Abstract
The human tumor necrosis factor alpha (TNF-α) gene is rapidly activated in response to multiple signals of stress and inflammation. We have identified transcription factors present in the TNF-α enhancer complex in vivo following ionophore stimulation (ATF-2/Jun and NFAT) and virus infection (ATF-2/Jun, NFAT, and Sp1), demonstrating a novel role for NFAT and Sp1 in virus induction of gene expression. We show that virus infection results in calcium flux and calcineurin-dependent NFAT dephosphorylation; however, relatively lower levels of NFAT are present in the nucleus following virus infection as compared to ionophore stimulation. Strikingly, Sp1 functionally synergizes with NFAT and ATF-2/c-jun in the activation of TNF-α gene transcription and selectively associates with the TNF-α promoter upon virus infection but not upon ionophore stimulation in vivo. We conclude that the specificity of TNF-α transcriptional activation is achieved through the assembly of stimulus-specific enhancer complexes and through synergistic interactions among the distinct activators within these enhancer complexes.
The human tumor necrosis factor alpha (TNF-α) gene is expressed in a variety of cell types in response to several different signal transduction pathways (for a review, see reference 2). In most cell types, TNF-α is not expressed prior to cellular stimulation. However, diverse extracellular stimuli, including exposure to calcium ionophore or antigen, virus infection, or bacterial lipopolysaccharide, can induce TNF-α gene expression. The common end point of these diverse signal transduction pathways is the activation of TNF-α gene transcription.
In activated T cells, TNF-α gene induction requires a cyclic AMP response element (CRE), which binds ATF-2/Jun, and two NFAT-binding sites, the −76-NFAT and κ3-NFAT sites (13, 28, 44). The nuclear translocation of NFAT proteins, which have been implicated in the regulation of a number of cytokine genes (for reviews, see references 7 and 37), requires the calcium-dependent phosphatase calcineurin. Nuclear translocation of NFAT proteins can be blocked by agents that inhibit the activity of calcineurin, such as the immunosuppressant drugs cyclosporin A (CsA) and FK506 (10).
TNF-α gene expression is also highly inducible in B cells activated through their antigen receptor or by calcium ionophore, and this induction is blocked by CsA (4, 14). However, in activated B cells, which have relatively lower levels of NFAT proteins than T cells do, TNF-α gene regulation does not require the κ3 element. Rather, induction of the gene depends upon the CRE and the high-affinity −76-NFAT site. Thus, the TNF-α gene is regulated in a cell type-specific manner in response to the same extracellular signal (45).
Infection of a cell by RNA or DNA viruses also induces TNF-α gene transcription (1, 51). Sendai virus, a single-stranded RNA paramyxovirus, is a prototypic inducer of the antiviral response (reviewed in reference 49). Infection of a variety of cell types by Sendai virus, including monocytes, T and B cells, and fibroblasts (11, 12, 14), results in the activation of the TNF-α gene and other virus-inducible genes.
Here we show that activation of TNF-α gene transcription by virus infection requires a unique combination of transcriptional activators and regulatory elements, which differs from the combination required for ionophore induction of the gene. We demonstrate that virus infection leads to intracellular calcium flux followed by NFAT dephosphorylation and translocation to the nucleus but to levels lower than those achieved after ionophore stimulation. Moreover, analysis of in vivo protein-DNA interactions reveals that stimulation by calcium ionophore leads to the formation of a TNF-α enhancer complex containing ATF-2/Jun and NFAT, while virus infection results in the recruitment of a unique TNF-α enhancer complex that contains ATF-2/Jun, NFAT, and Sp1. We also find that Sp1 functionally synergizes with NFAT, consistent with its requirement in activation of TNF-α gene expression under conditions of lower levels of NFAT following virus infection. Thus, the selective and inducer-specific recruitment of nucleoprotein-DNA complexes to the TNF-α promoter provides direct evidence for a general mechanism by which a single gene may be controlled in response to different extracellular stimuli.
MATERIALS AND METHODS
Cell culture, activation, and transfection.
The Ar-5 T cell clone, L929 cells, and A20 cells were grown and transfections were performed by using DEAE-dextran as previously described (11, 13, 24, 45). Thirty-six hours after transfection, cells were activated with Sendai virus (SPAFAS; Cantell strain) at a final concentration of 300 hemagglutinin (HA) units/ml or ionomycin (Calbiochem; 1 μM) or ionomycin and phorbol 12-myristate 13-acetate (PMA; Calbiochem; 200 nM) and harvested approximately 16 h later. Where indicated, cells were treated for 10 min with 1 μM CsA (Sandoz) before the addition of virus or ionomycin. Chloramphenicol acetyltransferase (CAT) assays were performed as previously described (13). Quantification of the conversion of [14C]chloramphenicol to its acetylated forms was obtained with a Betagen (Waltham, Mass.) Betascope. As a transfection control, the cytomegalovirus (CMV) β-galactosidase (β-Gal) plasmid (pCMVβ; Clontech) was cotransfected in all cases and extracts were normalized to β-Gal activity prior to performance of CAT assays. Differences in TNF-α reporter gene induction ratios between Fig. 1 and 2B resulted from the use of different antigen-specific stimulations of the Ar-5 T-cell clone in these experiments. SL2 cells were maintained and transfected as previously described, and Hsp β-Gal was used as a control for transfection efficiency (30, 41).
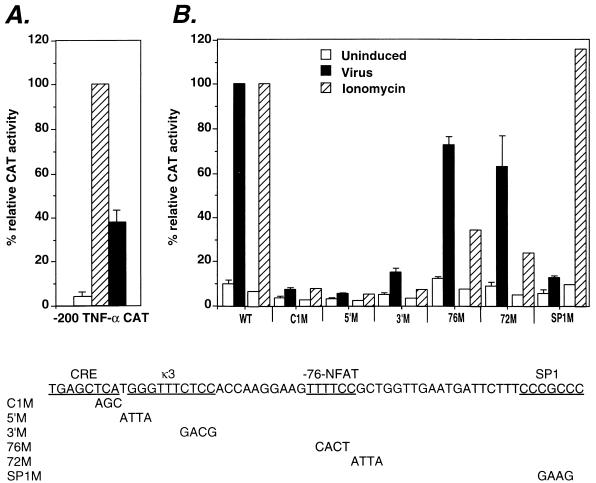
FIG. 1.
