Abstract
Glycoprotein K (gK) of pseudorabies virus (PrV) has recently been identified as a virion component which is dispensable for viral entry but required for direct cell-to-cell spread. Electron microscopic data suggested a possible function of gK in virus egress by preventing immediate fusion of released virus particles with the plasma membrane (B. G. Klupp, J. Baumeister, P. Dietz, H. Granzow, and T. C. Mettenleiter, J. Virol. 72:1949–1958, 1998). For more detailed analysis, a PrV mutant with a deletion of the UL53 (gK) open reading frame (ORF) from codons 48 to 275 was constructed, and the protein was analyzed with two monoclonal antibodies directed against PrV gK. The salient findings of this report are as follows. (i) From the PrV UL53 ORF, a functional gK is translated only from the first in-frame methionine. From the second in-frame methionine, a nonfunctional product is expressed which is not incorporated into virions. (ii) When constitutively expressed in a stable cell line without other viral proteins, gK is only incompletely processed. After superinfection with gK-deletion mutants, proper processing is restored and mature gK is incorporated into virions. (iii) The UL20 gene product is specifically required for processing of gK. gK is not correctly processed in a UL20 deletion mutant of PrV, and superinfection of gK-expressing cells with PrV-UL20− does not restore processing. However, all other known structural viral glycoproteins appear to be processed normally in PrV-UL20−-infected cells. (iv) Coexpression of gK and UL20 restored gK processing at least partially. Thus, our data show that the UL20 gene product is required for proper processing of PrV gK.
Herpesvirus glycoproteins, which are localized in the virion envelope, play a major role in virus entry by mediating attachment of virions to cell-surface receptors and fusion of the viral envelope with the plasma membrane during penetration. They are also involved in virus egress and direct cell-to-cell spread (36). In addition, they represent prominent targets for the host's immune response (29). For Pseudorabies virus (PrV), a member of the Varicellovirus genus of the Alphaherpesvirinae, 11 glycoproteins have been described which are designated as gB, gC, gD, gE, gG, gH, gI, gK, gL, gM, and gN (30). Homologs for these proteins have also been found in other alphaherpesviruses.
Mature glycoprotein K (gK) of PrV is a hydrophobic protein of 36 kDa which is present in virions and contains both high-mannose as well as complex forms of N-linked glycans (25). The protein is conserved among the alphaherpesviruses, and homologous proteins or open reading frames (ORFs) have also been described for herpes simplex virus types 1 and 2 (HSV-1 and -2), bovine herpesvirus 1, equine herpesvirus 1, varicella-zoster virus (VZV), infectious laryngotracheitis virus, and Marek's disease virus (12, 17, 20, 32, 34, 35, 39). All deduced gK homologous proteins are characterized by the presence of four hydrophobic domains of sufficient length to be membrane spanning. The secondary structure of HSV-1 gK has been determined by in vitro translation and processing of mutant proteins followed by protease digestion (31). The results indicated that only three of the four predicted hydrophobic domains indeed span the lipid bilayer. The third hydrophobic domain probably forms a loop which is anchored by the second and fourth hydrophobic domains.
First studies indicated that HSV-1 gK is involved in cell fusion, since various mutations leading to a syncytial phenotype mapped to the UL53 gene which encodes gK (5, 7, 13, 33). These syn mutations are preferentially located in the proposed ectodomains of gK (31). Whereas HSV-1 gK had so far only been detected in the endoplasmic reticulum and nuclear membranes of infected cells (14), it has been found in virions of PrV and VZV (25, 32). Detailed studies of HSV-1, PrV, and VZV demonstrated an important role for gK in virus replication (8, 15, 25, 32). Only small plaques or foci of infected cells were observed after infection of noncomplementing cells with gK mutants of HSV-1 and PrV (14, 25), and productive viral replication was prevented by deletion of VZV gK (32). Generally, the yield of gK mutants from noncomplementing cells is decreased compared to that of wild-type viruses, although final titers may depend on the cell type and particular virus mutant (14, 16).
HSV-1 and PrV gKs have been implicated in virus morphogenesis and egress. In cells infected with HSV-1 F-gKβ, which contains a lacZ expression cassette within the UL53 ORF numerous nonenveloped capsids were found in the cytoplasm (14), whereas in cells infected with a gK-deletion mutant of strain KOS, enveloped virions appeared to accumulate in the cytoplasm (16). These findings pointed to an important role for HSV-1 gK in nucleocapsid envelopment and efficient transport of enveloped virions to the extracellular space. Ultrastructural studies of a PrV gK mutant also revealed a defect in virus egress. Whereas only a few enveloped virions were detected outside of noncomplementing cells infected with the mutant virus, numerous nucleocapsids were observed just underneath the plasma membrane as well as fusion events which were interpreted as immediate entry of virions into the cell they just left (25). Thus, it was postulated that gK prevents entry of released virus particles by inhibition of fusion between the virion envelope and cytoplasmic membrane of infected cells (25).
Based on the model of gK topology (31), different functional domains for gK involved in nucleocapsid envelopment, membrane fusion, virus replication, and egress have been proposed (8). Effects on virus yield, plaque formation, and virus envelopment were tentatively assigned to amino-terminal portions of gK, comprising the N terminus, a cytoplasmic loop, and the second extracellular domain. The cytoplasmic tail was dispensable for virus replication and egress. In addition, comparison of gK sequences of various alphaherpesviruses identified conserved cysteine-rich and tyrosine-based motifs. Mutation of these motifs identified two domains of HSV-1 gK—the first cytoplasmic domain and the extracellular loop—which are crucial for virus replication and egress, while the N terminus was correlated with aberrant nucleocapsid envelopment and membrane fusion.
Besides gK, other putative multiply-membrane-spanning proteins include the product of the UL20 gene. The presumably nonglycosylated HSV-1 UL20 protein was detected in nuclear membranes and Golgi-derived vesicles of infected cells as well as in purified virions (38). Deletion of the HSV-1 UL20 gene affects viral egress in a cell-type-specific manner which correlates with fragmentation of the Golgi apparatus (1). After infection of Vero cells with an HSV-1 UL20-deletion mutant, virus particles accumulated in the perinuclear space (1, 2). Moreover, smaller amounts of viral glycoproteins gC and gD were found in the plasma membrane, and immature forms of both glycoproteins were present in enveloped virions, which indicates that the HSV-1 UL20 protein contributes to processing and transport of viral glycoproteins gC and gD from the trans-Golgi area to the plasma membrane (1). The PrV UL20 protein was also implicated in virus egress (10). However, in ultrastructural analyses, mutation of the UL20 protein led to accumulation of enveloped virions in large vesicles in the cytoplasm of infected Vero cells and failure of release of these virions into the extracellular space. No obvious alterations in maturation of glycoproteins expressed in Vero cells infected with the PrV-UL20− mutant were observed, but not all glycoproteins were investigated (10). Here we show that the absence of the UL20 protein specifically affects processing of gK, which indicates there is an intimate connection between these two multiply-membrane-spanning virion proteins.
