Abstract
The viral membrane proteins M and E are the minimal requirements for the budding of coronavirus particles. Since the E protein occurs in particles only in trace amounts, the lateral interactions between the M proteins apparently generate the major driving force for envelope formation. By using coimmunoprecipitation and envelope incorporation assays, we provide extensive evidence for the existence of such M-M interactions. In addition, we determined which domains of the M protein are involved in this homotypic association, using a mutagenetic approach. Mutant M proteins which were not able to assemble into viruslike particles (VLPs) by themselves (C. A. M. de Haan, L. Kuo, P. S. Masters, H. Vennema, and P. J. M. Rottier, J. Virol. 72:6838–6850, 1998) were tested for the ability to associate with other M proteins and to be rescued into VLPs formed by assembly-competent M proteins. We found that M proteins lacking parts of the transmembrane cluster, of the amphipathic domain, or of the hydrophilic carboxy-terminal tail, or M proteins that had their luminal domain replaced by heterologous ectodomains, were still able to associate with assembly-competent M proteins, resulting in their coincorporation into VLPs. Only a mutant M protein in which all three transmembrane domains had been replaced lost this ability. The results indicate that M protein molecules interact with each other through multiple contact sites, particularly at the transmembrane level. Finally, we tested the stringency with which membrane proteins are selected for incorporation into the coronavirus envelope by probing the coassembly of some foreign proteins. The observed efficient exclusion from budding of the vesicular stomatitis virus G protein and the equine arteritis virus M protein indicates that envelope assembly is indeed a highly selective sorting process. The low but detectable incorporation of CD8 molecules, however, demonstrated that this process is not perfect.
Enveloped viruses acquire their lipid membranes by the budding of the viral nucleocapsid (NC) through cellular membranes. Although little is known about the molecular details of this process, it has become clear that the roles played by the viral membrane proteins in the formation of the viral envelope vary tremendously among different viruses. At one extreme, these proteins are not required at all. Viruses such as rhabdoviruses and retroviruses bud normally in the absence of their glycoproteins to form the characteristic bullet-shaped and rounded particles, respectively. At the other extreme, the viral membrane proteins are all that is required for envelope formation. Here, these proteins have the capacity by themselves to carry out the budding of particles devoid of an NC. While such “empty” particles are often smaller than authentic virions—subviral particles have been demonstrated for flaviviruses (1, 26, 37, 47) and hepadnaviruses (41, 49)—their dimensions can perfectly match those of normal virions, as we and others have observed for coronaviruses (4, 56). Intermediate between these extremes are the many viruses for which the membrane proteins are essential but not sufficient to form the viral envelope. Here, internal components are also required: they act together with the membrane proteins to accomplish the budding. In this category, alphaviruses are the best-studied examples. (For a recent review of the topic, see reference 19.)
As for large biological complexes in general, molecular interactions between the structural components generate the free energy that drives virus assembly. In view of the widely differing roles of the viral membrane proteins in budding, the significance of the interactions between these proteins is also likely to vary greatly. Thus, while associations between the envelope glycoprotein trimers of retroviruses may be weak or even absent, protein-protein interactions are probably crucial for coronaviruses. Unfortunately, information about such interactions is largely lacking, particularly due to the technical difficulties of obtaining ultrastructural data for these viruses, which for the nonenveloped viruses has proved so valuable. An exception is the alphaviruses: cryoelectron microscopy and image reconstruction of Semliki Forest virus (59) and Sindbis virus (53) revealed among others the icosahedral surface symmetry (T=4) of both their nucleocapsids and their envelopes, as well as the trimeric nature of their spikes. In addition, and more recently, the reconstruction of the Ross River virus particle (9) visualized the tight association between the heterodimeric subunits of neighboring spikes.
Coronaviruses carry three or four proteins in their envelopes. The M protein is the most abundant component; it is a type III glycoprotein consisting of a short amino-terminal ectodomain, three successive transmembrane domains, and a long carboxy-terminal domain on the inside of the virion (or in the cytoplasm) (44). The small E protein is a minor but essential viral component (4, 5, 17, 48, 56). In cells, it accumulates in and induces the coalescence of the membranes of the intermediate compartment (IC), giving rise to typical structures (43). A fraction of the proteins appear extracellularly in membranous structures of unknown identity (35). The trimeric spike (S) protein forms the characteristic viral peplomers. These peplomers are involved in virus-cell attachment and in virus-cell and cell-cell fusion (8). A subset of coronaviruses contains a hemagglutinin-esterase (HE) protein, which occurs as a disulfide-linked homodimer (6).
For assembly of the coronavirus envelope, only the M protein and the E protein are needed (4, 5, 11, 21, 56). Expression in cells of the genes coding for these proteins leads to the formation and release of viruslike particles (VLPs) similar in size and shape to authentic virions. The S protein is dispensable for the formation of these particles. This has now been demonstrated for mouse hepatitis virus (MHV) (5, 11, 56), transmissible gastroenteritis virus (4), and feline infectious peritonitis virus (21). Particularly in MHV, the E protein is only present in trace amounts; though essential for their formation, the protein is barely detectable in VLPs of this virus (56). Thus, the protein component of the envelopes of these particles essentially consists of M molecules. We hypothesize that the coronavirus membrane basically consists of a dense matrix of laterally interacting M proteins, which in some way requires the E protein for budding and in which the S and HE glycoproteins are incorporated, if available, by specific interactions with M (13, 39, 40, 56).
The existence of M-M interactions has already been inferred from data obtained using sucrose gradient analysis. When expressed on its own, the M protein was found in large heterogeneous complexes in the Golgi apparatus (31). The S protein, which by itself is transported to the plasma membrane (40), appeared to associate with these M protein complexes when coexpressed, resulting in its retention in the Golgi complex. Further support for the existence of M-M protein interactions came from our recent observation that assembly-incompetent M protein mutants could be rescued into VLPs (11).
In view of the presumed importance of M proteins for the formation of the coronavirus envelope, the present study was undertaken to provide convincing evidence for the occurrence of interactions between them. In addition, we analyzed which domains of the M molecule are involved in these interactions and investigated where in the cell association of M proteins takes place. Finally, we studied the accuracy with which the M protein framework is composed by analyzing the sorting of foreign membrane proteins.
MATERIALS AND METHODS
Cells, viruses, and antibodies.
Recombinant vaccinia virus encoding the bacteriophage T7 RNA polymerase (vTF7-3) and OST7-1 cells were obtained from B. Moss. OST7-1 cells (16) were maintained as monolayer cultures in Dulbecco's modified Eagle's medium containing 10% fetal calf serum, 100 IU of penicillin/ml, and 100 μg of streptomycin/ml (all from Life Technologies, Ltd., Paisley, United Kingdom). The hybridoma line OKT8 producing the OKT8 monoclonal antibody against human CD8 (anti-CD8) was purchased from ECACC (Salisbury, United Kingdom). The rabbit polyclonal MHV strain A59 antiserum (K134; anti-MHV) (45), the rabbit polyclonal vesicular stomatitis virus (VSV) antiserum (K114; anti-VSV) (57), and the rabbit polyclonal peptide serum raised against the 18 carboxy-terminal amino acids of MHV M (anti-Mc) (30) have been described earlier. The monoclonal antibody J1.3 against the amino terminus of MHV M (anti-MN) (52) was kindly provided by J. Fleming.
Expression vectors and site-directed mutagenesis.
