Abstract
During postembryonic development of Caenorhabditis elegans, the heterochronic gene lin-14 controls the timing of developmental events in diverse cell types. Three alternative lin-14 transcripts are predicted to encode isoforms of a novel nuclear protein that differ in their amino-terminal domains. In this paper, we report that the alternative amino-terminal domains of LIN-14 are dispensable and that a carboxy-terminal region within exons 9 to 13 is necessary and sufficient for in vivo LIN-14 function. A transgene capable of expressing only one of the three alternative lin-14 gene products rescues a lin-14 null mutation and is developmentally regulated by lin-4. This shows that the deployment of alternative lin-14 gene products is not critical for the ability of LIN-14 to regulate downstream genes in diverse cell types or for the in vivo regulation of LIN-14 level by lin-4. The carboxy-terminal region of LIN-14 contains an unusual expanded nuclear localization domain which is essential for LIN-14 function. These results support the view that LIN-14 controls developmental timing in C. elegans by regulating gene expression in the nucleus.
Development of multicellular organisms requires the precise spatial and temporal control of developmental events, including cell fate determination and cell division and differentiation. In Caenorhabditis elegans, the temporal control of stage-specific developmental events in the larva is controlled by heterochronic genes (2; reviewed in references 1, 4, 25). Mutations in heterochronic genes cause cells to adopt fates that are characteristic of earlier or later larval stages. For example, loss-of-function (lf) mutations of lin-14 cause so-called “precocious” phenotypes in which animals skip their L1-specific developmental programs and precociously express L2-specific programs. By contrast, strong lin-14 gain-of-function (gf) mutations cause “retarded” phenotypes wherein animals fail to execute later-stage developmental programs and instead reiterate L1 developmental programs (2, 3). Consistent with these mutant phenotypes, lin-14 activity progressively decreases during development due to a decrease in LIN-14 protein between the L1 and L2 stages. This temporal gradient of LIN-14 governs the proper sequence of stage-specific programs for the developing larva (17, 28).
The analysis of lin-14(lf) mutant phenotypes revealed that lin-14 controls diverse developmental events in a range of different cell types and cell lineages. These events include stage-specific division patterns in lateral hypodermal cell lineages (2), cell cycle progression and developmental commitment in vulva precursor cells (VPCs) (10), cell divisions in the intestine (30), lin-29-dependent lateral hypodermal seam cell differentiation (22), neuronal remodeling (11), and the stage-specific initiation and expression of the dauer larva developmental program (18). It is not known how LIN-14 carries out these various developmental roles. LIN-14 protein is widely expressed (23) and presumably interacts with cell-specific factors to regulate the timing of target gene activation and repression.
Molecular analysis of the lin-14 locus reveals a relatively complex genomic structure with 13 exons spanning over 20 kilobases (kb) producing at least three different transcripts, lin-14A, lin-14B1, and lin-14B2 (28). The predicted proteins encoded by these three transcripts are all novel proteins of approximately 540 amino acids that differ in their N-terminal sequences. Except for a putative amphipathic helix domain near its carboxy terminus, there are no recognized functional motifs in LIN-14. In addition, although LIN-14 is nuclear localized (23), which would be consistent with the hypothesis that LIN-14 may be a transcriptional regulator, there is as yet no direct evidence for a specific biochemical activity for LIN-14.
The existence of alternative LIN-14 isoforms raises the question of whether these different isoforms carry out different and perhaps cell-type-specific functions. To further understand how lin-14 controls developmental timing, we performed an in vivo structure and function analysis of LIN-14 protein. In this report, we show that the alternative amino-terminal lin-14 exons (exons 2 and 4) are dispensable for LIN-14 activity. We also show that domains necessary and sufficient for LIN-14 in vivo function lie in the carboxy-terminal half of the protein in a region well conserved among different nematode species. LIN-14 has an unusual expanded nuclear localization domain also contained in the carboxy-terminal region, supporting the idea that LIN-14 controls developmental timing by regulating gene activity in the nucleus. Our results suggest that LIN-14 carboxy-terminal domains carry out the primary function of controlling downstream gene activity, while amino-terminal lin-14 sequences play secondary roles, perhaps in modulating lin-14 activity in different developmental contexts.
MATERIALS AND METHODS
Worm strains.
Nematode strains were grown and maintained as previously described (31). All animals were grown at 20°C unless otherwise indicated. The following strains were used (items separated by commas): VT284 lin-14(ma135)/szT1, VT573 lin-4(e912); lin-14(n179ts), MT1397 lin-14(n179ts), VT499 lin-4(e912)vab-9(e1744)/mnC1, VT885 lin-14(n179ts); maEx166, VT886 lin-14(ma135); maEx167, VT887 lin-14(n179ts); maEx168, VT888 lin-14(ma135); maEx168, VT889 lin-4(e912) vab-9(e1744); lin-14(n179ts); maEx166, VT889 lin-4(e912) vab-9(e1744); lin-14(ma135); maEx166, and VT890 lin-4(e912) vab-9(e1744); lin-14(ma135); maEx168.
Germ line transformation.
Worms were transformed by microinjection as previously described (9). maEx166 was generated by coinjecting p14GFP (pVT333G, 25 μg/ml) and rol-6(su1006) (pRF4, 150 μg/ml [20]). maEx167 was generated by coinjecting p14GFP (pVT333G, 25 μg/ml) and col-19::GFP (pVT301, 125 μg/ml). maEx168 was generated by coinjecting p14B2GFP (pVT389, 25 μg/ml) and col-19::GFP (pVT301, 125 μg/ml). The col-19 promoter is expressed only in adult hypodermal cells (19). All the deletion constructs D1 to D10 were coinjected at the concentrations indicated in Table 3, with col-19::GFP (pVT301, 150 μg/ml) as a transgenic marker. When lin-14(n179ts) was used as the host strain, transgenic lines were established at 15°C.
TABLE 3.
In vivo localization and genetic activity of lin-14 deletion constructs
| Genotype and constructa | Exons in transgene (encoded amino acids)b | Degree of NLc | % of seam cells making adult alae at L3 moltd
|
|||
|---|---|---|---|---|---|---|
| 25°Ce | 15°Cf
|
|||||
| GFP− | GFP+ | Diff | ||||
| lin-14(n179ts) | None | NA | 99 (770) | 13 (770) | ||
| lin-14(n179ts); D3::GFP | 5–8* (aa 66–283) | − | 100 (156) | 14 (364) | 27 (325) | 13 |
| 99 (208) | 12 (195) | 19 (260) | 7 | |||
| lin-14(n179ts); D5::GFP | 9–13 (aa 292–535) | + | 26 (323) | 10 (78) | 0 (195) | −10 |
| 27 (273) | 10 (78) | 0 (165) | −10 | |||
| lin-14(ma135); D5::GFP | 9–13 (aa 292–535) | + | 40 (192) | ND | ND | |
| lin-14(n179ts); D6::GFP | 10–13 (aa 328–535) | + | 100 (106) | 23 (120) | 63 (317) | 40 |
| 98 (95) | 16 (179) | 48 (330) | 32 | |||
| lin-14(n179ts); D7::GFP | 11–13 (aa 400–535) | − | 98 (377) | 7 (104) | 0 (117) | −7 |
| 97 (195) | 10 (221) | 11 (208) | 1 | |||
| lin-14(n179ts); D8::GFP | 9–11* (aa 292–436) | − | 95 (312) | 16 (260) | 7 (260) | −9 |
| 92 (208) | 11 (221) | 10 (169) | −1 | |||
| lin-14(n179ts); D9::GFP | 8*–12* (aa 284–466) | ++ | 62 (225) | ND | ND | |
| 56 (231) | ||||||
| lin-14(n179ts); D10::GFP | 9(n179ts)–13 (aa 292–535) | + | 97 (351) | 10 (182) | 3 (211) | −7 |
| 95 (325) | 21 (143) | 12 (286) | −9 | |||
Transgenic animals were homozygous mutants for either lin-14(n179ts) (strain MT1397) or lin-14(ma135) [homozygotes segregated from VT284 lin-14(ma135); szT1] and also contain an extrachromosomal array carrying the indicated LIN-14 deletion construct (see Fig. 1D and 4). lin-14(n179ts) animals were also assayed in an untransformed line (first row). The transgenic lines were generated by injecting each construct at a concentration of 20 ng/μl, except for D6 (5 ng/μl) and D7 (40 ng/μl).
See Fig. 1D and 4. ∗, construct does not contain the full length of this exon (for details, see Fig. 1D and text).
