Abstract
It was previously shown that the human U1A protein, one of three U1 small nuclear ribonucleoprotein-specific proteins, autoregulates its own production by binding to and inhibiting the polyadenylation of its own pre-mRNA. The U1A autoregulatory complex requires two molecules of U1A protein to cooperatively bind a 50-nucleotide polyadenylation-inhibitory element (PIE) RNA located in the U1A 3′ untranslated region. Based on both biochemical and nuclear magnetic resonance structural data, it was predicted that protein-protein interactions between the N-terminal regions (amino acids [aa] 1 to 115) of the two U1A proteins would form the basis for cooperative binding to PIE RNA and for inhibition of polyadenylation. In this study, we not only experimentally confirmed these predictions but discovered some unexpected features of how the U1A autoregulatory complex functions. We found that the U1A protein homodimerizes in the yeast two-hybrid system even when its ability to bind RNA is incapacitated. U1A dimerization requires two separate regions, both located in the N-terminal 115 residues. Using both coselection and gel mobility shift assays, U1A dimerization was also observed in vitro and found to depend on the same two regions that were found in vivo. Mutation of the second homodimerization region (aa 103 to 115) also resulted in loss of inhibition of polyadenylation and loss of cooperative binding of two U1A protein molecules to PIE RNA. This same mutation had no effect on the binding of one U1A protein molecule to PIE RNA. A peptide containing two copies of aa 103 to 115 is a potent inhibitor of polyadenylation. Based on these data, a model of the U1A autoregulatory complex is presented.
The U1 small nuclear ribonucleoprotein (snRNP) particle is the most abundant member of the spliceosomal snRNPs. Human U1 snRNP is comprised of 10 proteins and the 164-nucleotide U1 small nuclear RNA (U1RNA) and is required for splicing of pre-mRNA (38). One of the U1 snRNP-specific proteins, the U1A protein, contains two evolutionarily conserved RNA recognition motifs (RRMs) characteristic of a large family of proteins involved in the biosynthesis of cellular RNA (reviewed in reference 37). The signature motifs for the RRM family consist of two ribonucleoprotein (RNP) sequences, RNP1 and RNP2, which are the most conserved features of this family. The N-terminal RRM of U1A is, together with some flanking amino acids, necessary and sufficient for binding to the loop part of stem-loop 2 (SL2) sequence AUUGCAC of U1RNA (22, 27, 28). The structure of the N-terminal RRM of the U1A protein (amino acids [aa] 2 to 95) has been solved both by X-ray crystallography and by nuclear magnetic resonance (NMR) and consists of a β1α1β2β3α2β4 structure in which the β strands form a sheet with the highly conserved RNP1 and RNP2 motifs located in the two central β strands, β3 and β1, respectively (14, 23). An additional α helix (helix 3; hereafter referred to as helix C) is present when a longer fragment of the U1A protein is analyzed (aa 2 to 102; reference 15). Using both NMR and X-ray crystallography, the structure of U1A aa 2 to 98 in complex with SL2 of U1RNA has also been solved (1, 2, 15, 24). In this structure, the RNA loop lies across the β sheet, fitting into a groove formed between loop 3 (connecting β2 and β3) and the C-terminal portion of the RRM domain. In spite of intensive investigation, the C-terminal RRM (aa 202 to 283) of U1A does not seem to have any affinity for RNA (21).
The U1 snRNP particle is involved in the first step of spliceosome formation, in which it binds to the 5′ splice site of the pre-mRNA (reviewed in reference 18). It is possible that U1A is not essential for the splicing reaction, since in vitro splicing can still proceed in the absence of U1A. It has been suggested, however, that the U1A protein might play an important role in 5′ and 3′ splice site communication (33). In vertebrates, the U1A protein is able to regulate the polyadenylation of U1A pre-mRNA, thereby regulating its own expression level (4). The 3′ untranslated region (UTR) of the human U1A pre-mRNA contains a 50-nucleotide region, designated the polyadenylation-inhibitory element (PIE) RNA, whose sequence and structure are conserved in vertebrates. Located within the PIE RNA are two stretches of seven unpaired nucleotides designated loops 1 and 2, each being able to bind one molecule of U1A protein. Although one of the loops, when studied in isolation, has 27-fold lower affinity for U1A than the other, it was demonstrated that two molecules of U1A bind with high affinity (Kd, ∼0.1 nM) to PIE RNA, indicative of cooperative RNA binding (4, 35). The resulting (U1A)2-PIE RNA complex inhibits addition of the poly(A) tail to the U1A pre-mRNA by specifically inhibiting the enzyme poly(A) polymerase (PAP) (10). Inhibition of polyadenylation requires both the C-terminal 20 residues of PAP and aa 103 to 115 of U1A.
A model has been proposed in which the U1A autoregulatory complex inhibits PAP by bringing two copies of U1A aa 103 to 115 into close proximity (11, 34). In support of this model, it was found that two molecules, but not one molecule, of U1A bound to PIE RNA inhibit PAP. Likewise, a monomeric peptide consisting of U1A aa 103 to 115 is unable to inhibit PAP; however, upon increasing its local concentration by conjugation to bovine serum albumin (BSA), this same peptide becomes a potent inhibitor of PAP (11). PAP inhibition by the BSA-peptide conjugate does not require PIE RNA, suggesting that the main role of PIE RNA in PAP inhibition is to bring the two U1A proteins into close proximity. Indeed, the unusual secondary structure of PIE RNA is not essential for inhibition (11, 34). Independent of the biochemical analysis, the determination of the structure by NMR of one molecule of U1A (aa 2 to 98) bound to PIE RNA (1) has also led to the proposal (based on modeling) that the two PIE-RNA-bound U1A proteins make extensive protein-protein interactions throughout the N-terminal 100 residues (16).
Here we show, by using both the yeast two-hybrid system and in vitro assays, that U1A is able to homodimerize in the absence of RNA sequences that specifically bind U1A (i.e., SL2 of U1RNA and PIE RNA). Dimerization requires two regions, both located in the N-terminal 115 residues. Mutations of the second region (aa 103 to 115) which abolish dimerization also result in either reduction or complete loss of cooperative binding to PIE RNA but with no effect on U1A binding as a monomer to PIE RNA. These same mutations also result in loss of U1A's ability to inhibit polyadenylation. A dimeric form of a peptide containing these residues also inhibits polyadenylation. A model integrating these results will be presented explaining how the U1A autoregulatory complex functions.
MATERIALS AND METHODS
cDNAs and plasmids.
All of the wild-type and mutant U1A proteins and mRNAs used in this study were encoded by cDNAs lacking the U1A binding site (previously designated ΔB1/2) located in the 3′ UTR; i.e., the cDNAs lack the PIE RNA sequence. The plasmid used to produce the U1A(52/53) mutant lacking PIE RNA was constructed by inserting the NcoI/BglII fragment of the U1A(52/53) plasmid (3) into an NcoI/BglII-digested U1AΔB1/2 plasmid (4). The mutant U1A(52/53, 106/108) and U1A(52/53, 110/112) plasmids contain, in addition to substitutions at positions 52 and 53, substitutions at positions 106 to 108 (Arg-Lys-Arg is changed to Gly-Ser-Ile) and 110 to 112 (Lys-Arg-Lys is changed to Gly-Ser-Ile), respectively. These two mutant plasmids were produced in a three-step PCR using U1A(52/53) as a template in essentially the same manner as described before (25), with the exception that 20 instead of 10 cycles of amplification were performed in the second and third steps. The resulting fragment was cloned into the EcoRI site of pGEM-3Zf(+) and then moved to a plasmid lacking the PIE RNA in the 3′ UTR as described above using NcoI/BglII. To produce U1A104–282, a cDNA encoding U1A(102/103) was digested with BamHI/HindIII, and the resulting fragment was cloned into the BamHI/HindIII site of U1A(s2/3) and then moved to a plasmid lacking the PIE RNA in the 3′ UTR (3).
Cloning of fusion proteins for the yeast two-hybrid assay.
