Abstract
Productive high-titer infection by human immunodeficiency virus type 1 (HIV-1) requires the activation of target cells. Infection of quiescent peripheral CD4 lymphocytes by HIV-1 results in incomplete, labile reverse transcripts and lack of viral progeny formation. An interplay between Tat and p53 has previously been reported, where Tat inhibited the transcription of the p53 gene, which may aid in the development of AIDS-related malignancies, and p53 expression inhibited HIV-1 long terminal repeat transcription. Here, by using a well-defined and -characterized stress signal, gamma irradiation, we find that upon gamma irradiation, HIV-1-infected cells lose their G1/S checkpoints, enter the S phase inappropriately, and eventually apoptose. The loss of the G1/S checkpoint is associated with a loss of p21/Waf1 protein and increased activity of a major G1/S kinase, namely, cyclin E/cdk2. The p21/Waf1 protein, a known cyclin-dependent kinase inhibitor, interacts with the cdk2/cyclin E complex and inhibits progression of cells into S phase. We find that loss of the G1/S checkpoint in HIV-1-infected cells may in part be due to Tat's ability to bind p53 (a known activator of the p21/Waf1 promoter) and sequester its transactivation activity, as seen in both in vivo and in vitro transcription assays. The loss of p21/Waf1 in HIV-1-infected cells was specific to p21/Waf1 and did not occur with other KIP family members, such as p27 (KIP1) and p57 (KIP2). Finally, the advantage of a loss of the G1/S checkpoint for HIV-1 per se may be that it pushes the host cell into the S phase, which may then allow subsequent virus-associated processes, such as RNA splicing, transport, translation, and packaging of virion-specific genes, to occur.
In mammalian cells, passage from G0 to G1 is controlled by cyclin-dependent kinases cdks that are regulated by D-type cyclins. D-type cyclins (D1, D2, and D3) act as growth factor sensors, are induced as part of the delayed early response to growth factor stimulation, and assemble with cdk4 and cdk6 in a growth factor-dependent way. D-type cyclins are labile proteins, and because their holoenzyme activity decays rapidly, cells deprived of mitogens exit the cycle and enter G0. However, mammalian cells are only rarely cycling and spend a considerable fraction of their life spans in a resting phase, termed G0, with a DNA content equal to that of a G1 cell. Recruitment of resting cells into the cell cycle is controlled by a circuitry that interacts with the cell cycle machinery (16, 43).
p53 induction is translated into inhibition of cell cycle progression by two different mechanisms. In the first mode of action, p53 acts as a transcriptional activator and induces the transcription of genes with p53 response elements. Such genes include p21/Waf1, a known cdk inhibitor (cdkI) (13, 29). Interaction of this protein with the cdk2/cyclin E complex is thought to inhibit progression of cells into S phase (6, 33). There is an excellent correlation between p53 status, capability for G1 arrest, and p21/Waf1 induction after treatment with DNA-damaging agents (7, 42). The main target for transactivation is the p21 gene; synthesis of the p21 protein is the cause of G1/S-phase arrest due to the inhibition of cdks. As a consequence, the presence of p21/Waf1 protein prevents phosphorylation of Rb. Therefore, physical or functional deletion of the p21/Waf1 gene product results in a loss of the G1/S checkpoint (44).
Mice lacking p21/Waf1 (p21−/− embryonic fibroblasts) are deficient in the ability to arrest in G1 in response to DNA damage and nucleotide pool perturbation. p21−/− cells also exhibit a significant growth alteration in vitro, achieving a saturation density as high as that observed in p53−/− cells. In contrast, other aspects of p53 function, such as thymocytic apoptosis and the mitotic spindle checkpoint, appear normal (8).
The effect of p21/Waf1 on various purified cdks has also been explored. p21/Waf1 effectively inhibits the cdk2, cdk3, cdk4, and cdk6 kinases (Ki, 0.5 to 15 nM) but is much less effective toward cdc2/cyclin B (Ki, approximately 400 nM) and cdk5/p35 (Ki, >2 mM) and does not associate with cdk7/cyclin H. Thus, p21/Waf1 is not a universal inhibitor of cdks but displays selectivity for G1/S cyclin-cdk complexes (14).
Productive infection by human immunodeficiency virus type 1 (HIV-1) requires the activation of target cells. Infection of quiescent peripheral CD4 lymphocytes by HIV-1 results in incomplete, labile reverse transcripts. It has been shown that optimal completion of reverse transcription takes place in late G1 phase when highly purified T cells which contain G1b-phase cells are used (24). Along the same lines, activation with the alpha CD3 molecule alone resulted in cell cycle progression into only G1a and incomplete HIV-1 reverse transcription. However, costimulation through the CD28 receptor and transition into G1b was required to efficiently complete the reverse-transcription process (25).
Tat (transactivator of transcription) is essential for HIV-1 replication in vivo and in vitro. Tat-(65-80), an RGD-containing domain, has been shown to regulate the proliferative functions of a variety of cell lines, including a human adenocarcinoma cell line, A549. Treatment with Tat-(65-80) has been shown to reduce the p53 gene product fivefold, whereas c-fos gene transcription increased sevenfold at 0.5 h posttreatment and subsequently declined to baseline levels at 8 h. Therefore, it was suggested that Tat-(65-80) can modulate growth-related genes in cells (12). A similar result has also been reported in which Tat inhibits the transcription of p53, and the downregulation of p53 by Tat may be involved in the development of AIDS-related malignancies (26). The interaction between Tat and p53 seems specific, since, with a lambda cI repressor system, Tat protein specifically interacts with the human p53 protein via the p53 dimerization domain. Two alternative biological consequences have been proposed as a result of Tat-p53 interaction: (i) Tat interaction with p53 may inactivate p53 regulatory functions, thus producing cell transformation, or (ii) Tat interaction favors the formation of p53 dimers, thus leading the cell towards apoptosis (27).
The inhibition effect of p53 on the HIV-1 viral long terminal repeat (LTR) has been reported. The p53 inhibition is observed both at transfection and in in vitro transcription assays. In addition, the Sp1 and the TATA box sites of the HIV-1 LTR were shown to be the primary sites involved in the p53-induced inhibition observed on this viral promoter (9).
We have been interested in the question of the various cell cycle checkpoints and their relationship to post-integration events in HIV-1-infected cells. The rationale came from the reported observations stated above, especially the Tat-p53 interaction, which we have also found in a two-hybrid system, and its functional consequences in a virus-infected cell. We therefore reasoned that the Tat-p53 physical interaction might play a role in manipulating the host cell cycle checkpoints. By using a well-defined and characterized stress signal, gamma irradiation, we found that HIV-1-infected cells lose their G1/S checkpoints, inappropriately enter the S phase, and eventually apoptose. The loss of this checkpoint is associated with loss of p21/Waf1 protein and increased activity of a major G1/S kinase, namely, cyclin E/cdk2. Finally, the advantage of loss of the G1/S checkpoint for HIV-1 is shown to be associated with an increase in viral transcription and progeny formation.
MATERIALS AND METHODS
Expression vectors, antibodies, and protein purification.
Wild-type Tat protein was overexpressed in bacteria and purified as described previously (2). p53 was expressed from baculovirus and purified as described previously (28). Glutathione-S-transferase–TATA-binding protein (GST-TBP), GST-Rb (Z51), and GST-Tat were expressed in Escherichia coli DH5α, induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h, pelleted, washed with phosphate-buffered saline (PBS), lysed with PBS plus 0.1% NP-40, sonicated, and bound to GST beads (10% slurry) overnight. The beads were washed the next day (three times) with the lysis buffer and run on sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) for quality prior to use (19, 20). Histadine-tagged proteins were purified by nickel affinity chromatography (Qiagen), followed by cation-exchange fast protein liquid chromatography (HiTrap SP; Amersham Pharmacia Biotech) (18).
Plasmids (pCMV-p53 and G5 p53-CAT) for lymphocyte transfection have been described elsewhere (32). A supercoiled double-banded cesium chloride plasmid, p53/G-Free, was used for in vitro transcription and has been described previously (31). HIV-LTR CAT (wild type) and TAR mutant (TM26; 37, 38) plasmids were electroporated into CEM (12D7) cells and scored for chloramphenicol acetyltransferase (CAT) activity (17). Wild-type Tat and mutant 41 (pGEM-Tat 41) were generous gifts of Andy Rice.
Various antibodies were used for Western blots at 1:1,000 dilution, which were incubated overnight, washed the next day, and detected with 125I-protein G (Amersham). The antibodies used were p53 (Ab-1; Calbiochem), p21/Waf1 (C-19; Santa Cruz), Kip2 p57 (C-20; Santa Cruz), cyclin E (M-20; Santa Cruz), cdk2 (H-298; Santa Cruz), p27 (N-20; Santa Cruz), and TBP (a generous gift of Nancy Thompson). Antibodies against HIV-1 viral proteins were from the National Institutes of Health AIDS research and reference program (operated by McKesson BioServices, Rockville, Md.).
In vitro transcription assay.
