Abstract
Rubella virus nonstructural proteins, translated from input genomic RNA as a p200 polyprotein and subsequently processed into p150 and p90 by an intrinsic papain-like thiol protease, are responsible for virus replication. To examine the effect of p200 processing on virus replication and to study the roles of nonstructural proteins in viral RNA synthesis, we introduced into a rubella virus infectious cDNA clone a panel of mutations that had variable defective effects on p200 processing. The virus yield and viral RNA synthesis of these mutants were examined. Mutations that completely abolished (C1152S and G1301S) or largely abolished (G1301A) cleavage of p200 resulted in noninfectious virus. Mutations that partially impaired cleavage of p200 (R1299A and G1300A) decreased virus replication. An RNase protection assay revealed that all of the mutants synthesized negative-strand RNA as efficiently as the wild type does but produced lower levels of positive-strand RNA. Our results demonstrated that processing of rubella virus nonstructural protein is crucial for virus replication and that uncleaved p200 could function in negative-strand RNA synthesis, whereas the cleavage products p150 and p90 are required for efficient positive-strand RNA synthesis.
Rubella virus (RV), the sole member of the Rubivirus genus of the Togaviridae family (8), contains a single positive-strand RNA genome of 9,762 nucleotides (nt) (23). There are two large open reading frames (ORFs) in the genome. The 5′-proximal nonstructural protein (NSP) ORF (from nt 41 to nt 6388) encodes NSPs involved in viral RNA synthesis, and the 3′-proximal structural protein (SP) ORF (from nt 6512 to nt 9701) encodes SPs required for virus assembly (5, 6, 9, 27). These NSPs are first translated from the input genomic RNA as a 200-kDa polyprotein (p200) which undergoes a single proteolytic cleavage by its own intrinsic protease activity into the N-terminal product with a molecular mass of 150 kDa (p150) and the 90 kDa C-terminal product (p90) (1, 4, 7, 20). The RV NSPs are responsible for RNA replication, which is initiated by the synthesis of a full-length negative strand complementary to the genomic 40S positive-strand RNA. This negative strand then serves as the template for the synthesis of new positive-strand genomic RNA and of a subgenomic RNA which is initiated at an internal site in the negative-strand RNA (9, 27).
The functions of RV NSPs are poorly understood. Most of our knowledge of their roles in viral RNA replication was either derived from sequence analysis or deduced from studies on alphaviruses (reviewed in reference 26), members of the only other genus of the Togaviridae family. Computer-assisted alignment predicted four conserved enzyme motifs in the RV NSP sequence, ordered from the N terminus to the C terminus as methyltransferase, protease, helicase, and RNA-dependent RNA polymerase domains (9, 14). The methyltransferase and protease domains are located on the N and C termini of p150, respectively (11, 24). The helicase and RNA polymerase domains are on the N and C termini of p90, respectively (10, 13). The RV nonstructural protease is a papain-like cysteine protease (11). The region containing the protease domain has been studied in detail ((4, 18a, 20, 29). Its deduced catalytic dyad (C1152 and H1273) and the cleavage site (between G1301 and G1302) were demonstrated by site-directed mutagenesis (4, 20). We have recently mapped the region from V920 to P1296 as necessary for trans-cleavage activity of the RV nonstructural protease and showed that the region from V920 to G1020, although required for trans-cleavage activity, is dispensable for cis-cleavage activity (Liang et al., unpublished data).
By analogy with alphavirus replication, it is believed that RV NSPs, along with host factors, form active replication complexes to synthesize three RV-specific RNA species, a negative-strand genomic RNA, a 40S positive-strand genomic RNA, and a 24S subgenomic RNA (9, 26). However, the components of active replication complexes required for synthesis of distinct viral RNA species have not been characterized, nor have the roles of p200, p150, and p90 in viral RNA synthesis been studied. Effects of NSP processing on virus replication and RNA synthesis have not been examined.
In this study, we examined the roles of RV NSPs in virus replication and the synthesis of positive- and negative-strand genomic RNA and subgenomic RNA. An infectious, full-length cDNA clone of RV RNA (30) permitted the introduction of mutations at the protease catalytic site (C1152S) and around the cleavage site (R1299A, G1300A, G1301A, and G1301S). The constructed NSP cleavage mutations were examined for their effects on NSP processing, virus replication, and viral RNA synthesis. We present evidence that NSP cleavage is essential for virus replication and that the impaired virus replication in the cleavage mutants is due to defective synthesis of positive-strand RNA and not of negative-strand RNA. A hypothesis about the role of NSP cleavage in the synthesis of the three viral RNA species is proposed.
MATERIALS AND METHODS
Cells and viruses.
Vero cells were cultured in Eagle's minimum essential medium (MEM; Gibco BRL) supplemented with 5% fetal calf serum (FCS). BHK-21 cells were grown in MEM containing 10% FCS and 10% tryptose phosphate broth. RV (strain M33) was propagated in Vero cells.
Plasmid construction.
Standard recombinant DNA techniques were used to generate all constructs (19). The full-length infectious cDNA clone in which mutations were created, pBRM33, was based on RV strain M33 (30).
A panel of site-directed mutations was introduced into pBRM33 by PCR-mediated mutagenesis with primers containing the desired nucleotide changes (all of the primer sequences are given in Table 1). Generation of the C1152S and G1301S mutations has been described previously (18a). The full-length cDNA clones containing C1152S and G1301S were named pBRM33(C1152S) and pBRM33(G1301S). To construct the R1299A mutation, fusion PCR (30) was employed with pBRM33 DNA as the template and two pairs of primers, JSY-13 plus YL-16 and YL-15 plus JSY-12. The PCR product containing the R1299A mutation was used to replace the NheI-EcoRV fragment (nt 2803 to 4213) of pBRM33, generating plasmid pBRM33(R1299A). To facilitate mutagenesis, a silent mutation was introduced into pBRM33 to create a new XbaI site by changing CGG to AGA at nt 3935 to 3937. Fusion PCR was employed using pBRM33 DNA as the template and two paired primers, JSY-13 plus YL-14 and YL-13 plus JSY-12. The PCR product was used to replace the NheI-EcoRV fragment (nt 2803 to 4213) of pBRM33, and the resultant construct was named pBRM33-X. To construct the G1300A and G1301A mutations, PCR amplifications were performed using pBRM33-X as the template and mutagenic primers containing the desired mutations: YL-17 for mutation G1300A and YL-18 for mutation G1301A. The PCR products were used to replace the corresponding XbaI-EcoRV (nt 3933 to 4213) fragment of pBRM33-X. The constructs were named pBRM33(G1300A) and pBRM33(G1301A), respectively.
TABLE 1.