MATERIALS AND METHODS
Viruses and cells.
Virus mutants were derived from wild-type PrV strain Kaplan (PrV-Ka) (19). PrV-1112 carries a β-galactosidase expression cassette in the gG locus and exhibits growth properties similar to those of PrV-Ka (28). Mutant viruses PrV-UL20− (10), and PrV-UL3.5−, (9) as well as rescuant PrV-UL20R (10), have been described. Construction of PrV-gKβ (see Fig. 1C) has been described before (25). Viruses were grown on rabbit (RK13) or African green monkey (Vero) kidney cells for plaque assays, one-step growth kinetics, Western blot analysis, and immunofluorescence. Porcine kidney cells (PSEK) were used for virus propagation followed by virion purification. Cells were propagated in Eagle's minimum essential medium supplemented with 5 or 10% (RK13) fetal calf serum. Cotransfections were performed by calcium phosphate coprecipitation (11). RK13 cells were transfected with SuperFect (Qiagen, Hilden, Germany).
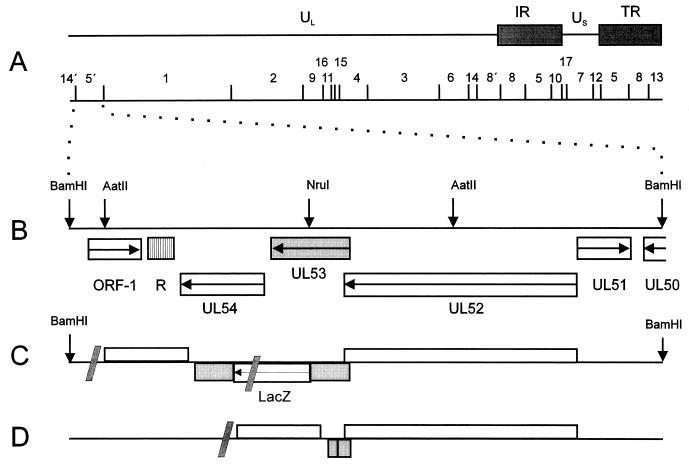
FIG. 1.
Construction of PrV-gK mutants. (A) A schematic map of the PrV genome with the BamHI restriction fragment map is depicted. The PrV genome consists of a unique long region (UL) and a unique short region (US); the latter is flanked by inverted repeats (IR, internal repeat; TR, terminal repeat). (B) Enlargement of the BamHI 5′ region. The locations of the identified ORFs, with transcriptional orientation indicated by arrows, are shown (R, region of reiterated sequences), and relevant restriction sites for cloning are indicated (3). BamHI 5′ genome organization for the UL52, UL53, and UL54 genes of the insertion mutant PrV-gKβ (C) (25) and gK deletion mutant PrV-ΔgKw (D). Shaded slashes in panels C and D indicate the figure is not drawn to scale.
Construction of PrV-ΔgKw.
For the construction of PrV-ΔgKw, a gK deletion mutant lacking most of the UL53 gene, a 4.2-kb AatII subfragment of the BamHI 5′ fragment was inserted into pUC19, cleaved with NruI (Fig. 1B), a singular restriction site within this subfragment, and digested with exonuclease Bal31 for bidirectional deletion of the UL53 ORF. The resulting plasmid, pUC19/gK/Bal31, carries a large deletion from codon 48 to codon 275 of the gK ORF (corresponding to nucleotides [nt] 4058 to 4743 of GenBank accession no. X87246) (Fig. 1D), as verified by sequencing. After cotransfection with genomic DNA of PrV-gKβ, recombinant viruses were screened for a white plaque phenotype. One single plaque isolate, designated as PrV-ΔgKw, was further characterized. The genotype was verified by restriction analysis and Southern blotting (data not shown).
Construction of cell lines.
For the construction of gK-expressing cell lines, the complete UL53 gene (nt 3918 to 4856) (3) and the UL53 gene from a second possible start codon (nt 3963 to 4856) (3) were amplified by PCR with Pfu polymerase (Promega, Mannheim, Germany). The forward primers, which in their 5′ extensions create a novel HindIII restriction site (underlined) were as follows: gK-1 (5′-ATAAGCTTCATGCTCCTCGGCGGGCGCC-3′; nt 3916 to 3936) and gK-2 (5′-AGAAGCTTGGTGATGGGCGCGTACGCCGG-3′; nt 3959 to 3979). The reverse primer, which adds an XbaI restriction site at the 3′ end of the amplified products (underlined), was gK-3 (5′-AGTCTAGACGTCCCCGCGCCGACCTTCATCC-3′; nt 4873 to 4851). The PCR product gK1-3 was inserted into HindIII- and XbaI-cleaved vector pcDNA3 (InVitroGen, Leek, The Netherlands), and the gK2-3 product was inserted into pRc/CMV (InVitroGen). The correct sequences of gK1-3 and gK2-3 were confirmed by direct sequencing with the T7 DNA sequencing kit (U.S. Biochemicals, Cleveland, Ohio). Plasmids were used for transfection of RK13 cells by SuperFect (Qiagen, Hilden, Germany). Transfectants were selected with 500 μg of Geneticin (Life Technologies, Eggenstein, Germany) per ml and tested for gK expression by indirect immunofluorescence (data not shown), Western blotting, and their ability to complement PrV gK mutants in trans. One cell clone each was selected and designated RK13-gK1-3 (complete gK ORF) and RK13-gK2-3 (gK from the second possible translation start).
For transient transfection of RK13-gK1-3 cells, a UL20 expression plasmid, designated pcDNA3-UL20, was constructed. To this end, the UL20 gene was amplified by PCR with a forward primer, which creates an EcoRI restriction site at the 5′ end of the amplified product (underlined) 5′-CACAGAATTCGCGGCGCGGGGATGGAGGAC-3′ (nt 6673 to 6692 of GenBank accession no. L00676) (21, 23); and a reverse primer, which adds an XhoI restriction site at the 3′ end of the amplified product (underlined), 5′-CACACTCGAGGTCGCTGGGGAGCAGGGGGG-3′ (nt 7219 to 7200). After restriction enzyme cleavage, the PCR product was cloned into EcoRI- and XhoI-cleaved vector pcDNA3.
Plaque assay.
Plaque formation was analyzed after titration of virus mutants on various cell lines 2 days after infection under a methylcellulose overlay. Thereafter, cells were fixed and stained with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) as described elsewhere (25). Although PrV-ΔgKw does not express β-galactosidase, the same procedure was used to document unstained plaques or foci of infected cells.
