Abstract
Rel and IκB protein families form a complex cellular regulatory network. A major regulatory function of IκB proteins is to retain Rel proteins in the cell cytoplasm. In addition, IκB proteins have also been postulated to serve nuclear functions. These include the maintenance of inducible NF-κB-dependent gene transcription, as well as termination of inducible transcription. We show that IκBα shuttles between the nucleus and the cytoplasm, utilizing the nuclear export receptor CRM1. A CRM1-binding export sequence was identified in the N-terminal domain of IκBα but not in that of IκBβ or IκBɛ. By reconstituting major aspects of NF-κB–IκB sequestration in yeast, we demonstrate that cytoplasmic retention of p65 (also called RelA) by IκBα requires Crm1p-dependent nuclear export. In mammalian cells, inhibition of CRM1 by leptomycin B resulted in nuclear localization of cotransfected p65 and IκBα in COS cells and enhanced nuclear relocation of endogenous p65 in T cells. These observations suggest that the main function of IκBα is that of a nuclear export chaperone rather than a cytoplasmic tether. We propose that the nucleus is the major site of p65-IκBα association, from where these complexes must be exported in order to create the cytoplasmic pool.
The NF-κB family of transcription factors consists of proteins that share a domain of approximately 300 amino acids known as the Rel homology domain (RHD) (10, 18). The RHD is required for sequence-specific DNA binding and also mediates protein-protein interactions. Homotypic interactions between RHDs generates a complex array of homo- and heterodimeric NF-κB-related proteins in cells, with the term NF-κB usually referring to the p50-p65 heterodimer. The RHDs also interact with other structural motifs, including ankyrin domains found in the family of IκB proteins (29, 31). Interactions between RHD and IκB proteins results in inhibition of DNA binding and retention of Rel complexes in the cytoplasm. Signals that induce NF-κB lead to the phosphorylation of IκB proteins, which are then targeted for ubiquitination and proteasome-mediated degradation. Rel proteins are thereby released to translocate to the nucleus, bind DNA, and activate gene expression. IκB proteins are therefore central regulators of NF-κB function.
The IκB proteins are a family of functionally diverse molecules. IκBα, IκBβ, and IκBɛ are the most similar, to the extent that they all interact with p65 (also known as RelA) or c-Rel to inhibit DNA binding and are targeted by signal induced phosphorylation for degradation (29, 31). p100 and p110, which are the precursors of RHD-containing p50 and p52 proteins, also contain at their C termini multiple ankyrin repeats that serve IκB-like functions by intramolecularly inhibiting DNA binding by the respective N-terminal RHDs. However, it is unclear whether these IκB proteins are targeted for signal induced degradation. Finally, the protooncogene bcl-3 contains multiple ankyrin domains and looks IκB-like, yet it does not inhibit DNA binding by Rel proteins and has been proposed to be a transcriptional activator in association with nuclear p50 or with p52 (16). The existence of functional differences amongst the IκB proteins is underscored by the differing phenotypes of the genetic deletion of individual IκB genes. IκBα knockout has the most severe phenotype, with the null mice dying within a week of birth (4, 14), whereas bcl-3-deficient mice have defects in germinal center formation (8, 26).
Though identified as inhibitors of NF-κB whose main function is to retain NF-κB in a non-DNA binding form in the cytoplasm, IκB proteins have been proposed to regulate NF-κB in several other ways. For example, IκBα has been shown to contain an unconventional nuclear localization signal (24) as well as a leucine-rich nuclear export sequence (NES) (2, 21). Taken together with earlier reports of transient IκBα presence in the nucleus, it has been proposed that IκBα may be involved in the removal of NF-κB from the nucleus (2, 33). Some evidence in favor of this model has been obtained with Xenopus oocytes, in which IκBα microinjected into the nucleus enhanced p65-RelA loss (2). Similarly, IκBβ has also been proposed to have a nuclear function; unphosphorylated IκBβ has been shown to interact with nuclear NF-κB without inhibiting DNA binding (22, 28, 30). Souyang et al. (28) suggested that NF-κB–IκBβ complexes may protect the transcription factor from being down-regulated by other IκB proteins, thereby leading to long-term NF-κB activation. Despite the differences, replacement of the IκBα gene by the IκBβ gene does not result in the severe phenotype of IκBα-null mice, suggesting that the two IκB proteins can functionally substitute for each other (5).
In this paper, we examined the mechanism of cytoplasmic retention of Rel proteins by IκBα. In a yeast model we show that IκBα shuttles between the cytoplasm and the nucleus, utilizing the nuclear export receptor, Crm1p. In contrast, IκBβ and IκBɛ were not shuttling proteins in this assay. A functional NES was mapped to the N-terminal domain of IκBα that precedes the first ankyrin domain. Mutations in the previously identified C-terminal NES did not affect protein shuttling in our assays. Second, cytoplasmic tethering of p65 in yeast also required nuclear export mediated by IκBα. These observations were extended to mammalian cells; in transfected COS cells cytoplasmic localization of p65 by IκBα was blocked, and increased nuclear p65 was detected in unactivated T cells, by inhibiting CRM1-dependent nuclear export. Our observations suggest that the nucleus is the major site of p65-IκBα association, and the export chaperone property of IκBα is required for cytoplasmic sequestration of this complex.
MATERIALS AND METHODS
Cell lines and strains.
Yeast strains used in this study are listed in Table 1. Yeast strains were generally grown in synthetic medium with the appropriate amino acid and nitrogen base supplement.
TABLE 1.
Strains and plasmids
| Strain or plasmid | Source and/or reference | Genotype or description |
|---|---|---|
| Strains | ||
| W303 | M. Rosbash (20) | MATa ade2-1 ura3-1 trp1.1 leu2-3,112 his3-11,15 |
| crm1.1 | L. Davis (20, 32) | MATa crm1-1 ade2 ura3 trp1 leu2 his3 |
| CRM1+ | This study | MATa crm1-1 ade2 ura3 trp1 leu2 his3; pLDB391 (CRM1 LEU2) integrated at CRM1 |
| EGY48 | M. Rosbash | MATα his3 ura3 trp13 lexA-op-LEU2; pSH18-34 (URA3 6lexA-op-lacZ) |
| RFY206 | M. Rosbash | MATa his3 leu2 trp1 lys2 ura3; pSH18-34 (URA3 6lexA-op-lacZ) |
| Yeast plasmids | ||
| pLDB391 | L. Davis (32) | XhoI-AatII fragment containing CRM1 ORF cloned into HindIII site (blunt ended) of pRS305 (LEU2) |
| pCGF-1A (pPS808) | P. Silver (13) | pPS293-based vector, 2μm, GFP under GAL promoter control, URA3, Ampr |
| pGFP | This study | pCGF-1A with terminator sequence from ADH in SalI and SphI sites |
| pGFP-IκBα | This study | Full-length human IκBα cDNA was cloned in frame after GFP in BamHI and XbaI sites of pGFP vector |
| pGFP-IκBαNEc | This study | Full-length human IκBα cDNA with leucine-to-alanine mutations at residues 272, 274, and 277 (L234) was cloned in frame after GFP |
| pGFP-NΔ32IκBα | This study | Human IκBα cDNA encoding residues from the number indicated after Δ (e.g., residue 32 in pGFP-NΔ32IκBα) to the end of the protein (residue 317) was cloned in frame after GFP |
| pGFP-NΔ42IκBα | ||
| pGFP-NΔ55IκBα | ||
| pGFP-NΔ59IκBα | ||
| pGFP-NΔ32IκBαNEc | This study | Same as above, except that the constructs contain L234 mutations at the C terminus |
| pGFP-NΔ42IκBαNEc | ||
| pGFP-NΔ55IκBαNEc | ||
| pGFP-NΔ59IκBαNEc | ||
| pGFP-NΔ71IκBαNEc | ||
| pGFP-NΔ84IκBαNEc | ||
| pGFP-IκBα-LIL3A49 | This study | Full-length human IκBα cDNA with leucine- or isoleucine-to-alanine mutations at residues 49, 52, and 54 was cloned in frame after GFP |
| pGFP-IκBαNEc-LIL3A49 | This study | Same as above, except that the construct contains L234 mutation at the C terminus |