All of the U1A-derived, yeast two-hybrid plasmids had a mutated PIE RNA sequence which inactivated U1A binding. Fusions to the LexA DNA-binding domain were constructed by subcloning of either the EcoRI fragment of U1C (32) or the NcoI/EcoRI fragments of U1AΔB1/2 and U1AΔB1/2(52/53) into pEG202. Fusions to the B42 activation domain were constructed by subcloning into pJG4-5 using EcoRI for U1C and U2B" (13) or NcoI/EcoRI for U1A, U1A(52/53), (U1AΔB1/2(52/53), U1AΔB1/2, U1A1–101, U1AΔB1/2104–282, U1AΔB1/2(52/53, 106/108), U1AΔB1/2(52/53, 110/112), U2B"1–98AΔB1/2, U2B"1–145AΔB1/2, U1A1–101B", and U1A1–202B".
Yeast two-hybrid assay.
We used the Brent yeast two-hybrid system described by Guyris and coworkers (12) to study protein-protein interactions. EGY48 yeast cells (trp1, his3, ura3) were grown and transformed with the following vectors: (i) pEG202 (36), encoding the DNA binding domain of LexA and a HIS3 selectable marker; (ii) pJG4-5 (36), encoding the activation domain of B42 and a TRP1 selectable marker; and (iii) pSH18-34, containing the lacZ gene under the control of the lexA operator and a URA3 selectable marker. Transformants were selected on glucose plates lacking histidine, uracil, and tryptophan, and several colonies were streaked and grown in medium lacking histidine, uracil, and tryptophan. Exponentially growing cells were harvested (optical density at 600 nm [OD600], 0.5 to 1.0/ml), and 1 ml of yeast cells was pelleted (1 min at 10,000 × g) and resuspended in 250 μl of buffer Z (100 mM NaPO4 [pH 7.2], 10 mM KCl, 1 mM MgSO4, 0.36% β-mercaptoethanol) and 100 μl of double-distilled H2O saturated with ether. After mixing and spinning (1 min at 10,000 × g), the ether was evaporated (30 min, 20°C). Subsequently, 100 μl of extract was transferred to a 96-well plate and incubated for 5 min at 30°C and 100 μl of o-nitrophenyl-β-d-galactopyranoside (4 mg/ml in buffer Z) was added to start the assay. The activity was followed during the course of the reaction (generally, 60 min) by determining the OD405 every 60 s in a microplate scanner (Biotek Instruments Ceres 900CUV). The amount of protein present in the β-galactosidase (β-gal) assay was established by addition of 100 μl of Bradford reagens (Bio-Rad) to 10 μl of extract and measurement of OD595. The β-gal activity was determined as follows: X · 1/OD595. The linear part of the obtained OD405 curve was used to determine X, which was defined as the increase in the measured OD per unit of time in which absorption was measured. At least three independent colonies were analyzed for each pair of constructs. Independently of β-galactosidase activity, Western blotting was performed to monitor the efficiency of expression of U1A, U2B", and the chimeric proteins.
Recombinant U1A proteins and bovine PAP.
cDNAs encoding U1A(106/108) and U1A(110/112) containing a C-terminal tag of six histidines were constructed in PCRs using U1A(52/53, 106/108) and U1A(52/53, 110/112) as templates. U1A(scrambled), in which the sequence ERDRKREKRK (aa 103 to 112) was mutated into KKRRREDREK, was constructed using crossover PCR using appropriate primers. The histidine-tagged U1A proteins were purified from Escherichia coli using Ni2+-nitrilotriacetic acid and MonoS chromatography (11). Histidine-tagged bovine PAP was purified as described before (11).
Gel shift assays.
Electrophoretic mobility shift assays (EMSAs) were performed as described before (35). Briefly, either a 32P-end-labeled SL2 RNA oligonucleotide or uniformly labeled PIE RNA was added to the U1A protein in a mixture containing 10 mM Na-HEPES (pH 7.4), 50 mM KCl, 5% glycerol, 3 μg of BSA, 4 U of rRNasin (Promega), 1 mM MnCl2, and 2 μg of competitor tRNA at room temperature. The 15-μl reaction mixture was immediately loaded on a 7% (60:1) native polyacrylamide gel using 1× Tris-borate-EDTA as the running buffer. Gels containing the SL2 RNA oligonucleotide were electrophoresed for 1.5 h at 20 V/cm, while gels containing PIE RNA were electrophoresed for 2.5 to 3 h.
In vitro dimerization assay.
A 400-ng sample (3 to 5 μl) of recombinant U1A(his) in phosphate-buffered saline–250 mM imidazole (17) was incubated with in vitro-translated, 35S-labeled U1A proteins in a final volume of 20 μl of buffer containing 20 mM HEPES-KOH (pH 7.9), 100 mM KCl, 1 mM MgCl2, and 0.05% NP-40. After a 30-min incubation, 10 μl of packed protein A-agarose beads coupled to polyclonal anti-(his)6 antibodies (Biozym) were added together to the binding reaction mixture along with 300 μl of IPP100 (10 mM Tris [pH 8.0], 100 mM NaCl, 0.05% NP-40) and the mixture was rotated for 90 min at 4°C. The beads were washed three times with IPP100 and then resuspended in sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis (PAGE) sample buffer, and the bound proteins were separated on an SDS–10% polyacrylamide gel.
Determination of Kds.
Gel shifts were quantified with a PhosphorImager, and the Kds of monomer and dimer binding to PIE RNA or the Kd of monomer binding to U1 RNA was calculated as previously described (9, 35). Because U1A protein was sufficiently in excess of PIE RNA or U1RNA, we could assume that [U1A]free = [U1A]total. The Kd of monomer binding to U1RNA is the fraction of U1 RNA shifted by U1A divided by [U1A]total. The Kd of monomer binding to PIE RNA is the fraction of PIE RNA shifted both as a monomer and as a dimer divided by [U1A]total. The Kd of dimer binding to PIE RNA is the fraction of PIE RNA shifted as a dimer divided by [U1A]total.
Polyadenylation assay and peptides.
The polyadenylation assay was performed as described before (10, 11). Reactions proceeded for 30 min at 37°C and were followed by phenol-chloroform extraction and ethanol precipitation. Polyadenylated RNAs were then separated by denaturing PAGE and visualized by autoradiography. Peptides were purchased from Research Genetics. The homodimerization of the monomeric peptide via N-terminal cysteine disulfide bond formation was performed under reducing conditions, and the resulting dimeric peptide purified by reverse-phase chromatography.
RESULTS
The human U1A protein homodimerizes in the yeast two-hybrid system.
In the crystal structure of the free human U1A protein (aa 2 to 95), two U1A molecules appeared to interact through a hydrophobic surface at the side opposite the RNA binding surface (23). Paradoxically, no dimerization of U1A in solution has been detected (2), although cooperative binding of the two U1A proteins to PIE RNA, indicating a possible direct interaction between the two U1A proteins within the autoregulatory complex, has been demonstrated (35). To determine whether U1A protein could dimerize, the yeast two-hybrid system that allows detection of protein-protein interactions in vivo was employed (6). In this assay, two proteins are fused to either the lexA DNA binding domain, the so-called bait, or the B42 activation domain, the so-called prey. If the two fusion proteins interact, the lacZ gene from the reporter plasmid is transcribed. By determining the β-gal activity in a quantitative assay, the efficiency of interaction between bait and prey fusion proteins can be measured. Because PIE RNA binds with high affinity to two human U1A molecules, its presence in yeast cells would artifactually promote U1A protein dimerization in vivo. Therefore, all of our U1A constructs contained a mutated PIE RNA sequence previously shown to inactivate U1A binding (4).
