Abstract
To explore the dynamics of snRNP structure and function, we have studied Cus1p, identified as a suppressor of U2 snRNA mutations in budding yeast. Cus1p is homologous to human SAP145, a protein present in the 17S form of the human U2 snRNP. Here, we define the Cus1p amino acids required for function in yeast. The segment of Cus1p required for binding to Hsh49p, a homolog of human SAP49, is contained within an essential region of Cus1p. Antibodies against Cus1p coimmunoprecipitate U2 snRNA, as well as Hsh155p, a protein homologous to human SAP155. Biochemical fractionation of splicing extracts and reconstitution of heat-inactivated splicing extracts from strains carrying a temperature-sensitive allele of CUS1 indicate that Cus1p and Hsh155p reside in a functional, high-salt-stable complex that is salt-dissociable from U2 snRNA. We propose that Cus1p, Hsh49p, and Hsh155p exist in a stable protein complex which can exchange with a core U2 snRNP and which is necessary for U2 snRNP function in prespliceosome assembly. The Cus1p complex shares functional as well as structural similarities with human SF3b.
Pre-mRNA splicing is catalyzed by a large ribonucleoprotein complex called the spliceosome. Several RNA molecules and many proteins are essential for splicing, assisting in spliceosome assembly, spliceosome activation, and conformational rearrangements before the actual transesterification reactions occur. An ordered assembly pathway for the construction of the spliceosome, including numerous points at which ATP hydrolysis is required, provide a rich sequence of biochemical events that is carried along in part by the action of splicing proteins (for reviews, see references 29 and 39).
Proteins that act during splicing have been identified by both biochemical and genetic means. In mammalian systems, splicing proteins have operationally been classified as small nuclear ribonucleoprotein particle (snRNP) proteins (that remain associated with a particular snRNA during biochemical fractionation) or splicing factors (that are transiently associated with snRNAs) (29, 39). Upon closer inspection, a number of splicing proteins cannot be classified by this simple operational distinction. For example, two multimeric protein factors required for prespliceosome assembly in mammalian cell extracts, SF3a and SF3b, can be separated from snRNAs and purified as discrete protein complexes that function in prespliceosome assembly (13, 28). Surprisingly, SF3a and SF3b subunits comprise seven of the nine salt-dissociable proteins identified in the 17S form of the U2 snRNP, the form that is recruited to the pre-mRNA during spliceosome assembly (8, 12, 30). The SF3a and SF3b proteins are also found in preparations of assembled spliceosomes, and most can be cross-linked to regions near the pre-mRNA branch point within the assembled spliceosome (10, 20, 38). The SF3a polypeptides are SAP61, SAP62, and SAP114, and the four known SF3b polypeptides are SAP49 SAP130, SAP145, and SAP155 (reviewed in reference 29). Thus, SF3a and SF3b are protein complexes with biochemical characteristics of splicing factors but which associate specifically with the free U2 snRNP under splicing conditions and remain as part of the spliceosome after U2 snRNP has been recruited. The SF3b proteins are also associated with the minor spliceosome, which has the U12 snRNP in place of U2 snRNP (44).
In contrast, Saccharomyces cerevisiae splicing factors have been discovered mainly through genetic approaches but have led independently to a similar set of proteins (29, 39). For example, PRP9 and PRP11 were identified among Hartwell's original temperature-sensitive mutations (originally called RNA9 and RNA11 [24]), and PRP21 was identified as a suppressor of prp9-1 (17), as well as in a search for temperature-sensitive splicing mutations (4, 40). The products of these three genes are similar to human SF3a subunits: Prp21p corresponds to SAP114, Prp9p corresponds to SAP61, and Prp11p corresponds to SAP62 (29, 39). Identification of yeast proteins similar to human SF3b subunits has occurred more recently, aided by the completion of the yeast genome. Cus1p was identified genetically by its ability to suppress U2 snRNA mutations and is similar to human SAP145 (19, 43). Hsh49p is an essential yeast protein homologous to SAP49 (26). A yeast protein similar to SAP155, which we refer to here as Hsh155p, has been identified (18, 37, 41). Rse1p is a conserved protein associated with the U2 snRNP (16) and is structurally related to human SAP130 (16, 18). The sequence similarities between the yeast proteins and their mammalian counterparts, as well as several examples of parallel protein-protein interactions (26, 29), have led to the hypothesis that these proteins function in a similar fashion in both yeast and mammals. Although numerous protein-protein interactions (18), phosphorylation events (41), and protein-RNA cross-links (19) have been described, exactly how human SF3b promotes spliceosome assembly and splicing remains mysterious.
In this study, we focus on Cus1p and provide evidence that the minimum portion of Cus1p required for viability in yeast contains the region of significant homology to human SAP145. The part of Cus1p required for binding to Hsh49p is contained within this essential conserved region but does not include the amino acid altered in the U2 suppressor protein Cus1-54p. Cus1p is physically associated with a fraction of U2 snRNA in splicing extracts and is efficiently associated with Hsh155p, the yeast protein similar to SAP155. Pre-mRNA becomes detectably associated with Cus1p before the first step of splicing and remains associated through the second step. Biochemical complementation studies using heat-inactivated splicing extracts from a cus1 temperature-sensitive mutant demonstrate that Cus1p functions as part of a protein complex which includes Hsh155p. This Cus1p complex dissociates from the U2 snRNP in a salt-sensitive fashion. These experiments reveal significant functional similarities between the Cus1p complex and human SF3b and suggest that the roles of these proteins in the function of the branch point binding snRNPs are similar in the yeast and human major and minor spliceosomes.
MATERIALS AND METHODS
Plasmid constructs.
All recombinant DNA manipulations were carried out as described previously (6). Yeast manipulations were also carried out in accordance with standard techniques (21). To create the CUS1 deletion series clones, an NdeI site was generated at the Cus1p start codon by site-directed mutagenesis (31) to make pRS314-CUS1Nde. Oligonucleotides containing NdeI (N termini) or XhoI (C termini) restriction sites were used to produce deletions of CUS1 with start and stop codons at the desired positions by PCR. These plasmids were transformed into a cus1::HIS3 knockout strain carrying wild-type CUS1 on a URA3 plasmid (43). Transformants that arose on synthetic complete dextrose medium (SCD)-His-Trp plates were replica plated onto 5-fluoroorotic acid (5-FOA)-Trp-His to check viability at different temperatures.
Temperature-sensitive cus1 allele.
A fragment spanning the open reading frame of CUS1 was amplified by mutagenic PCR (14). The PCR product was cotransformed into the cus1::HIS3 knockout strain along with a gapped linear fragment carrying pRS314-CUS1 without a coding region, in order that functional pRS314-CUS1 could only be regenerated by incorporation of a mutagenized copy of the CUS1 coding region. Transformants were replica plated onto 5-FOA to select for cells that had lost the URA3 plasmid. Cells that grew on 5-FOA were replica plated at different temperatures. DNA was recovered from colonies unable to grow at 34°C, and temperature-sensitive phenotypes were verified by retransformation and testing for temperature-sensitive in vitro splicing defects. One plasmid, designated cus1-3, carries 14 mutations in the CUS1 coding region that predict 10 amino acid substitutions (see Fig. 1A).