Preparation of MAbs against gK.
A bacterial fusion protein, pGEX-Bst XI/XhoI, encompassing codons 64 to 196 of the UL53 ORF fused to the glutathione S-transferase gene (25), was used for preparation of monoclonal antibodies (MAbs). Twelve-week-old mice were immunized intramuscularly with 100 μg of purified fusion protein mixed with complete Freund's adjuvant (Sigma, Deisenhofen, Germany). Subsequent immunizations were made with incomplete Freund's adjuvant after 6 weeks and, thereafter, repeated seven times in 4-week intervals. Four days prior to fusion, a booster immunization was applied. Hybridoma supernatants were differentially screened by Western blotting with purified virion preparations of PrV-Ka and PrV-gKβ. Indirect immunofluorescence was performed with PrV-Ka-infected cells. The resulting MAb D4-1 was selected for Western blotting; MAb b7-b6 was selected for indirect immunofluorescence (data not shown).
Western blot analysis of cell lysates and virions.
Cell lysates were harvested 24 or 48 h after infection or transfection. After centrifugation at 14,000 rpm for 1 min in an Eppendorf centrifuge, cells were washed twice with phosphate-buffered saline (PBS), resuspended in 100 μl of PBS, and mixed with the same volume of sample buffer. Virions were purified as described previously (24). Briefly, cells were infected and incubated until complete CPE developed. Medium was removed and cells were lysed in PBS by three freeze (−70°C) and thaw (37°C) cycles. Cellular material was removed by low-speed centrifugation, and the resulting supernatant was combined with the virus-containing medium. Virions were then purified by sucrose gradient centrifugation. Twenty microliters of cell lysate or 10 μg of purified virions was separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (27), electrotransferred onto polyvinylidene difluoride membrane (PVDF) (Immobilon-P; Millipore, Eschborn, Germany) (37), and reacted for 1 h with monoclonal or polyclonal antibodies against PrV glycoproteins diluted in PBS as follows: b43-b5 (anti-gB), 1:1,000; B16-c8 (anti-gC), 1:100; A9-b15 (anti-gE) 1:100 (26); D4-1 (anti-gK), 1:50 (this study); Vacc-gD 1:1,000 and glutathione S-transferase (GST)-gH 1:50,000 (24); GST-gI 1:50,000 (4), GST-gL 1:1,000 (22), anti-gM peptide serum 1:5,000 (6), and Baculo-gN 1:500 (18). After incubation with peroxidase-conjugated secondary antibody (Dianova, Hamburg, Germany), bound antibody was visualized by enhanced chemiluminescence (ECL Western blot system; Amersham, Braunschweig, Germany) and detected on X-ray film.
Glycosidase digestions.
Deglycosylation studies were performed as described elsewhere (25).
Tunicamycin treatment.
To inhibit N glycosylation, tunicamycin (Sigma, Deisenhofen, Germany) at a concentration of 20 μg per ml was added during infection of RK13 cells at a multiplicity of infection (MOI) of 1. After 24 h at 37°C, infected cells were harvested, and cell lysates were analyzed by Western blotting.
One-step growth analysis.
For analysis of growth behavior, RK13 and RK13-gK1-3 cells were infected with PrV-Ka, PrV-gKβ, and PrV-ΔgKw at an MOI of 5. After 1 h at 4°C, the inoculum was removed, prewarmed medium was added, and virus was allowed to penetrate for 1 h at 37°C. The remaining extracellular virus was inactivated by low-pH treatment. Immediately thereafter and after 4, 8, 12, 24, and 34 h, supernatant and cells were harvested separately and titrated. The cell pellet was treated by low pH to inactivate cell-associated but extracellular infectious virus. Titers of extra- and intracellular virus progeny were added, and average values and standard deviations of two independent experiments were calculated.
RESULTS
Construction and phenotypic characterization of PrV gK mutants.
Recently, a PrV gK mutant, PrV-gKβ, has been characterized in which the UL53 ORF had been interrupted after codon 164 by insertion of a gG-lacZ expression cassette (Fig. 1C) (25). Since in this mutant the expression of a truncated N-terminal gK fragment of 164 amino acids was still possible, another mutant, PrV-ΔgKw, was constructed by bidirectional exonuclease digestion beginning at a single NruI restriction site located in the middle of the UL53 gene (Fig. 1B). This mutant carries a large deletion in the UL53 ORF, starting after codon 47 and extending until codon 275 (Fig. 1D).
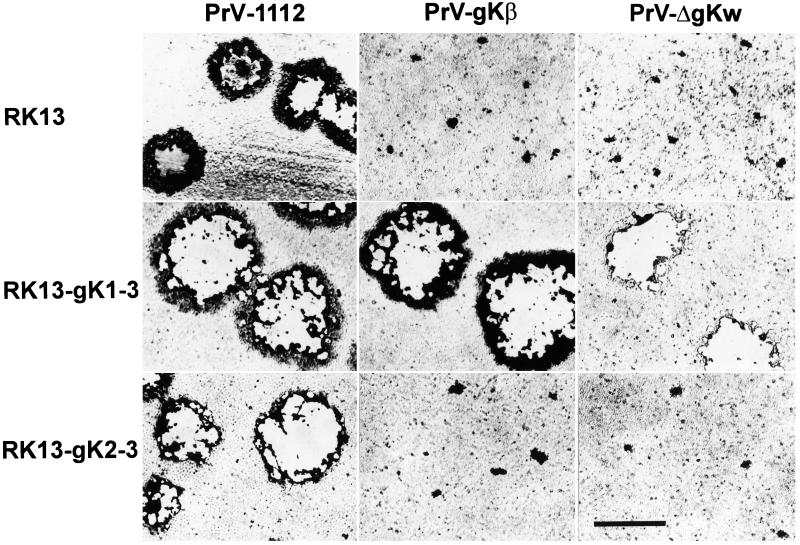
To analyze the growth properties of the gK mutants, plaque assays on normal RK13 and gK-expressing cells were performed. Two days after infection, plaques or foci of infected cells were analyzed. As shown in Fig. 2, PrV-1112 forms similar plaques on all cell lines tested. In contrast, the gK-negative mutants were unable to form plaques on parental RK13 cells, but were capable of plaque formation on RK13-gK1-3 cells which constitutively express gK from the first methionine. RK13-gK2-3 cells which express gK from the second in-frame methionine did not complement plaque formation in any gK mutant. These data confirm the important role of gK in plaque formation of PrV and indicate that functional gK is expressed only from the first methionine in the UL53 ORF.
FIG. 2.
