Abstract
Aging is a time-dependent complex biological process of organisms with gradual deterioration of the anatomical and physiological functions. The role of gut microbiota is inevitable in the aging process. Probiotic interventions improve gut homeostasis and support healthy aging by enhancing beneficial species and microbial biodiversity in older adults. The present preliminary clinical trial delves into the impact of an 8-week Lactobacillus rhamnosus intervention (10 × 109 CFU per day) on the glycaemic index, lipid profile, and microbiome of elderly subjects. Body weight, body fat, fasting blood glucose, total cholesterol, triglyceride, high-density lipoprotein, and low-density lipoprotein (LDL) are assessed at baseline (Week 0) and after treatment (Week 8) in placebo and probiotic groups. Gaussian regression analysis highlights a significant improvement in LDL cholesterol in the probiotic group (p = 0.045). Microbiome analysis reveals numeric changes in taxonomic abundance at various levels. At the phylum level, Proteobacteria increases its relative frequency (RF) from 14.79 ± 5.58 at baseline to 23.46 ± 8.02 at 8 weeks, though statistically insignificant (p = 0.100). Compared to the placebo group, probiotic supplementations significantly increased the proteobacteria abundance. Genus-level analysis indicates changes in the abundance of several microbes, including Escherichia-Shigella, Akkermansia, and Bacteroides, but only Butyricimonas showed a statistically significant level of reduction in its abundance. Probiotic supplementations significantly altered the Escherichia-Shigella and Sutterella abundance compared to the placebo group. At the species level, Bacteroides vulgatus substantially increases after probiotic treatment (p = 0.021). Alpha and beta diversity assessments depict subtle shifts in microbial composition. The study has limitations, including a small sample size, short study duration, single-strain probiotic use, and lack of long-term follow-up. Despite these constraints, the study provides valuable preliminary insights into the multifaceted impact of L. rhamnosus on elderly subjects. Further detailed studies are required to define the beneficial effect of L. rhamnosus on the health status of elderly subjects.
Keywords: Lactobacillus rhamnosus, aging, gut microbiota, probiotics, lipid profile, microbiome
1. Introduction
Aging is cellular senescence, a complex biological process with progressive decline in anatomical and physiological functions of multiple cells and tissues. The irreversible aging process is genetically marked and regulated by genetic and environmental factors [1]. The accumulated cellular senescence effects during aging produce damage and disturb somatic maintenance, which could cause certain cellular and molecular distractions leading to genetic instability, epigenetic alterations, mitochondrial dysfunction, proteostasis loss, stem cell exhaustion, cellular senescence, deregulation of nutrient sensing, and so on [2]. The intestine has been recognized as a crucial organ because age-related pathologies are associated with gut microbiota imbalances and gut-associated immune systems. The gut microbiota forms a biological ecosystem with trillions of bacteria and fungi and maintains the host’s health and energy homeostasis. The composition of gut microbiota is relatively stable throughout adult life, designs the health conditions of the host by balancing the pathogens, nutritional status, and energy expenditure, and substantially changes with aging and results in age-associated diseases [3]. The microbial imbalance causes cellular senescence, wherein the cells undergo irreversible growth arrest and accumulate senescent cells during aging due to the development of a senescent-associated secretory phenotype. This comprises the secretion of pro-inflammatory cytokines, growth factors, chemokines, proteases, and extracellular matrix components [4]. The accumulated senescent cells in the skin cause dermatological disorders mediated by the gut–skin axis influenced by the gut microbiota and their metabolites [4]. The mutual relationship between the gut microbiota and the human host helps to protect against the colonization of pathogenic bacteria, metabolic activities, synthesizing bioactive peptide compounds, vitamins, and hormones, and managing the immune system [5,6].
The gut microbiota is stable throughout adulthood and fluctuates with aging and disease conditions [3]. The gut microbial composition changes in the elderly population compared to adults due to various factors such as the aging process, nutritional habits, lifestyle, stress, reduced intestinal functions [3,7], changes in immune responses, lack of physical activity, infections, use of medications, and hospitalization [8,9,10]. Generally, the gut microbiota in adults mostly comprises Firmicutes and Bacteroides and smaller populations of Actinobacteria, Proteobacteria, and Verrucomicrobia [11]. Aged peoples’ gut microbiota is abundant with Bacteroides and Proteobacteria with a decrease in Firmicutes and Bifidobacteria [9,12,13]. Elderly subjects have reduced bacterial diversity and beneficial bacteria. Also, the elderly subjects have lower Bifidobacteria and Lactobacilli levels and increased Enterobacteriaceae and Clostridia levels than young subjects [14,15]. Older adults showed a reduced proportion of butyrate-producing Clostridium cluster XIVa, Roseburia and Ruminococcus [16]. In long-living people, their intestinal microbiota abundantly presented with Akkermansia, Bifidobacterium, and Christensenellaceae [17,18]. In elderly subjects, the gut microbial composition changes, and the microbial diversity is reduced because of various factors such as the accumulation of pro-inflammatory microbes and reduced beneficial microbes [19], aging, nutritional and lifestyle changes, decreased gut functions, and stress [3]. The age-dependent changes in the gut microbiota mean the gradual changes in the microbial diversity involving the decline in the abundance of core dominant microbial species like Bifidobacteria counts and the ratio of Firmicutes to Bacteroides and the increase in the abundance of sub-dominant microbial species like Proteobacteria [17,20].
During aging, the changes in lipid metabolism and their metabolite levels cause an increase in body adiposity. Excess adiposity can cause lipid toxicity, leading to cardiovascular diseases, arthritis, cancer, type 2 diabetes, and Alzheimer’s disease [21]. In humans, with an increase in age, the blood glyceride levels tend to increase, and the white blood lysophosphatidylcholine levels decrease. Human longevity is associated with specific sphingolipid and phospholipid blood profile changes. Furthermore, certain lipids and lipid-related compounds have been found to change depending on age. Lipid-related interventions in various model organisms are capable of modulating the lifespan. It shows that lipid metabolism is linked to the aging process and may enhance the health span. Blood lipids and lipid-related molecules can be biomarkers for human aging studies [22]. Lactic acid bacteria (LAB) and bifidobacteria are found commonly in the gut of humans, with potential physiological benefits, such as enhancing gut function and increasing the uptake of micronutrients, lowering cholesterol, and protecting the gastrointestinal tract from infection by modulating immune functions [23]. It is well-known that an individual’s gut microbiota is shaped right from the gestational stage. Increasing evidence states that probiotic interventions and dietary patterns significantly influence or modulate age-associated changes in gut microbiota and immune functions, promoting healthy aging outcomes [24]. Although several clinical studies evaluated every aspect of aging consequences, anti-aging, or healthy aging properties, the effect of each probiotic strain widely varies on gut composition, immune function, and metabolite synthesis. Certain other studies investigated the involvement of probiotics in gut health, cognitive functions, lipid metabolism, and other biomarkers. In this study, we evaluated the impact of Lactobacillus rhamnosus interventions on glycaemic indexes, lipid profiles, and differences in the microbial diversity of healthy Thai elderly subjects.
2. Materials and Methods
The study protocols were approved by the Ethical Committee of Ubon Ratchathani University (Code: UBU-REC-44/2564). The participants’ consent was obtained before the study.
2.1. Study Group
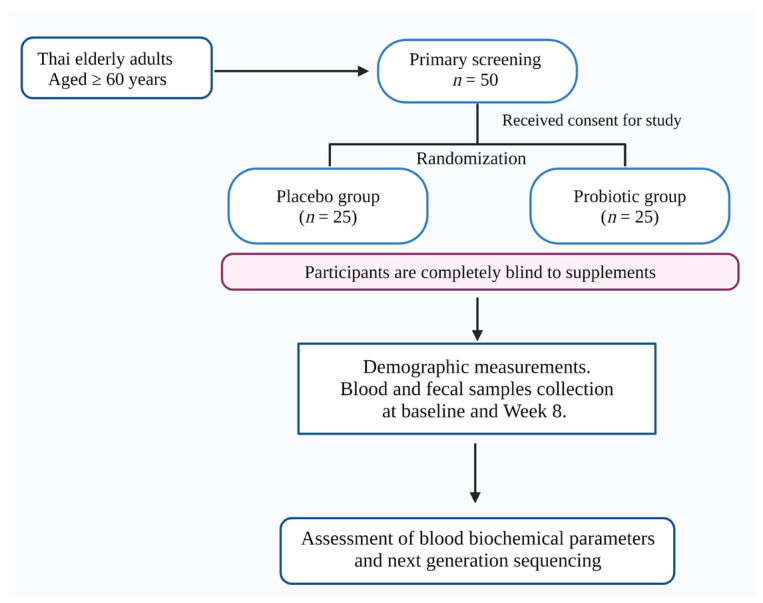
Thai elderly adults aged ≥ 60 who were willing to participate and complete the study were included. Participants under any other medications or taking probiotics in the previous 2 weeks were omitted from the study. After this primary screening, participants (n = 50; male =12; female = 38) were included in the study. The participants were randomly allotted into placebo (n = 25) and probiotic (n = 25) groups. Participants were completely blind to the supplements. The samples (blood and fecal) were gathered from the study subjects at baseline (week 0) and after 8 weeks. Participants were asked to follow the allocated follow-up visits without absence. The changes in the glycemic, lipid parameters and microbial composition were studied. The study protocol is illustrated in Figure 1.
Figure 1.
The illustration shows the study protocol (created using Biorender.com; accessed on 4 March 2024).
2.2. Probiotics Supplementation
Aluminum foil sachets containing 10 × 109 CFU of Lactobacillus rhamnosus received from Lactomason Co., Ltd. (Gyeongsangnam-do, Republic of Korea) were provided to the participants in the probiotic group. The placebo group participants were given a 10 g cornstarch sachet. All participants were instructed to take the supplement regularly by dissolving the contents of one sachet in a glass of water before breakfast. Instructions were given to store the sachet in a dry place at 2–8 °C. Participants were encouraged not to change their physical activities, nutrition, or lifestyle. The participants were advised to avoid the intake of any other probiotics, dietary supplements, or fermented food throughout the study.