Virus induction of TNF-α gene requires the CRE/κ3 and SP1 sites. (A) Relative activation levels of the −200 TNF-α–CAT fusion construct by ionophore and virus. The Ar-5 T-cell clone was transfected with the −200 TNF-α promoter–CAT reporter construct. Cells were mock stimulated (Uninduced) or stimulated with ionomycin (Ionomycin) or Sendai virus (Virus). CAT assays were performed, and the percent conversion of [14C]chloramphenicol to its acetylated forms was quantified. Within individual experiments, the results were normalized to the −200 TNF-α–CAT ionomycin-induced level (100%) and then averaged and plotted in the displayed histogram. Standard deviation is represented by the error bars. All cells were cotransfected with the cytomegalovirus β-Gal plasmid, and β-Gal assays were performed to control for transfection efficiency. The figure shows the results of three independent experiments. (B) Relative activities of human TNF-α–CAT fusion constructs containing mutations in the CRE, κ3-NFAT, −76-NFAT, or SP1 site in virus-stimulated Ar-5 T cells. Ar-5 T cells were transfected with the wild-type −200 TNF-α promoter–CAT reporter (WT) and mutated −200 TNF-α promoter–CAT reporter genes (C1M, 5′M, 3′M, 76M, 72M, SP1M), and CAT assays were performed and quantified as described above. The figure shows the results of three independent experiments. Previously published results showing the effect of these mutations upon ionomycin induction of the gene (13, 44, 45) are included in the figure for comparison. Each ionomycin set is independently normalized to the −200 WT ionomycin-induced level (100%) on the same graph as the virus set, which is independently normalized to the −200 WT virus-induced level (100%).
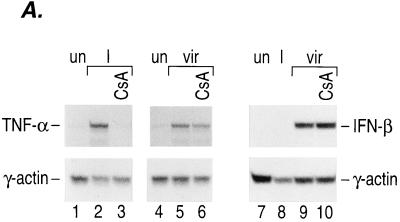
FIG. 2.
Calcineurin is involved in virus-inducible TNF-α gene induction. (A) Induction of TNF-α mRNA by ionophore and virus in Ar-5 T cells. An autoradiogram of an RNase protection assay mapping TNF-α and γ-actin mRNAs is shown. Ar-5 cells were stimulated with ionomycin (I) for 30 min or with Sendai virus (vir) for 2 h in the presence or absence of CsA. The γ-actin probe was made to have a specific activity that was one-fifth the specific activity of the mouse TNF-α probe. (B) CsA inhibits virus induction of TNF-α CAT reporter activity. Ar-5 T cells, A20 B cells, or L929 fibroblasts were transfected with the −200 TNF-α–CAT reporter gene. Twenty-four hours after transfection, the cells were mock induced (Uninduced) or stimulated with Sendai virus (Virus) in the presence or absence of CsA as indicated in the figure. CAT assays were performed and quantified as described in the legend to Fig. 1. The figure shows the results of three independent experiments. (C) Calcineurin augments virus induction of TNF-α. Ar-5 T cells, A20 B cells, or L929 fibroblasts were transfected with the −200 TNF-α–CAT reporter gene and a plasmid that constitutively expresses the catalytic subunit of calcineurin (ΔCAM) or a control plasmid (SR2α) as indicated in the figure. Twenty-four hours after transfection, the cells were either mock induced (Uninduced) or stimulated with Sendai virus (Virus) as described for panel B. CAT assays were performed and quantified as described in the legend to Fig. 1. The figure shows the results of three independent experiments for Ar-5 and A20 cells and a representative experiment for L929 cells.
RNA analysis.
RNA was prepared from Ar-5 cells, and 32P-labeled RNA probes were prepared from SP6 γ-actin and a murine TNF-α probe. RNase protection assays were performed as described previously (13) and quantified with a PhosphorImager (Molecular Dynamics).
Plasmids.
The TNF-α–CAT wild-type and mutant promoter constructs have been described previously (13, 44). The synthetic multimer constructs −39 TNF-α–CAT, κ3(L)1, κ3(L)2, and (3′M)2 have all been described before (45). Two copies of the CRE/κ3 site bearing the C1 mutation [(C1)2] or three copies of the oligonucleotide spanning positions −85 to −59 containing the −76-NFAT site [(−76-NFAT)3] were cloned upstream of the −39 TNF-α–CAT reporter gene as previously described (45). The pPAC-Sp1, pPAC-ATF-2, and pPAC-c-jun constructs have been described previously (30, 41). The pPAC-NFATp S>A vector was constructed as follows. The HindIII fragment of pREP4-NFATp (a gift from Tim Hoey [20]) was subcloned into the HindIII site of pcDNA3 (Invitrogen). The BspEI-DraIII fragment of pREP4-NFATp was then subcloned into the BspEI-EcoRV sites of the resulting plasmid. Serine-to-alanine changes from positions 168 to 177 were introduced by PCR to induce constitutive nuclear localization (3). This fragment was then subcloned into pPAC2 (a gift from Jeremiah Hagler), which contains the Drosophila actin promoter and Drosophila Hsp70 polyadenylation site subcloned into pBluescript (Stratagene).
Electrophoretic mobility shift assays (EMSA) and Western blot analysis.
Nuclear extracts were prepared from Ar-5 cells stimulated for 2 h with Sendai virus or for 30 min with ionomycin (1 μM) as previously described (13). Where indicated, cells were treated with 1 μM CsA for 10 min before the addition of the stimulus.
EMSA were performed with approximately 5 μg of protein, and antibody competition assays were performed as described previously (45) with an antibody specific for NFATp (anti-67.1; a gift from Anjana Rao) or specific for NFATc (Affinity Bioreagents).
For Western blot analysis, after nuclear extract preparation, 15 μg of protein was combined with an equal volume of 2× Laemmli sample buffer and boiled for 5 min in reducing sample buffer. The lysates were analyzed by sodium dodecyl sulfate–6% polyacrylamide gel electrophoresis followed by Western blot analysis with the anti-NFATp antibody as previously described (26).
Calcium imaging.
Ar-5 T cells were plated on coverslips and incubated for 1 h at room temperature in Dulbecco's modified Eagle medium–Ham's F-12 medium without phenol red (Sigma) with 2.5 μM Fura-2 AM in 0.03% plurionic F-127 (Molecular Probes). Coverslips with Fura-2-loaded cells were placed in a glass-bottom petri dish containing external saline. The dish was maintained at 37°C for the duration of the experiment. Calibrated calcium values were determined at a frequency of 0.5 Hz by ratiometric analysis as previously described (16).
Formaldehyde cross-linking and chromatin immunoprecipitation.
Assays were performed essentially as described previously (34, 50). Ar-5 T cells, A20 B cells, and L929 fibroblasts (∼2 × 108 cells) were treated with Sendai virus, 1 μM ionomycin, or 1 μM ionomycin and 200 nM PMA along with control samples for 3 h as indicated in Fig. 5 and then treated with formaldehyde (1% final concentration) for 30 min at 37°C. Cells were harvested, and chromatin was sonicated, extracted, and purified, followed by immunoprecipitation with anti-c-jun, anti-Sp1 (Santa Cruz Biotechnology), anti-ATF-2 (8), anti-NFATc (Affinity Bioreagents), or anti-NFATp (a gift from Tim Hoey). The anti-NFATp antibody is a rabbit polyclonal antibody raised against a fragment of recombinant human NFATp containing amino acids 220 to 318 (19). Immunoprecipitated DNA was then amplified by PCR with primers flanking the murine TNF-α promoter. Cycles of the PCRs were titrated to ensure that amplification was in the linear range. Radiolabeled primers were included in PCRs to visualize PCR products by autoradiography; products were resolved on 5% nondenaturing polyacrylamide gels.
