Abstract
To examine the cell fusion activity of hepatitis C virus (HCV) envelope proteins (E1 and E2), we have established a sensitive cell fusion assay based on the activation of a reporter gene as described previously (O. Nussbaum, C. C. Broder, and E. A. Berger, J. Virol. 68:5411–5422, 1994). The chimeric HCV E1 and E2 proteins, each consisting of the ectodomain of the E1 and E2 envelope protein and the transmembrane and cytoplasmic domains of the vesicular stomatitis virus G glycoprotein, were expressed on the cell surface. Cells expressing the chimeric envelope proteins and T7 RNA polymerase were cocultured with the various target cell lines transfected with a reporter plasmid encoding the luciferase gene under the control of the T7 promoter. After cocultivation, the cell fusion activity was determined by the expression of luciferase in the cocultured cells. The induction of cell fusion requires both the chimeric E1 and E2 proteins and occurs in a low-pH-dependent manner. Although it has been shown that HCV E2 protein binds human CD81 (P. Pileri, Y. Uematsu, S. Campagnoli, G. Galli, F. Falugi, R. Petracca, A. J. Weiner, M. Houghton, D. Rosa, G. Grandi, and S. Abrignani, Science 282:938–941, 1998), the expression of human CD81 alone is not sufficient to confer susceptibility to cell fusion in the mouse cell line. Treatment of the target cells with pronase, heparinase, or heparitinase reduced the cell fusion activity induced by the chimeric envelope proteins. These results suggest (i) that both HCV E1 and E2 proteins are responsible for fusion with the endosomal membrane after endocytosis and (ii) that certain protein molecules other than human CD81 and some glycosaminoglycans on the cell surface are also involved in the cell fusion induced by HCV.
Hepatitis C virus (HCV) is the major causative agent of non-A, non-B hepatitis throughout the world (9, 38). The majority of patients infected with HCV are unable to clear the virus, and many of these patients eventually develop chronic liver diseases. The spectrum of diseases caused by persistent HCV infection extends from chronic hepatitis to cirrhosis and finally to hepatocellular carcinoma. While the incidence of HCV infection has been remarkably diminished by the introduction of an efficient blood-screening system, HCV has already infected more than 100 million people worldwide (29).
HCV is a positive-stranded RNA virus with a genome size of approximately 9.4 kb; it contains a large open reading frame encoding a precursor polyprotein of approximately 3,000 amino acids (10, 34). Cleavages of this polyprotein are co- and posttranslational and generate at least 11 viral proteins, including two glycoproteins, E1 and E2 (27, 61, 63). The nonstructural proteins are processed by NS2 and NS3 proteases (5, 27, 65), whereas processing of the capsid protein and the two membrane glycoproteins, E1 and E2, is mediated by a host signal peptidase(s) (27, 28). Although some cell lines exhibit limited replication of HCV (33, 51, 62), studies of the infection mechanisms of HCV have been hampered by the lack of an efficient, reliable cell culture system that supports the full replication of HCV. The E1 and E2 proteins are putative virion envelope glycoproteins that contain 5 or 6 and 11 N-linked glycosylation sites, respectively (49). They are transmembrane proteins and associate with each other to form a stable, noncovalently linked heterodimer (18, 46, 58). The carboxyl-terminal deletion that removes the hydrophobic regions of these proteins results in translocation onto the cell surface, suggesting that E1 and E2 proteins have signals to retain these proteins in the endoplasmic reticulum membrane (12, 13, 46, 50). The extracellular domains of the E1 and E2 glycoproteins are thought to play an important role in the interactions between the virus and its receptor(s). Recently, human CD81 (hCD81) has been shown to be a binding receptor for the HCV E2 protein (57), and a correlation between the development of an antibody capable of inhibiting the interaction of E2 protein with hCD81 and recovery from chronic hepatitis C has been demonstrated (32).
To examine the initial step of HCV infection in more detail, we established a highly sensitive cell fusion assay system. The chimeric HCV envelope proteins each consists of (i) a signal sequence and transmembrane and cytoplasmic domains of vesicular stomatitis virus (VSV) G glycoprotein and (ii) the ectodomain of the HCV E1 or E2 envelope protein expressed on the cell surface. Cell fusion activity was determined by the reporter gene activation method (54). Characterization of the cell fusion induced by the chimeric HCV envelope proteins, including optimal pH, host range, and the surface molecules on the target cells responsible for the fusion, will be discussed.
MATERIALS AND METHODS
Construction of expression plasmids.
The HCV cDNA (genotype 1b) used in this study was originally isolated from a blood sample of an HCV carrier (NIHJ1) (2). The infectivity of the blood was shown by incidental infection of a human and by experimental infection of chimpanzees. To construct expression plasmids encoding chimeric HCV envelope proteins, pCAGVSV, encoding a signal sequence, a transmembrane domain, and the cytoplasmic tail of the VSV G protein was generated. The signal sequence gene of the G protein of VSV (serotype Indiana) was synthesized by the following oligonucleotides: 5′-AATTCTGACA CTATGAAGTGCCTTTTGTACTTAGCCTTTTTATTCATTGGGGTGAAT TGC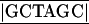 T-3′ (top strand) and 5′-CTAGA
T-3′ (top strand) and 5′-CTAGA GCAATTCACCC CAATGAATAAAAAGGCTAAGTACAAAAGGCACTTCATAGTGTCAG-3′ (bottom strand), both of which contain an additional EcoRI site (underlined) at the 5′ end and an additional NheI site (box) and XbaI site (boldface) at the 3′ end. The transmembrane and cytoplasmic regions of the VSV G protein were amplified by PCR using pGL-1 (59) as a template and the following primers: sense primer, 5′-AAATCTAGAAAAAGCTCTATTGCCTCTTTTTTCT-3′, containing an additional XbaI site (underlined); antisense primer, 5′-AAA
GCAATTCACCC CAATGAATAAAAAGGCTAAGTACAAAAGGCACTTCATAGTGTCAG-3′ (bottom strand), both of which contain an additional EcoRI site (underlined) at the 5′ end and an additional NheI site (box) and XbaI site (boldface) at the 3′ end. The transmembrane and cytoplasmic regions of the VSV G protein were amplified by PCR using pGL-1 (59) as a template and the following primers: sense primer, 5′-AAATCTAGAAAAAGCTCTATTGCCTCTTTTTTCT-3′, containing an additional XbaI site (underlined); antisense primer, 5′-AAA GAATTCAT CTGTTGTGCAGGATTTGAGTTACT-3′, containing an additional EcoRI site (underlined) and HindIII site (box). The signal sequence and the amplified fragment digested with XbaI and HindIII were inserted into the EcoRI and HindIII sites of pUC19. After amplification in Escherichia coli, the VSV G protein gene sequences were cut out by digestion with EcoRI and inserted into the pCAGGS vector, which has the CAG promoter consisting of the cytomegalovirus (CMV) immediate-early enhancer, the chicken β-actin promoter, and the rabbit β-globin polyadenylation signal (53). The CAG promoter can be utilized in a wide variety of mammalian cell lines and exhibits stronger expression than the CMV promoter (53). Coding sequences for ectodomains of HCV envelope proteins (E1, amino acids [aa] 192 to 340; E2, aa 384 to 711) were amplified by PCR using pNIHJ1 as a template after the addition of NheI and XbaI sites at the 5′ and 3′ ends, respectively. The ectodomain-encoding sequences of the amplified genes were excised by NheI and XbaI digestion and inserted into the same sites of pCAGVSV. The resulting plasmids, pCAV340V and pCAV711V, carried the genes encoding the chimeric HCV E1 and E2 proteins, respectively under the control of the CAG promoter (Fig. 1A). Plasmids encoding the signal sequence of the VSV G protein and an authentic E1 (aa 192 to 383; pCAV383) or E2 (aa 384 to 809; pCAV809) protein under the control of the CAG promoter were also constructed. These expression plasmids were verified by DNA sequencing. Four plasmids encoding VSV G, HCV C, E1, E2 (aa 1 to 810), HCV E1, and E2 (aa 155 to 810) proteins and β-galactosidase, respectively, under the control of the CAG promoter were also constructed and used as controls.