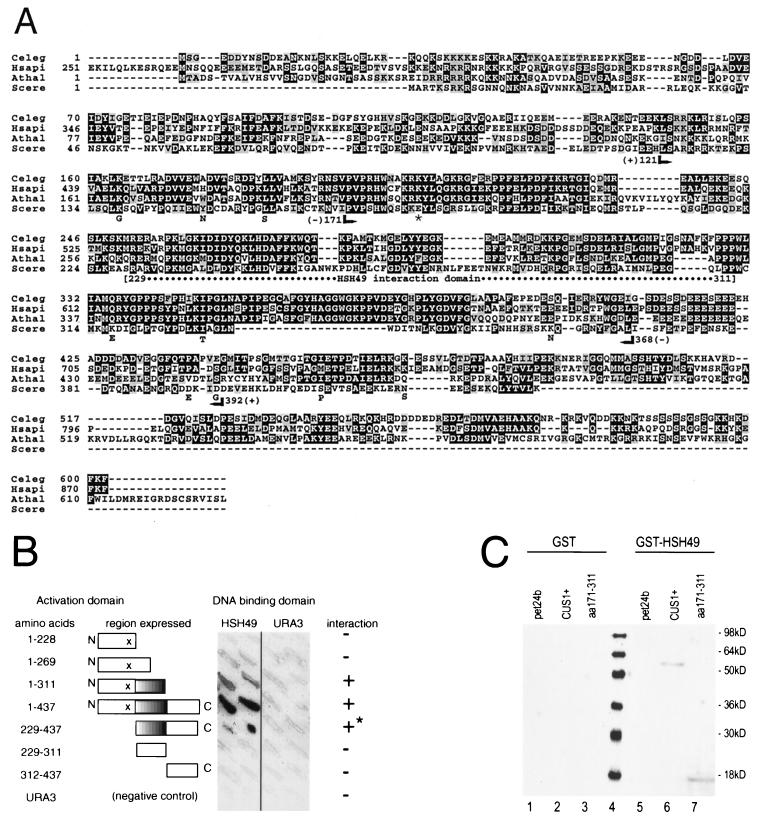
FIG. 1.
Structure and function of Cus1p. (A) Alignment of Cus1p amino acid sequence with those of Cus1p/SAP145 family members from metazoans. Scere, yeast Cus1p; Celeg, C. elegans (GenBank accession no. AAB69931); Hsapi, human SAP145 (Swissprot accession no. Q13435) (19); Athal, Arabidopsis (EMBL accession no. CAB36810). Black boxes indicate identity; grey boxes indicate similarity. Searches used BLAST (3), and alignment was performed by CLUSTALW (25). Positions of the smallest viable deletions (+) and the largest lethal deletions (−) are indicated below the yeast sequence. An asterisk marks the position of the Cus1-54p suppressor mutation E181K. Letters below the yeast sequence indicate predicted amino acid substitutions in the cus1-3 allele. The Hsh49p interaction domain is indicated between positions 229 and 311. (B) An 82-amino-acid region of Cus1p is necessary for the interaction with Hsh49p in the two-hybrid system. Amino-terminal deletions of Cus1p to amino acid 229 (229–437) can interact with Hsh49p, although the deletions produce a dominant negative growth defect (asterisk). Carboxy-terminal deletions to amino acid 311 (1–311) also support an interaction. Interaction is indicated by blue color (shown as dark shading on the figure) accumulating on the yeast patches after application of X-Gal. The 229–311 region of Cus1p alone is unable to support this interaction in this system (229–311). None of the constructs interacted with the Ura3p control. See panel A for position of the deletion end points. (C) Cus1p and a Cus1p fragment containing amino acids 171 to 311 bind Hsh49p. Pulldown studies were performed with GST or GST-Hsh49p and either recombinant Cus1p or a region of Cus1p containing the amino acids between positions 117 and 311. Recombinant extracts were mixed in a pairwise manner and then purified with glutathione agarose. Associated proteins were eluted with reduced glutathione, run on a 10% polyacrylamide gel, transferred, and blotted with anti-Cus1p antibody. Pairwise combinations are as follows: lane 1, GST plus pET24b (empty vector); lane 2, GST plus Cus1p; lane 3, GST plus Cus1p amino acids 171 to 311 (aa171–311); lane 4, GST-Hsh49p plus pET24b; lane 5, GST-Hsh49p plus Cus1p; lane 6, GST-Hsh49p plus Cus1p aa171–311. See panel A for position of the deletion end points.
Two-hybrid assay.
The pACT2-CUS1 fusion plasmid, the pAS2-HSH49 fusion plasmid, and the URA3 control plasmids were as described previously (26). Deletions of CUS1 within the pACT2-CUS1 plasmid were made by utilization of restriction sites that maintained the open reading frame of the fusion. Pairs of constructs were cotransformed into yeast strain Y190 (7, 23). Multiple transformants from each pairwise combination of plasmids were streaked on a grid, grown and overlaid with 0.5% agarose–0.5 M NaPO4 (pH 7.0)–0.1% sodium dodecyl sulfate (SDS)–2% dimethyl formamide–0.2% (wt/vol) X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside), and incubated at 37°C.
Protein-protein interactions.
Regions of Cus1p that interacted with Hsh49p in two-hybrid assays were cloned into pET24b (Novagen). Expression of these Cus1p fragments in Escherichia coli BL21 was confirmed by Western blot analysis (22). pGEX2t-Hsh49p was subcloned as previously described (26). The proteins were extracted after induction and incubation at 30°C for 3 h as previously described (26). Expression of glutathione S-transferase (GST)-Hsh49p and GST was checked by Western blot analysis utilizing anti-GST monoclonal antibody (gift from Santa Cruz Biotechnology). The in vitro protein-protein interaction assay was performed essentially as described previously (26).
Protein purification and polyclonal antibody production.
The CUS1 coding sequence was amplified with an NdeI site at the start codon and cloned into pET24b to produce an untagged protein. The plasmid was transformed into the E. coli strain BL21, and colonies were screened for optimal protein production. Cultures were grown to an A595 of 0.6 and induced with 10 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h at 37°C. Cells were pelleted by centrifugation and resuspended in 0.1 volume of HK100 (0.05 M HEPES [pH 7.5], 100 mM KCl) buffer. Triton-X (to 0.1%) and lysozyme (to 0.1 mg/ml) were added, and the mixture was incubated at 30°C for 20 min, put on ice, sonicated, and centrifuged at 10,000 rpm for 10 min at 4°C in a Sorvall SS-34 rotor. The lysate was loaded onto cation-exchange resin Bio-Rex70 (Bio-Rad), washed with 5 column volumes of HK100 and then 2 volumes of HK400 (0.05 M HEPES [pH 7.5], 400 mM KCl), and eluted with HK700 (0.05 M HEPES [pH 7.5], 700 mM KCl). Approximately 90% purification was achieved with this protocol. Protein was concentrated with Centriprep30s (Amicon) and loaded onto a 10% denaturing Tris-glycine acrylamide gel. Side strips of the gel were stained with Coomassie blue dye to locate the protein. The strip of gel containing Cus1p was cut out, and protein was electroeluted and injected into a rabbit (22). Polyclonal anti-Cus1p antibody was affinity purified by binding rabbit serum to denatured recombinant Cus1p on nitrocellulose strips, washing, and eluting bound antibody with glycine (22). The antibody was determined to be specific for Cus1p through detection of recombinant Cus1p, native yeast Cus1p, and Cus1p-His6 in whole-cell extracts of yeast by Western blotting (data not shown).
Immunoprecipitation and Western blotting.
Immunoprecipitation experiments were performed essentially as described before (42). Protein A-Sepharose was preswollen in HK150 (0.05 M HEPES [pH 7.5], 150 mM KCl) or HK200 (0.05 M HEPES [pH 7.5], 200 mM KCl) buffer overnight at 4°C, and 400 μl of a 50% slurry was incubated with preimmune serum or anti-Cus1p serum for 3 h at 4°C. Beads were washed three times with HK150 with 5-min incubations on ice between washes. Aliquots of 50% slurry (40 μl) were incubated with 100 μl of yeast splicing extract or yeast protein preparation (6) and incubated at 4°C for at least 3 h. Beads were collected and washed three times in HK150 as described above. Proteins were eluted by suspension in 20 μl of SDS loading dye and separated on SDS–10% polyacrylamide gels.
Western blots (22) were blocked in 3% bovine serum albumin in Tris-buffered saline–Tween (TBST) for 30 min at room temperature, probed with primary antibody in the same solution for 1 h at room temperature and washed in TBST for 30 min, and then secondary antibody was added for 1 h at room temperature. Blots were washed in TBST, and proteins were visualized with 5-bromo-4-chloro-3-indolyl phosphate–nitroblue tetrazolium (NBT-BCIP). Anti-Cus1p antibody was used at a dilution of 1:10,000 for detection of Cus1p. Monoclonal antibody 9E10 directed against the myc epitope (gift from Santa Cruz Biotechnology) was used at a dilution of 1:2,000 for detection of Hsh155-Mycp.