All of the expression vectors used contain the genes under control of bacteriophage T7 transcription regulatory elements. Expression construct pTM5ab contains the MHV strain A59 open reading frames 5a and 5b, the latter coding for the E protein, in pTUG31 (56, 58). The construction of M genes coding for the mutant proteins ΔC, Δ(a+b) and Δ(b+c) (30), A2A3 and Δ18 (11), and M-KK and 3AT5-KK (12) has been described before (Fig. 1). Also, the constructs encoding the VSV G protein (58) and the equine arteritis virus (EAV) hybrid protein M+9A have been described before (12). The latter protein has an insertion of 9 amino acids, corresponding to the MHV M amino-terminal sequence (residues S2 to P10), behind the initiating methionine of EAV M. The construct coding for the MHV M protein ΔLT, which has a deletion of 5 amino acids (ΔL108 to T112), was fortuitously obtained during the construction of the gene coding for the mutant M protein SapΔ1 (13). To make the M gene encoding the mutant protein ΔRK, which lacks amino acids R188 through K207, pLITMUS38 (New England Biolabs) containing the gene coding for the M protein Sap (13) was digested with BssHII and StyI, treated with mung bean nuclease (Pharmacia), and religated. The construct was treated with BamHI, and the resulting fragment was cloned into expression vector pTUG3. In hybrid protein VGM, the amino-terminal ectodomain of MHV M was replaced by that of VSV G. In order to make the construct encoding this protein, an SstI restriction site was engineered in the MHV M gene by PCR mutagenesis using primers 891 (5′-GTTCAGAGCTCTAAGGAATGGAACTTCTCG-3′) and 746 (5′-CGTCTAGATTAGGTTCTCAACAATGCGG-3′), corresponding to the region coding for the carboxy-terminal part of the ectodomain (and introducing the SstI restriction site) and the 3′ end of the MHV M gene, respectively. The PCR product obtained was cloned into the pNOTA/T7 shuttle vector (5 prime→3 prime, Inc.) and subsequently excised from the plasmid with BamHI and cloned into pTUG3, resulting in construct pTUG3MSacI. The fragment encoding the VSV G ectodomain was excised from pSV045R-ts (18) (a kind gift from J. K. Rose) by using XhoI and SstI and cloned into pTUG3MSacI treated with the same enzymes, resulting in expression construct pTUG3VGM. Plasmids S83 and S84 were a kind gift from S. Munro (38). Plasmid S83 encodes a human CD8 protein, in which the cytoplasmic tail has been replaced by four foreign amino acids (KRLK), while plasmid S84 encodes human CD8 protein which contains the CD8 cytoplasmic tail starting with these 4 amino acids. The sequence coding for KRLK contains an AflII site which facilitates the exchange of cytoplasmic tails. The expression cassettes of plasmids S83 and S84 were excised by using HindIII and XbaI and cloned into pNOTA/T7 treated with the same enzymes, resulting in expression vectors pNOTACD8tr and pNOTACD8, respectively. The construct encoding hybrid protein CD8Mc contains the sequence encoding the extracellular and transmembrane domains of CD8 followed by the MHV M cytoplasmic domain sequence starting with the codon for residue S105. In order to make this construct, an AflII restriction site was engineered in the MHV M gene by PCR mutagenesis using primers 586 (5′-GTATTTTCTTAAGAGCATTAGGTG-3′) and 495 (5′-TTAGATTCTCAACAATGCGG-3′), corresponding to the region coding for the amino-terminal part of the cytoplasmic domain (and introducing the AflII site) and the 3′ end of the MHV M gene, respectively. The PCR product obtained was cloned into the pNOTA/T7 vector and subsequently excised using AflII and XbaI and cloned into pNOTACD8tr treated with the same enzymes, resulting in pNOTACD8Mc. In hybrid protein CD8ΔN, the amino-terminal ectodomain of MHV M was replaced by that of CD8. The region encoding the MHV M transmembrane and cytoplasmic domains was excised from pTZ19RMΔN (30) by using PvuII and BamHI and cloned into pNOTACD8tr treated with EcoRV and BamHI, resulting in pNOTACD8ΔN. All constructs were verified by sequencing.
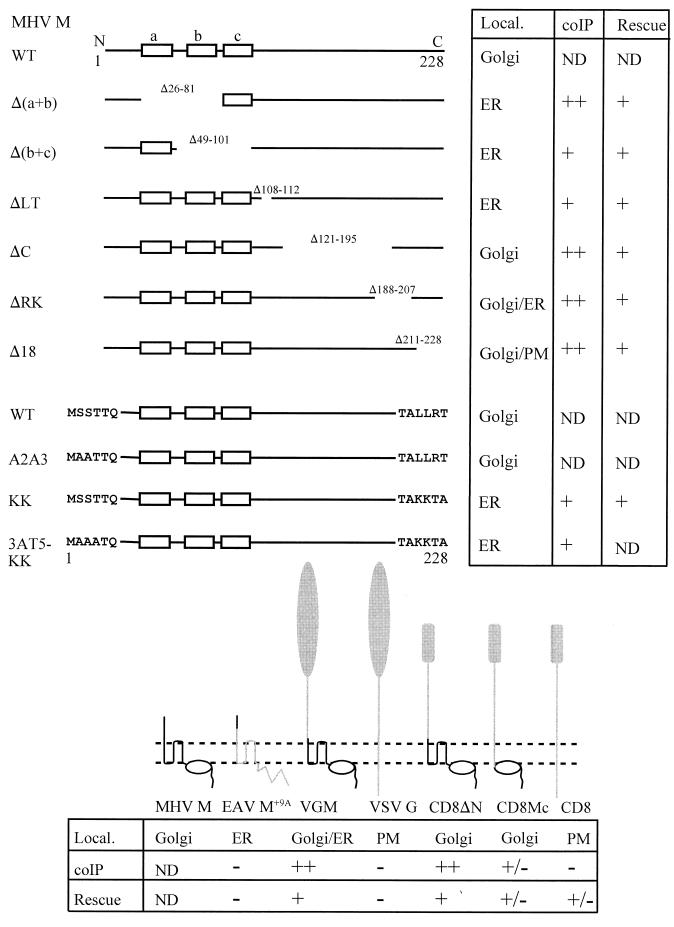
FIG. 1.
Overview of mutant M proteins. A schematic linear representation of the M protein, with its three transmembrane domains (a, b, and c) indicated, is shown on top. Mutant proteins with deletions in the transmembrane region [Δ(a+b) and Δ(b+c)], the amphipathic domain (ΔLT, ΔC, and ΔRK), and the extreme carboxy terminus (Δ18) are depicted at the top. Gaps represent deletions; the numbers indicate the deleted amino acids. Mutant proteins with amino acid substitutions in the amino terminus and/or carboxy terminus (A2A3, KK, and 3AT5-KK) are also depicted. The six amino-terminal and carboxy-terminal residues are shown. Below, the membrane structures of MHV M, EAV M+9A, VSV G, and CD8, as well as their chimeric forms, are drawn. The black lines represent amino acid sequences derived from MHV M; the oval symbolizes the amphipathic domain. The gray lines and symbols designate sequences derived from EAV M, VSV G, or CD8. The intracellular localization of the mutant proteins (Local.), their abilities to coimmunoprecipitate indicator M proteins (coIP), and their abilities to become incorporated into VLPs when coexpressed with M protein A2A3 (Rescue) are indicated at the upper right and bottom. Golgi, ER, and PM indicate localization of the proteins in the Golgi complex, in the ER, and in the plasma membrane, respectively (references 11, 12, and 13 and data not shown). The semiquantitative scores ++, +, +/−, and − indicate efficient, moderately efficient, inefficient, and no coIP of the indicator proteins M-Δ18 (for 3AT5-KK) and M-A2A3 (for the others). The semiquantitative scores +, +/−, and − indicate efficient, inefficient, and no rescue of the M proteins into VLPs as determined by immunoisolation of intact VLPs. ND, not determined. Pulse-chase analysis demonstrated that the stabilities of all mutant M proteins were similar to that of WT M, with the exception of the M protein ΔRK, which was slightly less stable.
Metabolic labeling and immunoprecipitation.
Subconfluent monolayers of OST7-1 cells in 10-cm2 tissue culture dishes were inoculated with vTF7-3 (t = 0 h) and subsequently transfected 1 h later with plasmid DNA by using lipofectin (Life Technologies) as described previously (11). At t = 2 h, the cells were placed at 32°C. At t = 4.5 h, the cells were washed with phosphate-buffered saline and starved for 30 min in cysteine- and methionine-free modified Eagle's medium containing 10 mM HEPES, pH 7.2, and 5% dialyzed fetal calf serum. The medium was then replaced by 600 μl of similar medium containing 100 μCi of 35S in vitro cell-labeling mixture (Amersham), and the cells were labeled for the indicated time periods. In some experiments, the radioactivity was chased by incubating the cells with culture medium containing 2 mM methionine and 2 mM cysteine for 2 h. Proteins were immunoprecipitated from cell lysates as described before (40). Culture media were prepared for immunoprecipitation (IP) in the presence or absence of detergents by addition of 1/4 volume of five-times-concentrated lysis buffer or by addition of 2.5 volumes of TEN buffer consisting of 40 mM Tris-HCl (pH 7.6), 50 mM NaCl, and 1 mM EDTA, respectively. The immune complexes were adsorbed to Pansorbin cells (Calbiochem) for 30 min at 4°C and were subsequently collected by low-speed centrifugation. The pellets were washed three times by resuspension and centrifugation using 50 mM Tris-HCl (pH 8.0)–62.5 mM EDTA–0.5% Nonidet P-40–0.5% Na-deoxycholate or TEN buffer. The final pellets were suspended in electrophoresis sample buffer. The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis in 15% polyacrylamide gels.
RESULTS
Demonstration of M-M interaction.