NL, nuclear localization. Results from Fig. 4. −, 1.00 to 1.49; +, 1.50 to 4.00; ++, >4.00. (See Fig. 4J for units.) NA, not applicable.
Seam cell fates were scored as described previously (2) and in Materials and Methods. For each construct, except for D5 in lin-14(ma135) animals, two independent transgenic lines were scored (see Materials and Methods).
At 25°C, essentially 100% of lateral hypodermal seam cells in lin-14(n179ts) animals produce adult lateral alae at the L3 molt, one stage earlier than in the wild type. n, the total number of seam cells scored. Transgenic animals were identified by their GFP fluorescence.
The precocious phenotype of lin-14(n179ts) at 15°C is variable, so in each individual case, animals carrying the transgene (GFP+) and animals lacking the transgene (GFP−) were scored for precocious phenotype in seam cells. The occurrence of rescue or dominant negative phenotype was then assessed by subtracting the data for GFP+ animals from the data for GFP− animals. Diff = GFP+ − GFP−. n, total number of seam cells scored.
Plasmid constructions.
A 5.7-kb EcoRI fragment from cosmid KE7 containing lin-14 exons 4 to 13 plus about 1.7 kb of the lin-14 3′ untranslated region (3′UTR) was first cloned into the EcoRI site of pBluescript to make pB14R. An internal EcoRI site within this fragment was eliminated by partial digestion and end-filling during the cloning. The green fluorescent protein (GFP) coding sequence was cut from pPD95.02 with AgeI and SmaI and then blunted and cloned into a blunted EcoNI site of pB14R (with GFP sequences in frame with the carboxy end of lin-14 coding sequence) to make pB14RGFP. p14GFP was made by inserting a 12.8-kb KpnI-SalI fragment from cosmid KE7 into the corresponding sites of pB14RGFP (see Fig. 1). p14B2GFP was made in two steps. First, the 1.89-kb BsiWI-AgeI sequence which contains exon 2 was replaced with a 760-bp PCR fragment from BsiWI to a downstream sequence near a PacI site, which is about 400 bp upstream of exon 2. Second, exon 4 was then deleted in p14B2GFP by deleting a 514-bp PacI-ClaI fragment that contains exon 4.
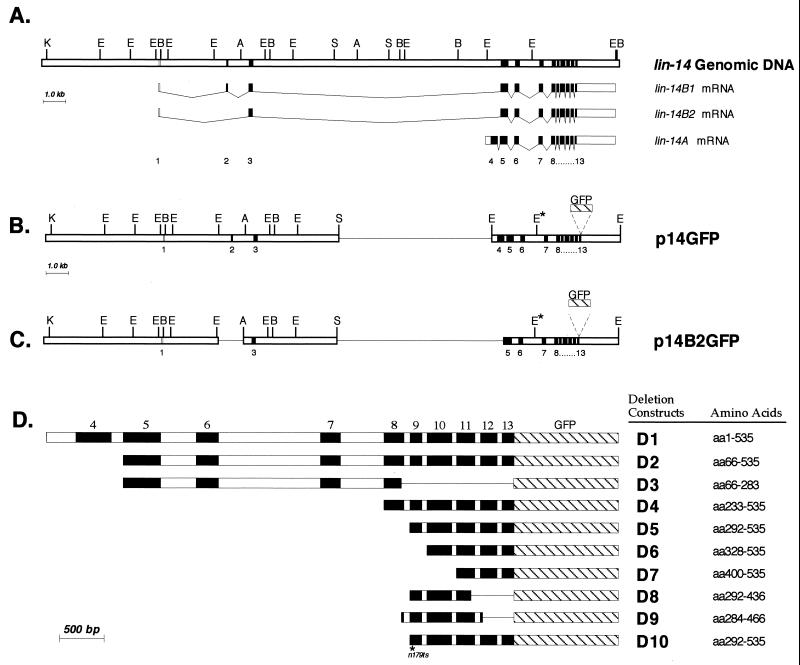
FIG. 1.
Genomic organization of wild-type lin-14 and lin-14 deletion constructs. (A) lin-14 intron-exon structure of the three identified lin-14 transcripts, lin-14B1, lin-14B2, and lin-14A (adapted from reference 28). Restriction enzyme map and intron-exon spacing are drawn to scale. Open box in exon 4 indicates that the 5′ end of the exon 4 open reading frame is undetermined. A, AgeI; B, BglII; E, EcoRI; K, KpnI; S, SalI. (B and C) LIN-14 deletion-GFP fusion constructs p14GFP and p14B2GFP. A single line represents a region that is deleted. E∗, EcoRI site that was eliminated (see Materials and Methods). (D) Truncated lin-14 constructs expressed from the col-10 promoter. col-10 promoter sequences and 3′UTR sequence of lin-14 are not shown. Open boxes indicate introns and filled boxes indicate exons. GFP sequence is represented by the hatched box (artificial introns in GFP are not shown). Amino acids are numbered according to Wightman et al. (28). D10 contains the lin-14(n179ts) mutation (R303G in exon 9; B. Reinhart and G. Ruvkun, personal communication; see Materials and Methods).
A 1.2-kb BglII-BamHI col-10 sequence without an ATG start codon was inserted into the BamHI site of pB14RGFP to make deletion construct D1. For constructs D2 to D9, the same col-10 sequence, but with an in-frame ATG codon, was used. Deletion constructs D2 to D10 were generated by PCR amplification of portions of the lin-14 coding sequence by using pB14R as a template. To make D10, exon 9 in D5 was replaced with DNA carrying the n179 point mutation which was PCR amplified from lin-14(n179ts) genomic DNA, subcloned, and verified by sequencing.
Microscopy and photography.
Images of live animals anesthetized with 1 mM levamisole were captured with an Optronics DE1750 integrating color CCD video camera and a Scion CG-7 RGB video capture board on a Power Macintosh 8500AV. GFP fluorescence images were taken with exposures ranging from 1/4 to 2 s, unless stated otherwise. The digital images were processed with Adobe Photoshop.
Scoring the rescuing activity of lin-14 constructs.
For each construct, multiple transgenic lines were generated and one or two of the lines that displayed the most improved general appearance and the highest level of GFP expression were analyzed in detail for the rescue of heterochronic phenotypes. To score the rescue of lin-14(n179ts), animals were grown at 25°C. The number of seam cells at the end of the L1, the timing of adult alae formation, and the timing of VPC divisions were scored as previously described (2). For scoring the formation of precocious dauer larvae, plates were inoculated with 100 or more transgenic adults and their progeny were allowed to exhaust the food supply. Dauer larvae were collected by 1% sodium dodecyl sulfate selection (18), and L1 dauer larvae were identified by the presence of dauer alae on animals with body size and a number (n = 12) of gonadal nuclei consistent with developmental arrest at the L1 molt (18).
Efficiency of LIN-14::GFP nuclear localization.
CCD images were captured in RGB format, and the green channel was selected with Adobe Photoshop and transferred to an NIH Image file as a grayscale image. Using NIH Image, the area of each nucleus and the area of each cytoplasm were separately circumscribed and their mean light intensity was measured. The data of mean intensity were further normalized to a 1-s exposure according to the camera's measured exposure calibration. Background light intensity was also measured and normalized similarly and subtracted from the nuclear and cytoplasmic intensities. For any given deletion construct, the GFP fluorescence intensity of at least 20 nuclei and their surrounding cytoplasm were measured.
RESULTS
A LIN-14::GFP fusion construct that can fully rescue a lin-14 null mutation.
lin-14 sequences span approximately 20 kb of genomic DNA (28) (Fig. 1A). To analyze the functional organization of LIN-14 protein, a modified lin-14 genomic construct, p14GFP, was constructed (Fig. 1B). p14GFP encompasses lin-14 genomic DNA from 5.2 kb upstream of exon 1 to about 500 bp downstream of the polyadenylation signal, with a deletion of approximately 7 kb of intron sequence between exon 3 and exon 4 (Fig. 1B; see Materials and Methods). p14GFP also contains a GFP sequence inserted in frame at the LIN-14 carboxy terminus (see Materials and Methods) to enable the detection of LIN-14::GFP fusion protein in transgenic worms.
p14GFP was transformed into lin-14(ma135) animals, a lin-14 genetic null (18; V. Ambros, unpublished data) and stable transgenic lines were established. The anatomical pattern of expression of p14GFP from the transgenic extrachromosomal array maEx167 is consistent with that of the endogenous LIN-14 protein as previously determined by LIN-14 antibody staining (23) (see below). LIN-14::GFP was nuclear localized, as is the case for endogenous LIN-14 (23) (see below).