Immunoprecipitated snRNA was isolated by the addition of 200 μl of RNA elution buffer (0.3 M Na acetate [pH 5.2], 0.2% SDS, 1 mM EDTA, 10 μg of proteinase K/ml) to the beads and incubation at 65°C for 10 min, followed by phenol extraction and ethanol precipitation. Primer extension was performed as described previously (45) with oligonucleotides complementary to U1, U2, U4, U5, and U6 snRNAs.
Association of Cus1p with pre-mRNA was evaluated by coimmunoprecipitation of pre-mRNA with anti-Cus1p antibody. Splicing reaction mixtures were incubated for 20 min at 23°C and added to protein A-Sepharose prebound to anti-Cus1p antibodies or preimmune serum from the same rabbit as described previously (42).
Splicing extract preparation, splicing, and complex formation.
Extract preparation was performed as previously described (43). Strains were grown at 26°C until saturation, diluted to an A600 of 0.05, and grown to an A600 of 2.8 to 3.5. Splicing assays (43) and oligonucleotide ablation (33, 42) were performed as previously described.
Fractionation of yeast splicing extracts.
Chromatography of splicing extracts on a Superose6 gel filtration column (Pharmacia) was done as follows: 500 μl of splicing extract was run through a Spin-X 100 column to remove aggregated proteins. This filtrate was passed over the Superose6 column with a flow rate of 0.2 ml of buffer D150 or D500 (buffer D with 150 or 500 mM KCl) per ml, with collection volumes of 0.5 ml. Fractions were concentrated to 200 μl with Microcon10 centrifugal concentrators (Amicon). Even-numbered fractions were assayed for Cus1p and Hsh155-mycp by gel electrophoresis and Western blotting. To test for Cus1p activity, an aliquot of splicing extract from the temperature-sensitive cus1-3 strain was heat inactivated at 36°C for 1 h and placed on ice. Then, 2 μl of each fraction was added to 2 μl of heat-inactivated cus1-3 extract in a 10-μl splicing reaction volume and incubated for 30 min at room temperature. Extracts were derived from wild-type strain HI227 (43), the cus1-3 strain described above, and a derivative of strain EJ101 (27) carrying Hsh155-mycp, the product of an allele of HSH155 (also called ySAP155) with 13 C-terminal copies of the myc tag. This last strain was constructed by the PCR tagging method of Longtine et al. (32).
RESULTS
Essential Cus1p amino acids correspond to those shared among Cus1/SAP145 family members.
Cus1p is similar to SAP145 from humans (19, 43), as well as proteins predicted from cDNAs derived from Caenorhabditis elegans and Arabidopsis thaliana (Fig. 1A). The region of homology between the yeast and metazoan proteins comprises approximately 200 amino acids and is about 43% identical and 65% similar at positions shared by all. To explore the relationship between this structural conservation and the functional requirement for Cus1p in growth and splicing (43), we defined the region of CUS1 required for viability in yeast. A series of partial CUS1 deletions was tested for their ability to rescue a lethal cus1::HIS3 knockout allele (Fig. 1A and data not shown). Whereas an N-terminal deletion to amino acid 121 rescues a lethal CUS1 disruption, further deletion to position 171 does not. Deletion of the carboxy-terminal 40 amino acids (to residue 392) rescues the cus1::HIS3 disruption, but deletion past residue 368 is lethal. Nearly half of the last 59 residues of Cus1p are acidic (23 are E or D) (Fig. 1A). The nonlethal deletion to position 392 removes 12 of these. The lethal deletion to position 368 removes additional glutamate residues that are shared among all family members (Fig. 1A). Therefore, the elements of Cus1p required for normal growth are contained within amino acids 121 to 392 and correspond to the most highly conserved region of Cus1p. We conclude that the conserved elements of Cus1p structure are required for function in yeast and likely in metazoans as well.
A distinct region of Cus1p is required for the interaction with Hsh49p.
Full-length Cus1p binds Hsh49p both in vivo and in vitro (26). To map the region of Cus1p required for the interaction with Hsh49p, we used the two-hybrid system (7, 11, 34) and verified our findings by an in vitro protein binding assay (see below). GAL4-DNA binding domain–Hsh49p fusion protein was expressed with different segments of CUS1 fused to the GAL4 activation domain in yeast carrying a GAL-regulated lacZ gene, and β-galactosidase activity was assayed on plates (Fig. 1B). The derivatives containing amino acids 1 to 311 or 229 to 437 of Cus1p are sufficient for the interaction with Hsh49p (Fig. 1B; region marked on Fig. 1A), suggesting that the 82 amino acids between positions 229 and 311 represent the Hsh49p interaction domain of Cus1p. Although neither construct expressed separately or with other constructs produced a growth defect, the specific pair of two-hybrid plasmids expressing the GAL4 DNA binding domain–full-length HSH49 fusion and the GAL4 activation domain fused to amino acids 229 to 437 of Cus1p demonstrated a severe dominant negative growth defect (and produced strong lacZ reporter expression) (Fig. 1B). The construct containing only amino acids 229 to 311 failed to support the two-hybrid interaction with Hsh49p, suggesting that either the Hsh49p interaction region of Cus1p requires the support of other Cus1p elements or that this particular fusion protein is unstable or incorrectly folded. The region of Cus1p altered in the Cus1-54p suppressor protein (position 181) is not required for interaction with Hsh49p (Fig. 1B), suggesting that the mechanism of suppression does not directly involve changes in association of these subunits with each other.
To confirm that the two-hybrid results reflect physical interactions, we tested the ability of recombinant derivatives of Cus1p to bind a GST-Hsh49p protein in vitro. Since expression of a Cus1p fragment representing residues 229 to 311 was not detectable in E. coli, we used a slightly larger segment spanning residues 171 to 311 (Fig. 1A). Association of Cus1p with Hsh49p was assessed by binding to glutathione agarose loaded with either control GST (Fig. 1C, left) or the GST-Hsh49p fusion (Fig. 1C, right) and detected by Western blotting of bound material probed with an anti-Cus1p antibody (see Materials and Methods). Proteins representing full-length Cus1p (lanes 2 and 6) or the Cus1p region spanning residues 171 to 311 (lanes 3 and 7) were not detected when incubated with GST (lanes 1 to 3) but were readily detected after incubation with GST-Hsh49p (lanes 6 and 7). This demonstrates that the region of Cus1p containing amino acids 171 to 311 is sufficient for the interaction with Hsh49p, and we conclude that this region contains the Cus1p element required for binding to Hsh49p. This region is conserved in the metazoan members of the Cus1/SAP145 family (Fig. 1A), suggesting that the structure of the interface between these two subunits is conserved. In Hsh49, this interaction involves predominantly the first of two RNA recognition motifs (26).
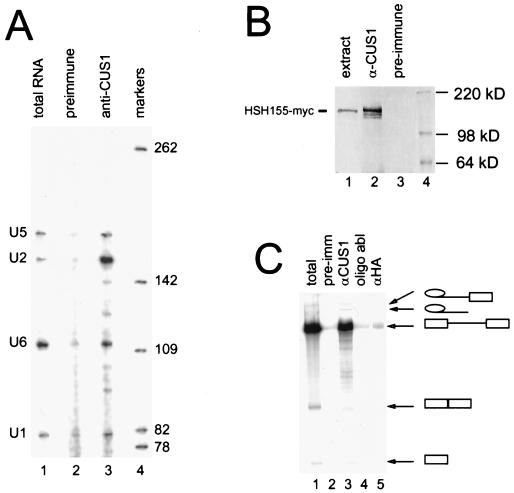
A fraction of U2 snRNA is bound to Cus1p in splicing extracts.
As a suppressor of a cold-sensitive U2 snRNA mutation, Cus1p has clear functional association with U2 RNA. To determine whether Cus1p is physically associated with U2 or other snRNAs, yeast extracts were immunoprecipitated with anti-Cus1p antibody and examined for the presence of snRNAs by primer extension (Fig. 2A). Only a fraction of U2 snRNA is associated with Cus1p in splicing extracts (Fig. 2A, lane 3), and the association is specific, since preimmune serum did not precipitate U2 snRNA (lane 2). The amount of U2 snRNA in the precipitate is less than 10% of that present in the extract, even though the antibody recovers >90% of the input Cus1p as determined by Western blotting of the immunosupernatant (data not shown). This indicates that only a fraction of total U2 snRNA is immunoprecipitable with Cus1p antibody at 150 mM KCl. Anti-Cus1p antibody also reproducibly precipitates small amounts of U1, U5, and U6 that are not recovered in the preimmune control (compare lanes 2 and 3). This is consistent with the presence of endogenous spliceosomal complexes in the extract and the fact that anti-Cus1p recognizes the spliceosome (see below).