PCR primers used in the generation of mutants
| Primer | Polaritya | Nucleotide positionb | Amino acid change | Sequencec |
|---|---|---|---|---|
| PCR | ||||
| JSY-13 | + | 2770–2790 | 5′ GCTGCTCGAGCGCGCCTACCG 3′ | |
| JSY-12 | − | 4220–4240 | 5′ GTAGGTGGCGGCGTTCTTGAT 3′ | |
| Mutagenic | ||||
| JSY-2 | + | 3482–3508 | C1152S | 5′ GACCCAAACACCAGCTGGCTCCGCGCC 3′ |
| JSY-3 | − | 3482–3508 | C1152S | 5′ GGCGCGGAGCCAGCTGGTGTTTGGGTC 3′ |
| JSY-4 | + | 3929–3955 | G1301S | 5′ CTGTCTCGGGGCAGCGGCACTTGTGCC 3′ |
| JSY-5 | − | 3929–3955 | G1301S | 5′ GGCACAAGTGCCGCTGCCCCGAGACAG 3′ |
| YL-15 | + | 3920–3952 | R1299A | 5′ GCGGTCCCCCTGTCTGCAGGCGGCGGCACTTGT 3′ |
| YL-16 | − | 3920–3952 | R1299A | 5′ ACAAGTGCCGCCGCCTGCAGACAGGGGGACCGC 3′ |
| YL-13 | + | 3920–3952 | 5′ GCGGTCCCCCTGTCTAGAGGCGGCGGCACTTGT 3′ | |
| YL-14 | − | 3920–3952 | 5′ ACAAGTGCCGCCGCCTCTAGACAGGGGGACCGC 3′ | |
| YL-17 | + | 3941–3955 | G1300A | 5′ CTGTCTAGAGCAGGCGGCACTTGTGCC 3′ |
| YL-18 | + | 3948–3958 | G1301A | 5′ CTGTCTAGAGGCGCAGGCACTTGTGCCGCC 3′ |
Polarity of primers on M33 genome. +, forward; −, backward.
Positions of primers on M33 genome.
Sequences of primers. The mutated nucleotides are in boldface. The restriction enzyme XbaI sites are underlined.
All PCRs were carried out in 25 cycles of 98°C for 30 s, 50°C for 2 min, and 70°C for 2 min using either 2.5 U of ExTaq temperature-stable DNA polymerase (TaKaRa LA PCR kit) or Native Pfu DNA polymerase (Strategene) in buffers provided by the manufacturers and supplemented with 10% dimethyl sulfoxide. The resulting PCR fragments were purified with a QIAquick Spin PCR purification kit (QIAgen).
In vitro transcription.
The cDNA clones were linearized at the unique HindIII site and transcribed with SP6 RNA polymerase (Promega) in the presence of a cap analog, 7mG5′ppp5′G (Promega), using the protocol recommended by the manufacturer.
In vitro translation and determination of the NSP processing ratio.
In vitro translation was performed in a system described by Liang et al. (18a)). Briefly, the 50-μl reaction mixtures containing nuclease-treated rabbit reticulocyte lysate (Promega), an amino acid mixture minus methionine, RNasin (RNase inhibitor), and RNA transcripts in the presence of [35S]methionine (NEN) at 400 μCi/ml were incubated at 30°C for the indicated time. Radiolabeled proteins were visualized by fluorescence autoradiography after sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis analysis. The processing of wild-type (WT) or mutant NSPs was studied by time course analysis of in vitro translation reactions. The processing ratio of each construct was calculated as the percentage of cleavage products in the total proteins and plotted against the incubation time as described by Liang et al. (18a).
RNA transfection.
About 20 μg of in vitro-transcribed RNA (in a 20-μl transcription reaction) and 10 μg of Lipofectin (Gibco BRL) were suspended in 0.5 ml of FCS-free MEM and incubated for 20 min at room temperature. The formed Lipofectin-RNA mixtures were applied to a Vero cell monolayer (in a 35-mm-diameter dish) which had been washed with FCS-free MEM twice. After incubation for 2 to 3 h at 37°C, the mixtures were removed and replaced with culture medium. At day 6 posttransfection, culture fluids were harvested and the virus released into the culture medium was quantitated by plaque assay on Vero cells.
BHK-21 cells were transfected by electroporation as described previously (30). BHK-21 cells were harvested by trypsin treatment and washed twice with cold phosphate-buffered saline (without Ca2+ and Mg2+) and resuspended at a concentration of 107/ml. A 0.5-ml sample of the cell suspension was mixed with about 20 μg of in vitro-transcribed RNA (in a 20-μl transcription reaction mixture) and transferred to a 2-mm-diameter cuvette. Electroporation utilized two consecutive 1.5-kV, 250-μF pulses with a Gene-Pulser (Bio-Rad). The cells were diluted with culture medium and distributed among four 35-mm-diameter dishes. Culture fluids were collected at 48 h postelectroporation, and the released virus particles were quantitated by plaque assay on Vero cells.
Plaque assay and virus growth analysis.
For viral plaque assay, Vero cells infected by a serially diluted virus stock were overlaid with 0.5% agarose in MEM containing 5% FCS, incubated at 35°C for 6 or 8 days, and stained with 5% neutral red diluted in MEM supplemented with 5% FCS.
For virus growth rate analysis, Vero cell monolayers (35-mm-diameter dish) were transfected with WT or mutant RNA mediated by Lipofectin as described above. After removal of the RNA-Lipofectin mixtures, the cells were washed with PBS, overlaid with fresh medium, and incubated at 37°C. The culture medium was harvested and replaced with fresh medium every 24 h. The released virus was quantitated by plaque assay.
RPA.
An RNase protection assay (RPA) was employed to analyze the synthesis of virus-specific RNAs during virus replication. For synthesis of a plus or minus polarity RNA probe in vitro, a DNA fragment (nt 6323 to 6623) of pBRM33, representing the region covering the subgenomic RNA initiation site (nt 6436), was separately cloned into vector pSPT18 or pSPT19 (Pharmacia Biotech) at the EcoRI and XbaI sites to make construct pSPT18-pb or pSPT19-pb. A 328-bp minus polarity RNA probe (pb18), synthesized with SP6 RNA polymerase from EcoRI-linearized pSPT18-pb, can protect 301-nt positive-strand genomic RNA and 188-nt subgenomic RNA. A 328-nt plus polarity RNA probe (pb19), synthesized with SP6 RNA polymerase from HindIII-linearized pSPT19-bp, can protect 301-nt RV negative-strand genomic RNA. The 35S-labeled RNA probe was synthesized with SP6 polymerase in a 20-μl in vitro transcription reaction mixture containing buffer (provided by the manufacturer); RNasin; 0.5 mM each ATP, GTP, and UTP; 12.5 μM CTP; and [α-35S]CTP (NEN) at 2.5 mCi/ml. After incubation for 90 min at 37°C, 2 μl of DNase I (7,500 U/ml; Pharmacia Biotech) was added to the reaction mixture and it was incubated for a further 15 min. The probe was precipitated with ethanol after phenol-chloroform extraction and resuspended in H2O.