| pGFP-IκBαNEc-LI2A78 | This study | Full-length human IκBα cDNA with leucine- or isoleucine-to-alanine mutations at residues 78, 82 (LI2A78), 272, 274, and 277 (L234) was cloned in frame after GFP |
| pGFP-IκBαNEc-LLII4A78 | This study | Full-length human IκBα cDNA with leucine- or isoleucine-to-alanine mutations at residues 78, 80, 82, 83 (LLII4A), 272, 274, and 277 (L234) was cloned in frame after GFP |
| pGFP-IκBβ | This study | Full-length mouse IκBβ cDNA was cloned in frame after GFP in BamHI and XbaI sites of pGFP vector |
| pGFP-IκBɛ | This study | Full-length mouse IκBɛ cDNA was cloned in frame after GFP in BamHI and XbaI sites of pGFP vector |
| pGFP-p65 | This study | Full-length mouse p65 cDNA was cloned in frame after GFP in BamHI and XbaI sites of pGFP vector |
| p424 (pLDB229) | L. Davis | 2μm, GALL promoter, CYC1 terminator, TRP1, Ampr |
| pGAL1 | This study | GAL1 promoter of p424 was replaced with GAL1 promoter from pCGF-1A |
| pGAL1.HAIκBα | This study | An HA tag (YPYDVPDYA) was created in the 5′ primer that was used for cloning IκBα by PCR. The full-length HA-tagged IκBα cDNA was inserted in BamHI and EcoRI sites of pGAL1 |
| pJG 4-5 | M. Rosbash | 2μm, GAL promoter, ADH terminator, TRP1, Ampr; A cassette containing SV40 NLS, the acid blob B42, and the HA epitope tag was inserted after GAL promoter (NL-B42-HA) |
| pJG-Rev | M. Rosbash | Full-length Rev cDNA was inserted after NL-B42-HA cassette |
| pEG202 | M. Rosbash | 2 μm, ADH promoter, ADH terminator, HIS3, Ampr; a LexA DNA binding domain was inserted after ADH promoter |
| pEG202-CRM1 | M. Rosbash | Full-length cDNA encoding yeast CRM1 was cloned in frame after LexA |
| pJG-IκBα | This study | Full-length human IκBα was inserted in frame at EcoRI and XhoI sites after NL-B42-HA cassette |
| pJG-IκBαNEc5A | This study | Full-length human IκBα with leucine- or isoleucine-to-alanine mutations at residues 265, 269, 272, 274, and 277 was inserted after NL-B42-HA cassette |
| pJG-IκBαΔNEc | This study | Same as above, except that the C-terminal putative NES from residues 265 to 277 (IQQQLGQLTLE) was deleted |
| pJG-IκBα-LIL3A49 | This study | Full-length human IκBα with leucine-to-isoleucine-to-alanine mutations at 49, 52, and 54 (putative N-terminal NES) was cloned in frame after NL-B42-HA cassette |
| pJG-IκBαNEc-LIL3A49 | This study | Same as above, except that in addition to the mutations at the putative N-terminal NES, putative C-terminal NES was mutated (L234) |
| pJG-α60 | This study | A cDNA encoding human IκBα from amino acid residues 1 to 60 was inserted after NL-B42-HA cassette |
| pJG-α60-LIL3A49 | This study | Same as above, except that the putative N-terminal NES was mutated |
| pJG-α73 | This study | A cDNA encoding human IκBα from amino acid residues 1 to 73 was inserted after NL-B42-HA cassette |
| pJG-α73-LIL3A49 | This study | Same as above, except that the putative N-terminal NES was mutated |
| pJG-β56 | This study | A cDNA encoding mouse IκBβ from residues 1 to 56, right before the first ankyrin repeat was inserted after NL-B42-HA cassette |
| pJG-ɛ122 | This study | A cDNA encoding mouse IκBɛ from residues 1 to 122, right before the first ankyrin repeat was inserted after NL-B42-HA cassette |
| pYEX-BX | Clontech | 2μm, pCUP1, URA3, LEU2-d, Ampr |
| pYEX-BX-IκBα | This study | A HA-tagged, full-length human IκBα was inserted into BamHI and EcoRI sites of pYEX-BX, under the control of a copper-inducible promoter |
| pCu.IκBα | This study | A HindIII (fill in)-EcoRI fragment containing HA-IκBα and the copper-induced promoter was excised from pYEX-BX-IκBα and cloned into SacI (fill in) and EcoRI sites of p424 |
| Mammalian plasmids | ||
| pEGFP.C3 | Clontech | SV40 ori, pUC ori, pCMV, EGFP, SV40 poly(A), Ampr |
| pGFP-p65 | This study | Full-length mouse p65 cDNA was inserted in frame behind GFP into XhoI and EcoRI sites of pEGFP.C3 |
| pGFP-IκBα | This study | Full-length human IκBα cDNA was cloned in frame after GFP |
| pGFP-IκBαNEc | This study | Same as above, except that the construct contains L234 mutations |
| pGFP-IκBαNEc-LIL3A49 | This study | Same as above, except that the construct also contains the N-terminal mutations |
| pGFP-IκBα-LIL3A49 | This study | Same as above, except that the construct does not contain L234 mutations |
| pGFP-IκBβ | This study | Full-length mouse IκBβ cDNA was cloned in frame after GFP |
| pGFP-IκBɛ | This study | Full-length mouse IκBɛ cDNA was cloned in frame after GFP |
| pCDNA3 | Clontech | SV40 ori, ColE1 ori, pCMV, BGH poly(A), Ampr |
| pCDNA3.HA-IκBα | This study | Full-length human IκBα with the HA tag was cloned into pCDNA3 |
ORF, open reading frame; SV40, simian virus 40; BGH, bovine growth hormone; EGFP, enhanced GFP; ADH, alcohol dehydrogenase.
D5h3 T hybridoma cells were grown in Dulbecco modified Eagle medium (DMEM) (GIBCO BRL) supplemented with 10% heat-inactivated fetal bovine serum, 50 μM β-mercaptoethanol, 50 U of penicillin per ml, and 50 μg of streptomycin per ml. COS cells were cultured in DMEM medium with 10% newborn calf serum and the above supplements. BOSC 23 cells were cultured in DMEM medium with 10% heat-inactivated fetal bovine serum, 50 U of penicillin per ml, and 50 μg of streptomycin per ml.
The plasmids used in this study were confirmed by sequencing, and expression of proteins was verified by Western blot analysis. The transcriptional activities of the fusion proteins green fluorescent protein (GFP)-p65 and hemagglutinin (HA)-IκBα were checked in yeast and COS cells with NF-κB enhancer-dependent, LacZ, and chloramphenicol acetyltransferase reporter genes, respectively.
Transformation and transfection.
Yeast expression plasmids and linear plasmid pLDB391 (Crm1p expression vector) were introduced into yeast by lithium acetate transformation (11). The transformed cells were then selected with synthetic complete medium lacking the appropriate nutrient.
COS cell transfection was done by the calcium phosphate method as previously described (19). The amount of plasmid added was equalized with a carrier plasmid in each sample. The medium was changed 12 h after transfection, and leptomycin B (LMB) was added 4 h prior to harvest.
Cytoplasmic and nuclear extracts.
The procedures for making cytoplasmic and nuclear extracts from D5h3 T cells have been described previously (12). Briefly, cells were washed with phosphate-buffered saline (PBS) and the cytoplasmic extracts were obtained by resuspending the pellets in hypotonic buffer A (10 mM Tris [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.5 mM dithiothreitol (DTT), 0.1% NP-40). Nuclei were collected by centrifugation, and nuclear proteins were extracted in buffer C (20 mM HEPES [pH 7.9], 420 mM NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 20% glycerol). To make nuclear extract from COS cells, streptolysin O (Sigma) in buffer S (115 mM potassium acetate [pH 7.3], 25 mM HEPES [pH 7.4], 2.5 mM MgCl2) was used to lyse the cytoplasmic membrane (23). The nuclei were then solubilized in buffer T (30 mM Tris [pH 8.6], 150 mM NaCl, 2 mM EDTA, 2% Triton X-100). Contamination of cytoplasmic proteins in nuclear extracts was estimated by α-tubulin with Western blotting.
Yeast whole-cell extracts for Western blotting were prepared by trichloroacetic acid (TCA) method. Cells were pelleted, washed, and disrupted with 50% TCA and acid-washed glass beads (425 to 500 μm) using a glass bead beater at 4°C. The TCA-precipitated proteins were washed twice with water to remove residual TCA and then boiled in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer prior to electrophoresis. Yeast whole-cell extracts for immunoprecipitation were prepared with glass bead disruption buffer (20 mM Tris-Cl [pH 7.9], 10 mM MgCl2, 1 mM EDTA, 5% glycerol, 1 mM DTT, 0.3 M ammonium sulfate). The procedure has been described previously (3).
Western blot analysis.
