Abstract
Anthracnose, caused by the fungus Colletotrichum lindemuthianum, poses a significant and widespread threat to the common bean crop. The use of plant genetic resistance has proven to be the most effective strategy for managing anthracnose disease. The Amendoim Cavalo (AC) Andean cultivar has resistance against multiple races of C. lindemuthianum, which is conferred by the Co-AC gene. Fine mapping of this resistance gene to common bean chromosome Pv01 enabled the identification of Phvul.001G244300, Phvul.001G244400, and Phvul.001G244500 candidate genes for further validation. In this study, the relative expression of Co-AC candidate genes was assessed, as well as other putative genes in the vicinity of this locus and known resistance genes, in the AC cultivar following inoculation with the race 73 of C. lindemuthianum. Gene expression analysis revealed significantly higher expression levels of Phvul.001G244500. Notably, Phvul.001G244500 encodes a putative Basic Helix–Loop–Helix transcription factor, suggesting its involvement in the regulation of defense responses. Furthermore, a significant modulation of the expression of defense-related genes PR1a, PR1b, and PR2 was observed in a time-course experiment. These findings contribute to the development of improved strategies for breeding anthracnose-resistant common bean cultivars, thereby mitigating the impact of this pathogen on crop yields and ensuring sustainable bean production.
Keywords: candidate gene expression, common bean–anthracnose interaction, plant defense genes
1. Introduction
The common bean (Phaseolus vulgaris L.) holds the distinction of being the most widely consumed legume in human diets [1]. Known for its affordability, the common bean seeds serve as a crucial source of protein, dietary fiber, complex carbohydrates, and other essential nutrients, particularly for low-income populations in Africa and Latin America [2,3]. However, the productivity and quality of common bean crops are significantly threatened by Colletotrichum lindemuthianum (Sacc. and Magnus) Briosi and Cavara, a hemibiotrophic ascomycete fungus that causes anthracnose (ANT) [4,5]. This pathogen represents one of the most severe, widespread, and recurring threats to the common bean cultivation. Under favorable environmental conditions, ANT can lead to reduced seed quality and significant yield losses [5,6].
Efforts to control C. lindemuthianum have primarily relied on genetic resistance, as the pathogen exhibits high genetic variability that challenges conventional breeding programs [4,5]. Unfortunately, the pathogen has shown a remarkable ability to overcome cultivated plant resistance through coevolution, rendering previously resistant cultivars to become susceptible over time [7,8]. The use of resistant cultivars remains the most effective and environmentally friendly approach for managing C. lindemuthianum in common bean cultivation [9]. These cultivars offer a cost-effective and user-friendly solution. However, developing cultivars that are resistant to the diverse range of physiological races of C. lindemuthianum poses significant challenges [7].
Anthracnose (ANT) resistance in common beans is conferred by independent loci known as ‘Co.’ These resistance loci have been mapped to various chromosomes of the common bean genome, often clustering in disease-resistance regions [10]. Among the identified resistance loci in the Andean genetic pool, many have been mapped to chromosome Pv01. Notable alleles include Co-1, Co-12, Co-3, Co-14, Co-15 and Co-1HY at the Co-1 locus [11,12,13,14], Co-x [15,16], Co-Pa [17], CoPv01CDRK [18], and Co-AC [19], all located at the end of Pv01.
The identification and molecular characterization of novel resistance genes plays a crucial role in breeding for disease resistance [20,21]. Gene expression analysis has proven to be a valuable tool in understanding the resistance response of common bean genotypes SEL 1308 and T9576R to the race 73 of C. lindemuthianum [22,23]. Several defense-related genes, including those encoding pathogenesis-related proteins (PR) such as PR1a, PR1b, and PR2, were found to be spatially and temporally induced by the pathogen [22,23,24]. Notably, a potential gene associated with the Co-12 locus was identified [23]. Similarly, changes in the expression levels of PR1a, PR1b, and PR2 genes were reported during incompatible interactions with race 2 of C. lindemuthianum [25].
In the evaluation of expression levels of candidate genes for the Co-12 allele of the Co-1 gene in response to race 73 of C. lindemuthianum, the gene Phvul.001G243800 exhibited high induction, suggesting its potential as a candidate gene for the Co-12 allele of the Co-1 gene [23]. Additionally, through fine mapping in the Hongyundou genotype, four candidate genes were identified for the Co-1HY allele. Among them, Phvul.001G243600 and Phvul.001G243700 showed higher induction in response to race 81 of C. lindemuthianum, indicating their potential as candidate genes for the Co-1HY locus [13]. Moreover, in a genetic study focusing on the anthracnose resistance Co-x gene, a gene called KTR2/3 was found within a CRINKLY4 kinase cluster located between the Phvul.001G243600 and Phvul.001G243700 genes. Gene expression analysis revealed that KTR2/3 was induced by strain 100 (race 3993) of C. lindemuthianum, and its transient expression in susceptible genotype BAT93 resulted in increased resistance to the pathogen [16]. The authors suggested that this gene may act as a decoy involved in indirectly recognizing fungal effectors [16]. These findings highlight the significance of gene expression analysis in uncovering potential resistance genes and their roles in the common bean’s defense response to C. lindemuthianum.
Through fine mapping, five candidate genes for the CoPv01CDRK locus, which confer resistance to anthracnose and angular leaf spot in the CDRK common bean cultivar, were identified [18]. Among these candidate genes, gene expression analysis revealed that Phvul.001G246300, potentially encoding an Abscisic Acid Receptor (PYL5), exhibited the highest responsiveness to both pathogens, making it the primary candidate gene for the CoPv01CDRK locus [26].
Another noteworthy common bean cultivar, AC, is an Andean landrace collected in the state of Santa Catarina, Brazil [19]. AC possesses the Co-AC gene that confers resistance to races 2, 7, 9, 19, 23, 39, 55, 65, 73, 89, 1545, 2047, and 3481 of C. lindemuthianum [27]. Through fine mapping, the Co-AC gene was located at the end of the Pv01 chromosome within a genomic region containing three candidate genes: Phvul.001G244300, Phvul.001G244400, and Phvul.001G244500 [19].
Candidate genes located within the Co-AC loci on Pv01 demonstrate unique expression patterns when inoculated with race 73 of C. lindemuthianum in the AC cultivar. The primary aim of this study was to investigate the dynamic expression profiles of Co-AC candidate genes Phvul.001G244300, Phvul.001G244400, and Phvul.001G244500 within the AC cultivar upon exposure to C. lindemuthianum race 73. The present study hypothesizes that each of the candidate genes that overlap with the Co-AC loci on Pv01 exhibits distinct expression patterns in response to inoculations with race 73 of C. lindemuthianum in the AC cultivar. The objective of this study was to investigate the relative expression patterns of the Co-AC candidate genes and other disease-resistance genes in the AC cultivar in response to C. lindemuthianum race 73, employing quantitative real-time PCR. Specifically, the focus was to gain insights into their potential roles in the plant’s defense mechanisms against this pathogen, contributing to a deeper understanding of disease resistance in common beans.