FIG. 2.
Anti-Cus1p antibody coimmunoprecipitates U2 snRNA, Hsh155p, and the intermediates and products of splicing. (A) Anti-Cus1p antibody coimmunoprecipitates U2 snRNA. Splicing extracts were incubated with prebound anti-Cus1p serum or preimmune serum. Bound RNA was washed and eluted. Primer extension analysis with primers complementary to U1, U2, U5, and U6 snRNAs was performed. Lane 1, total RNA; lane 2, preimmune serum; lane 3, anti-Cus1p; lane 4, end-filled Sau3A fragments of pUC13 as markers. Extension products from the U snRNAs are indicated at the left. (B) Anti-Cus1p antibodies specifically coimmunoprecipitate Hsh155p, the yeast homologue of the human SF3b subunit SAP155. Yeast splicing extract was made from a strain with Hsh155p tagged with the myc epitope. Extract was incubated with protein A-Sepharose-bound anti-Cus1p serum or preimmune serum and washed, and associated proteins were removed and loaded on a 10% polyacrylamide gel, transferred, and blotted with monoclonal anti-myc antibody. Lane 1, total extract (20% of input for lanes 2 and 3); lane 2, anti-Cus1p antibody; lane 3, preimmune serum; lane 4, protein markers. (C) Anti-Cus1p antibodies coimmunoprecipitate pre-mRNA substrate, intermediates, and products of splicing. Splicing reaction mixtures were incubated either with (lane 4) or without (lanes 1 to 3 and 5) preincubation of extract with a U2 snRNA-complementary oligonucleotide. Reaction mixtures were immunoprecipitated with anti-Cus1p serum (lanes 3 and 4), preimmune serum (lane 2), or anti-hemagglutinin antibody (lane 5). Bound RNA was then extracted and run on a 6% denaturing acrylamide gel. Lane 1 contains RNA from 20% of the input used in the immunoprecipitations for the other lanes.
To determine whether the number of Cus1p molecules might be limiting for the immunoprecipitation of U2 snRNA, we used Western blotting and known amounts of recombinant Cus1p to estimate the number of Cus1p molecules per yeast cell (data not shown). This analysis indicates that there are approximately 2,000 molecules of Cus1p per cell. To estimate the number of U2 snRNA molecules in the cell, we compared the ratio of a U2 snRNA primer extension product from a known number of cells to a known amount of a synthetic (T7) mutant U2 snRNA able to produce a different primer extension product (C121U [5]). This indicates that there are approximately 500 molecules of U2 snRNA in a yeast cell, a number that is consistent with a previous general estimate of snRNA content by Northern blotting (35). Given this, we can at least conclude that limiting Cus1p does not explain the inefficient precipitation of U2 snRNA. We suggest that the specific interaction between Cus1p and U2 snRNA may be restricted to a functional subset of molecules, their association may be salt labile, or both.
A substantial fraction of Hsh155p is associated with Cus1p in splicing extracts.
Identification of a yeast homolog of SAP155 (37, 41) prompted us to ask whether this protein is associated with Cus1p. Although called ySAP155 or ySAP110 by other workers, the designation SAP is in use for other yeast genes not involved in splicing. We refer to this gene as HSH155 (for human SAP homolog 155). The gene is essential (18); however, a strain carrying an HSH155 gene with 13 C-terminal copies of the myc epitope is viable, and splicing extracts made from the tagged strain are active (data not shown).
To determine whether Hsh155p is associated with Cus1p, we immunoprecipitated splicing extracts from the strain carrying the HSH155-myc gene with anti-Cus1p antibody and evaluated the presence of tagged Hsh155p by Western blotting with anti-Myc antibody (Fig. 2B). To evaluate the fraction of myc-tagged Hsh155p associated with Cus1p, we used an aliquot of the total extract equivalent to 20% of that used for the immunoprecipitation (lane 1). Comparison of this signal to that observed in the anti-Cus1p immunoprecipitate (lane 2) indicates that more than half and possibly all of the input myc-tagged Hsh155p is immunoprecipitated by the anti-Cus1p antibody. Because a much larger fraction of input Hsh155-myc protein than U2 RNA is associated with Cus1p, it seems likely that the interaction between the proteins does not require U2 RNA. The interaction is specific, since immunoprecipitation with preimmune serum did not result in coimmunoprecipitation of Hsh155-mycp (Fig. 2B, lane 3). The association of Cus1p with Hsh155p (Fig. 2B), together with the interaction between Cus1p and Hsh49p (Fig. 1B and C) (26), significantly extends the parallels in subunit composition between the yeast and mammalian SF3b complexes.
Cus1p is associated with intermediates and products of splicing.
To explore the association of Cus1p with the pre-mRNA during splicing, we immunoprecipitated splicing reaction mixtures containing radiolabeled pre-mRNA (Fig. 2C). Cus1p is associated with the pre-mRNA substrate, the splicing intermediates (free exon 1 and lariat intermediate), and the splicing products (spliced exons and lariat intron), as demonstrated by the coimmunoprecipitation of these RNAs with Cus1p (Fig. 2C, lane 3). This association is specific, since immunoprecipitation with preimmune serum (Fig. 2C, lane 2) or antihemagglutinin antibody (Fig. 2C, lane 5) did not efficiently precipitate pre-mRNA or any intermediates or products of splicing. Oligonucleotide ablation of U2 snRNA in the splicing reactions prior to immunoprecipitation demonstrated that interaction of Cus1p with pre-mRNA is dependent upon the presence of U2 snRNA (Fig. 2C, lane 4). These data indicate that Cus1p is accessible to the antibody in spliceosomes at various stages of the splicing reaction. The immunoprecipitation of intermediates and products of splicing by anti-Cus1p antibodies is in contrast to similar experiments with Prp9p, Prp11p, and Prp21p. These homologs of mammalian SF3a components have been demonstrated to coimmunoprecipitate only pre-mRNA and have been considered to be destabilized or lost from the spliceosome or shielded from antibodies before the first step of splicing (1, 2, 4, 36, 42).
Splicing in heat-inactivated temperature-sensitive Cus1p extracts can be rescued by extracts depleted of U2 snRNA.
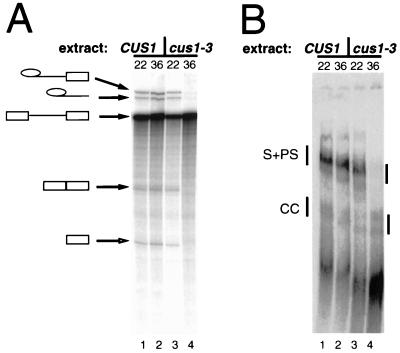
To explore the biochemical role of Cus1p in splicing, we began to develop assays that depend on Cus1p function. Extracts made after repression of Cus1p expression using a GAL:CUS1 gene do not support splicing complex formation in vitro, and a small but reproducible rescue of prespliceosome formation occurs after addition of partially purified Cus1p-containing fractions from yeast extracts (43). Because splicing is poorly reconstituted in a genetically depleted Cus1p extract, we were concerned that secondary effects of CUS1 repression might hinder efforts to reconstitute Cus1p activity. To circumvent this difficulty, we isolated temperature-sensitive Cus1p mutants and made splicing extracts that could be heat inactivated (Fig. 3). Preheating of extracts derived from one temperature-sensitive mutant, the cus1-3 mutant, specifically inactivated splicing (Fig. 3A, lane 4; see Fig. 1A for sequence changes in the cus1-3 mutant) and prespliceosome formation (Fig. 3B, lane 4). Commitment complexes (Fig. 3B) still appear in the heated mutant extracts, consistent with a role for Cus1p after commitment complex formation but before or during prespliceosome formation (43).
FIG. 3.