GAATTCAT CTGTTGTGCAGGATTTGAGTTACT-3′, containing an additional EcoRI site (underlined) and HindIII site (box). The signal sequence and the amplified fragment digested with XbaI and HindIII were inserted into the EcoRI and HindIII sites of pUC19. After amplification in Escherichia coli, the VSV G protein gene sequences were cut out by digestion with EcoRI and inserted into the pCAGGS vector, which has the CAG promoter consisting of the cytomegalovirus (CMV) immediate-early enhancer, the chicken β-actin promoter, and the rabbit β-globin polyadenylation signal (53). The CAG promoter can be utilized in a wide variety of mammalian cell lines and exhibits stronger expression than the CMV promoter (53). Coding sequences for ectodomains of HCV envelope proteins (E1, amino acids [aa] 192 to 340; E2, aa 384 to 711) were amplified by PCR using pNIHJ1 as a template after the addition of NheI and XbaI sites at the 5′ and 3′ ends, respectively. The ectodomain-encoding sequences of the amplified genes were excised by NheI and XbaI digestion and inserted into the same sites of pCAGVSV. The resulting plasmids, pCAV340V and pCAV711V, carried the genes encoding the chimeric HCV E1 and E2 proteins, respectively under the control of the CAG promoter (Fig. 1A). Plasmids encoding the signal sequence of the VSV G protein and an authentic E1 (aa 192 to 383; pCAV383) or E2 (aa 384 to 809; pCAV809) protein under the control of the CAG promoter were also constructed. These expression plasmids were verified by DNA sequencing. Four plasmids encoding VSV G, HCV C, E1, E2 (aa 1 to 810), HCV E1, and E2 (aa 155 to 810) proteins and β-galactosidase, respectively, under the control of the CAG promoter were also constructed and used as controls.
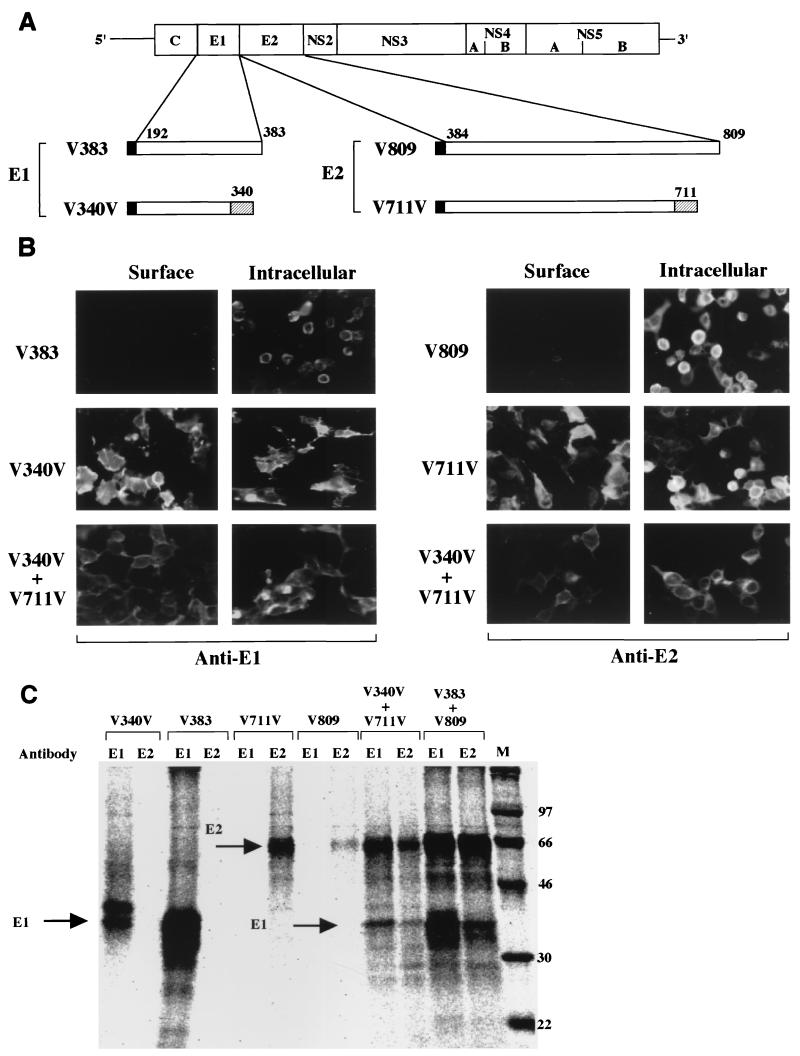
FIG. 1.
Expression of chimeric HCV envelope proteins. (A) Structures of HCV envelope genes used for expression. Solid bar in N terminus, signal sequence of VSV G protein. V383 and V809 are cDNA clones encoding full-length E1 and E2 envelope proteins, respectively. V340V and V711V encode chimeric envelope constructs, each of which has a deletion in the C terminus and possesses transmembrane and cytoplasmic domains of the VSV G protein (hatched bars). All these cDNAs were inserted into the pCAGGS vector under the control of the CAG promoter (53). (B) Immunofluorescence micrographs of 293T cells transfected with the expression plasmids encoding the full-length or chimeric HCV envelope proteins. Cell surface (left) and intracellular (right) expression of the envelope proteins are indicated. (C) Immunoprecipitation of cells expressing the full-length or chimeric envelope proteins. Cells transfected with the expression plasmids were labeled with [35S]methionine and immunoprecipitated with anti-E1 or -E2 monoclonal antibodies. In cells expressing one of the full-length or chimeric proteins, each envelope protein was precipitated by a specific antibody. In cells coexpressing full-length or chimeric E1 and E2 envelope proteins, both E1 and E2 proteins were coprecipitated by either of the antibodies. Molecular mass markers (kilodaltons) are at the right.
Cells.
CHO, Huh7, FLC4, Hep G2, 293T, BRL3A, MA104, COS7, HeLa, BHK21, NMuLi, and NIH 3T3 cell lines were maintained in Dulbecco's modified Eagle's medium (DMEM; Gibco BRL, Gaithersburg, Md.) containing 2 mmol of l-glutamine, penicillin (50 IU/ml), and streptomycin (50 μg/ml) and supplemented with 10% fetal bovine serum (FBS). Hep G2 cells were purchased from Dainippon Pharmaceutical Co., Ltd., Osaka, Japan. Huh7 cells were a gift from the Japanese Cancer Research Resources Bank-Cell, Tokyo, Japan. BRL3A and NMuLi cells were purchased from the American Type Culture Collection. NIH 3T3 cells constitutively expressing hCD81 were kindly provided by S. Abrignani (IRIS, Chiron, Siena, Italy).
Expression of the chimeric HCV proteins.