Extracts (10 μg) were separated by SDS–10% PAGE, and proteins were then transferred to enhanced chemiluminescence hybond nitrocellulose membrane (Amersham). Equal loading of each sample was confirmed with Ponceau S staining (Sigma). Anti-c-Rel, anti-p65, and anti-IκBα (Santa Cruz Biotechnology) as well as anti-α-tubulin (ICN Biochemical Inc.) were each used at a dilution of 1:1,000. After incubating with the primary antibody for 1 h at room temperature, filters were washed and incubated with peroxidase-conjugated anti-rabbit immunoglobulin (Ig) (Amersham) or anti-mouse Ig (Jackson ImmunoResearch Lab. Inc.) at a dilution of 1:2,000. The chemiluminescence signal was detected using SuperSignal substrate according to the manufacturer's specification (Pierce).
Immunostaining.
Cells were fixed with 4% paraformaldehyde in PBS for 10 min and permeabilized with 0.2% Triton X-100 in PBS for 10 min at room temperature. Blocking was done with 1 mg of bovine serum albumin per ml in PBS and then with 5% normal rabbit serum in PBS (Jackson ImmunoResearch Lab. Inc.) for 30 min. After blocking, cells were incubated with mouse monoclonal anti-HA (Clone 116B12) (Berkeley Antibody Company) at a 1:1,000 dilution in PBS containing 5% normal rabbit serum for 45 min. Cells were then washed several times with PBS and incubated with lissamine rhodamine-conjugated anti-mouse IgG (heavy plus light chains) (Jackson ImmunoResearch Lab. Inc.) at a 1:200 dilution for 45 min. After incubation with the secondary antibody, cells were washed four times with PBS before mounting with Fluoromount (Fisher Scientific), sealed with nail polish, and observed by fluorescence microscopy.
Fluorescence microscopy.
To localize the subcellular localization of GFP in yeast, cells were grown in synthetic complete medium with 2% glucose lacking the appropriate amino acid. Cells were then shifted to raffinose-containing medium and grown to early log phase before inducing with 2% galactose or 0.5 mM copper.
The subcellular localization of GFP in COS cells was determined 40 h after transfection. The GFP signals in living cells or the immunofluorescence signals were observed by fluorescence microscopy (Axiophot; Zeiss) with a GFP generic long pass filter.
Yeast interaction mating assay.
The procedures have been described previously (7). Briefly, EGY48 yeast strains containing the fish plasmids (pJG-) were plated on Ura-Trp dropout minimal plates. RFY206 yeast strains containing the bait plasmids (pEG202-) were plated on Ura-His dropout minimal plates. Before mating, the yeast strains were streaked on yeast extract-peptone-dextrose plates and incubated at 30°C for a day. The EGY48 yeast strains were then replicated perpendicularly to RFY206 strains on Ura-Trp-His dropout plates with 1% raffinose and 2% galactose. Photographs were taken after 2 to 3 days of incubation at 30°C.
RESULTS
IκBα is a shuttling protein.
The properties of individual Rel or IκB proteins are often difficult to evaluate in the complex milieu of mammalian cells, where several of these proteins are simultaneously expressed. Yeast cells do not contain any known Rel or IκB proteins, and the cytoplasmic tethering of p65-RelA by IκBα has been reconstituted in these cells (6). Therefore, we investigated the properties of IκB proteins in yeast. IκBα, IκBβ, and IκBɛ were tagged at the N terminus with GFP and expressed from a galactose-inducible promoter. Subcellular location was monitored by fluorescence microscopy.
Several observations suggest that IκBα may interact with the nuclear export machinery. First, IκBα has been shown to contain two leucine-rich sequence motifs that are reminiscent of the recognition sites of the nuclear export receptor Crm1p (2, 21); one of these elements has been shown to bind nuclear export receptor Crm1p in vitro and serve as an export sequence when attached to pyruvate kinase (21). Second, IκBα microinjected into Xenopus oocytes nuclei decreases nuclear p65-RelA, though this may be due to inhibition of DNA binding and consequent loss of the protein from the nucleus (2). Third, IκBα regulation of v-Rel distribution in transfected cells has been shown to be sensitive to LMB (24), a drug that blocks CRM1 activity (9, 15). To obtain functional evidence for interaction of IκBα with the nuclear export machinery, we compared the subcellular distribution of IκB proteins in the crm1-1 yeast strain, which is defective for Crm1p-mediated export, (20, 32), to the same strain transformed with a wild-type CRM1 gene (CRM1+). In CRM1+ cells, GFP-IκBα was present in both the nucleus and the cytoplasm, whereas GFP-IκBβ and GFP-IκBɛ were located predominantly in the cytoplasm (Fig. 1A, left panels). Western blot analyses showed that proteins of appropriate sizes were expressed in all transformants (data not shown).
FIG. 1.
Nucleocytoplasmic shuttling of IκB. (A) GFP or GFP-IκB fusion proteins were expressed from a galactose-inducible promoter in two yeast strains. crm1-1 has a mutation in the CRM1 gene that encodes a nuclear export receptor, and the strain is consequently defective for nuclear export. These cells were transformed with a vector that constitutively expresses a WT CRM1 gene and serve as an isogenic WT control (CRM1+). Cells were induced for 3 h with galactose, and GFP expression was monitored by fluorescence microcopy. (B) GFP-IκB fusion proteins, as indicated, were expressed in mammalian BOSC 23 cells by transcient transfection. Forty hours after transfection, half of the cells were treated with LMB (+LMB) and, after an additional 4 h, fixed for fluorescent visualization. The second and fourth columns show DAPI-stained nuclei; GFP fluorescence in only a subset of cells reflects the reduced (less than 100%) efficiency of transient transfection.
In contrast, GFP-IκBα was located predominantly in the nucleus of crm1-1 cells, whereas the distribution of GFP-IκBβ or GFP-IκBɛ or GFP itself did not change significantly compared to that observed with CRM1+ cells (Fig. 1A, right panels). The substantial redistribution of IκBα to the nucleus in crm1-1 strain suggests that most of the cellular IκBα transits through the nucleus and requires active export for its cytoplasmic localization. This is to be distinguished from earlier interpretations that overexpressed IκBα spills over from the cytoplasm into the nucleus. We propose that the mixed cytoplasm and nuclear distribution of overexpressed IκBα is probably the result of saturating nuclear export, import, or both. We conclude that IκBα, but not IκBβ or IκBɛ, transits through the nucleus before residing in the cytoplasm.
The subcellular distribution of IκB proteins was also examined in transiently transfected mammalian BOSC23 cells using GFP fluorescence (Fig. 1B). The involvement of CRM1 was evaluated by using LMB, a specific inhibitor of CRM1-dependent export (9, 15). In the presence of LMB, GFP-IκBα distribution shifted to being predominantly nuclear, compared to being predominantly cytoplasmic in untreated cells (Fig. 1B). However, GFP-IκBβ and GFP-IκBɛ subcellular distribution was the same in the presence and absence of LMB (Fig. 1B). These observations are consistent with those in yeast and indicate that IκBα, but not IκBβ or IκBɛ, utilizes CRM1 to shuttle between the nucleus and the cytoplasm.
CRM1-responsive sequence in N terminus of IκBα.
IκBα has been previously shown to contain a CRM1-dependent nuclear export sequence located just after the sixth ankyrin repeat (Fig. 2A). However, an IκBα derivative that was mutated in this motif (IκBαNEc) redistributed to the nucleus in crm1-1 cells just like the wild-type protein (Fig. 2B), indicating the presence of at least one other Crm1p-interacting export sequence. It was possible that the functional NES(s) lay within the ankyrin domain; for example, one such sequence has been proposed to be located in ankyrin 2 (24). Because mutations within ankyrin domains were more likely to affect IκBα structure and function in other ways, we first sought the putative NES in the N-terminal domain that precedes ankyrin repeat 1. N-terminal truncation mutants of IκBα, as indicated in Fig. 2A, were generated in the context of a wild-type (WT) IκBα gene or one containing a mutation in the C-terminal NES (NEc). These IκBα derivatives were expressed as GFP fusion proteins in CRM1+ cells, followed by fluorescent visualization. Full-length IκBα and the first two deletion mutants were found both in the nucleus and the cytoplasm (Fig. 2C, top three panels, only the data in the context of the NEc mutation are shown). In contrast, NΔ55 and NΔ59 proteins were located predominantly in the nucleus (Fig. 2C, bottom two panels). A similar distribution pattern was observed when the truncations were assayed in the context of a protein that was not mutated in the C-terminal NES (summarized in Fig. 2A). We concluded that a peptide motif between residues 42 and 55 is necessary for cytoplasmic location of IκBα.
FIG. 2.