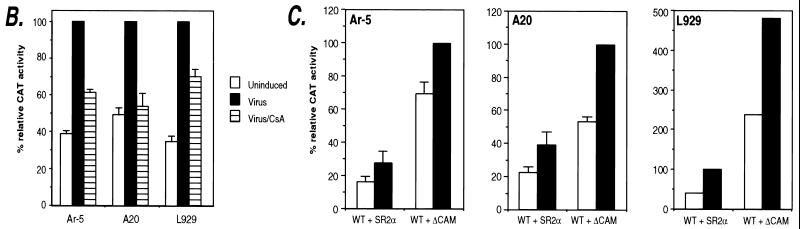
FIG. 5.
Association of specific transcriptional activator proteins with the TNF-α promoter in vivo upon ionophore stimulation and virus induction. Ar-5 T cells (A), A20 B cells (B), and L929 fibroblasts (C) were unstimulated (−) or stimulated with Sendai virus (V), ionomycin (I), or PMA plus ionomycin (P/I) for 3 h as indicated. The cells were then treated with formaldehyde to cross-link protein to DNA, and chromatin was isolated and purified and immunoprecipitated with the indicated antibodies as described previously (34, 50). After reversal of the cross-links, the DNA was amplified by PCR with primers to TNF-α (−210 to +36) (PCRs, 20 cycles of 1 min each at 94, 70, and 72°C). A range of PCR cycles was performed to ensure that the products visualized were in the linear range of amplification. Control amplification using genomic DNA (gDNA) and water (H20) are shown along with a 123-bp marker (Life Technologies; m.w. marker). The high levels of constitutive binding of c-jun to the TNF-α promoter in Ar-5 cells in the chromatin immunoprecipitation assay are consistent with studies demonstrating that c-jun homodimers can bind the TNF-α CRE (44). Although low relative increases in levels of amplified DNA were observed in the NFATp immunoprecipitations, the antibody used was the only one of those tested that was suitable for the chromatin immunoprecipitation assay, and results with Ar-5 and A20 cells were consistent.
RESULTS
The CRE/κ3 and Sp1 sites are required for virus induction of TNF-α gene expression.
To identify the TNF-α promoter sequences required for the transcriptional activation of the TNF-α gene by virus, we first transfected Ar-5 T cells with a TNF-α CAT reporter gene containing either 1045 or 200 nucleotides upstream of the TNF-α mRNA cap site. Consistent with our studies of multiple cell types with a variety of inducers (5, 11, 13, 14, 45), 200 nucleotides upstream of the start site of transcription are sufficient for maximal inducibility of the gene by virus and ionophore in T cells (47). Notably, when the levels of TNF-α reporter gene induction were quantified, the levels reached after virus stimulation were approximately 40% of the levels achieved after ionophore stimulation (Fig. 1A).
To identify the TNF-α promoter elements required for virus induction of the gene, we transfected Ar-5 T cells with TNF-α–CAT reporter constructs bearing mutations in different regulatory elements. Figure 1B shows that mutation of either the CRE (C1M), the κ3-NFAT site (5′M and 3′M), or the Sp1 (SP1M) site significantly reduced virus induction of the gene. The CRE, κ3-NFAT, and −76-NFAT sites were previously shown to be required for ionophore induction (13, 44, 45). Thus, induction of TNF-α gene expression by virus, in contrast to ionophore, requires an intact Sp1 site (Fig. 1B). Furthermore, while the −76-NFAT site is critical for induction by ionophore, it is required only to achieve maximal levels of induction by virus (Fig. 1B). Thus, distinct combinations of regulatory elements are required for activation of the TNF-α gene by virus and by ionophore.
CsA inhibits virus induction of TNF-α.
To further investigate activation of the TNF-α gene by virus, we performed quantitative RNase protection assays with RNA from cells stimulated with virus or with ionomycin in the presence or absence of the calcineurin inhibitor CsA. Consistent with previous results (13), Fig. 2A shows that in T cells, TNF-α transcription is stimulated by ionomycin and this induction can be blocked by pretreatment of the cells with CsA (Fig. 2A, lanes 1 to 3). Concordant with our transfection analysis, virus also stimulated TNF-α gene expression in Ar-5 T cells to approximately 50% of the levels reached with ionophore stimulation of the cells (Fig. 2A, lanes 4 and 5). Remarkably, virus induction of TNF-α mRNA levels was partially blocked (an average of 30% in three independent experiments) by pretreatment of the cells with CsA (lane 6). In contrast, virus induction of beta interferon (IFN-β) transcription was not blocked by CsA (Fig. 2A, lanes 7 to 10).
We also studied the effect of CsA upon TNF-α gene transcription in transient-transfection experiments. We note that in B cells and fibroblasts, as in T cells, 200 nucleotides upstream of the start site of transcription are sufficient for maximal inducibility of the gene by virus (11, 45, 48). Consistent with the RNase protection analysis in Ar-5 T cells, virus induction of the −200 TNF-α–CAT reporter gene was partially inhibited by CsA in Ar-5 T cells and L929 fibroblasts and blocked by CsA in A20 B cells (Fig. 2B). Thus, calcineurin phosphatase activity is required for full activation of the TNF-α gene by virus in all three cell types. These results indicate that virus induction of TNF-α, unlike that of IFN-β, involves the phosphatase activity of calcineurin.
Additional evidence for the involvement of calcineurin in virus induction of TNF-α was provided by cotransfecting a constitutively active form of calcineurin (ΔCAM) (33) along with the TNF-α–CAT reporter gene. Previous studies demonstrated that ΔCAM induced transcription of the cotransfected −200 TNF-α–CAT reporter gene in Ar-5 T cells in the absence of any other cellular stimulation (15). As shown in Fig. 2C, ΔCAM augments virus-induced levels of transcription of the TNF-α promoter as well as basal TNF-α promoter activity in Ar-5 T cells, A20 B cells, and L929 fibroblasts. Taken together, the results of these experiments are consistent with calcineurin acting as an intermediate in the signal transduction pathway stimulated by virus. Furthermore, given that CsA inhibits NFAT translocation to the nucleus (10), these experiments support a role for NFAT proteins in the regulation of TNF-α gene expression by virus and correlate with the involvement of cis-acting NFAT-binding elements in virus induction of TNF-α (Fig. 1B).
Virus infection induces dephosphorylation of NFAT and a rise in intracellular calcium levels.
