Abstract
The Borrelia burgdorferi SpoVG protein has previously been found to be a DNA- and RNA-binding protein. To aid in the elucidation of ligand motifs, affinities for numerous RNAs, ssDNAs, and dsDNAs were measured and compared. The loci used in the study were spoVG, glpFKD, erpAB, bb0242, flaB, and ospAB, with particular focus on the untranslated 5′ portion of the mRNAs. Performing binding and competition assays yielded that the 5′ end of spoVG mRNA had the highest affinity while the lowest observed affinity was to the 5’ end of flaB mRNA. Mutagenesis studies of spoVG RNA and ssDNA sequences suggested that the formation of SpoVG-nucleic acid complexes are not entirely dependent on either sequence or structure. Additionally, exchanging uracil for thymine in ssDNAs did not affect protein-nucleic acid complex formation.
Keywords: SpoVG, RNA binding protein, DNA binding protein, Borrelia burgdorferi, Electrophoretic mobility shift assay (EMSA)
1. Introduction
SpoVG proteins are conserved across diverse groups of bacteria, including Firmicutes, Spirochaetota, and Myxococcota (Delta-proteobacteria) [1]. The protein’s nomenclature derives from early studies of Bacillus spp., where it was observed that mutations within the spoVG locus impaired sporulation at stage V [2]. The SpoVG homologues of Borrelia burgdorferi and other non-sporulating bacterial species affect expression of a broad range of genes, and spoVG mutants exhibit physiological defects [3–8]. Our group has studied SpoVG proteins from B. burgdorferi, Staphylococcus aureus, and Listeria monocytogenes, and found that they exhibit affinities for specific DNA sequences [1]. Subsequent studies by Burke and Portnoy found that L. monocytogenes SpoVG exhibits a substantially greater affinity for single-stranded RNAs than for cognate double-stranded DNAs (dsDNAs), and hypothesized that SpoVG functions primarily as an RNA-binding protein [3]. Evaluation of the B. burgdorferi homologue found a similarly greater affinity for RNAs [4].
The Lyme disease spirochete, B. burgdorferi, is a particularly useful model for studies of gene and protein regulation in a vector-borne pathogen [9–11]. In addition to being the cause of a significant human disease, B. burgdorferi infects both vertebrates and ticks [12]. The Lyme spirochete must, therefore, possess mechanisms to accurately determine which host it is in, and produce proteins and other essential factors for each stage of its infectious cycle. B. burgdorferi must also recognize when a tick is feeding, in order to undergo the physiological changes that are necessary for transmission into the vertebrate host. Our group’s previous data indicate that B. burgdorferi SpoVG affects expression levels of numerous proteins and affects bacterial physiology, apparently through its activities as an RNA- and/or DNA-binding protein [4]. To gain further insights into SpoVG function, we identified high- and low-affinity ligands of B. burgdorferi SpoVG, which were then employed to gain insights on the ability of SpoVG to bind single-stranded and double-stranded nucleic acids.
2. Materials and methods
2.1. Purification of recombinant proteins
Polyhistidine-tagged B. burgdorferi SpoVG was purified essentially as described previously [1,4]. Briefly, Escherichia coli Rosetta II (Invitrogen, MA) was transformed with pBLJ132, which consists of spoVG cloned into pET101 [1]. E. coli were then grown to an OD600 of at least 1.0 in Super Broth (32 g Tryptone, 20 g Yeast Extract, and 5 g NaCl per liter), and recombinant SpoVG expression was induced for 1h by adding isopropyl-β-d-thiogalactopyranoside (IPTG) to a final concentration of 1 mM. Bacteria were harvested by centrifugation at 5400×g for 30 min and frozen at −80 °C until needed. Resuspended cells were lysed by sonication with the addition of B-PER bacterial protein extraction reagent to 2% v/v (Thermo-Fisher, MA). Recombinant proteins were purified using MagneHis nickel particles (Promega, WI), then dialyzed against EMSA buffer (50 mM Tris-HCl, 25mMKCl, 10% glycerol (v/v), 0.01% Tween 20, 100 nM dithiothreitol (DTT), and 1 mM phenylmethanesulfonyl fluoride (PMSF)). Proteins were concentrated using 10 kDa Amicon centrifugal units (MilliporeSigma, MA) and aliquots were stored at −80 °C until needed. Protein purity and concentration were assessed by SDS-PAGE, Quick Start Bradford protein assay (Bio-Rad), and bicinchoninic acid assay (BCA) (Thermo-Fisher, MA).
2.2. Electromobility shift assay (EMSA)
Fluorescently tagged and untagged DNA and RNA oligonucleotides were synthesized by Integrated DNA Technologies (IDT, IA). The sequences of oligonucleotides used in this study are listed in Table 1 and Supplemental Table 1. DNA and RNA probes were tagged on the 5’ end with an IRDye 800 fluorescent tag (LI-Cor, NE) or an Alexa Fluor 488, respectively. Double-stranded DNA (dsDNA) probes and competitors were produced from pairs of separately-synthesized complimentary oligonucleotides by mixing equal molar concentrations, heating to 95 °C, and slowly cooling to room temperature.
Table 1.