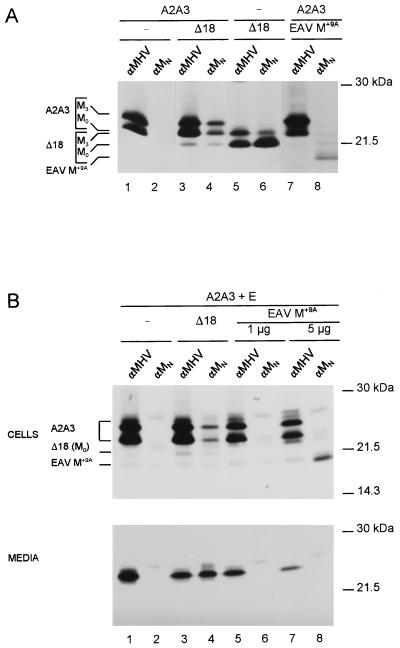
The monoclonal antibody J1.3 is directed against the amino terminus of the MHV M protein (52). Fine mapping of the epitope recognized by this antibody—designated anti-MN—enabled us to develop a coimmunoprecipitation (coIP) assay for the detection of interactions between the M molecules. We found out recently (11) that recognition of this epitope by the antibody is critically dependent on the presence of the serine residues at positions 2 and 3. A mutant of the M protein, named A2A3, in which these residues have been replaced by alanines, was not recognized by the monoclonal antibody. This mutant protein otherwise behaved identically to the wild-type (WT) M protein in every aspect studied, including its ability to assemble VLPs (11, 12). The coIP assay is thus based on the coexpression of the A2A3 protein with mutant M proteins carrying an intact anti-MN epitope: association of the proteins is monitored by the coprecipitation of A2A3 M molecules by the monoclonal antibody. The assay is demonstrated in Fig. 2A. In this experiment, the A2A3 M gene was either expressed alone or in combination with the gene encoding the carboxy-terminal deletion mutant MΔ18 (Fig. 1) or with the gene encoding a control protein, the chimeric EAV protein M+9A (12). This protein consists of the EAV M protein extended at its extreme amino terminus by inserting the 9-residue amino-terminal sequence of MHV M (residues S2 to P10). As a result of this extension, the EAV protein acquired the epitope recognized by the MHV-specific antibody anti-MN. The EAV M protein is a triple-membrane-spanning protein with a topology similar to that of the MHV M protein but is slightly smaller (15). The genes were expressed in OST7-1 cells by using the vTF7-3 expression system. The cells were labeled for 2 h with 35S-labeled amino acids starting at 5 h postinfection. Cell lysates were prepared and subjected to IP with either an anti-MHV serum or the monoclonal antibody anti-MN. Mutants A2A3 and Δ18 were well expressed both in the single expression and in the coexpressions, as was demonstrated by IP using the anti-MHV serum (Fig. 2A, lanes 1, 3, 5, and 7). The A2A3 protein appeared as the well-known set of O-glycosylated forms described before (29, 54), with the unglycosylated form (M0) and the Golgi-modified form containing galactose and sialic acid (M3) being the most prominent species. The M protein mutant Δ18 also becomes O glycosylated normally (11). Its M0 form runs slightly faster in the gel than A2A3, while its M3 form comigrates with the unglycosylated form (Fig. 2A, cf. lanes 1 and 5). Importantly, the mutant protein A2A3 was clearly not recognized by anti-MN, in contrast to the M protein Δ18, as seen after single expression (Fig. 2A, lanes 2 and 6). Analysis of the lysate from cells expressing protein A2A3 and the M mutant Δ18 revealed the formation of M-M complexes. The anti-MHV serum precipitated both proteins A2A3 and Δ18 (lane 3). The monoclonal antibody anti-MN not only precipitated protein MΔ18 but also the glycosylated forms of A2A3 (lane 4). Analysis of the lysate from cells expressing protein A2A3 and the EAV M protein revealed the specificity of the assay. While the anti-MHV serum only precipitated protein A2A3 (lane 7), the monoclonal antibody only precipitated the EAV M+9A protein (lane 8). No coIP was observed. As another control for the specificity of the interactions measured, lysates of cells singly expressing the M protein mutants A2A3 and Δ18 were pooled and subsequently processed for IP using monoclonal anti-MN. No coIP was observed from the pooled lysates (not shown).
FIG. 2.
Demonstration of M-M interaction. Genes coding for the mutant M proteins A2A3 and Δ18, the chimeric protein EAV M+9A, and the E protein were expressed in OST7-1 cells in various combinations, as indicated above each lane (−, absent), by using the vTF7-3 expression system. For the plasmid encoding protein A2A3, 5 μg was transfected, while for the plasmid encoding the E protein, 1 μg was used (A and B); 5 μg of the plasmid encoding the M protein Δ18 was used for panel A, and 1 μg was used for panel B, while 3 μg of the plasmid encoding the chimeric protein EAV M+9A was used for panel A and 1 or 5 μg was used for panel B. Cells were labeled for 2 (A) or 3 (B) h. Cell lysates were prepared and subjected to IP with either the anti-MHV serum (αMHV) or the monoclonal antibody to the amino terminus of M (αMN). When the E protein was coexpressed (B), culture media were also collected and processed for IP or for affinity isolation of VLPs. The affinity isolations were performed by using the monoclonal antibody anti-MN in the absence of detergents. The positions of the different proteins are indicated at the left, while the molecular mass markers are at the right. Only the relevant parts of the gels are shown.
Our second assay for the detection of M-M interactions was based on the VLP assembly system. To demonstrate and validate this approach, we analyzed the coincorporation of the mutant proteins MΔ18 and EAV M+9A into VLPs assembled from protein A2A3. Earlier we showed that protein A2A3, when coexpressed with the E protein, is assembled into VLPs as efficiently as WT M protein, while both mutant MΔ18 (11) and EAV M+9 (unpublished data) proteins failed to be. In the experiment shown in Fig. 2B, the E protein gene is coexpressed with the genes coding for the mutant proteins A2A3, Δ18, and EAV M+9A in a way similar to that described above except that the cells were labeled for 3 h. The cells and culture media were collected separately and processed for IP with the anti-MHV serum and with the monoclonal antibody anti-MN. Analysis of the cell lysates (Fig. 2B, top) revealed that the coexpression of the E protein gene did not affect the coIP results (cf. Fig. 2A). Again, protein A2A3 was not recognized by monoclonal anti-MN antibody (lane 2) and was coprecipitated when coexpressed with protein MΔ18 (lane 4) but not with control protein EAV M+9A (lanes 6 and 8). Due to the longer labeling time used in this experiment to allow detection of released VLPs, some more background bands were observed around the M protein bands. The E protein was not resolved with the antibodies used. Analysis of the culture media by the normal IP procedure (i.e., using detergents) with the anti-MHV serum showed that all combinations of plasmids had been productive in VLP formation (Fig. 2B, bottom, lanes 1, 3, 5, and 7). By carrying out the precipitations on the media with anti-MN in the absence of detergents, an immunoisolation of intact VLPs was performed. As expected, these VLPs could not be affinity isolated with the monoclonal anti-MN antibody when only the M mutant A2A3 had been coexpressed with the E protein (lane 2). The additional expression of MΔ18 protein, however, enabled isolation of the A2A3-based VLPs (lane 4), apparently due to the coincorporation of the truncated M protein. VLPs could not be affinity isolated after coexpression of EAV M+9A, indicating that this protein is not incorporated (lanes 6 and 8). The combined results demonstrate the specificity and consistency of the two assays in detecting interactions between M molecules.
Mapping of M protein domains involved in homotypic interactions.
The assays were subsequently used to investigate the involvement of different domains of the M molecule in M-M interactions. To this end, a number of M protein mutants were evaluated (Fig. 1). Mutant proteins Δ(a+b) and Δ(b+c) have a deletion of the first and second transmembrane domains and of the second and third transmembrane domains, respectively, resulting in M proteins with only the third or only the first transmembrane domain left. With their amino termini in the lumen and their carboxy termini in the cytoplasm, these proteins have the same membrane topology as WT M (30). When expressed, they appear mainly in an unglycosylated form, which is indicative of their inefficient transport out of the endoplasmic reticulum (ER), as we verified by immunofluorescence (not shown). The mutant proteins ΔLT, ΔC, and ΔRK each lack a different part of the amphipathic domain which encompasses the region of the M molecule between the transmembrane cluster and the approximately 20-residue hydrophilic carboxy-terminal tail. The disposition of the amphipathic domain has not yet been resolved. While the M protein mutant ΔLT does not become glycosylated and localizes to the ER, the mutant proteins ΔC and ΔRK acquire O-linked sugars, which is indicative of their transport to the Golgi complex (references 11, 12 and unpublished results). Furthermore, we also tested a mutant M protein with an ER retrieval signal (M-KK). This protein carries a cytoplasmic KKXX ER retrieval and retention signal (2, 24) which localizes it to the ER. While this protein can become O glycosylated under artificial conditions (e.g., during treatment with brefeldin A [BFA]), no trace of glycosylation can be detected in standard pulse-chase experiments even after 3 h of chase (12). Apparently, the protein is either retained very efficiently in the ER or rapidly retrieved from pre-Golgi compartments, where no O glycosylation takes place (12). The mutant protein Δ18 was used as a positive control. Importantly, all these mutant proteins were found to be deficient in VLP assembly when coexpressed with the E protein gene (reference 11 and unpublished data).