2. Results
2.1. Phenotypic Evaluation of the Cultivars
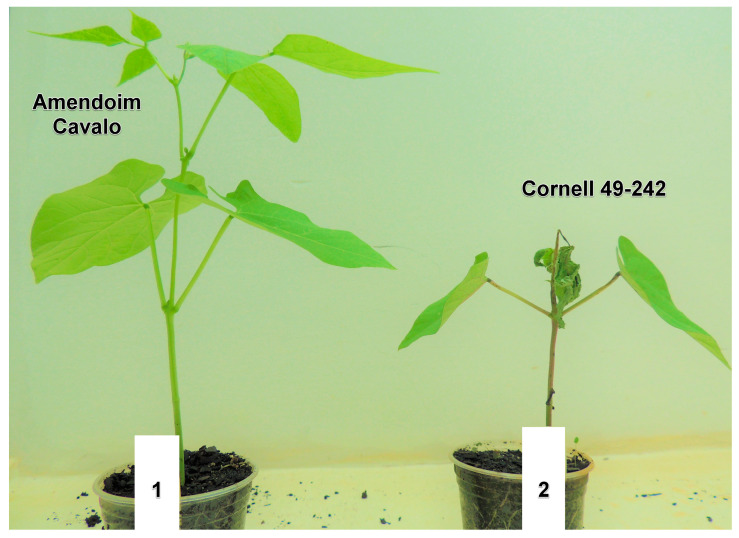
Inoculation of C. lindemuthianum race 73 on the resistant cultivar AC and the susceptible cultivar Cornell 49-242 resulted in disease development exclusively in the susceptible cultivar (Figure 1). Disease symptoms manifested as small water-soaked lesions on the underside of the leaves and small sunken lesions on the stems, ultimately leading to plant mortality. Notably, symptom expression occurred only after 72 h post-inoculation (hpi), indicating the hemibiotrophic nature of the fungus. Conversely, no symptoms or hypersensitive responses were observed in the resistant cultivar.
Figure 1.
Disease reaction of the resistant cultivar Amendoim Cavalo (1) and susceptible cultivar Cornell 49-242 (2) after 72 h post-inoculation with race 73 of Colletotrichum lindemuthianum.
2.2. Differential Expression of Candidate in the Amendoim Cavalo Cultivar Inoculated with Race 73 of C. lindemuthianum
Aiming to identify the molecular mechanisms underlying Co-AC resistance, the relative expression of the following candidate genes was assessed: KTR2/3, Phvul.001G243800, Phvul.001G244300, Phvul.001G244400, Phvul.001G244500, Phvul.001G245300, and Phvul.001G246300. Additionally, the expression of defense genes PR1a, PR1b, and PR2 were evaluated as markers for resistance upon C. lindemuthianum race 73 inoculation. These evaluated genes with functional annotation are described in Table S1.
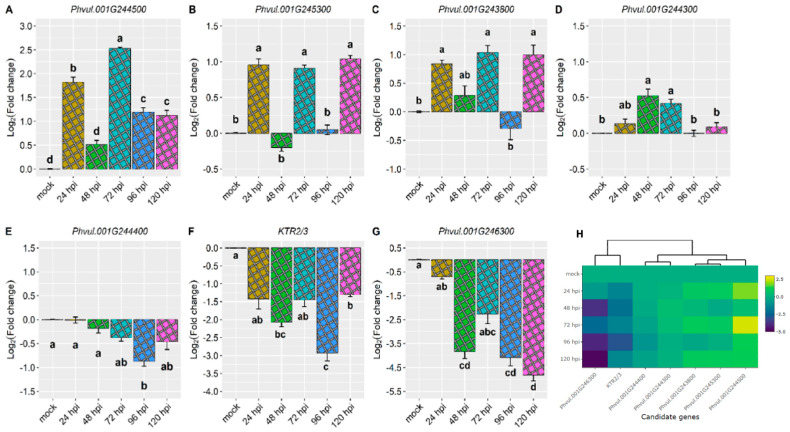
Phvul.001G244500 exhibited the most significant response to the pathogen, showing a 2.5-fold change at 72 h post-inoculation (hpi). Additionally, an approximately 1.8-fold increase was observed at 24 hpi, while increases higher than 1.0-fold were observed at 96 and 120 hpi (Figure 2A,H and Table 1). The gene Phvul.001G245300 displayed the second-highest response to the pathogen, being induced at 24, 72, and 120 hpi with an average increase of 0.90-fold (Figure 2B,H and Table 1). The gene Phvul.001G243800 was also induced at 24, 72, and 120 hpi, albeit with a relatively small average increase of 0.9-fold (Figure 2C,H and Table 1).
Figure 2.
Relative expression of candidate genes (A) Phvul.001G244500, (B) Phvul.001G245300, (C) Phvul.001G243800, (D) Phvul.001G244300, (E) Phvul.001G2444400, (F) KTR2/3, (G) Phvul.001G246300 in the Amendoim Cavalo at 24, 48, 72, 96, and 120 h post-inoculation (hpi) with the race 73 of C. lindemuthianum and mock. The results are presented as logarithmic base 2 of the fold change of gene expression. Means with the same letter for each gene are not significantly different at the 5% significance level, using the Alexander–Govern test. (H) Heatmap of the relative expression of candidate genes for the Co-AC and genes proximal to this locus in the Amendoim Cavalo cultivar. Yellow shading indicates higher expression, and dark blue shading has lower expression than reference genes.
Table 1.
Summary table of mean relative gene expression (Log2(fold change)) of Co-AC candidate genes and pathogenesis-related genes in response to C. lindemuthianum race 73 in Amendoim Cavalo cultivar.
| Gene | Gene Model | C. lindemuthianum Race 73 | ||||
|---|---|---|---|---|---|---|
| 24 hpi | 48 hpi | 72 hpi | 96 hpi | 120 hpi | ||
| Co-x | KTR2/3 | −1.4 | −2.1 | −1.4 | −2.9 | −1.3 |
| Co-1 | Phvul.001G243800 | 0.8 | 0.3 | 1.0 | −0.3 | 1.0 |
| Co-AC | Phvul.001G244300 | 0.1 | 0.5 | 0.4 | 0.0 | 0.1 |
| Phvul.001G244400 | 0.0 | −0.2 | −0.4 | −0.9 | −0.5 | |
| Phvul.001G244500 | 1.8 | 0.5 | 2.5 | 1.2 | 1.1 | |
| CoPv01CDRK | Phvul.001G245300 | 1.0 | −0.2 | 0.9 | 0.0 | 1.0 |
| Phvul.001G246300 | −0.7 | −3.8 | −2.3 | −4.1 | −4.8 | |
| Pathogenesis-related genes | Phvul.003G109100 (PR1a) | −0.9 | −1.6 | 1.4 | 2.3 | 1.8 |
| Phvul.006G196900 (PR1b) | −1.6 | −2.0 | 1.4 | 3.8 | 3.9 | |
| Phvul.009G256400 (PR2) | −1.2 | −1.0 | 0.6 | 1.2 | 1.8 | |
The expression of the Phvul.001G244300 gene was induced by the pathogen at 48 and 72 hpi, showing a small increase of 0.4-fold (Figure 2D,H and Table 1). Conversely, the Phvul.001G244400 gene was downregulated only at 96 hpi, with a reduction of 0.9-fold (Figure 2E,H and Table 1). The KTR2/3 and Phvul.001G246300 genes exhibited downregulation at 48, 96, and 120 hpi (Figure 2F–H and Table 1).
The Phvul.001G244500 gene was the most responsive candidate gene to the pathogen, particularly at 72 hpi. Additionally, the Phvul.001G246300 candidate gene for CoPv01CDRK and the KTR2/3 candidate gene for Co-x were significantly downregulated in the AC cultivar upon inoculation with the race 73 of C. lindemuthianum. This suggests that different candidate genes are expressed in each anthracnose-resistant cultivar (Figure 2 and Table 1).