Heat-treated cus1-3 extract is blocked in prespliceosomeformation and splicing. (A) The cus1-3 extract is temperature sensitive for splicing. cus1-3 or wild-type (CUS1) extracts were preincubated at 22 or 36°C for 1 h and then incubated with radiolabeled actin pre-mRNA at 22°C for 20 min. RNA was extracted and run on a 6% denaturing acrylamide gel. Lane 1, wild-type extract treated at 22°C; lane 2, wild-type extract treated at 36°C; lane 3, cus1-3 extract treated at 22°C; lane 4, cus1-3 extract treated at 36°C. (B) The cus1-3 extract is temperature sensitive for prespliceosome formation. Reactions were set up as described for panel A, incubated, and loaded onto a nondenaturing 3.5% acrylamide–0.5% agarose gel. Lane 1, wild-type extract treated at 22°C; lane 2, wild-type extract treated at 36°C; lane 3, cus1-3 extract treated at 22°C; lane 4, cus1-3 extract treated at 36°C. CC, commitment complexes.
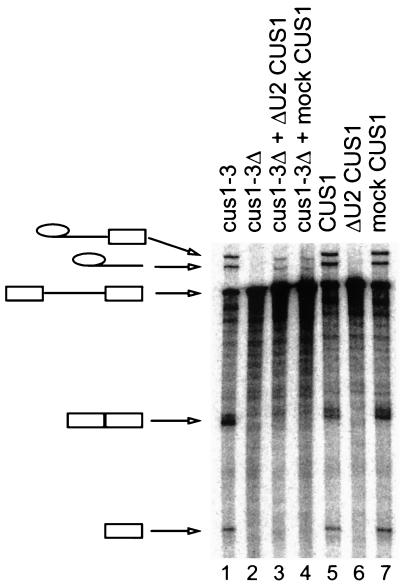
The addition of recombinant Cus1p alone does not rescue splicing or prespliceosome formation in the heat-inactivated cus1-3 extract (data not shown). This suggests that execution of Cus1p function requires additional factors, for example, the other yeast SF3b-like proteins, which may not freely diffuse from inactivated cus1-3p-containing mutant complexes. To determine whether the addition of wild-type Cus1p in some form could rescue heat-inactivated cus1-3 extracts, we prepared wild-type extracts depleted of functional U2 snRNA by oligonucleotide-directed RNase H digestion of U2 RNA (Fig. 4). Such extracts do not splice (Fig. 4, lane 6) because they lack intact U2 snRNA (33); however, they can reconstitute splicing when mixed with heat-inactivated cus1-3 extracts (lane 3). Complementation is not efficient, as indicated by the reduced levels of the lariat product and lariat-exon 2 intermediate migrating more slowly than the pre-mRNA substrate. This inefficiency may be due to a loss or dilution of a general splicing factor or to the formation of denatured Cus1p that may inhibit splicing, because the same low efficiency is observed with wild-type extracts treated with an irrelevant oligonucleotide (mock treated) (Fig. 4, lane 4). Despite this, extracts containing disintegrated U2 snRNPs retain significant Cus1p activity that can exchange with that of the intact U2 snRNA from the cus1-3 extract. Together with the efficient association of Hsh155p with Cus1p (Fig. 2), this result suggests that yeast Cus1p activity exists in a complex with Hsh155p that can exchange with U2 snRNA under splicing conditions.
FIG. 4.
Reconstitution of splicing in heat-inactivated cus1-3 extracts does not require U2 snRNA. Splicing in heat-inactivated cus1-3 extracts alone or with the addition of wild-type (CUS1) extracts depleted of U2 RNA by oligonucleotide-RNase H depletion is shown. Lane 1, cus1-3 extract treated at 22°C; lane 2, cus1-3 extract treated at 36°C; lane 3, cus1-3 extract treated at 36°C plus U2-depleted wild-type extract; lane 4, cus1-3 extract treated at 36°C plus mock-depleted wild-type extract; lane 5, untreated wild-type extract; lane 6, U2-depleted wild-type extract; lane 7, mock-depleted wild-type extract.
Cus1p complementing activity is associated with Hsh155p and U2 snRNA under splicing conditions.
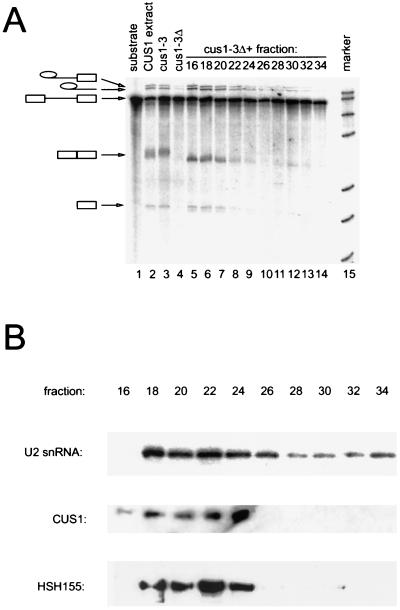
To study the properties of the factor that complements Cus1p function in vitro, we fractionated splicing extracts derived from the Hsh155-myc-tagged strain by gel exclusion fast protein liquid chromatography in a buffer containing 150 mM KCl (Fig. 5). Fractions were added to heat-inactivated cus1-3 splicing extract and assayed for complementation of splicing (Fig. 5A). Most of the rescuing activity chromatographed as a broad peak (Fig. 5A, lanes 5 to 8), with an apparent native molecular mass of 669 kDa to 2 MDa as estimated by comparing its Kav determined from the elution volume with the values obtained for several globular protein calibration standards and by the known upper limit of separation for the Superose6 column (2 MDa). Western blot analysis of these fractions using the anti-Cus1p antibody demonstrates that they contain Cus1p (Fig. 5B, fractions 18 to 24). These same fractions were also blotted and probed with the anti-myc antibody in order to detect Hsh155-mycp (Fig. 5B). Hsh155-mycp was also found in the fractions that reconstitute splicing (fractions 18 to 24). The Cus1p antibody is barely able to detect the protein at the levels we recover from the column fractions (data not shown), and only peak fractions have detectable levels of protein. The level of Cus1p in the weakly complementing fractions of Fig. 5 (28 and 30) is below the level of detection of the antibody. The anti-myc antibody that recognizes epitope tag at the C terminus of Hsh155-mycp is much more sensitive, so some side fractions may appear to contain 155 but not Cus1p. Fraction 16 is problematic, because it appears to contain aggregated protein, including nucleases and proteases that differentially influence the recovery and analysis of RNA and tagged protein in different experiments (data not shown). RNA was extracted from the fractions and assayed by primer extension for the presence of U2, which is also present in fractions where the reconstituting activity is found (Fig. 5B, lanes 18 to 26). Given the large size of this complex, and the identity of components found in these fractions, it seems possible that the rescuing factor in this experiment is the U2 snRNP itself.
FIG. 5.
Rescue of heat-treated cus1-3 extracts by large complexes containing U2 RNA, Cus1p, and Hsh155p under low-salt conditions. (A) Fractionation of Cus1p activity at 150 mM KCl. Hsh155p-myc extract was separated over a Superose6 sizing column in the presence of 150 mM KCl, added to heat-inactivated cus1-3 extracts, and assayed for splicing. Lane 1, pre-mRNA substrate; lane 2, wild-type extract; lane 3, cus1-3 extract at 22°C; lane 4, cus1-3 extract at 36°C; lanes 5 to 14, heat-inactivated cus1-3 extract incubated with fractionated Hsh155p-Myc extract from even-numbered fractions; lane 15, markers. (B) Profiles of U2 snRNA, Cus1p, and Hsh155p in Hsh155p-myc extract fractionated at 150 mM KCl. RNA was extracted from even-numbered fractions and used in primer extension analysis with an oligonucleotide complementary to U2 snRNA. Cus1p and Hsh155p in Hsh155p-myc extract fractionated at 150 mM KCl were detected by using even-numbered fractions loaded onto 10% acrylamide gels, transferred, and blotted with anti-Cus1p antibodies or anti-myc antibodies.