2.3. Demographic Assessments
After assigning the participants to respective groups, the sociodemographic characteristics, including age, gender, smoking, alcohol drinking, and body weight, were recorded. Body and visceral fat were manually noted using an electronic scale (Picooc®, Model S1 Pro, Beijing, China) (Table 1). The blood parameters, including triglyceride, fasting blood glucose (FBS), total cholesterol (TC), high-density lipoprotein-cholesterol (HDL-C), and low-density lipoprotein-cholesterol (LDL-C), were determined from blood using the automated machine at AMS Clinical Service Center, Chiang Mai University, Chiang Mai, Thailand.
Table 1.
Sociodemographic data of the study participants.
| No. | Variables | Group | p-Value | |
|---|---|---|---|---|
| Placebo (n = 25) | Probiotic (n = 25) | |||
| 1 | Age | 64.96 ± 0.86 | 63.00 ± 1.09 | 0.165 a |
| 2 | Male, n (%) | 3 (12.00) | 9 (36.00) | 0.095 b |
| Female, n (%) | 22 (88.00) | 16 (64.00) | ||
| 3 | Smoking | 0.490 b | ||
| No, n (%) | 25 (100.00) | 23 (92.00) | ||
| Yes, n (%) | 0 (0.00) | 2 (8.00) | ||
| 4 | Alcohol drinking | 0.235 b | ||
| No, n (%) | 25 (100.00) | 22 (88.00) | ||
| Yes, n (%) | 0 (0.00) | 3 (12.00) | ||
| 5 | Body weight (kg) | 63.80 ± 2.29 | 59.40 ± 2.17 | 0.170 a |
| 6 | Body fat (%) | 32.77 ± 1.41 | 27.76 ± 1.45 | 0.036 *a |
| 7 | Visceral fat (%) | 15.18 ± 0.57 | 13.55 ± 1.49 | 0.060 c |
| 8 | FBS (mg/dL) | 106.12 ± 8.77 | 104.00 ± 5.97 | 0.843 a |
| 9 | TC (mg/dL) | 211.68 ± 9.41 | 209.12 ± 8.55 | 0.841 a |
| 10 | TG (mg/dL) | 134.48 ± 11.20 | 157.36 ± 12.49 | 0.179 a |
| 11 | HDL (mg/dL) | 53.04 ± 1.95 | 52.72 ± 3.09 | 0.931 a |
| 12 | LDL (mg/dL) | 131.08 ± 8.35 | 120.01 ± 6.22 | 0.293 a |
FBS: fasting blood sugar; TC: total cholesterol; TG: triglycerides; HDL: high-density lipoprotein; LDL: low-density lipoprotein. Data are mean ± SE. * indicates the significant difference in p-value at a 95% confidence interval. a p-value from the independent t-test, b p-value from Fischer’s exact test, c p-value from the Mann–Whitney U test.
2.4. Next-Generation Sequencing (NGS)
The fecal genomic DNA was isolated, and DNA sequencing was conducted by the Omics Sciences and Bioinformatics Center, Faculty of Science, Chulalongkorn University, Thailand [25]. Due to the lack of quality DNA, we conducted NGS for only 12 samples each for the placebo and probiotic groups.
2.5. Statistical Analyses
The data are shown as the mean ± standard error of the mean (SEM) for continuous outcomes or as an absolute number and percentage for categorical outcomes. A paired t-test was performed for normally distributed data. The Wilcoxon signed-rank test analyzed the skewed data. The differentiation of outcomes between the placebo and probiotic groups was compared using a Mann–Whitney U test. The p-value < 0.05 was set as significant (two-tailed). The STATA version 15.1 for Windows (StataCorp, College Station, TX, USA) was utilized for statistical analysis.
3. Results
3.1. Changes in Biochemical Parameters
The basic parameters like body weight, body fat, visceral fat, and blood biochemical parameters like FBS, TC, TG, HDL, and LDL were measured at baseline and week 8 of the study. There were no significant differences in glycaemic or lipid parameters between the baseline and after treatment in the placebo group (Table 2). The marginal differences were observed in FBS, TG, HDL, and LDL after treatment in probiotic groups. These changes can be considered the initial state of functional changes due to the probiotic intervention. However, more duration is required to achieve significant effects in these parameters.
Table 2.
Biochemical parameters at baseline (week 0) and treatment (week 8) in the placebo and probiotic groups.
| Parameters | Placebo (n = 25) | p-Value | Probiotic (n = 25) | p-Value | ||
|---|---|---|---|---|---|---|
| Baseline (P-Week 0) |
Treatment (P-Week 8) |
Baseline (T-Week 0) |
Treatment (T-Week 8) |
|||
| Body weight (kg) | 63.80 ± 2.29 | 63.74 ± 2.30 | 0.884 a | 59.40 ± 2.17 | 58.73 ± 2.11 | 0.058 a |
| Body fat (%) | 32.77 ± 1.41 | 34.95 ± 1.53 | 0.127 a | 27.76 ± 1.45 | 27.28 ± 1.67 | 0.657 a |
| Visceral fat (%) | 15.18 ± 0.57 | 15.27 ± 0.66 | 0.402 b | 13.55 ± 1.49 | 12.09 ± 1.13 | 0.098 b |
| FBS (mg/dL) | 106.12 ± 8.77 | 109.92 ± 6.95 | 0.504 a | 104.00 ± 5.97 | 106.96 ± 5.84 | 0.375 a |
| TC (mg/dL) | 211.68 ± 9.41 | 201.96 ± 11.98 | 0.178 a | 209.12 ± 8.55 | 194.76 ± 7.63 | 0.055 a |
| TG (mg/dL) | 134.48 ± 11.20 | 149.80 ± 13.62 | 0.184 a | 157.36 ± 12.49 | 149.48 ± 12.62 | 0.463 a |
| HDL (mg/dL) | 53.04 ± 1.95 | 53.52 ± 3.55 | 0.858 a | 52.72 ± 3.09 | 56.28 ± 3.46 | 0.137 a |
| LDL (mg/dL) | 131.08 ± 8.35 | 128.42 ± 8.46 | 0.728 a | 120.01 ± 6.22 | 114.57 ± 8.00 | 0.439 a |
P-week 0: Placebo-week 0; P-week 8: Placebo-week 8; T-week 0: Treatment-week 0; T-week 8: Treatment-week 8; Baseline-Week 0; Treatment-Week 8; TC: total cholesterol; TG: triglycerides; HDL: high-density lipoprotein; LDL: low-density lipoprotein. Data are mean ± SE. a p-value from Paired t-test, b p-value from the Wilcoxon signed-rank test.
The statistical comparison of the considered parameters between the treatment and placebo groups was detailed (Table 3). The body weight (reduced; placebo (−0.06); probiotic (−0.68)), body fat (reduced; probiotic (−0.48)), visceral fat (reduced; probiotic (−1.45)), FBS (reduced; probiotic (−2.96)), and TC (reduced; probiotic (−14.36)), TG (reduced; probiotic (−7.88)), and LDL (reduced; probiotic (−5.44)) values were changed after the treatment in the placebo and probiotic groups, but not statistically significant (Table 3).
Table 3.
The differentiation of biochemical parameters between the placebo and probiotic groups at the end of the study.
| Variables | Difference | p-Value * | |
|---|---|---|---|
| Placebo (n = 25) | Probiotic (n = 25) | ||
| Body weight (kg) | −0.06 | −0.68 | 0.252 |
| Body fat (%) | 2.17 | −0.48 | 0.163 |
| Visceral fat (%) | 0.09 | −1.45 | 0.104 |
| FBS (mg/dL) | 3.80 | −2.96 | 0.734 |
| TC (mg/dL) | −9.72 | −14.36 | 0.869 |
| TG (mg/dL) | 15.32 | −7.88 | 0.479 |
| HDL (mg/dL) | 0.48 | 3.56 | 0.214 |
| LDL (mg/dL) | −2.66 | −5.44 | 0.823 |
FBS: Fasting blood sugar; TC: total cholesterol; TG: triglycerides; HDL: high-density lipoprotein; LDL: low-density lipoprotein. Data are mean. * p-value from the Mann–Whitney U test.
L. rhamnosus supplementation for 8 weeks substantially improved LDL (−69.85 to −0.79; p = 0.045) (Table 4). Besides LDL, no other parameters showed significant differences after 8 weeks of study.
Table 4.
Gaussian regression analysis of the probiotic treatment group after 8 weeks of study.
| Parameters | Coefficient | 95% Confidence Interval | p-Value |
|---|---|---|---|
| Body weight (kg) | −0.65 | −2.24 to 0.94 | 0.409 |
| Body fat (%) | −2.48 | −6.48 to 1.52 | 0.214 |
| Visceral fat (%) | −0.39 | −2.14 to 1.36 | 0.650 |
| FBS (mg/dL) | −5.19 | −22.15 to 11.78 | 0.536 |
| Total cholesterol (mg/dL) | −4.99 | −28.91 to 18.93 | 0.675 |
| Triglyceride (mg/dL) | −25.07 | −74.60 to 24.47 | 0.308 |
| HDL (mg/dL) | 9.76 | −2.43 to 21.94 | 0.112 |
| LDL (mg/dL) | −35.32 | −69.85 to −0.79 | 0.045 * |
FBS: Fasting blood sugar; TC: total cholesterol; TG: triglycerides; HDL: high-density lipoprotein; LDL: low-density lipoprotein. Data are mean. * indicates the significant difference in p-value at a 95% confidence interval.
3.2. Microbiome Analysis
The valid sequences were identified by matching the raw sequences with the corresponding barcode. The raw-sequence tags underwent analysis using QIIME 2™. Following chimera detection, the sequences were grouped into operational taxonomic units (OTUs) at a 97% sequence identity threshold. Quality assessment of pair-end reads was conducted by examining information in FASTQ files using DADA2. Reads failing to meet the default QIIME 2™ threshold values were filtered out, including a minimum quality score of 25, minimum/maximum length requirements of 200/1000, prohibition of ambiguous bases, and absence of mismatches in the primer sequence. The details of processed sequence reads are detailed in Table 5.