5′-3′ sequences of dsDNA, ssDNA and RNA used throughout. Red highlighted nucleotides in the ssDNA and RNA sections deviate from the spoVG probe. ss22 and ss23 have a deoxyribose backbone with uracil bases instead thymine.
| Name | Sequence |
|---|---|
| dsDNA | |
| SpoVG DNA | AGTTATGTACTTTTTGCGGGAGGCTTATAA |
| erpAB DNA | CTTATGGAGAAATTTATGAATAAGAAAATGAAA |
| glpFKD DNA | ATTAAATATAATTTTAATAAGGCTTTTATTAGAAAAATTAAT |
| bb0242 DNA | GTATTCAAAAAATAAACTGTCTAAACCTTTTGAAAAGG |
| OSpAB DNA | GTATTAAGTTATATTAATATAAAAGGAGAATATATT |
| flaB DNA | AGGCAAAAGGATTTGCCAAAGTCAGAAATT |
| ssDNA | |
| spoVG CD | TATGTACTTTTTGCGGGAGGCTTATAA |
| SpoVG NC | TTATAAGCCTCCCGCAAAAAGTACATA |
| ss01 | AATGTACTTTTTGCGGGAGGCTTATAA |
| ss02 | GATGTACTTTTTGCGGGAGGCTTATAA |
| ss03 | TATGTACAAAAAGCGGGAGGCTTATAA |
| ss04 | TATGTACCCCCCGCGGGAGGCTTATAA |
| ss05 | TATGTACGGGGGGCGGGAGGCTTATAA |
| ss06 | TATGTACTTTTTGCGGGTGGCTTATAA |
| ss07 | TATGTACTTTTTGCGGGTCCGAAATAA |
| ss08 | TATGTACTTTTTGCGGGGAATCCATAA |
| ss09 | GTACTTTTTGCGGGAGGCTTATAA |
| ss10 | CTTTTTGCGGGAGGCTTATAA |
| ss11 | TATGTACATTTTGCGGGAGGCTTATAA |
| ss12 | TATGTACTTATTGCGGGAGGCTTATAA |
| ss13 | TATGTACTTTTAGCGGGAGGCTTATAA |
| ss14 | TATGTACAATTTGCGGGAGGCTTATAA |
| ss15 | TATGTACTAATTGCGGGAGGCTTATAA |
| ss16 | TATGTACTTAATGCGGGAGGCTTATAA |
| ss17 | TATGTACTTTAAGCGGGAGGCTTATAA |
| ss18 | TATGTACAAATTGCGGGAGGCTTATAA |
| ss19 | TATGTACTAAATGCGGGAGGCTTATAA |
| ss20 | TATGTACTTAAAGCGGGAGGCTTATAA |
| ss21 | TATGTACAAAAAGCGGGTCCGAAATAA |
| ss22 | TATGTACUUUUUGCGGGAGGCTTATAA |
| ss23 | UAUGUACUUUUUGCGGGAGGCUUAUAA |
| RNA | |
| SpoVG RNA | UAUGUACUUUUUGCGGGAGGCUUAUAA |
| erpAB RNA | AUGGAGAAAUUUAUGAAUAAGAAAAUGAAA |
| glpFKD RNA | AUAAUUUUAAUAAGGCUUUUAUUAGAAAAAUUAAU |
| bb0242 RNA | GUAUUCAAAAAAUAAACUGUCUAAACCUUUUGAAAAGG |
| ospAB RNA | GUAUUAAGUUAUAUUAAUAUAAAAGGAGAAUAUAUU |
| flaB RNA | AGGCAAAAGGAUUUGCCAAAGUCAGAAAUU |
| RNA-03 | UAUGUACAAAAAGCGGGAGGCUUAUAA |
| RNA-17 | UAUGUACUUUAAGCGGGAGGCUUAUAA |
| RNA-20 | UAUGUACUUAAAGCGGGAGGCUUAUAA |
EMSAs were performed as previously described [1,4]. Unless stated otherwise, purified proteins were added to 100 nM of labeled nucleic acid probes and incubated at room temperature for 5–15 min. Unlabeled nucleic acid competitors were added prior to the addition of protein. EGTA was added to the indicated reactions to inhibit RNAse activity in protein aliquots. Poly-dI-dC was added to indicated gels as a non-specific competitor. One-sixth volume of EMSA loading dye (15 mg/mL Ficol 400, 0.8 mg/mL Orange G) was added. Electrophoresis was performed using pre-run 6% TBE gels (Invitrogen, MA) at 100-V in 0.5x TBE buffer. Gels were imaged and analyzed via densitometry with a ChemiDoc MP and Image-Lab software, respectively (Bio-Rad, CA). Lanes and bands were added manually according to the strongest free probe and the strongest shifted band. Equivalent bands were then added across the entire EMSA.’ Background noise was calculated and accounted for by the Image-Lab software. Percent shifted was graphed using GraphPad Prism 9 (Dotmatics, MA).
2.3. Calculation of apparent KD and IC50 values
For direct binding assays, the fraction of bound complex was fit to a one-site saturation ligand binding model (Fraction Bound = Bmax SpoVG/(Kd + SpoVG)) where Bmax is the maximum fraction bound and KD is the apparent dissociation equilibrium constant. [13]. It should be noted that this estimate of the binding constant is limited by the high concentration of nucleic acid in the binding assays which likely puts acids in the “Titration Regime” but can still be used to determine relative affinities of one sequence to another [13]. In competition assays, data were fit globally to determine the concentration of inhibitor resulting in 50% of the complex being disrupted (IC50) to the equation fraction bound to labeled probe = m – (m-b)/(1+IC50/[C]) where m is the maximum signal, b is the minimum (background) signal, [C] is the concentration of the unlabeled nucleic acid competitor, and IC50 is the concentration of the competitor that results in 50% of the complex being disrupted.
3. Results
3.1. SpoVG is a site-specific RNA- and DNA-binding protein
We previously reported that B. burgdorferi SpoVG can bind to the 5′ ends of the borrelial spoVG and glpFKD mRNAs, and then cognate dsDNAs [4]. It was also found that SpoVG binds within the transcript of a small gene of unknown function, bb0242, that is located between the glpK and glpD genes [4]. The present studies delved further into the interactions between SpoVG and other nucleic acid sequences to assess relative affinities and mechanisms of binding. Noting that spoVG and the glpFKD operon are highly expressed during tick colonization but not during mammalian infection, we investigated the ability of SpoVG to bind other borrelial sequences; specifically the 5’ ends of erpAB (which is repressed during tick colonization but highly expressed during mammalian infection), ospAB (which is highly expressed during tick colonization but repressed in mammals), and flaB (which is constitutively expressed throughout the borrelial tick-mammal cycle) [4,12,14–18]. Both RNA and dsDNA were examined for each target.