As shown in Tables 1 and 2, lin-14(ma135); maEx167 animals were fully rescued for all defects scored, including precocious L1 cell lineage patterns in the V lineage, precocious seam cell differentiation, precocious vulva development, and precocious initiation of dauer larval development (Tables 1 and 2).
TABLE 1.
Rescue of lin-14(lf) phenotypes by p14GFP and p14B2GFP transgenes
| Straina | No. of seam cells at L1 molt (n)b | % adult alae at L3 molt (n)c | % VPC divisions at L2 molt (n)d | % egl(−) adults (n)e | No. of intestinal cells at L2 molt (n)f | % Alae formation at L4 molt (n)g |
|---|---|---|---|---|---|---|
| Wild type | 6.0 ± 0.0 (20) | 0.0 (585) | 0.0 (69) | 0.0 (100) | 33.2 ± 1.3 (21) | 0.0 (455) |
| lin-14(ma135) | 11.2 ± 0.9 (15) | 100.0 (312) | 100.0 (45) | 100.0 (100) | 19.7 ± 0.6 (17) | NA |
| lin-14(n179ts) @ 25°C | 10.5 ± 0.8 (11) | 99.6 (770) | 54.4 (57) | 100.0 (110) | 20.2 ± 1.4 (19) | NA |
| VT886 (ma135; maEx167) | 6.0 ± 0.0 (10) | 8.3 (325) | 12.4 (57) | 7.5 (120) | 29.0 ± 3.4 (19) | 0.4 (260) |
| VT887 @25°C (n179; maEx168) | 6.2 ± 0.4 (12) | 2.0 (546) | 0.0 (36) | 13.6 (110) | 30.0 ± 2.4 (12) | 1.3 (234) |
| VT888 (ma135; maEx168) | 6.0 ± 0.0 (15) | 0.3 (338) | 3.3 (60) | 11.7 (120) | 31.1 ± 2.2 (21) | 6.8 (585) |
Transgenic animals were identified by their GFP fluorescence.
In lin-14(lf) mutant animals, lateral hypodermal V cells in the L1 stage express an L2-specific division pattern in place of the L1-specific division pattern and consequently will contain up to 12 seam cells at each side of the animal, instead of six, by the end of the L1 stage. Rescue of this L1 phenotype was measured here by the number of seam cells at the L1 molt on one side of the animal ± standard error of the mean. n, number of animals scored.
lin-14(lf) animals undergo only three larval molts and synthesize adult cuticle precociously at the third molt. The adult alae scored here are a cuticle structure that is specifically made by seam cells during their synthesis of adult cuticle (2). n, total number of seam cells scored.
In lin-14(lf) hermaphrodites, VPCs P5.p, P6.p, and P7.p divide and produce vulva cells one stage earlier than normal, at the L2 instead of the L3 stage. n, total number of VPCs from among P5.p, P6.p, and P7.p that were scored for precocious divisions at the L2 molt.
The precocious vulva development contributes to the egg-laying defects [egl(−)] in lin-14(lf) adults. To score the egl(−) phenotype, late L4 animals were picked onto one plate, and 24 to 48 h later, the number of egl(−) animals were counted by their “egg-bag” phenotype. n, total number of animals.
lin-14 activity is required for intestinal cells to execute a round of divisions in the late L1 stage (30). In the wild type, many of the 20 intestinal nuclei divide at the end of the L1 stage, resulting in a final complement of approximately 33 total intestinal nuclei (31). In lin-14(ma135) animals and in lin-14(n179ts) animals at 25°C, none of the intestinal nuclei divides and the total number of nuclei after the L1 stage remains at 20. n, total number of animals scored. Data are means ± standard errors of the mean.
Overexpression of LIN-14 late in development leads to retarded larval fates, including the production of larval cuticle instead of adult cuticle at the L4 molt, revealed by the absence of adult-specific lateral alae. n, number of seam cells scored for the production of adult lateral alae at the L4 molt.
TABLE 2.
Rescue of precocious dauer formation by p14GFP and p14B2GFP in lin-14(n179ts) and lin-14(ma135) mutants
| Straina | Cultureb | % Dauer larvae arrested at L1 molt (n)
|
|
|---|---|---|---|
| GFP− | GFP+ | ||
| VT886 (ma135; maEx167) | 886-1 | 85 (148) | 8 (116) |
| 886-2 | 70 (67) | 2 (82) | |
| VT887 @ 25°C (n179; maEx168) | 887-1 | 67 (21) | 1 (101) |
| 887-2 | 69 (150) | 0 (63) | |
| VT888 (ma135; maEx168) | 888-1 | 81 (100) | 14 (131) |
| 888-2 | 76 (66) | 12 (67) | |
These transgenic lines carry extrachromosomal arrays (maEx167 [p14GFP] or maEx168 [p14B2GFP]) that are frequently lost at meiosis, generating a mixture of nontransgenic (GFP−) and transgenic (GFP+) animals. Dauer larvae were harvested from starved plate cultures and separated into nontransgenic (GFP−) and transgenic (GFP+) classes. The percentage of the dauer larvae in each class that were arrested at the L1 molt was determined as described in Materials and Methods. n, total number of dauer larvae scored; all dauer larvae not arrested at the L1 molt were arrested at the L2 molt, as in the wild type.
For each strain, two starved plates were scored separately.
Alternative exons 2 and 4 are not required for lin-14 activity.
p14GFP should be capable of making all three lin-14 transcripts (lin-14A, -B1, and -B2) since it contains the alternative exons 2 (which is B1 specific) and 4 (which is A specific). To test whether lin-14 products containing those alternative exons are required for lin-14 function, we examined the rescuing activity of a construct (p14B2GFP) that is missing exons 2 and 4. p14B2GFP was generated by the deletion from p14GFP of an approximately 1.0-kb DNA sequence around (and including) exon 2 and the deletion of a 500-bp DNA sequence containing exon 4 (Fig. 1C). While it is impossible for p14B2GFP to make either lin-14A or lin-14B1 mRNA, GeneFinder (9, 27) predicts that this construct should produce the lin-14B2 transcript (data not shown).
The overall expression pattern of GFP from maEx168 is similar to that from maEx166 (see below) and maEx167, indicating that no tissue-specific promoter sequences were affected by the p14B2GFP deletions (data not shown). maEx168, an extrachromosomal array carrying p14B2GFP, was tested for its ability to rescue lin-14(n179ts). n179 is a temperature-sensitive (ts) allele of lin-14, and at 25°C, lin-14(n179ts) animals exhibit the lin-14(lf) phenotype (3), though the phenotype is somewhat less severe than that of lin-14(ma135) (see Table 1). As summarized in Table 1, maEx168 fully rescues lin-14(n179ts) for all the phenotypes tested.
The temperature-sensitive lin-14(n179ts) mutation does not completely eliminate lin-14 activity at 25°C. To exclude the possibility that p14B2GFP might rescue lin-14(n179ts) by boosting the weak residual activity of the temperature-sensitive LIN-14 protein, we crossed maEx168 into animals carrying the non-temperature-sensitive allele lin-14(ma135) and found efficient rescue of lin-14(ma135) precocious defects (Table 1). These results provide strong evidence that neither exon 2 or exon 4 is required for lin-14 activity and that a single predicted product of lin-14 is sufficient for LIN-14 function in different cell types.
lin-4-dependent developmental down regulation of LIN-14::GFP expression from transgenes.
The successive execution of larval stage-specific developmental programs requires the temporal down regulation of lin-14 activity between the L1 and L2 stages. We found that the effective level of LIN-14::GFP expressed from maEx167 is within the normal range (in terms of lin-14 activity) and appears to be down regulated satisfactorily to achieve the normal sequence of larval developmental events. The phenotype of maEx167 animals showed no significant evidence of LIN-14 overexpression, as seam cells made adult cuticle at the L4 molt and the timing of vulva development was normal (Table 1).