For RNA analysis, total cytoplasmic RNAs were extracted with TRIzol reagent (GIBCO BRL) at the times posttransfection indicated in Results. Negative-strand RNA was analyzed by a two-cycle RPA essentially as described by Novak and Kirkegaard (22). Approximately 20 μg of cytoplasmic RNA was incubated with 20 ng of RNA probe pb19 (approximately 1011 molecules) in 30 μl of hybridization buffer (40 mM PIPES, 400 mM NaCl, 1 mM EDTA, 80% deionized formamide; pH 6.4) overnight at 55°C. RNase digestion was for 60 min at 30°C in an RNase mixture (300 mM sodium acetate, 10 mM Tris-HCl [pH 7.5]; 5 mM EDTA; RNase A at 10 μg/ml, RNase T1 at 70 U/ml). The reaction mixture was treated with SDS-proteinase K for 15 min at 37°C, extracted with phenol-chloroform, and ethanol precipitated with tRNA at 5 μg/ml. Samples were resuspended in 30 μl of hybridization buffer, and 106 cpm of 35S-labeled RNA probe pb19 was added. The samples were denatured at 85°C for 5 min, hybridized overnight at 55°C, and subjected to RNase treatment as described above. The digestion products were analyzed on a 5% polyacrylamide–urea gel, which was fixed in 7% acetic acid, infiltrated with Enhancer (DuPont), dried, and exposed to X-ray film. Positive-strand genomic and subgenomic RNAs were analyzed by a single-round RPA. A 2-μg sample of total cytoplasmic RNAs was hybridized with 106 cpm of 35S-labeled RNA probe pb18 overnight at 55°C. The samples were treated as described above.
Image analysis.
Image analysis was performed on a personal computer using the Scion Image program for Windows (Beta 3b) (http://www.scioncorp.com/frames/fr_download_now.htm), the personal computer version of the public domain NIH Image program (developed at the U.S. National Institutes of Health and available on the internet at http://rsb.info.nih.gov/nih-image/).
RESULTS
Construction of mutations.
In order to study the effects of RV NSP processing on virus replication, the RV genome was altered by mutations that affect only NSP processing while avoiding any other possible structural or functional alterations in the NSP or in the viral genome. A panel of site-directed mutations was generated by PCR mutagenesis. These included catalytic-site mutation C1152S and cleavage site mutations G1301S, R1299A, G1300A, and G1301A. Most of the introduced mutations are conservative alterations, such as C to S and G to A, and thus are unlikely to affect the overall structure of the protein or functions of NSP other than proteolytic processing. To facilitate the process of mutagenesis for construction of the G1300A and G1301A mutations, a silent mutation was introduced into the RV infectious cDNA clone pBRM33 (30) to create a new XbaI site at nt 3935. The resultant cDNA clone was named pBRM33-X. In terms of virus growth, plaque size, and specific infectivity, pBRM33-X was indistinguishable from pBRM33 (data not shown). Amplified PCR fragments containing the desired mutations were reintroduced into pBRM33 or pBRM33-X, and the respective cDNA clones were named after their mutations: pBRM33(C1152S), pBRM33 (G1301S), pBRM33(R1299A), pBRM33(G1300A), and pBRM33(G1301A). The plasmid constructs encoding these mutations are listed in Table 2.
TABLE 2.
Mutations created at the catalytic site and around the cleavage site
| Mutation | Catalytic site Around cleavage sitea |
|---|---|
| ↓ | |
| None (WT) | C1152R1299-G1300-G1301-G1302 |
| C1152S | S |
| R1299A | A |
| G1300A | A |
| G1301A | A |
| G1301S | S |
Cleavage occurs after G1301, as indicated by the vertical arrow.
Effects of mutations on NSP processing.
To determine whether the p200 polyprotein itself can function in RNA replication, we constructed a panel of cleavage-defective mutations. The effect of mutations on NSP processing was determined using time course analysis of in vitro translation reactions programmed with full-length RNA transcripts from cDNA clones as described by Liang et al. (18a). The extent of NSP processing differed greatly among the WT and mutant RNAs (Fig. 1). To compare their cleavage efficiencies more precisely, the processing ratio, calculated as the percentage of cleavage products in the total proteins, was assessed for each mutant and plotted against the incubation time (Fig. 2). From both gel analysis and the calculated processing ratio, WT NSP processing (Fig. 1A) was almost complete at 3 h of incubation (Fig. 2). The NSP processing of the G1300A mutant was slightly delayed and decreased (Fig. 1B and 2); its processing ratio at 3 h was 75% of the WT level (Fig. 2). Mutation R1299A substantially impaired NSP processing, since the cleavage products were detected only after 3 h of incubation (Fig. 1C) and the cleavage ratio at 3 h was approximately 20% of the WT level (Fig. 2). Mutation G1301A resulted in minimally detectable cleavage of p200, with a minute amount of p90 detected after a 3-h incubation time (Fig. 1D). Mutations C1152S and G1301S abolished NSP processing completely (Fig. 1E and F). Thus, our generated mutations either abolished (C1152S and G1301S) or blocked (R1299A, G1300A, and G1301A) NSP processing to various degrees.
FIG. 1.
Effects of mutations on NSP processing in an in vitro translation system. Full-length RV RNAs containing site-directed mutations were generated from corresponding HindIII-linearized cDNA templates with SP6 RNA polymerase. In vitro translation reaction mixtures containing rabbit reticulocyte lysates were prepared at 30°C. Aliquots were removed at the indicated times, and the protein products were resolved by SDS-polyacrylamide gel electrophoresis. Panels: A, WT NSP; B, G1300A mutant NSP; C, R1299A mutant NSP; D, G1301A mutant NSP; E, C1152S mutant NSP; F, G1301S mutant NSP. Positions of molecular mass markers and protein products are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
FIG. 2.
Comparison of the processing ratios of WT and mutant NSPs. Protein bands of p200 and the p150 and p90 cleavage products were each quantitated at the indicated times using SCImage software as described in Materials and Methods. The cleavage ratio was calculated as the percentage of cleaved products in the total proteins and plotted against incubation time. Symbols: ⧫, WT NSP; ■, G1300A mutant NSP; ▴, R1299A mutant NSP; ○, G1301A mutant NSP; ●, C1152S and G1301S mutant NSPs.