The G-free DNA templates used in the in vitro transcription assays were amplified in E. coli and double CsCl2 purified prior to use. For the in vitro transcription reactions, preincubation with extract was done at 30°C for 30 min, followed by the addition of 2 μl of [α-32P]UTP (Amersham Pharmacia Biotech) (400 Ci/mmol) and incubation at 30°C for 60 min. Reaction mixtures contained CEM (12D7) whole-cell extract (20 μl; 14 μg/μl), 1.0 μg of supercoiled DNA, 300 ng of Tat protein, and 200 ng of p53 protein (pretreated with Pab421 antibody [31]) in a total volume of 65 μl. Transcription buffer (31 μl/reaction) contained 3 μl of 20% polyethylene glycol (molecular weight, 6000), 3 μl of 50 mM MgCl2, 3 μl of 1 mM dithiothreitol (DTT), 1 μl of 0.2 M creatine phosphate (Boehringer Mannheim), 1.5 μl of 50 mM ATP-CTP, 1 μl of 20 mM 3′-O-methylguanosine 5′-triphosphate (Amersham Pharmacia Biotech), 20 U of RNase T1 (100 U/μl; Boehringer Mannheim), and 18.5 μl of buffer D containing a final concentration of 20 mM HEPES (pH 7.9), 100 mM KCl, 12.5 mM MgCl2, 0.1 mM EDTA, 17% glycerol, and 1 mM DTT. Samples were processed and loaded on a 4% denaturing urea PAGE gel and exposed to a PhosphorImager cassette (Molecular Dynamics) overnight.
Lymphocyte transfection.
Lymphocyte [CEM (12D7)] cells were grown to mid-log phase and were processed for protein electroporation according to a previously published procedure (17). Only one modification was introduced, in which the cells were electroporated at 230 V and plated in 10 ml of complete RPMI 1640 medium for 18 h prior to harvest and CAT assay.
Microscale preparation of extracts.
Cytoplasmic fractions were obtained according to the following procedure. Cells were harvested at 1,000 rpm (Sorval) at 4°C for 10 min. The cell pellet was resuspended in 10 volumes of cold PBS without Ca2+ or Mg2+. The cell pellet was then washed (twice) in 0.1 volume of cold buffer A (10 mM HEPES [pH 7.9 at 4°C], 1.5 mM MgCl2, 10 mM KCl, and 0.5 mM DTT) and pelleted as described above. The washed cell pellet was resuspended in 60 μl of cold buffer C (20 mM HEPES [pH 7.9], 25% [vol/vol] glycerol, 0.42 M NaCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM phenylmethylsulfonyl fluoride [PMSF] [added fresh in isopropanol], and 0.5 mM DTT) plus 0.1% NP-40 per 107 cells, incubated on ice for 10 min, mixed briefly (10 s) by vortexing, and spun at 10,000 rpm at 4°C for 10 min in an Eppendorf microcentrifuge. The lysed cell supernatant was retained, diluted with 120 μl of cold modified buffer D (20 mM HEPES [pH 7.9], 20% [vol/vol] glycerol, 0.05 M KCl, 0.2 mM EDTA, 0.5 mM PMSF, and 0.5 mM DTT) per 107 cells, and stored at −70°C as 100-μl aliquots. Protein concentration determinations were made prior to storage.
To prepare nuclear extracts, cells were collected and washed once with PBS (without Ca2+ and Mg2+) and once with 200 μl of ice-cold buffer A. The cells were lysed in 200 μl of buffer A by gently passing the cell suspension through a 28-gauge needle. This procedure is done with the tube containing the cells submerged in ice. The nuclei were collected by pelleting the suspension for 30 s in an Eppendorf microcentrifuge, and the supernatant was kept for further analysis. Crude nuclei were extracted with ice-cold buffer C (20 mM HEPES [pH 7.9], 25% [vol/vol] glycerol, 420 mM KCl, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM DTT, 0.5 mM PMSF), 60 μl for 100 μl of cell pellet, for at least 15 min on ice. An equal volume of buffer D (20 mM HEPES [pH 7.9], 20% [vol/vol] glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT) was added, and the mixture was spun in an Eppendorf microcentrifuge for at least 10 min at 4°C. The supernatants were collected, and their volumes were measured. Generally, nuclear or cytoplasmic extracts from 48-h ACH2 cells had to be trichloroacetic acid precipitated prior to being loaded onto a gel (see Fig. 2B, 48 h). The protein concentration for each preparation was determined by using the Bio-Rad protein assay kit.
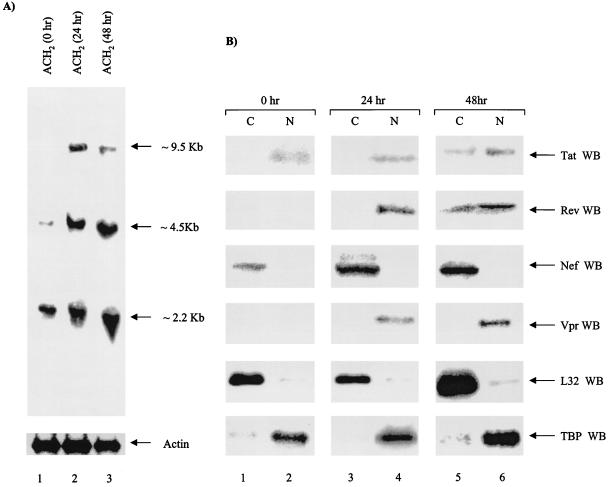
FIG. 2.
Gene expression and translation of HIV-1-infected cells following gamma irradiation. (A) Twenty micrograms of total RNA was separated on an RNA-formaldehyde-agarose gel, transferred, and probed with full-length labeled HIV-1 genomic RNA messages corresponding to a 2.2-kb collection of doubly spliced regulatory proteins, such as Tat, Nef, Rev, and Vpr. The ∼4.5-kb messages correspond to the Gag, Pol, and Env singly spliced messages, and the ∼9.5-kb messages represent full genomic RNA that is packaged into the virion. The blot was later stripped and used to probe with a 40-mer anti-sense actin probe (bottom). (B) Both cytoplasmic (50-μg) and nuclear (20-μg) extracts were processed from ACH2 cells and Western blotted for viral Tat, Nef, Rev, Vpr, cytoplasmic (L32) (C), and nuclear (TBP) (N) proteins. Forty-eight-hour samples were trichloroacetic acid precipitated prior to separation on a 4 to 20% Tris-glycine gel. WB, Western blotting.
Immunoprecipitation and immunoblotting.
Cells grown in culture were spun at 10,000 × g for 15 min. The supernatants were discarded, and the pellets were washed twice with 25 ml of PBS without Ca2+ or Mg2+. The pelleted cells were lysed with 1 ml of lysis buffer containing 50 mM Tris-Cl (pH 7.4), 120 mM NaCl, 5 mM EDTA, 0.5% NP-40, 50 mM NaF (phosphotyrosine phosphatase inhibitor), 1 mM DTT, and 1 mM PMSF. The cells were incubated on ice for 15 min and mixed gently every 5 min. The cells were then transferred to an Eppendorf tube and microcentrifuged at 4°C for 10 min. Protein concentrations in the lysates were determined with the bicinchoninic acid protein assay kit (Bio-Rad Laboratories). A total of 2 mg of cellular proteins with 50 μl of rabbit anti-human cyclin E antibody C-17 (Santa Cruz Biotechnology) were used for immunoprecipitation. They were mixed for 12 to 14 h at 4°C, and the next day, 150 μl of 30% Protein G Plus Protein Agarose beads (catalog no. IP05; Calbiochem) was used in TNE 50 plus 0.1% NP-40 buffer and mixed at 4°C for 3 h. The samples were microcentrifuged for 10 min at 4°C, and the supernatants were discarded. The agarose beads were washed three times with TNE 50 plus 0.1% NP-40, gently vortexed, and pelleted. To the pellets, 20 μl of 2× Tris-glycine–SDS sample buffer was added, and the solution was heated at 95°C for 5 min and separated on 4 to 20% Tris-glycine gels (NOVEX, Inc.) at 200 V for 60 min. The proteins were then transferred to nylon-reinforced nitrocellulose membranes (Immobilon-P transfer membranes; Millipore Corp.) overnight at 0.08 A. Following the transfer, the blots were blocked with 5% nonfat dry milk in 50 ml of TNE 50 plus 0.1% NP-40 for 30 min and washed twice with 25 ml of TNE 50 plus 0.1% NP-40 at 4°C. After the wash was discarded, the blots were probed with a 1:1,000 dilution of rabbit anti-human cdk2 (H-298) (catalog no. sc-748; Santa Cruz Biotechnology), rabbit anti-human cdk4 (H-303) (catalog no. sc-749; Santa Cruz Biotechnology), or rabbit anti-human cdk6 (H-96), (catalog no. sc-7180; Santa Cruz Biotechnology). The blots were probed for a period of 12 to 14 h at 4°C, followed by two washes with 25 ml of TNE 50 plus 0.1% NP-40. After the washes, the blots were treated with 10 ml of 125I-protein G (50 μl) (catalog no. IM. 244; Amersham) in TNE 50 plus 0.1% NP-40 for a period of 2 h at 4°C. Finally, the blots were washed twice in 25 ml of TNE 50 plus 0.1% NP-40 and placed on a PhosphorImager cassette for further analysis. For straight Western blots, a total of 25 to 50 μg of cellular proteins were run on 4 to 20% Tris-glycine gels, transferred, and blotted with a 1:1,000 dilution of cyclin E or cdk2 antibody.