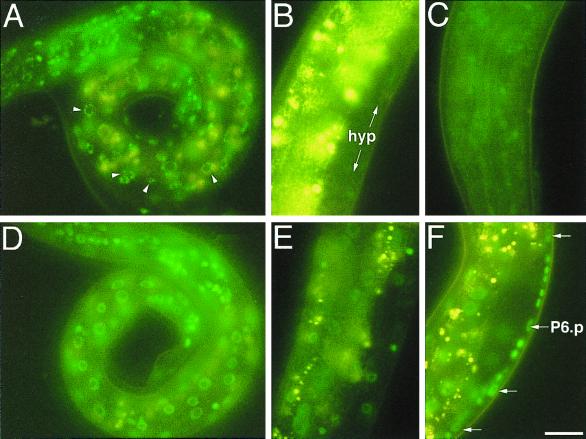
The temporal down regulation of lin-14 activity is executed at the posttranscriptional level. This down regulation requires the product of the heterochronic gene lin-4, which encodes a small 22-nucleotide RNA with sequence complementarity to seven elements in lin-14 3′UTR (17, 29). To confirm that LIN-14::GFP expression from transgenic p14GFP is down regulated by lin-4, LIN-14::GFP expression was monitored in wild-type and lin-4(e912) backgrounds, maEx166, a transgenic array similar to maEx167, was generated by coinjecting p14GFP along with pRF4 (rol-6(su1006)) (20). In wild-type maEx166 animals, the temporal profile of LIN-14::GFP expression faithfully reflects the results previously obtained by LIN-14 antibody staining of the wild type (23). maEx166 animals displayed the highest LIN-14::GFP expression in the nuclei of late embryos just before hatching and of newly hatched L1 animals (Fig. 2A). This high level of LIN-14::GFP expression starts to decrease in hypodermal and intestinal nuclei and in the nuclei of ventral nerve neurons during the L1 stage and becomes undetectable in the L2 stage (Fig. 2B). The only exception is a subset of neurons in the head in which LIN-14::GFP remains detectable as late as the L3 or L4 stage (Fig. 2C). The expression of LIN-14B2::GFP from maEx168 showed a temporal down regulation essentially indistinguishable from that of LIN-14::GFP expressed from maEx166 or maEx167 (data not shown).
FIG. 2.
lin-4-dependent down regulation of LIN-14::GFP from maEx166. (A) Newly hatched transgenic L1 larva carrying maEx166, which was generated by injection of p14GFP (Fig. 1). High levels of green LIN-14::GFP fluorescence is evident in hypodermal cells, muscle cells, intestinal cells, and neurons. Arrowheads point to nuclei that show punctuate LIN-14::GFP fluorescence typical of this construct. The yellow fluorescence seen in these images is due to the autofluorescence from the intestine. (B) LIN-14::GFP expression in maEx166 animals becomes virtually undetectable in hypodermal cells (hyp) by the L3 stage. Yellow signal is (non-GFP) intestinal autofluorescence. (C) In maEx166 animals, LIN-14::GFP expression in the head neurons remains detectable in some L3 animals. (D) lin-4(e912); maEx166 L1 larvae display fluorescence levels approximately equal to that of lin-4(+); maEx166 L1 larvae. (E) At the L3 stage, high levels of LIN-14::GFP expression persists in hypodermal cells and in intestinal cells of lin4(e912); maEx166 L3 larvae. (F) Expression of LIN-14::GFP in VPC (Pn.p) cells is easily detectable in this lin-4(e912); maEx166 L2 larva [but is undetectable in similarly staged lin-4(+); maEx166 L2 larvae; not shown]. Animals in B, E, and F are all oriented anterior down and ventral side to the right. Bar, 3 μm.
To test whether the down regulation of LIN-14::GFP from maEx166 or LIN-14B2::GFP from maEx168 is lin-4 dependent, we crossed maEx166 and maEx168 into a lin-4(e912) background. In lin-4(e912); maEx166 animals, LIN-14::GFP expression was poorly down regulated compared to the wild type and was easily detectable in hypodermal and intestinal nuclei in animals at stages as late as the L3 or L4 (Fig. 2D to F). This indicates that down regulation of LIN-14::GFP from maEx166 requires lin-4 activity, as is the case for LIN-14. Similarly, down regulation of LIN-14B2 was not evident in lin-14(e912); maEx168 animals. These results indicate that the 7 kb of intron sequences deleted in p14GFP and the additional sequences deleted from around exons 2 and 4 in p14B2GFP (Fig. 1) are dispensable for LIN-14 expression and temporal regulation by lin-4.
Interestingly, although wild-type VPCs display no detectable levels of endogenous LIN-14 (23, 28) or LIN-14::GFP (data not shown), LIN-14::GFP was easily detected in VPCs during the late L1 to L2 stage in lin-4(e912) animals (Fig. 2F). The expression of LIN-14 in lin-4(e912) VPCs is consistent with the retarded vulva phenotype of lin-14(e912) animals and the critical role of LIN-14 down regulation in regulating the competence and cell cycle progression of VPCs (10, 12).
Exons 9 to 13 of lin-14 are sufficient for lin-14 activity in lateral hypodermal cells.
To further delineate the parts of LIN-14 protein that are necessary and sufficient for its in vivo function, a series of deletions of the LIN-14::GFP fusion protein were constructed (Fig. 1D). In these experiments, LIN-14 deletion constructs were expressed from a simplified expression vector under the control of the C. elegans col-10 promoter. col-10 is specifically expressed in hypodermal cells (19), and thus the rescuing activity of LIN-14 deletion constructs was scored in the hypodermis. The assays were performed in a lin-14(n179ts) background because the temperature-sensitive phenotype of lin-14(n179ts) allows efficient transformation at permissive temperature (15°C) and a convenient assay for rescue simply by transferring the transgenic animals to nonpermissive temperature (25°C). It should be noted that although the injection conditions were similar for each construct (see Materials and Methods), the expression levels of these truncated LIN-14 proteins in general were greater than the LIN-14::GFP levels observed from maEx167 and maEx168 (see Fig. 2 and 4). In particular, the expression level of LIN-14D3::GFP was very high (see Fig. 4A).
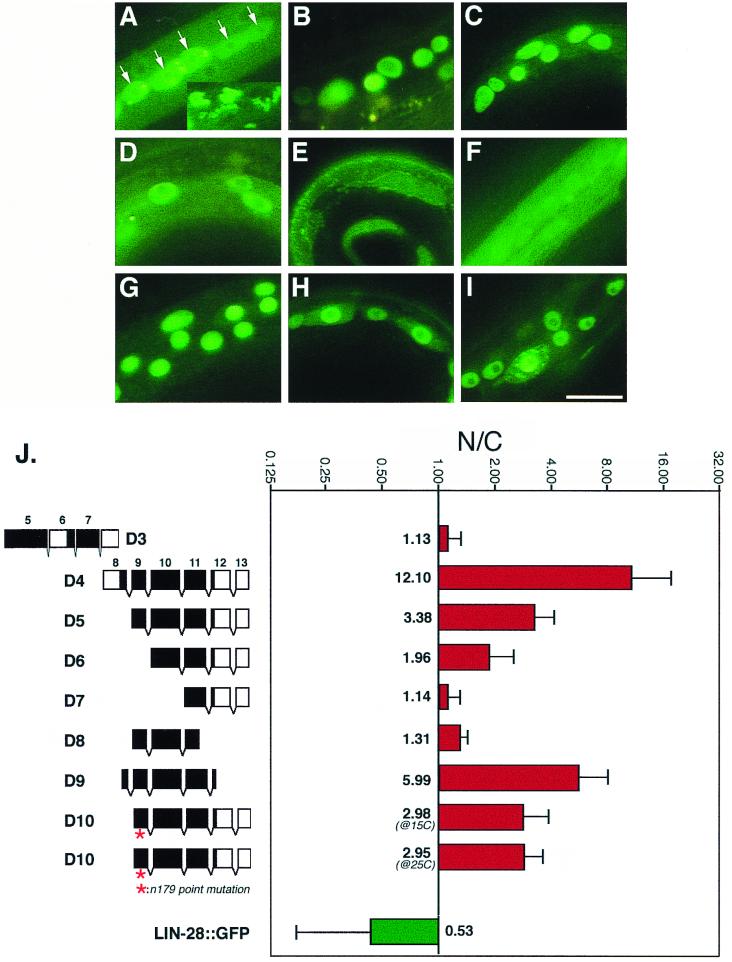
FIG. 4.