Effects of mutations on virus growth.
In order to examine the effects of mutations on RV replication, WT or mutant RNA, transcribed from respective full-length cDNA clones, was used to transfect either Vero or BHK-21 cells. The transfected Vero cells were incubated for 5 days and assayed for infectious virus released into the culture medium (Table 3). Infectious virus particles could be harvested from Vero cells transfected by the WT RNA and those with the G1300A and R1299A mutations, yielding virus titers of 3.4 × 106, 1.2 × 106, and 1.0 × 103 PFU/ml, respectively. In contrast, no plaques were detected in the medium containing RNAs with the mutations G1301A, G1301S, and C1152S, indicating that they are noninfectious (Table 3). Transfected Vero cells were also analyzed for the production of RV-specific SPs by immunoprecipitation with human anti-RV serum. RV SPs were readily detectable for the WT and the G1300A mutant but in a substantially lower quantity for the R1299A mutant. No RV SPs could be detected from cells transfected by the G1301A, G1301S, and C1152S RNAs (data not shown), indicating that they are defective in replication. To confirm that the different amounts of virus produced by the WT and mutant RNAs are not dependent on the host cells used, we also transfected BHK-21 cells with RNA transcripts by electroporation. At 48 h postelectroporation, the culture medium was collected and the released virus particles were quantitated. Consistent with our results obtained with Vero cells, infectious virus particles were only detected from BHK-21 cells transfected with the WT, G1300A, and R1299A RNAs. The virus titers were 1.5 × 107, 6 × 106, and 5 × 103 PFU/ml, respectively. Again, no infectious virus could be harvested from BHK-21 cells transfected with transcripts of G1301A, C1152S, or G1301S mutant RNA (Table 3). RNA transfection of BHK-21 cells by electroporation resulted in higher virus titers than that of Vero cells using Lipofectin, most likely due to the higher transfection efficiency of electroporation. The results obtained with the two cell types are comparable. The amount of virus produced from each infectious RNA (WT, G1300A, or R1299A) varies with its NSP-processing efficiency. In both cases, WT RNA, having the most efficient NSP processing, produced the highest virus titer. The G1300A mutant RNA, with 75% of the WT level of NSP processing, produced viruses at 30 to 40% of the WT level. The R1299A mutant RNA, with NSP processing at 20% of the WT level, released viruses at a level 2 × 103 to 3 × 103-fold lower than that of the WT. The G1301A mutant RNA, with minimally detectable NSP processing in vitro (processing ratio of less than 10% at 5 h of incubation), and the C1152S and G1301S mutant RNAs, abolishing NSP processing completely, released no infectious virus particles in either Vero or BHK-21 cells and thus are effectively lethal. In addition to the differences in virus titer, the WT and mutant RNAs also have different plaque phenotypes. The WT and G1300A mutant RNAs produced large, clear plaques at day 6 postinfection, while the R1299A RNA resulted in tiny, unclear plaques only after day 8 postinfection (data not shown).
TABLE 3.
Effects of RV NSP cleavage mutants on virus replication
| RV | Virus titera (PFU/ml) at:
|
Plaque phenotype on Vero cellsb | |
|---|---|---|---|
| Day 5 posttransfection of Vero cells | 48 h posttransfection of BHK-21 cells | ||
| WT | 3.4 × 106 | 1.5 × 107 | Big, clear, 6 dpi |
| G1300A mutant | 1.2 × 106 | 6 × 106 | Big, clear, 6 dpi |
| R1299A mutant | 1 × 103 | 5 × 103 | Tiny, unclear, 8 dpi |
| G1301A mutant | 0 | 0 | None |
| G1301S mutant | 0 | 0 | None |
| C1152S mutant | 0 | 0 | None |
The virus titers, determined by plaque assay on Vero cells, are the means of at least two independent experiments. Culture medium was tested without dilution and at 1:10 and 1:100 dilutions for the plaque assay. 0, no plaque formation was observed.
Vero cells infected by WT or mutant virus initiated by RNA transfection were overlaid with agarose medium and incubated for 6 days (for the WT and G1300A mutant) or 8 days (for the R1299A mutant) before being stained with neutral red. dpi, days postinfection.
To further analyze the influences of NSP cleavage on virus replication, growth rates were determined for the WT and infectious mutant (R1299A and G1300A) RNAs. Vero cells were transfected with the respective full-length RNAs mediated by Lipofectin. Culture medium was harvested every 24 h and replaced with fresh medium. Virus titers in the culture medium were quantitated by plaque assay on Vero cells and are shown in Fig. 3. For the WT, the amount of virus produced was about 3 × 103 PFU/ml at day 1 and reached a peak of 5 × 106 PFU/ml at day 4. The G1300A mutant had growth kinetics similar to those of the WT but yielded a 10-fold lower amount of released virus (2 × 102 PFU/ml) at day 1 and a 3-fold lower amount (1.6 × 106 PFU/ml) at day 4. The R1299A mutant virus was not detectable until after day 3, and its titer at day 5 was 2 × 103-fold lower than that of the WT. The failure to detect R1299A virus plaques before day 3 posttransfection may be due to their small size at early stages of infection. Our results demonstrate that NSP cleavage plays an important role in virus replication.
FIG. 3.
Growth curves of the WT and G1300A and R1299A mutant viruses. Vero cells in 35-mm-diameter dishes were transfected with WT or mutant RNAs mediated by Lipofectin for 2 h at 37°C. The cells were overlaid with culture medium after removal of the Lipofectin-RNA mixtures. The culture medium was changed every 24 h, and the virus particles released into the medium were quantitated by plaque assay. The results shown are the means of at least two independent experiments. Symbols: ■, WT RV; ▴, G1300A mutant; □, R1299A mutant.
Effects of mutations on viral RNA synthesis.
The reduction in virus yield due to defects in NSP cleavage presumably occurred at the level of viral RNA synthesis. To determine at which step(s) RNA synthesis is impaired in the mutant viruses, we examined the synthesis of three viral RNA species in the WT and NSP cleavage mutant viruses at the early stage of virus replication using an RPA.