Deletion analysis of IκBα to identify a functional NES. (A) Schematic representation of IκBα (top line) showing the relative locations of the six ankyrin repeats, the C-terminal PEST domain, and a proposed C-terminal NES. The amino acid sequence of the C-terminal NES is shown, and mutations that alter three leucines to alanines are indicated. This combination of mutations was previously shown to inactivate the NES and is referred to as NEc in our assays. The lower part of the figure shows an expanded view of the N-terminal and first ankyrin domains of IκBα with the positions of several N-terminal truncations used in this study. Note that all deletion mutants were tested with the rest of the protein either intact or containing the NEc mutation. The columns on the right summarize the subcellular distribution of these IκBα derivatives in CRM1+ cells. C, either cytoplasmic or mixed cytoplasmic and nuclear location; N, nuclear expression; nd, not determined. Representative data on the basis of which these conclusions are drawn are shown in panel C. (B) Mutation of the C-terminal NES does not affect IκBα localization. GFP fusion proteins containing a WT IκBα gene or the NEc mutation were expressed in crm1-1 (Crm1p mutant) or CRM1+ (reconstituted WT) cells and visualized by fluorescence. (C) Subcellular location of N-terminal truncation mutants of IκBα in CRM1+ cells. Results shown are representative of at least three independent experiments.
Examination of the sequence in this region showed a leucine-containing hydrophobic patch that could be an export motif (Fig. 3A). Similar leucine-rich patches, such as the sequence highlighted in the first ankyrin repeat, are also present elsewhere in the IκBα molecule (Fig. 3A). To determine if either or both motifs were required for cytoplasmic location, these sequences were mutated in the context of the WT IκBα or one that contains a mutated C-terminal NES. GFP fusion protein versions of these derivatives were expressed in CRM1+ cells and visualized by fluorescence microscopy. Alteration of the two leucines and one isoleucine in the N-terminal domain to three alanines (LIL3A) changed the subcellular distribution of the protein to being predominantly nuclear (Fig. 3B). This was regardless of whether the C-terminal NES was mutated or not. However, both mutations in the LHLAII motif in ankyrin 1 behaved like the WT protein with respect to subcellular distribution in CRM1+ cells (data not shown). These observations suggest that the N-terminal sequence LQEIRL is required for cytoplasmic location of IκBα; furthermore, its function cannot be substituted by other leucine-rich sequences in IκBα, including the C-terminal NES.
FIG. 3.
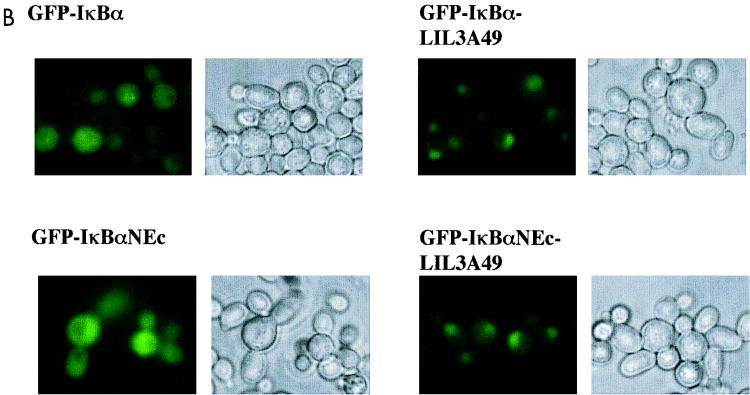
Point mutational analysis of a putative N-terminal NES in IκBα. (A) Schematic representation of the N-terminal and first ankyrin domains of IκBα. The sequence shown above the N-terminal domain includes residues 45 to 58 of human IκBα, whose deletion in NΔ55 (Fig. 2) makes IκBα constitutively nuclear. The indicated residues were changed to alanines to generate the mutant referred to as LIL3A49 (the first leucine is residue 49). The sequence above the ankyrin domain shows another leucine- or isoleucine-rich motif comprised of residues 76 to 85. Mutations LI2A78 and LLII4A78 alter the first leucine and isoleucine, or both leucines and isoleucines, respectively. Mutations were in the context of full-length (F.L.) WT IκBα or a derivative mutated at the C-terminal NES (NEc). Columns on the right summarize the subcellular location of mutants. C, either cytoplasm or cytoplasm plus nuclear; N, nuclear localization. Representative data in yeast and mammalian cells are shown in panels B and C, respectively. (B) Subcellular distribution of GFP-IκBα derivatives indicated in CRM1+ yeast strain. Data shown are representative of at least three independent experiments. (C) Subcellular distribution of GFP-IκBα derivatives in BOSC 23 mammalian cells visualized after transient transfection with appropriate expression vectors as indicated. DAPI staining was used to visualize nuclei; GFP fluorescence is present in a subset of cells because not all cells pick up transfected DNA.
To extend these observations, we compared the subcellular distribution of these IκBα derivatives in mammalian cells. GFP-IκBα and GFP-IκBαNEc, which were located in the cytoplasm of transiently transfected BOSC 23 cells (Fig. 3C, left panels) could be driven to the nucleus by treating the cells with LMB (Fig. 1B shows an example of location in LMB-treated cells), indicating that cytoplasmic location of both proteins was the result of active nuclear export. In contrast, LIL3A-mutated IκBα in the WT or NEc context was located primarily in the nucleus even in the absence of LMB treatment (Fig. 3C, right panels). The LQEIRL motif is therefore necessary for cytoplasmic location of IκBα in yeast as well as mammalian cells.
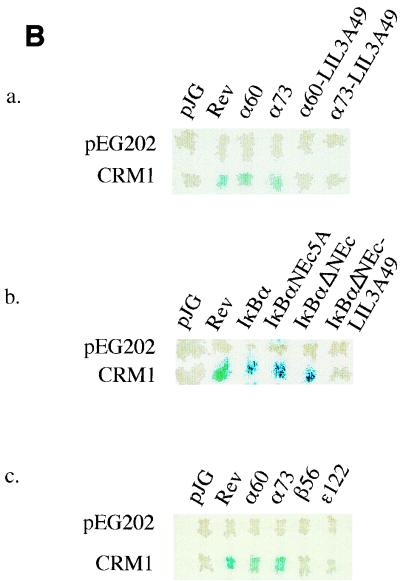
The simplest interpretation of the similarity of the subcellular distribution of IκBα in CRM1-inhibited (crm1-1) cells and in those with the LIL3A mutation is that the LQEIRL motif is a CRM1-dependent NES. An alternate possibility that we could not rule out from the experiments described above was that the sequence served as a cytoplasmic tether and was not involved in nuclear export. To gain additional insight, we tested whether this IκBα sequence bound CRM1 in a yeast two-hybrid assay. This assay has been previously used to examine interactions between CRM1 and its substrates (20). IκBα derivatives fused to a transcription activation domain in the vector pJG4-5 (Fig. 4A) were used to transactivate β-galactosidase expression by interacting with a LexA DNA binding domain-CRM1 fusion. The DNA binding domain of LexA did not interact with any of the fusion proteins in pJG4-5 (Fig. 4B, top rows). Interaction with CRM1 was evident when the fish contained Rev sequences (positive control) or two fragments derived from the N terminus of IκBα (α60 and α73) that contained the LQEIRL motif identified above. The LIL3A mutation in the context of either fragment abolished CRM1 interaction (Fig. 4Ba). These observations strengthen the idea that the newly identified motif is an NES and not a cytoplasmic tether.
FIG. 4.
CRM1 binding by IκB proteins. (A) Schematic representation of plasmids used in yeast two-hybrid assays to study IκB-CRM1 interactions. A fusion protein consisting of the LexA DNA binding domain and yeast Crm1p serves as the bait. IκBα transactivation domain fusion proteins were expressed in the vector pJG4-5. Plasmids are denoted by the features of the IκB portions of the fusion proteins. IκBα, full-length WT IκBα; IκBαNEc5A, full-length IκBα with a 5-alanine substitution in the C-terminal NES; IκBαΔNEc, full-length IκBα with a 12-amino-acid deletion of the C-terminal NES; IκBα-LIL3A49, full-length IκBα carrying the LIL3A mutation (Fig. 3A) in the N-terminal NES; IκBαNEc-LIL3A49, full-length IκBα carrying the LIL3A mutation and the NEc mutation (Fig. 2A); α60, first 60 amino acids from IκBα; α73, first 73 amino acids from IκBα; α60LIL3A49 and α73LIL3A49, LIL3A mutations in the context of α60 and α73, respectively; β56, first 56 amino acids of murine IκBβ; ɛ122, first 122 amino acids of murine IκBɛ. (B) IκB-Crm1p interaction using the two-hybrid assay. Bait plasmids (pEG202 or CRM1) were transformed into the RFY206 (MATa) yeast strain, and fish plasmids were transformed into the EGY48 (MATα) yeast strain. β-Galactosidase activity was assayed in diploids generated after mating fish- and bait-containing transformants.