Of the tested probes and competitors, SpoVG exhibited the greatest affinity for the spoVG mRNA 5′ end, henceforth referred to as spoVGRNA (Fig. 1 and Fig. 2). This high affinity binding site was further tested by competition EMSAs using an unlabeled spoVGRNA as a competitor (Fig. 1A and Supp. Fig. 1). No intermediate shift was observed indicating that there was no formation of a dsRNA probe. The protein-RNA complex shift stayed above 50% until 1 μM (10x) of competitor was added. Addition of up to 100 ng/μL poly-dI-dC did not result in an appreciable difference of SpoVG binding to the spoVGRNA (Fig. 1B), further indicating that binding of SpoVG to spoVGRNA is specific. SpoVG also bound to the glpFKDRNA, bb0242RNA, and erpABRNA probes, but EMSAs did not approach saturation in the tested ranges of SpoVG protein concentrations, indicating that the affinity of SpoVG for those three RNAs is weaker than spoVGRNA (Fig. 2).. (Insert Figs. 1 and 2)
Fig. 1. spoVG RNA is a SpoVG high affinity binding site.
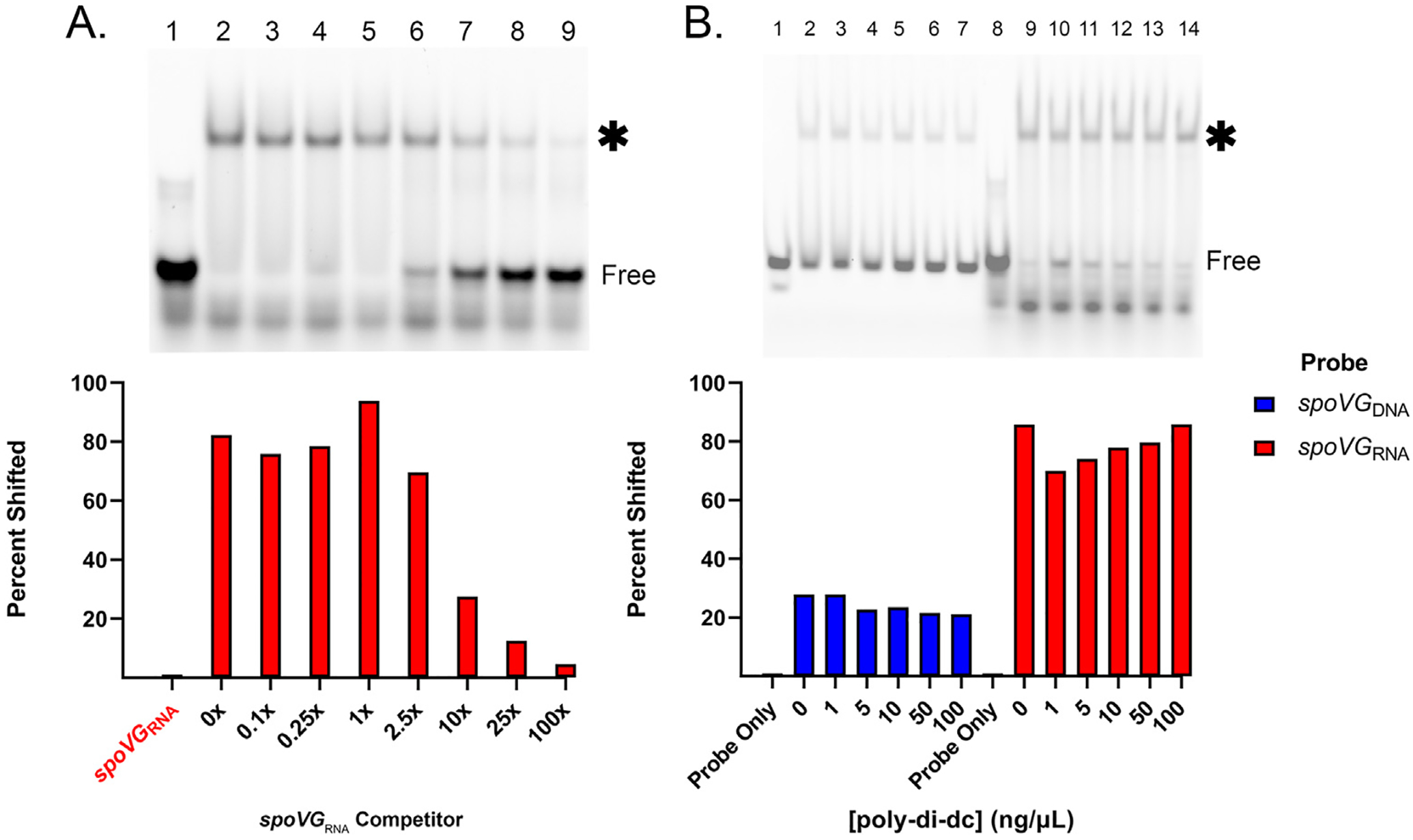
A. Competition assay using 100 nM labeled spoVGRNA. Free probe is indicated, and the shifted probe is indicated by an asterisk. SpoVG concentration was 500 nM in lanes 2–9. Unlabeled spoVGRNA was added at 10 nM, 25 nM, 100 nM, 250 nM, 1 μM, 2.5 μM, and 10.0 μM in lanes 3–9 respectively. EGTA was added to 5 mM. This assay was done in triplicate (Sup. Fig. 1). B. Competition assay using labeled spoVGDNA and spoVGRNA probe against poly-dI-dC. Lanes 1–7 included 100 nM spoVGDNA probe. Lanes 8–14 included 100 nM spoVGRNA probe. SpoVG was added to lanes 2–7 at 10 μM and lanes 9–14 at 100 nM. Poly-dI-dC was added in at 1,5,10,50, and 100 ng/μL in lanes 3–7 and 10–14 respectively.
Fig. 2. SpoVG has a higher binding affinity for RNA than DNA.