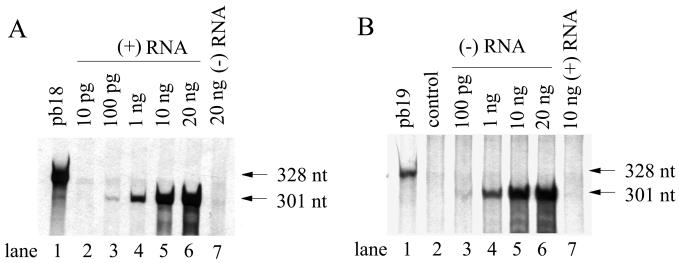
To evaluate the sensitivity of the RPA for detection of positive-strand RV RNA, various amounts of positive-strand RV RNA, transcribed in vitro from pBRM33, were subjected to an RPA using 106 cpm of 35S-labeled probe pb18, which contains 301 nt of the RV sequence and 27 nt of the vector sequence. The negative-polarity RV RNA, transcribed from a cDNA clone encoding an RV genome of reverse polarity, was also used as a negative control. As shown in Fig. 4A, a protected band of 301 nt was present in reaction mixtures containing 100 pg (lane 3), 1 ng (lane 4), 10 ng (lane 5), and 20 ng (lane 6) of positive-strand RNA but was absent in reaction mixtures containing 10 pg of positive-strand RNA (lane 2) and 20 ng of negative-strand genomic RNA (lane 7). These data indicate that this assay is strand specific and sensitive enough to detect at least 100 pg of positive-strand RV RNA (approximately 107 molecules). Furthermore, quantitative analysis of the protected probe suggested that the signal was proportional to the amount of positive-strand RNA used.
FIG. 4.
Sensitivity of RPA for detection of both positive- and negative-strand RV RNAs. RPA reactions were carried out as described in Materials and Methods. The full-length RV RNAs of either positive or negative polarity used in this experiment were transcribed in vitro from the respective RV cDNA clone, pBRM33 or pBRNM33, encoding a full-length RV genome downstream of the SP6 RNA polymerase promoter in either the forward or the backward orientation. (A) Standard RNase protection reactions were performed on 10 pg, 100 pg, 1 ng, 10 ng, and 20 ng (lanes 2 to 6) of positive-strand RV RNAs in the presence of 106 cpm of 35S-labeled probe pb18. A reaction mixture containing 20 ng of negative-strand RV RNA (lane 7) was also included as a negative control (lane 7). The products of RPA reactions along with 2 × 103 cpm of pb18 (lane 1) were resolved on a 5% polyacrylamide–7 M urea gel, which was treated with Enhancer (DuPont), dried, and exposed to X-ray film. (B) Two-cycle RNase protection reactions were performed with various amounts of negative-strand RV RNA. Negative-strand RV RNA was hybridized with 10 ng of transcribed probe pb19 and subjected to the first-cycle RPA reaction. The products of the first-cycle RPA reaction were hybridized with 106 cpm of 35S-labeled pb19 and subjected to the second-round RPA reaction. To examine the strand specificity of probe pb19, 10 ng of positive-strand RV RNA (lane 7) was analyzed in parallel. The control reaction mixture contained no RV RNA (lane 2). Lanes 3 to 6 represent reaction mixtures containing 100 pg, 1 ng, 10 ng, and 20 ng of negative-strand RV RNA, respectively. Lane 1, pb19. The autoradiographs were exposed for 1 day. The positions of the 328- and 301-nt bands are indicated. Images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
In virus-infected cells, RV negative-strand genomic RNA exists mostly as a double-stranded intermediate form with positive-strand RNA present in large molar excess. To prevent interference between the probe and negative-strand RNA by the large molar excess of positive-strand RNA, a two-cycle RPA was employed to detect negative-strand genomic RNA. To determine the sensitivity of the two-cycle RPA for negative-strand RNA, various amounts of negative-strand genomic RNA, transcribed in vitro from a cDNA clone encoding an RV genome of reverse polarity, were hybridized with 20 ng of unlabeled RNA probe pb19 (complementary to the sequence of pb18) in the first-cycle RPA. The products were subsequently hybridized with 106 cpm of 35S-labeled pb19 and subjected to a second cycle (Fig. 4B). The probe pb19 is negative strand specific, since no signal band was observed in the reaction mixture containing 10 ng of positive-strand genomic RNA (lane 7). The 301-nt signal band was apparent in a reaction mixture containing 1 ng (lane 4), 10 ng (lane 5), or 20 ng (lane 6) of negative-strand genomic RNA. A longer exposure (3 days) also detected the existence of this band in a reaction mixture containing 100 pg of negative-strand RNA (lane 3). Therefore, this two-cycle RPA is sensitive enough to detect more than 100 pg of negative-strand genomic RNA (approximately 107 RNA molecules). The signal intensity was proportional to the amount of negative-strand RNA.
We then examined viral RNA synthesis in BHK-21 cells transfected by electroporation with WT or mutant RNA transcripts. We consider virus-infected cells a less ideal system for the analysis of viral RNA synthesis because (i) revertants or second-site mutations may exist in virus stocks, (ii) the system might be affected by the early steps of virus entry prior to viral RNA synthesis (e.g., virus entry and nucleocapsid uncoating), (iii) the percentage of cells initially infected by RV is quite low (10 to 20%) (9, 12), and (iv) studies of noninfectious mutants are impossible. In contrast, electroporation of viral RNA transcripts into BHK-21 cells provides an advantageous system for the analysis of RNA synthesis at an early stage of virus replication. This system bypasses the steps of virus entry and nucleocapsid disassembly; has a high efficiency of transfection, allowing detection of low levels of negative-strand RNA; and allows studies of noninfectious mutants.