Cell culture.
ACH2 (10) and 8E5 (36) cells are both HIV-1-infected cells, with integrated wild-type single-copy virus (ACH2) and an integrated single-copy reverse transcriptase-defective virus (8E5) in CEM (12D7) cells (5). The CEM (12D7) T-cell is the parental cell for both ACH2 and 8E5 cells. CEM (Vector) and CEM (Tat) cells have been described elsewhere (22). These and other cell lines were cultured at 37°C up to 105 cells per ml in RPMI 1640 medium containing 10% fetal bovine serum (FBS) treated with a mixture of 1% streptomycin, penicillin antibiotics, and 1% l-glutamine (Gibco/BRL).
cdk assays.
Twenty million T cells were cultured to the mid-log phase of growth and lysed in a buffer containing 150 mM NaCl, 50 mM HEPES (pH 7.5), 1 mM EDTA, 2.5 mM EGTA, 1 mM DTT, 0.1% Tween-20, 100 μM Na3VO4, 1 mM NaF, 30 nM aprotinin, 500 nM leupeptin, 100 μM PMSF, 10 mM beta-glycerophosphate, and 1 mM sodium pyrophosphate. The kinase activity of the immunoprecipitated anti-cyclin E complexes from the cell lysis were assessed by the transfer of phosphate from [γ-32P]ATP to truncated recombinant GST-Rb (Z51) protein in a reaction buffer consisting of 50 mM HEPES (pH 7.5), 10 mM MgCl2, 1 mM DTT, 2.5 mM EGTA, 10 mM beta-glycerophosphate, 100 μM Na3VO4, 1 mM NaF, 20 μM ATP, 200 ng of the substrate GST-Rb protein (eluted from glutathione beads), and 10 μCi of [γ-32P]ATP (specific activity, 11 Ci/mmol; ICN Biochemical). The reactions were performed for 30 min at 30°C and stopped by the addition of SDS sample buffer. The samples were boiled for 5 min at 65°C, and the proteins were separated on a 4 to 20% Tris-glycine gel. The gels were autoradiographed, and bands were counted on a Molecular Dynamics PhosphorImager plate.
For histone H1 kinase activity, cells were cultured to the mid-log phase of growth, treated, and processed at various time points by lysis in a buffer containing 250 mM NaCl, 50 mM Tris (pH 7.4), 5 mM EDTA, 0.1% Nonidet P-40, 100 μM Na3VO4, 50 mM NaF, 30 nM aprotinin, and 500 nM leupeptin. Immunoprecipitated associated complexes were assessed by the transfer of phosphate from [γ-32P]ATP (specific activity, 11 Ci/mmol) to histone H1 (10 μg; Boehringer Mannheim) in a reaction buffer consisting of 50 mM Tris (pH 7.4), 10 mM MgCl2, 1 mM DTT, and 144 μM ATP (40 μCi of [γ-32P]ATP). The reactions were performed for 15 min at 30°C and stopped by the addition of SDS sample buffer. The samples were boiled for 5 min at 95°C, and the proteins were separated on a 4 to 20% Tris-glycine gel. One U of cdk2-associated activity/min was defined as the incorporation of 1 pmol of phosphate/min into the substrate.
Northern blotting.
Total cellular RNA was extracted using the RNAZol reagent (Gibco/BRL). Total RNA (20 μg) was run on a 1% formaldehyde-agarose gel overnight at 75 V, transferred onto a 0.2-μm nitrocellulose membrane (Millipore Inc.), UV cross-linked, and hybridized overnight at 42°C with end-labeled 32P-HIV full-genomic RNA (Loftsrand, Gaithersburg, Md.). The next day, the membrane was washed two times for 15 min each time with 10 ml of 0.2% SDS–2× SSC [1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate]) at 37°C, exposed, and counted on a PhosphorImager cassette (Molecular Dynamics).
Gamma irradiation of cells.
Gamma irradiation was performed in a J. L. Shepherd and Associates Mark I irradiator, model 68A, utilizing a pair of 6,000-Ci 137Cs sources in type 6810 capsules. Dosage values were initially calibrated by J. L. Shepherd and Associates and were updated annually by the Office of Radiation Safety of the University of Medicine and Dentistry of New Jersey (UMDNJ) with a Thomson and Nielsen Electronics Ltd. irradiator dosimeter, model MD-10, and high-dose sensors according to the instructions in the manual.
A 4-in. turntable on a base was used at position 3 of the irradiator chamber. The center of position 3 was located 8.2 in. from the sources. At this position, a uniform dose was given from 4.3 to 9.7 in. from the floor of the irradiator chamber and at a 2-in. radius from the center of the turntable with no attenuators used. Four individual 25-ml tissue culture flasks (5 ml of cells laid flat) fit into the uniform-dosage area. In this position, irradiation of about 1 min is calculated to give a dosage of 770 rads, or 7.7 Gy.
Cultures were grown prior to irradiation in a 37°C CO2 incubator under normal-serum or serum-starved conditions (RPMI 1640 or Dulbecco's modified Eagle's medium from Quality Biological, Inc.). The cultures were removed, irradiated, and then returned to 37°C to incubate for 24 or 48 h before being processed for fluorescence-activated cell sorter (FACS) analysis or cell extraction procedures. When cells were grown under serum-starved (1% FBS) conditions for 3 days, at time zero, 1/10 volume of fetal calf serum (from Atlanta Biologicals) was added immediately following gamma irradiation, and then incubation was continued for 24 or 48 h.
Cell cycle block and analysis.
Cells for all experiments were grown to mid-log phase, washed, and kept in complete medium with 1% FBS for 3 days. They were then treated with gamma irradiation, 10% serum was added, and then they were incubated for various times (i.e., 0, 24, and 48 h). For FACS analysis, the cells were removed from medium at each time point, washed with PBS without Mg2+ or Ca2+, fixed with 70% ethanol, and stained with a cocktail of PI buffer (PBS with Ca2+ and Mg2+, RNase A [10 μg/ml], NP-40 [0.1%], and propidium iodide [50 μg/ml]) followed by cell-sorting analysis. The FACS analyses for CEM, ACH2, and 8E5 cells were all performed at the same time after the collection of cells. At least three independent experiments, at different times (Fig. 1), were performed for each cell type. FACS data was acquired on a Becton Dickinson FACScaliber with a 488-nm argon laser. Acquisition was done with CELLQuest software (Becton Dickinson), and analysis was performed on a Macintosh computer with ModFit LT software (Verity Software House, Inc.). The gates used for FACS analysis were at channel 200 for FL2 width versus FL2 area, with a doublet discriminator. CELLQuest acquisition was for 1,024-channel documents, and the ModFit analysis was done for 250-channel documents. Apoptosis, G1, S, and G2/M peaks are shown in Fig. 1. We normally gate on live cells for flow cytometry, but the cell cycle parameters were calculated after the apoptotic cells were additionally gated out. More information on gating will be provided upon request.
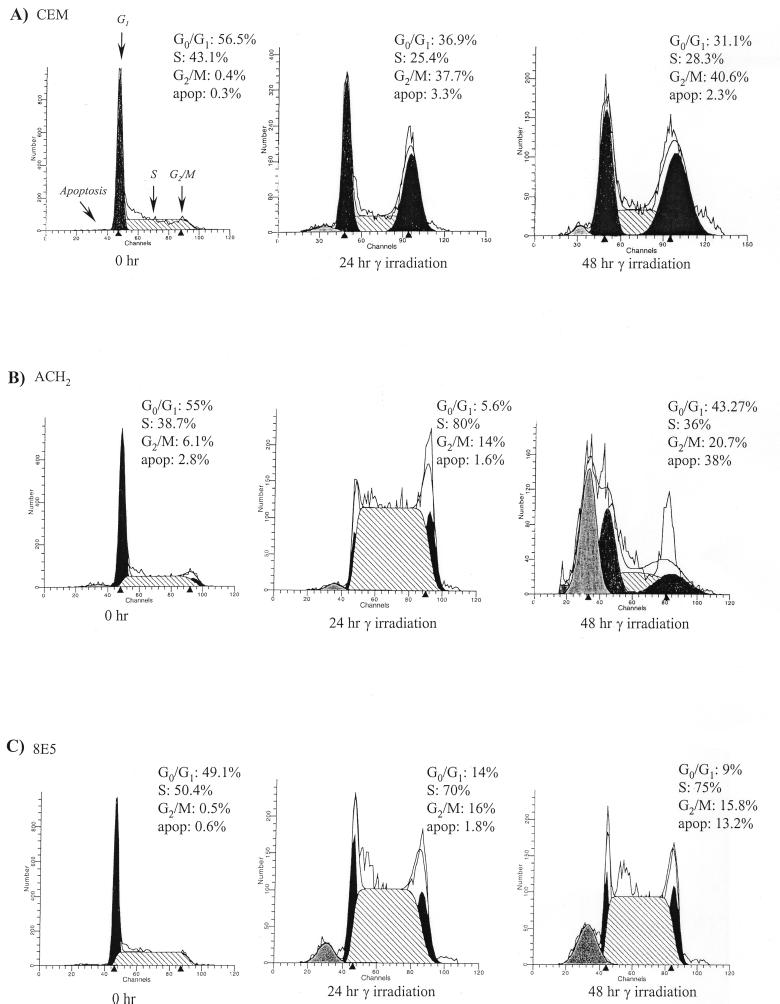
FIG. 1.