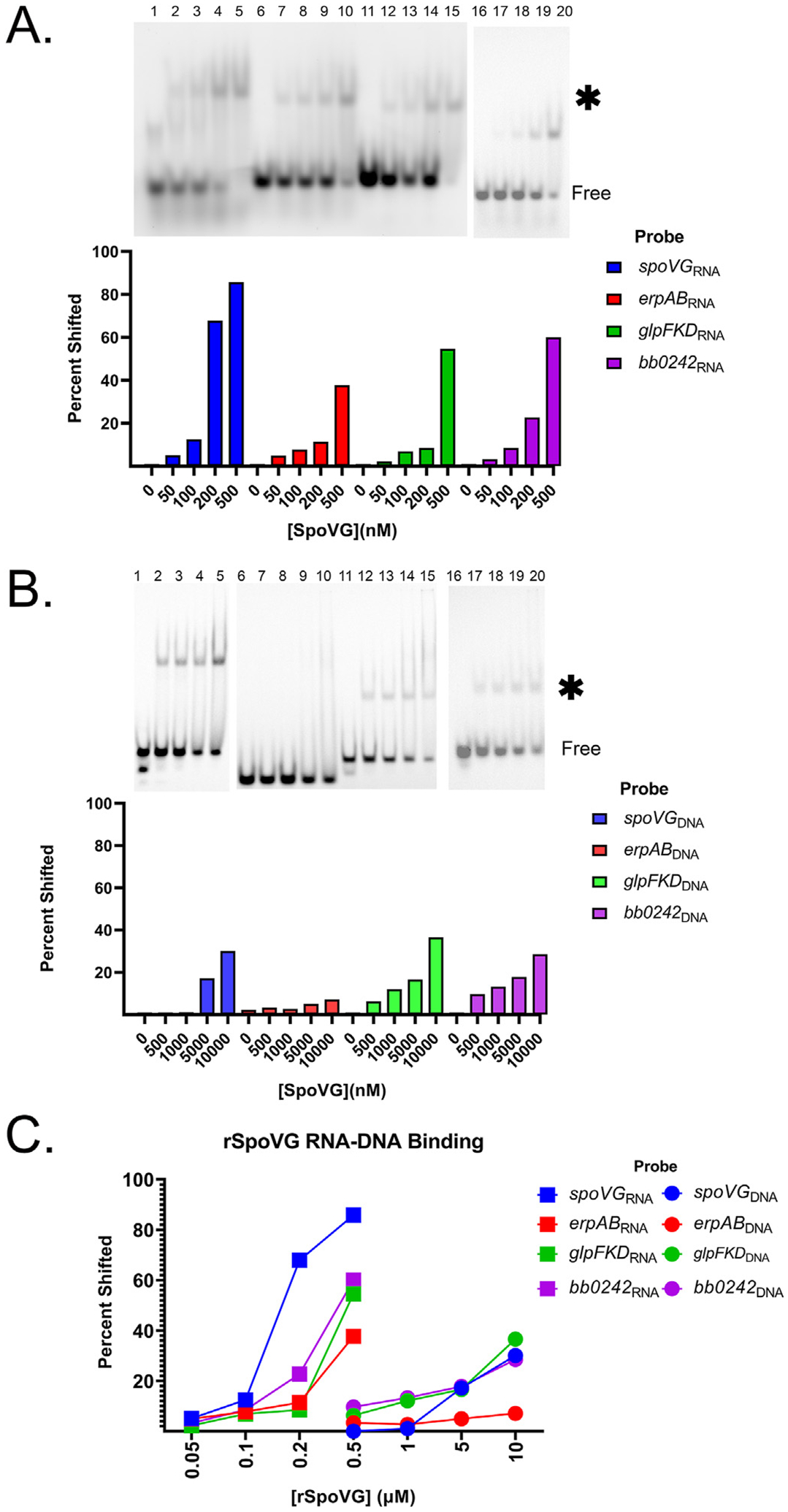
A. EMSA using labeled spoVGRNA, erpABRNA, glpFKDRNA, and bb0242RNA at 100 nM from lanes 1–5, 6–10, and 11–15, 16–20 respectively. Free probe is indicated, and the shifted probe is indicated by an asterisk. SpoVG was added to lanes 2–5, 7–10, 12–15, and 17–20 at 50 nM, 100 nM, 200 nM, and 500 nM. B. EMSAs using labeled spoVGDNA, glpFKDDNA, erpABDNA, and bb0242DNA at 100 nM from 1 to 5, 6–10, 11–15, 16–20 respectively. SpoVG was added to lanes 2–5, 7–10, 12–15, and 17–20 at 500 nM, 1 μM, 5 μM, and 10 μM respectively. C. Graph representing the percentage of 100 nM probe shifted due to SpoVG binding from the EMSAs in A and B.
Our initial studies of B. burgdorferi and S. aureus SpoVG found that both proteins preferentially bound to certain dsDNA sequences [1]. In contrast, a study of the L. monocytogenes orthologue with a variety of DNA probes led those researchers to conclude that DNA binding appeared to be non-specific [3]. To build upon those observations, we assessed the affinities of B. burgdorferi SpoVG for the above-described borrelial sequences as dsDNA. Affinities for all tested dsDNAs were significantly lower than their cognate RNAs (Fig. 2). The relative affinity of SpoVG to the spoVGDNA probe was over 100x weaker than for spoVGRNA, with addition of 10 μM SpoVG protein yielding approximately 30% shifted probe (Fig. 2B). This difference signifies that a substantially higher concentration of SpoVG is required to form a complex with SpoVGDNA dsDNA compared to SpoVGRNA.
These results were highlighted by performing EMSAs of the spoVGDNA, spoVGRNA, glpFKDDNA, and glpFKDRNA probes (Supp. Fig. 2). EMSAs with spoVG RNA achieved close to 100% shift at 200 nM compared to the unshifted dsDNA at 200 nM concentration (Supp. Fig. 3A). This was also observed with the glpFKDRNA. Saturation of the glpFKDRNA probe occurred between 1 μM and 10 μM SpoVG, whereas the glpFKDDNA probe showed almost no detectable shift at that concentration. While SpoVG had an inherently weaker interaction with the DNAs tested than the RNAs, there were still substantial differences when comparing affinities for the DNA probes out of the four tested. SpoVG had the lowest relative affinity for the erpABDNA probe (Fig. 2B and C).