Table 5.
The 16s rRNA amplicon sequences of the placebo (Baseline: PW0, After-treatment: PW8) and probiotics (Baseline: TW0, After-treatment: TW8) groups.
| Sample-ID | Input | Filtered | Denoised | Merged | Non-Chimeric |
|---|---|---|---|---|---|
| Placebo group | |||||
| PW0-1-02 | 228,670 | 156,586 | 155,663 | 152,508 | 84,553 |
| PW0-1-13 | 76,716 | 50,519 | 49,906 | 48,821 | 41,576 |
| PW0-1-15 | 121,423 | 88,254 | 87,612 | 85,709 | 52,312 |
| PW0-1-24 | 65,607 | 49,262 | 48,994 | 48,202 | 28,559 |
| PW0-1-35 | 72,981 | 50,936 | 50,576 | 49,171 | 36,557 |
| PW0-1-36 | 121,096 | 82,249 | 81,562 | 79,976 | 68,897 |
| PW0-1-39 | 48,332 | 34,305 | 34,012 | 33,225 | 25,337 |
| PW0-1-42 | 83,999 | 62,262 | 61,820 | 60,451 | 40,175 |
| PW0-1-43 | 62,702 | 43,389 | 42,818 | 41,423 | 30,313 |
| PW0-1-63 | 64,517 | 54,975 | 54,789 | 53,894 | 35,429 |
| PW0-1-69 | 69,463 | 57,120 | 56,965 | 56,297 | 42,940 |
| PW0-1-78 | 82,033 | 67,793 | 67,539 | 66,816 | 48,736 |
| PW8-2-63 | 34,397 | 14,825 | 14,708 | 14,531 | 11,610 |
| PW8-2-69 | 41,790 | 19,358 | 19,147 | 18,877 | 13,859 |
| PW8-2-78 | 54,624 | 25,338 | 25,053 | 24,421 | 18,908 |
| PW8-3-02 | 99,997 | 68,321 | 67,637 | 66,196 | 46,325 |
| PW8-3-13 | 65,990 | 46,652 | 46,327 | 45,632 | 36,879 |
| PW8-3-15 | 50,672 | 32,262 | 31,961 | 31,489 | 24,307 |
| PW8-3-24 | 128,327 | 93,300 | 92,849 | 90,848 | 70,533 |
| PW8-3-35 | 50,172 | 36,302 | 35,955 | 34,840 | 25,906 |
| PW8-3-36 | 88,839 | 64,988 | 64,507 | 63,085 | 51,453 |
| PW8-3-39 | 84,642 | 60,545 | 60,065 | 59,023 | 44,259 |
| PW8-3-42 | 264,836 | 208,317 | 207,379 | 203,133 | 145,732 |
| PW8-3-43 | 100,425 | 74,396 | 73,846 | 72,279 | 52,322 |
| Probiotic group | |||||
| TW0-1-22 | 138,660 | 101,667 | 101,218 | 99,829 | 74,148 |
| TW0-1-23 | 108,785 | 78,702 | 77,922 | 76,382 | 51,645 |
| TW0-1-26 | 148,589 | 111,060 | 110,479 | 108,555 | 71,637 |
| TW0-1-30 | 145,969 | 108,395 | 107,827 | 106,324 | 79,072 |
| TW0-1-33 | 124,551 | 100,196 | 99,962 | 99,237 | 62,441 |
| TW0-1-4 | 67,800 | 48,002 | 47,444 | 46,493 | 36,199 |
| TW0-1-101 | 92,779 | 77,626 | 77,267 | 76,435 | 65,627 |
| TW0-1-28 | 123,207 | 98,275 | 97,849 | 95,999 | 57,353 |
| TW0-1-44 | 93,555 | 76,108 | 75,727 | 74,927 | 57,845 |
| TW0-1-64 | 79,342 | 63,330 | 63,010 | 62,273 | 45,977 |
| TW0-1-87 | 80,502 | 65,053 | 64,689 | 63,708 | 46,194 |
| TW0-1-93 | 85,017 | 67,246 | 67,043 | 66,112 | 54,043 |
| TW8-2-101 | 29,904 | 15,109 | 14,925 | 14,678 | 13,074 |
| TW8-2-28 | 37,619 | 18,604 | 18,249 | 17,977 | 14,104 |
| TW8-2-44 | 39,047 | 19,011 | 18,854 | 18,650 | 15,916 |
| TW8-2-64 | 35,484 | 17,170 | 17,017 | 16,849 | 13,546 |
| TW8-2-87 | 29,232 | 13,553 | 13,405 | 13,298 | 9193 |
| TW8-2-93 | 51,548 | 25,060 | 24,826 | 24,404 | 19,697 |
| TW8-3-22 | 80,475 | 57,923 | 57,475 | 56,476 | 49,362 |
| TW8-3-23 | 88,815 | 66,880 | 66,592 | 65,661 | 50,791 |
| TW8-3-26 | 240,804 | 181,521 | 180,569 | 177,129 | 145,464 |
| TW8-3-30 | 87,286 | 64,381 | 64,027 | 63,254 | 50,862 |
| TW8-3-33 | 137,037 | 94,961 | 94,181 | 92,399 | 81,152 |
| TW8-3-4 | 128,895 | 93,093 | 92,460 | 90,949 | 76,221 |
Changes in the phylum, genus, and species diversity between and within (baseline vs. after treatment) the placebo and probiotics group samples were compared. PW0 and PW8 indicate the baseline and after-treatment samples of the placebo group, respectively. TW0 and TW8 indicate the baseline and after-treatment samples of the probiotic group, respectively.
3.2.1. Alpha Diversity
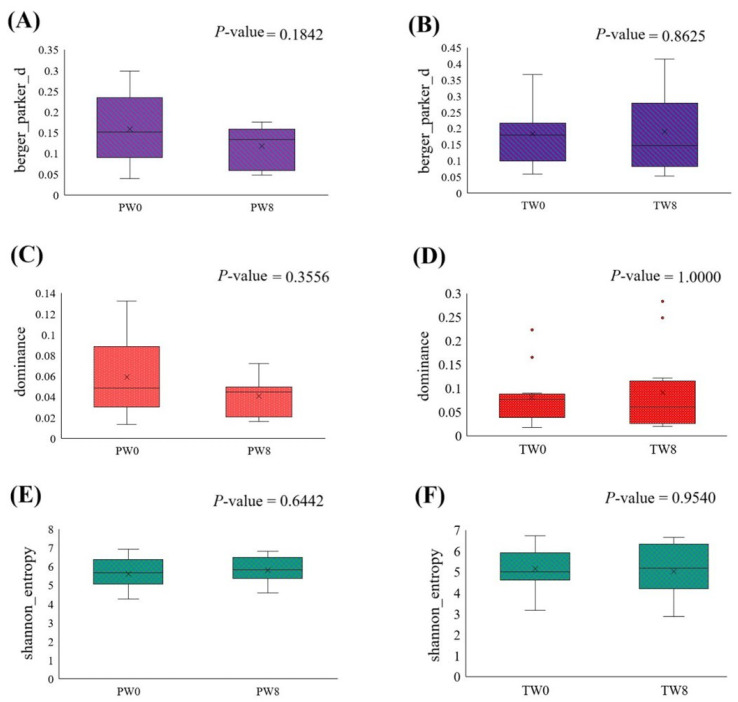
The Shannon diversity index was calculated to evaluate substantial group variations and analyzed using the Kruskal–Wallis (pairwise) test. Berger–Parker analysis was carried out to measure dominance within the community and the proportion of individuals belonging to the most abundant species in the sample. There were no significant changes observed in Berger–Parker analysis (Figure 2A,B), dominance metrics (Figure 2C,D), or Shannon entropy analysis (Figure 2E,F) after treatment in placebo and probiotic groups compared to the respective baseline. The results indicate that, despite observing numerical changes, there was no alteration in microbial abundance or consistency following the treatments.
Figure 2.
Alpha diversity estimation between the baseline and post-treatment of placebo (PW0 and PW8) and probiotic groups (TW0 and TW8). The results of Berger–Parker (A,B) analysis, dominance metrics (C,D), and Shannon entropy (E,F).
3.2.2. Beta Diversity
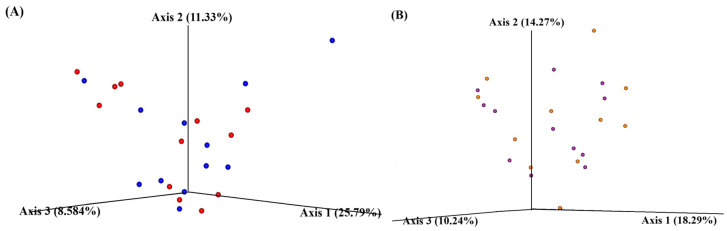
Principal coordinate analysis (PCoA) was employed to assess the relationships between samples, with visualization facilitated by QIIME 2™ View. PCoA plots were generated based on the first three principal coordinates, annotated according to their variance. In the placebo group, 25.79, 11.33, and 8.58% microbial variations were observed in axes 1, 2, and 3, respectively. Similarly, microbial variations of 18.29, 14.27, and 10.24% were observed in axes 1, 2, and 3, respectively, in the probiotic group. The scattered dots, representing baseline and after-treatment samples, indicated that microbial diversity changed after treatments (Figure 3).
Figure 3.
Beta diversity of placebo (A) and probiotic (B) group. The circle dots indicate the outlier samples. The red and blue dots indicate the baseline and treatment samples of the placebo group. The purple and orange dots indicate the baseline and treatment samples of the probiotic group.