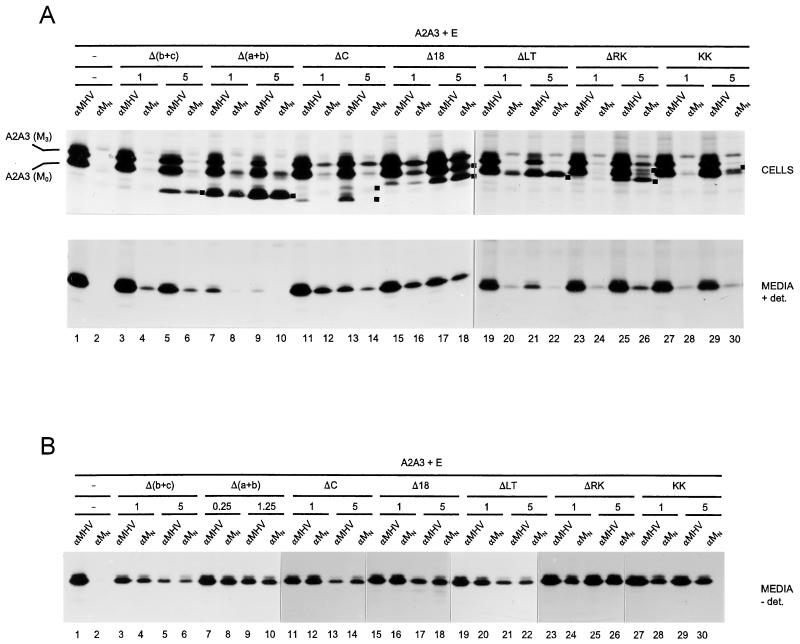
Each of these mutant M genes was expressed together with genes encoding the M protein mutant A2A3 and the E protein in two different concentrations, as in the previous experiment. The coIP assay was performed both on the cell lysates and on the culture media, as shown in Fig. 3A. As is clear from the analysis of the cell lysates (Fig. 3A, top), protein A2A3 was well expressed in all combinations; it was not precipitated by monoclonal anti-MN antibody when expressed only with the E protein (lane 2) but appeared when the mutant protein Δ18 was additionally coexpressed, particularly at the higher expression level of this mutant (lanes 16 and 18). Consistent with their transmembrane deletions, the mutant proteins Δ(b+c) and Δ(a+b) migrate faster in the gel than protein A2A3. Upon coexpression of these mutant proteins with A2A3 and E protein, protein Δ(b+c) appeared to coprecipitate only low levels of protein A2A3 (lanes 4 and 6), while the mutant protein Δ(a+b) clearly precipitated the M0 form of A2A3 as well as low levels of its glycosylated species (lanes 8 and 10). Coexpression of the mutant protein ΔC, which also migrates ahead of protein A2A3, also resulted in coprecipitation of the latter protein (lanes 12 and 14), even though—for reasons not understood—protein ΔC itself was not efficiently precipitated with the monoclonal antibody. The ER-retained mutant protein ΔLT has approximately the same electrophoretic mobility as the M0 form of protein A2A3. After coexpression of the mutant proteins ΔLT and A2A3, coprecipitation of small amounts of the glycosylated A2A3 species was observed (lanes 20 and 22). The unglycosylated form of the mutant ΔRK protein migrates slightly faster in the gel than that of protein A2A3, and the same is true of their glycosylated forms. Hence, the M3 form of the ΔRK protein runs in between the M0 and M3 forms of protein A2A3. Protein A2A3 was clearly coprecipitated with the mutant protein ΔRK (lanes 24 and 26). This coprecipitation was more pronounced at the higher expression level of the mutant ΔRK protein but was not as efficient as with protein Δ18. The M protein with the ER retrieval signal (M-KK) runs at a slightly higher position in the gel than the M0 form of protein A2A3. Coexpression of the protein A2A3 did not induce glycosylation of the mutant protein M-KK (not shown; see below), indicating that the transport-competent M proteins are not able to ferry the M proteins with the ER retention and retrieval signal to the Golgi complex. Upon coexpression of proteins M-KK and A2A3, small amounts of glycosylated forms of A2A3 were coprecipitated in addition to unglycosylated A2A3 protein (lanes 28 and 30). An explanation for the apparent association between glycosylated A2A3 and unglycosylated M-KK is that M proteins that have acquired Golgi modifications are able to return to pre-Golgi compartments, where they can subsequently interact with M proteins carrying an ER retention and retrieval signal. Since similar results were obtained in the absence of the E protein (not shown), the IP of glycosylated M proteins by the monoclonal antibody anti-MN does not result from VLPs which have not yet been secreted. The combined results demonstrate that all the M proteins tested were able to associate with the indicator protein A2A3 but with different efficiencies. It appeared that transport-competent proteins, such as the cytoplasmic deletion mutants Δ18, ΔRK, and ΔC, were more efficient than those that were not (M-KK and ΔLT) or were very poorly [Δ(b+c) and Δ(a+b)] transported to the Golgi complex. Whether this correlation results only from differences in localization or is a reflection of the transport-incompetent proteins being unable to pass the ER quality control and to become available for interaction with A2A3 protein is not clear.
FIG. 3.
Mapping of M protein domains involved in homotypic interaction. The genes encoding the M protein A2A3 and the E protein were coexpressed together with different amounts of mutant M genes as described in the legend to Fig. 2. The different combinations are indicated above the gel, as are the amounts (in micrograms) of the mutant M plasmids transfected (−, absent). Cells were labeled for 3 h. (A) The cells and culture media were collected separately and subjected to IP with anti-MHV (αMHV) and anti-MN (αMN) antibodies in the presence of detergents. The positions of the mutant M proteins in the gel are indicated by black squares at the right sides of the lanes. The positions of the M0 and M3 forms of protein A2A3 are also indicated. (B) In an otherwise-identical experiment, the culture media were analyzed in the absence of detergents to allow isolation of intact VLPs.
In the lower half of Fig. 3A, the results of the IPs—done in the presence of detergents—on the corresponding culture media are shown. The observations with the anti-MHV serum reveal that all combinations were productive in VLP formation but that the coexpression of the assembly-incompetent mutant M proteins inhibited formation of the A2A3-based particles, in a concentration-dependent manner, as we have observed before (11). Exceptions were the mutant proteins ΔRK and M-KK, which appeared not to interfere (lanes 23, 25, 27, and 29). The IPs with the anti-MHV antibodies showed that these mutant proteins were barely or not detectable directly in the VLPs. Indirectly, however, through their coIP of the A2A3 protein, their coincorporation into VLPs was evident in all cases, and the extent to which this occurred was generally consistent with the level of coIP observed with the cell lysates. The poor formation of VLPs observed in the presence of the mutant protein Δ(a+b) (lanes 7 and 9) was probably due to its relatively high expression level and, consequently, its stronger interference with the expression of protein A2A3 and with the assembly process.
In a separate experiment, the incorporation of the mutant M protein into VLPs was further confirmed by the immunoisolation assay. The same amounts of plasmid DNA encoding the different proteins were transfected, except for mutant Δ(a+b), for which a four-times-smaller amount of plasmid DNA was used in view of the considerations just mentioned. The analysis of the IPs performed in the absence of detergents are shown in Fig. 3B. Clearly, all the mutant proteins were coincorporated into VLPs. The amount of particles produced in the presence of Δ(a+b) protein was largely increased. The high sensitivity of this assay is illustrated by the observation that in all combinations tested, similar levels of protein A2A3 were (co)immunoprecipitated with monoclonal anti-MN antibody as with the anti-MHV serum.
Replacement of the MHV M ectodomain.
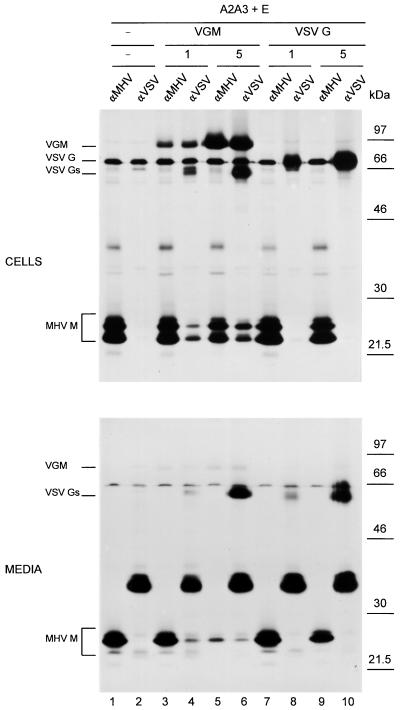
The MHV M protein contains a short (25 amino acids) ectodomain that is located in the lumens of intracellular organelles or on the outside of the virion. Mutations in this domain render the protein deficient in VLP assembly (11). In the present study, we evaluated whether replacement of the MHV M ectodomain by heterologous ectodomains affects M-M interactions. In the experiment shown in Fig. 4, we tested the hybrid protein VGM, which contains the ectodomain of the VSV G protein (Fig. 1). WT VSV G protein was included as a control. When coexpressed with the E protein, neither the hybrid protein nor the VSV G protein was productive in VLP assembly (not shown). It has been established that transport of VSV G protein out of the ER requires the formation of G protein trimers (27). Immunofluorescence analysis indicated the VGM hybrid protein to be located in the ER and the Golgi compartment (not shown). The oligomeric state of this protein was not studied.
FIG. 4.