The relative affinities of SpoVG for the ospABRNA and flaBRNA were determined by use of unlabeled RNAs as competitors against labeled spoVGRNA or glpFKDRNA probes. When using the spoVGRNA probe, addition of 100x excess of the unlabeled ospABRNA reduced the shifted protein-RNA complex by approximately 18% (IC50 = 5.9±4.6 × 10–5 M), while 100x excess of the unlabeled flaBRNA did not detectably affect the SpoVG-spoVGRNA complex (Fig. 3A and B). When labeled glpFKDRNA was used as a probe, addition of 25x excess of unlabeled flaBRNA led to a 20% reduction in the SpoVG-glpFKDRNA complex (Fig. 3C). In contrast, 25x excess of unlabeled glpFKDRNA, erpABRNA, or ospABRNA reduced binding to the probe by approximately 80%, and unlabeled spoVGRNA virtually eliminated the SpoVG-glpFKDRNA complex (Fig. 3C). The data above indicate that SpoVG has a substantially greater affinity for RNA compared to DNA, but that this affinity differs based on the sequence of the nucleic acid.
Fig. 3. SpoVG is a specific RNA binding protein.
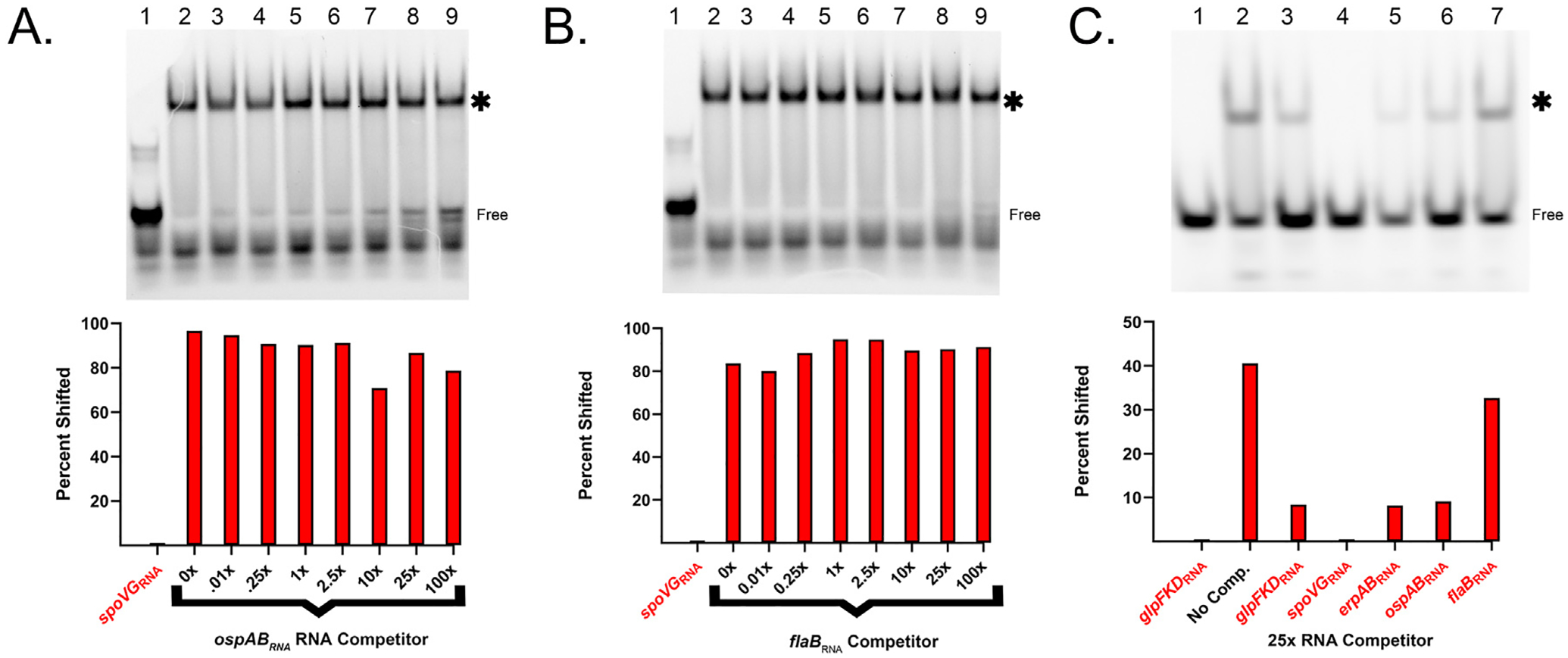
A. Competition assay using 100 nM labeled spoVGRNA. Free probe is indicated, and the shifted probe is indicated by an asterisk. The SpoVG concentration was 500 nM in lanes 2–9. Unlabeled ospABRNA was added at 10 nM, 25 nM, 100 nM, 250 nM, 1 μM, 2.5 μM, and 10.0 μM in lanes 3–9 respectively. EGTA was added to 5 mM. B. Competition assay using unlabeled flaBRNA and 100 nM labeled spoVGRNA. Lane values match (A), except unlabeled flaBRNA was used instead of ospABRNA. C. Competition assay using 100 nM labeled glpFKDRNA (lanes 8–14). SpoVG concentration was 500 nM. Competitors were added at a concentration of 2.5 μM in the following sequence glpFKDRNA, spoVGRNA, erpABRNA, bb0242RNA, and flaBRNA. The gamma of entire image was increased to more accurately visualize differences in competition.