To determine whether additional CRM1 interacting sequences were located within IκBα, we examined the interaction of IκBα derivatives that contained all the ankyrin domains (Fig. 4Bb). Full-length IκBα associated with CRM1, as did versions of IκBα that were mutated or deleted, in the C-terminal NES (labeled NEc5A or ΔNEc). However, an IκBα derivative carrying the LIL3A mutation no longer associated with CRM1. We conclude that ankyrin domains of IκBα do not contain strong CRM1-binding motifs and propose that nuclear export of IκBα is determined by the N-terminal LQEIRL motif.
The observation that subcellular location of IκBβ or IκBɛ was not affected by inhibiting CRM1 function suggested that there were no CRM1-binding motifs in these proteins. We tested the ability of the N-terminal domains of IκBβ and IκBɛ to bind CRM1 in the yeast assay. Whereas two fragments from the N terminus of IκBα scored positive in this assay, similar regions of IκBβ and IκBɛ did not interact with CRM1 (Fig. 4Bc) confirming the prediction of the cellular assays.
Role of nuclear export in cytoplasmic sequestration.
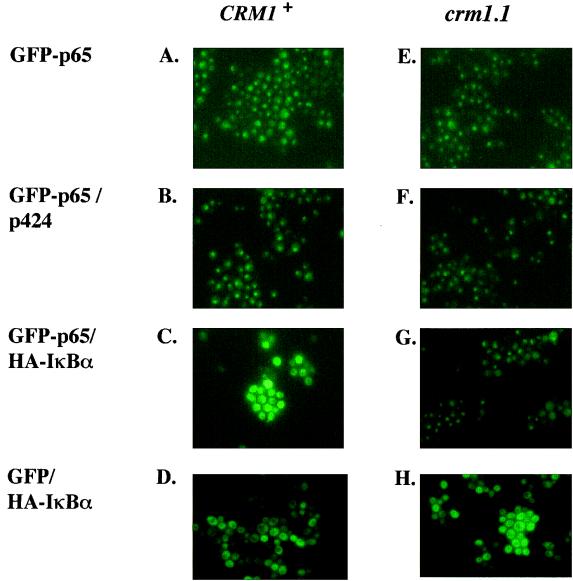
Epinat et al. have previously shown that coexpression of p65 and IκBα in yeast results in cytoplasmic retention of the Rel protein (6). To investigate the role of nuclear export in this process, we coexpressed a GFP-tagged p65 and IκBα in the export-deficient crm1-1 yeast strain or in crm1-1 cells reconstituted with a WT CRM1 gene. In the absence of IκBα, GFP-p65 expression was exclusively nuclear in either yeast strain (Fig. 5A, B, E, and F). Coexpression with IκBα led to relocation of GFP-p65 to the cytoplasm in the reconstituted CRM1+ cells (Fig. 5C), but not in crm1-1 cells (Fig. 5G). The subcellular distribution of GFP-p65 was not affected by the empty expression vector used to express IκBα (Fig. 5B and F), nor was the distribution of GFP alone affected by IκBα (Fig. 5D and H). These observations suggest that cytoplasmic retention of p65 by IκBα requires active nuclear export.
FIG. 5.
Cytoplasmic retention of p65 by IκBα requires nuclear export. GFP or GFP-p65 was expressed from a galactose-inducible promoter in the yeast strains as indicated in the legend to Fig. 1. The IκBα gene was tagged with a 9-amino-acid epitope from the influenza virus HA (HA-IκBα) and expressed from the vector p424, which also contains a galactose-inducible promoter. Single or double transformants, as indicated, were induced with galactose for 3 h, and GFP expression was monitored by fluorescence microscopy. Results shown are from one of three independent experiments.
In this experiment both GFP–p65 and HA-IκBα were transcribed from galactose inducible promoters, with the idea that both proteins would be expressed together and, presumably, retained in the cytoplasm. The observation that p65 was localized to the nucleus in crm1-1 cells under these conditions suggested that a significant proportion of the coexpressed p65 and IκBα made its way to the nucleus and remained there in the absence of Crm1p-dependent export. This could be because coordinately synthesized p65 and IκBα did not find each other before p65 translocated to the nucleus or because p65 synthesis preceded that of IκBα and resulted in its nuclear localization before associating with IκBα. In either case, cytoplasmic localization would be the result of export in Crm1p-containing cells.
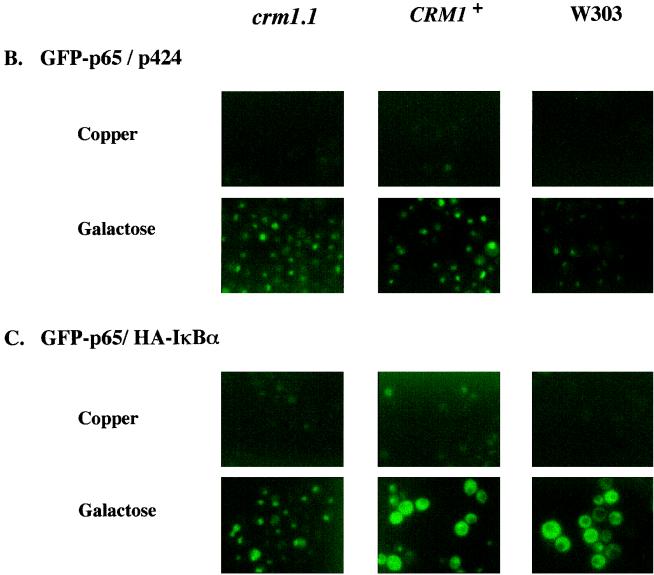
To minimize nuclear translocation of p65, we established ongoing IκBα synthesis prior to p65 expression. Towards this goal, IκBα was expressed from a copper-inducible promoter (17) and GFP-p65 was expressed from a galactose-inducible promoter in crm1-1 and CRM1+ strains, and a WT yeast strain, W303. The tight regulation of p65 in glucose medium ensured that IκBα protein was evident before p65 (Fig. 6A). IκBα expression was detected by immunoblotting even before treatment with copper because of leakiness in this promoter; however, higher levels of IκBα were apparent after copper treatment (Fig. 6A, lanes 2, 5, and 8). During this time, p65 expression could not be detected (Fig. 6A, lanes 1, 2, 4, 5, 7, and 8). After treatment with copper, the cells were shifted to galactose- and copper-containing medium that resulted in GFP-p65 expression (Fig. 6A, lanes 3, 6, and 9). GFP-p65 fluorescence was detected primarily in the nucleus in the absence of IκBα in all strains (Fig. 6B). When IκBα was expressed first, followed by GFP-p65, cytoplasmic fluorescence was evident in most CRM1+ and W303 cells, indicating that GFP-p65 was retained in the cytoplasm (Fig. 6C). However, even when IκBα was expressed first, most of the GFP-p65 was located in the nucleus of crm1-1 cells (Fig. 6C). These observations suggest that cytoplasmic localization of p65 in the presence of ongoing IκBα synthesis requires nuclear export.
FIG. 6.
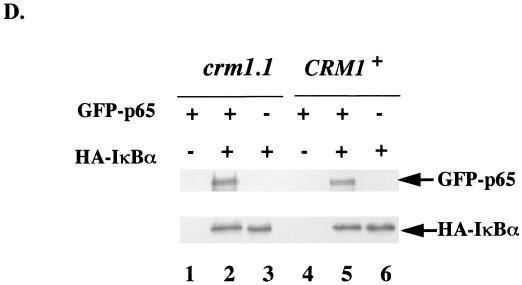
Sequential induction of IκBα and GFP-p65. (A) HA-IκBα gene was cloned into an expression vector that contains a copper-inducible promoter. GFP-p65 was expressed from the galactose-inducible promoter. Double transformants in different yeast strains were treated first with copper to induce IκBα expression and then with galactose to induce GFP-p65 expression. Whole-cell extracts were prepared from double transformants that were not treated either with inducing agent (lanes 1, 4, and 7), or were treated with 0.5 mM copper sulfate for 1 h (lanes 2, 5, and 8) or with copper sulfate for 1 h followed by galactose and copper for an additional 2.5 h (lanes 3, 6, and 9). GFP-p65 and IκBα were detected by immunoblotting after separation of the extracts by SDS-PAGE. crm1-1 and CRM1+ strains were defined in the legend to Fig. 1; W303 represents another WT strain. Results shown are from one of three independent experiments. (B and C) Fluorescent visualization of GFP-p65 localization in single and double transformants, respectively, as noted on the left of the panels. p424 is an empty expression vector. Yeast strains used are indicated on the top. Results shown are from one of three independent experiments. (D) Nuclear association of GFP-p65 and HA-IκBα in crm1-1 cells. CRM1+ and crm1-1 cells transformed with expression vectors described for panel A were induced (+) to express GFP-p65 alone, HA-IκBα alone, or both together as indicated. Whole-cell extracts were first incubated with anti-IκBα antibodies, and then the immunoprecipitate was fractionated by SDS-PAGE. Proteins were transferred to nitrocellulose filters which were probed with anti-p65 and anti-IκBα anti-sera. The immunoblots were visualized by chemiluminescence.