Replacement of the MHV M ectodomain by the VSV G ectodomain. Genes encoding the M protein A2A3 and the E protein were expressed together or in combination with genes encoding VSV G or the hybrid protein VGM, as described in the legend to Fig. 2. The amounts of the plasmids encoding VGM and VSV G that were transfected are indicated (in micrograms; −, absent). Cells were labeled for 3 h. The cells and media were collected separately and subjected to IP by using the anti-MHV serum (αMHV) or the anti-VSV serum (αVSV). When IP on the medium was performed with the anti-VSV serum, no detergents were added to allow affinity isolation of VLPs. The positions of the different proteins are indicated at the left, while the molecular mass markers are at the right.
The genes coding for VGM and VSV G were coexpressed with the A2A3 and E protein genes, as described above. Cell lysates and culture media were subjected to IP with the anti-MHV serum and with an anti-VSV serum. Analysis of the cell lysates (Fig. 4, top) showed that the A2A3 protein was well expressed in all combinations, although the expression level was somewhat decreased when a large amount of VGM plasmid DNA was cotransfected (Fig. 4, lane 5). The A2A3 protein was not recognized by the anti-VSV serum, as it was not precipitated after coexpression with the E protein only (lane 2). The hybrid protein VGM (apparent molecular mass, 89 kDa) was recognized by both the anti-MHV serum and by the anti-VSV serum. In addition, the anti-VSV, but not the anti-MHV, serum precipitated a protein with an apparent mass of 62 kDa. The anti-VSV serum clearly coprecipitated the A2A3 protein, and this coIP was more pronounced at the higher VGM protein expression level (lanes 4 and 6). The VSV G protein (apparent mass, 70 kDa) was well expressed; its analysis was somewhat obscured by the precipitation of a (probably vaccinia virus-related) background protein with the same electrophoretic mobility. No trace of the A2A3 protein was found to be coprecipitated with the VSV G protein (lanes 8 and 10).
The IPs performed on the culture media were done in the presence (Fig. 4, lane αMHV) or in the absence (lane αVSV) of detergents. As inferred from the appearance of the A2A3 protein, VLPs were formed in all plasmid combinations, although to a lesser extent when the large amount of VGM plasmid DNA had been cotransfected (lane 5). The VGM protein (lanes 4 and 6) but not the VSV G protein (lanes 8 and 10) coprecipitated A2A3 protein, indicating that the hybrid protein, but not VSV G, was incorporated into VLPs. Although the amount of VLPs released decreased with the higher VGM expression level, relatively more A2A3 protein was coprecipitated, indicating that the rescue of VGM into VLPs was more efficient. After prolonged exposure of the gel to the film, the VGM protein itself became visible (not shown). The anti-VSV serum also precipitated the 62-kDa protein, from culture media of both cells expressing the VGM protein and cells producing the VSV G protein. This protein, which was not observed when the anti-MHV serum was used, apparently corresponds to the 62-kDa protein observed in the cell lysates. It most likely represents a soluble form of the VSV G (hybrid) protein that has been observed before in VSV-infected cells (20, 22). When precipitations on the media were performed in the absence of detergents, an intense background band was observed with a mass between 30 and 46 kDa. This background band was also observed when other antibodies were used.
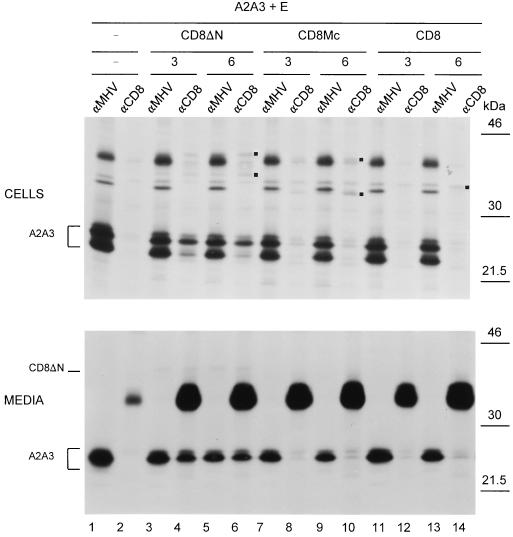
In order to study the effect of the ectodomain replacement in more detail, we also prepared two chimeric CD8 constructs (Fig. 1). In the CD8ΔN protein, the MHV M ectodomain was replaced by that of the CD8 protein, while in the CD8Mc protein, both ecto- and transmembrane domains were replaced, yielding a CD8 protein having the cytoplasmic domain of the M protein. It is of note that, while the VSV G protein naturally oligomerizes into noncovalently linked trimers, CD8 forms disulfide-linked dimers. Coexpression studies revealed that both the CD8ΔN and the CD8Mc proteins were deficient in VLP assembly (not shown). The genes encoding CD8ΔN and CD8Mc, as well as CD8, were each coexpressed with the genes coding for the A2A3 protein and the E protein, as before. Cell lysates and culture media were subjected to IP by using the anti-MHV serum and a monoclonal antibody to CD8 (OKT8; here designated anti-CD8). Analysis of the cell lysates (Fig. 5, top) showed that mutant protein A2A3 was well expressed in all combinations and not precipitated by the anti-CD8 antibody when coexpressed with the E protein only (lane 2). The expression levels of the CD8 (hybrid) proteins were very low compared to that of the A2A3 protein, but prolonged exposure times, necessary for their visualization, revealed that the different hybrid proteins were expressed similarly. Because these levels were much higher when the CD8 proteins were expressed singly, we assume that the effect is somehow caused by interference of the constructs. Cloning of the CD8 expression cassettes into another plasmid did not improve their expression levels. All CD8-derived proteins became glycosylated, which is indicative of their transport to the Golgi complex. Analysis in nonreducing gels demonstrated that both CD8 hybrid proteins occurred as dimers (not shown). The coIP assay revealed that the A2A3 protein was precipitated quite efficiently with the CD8ΔN protein (lanes 4 and 6), while its coprecipitation was dramatically decreased with the CD8Mc protein (lanes 8 and 10) and absent with the CD8 protein (lanes 12 and 14). The IPs on the culture media were again performed in the presence (lane αMHV) or in the absence (lane αCD8) of detergents. The analyses demonstrated that all combinations resulted in the production of VLPs (Fig. 5, bottom). Mutant protein CD8ΔN was clearly incorporated into VLPs as judged from coprecipitation of the M protein mutant (lanes 4 and 6). The fusion protein itself was indeed visible after prolonged exposure of the film to the gel (not shown). Both the CD8Mc and the CD8 proteins were incorporated very inefficiently, but due to the extreme sensitivity of the assay, some inclusion could be detected through the coIP of the A2A3 protein (lanes 10 and 14). Their levels of incorporation seemed to increase with higher levels of expression (Fig. 5 and data not shown).
FIG. 5.
Replacement of the MHV M ecto- and transmembrane domains by the corresponding domains of CD8. Genes encoding the M protein A2A3 and the E protein were expressed together or in combination with genes encoding either CD8, hybrid protein CD8Mc, or hybrid protein CD8ΔN, as described in the legend to Fig. 2. The amounts of the CD8 constructs transfected are indicated (in micrograms; −, absent). Cells were labeled for 3 h. The cells and media were collected separately and subjected to IP by using the anti-MHV serum (αMHV) or a monoclonal antibody directed against CD8 (αCD8). When IP on the medium was performed with the anti-CD8 antibodies, detergents were omitted to allow isolation of intact VLPs. The positions of the unglycosylated (CD8ΔN and CD8Mc) forms and of the fully glycosylated (CD8ΔN, CD8Mc, and CD8) forms of the CD8 (hybrid) proteins are indicated by black squares at the right of the lanes. The position of M protein A2A3 is also indicated. The molecular mass markers are at the right.
The results indicate that the ectodomain of MHV M can be replaced by the ectodomain of VSV G or that of CD8 without much loss of M-M interaction. These hybrid proteins were also incorporated into VLPs, the hybrid protein containing the CD8 ectodomain being more efficient than the one with the VSV G ectodomain. Additional substitution of the transmembrane domains, as in the CD8Mc protein, reduced the interaction with the M protein to background level: the extent of incorporation into VLPs was similar to that of the control protein CD8. The other control protein, VSV G, did not associate with MHV M protein and was not incorporated into VLPs.
M proteins interact in pre-Golgi compartments.
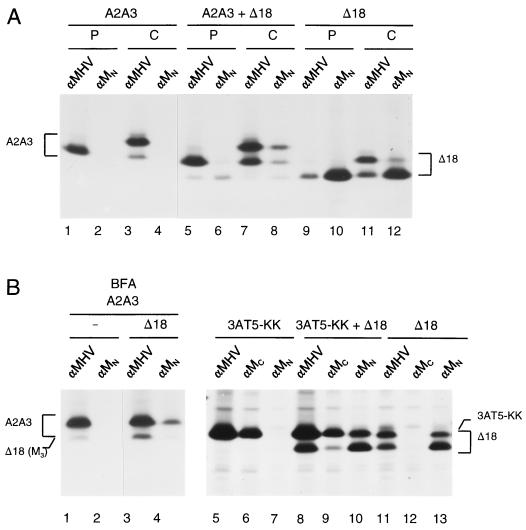
The results obtained with the ER-retained M proteins suggest that M proteins interact already in the ER. To further study the kinetics and first site of these interactions, we expressed the genes encoding the mutant M proteins A2A3 and Δ18 individually and together and carried out a pulse-chase experiment in which we labeled the cells for 15 min followed by a 105-min chase. As shown in Fig. 6A, both proteins were mainly present in their unglycosylated forms (M0) immediately after the labeling (lanes 1 and 9), while during the chase the slower-migrating (Golgi-modified) M protein species appeared (lanes 3 and 11). The analysis of the lysates from cells coexpressing the two proteins revealed that little association had occurred during the labeling period, as hardly any of the unglycosylated (pre-Golgi) A2A3 protein appeared in the precipitate prepared with the anti-MN antibody (lane 6). During the chase, a significant fraction of the proteins had associated, as illustrated by the efficient coIP of the glycosylated (Golgi-modified) A2A3 forms by the monoclonal antibody (lane 8).