Plaque formation of PrV-gK mutants under plaque assay conditions. Normal RK13 cells and constitutively gK-expressing RK13-gK1-3 and RK13-gK2-3 cells were infected with PrV-1112, PrV-gKβ, and PrV-ΔgKw for 2 days. The cells were then fixed, stained, and photographed. Bar, 1 mm.
One-step growth kinetics of PrV gK mutants.
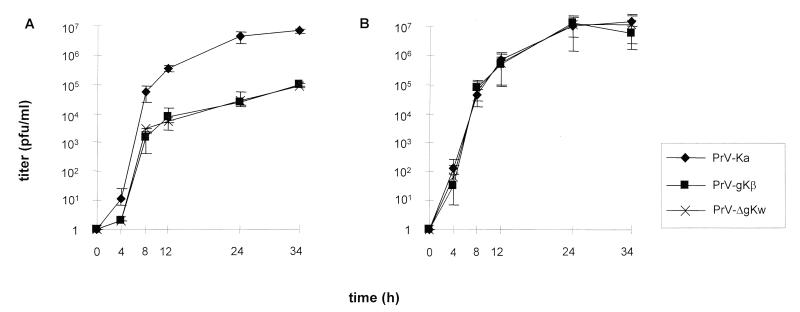
To further investigate replication of PrV gK mutants, one-step growth kinetics were assayed on noncomplementing RK13 (Fig. 3A) and complementing RK13-gK1-3 (Fig. 3B) cells after infection with PrV-Ka, PrV-gKβ, and PrV-ΔgKw at an MOI of 5. Virus progeny was titrated on complementing cells, and average values as well as standard deviations of two independent experiments were calculated. The interruption or deletion of the UL53 ORF resulted in a significant growth defect on RK13 cells at all times postinfection compared to PrV-Ka. Final viral titers of PrV-gKβ and PrV-ΔgKw were reduced by about 50-fold. On complementing RK13-gK1-3 cells, replication of PrV-gKβ and PrVΔgKw was comparable to that of PrV-Ka and resulted in nearly identical final titers.
FIG. 3.
One-step-growth kinetics. RK13 (A) and RK13-gK1-3 (B) cells were infected at an MOI of 5 with PrV-Ka, PrV-gKβ, and PrV-ΔgKw. After the indicated times, supernatant and cells were harvested and titrated on complementing cells. The calculated virus titers of supernatant and cells were added. The mean values with standard deviations of two independent experiments are shown.
Analysis of constitutively gK-expressing cell lines.
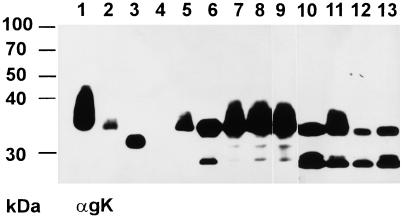
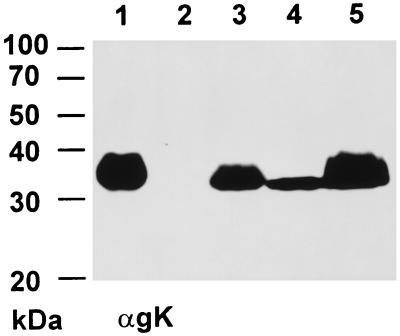
Two constitutively gK-expressing cell lines were constructed. RK13-gK1-3 cells expressed the complete gK, whereas RK13-gK2-3 cells were only able to express a gK from a second possible translational start codon. To investigate the expression products in detail, lysates of RK13-gK1-3 and RK13-gK2-3 cells infected with PrV-Ka, PrV-gKβ, and PrV-ΔgKw or mock-infected cells were separated by PAGE under reducing conditions, blotted onto PVDF membrane, and probed with MAb D4-1 directed against PrV gK. As shown in Fig. 4, in purified virion preparations of PrV-Ka, MAb D4-1 recognized the glycosylated ca. 36-kDa mature gK, which appeared as a broad band indicative of glycosylated proteins (Fig. 4, lane 1), as well as a 34-kDa form after treatment with endo-β-N-acetylglucosaminidase H (endo H) (Fig. 4, lane 2) and a 32-kDa form after PNGase F digest (Fig. 4, lane 3).
FIG. 4.
Western blot analysis of constitutively gK-expressing cell lines. Proteins were separated by 10% PAGE, blotted onto PVDF membrane, and incubated with a MAb against gK (αgK). Samples were purified PrV virions (lane 1) after incubation with endo H (lane 2) or PNGase F (lane 3); lysates of noninfected RK13 cells (lane 4) or RK13 cells after infection with PrV-Ka (lane 5); lysate of RK13-gK1-3 cells (lane 6) infected with PrV-Ka (lane 7), PrV-gKβ (lane 8), or PrV-ΔgKw (lane 9); or cell lysate of RK13-gK2-3 cells (lane 10) infected with PrV-Ka (lane 11), PrV-gKβ (lane 12), or PrV-ΔgKw (lane 13). The locations of molecular mass markers are indicated on the left.
Surprisingly, expressed proteins detected by the anti-gK MAb in RK13-gK1-3 and RK13-gK2-3 cells were different from gK found in purified virions. As shown in Fig. 4, lane 6, a major protein with a molecular mass of around 34 kDa and a minor expression product of 28 kDa were detected in RK13-gK1-3 cells, whereas RK13-gK2-3 cells expressed a protein of 33 kDa and two smaller products of 27 and 28 kDa (Fig. 4, lane 10). Notably, these protein bands all lacked the “smeary” appearance found in mature gK. Interestingly, after infection of RK13-gK1-3 cells with wild-type PrV (Fig. 4, lane 7) or any of the gK mutants (Fig. 4, lanes 8 and 9), a broad protein band of ca. 36 kDa was recognized as the main protein which appeared comparable to gK detected in PrV-Ka-infected RK13 cells (Fig. 4, lane 5) and purified PrV-Ka virions (Fig. 4, lane 1). In contrast to RK13-gK1-3 cells, the glycosylated ca. 36-kDa form of gK could only be recognized in RK13-gK2-3 cells infected with PrV-Ka (Fig. 4, lane 11), but not after infection with any of the gK mutants (Fig. 4, lanes 12 and 13). The smaller protein species present in these cell lines may represent precursor forms, since a gK precursor protein with an apparent molecular mass of ca. 28 kDa has been identified in earlier in vitro translation studies (J. Baumeister et al., unpublished observations). The alteration in the gK pattern after infection of RK13-gK1-3 cells may indicate that gK is not completely processed in these cells when expressed alone. Indirect immunofluorescence analysis of both cell lines could not clarify whether gK was expressed on the cell surface, since the MAb recognized gK only in permeabilized cells. Here a diffuse, equally distributed cytoplasmic fluorescence was detected for both cell lines (not shown).