293T cells (8 × 105 cells in a 35-mm-diameter plate) were transfected with 1.0 μg of plasmids in the presence of Trans IT-LT1 (Mirus Co., Madison, Wis.) and Opti-MEM (Gibco BRL) and incubated at 37°C for 36 h. Expression of the HCV proteins was analyzed by indirect immunofluorescence assay and immunoprecipitation analysis. The cells were washed with phosphate-buffered saline (PBS) and fixed with 3% paraformaldehyde in PBS for 20 min at room temperature. These fixed cells were then permeabilized with 0.2% Triton X-100 for 3 min at room temperature and blocked with a nonfat milk solution (Block Ace; Yukijirushi Co., Sapporo, Japan). The cells were incubated with anti-HCV E1 or E2 monoclonal antibodies (46), which were provided by M. Kohara (The Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan) and which were diluted 1:200 in PBS for 60 min at 37°C. The cells were further incubated with fluorescein isothiocyanate-conjugated goat anti-mouse immunoglobulin G (TAGO, Burlingame, Calif.) diluted 1:500 in PBS. Observations were made through the use of fluorescence microscopy (TE300; Nikon, Tokyo, Japan). For immunoprecipitation, 293T cells transfected with the expression plasmids were incubated for 24 h and labeled with 20 μCi of Tran 35S-label (ICN, Irvine, Calif.) for 4 h after 2 h of starvation with medium deficient in FBS, methionine, and cysteine. The cells were washed twice with PBS and dissolved in 400 μl of TNE buffer (10 mM Tris-HCl [pH 7.8], 150 mM NaCl, 1 mM EDTA, 1% NP-40, 10 μg of aprotinin per ml). Cell lysate (100 μl) was diluted with 900 μl of TNE buffer and incubated with 0.5 μl of the anti-E1 or -E2 monoclonal antibody and 20 μl of protein A-Sepharose (Pharmacia) (50% [vol/vol] suspension in TNE buffer) for 1 h at 4°C with rotation. After centrifugation at 8,000 × g for 10 s at 4°C, the pellets were washed twice with TNE buffer. The immunoprecipitates were analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis.
Cell fusion assay of 293T cells transiently expressing the chimeric HCV envelope proteins.
293T cells (8 × 105 cells in a 35-mm-diameter plate) were transfected with expression plasmids encoding the chimeric envelope proteins or HCV structural proteins (0.8 μg), VSV G protein (0.25 μg), or β-galactosidase (0.8 μg) together with two reporter plasmids, pT7EMCVLuc (1.0 μg) (4) and pRL-CMV (0.1 μg) (Promega, Madison, Wis.) by Trans IT-LT1 (Mirus). To avoid severe cytopathic effects (CPE) due to the expression of VSV G protein, a smaller amount of the expression plasmid for the VSV G protein was used for transfection. pT7EMCVLuc has a firefly luciferase gene under the control of the T7 promoter, and pRL-CMV has a sea pansy luciferase gene under the control of the CMV promoter; expression of the latter gene was used as an internal standard for transfection. The target cell lines (2 × 105 cells per well in a 24-well plate) of Hep G2 and FLC4 were infected with a replication-deficient vaccinia virus recombinant expressing T7 RNA polymerase (DIsT7; Ishii et al., unpublished data) at a multiplicity of infection of 2.5 and incubated for 12 h. We utilized this virus to prevent the extensive CPE caused by replication of the vaccinia virus, which can interfere with a long-term analysis of the expressed products (69). Between 36 and 48 h after transfection, the 293T cells were treated with 0.05% EDTA in PBS and suspended in DMEM containing 10% FBS. The 293T cells (2 × 105 cells per well) were overlaid onto the target cells and incubated for 5 h. The cocultured cells were bathed in PBS at pH 5.0 for 2 min at 37°C and then were incubated with DMEM containing 10% FBS for 5 h. The cell fusion activity was quantitatively determined by measuring firefly luciferase gene expression in the lysates of the cocultured cells after it was standardized with sea pansy luciferase gene expression. Firefly and sea pansy luciferase activities were determined by the dual-luciferase reporter assay system (Promega) as described previously (4). Relative light units were measured with a luminometer (Berthold, Wildbad, Germany).
Cell fusion assay of CHO cell lines constitutively expressing HCV envelope proteins.
CHO cell lines constitutively expressing the chimeric HCV envelope proteins (Matsuura et al., submitted for publication) (2 × 104 cells per well in a 96-well plate) were transfected with an expression plasmid carrying the T7 RNA polymerase gene under the control of the CAG promoter (Ishii et al., unpublished data) (pCAGT7; 0.05 μg/well) and incubated for 24 h. The target cell lines (8 × 105 cells in a 35-mm-diameter plate) were transfected with pT7EMCVLuc (1.0 μg) together with pRL-CMV (0.1 μg). Twenty-four hours after transfection, the target cells were trypsinized, suspended in DMEM containing 10% FBS, and cultured for 8 h in suspension. The target cells (2 × 104 cells per well) were overlaid onto the CHO cell lines expressing T7 RNA polymerase, cocultured for 5 h, bathed in PBS at pH 5.0 for 2 min at 37°C, and incubated in DMEM containing FBS for 5 h. The cell fusion activity was determined as described above.
Chemical and enzymatic modification of cells.
Hep G2 cells were transfected with the reporter plasmids and suspended in serum-free DMEM. Sodium periodate, pronase, phospholipase C, or glycosaminoglycan (GAG) lyases (heparinase, heparitinase, hyaluronidase, and keratanase) were added to the cells in various concentrations, and the cells were incubated for 1 h. To stop the treatment, an equal volume of DMEM containing 10% FBS was added, and the cells (2 × 104 cells) were cocultured with the same number of CHO cells (in a 96-well plate) either constitutively expressing both the chimeric E1 and E2 proteins or transfected with the expression plasmid encoding the VSV G protein, as described previously. To avoid extensive CPE due to expression of the VSV G protein, CHO cells (2 × 104 cells per well in a 96-well plate) were transfected with 0.02 μg of the expression plasmid. After 5 h of cocultivation, cells were bathed in PBS, pH 5.0, for 2 min at 37°C, and the cell fusion activity was determined as described above.
Reagents.
Phospholipase C (Bacillus cereus grade I) was obtained from Boehringer GmbH, Mannheim, Germany. Pronase and sodium periodate were obtained from Sigma-Aldrich, Tokyo, Japan. Heparitinase (Flavobacterium heparinum), heparinase (F. heparinum), hyaluronidase (Streptomyces hyalurolyticus), and keratanase (Pseudomonas sp.) were kindly provided by Seikagaku Corporation, Tokyo, Japan.
RESULTS
Characterization of chimeric HCV envelope proteins.
To express HCV envelope proteins on the cell surface, we generated chimeric HCV envelope constructs (V340V and V711V) encoding the ectodomains of the E1 or E2 protein and the signal sequence, transmembrane region, and cytoplasmic tail of the VSV G protein (Fig. 1A). As negative controls, we also prepared cDNA clones encoding the authentic E1 and E2 proteins (V383 and V809). To normalize the expression levels of these envelope proteins, we utilized the signal sequence of the VSV G protein instead of the authentic anchor/signal sequences of HCV proteins. 293T cells transfected with the expression plasmids encoding authentic or chimeric HCV envelope proteins were examined by immunofluorescence analysis to determine the localization of the proteins (Fig. 1B). The authentic E1 and E2 proteins expressed by transfection with pCAV383 or pCAV809 were detected in the cytoplasm, but these proteins were not translocated onto the cell surface. In contrast, the chimeric E1 and E2 proteins expressed by transfection with pCAV340V or pCAV711V were efficiently translocated onto the cell surface. Although it has been previously reported that the folding of the E1 protein is dependent on the coexpression of the E2 protein (48), the chimeric E1 protein in this study was efficiently expressed and translocated onto the cell surface without coexpression of the chimeric E2 protein.
To examine the interaction of the HCV envelope proteins, cells transfected with the expression plasmids were labeled with [35S]methionine and immunoprecipitated with the specific monoclonal antibodies. Cells expressing the authentic and chimeric E1 or E2 proteins were precipitated by either of the specific antibodies. In cells coexpressing the authentic or chimeric E1 and E2 proteins, coprecipitation of E1 and E2 proteins by each antibody was observed. The expression level of the chimeric envelope proteins in cells cotransfected with the expression plasmids was lower than that in cells transfected with either of the expression plasmids (Fig. 1C). The ectodomain but not the transmembrane or cytoplasmic region of VSV G protein was responsible for the oligomerization of the protein (14). Therefore, the coprecipitation of the chimeric E1 and E2 proteins suggests that the ectodomains of the E1 and E2 proteins of HCV are sufficient for heterodimer formation.