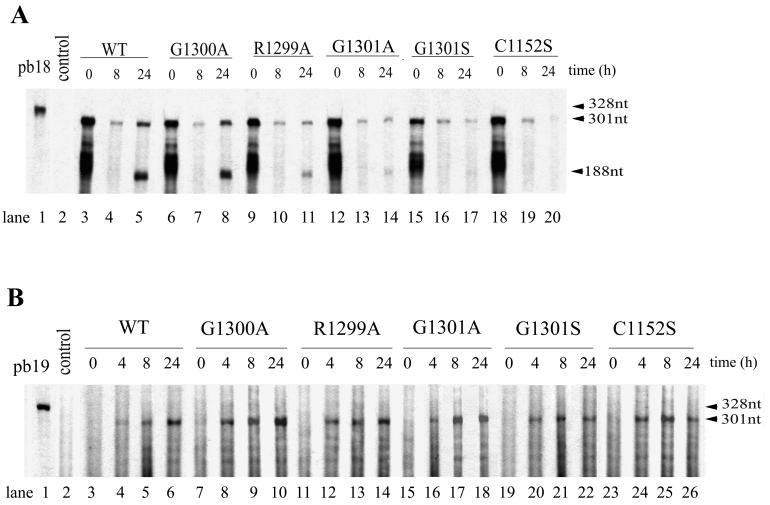
To determine RNA synthesis in WT and mutant viruses, the respective viral RNAs transcribed in vitro were used to transfect BHK-21 cells by electroporation. At the indicated times, total cellular RNA was extracted and subjected to an RPA. A 2-μg sample of RNA was used for positive-strand RNA detection, and 20 μg was used for negative-strand RNA detection. At 0 h postelectroporation, the 301-nt protected fragment representing the positive-strand genomic RNA was apparent for all constructs (Fig. 5A, lanes 3, 6, 9, 12, 15, and 18), representing the input genomic RNA transfected into cells. By 8 h postelectroporation, the intensity of the 301-nt band decreased to a low level (Fig. 5A, lanes 4, 7, 10, 13, 16, and 19), suggesting that the input genomic RNA had mostly been degraded at that time. At 24 h postelectroporation, accumulation of both the protected 301-nt fragment and the 188-nt fragment (representing subgenomic RNA) was apparent for the WT and the G1300A mutant RNA (Fig. 5A, lanes 5 and 8). Much less of these bands was found with the R1299A mutant RNA (Fig. 5A, lane 11). The G1301S and C1152S mutant RNAs exhibited decreased levels of the 301-nt band (compared with the amounts detected at 8 h; Fig. 5A, lanes 17 and 20). Subgenomic RNA represented by the 188-nt fragment was scarcely detectable for the two mutants. Thus, they show no evidence of production of positive-strand RNA. The G1301A mutant showed the presence of low levels of the 301-nt band with little increase at 24 h over the level found at 8 h. By 24 h, a trace of the 188-nt fragment was detected (Fig. 5A, lane 14), indicating some slight synthesis of positive-strand RNA. These results confirm that the more-infectious constructs produce more positive-strand RNAs and the noninfectious ones produce little (G1301A) or no (G1301S and C1152S) positive strands. Detailed quantitation of the amount of positive-strand RNA is presented below.
FIG. 5.
RNA analysis of WT and mutant constructs. (A) Positive-strand RNA analysis. BHK-21 cells were electroporated with RNAs transcribed in vitro from a cDNA clone containing the WT sequence (lanes 3 to 5) or the G1300A (lanes 6 to 8), R1299A (lanes 9 to 11), G1301A (lanes 12 to 14), G1301S (lanes 15 to 17), or C1152S (lanes 18 to 20) mutant sequence. At indicated times postelectroporation (0, 8, and 24 h), total cytoplasmic RNAs were extracted using TRIzol reagent and subjected to an RPA using 35S-labeled probe pb18, which was loaded in lane 1. BHK-21 cells electroporated with no RV RNA served as a control (lane 2). The autoradiograph was exposed for 1 day. (B) Negative-strand genomic RNA analysis. Total cytoplasmic RNAs extracted at 0, 4, 8, and 24 h postelectroporation of BHK-21 cells transfected with the WT virus (lanes 3 to 6) or the G13000A (lanes 7 to 10), R1299A (lanes 11 to 14), G1301A (lanes 15 to 18), G1301S (lanes 19 to 22), or C1152S (lanes 23 to 26) mutant virus, respectively, were subjected to a two-cycle RPA as described in Materials and Methods. The 35S-labeled probe pb19 was loaded in lane 1. BHK-21 cells electroporated with no RV RNA served as a control (lane 2). The autoradiograph was exposed for 2 days. The positions of the 328, 301, and 188-nt bands are indicated. The images were scanned using a UMAX Astra 1220U scanner with Adobe Photoshop 5.0 software.
Figure 5B shows the levels of negative-strand RNA produced by the constructs. At 0 h postelectroporation, no protected negative-strand RNA fragment (301 nt) was observed for any construct (Fig. 5B, lanes 3, 7, 11, 15, 19, and 23). By 4 h postelectroporation, all constructs had produced detectable negative-strand RNA (Fig. 5B, lanes 4, 8, 12, 16, 20, and 24), with the WT showing the lowest level. Negative-strand RNA continued to accumulate at 8 h in all of the constructs (Fig. 5B, lanes 5, 9, 13, 17, 21, and 25). At 24 h, the amount of negative-strand RNA had further increased in the WT and the G1300A and R1299A mutants (Fig. 5B, lanes 6, 10, and 14) but had not increased (or even decreased) in the G1301A, G1301S, and C1152S mutants (Fig. 5B, lanes 18, 22, and 26). The last three mutants are those that showed little or no positive-strand RNA synthesis (see above), suggesting that the absence of positive-strand RNA accumulation prevents continued synthesis of negative-strand RNA or allows its degradation. The increased amount of negative-strand RNA in the WT, G1300A, and R1299A viruses at 24 h may mean that the presence of newly synthesized positive-strand RNA allows negative-strand RNA to accumulate further. Whether this occurs in new cells from reinfection or in the same cells is unclear. In general, all of the constructs, including those without synthesis of positive-strand RNA, produced negative-strand RNA at early stages of infection.
To compare the efficiencies of RNA production between the WT and mutant viruses more precisely, we assessed the amounts of negative-strand RNA, positive-strand genomic RNA, and subgenomic RNA for all of the constructs and normalized the results for the mutants against those for the WT. The molar ratio of subgenomic RNA to positive-strand genomic RNA was also calculated for the WT and the G1300A and R1299A mutant viruses. The results are summarized in Table 4. Negative-strand RNA was compared at 4 and 8 h, because these are the points when synthesis of negative-strand RNA was not complicated by new synthesis of positive-strand RNA or by reinfection. In general, the mutants produced significantly more negative-strand RNA than did the WT at 4 h and almost the same amount as the WT at 8 h. We also compared the amount of negative-strand RNA between the WT and G1301S viruses every 2 h after electroporation. The results confirmed the above observations, with the G1301S mutant producing substantially more negative-strand RNA than the WT at 2 and 4 h. They produced similar levels of negative-strand RNA by 8 h (data not shown). How the WT differs from the mutants at early stages is unclear. One explanation might be that more efficient processing of WT NSP decreases the available negative-strand RNA replication complex. However, the widely varied NSP cleavage ratios among the mutants (0 to 75% cleavage ratios) did not result in correlated production of negative-strand RNA at 4 h. A likely reason is that a limiting host factor(s) might define the number of replication complexes formed, so further increased amounts of p200 had no effect on the levels of replication complexes available. We propose that at 4 h, WT p200 has not saturated the limited host factor(s) in the formation of the negative-strand RNA replication complex. Increased amounts of p200 in G1300A, due to delayed and less-efficient NSP cleavage, result in higher production of negative strands. Yet further increased amounts of p200 in other mutants would not further increase negative-strand RNA synthesis because such a host factor(s) is limited. At 8 h, all of the constructs produced similar amount of negative strands, suggesting that they contained comparable negative-strand RNA replication complexes, possibly limited by a host factor(s).
TABLE 4.