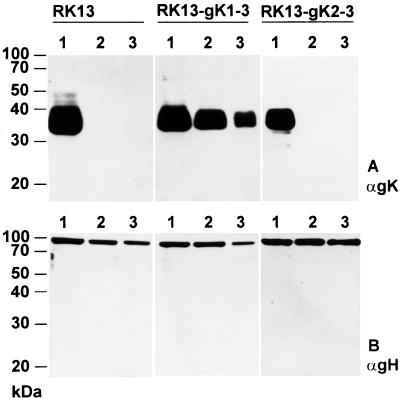
gK is a structural component of virions of gK mutants grown on RK13-gK1-3 cells, but not on RK13-gK2-3 cells.
To test for incorporation of gK expressed from the transgenic cell lines into virions, Western blot analyses of purified virions were performed. RK13, RK13-gK1-3, and RK13-gK2-3 cells were infected with PrV-Ka, PrV-gKβ, and PrV-ΔgKw at an MOI of 1, cells and supernatant were harvested 2 days after infection, and virions were purified. Proteins were separated by SDS-PAGE, blotted, and incubated with anti-gK MAb D4-1 (Fig. 5A). For control, the blot was then stripped and reprobed with a polyclonal anti-gH serum (Fig. 5B). The ca. 36-kDa mature gK was recognized in PrV-Ka virions grown on RK13, RK13-gK1-3, and RK13-gK2-3 cells (Fig. 5A, lanes 1). A protein with similar electrophoretic mobility was also detected in all virion preparations of gK mutants which had been propagated on RK13-gK1-3 cells (Fig. 5A, middle panel, lanes 2 and 3), whereas in virions from gK mutants propagated on RK13-gK2-3 cells, no gK was recognized (Fig. 5A, right panel, lanes 2 and 3). In contrast, the 95-kDa gH was detected similarly in all virion preparations independent of the cell line used for propagation. These results show that the protein expressed from a second in-frame methionine in the UL53 ORF is not incorporated into virions. Therefore, the first translation start codon appears to be used for correct translation of PrV gK.
FIG. 5.
Identification of gK in virions propagated on different gK-expressing cell lines. Purified virions of PrV-Ka (lanes 1), PrV-gKβ (lanes 2), and PrV-ΔgKw (lanes 3) were isolated from RK13, RK13-gK1-3, and RK13-gK2-3 cells and analyzed by Western blotting after separation in a 10% polyacrylamide gel. The blot was incubated with a MAb against gK (A [αgK]) then stripped and probed with a gH-specific rabbit serum [αgH]). The locations of molecular mass markers are indicated on the left.
Identification of another viral protein that is involved in the correct processing of gK.
Obviously, the gK protein as expressed from RK13-gK1-3 cells migrates with an electrophoretic mobility which is different from that of mature gK as detected after infection. To check whether another viral protein is required for efficient glycosylation of gK, cell lysates and purified virion preparations of a multitude of PrV glycoprotein mutants available in our laboratory (mutants lacking gB, gC, gD, gE, gG, gH, gI, gL, gM, or gN, as well as several double and triple mutants) were tested for expression of gK by Western blotting (not shown). The blots clearly demonstrated that the mature 36-kDa form of gK was expressed by all virus mutants tested. These findings indicated that other glycoproteins of PrV seemed not to be involved in the processing of gK. Furthermore, cell lysates of two virus mutants with defects in virus egress, PrV-UL20− and PrV-UL3.5−, were analyzed. As shown in Fig. 6, lane 4, the gK signal detected in PrV-UL20−-infected cells was smaller than and not as diffuse as the signal normally observed for gK. In PrV-UL3.5−-infected cells (Fig. 6, lane 5) gK was comparable to the gK present in purified PrV-Ka virions (Fig. 6, lane 1) and infected cell lysates (Fig. 6, lane 3). This indicates that the UL20 gene product of PrV may be involved in the processing of gK.
FIG. 6.
Expression of gK in two different PrV mutants with defects in the virus egress pathway. Noninfected RK13 cells (lane 2) or RK13 cells infected with PrV-Ka (lane 3), PrV-UL20− (lane 4), or PrV-UL3.5− (lane 5) were harvested 24 h after infection and separated by 10% PAGE. Purified PrV-Ka virions (lane 1) were used as a positive control. Expression of gK was detected by the anti-gK MAb (αgK). The locations of molecular mass markers are indicated on the left.
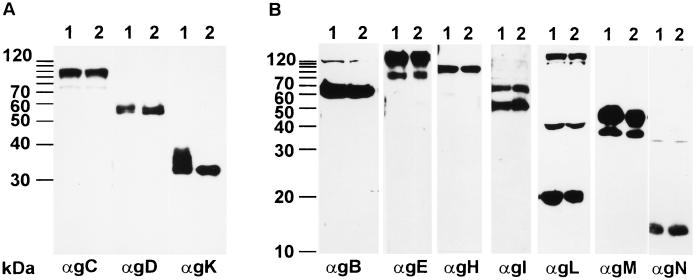
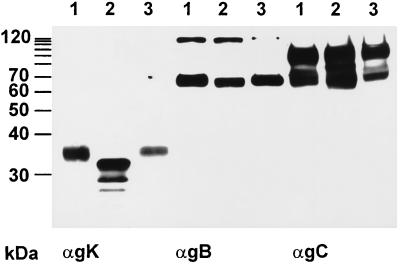
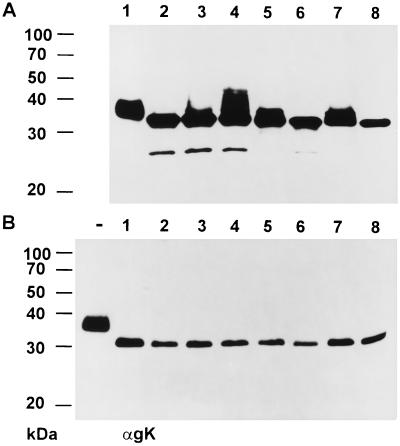
To analyze whether expression and/or modification of other glycoproteins is altered in PrV-UL20−-infected cells, all known structural glycoproteins of PrV were analyzed in PrV-UL20−-infected RK13 cell lysates by Western blotting with a panel of monoclonal and polyclonal antibodies (Fig. 7). The experiment clearly showed that the amounts and apparent molecular weights of the other glycoproteins expressed in PrV-UL20− infected cells (Fig. 7, lanes 2) were identical to those of proteins of PrV-Ka-infected cells (Fig. 7, lanes 1). Only the appearance of gK was altered in PrV-UL20−-infected cells. Moreover, the incompletely processed form of gK was also detectable in purified PrV-UL20− virions (Fig. 8, lane 2), whereas the corresponding UL20 rescue mutant showed a mature gK (Fig. 8, lane 3). gB and gC, used as controls, were unaltered (Fig. 8).