3.2. Effects of nucleic acid sequence on SpoVG-binding
As the SpoVG protein appears to have the strongest affinity for the spoVGRNA probe, we made a series of sequence variants to determine whether the sequence or structure of the RNA was important in recognition and affinity of the protein to the nucleic acid. EMSAs were undertaken using the labeled spoVGRNA probe and numerous unlabeled variant nucleic acids as competitors (Table 1). As a cost saving first approach, ssDNA competitors were used for the initial analyses. Sequence variations in competitors included changing the 5′ nucleotide, removing 3 or 6 nucleotides from the 5’ end, altering the run of five consecutive thymines, and altering an AGGCT sequence near the 3′ end (Table 1).
Of the 21 tested unlabeled ssDNAs, only a subset of the oligonucleotides that disrupted the five-thymine run had appreciable impacts on competition for SpoVG (Fig. 4A and B). Competition of the spoVGRNA with nucleic acid fragments that replace the sequence TTTTT in the unlabeled probe with CCCCC (ss04) or GGGGG (ss05) did not appreciably affect their ability to compete with the wild type spoVG sequence (Fig. 4A). However, replacing the run of thymines in the sequence with adenines in some positions: (AAAAA) (ss03), TTTAA (ss17), or AAATT (SS18), or with Us (ss21) resulted in reduced competition. Lastly, when combining the changes of the TTTTT sequence to AAAAA with deletion of the AGGCT sequence (ss21), the competition was reduced but was similar to that of the AAAAA (ss03) change only.
Fig. 4. Mutagenesis of spoVG ssDNA and RNA.
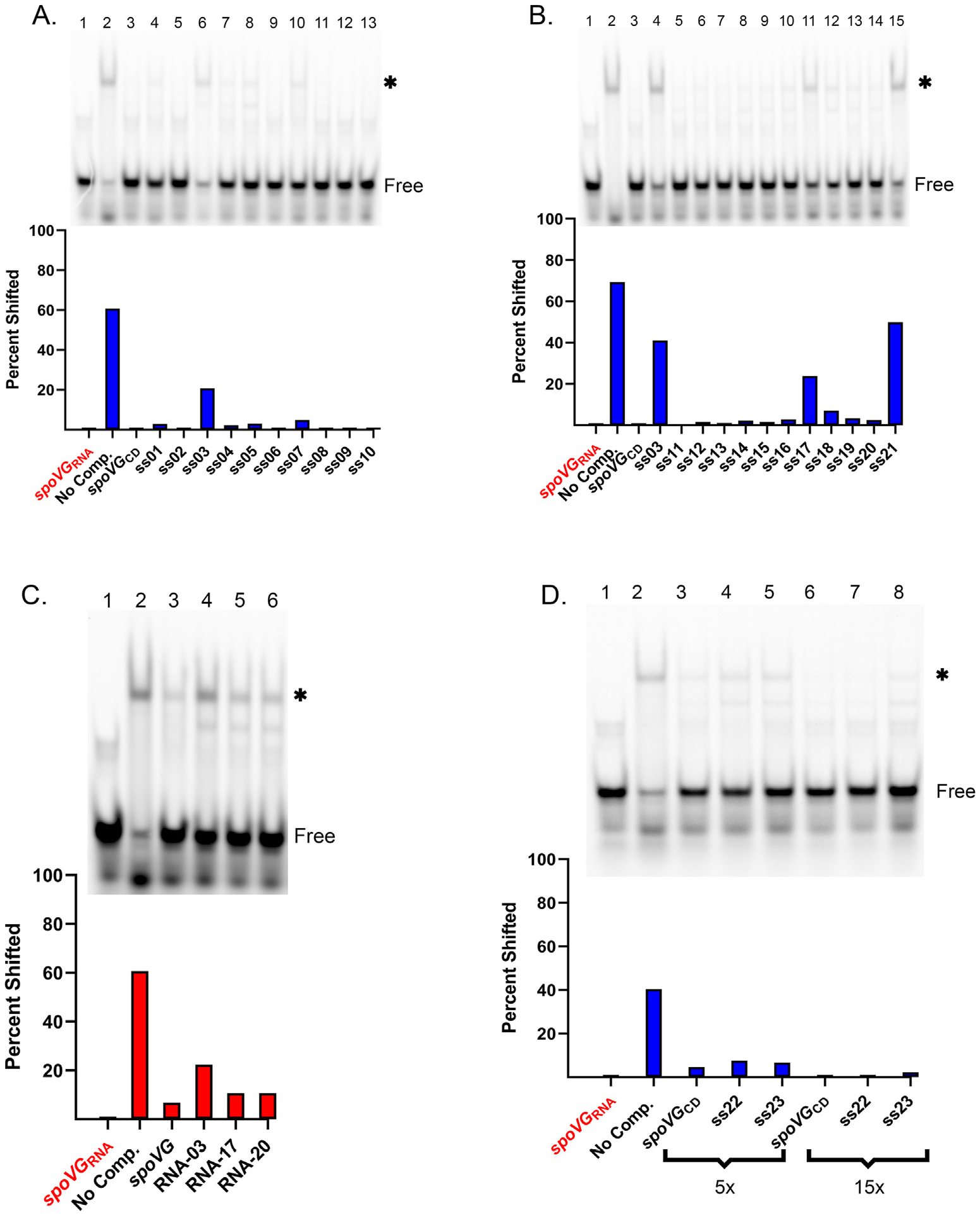
A. EMSA using 100 nM spoVGRNA probe and 100 nM of SpoVG in lanes 2–13. ssDNA competitors were added to lanes 3–13 at 5 μM. Free probe is indicated, and the shifted probe is indicated by an asterisk. Competitors used were spoVG + through ss10 (Table 1 and Supp. Table 1). B. EMSA using 100 nM spoVGRNA probe and 100 nM of SpoVG in lanes 2–15. Competitors at 5 μM were added to lanes 3–15. Competitors were spoVGCD, ss03, followed by ss11 through ss21 (Table 1). C. EMSA using 100 nM spoVGRNA probe with 100 nM SpoVG added to lanes 2–6. Unlabeled spoVGRNA, RNA-03, RNA-17, and RNA-20 were added at 1.5 μM to lanes 3–6 respectively. D. EMSA using 100 nM of spoVGRNA probe and 100 nM of SpoVG added to lanes 2–8. ssDNA competitors consisted of spoVGCD, ss22, and ss23 at 500 nM in lanes 3–5 and 2 μM in lanes 6–8.