Nuclear localization efficiency of various truncated LIN-14::GFP proteins. (A to I) Hypodermal cells expressing LIN-14D3::GFP (A) LIN-14D4::GFP (B), LIN-14D5::GFP (C), LIN-14D6::GFP (D), LIN-14D7::GFP (E), LIN-14D8::GFP (F), LIN-14D9::GFP (G), LIN-14D10::GFP (at 15°C) (H), and LIN-14D10::GFP (at 25°C) (I). Animals were at developmental stages ranging from L1 to L3. (E and F) Unlocalized LIN-14D7::GFP and LIN-14D8::GFP appear as a uniform green fluorescence, while in all other panels except A; the green fluorescent spots are nuclei in which LIN-14::GFP is localized. Nucleoli are relatively free of LIN-14::GFP staining and appear as darker spots within the nuclei. The nuclear localization patterns of LIN-14D1::GFP and LIN-14D2::GFP (data not shown) are similar to that of LIN-14D4::GFP. (A) Lateral hypodermal seam cells (arrow) are filled with unlocalized LIN-14D3::GFP and show a higher level of GFP fluorescence than surrounding hypodermal cells, and the inset shows the unidentified GFP-containing inclusions that are frequently evident in animals expressing LIN-14D3::GFP. We have not determined whether the structures are extracellular or intracellular. The image was captured with relatively short exposures (1/30 or 1/60 s) so the actual level of GFP fluorescence is much higher than in the rest of panels, which were taken at exposures ranging from 1/4 to 2 s. (J) Quantitative assay of the nuclear localization efficiency of truncated LIN-14::GFP proteins. In the diagram on the left, black regions in exon boxes represent the amino acid sequences that are relatively well conserved among LIN-14 proteins from different nematode species (see Fig. 3). GFP sequences are not shown, and the intron spaces in D3 are not drawn in scale. N/C, nuclear-to-cytoplasmic ratio of GFP fluorescence plotted in a log scale (see Materials and Methods). LIN-28::GFP is a cytoplasmically localized protein (4) used here as a control. Bar, 5 μm.
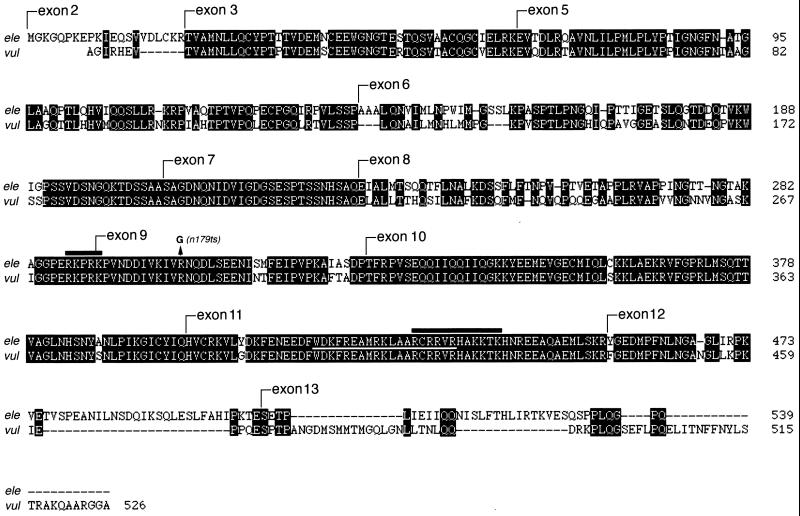
Surprisingly, although lin-14 contains a total of 13 exons, we found that a deletion construct containing only exons 9 to 13 (Fig. 1D, construct D5) rescued lin-14(n179ts) at 25°C (Table 3 and data not shown). Similarly, deletion construct D9 (containing exons 8 through 12) rescued lin-14(n179ts), though not as strongly as D5 (Table 3). This suggests that sequences sufficient for lin-14 activity are contained within exons 9 to 12, although some of exon 13 may also contribute. Sequences in exon 9 appear to be critical for lin-14 activity, as construct D6 (containing exons 10 to 13; see Fig. 1D) showed no rescuing activity (Table 3). Construct D8 (containing exons 9 and 10 and part of exon 11; see Fig. 1D) also showed no rescuing activity, indicating that sequences in the exon 11-to-12 interval are essential. These findings indicate that sequences contained within a carboxy-terminal region of LIN-14 encoded by exons 9 to 13 are both necessary and sufficient for lin-14 activity, as assayed by rescue of lin-14(n179ts) defects in the hypodermis. Exons 9 to 12 corresponds to a part of LIN-14 where the amino acid sequence is well conserved among the nematode species C. elegans, Caenorhabditis vulgaris (Fig. 3), and Caenorhabditis briggsae (B. Reinhart and G. Ruvkun, personal communication), consistent with the functional activity of this region of the protein.
FIG. 3.
Amino acid sequence alignment between LIN-14 proteins from C. elegans (ele) and C. vulgaris (vul). Identical amino acids are labeled in black boxes, and dashes indicates gaps generated by the aligning program GeneInspector. The start position of each exon in C. elegans LIN-14 is labeled. Black lines highlight the potential consensus sequences for nuclear localization activity (15). The white line between the two sequences within exon 11 indicates the putative amphipathic helix motif (28). Also shown is the n179 point mutation (R303G) (B. Reinhart and G. Ruvkun, personal communication). Note that sequences shown here are equivalent to the predicted lin-14B1 product (see Fig. 1A) and the very amino-terminal end of the C. vulgaris LIN-14 sequence is undetermined. C. vulgaris sequence was determined from cDNA.
To exclude the possibility that the rescuing activity of the above constructs depends on endogenous residual activity of LIN-14(n179ts) protein, for example, by synergizing with or stabilizing the temperature-sensitive LIN-14 protein, we tested the rescuing activity of LIN-14D5::GFP in lin-14(ma135) animals, which carry a non-temperature-sensitive null allele of lin-14. In lin-14(ma135) animals carrying a transgenic array of construct D5, 60.4% of seam cells are rescued from making precocious adult cuticle at the L3 stage. These data are comparable to the rescuing activity of LIN-14D5::GFP protein in lin-14(n179ts) (Table 3, ∼73%), demonstrating that the product of exons 9 to 13 alone supplies lin-14 activity.
Truncated LIN-14::GFP fusion proteins, particularly D4 to D10, are generally not well down regulated during larval development. Unlike animals transformed with full-length LIN-14::GFP constructs (see above), animals carrying truncated constructs often exhibit bright fluorescence after the L1 stage (see Fig. 5). Truncated rescuing constructs can also cause retarded phenotypes of varying strengths, although we have not quantitatively compared the truncated constructs to the full-length constructs with respect to retarded phenotypes (data not shown).
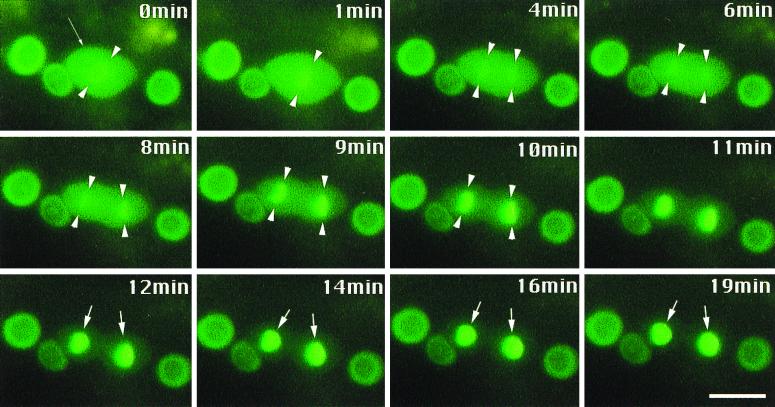
FIG. 5.
LIN-14D4::GFP is efficiently localized to nuclear material during mitosis. A dividing cell in a transgenic lin-14(n179ts) at the late L2 stage expressing LIN-14D4::GFP at 20°C. Small arrow in 0 min image points to a hypodermal cell undergoing mitosis, and the arrowheads in this and subsequent frames highlight the mitotic chromosomes. Larger arrows point to new sister nuclei. LIN-14D4::GFP is completely nuclear localized in surrounding interphase hypodermal cells. Bar, 3 μm.
Partial dominant negative activity of truncated LIN-14 products.
lin-14(n179ts) at permissive temperature (15°C) provides a sensitized genetic background for testing the potential antimorphic (“dominant negative”) activity of the truncated LIN-14 proteins produced by these deletion constructs. We have found that at 15°C, approximately 13% of seam cells in n179 animals express precocious adult cuticle (Table 3), and this weak precocious phenotype indicates that at 15°C, the level of lin-14 activity in lin-14(n179ts) animals might be slightly below the threshold required for wild-type seam cell development. Such a leaky precocious phenotype of lin-14(n179ts) would be enhanced in the presence of a truncated LIN-14 protein which is able to interfere with endogenous lin-14 function.
lin-14 deletion constructs were tested for their ability to either rescue or enhance the leaky precocious phenotype of lin-14(n179ts) animals at 15°C. As shown in Table 3, rescuing constructs clearly prevent the formation of precocious adult cuticle in lin-14(n179ts) animals at 15°C, but one of the nonrescuing constructs, D6 (which contains exons 10 to 13), enhances the leaky precocious phenotype. In lin-14(n179ts) animals carrying the D6 transgenic array, more seam cells express precocious adult cuticle at 15°C than in lin-14(n179ts) animals (Table 3). This antimorphic, or partial dominant negative, activity of LIN-14D6::GFP protein requires exon 10, since a further truncated construct (D7) containing only exons 11 to 13 does not modify the lin-14(n179ts) phenotype. The antimorphic activity of LIN-14D6::GFP seems to require the sensitized lin-14 hypomorphic genetic background provided by lin-14(n179ts), since no such activity was observed in a wild-type background.