FIG. 7.
Analysis of glycoproteins in PrV-UL20−-infected cells. Lysates of RK13 cells infected with PrV-Ka (lanes 1) and the PrV-UL20− mutant (lanes 2) were separated by 10% (A) or 12% (B) PAGE and blotted. Thereafter, the membrane was incubated with monoclonal or polyclonal antibodies (α) against PrV glycoproteins as indicated at the bottom. The locations of molecular mass markers are indicated to the left of each panel.
FIG. 8.
Incorporation of incompletely processed gK into PrV-UL20− virions. Purified PrV-Ka (lanes 1), PrV-UL20− (lanes 2), and PrV-UL20R (lanes 3) virions were lysed and analyzed by Western blotting after separation in a 10% polyacrylamide gel. The blot was incubated with MAbs against gK (αgK), gB (αgB), or gC (αgC).
To exclude the possibility that the smaller form of gK expressed in PrV-UL20−-infected cells is due to differences in the protein backbone, infected cell lysates were analyzed by Western blotting before and after tunicamycin treatment. After inhibition of N glycosylation, the same gK-specific signal was recognized in the cell lysate of PrV-Ka- and PrV-UL20−-infected cells (data not shown).
Coexpression with UL20 partially restores gK processing.
To demonstrate that the UL20 gene product is required for processing of gK, RK13-gK1-3 cells were transfected with pcDNA3-UL20 and analyzed by Western blotting 24 or 48 h after transfection. As shown in Fig. 9A, an increase in the apparent molecular mass of cellularly expressed gK (Fig. 9, lane 2) indicative of processing was first observed 24 h after transfection (Fig. 9, lane 3), which grew more pronounced 48 h after transfection (Fig. 9, lane 4). Thus, coexpression of UL20 at least partially restored processing of gK. For comparison, mature gK present in purified virions is shown in Fig. 9A, lane 1. As controls, lysates of RK13-gK1-3 cells (Fig. 9A, lanes 5 and 6) or RK13 cells (Fig. 9A, lanes 7 and 8) infected with PrV-Ka (Fig. 9A, lanes 5 and 7) or PrV-UL20− (Fig. 9A, lanes 6 and 8) were probed with the anti-gK MAb. After PNGase F digestion (Fig. 9B, lanes 1 to 8), the molecular masses of gK were identical at 32 kDa in all samples. This indicates that the observed processing of gK in the presence of UL20 is effected by N-linked glycosylation. When gK-expressing RK13-gK1-3 cells were first transfected with the UL20 expression plasmid and then infected with UL20-negative PrV, fully processed gK was detected (data not shown).
FIG. 9.
Western blot analysis of gK expressed in RK13-gK1-3 cells after transient expression of UL20. Proteins of purified PrV-Ka virions (lane 1), lysate of RK13-gK1-3 cells (lane 2), and RK13gK1-3 cells transfected with pcDNA3-UL20 24 h (lane 3) or 48 h (lane 4) after transfection were separated in an SDS–10% polyacrylamide gel and probed with the anti-gK MAb (αgK). As controls, lysates of RK13-gK1-3 cells infected with PrV-Ka (lane 5) or PrV-UL20− (lane 6) and of RK13 cells infected with PrV-Ka (lane 7) or PrV-UL20− (lane 8) after incubation with the anti-gK MAb are included. (A) Samples without PNGase F digestion. (B) Samples after PNGase F digestion. In panel B, lane − is an undigested control of PrV virion proteins. The locations of molecular mass markers are indicated on the left.
DISCUSSION
In this report, we show a specific effect on maturation of gK by inactivation of the UL20 gene. Both UL20 and gK play a role in virus egress from infected cells. Whereas in cells infected with PrV gK mutants, virion morphogenesis appears to proceed normally until release of virions into the extracellular space (25), in cells infected with PrV-UL20−, enveloped virions were shown to accumulate in huge vacuoles and are only inefficiently released (10). Thus, the UL20 protein seems to be functionally required before gK function is needed. So far, the nature of the interplay between UL20 and gK is unclear. Both are membrane proteins (12, 31, 38). HSV-1 gK has been shown to span the membrane three times, and the UL20 protein also contains hydrophobic domains of sufficient length to traverse the lipid bilayer up to four times. Thus, both seem to be intimately associated with membranes. Since we do not yet have serologic reagents to detect the PrV UL20 protein, it is unclear whether the observed dependence of gK processing on the UL20 protein is associated with the formation of a physical complex between the two polypeptides. However, it is clear from our results that the effect on glycoprotein maturation by the UL20 protein is specific for gK. None of the other known viral structural glycoproteins seems to be affected. Surprisingly, the incompletely processed form of gK is incorporated into virions in the absence of the UL20 protein, indicating (i) that proper processing of gK is not required for virion incorporation and (ii) that the UL20 protein is required for maturation, but not for virion localization of gK. A possible explanation for the effect on maturation is that by interaction with gK, the UL20 protein mediates retention of a conformation of gK which renders it amenable for processing. In the absence of the UL20 protein, gK could assume a different conformation which does not allow processing to occur. Using our MAbs, we are currently trying to analyze this phenomenon.
Our data also show that functional gK is only expressed from the first in-frame methionine present in the UL53 ORF (3). Cells constitutively expressing gK from this start codon complement insertion (PrV-gKβ) and deletion (PrV-ΔgKw) mutants with regard to plaque size and one-step growth, and the cellularly expressed gK is properly processed and incorporated into virions. In contrast, from the second in-frame methionine, a gK form is expressed which, although migrating in SDS-PAGE similarly to gK expressed from the first methionine, is not properly processed and is not incorporated into virions. Also, these cells do not complement plaque formation of either of the gK mutants.
Studies of PrV gK were greatly facilitated with the isolation of two MAbs which recognize gK. These are the first MAbs described which are specific for herpesvirus gK proteins. Our analyses confirm that mature ca. 36-kDa virion gK contains ca. 2 kDa of endo H-sensitive carbohydrates and an additional 2 kDa of PNGase F-sensitive N-linked glycans. Interestingly, in RK13-gK1-3 cells, a 34-kDa species which comigrates with the endo H-digested virion gK is expressed, which is then apparently correctly processed after superinfection by wild-type PrV and either of the gK mutants and is incorporated into virions. An only slightly smaller protein of 33 kDa is present in RK13-gK2-3 cells; however, it is not processed after superinfection with gK-deletion mutants and is not present in virions propagated on these cells. Thus, the product expressed from the second in-frame methionine within UL53 is detected by the MAb in cell lysates, but is not functional. In these cells, a prominent, second 28-kDa form of gK is recognized by the MAb which may represent a stable breakdown product of the 33-kDa gK.