Comparisons of the relative amounts of RNAs produced by WT and mutant RVsa
| Construct | Negative-strand RNAb
|
Positive-strand RNAc
|
SG/G molar ratiod | ||
|---|---|---|---|---|---|
| 4 h | 8 h | Genomic | Subgenomic | ||
| WT | 1 | 1 | 1 | 1 | 4.70 ± 1.00 |
| G1300A | 2.54 ± 0.37 | 1.13 ± 0.40 | 0.88 ± 0.13 | 0.82 ± 0.08 | 4.45 ± 1.20 |
| R1299A | 2.05 ± 0.50 | 1.41 ± 0.35 | 0.19 ± 0.13 | 0.24 ± 0.10 | 5.15 ± 1.06 |
| G1301A | 2.21 ± 1.11 | 1.39 ± 0.37 | NAe | NA | NA |
| G1301S | 2.36 ± 0.52 | 0.97 ± 0.11 | NA | NA | NA |
| C1152S | 1.90 ± 0.57 | 1.43 ± 0.33 | NA | NA | NA |
The values shown are the results of at least two independent experiments.
The amount of negative-strand RNA at the respective time point (4 or 8 h postelectroporation) was assessed as described in Materials and Methods and normalized against that of WT (value of 1.0).
Positive-strand RNA, either genomic or subgenomic, produced at 24 h postelectroporation was assessed and normalized against that of the WT (value of 1.0).
The SG/G molar ratio is the calculated molar ratio of subgenomic RNA to genomic RNA.
NA, data not available.
It was assumed that the G1301S and C1152S mutants synthesized no positive-strand RNA. The average density of the 301-nt band for the two at 24 h was taken to represent the level of input genomic RNA present in other mutants and was subtracted from their measured values. Although the G1301A mutant produced a low level of subgenomic RNA and possibly some genomic RNA as well (Fig. 5A), these were not subjected to quantitation or molar-ratio calculation because of the likely presence of background genomic RNA. It is evident that all of the mutants produced less positive-strand RNA than did the WT (Table 4). Positive-strand RNA levels in mutants are in accord with their respective NSP-processing efficiencies and virus yields. The more NSP cleavage is impaired, the less positive-strand RNA is produced. The slightly impaired G1300A mutant produced 88% of the level of genomic RNA and 82% of the subgenomic RNA produced by the WT. The substantially impaired R1299A mutant transcribed 19% of genomic RNA and 24% of subgenomic RNA produced by the WT. Production of the genomic and subgenomic positive-strand RNAs was almost equally lowered. The molar subgenomic/genomic RNA ratios, ranging from 4.5 to 5.2 for the mutants, did not vary dramatically from that of the WT. These results indicate that NSP cleavage may not be the cause for the differential production of subgenomic and genomic RNAs in virus-infected cells.
DISCUSSION
Cleavage of p200 is essential for virus replication.
In RNA viruses, nonstructural-polyprotein processing is temporally regulated such that the ratio of polyprotein to cleavage products changes over the course of infection. Nonstructural-polyprotein processing has been demonstrated to be essential for virus replication in alphavirus (25), the flavivirus yellow fever virus (2, 3, 21), and bovine viral diarrhea virus (28) by examining the effects of mutations that inactivate the protease function or block the cleavage site with the use of infectious cDNA clones. In this study, we generated a panel of cleavage mutants and demonstrated that the cleavage of nonstructural polyprotein p200 is essential for RV replication. The effects on RV replication were found to correlate with the efficiency of p200 polyprotein processing. Mutations that completely (G1301S and C1152S) or nearly (G1301A) abolished p200 cleavage shut down virus replication (Table 3). A mutation with a minor influence on NSP processing (G1300A) produced infectious virus with a growth rate decreased by 3- to 10-fold. A mutation with a profound effect on NSP cleavage (R1299A) produced a viable virus but with a growth rate lowered by 2,000- to 3,000-fold. Examination of RNA synthesis suggested that defective production of positive-strand RNA in the mutants, including both positive-strand genomic RNA and subgenomic RNA, may be the cause for reduced virus production (Table 4). More-infectious constructs produced more positive-strand RNAs than less-infectious ones, with noninfectious constructs producing little or none. However, given its 20% p200-processing level and 20% positive-strand genomic RNA, the R1299A mutant produced an unexpectedly lower virus yield (103-fold lower than that of the WT). Although alternative explanations may exist, a likely reason is reinfection. The positive-strand RNA level was compared at 24 h postelectroporation, whereas virus yields were compared at 48 h (BHK-21 cells) or 5 days (Vero cells) posttransfection. It also seems that productive release of infectious virus particles may require a threshold level of positive-strand RNA synthesis. For example, the G1301A mutant is noninfectious by plaque assay although a minimal level of positive-strand RNA could be detected by RPA. We believe that these mutations act by impairing p200 cleavage rather than by directly affecting the activity of the RNA replicase. First, the introduced mutations, except for R1299A, are conserved substitutions. Second, the mutations at cleavage sites are unlikely to be important for replicase activity. Third, for all of the mutants, there is a good correlation among p200 cleavage efficiency, virus production, and viral RNA synthesis.
Uncleaved polyprotein p200 can produce negative-strand RNA, whereas cleavage products from p200 are required for efficient positive-strand RNA synthesis.
We have shown that all of the mutants, including the noninfectious, cleavage-defective G1301S and C1152S mutants, accumulated negative-strand RNA as efficiently as the WT did at 8 h (Fig. 5B and Table 4), suggesting that uncleaved p200 is sufficient to produce negative-strand RNA from the input genomic RNA. Interestingly, mutants even produced more negative-strand RNA at 4 h than did the WT, providing further evidence for the role of p200 in negative-strand RNA synthesis. However, the amount of negative-strand RNA did not increase proportionally to the amount of p200 among mutants, suggesting that a limiting host factor(s) may also play a role in regulating the number of replication complexes for negative-strand RNA synthesis.
The capacity to synthesize positive-strand RNA differed greatly between the WT and the mutants. All of the mutants produced lower levels of positive-strand RNA, both genomic and subgenomic RNAs, than the WT. Mutants more defective in cleaving p200 produced less positive-strand RNA (Fig. 5A and Table 4). The cleavage-defective G1301S and C1152S mutants showed accumulation of positive-strand RNA barely detected by RPA (Fig. 5A, lane 8). This suggests that cleavage products from p200 (i.e., p150 and p90) are responsible for efficient synthesis of both positive-strand genomic RNA and subgenomic RNA. In view of the limited sensitivity of the RPA used in this study, the possibility of inefficient synthesis of positive-strand RNA by uncleaved p200 cannot be ruled out. For the two infectious (G1300A and R1299A) mutants, the molar ratios of subgenomic RNA to positive-strand genomic RNA were not significantly different from that of the WT, indicating that p200 cleavage does not contribute to the differential synthesis of positive-strand genomic and subgenomic RNAs.