2.3. Expression Profile of Defense Genes in the Amendoim Cavalo Cultivar Inoculated with Race 73 of C. lindemuthianum
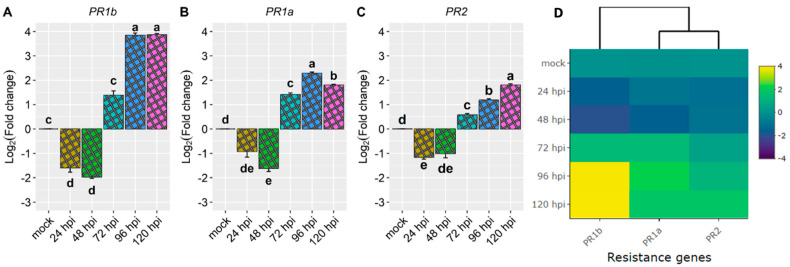
Concerning the PR genes, namely PR1b (Phvul.006G196900), PR2 (Phvul.009G256400), and PR1a (Phvul.003G109100), their induction was observed only starting from 72 h post-inoculation (hpi), with PR1b displaying notable activation between 96 and 120 hpi (refer to Figure 3 and Table 1). These findings suggest that the initiation of the resistance response may be attributed to the candidate gene Phvul.001G244500. Furthermore, by 72 hpi, additional defense genes appear to contribute to the activation of the resistance response.
Figure 3.
Relative expression of plant defense genes (A) Phvul.006G196900 (PR1b), (B) Phvul.003G109100 (PR1a), and (C) Phvul.009G256400 (PR2) in the common bean cultivar Amendoim Cavalo at 24, 48, 72, 96, and 120 h post-inoculation (hpi) with the race 73 of C. lindemuthianum and mock. The results are presented as logarithmic base 2 of the fold change of gene expression. Means with the same letter for each gene are not significantly different at the 5% significance level, using the Alexander–Govern test. (D) Heatmap of the relative expression of plant defense genes Phvul.006G196900 (PR1b), Phvul.003G109100 (PR1a), and Phvul.009G256400 (PR2) in the common bean cultivar Amendoim Cavalo Yellow shading indicates higher expression and dark blue shading lower expression than that of reference genes.
Among them, PR1b (Phvul.006G196900) stood out as the most responsive to the pathogen, exhibiting a substantial increase in expression from 72 to 96 hpi and a remarkable 3.9-fold increase at 120 hpi (Figure 3A,D and Table 1). The gene PR1a (Phvul.003G109100) showed a moderate level of expression and response upon exposure to the pathogen. Remarkably, there was a 2.3-fold increase in expression observed at 96 hpi (Figure 3B,D and Table 1). The expression pattern of the PR2 (Phvul.009G256400) gene mirrored that of PR1a. It experienced downregulation at 24 and 48 hpi, with a one-fold reduction, but demonstrated increased expression levels at 72, 96, and 120 hpi with fold changes of 0.6, 1.2, and 1.8, respectively (Figure 3C,D and Table 1).
3. Discussion
Gene expression analysis plays a crucial role in understanding the genetic basis of disease resistance and can aid in the identification of effective resistance genes for plant breeding and molecular studies within specific pathosystems. In this study, the resistance response to C. lindemuthianum in the AC cultivar was investigated, focusing on the relative expression of candidate genes associated with resistance genes, namely Co-AC [19], CoPv01CDRK [18], Co-x [15,16], and the Co-12 allele for the Co-1 locus [23] (Figure 2). Additionally, the relative expression of disease-resistance genes PR1a, PR1b, and PR2 was examined in the same pathosystem (Figure 3).
This study focused on a genomic region spanning 250 Kb at the end of Pv01, which encompasses the candidate genes for Co-AC and potential genes closely linked to this locus (Figure S1). Notably, distinct expression patterns among the candidate genes of the Co-AC resistance gene in the AC common bean cultivar were observed. Among these candidate genes (Phvul.001G244300, Phvul.001G244400, and Phvul.001G244500), Phvul.001G244500 displayed the most pronounced responsiveness to race 73 of C. lindemuthianum, particularly at 72 hpi, with a 2.5-fold change in gene expression (Figure 2 and Figure 3 and Table 1).
These findings highlight the potential role of the Phvul.001G244500 gene, which encodes a Basic Helix–Loop–Helix (bHLH) transcription factor, in regulating defense processes against C. lindemuthianum race 73. Proteins containing the Basic Helix–Loop–Helix domain are known to regulate the expression of their target genes, which are involved in many physiological processes and have a broad range of functions in biosynthesis, metabolism, and transduction of plant hormones [28]. Newly, bHLHs were observed differentially expressed in rose petals upon disease infection, suggesting candidate genes that regulate the response of rose plants to Botrytis cinerea [29].
Unlike the robust induction of Phvul.001G244500 in the AC cultivar, a previous study showed a low expression pattern of this candidate gene in the California Dark Red Kidney (CDRK) cultivar inoculated with C. lindemuthianum race 73. Lovatto [26] et al. (2023) reported that Phvul.001G244500 displayed only a slight induction at 120 hpi, with less than a one-fold change in gene expression. Phvul.001G244500 is a strong candidate for the Co-AC resistance gene in the AC cultivar, while it was not responsive in the CDRK cultivar.
Phvul.001G245300 expression in AC cultivar was the second most induced gene, although at lower levels, with approximately 1.0-fold change at 24, 72, and 120 hpi. This observation suggests that Phvul.001G245300 may function in a secondary layer of the resistance response. Phvul.001G245300 encodes a putative Leucine-Rich Repeat Protein Kinase (LRR-Kinase), and proteins encoding LRR and Kinase domains are known to be expressed by resistance genes [10].
The AC cultivar Phvul.001G243800 showed only minor increases in expression levels at 24, 72, and 120 hpi (approximately one-fold change). In contrast, the Phvul.001G244500 gene exhibited a substantial 2.5-fold change in gene expression (Figure 2 and Figure 3 and Table 1). These results suggest that Phvul.001G243800 has a limited effect on the resistance response in the AC cultivar. The Phvul.001G243800 gene, which is a candidate for the Co-12 allele of the Co-1 locus, was found to be highly induced at 72 hpi in the T9576R genotype inoculated with race 73 of C. lindemuthianum [23]. Therefore, besides the allelism test, this candidate gene expression study corroborates that different genes confer resistance to the same pathogen in each cultivar once each resistant cultivar expresses high levels of different candidate genes.
The present study demonstrated a different pattern for the KTR2/3 gene in the AC cultivar inoculated with race 73 of C. lindemuthianum. It was consistently downregulated at 48, 96, and 120 hpi, indicating that this gene may not trigger the resistance response in this specific pathosystem. Conversely, the KTR2/3, a candidate gene for the Co-x gene, was upregulated at 24 hpi in the JaloEEP558 cultivar [16]. Again, this corroborates that different genes confer resistance to the same pathogen in each cultivar.
In this study, the Phvul.001G246300 gene in the AC cultivar was downregulated, suggesting that it may not be the responsive resistance gene in this specific pathosystem. On the other hand, in the CDRK cultivar, the Phvul.001G246300 gene showed the highest responsiveness to both C. lindemuthianum race 73 and P. griseola race 63-39, indicating that the CoPv01CDRK resistance gene confers resistance to both diseases in common bean. Therefore, the expression levels of candidate genes of both resistant cultivars indicate that different genetic resistances are involved in each cultivar.
Taken together, it is possible to identify distinct roles of the genes Phvul.001G244500, Phvul.001G243800, KTR2/3, and Phvul.001G246300 involved in the resistance response to race 73 of C. lindemuthianum in different common bean cultivars. The contrasting expression patterns emphasize the complexity of resistance mechanisms and highlight the importance of the candidate gene Phvul.001G244500 for Co-AC in conferring robust resistance.