FIG. 6.
M proteins interact in pre-Golgi compartments. Genes encoding the M proteins A2A3, Δ18, and 3AT5-KK were expressed as described in the legend to Fig. 2. The different combinations are indicated at the top. When indicated, BFA (6 μg/ml) was present from t = 3 h. Cells were pulse-labeled for 15 min (P) followed by a 105-min chase (C) (panel A) or were labeled for 2 h (panel B). The positions of the different proteins are indicated. −, absent.
Due to their similar electrophoretic mobilities, the glycosylated form of the Δ18 protein and the unglycosylated form of the A2A3 protein could not be discriminated in the gel. Hence, the pulse-chase experiment did not reveal whether M-M association starts in pre-Golgi compartments. In one approach to study this issue, we made use of BFA. This drug blocks the exit of newly synthesized proteins from the ER and causes a rapid distribution of Golgi enzymes to the ER (32, 33). In the presence of BFA, MHV M proteins are rapidly O glycosylated and completely converted into the M3 form (29). In the experiment shown in Fig. 6B (left), we again coexpressed the A2A3 and Δ18M proteins and performed labeling in the presence of BFA. The IPs with anti-MHV and anti-MN antibodies showed that the proteins had each been efficiently glycosylated. The A2A3 protein was not precipitated by monoclonal antibody anti-MN when expressed alone (lane 2) but was clearly coprecipitated with mutant protein Δ18, as was demonstrated by the prominent appearance of its glycosylated form (lane 4). Obviously, transport out of the ER is not a prerequisite for M proteins to associate.
In another approach, we took advantage of the availability of the mutant M protein 3AT5-KK (12). The replacement of the serine and threonine residues at positions 2, 3, and 4 by alanines in this M protein has destroyed the epitope recognized by the anti-MN antibody. Thus, only the threonine at position 5 of the amino-terminal hydroxyl amino acid cluster has remained, which appeared to be sufficient for the O glycosylation of the M protein. In addition to these changes, the 3AT5-KK polypeptide carries at its carboxy terminus the KKXX retrieval and retention signal, which we showed to be functional (12). We expressed the mutant M protein alone and together with the Δ18 M protein and performed a 2-h radiolabeling. For the IPs, we again used the anti-MHV and anti-MN antibodies, as well as a rabbit anti-peptide serum directed against the extreme carboxy terminus of MHV M (anti-Mc). As is clear from the analyses of the single expressions shown in Fig. 6B (right), the latter antiserum recognized the 3AT5-KK protein (lane 6) but, as predicted, not the truncated Δ18 protein (lane 12), while the converse was true for the anti-MN antibody (lane 7 and 13). Moreover, no trace of glycosylation of the 3AT5-KK protein was observed, demonstrating its tight ER retention. After coexpression of the two proteins, the anti-MC antibodies did precipitate the unglycosylated form of the truncated Δ18 protein, apparently as a result of an interaction with the other M protein in the ER. The apparent coIP of 3AT5-KK protein with Δ18 protein by the anti-MN antibodies supported this interpretation, although the picture was obscured by the comigration of the former protein with the glycosylated form of the truncated M protein.
DISCUSSION
The formation of progeny virions in coronavirus-infected cells involves two main processes, assembly of the helical nucleocapsids and of the viral envelopes. These processes are spatially separated, occurring in the cytoplasm and in intracellular membranes, respectively, and they apparently take place independently of each other. Obviously, the M protein is the key player in virion assembly, as it not only directs envelope formation but in addition provides the matrix to which the NC can attach for budding. The molecular interactions between the M molecules are most likely essential to the functioning of the M protein. Here we provide direct evidence for such interactions, which appear to occur through multiple contact sites and which generate a framework in the membrane from which foreign proteins are selectively excluded.
For the study of M-M interactions, we established two assays, both based on a mutant M protein named A2A3. The protein behaved like WT M in all relevant respects but was immunologically distinguishable due to the lack of an epitope caused by two subtle mutations. This property allowed its use as a reporter in the coIP assay after coexpression with other mutant M proteins. In the VLP incorporation assay, it enabled the sensitive detection of mutant M proteins coassembled into A2A3 protein-based particles. The two assays clearly demonstrated the existence of M-M interactions. In addition, the coprecipitation of relatively large amounts of reporter protein by only trace amounts of bait protein indicated the occurrence of large M protein complexes. This result confirmed our earlier observation obtained by sucrose-gradient analysis of singly expressed proteins (31). In these studies, we found that WT M protein accumulated in Golgi membranes in large heterogeneous complexes consisting of up to 40 M molecules. Somewhat smaller complexes appeared when a cytoplasmic tail truncation mutant was analyzed.
The involvement of the different domains of the M molecule in M-M interaction was investigated by evaluating several M protein mutants. Strikingly, mutant M proteins with deletions in the transmembrane domains, in the amphipathic domain, or in the carboxy-terminal hydrophilic tail or hybrid M proteins with heterologous ectodomains were still able to interact with the reporter M molecules, resulting in their incorporation into envelope particles. Only when all three transmembrane domains had been replaced by a heterologous transmembrane domain were interactions with A2A3 M protein and subsequent incorporation into particles severely reduced. Apparently, the M molecules interact with each other through multiple contact sites along the polypeptides. These sites may not be limited to the transmembrane region. For instance, while the replacement of the MHV M ectodomain by heterologous ectodomains hardly affected M-M interaction, involvement of this ectodomain in homotypic interactions cannot be excluded. Rather, such interactions have actually been demonstrated for the M protein of the human coronavirus 229E, where a cysteine residue in the short ectodomain gives rise to the formation of disulfide-linked homodimers (3). Thus, interactions (though generally noncovalent) between ectodomains may be a common feature of coronavirus M proteins.
An important conclusion from our observations is that M-M interactions are essential for coronavirus envelope assembly but that they are not sufficient. While the different mutant M proteins studied were each able to associate with the A2A3 protein, none of them was able by itself to assemble into particles when coexpressed with the E protein (reference 11 and data not shown). Obviously, additional requirements have to be met. We hypothesize that the full complement of interactions between the M molecules is required for efficient particle formation. Conceivably, the interactions at the various contact sites along the M polypeptides provide the free energy needed to generate and stabilize membrane curvature. In this respect, the E protein is unlikely to contribute significantly due to its numerical underrepresentation. In addition, the M proteins may need to interact with viral (E) and host proteins.
Associations between the M proteins appeared to take place in early (i.e., pre-Golgi) compartments. This is not surprising, since coronaviruses are assembled at the membranes of the IC (25, 28, 54). Consistently, the M protein has also been shown to engage in its interactions with the other viral membrane proteins (S and HE) in early compartments, most likely in the ER (13, 39, 40). The resulting higher-order complexes are thought to be maintained primarily by the M-M interactions (40). When expressed alone, the M protein accumulates in the Golgi compartment (25, 29, 46). However, coexpressed ER-retained mutant M proteins were found to interact with O-glycosylated—i.e., Golgi-modified—M molecules. This implies that M proteins recycle from the Golgi complex back to early compartments. Recycling of Golgi-resident membrane proteins is not without precedent. Both Golgi-resident glycosyltransferases (50) and proteins equipped with Golgi-targeting signals (10) have been shown to recycle through the ER. Furthermore, an inhibitor of sphingolipid synthesis shifted the steady-state distribution of infectious bronchitis virus M protein from the Golgi complex to the ER, suggesting that this M protein is at least in part localized by retrieval mechanisms (34). Interesting as this recycling process may be by itself, relocation of M proteins back to the ER and IC is probably functionally important in coronavirus-infected cells. Retrograde transport of escaped M molecules offers these proteins another opportunity to become assembled into progeny virions. In addition, recycling may provide a clearance mechanism to prevent saturation of the Golgi system with M molecules and subsequent impaired passage of progeny virions on their way out of the cell.
Our studies with the ER-retained M protein mutants not only show that the M proteins interact in early compartments; the observation that ER-retained M proteins can be rescued into VLPs also indicates that VLP budding can occur in these compartments, as is the case for coronavirions. It is not yet clear which factors determine the site of budding. Neither WT M proteins nor M-S complexes are retained by themselves in the budding compartment (25, 29, 40). A good candidate for controlling the site of budding is the E protein. When this protein is expressed independently, it appears to accumulate in membranes of the ER and IC (43). Thus, through its interaction with the M protein in infected cells, the E protein might be able to retain the M and M-S complexes in these early compartments where the viral particles are formed.