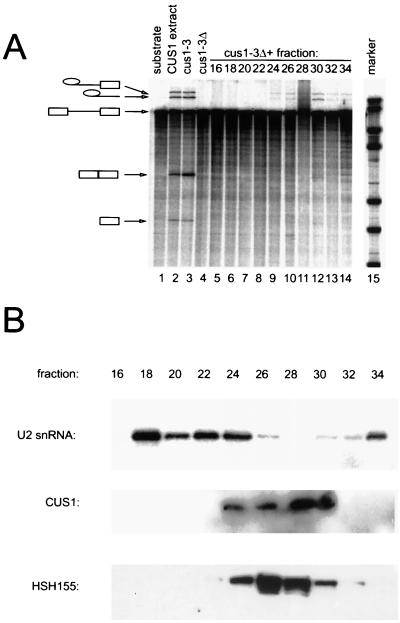
Cus1p-complementing activity remains associated with Hsh155p but not U2 snRNA under high-salt conditions.
Although most of the complementing activity migrates as a large complex in 150 mM KCl, in some experiments a small amount of complementing activity could be observed in fractions 28 to 30 (Fig. 5A, lanes 11 and 12). This activity is very weak and migrates with an apparent mass of between 232 and 669 kDa. Although not present in sufficient amounts to be detectable on Western blots by our anti-Cus1p antibody, the more avid anti-myc monoclonal antibody detects small amounts of Hsh155-mycp in these fractions. To test the idea that these fractions represent Cus1p-containing complexes that dissociate from U2 snRNP, and to evaluate the salt sensitivity of the association of the Cus1p complex with U2 RNA, splicing extracts from the Hsh155-mycp strain were fractionated in the presence of 500 mM KCl (Fig. 6). Under these conditions, the majority of the complementing activity is present in fractions 26 to 30, but splicing is inefficient. (The mRNA product and exon 1 intermediates are obscured. Refer to lariat intermediate and product.) This activity has an apparent mass of 232 to 669 kDa (Fig. 6A, lanes 9 to 14), similar to the very minor peak observed under low-salt conditions. This value is larger than that expected for Cus1p alone (50 kDa), providing biochemical evidence for a salt-resistant complex containing Cus1p activity. Indeed, Western blot analysis of these fractions shows that Cus1p and Hsh155-mycp fractionate with the reconstituting activity (Fig. 6B, fractions 24 to 30). Because the amounts of Cus1p in the peak fractions are near the detection limit of our anti-Cus1p antibody, quantitation of the Cus1p signal is not reliable. Furthermore, because the anti-myc antibody is much more sensitive, the signals arising from the two proteins cannot be compared directly. Because of this, we can only qualitatively assign Cus1p to a subset of fractions containing Hsh155-mycp, and in every case all fractions that contain abundant Hsh155-mycp also contain Cus1p. Although this suggests that the two proteins are together in a single complex, it is formally possible that they are each in different complexes that comigrate on the column. Analysis of the RNA in these fractions shows that most of the U2 snRNA still migrates in the larger fractions (Fig. 6B, fractions 18 to 24), consistent with the very large mass of yeast U2 snRNA (387 kDa) plus additional proteins (15). This result indicates that in 500 mM KCl, the cus1-3p-complementing activity is no longer associated with the bulk of U2 snRNA. We conclude that increasing the salt concentration from 0.15 to 0.5 M releases a functional protein complex containing Cus1p and Hsh155p from a larger complex that may represent a form of the U2 snRNP.
FIG. 6.
Rescue of heat-treated cus1-3 extracts by smaller complexes containing Cus1p and Hsh155p under high-salt conditions. (A) Rescue of splicing in heat-inactivated splicing extracts by sizing fractions separated at 500 mM KCl. Hsh155p-myc extract was separated over a Superose6 sizing column in the presence of 500 mM KCl, added to heat-inactivated cus1-3 extracts, and assayed for splicing. Lane 1, pre-mRNA; lane 2, wild-type extract; lane 3, cus1-3 extract at 22°C; lane 4, cus1-3 extract at 36°C; lanes 5 to 14, heat-inactivated cus1-3 extract incubated with fractionated Hsh155p-myc extract from even-numbered fractions; lane 15, markers. (B) Profiles of U2 snRNA, Cus1p, and Hsh155p in Hsh155p-myc extract fractionated at 500 mM KCl. RNA was extracted from even-numbered fractions and used in primer extension analysis with an oligonucleotide complementary to U2 snRNA. Cus1p and Hsh155p in Hsh155p-myc extract fractionated at 500 mM KCl were detected by using even-numbered fractions loaded onto 10% acrylamide gels, transferred, and blotted with anti-Cus1p antibodies or anti-Myc antibodies.
DISCUSSION
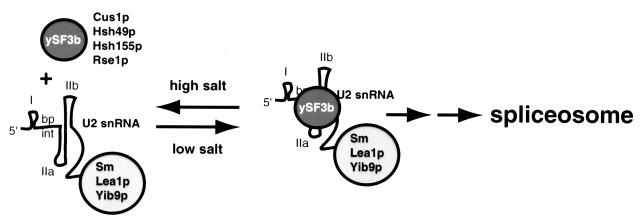
Understanding snRNP function will require knowing how snRNP proteins and splicing factors interact with RNA during spliceosome assembly and splicing. Building on the identification of the splicing factor Cus1p in a U2 RNA suppressor screen (43), we found that the most widely conserved parts of Cus1p are those essential for function in yeast (Fig. 1A). An 82-amino-acid region within the essential part of Cus1p is sufficient for binding to another evolutionarily conserved splicing factor, Hsh49p (Fig. 1B). Anti-Cus1p antibodies coimmunoprecipitate U2 snRNA as well as pre-mRNA and the intermediates and products of the splicing reaction, demonstrating that Cus1p is present in the spliceosome and accessible to antibodies throughout the splicing process (Fig. 2A and C). A new interaction between Cus1p and Hsh155p, the homolog of human SAP155 (Fig. 2B), establishes an important functional parallel between the structurally similar yeast and human proteins. Gel exclusion chromatography of splicing extracts under different ionic conditions shows that functional Cus1p-complementing activity contains both Cus1p and Hsh155p, suggesting that rescue is mediated by a Cus1p complex that also contains Hsh155p (Fig. 5 and 6). Considering the binding between Hsh49p and Cus1p (Fig. 1B and C) (26), and the association of Cus1p with Hsh155p (Fig. 2B), we conclude that a complex of yeast proteins similar to mammalian SF3b is required for the association of the U2 snRNP with the assembling spliceosome (Fig. 7). The dynamic salt-sensitive behavior of this protein complex with respect to its association with U2 snRNA also seems similar to that observed for mammalian SF3b (8, 12), suggesting that the underlying features of the splicing apparatus that lead to this phenomenon are conserved as well.
FIG. 7.
A model for salt-labile interaction of Cus1p-containing complexes (ySF3b) during activation of the U2 snRNP for intron recognition and spliceosome assembly in yeast. I, IIa, and IIb, conserved stem-loops of U2 snRNA; Sm, core Sm protein complex of the snRNP. Other individual proteins are named by their gene names, followed by a lowercase p. Proteins that remain associated with U2 in high salt are described in reference 15.
Cus1p function in splicing.
Cus1p was identified as a suppressor of U2 snRNA mutations in yeast. A glutamate at position 181 in the wild-type protein is changed to a lysine, somehow accelerating the rate of prespliceosome formation at low temperature in extracts containing mutant U2 RNA (43). Extracts made from strains in which the expression of Cus1p is repressed have been difficult to complement; however, extracts made from the temperature-sensitive cus1-3 strain could be heat inactivated (Fig. 3) and complemented with splicing extract fractions of different types (Fig. 4 to 6). The defect in the heat-inactivated temperature-sensitive extracts (failure to form prespliceosomes) (Fig. 3) is the same as that observed in the genetically depleted extract (43), and thus both results indicate that the earliest function of Cus1p in the splicing pathway is at the step of U2 snRNP addition to the commitment complex to form the prespliceosome. Since Cus1p remains associated with the spliceosome throughout splicing (Fig. 2), there could be additional functions of Cus1p at later steps of splicing.