PR proteins are induced by phytopathogens as well as defense-related signaling molecules. They are the key components of the plant’s innate immune system, especially systemic acquired resistance (SAR), and are widely used as diagnostic molecular markers of defense signaling pathways [30]. Plant resistance to pathogens involves the activation of genes encoding PR proteins, which are categorized into 17 families and are known to accumulate following pathogen infection in various plant species [31]. PR1 genes, a subset of the PR family, are commonly used as markers for systemic acquired resistance [32]. However, the understanding of PR1 genes remains limited, with only a small proportion having been studied thus far [33].
In this current investigation, it was noted that among the tested defense resistance genes, PR1b displayed the highest level of responsiveness to race 73 of C. lindemuthianum in the AC cultivar, particularly evident at 96 and 120 hpi (refer to Figure 3). Interestingly, PR1b also exhibited the most conspicuous induction in the CDRK cultivar upon inoculation with race 73 of C. lindemuthianum, predominantly at 120 hpi [25].
Theoretical considerations propose that PR1b encodes a PR1-like protein, typically secreted into the extracellular spaces of plant leaves, as a response to pathogen infection [34]. In Arabidopsis thaliana, a homolog of PR1b plays a crucial role in defense responses against necrotrophic pathogens, mediated by methyl jasmonate and ethylene while being repressed by salicylic acid [35].
In this investigation, PR1a demonstrated significant upregulation in the AC cultivar when inoculated with race 73 of C. lindemuthianum, particularly peaking between 72 and 120 hpi, with a notable zenith at 96 hpi (refer to Figure 3 and Table 1). Likewise, in the CDRK cultivar subjected to the same inoculation, PR1a exhibited its highest induction at 72 and 96 hpi [26]. The upregulation of PR1a was also evident in the SEL 1308 cultivar following inoculation with race 73 of C. lindemuthianum [22,24], as well as in the T9576R common bean when exposed to race 73 of C. lindemuthianum [23], and in the ‘Naz’ cultivar during inoculation with race 2 of C. lindemuthianum [25].
PR1a is postulated to encode a PR protein featuring the Bet v I domain [36]. A recent transcriptome study, delving into the incompatible interaction between strain C531 and the BAT93 cultivar, underscored the pivotal role of PR10/Bet v I in conferring disease resistance in common beans [37].
In this study involving the AC cultivar inoculated with race 73 of C. lindemuthianum, the expression of PR2 exhibited a modest repression at 24 hpi, followed by a substantial induction from 72 hpi, reaching a notable peak at 120 hpi. Similarly, in the CDRK cultivar, PR2 displayed an upregulation in response to race 73 of C. lindemuthianum, particularly between 72 and 96 hpi [26]. Examining the SEL 1308 cultivar, PR2 was also observed to be upregulated following inoculation with race 73 of C. lindemuthianum [22,24]. In the case of the Naz cultivar, known for its resistance to race 2 of C. lindemuthianum, an upregulation of PR2 was noted from 48 hpi onwards [25]. In the T9576R genotype subjected to inoculation with race 73 of C. lindemuthianum, PR2 exhibited upregulation at all evaluated time points except at 96 h post-inoculation [23].
Hypothetically, it is suggested that PR2 encodes a 1,3-beta-glucan endohydrolase [38]. Consequently, PR2 may play a pivotal role in the resistance response by actively degrading fungal cell walls, potentially triggering a plant’s pattern-triggered immunity (PTI) [39,40].
Gene expression analysis offers valuable insights into the role and interaction of these genes in mounting an effective resistance response. Moreover, it provides additional knowledge necessary for the identification of promising genes for utilization in plant breeding programs. The most expressed candidate genes can be validated by phenotypic evaluation of the same cultivar with candidate gene knockout and ultimately developing molecular markers for enhanced selection and incorporation of effective genes into breeding strategies.
It is crucial to emphasize that the Andean AC cultivar is resistant to 13 out of the 15 Colletotrichum lindemuthianum races assessed [19]. Additionally, this cultivar exhibits commendable agronomic traits. Previous studies conducted by Vidigal Filho and colleagues (2020) [3] demonstrated that the same cultivar displayed a broad spectrum of resistance, covering four distinct races of C. lindemuthianum.
These findings contribute valuable knowledge regarding the genetic mechanisms underlying resistance to C. lindemuthianum in the AC cultivar and provide insights into potential genes involved in the resistance response to anthracnose in common beans. These results have implications for future research and can aid in the development of effective strategies for anthracnose resistance in common bean breeding programs. The characterization of a specific candidate gene, Phvul.001G244500, broadens the understanding of the defense networks activated in response to pathogen infection. Additionally, PR1a, PR1b, and PR2 would play significant roles in the defense responses of different common bean cultivars against C. lindemuthianum.
4. Conclusions
The observed upregulation of candidate genes in the incompatible interaction may signify the host’s response to counterbalance the compromised resistance caused by pathogen infection. This research has successfully pinpointed the candidate gene Phvul.001G244500 as an effective defense mechanism against the specific race 73 of C. lindemuthianum. Moreover, the involvement of defense genes PR1a, PR1b, and PR2 in the resistance response was demonstrated, with a particular emphasis on PR1b. This study has yielded invaluable insights into the genetic underpinnings of resistance to C. lindemuthianum race 73 within the AC cultivar. These discoveries significantly bolster our capacity to develop more effective strategies for breeding anthracnose-resistant common bean cultivars. By incorporating this resistance gene into breeding programs, these findings can enhance the resilience of common bean crops against this devastating pathogen, contributing to sustainable agriculture and food security.
5. Materials and Methods
5.1. Plant Material and Growth Conditions
The experiment was performed in a completely randomized design. Seedlings of the resistant AC and the susceptive Cornell 49-242 cultivars were inoculated with race 73 of C. lindemuthianum, and relative expression of ten genes only in AC cultivar was evaluated at 24, 48, 72, 96, and 120 hpi and in the mock. Three biological replicates (plants) were collected for each experimental condition evaluated, and for each biological replicate, three technical replicates (qPCR reactions) were performed in each experiment. The experiment was conducted at the Núcleo de Pesquisa Aplicada à Agricultura (Nupagri) at the Universidade Estadual de Maringá (UEM) in Maringá, Paraná, Brazil (latitude 23° 26′8″ S, longitude 51° 53′42″ W). Briefly, seeds were planted in plastic trays filled with a commercial substrate, MecPlant (MEC PREC—Ind. Com Ltd., Telemaco Borba, Brazil), that had been previously sterilized and fertilized. The seedlings were grown in greenhouses under natural light at a temperature of 25 °C until the first trifoliate leaf growth stage [8].
5.2. Pathogenesis Assay
Monosporic cultures of C. lindemuthianum were prepared following the methodologies described by Mathur et al. [41]. The inoculum was produced on a medium comprising green common bean pods incubated at 22 ± 2 °C without light for 14 days. Conidiospore quantification was conducted using a hemacytometer under an optical microscope. The plants were sprayed with a conidiospore suspension prepared in distilled water and Tween 20® (0.01%) at an approximate concentration of 1.2 × 106 mL−1. Inoculation was carried out by spraying the suspension onto the plants using a manual pressurized pump sprayer. For the negative control (mock), plants were sprayed only with distilled water and Tween 20® (0.01%). After inoculation, the plants were placed in a mist chamber at a temperature of 22 ± 2 °C, a photoperiod of 12 h, and ≥95% relative humidity for 72 h. Subsequently, the plants were transferred to a growth chamber with a temperature of 22 ± 2 °C and a photoperiod of 12 h for the duration of the experiment. Anthracnose symptoms were evaluated using the 1-to-9 disease severity scales proposed by Pastor-Corrales et al. [8]. Plants with disease reaction scores between 1 and 3 were considered resistant, whereas plants with scores from 4 to 9 were considered susceptible.