Cell fusion assay using 293T cells transiently expressing HCV envelope proteins.
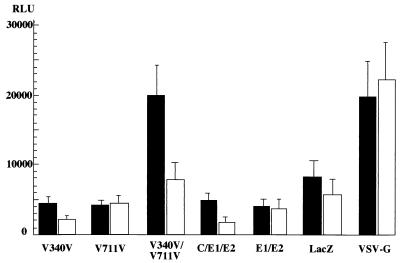
To examine the cell fusion activity of the HCV envelope proteins, we established a highly sensitive and quantitative reporter gene activation method after modifying the original method (54). We used the luciferase gene instead of the β-galactosidase gene as a reporter to enhance the sensitivity and an internal standard to normalize the transfection efficiency (55). 293T cells were transfected with expression plasmids encoding the chimeric HCV envelope proteins, VSV G protein, authentic HCV structural proteins, or β-galactosidase together with a reporter plasmid carrying a firefly luciferase gene under the control of the T7 promoter. As an internal standard, a plasmid carrying a sea pansy luciferase gene under the control of the CMV promoter was used. The target cell lines of Hep G2 and FLC4 were infected with DIsT7, which expresses T7 RNA polymerase, and incubated for 12 h. The 293T cells were overlaid onto the target cells and incubated for 5 h. The cocultured cells were treated with medium having an acidic pH for 2 min and incubated in DMEM containing FBS for a further 5 h. The cell fusion activity was then determined by the expression of firefly luciferase in the lysates of the cocultured cells after standardization with sea pansy luciferase (Fig. 2). Although the fusion activity of cells expressing both the chimeric E1 and E2 proteins was slightly higher than that of cells expressing LacZ, it was three to four times higher than that of cells expressing either of the chimeric envelope proteins. Cells expressing authentic HCV proteins (all of the HCV structural proteins or both the E1 and E2 proteins) or β-galactosidase exhibited a low level of activity. Cells expressing the VSV G protein exhibited a high level of fusion activity. These results indicate that both the chimeric E1 and E2 proteins, which are expressed on the cell surface, are required for cell fusion.
FIG. 2.
Cell fusion assay of 293T cells transiently expressing the chimeric HCV envelope proteins. 293T cells transfected with expression plasmids encoding the chimeric HCV envelope proteins (V340V and/or V711V), authentic HCV structural proteins (C, E1, and E2 or E1 and E2), β-galactosidase (LacZ), or VSV G protein together with two reporter gene plasmids were cocultured with human hepatoma cell lines Hep G2 (solid bars) or FLC4 (open bars) expressing T7 RNA polymerase. The relative luciferase activity (relative light units [RLU]) after normalization (firefly luciferase activity/sea pansy luciferase activity × 106) is shown. The relative activities were determined by at least three independent experiments, each of which was conducted with triplicate samples. Vertical lines, standard deviations.
Cell fusion assay of CHO cell lines constitutively expressing HCV envelope proteins.
Although the cell fusion assay using 293T cells proved to be useful, the expression of the proteins depended on the transfection efficiency of the expression plasmids, and this was not always stable. Therefore, we established CHO cell lines constitutively expressing the chimeric HCV envelope proteins, and these cell lines were used for further study. The expression of the HCV envelope glycoproteins on the surface was confirmed by immunofluorescence and fluorescence-activated cell sorter analyses (Matsuura et al., submitted).
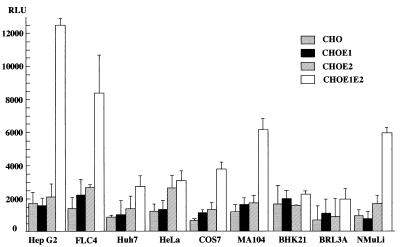
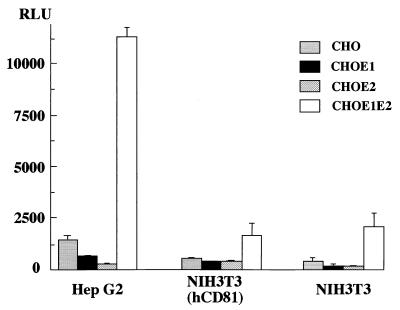
To examine the cell fusion activity of the chimeric HCV envelope proteins with various mammalian cell lines, the CHO cell line constitutively expressing the chimeric HCV proteins was cocultured with various target cell lines (Fig. 3). The target cells transfected with the reporter plasmids were trypsinized 24 h after transfection and incubated for 8 h in suspension in DMEM containing 10% FBS. They were then overlaid onto the CHO cell lines, which were transfected with pCAGT7, which carries the T7 RNA polymerase gene under the control of the CAG promoter (53). After 5 h of cocultivation and 2 min of low-pH exposure, the cocultured cells were incubated in DMEM containing 10% FBS for a further 5 h, and the cell fusion activity was determined as described above. Levels of expression of sea pansy luciferase, which was used as an internal standard for the transfection of reporter plasmids, among the target cell lines examined were similar (data not shown). The CHO cell line expressing both the E1 and E2 proteins consistently exhibited higher fusion activity than those expressing either of the chimeric envelope proteins and parental CHO cells. Among the cell lines examined, human hepatoma cell line Hep G2 exhibited the highest fusion activity. Human hepatoma cell line FLC4, monkey kidney cell lines MA104 and COS7, and murine liver cell line NMuLi also exhibited a relatively high level of fusion activity. In contrast, human hepatoma cell line Huh7, human cervix cancer cell line HeLa, hamster kidney cell line BHK, and rat liver cell line BRL3A showed little cell fusion activity. These results confirm the data obtained from the transient expression, suggesting that both the E1 and E2 proteins are required for efficient cell fusion.
FIG. 3.
Cell fusion assay of CHO cell lines expressing the chimeric HCV envelope proteins with various mammalian cell lines. CHO cell lines expressing either or both of the chimeric E1 and E2 proteins (CHOE1, CHOE2, and CHOE1E2, respectively) and the parent cell line (CHO) were cocultured with various cell lines. Cell fusion activity was determined as described for Fig. 2. RLU, relative light units.
Low-pH dependency in cell fusion.
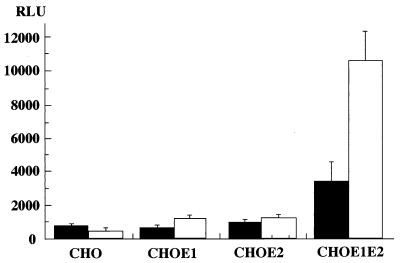
Flaviviruses have been shown to require low pH for cell fusion (66). To confirm the pH-dependent cell fusion of HCV envelope proteins, we examined the cell fusion activity with low-pH exposure (from pH 4.8 to 6.4) for 2 min after cocultivation with Hep G2 cells and found that treatment with medium at pH 5.0 resulted in the highest activity (data not shown). The CHO cell line expressing both of the HCV envelope proteins exhibited three-times-higher activity of fusion to Hep G2 cells after an exposure of pH 5.0 than those not treated (Fig. 4). Other cell lines expressing either of the chimeric envelope proteins and parent CHO cells showed a low level of cell fusion irrespective of the low-pH treatment. The enhancement of cell fusion activity by low-pH treatment suggests that HCV enters the target cell via the endosome, where the envelope proteins undergo conformational change into a fusion-competent state.