3.2.3. Taxonomical Allocations and Determinations
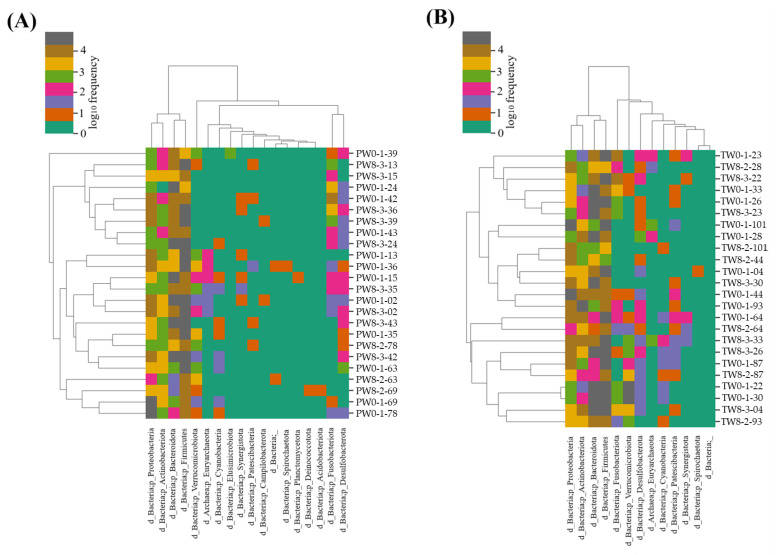
A heat map was generated to illustrate the predicted bacterial taxonomy in the placebo and probiotic samples (Figure 4A,B).
Figure 4.
The heat-map represents the taxonomy estimated for the placebo (A) and probiotic (B) samples after week 0 and week 8 of treatment. The diversity was represented with log10 frequency.
The changes in microbial abundance after probiotic treatment are detailed in Table 6. There were numerical changes in the RF of the microbial abundance at the phylum level, but they were not statistically significant. Similarly, non-significant alteration of microbial abundance was observed at the genus and species level, except Butyricimonas (p = 0.017) and Bacteroides vulgatus (p = 0.021) (Table 6).
Table 6.
The statistical differences in the phylum, genus, and species between the baseline and after-treatment samples in the probiotic group.
| Taxonomy | Baseline (TW0) | After Treatment (TW8) | p-Value |
|---|---|---|---|
| Phyla | |||
| Proteobacteria | 14.79 ± 5.58 | 23.46 ± 8.02 | 0.100 |
| Verrucomicrobiota | 5.54 ± 3.71 | 2.39 ± 1.84 | 0.475 |
| Bacteroidota | 27.77 ± 7.90 | 24.61 ± 6.31 | 0.938 |
| Actinobacteriota | 9.27 ± 4.00 | 6.66 ± 1.84 | 0.875 |
| Firmicutes | 41.09 ± 6.81 | 41.48 ± 6.57 | 0.754 |
| Fusobacteriota | 1.32 ± 0.80 | 1.27 ± 0.80 | 0.969 |
| Desulfobacterota | 0.19 ± 0.07 | 0.11 ± 0.05 | 0.388 |
| Patescibacteria | 0.04 ± 0.02 | 0.01 ± 0.004 | 0.254 |
| Genera | |||
| Escherichia-Shigella | 13.71 ± 5.90 | 21.90 ± 8.63 | 0.117 |
| Akkermansia | 5.63 ± 3.77 | 2.49 ± 1.90 | 0.751 |
| Bacteroides | 16.81 ± 6.76 | 18.21 ± 4.90 | 0.272 |
| Bifidobacterium | 5.80 ± 3.86 | 4.30 ± 1.52 | 0.610 |
| Phascolarctobacterium | 4.66 ± 1.87 | 2.66 ± 1.15 | 0.724 |
| Prevotella | 9.58 ± 3.40 | 4.96 ± 2.78 | 0.609 |
| Faecalibacterium | 2.85 ± 0.74 | 5.75 ± 1.96 | 0.182 |
| Blautia | 5.52 ± 2.78 | 3.77 ± 2.17 | 0.239 |
| Collinsella | 1.70 ± 0.61 | 2.22 ± 0.77 | 0.388 |
| Weissella | 3.89 ± 3.70 | 0.07 ± 0.03 | 0.308 |
| Subdoligranulum | 0.79 ± 0.32 | 1.32 ± 0.48 | 0.638 |
| Agathobacter | 0.41 ± 0.13 | 1.13 ± 0.41 | 0.100 |
| Romboutsia | 0.52 ± 0.21 | 0.39 ± 0.14 | 1.000 |
| Roseburia | 0.72 ± 0.22 | 0.99 ± 0.55 | 0.784 |
| Alistipes | 0.45 ± 0.17 | 0.42 ± 0.36 | 0.325 |
| Paraprevotella | 2.05 ± 1.17 | 2.79 ± 2.40 | 0.305 |
| Streptococcus | 1.43 ± 0.90 | 1.38 ± 0.87 | 0.433 |
| Fusobacterium | 1.14 ± 0.85 | 0.38 ± 0.16 | 0.969 |
| Slackia | 0.84 ± 0.37 | 1.76 ± 1.12 | 0.906 |
| UCG 002 | 0.69 ± 0.17 | 1.14 ± 0.28 | 0.410 |
| Dorea | 1.53 ± 1.24 | 1.09 ± 0.53 | 0.255 |
| Lactobacillus | 0.08 ± 0.02 | 0.39 ± 0.16 | 0.784 |
| Monoglobus | 0.05 ± 0.03 | 1.31 ± 1.18 | 0.365 |
| Parasutterella | 0.57 ± 0.43 | 0.55 ± 0.42 | 0.184 |
| Enterococcus | 0.22 ± 0.08 | 0.64 ± 0.33 | 0.723 |
| Butyricicoccus | 0.91 ± 0.74 | 0.08 ± 0.04 | 0.388 |
| Olsenella | 0.20 ± 0.11 | 0.72 ± 0.32 | 0.305 |
| CAG 352 | 0.81 ± 0.39 | 0.30 ± 0.15 | 0.076 |
| Holdemanella | 0.33 ± 0.17 | 0.35 ± 0.11 | 0.145 |
| Fusicatenibacter | 0.45 ± 0.17 | 0.42 ± 0.36 | 0.583 |
| Parabacteroides | 0.66 ± 0.22 | 0.61 ± 0.21 | 0.969 |
| Coprococcus | 0.28 ± 0.14 | 0.69 ± 0.27 | 0.383 |
| Lachnoclostridium | 0.50 ± 0.15 | 0.30 ± 0.12 | 0.153 |
| Barnesiella | 0.38 ± 0.15 | 0.07 ± 0.04 | 0.178 |
| Odoribacter | 0.32 ± 0.10 | 0.17 ± 0.10 | 0.267 |
| Atopobium | 0.52 ± 0.51 | 0.06 ± 0.05 | 0.344 |
| Megamonas | 0.85 ± 0.35 | 0.39 ± 0.27 | 0.268 |
| Clostridia UCG 014 | 0.09 ± 0.06 | 0.49 ± 0.37 | 0.767 |
| Klebsiella | 1.32 ± 0.63 | 0.48 ± 0.21 | 0.289 |
| Ruminococcus | 0.38 ± 0.24 | 0.24 ± 0.12 | 0.969 |
| Erysipelotrichaceae UCG 003 | 0.31 ± 0.13 | 0.08 ± 0.04 | 0.234 |
| Enterobacteriaceae | 0.35 ± 0.18 | 0.19 ± 0.12 | 0.456 |
| Anaerostipes | 0.08 ± 0.03 | 0.38 ± 0.16 | 0.132 |
| Enterobacter | 0.21 ± 0.07 | 0.31 ± 0.20 | 0.505 |
| Veillonella | 0.37 ± 0.15 | 1.25 ± 1.14 | 0.326 |
| Flavonifractor | 0.10 ± 0.04 | 0.13 ± 0.10 | 1.000 |
| UBA1819 | 0.09 ± 0.03 | 0.13 ± 0.07 | 0.875 |
| Lachnospiraceae UCG 010 | 0.13 ± 0.09 | 0.04 ± 0.02 | 0.692 |
| Hungatella | 0.15 ± 0.12 | 0.15 ± 0.09 | 0.966 |
| Butyricimonas | 0.18 ± 0.05 | 0.06 ± 0.04 | 0.017 * |
| Bilophila | 0.11 ± 0.06 | 0.11 ± 0.06 | 0.784 |
| Lachnospiraceae | 0.23 ± 0.06 | 0.26 ± 0.13 | 0.724 |
| Sutterella | 0.10 ± 0.04 | 0.15 ± 0.11 | 1.000 |
| Species | |||
| Bacteroides stercoris | 10.31 ± 7.83 | 1.96 ± 1.01 | 0.422 |
| Bacteroides vulgatus | 12.34 ± 3.94 | 28.81 ± 8.31 | 0.021 * |
| Bacteroides fragilis | 3.26 ± 1.61 | 13.96 ± 7.30 | 0.222 |
| Bacteroides uniformis | 10.70 ± 3.76 | 5.32 ± 2.28 | 0.365 |
| Lachnospiraceae NK4A136 group | 4.77 ± 2.02 | 6.29 ± 2.67 | 0.634 |
| Eubacterium hallii group | 12.40 ± 5.98 | 12.80 ± 5.46 | 0.610 |
| Ruminococcus torques group | 7.95 ± 2.23 | 6.66 ± 2.75 | 0.326 |
| Slackia isoflavoniconvertens | 7.76 ± 4.49 | 3.97 ± 1.77 | 0.579 |
| Bacteroides massiliensis | 6.64 ± 3.71 | 1.14 ± 0.62 | 0.148 |
| Eubacterium eligens group | 2.30 ± 0.96 | 1.12 ± 0.47 | 0.222 |
| Incertae sedis | 0.66 ± 0.22 | 0.56 ± 0.29 | 0.555 |
| Eubacterium ramulus | 0.48 ± 0.21 | 0.95 ± 0.42 | 0.969 |
| Parabacteroides distasonis | 1.49 ± 0.61 | 0.88 ± 0.49 | 0.422 |
The Wilcoxon signed-rank test was used to determine the statistical significance. * Statistically significant if p ≤ 0.05.