5.3. RNA Extraction
For sample collection and total RNA extraction, leaf samples weighing approximately 100 ± 10 mg from the AC cultivar inoculated with race 73 of C. lindemuthianum were collected at 24, 48, 72, 96, and 120 h post-inoculation (hpi), as well as from the mock control. The leaf samples were immediately submerged in liquid nitrogen (N2) and stored at −80 °C until further processing. Total RNA extraction was performed by macerating the tissue and adding 1000 µL of TRIzol® (Invitrogen™, Waltham, MA, USA) to each microtube. The subsequent steps for RNA extraction and isolation followed the manufacturer’s recommendations. The precipitated total RNA pellets were washed with 70% ethanol (EtOH) and then suspended in RNase-free H2O.
The integrity of the total RNA was assessed by electrophoresis on a 1% m/v agarose gel, run for 80 min at 80 volts, at 5 °C, and in the absence of light. To assess the quality and quantity of total RNA, a spectrophotometer (FEMTO 700STM) was used to measure absorbance at 230 nm, 240 nm, 260 nm, and 280 nm. The following absorbance ratios were used to determine RNA purity: A260/A230 between 1.9 and 2.4, A260/A240 ≥ 1.4, and A260/A280 between 1.8 and 2.2. The concentration of total RNA was calculated using the formula [RNA] (ng µL−1) = A260nm × 40 × 100 [42]. Total RNA samples that met the purity criteria and exhibited no visual signs of degradation were subjected to DNase I treatment using DNase ITM (Invitrogen™, Waltham, MA, USA) to remove any residual DNA. The purification reaction was carried out using 1 µg of total RNA, following the manufacturer’s instructions.
5.4. Reverse Transcription (cDNA Synthesis)
The cDNA synthesis was carried out using the ‘Superscript® IV First-Strand Synthesis System’ kit (Invitrogen™, Waltham, MA, USA) following the manufacturer’s protocol. The total volume of cDNA synthesis reaction was 20 µL with the following components: 1 µg of total RNA, primer-oligo d(T) (2.5 µM), dNTP mix (0.5 mM each), First-Strand Buffer (1X), DL-dithiothreitol (5 mM), ribonuclease inhibitor (2 U µL−1), MMLV-RT (10 U µL−1) and RNase-free water. Initially, total RNA, primer-oligo d(T), dNTP mix, and RNase-free water (to 13 µL) were added to the reaction. The samples were incubated in a thermocycler (Applied Biosystems® Veriti® 96-Well Fast Thermal Cycler, Waltham, MA, USA) at 65 °C for 5 min, followed by 4 °C for 1 min. Then, the First-Strand Buffer, DL-dithiothreitol, ribonuclease inhibitor, and 1 MMLV-RT were added to the reaction. The samples were incubated at 55 °C for 10 min for cDNA synthesis activation, followed by 80 °C for 10 min to inactivate the reaction. To remove residual RNA after cDNA synthesis, 1 µL of Escherichia coli RNase H was added, and the samples were incubated at 37 °C for 20 min. The cDNA synthesis product (20 µL) was diluted 1:100 for qPCR analysis. To assess the cDNA synthesis efficiency, positive control was included, in which HeLa-S3 RNA (10 ng) was used instead of total RNA.
For the control of cDNA synthesis, the PCR reaction was conducted using the following components: 5 µL of PCR buffer (10X), 2 µL of MgCl2 (50 mM), 1 µL of dNTP Mix (10 mM), 1 µL of sense primer (10 µM), 1 µL of antisense primer (10 µM), 2 µL of cDNA for the positive control, and 2 µL of ultrapure H2O for the negative control. Additionally, 0.2 µL of Taq PlatinumTM DNA polymerase (Invitrogen™, Waltham, MA, USA) and 37.8 µL of ultrapure H2O were included in the reaction mixture. The PCR amplification was performed under the following thermocycling conditions: an initial denaturation step at 94 °C for 2 min, followed by 35 cycles of denaturation at 94 °C for 15 s, annealing at 55 °C for 30 s, and synthesis at 68 °C for 1 min. After completion of the PCR reaction, both the positive and negative controls were subjected to electrophoretic analysis using a 1.5% (w/v) agarose gel. The expected fragment size of approximately 353 bp was observed in the positive control lane, confirming the successful amplification, while no fragment was detected in the negative control lane, validating the absence of non-specific amplification as indicated by the manufacturer’s instructions.
5.5. Target Genes and Primer Design
The candidate genes selected for expression analysis in the AC cultivar were associated with resistance to race 73 of C. lindemuthianum. Specifically, the genes Phvul.001G244300, Phvul.001G244400, and Phvul.001G244500 were identified within the Co-AC locus of the AC cultivar [19]. Additionally, the gene Phvul.001G246300, located within the CoPv01CDRK loci, exhibited significant responsiveness in the CDRK cultivar when inoculated with race 73 of C. lindemuthianum [26]. The gene Phvul.001G245300 is located in close proximity to the genomic region of the CDRK cultivar. The inclusion of the Phvul.001G243800 gene was based on its induction in the near-isogenic line T9576R, possessing the Co-12 resistance allele, when inoculated with race 73 of C. lindemuthianum [23]. The KTR2/3 candidate gene for Co-x in the Jalo EEP558 cultivar was also evaluated due to its induction in response to race 3993 of C. lindemuthianum [16]. Furthermore, the well-known plant defense genes Phvul.003G109100 (PR1a), Phvul.006G196900 (PR1b), and Phvul.009G256400 (PR2) were included in the analysis [22,23,24]. To standardize gene expression levels, the reference genes Phvul.008G011000 (actin—ACT) and Phvul.001G133200 (insulin-degrading enzyme—IDE) [43] were used. ACT had previously been validated for quantifying the relative expression of candidate genes in studies [23,25], while IDE’s validation was previously established by Oblessuc et al. [24]. Both genes have been utilized as reference genes for quantifying the relative expression of resistance genes against ANT in studies [16,43]. For normalization purposes, the reference genes Phvul.008G011000 (ACT) and Phvul.001G133200 (IDE) were used [43].
To obtain the coding sequences (CDS) and DNA sequences of the target genes, the common bean (P. vulgaris L.) genome available at v1.2 Phytozome [44] was accessed. Primer design for the qPCR assay was performed using the ‘Primer-Blast web tool’ [45] with the following specifications: primer size between 18 and 24 base pairs (bp), melting temperature between 59 and 61 °C, amplicon size between 80 and 160 bp, and, when possible, the primer pair should be separated by at least one intron in the corresponding genomic DNA sequence. Primer dimers and secondary structures were assessed using Gene Runner software (version 6.5.52), the ‘Multiple Prime Analyzer’ web tool (Thermo Fisher Scientific, Waltham, MA, USA): https://bit.ly/34kZpnP, accessed on 11 May 2020), and ‘The Sequence Manipulation Suite’ web tool [46]. The secondary structure of the amplicons was verified using ‘The Mfold Web Server’ platform [47] with coding sequences obtained from v1.2 Phytozome. All primer designs and in silico validation procedures not explicitly mentioned followed established literature recommendations [48,49]. Table S2 provides the primer sequences for each candidate gene evaluated, with the primers for the KTR2/3 gene obtained from Richard et al. [16].