FIG. 4.
pH dependency of cell fusion. CHO cell lines expressing either or both of the chimeric E1 and E2 proteins (CHOE1, CHOE2, and CHOE1E2, respectively) and the parent cell line (CHO) were cocultured with Hep G2 cells. Cell fusion activity was determined after low-pH treatment (open bars) or without the treatment (solid bars). Cell fusion activity was determined as described for Fig. 2. RLU, relative light units.
Effect of hCD81 expression on cell fusion.
Recently, hCD81 has been shown to be a binding receptor of the E2 protein (57). hCD81 is a member of the tetraspanin family, which traverses the membrane four times. To examine the role of hCD81 in the cell fusion of HCV, mouse cell line NIH 3T3 constitutively expressing hCD81 was examined for its fusion activity with the CHO cell lines expressing either or both of the chimeric E1 and E2 proteins. Even though its fusion activity is much lower than that of the Hep G2 cell line, the NIH 3T3 cell line shows four- to fivefold higher activity of fusion to CHO cells expressing both of the chimeric envelope proteins than cell lines expressing either of the envelope proteins and the parental CHO cells. However, no augmentation was observed in the NIH 3T3 cell line by expression of hCD81, as shown in Fig. 5. Indeed, the Hep G2 cell line shows the highest fusion activity, but the expression level of hCD81 in this cell line is much lower than that in other human cell lines examined (see Discussion). These results suggest that a molecule other than hCD81 might be required for HCV cell fusion.
FIG. 5.
Effect of hCD81 expression on cell fusion. CHO cell lines expressing either or both of the chimeric E1 and E2 proteins (CHOE1, CHOE2, and CHOE1E2, respectively) and the parent cell line (CHO) were cocultured with the NIH 3T3 cell line constitutively expressing hCD81 or the parent cell line. As a positive control, Hep G2 cells were used. Cell fusion activity was determined as described for Fig. 2. RLU, relative light units.
Effects of chemical and enzymatic modifications of Hep G2 cells on cell fusion activity.
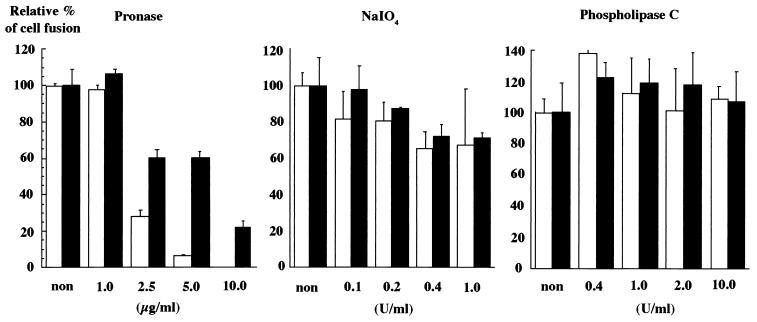
To determine the role of cell surface proteins, carbohydrates, and phospholipids on the target cells in the cell fusion mediated by HCV envelope proteins, Hep G2 cells were treated with various concentrations of pronase, sodium periodate, or phospholipase C (Fig. 6). When cells were treated with pronase, the fusion activity of the cell was reduced in a dose-dependent manner. The reduction in the fusion activity of Hep G2 cells in response to the treatment was much more evident in the CHO cell lines expressing HCV envelope proteins than in those expressing the VSV G protein. On the other hand, treatment with phospholipase C or sodium periodate resulted in no clear reduction in fusion activity.
FIG. 6.
Effects of chemical modifications of Hep G2 cells on cell fusion. CHO cell lines constitutively expressing both the chimeric E1 and E2 proteins (open bars) or transiently expressing the VSV G protein (solid bars) were cocultured with Hep G2 cells treated with various concentrations of pronase, sodium periodate, or phospholipase C. The relative cell fusion activities were determined by at least three independent experiments, each of which was conducted with triplicate samples. Vertical lines, standard deviations.
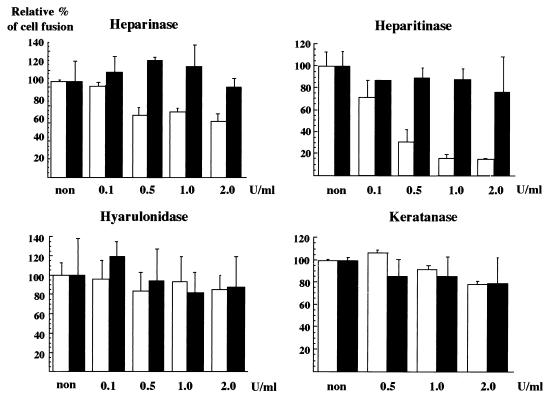
GAGs have been shown to be important in the cell surface binding of a number of microorganisms (60). In dengue virus, for example, a highly sulfated heparan sulfate has been shown to effectively prevent the infection of target cells (8). To assess the role of cell surface GAGs on the cell fusion of HCV, Hep G2 cells were treated with various GAG lyases, as shown in Fig. 7. The treatment of Hep G2 cells with heparinase or heparitinase reduced the fusion activity, whereas no effect was observed in response to treatment with keratanase or hyaluronidase. In particular, heparitinase treatment reduced by more than 80% the fusion activity induced by the HCV envelope proteins but had no effect on the cell fusion by the VSV G protein. These results suggest that protein molecules and certain GAGs on the surface of Hep G2 cells play an important role in cell fusion.
FIG. 7.
Effects of GAG lyase treatments of Hep G2 cells on cell fusion. CHO cell lines constitutively expressing both the chimeric E1 and E2 proteins (open bars) or transiently expressing VSV G protein (solid bars) were cocultured with Hep G2 cells treated with various concentrations of heparinase, heparitinase, hyaluronidase, or keratanase. The relative cell fusion activities were determined as described for Fig. 6. non, no treatment.
DISCUSSION
Most enveloped viruses invade host cells by attachment to cell surface receptors, followed by fusion of the viral membrane to the host membrane either at neutral pH or at low pH in endocytic vesicles (67). In this context, fusion events may well be divided into two classes, low pH dependent and pH independent (44, 66, 68). Orthomyxovirus, e.g., influenza virus (30), togavirus, e.g., Semliki Forest virus (43), rhabdovirus, e.g., VSV (45), flavivirus, e.g., West Nile virus (35), and bunyavirus, e.g., LaCrosse virus (26), and poxvirus, e.g., vaccinia virus (25), have been shown to require low pH for cell fusion. They enter cells by an endocytotic pathway that exploits a normal cellular pathway for the uptake of materials bound to cell surface receptors. In an endosome, the low pH induces conformational changes of the virus envelope protein to a fusion-active state and causes the exposure of a hydrophobic portion of the viral envelope protein. This hydrophobic region promotes fusion with the vesicle membrane. On the other hand, retrovirus, e.g., human immunodeficiency virus (HIV) (47), herpesvirus, e.g., herpes simplex virus (7), and paramyxovirus, e.g., Sendai virus (20), enter the host cells in a pH-independent fashion. Binding of the virus to the cell surface receptor leads to fusion between the virion lipid envelope and the cell plasma membrane.
Because HCV is a flavivirus, we established a cell fusion assay system that is triggered by low pH. Although a weak cell fusion was observed without low-pH treatment (Fig. 4), low-pH exposure was shown to enhance the cell fusion mediated by the chimeric HCV E1 and E2 proteins. Although we could demonstrate the cell fusion activity of the chimeric HCV envelope proteins by the reporter gene activation method, syncytium formation of transfected cells was not detected by microscopy. Recently, Flint et al. (22) have shown that a chimeric HCV E2 protein consisting of the ectodomain of E2, the transmembrane, and the cytoplasmic regions of influenza virus hemagglutinin expressed on the cell surface undergoes conformational change at low pH. No syncytium formation was observed in their system either (22). Syncytium formation is known to be dependent on the density of the fusion protein on the cell surface (15). A possible explanation for the lack of syncytium formation by the HCV envelope proteins might be that the chimeric HCV envelope proteins are not sufficient to quantitatively induce syncytia or that HCV envelope proteins per se may possess a little fusion activity.