We used immunoprecipitation assays to determine whether GFP-p65 and HA-IκBα were complexed in the nucleus of crm1-1 cells. The proteins were induced, individually or together, in crm1-1 and CRM1+ cells. Anti-IκBα antibody was used to immunoprecipitate HA-IκBα from whole-cell lysates, and associated p65 was detected by immunoblotting after fractionation of the precipitate by SDS-PAGE. The membranes were also probed with anti-IκBα antibody. GFP-p65 was only detected when coexpressed with HA-IκBα (Fig. 6D, compare lanes 1 and 2 or 4 and 5). Levels of HA-IκBα, or the efficiency of immunoprecipitation, were unchanged in the presence or absence of GFP-p65 (Fig. 6D, compare lanes 2 and 3 or 5 and 6). Importantly, comparable levels of GFP-p65 were associated with IκBα in crm1-1 and CRM1+ strains, though the complex is predominantly nuclear in crm1-1 cells and predominantly cytoplasmic in the CRM1+ cells (Fig. 6C). These observations indicate that mutation of CRM1 does not affect p65-IκBα protein association; more likely it affects the translocation of the p65-IκBα complex from the nucleus to the cytoplasm.
Cytoplasmic sequestration in mammalian cells.
To extend these observations to mammalian cells, COS cells were transiently transfected with a GFP-p65 expression vector in the presence or absence of an IκBα expression vector, and nuclear export via CRM1 was blocked by treating cells with the CRM1 inhibitor LMB (9, 15). As expected, GFP-p65 was exclusively nuclear when expressed in the absence of IκBα (Fig. 7A) and substantially cytoplasmic in the presence of coexpressed IκBα (Fig. 7C). Immunofluorescence using anti-HA antibodies showed that IκBα localization closely paralleled that of GFP-p65. We found that GFP-p65 transactivated κB-dependent reporter at a level comparable to that seen with WT p65; furthermore, GFP-p65 dependent transactivation was efficiently suppressed by coexpressed IκBα (data not shown). These observations suggest that the DNA binding and transcription activation characteristics of GFP-p65 are similar to those of p65, and the observations validate the subcellular distribution studies shown in Fig. 7. To determine the contribution of export to IκBα induced redistribution of p65, we treated transfected COS cells with LMB for 2 to 4 h prior to fixation and fluorescent visualization. In cells transfected only with GFP-p65, LMB treatment did not alter the nuclear expression of this protein (Fig. 7B). However, in cells that coexpressed GFP-p65 and IκBα, both proteins were present in the nuclei of LMB-treated cells at levels significantly higher than those in untreated cells (compare Figs. 7C and D). We conclude that cytoplasmic retention of p65 by IκBα requires active nuclear export.
FIG. 7.
Cytoplasmic retention of p65 by IκBα in transfected COS cells is sensitive to LMB. (A) COS cells were transfected with GFP-p65 and HA-tagged IκBα expression vectors singly, or in combinations, as noted on the left. LMB (10 ng/ml) was added to the transfected cells 36 h after transfection (indicated on the left as +LMB). Subcellular localization of GFP-p65 was detected 40 h after transfection. HA-IκBα was detected after staining fixed cells with Texas red-conjugated anti-HA antibodies. Results shown are from one of three independent experiments. (B) Nuclear association of GFP-p65 and HA-IκBα in COS cells. COS cells were transiently transfected with GFP-p65 and HA-IκBα expression vectors as indicated. Half the cells were treated with LMB for the last 4 h, and nuclear extracts were prepared as described in Materials and Methods. Nuclear extracts were first treated with anti-p65 antiserum, and then the immunoprecipitate was fractionated by SDS-PAGE and the proteins were transferred to nitrocellulose filters. The filters were probed with anti-p65, anti-IκBα, or anti-α-tubulin and visualized by chemiluminescence.
Colocalization of GFP-p65 and HA-IκBα in the nucleus of LMB-treated cells suggests that the two proteins are associated. This was confirmed by coimmunoprecipitation assays. Nuclear extracts from COS cells transfected with expression vectors for GFP-p65 and IκBα, with or without LMB treatment, were immunoprecipitated with anti-p65 antibodies, and the precipitate was probed with anti-IκBα antibody after separation by SDS-PAGE. In the absence of LMB, nuclear p65 level was reduced in cells that coexpressed IκBα (Fig. 7E, compare lanes 1 and 2), but very little IκBα could be detected in the nucleus. The lower level of nuclear p65 is most likely due to cytosolic localization of the protein by IκBα. Several factors contribute to the incomplete depletion of nuclear p65 (Fig. 7E, lane 2). First, not all cells coexpress IκBα and p65, and second, we detected some degree of cytosolic contamination in the nuclear extract preparation, as evidenced by the presence of α-tubulin (Fig. 7E, bottom panel). When the cells were treated with LMB, the levels of nuclear GFP-p65 were similar regardless of whether IκBα was coexpressed (Fig. 7G, lanes 3 and 4), consistent with the fluorescent visualization that shows both proteins to be predominantly nuclear. More importantly, p65-associated IκBα was easily detected in nuclear extracts from cells in which both proteins were coexpressed (Fig. 7E, lane 4). Given the particularly low cytoplasmic contamination in these extracts (the lowest α-tubulin levels are shown in Fig. 7E, lane 4), we conclude that nuclear p65 and IκBα form a complex in LMB-treated cells. These observations further strengthen the view that cytosolic sequestration requires active export of NF-κB–IκB complexes from the nucleus.
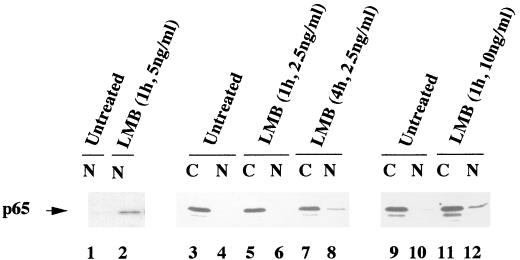
In the two previous experiments we showed that cytoplasmic localization of ectopically expressed p65 required the nuclear export receptor CRM1. To assess whether CRM1 was also required to maintain cytoplasmic p65 in untransfected cells, we treated D5h3 T hybridoma cells with LMB and assayed p65 levels in the nucleus by immunoblotting. p65 was not detected in nuclear extracts from untreated D5h3 cells (Fig. 8, lane 1), but 1 h of treatment with LMB resulted in accumulation of this protein in nuclei (Fig. 8, lane 2). The effect was also evident with a lower dose of LMB; however, longer times of treatment were required (Fig. 8, lanes 3 to 8). No significant difference was observed with a higher concentration of LMB (Fig. 8, lanes 9 to 12). Use of these nuclear extracts in electrophoretic mobility shift assays with a κB DNA probe did not reveal increased κB DNA binding activity (data not shown), suggesting that the nuclear p65 was associated with an IκB protein. Similar results were recently reported by Rodriguez et al. (23), who showed that treatment of HeLa cells with LMB resulted in elevated levels of nuclear p65. In both cases only a modest increase in nuclear p65 levels was observed after LMB treatment, compared to the more clearcut results in transfected COS cells. As described more fully in the Discussion, our interpretation of these observations is that the p65 detected in the nucleus reaches there due to ongoing disruption of NF-κB–IκBα complexes in the cytoplasm. We conclude that maintenance of p65 in the cytoplasm of unstimulated cells requires continuous retrieval of the nuclear protein (Fig. 9).
FIG. 8.
Effect of LMB treatment of unactivated T cells. D5h3 T hybridoma cells were treated with LMB, and p65 expression in nuclear (N) and cytoplasmic (C) extracts at different times was followed by immunoblotting. LMB concentrations and times of treatment are noted above the lanes. IκBα or IκBβ levels were assayed by immunoblotting of whole-cell extracts and did not change significantly in the presence or absence of LMB (data not shown). Results shown are from one of three independent experiments.
FIG. 9.