Table 7 details the changes in microbial abundance in the placebo group. The abundance of the phylum Actinobacteriota significantly (p = 0.023) increased after treatment. The abundance of Escherichia-Shigella (p = 0.050) was reduced, and the abundance of Collinsella (p = 0.045) and Sutterella (p = 0.047) increased significantly after treatment. No significant differences were reflected in species level in the placebo group (Table 7).
Table 7.
The statistical differences in the phylum, genus, and species between the baseline and after-treatment samples in the placebo group.
| Taxonomy | Baseline (PW0) | After Treatment (PW8) | p-Value |
|---|---|---|---|
| Phyla | |||
| Proteobacteria | 19.71 ± 6.45 | 7.66 ± 1.86 | 0.071 |
| Bacteroidota | 30.95 ± 8.51 | 24.05 ± 4.91 | 0.754 |
| Firmicutes | 44.75 ± 6.74 | 60.11 ± 2.45 | 0.136 |
| Actinobacteriota | 1.68 ± 0.46 | 4.35 ± 1.57 | 0.023 * |
| Fusobacteriota | 1.69 ± 1.17 | 0.80 ± 0.50 | 0.938 |
| Verrucomicrobiota | 1.02 ± 0.39 | 2.85 ± 2.24 | 0.609 |
| Desulfobacterota | 0.21 ± 0.09 | 0.18 ± 0.06 | 0.969 |
| Genera | |||
| Escherichia-Shigella | 16.30 ± 6.58 | 5.85 ± 2.14 | 0.050 * |
| Bacteroides | 14.37 ± 4.14 | 14.92 ± 3.88 | 1.000 |
| Faecalibacterium | 3.22 ± 0.83 | 3.46 ± 1.24 | 0.530 |
| Blautia | 6.07 ± 3.24 | 5.11 ± 0.88 | 0.182 |
| Prevotella | 14.65 ± 6.52 | 6.97 ± 2.54 | 0.609 |
| Agathobacter | 1.90 ± 0.83 | 0.57 ± 0.24 | 0.224 |
| Collinsella | 0.98 ± 0.29 | 3.62 ± 1.89 | 0.045 * |
| Subdoligranulum | 2.49 ± 1.04 | 1.20 ± 0.53 | 0.158 |
| Streptococcus | 2.13 ± 1.53 | 5.47 ± 3.56 | 0.695 |
| Roseburia | 1.33 ± 0.28 | 2.41 ± 0.83 | 0.195 |
| Phascolarctobacterium | 2.61 ± 0.55 | 3.78 ± 1.09 | 0.433 |
| Romboutsia | 0.46 ± 0.16 | 1.22 ± 0.51 | 0.081 |
| Klebsiella | 1.54 ± 0.59 | 0.63 ± 0.22 | 0.170 |
| Fusobacterium | 1.75 ± 1.21 | 0.92 ± 0.57 | 0.938 |
| Ruminococcus | 0.92 ± 0.33 | 1.11 ± 0.46 | 0.906 |
| Dorea | 0.64 ± 0.17 | 1.20 ± 0.53 | 0.254 |
| Akkermansia | 1.12 ± 0.43 | 3.92 ± 3.04 | 0.609 |
| Lachnoclostridium | 0.77 ± 0.14 | 1.28 ± 0.49 | 0.814 |
| UCG-002 | 1.50 ± 0.55 | 0.95 ± 0.44 | 0.289 |
| Fusicatenibacter | 0.68 ± 0.28 | 0.35 ± 0.25 | 0.209 |
| Enterobacter | 1.29 ± 0.60 | 0.09 ± 0.03 | 0.090 |
| Veillonella | 0.21 ± 0.08 | 4.91 ± 2.59 | 0.100 |
| Alistipes | 1.23 ± 0.54 | 0.96 ± 0.38 | 0.480 |
| CAG-352 | 0.39 ± 0.12 | 0.52 ± 0.22 | 0.969 |
| Enterobacteriaceae | 1.88 ± 1.10 | 0.35 ± 0.17 | 0.209 |
| Coprococcus | 0.70 ± 0.23 | 0.77 ± 0.22 | 0.410 |
| Sutterella | 0.49 ± 0.24 | 1.64 ± 0.99 | 0.047 * |
| Anaerostipes | 0.41 ± 0.18 | 0.64 ± 0.22 | 0.556 |
| Parabacteroides | 0.90 ± 0.29 | 1.04 ± 0.35 | 0.875 |
| Holdemanella | 0.68 ± 0.37 | 0.53 ± 0.32 | 0.906 |
| Butyricicoccus | 0.52 ± 0.14 | 1.03 ± 0.25 | 0.100 |
| Enterococcus | 0.15 ± 0.09 | 2.62 ± 1.83 | 0.057 |
| Lachnospira | 0.41 ± 0.20 | 0.22 ± 0.10 | 0.428 |
| Lachnospiraceae | 0.62 ± 0.24 | 1.04 ± 0.31 | 0.255 |
| Paraprevotella | 0.58 ± 0.39 | 0.42 ± 0.25 | 0.969 |
| Muribaculaceae | 0.38 ± 0.18 | 0.95 ± 0.54 | 0.937 |
| Haemophilus | 0.10 ± 0.05 | 0.26 ± 0.15 | 0.178 |
| Bifidobacterium | 0.13 ± 0.05 | 0.50 ± 0.22 | 0.170 |
| UBA1819 | 0.06 ± 0.02 | 0.10 ± 0.05 | 0.969 |
| Lachnospiraceae UCG010 | 0.16 ± 0.04 | 0.14 ± 0.05 | 0.692 |
| Odoribacter | 0.15 ± 0.06 | 0.17 ± 0.10 | 0.937 |
| Oscillibacter | 0.05 ± 0.01 | 0.13 ± 0.08 | 0.937 |
| Bilophila | 0.13 ± 0.10 | 0.07 ± 0.03 | 0.783 |
| Butyricimonas | 0.26 ± 0.12 | 0.15 ± 0.06 | 0.844 |
| Species | |||
| Bacteroides uniformis | 7.72 ± 2.21 | 5.48 ± 3.86 | 0.158 |
| Bacteroides vulgatus | 7.99 ± 2.23 | 8.17 ± 2.28 | 0.875 |
| Bacteroides plebeius | 5.44 ± 3.84 | 4.16 ± 2.49 | 0.937 |
| Lachnospiraceae NK4A136 group | 2.39 ± 1.72 | 3.78 ± 1.52 | 0.665 |
| Bacteroides coprophilus | 3.10 ± 2.96 | 2.03 ± 1.32 | 1.000 |
| Eubacterium hallii group | 11.47 ± 6.49 | 9.85 ± 5.06 | 0.754 |
| Ruminococcus gnavus group | 0.84 ± 0.38 | 7.56 ± 4.52 | 0.812 |
| Ruminococcus bicirculans | 1.81 ± 0.82 | 1.70 ± 0.80 | 0.906 |
| Lactobacillus salivarius | 7.42 ± 7.29 | 4.51 ± 4.36 | 0.902 |
| Prevotella stercorea | 3.32 ± 1.81 | 2.87 ± 1.50 | 0.475 |
| Prevotella copri | 1.55 ± 1.36 | 10.04 ± 4.89 | 0.475 |
| Ruminococcus torques group | 2.35 ± 0.48 | 3.51 ± 1.05 | 0.194 |
| Bacteroides stercoris | 4.60 ± 1.84 | 1.11 ± 0.59 | 0.194 |
| Parabacteroides merdae | 1.30 ± 0.43 | 0.90 ± 0.30 | 0.255 |
| Eubacterium eligens group | 1.28 ± 0.36 | 1.36 ± 0.63 | 0.433 |
| Parabacteroides distasonis | 2.05 ± 1.14 | 1.41 ± 0.81 | 0.692 |
| Ruminococcus gauvreauii group | 0.71 ± 0.36 | 1.75 ± 1.45 | 0.410 |
| Alistipes shahii | 0.87 ± 0.45 | 0.46 ± 0.23 | 0.194 |
The Wilcoxon signed-rank test was used to determine the statistical significance. * Statistically significant if p ≤ 0.05.
The comparison of placebo and probiotic groups after the treatment period indicated that proteobacteria (p = 0.015), Escherichia-Shigella (p = 0.024), Sutterella (p = 0.039), and Bacteroides vulgatus abundancy were altered significantly (Table 8).
Table 8.
The differentiation of microbiome between the placebo and probiotic groups at the end of the study.