In paramyxovirus, both fusion (F) and hemagglutinin-neuraminidase (HN) proteins are required for cell fusion. The HN protein mediates attachment of the virion to its cellular receptor, permitting the virus to bind to receptors, and also promotes fusion (40), while the fusion protein is known to mediate virus-cell fusion (11). In rubella virus, the internal hydrophobic region in the E1 protein plays a major role in the formation of the E1-E2 heterodimer and in low-pH-induced cell fusion (70). Semliki Forest virus fusion is mediated by E1, E2, and peripheral glycopolypeptide E3 (24), and pestivirus infection is initiated by the interaction of Erns and E2 (31). HCV E1 and E2 glycoproteins have been shown to form a noncovalent heterodimer, and their role(s) during HCV infection has been suggested in previous studies (16, 17, 27, 41, 46, 58). Recently, Lagging et al. have reported that the pseudotype VSVs possessing either of the chimeric HCV envelope proteins recover the ability to infect various mammalian cells, although they have not examined a pseudotype virus bearing both of the envelope proteins (39). On the other hand, we have demonstrated that a pseudotype VSV possessing both the E1 and E2 proteins exhibits a significantly higher infectivity than those possessing either of the HCV envelope proteins (Matsuura et al., submitted). In this study, we were able to demonstrate the requirement for both the chimeric E1 and E2 proteins and the augmentation by the low-pH treatment of the fusion activity of HCV envelope proteins. These results suggest that HCV enters the target cells via an endosomal pathway. In the endosome, a low-pH-dependent conformation change of the E1 and/or E2 proteins occurs, which then triggers membrane fusion and the entry of the nucleocapsid into the cytoplasm. Further study to identify specific regions in HCV envelope proteins responsible for receptor binding and membrane fusion is needed to understand the dynamic mechanism of HCV infection.
hCD81 has been shown to be a binding receptor of the E2 protein (57). However, there was no difference in cell fusion activity between the NIH 3T3 cell line expressing hCD81 and the parental cell line in this study. This result reminds us of cofactors such as human CCR5 and CXCR4 in HIV infection (3, 21). In HIV infection, three distinct entry mechanisms have been proposed: direct fusion between the viral and cellular membrane (47, 64), receptor-mediated endocytosis (56), and hCD4-independent entry (19). Although hCD81 is ubiquitously expressed on the surfaces of human cells (42), the expression level of hCD81 in Hep G2 cells is much lower than that in other human cell lines examined (data not shown), despite the fact that this cell line has the highest fusion activity. These results suggest that hCD81 alone is not sufficient to allow cell fusion and that another cofactor(s) might be required or that HCV cell fusion may occur in an hCD81-independent manner. Recently, Agnello et al. suggested the importance of the low-density lipoprotein (LDL) receptor for HCV entry (1). However, treatment of Hep G2 cells with an anti-LDL receptor antibody did not block cell fusion with CHO cells expressing both the chimeric E1 and E2 proteins (data not shown).
Treatment of Hep G2 cells with pronase, heparinase, or heparitinase reduced the levels of cell fusion activity, suggesting that certain protein molecules and GAGs on the cell surface play an important role in cell fusion. In dengue virus, a highly sulfated heparan sulfate has been shown to prevent the infection of target cells (8). GAGs are unbranched polysaccharides that are ubiquitously present on the cell surface, acquiring a net negative charge through N and O sulfation (36). GAG binding domains in the proteins are positively charged regions containing arginine and lysine (23). Interaction with the GAG molecule increases the population of the virus or the concentrations of the virus ligand in the vicinity of its entry receptor, enhancing the overall efficiency of infection. In Sindbis virus, it has been shown that adaptive mutation to a positive charge increases the attachment affinity (37). This model may also be applicable to HCV infection. However, it should be noted that additional receptors besides GAGs are involved in the binding and entry of many of these viruses. Sindbis virus has been shown to replicate on a GAG-deficient CHO cell line (6), and herpes simplex virus type 1 binds to heparan sulfate, followed by binding to a more specific protein receptor, TNF/NGF (52).
To our knowledge, this is the first report to demonstrate the fusion activity of HCV envelope proteins. Despite numerous efforts, there is no sufficient and specific treatment for patients infected with HCV. The cell fusion assay system described in this study might offer an efficient tool for future studies on cellular receptors for HCV, for the assignment of biological functions of the E1 and E2 envelope proteins in the binding, fusion, and penetration of HCV, and for the development of prophylactics and therapeutics for hepatitis C.
ACKNOWLEDGMENTS
We thank T. Shimoike, H. Tani, T. Someya, Y. W. Koh, and T. Tsutsumi for helpful discussions, S. Abrignani for providing NIH 3T3 cells, K. Suzuki for CHO cells, M. Kohara for anti-E1 and -E2 monoclonal antibodies, and S. Nagamori for FLC4 cells. We also thank Y. H. Suzuki and S. Ogawa for technical assistance and T. Mizoguchi for secretarial work.
S.T. is a research fellow of the Japanese Foundation of Viral Hepatitis Research. This work was supported by grants from Research on Advanced Medical Technology, Research on Emerging and Re-Emerging Infectious Diseases, and Research on Health Sciences Focusing on Drug Innovation of the Ministry of Health and Welfare and the Organization for Pharmaceutical Safety and Research of Japan.