Sorting of membrane proteins plays an important role in the assembly of virus envelopes. In coronaviruses the membrane proteins S and HE are specifically incorporated into the budding particle via lateral interactions with M proteins (13, 39, 40, 56). Foreign membrane proteins seem to be efficiently excluded. The effective segregation of the VSV G and the EAV M proteins from the budding VLP indeed indicates that envelope assembly is a very selective process. This process is, however, not perfect, as was apparent from the low but detectable level of incorporation of CD8 molecules into virus particles. Inclusion of foreign membrane proteins has been shown before when MHV pseudotypes containing the murine leukemia virus envelope determinants were observed after propagation of MHV in cells persistently infected with the leukemia virus (60). Other enveloped viruses exhibit different sorting stringencies, in keeping with their mechanisms of assembly. Thus, retroviruses—dependent for budding only on the Gag protein—are not very selective against foreign membrane proteins, allowing incorporation of substantial amounts of proteins of host and viral origin (19, 55). In contrast, the extensive and specific interactions between the spikes themselves and with the NC during the budding of alphaviruses seem to leave little room for other proteins to steal into particles (19, 51).
An intriguing question that remains is how the selectivity in the incorporation of membrane proteins in coronaviruses is realized. Our working hypothesis is that the M proteins form a molecular matrix, a geometric framework in which vacancies occur at regular positions. Budding does not require that these vacancies be filled by proteins, but the positions can be taken by the S and/or HE proteins via specific interactions with M. Foreign membrane proteins generally will not associate with the M protein and will thus not be taken into the matrix. Nonspecific incorporation may occur, though rarely, by accidental fit, as we observed for the CD8 protein. Interestingly, while the S protein occurs in the form of trimers (14), the HE protein is present in disulfide-linked homodimeric form (for a review, see reference 6). This suggests that there may actually be two types of vacancies, one for each oligomeric structure. How the E protein finds its way into the viral envelope is still enigmatic.
Like coronaviruses, hepadnaviruses and flaviviruses exhibit an NC-independent budding mechanism, which in these cases leads to formation of subviral particles. Interactions between the envelope proteins have also been demonstrated for these viruses. The hepadnavirus small envelope S protein, the single requirement for subviral particle formation (41, 49), forms disulfide-linked oligomers (23). It was found that secretion-deficient mutant S proteins can either retain secretion-competent S protein in the cell (36, 42) or be rescued into secreted particles (7). For flaviviruses, heterodimer formation between the envelope proteins E and prM is required to allow assembly and secretion of subviral particles (1). Clearly, we are only beginning to tackle the many fundamental questions regarding the generation of all these viruses.
ACKNOWLEDGMENT
These investigations were supported by the Council for Chemical Sciences of the Netherlands Organization for Scientific Research (CW-NWO).
REFERENCES
- 1.Allison S L, Stadler K, Mandl C W, Kunz C, Heinz F X. Synthesis and secretion of recombinant tick-borne encephalitis virus protein E in soluble and particulate form. J Virol. 1995;69:5816–5820. doi: 10.1128/jvi.69.9.5816-5820.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Andersson H, Kappeler F, Hauri H P. Protein targeting to endoplasmic reticulum by dilysine signals involves direct retention in addition to retrieval. J Biol Chem. 1999;274:15080–15084. doi: 10.1074/jbc.274.21.15080. [DOI] [PubMed] [Google Scholar]
- 3.Arpin N, Talbot P J. Molecular characterization of the 229E strain of human coronavirus. Adv Exp Med Biol. 1990;276:73–80. doi: 10.1007/978-1-4684-5823-7_11. [DOI] [PubMed] [Google Scholar]
- 4.Baudoux P, Carrat C, Besnardeau L, Charley B, Laude H. Coronavirus pseudoparticles formed with recombinant M and E proteins induce alpha interferon synthesis by leukocytes. J Virol. 1998;72:8636–8643. doi: 10.1128/jvi.72.11.8636-8643.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bos E C, Luytjes W, van der Meulen H V, Koerten H K, Spaan W J. The production of recombinant infectious DI-particles of a murine coronavirus in the absence of helper virus. Virology. 1996;218:52–60. doi: 10.1006/viro.1996.0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brian D A, Hogue B G, Kienzle T E. The coronavirus hemagglutinin esterase glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 165–179. [Google Scholar]
- 7.Bruss V, Ganem D. Mutational analysis of hepatitis B surface antigen particle assembly and secretion. J Virol. 1991;65:3813–3820. doi: 10.1128/jvi.65.7.3813-3820.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cavanagh D. The coronavirus surface glycoprotein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 73–113. [Google Scholar]
- 9.Cheng R H, Kuhn R J, Olson N H, Rossmann M G, Choi H K, Smith T J, Baker T S. Nucleocapsid and glycoprotein organization in an enveloped virus. Cell. 1995;80:621–630. doi: 10.1016/0092-8674(95)90516-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cole N B, Ellenberg J, Song J, DiEuliis D, Lippincott-Schwartz J. Retrograde transport of Golgi-localized proteins to the ER. J Cell Biol. 1998;140:1–15. doi: 10.1083/jcb.140.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.de Haan C A M, Kuo L, Masters P S, Vennema H, Rottier P J M. Coronavirus particle assembly: primary structure requirements of the membrane protein. J Virol. 1998;72:6838–6850. doi: 10.1128/jvi.72.8.6838-6850.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Haan C A M, Roestenberg P, de Wit M, de Vries A A F, Nilsson T, Vennema H, Rottier P J M. Structural requirements for O-glycosylation of the mouse hepatitis virus membrane protein. J Biol Chem. 1998;273:29905–29914. doi: 10.1074/jbc.273.45.29905. [DOI] [PubMed] [Google Scholar]
- 13.de Haan C A M, Smeets M, Vernooij F, Vennema H, Rottier P J M. Mapping of the coronavirus membrane protein domains involved in interaction with the spike protein. J Virol. 1999;73:7441–7452. doi: 10.1128/jvi.73.9.7441-7452.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Delmas B, Laude H. Assembly of coronavirus spike protein into trimers and its role in epitope expression. J Virol. 1990;64:5367–5375. doi: 10.1128/jvi.64.11.5367-5375.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elroy-Stein O, Moss B. Cytoplasmic expression system based on constitutive synthesis of bacteriophage T7 RNA polymerase in mammalian cells. Proc Natl Acad Sci USA. 1990;87:6743–6747. doi: 10.1073/pnas.87.17.6743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischer F, Stegen C F, Masters P S, Samsonoff W A. Analysis of constructed E gene mutants of mouse hepatitis virus confirms a pivotal role for E protein in coronavirus assembly. J Virol. 1998;72:7885–7894. doi: 10.1128/jvi.72.10.7885-7894.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gallione C J, Rose J K. A single amino acid substitution in a hydrophobic domain causes temperature-sensitive cell-surface transport of a mutant viral glycoprotein. J Virol. 1985;54:374–382. doi: 10.1128/jvi.54.2.374-382.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Garoff H, Hewson R, Opstelten D-J E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garreis-Wabnitz C, Kruppa J. Intracellular appearance of a glycoprotein in VSV-infected BHK cells lacking the membrane-anchoring oligopeptide of the viral G-protein. EMBO J. 1984;3:1469–1476. doi: 10.1002/j.1460-2075.1984.tb01998.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Godeke G-J, de Haan C A M, Rossen J W A, Vennema H, Rottier P J M. Assembly of spikes into coronavirus particles is mediated by the carboxy-terminal domain of the spike protein. J Virol. 2000;74:1566–1571. doi: 10.1128/jvi.74.3.1566-1571.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Graeve L, Garreis-Wabnitz C, Zauke M, Breindl M, Kruppa J. The soluble glycoprotein of vesicular stomatitis virus is formed during or shortly after the translation process. J Virol. 1986;57:968–975. doi: 10.1128/jvi.57.3.968-975.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huovila A P, Eder A M, Fuller S D. Hepatitis B surface antigen assembles in a post-ER, pre-Golgi compartment. J Cell Biol. 1992;118:1305–1320. doi: 10.1083/jcb.118.6.1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jackson M R, Nilsson T, Peterson P A. Identification of a consensus motif for retention of transmembrane proteins in the endoplasmic reticulum. EMBO J. 1990;9:3153–3162. doi: 10.1002/j.1460-2075.1990.tb07513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klumperman J, Krijnse Locker J, Meijer A, Horzinek M C, Geuze H J, Rottier P J M. Coronavirus M proteins accumulate in the Golgi complex beyond the site of virion budding. J Virol. 1994;68:6523–6534. doi: 10.1128/jvi.68.10.6523-6534.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Konishi E, Pincus S, Paoletti E, Shope R E, Burrage T, Mason P W. Mice immunized with a subviral particle containing the Japanese encephalitis virus prM/M and E proteins are protected from lethal JEV infection. Virology. 1992;188:714–720. doi: 10.1016/0042-6822(92)90526-u. [DOI] [PubMed] [Google Scholar]