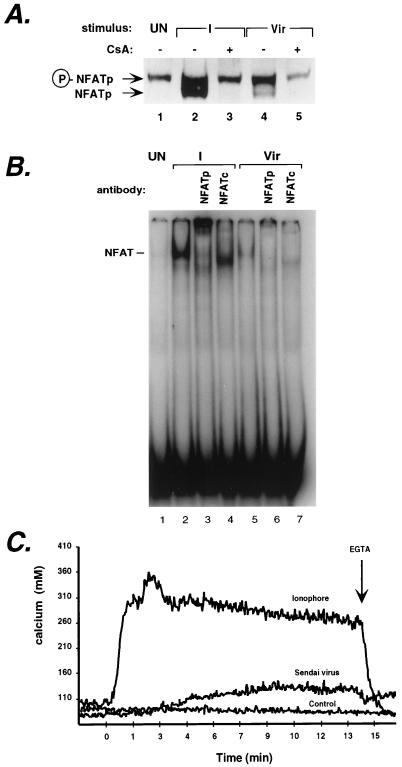
The experiments described above demonstrated that the TNF-α NFAT binding site (κ3) is required for virus induction of the TNF-α gene and that the −76-NFAT site is required for maximal transcriptional levels of virus-stimulated TNF-α gene expression in Ar-5 T cells. To investigate whether virus infection indeed leads to the dephosphorylation of NFAT, we carried out Western blot experiments using nuclear extracts prepared from unstimulated or virus-stimulated Ar-5 T cells and an antibody to NFATp. Using this technique, inhibition of calcineurin-dependent dephosphorylation of NFATp by the specific calcineurin inhibitor CsA can be monitored by observing a size shift in the NFATp detected in the immunoblot (37). In unstimulated cells, NFATp is constitutively present in the nucleus in its phosphorylated form (Fig. 3A, lane 1). However, ionophore induction of the cells leads to the appearance of the dephosphorylated form of NFATp, which appears as a faster-migrating band on the Western blot (Fig. 3A, lane 2) and is inhibited by CsA (lane 3). Strikingly, virus infection also leads to the dephosphorylation of NFATp, which is also inhibited by CsA (Fig. 3A, lanes 4 and 5).
FIG. 3.
Virus infection causes NFAT dephosphorylation and an increase in intracellular calcium levels. (A) Infection of Ar-5 T cells by Sendai virus causes dephosphorylation of NFATp. Nuclear extracts were prepared from Ar-5 T cells stimulated with ionomycin (I) for 30 min or with Sendai virus (Vir) for 2 h, in the presence or absence of CsA as indicated in the figure. Lysates were analyzed by sodium dodecyl sulfate–6% polyacrylamide gel electrophoresis followed by Western blot analysis with the anti-NFATp (67.1) antibody. After ionomycin or virus stimulation of the cells, phosphorylated NFATp (P-NFATp) is dephosphorylated (NFATp) and represented as a size shift in the gel. (B) Virus-inducible NFATp/c binds to the −76-NFAT site. An EMSA using nuclear extracts from unstimulated Ar-5 cells (UN) or cells stimulated with ionomycin (I) for 30 min or with Sendai virus (Vir) for 2 h as indicated in the figure was performed. An ionomycin- and virus-inducible complex binds to the oligonucleotide probe spanning positions −85 to −59 containing the −76-NFAT site. Antibodies to NFATp and to NFATc specifically react with the ionomycin- and virus-inducible complex, since they do not react with complexes bound to an Sp1 oligonucleotide probe (47). (C) Infection of Ar-5 T cells by Sendai virus causes an increase in intracellular calcium levels. Cells were exposed to either Sendai virus or ionomycin or a control carrier (allantoic fluid in which the Sendai virus is prepared) at 0 min. Extracellular calcium was later buffered to 0 mM by the addition of 4 mM EGTA (arrow).
Electrophoretic mobility shift assays with Ar-5 nuclear extracts demonstrated that the levels of NFATp in the nucleus induced by virus infection are lower than those induced by ionophore (Fig. 3B). This is consistent with the lower levels of dephosphorylated NFATp detected by Western blot analysis following virus infection in comparison to those after ionophore stimulation (Fig. 3A). This quantitative difference of NFATp activation is concordant with the quantitative difference of TNF-α gene induction stimulated by ionophore relative to virus treatment of the cells (Fig. 1A and 2A and see below).
The ability of the calcineurin inhibitor CsA to block virus-dependent NFAT dephosphorylation suggests that Sendai virus infection may effect gene activation at least in part through an increase in intracellular calcium. To examine this hypothesis, we performed ratiometric calcium imaging studies in Ar-5 T cells stimulated with ionomycin or Sendai virus. Remarkably, virus infection of Ar-5 T cells leads to an increase in intracellular calcium (Fig. 3C). The magnitude of this effect (an increase of approximately 40 nM) is not as strong as the effect elicited by treatment with calcium ionophore (an increase of approximately 200 nM). Thus, the more modest rise in calcium after virus stimulation correlates with the lower levels of dephosphorylated NFAT and the lower levels of TNF-α transcription after virus infection relative to ionophore stimulation of the cells. Treatment with 4 mM EGTA, which eliminates calcium influx by chelating extracellular calcium, reversed the effect of ionomycin, but not virus, indicating that virus infection results in the release of intracellular calcium stores (Fig. 3C).
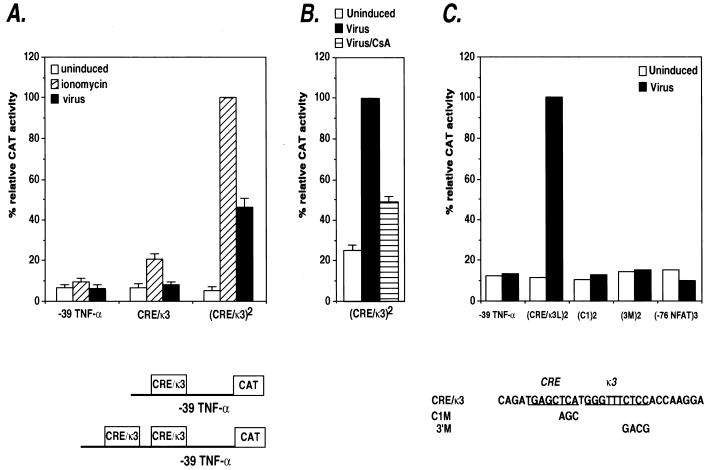
The functional significance of the quantitative difference in NFATp activation by ionophore as compared to virus was also investigated in transfection experiments using reporter constructs bearing one or two copies of the CRE/κ3 composite element placed upstream of the minimal −39 TNF-α promoter. Figure 4A demonstrates that although a single copy of the CRE/κ3 site is inducible by ionomycin, it is not activated by virus. However, two copies of the CRE/κ3 site are efficiently activated by virus (approximately 50% of the levels achieved following ionophore stimulation of the cells) (Fig. 4A), and this induction is sensitive to CsA (Fig. 4B). These results are in agreement with the data presented above (Fig. 3A) showing that ionophore stimulation leads to higher levels of nuclear localization of NFAT relative to the levels achieved after virus infection of the cells.
FIG. 4.
Inducer-specific regulation of the CRE/κ3 composite element by ionophore and virus. (A) Activation of synthetic TNF-α promoters containing the CRE/κ3 multimers by ionomycin or virus in Ar-5 T cells. Ar-5 cells were transfected with the truncated −39 TNF-α–CAT construct containing either one or two copies of the κ3(L) site. Twenty-four hours later, the cells were mock stimulated (uninduced) or stimulated with ionomycin (ionomycin) or Sendai virus (virus). Within the individual experiments displayed, the results were normalized to the induced level (100%) of the wild-type (CRE/κ3)2 multimer construct and then averaged and plotted in the displayed histogram. CAT assays were performed and quantified as described in the legend to Fig. 1. The figure shows the results of three independent experiments. (B) CsA inhibits virus activation of the CRE/κ3 site. Ar-5 cells were transfected with the κ3(L)2 synthetic promoter construct, stimulated with virus in the presence or absence of CsA, and analyzed and quantified as described in the legend to Fig. 1. The figure shows the results of three independent experiments. (C) Activation of synthetic TNF-α promoters containing CRE/κ3 or −76-NFAT multimers in Ar-5 T cells. Ar-5 cells were transfected with the truncated −39 TNF-α–CAT construct containing either two copies of the wild-type CRE/κ3 site or two copies of the CRE/κ3 site containing the 3′ mutation or the C1 mutation or with three copies of the −76-NFAT site as indicated in the figure. Twenty-four hours later, the cells were mock stimulated (Uninduced) or stimulated with Sendai virus (Virus) as indicated. The figure shows a representative experiment. The results were normalized to the induced level (100%) of the wild-type (CRE/κ3)2 multimer construct and then averaged and plotted in the displayed histogram.