5.6. Quantitative PCR (qPCR) and Data Analysis
The determination of PCR efficiency for each primer involved establishing a standard curve through a fivefold serial dilution, utilizing the cDNA pool as the template. This process incorporated three replicates at every dilution point [50,51]. The amplification efficiency was computed employing the equation E = [10(−1/slope)] − 1 [51], using the slope values derived from linear regression analysis. This analysis encompassed the log10-transformed cDNA concentrations on the x-axis and corresponding Cq values on the y-axis. The calculated amplification efficiency for each primer pair ranged from 0.92 to 1.09 while maintaining a coefficient of determination (R2) for the linear regression of at least 0.98 (Table S2).
The cDNA quantification reactions were conducted in the StepOnePlus™ real-time PCR system (Applied Biosystems™, Waltham, MA, USA; StepOnePlus™ Real-Time PCR Systems) using 96-well microplates [MicroAmp™ Fast 96-well Reaction Plate (0.1 mL)] sealed with MicroAmp™ Optical Adhesive Film. The total reaction volume was 10 µL, consisting of 3.4 µL of cDNA, 1.6 µL of forward and reverse primer mix (800 nM), and 5 µL of PowerUp™ SYBR™ Green Master Mix (Applied Biosystems™, Waltham, MA, USA). The thermocycling conditions included 50 °C for 2 min, 95 °C for 2 min, 40 cycles of 15 s at 95 °C, and 30 s at 60 °C.
After completing the cDNA quantitation reaction, a thorough assessment of target specificity was conducted through a dissociation curve analysis, employing the continuous melt curve setup as per the manufacturer’s specifications. Only samples demonstrating clear specificity in accordance with the dissociation curve were considered for subsequent analysis. Quantification cycle (Cq) values were extracted using StepOnePlus™ Software v2.3 (Applied Biosystems™, Waltham, MA, USA). The baseline was automatically established, and the threshold was set manually during the exponential phase of amplification. For all cDNA quantification reactions, a consistent threshold value of 0.7707 was applied.”
The genes Phvul.008G011000 (IDE) and Phvul.001G133200 (ACT) served as reference genes [43]. The arithmetic mean of quantification cycle (Cq) values [52] was computed for each experimental condition under consideration. Relative expression levels were determined by normalizing Cq values with the reference genes, employing the 2−ΔΔCT method [53,54]. Mean Cq values were derived for each gene in every experimental condition through the calculation based on three biological repetitions and three replicates (n = 3 × 3).
The investigation into the relative expression of candidate genes at the Co-AC locus and known disease-resistance genes was conducted in response to race 73 of C. lindemuthianum, spanning time points at 24, 48, 72, 96, and 120 h post-inoculation (hpi) in the AC cultivar. The calibrator condition for each gene was the relative expression observed in the mock (control, without pathogen). For data analysis and presentation of results, a logarithmic base 2 transformation was applied before statistical analysis. The Alexander-Govern test, with a significance level of 5%, was utilized to compare expression levels among experimental conditions. Pairwise comparisons of relative expression means at different time points for each gene were assessed, with significance levels adjusted using Bonferroni correction (p ≤ 0.05). These statistical analyses employed the ‘oneway.test’ [55] and ‘companion’ R packages.
All data and statistical analyses were conducted using R software (version 4.0.3) (R Core Team, Vienna, Austria, accessed on 14 June 2021), and plots were generated using the ggplot2 package [56] and R base. Error bars, representing the standard deviation of the means from three biological and three technical replicates (3 × 3), were incorporated into the visualizations. Heatmaps representing mean Cq values were generated using the ‘heatmaply’ R package, and the dendrogram was constructed based on the Euclidean distance measure and the average linkage function [50] among the relative expression values of the genes.
Acknowledgments
M. Lovatto, A.C. Calvi, E.A. Nascimento, and M. Vaz Bisneta are grateful for Grants or Scholarships from the National Council for Scientific and Technological Development (CNPq) and Capes. M.C. Gonçalves-Vidigal and P.S. Vidigal Filho are grateful for Grants from CNPq and Capes.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/plants13091245/s1; Table S1: Gene model and predicted functional annotation based on Phytozome; Table S2 Target genes, primers used, qPCR product size (amplicon), primer melting temperature (Tm), amplification efficiency (E) and coefficient of determination of linear regression (R²); Figure S1 Common bean chromosome Pv01 containing candidate genes for anthracnose resistance genes Co-x (KTR2/3), Co-1 (Phvul.001G243800), Co-AC (Phvul.001G244300, Phvul.001G244400, and Phvul.001G244500), and CoPv01CDRK (Phvul.001G245300 and Phvul.001G246300).
Author Contributions
Design and supervision of the research: P.S.V.F., M.C.G.-V. and M.M.; Conduct of the experiments: M.L., E.A.N. and A.C.C.; Analyzed the data: M.L., M.C.G.-V. and M.V.B.; Original draft of the manuscript: M.L.; Revision and editing of the manuscript: M.C.G.-V., T.A.S.G., M.V.B., P.S.V.F. and M.M. All authors have read and agreed to the published version of the manuscript.
Data Availability Statement
All data are presented within the article or in the Supplementary Materials.
Conflicts of Interest
The authors declare no conflicts of interest.
Funding Statement
Federal Funding Institution National Council for Scientific and Technological Development (CNPq) Grant/Award Number: 408472/2018-9; and from Coordination for the Improvement of Higher Education Personnel (Capes), Award number 88887.470337/2019-00.
Footnotes
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content.
References
- 1.Bitocchi E., Rau D., Bellucci E., Rodriguez M., Murgia M.L., Gioia T., Santo D., Nanni L., Attene G., Papa R. Beans (Phaseolus ssp.) as a model for understanding crop evolution. Front. Plant Sci. 2017;8:722. doi: 10.3389/fpls.2017.00722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vaz Patto M.C., Amarowicz R., Aryee A.N.A., Boye J.I., Chung H.-J., Martín-Cabrejas M.A., Domoney C. Achievements and Challenges in Improving the Nutritional Quality of Food Legumes. Crit. Rev. Plant Sci. 2015;34:105–143. doi: 10.1080/07352689.2014.897907. [DOI] [Google Scholar]
- 3.Vidigal Filho P.S., Gonçalves-Vidigal M.C., Vaz Bisneta M., Souza V.B., Gilio T.A.S., Calvi A.C., Lima L.R.L., Pastor-Corrales M.A., Melotto M. Genome-wide association study of resistance to anthracnose and angular leaf spot in Brazilian Mesoamerican and Andean common bean cultivars. Crop Sci. 2020;60:2931–2950. doi: 10.1002/csc2.20308. [DOI] [Google Scholar]
- 4.Padder B.A., Sharma P.N., Awale H.E., Kelly J.D. Colletotrichum lindemuthianum, the causal agent of bean anthracnose. J. Plant Pathol. 2017;99:317–330. doi: 10.4454/jpp.v99i2.3867. [DOI] [Google Scholar]