| Taxonomy | Placebo vs. Treatment | p-Value | |
|---|---|---|---|
| Phylum | |||
| Proteobacteria | −12.06 | 8.67 | 0.015 * |
| Bacteroidota | −6.90 | −3.15 | 0.773 |
| Firmicutes | 15.36 | 0.39 | 0.166 |
| Actinobacteriota | 2.67 | −2.61 | 0.564 |
| Fusobacteriota | −0.89 | −0.05 | 0.817 |
| Verrucomicrobiota | 1.84 | −3.15 | 0.384 |
| Desulfobacterota | −0.03 | −0.07 | 0.686 |
| Genus | |||
| Escherichia-Shigella | −10.45 | 8.20 | 0.024 * |
| Bacteroides | 0.55 | 1.40 | 0.326 |
| Faecalibacterium | 0.24 | 2.90 | 0.273 |
| Blautia | −0.96 | −1.76 | 0.106 |
| Prevotella | −7.68 | −4.61 | 0.908 |
| Agathobacter | −1.33 | 0.72 | 0.050 |
| Collinsella | 2.63 | 0.52 | 0.525 |
| Subdoligranulum | −1.29 | 0.53 | 0.133 |
| Streptococcus | 3.34 | 0.75 | 0.225 |
| Roseburia | 1.08 | 0.27 | 0.237 |
| Phascolarctobacterium | 1.18 | −1.99 | 0.419 |
| Romboutsia | 0.76 | −0.12 | 0.138 |
| Klebsiella | −0.91 | −0.83 | 0.977 |
| Fusobacterium | −0.83 | −0.05 | 0.817 |
| Ruminococcus | 0.19 | −0.14 | 0.862 |
| Dorea | 0.55 | 0.45 | 0.817 |
| Akkermansia | 2.80 | −3.14 | 0.487 |
| Lachnoclostridium | 0.50 | −0.20 | 0.453 |
| UCG-002 | −0.55 | 0.92 | 0.260 |
| Fusicatenibacter | −0.33 | 0.02 | 0.204 |
| Enterobacter | −1.19 | 0.10 | 0.355 |
| Veillonella | 4.70 | 0.88 | 0.106 |
| Alistipes | −0.28 | −0.25 | 1.000 |
| CAG-352 | 0.13 | 0.52 | 0.148 |
| Enterobacteriaceae | −1.53 | −0.16 | 0.419 |
| Coprococcus | 0.07 | 0.40 | 0.977 |
| Sutterella | 1.15 | 0.05 | 0.039 * |
| Anaerostipes | 0.23 | 0.30 | 0.470 |
| Parabacteroides | 0.13 | −0.04 | 0.908 |
| Holdemanella | −0.16 | −0.50 | 0.386 |
| Butyricicoccus | 0.51 | 0.43 | 0.326 |
| Enterococcus | 2.47 | −0.02 | 0.182 |
| Lachnospiraceae | 0.41 | 0.03 | 0.312 |
| Paraprevotella | −0.16 | −0.03 | 0.541 |
| Bifidobacterium | 0.37 | −1.50 | 0.795 |
| UBA1819 | 0.04 | 0.04 | 0.729 |
| Lachnospiraceae UCG-010 | −0.02 | −0.09 | 0.931 |
| Odoribacter | 0.03 | −0.15 | 0.309 |
| Bilophila | −0.06 | 0.00 | 0.862 |
| Butyricimonas | −0.11 | −0.12 | 0.271 |
| Species | |||
| Bacteroides uniformis | −2.24 | −5.38 | 0.908 |
| Bacteroides vulgatus | 0.18 | 16.47 | 0.033 * |
| Lachnospiraceae NK4A136 group | 1.39 | 1.52 | 1.000 |
| Bacteroides stercoris | −3.49 | −8.35 | 0.727 |
| Parabacteroides distasonis | −0.64 | −0.61 | 0.447 |
* Statistically significant if p ≤ 0.05.
4. Discussion
Age is a significant factor influencing the composition of the human intestinal microbiota, which changes throughout a person’s life [26,27,28,29]. These changes are influenced by factors such as physiological alterations in the gastrointestinal tract associated with aging, dietary habits specific to different countries, lifestyle choices, frailty conditions, antibiotic usage, and nutritional behaviors. Numerous studies have explored the impact of age-related physiological changes, lifestyle factors, and dietary habits on the gut microbiota [26,27,28,29,30]. Aging is accompanied by gut microbiome alterations that increase the various aging-associated diseases. It is difficult to conclude which factor initially contributed to this shift in the gut microbiome. However, lifestyle practices like increased medication, reduced mobility, and diet are found to manipulate the gut microbial composition. Understanding microbial manipulation in the elderly develops promising strategies to prevent age-associated diseases [31]. In the human body, gut microbiota acts as an important metabolic organ facilitating the metabolism of nutrients [32]. In addition to its association with metabolic diseases like obesity, liver diseases, intestinal diseases, neuropsychiatric diseases, diabetes, and cardiovascular diseases (CVD), the gut microbiota also acts as a reservoir for antibiotic-resistant genes [33]. The genotype of the host, diet, diseases, and age affect the composition and diversity of gut microbiota [34,35]. Therefore, employing microbial intervention to manipulate the gut microbiota represents an innovative strategy for impacting sleep and well-being [36]. Ingesting probiotics orally enables the restoration of functional activities within the gut microbiota [37].
Probiotic supplementation can protect gut integrity and restore its functions by initiating the growth of beneficial microbes, safeguarding the intestinal barrier, and positively modulating immune functions [1]. Costabile et al. investigated the effects of probiotic L. rhamnosus GG and pilus-deficient L. rhamnosus GG-PB12combined with soluble corn fiber on microbiota, immunity, metabolism, and blood lipids in healthy elder persons. Consumption of L. rhamnosus GG and prebiotics increased the natural killer cell activity compared to the baseline group. The fecal microbiota analysis showed that the synbiotic supplementation of L. rhamnosus GG with corn fiber and L. rhamnosus GG-PB12 with corn fiber significantly increased Parabacteroides. L. rhamnosus GG with corn fiber increased Ruminococcus, and Incertae Sedis and decreased the Oscillospira and Desulfovibrio. L. rhamnosus GG and corn fiber further reduced the total cholesterol and LDL. L. rhamnosus GG-PB12 with corn fiber treated volunteers showed a significant reduction in C-reactive protein compared to baseline. Thus, the dietary intervention with L. rhamnosus GG and corn fiber could positively enhance the immune response and microbial community [38]. The lipid status of an individual plays an important function in reducing the risk of CVD. Few observational studies suggested that hyperlipidemic subjects have a high risk of developing CVD. Additionally, it was found that reduced serum cholesterol, in turn, reduces the CVD risk. Clinical studies evaluated the beneficial effects of probiotic supplementation on serum lipid profiles [39,40,41]. Synbiotic interventions have been found to have more benefits in hypercholesterolemic patients than in normal people, and the reduction in total cholesterol and LDL levels is greater in the elderly than in younger individuals [38].
In this study, the difference between placebo and probiotic groups at the end of 8 weeks showed that the mean body weight reduction was almost the same. Reductions in visceral fat, FBS, TC, TG, and LDL were greater than placebo but not up to significant levels; simultaneously, there was a suggestive trend indicating improvement from baseline to week 8. However, after 8 weeks, this significance was reduced (p = 0.25) [36]. The difference in HDL level was increased 8 weeks after treatment (Table 3). Certain studies correlated that elevated serum TC, TG, and LDL and low HDL increase the risk for CVD [42,43,44]. Probiotics had no significant effects on the body, visceral fat, FBS, TC, TG, and HDL. The effect of probiotics on LDL depends on various factors, and a significant reduction in LDL was observed in the present study (Table 4). These results indicate that probiotic L. rhamnosus may improve the lipid profile when treated long or combined with other probiotics or prebiotic compounds.
In adults, Firmicutes are predominantly present in the gut, followed by Bacteroidetes [13]. Among the oldest adults, there is generally a decrease in Firmicutes and an increase in Bacteroidetes, which aligns with previous findings indicating a rise in the Firmicutes/Bacteroidetes ratio during adulthood followed by a decline in older age [45]. Vogt et al. [46] reported that Bacteroidetes showed an increase in abundance, whereas Firmicutes and the genus Bifidobacterium exhibited a decrease. Similarly, prior research has demonstrated that excessively high and excessively low Firmicutes/Bacteroidetes ratios can be linked to metabolic and gastrointestinal disorders [47]. These findings imply that a balanced distribution of these core phyla may signify good health and longevity, although specific environmental factors may partly influence this equilibrium. The abundance of Proteobacteria increased after L. rhamnosus supplementation; even though the changes were insignificant, it is noteworthy that L. rhamnosus supplementation could improve the microbiome positively in elderly subjects (Table 6). With the support of the evidence from the previous studies, proteobacteria, which is associated with increased gut inflammation and dysbiosis, was more abundant in older adults than in younger adults [48,49,50,51]. The Shannon–Wiener index considers fewer common species [37].
Following the treatment period, a comparison between the placebo and probiotic groups revealed significant alterations in the richness of Escherichia-Shigella (p = 0.024), Sutterella (p = 0.039), Agathobacter (p = 0.050), and Bacteroides vulgatus (p = 0.033) (Table 8). Laongkham et al. explained that the core gut microbiota of healthy Thai individuals comprises eleven species, including Firmicutes, Bacteroidetes, and Proteobacteria, shared by over 90% of subjects. Notably, Escherichia coli was found to be highly prevalent, especially among Thai elderly individuals. Age and PCA coordination were also correlated, particularly regarding the loading vector associated with the genus Escherichia/Shigella. However, no significant difference was observed in the abundance of this genus group between the adult and elderly groups [52]. After 8 weeks of probiotic intervention, non-significant changes were observed in the abundance of Akkermansia (p = 0.751), Prevotella (p = 0.609), and Bifidobacterium (p = 0.610) (Table 6). The alterations in Akkermansia, along with the changes in Bifidobacterium and the decline of Prevotella, have been proposed as biomarkers for PD [53].
In the case of the Agathobacter genus, López-García et al. mentioned that it did not meet statistical significance during the analysis. Still, there was a noticeable fluctuation in its occurrence throughout the clinical trial. In the Lactiplantibacillus pentosus-supplemented group, there was a rise in the average frequency of Agathobacter sequences from 4.67 to 4.98%. Conversely, there was a decrease in the placebo group from an initial frequency of 3.70% to a final frequency of 1.98% [54]. Similarly, in an investigation, nine healthy individuals were chosen to undergo a fasting regimen of approximately 17 h per day for 29 days. After the trial, no notable alterations in the measured values were reported. However, a noteworthy rise in the prevalence of both Akkermansia muciniphila and Bacteroides fragilis was reported [55].
Additionally, two clinical trials demonstrated a reduction in the abundance of B. vulgatus following the administration of probiotics [56,57]. Assessing intestinal microbiota composition poses a significant challenge due to its notable variability. This is primarily because modifications to commensal strains can occur within a short span of just a few days through changes in diet and lifestyle [58,59]. These findings underscore specific taxonomic changes within the probiotic group over the study period, emphasizing the importance of evaluating microbial dynamics at different taxonomic levels for a comprehensive understanding of the impact of probiotic intervention.
5. Limitations
The study has a few limitations that warrant consideration in interpreting its findings. The sample size of the study was relatively small, and the results may not be fully representative of the broader elderly population of Thailand. Individual variations among the study subjects may impact the generalizability of the observed effects. The study duration (8 weeks) was relatively short, with long-term effects. The sustainability of changes in glycaemic index, lipid profile, and microbiome may not be adequately captured within this timeframe.