The CRE and κ3 sites synergistically activate transcription.
To further dissect the functional cooperation between the ATF-2/Jun and NFAT subsites of the CRE-κ3 composite element in conferring virus inducibility, we investigated the role of each half-site in synthetic promoters bearing mutations in these sites. Mutation of the 3′ aspect of κ3 (3′M), which is required for the binding of NFAT proteins to the site, or mutation of the CRE (C1), which is required for ATF-2/Jun binding to the site, abrogated the ability of the CRE/κ3 site to act as a virus-inducible element (Fig. 4C). Interestingly, up to three copies of the higher-affinity −76-NFAT site, which binds NFAT without an AP-1 protein partner, does not confer virus inducibility (Fig. 4C) or ionophore inducibility (47) upon the truncated −39 TNF-α promoter. Thus, the transcriptional activity of the CRE/κ3 element requires synergistic interactions between the κ3 and CRE half-sites of the element.
ATF-2/Jun, Sp1, and NFAT proteins interact with the endogenous TNF-α promoter upon virus infection.
The transfection experiments described above (Fig. 1 and 4) demonstrate the importance of the CRE, NFAT, and Sp1 regulatory elements in the inducer-specific regulation of TNF-α by ionophore and virus infection in Ar-5 T cells. Consistent with these results, transfection experiments in A20 B cells and in L929 fibroblast cells confirmed the significance of the CRE, NFAT, and Sp1 regulatory elements in the inducer-specific regulation of TNF-α by virus (48).
To directly test whether the cognate transcriptional activator proteins bind to the CRE, NFAT, and Sp1 sites of the TNF-α promoter in these cell types in vivo, we performed formaldehyde cross-linking and chromatin immunoprecipitation experiments using specific antibodies. TNF-α promoter DNA was immunoprecipitated by the antibodies shown in Fig. 5 and then amplified by PCR. This provides a measure of the relative amounts of protein binding to the promoter in vivo before and after stimulation by ionophore or virus in Ar-5 T cells (Fig. 5A), by PMA and ionomycin or virus in A20 B cells (Fig. 5B), and by virus in L929 fibroblasts (Fig. 5C).
In Ar-5 T cells, both virus infection and ionophore stimulation induce binding of ATF-2 to the TNF-α promoter (Fig. 5A, lanes 4 to 6), whereas c-jun binding was constitutive and minimally inducible (lanes 7 to 9). Strikingly, virus infection but not ionophore stimulation results in the induction of Sp1 binding to the TNF-α promoter in Ar-5 cells (Fig. 5A, lanes 10 to 12). In addition, the binding of NFATp was induced by virus as well as by ionomycin in Ar-5 cells (Fig. 5A, lanes 13 to 15). Similar results were obtained in A20 B cells stimulated by PMA and ionomycin or by virus (Fig. 5B). We note that the detection of NFATp binding in unstimulated cells may reflect constitutive levels of NFATp in the nucleus (Fig. 3A) or persistent shuttling of NFAT between the nucleus and the cytoplasm (39).
TNF-α induction is selectively induced by virus and not by ionophore in L929 cells (11, 23). Consistent with the results obtained in the other cell lines, virus infection of L929 cells also resulted in the recruitment of ATF-2 and Sp1 to the TNF-α promoter (Fig. 5C, lanes 4 and 5 and lanes 8 and 9). In L929 cells, binding of c-jun was inducible (Fig. 5C, lanes 6 and 7). Furthermore, in L929 cells, which lack NFATp (23, 29), binding of NFATc to the TNF-α promoter was also strongly induced by virus infection (Fig. 5C, lanes 10 and 11). Consistent with the requirement for the Sp1 site in virus induction, we previously showed that a minimal TNF-α promoter containing the Sp1 site retains virus inducibility in L929 cells, whereas a minimal promoter with a compromised Sp1 site does not (11). Taken together, these results strongly correlate with the critical roles of the Sp1, CRE, and NFAT sites in the activation of the TNF-α gene by virus and with the involvement of Sp1 in the inducer-specific assembly of enhancer complexes on the TNF-α promoter.
Sp1 functionally synergizes with NFATp and ATF-2/c-jun.
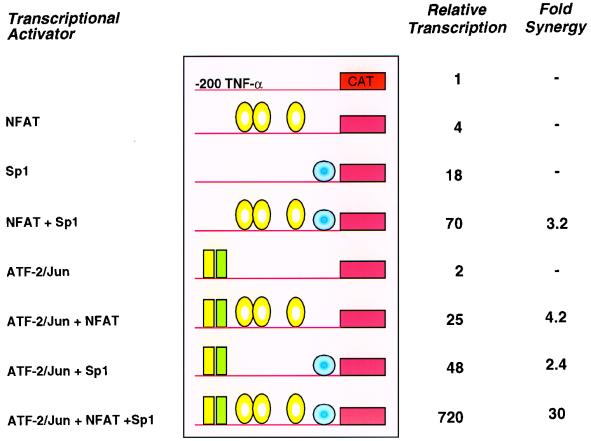
Given that Sp1 is recruited to the TNF-α promoter following virus infection, we next examined whether Sp1 could synergize with ATF-2/c-jun and NFATp in activation of the TNF-α promoter. We expressed these activators by transient transfection in Drosophila Schneider-2 (SL2) cells, which are devoid of these factors, and tested their effect upon the −200 TNF-α CAT reporter. Cotransfection of NFATp bearing serine-to-alanine mutations to induce constitutive nuclear localization (NFATp S>A) and Sp1 resulted in more than threefold synergistic induction of TNF-α-driven CAT activity (Fig. 6). Consistent with our mutational analyses (Fig. 1B and 4C), ATF-2/c-jun and NFATp also activate the TNF-α promoter in a synergistic fashion, to a level over fourfold higher than their additive effect (Fig. 6). Remarkably, when all four activators were cotransfected with the TNF-α–CAT reporter plasmid, the relative transcriptional reached was 30-fold higher than the additive transcription effect of all of the activators (Fig. 6). Quantitative DNase footprinting revealed that the binding of ATF-2/c-jun and NFATp (45) and the binding of Sp1 and NFATp or ATF-2/c-jun (46) is not cooperative, suggesting that the observed transcriptional synergy occurs at the level of recruitment of the basal transcription machinery (6). Thus, binding of Sp1 to the TNF-α promoter in the context of the enhancer complex, either by overexpression of Sp1 and the other transactivators or by virus-specific recruitment of Sp1, results in high levels of TNF-α gene transcription.