Whereas the inhibition of gK processing in cells infected with PrV-UL20− indicated that the UL20 protein is necessary for gK maturation, it did not exclude an indirect effect. However, transfection of RK13-gK1-3 cells with a UL20 expression plasmid at least partially restored gK processing, which shows a direct effect of UL20 on gK. The fact that gK processing appeared to be restored only partially may be explained by the transfection efficiency with the UL20 expression plasmid, which never reaches 100% of gK-expressing cells, and the timing of the expression of the two proteins which is certainly different in the transfected cell line from that in virus-infected cells. On the other hand, it cannot be excluded that yet another viral protein is required for complete processing of gK.
HSV-1 UL20 has been implicated in compensation for disruption of the Golgi apparatus late after infection, and UL20-deletion mutants of HSV-1 have been shown to exhibit defects in the processing of gC and gD (1). In PrV, except for gK, the processing of none of the other known structural viral glycoproteins appears to be affected, which cannot be explained by a more general disruption of the protein export pathway. In contrast, the specific effect of the UL20 deletion on gK indicates that both intimately cooperate.
Electron microscopical analyses of our first gK mutant, PrV-gKβ (25), indicated that, in the absence of gK, virions are released from infected cells, but immediately fuse with the cell they left. Thus, gK was postulated to play a role in interference. We tried to test this hypothesis by establishing the gK-expressing cell lines that are described in this report. However, these cells did not show any interference, which, with hindsight, may now be explained by the fact that these cells do not express mature gK in the absence of the UL20 protein. Therefore, we are currently trying to construct cells coexpressing gK and UL20, which are more likely to express functional gK.
Although in both HSV-1 and PrV, the UL20 and gK proteins are involved in virion maturation and egress, there are differences in the phenotypes displayed by the respective mutants. In HSV-1, deletion of UL20 resulted in accumulation of enveloped virions in the perinuclear space (2) and disruption of the Golgi apparatus with concomitant alteration of glycoprotein maturation (1), whereas in the absence of gK, enveloped virions accumulated in the cytoplasm (16). In PrV, deletion of UL20 resulted in accumulation of enveloped virions within intracytoplasmic vesicles (10). In the absence of gK, virion morphogenesis appears to proceed normally, but extracellular virions are found only rarely (25). Most strikingly, in cells infected with gK-negative PrV, numerous intracellular nucleocapsids were found close to the plasma membrane, and fusion events were observed which might represent entry stages of released virions. Since HSV-1 UL20 and gK-null mutants exhibited different phenotypes on different cells, indicating partial complementation by cellular factors (2, 16), we cannot exclude that the differences in phenotypes between HSV-1 and PrV are, at least partially, due to the difference in cell systems. However, from our data, we deduce that UL20 function precedes gK function, which would imply that a UL20-gK double mutant should exhibit essentially a UL20 phenotype. Our future goal is to isolate and analyze this mutant after establishment of doubly expressing cells.
ACKNOWLEDGMENTS
This work was supported by the Deutsche Forschungsgemeinschaft (Me 854/4-1) and the European Union (EEC-contract no. BMH4-CT97-2573).
REFERENCES
- 1.Avitabile E, Ward P L, Di Lazzaro C, Torrisi M R, Roizman B, Campadelli-Fiume G. The herpes simplex virus UL20 protein compensates for the differential disruption of exocytosis of virions and viral membrane glycoproteins associated with fragmentation of the Golgi apparatus. J Virol. 1994;68:7397–7405. doi: 10.1128/jvi.68.11.7397-7405.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baines J D, Ward P L, Campadelli-Fiume G, Roizman B. The UL20 gene of herpes simplex virus 1 encodes a function necessary for viral egress. J Virol. 1991;65:6414–6424. doi: 10.1128/jvi.65.12.6414-6424.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Baumeister J, Klupp B G, Mettenleiter T C. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J Virol. 1995;69:5560–5567. doi: 10.1128/jvi.69.9.5560-5567.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brack A R, Dijkstra J M, Granzow H, Klupp B G, Mettenleiter T C. Inhibition of virion maturation by simultaneous deletion of glycoproteins E, I, and M of pseudorabies virus. J Virol. 1999;73:5364–5372. doi: 10.1128/jvi.73.7.5364-5372.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Debroy C, Pederson N, Person S. Nucleotide sequence of a herpes simplex virus type 1 gene that causes cell fusion. Virology. 1985;145:36–48. doi: 10.1016/0042-6822(85)90199-0. [DOI] [PubMed] [Google Scholar]
- 6.Dijkstra J M, Visser N, Mettenleiter T C, Klupp B G. Identification and characterization of pseudorabies virus glycoprotein gM as a nonessential virion component. J Virol. 1996;70:5684–5688. doi: 10.1128/jvi.70.8.5684-5688.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dolter K E, Ramaswamy R, Holland T C. Syncytial mutations in the herpes simplex virus type 1 gK (UL53) gene occur in two distinct domains. J Virol. 1994;68:8277–8281. doi: 10.1128/jvi.68.12.8277-8281.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Foster T P, Kousoulas K G. Genetic analysis of the role of herpes simplex virus type 1 glycoprotein K in infectious virus production and egress. J Virol. 1999;73:8457–8468. doi: 10.1128/jvi.73.10.8457-8468.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fuchs W, Klupp B G, Granzow H, Rziha H-J, Mettenleiter T C. Identification and characterization of the pseudorabies virus UL3.5 protein, which is involved in virus egress. J Virol. 1996;70:3517–3527. doi: 10.1128/jvi.70.6.3517-3527.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fuchs W, Klupp B G, Granzow H, Mettenleiter T C. The UL20 gene product of pseudorabies virus functions in virus egress. J Virol. 1997;71:5639–5646. doi: 10.1128/jvi.71.7.5639-5646.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Graham F L, van der Eb A J. A new technique for the assay of infectivity of human adenovirus. Virology. 1973;52:456–467. doi: 10.1016/0042-6822(73)90341-3. [DOI] [PubMed] [Google Scholar]