Effect of gamma irradiation on HIV-infected and uninfected cells. The cells were grown to the mid-log phase of growth, serum starved for 3 days in 1% FBS, gamma irradiated, and processed at various time points for FACS analysis. (A) Parental CEM (12D7) cells; (B and C) HIV-1-integrated latently infected cells with wild-type (ACH2) and reverse transcription mutant (8E5) proviral genomes. Each panel shows cell cycle histogram profiles and percentages of cell numbers at various stages of the cell cycle. apop, percentage of cells that were apoptosing at each 0-, 24-, and 48-h time point. Apoptosis, G1, S, and G2/M peaks are indicated by arrows. Apoptotic cells are cells that have exited the cell cycle and are about to apoptose. We normally gate on live cells for flow cytometry, but the cell cycle parameters were calculated after the apoptotic cells were additionally gated out. More information on gating will be provided upon request.
RESULTS
We initially asked whether HIV-1-infected cells had any abnormalities in their cell cycle profiles when treated with gamma irradiation. Gamma irradiation has been one of the best-studied agents for inducing DNA damage in a variety of eukaryotic systems, ranging from yeast to humans, and its consequential effects on various pathways have been well characterized. These pathways, including evolutionarily conserved Chk1, ATR, and nuclear poly(ADP-ribose) polymerase and nonconserved p21, p53 and AbI, guard genomic integrity after DNA damage (21, 30, 34, 47).
Normally, when primary, untransformed cells are gamma irradiated, they stop at a time point called the G1/S checkpoint and do not proceed into S phase. This block at G1/S allows time to repair damaged DNA prior to S-phase entry. Transformed cells, as well as various in vitro cell lines, however, will stop at two checkpoints, one at G1/S and the other at G2/M. The proteins responsible for the G1/S and G2/M blocks are p21/Waf1 and 14-3-3 family members, respectively (3, 4). p21/Waf1 inhibits G1/S cyclin-cdk complexes and therefore stops entry into S phase, whereas a block at the G2/M checkpoint allows sufficient time for increase of cell mass and rescanning and repairing of damaged DNA prior to mitosis. The discrepancy between primary and transformed cell lines is that primary cells are mostly at G0/G1 (∼80 to 90%; i.e., human peripheral blood lymphocytes [PBLs]), whereas transformed cell lines have a G0/G1 population of about 40 to 60% and an S-phase population of 30 to 40%. It is the G0/G1 population that stops at G1/S and S-phase cells that stop at G2/M upon gamma irradiation.
Effect of gamma irradiation on HIV-1-infected cells.
To determine the effect of gamma irradiation on lymphocytes, we initially used various T-cell lines and performed FACS analysis after irradiation. A typical profile of treated cells within 48 h postirradiation is shown in Fig. 1A. CEM (12D7) lymphocytes are the parental host cells for two well-studied latent HIV-1-infected cells, namely, ACH2 and 8E5. As expected, upon irradiation of CEM parental cells, two populations blocked at G0/G1 and G2/M appeared within the first 48 h. These populations remained blocked until DNA damage was repaired and allowed reentry into the next phase of the cell cycle. We have also observed a similar pattern of G0/G1 and G2/M blocks in other commonly used T cells, namely, H9, Jurkat, and Molt-4 cells (data not shown). However, the profile of HIV-1-infected cells looked very different upon gamma irradiation. The results of such an experiment are shown in Fig. 1B and C. In both ACH2 and 8E5 cells, a loss of G0/G1 population, as well as an increase in S-phase cells, was observed within the first 24 h. This was in marked contrast to CEM parental cells, where only a twofold drop in G0/G1 cells was observed after 24 h (56.5% at 0 h versus 36.9% at 24 h). HIV-1-infected cells also showed a dramatic increase in apoptotic cells after 48 h post-gamma irradiation. This increase in apoptotic cells was absent in the parental cells (Fig. 1, CEM 48-h samples). We have therefore attributed the loss of the G0/G1 population and the massive apoptosis effect to the presence of the HIV-1 genome in ACH2 and 8E5 cells.
We next examined the effect of HIV-1 gene expression and subsequent translation in infected cells. We chose ACH2 cells over 8E5 cells for the continuation of these studies mainly because they produced wild-type HIV-1 particles and both were from the same parental line. Total RNA was extracted before and after gamma irradiation and was used for Northern blot analysis. A full-length 32P-RNA-labeled probe was used for detection of various-size HIV-1 RNA populations. Generally, a doubly spliced series of RNA molecules was observed in ACH2 cells prior to gamma irradiation. They represent the first wave of HIV-1 transcripts, which are doubly spliced messages that are then translated for the next wave of processes, such as transactivation (by Tat), RNA transport and splicing (by Rev), and other virus-associated steps. Therefore, these small messages code for regulatory proteins, such as Tat, Rev, Nef, Vpr, and other accessory molecules needed for proper progeny formation. When performing Northern blot analysis from ACH2 cells, we found that HIV-1 transcripts increase within the first 24 h and remain constant up to 48 h (Fig. 2A). In fact all the appropriate transcripts prior to the assembly of virions were made within the first 24 h, which correlated with loss of a G0/G1 population and increase of S-phase cells.
To determine which of the accessory proteins was present in ACH2 cells before and after gamma irradiation, we performed Western blots of nuclear and cytoplasmic fractions, using anti-Tat, -Rev, -Vpr, and -Nef antibodies. The results are shown in Fig. 2B, where only Tat protein was present in the nuclear fractions of untreated zero-hour samples and only Nef protein was present in the cytoplasmic fractions of untreated ACH2 cells. Two positive-control cellular proteins, L32 and TBP, were used as protein markers for the cytoplasmic and nuclear fractions, respectively. Taken together, these data imply that the accessory proteins, such as Tat, in the nucleus may contribute to the loss of the G1/S checkpoint observed in latent HIV-1-infected cells.
Effect of Tat on various stages of the cell cycle in lymphocytes.
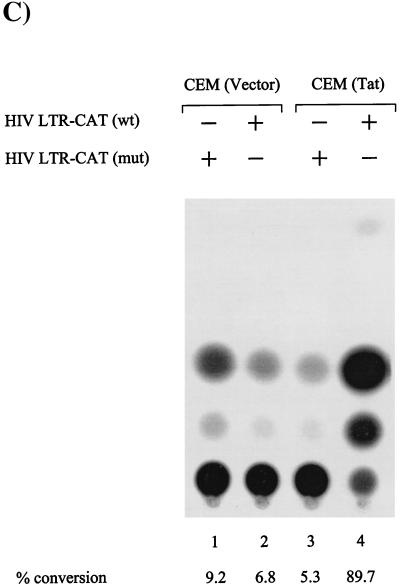
It has previously been shown that p53 protein is responsible for the G1/S and G2/M checkpoints in higher eukaryotic cells (47). Therefore, the sequestration of p53 protein by various cellular and/or viral proteins provides an important regulatory mechanism for abrogation of a given checkpoint (35, 41, 49). Along the same lines, HIV-1 Tat has been shown to bind to p53 (9, 26, 27) and is proposed to be important for the development of HIV-1-related malignancies (26). Interestingly, Tat expression has also been related to increased apoptosis in infected cells (40, 46). We therefore wished to examine whether Tat-expressing cells would show either loss of the G1/S checkpoint or apoptosis, as both are seen in ACH2 cells. We used two cell lines, CEM (Tat), containing a vector carrying the Tat gene, and CEM (Vector), containing only the backbone vector, and selected them in the presence of hygromycin (22). Upon gamma irradiation of these cells, there was a 10-fold drop in G0/G1 cells within the first 24 h in the CEM (Tat) cells compared to only a 2-fold drop among vector-transfected cells (Fig. 3A and B). However, no increase in apoptosis was observed at any time point following gamma irradiation in Tat-expressing cells. We have observed similar results with other T lymphocytes, such as H9-Tat and Jurkat-Tat cells (data not shown). To determine whether the Tat in CEM (Tat) cells was a functional protein, we transfected both a wild-type and a TAR mutant (TM26) reporter plasmid into these cells and scored for activated transcription using standard CAT assays (17, 19). As expected, the wild-type but not the mutant LTR-CAT construct was transactivated in CEM (Tat) cells (Fig. 3C). Taken together, these data suggest that loss of the G1/S checkpoint in ACH2 and CEM (Tat) cells is due to the presence of Tat protein and that the increase in apoptosis observed in irradiated ACH2 cells is due to other HIV regulatory or structural proteins.
FIG. 3.
Effect of Tat on G1/S checkpoint in CEM cells. Both CEM (Vector) and CEM (Tat) cells were grown in the presence of 100 μg of hygromycin/ml to the mid-log phase of growth. The cells were serum starved for 3 days prior to gamma irradiation. Following gamma irradiation, the cells were put in complete medium with 10% FBS and processed for FACS analysis at various time points. (A and B) Histograms and numbers of cells at various stages of the cell cycle. (C) Transfection of both cell types with either wild-type (wt) LTR-CAT or a TAR mutant (mut) (TM26) LTR-CAT in CEM (Vector) and CEM (Tat) cells. Five micrograms of each DNA was transfected (by electroporation) into the cells and processed 18 h later for CAT assay. Fifty micrograms of total protein was used for the CAT assay, and acetylated products were run on thin-layer chromatography, exposed on a PhosphorImager cassette and counted with Molecular Dynamics software. +, present; −, absent.