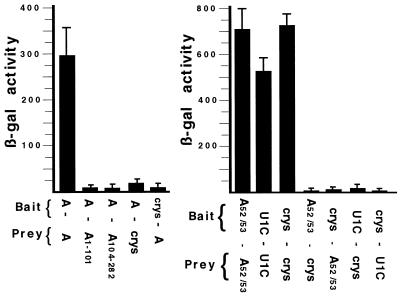
As shown in Fig. 1, U1A protein does homodimerize in yeast with an affinity similar to that of the positive controls represented in the right panel by homodimerization of either the U1 snRNP-specific U1C protein or the αBcrystallin protein (5, 17). U1A homodimerization was specific, since only low levels of β-gal activity were observed between U1A and αBcrystallin. However, these results did not distinguish whether U1A dimerization was based on direct protein-protein interactions or indirect interactions, for example, two U1A protein molecules binding to one of RNA. We would not expect RNA involvement, since it has been reported that the human U1A protein does not bind either yeast U1 RNA or the pre-mRNA for the yeast U1A homologue, the mud1 gene (19, 20). Because it was still a formal possibility that yeast cells contain an RNA with multiple, cryptic U1A binding sites that promote U1A dimerization, we also tested the U1A52/53 mutant, in which residues 52 and 53 were mutated to Gly and Ser, respectively. Compared to wild-type U1A, the U1A52/53 mutant has a severe (1,000-fold) reduction in affinity for either SL2 of vertebrate U1RNA or PIE RNA (4, 27). If U1A homodimerization is based on yeast cells containing single RNA molecules with multiple cryptic U1A binding sites (or, alternatively, nonspecific RNA binding), we predict that the U1A(52/53) mutant would dimerize less efficiently in yeast. Instead, just the opposite is observed in that the U1A(52/53) mutant homodimerized more efficiently than wild-type U1A protein and was specific since only low levels of β-gal activity were observed between U1A(52/53) and αBcrystallin. This observation is most likely explained by the fact that U1A(52/53) expression in yeast is more efficient than expression of wild-type U1A (Western blot; data not shown). The data presented below and the fact that U1A(52/53) homodimerizes at least as well as wild-type U1A support our view that homodimerization is primarily based on direct U1A protein-protein interactions and not on RNA binding. However, given the constraints of the two-hybrid assay, we cannot rule out contributions from nonspecific RNA binding.
FIG. 1.
Two-hybrid analysis of U1A. The two panels represent two independent experiments. Each histogram represents the mean value of three independent transformants, and standard deviations are indicated. The y axis is in β-gal units. Indicated below each histogram are the identities of the fusion proteins expressed in the cells and whether the fusion proteins come from the bait or the prey plasmid. (Left) Analysis of wild-type and deletion mutant forms of U1A. (Right) Analysis of U1A(52/53) mutant proteins. U1C is the human U1 snRNP-specific U1C protein, and crys is the αBcrystallin protein.
Two domains located in the N-terminal 115 aa residues are involved in homodimerization.
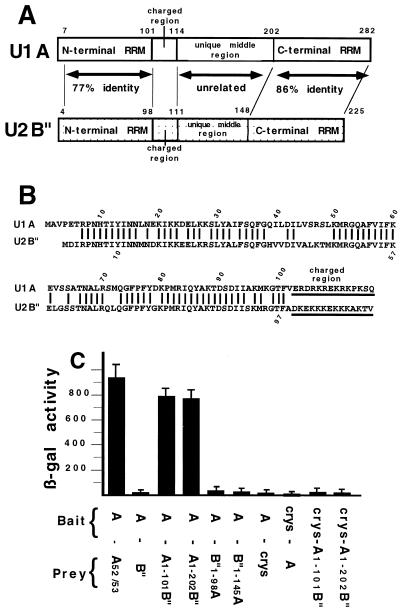
To identify which domains of U1A were involved in homodimerization the N-terminal RRM domain, U1A(1-101), and the C-terminal two-thirds of the protein, U1A(104-282), were cloned separately into the prey vector pJG4-5 and tested in the two-hybrid assay. Somewhat surprisingly, both U1A(1-101) and U1A(104-282) were unable to dimerize with wild-type U1A in the yeast two-hybrid system (left panel of Fig. 1) although both fusion proteins were expressed in the yeast cells (Western blot; data not shown). These results imply that these two constructs disrupted the dimerization domain or that more than one domain of U1A is necessary, although it is also possible that these deletion mutants were improperly folded. To test these possibilities, we attempted to test other truncated U1A protein constructs. Unfortunately, these truncated U1A proteins were not stably expressed in the yeast two-hybrid assay (J. Klein Gunnewiek, unpublished observations). This led us to take a different approach, which took advantage of our previous work using chimeric proteins consisting of human U1A and human U2B", a U2 snRNP-specific protein which is homologous to U1A (Fig. 2A and B). These chimeric proteins are known to fold properly in solution and can have either U1A or U2B" characteristics, depending on which type of domain is present (28). Indeed, these hybrid proteins have been extensively utilized for understanding both RNA-protein and protein-protein interactions that occur in the U1 and U2 snRNP particles (28, 29, 30). Several of these chimeric U1A/U2B" proteins were cloned into the prey vector pJB4-5 and tested for the ability to heterodimerize with U1A protein in the two-hybrid assay.
FIG. 2.
Two-hybrid analysis of U1A/U2B" chimeras. Structural features of U1A (31) and U2B" (13) and results of yeast two-hybrid analysis of chimeras of U1A and U2B". (A) The open box represents the U1A protein, and the speckled box represents the U2B" protein. Both the U1A and the U2B" proteins contain two RRMs separated by an unrelated middle region. The charged region which is found in both proteins is also indicated. (B) Sequence alignment of the N-terminal portion of the human U1A (above) and U2B" (below) proteins. Only identical residues are indicated by a line. The position of the charged region is also indicated. (C) Results of two-hybrid analysis of the chimeric U1A/U2B" proteins. Chimera U1A(1-101)-B" is aa 1 to 101 of U1A and aa 101 to 225 of U2B", chimera U1A(1-202)-B" is aa 1 to 202 of U1A and aa 148 to 225 of U2B", chimera B"(1-98)-U1A is aa 1 to 98 of U2B" and aa 104 to 282 of U1A, and chimera B" (1-145)-U1A is aa 1 to 145 of U2B" and aa 205 to 282 of U1A. The features of the histograms are as described in the legend to Fig. 1.
Somewhat surprisingly, given that the two proteins are so closely related, wild-type U2B" was unable to heterodimerize with the U1A protein (Fig. 2C). This result further confirmed the specificity of the two-hybrid approach. In contrast to U2B", the chimera U1A(1-101)-B", which contains the N-terminal 101 amino acids of U1A fused to the C-terminal part (aa 101 to 225) of U2B", was able to heterodimerize to wild-type U1A. Three conclusions could be drawn from this result. First, U1A residues 1 to 101 are required for dimerization even though they are not sufficient (Fig. 1). Second, residues 101 to 225 of U2B" were actively contributing to dimerization with U1A. Third, not any RRM can functionally substitute for the N-terminal RRM of U1A. Thus, at least two regions of U1A were needed for homodimerization, with one region being located in U1A residues 1 to 101 and the other being located in residues 102 to 282 but being able to be replaced with the corresponding residues in U2B". Note that these results do not distinguish whether the contribution to homodimerization of U1A's RRM domain is due to RNA-protein binding or to protein-protein interactions.
Testing of several other chimeric constructs demonstrated that residues 1 to 98 of U2B" are unable to functionally substitute for corresponding residues 1 to 101 in U1A. This result is all the more striking since this domain of B" is not only 77% identical in sequence to U1A (Fig. 2B) but is also known, based on its X-ray crystal structure, to fold into the same three-dimensional structure, in which the primary chain folds into a βαββαβ pattern (26). Thus, the contribution of aa 1 to 101 of U1A to homodimerization is mediated by only a few residues specific to U1A which are not found in U2B". As expected, none of these chimeric proteins bound the control protein, αBcrystallin. Similar results were obtained when U1A(52/53) was used as bait instead of U1A (data not shown). Although testing of other chimeric proteins suggested that the second region included aa 103 to 115 (data not shown), we could not be certain that the chimeric proteins accurately reflected interactions occurring during U1A-U1A homodimerization. Thus, we decided to return to analyzing only the U1A protein.