FIG. 6.
Sp1 synergizes with NFATp and ATF-2/c-jun. Drosophila SL2 cells were cotransfected with NFATp S>A (2.4 μg), Sp1 (100 ng), and ATF-2/c-jun (500 ng) in the combinations indicated along with Hsp–β-Gal (500 ng) and the −200 TNF-α–CAT reporter (500 ng). CAT activity normalized for β-Gal expression is shown for a representative experiment. The fold synergy of activation (i.e., fold activation greater than the additive effect of the individual activators) is noted. The experiment shown is representative of two independent experiments. The experiment testing the synergy between NFATp and Sp1 was performed five independent times. We note that increasing amounts (0.1 to 2.4 μg) of NFATp S>A cotransfected into SL2 cells with the TNF-α–CAT reporter gene resulted in up to a fourfold increase of TNF-α-driven CAT activity (31).
DISCUSSION
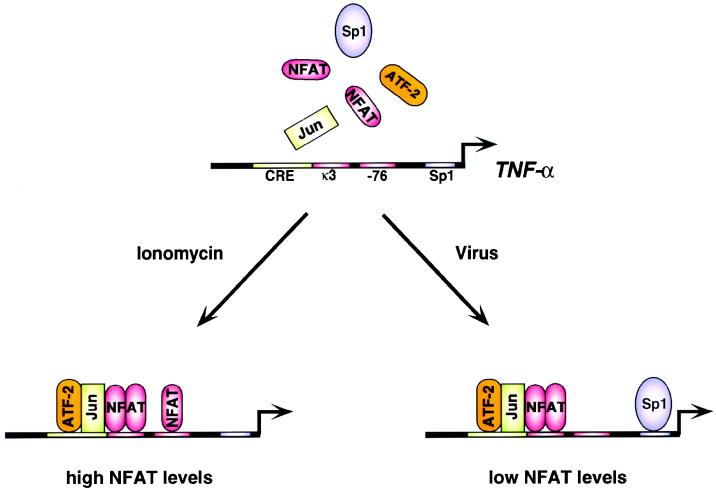
A critical question in eukaryotic gene regulation is how individual genes are specifically activated in response to a particular cellular stress. Here we present evidence for a general mechanism for this process: the assembly of distinct enhancer complexes on the same promoter in response to different inducers (see model in Fig. 7).
FIG. 7.
Inducer-specific assembly of distinct enhancer complexes on the TNF-α gene promoter. The cis-acting TNF-α promoter elements, transcription factors, and coactivators involved in the inducer-specific regulation of TNF-α by ionophore and virus in T cells are summarized schematically. The sites that are critical for activation of the promoter are indicated. The transcription factors which bind to the TNF-α promoter in vivo (NFAT, ATF-2/c-jun, and Sp1) following the indicated stimuli are shown.
Previous studies have shown that enhanceosome assembly is dependent upon the precise spatial arrangement of activator binding sites, which facilitate a unique pattern of protein-protein and protein-DNA interactions (reviewed in references 6, 36, and 42). The IFN-β promoter is a well-characterized example of inducible enhanceosome assembly. In contrast to TNF-α, which is induced by multiple stimuli, IFN-β is tightly regulated in response to a single specific stimulus, virus infection (reviewed in reference 27). Virus infection of a cell results in the coordinate assembly of a specific set of transcriptional activators on the IFN-β promoter, including NF-κB p50/p65, ATF-2/c-jun, HMG(I)Y, and interferon regulatory factors 3 and 7 (reviewed in reference 27). These activators bind to a fixed set of binding sites in its promoter and associate with the CREB-binding protein/p300 coactivators (25, 32, 50, 53). Thus, the IFN-β enhanceosome has a stringent requirement for a specific set of binding sites and cognate transcription factors, which are coordinately activated by virus.
Our experiments in Drosophila Schneider cells demonstrated that expression of ATF-2/c-jun, NFATp, and Sp1 results in synergistic rather than additive levels of transcription mediated by the TNF-α promoter, which is a critical feature of enhanceosome assembly (6). Spatial constraint upon the arrangement of transcription factor binding sites also plays an important role in the induction of TNF-α transcription; separation of the ATF-2/c-jun and NFATp binding sites at the CRE and κ3 by insertion of additional base pairs abolishes inducibility of the promoter in response to ionophore or virus (9). In contrast to IFN-β, however, in the case of TNF-α gene activation, a specific set of promoter binding sites are recognized by different activator complexes in response to either virus or ionophore. In response to calcium influx, the CRE and the κ3 and −76-NFAT sites are required (13, 44, 45). In response to virus infection, however, the κ3-NFAT site but not the −76- NFAT site is required and the Sp1 site is critical for transcriptional induction of the gene (Fig. 7). Thus, stimulus-specific enhancer complexes assemble on the TNF-α promoter, which use distinct sets of binding sites and cognate proteins.
Our experiments have also established that virus infection causes an increase in intracellular calcium levels, calcineurin-dependent NFAT dephosphorylation, and binding of NFAT to the TNF-α promoter in vivo, thus establishing NFAT as a transcription factor involved in virus-mediated gene regulation. The TNF-α promoter contains multiple sites which bind NFAT with different affinities and which are utilized in a cell type-specific manner (13, 44, 45). Here we have shown that the level of dephosphorylated NFAT in the nucleus after virus infection is lower than that achieved after ionophore stimulation. We have also demonstrated dramatic transcriptional synergy between NFATp, ATF-2/c-jun, and Sp1 in Drosophila SL2 cells. Our functional analysis indicates that under conditions of low NFAT levels such as occur after virus infection, NFAT functionally synergizes with ATF-2/Jun at the composite CRE/κ3 site and the Sp1 binding site is critical for TNF-α gene induction. By contrast, under conditions of high NFAT levels such as occur after ionophore stimulation, TNF-α transcription requires the −76-NFAT site in addition to κ3 but is independent of the Sp1 binding site. Taken together, our data are consistent with a model in which functional synergy between Sp1 and NFAT and ATF-2/Jun is required for transcriptional activation of TNF-α under conditions of low NFAT levels following virus infection.
The TNF-α CRE site is an element that integrates signals from distinct signal transduction pathways reminiscent of the serum response element (SRE) of the c-fos promoter (18, 35). In the case of TNF-α, activated binding of ATF-2/Jun to the CRE is the target of diverse signal transduction pathways. TNF-α gene activation following antigen or calcium stimulation of lymphocytes (44, 45) (Fig. 5A), FcɛRI engagement in mast cells (17), and TNF receptor engagement of fibroblasts (5) all involve the CRE site. As well, the CRE is critical for TNF-α gene activation upon lipopolysaccharide stimulation of monocytes (43, 52) and virus infection of multiple cell types (Fig. 2A and 5). Thus, these experiments provide a striking example of integration of diverse signal transduction pathways at a particular DNA element, the TNF-α CRE.