Mechanism of Tat-mediated loss of G1/S checkpoint.
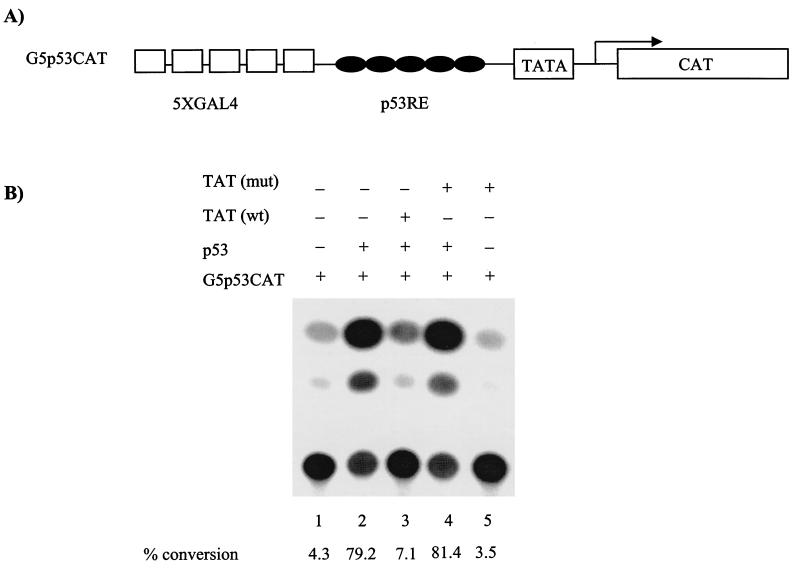
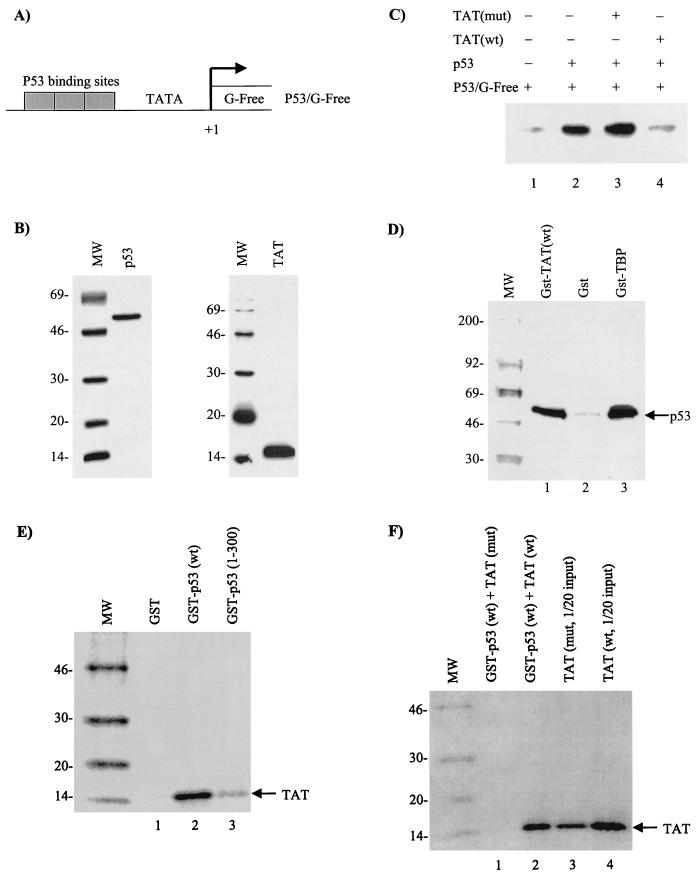
To determine the mechanism of loss of the G1/S checkpoint by Tat and its relationship with p53, we decided to perform two sets of in vivo and in vitro experiments. First, we performed transfections into CEM cells using a p53-responsive reporter, p53 activator, and Tat plasmids. The results of such an experiment are shown in Fig. 4. The plasmid G5p53CAT was responsive to a p53 activator construct (Fig. 4B, compare lanes 1 and 2). Furthermore, in agreement with previously published results (9, 26), wild-type but not mutant Tat was able to downregulate the effect of p53 in transfections (Fig. 4B, compare lanes 3 to 5). Transfection experiments score for transcription, message half-life, nuclear transport, and translation events. We wondered whether the observed effect of Tat and p53 was at the level of transcription and/or other subsequent events stated above. To find out, we performed in vitro transcription analysis with Tat and p53 using a supercoiled p53-responsive G-free cassette. The results are shown in Fig. 5C, where purified p53 was able to activate the p53/G-Free plasmid by a fourfold increase (1,250 counts for basal transcription versus 5,375 counts for activated transcription). In the presence of wild-type but not mutant Tat, the p53 activation is nearly abolished (Fig. 5, compare lanes 2 to 4). To further show that Tat indeed complexes with p53 as reported previously (27), we performed GST pull-down assays using GST-Tat and radiolabeled p53. The results of such an experiment are shown in Fig. 5D, where p53 binds as efficiently to Tat as to TBP. We next performed other control experiments, where the GST-p53 wild type or a mutant GST-p53 (1-300) was incubated with 35S-labeled wild-type Tat or a Tat mutant 41 (pGEM-Tat 41; a generous gift of A. Rice), and found that Tat binds to the C-terminal domain of p53 (Fig. 5E, lanes 2 and 3) and that the Tat mutant 41 was not able to bind to p53 (Fig. 5F, lanes 2 and 3) under 150 mM salt wash conditions. Collectively, these experiments imply that Tat may indeed bind to p53 and sequester its transactivation capability on p53-responsive elements.
FIG. 4.
In vivo effect of Tat on p53-activated transcription. CEM (12D7) cells were grown to mid-log phase and transfected with a reporter (G5P53CAT) and activator (CMV-p53) in the presence (+) or absence (−) of Tat expression vector. (A) Diagram of the reporter gene used for transfection studies, which contains five GAL4 binding sites and five DNA-responsive elements. (B) Transfection experiment in CEM cells using electroporation method. The samples were processed similarly to those in Fig. 3C.
FIG. 5.
In vitro analysis of Tat on P53-activated transcription. In vitro transcription assays were performed with a supercoiled G-free cassette DNA (P53/G-Free), CEM (12D7) whole-cell extract, and purified p53 and Tat proteins. (A) Diagram of the reporter G-Free plasmid. (B) Coomassie blue stain of p53 and Tat proteins used for in vitro transcription assays. (C) In vitro transcription with p53 (lane 2 was treated with monoclonal antibody PAb 421 to increase DNA binding) in the presence (+) or absence (−) of either mutant (lane 3, Tat 41 mutant [25]) (mut) or wild-type (lane 4) (wt) Tat. Neither wild-type nor mutant Tat protein alone activated P53/G-Free transcription (data not shown). (D) In vitro binding of GST-TBP (positive control), GST, or GST-Tat to 35S-labeled p53 (TNT; Promega). Samples were bound overnight and washed the next day, and bound p53 proteins were run on a 4 to 20% Tris-glycine gel, dried, and exposed to a PhosphorImager cassette. (E) Binding of 35S-labeled Tat to GST-p53 wild type or a mutant containing p53 residues 1 to 300. As previously reported (27), Tat binds to the C-terminal domain of p53. (F) Binding of either the wild-type or a 41 mutant of Tat to GST-p53. The experiments shown in panels E and F were performed in the presence of 50 μg of ethidium bromide/ml (to eliminate nonspecific binding to nucleic acids) and washed with TNE150 plus 0.1% NP-40 buffer prior to being loaded on SDS-PAGE.
Effect of cdkIs in HIV-1-infected cells.
cdks are generally active at specific stages of the cell cycle when bound to specific cyclin partners. The cyclin-cdk complexes are subject to inhibition by cdkIs, which sequester the enzymatic functions of cyclin-cdk complexes, thereby stopping cells at specific checkpoints. The G1 phase of the cell cycle contains two sets of inhibitors, the INK and KIP family members for early G1 and late G1 phases, respectively. The INK family members consist of p16(INK4A), p15(INK4B), p18(INK4C), and p19(INK4D), and they mainly inhibit early G1 kinases, such as cyclin D1 to -3/cdk4 to -6 (1, 11). The CIP/KIP family members are p21/Waf1/CIP1, p27(KIP1), and p57(KIP2), and they inhibit some early G1 kinases (i.e., p27 association with cyclin D1/cdk complex) but mainly the late G1/S checkpoint kinase, namely, cyclin E/cdk2 (48).