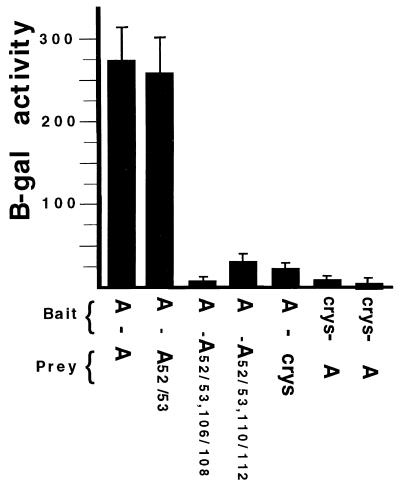
Both the U1A and U2B" proteins contain a middle region which separates the two highly-related RRM domains (Fig. 2A). Although both middle regions are, for the most part, unrelated in sequence, we noted that both contain a short stretch of 14 charged residues (Fig. 2B). Results from a systematic analysis of the U1A autoregulatory complex suggested that these charged residues form a homodimerization surface which would be essential for inhibition of polyadenylation (11). To investigate this, three residues (aa 106 to 108) in this region were mutated from Arg-Lys-Arg to Gly-Ser-Ile in the context of the U1A(52/53) mutant construct to produce the construct U1A(52/53, 106/108). Two-hybrid analysis of this mutant (Fig. 3) demonstrated that it could not dimerize with the wild-type U1A protein. Mutation of three adjacent residues (aa 110 to 112) from Lys-Arg-Lys to Gly-Ser-Ile, producing construct U1A(52/53, 110/112), also resulted in loss of dimerization activity (Fig. 3). In summary, the two-hybrid data supported our conclusion that U1A homodimerization requires two regions, the first (aa 1 to 101) being more constrained in its sequence content than the second (aa 103 to 115).
FIG. 3.
Two-hybrid analysis of U1As mutated in the charged domain. The features of the histograms are as described in the legend to Fig. 1.
Homodimerization is also observed in vitro.
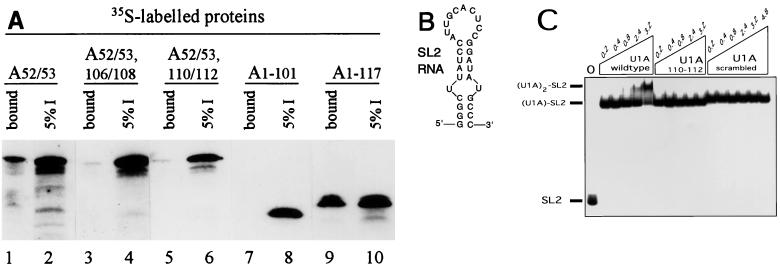
Because the two-hybrid approach is inherently limited in being able to show direct protein-protein interactions, we decided to utilize in vitro approaches. In one type of approach, we incubated bacterially expressed and highly purified U1A(his) (containing a C-terminal histidine tag) with in vitro-translated, 35S-labeled U1A protein. The incubation mixture was then subjected to selection using agarose beads coupled to a polyclonal antibody specific to the histidine tag. After extensive washing of the beads, the tightly bound proteins were eluted in SDS buffer and analyzed by SDS-PAGE and autoradiography. The appearance of 35S-labeled U1A protein in the bound fraction would indicate that it physically interacts with the unlabeled U1A(his) protein. Two controls that were employed in the two-hybrid analysis were used here: (i) the U1A(52/53) mutant mRNA, to ensure that the 35S-labeled U1A protein was not being coselected by RNA binding, and (ii) U1A mRNA that lacked the PIE RNA sequence in the 3′ UTR. The results shown in lanes 1 and 2 of Fig. 4A indicate that the two U1A proteins interact. Although this interaction is rather inefficient, it requires the same domains identified in vivo since the mutant constructs U1A(52/53, 106/108) and U1A(52/53, 110/112) showed a marked reduction in binding to the unlabeled U1A(his) protein (Fig. 4A, lanes 3 to 6). To determine the minimal fragment size of U1A needed for interaction to occur, we tested two N-terminal fragments. As shown in lanes 7 to 10, and consistent with the results in Fig. 1, U1A(1-101) could not interact with wild-type U1A(his). In contrast, U1A(1-117) was able to bind to U1A(his), indicating that the N-terminal 117 residues contain all of the determinants needed to dimerize with the wild-type U1A protein. Finally, we noted that although RNAs containing high-affinity U1A binding sites were absent in these assays, RNase treatment of these complexes, while not changing the specificity of these interactions, did reduce the overall efficiency of interaction (data not shown). Additionally, as expected, none of the 35S-labeled proteins bound to the agarose beads in the absence of the U1A(his) protein (data not shown).
FIG. 4.
U1A homodimerizes in vitro using the same domains as in vivo. (A) Results of histidine tag coselection experiments using an anti-his polyclonal antibody coupled to agarose beads. A 13-pmol (400-ng) sample of recombinant U1A(his) (histidine tagged) was incubated with 35S-labeled mutant U1A proteins (as indicated above the autoradiogram); this was followed by coselection with anti-his antibody coupled to agarose beads. After extensive washing of the beads, the bound proteins were eluted in SDS buffer and analyzed by SDS-PAGE and autoradiography. The even-numbered lanes represent 5% of the total input of 35S-labeled protein, and the odd-numbered lanes represent 50% of the bound proteins that were eluted. (B) Sequence of the SL2 RNA oligonucleotide used in the gel shift experiments in panel C. (C) EMSA of wild-type and mutant U1A proteins binding to the SL2 RNA oligonucleotide. Each lane contains 1 nM 32P-end-labeled SL2 RNA oligonucleotide. Lanes 2 to 17 contain, in addition, increasing micromolar concentrations of wild-type U1A (lanes 2 to 6), U1A(110/112) mutant protein (lanes 7 to 11), or U1A(scrambled) mutant protein (lanes 12 to 17), as indicated above the autoradiogram. On the left are indicated the positions of the unbound SL2 RNA, the (U1A)1-SL2 RNA complex, and the (U1A)2-SL2 RNA complex.
Recombinant, purified U1A also dimerizes.
Because the in vitro assay used in Fig. 4A contains RNA and protein present in the wheat germ extract, we next asked whether recombinant, purified U1A would homodimerize in vitro. Using an EMSA, it has been shown that recombinant U1A protein efficiently and specifically binds to labeled RNA oligonucleotides containing the sequence of stem-loop 2 of U1RNA (27). As shown in Fig. 4C, the 32P-labeled SL2 RNA oligonucleotide (the sequence is given in Fig. 4B) is completely bound when 0.2 μM wild-type or mutant U1A is present. Further addition of wild-type U1A eventually leads to a second U1A molecule binding; however, only when 3.2 μM U1A is added is all of the (U1A)1-SL2 RNA complex shifted to (U1A)2-SL2 RNA (lanes 2 to 6). Thus, binding of the second U1A molecule is very inefficient, yielding an approximate Kd of interaction in the 10 μM range.
Based on the homodimerization analysis already presented, we would predict that the second U1A molecule enters the (U1A)1-SL2 RNA complex primarily by protein-protein interactions rather than by nonspecifically binding to an exposed portion of the SL2 RNA. To test this prediction, we performed the same EMSA analysis with identical concentrations of the U1A(110/112) mutant construct. As seen in lanes 7 to 11 of Fig. 4C, this mutant, which was defective in homodimerization, is also unable to bind as two molecules to SL2 RNA even though it can readily bind as one molecule. Under these same conditions, the other homodimerization mutant construct U1A(106/108) was also unable to bind as two molecules to SL2 RNA (data not shown). An additional U1A mutant protein, U1A(scrambled), in which the sequence of residues ERDRKREKRK, from aa 103 to aa 112, was scrambled to become KKRRREDREK, was also unable to bind as two molecules to SL2 RNA (Fig. 4C, lanes 12 to 17). Furthermore, two other recombinant U1A proteins, consisting of either just the N-terminal RRM domain (aa 1 to 101) or just the C-terminal RRM domain (aa 200 to 282), were also unable to bind as two molecules to SL2 RNA (data not shown). Note that these same results were obtained when the EMSA was performed at 4°C, which can, in some cases, stabilize weak protein-protein interactions (data not shown). Taken together, these data argue in favor of the view that the second U1A molecule binds the (U1A)1-SL2 RNA complex primarily by protein-protein interactions that depend on aa 103 to 115. Therefore, we conclude that the charged region of U1A, from aa 103 to aa 115, is required for homodimerization both in the yeast two-hybrid system and in two types of in vitro assays.