Model for nuclear export-dependent cytoplasmic sequestration of NF-κB in unstimulated cells. Two central interpretations of our observations are pictorially represented in this figure. Numbers indicate steps, as explained below. First, cytoplasmic NF-κB–IκBα complexes are actively retained in the cytoplasm. We think this is needed because p65 can leak into the nucleus in the absence of stimulation, for example, because of constitutive turnover of IκBα (step 1). A Rel protein that is released due to constitutive degradation of its associated IκBα could, in principle, meet up with a newly synthesized IκBα molecule and be held back in the cytoplasm. We suggest that this does not happen to any significant extent, and the released p65 protein migrates to the nucleus (step 2). The second tenet of our model is that IκBα and p65 do not associate in the cytoplasm. Thus, newly synthesized IκBα translocates independently to the nucleus (step 3). Similarly, transiently released or newly synthesized p65 also migrates to the nucleus (step 4). The two proteins associate in the nucleus (step 5), from where the p65-IκBα complex is then exported out by CRM1 to maintain the cytosolic pool in unstimulated cells (step 6).
DISCUSSION
Using GFP-tagged IκB proteins, we found that the subcellular location of IκBα, but not IκBβ or IκBɛ, depended upon the nuclear export receptor, CRM1. In a yeast strain that contained a mutated CRM1 gene, GFP-IκBα was located predominantly in the nucleus, indicating that active export was required for its cytoplasmic localization. Our observations directly demonstrate that IκBα is a shuttling protein, which is presumably due to the presence of nuclear localization and nuclear export sequences within this polypeptide. However, mutation of the previously identified C-terminal NES did not affect CRM1-dependent subcellular localization of IκBα, prompting us to look for another sequence that regulated IκBα location. A leucine-rich region in the second ankyrin repeat has been shown to affect v-Rel localization (24). Because this sequence contacts p65 in the p65-IκBα complex, and structural integrity of the ankyrin motifs is critical for IκBα function, we did not alter it. Instead, mutational studies showed that the sequence LQEIRL, in the N-terminal domain of IκBα, was essential for CRM1-dependent IκBα shuttling. This motif was also shown to bind CRM1 protein, suggesting that it is a functional NES. Functional consequences of IκBα shuttling are further discussed below.
IκBβ and IκBɛ do not contain CRM1 binding motifs at their N termini, and their cellular location is not affected by CRM1. The differences between IκBα and the other two IκB proteins indicate that they are not shuttling proteins, at least via the CRM1 pathway and suggest that the three related polypeptides may have different biological functions. Recently, Cheng et al. (5) showed that replacing the IκBα gene with the IκBβ gene rescued the neonatal lethality observed in IκBα-deficient mice. These observations were interpreted to mean that IκBα and IκBβ were functionally similar, and the inability of IκBβ to rescue mice lacking IκBα was due to inappropriate gene regulation. It is possible that the functional differences between IκB family members will be reflected in more subtle cellular assays. Alternatively, our present studies do not rule out the possibility that IκBβ and IκBɛ are also shuttling proteins but that they use an exportin different from CRM1.
Two major biological functions have been ascribed to IκBα: the tethering of Rel proteins in the cytoplasm and the removal of induced Rel proteins from the nucleus. The tethering function is well supported by the observations that all cells contain non-DNA binding NF-κB–IκBα complexes in the cytoplasm, that IκBα degradation is required for nuclear translocation of NF-κB, and that NF-κB is constitutively nuclear in IκBα-deficient mice. Down-regulation function has been inferred from the observation that IκBα transiently appears in the nucleus of HeLa cells after removal of an NF-κB inducing TNFα signal (1). More recently, identification of a leucine-rich NES in IκBα (21) and the demonstration that subcellular distribution of v-Rel is sensitive to LMB (24) have provided additional evidence in favor of the down-regulation hypothesis. In this paper, we directly demonstrate that the shuttling property of IκBα is required for cytoplasmic retention of p65. The requirement of a viable CRM1-dependent export pathway for cytoplasmic localization of p65 by IκBα was unexpected, because IκBα is believed to be a cytoplasmic tether of Rel proteins. If so, coexpressed p65 and IκBα should have formed complexes in the cytoplasm and stayed there. However, we found this was not the case, even when IκBα synthesis was established prior to GFP-p65 expression. The simplest interpretation of these observations is that IκBα is not a cytoplasmic tether as is generally assumed; rather, the main function of IκBα is that of a nuclear export chaperone. In resting cells, this leads to localization of p65 to the cytoplasm.
In yeast, where the properties of proteins could be studied individually, we found that IκBα shuttled continuously between the nucleus and the cytoplasm. The requirement of nuclear export for cytoplasmic retention of NF-κB–IκBα complexes raised the question whether NF-κB–IκB complexes also shuttled continuously. The predominant cytosolic location of NF-κB–IκBα complexes in unstimulated cells could then be explained by the greater efficiency of IκBα-mediated export compared to nuclear localization signal (NLS)-dependent import. Two previous studies shed light on this question. Sachdev and Hannink (24) noted that v-Rel protein rapidly accumulated in the nucleus of chicken embryo fibroblasts treated with LMB. In the same cells, c-Rel localization was unaffected by LMB. The authors proposed that v-Rel–IκBα complexes shuttled between the nucleus and the cytoplasm because weak interactions between v-Rel and IκBα exposed the v-Rel NLS for nuclear import of the complex. These observations showed that net cytosolic distribution of Rel-IκBα complexes could be maintained despite continuous shuttling; however, all Rel-IκBα complexes did not behave identically. In the second study Rodriguez et al. (23) recently concluded that the portion of NF-κB complexed to IκBα in HeLa cells shuttled continuously, based on the observation that treatment of HeLa cells with LMB for 30 min resulted in increased nuclear p65 and IκBα expression. Their proposal of shuttling was strengthened by the demonstration that preexisting IκBα also accumulated in the nucleus. These observations suggest that p65-IκBα complexes can shuttle, whereas c-Rel–IκBα complexes cannot.
In D5h3 T cells, although LMB treatment increased nuclear p65, only a small fraction of the cellular p65 was found in the nucleus. Furthermore, nuclear c-Rel levels did not change significantly under these conditions (data not shown). Our interpretation of these observations is that there is a continuous leak of Rel proteins to the nucleus due to, for example, constitutive IκBα breakdown in unstimulated cells (Fig. 9). We suggest that retrieval of these molecules from the nucleus (in order to maintain the cytosolic store) requires IκBα and its export chaperone characteristics. When retrieval is blocked by LMB, nuclear accumulation of p65-RelA results. In this model we envisage that p65-IκBα complexes do not shuttle continuously, probably because the nuclear localization sequence of p65 is not accessible when it is associated with IκBα. Our model accounts for the low levels of nuclear accumulation by Rel proteins in LMB-treated cells seen in all three studies, yet emphasizes the role of IκBα-dependent export in maintaining the cytosolic pool of NF-κB–IκBα complexes in resting cells.
IκBα has been generally considered to be a cytoplasmic tether of Rel proteins, because its association with Rel proteins hides their nuclear localization signals and thereby prevents nuclear entry. However, we found in both yeast and COS cells that cytoplasmic sequestration of p65 by IκBα required CRM1-dependent nuclear export. Because we think it unlikely that p65-IκBα complexes shuttle continuously, this observation raises the question of why simple cytoplasmic tethering of p65 by IκBα does not occur. That is, why are p65-IκBα complexes not held back in the cytoplasm, as was previously believed, and why do they instead require nuclear export to create the cytoplasmic pool? A fundamental assumption of the tethering model is that p65 and IκBα associate in the cytoplasm. We propose an alternative possibility that newly synthesized p65 and IκBα (which will create the cytoplasmic pool) do not complex in the cytoplasm (Fig. 9). This could be because both proteins translocate independently to the nucleus, which is their default cellular location, and active export is required to bring the complex out to the cytoplasm. Furthermore, uncomplexed IκBα has been shown to be very unstable (27), making it unlikely that there is ever a pool of IκBα (27) in the cell cytoplasm awaiting the synthesis of Rel proteins in order to retain them in the cytoplasm. Therefore, we propose that most cellular p65-IκBα complexes are formed in the nucleus (Fig. 9). Once formed, nuclear p65-IκBα complexes are exported to the cytoplasm using the chaperone properties of IκBα (Fig. 9, step 6). This leads to a net accumulation of p65-IκBα complexes in the cytoplasm, as the complex cannot reenter the nucleus because the NLS on the Rel protein is hidden by IκBα.
ACKNOWLEDGMENTS
LMB used in this study was kindly provided by M. Yoshida. We thank M. Rosbash for comments on the manuscript, Phil Gnatowski for help in the preparation of the manuscript, and Zaira Garcea for enthusiastic assistance in some of these experiments during the summer of 1998.
This work was supported by NIH grants to R.S. (AI 41035) and L.D. (GM54768).
ADDENDUM IN PROOF
The N-terminal NES in IκBα has also been identified by Johnson et al. (EMBO J. 18:6682–6693, 1999).