Cus1p binds the pre-mRNA substrate in active splicing extracts, and this binding is dependent on intact U2 snRNA (Fig. 2). This argues that Cus1p does not act by binding to the pre-mRNA before it becomes associated with U2, although the possibilities that Cus1p binds pre-mRNA in an unstable fashion and that U2 snRNP is required to stabilize that interaction cannot strictly be ruled out. Based on gel exclusion chromatography in high-salt conditions, the protein that rescues heat-inactivated cus1-3 splicing extracts is much larger than the 50 kDa expected for monomeric Cus1p, a result consistent with the expectation that Cus1p is a subunit of a protein complex. Since other yeast U2 snRNP proteins remain associated with U2 snRNA in up to 0.3 M KCl (15), it seems likely that the Cus1p complex dissociates under high-salt conditions from a core U2 snRNP containing the high-salt-stable proteins and reassociates at lower ionic strengths in order to participate in spliceosome assembly (Fig. 7). Precisely how Cus1p helps U2 snRNP assemble into the complex and especially how the Cus1-54p suppressor protein enables mutant U2 RNA to participate in this reaction at a normally nonpermissive temperature remain to be determined.
Other proteins in the Cus1p complex.
Cus1p interacts with a number of other proteins. Hsh49p binds Cus1p in vitro, through the first RNA recognition motif of Hsh49p (26), and through amino acids 229 to 311 of Cus1p (Fig. 1). Although we have not provided direct evidence that Hsh49 is present in the fractions that rescue heat-inactivated cus1-3 extracts in vitro, the robust nature of the protein-protein interaction suggests that binding of these two proteins to each other is important for their function, and in both proteins the minimum binding domain includes essential amino acids. Thus, Hsh49p is a likely component of the functional Cus1p complex.
Anti-Cus1p antibodies coimmunoprecipitate nearly all of the tagged Hsh155p present in splicing extracts (Fig. 2B), indicating that most if not all of Hsh155p is bound directly or indirectly to Cus1p, at least at 150 mM NaCl. All rescuing fractions contain both Cus1p and Hsh155p (Fig. 5 and 6), consistent with the coimmunoprecipitation. Taken together, the coimmunoprecipitation and cofractionation results support the interpretation that Hsh155p is part of the Cus1p complex (Fig. 7). Because we have not yet shown that we can coimmunoprecipitate Cus1p and Hsh155-mycp from the complementing fractions of the 0.5 M KCl fractionation, we cannot discern whether the two proteins are together in a single complex or in two different, similarly sized complexes. Future work will address the composition of the functional Cus1p complex in these fractions with respect to the presence and direct physical association of Hsh155p, Rse1p, and Hsh49p with Cus1p and each other. Since the human homologues of these three yeast proteins form part of the SF3b complex in mammals, we propose that the Cus1p complex we observe (Fig. 5 and 6) is equivalent to yeast SF3b. The yeast splicing factor Rse1p is equivalent to mammalian SAP130, the fourth subunit of mammalian SF3b (16, 18, 44). Rse1p is associated with U2 snRNA in a salt-sensitive fashion and is required for prespliceosome formation, properties expected of a yeast SF3b subunit (16). The sum total of the predicted masses of the proposed yeast SF3b subunits Cus1p, Hsh49p, Hsh155p, and Rse1p is about 336 kDa, a value consistent with the migration of cus1-3p-complementing activity by gel exclusion chromatography in 0.5 M KCl (Fig. 6). The limited resolution of the analysis thus far does not allow us to determine whether other proteins are also present in the functional Cus1p complex.
Relationship of the Cus1p complex to mammalian SF3b.
The splicing factor SF3b binds to 12S U2 snRNP to create the 15S U2 snRNP, which is then competent to bind the SF3a complex to create the 17S U2 snRNP, the form that participates in spliceosome assembly (8, 12, 30). Human SF3b (hSF3b) contains four major proteins: SAP155, SAP130, SAP145, and SAP49 (29). The corresponding proteins of ySF3b are Hsh155p, Rse1p, Cus1p, and Hsh49p (references 16, 19, 26, 29, 37, 41, and 43 and present study). Similarity in the amino acid sequences between the corresponding human and yeast proteins varies for the different subunits and ranges from 50 to 25% identity and 70 to 40% similarity, indicating that some of the subunits are more constrained than others to maintain the function of the complex. The human 17S U2 snRNP forms under low-salt conditions and contains nine additional proteins that the high-salt-stable 12S U2 snRNP lacks, and four of these represent the SF3b complex (8). In yeast extracts, complexes containing at least Cus1p, Hsh155p, and U2 snRNA (Fig. 2) or Rse1p and U2 snRNA (16) are detectable under low (150 mM)-salt conditions, whereas under high-salt conditions, the U2 RNA cannot be detected. Furthermore, the functional Cus1p complex migrates away from U2 snRNA on gel exclusion columns under high-salt conditions (Fig. 6) but must depend on U2 snRNA to participate in spliceosome assembly (Fig. 2) during active splicing in reconstituted extracts (Fig. 6). Thus, the association of SF3b with U2 snRNA is salt labile in both systems. Once assembled into spliceosomes, the hSF3b proteins remain near the pre-mRNA sequences until after the second catalytic step of splicing, as assayed by cross-linking (9, 19, 38). In yeast, Cus1p remains bound to pre-mRNA until after the second step, as assayed by coimmunoprecipitation of substrate RNA (Fig. 2C). Cross-linking of Hsh155p to sequences near the branch site of a yeast pre-mRNA is also observed following prespliceosome formation (D. S. McPheeters and P. Muhlenkamp, unpublished data). Thus, in both systems, the SF3b subunits appear to become integral components of the spliceosome once the U2 snRNP is recruited to the branch point.
Salt sensitivity may seem simply a convenient phenomenon by which to categorize the behavior of protein factors; however, its conservation in two diverse systems may betray a deeper biological significance. A specific association between factors that can be broken by raising the salt slightly above physiological levels may indicate that the association itself may be physiologically dynamic. If such an association can be controlled by small shifts in ionic strength, then it may be controlled physiologically by protein modification, for example, phosphorylation. If there is a functional reason for controlling the association of SF3b with U2 snRNP, it is sufficiently important to have been conserved in evolution. Future experiments to address how and why the association of SF3b with U2 snRNP may be controlled are needed.
ACKNOWLEDGMENTS
We thank Haller Igel for sequencing the cus1-3 allele. We also thank Rhonda Perriman for helpful technical suggestions and Rhonda Perriman and Marc Spingola for critical comments on the manuscript.
This work was supported by a Department of Education Graduate Fellowship to M.H.P. and by National Institutes of Health grants GM40478 (to M.A.) and GM52310 (to D.S.M.).