Cooperative RNA binding requires one of the homodimerization domains.
It has been previously demonstrated that the Kds of the two individual binding sites on PIE RNA for U1A differ by about 27-fold (35). This was done by measuring the Kd of each site in the absence of the other site which was inactivated by mutation. However, when both binding sites are present, as is the case for wild-type PIE RNA, the second molecule of U1A binds with nearly the same affinity as the first molecule, indicating that the two U1A proteins cooperatively bind, suggesting a direct interaction between the two RNA-bound U1A proteins. Thus, the presence of the first RNA-bound U1A protein raises the affinity of binding of the second U1A protein by roughly 27-fold. To determine whether the homodimerization regions identified as described above were responsible for cooperative RNA binding, we measured the Kds of U1A proteins mutated in these regions to determine whether the binding affinity of the second U1A molecule for PIE RNA was affected. Unfortunately, we could not analyze mutations in the N-terminal domain (aa 1 to 101) of U1A since they are known to severely affect the binding of one molecule of U1A to PIE RNA (as well as to SL2 of the U1RNA). In contrast, we could analyze the mutations in aa 103 to 115 since they do not affect the binding of one molecule of U1A to PIE RNA.
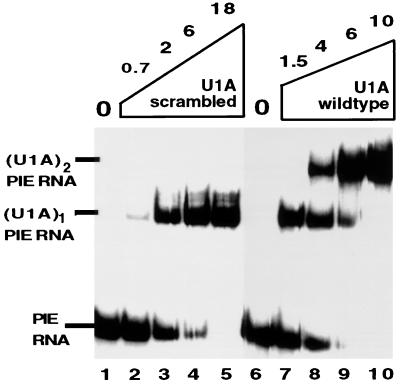
Table 1 summarizes the Kds that were determined for the binding of the wild-type and mutant U1A proteins to both the PIE and U1RNAs. All three mutant proteins showed no statistically significant defect in binding of one molecule of U1A to either U1 or PIE RNA compared with wild-type U1A. In contrast, all three mutant proteins showed a statistically significant reduction in the ability to bind as two molecules to PIE RNA compared to wild-type U1A, indicating they were defective in cooperative binding to PIE RNA. The most severe defect was seen with the U1A(scrambled) mutant protein, which had a 34-fold reduction in its Kd of binding as two molecules to PIE RNA. Figure 5 gives an example of the EMSA data for the U1A(scrambled) mutant protein compared to wild-type U1A. Both proteins bind with similar affinity as a monomer to PIE RNA; however, U1A(scrambled) is severely impaired for binding as two molecules to PIE RNA. Increasing the U1A(scrambled) concentration to 300 nM eventually results in significant dimer binding (data not shown). Thus, within the statistical limits of these measurements, we conclude that U1A(scrambled) has a complete loss of cooperative binding to PIE RNA while the other two mutant proteins showed a modest (twofold), but statistically significant, reduction in cooperative binding.
TABLE 1.
Kds of wild-type and mutant U1A binding to U1 RNA or to PIE RNA
| RNA substrate | Mean Kd of U1A protein ± SEMa
|
|||
|---|---|---|---|---|
| Wild type | 106/108 mutant | 110/112 mutant | Scrambled | |
| U1 RNA monomer | 5 ± 1.5 | 5 ± 2 | 5 ± 1.7 | 6 ± 1.8 |
| PIE RNA monomer | 9 ± 1.9 | 10 ± 2.1 | 10 ± 1.9 | 11 ± 2 |
| PIE RNA dimer | 11 ± 1.8 | 20 ± 4.2 | 27 ± 5 | 310 ± 55 |
All Kds are expressed as 10−11 molar and are rounded to the nearest whole digit. All Kds were determined by EMSA.
FIG. 5.
The U1A(scrambled) mutant protein has lost cooperative binding to PIE RNA. EMSA of wild-type U1A and U1A(scrambled) mutant protein binding to 32P-labeled PIE RNA. Each lane contains 0.5 nM 32P-labeled PIE RNA. Lanes 2 to 5 and 7 to 10 contain, in addition, increasing nanomolar concentrations of recombinant wild-type U1A or U1A(scrambled), as indicated above the autoradiogram. On the left are indicated the positions of PIE RNA, the (U1A)1-PIE RNA complex, and the (U1A)2-PIE RNA complex. Note that the amounts of U1A used here are ∼1,000-fold less than those used for Fig. 4C.
Inhibition of polyadenylation requires one of the homodimerization domains.
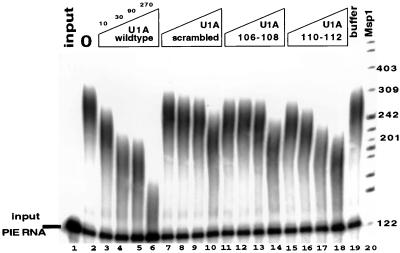
It has previously been shown that aa 103 to 115 of U1A are required for inhibition of PAP (11); however, an analysis of finer-scale mutant proteins was not done at that time. To determine whether the homodimerization-cooperativity region identified above was also important for inhibition of polyadenylation and PAP, we analyzed these same U1A mutants in polyadenylation assays. Figure 6 gives the results of this analysis in which poly(A) assays with recombinant bovine PAP were performed as described before (11). Inhibition of PAP activity was observed with 10 nM wild-type U1A. However, all three U1A mutant proteins showed a marked defect in being able to inhibit PAP activity. The U1A(scrambled) and U1A(106/108) proteins were the most defective, whereas U1A(110/112) showed a partial defect in the ability to inhibit PAP, although it was still less efficient than wild-type U1A. Interestingly, the three mutant proteins have somewhat different Kds of binding as a dimer to PIE RNA compared to their ability to inhibit polyadenylation. This may indicate that different residues of this region make different contributions to RNA binding or polyadenylation inhibition, although other explanations cannot be excluded. For example, RNA binding itself could alter the conformation of U1A such that the mutant regions 106 to 108 and 110 to 112 become less effective in dimerization. A more extensive mutagenic analysis of this region, along with determination of the atomic structure of the complex, will likely resolve this issue. Regardless, these results demonstrate that U1A residues involved in U1A dimerization and cooperative RNA binding are also important for inhibition of polyadenylation. Note that compared to Fig. 5, more U1A (three- to fivefold) is needed for significant inhibition of PAP than is needed to bind PIE RNA. This observation is consistent with our previous results (10, 11) and is because (i) inhibition of PAP will occur when most of the PIE RNA is bound by two molecules of U1A and (ii) twice as much PIE RNA is present in the polyadenylation reactions than in Fig. 5.
FIG. 6.
The U1A dimerization domain (aa 103 to 115) is also needed for PAP inhibition. Shown is the inhibition of PAP by either wild-type U1A (lanes 3 to 6), U1A(scrambled) (lanes 7 to 10), U1A(106/108) (lanes 11 to 14), or U1A(110/112) (lanes 15 to 18). Above the autoradiogram is indicated the nanomolar concentration of U1A present in each lane. Polyadenylated RNAs were separated on a denaturing polyacrylamide gel. Each lane contains polyadenylation buffers and 1 nM 32P-labeled PIE RNA incubated either in the absence (lane 1) or in the presence (lanes 2 to 19) of 5 nM recombinant bovine PAP. Lane 19 contains U1A storage buffer in place of U1A protein as a control. Lane 20 is a 32P-end-labeled MspI digest of pBR322, and the sizes of the bands are indicated in nucleotides.