REFERENCES
- 1.Agnello V, Abel G, Elfahal M, Kight G B, Zhang Q-X. Hepatitis C virus and other flaviviridae viruses enter cells via low density lipoprotein receptor. Proc Natl Acad Sci USA. 1999;96:12766–12771. doi: 10.1073/pnas.96.22.12766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aizaki H, Aoki Y, Harada T, Ishii K, Suzuki T, Nagamori S, Toda G, Matsuura Y, Miyamura T. Full-length complementary DNA of hepatitis C virus genome from an infectious blood sample. Hepatology. 1998;27:621–627. doi: 10.1002/hep.510270242. [DOI] [PubMed] [Google Scholar]
- 3.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 4.Aoki Y, Aizaki H, Shimoike T, Tani H, Ishii K, Saito I, Matsuura Y, Miyamura T. A human liver cell line exhibits efficient translation of HCV RNAs produced by a recombinant adenovirus expressing T7 RNA polymerase. Virology. 1998;250:140–150. doi: 10.1006/viro.1998.9361. [DOI] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Byrnes A P, Griffin D E. Binding of Sindbis virus to cell surface heparan sulfate. J Virol. 1998;72:7349–7356. doi: 10.1128/jvi.72.9.7349-7356.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campadelli-Fiume G, Avitabile E, Fini S, Stirpe D, Arsenakis M, Roizman B. Herpes simplex virus glycoprotein D is sufficient to induce spontaneous pH-independent fusion in a cell line that constitutively expresses the glycoprotein. Virology. 1988;166:598–602. doi: 10.1016/0042-6822(88)90533-8. [DOI] [PubMed] [Google Scholar]
- 8.Chen Y, Maguire T, Hileman R E, Fromm J R, Esko J D, Linhardt R J, Marks R M. Dengue virus infectivity depends on envelope protein binding to target cell heparan sulfate. Nat Med. 1997;3:866–871. doi: 10.1038/nm0897-866. [DOI] [PubMed] [Google Scholar]
- 9.Choo Q-L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 10.Choo Q-L, Richman K H, Han J H, Berger K, Lee C, Dong C, Gallegos C, Coit D, Medina-Selby A, Barr P J, Weiner A J, Bradley D W, Kuo G, Houghton M. Genetic organization and diversity of the hepatitis C virus. Proc Natl Acad Sci USA. 1991;88:2451–2455. doi: 10.1073/pnas.88.6.2451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Choppin P W, Scheid A. The role of viral glycoproteins in adsorption, penetration, and pathogenicity of viruses. Rev Infect Dis. 1980;2:40–61. doi: 10.1093/clinids/2.1.40. [DOI] [PubMed] [Google Scholar]
- 12.Cocquerel L, Duvet S, Meunier J C, Pillez A, Cacan R, Wychowski C, Dubuisson J. The transmembrane domain of hepatitis C virus glycoprotein E1 is a signal for static retention in the endoplasmic reticulum. J Virol. 1999;73:2641–2649. doi: 10.1128/jvi.73.4.2641-2649.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cocquerel L, Meunier J C, Pillez A, Wychowski C, Dubuisson J. A retention signal necessary and sufficient for endoplasmic reticulum localization maps to the transmembrane domain of hepatitis C virus glycoprotein E2. J Virol. 1998;72:2183–2191. doi: 10.1128/jvi.72.3.2183-2191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Crise B, Ruusala A, Zagouras P, Shaw A, Rose J K. Oligomerization of glycolipid-anchored and soluble forms of the vesicular stomatitis virus glycoprotein. J Virol. 1989;63:5328–5333. doi: 10.1128/jvi.63.12.5328-5333.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Danieli T, Pelletier S L, Henis Y I, White J M. Membrane fusion mediated by the influenza virus hemagglutinin requires the concerted action of at least three hemagglutinin trimers. J Cell Biol. 1996;133:559–569. doi: 10.1083/jcb.133.3.559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Deleersnyder V, Pillez A, Wychowski C, Blight K, Xu J, Hahn Y S, Rice C M, Dubuisson J. Formation of native hepatitis C virus glycoprotein complexes. J Virol. 1997;71:697–704. doi: 10.1128/jvi.71.1.697-704.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dubuisson J, Hsu H H, Cheung R C, Greenberg H B, Russell D G, Rice C M. Formation and intracellular localization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia and Sindbis viruses. J Virol. 1994;68:6147–6160. doi: 10.1128/jvi.68.10.6147-6160.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubuisson J, Rice C M. Hepatitis C virus glycoprotein folding: disulfide bond formation and association with calnexin. J Virol. 1996;70:778–786. doi: 10.1128/jvi.70.2.778-786.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 20.Fan D P, Sefton B M. The entry into host cells of Sindbis virus, vesicular stomatitis virus, and Sendai virus. Cell. 1978;15:985–992. doi: 10.1016/0092-8674(78)90282-9. [DOI] [PubMed] [Google Scholar]
- 21.Feng Y, Broder C C, Kennedy P E, Berger E A. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science. 1996;272:872–887. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 22.Flint M, Thomas J M, Maidens C M, Shotton C, Levy S, Barclay W S, McKeating J A. Functional analysis of cell surface-expressed hepatitis C virus E2 glycoprotein. J Virol. 1999;73:6782–6790. doi: 10.1128/jvi.73.8.6782-6790.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Flynn S J, Ryan P. A heterologous heparin-binding domain can promote functional attachment of a pseudorabies virus gC mutant to cell surfaces. J Virol. 1995;69:834–839. doi: 10.1128/jvi.69.2.834-839.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Garoff H, Wilschut J, Liljeström P, Wahlberg J M, Bron R, Suomalainen M, Smyth J, Salminen A, Barth B U, Zhao H, Forsell K, Ekström M. Assembly and entry mechanisms of Semliki Forest virus. Arch Virol (Suppl) 1994;9:329–338. doi: 10.1007/978-3-7091-9326-6_33. [DOI] [PubMed] [Google Scholar]
- 25.Gong S C, Lai C F, Esteban M. Vaccinia virus induces cell fusion at acid pH and this activity is mediated by the N-terminus of the 14-kDa virus envelope protein. Virology. 1990;178:81–91. doi: 10.1016/0042-6822(90)90381-z. [DOI] [PubMed] [Google Scholar]
- 26.Gonzalez-Scarano F. La Crosse virus G1 glycoprotein undergoes a conformational change at the pH of fusion. Virology. 1985;140:209–216. doi: 10.1016/0042-6822(85)90359-9. [DOI] [PubMed] [Google Scholar]
- 27.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hijikata M, Kato N, Ootsuyama Y, Nakagawa M, Shimotohno K. Gene mapping of the putative structural region of the hepatitis C virus genome by in vitro processing analysis. Proc Natl Acad Sci USA. 1991;88:5547–5551. doi: 10.1073/pnas.88.13.5547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Houghton M, Weiner A, Han J, Kuo G, Choo Q-L. Molecular biology of the hepatitis C viruses: implications for diagnosis, development and control of viral disease. Hepatology. 1991;14:381–388. [PubMed] [Google Scholar]
- 30.Huang R T C, Rott R, Klenk H D. Influenza viruses cause hemolysis and fusion of cells. Virology. 1981;110:243–247. doi: 10.1016/0042-6822(81)90030-1. [DOI] [PubMed] [Google Scholar]
- 31.Hulst M M, Moormann R J M. Inhibition of pestivirus infection in cell culture by envelope proteins Erns and E2 of classical swine fever virus: Erns and E2 interact with different receptors. J Gen Virol. 1997;78:2779–2787. doi: 10.1099/0022-1317-78-11-2779. [DOI] [PubMed] [Google Scholar]
- 32.Ishii K, Rosa D, Watanabe Y, Katayama T, Harada H, Wyatt C, Kiyosawa K, Aizaki H, Matsuura Y, Houghton M, Abrignani S, Miyamura T. High titers of antibodies inhibiting the binding of envelope to human cells correlate with natural resolution of chronic hepatitis C. Hepatology. 1998;28:1117–1120. doi: 10.1002/hep.510280429. [DOI] [PubMed] [Google Scholar]
- 33.Ito T, Mukaigawa J, Zuo J, Hirabayashi Y, Mitamura K, Yasui K. Cultivation of hepatitis C virus in primary hepatocyte culture from patients with chronic hepatitis C results in release of high titre infectious virus. J Gen Virol. 1996;77:1043–1054. doi: 10.1099/0022-1317-77-5-1043. [DOI] [PubMed] [Google Scholar]