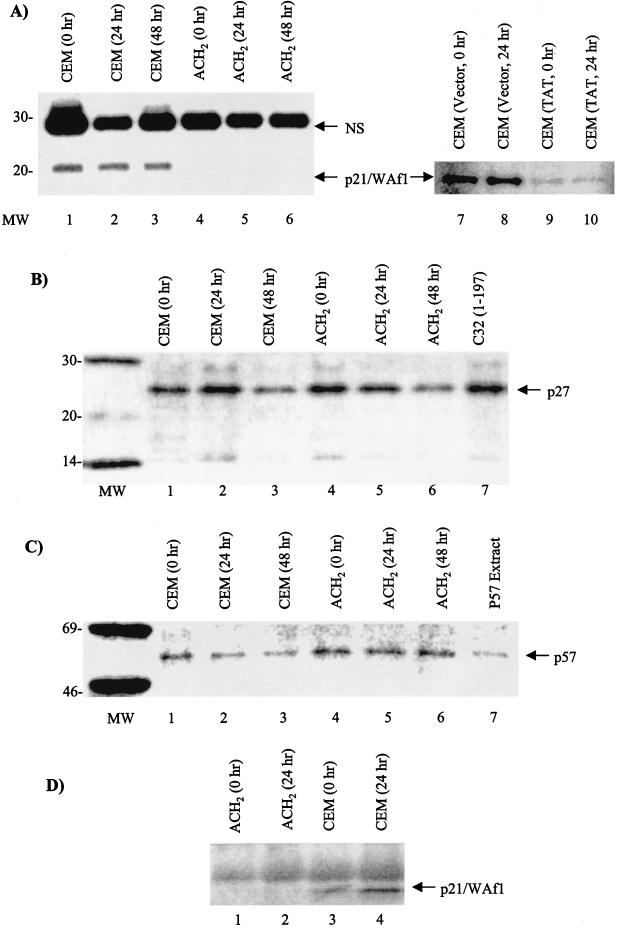
We therefore wished to examine the levels of late G1 cyclin inhibitors in infected and uninfected cells. Upon Western blot analysis, we observed the absence of p21/Waf1 in ACH2 cells before or after gamma irradiation (Fig. 6A, p21/Waf1 WB). Interestingly, p21/Waf1 levels were constant in CEM parental cells even 24 or 48 h post-gamma irradiation. However, similar levels of p27 and p57 inhibitors were present in infected and uninfected cells (Fig. 6B and C). A similar result was also obtained using the CEM vector control and the Tat-containing cell line, where p21/Waf1 levels were decreased in the Tat-containing cells (Fig. 6A, lanes 7 to 10). When we looked at the localization of p21/Waf1 protein in uninfected cells, we found that more of p21/Waf1 protein is localized in the nucleus following gamma irradiation (Fig. 6D, lanes 3 and 4), implying that transport of p21/waf1 into the nucleus may be a rate-limiting step as opposed to a transcriptional increase in these cells. Western blot analysis of early INK family members showed no change of cdkIs in infected or uninfected cells (data not shown). Collectively, these data imply that, in ACH2 cells, loss of the G1/S checkpoint is attributed to lack of cdkI p21/Waf1 expression.
FIG. 6.
Late-G1-phase cdkI KIP family members in infected and uninfected cells. Both CEM and ACH2 cells were gamma irradiated, and nuclear extracts were processed at various time points for cdkI levels. (A, B, and C) Western blots with anti-p21 (Waf1) monoclonal antibody (MAb) (A), anti-p27 MAb (B), and anti-p57 MAb (C). All samples were processed on a 10 to 20% Tricine gel (Novex), transferred to a polyvinylidene difluoride membrane, and Western blotted with appropriate antibodies. Antigen-antibody-associated complexes were detected with 125I-protein G (Amersham). The samples in panel A, lanes 7 to 10, were processed from CEM vector control and CEM Tat-expressing cells. (D) Nuclear extracts from CEM and ACH2 after gamma irradiation and Western blotting for p21/Waf1 protein.
Functional activation of G1/S kinase and cyclin E/cdk2 in HIV-1-infected cells.
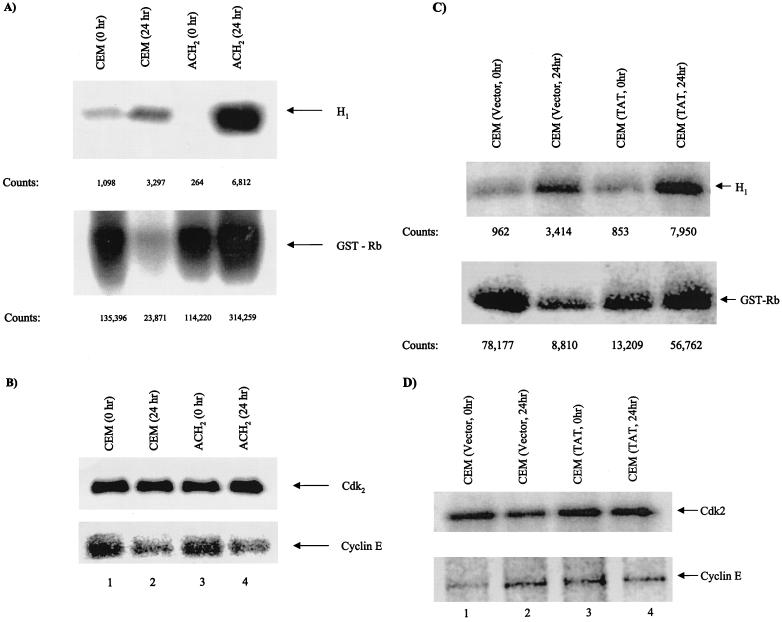
Next, we examined the effect of cyclin E/cdk2 kinase activity in infected and uninfected cells. We reasoned that if ACH2 cells have no p21/Waf1 present, they should have higher cyclin E/cdk2 kinase activity. We performed kinase assays with immunoprecipitated cyclin E/cdk2 complex from both CEM and ACH2 cells. The results of such an experiment are shown in Fig. 7A, where cyclin E/cdk2 immunoprecipitate was capable of a better phosphorylation of both histone H1 and Rb substrates post-gamma irradiation in HIV-1-infected cells. The cyclin E and cdk2 protein levels were similar before and after gamma irradiation (Fig. 7B). A similar result was also obtained in Tat-expressing cells, where higher levels of histone H1 and Rb phosphorylation were apparent 24 h post-gamma irradiation (Fig. 7C). The two cell types contained similar levels of cyclin E and cdk2, indicating that the change in phosphorylation pattern is not simply due to changes in expression levels of these cyclin-cdk complexes (Fig. 7D). It is interesting to note that both CEM and CEM (Vector) cells show lower levels of Rb phosphorylation 24 h post-gamma irradiation, perhaps due not so much to the p21/Waf1 transcriptional upregulation but simply to an active transport of p21/Waf1 into the nucleus, as seen in Fig. 6D. Taken together, loss of p21/Waf1 in infected cells may allow the cyclin E/cdk2-associated complex to push cells inappropriately into S phase and thus promote apoptosis.
FIG. 7.
Immunoprecipitation of cyclin E-associated complex in both infected and uninfected cells. Infected and uninfected cell extracts from gamma-irradiated cells were treated for immunoprecipitation with polyclonal rabbit anti-cyclin E antibody at 4°C. The immune complexes were pelleted the next day with protein A plus protein G beads, washed, and used for kinase assays with histone H1 or GST-Rb as substrates. (A) Labeled products resolved by SDS–4 to 20% PAGE and their corresponding counts with the Molecular Dynamics PhosphorImager software. (B) Straight Western blots for cdk2 and cyclin E in infected and uninfected cells. (C and D) Similar to panels A and B except that CEM (Vector) and CEM (Tat) were used as the starting materials for immunoprecipitations.
DISCUSSION
HIV-1 establishes latent infection of a certain population of CD4+ host cells, which could be long-term reservoirs for HIV-1. The expression of viral genes in such long-term-infected cells is strongly regulated by the cellular status, such as the phase of the cell cycle or the stage of cell differentiation. It has been demonstrated that activation of HIV-1 by NF-κB was induced by phorbol myristate acetate treatment during late G1 to S but not after entering G2 phase, indicating that the transcriptional factor(s) involved in viral gene expression is also largely regulated by the host cell cycle (45).
It has been proposed that retroviruses establish productive infection only in proliferating cells, such as T cells. Macrophages, however, are often considered to be nonproliferating in vitro yet are susceptible to HIV-1 infection. Treatment of monocyte-derived macrophages with aphidicolin, a specific inhibitor of DNA polymerase alpha and delta which arrests cells in G1/S phase of the cell cycle, also inhibited DNA synthesis but did not prevent establishment of productive infection, which is completely analogous to observations in T cells (39). More specifically, it has been demonstrated that completion of reverse transcription in macrophages inoculated with the HIV-1 Ba-L variant, and a macrophage-tropic ADA strain and two primary macrophage-tropic HIV-1 variants isolated from cerebrospinal fluid and from bronchoalveolar lavage from patients with AIDS, is dependent on cellular conditions that coincide with cell proliferation. Therefore, HIV-1 replication is restricted to cells with proliferative potential (23). Collectively, these data suggest that most if not all HIV-1 subtypes require the presence of an environment where cells either are doubling or are simply able to go from early G1 to late G1 phase of the cell cycle (i.e., macrophages).
It has been shown that the cdkI p21 (Sdi1, Cip1, and Waf1) binds to and inactivates all cyclin E/cdk2 complexes in cells that are under stress, such as gamma irradiation. Interestingly, it has recently been shown that in the senescent-cell-cycle-arrested cells, p21/Waf1 expression occurs prior to the accumulation of the cdk4-cdk6 inhibitor p16 (Ink4a), suggesting that p21 may also be an earlier upstream inhibitor than INK family members (42). However, the mechanism of inhibition, and therefore the blocking of cells at G1/S, is generally attributed to lack of complete Rb phosphorylation and the subsequent release of E2F family members.