Based on above results, three unlabeled RNAs were synthesized for use as EMSA competitors: RNA-03 changed the run of five uracils (UUUUU) to adenines (AAAAA), RNA-17 changed the run to UUUAA, and RNA-20 changed it to UUAAA (Table 1). When added to EMSAs at 15-fold excess, each of these mutant competitors was slightly less effective at competing for SpoVG than the spoVGRNA sequence (Fig. 4C). RNA-03 saw a 29% decrease in competition, whereas RNA-17 and RNA-20 saw a 7% decrease in competition compared to the wild type sequence. All RNAs effectively competed for SpoVG when added at 50x excess whereas the ssDNA counter-parts, ss03 and ss17, were less effective than spoVGCD (Table 1 and Fig. Supp. 3A). In contrast, 50x excess of unlabeled glpFKDRNA competitor did not compete nearly as well as any tested spoVG mutant RNA sequence. (Fig. Supp. 3B). Altogether, this suggests that SpoVG differentially interacts with additional aspects of the spoVGRNA that are not present in the glpFKDRNA.
Due to the observed differential affinities above between the RNA probes derived from the start of transcription and their cognate dsDNAs, we performed competition assays comparing the coding (spoVGCD) and non-coding (spoVGNC) strands of DNA. EMSAs were performed in which the two ssDNAs competed against labeled erpABRNA, which was used to inhibit possible base pairing. This competition EMSA indicated that spoVGNC better competed against erpABRNA than spoVGCD (Supp. Fig. 4A and B). A substantial difference was observed when competitors were added at 100 nM concentration: spoVGNC was able to compete away 62% of the SpoVG-erpABRNA complex compared to spoVGCD, which only competed away 12%.
3.3. Differences of uracil vs thymine alone can not explain the differences in RNA and ssDNA binding
The differential competition of the SpoVG derived ssDNA and RNA sequences lead us to further investigate whether the presence of uracils in RNA and thymines in ssDNA contributed to differences in SpoVG affinity. To that end, we produced two ssDNAs based on the spoVGCD sequence: competitor ss22 replaced only the TTTTT with UUUUU, whereas competitor ss23 replaced every thymine in the sequence with uracil (Table 1). Competition analysis using the altered ssDNA sequences did not result in more effective competition than the original SpoVG ssDNA sequence at 5x and 15x competitor concentrations (Fig. 4D).
4. Discussion
B. burgdorferi SpoVG is a sequence-specific RNA- and DNA-binding protein, exhibiting binding constants of 5.9±2.1 × 10–8 M from direct binding assays and with an apparent IC50 of 1.033 × 10–6 M ± 2.7 × 10–7 for spoVGRNA. Binding affinities for tested dsDNAs appear to be over 100-times weaker than for cognate RNAs, yet selectivity and differential affinity for certain dsDNA sequences was observed.
Analyses with the high-affinity RNA sequence derived from the spoVG start of transcription provided insights on the nature of those interactions. This RNA contains a run of five uracils, UUUUU, which, when changed to adenines, AAAAA, substantially affected SpoVG binding. Two additional base replacements, UUUUU to UUUAA or UUAAA, also led to decreases in SpoVG-RNA complex formation. Note that two other RNAs that SpoVG bound with high affinity also contains runs of uracils: the glpFKDRNA probe contains two poly-U tracts, UUUUAAU and UUUUAUU, and the erpABRNA probe contains one poly-U tract, UUUAU. However, ospABRNA contains runs of only two sequential uracils, UU, and the flaBRNA contains a single uracil tract of UUU. These results suggest that a poly-U tract may play a role in SpoVG binding. Noting that unlabeled glpFKDRNA was less effective than any of the mutated spoVG RNAs, it is evident that additional features have important impacts on complex formation.
Having observed that spoVG ssDNA and RNA sequence variants showed a wide variation in their abilities to bind SpoVG, we used RNA structure v6.4 to predict the secondary structures of these sequences and the other natural substrates that were observed (Supp. Table 1) [19]. A majority of the constructs that were the least effective competitors e.g. (SS-03 and RNA-03, SS-17 and RNA-17, SS-20 and RNA-20) are predicted to share a secondary structure where the nucleic acid has a stem of 4 or 5 base pairs and a loop with a sequence GGAG. Lacking a 2′ hydroxyl group, the RNA sequences may fold into a more A-form helix compared to the same sequence of DNA, providing a possible rationale for why SpoVG prefers RNA over DNA. This seems particularly likely in that replacing thymines with uracils (ss22 and ss23) was insufficient to increase affinity of the SpoVG protein for these sequences. Yet, sequence ss21, which is predicted to have a different structure, with variations in both the stem and the loop, was still an efficient competitor, suggesting that there are likely other determinants to binding.
The greater affinity of SpoVG for RNA over cognate ssDNA containing uracil implies that SpoVG is more likely to form complexes with RNA rather than DNA either through direct interactions with the backbone or because of RNA’s preferred tertiary folds compared to those of DNA. Previous site-directed mutagenesis of SpoVG revealed that two domains are involved with nucleic acid-binding [1]. Residues in the sole alpha helix of the protein confer nucleic acid sequence preference. Two positively charged residues (argi-nine or lysine) in a separate loop are conserved across all bacterial species and changing either residue to an alanine eliminated nucleic acid binding. Taken together, these results suggest that the charged residues of the loop may bind to the sugar-phosphate backbone and may be better positioned to interact with ribose rather than deoxyribose.
We acknowledge that the sequences used in this study were short fragments of the 5’ end of mRNAs and lack the complexity of full-length mRNAs of coding genes. We are presently examining the B. burgdorferi transcriptome by RNA immunoprecipitation -sequencing (RIP-Seq) to identify additional high-affinity SpoVG binding sites, which can then be compared to the binding sites herein, to provide an enhanced rational approach to characterizing RNA features that are involved with SpoVG-binding.