Additionally, the exclusive use of a single strain, L. rhamnosus, as the probiotic intervention limits the understanding of potential synergistic effects that may arise from combining multiple strains. Furthermore, the study employed a single dosage of the probiotic, and exploring the impact of different dosage levels could provide insights into the dose–response relationship and optimal supplementation levels. The absence of follow-up research beyond the 8-week intervention period is another limitation, as longer-term assessments could reveal whether the observed effects persist or diminish over time.
6. Conclusions
The 8-week supplementation of L. rhamnosus improved the glycaemic index and lipid profile positively, but statistical significance was not observed. Gaussian regression analysis indicated that the probiotic supplementation significantly reduced the LDL level in the elderly subjects. Microbiome analysis revealed taxonomic shifts in both the placebo and probiotic groups. The results revealed that L. rhamnosus supplementation did not significantly affect the microbiome of the healthy elderly subjects. The study, however, is limited by its small sample size, short duration, and the use of a single probiotic strain. Future research with larger cohorts, extended study periods, and different probiotic formulations must confirm the findings. Addressing these limitations will contribute to a more comprehensive understanding of the potential benefits and mechanisms underlying probiotic interventions in the elderly population.
Acknowledgments
The authors gratefully acknowledge the Faculty of Pharmacy and Chiang Mai University, Chiang Mai, Thailand, for their support. T.C. wishes to acknowledge the support of the Postdoctoral Fellowship from Mae Fah Luang University, Thailand.
Author Contributions
Conceptualization, C.C., B.S.S., W.R., S.S., P.K. and P.S.; methodology, C.C., S.K., W.R. and T.C.; software, S.T., M.B., S.K. and B.S.S.; validation, C.C., M.B., P.K., W.R. and S.S.; formal analysis, T.C., S.T., M.B., P.K., P.F., W.R. and C.C.; investigation, S.K., S.S., S.T., W.R. and C.C.; resources, C.C.; data curation, S.T., M.B., T.C., P.F. and S.K.; writing—original draft preparation, B.S.S., S.T., C.C., P.K. and N.S.; writing—review and editing, B.S.S., S.T., C.C., P.K., P.S., W.R. and N.S.; supervision, W.R.; project administration, S.P. and C.C.; funding acquisition, C.C. All authors have read and agreed to the published version of the manuscript.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
This research was funded by Fundamental Fund Research-2024, Chiang Mai University, Chiang Mai, Thailand.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Landete J.M., Gaya P., Rodríguez E., Langa S., Peirotén Á., Medina M., Arqués J.L. Probiotic Bacteria for Healthier Aging: Immunomodulation and Metabolism of Phytoestrogens. Biomed. Res. Int. 2017;2017:5939818. doi: 10.1155/2017/5939818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.López-Otín C., Blasco M.A., Partridge L., Serrano M., Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–1217. doi: 10.1016/j.cell.2013.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakshminarayanan B., Stanton C., O’Toole P.W., Ross R.P. Compositional dynamics of the human intestinal microbiota with aging: Implications for health. J. Nutr. Health. Aging. 2014;18:773–786. doi: 10.1007/s12603-014-0549-6. [DOI] [PubMed] [Google Scholar]
- 4.Boyajian J.L., Ghebretatios M., Schaly S., Islam P., Prakash S. Microbiome and Human Aging: Probiotic and Prebiotic Potentials in Longevity, Skin Health and Cellular Senescence. Nutrients. 2021;13:4550. doi: 10.3390/nu13124550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guarner F., Malagelada J.R. Gut flora in health and disease. Lancet. 2003;361:512–519. doi: 10.1016/S0140-6736(03)12489-0. [DOI] [PubMed] [Google Scholar]
- 6.Bäckhed F., Ley R.E., Sonnenburg J.L., Peterson D.A., Gordon J.I. Host-bacterial mutualism in the human intestine. Science. 2005;307:1915–1920. doi: 10.1126/science.1104816. [DOI] [PubMed] [Google Scholar]
- 7.Hickson M. Malnutrition and ageing. Postgrad. Med. J. 2006;82:2–8. doi: 10.1136/pgmj.2005.037564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tiihonen K., Ouwehand A.C., Rautonen N. Human intestinal microbiota and healthy ageing. Ageing. Res. Rev. 2010;9:107–116. doi: 10.1016/j.arr.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 9.Claesson M.J., Cusack S., O’Sullivan O., Greene-Diniz R., de Weerd H., Flannery E., Marchesi J.R., Falush D., Dinan T., Fitzgerald G., et al. Composition, variability, and temporal stability of the intestinal microbiota of the elderly. Proc. Natl. Acad. Sci. USA. 2011;108:4586–4591. doi: 10.1073/pnas.1000097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Claesson M.J., Jeffery I.B., Conde S., Power S.E., O’Connor E.M., Cusack S., Harris H.M., Coakley M., Lakshminarayanan B., O’Sullivan O., et al. Gut microbiota composition correlates with diet and health in the elderly. Nature. 2012;488:178–184. doi: 10.1038/nature11319. [DOI] [PubMed] [Google Scholar]
- 11.Eckburg P.B., Bik E.M., Bernstein C.N., Purdom E., Dethlefsen L., Sargent M., Gill S.R., Nelson K.E., Relman D.A. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rondanelli M., Giacosa A., Faliva M.A., Perna S., Allieri F., Castellazzi A.M. Review on microbiota and effectiveness of probiotics use in older. World J. Clin. Cases. 2015;3:156–162. doi: 10.12998/wjcc.v3.i2.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Odamaki T., Kato K., Sugahara H., Hashikura N., Takahashi S., Xiao J.Z., Abe F., Osawa R. Age-related changes in gut microbiota composition from newborn to centenarian: A cross-sectional study. BMC Microbiol. 2016;16:90. doi: 10.1186/s12866-016-0708-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Woodmansey E.J. Intestinal bacteria and ageing. J. Appl. Microbiol. 2007;102:1178–1186. doi: 10.1111/j.1365-2672.2007.03400.x. [DOI] [PubMed] [Google Scholar]
- 15.Hopkins M.J., Macfarlane G.T. Changes in predominant bacterial populations in human faeces with age and with Clostridium difficile infection. J. Med. Microbiol. 2002;51:448–454. doi: 10.1099/0022-1317-51-5-448. [DOI] [PubMed] [Google Scholar]
- 16.Mäkivuokko H., Tiihonen K., Tynkkynen S., Paulin L., Rautonen N. The effect of age and non-steroidal anti-inflammatory drugs on human intestinal microbiota composition. Br. J. Nutr. 2010;103:227–234. doi: 10.1017/S0007114509991553. [DOI] [PubMed] [Google Scholar]
- 17.Biagi E., Franceschi C., Rampelli S., Severgnini M., Ostan R., Turroni S., Consolandi C., Quercia S., Scurti M., Monti D., et al. Gut Microbiota and Extreme Longevity. Curr. Biol. 2016;26:1480–1485. doi: 10.1016/j.cub.2016.04.016. [DOI] [PubMed] [Google Scholar]
- 18.Kong F., Hua Y., Zeng B., Ning R., Li Y., Zhao J. Gut microbiota signatures of longevity. Curr. Biol. 2016;26:R832–R833. doi: 10.1016/j.cub.2016.08.015. [DOI] [PubMed] [Google Scholar]
- 19.Du Y., Gao Y., Zeng B., Fan X., Yang D., Yang M. Effects of anti-aging interventions on intestinal microbiota. Gut Microbes. 2021;13:1994835. doi: 10.1080/19490976.2021.1994835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bischoff S.C. Microbiota and aging. Curr. Opin. Clin. Nutr. Metab. Care. 2016;19:26–30. doi: 10.1097/MCO.0000000000000242. [DOI] [PubMed] [Google Scholar]
- 21.Chung K.W. Advances in Understanding of the Role of Lipid Metabolism in Aging. Cells. 2021;10:880. doi: 10.3390/cells10040880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson A.A., Stolzing A. The role of lipid metabolism in aging, lifespan regulation, and age-related disease. Aging Cell. 2019;18:e13048. doi: 10.1111/acel.13048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chiang B.L., Sheih Y.H., Wang L.H., Liao C.K., Gill H.S. Enhancing immunity by dietary consumption of a probiotic lactic acid bacterium (Bifidobacterium lactis HN019): Optimization and definition of cellular immune responses. Eur. J. Clin. Nutr. 2000;54:849–855. doi: 10.1038/sj.ejcn.1601093. [DOI] [PubMed] [Google Scholar]
- 24.Sivamaruthi B.S., Kesika P., Chaiyasut C. A review on anti-aging properties of probiotics. Int. J. Appl. Pharm. 2018;10:23–27. doi: 10.22159/ijap.2018v10i5.28249. [DOI] [Google Scholar]
- 25.Chaiyasut C., Sirilun S., Juntarachot N., Tongpong P., Ouparee W., Sivamaruthi B.S., Peerajan S., Waditee-Sirisattha R., Prombutara P., Klankeo P., et al. Effect of Dextranase and Dextranase-and-Nisin-Containing Mouthwashes on Oral Microbial Community of Healthy Adults-A Pilot Study. Appl. Sci. 2022;12:1650. doi: 10.3390/app12031650. [DOI] [Google Scholar]
- 26.Mueller S., Saunier K., Hanisch C., Norin E., Alm L., Midtvedt T. Differences in fecal microbiota in diferent European study populations in relation to age, gender, and country: A cross-sectional study. Appl. Environ. Microbiol. 2006;72:1027–1033. doi: 10.1128/AEM.72.2.1027-1033.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Biagi E., Nylund L., Candela M., Ostan R., Bucci L., Pini E., Nikkïla J., Monti D., Satokari R., Franceschi C., et al. Through ageing, and beyond: Gut microbiota and infammatory status in seniors and centenarians. PLoS ONE. 2010;5:e10667. doi: 10.1371/annotation/df45912f-d15c-44ab-8312-e7ec0607604d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao L., Qiao X., Zhu J., Zhang X., Jiang J., Hao Y., Ren F. Correlations of fecal bacterial communities with age and living region for the elderly living in Bama, Guangxi. China. J. Microbiol. 2011;49:186–192. doi: 10.1007/s12275-011-0405-x. [DOI] [PubMed] [Google Scholar]