We have investigated the effect of the G1/S checkpoint in latent HIV-1-infected cells. We found that gamma-irradiated ACH2 and 8E5 cells showed a loss of G0/G1 population within the first 24 h, increase of singly spliced RNA, and a dramatic increase of apoptotic cells after 48 h. This result is intriguing in that HIV basal transcription and transactivation may be phenomena of the mid-to-late G1 phase of the cell cycle. At the same time, Tat may directly affect the host cell cycle machinery at the G1/S checkpoint by sequestering p53, increasing Rb phosphorylation, and pushing cells into S phase. Subsequently, the host cell may sense the inappropriate entry into S phase and turn on its apoptotic machinery.
To our surprise, when we looked at the apoptosis pattern of Tat-expressing cells, we observed no increase of apoptosis in lymphocytes after gamma irradiation. Similar results were also seen in Jurkat-Tat, H9-Tat, THP-1–Tat, and HeLa-Tat cells compared to their parental counterparts, Jurkat, H9, THP-1, and HeLa cells, respectively (data not shown). In fact, we have always observed loss of a G0/G1 population in Tat-expressing cells and no apparent apoptosis within 48 h post-gamma irradiation. We therefore believe that the massive apoptosis seen in ACH2 or 8E5 cells is due to HIV-1 open reading frames other than that for Tat. Current preliminary results indicate that Env protein synthesis alone may be responsible for a majority of the apoptosis in these cells (F. Santiago and S. Chong, unpublished results). Future in-depth experiments with various Tats and Envs, from clades A to O, will determine whether the loss of G0/G1 population by Tat and apoptosis, possibly by Env, are general phenomena of all HIV-1 strains.
Loss of the G1/S checkpoint by Tat-expressing cells in HIV-1 latent cells is intriguing in light of reports that Tat functionally and physically interacts with p53 (Fig. 8). As reported previously, Tat-p53 functional interactions may allow the development of AIDS-related malignancies (26). This suggestion is plausible, since free Tat protein is able to act as a mitogen and can activate the transcription of a number of proliferative cytokines (15). The physical interaction of Tat and p53 may also have additional roles, one of which is indicated by the observation that p53 inhibits HIV-1 LTR gene expression in the absence of Tat (9). The p53-induced repression of the LTR could possibly be relieved by the sequestration of p53 protein at the G1/S checkpoint by Tat protein. This interaction would allow LTR gene expression and transactivation to take place. Simultaneously, Tat-p53 physical interaction can alter the gene expression of p53-responsive promoters, such as p21/Waf1. Downregulation of a cdkI, such as p21/Waf1, may account for the observed loss of the G1/S checkpoint.
FIG. 8.
Proposed model of Tat-p53 interaction and its functional consequences for p21/Waf1 levels. Upon DNA damage, the primary effect of p53 response at G1/S is upregulation of the p21/Waf1 gene product and its subsequent binding to the cyclin-cdk complex. The functional consequence of the p21-cyclin-cdk complex is to stop cells at the G1/S border prior to entry of the cells into the S phase. In HIV-infected cells, Tat inactivates p53 function, thus downregulating p21 expression and allowing loss of the G1/S checkpoint.
We also attempted to observe whether a similar loss of the G1/S checkpoint could occur in PBL samples from patients with HIV-1 infection. Using available patient samples (UMDNJ cohort group), we were unable to establish a semipurified baseline set of infected CD4+ cells from 10 patients with AIDS at various stages of disease development. The major inconsistency in experiments with patient PBLs had been that there were no available methods to purify infected CD4+ cells from patients who were under treatment with zidovudine and protease inhibitors (data not shown).
Loss of a checkpoint in HIV-1-infected cells has profound consequences for the virus. This may be a mechanism for the virus to push cells into S phase, which may be needed for subsequent virus-associated processes, such as RNA splicing, transport, translation, and packaging of virions. If so, then HIV-1 could serve as a model system to study many similar cellular pathways, such as basal transcription, activation, etc., in a cell cycle-dependent manner. Thus, retroviruses could serve as in vivo functional molecular probes, allowing us to better understand basic cellular machineries in higher eukaryotic cells.
ACKNOWLEDGMENTS
E. Clark and F. Santiago contributed equally to this work.
We thank Marlene Healey for the critical initial observations of loss of p21/Waf1 protein in infected cells and Ebony Brooks for assistance in preparing the manuscript.
This work was supported by NIH grants AI42524 and 13969 and in part by grant AI43894 to F.K.
REFERENCES
- 1.Biggs J R, Kraft A S. Inhibitors of cyclin-dependent kinase and cancer. J Mol Med. 1995;73:509–514. doi: 10.1007/BF00198902. [DOI] [PubMed] [Google Scholar]
- 2.Bohan C A, Kashanchi F, Ensoli B, Buonaguro L, Boris-Lawrie K A, Brady J N. Analysis of Tat transactivation of human immunodeficiency virus transcription in vitro. Gene Expr. 1992;2:391–407. [PMC free article] [PubMed] [Google Scholar]
- 3.Brugarolas J, Chandrasekaran C, Gordon J I, Beach D, Jacks T, Hannon G J. Radiation-induced cell cycle arrest compromised by p21 deficiency. Nature. 1995;377:552–557. doi: 10.1038/377552a0. [DOI] [PubMed] [Google Scholar]
- 4.Bunz F, Dutriaux A, Lengauer C, Waldman T, Zhou S, Brown J P, Sedivy J M, Kinzler K W, Vogelstein B. Requirement for p53 and p21 to sustain G2 arrest after DNA damage. Science. 1998;282:1497–1501. doi: 10.1126/science.282.5393.1497. [DOI] [PubMed] [Google Scholar]
- 5.Chang I J, McNulty E, Martin M. Human immunodeficiency viruses containing heterologous enhancer/promoters are replication competent and exhibit different lymphocyte tropisms. J Virol. 1993;67:743–752. doi: 10.1128/jvi.67.2.743-752.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chen J, Saha P, Kornbluth S, Dynlacht B D, Dutta A. Cyclin-binding motifs are essential for the function of p21CIP1. Mol Cell Biol. 1996;16:4673–4682. doi: 10.1128/mcb.16.9.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffman F D, Studzinski G P. Differentiation-related mechanisms which suppress DNA replication. Exp Cell Res. 1999;248:58–67. doi: 10.1006/excr.1999.4457. [DOI] [PubMed] [Google Scholar]
- 8.Deng C, Zhang P, Harper J W, Elledge S J, Leder P. Mice lacking p21CIP1/WAF1 undergo normal development, but are defective in G1 checkpoint control. Cell. 1995;82:675–684. doi: 10.1016/0092-8674(95)90039-x. [DOI] [PubMed] [Google Scholar]
- 9.Duan L, Ozaki I, Oakes J W, Taylor J P, Khalili K, Pomerantz R J. The tumor suppressor protein p53 strongly alters human immunodeficiency virus type 1 replication. J Virol. 1994;68:4302–4313. doi: 10.1128/jvi.68.7.4302-4313.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Duh E J, Maury W J, Folks T M, Fauci A S, Rabson A B. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA. 1989;86:5974–5978. doi: 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Drexler H G. Review of alterations of the cyclin-dependent kinase inhibitor INK4 family genes p15, p16, p18 and p19 in human leukemia-lymphoma cells. Leukemia. 1998;12:845–859. doi: 10.1038/sj.leu.2401043. [DOI] [PubMed] [Google Scholar]
- 12.el-Solh A, Kumar N M, Nair M P, Schwartz S A, Lwebuga-Mukasa J S. An RGD containing peptide from HIV-1 Tat-(65-80) modulates protooncogene expression in human bronchoalveolar carcinoma cell line, A549. Immunol Investig. 1997;26:351–370. doi: 10.3109/08820139709022692. [DOI] [PubMed] [Google Scholar]
- 13.Gartel A L, Serfas M S, Tyner A L. p21—negative regulator of the cell cycle. Proc Soc Exp Biol Med. 1996;213:138–149. doi: 10.3181/00379727-213-44046. [DOI] [PubMed] [Google Scholar]
- 14.Harper J W, Elledge S J, Keyomarsi K, Dynlacht B, Tsai L H, Zhang P, Dobrowolski S, Bai C, Connell-Crowley L, Swindell E, et al. Inhibition of cyclin-dependent kinases by p21. Mol Biol Cell. 1995;6:387–400. doi: 10.1091/mbc.6.4.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hofman F M, Wright A D, Dohadwala M M, Wong-Staal F, Walker S M. Exogenous tat protein activates human endothelial cells. Blood. 1993;82:2774–2780. [PubMed] [Google Scholar]