A peptide containing two copies of the homodimerization domain is a potent inhibitor of PAP.
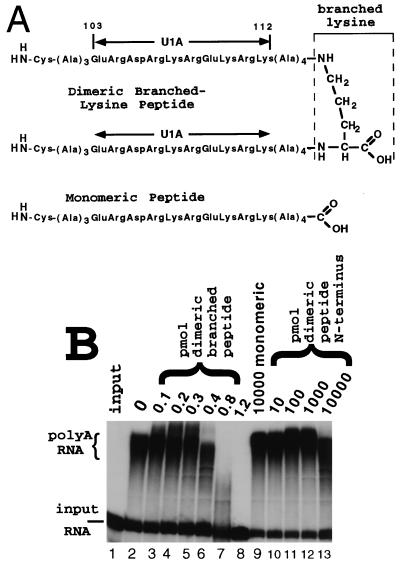
Previous biochemical and NMR structural analyses of the U1A autoregulatory complex led to the hypothesis that the binding of two U1A proteins to PIE RNA juxtaposes two copies of U1A residues 103 to 115, resulting in the formation of a novel protein surface able to inhibit PAP (10, 11, 34). Indirect support of this hypothesis came from the observation that a monomeric peptide corresponding to aa 103 to 115 became a potent inhibitor of polyadenylation only when it was conjugated to BSA (11). This observation was not conclusive, however, since it was estimated that, on average, 10 to 15 peptide molecules were conjugated to every BSA molecule. To devise a more rigorous test of this hypothesis, we used a chemically synthesized peptide containing two tandem copies of aa 103 to 112 held together by a “branched” lysine (the structure is shown in Fig. 7A), which was the first residue in the synthesis. Note that chemical synthesis proceeds from the carboxy terminus to the amino terminus. As shown in Fig. 7B, the monomeric form of this peptide did not inhibit PAP even when used at 10,000-fold stoichiometric excess over the enzyme (lane 9). In contrast, the dimeric branched peptide was a potent inhibitor of PAP in that a 2:1 stoichiometry was sufficient for partial inhibition and a 12:1 ratio resulted in complete inhibition (lanes 3 to 8). An additional control consisting of the same peptide sequence but homodimerized by cysteine disulfide bridging of the N termini was also unable to inhibit PAP, even when used at 10,000-fold stoichiometric excess (lanes 10 to 13). Taken together, these results suggest that the active form of the U1A dimerization interface found in the U1A autoregulatory complex consists of two peptides of U1A lying approximately side by side in a parallel, instead of antiparallel, orientation. These results led us to propose the model shown in Fig. 8, which is discussed in more detail below.
FIG. 7.
Two copies of the aa 103 to 115 dimerization domain are sufficient to inhibit PAP. (A) Schematic of the structure of the monomeric peptide and the dimeric branched-lysine peptide used in the polyadenylation assay shown in panel B. Note that the monomeric peptide has an N-terminal cysteine to allow homodimerization by disulfide bond formation under reducing conditions. (B) The polyadenylation assay is as described in the legend to Fig. 6, except that the chemically synthesized peptides shown in panel A were used in place of U1A. Lanes 1 and 2 are as in Fig. 6. Lanes 2 to 13 contain 5 nM recombinant PAP. Lanes 3 to 8 contain increasing amounts of the dimeric branched-lysine peptide. Lane 9 contains 10,000 pmol of the monomeric peptide. Lanes 10 to 13 contain increasing amounts of the homodimeric peptide covalently linked N terminus to N terminus via cysteine disulfide bonds. The amount of peptide used in each lane is indicated in picomoles above the autoradiogram.
FIG. 8.
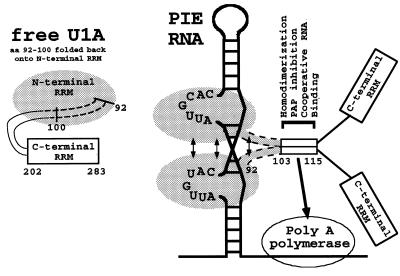
The U1A dimerization model incorporates results from a number of publications, including this one, as described in the text. Shown is a schematic of the structure of free U1A protein in order to illustrate that aa 92 to 102 are in a different conformation when the protein is RNA bound. Also shown are two U1A proteins in complex with PIE RNA. The region containing amino acids 92 to 115 of both U1A proteins is involved in protein-protein interactions, as indicated by the arrows. The dimerization of aa 103 to 113 is sufficient to create a surface that promotes both cooperative binding to PIE RNA and binding to and inhibition of vertebrate PAP. Note that the two copies of aa 92 to 115 are deliberately aligned in parallel to each other. In addition, stabilizing protein-protein interactions are present within the N-terminal 100 residues, as indicated by arrows.
DISCUSSION
We have presented a systematic analysis of the human U1A protein and the U1A autoregulatory complex which provides the first direct experimental data in support of predictions previously made by several laboratories (including our own) that protein-protein interactions between the N-terminal regions (aa 1 to 115) of the two U1A proteins would form the basis for cooperative binding to PIE RNA and for inhibition of polyadenylation. The work also uncovers some unexpected features of how the U1A autoregulatory complex functions. For example, U1A homodimerization both in vitro and in vivo was unexpected, as was the importance of the contribution of aa 103 to 115 to cooperative RNA binding. Additionally, the finding that a dimeric peptide inhibits polyadenylation when oriented in parallel to itself was unexpected and underscored the specificity of interactions present in the U1A autoregulatory complex.
Two regions of U1A have been identified which are required for homodimerization both in vivo and in vitro. One of these two regions, aa 103 to 115, is also required for cooperative binding to PIE RNA and for inhibition of PAP, both functions being used by U1A for autoregulation. We also demonstrate that the entire U1A autoregulatory complex can be functionally replaced, in terms of inhibition of PAP, by a small dimeric peptide containing two copies of U1A aa 103 to 112 linked at the C termini. The inhibition is specific, since a related dimeric peptide in which the two copies are linked via their N termini is unable to inhibit PAP. The dramatic difference in activity between these two dimeric peptides which are identical in sequence supports our view that conformational constraints imposed by the unusual architecture of this autoregulatory complex play a key role in its functioning. Thus, although we do not know the precise orientation of the two copies of U1A aa 103 to 115, our results are consistent with the idea that the active site of the U1A autoregulatory complex contains just two copies of aa 103 to 112, with one copy lying roughly parallel with the other.
Independent of the biochemical efforts to understand the mechanics of the U1A autoregulatory complex, the atomic structure of part of this complex was solved by both NMR and X-ray crystallography (1, 2, 16). Although, the structural determination used a truncated form of U1A (aa 2 to 98) that terminated just at the N-terminal boundary of aa 103 to 115, a number of features of this structure allowed predictions to be made, based on computer modeling, about the mechanics of the U1A autoregulatory complex. The results presented here provide direct biochemical evidence in support of these predictions and uncover some unpredicted features which are integrated into the model described below.
Model of the U1A autoregulatory complex.