Given that both the TNF-α and IFN-β genes are coinduced by virus in multiple cell types (11, 12), it is notable that both genes contain CRE sites, which bind ATF-2/Jun proteins (8, 44). ATF-2/Jun proteins become transcriptionally active upon phosphorylation by the p38 and JNK members of the mitogen-activated protein kinase family of protein kinases (reviewed in reference 38), and JNK activity can be augmented by increasing levels of intracellular calcium (40). Thus, together with calcium-mediated pathways and calcineurin, JNK and/or p38 are likely to be involved in virus-mediated signal transduction and gene regulation.
Using chromatin immunoprecipitation assays, we have shown that Sp1 binding to the TNF-α promoter is virus inducible in vivo in three different cell types. The involvement of Sp1 in virus-mediated induction of TNF-α gene expression is particularly striking because it demonstrates that a protein associated with the regulation of constitutively expressed housekeeping genes (21) is recruited in an inducible fashion to the promoter of an immediate early-response cytokine gene. It is interesting to note that the Sp1 site in the TNF-α promoter varies from the Sp1 consensus binding site sequence (22) and is a relatively weak Sp1 binding site (46). This is consistent with the idea that Sp1 binding to the TNF-α promoter element site is inducible rather than constitutive.
In summary, we have shown that distinct enhancer complexes form on different activator recognition sites on the TNF-α promoter in response to distinct extracellular stimuli. The specificity of TNF-α transcriptional activation is achieved through synergistic interactions among specific activators in these enhancer complexes. The differential transcription of genes in temporal, spatial, and signaling-specific patterns is at the heart of the development and cellular regulation of complex organisms. We anticipate that the differential assembly of enhanceosomes is a general mechanism by which a gene may be controlled in a temporal and tissue-specific manner.
ACKNOWLEDGMENTS
We are indebted to Tim Hoey for the generous gift of an unpublished NFATp antibody. We are grateful to Lori Daniels for laboratory assistance, Ann Corbett for help with manuscript preparation, and Judith Grisham for editorial assistance. We also thank Anjana Rao and Jeremiah Hagler for gifts of reagents.
This work was supported by an Established Investigator Award from the American Heart Association to A.E.G. and by grants from the National Institutes of Health to D.T. (GM54605) and T.M. (AI20642).
J.V.F. and A.M.U. contributed equally to this work.
Footnotes
Dedicated to the memory of Mauricio X. Zuber and Ernest G. Peralta.
REFERENCES
- 1.Aderka D, Holtmann H, Toker L, Hahn T, Wallach D. Tumor necrosis factor induction by Sendai virus. J Immunol. 1986;136:2938–2942. [PubMed] [Google Scholar]
- 2.Aggarwal B B, Puri R K. Human cytokines: their role in disease and therapy. Cambridge, Mass: Blackwell Science; 1995. [Google Scholar]
- 3.Beals C R, Clipstone N A, Ho S N, Crabtree G R. Nuclear localization of NF-ATc by a calcineurin-dependent, cyclosporin-sensitive intramolecular interaction. Genes Dev. 1997;11:824–834. doi: 10.1101/gad.11.7.824. [DOI] [PubMed] [Google Scholar]
- 4.Boussiotis V A, Nadler L M, Strominger J L, Goldfeld A E. Tumor necrosis factor α is an autocrine growth factor for normal human B cells. Proc Natl Acad Sci USA. 1994;91:7007–7011. doi: 10.1073/pnas.91.15.7007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brinkman B M N, Telliez J-B, Schievella A R, Lin L-L, Goldfeld A E. Engagement of TNF receptor 1 leads to ATF-2 and p38 MAP kinase-dependent TNF-α gene expression. J Biol Chem. 1999;274:30882–30886. doi: 10.1074/jbc.274.43.30882. [DOI] [PubMed] [Google Scholar]
- 6.Carey M. The enhanceosome and transcriptional synergy. Cell. 1998;92:5–8. doi: 10.1016/s0092-8674(00)80893-4. [DOI] [PubMed] [Google Scholar]
- 7.Crabtree G R. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–614. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- 8.Du W, Thanos D, Maniatis T. Mechanisms of transcriptional synergism between distinct virus-inducible enhancer elements. Cell. 1993;74:887–898. doi: 10.1016/0092-8674(93)90468-6. [DOI] [PubMed] [Google Scholar]
- 9.Falvo, J. V., and A. E. Goldfeld. Unpublished data.
- 10.Flanagan W M, Corthesy B, Bram R J, Crabtree G R. Nuclear association of a T-cell transcription factor blocked by FK-506 and cyclosporin A. Nature. 1991;352:803–807. doi: 10.1038/352803a0. [DOI] [PubMed] [Google Scholar]
- 11.Goldfeld A E, Doyle C, Maniatis T. Human tumor necrosis factor α gene regulation by virus and lipopolysaccharide. Proc Natl Acad Sci USA. 1990;87:9769–9773. doi: 10.1073/pnas.87.24.9769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goldfeld A E, Maniatis T. Coordinate viral induction of tumor necrosis factor α and interferon β in human B cells and monocytes. Proc Natl Acad Sci USA. 1989;86:1490–1494. doi: 10.1073/pnas.86.5.1490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goldfeld A E, McCaffrey P G, Strominger J L, Rao A. Identification of a novel cyclosporin-sensitive element in the human tumor necrosis factor α gene promoter. J Exp Med. 1993;178:1365–1379. doi: 10.1084/jem.178.4.1365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goldfeld A E, Strominger J L, Doyle C. Human tumor necrosis factor α gene regulation in phorbol ester stimulated T and B cell lines. J Exp Med. 1991;174:73–81. doi: 10.1084/jem.174.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Goldfeld A E, Tsai E, Kincaid R, Belshaw P J, Schrieber S L, Strominger J L, Rao A. Calcineurin mediates human tumor necrosis factor α gene induction in stimulated T and B cells. J Exp Med. 1994;180:763–768. doi: 10.1084/jem.180.2.763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grynkiewicz G, Poenie M, Tsien R Y. A new generation of Ca2+ indicators with greatly improved fluorescence properties. J Biol Chem. 1985;260:3440–3450. [PubMed] [Google Scholar]
- 17.Hata D, Kawakami Y, Inagaki N, Lantz C S, Kitamura T, Khan W N, Maeda-Yamamoto M, Miura T, Han W, Hartman S E, Yao L, Nagai H, Goldfeld A E, Alt F W, Galli S J, Witte O N, Kawakami T. Involvement of Bruton's tyrosine kinase in FcɛRI-dependent mast cell degranulation and cytokine production. J Exp Med. 1998;187:1235–1247. doi: 10.1084/jem.187.8.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hill C S, Wynne J, Treisman R. The Rho family GTPases RhoA, Rac1, and CDC42Hs regulate transcriptional activation by SRF. Cell. 1995;81:1159–1170. doi: 10.1016/s0092-8674(05)80020-0. [DOI] [PubMed] [Google Scholar]
- 19.Hoey, T. Personal communication.