REFERENCES
- 1.Abovich N, Legrain P, Rosbash M. The yeast PRP6 gene encodes a U4/U6 small nuclear ribonucleoprotein particle (snRNP) protein, and the PRP9 gene encodes a protein required for U2 snRNP binding. Mol Cell Biol. 1990;10:6417–6425. doi: 10.1128/mcb.10.12.6417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Abovich N, Liao X C, Rosbash M. The yeast MUD2 protein: an interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- 3.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 4.Arenas J E, Abelson J N. The Saccharomyces cerevisiae PRP21 gene product is an integral component of the prespliceosome. Proc Natl Acad Sci USA. 1993;90:6771–6775. doi: 10.1073/pnas.90.14.6771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ares M, Jr, Igel A H. Lethal and temperature-sensitive mutations and their suppressors identify an essential structural element in U2 small nuclear RNA. Genes Dev. 1990;4:2132–2145. doi: 10.1101/gad.4.12a.2132. [DOI] [PubMed] [Google Scholar]
- 6.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1987. [Google Scholar]
- 7.Bai C, Elledge S J. Gene identification using the yeast two-hybrid system. Methods Enzymol. 1996;273:331–347. doi: 10.1016/s0076-6879(96)73029-x. [DOI] [PubMed] [Google Scholar]
- 8.Behrens S E, Tyc K, Kastner B, Reichelt J, Luhrmann R. Small nuclear ribonucleoprotein (RNP) U2 contains numerous additional proteins and has a bipartite RNP structure under splicing conditions. Mol Cell Biol. 1993;13:307–319. doi: 10.1128/mcb.13.1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bennett M, Michaud S, Kingston J, Reed R. Protein components specifically associated with prespliceosome and spliceosome complexes. Genes Dev. 1992;6:1986–2000. doi: 10.1101/gad.6.10.1986. [DOI] [PubMed] [Google Scholar]
- 10.Bennett M, Pinol-Roma S, Staknis D, Dreyfuss G, Reed R. Differential binding of heterogeneous nuclear ribonucleoproteins to mRNA precursors prior to spliceosome assembly in vitro. Mol Cell Biol. 1992;12:3165–3175. doi: 10.1128/mcb.12.7.3165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brent R, Finley R L., Jr Understanding gene and allele function with two-hybrid methods. Annu Rev Genet. 1997;31:663–704. doi: 10.1146/annurev.genet.31.1.663. [DOI] [PubMed] [Google Scholar]
- 12.Brosi R, Groning K, Behrens S E, Luhrmann R, Kramer A. Interaction of mammalian splicing factor SF3a with U2 snRNP and relation of its 60-kD subunit to yeast PRP9. Science. 1993;262:102–105. doi: 10.1126/science.8211112. [DOI] [PubMed] [Google Scholar]
- 13.Brosi R, Hauri H P, Kramer A. Separation of splicing factor SF3 into two components and purification of SF3a activity. J Biol Chem. 1993;268:17640–17646. [PubMed] [Google Scholar]
- 14.Cadwell R C, Joyce G F. Mutagenic PCR. PCR Methods Appl. 1994;3:S136–S140. doi: 10.1101/gr.3.6.s136. [DOI] [PubMed] [Google Scholar]
- 15.Caspary F, Seraphin B. The yeast U2A′/U2B complex is required for pre-spliceosome formation. EMBO J. 1998;17:6348–6358. doi: 10.1093/emboj/17.21.6348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Caspary F, Shevchenko A, Wilm M, Seraphin B. Partial purification of the yeast U2 snRNP reveals a novel yeast pre-mRNA splicing factor required for pre-spliceosome assembly. EMBO J. 1999;18:3463–3474. doi: 10.1093/emboj/18.12.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chapon C, Legrain P. A novel gene, spp91-1, suppresses the splicing defect and the pre-mRNA nuclear export in the prp9-1 mutant. EMBO J. 1992;11:3279–3288. doi: 10.1002/j.1460-2075.1992.tb05406.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Das B K, Xia L, Palandjian L, Gozani O, Chyung Y, Reed R. Characterization of a protein complex containing spliceosomal proteins SAPs 49, 130, 145, and 155. Mol Cell Biol. 1999;19:6796–6802. doi: 10.1128/mcb.19.10.6796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gozani O, Feld R, Reed R. Evidence that sequence-independent binding of highly conserved U2 snRNP proteins upstream of the branch site is required for assembly of spliceosomal complex A. Genes Dev. 1996;10:233–243. doi: 10.1101/gad.10.2.233. [DOI] [PubMed] [Google Scholar]
- 20.Gozani O, Patton J G, Reed R. A novel set of spliceosome-associated proteins and the essential splicing factor PSF bind stably to pre-mRNA prior to catalytic step II of the splicing reaction. EMBO J. 1994;13:3356–3367. doi: 10.1002/j.1460-2075.1994.tb06638.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guthrie C, Fink G. Guide to yeast genetics and molecular biology. San Diego, Calif: Academic Press, Inc.; 1991. [Google Scholar]
- 22.Harlow E, Lane D. Antibodies: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1988. [Google Scholar]
- 23.Harper J W, Adami G R, Wei N, Keyomarsi K, Elledge S J. The p21 Cdk-interacting protein Cip1 is a potent inhibitor of G1 cyclin-dependent kinases. Cell. 1993;75:805–816. doi: 10.1016/0092-8674(93)90499-g. [DOI] [PubMed] [Google Scholar]
- 24.Hartwell L H. Macromolecule synthesis in temperature-sensitive mutants of yeast. J Bacteriol. 1967;93:1662–1670. doi: 10.1128/jb.93.5.1662-1670.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Higgins D G, Thompson J D, Gibson T J. Using CLUSTAL for multiple sequence alignments. Methods Enzymol. 1996;266:383–402. doi: 10.1016/s0076-6879(96)66024-8. [DOI] [PubMed] [Google Scholar]
- 26.Igel H, Wells S, Perriman R, Ares M., Jr Conservation of structure and subunit interactions in yeast homologues of splicing factor 3b (SF3b) subunits. RNA. 1998;4:1–10. [PMC free article] [PubMed] [Google Scholar]
- 27.Jones E W. Tackling the protease problem in Saccharomyces cerevisiae. Methods Enzymol. 1991;194:428–453. doi: 10.1016/0076-6879(91)94034-a. [DOI] [PubMed] [Google Scholar]
- 28.Kramer A. Presplicing complex formation requires two proteins and U2 snRNP. Genes Dev. 1988;2:1155–1167. doi: 10.1101/gad.2.9.1155. [DOI] [PubMed] [Google Scholar]
- 29.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 30.Kramer A, Gruter P, Groning K, Kastner B. Combined biochemical and electron microscopic analyses reveal the architecture of the mammalian U2 snRNP. J Cell Biol. 1999;145:1355–1368. doi: 10.1083/jcb.145.7.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kunkel T A, Bebenek K, McClary J. Efficient site-directed mutagenesis using uracil-containing DNA. Methods Enzymol. 1991;204:125–139. doi: 10.1016/0076-6879(91)04008-c. [DOI] [PubMed] [Google Scholar]
- 32.Longtine M S, McKenzie A R, Demarini D J, Shah N G, Wach A, Brachat A, Philippsen P, Pringle J R. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 33.McPheeters D S, Fabrizio P, Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes Dev. 1989;3:2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- 34.Phizicky E M, Fields S. Protein-protein interactions: methods for detection and analysis. Microbiol Rev. 1995;59:94–123. doi: 10.1128/mr.59.1.94-123.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Riedel N, Wise J A, Swerdlow H, Mak A, Guthrie C. Small nuclear RNAs from Saccharomyces cerevisiae: unexpected diversity in abundance, size, and molecular complexity. Proc Natl Acad Sci USA. 1986;83:8097–8101. doi: 10.1073/pnas.83.21.8097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ruby S W, Chang T H, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- 37.Schmidt-Zachmann M S, Knecht S, Kramer A. Molecular characterization of a novel, widespread nuclear protein that colocalizes with spliceosome components. Mol Biol Cell. 1998;9:143–160. doi: 10.1091/mbc.9.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staknis D, Reed R. Direct interactions between pre-mRNA and six U2 small nuclear ribonucleoproteins during spliceosome assembly. Mol Cell Biol. 1994;14:2994–3005. doi: 10.1128/mcb.14.5.2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Staley J P, Guthrie C. Mechanical devices of the spliceosome: motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- 40.Vijayraghavan U, Company M, Abelson J. Isolation and characterization of pre-mRNA splicing mutants of Saccharomyces cerevisiae. Genes Dev. 1989;3:1206–1216. doi: 10.1101/gad.3.8.1206. [DOI] [PubMed] [Google Scholar]
- 41.Wang C, Chua K, Seghezzi W, Lees E, Gozani O, Reed R. Phosphorylation of spliceosomal protein SAP 155 coupled with splicing catalysis. Genes Dev. 1998;12:1409–1414. doi: 10.1101/gad.12.10.1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wells S E, Ares M., Jr Interactions between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wells S E, Neville M, Haynes M, Wang J, Igel H, Ares M., Jr CUS1, a suppressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- 44.Will C L, Schneider C, Reed R, Luhrmann R. Identification of both shared and distinct proteins in the major and minor spliceosomes. Science. 1999;284:2003–2005. doi: 10.1126/science.284.5422.2003. [DOI] [PubMed] [Google Scholar]
- 45.Yan D, Perriman R, Igel H, Howe K J, Neville M, Ares M., Jr CUS2, a yeast homolog of human Tat-SF1, rescues function of misfolded U2 through an unusual RNA recognition motif. Mol Cell Biol. 1998;18:5000–5009. doi: 10.1128/mcb.18.9.5000. [DOI] [PMC free article] [PubMed] [Google Scholar]