- 5.Nunes M.P.B.A., Gonçalves-Vidigal M.C., Martins V.S.R., Xavier L.F.S., Valentini G., Vaz Bisneta M., Vidigal Filho P.S. Relationship of Colletotrichum lindemuthianum races and resistance loci in the Phaseolus vulgaris L. genome. Crop Sci. 2021;61:3877–3893. doi: 10.1002/csc2.20601. [DOI] [Google Scholar]
- 6.Singh S.P., Schwartz H.F. Breeding common bean for resistance to diseases: A review. Crop Sci. 2010;50:2199–2223. doi: 10.2135/cropsci2009.03.0163. [DOI] [Google Scholar]
- 7.Kelly J.D., Afanador L., Cameron L.S. New races of Colletotrichum lindemuthianum in Michigan and implications in dry bean resistance breeding. Plant Dis. 1994;78:892–894. doi: 10.1094/PD-78-0892. [DOI] [Google Scholar]
- 8.Pastor-Corrales M.A., Otoya M.M., Molina A., Singh S.P. Resistance to Colletotrichum lindemuthianum isolates from middle America and Andean South America in different common bean races. Plant Dis. 1995;79:63–67. doi: 10.1094/PD-79-0063. [DOI] [Google Scholar]
- 9.Pacheco L.M., Berrouet K.V., Yepes M.S., Sánchez P.G., Montoya M.M. Detección por PCR de Colletotrichum lindemuthianum en cultivos y semillas de frijol en Antioquia, Colombia. Acta Agrómica. 2014;63:377–387. doi: 10.15446/acag.v63n4.42035. [DOI] [Google Scholar]
- 10.Vaz Bisneta M., Gonçalves-Vidigal M.C. Integration of anthracnose resistance loci and RLK and NBS-LRR-encoding genes in the Phaseolus vulgaris L. genome. Crop Sci. 2020;60:2901–2918. doi: 10.1002/csc2.20288. [DOI] [Google Scholar]
- 11.Gonçalves-Vidigal M.C., Cruz A.S., Garcia A., Kami J., Vidigal Filho P.S., Sousa L.L., McClean P., Gepts P., Pastor-Corrales M.A. Linkage mapping of the Phg-1 and Co-14 genes for resistance to angular leaf spot and anthracnose in the common bean cultivar AND 277. Theor. Appl. Genet. 2011;122:893–903. doi: 10.1007/s00122-010-1496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuiderveen G.H., Padder B.A., Kamfwa K., Song Q., Kelly J.D. Genome-wide association study of anthracnose resistance in Andean beans (Phaseolus vulgaris) PLoS ONE. 2016;11:e0156391. doi: 10.1371/journal.pone.0156391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen M., Wu J., Wang L., Mantri N., Zhang X., Zhu Z., Wang S. Mapping and genetic structure analysis of the anthracnose resistance locus Co-1HY in the common bean (Phaseolus vulgaris L.) PLoS ONE. 2017;12:e0169954. doi: 10.1371/journal.pone.0169954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lima L.R.L., Gonçalves-Vidigal M.C., Vaz Bisneta M., Valentini G., Vidigal Filho P.S., Martins V.S.R., Souza T.L.P.O. Genetic fine-mapping of anthracnose disease-resistance allele Co-14 present in the Andean common bean cultivar AND 277. Crop Sci. 2023;63:750–763. doi: 10.1002/csc2.20905. [DOI] [Google Scholar]
- 15.Richard M.M.S., Pflieger S., Sevignac M., Thareau V., Blanchet S., Li Y., Jackson S.A., Jackson S.A., Geffroy V. Fine mapping of Co-x, an anthracnose resistance gene to a highly virulent strain of Colletotrichum lindemuthianum in common bean. Theor. Appl. Genet. 2014;127:1653–1666. doi: 10.1007/s00122-014-2328-5. [DOI] [PubMed] [Google Scholar]
- 16.Richard M.M.S., Gratias A., Diaz J.C.A., Thareau V., Pflieger S., Meziadi C., Blanchet S., Marande W., Bitocchi E., Papa R., et al. A common bean truncated CRINKLY4 kinase controls gene-for-gene resistance to the fungus Colletotrichum lindemuthianum. J. Exp. Bot. 2021;72:3569–3581. doi: 10.1093/jxb/erab082. [DOI] [PubMed] [Google Scholar]
- 17.Lima-Castro S.A., Gonçalves-Vidigal M.C., Gilio T.A.S., Lacanallo G.F., Valentini G., da Silva Ramos Martins V., Song Q., Galván M.Z., Hurtado-Gonzales O.P., Pastor-Corrales M.A. Genetics and mapping of a new anthracnose resistance locus in Andean common bean Paloma. BMC Genom. 2017;18:306. doi: 10.1186/s12864-017-3685-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonçalves-Vidigal M.C., Gilio T.A.S., Valentini G., Vaz Bisneta M., Vidigal Filho P.S., Song Q., Oblessuc P.R., Melotto M. New Andean source of resistance to anthracnose and angular leaf spot: Fine-mapping of disease-resistance genes in California Dark Red Kidney common bean cultivar. PLoS ONE. 2020;15:e0235215. doi: 10.1371/journal.pone.0235215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gilio T.A.S., Hurtado-Gonzales O.P., Gonçalves-Vidigal M.C., Valentini G., Elias J.C.F., Song Q., Pastor-Corrales M.A. Fine mapping of an anthracnose-resistance locus in Andean common bean cultivar Amendoim Cavalo. PLoS ONE. 2020;15:e0239763. doi: 10.1371/journal.pone.0239763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li W., Deng Y., Ning Y., He Z., Wang G.-L. Exploiting broad-spectrum disease resistance in crops: From molecular dissection to breeding. Annu. Rev. Plant Biol. 2020;71:575–603. doi: 10.1146/annurev-arplant-010720-022215. [DOI] [PubMed] [Google Scholar]
- 21.Kankanala P., Nandety R.S., Mysore K.S. Genomics of plant disease resistance in legumes. Front. Plant Sci. 2019;10:1345. doi: 10.3389/fpls.2019.01345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Borges A., Melotto M., Tsai S.M., Caldas D.G.G. Changes in spatial and temporal gene expression during incompatible interaction between common bean and anthracnose pathogen. J. Plant Physiol. 2012;169:1216–1220. doi: 10.1016/j.jplph.2012.04.003. [DOI] [PubMed] [Google Scholar]
- 23.Mahiya-Farooq, Padder B.A., Bhat N.N., Shah M.D., Shikari A.B., Awale H.E., Kelly J.D. Temporal expression of candidate genes at the Co-1 locus and their interaction with other defense related genes in common bean. Physiol. Mol. Plant Pathol. 2019;108:101424. doi: 10.1016/j.pmpp.2019.101424. [DOI] [Google Scholar]
- 24.Oblessuc P.R., Baroni R.M., Garcia A.A.F., Chioratto A.F., Carbonell S.A.M., Camargo L.E.A., Benchimol L.L. Mapping of angular leaf spot resistance QTL in common bean (Phaseolus vulgaris L.) under different environments. BMC Genet. 2012;13:50. doi: 10.1186/1471-2156-13-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Shams E., Javan-Nikkhah M., Mirzadi Gohari A. Dissecting molecular events and gene expression signatures involved in Colletotrichum lindemuthianum-Phaseolus vulgaris pathosystem in compatible and incompatible interactions. Eur. J. Plant Pathol. 2020;156:925–937. doi: 10.1007/s10658-020-01944-8. [DOI] [Google Scholar]
- 26.Lovatto M., Gonçalves-Vidigal M.C., Vaz Bisneta M., Calvi A.C., Mazucheli J., Vidigal Filho P.S., Miranda E.G.R., Melotto M. Responsiveness of candidate genes on CoPv01CDRK/PhgPv01CDRK loci in common bean challenged by anthracnose and angular leaf spot pathogens. Int. J. Mol. Sci. 2023;24:16023. doi: 10.3390/ijms242216023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nanami D.S.Y., Gonçalves-Vidigal M.C., Castro S.A.L., Frias A.A.T., Vidigal Filho P.S., Elias H.T. Characterization of genetic resistance in Andean common bean cultivar Amendoim Cavalo to Colletotrichum lindemuthianum. Agron. Sci. Biotechnol. 2017;3:43–52. doi: 10.33158/ASB.2017v3i1p43. [DOI] [Google Scholar]