- 16.Iliakis G. Cell cycle regulation in irradiated and nonirradiated cells. Semin Oncol. 1997;24:602–615. [PubMed] [Google Scholar]
- 17.Kashanchi F, Duvall J F, Brady J N. Electroporation of viral transactivator proteins into lymphocyte suspension cells. Nucleic Acids Res. 1992;20:4673–4674. doi: 10.1093/nar/20.17.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kashanchi F, Duvall J F, Kwok R P, Lundblad J R, Goodman R H, Brady J N. The coactivator CBP stimulates human T-cell lymphotrophic virus type I Tax transactivation in vitro. J Biol Chem. 1998;273:34646–34652. doi: 10.1074/jbc.273.51.34646. [DOI] [PubMed] [Google Scholar]
- 19.Kashanchi F, Khleif S N, Duvall J F, Sadaie M R, Radonovich M F, Cho M, Martin M A, Chen S Y, Weinmann R, Brady J N. Interaction of human immunodeficiency virus type 1 Tat with a unique site of TFIID inhibits negative cofactor Dr1 and stabilizes the TFIID-TFIIA complex. J Virol. 1996;70:5503–5510. doi: 10.1128/jvi.70.8.5503-5510.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kashanchi F, Piras G, Radonovich M F, Duvall J F, Fattaey A, Chiang C M, Roeder R G, Brady J N. Direct interaction of human TFIID with the HIV-1 transactivator tat. Nature. 1994;367:295–299. doi: 10.1038/367295a0. [DOI] [PubMed] [Google Scholar]
- 21.Kastan M B, Kuerbitz S. Control of G1 arrest after DNA damage. Environ Health Perspect. 1993;101:55–58. doi: 10.1289/ehp.93101s555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kira T, Merin J P, Baba M, Shigeta S, Okamoto T. Anti-Tat MTT assay: a novel anti-HIV drug screening system using the viral regulatory network of replication. AIDS Res Hum Retroviruses. 1995;11:1359–1366. doi: 10.1089/aid.1995.11.1359. [DOI] [PubMed] [Google Scholar]
- 23.Kootstra N A, Schuitemaker H. Proliferation-dependent replication in primary macrophages of macrophage-tropic HIV type 1 variants. AIDS Res Hum Retroviruses. 1998;14:339–345. doi: 10.1089/aid.1998.14.339. [DOI] [PubMed] [Google Scholar]
- 24.Korin Y D, Zack J A. Nonproductive human immunodeficiency virus type 1 infection in nucleoside-treated G0 lymphocytes. J Virol. 1999;73:6526–6532. doi: 10.1128/jvi.73.8.6526-6532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Korin Y D, Zack J A. Progression to the G1b phase of the cell cycle is required for completion of human immunodeficiency virus type 1 reverse transcription in T cells. J Virol. 1998;72:3161–3168. doi: 10.1128/jvi.72.4.3161-3168.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li C J, Wang C, Friedman D J, Pardee A B. Reciprocal modulations between p53 and Tat of human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1995;92:5461–5464. doi: 10.1073/pnas.92.12.5461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Longo F, Marchetti M A, Castagnoli L, Battaglia P A, Gigliani F. A novel approach to protein-protein interaction: complex formation between the p53 tumor suppressor and the HIV Tat proteins. Biochem Biophys Res Commun. 1995;206:326–334. doi: 10.1006/bbrc.1995.1045. [DOI] [PubMed] [Google Scholar]
- 28.Lu H, Levine A J. Human TAFII31 protein is a transcriptional coactivator of the p53 protein. Proc Natl Acad Sci USA. 1995;92:5154–5158. doi: 10.1073/pnas.92.11.5154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mantel C, Braun S E, Reid S, Henegariu O, Liu L, Hangoc G, Broxmeyer H E. p21(cip-1/waf-1) deficiency causes deformed nuclear architecture, centriole overduplication, polyploidy, and relaxed microtubule damage checkpoints in human hematopoietic cells. Blood. 1999;93:1390–1398. [PubMed] [Google Scholar]
- 30.Masutani M, Nozaki T, Wakabayashi K, Sugimura T. Role of poly(ADP-ribose) polymerase in cell-cycle checkpoint mechanisms following gamma-irradiation. Biochimic. 1995;77:462–465. doi: 10.1016/0300-9084(96)88161-2. [DOI] [PubMed] [Google Scholar]
- 31.Mundt M, Hupp T, Fritsche M, Merkle C, Hansen S, Lane D, Groner B. Protein interactions at the carboxyl terminus of p53 result in the induction of its in vitro transactivation potential. Oncogene. 1997;15:237–244. doi: 10.1038/sj.onc.1201174. [DOI] [PubMed] [Google Scholar]
- 32.Muralidhar S, Doniger J, Mendelson E, Araujo J C, Kashanchi F, Azumi N, Brady J N, Rosenthal L J. Human cytomegalovirus mtrII oncoprotein binds to p53 and down-regulates p53-activated transcription. J Virol. 1996;70:8691–8700. doi: 10.1128/jvi.70.12.8691-8700.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ogryzko V V, Wong P, Howard B H. WAF1 retards S-phase progression primarily by inhibition of cyclin-dependent kinases. Mol Cell Biol. 1997;17:4877–4882. doi: 10.1128/mcb.17.8.4877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perry M E, Levine A J. Tumor-suppressor p53 and the cell cycle. Curr Opin Genet Dev. 1993;3:50–54. doi: 10.1016/s0959-437x(05)80340-5. [DOI] [PubMed] [Google Scholar]
- 35.Pise-Masison C A, Choi K S, Radonovich M, Dittmer J, Kim S I, Brady J N. Inhibition of p53 transactivation function by the human T-cell lymphotropic virus type 1 Tax protein. J Virol. 1998;72:1165–1170. doi: 10.1128/jvi.72.2.1165-1170.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Quillent C, Dumey N, Dauguet C, Clavel F. Reversion of a polymerase-defective integrated HIV-1 genome. AIDS Res Hum Retroviruses. 1993;9:1031–1037. doi: 10.1089/aid.1993.9.1031. [DOI] [PubMed] [Google Scholar]
- 37.Rounseville M P, Kumar A. Binding of a host cell nuclear protein to the stem region of human immunodeficiency virus type 1 trans-activation-responsive RNA. J Virol. 1992;66:1688–1694. doi: 10.1128/jvi.66.3.1688-1694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rounseville M P, Lin H C, Agbottah E, Shukla R R, Rabson A B, Kumar A. Inhibition of HIV-1 replication in viral mutants with altered TAR RNA stem structures. Virology. 1996;216:411–417. doi: 10.1006/viro.1996.0077. [DOI] [PubMed] [Google Scholar]
- 39.Schuitemaker H, Kootstra N A, Fouchier R A, Hooibrink B, Miedema F. Productive HIV-1 infection of macrophages restricted to the cell fraction with proliferative capacity. EMBO J. 1994;13:5929–5936. doi: 10.1002/j.1460-2075.1994.tb06938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seve M, Favier A, Osman M, Hernandez D, Vaitaitis G, Flores N C, McCord J M, Flores S C. The human immunodeficiency virus-1 Tat protein increases cell proliferation, alters sensitivity to zinc chelator-induced apoptosis, and changes Sp1 DNA binding in HeLa cells. Arch Biochem Biophys. 1999;361:165–172. doi: 10.1006/abbi.1998.0942. [DOI] [PubMed] [Google Scholar]
- 41.Steegenga W T, Shvarts A, Riteco N, Bos J L, Jochemsen A G. Distinct regulation of p53 and p73 activity by adenovirus E1A, E1B, and E4orf6 proteins. Mol Cell Biol. 1999;19:3885–3894. doi: 10.1128/mcb.19.5.3885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stein G H, Drullinger L F, Soulard A, Dulic V. Differential roles for cyclin-dependent kinase inhibitors p21 and p16 in the mechanisms of senescence and differentiation in human fibroblasts. Mol Cell Biol. 1999;19:2109–2117. doi: 10.1128/mcb.19.3.2109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stein G H, Dulic V. Origins of G1 arrest in senescent human fibroblasts. Bioessays. 1995;17:537–543. doi: 10.1002/bies.950170610. [DOI] [PubMed] [Google Scholar]
- 44.Szumiel I. Monitoring and signaling of radiation-induced damage in mammalian cells. Radiat Res. 1998;150:S92–S101. [PubMed] [Google Scholar]
- 45.Tobiume M, Fujinaga K, Kameoka M, Kimura T, Nakaya T, Yamada T, Ikuta K. Dependence on host cell cycle for activation of human immunodeficiency virus type 1 gene expression from latency. J Gen Virol. 1998;79:1363–1371. doi: 10.1099/0022-1317-79-6-1363. [DOI] [PubMed] [Google Scholar]
- 46.Wang P, Barks J D, Silverstein F S. Tat, a human immunodeficiency virus-1-derived protein, augments excitotoxic hippocampal injury in neonatal rats. Neuroscience. 1999;88:585–597. doi: 10.1016/s0306-4522(98)00242-5. [DOI] [PubMed] [Google Scholar]
- 47.Westphal C H. Cell-cycle signaling: Atm displays its many talents. Curr Biol. 1997;7:R789–R792. doi: 10.1016/s0960-9822(06)00406-4. [DOI] [PubMed] [Google Scholar]
- 48.Xu X, Nakano T, Wick S, Dubay M, Brizuela L. Mechanism of Cdk2/Cyclin E inhibition by p27 and p27 phosphorylation. Biochemistry. 1999;38:8713–8722. doi: 10.1021/bi9903446. [DOI] [PubMed] [Google Scholar]
- 49.Zimmermann H, Degenkolbe R, Bernard H U, O'Connor M J. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–6219. doi: 10.1128/jvi.73.8.6209-6219.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]