- 29.Yatsunenko T., Rey F.E., Manary M.J., Trehan I., Dominguez-Bello M.G., Contreras M. Human gut microbiome viewed across age and geography. Nature. 2012;486:222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hayashi H., Sakamoto M., Kitahara M., Benno Y. Molecular analysis of fecal microbiota in elderly individuals using 16S rDNA library and T-RFLP. Microbiol. Immunol. 2003;47:557–570. doi: 10.1111/j.1348-0421.2003.tb03418.x. [DOI] [PubMed] [Google Scholar]
- 31.Hutchinson A.N., Bergh C., Kruger K., Sűsserová M., Allen J., Améen S., Tingö L. The Effect of Probiotics on Health Outcomes in the Elderly: A Systematic Review of Randomized, Placebo-Controlled Studies. Microorganisms. 2021;9:1344. doi: 10.3390/microorganisms9061344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liu Y.H., Kang L. Progress in researches on gut microbiota and metabolism. Chin. J. Microecol. 2013;3:362–364+367. [Google Scholar]
- 33.Sommer M.O., Dantas G. Antibiotics and the resistant microbiome. Curr. Opin. Microbiol. 2011;14:556–563. doi: 10.1016/j.mib.2011.07.005. [DOI] [PubMed] [Google Scholar]
- 34.Gajer P., Brotman R.M., Bai G., Sakamoto J., Schütte U.M., Zhong X., Koenig S.S., Fu L., Ma Z.S., Zhou X., et al. Temporal dynamics of the human vaginal microbiota. Sci. Transl. Med. 2012;4:132ra52. doi: 10.1126/scitranslmed.3003605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.David L.A., Maurice C.F., Carmody R.N., Gootenberg D.B., Button J.E., Wolfe B.E., Ling A.V., Devlin A.S., Varma Y., Fischbach M.A., et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505:559–563. doi: 10.1038/nature12820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Patterson E., Tan H.T.T., Groeger D., Andrews M., Buckley M., Murphy E.F., Groeger J.A. Bifidobacterium longum 1714 improves sleep quality and aspects of well-being in healthy adults: A randomized, double-blind, placebo-controlled clinical trial. Sci. Rep. 2024;14:3725. doi: 10.1038/s41598-024-53810-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vicariotto F., Malfa P., Viciani E., Dell’Atti F., Squarzanti D.F., Marcante A., Castagnetti A., Ponchia R., Governini L., De Leo V. Efficacy of Lactiplantibacillus plantarum PBS067, Bifidobacterium animalis subsp. lactis BL050, and Lacticaseibacillus rhamnosus LRH020 in the Amelioration of Vaginal Microbiota in Post-Menopausal Women: A Prospective Observational Clinical Trial. Nutrients. 2024;16:402. doi: 10.3390/nu16030402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Costabile A., Bergillos-Meca T., Rasinkangas P., Korpela K., de Vos W.M., Gibson G.R. Effects of Soluble Corn Fiber Alone or in Synbiotic Combination with Lactobacillus rhamnosus GG and the Pilus-Deficient Derivative GG-PB12 on Fecal Microbiota, Metabolism, and Markers of Immune Function: A Randomized, Double-Blind, Placebo-Controlled, Crossover Study in Healthy Elderly (Saimes Study) Front. Immunol. 2017;8:1443. doi: 10.3389/fimmu.2017.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sabatine M.S., Wiviott S.D., Im K., Murphy S.A., Giugliano R.P. Efficacy and Safety of Further Lowering of Low-Density Lipoprotein Cholesterol in Patients Starting With Very Low Levels: A Meta-analysis. JAMA Cardiol. 2018;3:823–828. doi: 10.1001/jamacardio.2018.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chen Z.Y., Jiao R., Ma K.Y. Cholesterol-lowering nutraceuticals and functional foods. J. Agric. Food Chem. 2008;56:8761–8773. doi: 10.1021/jf801566r. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu M., Hashiquchi M., Shiga T., Tamura H.O., Mochizuki M. Meta analysis: Effects of probiotic supplementation on lipid profiles in normal to mildly hypercholesterolemic individuals. PLoS ONE. 2015;10:e0139795. doi: 10.1371/journal.pone.0139795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gould A.L., Davies G.M., Alemao E., Yin D.D., Cook J.R. Cholesterol reduction yields clinical benefits: Meta-analysis including recent trials. Clin. Ther. 2007;29:778–794. doi: 10.1016/j.clinthera.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 43.Law M.R., Wald N.J., Rudnicka A.R. Quantifying effect of statins on low density lipoprotein cholesterol, ischaemic heart disease, and stroke: Systematic review and meta-analysis. BMJ. 2003;326:1423–1429. doi: 10.1136/bmj.326.7404.1423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.De Caterina R., Scarano M., Marfisi R., Lucisano G., Palma F., Tatasciore A., Marchioli R. Cholesterol-lowering interventions and stroke: Insights from a meta-analysis of randomized controlled trials. J. Am. Coll. Cardiol. 2010;55:198–211. doi: 10.1016/j.jacc.2009.07.062. [DOI] [PubMed] [Google Scholar]
- 45.Mariat D., Firmesse O., Levenez F., Guimarăes V., Sokol H., Doré J., Corthier G., Furet J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009;9:123. doi: 10.1186/1471-2180-9-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vogt N.M., Kerby R.L., Dill-McFarland K.A., Harding S.J., Merluzzi A.P., Johnson S.C., Carlsson C.M., Asthana S., Zetterberg H., Blennow K., et al. Gut microbiome alterations in Alzheimer’s disease. Sci. Rep. 2017;7:13537. doi: 10.1038/s41598-017-13601-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walters W.A., Xu Z., Knight R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014;588:4223–4233. doi: 10.1016/j.febslet.2014.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rizzatti G., Lopetuso L., Gibiino G., Binda C., Gasbarrini A. Proteobacteria: A common factor in human diseases. BioMed Res. Int. 2017;2017:9351507. doi: 10.1155/2017/9351507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kim B.S., Choi C.W., Shin H., Jin S.P., Bae J.S., Han M., Seo E.Y., Chun J., Chung J.H. Comparison of the gut microbiota of centenarians in longevity villages of South Korea with those of other age groups. J. Microbiol. Biotechnol. 2019;29:429–440. doi: 10.4014/jmb.1811.11023. [DOI] [PubMed] [Google Scholar]
- 50.Wu L., Zeng T., Zinellu A., Rubino S., Kelvin D.J., Carru C. A Cross-Sectional Study of Compositional and Functional Profiles of Gut Microbiota in Sardinian Centenarians. mSystems. 2019;4:e00325-19. doi: 10.1128/mSystems.00325-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yu X., Wu X., Qiu L., Wang D., Gan M., Chen X., Wei H., Xu F. Analysis of the intestinal microbial community structure of healthy and long-living elderly residents in Gaotian Village of Liuyang City. Appl. Microbiol. Biotechnol. 2015;99:9085–9095. doi: 10.1007/s00253-015-6888-3. [DOI] [PubMed] [Google Scholar]
- 52.Laongkham O., Nakphaichit M., Nakayama J., Keawsompong S., Nitisinprasert S. Age-related changes in the gut microbiota and the core gut microbiome of healthy Thai humans. 3 Biotech. 2020;10:276. doi: 10.1007/s13205-020-02265-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal-Martinez G., Chin B., Camarillo C., Herrera G.V., Yang B., Sarosiek I., Perez R.G. A Pilot Microbiota Study in Parkinson’s Disease Patients versus Control Subjects, and Effects of FTY720 and FTY720-Mitoxy Therapies in Parkinsonian and Multiple System Atrophy Mouse Models. J. Parkinsons Dis. 2020;10:185–192. doi: 10.3233/JPD-191693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.López-García E., Benítez-Cabello A., Arenas-de Larriva A.P., Gutierrez-Mariscal F.M., Pérez-Martínez P., Yubero-Serrano E.M., Garrido-Fernández A., Arroyo-López F.N. Oral intake of Lactiplantibacillus pentosus LPG1 Produces a Beneficial Regulation of Gut Microbiota in Healthy Persons: A Randomized, Placebo-Controlled, Single-Blind Trial. Nutrients. 2023;15:1931. doi: 10.3390/nu15081931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ozkul C., Yalinay M., Karakan T. Islamic fasting leads to an increased abundance of Akkermansia muciniphila and Bacteroides fragilis group: A preliminary study on intermittent fasting. Turk. J. Gastroenterol. 2019;30:1030–1035. doi: 10.5152/tjg.2019.19185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nicolucci A.C., Hume M.P., Martínez I., Mayengbam S., Walter J., Reimer R.A. Prebiotics reduce body fat and alter intestinal microbiota in children who are overweight or with obesity. Gastroenterology. 2017;153:711–722. doi: 10.1053/j.gastro.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 57.Dewulf E.M., Cani P.D., Claus S.P., Fuentes S., Puylaert P.G., Neyrinck A.M., Bindels L.B., de Vos W.M., Gibson G.R., Thissen J.P., et al. Insight into the prebiotic concept: Lessons from an exploratory, double blind intervention study with inulin-type fructans in obese women. Gut. 2013;62:1112–1121. doi: 10.1136/gutjnl-2012-303304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ciobârcă D., Cătoi A.F., Copăescu C., Miere D., Crișan G. Bariatric Surgery in Obesity: Effects on Gut Microbiota and Micronutrient Status. Nutrients. 2020;12:235. doi: 10.3390/nu12010235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alhusain F. Microbiome: Role and Functionality in Human Nutrition Cycle. Saudi Med. J. 2021;42:146–150. doi: 10.15537/smj.2021.2.25587. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The original contributions presented in the study are included in the article, further inquiries can be directed to the corresponding author.