- 12.Hutchinson L, Goldsmith K, Snoddy D, Ghosh H, Graham F L, Johnson D C. Identification and characterization of a novel herpes simplex virus glycoprotein, gK, involved in cell fusion. J Virol. 1992;66:5603–5609. doi: 10.1128/jvi.66.9.5603-5609.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hutchinson L, Graham F L, Cai W, Debroy C, Person S, Johnson D C. Herpes simplex virus (HSV) glycoproteins B and K inhibit cell fusion induced by HSV syncytial mutants. Virology. 1993;196:514–531. doi: 10.1006/viro.1993.1507. [DOI] [PubMed] [Google Scholar]
- 14.Hutchinson L, Johnson D C. Herpes simplex virus glycoprotein K promotes egress of virus particles. J Virol. 1995;69:5401–5413. doi: 10.1128/jvi.69.9.5401-5413.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hutchinson L, Roop-Beauchamp C, Johnson D C. Herpes simplex virus glycoprotein K is known to influence fusion of infected cells, yet is not on the cell surface. J Virol. 1995;69:4556–4563. doi: 10.1128/jvi.69.7.4556-4563.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jayachandra S, Baghian A, Kousoulas K G. Herpes simplex virus type 1 glycoprotein K is not essential for infectious virus production in actively replicating cells but is required for efficient envelopment and translocation of infectious virions from the cytoplasm to the extracellular space. J Virol. 1997;71:5012–5024. doi: 10.1128/jvi.71.7.5012-5024.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson M A, Prideaux C T, Kongsuwan K, Tyack S G, Sheppard M. ICP27 immediate early gene, glycoprotein K (gK) and DNA helicase homologues of infectious laryngotracheitis virus (gallid herpesvirus 1) SA-2 strain. Arch Virol. 1995;140:623–634. doi: 10.1007/BF01309954. [DOI] [PubMed] [Google Scholar]
- 18.Jöns A, Dijkstra J M, Mettenleiter T C. Glycoproteins M and N of pseudorabies virus form a disulfide-linked complex. J Virol. 1998;72:550–557. doi: 10.1128/jvi.72.1.550-557.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaplan A S, Vatter A E. A comparison of herpes simplex and pseudorabies viruses. Virology. 1959;7:394–407. doi: 10.1016/0042-6822(59)90068-6. [DOI] [PubMed] [Google Scholar]
- 20.Khadr A, Tikoo S K, Babiuk L A, van Drunen Littel-van den Hurk S. Sequence and expression of a bovine herpesvirus-1 gene homologous to the glycoprotein K-encoding gene of herpes simplex virus-1. Gene. 1996;168:189–193. doi: 10.1016/0378-1119(95)00776-8. [DOI] [PubMed] [Google Scholar]
- 21.Klupp B G, Kern H, Mettenleiter T C. The virulence-determining genomic BamHI fragment 4 of pseudorabies virus contains genes corresponding to the UL15 (partial), UL18, UL19, UL20, and UL21 genes of herpes simplex virus and a putative origin of replication. Virology. 1992;191:900–908. doi: 10.1016/0042-6822(92)90265-q. [DOI] [PubMed] [Google Scholar]
- 22.Klupp B G, Baumeister J, Karger A, Visser N, Mettenleiter T C. Identification and characterization of a novel structural glycoprotein in pseudorabies virus, gL. J Virol. 1994;68:3868–3878. doi: 10.1128/jvi.68.6.3868-3878.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klupp B G, Lomniczi B, Visser N, Fuchs W, Mettenleiter T C. Mutations affecting the UL21 gene contribute to avirulence of pseudorabies virus vaccine strain Bartha. Virology. 1995;212:466–473. doi: 10.1006/viro.1995.1504. [DOI] [PubMed] [Google Scholar]
- 24.Klupp B G, Fuchs W, Weiland E, Mettenleiter T C. Pseudorabies virus glycoprotein L is necessary for virus infectivity but dispensable for virion localization of glycoprotein H. J Virol. 1997;71:7687–7695. doi: 10.1128/jvi.71.10.7687-7695.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Klupp B G, Baumeister J, Dietz P, Granzow H, Mettenleiter T C. Pseudorabies virus glycoprotein gK is a virion structural component involved in virus release but is not required for entry. J Virol. 1998;72:1949–1958. doi: 10.1128/jvi.72.3.1949-1958.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Klupp B G, Mettenleiter T C. Glycoprotein gL-independent infectivity of pseudorabies virus is mediated by a gD-gH fusion protein. J Virol. 1999;73:3014–3022. doi: 10.1128/jvi.73.4.3014-3022.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature (London) 1979;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Mettenleiter T C, Rauh I. A glycoprotein gX-β-galactosidase fusion gene as insertional marker for rapid identification of pseudorabies virus mutants. J Virol Methods. 1990;30:55–66. doi: 10.1016/0166-0934(90)90043-f. [DOI] [PubMed] [Google Scholar]
- 29.Mettenleiter T C. Immunobiology of pseudorabies (Aujeszky's disease) Vet Immunol Immunopathol. 1996;54:221–229. doi: 10.1016/s0165-2427(96)05695-4. [DOI] [PubMed] [Google Scholar]
- 30.Mettenleiter T C. Aujeszky's disease (pseudorabies) virus: the virus and molecular pathogenesis—state of the art, June 1999. Vet Res. 2000;31:99–115. doi: 10.1051/vetres:2000110. [DOI] [PubMed] [Google Scholar]
- 31.Mo C, Holland T C. Determination of the transmembrane topology of herpes simplex virus type 1 glycoprotein K (gK) J Biol Chem. 1997;272:33305–33311. doi: 10.1074/jbc.272.52.33305. [DOI] [PubMed] [Google Scholar]
- 32.Mo C, Suen J, Sommer M, Arvin A. Characterization of varicella-zoster virus glycoprotein K (open reading frame 5) and its role in virus growth. J Virol. 1999;73:4197–4207. doi: 10.1128/jvi.73.5.4197-4207.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pogue-Geile K L, Spear P G. The single base pair substitution responsible for the syn phenotype of herpes simplex virus type 1, strain mp. Virology. 1987;157:67–74. doi: 10.1016/0042-6822(87)90314-x. [DOI] [PubMed] [Google Scholar]
- 34.Ramaswamy R, Holland T C. In vitro characterization of the HSV-1 UL53 gene product. Virology. 1992;186:579–587. doi: 10.1016/0042-6822(92)90024-j. [DOI] [PubMed] [Google Scholar]
- 35.Ren D, Lee L F, Coussens P M. Identification and characterization of Marek's disease virus genes homologous to ICP27 and glycoprotein K of herpes simplex virus-1. Virology. 1994;204:242–250. doi: 10.1006/viro.1994.1528. [DOI] [PubMed] [Google Scholar]
- 36.Spear P G. Entry of alphaherpesviruses into cells. Semin Virol. 1993;4:167–180. [Google Scholar]
- 37.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ward P L, Campadelli-Fiume G, Avitabile E, Roizman B. Localization and putative function of the UL20 membrane protein in cells infected with herpes simplex virus 1. J Virol. 1994;68:7406–7417. doi: 10.1128/jvi.68.11.7406-7417.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhao Y, Holden V R, Harty R N, O'Callaghan D J. Identification and transcriptional analyses of the UL3 and UL4 genes of equine herpesvirus 1, homologs of the ICP27 and glycoprotein K genes of herpes simplex virus. J Virol. 1992;66:5363–5372. doi: 10.1128/jvi.66.9.5363-5372.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]