Figure 8 presents an updated model of the U1A autoregulatory complex which incorporates both the biochemical analysis and the structural data and its consequent predictions. First, both U1A proteins lie on the same side of PIE RNA. Although this is consistent with the RNA geometry and was pointed out by van Gelder et al. (35), the details of the orientation and the intermolecular interactions of helices and beta sheets of the two U1A proteins could not be foreseen at that time. Second, the extreme C terminus of the U1A fragment used in the structural work (helix C; aa 92 to 98) undergoes a conformational change, shifting its position by 135° upon binding to PIE RNA (2). This conformational change was also observed when U1A binds to the highly related RNA derived from SL2 of U1RNA (24). This conformational change allows additional intermolecular protein-protein interactions to occur between the two RNA-bound U1A proteins with the most-extensive interactions occurring between helix C of one U1A molecule and the corresponding helix C of the second. Although not directly predicted at that time, it was clearly plausible that the helix C-helix C homodimeric interactions could extend in the C-terminal direction up to aa 115, thereby including the region studied in this work. Third, the conformational change in PIE RNA also contributes to the functioning of the autoregulatory complex. Recent analysis of the global structure of both free and U1A-bound PIE RNAs indicates the existence of a strong bend in the helical axis of the RNA (7, 8) which would bring the two U1A proteins into even closer proximity. Although this analysis identified a possible conformational change in PIE RNA upon U1A binding which could form the basis for the homodimerization reported here, we think this unlikely since homodimerization was also observed in the absence of PIE RNA (both in yeast and in vitro upon binding to SL2 RNA). Resolution of this issue will come with additional data. Thus, both the geometry of PIE RNA binding and the intrinsic bending of PIE RNA constrain the two U1A proteins to lie on the same side of the complex. This and the conformational change in helix C combine to promote homodimeric interactions only in the RNA-bound form.
Role of RNA in homodimerization.
U1A is known to exist predominately as a monomer in solution, even at high concentrations (2), but upon addition of RNA containing a single U1A binding site (Fig. 4C), a second molecule of U1A binds via intermolecular interactions between the two aa 102-to-115 regions. This is readily explainable by the conformational change observed in these residues leading to exposure of a hydrophobic patch, thereby promoting dimerization (2). As discussed above, several reports have indicated that PIE RNA also undergoes a conformational change resulting in bending of the helical axis of the RNA (7, 8). Indeed, it has been demonstrated that the central stems separating the two bound U1As is twisted upon protein binding, which is consistent with the fact that U1A stabilizes and extends the helical stem of the RNA to which it is bound. It remains to be determined whether such a conformational change would occur before or upon binding of the second U1A molecule. Thus, it is conceivable that binding of one molecule of U1A stabilizes PIE RNA in a conformation that has higher affinity for the second U1A. It is expected that the structure of this complex at atomic resolution will be solved in the near future, which will undoubtedly go far toward answering these questions.
U1A protein does not homodimerize in the complete absence of RNA; however, our data do not clarify what role low-affinity binding to RNA plays in homodimerization. It is somewhat paradoxical that the two hybrid and in vitro selection assays, which contain uncharacterized low-affinity sites, yielded results in full agreement with those of the other in vitro assays using the high-affinity binding sites. Several explanations are possible, including the possibility that U1A binding to weak RNA sites results in a conformational change in helix C which promotes homodimerization. An alternative possibility is that loading of multiple U1As onto RNA molecules with weak sites could increase the local concentration sufficiently to support homodimerization. Further experiments will distinguish between these possibilities.
Lessons for RRM-containing proteins.
At the molecular level, the U1A autoregulatory system is one of the best-understood examples of regulated RNA processing in metazoans and contains a number of features that are used in more-complicated regulatory systems. U1A was the first protein in the RRM family for which the structure of both the free protein and the RNA-RRM complex was known. The beta sheet of the RRM provides a folding platform for the RNA by extending the double-helical structures of the bound RNA. An “induced-fit” type of binding occurs as the PIE RNA structure bends upon binding U1A, and in turn, U1A also undergoes a large conformational change (helix C) upon binding. This results in a high level of specificity and affinity in the formation of the complex.
The family of proteins containing one or more RRM domains includes well over 1,000 members and contains examples of genes present in the diverse array of macromolecular machinery that utilizes and synthesizes RNA, as well as genes that regulate that machinery. We expect that biochemical, biophysical, and structural investigations into the underlying mechanics of the U1A autoregulatory system will provide insights and well-defined examples for investigations of these more complex systems. It is notable that pre-mRNA, represented by PIE RNA, can impose architectural constraints on RNP complexes, resulting in the generation of new interactions and binding specificities. The large conformational changes induced by pre-mRNA binding also add to the potential to generate new regulatory functions, and that in U1A is all the more striking since it involves only 14 residues. Furthermore, the U1A pre-mRNA recruits proteins into the complex based both on direct, high-affinity RNA-protein interactions and on lower-affinity interactions in which cooperative RNA binding plays a major role. This is reminiscent of the mechanical workings of the spliceosome and the cleavage-polyadenylation complex, which assemble onto very long pre-mRNAs by combining a number of low-affinity interactions that ultimately lead to the exquisitely precise joining of exons and to 3′-end processing. Thus, it is reasonable to expect that these elements of RNA recognition and RNA-protein reorganization used in the relatively simple case of the U1A autoregulatory complex will also be utilized by other RRM family members in these more complicated systems.
ACKNOWLEDGMENTS
We thank Wilbert Boelens for helpful discussion and assistance with the two-hybrid assay and Ger Pruijn, Bom Ko, Luca Varani, and Gabriele Varani for critical reading of the manuscript. We also thank Luca Varani and Gabriele Varani for sharing data prior to publication.
This work was supported by SON grant 331-013 from the Netherlands Foundation for Chemical Research with financial aid from the Netherlands Organization for Scientific Research (NWO) and by NIH grant 1R01-GM57286 to S.I.G.
REFERENCES
- 1.Allain F H-T, Gubser C C, Howe P W A, Nagai K, Neuhaus D, Varani G. Specificity of ribonucleoprotein interaction determined by RNA folding during complex formation. Nature. 1996;380:646–650. doi: 10.1038/380646a0. [DOI] [PubMed] [Google Scholar]
- 2.Avis J M, Allain F H-T, Howe P, Varani G, Nagai K, Neuhaus D. Solution structure of the N-terminal RNP domain of U1A protein: the role of the C-terminal residues in structure stability and RNA binding. J Mol Biol. 1996;257:398–411. doi: 10.1006/jmbi.1996.0171. [DOI] [PubMed] [Google Scholar]
- 3.Boelens W, Scherly D, Jansen E J R, Kolen K, Mattaj I W, van Venrooij W J. Analysis of in vitro binding of U1-A protein mutants to U1 snRNA. Nucleic Acids Res. 1991;19:4611–4618. doi: 10.1093/nar/19.17.4611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Boelens W C, Jansen E J R, van Venrooij W J, Stripecke R, Mattaj I W, Gunderson S I. The human U1 snRNP-specific U1A protein inhibits polyadenylation of its own pre-mRNA. Cell. 1993;72:881–892. doi: 10.1016/0092-8674(93)90577-d. [DOI] [PubMed] [Google Scholar]