- 20.Hoey T, Sun Y L, Williamson K, Xu X. Isolation of two new members of the NF-AT gene family and functional characterization of the NF-AT proteins. Immunity. 1995;2:461–472. doi: 10.1016/1074-7613(95)90027-6. [DOI] [PubMed] [Google Scholar]
- 21.Holler M, Westin G, Jiricny J, Schaffner W. Sp1 transcription factor binds DNA and activates transcription even when the binding site is CpG methylated. Genes Dev. 1988;2:1127–1135. doi: 10.1101/gad.2.9.1127. [DOI] [PubMed] [Google Scholar]
- 22.Kadonaga J T, Jones K A, Tjian R. Promoter-specific activation of RNA polymerase transcription by Sp1. Trends Biochem Sci. 1986;11:20–23. [Google Scholar]
- 23.King, H. C., and A. E. Goldfeld. Unpublished data.
- 24.Kubo M, Ransom J, Webb D, Hashimoto Y, Tada T, Nakayama T. T-cell subset-specific expression of the IL-4 gene is regulated by a silencer element and STAT6. EMBO J. 1997;16:4007–4020. doi: 10.1093/emboj/16.13.4007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin R, Heylbroeck C, Pitha P M, Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;18:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Luo C, Burgeon E, Carew J A, McCaffrey P G, Badalian T M, Lane W S, Hogan P G, Rao A. Recombinant NFAT1 (NFATp) is regulated by calcineurin in T cells and mediates transcription of several cytokine genes. Mol Cell Biol. 1996;16:3955–3966. doi: 10.1128/mcb.16.7.3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maniatis T, Falvo J V, Kim T H, Kim T K, Lin C H, Parekh B S, Wathelet M G. Structure and function of the interferon-β enhanceosome. Cold Spring Harbor Symp Quant Biol. 1998;63:609–620. doi: 10.1101/sqb.1998.63.609. [DOI] [PubMed] [Google Scholar]
- 28.McCaffrey P G, Goldfeld A E, Rao A. The role of NFATp in cyclosporin A-sensitive tumor necrosis factor-α gene transcription. J Biol Chem. 1994;269:30445–30450. [PubMed] [Google Scholar]
- 29.McCaffrey P G, Luo C, Kerppola T K, Jain J, Badalian T M, Ho A M, Burgeon E, Lane W S, Lambert J N, Curran T, Verdine G L, Rao A, Hogan P G. Isolation of the cyclosporin-sensitive T cell transcription factor NFATp. Science. 1993;262:750–754. doi: 10.1126/science.8235597. [DOI] [PubMed] [Google Scholar]
- 30.Merika M, Orkin S H. Functional synergy and physical interactions of the erythroid transcription factor GATA-1 with the Kruppel family proteins Sp1 and EKLF. Mol Cell Biol. 1995;15:2437–2447. doi: 10.1128/mcb.15.5.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Merika, M., and D. Thanos. Unpublished data.
- 32.Merika M, Williams A J, Chen G, Collins T, Thanos D. Recruitment of CBP/p300 by the IFNβ enhanceosome is required for synergistic activation of transcription. Mol Cell. 1998;1:277–287. doi: 10.1016/s1097-2765(00)80028-3. [DOI] [PubMed] [Google Scholar]
- 33.O'Keefe S J, Tamura J, Kincaid R L, Tocci M J, O'Neill E A. FK-506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature. 1992;357:692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- 34.Orlando V, Strutt H, Paro R. Analysis of chromatin structure by in vivo formaldehyde cross-linking. Methods. 1997;11:205–214. doi: 10.1006/meth.1996.0407. [DOI] [PubMed] [Google Scholar]
- 35.Price M A, Cruzalegui F H, Treisman R. The p38 and ERK MAP kinase pathways cooperate to activate ternary complex factors and c-fos transcription in response to UV light. EMBO J. 1996;15:6552–6563. [PMC free article] [PubMed] [Google Scholar]
- 36.Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–577. doi: 10.1038/386569a0. [DOI] [PubMed] [Google Scholar]
- 37.Rao A, Luo C, Hogan P G. Transcription factors of the NFAT family: regulation and function. Annu Rev Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 38.Schaeffer H J, Weber M J. Mitogen-activated protein kinases: specific messages from ubiquitous messengers. Mol Cell Biol. 1999;19:2435–2444. doi: 10.1128/mcb.19.4.2435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shibasaki F, Price E R, Milan D, McKeon F. Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature. 1996;382:370–373. doi: 10.1038/382370a0. [DOI] [PubMed] [Google Scholar]
- 40.Su B, Jacinto E, Hibi M, Kallunki T, Karin M, Ben-Neriah Y. JNK is involved in signal integration during costimulation of T lymphocytes. Cell. 1994;77:727–736. doi: 10.1016/0092-8674(94)90056-6. [DOI] [PubMed] [Google Scholar]
- 41.Thanos D, Maniatis T. Virus induction of human IFNβ gene expression requires the assembly of an enhanceosome. Cell. 1995;83:1091–1100. doi: 10.1016/0092-8674(95)90136-1. [DOI] [PubMed] [Google Scholar]
- 42.Tjian R, Maniatis T. Transcriptional activation: a complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- 43.Tsai, E. Y., and A. E. Goldfeld. Unpublished data.
- 44.Tsai E Y, Jain J, Pesavento P A, Rao A, Goldfeld A E. Tumor necrosis factor alpha gene regulation in activated T cells involves ATF-2/Jun and NFATp. Mol Cell Biol. 1996a;16:459–467. doi: 10.1128/mcb.16.2.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tsai E Y, Yie J, Thanos D, Goldfeld A E. Cell-type-specific regulation of the human tumor necrosis factor alpha gene in B cells and T cells by NFATp and ATF-2/JUN. Mol Cell Biol. 1996b;16:5232–5244. doi: 10.1128/mcb.16.10.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tsytsykova, A. V., and A. E. Goldfeld. Unpublished data.
- 47.Uglialoro, A. M., and A. E. Goldfeld. Unpublished data.
- 48.Uglialoro, A. M., H. C. King, and A. E. Goldfeld. Unpublished data.
- 49.Vilcek J, Sen G. Interferons and other cytokines. In: Fields B, Knipe D M, Howley P M, editors. Virology. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 375–399. [Google Scholar]
- 50.Wathelet M G, Lin C H, Parekh B S, Ronco L V, Howley P M, Maniatis T. Virus infection induces the assembly of coordinately activated transcription factors on the IFN-β enhancer in vivo. Mol Cell. 1998;1:507–518. doi: 10.1016/s1097-2765(00)80051-9. [DOI] [PubMed] [Google Scholar]
- 51.Wong G H, Goeddel D V. Tumour necrosis factors α and β inhibit virus replication and synergize with interferons. Nature. 1986;323:819–822. doi: 10.1038/323819a0. [DOI] [PubMed] [Google Scholar]
- 52.Yao J, Mackman N, Edgington T S, Fan S T. Lipopolysaccharide induction of the tumor necrosis factor-α promoter in human monocytic cells. Regulation by Egr-1, c-Jun, and NF-κB transcription factors. J Biol Chem. 1997;272:17795–17801. doi: 10.1074/jbc.272.28.17795. [DOI] [PubMed] [Google Scholar]
- 53.Yoneyama M, Suhara W, Fukuhara Y, Fukuda M, Nishida E, Fujita T. Direct triggering of the type I interferon system by virus infection: activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17:1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]