REFERENCES
- 1.Arenzana-Seisdedos P, Thompson J, Rodriguez M, Bachelerie F, Thomas D, Hay R T. Inducible nuclear expression of newly synthesized IκBα negatively regulates DNA-binding and transcriptional activities of NF-κB. Mol Cell Biol. 1995;15:2689–2696. doi: 10.1128/mcb.15.5.2689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Arenzana-Seisdedos P, Turpin P, Rodriguez M, Thomas D, Hay R T, Virelizier J L, Dargemont C. Nuclear localization of IκBα promotes active transport of NF-κB from the nucleus to the cytoplasm. J Cell Sci. 1997;110:369–378. doi: 10.1242/jcs.110.3.369. [DOI] [PubMed] [Google Scholar]
- 3.Ausubel F M, Brent R, Kingston R E, Moore D D, Sezdman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. Vol. 2. New York, N.Y: John Wiley & Sons; 1987. Preparation of protein extracts from yeast; p. 13.13.4. [Google Scholar]
- 4.Beg A A, Sha W C, Bronson R T, Baltimore D. Constitutive NF-κB activation, enhanced granulopoiesis, and neonatal lethality in IκBα-deficient mice. Genes Dev. 1995;9:2736–2746. doi: 10.1101/gad.9.22.2736. [DOI] [PubMed] [Google Scholar]
- 5.Cheng J D, Ryseck R P, Attar R M, Dambach D, Bravo R. Functional redundancy of the nuclear factor κB inhibitors IκBα and IκBβ. J Exp Med. 1998;188:1055–1062. doi: 10.1084/jem.188.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Epinat J C, Whiteside S T, Rice N R, Israel A. Reconstitution of the NFκB system in Saccharomyces cerevisiae for isolation of effectors by phenotype modulation. Yeast. 1997;13:599–612. doi: 10.1002/(SICI)1097-0061(19970615)13:7<599::AID-YEA109>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 7.Finley R L, Jr, Brent R. Two-Hybrid analysis of genetic regulatory networks. In: Bartel P L, Fields S, editors. The yeast two-hybrid system. New York, N.Y: Oxford University Press; 1997. pp. 197–214. [Google Scholar]
- 8.Franzoso G, Carlson L, Scharton-Kersten T, Shores E W, Epstein S, Grinberg A, Tran T, Shacter F, Leonardi A, Anver M, Love P, Sher A, Siebenlist U. Critical roles for the Bcl-3 oncoprotein in T cell-mediated immunity, splenic microarchitecture and germinal center reactions. Immunity. 1997;6:479–490. doi: 10.1016/s1074-7613(00)80291-5. [DOI] [PubMed] [Google Scholar]
- 9.Fukuda M, Ano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nichida E. CRM1 is responsible for intracellular transport mediated by the nuclear export signal. Nature. 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 10.Grimm S, Baeuerle P A. The inducible transcription factor NFκB: structure-function relationship of its protein subunits. Biochem J. 1993;290:297–308. doi: 10.1042/bj2900297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ito H, Fukuda Y, Murata K, Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983;153:163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jamieson C, McCaffrey P G, Rao A, Sen R. Physiologic activation of T cells via the T cell receptor induces NFκB. J Immunol. 1991;147:415–420. [PubMed] [Google Scholar]
- 13.Kahana J A, Silver P A. Use of the A. Victoria Green fluorescent protein to study protein dynamics in vivo. Curr Protocols Mol Biol. 1996;1(Suppl. 34):9.7.22–9.7.28. doi: 10.1002/0471142727.mb0907cs34. [DOI] [PubMed] [Google Scholar]
- 14.Klement J F, Rice N R, Car B D, Abbondanzo S J, Powers G D, Bhatt H, Chen C H, Rosen C A, Stewart C L. IκBα deficiency results in a sustained NF-κB response and severe widespread dermatitis in mice. Mol Cell Biol. 1996;16:2341–2349. doi: 10.1128/mcb.16.5.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Leptomycin B inhibition of signal-mediated nuclear export by direct binding to CRM1. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 16.Lenardo M, Siebenlist U. Bcl-3-mediated nuclear regulation of the NFκB trans-activating factor. Immunol Today. 1994;15:145–147. doi: 10.1016/0167-5699(94)90308-5. [DOI] [PubMed] [Google Scholar]
- 17.Macreadie I G, Jagadish M N, Azad A A, Vaughan P R. Versatile cassettes designed for the copper inducible expression of proteins in yeast plasmid. Plasmid. 1989;21:147–150. doi: 10.1016/0147-619x(89)90059-0. [DOI] [PubMed] [Google Scholar]
- 18.May M J, Ghosh S. Rel/NF-κB and IκB proteins: an overview. Semin Cancer Biol. 1997;8:63–73. doi: 10.1006/scbi.1997.0057. [DOI] [PubMed] [Google Scholar]
- 19.Nelsen B, Tian G, Erman B, Gregoire J, Maki R, Graves B, Sen R. Regulation of lymphoid-specific immunoglobulin μ heavy chain gene enhancer by ETS-domain proteins. Science. 1993;261:82–86. doi: 10.1126/science.8316859. [DOI] [PubMed] [Google Scholar]
- 20.Neville M, Stutz F, Lee L, Davis L I, Rosbash M. The importin-β family member Crm1p bridges the interaction between Rev and the nuclear pore complex during nuclear export. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 21.Ossareh-Nazari B, Bachelerie F, Dargemont C. Evidence for a role of CRM1 in signal-mediated nuclear protein export. Science. 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 22.Phillips R J, Ghosh S. Regulation of IκBβ in WEHI 231 mature B cells. Mol Cell Biol. 1997;17:4390–4396. doi: 10.1128/mcb.17.8.4390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodriguez M S, Thompson J, Hay R T, Dargemont C. Nuclear retention of IκBα protects it from signal-induced degradation and inhibits nuclear factor κB transcriptional activation. J Biol Chem. 1999;274:9108–9115. doi: 10.1074/jbc.274.13.9108. [DOI] [PubMed] [Google Scholar]
- 24.Sachdev S, Hannink M. Loss of IκBα-mediated control over nuclear import and DNA binding enables oncogenic activation of c-Rel. Mol Cell Biol. 1998;18:5445–5456. doi: 10.1128/mcb.18.9.5445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sachdev S, Hoffmann A, Hannink M. Nuclear localization of IκBα is mediated by the second ankyrin repeat: the IκBα ankyrin repeats define a novel class of cis-acting nucleas import sequences. Mol Cell Biol. 1998;18:2524–2534. doi: 10.1128/mcb.18.5.2524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schwarz E M, Krimpenfort P, Berns A, Verma I M. Immunological defects in mice with a targeted disruption in Bcl-3. Genes Dev. 1997;11:187–197. doi: 10.1101/gad.11.2.187. [DOI] [PubMed] [Google Scholar]
- 27.Scott M L, Fujita T, Liou H-C, Nolan G P, Baltimore D. The p65 subunit of NF-κB regulates IκB by two distinct mechanisms. Genes Dev. 1993;7:1266–1276. doi: 10.1101/gad.7.7a.1266. [DOI] [PubMed] [Google Scholar]
- 28.Souyang H, Phillips R, Douglas I, Ghosh S. Role of unphosphorylated, newly synthesized IκBβ in persistent activation of NF-κB. Mol Cell Biol. 1996;16:5444–5449. doi: 10.1128/mcb.16.10.5444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Verma I M, Stevenson J K, Schwartz E M, Van Antwerp D, Miyamoto S. Rel/NF-κB/IκB family: intimate tales of association and dissociation. Genes Dev. 1998;9:2723–2735. doi: 10.1101/gad.9.22.2723. [DOI] [PubMed] [Google Scholar]
- 30.Weil R, Whiteside S T, Israel A. Control of NFκB activity by the IκBβ inhibitor. Immunobiology. 1997;198:14–23. doi: 10.1016/s0171-2985(97)80023-x. [DOI] [PubMed] [Google Scholar]
- 31.Whiteside S T, Israel A. IκB proteins: structure, function and regulation. Semin Cancer Biol. 1997;8:75–82. doi: 10.1006/scbi.1997.0058. [DOI] [PubMed] [Google Scholar]
- 32.Yan C, Lee L H, Davis L I. Crm1p mediates regulated nuclear export of a yeast AP-1-like transcription factor. EMBO J. 1998;17:7416–7429. doi: 10.1093/emboj/17.24.7416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zabel U, Henkel T, Silver M S, Baeuerle P A. Nuclear uptake control of NFκB by MAD-3, and IκB protein present in the nucleus. EMBO J. 1993;12:201–211. doi: 10.1002/j.1460-2075.1993.tb05646.x. [DOI] [PMC free article] [PubMed] [Google Scholar]