- 5.Boelens W C, Croes Y, de Ruwe M, de Reu L, De Jong W W. Negative charges in the C-terminal domain stabilize the alpha-βcrystallin complex. J Biol Chem. 1998;273:28085–28090. doi: 10.1074/jbc.273.43.28085. [DOI] [PubMed] [Google Scholar]
- 6.Fields S, Song O K. A novel genetic system to detect protein-protein interactions. Nature. 1989;340:245–246. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 7.Grainger R J, Murchie A I H, Norman D G, Lilley D M J. Severe axial bending of RNA induced by the U1A binding element present in the 3′ untranslated region of the U1A mRNA. J Mol Biol. 1997;273:84–92. doi: 10.1006/jmbi.1997.1289. [DOI] [PubMed] [Google Scholar]
- 8.Grainger R J, Norman D G, Lilley D M J. Binding of U1A protein into the 3′ untranslated region of its pre-mRNA. J Mol Biol. 1999;288:585–594. doi: 10.1006/jmbi.1999.2717. [DOI] [PubMed] [Google Scholar]
- 9.Gunderson S I, Chapman K A, Burgess R R. Interactions of T7 RNA polymerase with T7 late promoters measured by footprinting with methidiumpropyl-EDTA-iron (II) Biochemistry. 1987;26:1539–1546. doi: 10.1021/bi00380a007. [DOI] [PubMed] [Google Scholar]
- 10.Gunderson S I, Beyer K, Martin G, Keller W, Boelens W C, Mattaj I W. The human U1A snRNP protein regulates polyadenylation via a direct interaction with poly(A) polymerase. Cell. 1994;76:531–541. doi: 10.1016/0092-8674(94)90116-3. [DOI] [PubMed] [Google Scholar]
- 11.Gunderson S I, Vagner S, Polycarpou-Schwarz M, Mattaj I W. Involvement of the carboxyl terminus of poly(A) polymerase in U1A autoregulation and in the coupling of splicing and polyadenylation. Genes Dev. 1997;11:761–773. doi: 10.1101/gad.11.6.761. [DOI] [PubMed] [Google Scholar]
- 12.Guyris J, Golemis E, Chertkov H, Brent R. Cdi1, a human G1 and S phase protein phosphatase that associates with Cdk2. Cell. 1993;75:791–803. doi: 10.1016/0092-8674(93)90498-f. [DOI] [PubMed] [Google Scholar]
- 13.Habets W J, Sillekens P T G, Hoet M H, Schalken J A, Roebroek A J M, Leunissen J A M, van de Ven W J M, van Venrooij W J. Analysis of a cDNA clone expressing a human autoimmune antigen: full length sequence of the U2 small nuclear RNA associated B" antigen. Proc Natl Acad Sci USA. 1987;84:2421–2425. doi: 10.1073/pnas.84.8.2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffman D W, Query C C, Golden B L, White S W, Keene J D. RNA-binding domain of the A protein component of the U1 small nuclear ribonucleoprotein analyzed by NMR spectroscopy is structurally similar to ribosomal proteins. Proc Natl Acad Sci USA. 1991;88:2495–2499. doi: 10.1073/pnas.88.6.2495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Howe P, Nagai K, Neuhaus D, Varani G. NMR studies of U1 snRNA recognition by the N-terminal RNP domain of the human U1A protein. EMBO J. 1994;13:3873–3881. doi: 10.1002/j.1460-2075.1994.tb06698.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jovine L, Oubridge C, Avis J M, Nagai K. Two structurally different RNA molecules are bound by the spliceosomal protein U1A using the same recognition strategy. Structure. 1996;4:621–631. doi: 10.1016/s0969-2126(96)00066-4. [DOI] [PubMed] [Google Scholar]
- 17.Klein Gunnewiek J M T, Van Aarssen Y, Wassenaar R, Legrain P, van Venrooij W J, Nelissen R L H. Homodimerization of the human U1 snRNP-specific protein C. Nucleic Acids Res. 1995;23:4864–4871. doi: 10.1093/nar/23.23.4864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 19.Kretzner L, Krol A, Rosbash M. Saccharomyces cerevisiae U1 small nuclear RNA secondary structure contains both universal and yeast specific domains. Proc Natl Acad Sci USA. 1990;87:851–855. doi: 10.1073/pnas.87.2.851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liao X C, Tang J, Rosbash M. An enhancer screen identifies a gene that encodes the yeast U1 snRNP A protein: implications for snRNP protein function in pre-mRNA splicing. Genes Dev. 1993;7:419–428. doi: 10.1101/gad.7.3.419. [DOI] [PubMed] [Google Scholar]
- 21.Lu J, Hall K B. An RBD that does not bind RNA: NMR secondary structure determination and biochemical properties of the C-terminal RNA binding domain from the human U1A protein. J Mol Biol. 1995;247:739–752. doi: 10.1006/jmbi.1995.0177. [DOI] [PubMed] [Google Scholar]
- 22.Lutz-Freyermuth C, Query C C, Keene J D. Quantitative determination that one of two potential RNA-binding domains of the A protein component of the U1 small nuclear ribonucleoprotein complex binds with high affinity to stem-loop II of U1 RNA. Proc Natl Acad Sci USA. 1990;87:6393–6397. doi: 10.1073/pnas.87.16.6393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nagai K, Oubridge C, Jessen T H, Li J, Evans P R. Crystal structure of the RNA-binding domain of the U1 small nuclear ribonucleoprotein A. Nature. 1990;348:515–520. doi: 10.1038/348515a0. [DOI] [PubMed] [Google Scholar]
- 24.Oubridge C, Ito N, Evans P R, Teo C H, Nagai K. Crystal structure at 1.92 A resolution of the RNA-binding domain of the U1A spliceosomal protein complexed with an RNA hairpin. Nature. 1994;372:432–438. doi: 10.1038/372432a0. [DOI] [PubMed] [Google Scholar]
- 25.Picard V, Ersdal-Badju E, Lu A, Bock S C. A rapid and efficient one tube PCR-based mutagenesis technique using Pfu DNA polymerase. Nucleic Acids Res. 1994;22:2587–2591. doi: 10.1093/nar/22.13.2587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Price S R, Evans P R, Nagai K. Crystal structure of the splicesomal U2B"-U2A′ protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 27.Scherly D, Boelens W, van Venrooij W J, Dathan N A, Hamm J, Mattaj I W. Identification of the RNA binding segment of human U1A protein and definition of its binding site on U1 snRNA. EMBO J. 1989;8:4163–4170. doi: 10.1002/j.1460-2075.1989.tb08601.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scherly D, Boelens W, Dathan N A, van Venrooij W J, Mattaj I W. Major determinants of the specificity of interaction between small nuclear ribonucleoproteins U1A and U2B". Nature. 1990;345:502–506. doi: 10.1038/345502a0. [DOI] [PubMed] [Google Scholar]
- 29.Scherly D, Dathan N A, Boelens W, van Venrooij W J, Mattaj I W. The U2B" RNP motif as a site of protein-protein interaction. EMBO J. 1990;9:3675–3681. doi: 10.1002/j.1460-2075.1990.tb07579.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Scherly D, Kambach C, Boelens W, van Venrooij W J, Mattaj I W. Conserved amino acid residues within and outside of the N-terminal ribonucleoprotein motif of U1A small nuclear ribonucleoprotein involved in U1 RNA binding. J Mol Biol. 1991;219:577–584. doi: 10.1016/0022-2836(91)90651-l. [DOI] [PubMed] [Google Scholar]
- 31.Sillekens P T G, Habets W J, Beijer R P, van Venrooij W J. cDNA cloning of the human U1snRNA-associated A protein: extensive homology between U1 and U2 snRNP-specific proteins. EMBO J. 1987;6:3841–3848. doi: 10.1002/j.1460-2075.1987.tb02721.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sillekens P T G, Beijer R P, Habets W J, van Venrooij W J. Human U1 snRNP-specific C protein: complete cDNA and protein sequence and identification of a multigene family in mammals. Nucleic Acids Res. 1988;16:8307–8321. doi: 10.1093/nar/16.17.8307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tarn W Y, Steitz J A. Modulation of 5′ splice site choice in pre-messenger RNA by two distinct steps. Proc Natl Acad Sci USA. 1995;92:2504–2508. doi: 10.1073/pnas.92.7.2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Teunissen S W M, van Gelder C W G, van Venrooij W J. Probing the 3′ UTR structure of U1A mRNA and footprinting analysis of its complex with U1A protein. Biochemistry. 1997;36:1782–1789. doi: 10.1021/bi9623237. [DOI] [PubMed] [Google Scholar]
- 35.van Gelder C W G, Gunderson S I, Jansen E J R, Boelens W C, Polycarpou-Schwarz M, Mattaj I W, van Venrooij W J. A complex secondary structure in U1A pre-mRNA that binds two molecules of U1A protein is required for regulation of polyadenylation. EMBO J. 1993;12:5191–5200. doi: 10.1002/j.1460-2075.1993.tb06214.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Watson M A, Buckholz R, Weiner M P. Vectors encoding alternative antibiotic resistance for use in the yeast two-hybrid system. BioTechniques. 1996;21:255–259. doi: 10.2144/96212st02. [DOI] [PubMed] [Google Scholar]
- 37.Will C L, Behrens S E, Luhrmann R. Protein composition of mammalian spliceosomal snRNP. Mol Biol Rep. 1993;18:121–126. doi: 10.1007/BF00986766. [DOI] [PubMed] [Google Scholar]
- 38.Will C L, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]