LIN-14D3::GFP, which contains LIN-14 sequences from exons 5, 6, and 7 and to the beginning of exon 8 (Fig. 1D), shows very weak antimorphic activity in lin-14(n179ts) animals at 15°C (Table 3). The relatively low strength of the LIN-14D3::GFP negative activity, coupled with the extraordinarily high level of expression of this construct relative to that of D6, casts doubt on the specificity of the D3 phenotype.
LIN-14 has an extended nuclear localization domain.
To characterize sequences required for the nuclear localization of LIN-14, we examined the nuclear localization of truncated LIN-14::GFP proteins. The ratio of nuclear GFP fluorescence to cytoplasmic GFP fluorescence was measured in transgenic worms carrying various LIN-14::GFP constructs (see Materials and Methods). Exons 1 to 7 do not seem to contain any essential nuclear localization signals (NLS), since LIN-14D4::GFP which contains exons 8 to 13 is efficiently nuclear localized (Fig. 4B and J). Further NLS sequences seem to be contained between the end of exon 8 and the beginning of 12, since LIN-14D9::GFP is also nuclear localized (Fig. 4G and J). LIN-14D5::GFP, which contains exons 9 to 13, is still nuclear localized, but its localization efficiency is less than that of LIN-14D::GFP (Fig. 4C and J), suggesting that nuclear localization of LIN-14 is influenced by sequences in exon 8. Further deletion of sequences from either the N terminus or C terminus diminishes the nuclear localization of LIN-14 significantly (Fig. 4D, E, F, and J).
These results suggest that LIN-14 nuclear localization requires sequences at approximately the exon 8 and 9 border and also requires sequences in the exon 11 and 12 region. Thus, the LIN-14 NLS is either bipartite or extends over a region of LIN-14 from exon 8 to exon 12. Basic Arg-Lys clusters similar to a typical NLS consensus sequence are found at both ends of this region (Fig. 3), but the above results show that neither of these regions alone is sufficient to bring LIN-14 to the nucleus. Interestingly, LIN-14D10::GFP, which contains exons 9 to 13 with the n179 point mutation (B. Reinhart and G. Ruvkun, personal communication), is localized to nuclei (Fig. 4), despite have in no rescuing activity or antimorphic activity (Table 3). This suggests that the n179 mutation impairs a component of LIN-14 function other than nuclear localization. LIN-14 sequences required for nuclear localization could function by interacting directly with the nuclear localization machinery, or indirectly, via a nuclear-transported partner.
The behavior of LIN-14::GFP fusion proteins during cell division suggests that LIN-14 nuclear localization is rapid and efficient. For example, after premitotic nuclear envelope breakdown, the truncated LIN-14 fusion protein LIN-14D4::GFP appears uniformly distributed in the cytoplasm and then becomes reconcentrated in the daughter nuclear material during a brief period shortly after metaphase (Fig. 5). This observation suggests that the process of LIN-14 nuclear localization is very efficient and may involve interaction with some component of the mitotic apparatus. A small amount of the LIN-14D4::GFP fluorescence during metaphase appears to be associated with chromosomes, suggesting a possible chromatin binding activity of the truncated protein (Fig. 5). However, we did not observe a similar mitotic chromatin association of the full-length LIN-14::GFP and LIN-14B2::GFP proteins (data not shown). The relatively lower level of overall expression of LIN-14::GFP and LIN-14B2::GFP compared to that of the truncated LIN-14D4::GFP protein could account for the difficulty in detecting chromosome-associated fluorescence for the full-length fusion proteins. However, it is noteworthy that LIN-14::GFP and LIN-14B2::GFP fluorescence was not apparent anywhere within dividing hypodermal cells, yet was easily detectable before cell division and in daughter cell nuclei (data not shown). This suggests that full-length LIN-14 may be preferentially decreased in level in association with mitosis, while in contrast, the truncated LIN-14 proteins may be relatively stable during mitosis.
DISCUSSION
In this work, we report that two of the three previously identified alternative lin-14 gene products (LIN-14A and LIN-14B1) are dispensable for lin-14 activity. Although p14B2GFP is not capable of producing either LIN-14A or LIN-14B1, the rescuing activity of lin-14(ma135) by p14B2GFP is indistinguishable from that of p14GFP, which is predicted to produce all three alternative transcripts. Our deletion analysis identified domains of LIN-14 in the carboxy-terminal region that are necessary and sufficient for lin-14 activity in hypodermal cells. The observed antimorphic, or partial dominant negative, activity of truncated LIN-14D6::GFP is consistent with the hypothesis that protein-protein interactions are involved in LIN-14 activity. Finally, we characterized the nuclear localization domain of LIN-14 and found that it spans the essential functional domain of LIN-14. This carboxy-terminal region, comprising exons 9 to 13, includes exons that are well conserved among LIN-14 proteins from different nematode species, indicating that this region interacts with relatively well-conserved cellular components. These interacting components likely including nuclear localization machinery, other nuclear proteins, and regulatory partners acting with LIN-14 in the control of gene expression.
Alternative lin-14 products are not required for lin-14 function in diverse cell lineages.
Earlier observations suggested that at least one of the three LIN-14 isoforms may perform tissue-specific functions. A genomic DNA fragment containing only exons 4 to 13 specifically rescues a precocious intestinal cell lineage phenotype (30), and overexpression of LIN-14A in intestinal cells causes an apparent cell autonomous gain-of-function lin-14 phenotype (Y. Hong, unpublished data). During our analysis of LIN-14 deletion constructs, we observed intestinal specific promoter/enhancer elements in the region between exons 4 and 8 (Y. Hong, unpublished data). These observations suggested that lin-14A might be an intestine-specific product and raised the question as to whether different lin-14 isoforms perform distinct tissue-specific functions. However, those experiments did not test for whether other LIN-14 isoforms also function in intestinal lineages or, more generally, whether the multiple LIN-14 isoforms are required for full lin-14 function.
Our results suggest that the ability to produce LIN-14 isoforms with alternative amino-terminal exons is not of developmental significance, at least under standard culture conditions. p14B2GFP, which cannot make either LIN-14A or LIN-14B1 due to the deletion of the alternative exons 2 and 4, fully rescues a lin-14 null mutant for all defects we tested, including precocious intestinal cell lineages. These results show that exons 2 and 4 are dispensable for lin-14 function in diverse cell types. Although LIN-14A may be expressed specifically, or predominantly, in intestinal cells, apparently the LIN-14A-specific exon (exon 4) is not required for LIN-14 function, even in intestinal cells.
Our experiments did not assay for the rescue of the timing of DD neuron remodeling. However, Hallam and Jin (11) have shown that exons 4 to 13 expressed under the neuron-specific unc-25 promoter can rescue the DD neuron remodeling defects of lin-14(ma135) larvae. This result is consistent with our finding that exon 2 is dispensable for lin-14 activity, but since a construct consisting of only exons 9 to 13 has not been tested for effects of DD remodeling, the possibility remains that exons 4 to 8 could be involved in the regulation of DD remodeling.
It is not uncommon for genes involved in developmental regulation to encode multiple isoforms. In some cases, these isoforms carry out distinct functions, and in other cases, genetic isoforms are functionally equivalent. For example, the Drosophila sex determination gene doublesex (dsx) encodes two protein isoforms (generated by alternative splicing) that have opposite effects on sexual phenotype (6). The Drosophila homeotic genes Abdominal-B (abd-B), Antennapedia (Antp), labial (lab), proboscipedia (pb), and Ultrabithorax (Ubx) each encode multiple evolutionarily conserved protein isoforms, (references 5 and 26 and references within), although the functional differences among the isoforms varies from gene to gene. The two alternative products of Abd-B (Abd-Bm and Abd-Br) are less distinct in function than are the dsx isoforms; nonetheless, Abd-Bm and Abd-Br exhibit distinguishable expression patterns and developmental activities (14, 16). The developmental roles of the six Ubx protein isoforms produced via alternative splicing are less distinct. Although the various Ubx isoforms are expressed in different patterns (13, 21), a Ubx mutant that expresses a single isoform is essentially wild type except for minor abnormalities (7, 26). Thus, individual Ubx protein isoforms do not apparently encode functionally distinct tissue-specific products and the existence of the various isoforms seems to mainly serve the purpose of fine tuning Ubx activity in different tissues (26; reviewed in reference 8). Like the Ubx isoforms of Drosophila, the alternative lin-14 transcripts, lin-14B1 and lin-14A, encode products that are not substantially distinct in function.