- 27.Kreis T E, Lodish H F. Oligomerization is essential for transport of vesicular stomatitis viral glycoprotein to the cell surface. Cell. 1986;46:929–937. doi: 10.1016/0092-8674(86)90075-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krijnse Locker J, Ericsson M, Rottier P J M, Griffiths G. Characterization of the budding compartment of mouse hepatitis virus: evidence that transport from the RER to the Golgi complex requires only one vesicular transport step. J Cell Biol. 1994;124:55–70. doi: 10.1083/jcb.124.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krijnse Locker J, Griffiths G, Horzinek M C, Rottier P J M. O-glycosylation of the coronavirus M protein. Differential localization of sialytransferases in N- and O-linked glycosylation. J Biol Chem. 1992;267:14094–14101. doi: 10.1016/S0021-9258(19)49683-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Krijnse Locker J, Rose J K, Horzinek M C, Rottier P J M. Membrane assembly of the triple-spanning coronavirus M protein; individual transmembrane domains show preferred orientation. J Biol Chem. 1992;267:21911–21918. doi: 10.1016/S0021-9258(19)36699-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Krijnse Locker J M, Opstelten D-J E, Ericsson M, Horzinek C, Rottier P J M. Oligomerization of a trans-Golgi/trans-Golgi network retained protein occurs in the Golgi complex and may be part of its retention. J Biol Chem. 1995;270:8815–8821. doi: 10.1074/jbc.270.15.8815. [DOI] [PubMed] [Google Scholar]
- 32.Lippincott-Schwartz J, Donaldson J G, Schweizer A, Berger E G, Hauri H P, Yuan L C, Klausner R D. Microtubule-dependent retrograde transport of proteins into the ER in the presence of brefeldin A suggests an ER recycling pathway. Cell. 1990;60:821–836. doi: 10.1016/0092-8674(90)90096-w. [DOI] [PubMed] [Google Scholar]
- 33.Lippincott-Schwartz J, Yuan L C, Bonifacino J S, Klausner R D. Rapid redistribution of Golgi proteins into the ER in cells treated with brefeldin A: evidence for membrane cycling from Golgi to ER. Cell. 1989;56:801–813. doi: 10.1016/0092-8674(89)90685-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Maceyka M, Machamer C E. Ceramide accumulation uncovers a cycling pathway for the cis-Golgi network marker, infectious bronchitis virus M protein. J Cell Biol. 1997;139:1411–1418. doi: 10.1083/jcb.139.6.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Maeda J, Maeda A, Makino S. Release of coronavirus E protein in membrane vesicles from virus-infected cells and E protein-expressing cells. Virology. 1999;263:265–272. doi: 10.1006/viro.1999.9955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mangold C M, Streeck R E. Mutational analysis of the cysteine residues in the hepatitis B virus small envelope protein. J Virol. 1993;67:4588–4597. doi: 10.1128/jvi.67.8.4588-4597.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mason P W, Pincus S, Fournier M J, Mason T L, Shope R E, Paoletti E. Japanese encephalitis virus-vaccinia recombinants produce particulate forms of the structural membrane proteins and induce high levels of protection against lethal JEV infection. Virology. 1991;180:294–305. doi: 10.1016/0042-6822(91)90034-9. [DOI] [PubMed] [Google Scholar]
- 38.Munro S. An investigation of the role of transmembrane domains in Golgi protein retention. EMBO J. 1995;14:4695–4704. doi: 10.1002/j.1460-2075.1995.tb00151.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nguyen V-P, Hogue B G. Protein interactions during coronavirus assembly. J Virol. 1997;71:9278–9284. doi: 10.1128/jvi.71.12.9278-9284.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Opstelten D-J E, Raamsman M J, Wolfs K, Horzinek M C, Rottier P J M. Envelope glycoprotein interactions in coronavirus assembly. J Cell Biol. 1995;131:339–349. doi: 10.1083/jcb.131.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Patzer E J, Nakamura G R, Simonsen C C, Levinson A D, Brands R. Intracellular assembly and packaging of hepatitis B surface antigen particles occur in the endoplasmic reticulum. J Virol. 1986;58:884–892. doi: 10.1128/jvi.58.3.884-892.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Prange R, Nagel R, Streeck R E. Deletions in the hepatitis B virus small envelope protein: effect on assembly and secretion of surface antigen particles. J Virol. 1992;66:5832–5841. doi: 10.1128/jvi.66.10.5832-5841.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Raamsman M J, Krijnse Locker J, de Hooghe A, de Vries A A F, Griffiths G, Vennema H, Rottier P J M. Characterization of the coronavirus MHV-A59 small membrane protein E. J Virol. 2000;74:2333–2342. doi: 10.1128/jvi.74.5.2333-2342.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rottier P J M. The coronavirus membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 115–139. [Google Scholar]
- 45.Rottier P J M, Horzinek M C, van der Zeijst B A M. Viral protein synthesis in mouse hepatitis virus strain A59-infected cells: effect of tunicamycin. J Virol. 1981;40:350–357. doi: 10.1128/jvi.40.2.350-357.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rottier P J M, Rose J K. Coronavirus E1 glycoprotein expressed from cloned cDNA localizes in the Golgi region. J Virol. 1987;61:2042–2045. doi: 10.1128/jvi.61.6.2042-2045.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schalich J, Allison S L, Stiasny K, Mandl C W, Kunz C, Heinz F X. Recombinant subviral particles from tick-borne encephalitis virus are fusogenic and provide a model system for studying flavivirus envelope glycoprotein functions. J Virol. 1996;70:4549–4557. doi: 10.1128/jvi.70.7.4549-4557.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Siddell S G. The small-membrane protein. In: Siddell S G, editor. The Coronaviridae. New York, N.Y: Plenum Press; 1995. pp. 181–189. [Google Scholar]
- 49.Simon K, Lingappa V R, Ganem D. Secreted hepatitis B surface polypeptides are derived from a transmembrane precursor. J Cell Biol. 1988;107:2163–2168. doi: 10.1083/jcb.107.6.2163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Storrie B, White J, Rottger S, Stelzer E H, Suganuma T, Nilsson T. Recycling of golgi-resident glycosyltransferases through the ER reveals a novel pathway and provides an explanation for nocodazole-induced Golgi scattering. J Cell Biol. 1998;143:1505–1521. doi: 10.1083/jcb.143.6.1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Strauss E G. Mutants of Sindbis virus. III. Host polypeptides present in purified HR and ts103 virus particles. J Virol. 1978;28:466–474. doi: 10.1128/jvi.28.2.466-474.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taguchi F, Fleming J O. Comparison of six different murine coronavirus JHM variants by monoclonal antibodies against the E2 glycoprotein. Virology. 1989;169:233–235. doi: 10.1016/0042-6822(89)90061-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tooze J, Tooze S A, Fuller S D. Sorting of progeny coronavirus from condensed secretory proteins at the exit from the trans-Golgi network of AtT20 cells. J Cell Biol. 1987;105:1215–1226. doi: 10.1083/jcb.105.3.1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tooze S A, Tooze J, Warren G. Site of addition of N-acetyl-galactosamine to the E1 glycoprotein of mouse hepatitis virus-A59. J Cell Biol. 1988;106:1475–1487. doi: 10.1083/jcb.106.5.1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremblay M J, Fortin J F, Cantin R. The acquisition of host-encoded proteins by nascent HIV-1. Immunol Today. 1998;19:346–351. doi: 10.1016/s0167-5699(98)01286-9. [DOI] [PubMed] [Google Scholar]
- 56.Vennema H, Godeke G-J, Rossen J W A, Voorhout W F, Horzinek M C, Opstelten D-J E, Rottier P J M. Nucleocapsid-independent assembly of coronavirus-like particles by co-expression of viral envelope protein genes. EMBO J. 1996;15:2020–2028. doi: 10.1002/j.1460-2075.1996.tb00553.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vennema H, Heijnen L, Zijderveld A, Horzinek M C, Spaan W J. Intracellular transport of recombinant coronavirus spike proteins: implications for virus assembly. J Virol. 1990;64:339–346. doi: 10.1128/jvi.64.1.339-346.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vennema H, Rijnbrand R, Heijnen L, Horzinek M C, Spaan W J. Enhancement of the vaccinia virus/phage T7 RNA polymerase expression system using encephalomyocarditis virus 5′-untranslated region sequences. Gene. 1991;108:201–209. doi: 10.1016/0378-1119(91)90435-E. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vogel R H, Provencher S W, von Bonsdorff C H, Adrian M, Dubochet J. Envelope structure of Semliki Forest virus reconstructed from cryo-electron micrographs. Nature. 1986;320:533–535. doi: 10.1038/320533a0. [DOI] [PubMed] [Google Scholar]
- 60.Yoshikura H, Taguchi F. Mouse hepatitis virus strain MHV-S: formation of pseudotypes with a murine leukemia virus envelope. Intervirology. 1978;10:132–136. doi: 10.1159/000148978. [DOI] [PubMed] [Google Scholar]