- 34.Kato N, Hijikata M, Ootsuyama Y, Nakagawa M, Ohkoshi S, Sugimura T, Shimotohno K. Molecular cloning of the human hepatitis C virus genome from Japanese patients with non-A, non-B hepatitis. Proc Natl Acad Sci USA. 1990;87:9524–9528. doi: 10.1073/pnas.87.24.9524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kimura T, Ohyama A. Association between the pH-dependent conformational change of West Nile flavivirus E protein and virus-mediated membrane fusion. J Gen Virol. 1988;69:1247–1254. doi: 10.1099/0022-1317-69-6-1247. [DOI] [PubMed] [Google Scholar]
- 36.Kjellen L, Lindahl U. Proteoglycans: structures and interactions. Annu Rev Biochem. 1991;60:443–475. doi: 10.1146/annurev.bi.60.070191.002303. [DOI] [PubMed] [Google Scholar]
- 37.Klimstra W B, Ryman K D, Johnston R E. Adaptation of Sindbis virus to BHK cells selects for use of heparan sulfate as an attachment receptor. J Virol. 1998;72:7357–7366. doi: 10.1128/jvi.72.9.7357-7366.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kuo G, Choo Q-L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W-S, Kuo C, Berger K, Shuster F R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 39.Lagging L M, Meyer K, Owens R J, Ray R. Functional role of hepatitis C virus chimeric glycoproteins in the infectivity of pseudotyped virus. J Virol. 1998;72:3539–3546. doi: 10.1128/jvi.72.5.3539-3546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lamb R A, Kolakofsky D. Paramyxoviridae: the viruses and their replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1177–1204. [Google Scholar]
- 41.Lanford R E, Notvall L, Chavez D, White R, Frenzel G, Simonsen C, Kim J. Analysis of hepatitis C virus capsid, E1, and E2/NS1 proteins expressed in insect cells. Virology. 1993;197:225–235. doi: 10.1006/viro.1993.1583. [DOI] [PubMed] [Google Scholar]
- 42.Levy S, Todd S C, Maecker H T. CD81 (TAPA-1): a molecule involved in signal transduction and cell adhesion in the immune system. Annu Rev Immunol. 1998;16:89–109. doi: 10.1146/annurev.immunol.16.1.89. [DOI] [PubMed] [Google Scholar]
- 43.Marsh M, Helenius A. Adsorptive endocytosis of Semliki Forest virus. J Mol Biol. 1980;142:439–454. doi: 10.1016/0022-2836(80)90281-8. [DOI] [PubMed] [Google Scholar]
- 44.Marsh M, Helenius A. Virus entry into animal cells. Adv Virus Res. 1989;36:107–151. doi: 10.1016/S0065-3527(08)60583-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matlin K S, Reggio H, Helenius A, Simons K. Pathway of vesicular stomatitis virus entry leading to infection. J Mol Biol. 1982;156:609–631. doi: 10.1016/0022-2836(82)90269-8. [DOI] [PubMed] [Google Scholar]
- 46.Matsuura Y, Suzuki T, Suzuki R, Sato M, Aizaki H, Saito I, Miyamura T. Processing of E1 and E2 glycoproteins of hepatitis C virus expressed in mammalian and insect cells. Virology. 1994;205:141–150. doi: 10.1006/viro.1994.1629. [DOI] [PubMed] [Google Scholar]
- 47.McClure M O, Marsh M, Weiss R A. Human immunodeficiency virus infection of CD4-bearing cells occurs by a pH-independent mechanism. EMBO J. 1988;7:513–518. doi: 10.1002/j.1460-2075.1988.tb02839.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Michalak J P, Wychowski C, Choukhi A, Meunier J C, Ung S, Rice C M, Dubuisson J. Characterization of truncated forms of hepatitis C virus glycoprotein. J Gen Virol. 1997;78:2299–2306. doi: 10.1099/0022-1317-78-9-2299. [DOI] [PubMed] [Google Scholar]
- 49.Miyamura T, Matsuura Y. Structural proteins of hepatitis C. Trends Microbiol. 1993;1:229–231. doi: 10.1016/0966-842x(93)90137-g. [DOI] [PubMed] [Google Scholar]
- 50.Mizushima H, Hijikata M, Tanji Y, Kimura K, Shimotohno K. Analysis of N-terminal processing of hepatitis C virus nonstructural protein 2. J Virol. 1994;68:2731–2734. doi: 10.1128/jvi.68.4.2731-2734.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mizutani T, Kato N, Saito S, Ikeda M, Sugiyama K, Shimotohno K. Characterization of hepatitis C virus replication in cloned cells obtained from a human T-cell leukemia virus type 1-infected cell line, MT-2. J Virol. 1996;70:7219–7223. doi: 10.1128/jvi.70.10.7219-7223.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Montgomery R I, Warner M S, Lum B J, Spear P G. Herpes simplex virus-1 entry into cells mediated by a novel member of the TNF/NGF receptor family. Cell. 1996;87:427–436. doi: 10.1016/s0092-8674(00)81363-x. [DOI] [PubMed] [Google Scholar]
- 53.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–200. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 54.Nussbaum O, Broder C C, Berger E A. Fusogenic mechanisms of enveloped-virus glycoproteins analyzed by a novel recombinant vaccinia virus-based assay quantitating cell fusion-dependent reporter gene activation. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Okuma K, Nakamura M, Nakano S, Niho Y, Matsuura Y. Host range of human T-cell leukemia virus type I analyzed by a cell fusion-dependent reporter gene activation assay. Virology. 1999;254:235–244. doi: 10.1006/viro.1998.9530. [DOI] [PubMed] [Google Scholar]
- 56.Pauza C D, Price T M. Human immunodeficiency virus infection of T cells and monocytes proceeds via receptor-mediated endocytosis. J Cell Biol. 1988;107:959–968. doi: 10.1083/jcb.107.3.959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner A J, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 58.Ralston R, Thudium K, Berger K, Kuo C, Gervase B, Hall J, Selby M, Kuo G, Houghton M, Choo Q-L. Characterization of hepatitis C virus envelope glycoprotein complexes expressed by recombinant vaccinia viruses. J Virol. 1993;67:6753–6761. doi: 10.1128/jvi.67.11.6753-6761.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rose J K, Bergmann J E. Expression from cloned cDNA of cell-surface secreted forms of the glycoprotein of vesicular stomatitis virus in eucaryotic cells. Cell. 1982;30:753–762. doi: 10.1016/0092-8674(82)90280-x. [DOI] [PubMed] [Google Scholar]
- 60.Rostand K S, Esko J D. Microbial adherence to and invasion through proteoglycans. Infect Immun. 1997;65:1–8. doi: 10.1128/iai.65.1.1-8.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Selby M J, Choo Q-L, Berger K, Kuo G, Glazer E, Eckart M, Lee C, Chien D, Kuo C, Houghton M. Expression, identification and subcellular localization of the proteins encoded by the hepatitis C viral genome. J Gen Virol. 1993;74:1103–1113. doi: 10.1099/0022-1317-74-6-1103. [DOI] [PubMed] [Google Scholar]
- 62.Shimizu Y K, Iwamoto A, Hijikata M, Purcell R H, Yoshikura H. Evidence for in vitro replication of hepatitis C virus genome in a human T-cell line. Proc Natl Acad Sci USA. 1992;89:5477–5481. doi: 10.1073/pnas.89.12.5477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Shoji I, Suzuki T, Sato M, Aizaki H, Chiba T, Matsuura Y, Miyamura T. Internal processing of hepatitis C virus NS3 protein. Virology. 1999;254:315–323. doi: 10.1006/viro.1998.9540. [DOI] [PubMed] [Google Scholar]
- 64.Stein B S, Gowda S D, Lifson J D, Penhallow R C, Bensch K G, Engleman E G. pH-independent HIV entry into CD4-positive cells via virus envelope fusion to the plasma membrane. Cell. 1987;49:659–668. doi: 10.1016/0092-8674(87)90542-3. [DOI] [PubMed] [Google Scholar]
- 65.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.White J, Kielian M, Helenius A. Membrane fusion proteins of enveloped animal viruses. Q Rev Biophys. 1983;16:151–195. doi: 10.1017/s0033583500005072. [DOI] [PubMed] [Google Scholar]
- 67.White J M. Viral and cellular membrane fusion proteins. Annu Rev Physiol. 1990;52:675–697. doi: 10.1146/annurev.ph.52.030190.003331. [DOI] [PubMed] [Google Scholar]
- 68.White J M, Litman D R. Viral receptors of the immunoglobulin superfamily. Cell. 1989;56:725–728. doi: 10.1016/0092-8674(89)90674-0. [DOI] [PubMed] [Google Scholar]
- 69.Wyatt L S, Moss B, Rozenblatt S. Replication-deficient vaccinia encoding bacteriophage T7 RNA polymerase for transient gene expression in mammalian cells. Virology. 1995;210:202–205. doi: 10.1006/viro.1995.1332. [DOI] [PubMed] [Google Scholar]
- 70.Yang D, Hwang D, Qiu Z, Gillam S. Effects of mutations in the rubella virus E1 glycoprotein on E1-E2 interaction and membrane fusion activity. J Virol. 1998;72:8747–8755. doi: 10.1128/jvi.72.11.8747-8755.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]