Carboxy-terminal domains of LIN-14 are sufficient for lin-14 activity.
Functional domains of LIN-14 sufficient for in vivo lin-14 activity are contained in the carboxy-terminal exons 9 to 13, a region approximately half the length of the full LIN-14 protein. Because we wished to test LIN-14 constructs that lacked all amino-terminal protein sequences and because of uncertainty about the position of the LIN-14 initiation codon, we used a hypodermal-specific col-10 promoter to drive expression of amino-terminal LIN-14 deletion constructs. Thus, our experiments involving the rescuing activity of LIN-14 deletions D1 to D10 (Fig. 1) were restricted to hypodermal cells. Consistent with our conclusion that a product of exons 9 to 13 is sufficient for in vivo lin-14 function, the region including exons 9 to 12 is highly conserved in amino acid sequence between different nematode species C. elegans and C. vulgaris (Fig. 3) and between C. elegans and C. briggsae (B. Reinhart and G. Ruvkun, personal communication). Eight of 10 sequenced loss-of-function mutations are within exons 9 and 12 (B. Reinhart and G. Ruvkun, personal communication; R. Lee and V. Ambros, unpublished data), further supporting the conclusion that this region contains essential LIN-14 functional domains. Since we did not test the rescuing activity of LIN-14 protein derivatives with deletions internal to exons 9 to 13, we do not know the relative importance of all these sequences, particularly exons 10 and 11. The amino acid sequence of exon 13 and part of exon 12 is not well conserved evolutionarily (Fig. 3), suggesting that these unconserved carboxy-terminal sequences correspond to less critical LIN-14 protein sequences. Indeed, deletion construct D9, which lacks these unconserved sequences, displayed significant rescuing activity (Table 3).
The function of lin-14 exons 1 to 8 is unclear from our experiments but could be significant under certain circumstances. Relatively high amino acid sequence conservation between the C. elegans and C. vulgaris LIN-14 proteins is not confined only to the carboxy-terminal region sufficient for lin-14 activity (exons 9 to 12). Exon 3, parts of exons 5 and 6, and all of exon 7 are also well conserved, suggesting that these sequences perform conserved, and hence significant, functions. Although we observed that deletion of exons 2 and 4 had no detectable effect on the rescuing activity of a LIN-14 transgene under the conditions of our assay, it is possible that under other culture conditions, these exons may have significant roles in obtaining optimal LIN-14 levels in all the relevant cell types throughout development. Furthermore, it is noteworthy that in lateral hypodermal cells, the rescuing activity of LIN-14D5::GFP, which lacks exons 1 to 8, is somewhat lower than that of LIN-14::GFP or LIN-14B2::GFP, which contain additional N-terminal exons, even though the expression level of LIN-14D5::GFP, driven by col-10 promoter, appears at least as high as that of LIN-14::GFP or LIN-14B2::GFP in the hypodermis. Thus, in lateral hypodermal cells, sequences contained within the N-terminal domains of LIN-14, although relatively dispensable, may be required for optimum lin-14 activity.
The N-terminal domains of LIN-14 could affect LIN-14 stability and hence might play a role in the developmental down regulation of LIN-14 protein levels. Although col-10::lacZ reporter genes containing a lin-14 3′UTR shows lin-4-dependent down regulation (29), other reporter constructs containing lin-14 3′UTR, such as col-10::GFP, col-10::luciferase (which contain no LIN-14 coding sequences; Y. Hong, unpublished data), or the amino-truncated LIN-14::GFP constructs (particularly D4 to D10) are not efficiently down regulated. In contrast, full-length LIN-14::GFP and LIN-14B2::GFP display lin-4-dependent down regulation. We have not examined in detail what LIN-14 coding sequences predispose a LIN-14::GFP fusion to accurate down regulation or whether such sequences act on the level of the protein or the mRNA. It is possible that certain structural features of the LIN-14 amino-terminal region may significantly influence LIN-14 stability. These sequences could include PEST sequences contained in exons 6 and 7 (28) and/or other sequences in exons 2 to 5.
Finally, unlike the truncated LIN-14::GFP proteins that we tested, endogenous LIN-14 proteins (visualized by antibody staining), as well as the full-length transgenes LIN-14::GFP and LIN-14B2::GFP (visualized by GFP fluorescence), appear to decrease in quantity rapidly in conjunction with mitosis and then reappear in daughter cell nuclei (M. Hristova, Y. Hong, and V. Ambros, unpublished data). Further experiments are required to determine whether this dynamic LIN-14 behavior is cell cycle triggered and is mediated by specific N-terminal sequences of LIN-14.
LIN-14 has an unconventional nuclear localization domain.
LIN-14 nuclear localization appears to require sequences contained in an approximately 200-amino-acid region between the end of exon 8 and the beginning of exon 12 (Fig. 4). Since we did not test the nuclear localization activity of deletions internal to this region, our data do not distinguish between an NLS extending throughout the region and a bipartite LIN-14 NLS located at each end. Near the ends of the exon 8-to-12 region are clusters of basic amino acids (RKPRK, amino acids 288 to 292 near the exon 8 and 9 border, and RCRRVR, amino acids 420 to 425 in exon 11) that strongly resemble typical NLS sequences (15). Neither of these particular elements is sufficient to localize LIN-14 to the nucleus, but they could act together as a bipartite NLS. A split or extended nuclear localization signal in LIN-14 would be unusual, since the nuclear localization signals of DNA or RNA-binding nuclear proteins are usually confined to a single region of fewer than approximately 50 amino acids (reviewed in reference 15). LIN-14 could be transported to the nuclei by direct association with the nuclear transport machinery and/or by association with other nuclear proteins.
The nuclear localization of LIN-14 does not require the rescuing activity of LIN-14. LIN-14D10::GFP with the n179 point mutation is efficiently concentrated in nuclei, but does not rescue lin-14(lf) phenotypes. This result suggests that although the nuclear localization domain of LIN-14 physically overlaps with the essential functional domain of LIN-14, the NLS activity is genetically separable from other essential LIN-14 activities.
LIN-14 may function in a protein complex.
A nonfunctional truncation of LIN-14, LIN-14D6::GFP, which contains only exons 10 to 13, has antimorphic, or partial dominant negative, activity in a lin-14 hypomorphic background. This observation suggests that LIN-14 may function in a protein complex, perhaps as a homodimer, or as a heterodimer or heteromultimer with other proteins. Although it cannot be excluded that LIN-14D6::GFP may have acquired novel function that is unrelated to endogenous lin-14 activity, we interpret its antimorphic activity to indicate that the truncated LIN-14 product, LIN-14D6::GFP, lacks certain essential function(s) but still retains a protein-protein interaction domain that mediates the association of LIN-14 with itself or with other proteins. Within exon 11 there is a predicted amphipathic helix sequence (28) which could mediate protein-protein interactions via coil-coil binding (reviewed in reference 24). LIN-14D6::GFP contains this amphipathic domain and could interfere with the endogenous lin-14 activity by competing with functional LIN-14 protein for binding to partners. LIN-14D7::GFP, containing exons 11 to 13, does not show any detectable dominant negative phenotype, suggesting that sequences in exon 10 are also critical for the dominant negative activity of LIN-14D6::GFP.
The antimorphic activity of LIN-14D6::GFP was detected in lin-14(n179ts) animals, but not in a wild-type background. Other hypomorphic alleles, besides n179, were not tested as sensitized genetic backgrounds, so it is possible that LIN-14D6::GFP acts allele specifically. However, it is also plausible that n179 simply supplies a partially reduced level of lin-14 activity suitable for detecting the further reduction in lin-14 function resulting from the antimorphic activity of LIN-14D6::GFP.
These observations are consistent with the hypothesis that LIN-14 may function in conjunction with other, as-yet-unidentified proteins to regulate the expression of lin-14 target genes. LIN-14 controls, directly or indirectly, diverse developmental events in numerous cell types, suggesting that it may have many genetic targets. One such candidate target gene is the C. elegans cyclin-dependent kinase inhibitor cki-1, which is responsible for mediating the temporal control of cell cycle progression by lin-14 in VPCs (12). Presumably, cell type-specific responses to lin-14 activity, such as the VPC-specific activation of cki-1, would originate from the interaction of LIN-14 with cell-type-specific cofactors, such as proteins involved in pre-mRNA synthesis or processing. The hypothetical LIN-14 partners may be discovered by identifying mutations that modify the phenotypes of either lin-14(lf) or lin-14(gf) or by direct biochemical screening for interacting proteins.