- 28.Hao Y., Zong X., Ren P., Quian Y., Fu A. Basic Helix-Loop-Helix (bHLH) transcription factors regulate a wide range of functions in Arabidopsis. Int. J. Mol. Sci. 2021;22:7152. doi: 10.3390/ijms22137152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding C., Gao J., Zhang S., Jiang N., Su D., Huang X., Zhang Z. The Basic/Helix-Loop-Helix transcription factor family gene RcbHLH112 is a susceptibility gene in gray mould resistance of rose (Rosa chinensis) Int. J. Mol. Sci. 2023;24:16305. doi: 10.3390/ijms242216305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ali S., Ganai B.A., Kamili A.N., Bhat A.A., Mir Z.A., Bhat J.A., Tyagi A., Islam S.T., Mushtaq M., Yadav P., et al. Pathogenesis-related proteins and peptides as promising tools for engineering plants with multiple stress tolerance. Microbiol. Res. 2018;212:29–37. doi: 10.1016/j.micres.2018.04.008. [DOI] [PubMed] [Google Scholar]
- 31.Friesen T.L., Faris J.D. Characterization of Effector–Target Interactions in Necrotrophic Pathosystems Reveals Trends and Variation in Host Manipulation. Annu. Rev. Phytopathol. 2021;59:77–98. doi: 10.1146/annurev-phyto-120320-012807. [DOI] [PubMed] [Google Scholar]
- 32.Riviere M.-P., Marais A., Ponchet M., Willats W., Galiana E. Silencing of acidic pathogenesis-related PR-1 genes increases extracellular beta-(1→3)-glucanase activity at the onset of tobacco defence reactions. J. Exp. Bot. 2008;59:1225–1239. doi: 10.1093/jxb/ern044. [DOI] [PubMed] [Google Scholar]
- 33.Zhang Q., Guo N., Zhang Y., Yu Y., Liu S. Genome-wide characterization and expression analysis of pathogenesis-related 1 (PR-1) gene family in tea plant (Camellia sinensis (L.) O. Kuntze) in response to blister-blight disease stress. Int. J. Mol. Sci. 2022;23:1292. doi: 10.3390/ijms23031292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dixon D.C., Cutt J.R., Klessig D.F. Differential targeting of the tobacco PR-1 pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J. 1991;10:1317–1324. doi: 10.1002/j.1460-2075.1991.tb07650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santamaria M., Thomson C.J., Read N.D., Loake G.J. The promoter of a basic PR1-like gene, AtPRB1, from Arabidopsis establishes an organ-specific expression pattern and responsiveness to ethylene and methyl jasmonate. Plant Mol. Biol. 2001;47:641–652. doi: 10.1023/A:1012410009930. [DOI] [PubMed] [Google Scholar]
- 36.Walter M.H., Liu J.-W., Grand C., Lamb C.J., Hess D. Bean pathogenesis-related (PR) proteins deduced from elicitor-induced transcripts are members of a ubiquitous new class of conserved PR proteins including pollen allergens. Mol. Genet. Genom. 1990;222:353–360. doi: 10.1007/BF00633840. [DOI] [PubMed] [Google Scholar]
- 37.Alvarez-Diaz J.C., Laugé R., Delannoy E., Huguet S., Roux C.P.-L., Gratias A., Geffroy V. Genome-Wide Transcriptomic Analysis of the Effects of Infection with the Hemibiotrophic Fungus Colletotrichum lindemuthianum on Common Bean. Plants. 2022;11:1995. doi: 10.3390/plants11151995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Edington B.V., Lamb C.J., Dixon R.A. cDNA cloning and characterization of a putative 1,3-b-Dglucanase transcript induced by fungal elicitor in bean cell suspension cultures. Plant Mol. Biol. 1991;16:81–94. doi: 10.1007/BF00017919. [DOI] [PubMed] [Google Scholar]
- 39.de Jonge R., van Esse H.P., Kombrink A., Shinya T., Desaki Y., Bours R., van der Krol S., Shibuya N., Joosten M.H., Thomma B.P.H.J. Conserved fungal lysM effector ecp6 prevents chitin-triggered immunity in Plants. Science. 2010;329:953–955. doi: 10.1126/science.1190859. [DOI] [PubMed] [Google Scholar]
- 40.Barreto-Bergter E., Figueiredo R.T. Fungal glycans and the innate immune recognition. Front. Cell. Infect. Microbiol. 2014;4:145. doi: 10.3389/fcimb.2014.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mathur R.S., Barnett H.l., Lilly V.G. Sporulation of Colletotrichum lindemuthianum in culture. Phytopathology. 1950;40:104–114. [Google Scholar]
- 42.Farrell R.E. RNA Methodologies: Laboratory Guide for Isolation and Characterization. 5th ed. Elsevier; Amsterdam, The Netherlands: 2017. pp. 1–855. [Google Scholar]
- 43.Borges A., Tsai S.M., Caldas D.G.G. Validation of reference genes for RT-qPCR normalization in common bean during biotic and abiotic stresses. Plant Cell Rep. 2012;31:827–838. doi: 10.1007/s00299-011-1204-x. [DOI] [PubMed] [Google Scholar]
- 44.Goodstein D.M., Shu S., Howson R., Neupane R., Hayes R.D., Fazo J., Mitros T., Dirks W., Hellsten U., Putnam N., et al. Phytozome: A comparative platform for green plant genomics. Nucleic. Acids Res. 2012;40:D1178–D1186. doi: 10.1093/nar/gkr944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ye J., Coulouris G., Zaretskaya I., Cutcutache I., Rozen S., Madden T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012;13:134. doi: 10.1186/1471-2105-13-134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stothard P. The sequence manipulation suite: JavaScript programs for analyzing and formatting protein and DNA sequences. BioTechniques. 2000;28:1102–1104. doi: 10.2144/00286ir01. [DOI] [PubMed] [Google Scholar]
- 47.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bustin S.A., Benes V., Garson J.A., Hellemans J., Huggett J., Kubista M. The MIQE Guidelines: Minimum information for publication of quantitative real-time PCR Experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 49.Bustin S., Huggett J. qPCR primer design revisited. Biomol. Detect. Quantif. 2017;14:19–28. doi: 10.1016/j.bdq.2017.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Svec D., Tichopad A., Novosadova V., Pfaffl M.W., Kubista M. How good is a PCR efficiency estimate: Recommendations for precise and robust qPCR efficiency assessments. Biomol. Detect. Quantif. 2015;3:9–16. doi: 10.1016/j.bdq.2015.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rasmussen R. Quantification on the LightCycler. In: Meuer S., Wittwer C., Nakagawara K.-I., editors. Rapid Cycle Real-Time PCR. Springer; Berlin/Heidelberg, Germany: 2001. pp. 21–34. [DOI] [Google Scholar]
- 52.Schmittgen T.D., Livak K.J. Analyzing real-time PCR data by the comparative CT method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 53.Dag O., Dolgun A., Konar N.M. Onewaytests: An R Package for One-Way Tests in Independent Groups Designs. R J. 2018;10:175–199. doi: 10.32614/RJ-2018-022. [DOI] [Google Scholar]
- 54.R Core Team . R: A Language and Environment for Statistical Computing. R Foundation for Statistics Computing; Vienna, Austria: 2020. [(accessed on 14 June 2021)]. Available online: http://www.R-project.org/ [Google Scholar]
- 55.Wickham H. ggplot2: Elegant Graphics for Data Analysis. 2nd ed. Springer International Publishing; Cham, Switzerland: 2016. [DOI] [Google Scholar]
- 56.Galili T., O’Callaghan A., Sidi J., Sievert C. Heatmaply: An R package for creating interactive cluster heatmaps for online publishing. Bioinformatics. 2018;34:1600–1602. doi: 10.1093/bioinformatics/btx657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are presented within the article or in the Supplementary Materials.