In conclusion, the results of these studies indicate that there is definitive preferential affinity of SpoVG for certain RNAs and DNAs within the B. burgdorferi genome. Furthermore, SpoVG can bind to numerous sites throughout the transcriptome and genome. B. burgdorferi controls levels of SpoVG throughout the infection cycle [4]. Thus, differential expression of SpoVG will result in occupancy of greater or fewer binding sites, facilitating a variety of different phenotypes.
Supplementary Material
Acknowledgments
These studies were funded by US NIH grant R01 AI144126. We thank Tatiana Castro-Padovani and Jessamyn Moore for assistance and helpful comments on this manuscript.
Footnotes
Declaration of competing interest
The authors certify that they do not have any conflicts of interest with this study or report.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrc.2023.02.044.
References
- [1].Jutras BL, Chenail AM, Rowland CL, Carroll D, Miller MC, Bykowski T, Stevenson B, Eubacterial SpoVG homologs constitute a new family of site-specific DNA-binding proteins, PLoS One 8 (2013), e66683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Rosenbluh A, Banner CD, Losick R, Fitz-James PC, Identification of a new developmental locus in Bacillus subtilis by construction of a deletion mutation in a cloned gene under sporulation control, J. Bacteriol 148 (1981) 341–351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Burke TP, Portnoy DA, SpoVG is a conserved RNA-binding protein that regulates Listeria monocytogenes lysozyme resistance, virulence, and swarming motility, mBio 7 (2016), e00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Savage CR, Jutras BL, Bestor A, Tilly K, Rosa PA, Tourand Y, Stewart PE, Brissette CA, Stevenson B, Borrelia burgdorferi SpoVG DNA- and RNA-binding protein modulates the physiology of the Lyme disease spirochete, J. Bacteriol 200 (2018) e00033, 00018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Liu X, Zhang S, Sun B, SpoVG regulates cell wall metabolism and oxacillin resistance in methicillin-resistant Staphylococcus aureus strain N315, Anti-microb. Agents Chemother 60 (2016) 3455–3461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Schulthess B, Bloes DA, Berger-Bächi B, Opposing roles of σB and σB-controlled SpoVG in the global regulation of esxA in Staphylococcus aureus, BMC Microbiol. 24 (2012) 12–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Zhu Q, Liu B, Sun B, SpoVG modulates cell aggregation in Staphylococcus aureus by regulating sasC expression and extracellular DNA release, Appl. Environ. Microbiol 86 (2020) e00591, 00520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Huang Q, Zhang Z, Liu Q, Liu F, Liu Y, Zhang J, Wang G, SpoVG is an important regulator of sporulation and affects biofilm formation by regulating Spo0A transcription in Bacillus cereus 0–9, BMC Microbiol. 21 (2021) 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Stevenson B, Seshu J, Regulation of gene and protein expression in the Lyme disease spirochete, in: Adler B (Ed.), Spirochete Biology: the Post Genomic Era, Springer-Nature, Heidelberg, 2018, pp. 83–112. [DOI] [PubMed] [Google Scholar]
- [10].Samuels DS, Lybecker MC, Yang XF, Ouyang Z, Bourret TJ, Boyle WK, Stevenson B, Drecktrah D, Caimano MJ, Gene regulation and transcriptomics, in: Radolf JD, Samuels DS (Eds.), Lyme Disease and Relapsing Fever Spirochetes, Caister Academic Press, London, 2021, pp. 87–129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Stevenson B, Krusenstjerna AC, Castro-Padovani TN, Savage CR, Jutras BL, Saylor TC, The consistent tick-vertebrate infectious cycle of the Lyme disease spirochete enables Borrelia burgdorferi to control protein expression by monitoring its physiological status, J. Bacteriol 204 (2022), e0060621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Radolf JD, Caimano MJ, Stevenson B, Hu LT, Of ticks, mice, and men: understanding the dual-host lifestyle of Lyme disease spirochaetes, Nat. Rev. Microbiol 10 (2012) 87–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Jarmoskaite IA-O, AlSadhan I, Vaidyanathan PP, Herschlag DA-O, How to measure and evaluate binding affinities, Elife (2020) 9. LID - 10.7554/eLife.57264 [doi] LID - e57264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Miller JC, von Lackum K, Babb K, McAlister JD, Stevenson B, Temporal analysis of Borrelia burgdorferi Erp protein expression throughout the mammal-tick infectious cycle, Infect. Immun 71 (2003) 6943–6952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Arnold WK, Savage CR, Brissette CA, Seshu J, Livny J, Stevenson B, RNA-seq of Borrelia burgdorferi in multiple phases of growth reveals insights into the dynamics of gene expression, transcriptome architecture, and noncoding RNAs, PLoS One 11 (2016), e0164165, 10.1371/journal.pone.0164165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pal U, Yang X, Chen M, Bockenstedt LK, Anderson JF, Flavell RA, Norgard MV, Fikrig E, OspC facilitates Borrelia burgdorferi invasion of Ixodes scapularis salivary glands, J. Clin. Investig 113 (2004) 220–230, 10.1172/JCI19894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].He M, Ouyang Z, Troxell B, Xu H, Moh A, Piesman J, Norgard MV, Gomelsky M, Yang XF, Cyclic di-GMP is essential for the survival of the Lyme disease spirochete in ticks, PLoS Pathog. 7 (2011), e1002133, 10.1371/journal.ppat.1002133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Pappas CJ, Iyer R, Petzke MM, Caimano MJ, Radolf JD, Schwartz I, Borrelia burgdorferi requires glycerol for maximum fitness during the tick phase of the enzootic cycle, PLoS Pathog. 7 (2011), e1002102, 10.1371/journal.ppat.1002102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Reuter JS, Mathews DH, RNAstructure: software for RNA secondary structure prediction and analysis, BMC Bioinf. 11 (2010) 129, 10.1186/1471-2105-11-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