ACKNOWLEDGMENTS
We thank Eric Moss for first testing the rescuing activity of lin-14 exons 4 to 13 and his generous help on this project. We are grateful to the Sanger Center for providing cosmid KE7 and to Brenda Reinhart and Gary Ruvkun for sharing their data prior to publication. We also thank Eric Moss, Philip Olsen, and Richard Roy for helpful discussion. pPD series plasmids were kindly provided by Andrew Fire (personal communication), and their sequences can be obtained at ftp://ciw1.ciwemb.edu.
This work was supported by U.S. Public Health Service research grant GM34208 (V.A.). Some nematode strains used in this work were provided by the Caenorhabditis Genetics Center, which is funded by NIH National Center for Research Resources (NCRR).
REFERENCES
- 1.Ambros V. Heterochronic genes. In: Riddle D L, Blumenthal T, Meyer B J, Priess J R, editors. C. elegans II. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 501–518. [PubMed] [Google Scholar]
- 2.Ambros V, Horvitz H R. Heterochronic mutants of the nematode Caenorhabditis elegans. Science. 1984;226:409–416. doi: 10.1126/science.6494891. [DOI] [PubMed] [Google Scholar]
- 3.Ambros V, Horvitz H R. The lin-14 locus of Caenorhabditis elegans controls the time of expression of specific postembryonic developmental events. Genes Dev. 1987;1:398–414. doi: 10.1101/gad.1.4.398. [DOI] [PubMed] [Google Scholar]
- 4.Ambros V, Moss E M. Heterochronic genes and the temporal control of C. elegans development. Trends Genet. 1994;10:123–127. doi: 10.1016/0168-9525(94)90213-5. [DOI] [PubMed] [Google Scholar]
- 5.Bomze H M, Lopez A J. Evolutionary conservation of the structure and expression of alternatively spliced Ultrabithorax isoforms from Drosophila. Genetics. 1994;136:965–977. doi: 10.1093/genetics/136.3.965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burtis K C, Baker B S. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- 7.Busturia A, Vernos I, Macias A, Casanova J, Morata G. Different forms of Ultrabithorax proteins generated by alternative splicing are functionally equivalent. EMBO J. 1990;9:3551–3555. doi: 10.1002/j.1460-2075.1990.tb07565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Duncan I. How do single homeotic genes control multiple segment identities? Bioessays. 1996;18:91–94. doi: 10.1002/bies.950180203. [DOI] [PubMed] [Google Scholar]
- 9.Epstein H F, Shakes D C. Caenorhabditis elegans: modern biological analysis of an organism. Methods in cell biology. Vol. 48. San Diego, Calif: Academic Press, Inc.; 1995. [Google Scholar]
- 10.Euling S, Ambros V. Heterochronic genes control cell cycle progress and developmental competence of C. elegans vulva precursor cells. Cell. 1996;84:667–676. doi: 10.1016/s0092-8674(00)81045-4. [DOI] [PubMed] [Google Scholar]
- 11.Hallam S J, Jin Y. lin-14 regulates the timing of synaptic remodeling in Caenorhabditis elegans. Nature. 1998;395:78–82. doi: 10.1038/25757. [DOI] [PubMed] [Google Scholar]
- 12.Hong Y, Roy R, Ambros V. Developmental regulation of a cyclin-dependent kinase inhibitor controls postembryonic cell cycle progression in Caenorhabditis elegans. Development. 1998;125:3585–3597. doi: 10.1242/dev.125.18.3585. [DOI] [PubMed] [Google Scholar]
- 13.Kornfeld K, Saint R B, Beachy P A, Harte P J, Peattie D A, Hogness D S. Structure and expression of a family of Ultrabithorax mRNAs generated by alternative splicing and polyadenylation in Drosophila. Genes Dev. 1989;3:243–258. doi: 10.1101/gad.3.2.243. [DOI] [PubMed] [Google Scholar]
- 14.Kuziora M A. Abdominal-B protein isoforms exhibit distinct cuticular transformations and regulatory activities when ectopically expressed in Drosophila embryos. Mech Dev. 1993;42:125–137. doi: 10.1016/0925-4773(93)90002-f. [DOI] [PubMed] [Google Scholar]
- 15.LaCasse E C, Lefebvre Y A. Nuclear localization signals overlap DNA- or RNA-binding domains in nucleic acid-binding proteins. Nucleic Acids Res. 1995;23:1647–1656. doi: 10.1093/nar/23.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lamka M L, Boulet A M, Sakonju S. Ectopic expression of UBX and ABD-B proteins during Drosophila embryogenesis: competition, not a functional hierarchy, explains phenotypic suppression. Development. 1992;116:841–854. doi: 10.1242/dev.116.4.841. [DOI] [PubMed] [Google Scholar]
- 17.Lee R C, Feinbaum R L, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 18.Liu Z, Ambros V. Heterochronic genes control the stage-specific initiation and expression of the dauer larva developmental program in Caenorhabditis elegans. Genes Dev. 1989;3:2039–2049. doi: 10.1101/gad.3.12b.2039. [DOI] [PubMed] [Google Scholar]
- 19.Liu Z, Kirch S, Ambros V. The Caenorhabditis elegans heterochronic gene pathway controls stage-specific transcription of collagen genes. Development. 1995;121:2471–2478. doi: 10.1242/dev.121.8.2471. [DOI] [PubMed] [Google Scholar]
- 20.Mello C C, Kramer J M, Stinchcomb D, Ambros V. Efficient gene transfer in C. elegans: extrachromosomal maintenance and integration of transforming sequences. EMBO J. 1991;10:3959–3970. doi: 10.1002/j.1460-2075.1991.tb04966.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.O'Connor M B, Binari R, Perkins L A, Bender W. Alternative products from the Ultrabithorax domain of the bithorax complex. EMBO J. 1988;7:435–445. doi: 10.1002/j.1460-2075.1988.tb02831.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rougvie A E, Ambros V. The heterochronic gene lin-29 encodes a zinc finger protein that controls a terminal differentiation event in Caenorhabditis elegans. Development. 1995;121:2491–2500. doi: 10.1242/dev.121.8.2491. [DOI] [PubMed] [Google Scholar]
- 23.Ruvkun G, Giusto J. The Caenorhabditis elegans heterochronic gene lin-14 encodes a nuclear protein that forms a temporal developmental switch. Nature. 1989;338:313–319. doi: 10.1038/338313a0. [DOI] [PubMed] [Google Scholar]
- 24.Segrest J P, De Loof H, Dohlman J G, Brouillette C G, Anantharamaiah G M. Amphipathic helix motif: classes and properties. Proteins Struct Funct Genet. 1990;8:103–117. doi: 10.1002/prot.340080202. [DOI] [PubMed] [Google Scholar]
- 25.Slack F, Ruvkun G. Temporal pattern formation by heterochronic genes. Annu Rev Genet. 1997;31:611–634. doi: 10.1146/annurev.genet.31.1.611. [DOI] [PubMed] [Google Scholar]
- 26.Subramaniam V, Bomze H M, Lopez A J. Functional differences between Ultrabithorax protein isoforms in Drosophila melanogaster: evidence from elimination, substitution and ectopic expression of specific isoforms. Genetics. 1994;136:979–991. doi: 10.1093/genetics/136.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.The C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: a platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- 28.Wightman B, Burglin T R, Gatto J, Arasu P, Ruvkun G. Negative regulatory sequences in the lin-14 3′-untranslated region are necessary to generate a temporal switch during Caenorhabditis elegans development. Genes Dev. 1991;5:1813–1824. doi: 10.1101/gad.5.10.1813. [DOI] [PubMed] [Google Scholar]
- 29.Wightman B, Ha I, Ruvkun G. Posttranscriptional regulation of the heterochronic gene lin-14 by lin-4 mediates temporal pattern formation in C. elegans. Cell. 1993;75:855–862. doi: 10.1016/0092-8674(93)90530-4. [DOI] [PubMed] [Google Scholar]
- 30.Wightman B C. Post-transcriptional regulation of the C. elegans heterochronic gene lin-14. Ph.D. thesis. Boston, Mass: Harvard Medical School; 1992. [Google Scholar]
- 31.Wood W B The Community of C. elegans Researchers. The nematode Caenorhabditis elegans. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1988. [Google Scholar]