Our studies suggest a strong similarity between RV and alphavirus (reviewed in reference 26), a well-characterized positive-strand RNA virus genus, in NSP processing and viral RNA synthesis. Alphavirus NSP contains three cleavage sites, generating four cleavage products (nsP1 and nsP4) and a number of intermediates. A model for the composition of replication complexes and the temporal regulation of negative- and positive-strand RNAs has been proposed from several lines of study (15–18, 25). Three forms of replication complexes are involved in alphavirus replication: uncleaved P123 and nsP4 generate only negative-strand RNA; the complex composed of nsP1, P23, and nsP4 is active in the synthesis of both negative-strand RNA and 49S positive-strand genomic RNA; and the complex consisting of the final cleavage products nsP1, nsP2, and nsP3, and nsP4 produces only 49S positive-strand genomic RNA and subgenomic RNA. Cleavage at the 1/2 and 2/3 sites, respectively, switches the template preference of the replication complex from negative- to positive-strand RNA and also inactivates its capacity for negative-strand RNA synthesis, which explains the shutoff of negative-strand RNA synthesis after 4 to 6 h postinfection. In RV, p200 is cleaved into p150 and p90, giving a much simpler NSP composition. This work demonstrates that the replication complex composed of polyprotein p200 is active in negative-strand RNA synthesis but incapable of efficient positive-strand RNA synthesis, while cleavage of p200 is required for efficient positive-strand RNA synthesis. For both alphavirus and RV, cleavage of polyprotein or intermediates causes the switching of negative-strand to more-efficient positive-strand RNA synthesis. However, it has yet to be determined that synthesis of negative-strand RNA ceases after an early stage of RV replication. Our RNase protection assay results are complicated by reinfection, as well as the fast growth rate of BHK-21 cells. Studies using virus-infected Vero cells are under way. It also remains to be determined whether or not the cleaved products of RV NSP, p150 and p90, transcribe negative-strand RNA.
Our work provides the first experimental data demonstrating the relationships between RV NSP cleavage and virus replication, particularly between NSP cleavage and viral RNA synthesis. From our results and the studies of alphavirus replication, we hypothesize that uncleaved p200 forms the replication complex for negative-strand RNA synthesis and that cleavage of p200 into p150 and p90 converts the complex into one with the capacity for efficient positive-strand RNA synthesis. Whether p150 and p90 also produce negative-strand RNA remains to be investigated. It is of interest that p200 is capable of negative-strand RNA synthesis but not of positive-strand RNA synthesis. One possibility is that recognition of positive-strand RNA promoters or initiation of positive-strand RNA synthesis needs a specific component or conformation not present in p200 but generated by its cleavage. Positive-strand RNA viruses replicate through negative-strand intermediates, the regulatory mechanism of which is worthy of study. Previous studies of alphavirus and the present one of RV may indicate a possible mechanism of RNA replication for a group of viruses, namely, that the change from synthesis of negative-strand RNA to that of positive-strand RNA is mediated by NSP cleavage.
ACKNOWLEDGMENTS
This work was supported by a grant from the Medical Research Council of Canada. Y.L. is supported by a studentship from the British Columbia Children's Hospital Foundation. S.G. is an investigator of the British Columbia Children's Hospital Foundation.
We are grateful to J. Yao for technical assistance.
REFERENCES
- 1.Bowden D S, Westaway E G. Rubella virus structural and nonstructural proteins. J Gen Virol. 1984;65:933–943. doi: 10.1099/0022-1317-65-5-933. [DOI] [PubMed] [Google Scholar]
- 2.Chambers T J, Nestorowicz A, Amberg S M, Rice C M. Mutagenesis of yellow fever virus NS2B protein: effects on proteolytic processing. NS2B-NS3 complex formation, and viral replication. J Virol. 1993;67:6797–6807. doi: 10.1128/jvi.67.11.6797-6807.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chambers T J, Nestorowicz A, Rice C M. Mutagenesis of the yellow fever virus NS2B/3 cleavage site: determinants of cleavage site specificity and effects on polyprotein processing and viral replication. J Virol. 1995;69:1600–1605. doi: 10.1128/jvi.69.3.1600-1605.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chen J-P, Strauss J H, Strauss E G, Frey T K. Characterization of the rubella virus nonstructural protease domain and its cleavage site. J Virol. 1996;70:4707–4713. doi: 10.1128/jvi.70.7.4707-4713.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Clarke D M, Loo T W, Hui I, Chong P, Gillam S. Nucleotide sequence and in vivo expression of rubella virus 24S subgenomic mRNA encoding the structural proteins E1, E2, and C. Nucleic Acids Res. 1987;15:3041–3057. doi: 10.1093/nar/15.7.3041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dominguez G, Wang C Y, Frey T K. Sequence of the genome RNA of rubella virus: evidence for genetic rearrangement during Togavirus evolution. Virology. 1990;177:225–238. doi: 10.1016/0042-6822(90)90476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Forng R-Y, Frey T K. Identification of the rubella virus nonstructural proteins. Virology. 1995;206:843–853. doi: 10.1006/viro.1995.1007. [DOI] [PubMed] [Google Scholar]
- 8.Francki R I B, Fauquet C M, Knudson D L, Brown F, editors. Classification and nomenclature of viruses. Fifth report of the International Committee on Taxonomy of Viruses. Archives of Virology, suppl. 2. Vienna, Austria: Springer-Verlag; 1991. [Google Scholar]
- 9.Frey T K. Molecular biology of rubella virus. Adv Virus Res. 1994;44:69–160. doi: 10.1016/S0065-3527(08)60328-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorbalenya A E, Koonin E V, Wolf Y I. A new superfamily of putative NTP-binding domains encoded by genomes of small DNA and RNA viruses. FEBS Lett. 1990;262:145–148. doi: 10.1016/0014-5793(90)80175-i. [DOI] [PubMed] [Google Scholar]
- 11.Gorbalenya A E, Koonin E V, Lai M M-C. Putative papain-related thiol proteases of positive-strand RNA viruses. FEBS Lett. 1991;288:201–205. doi: 10.1016/0014-5793(91)81034-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hemphill M L, Forng R-Y, Abernathy E S, Frey T K. Time course of virus-specific macromolecular synthesis during rubella virus infection in Vero cells. Virology. 1988;162:65–75. doi: 10.1016/0042-6822(88)90395-9. [DOI] [PubMed] [Google Scholar]
- 13.Kamer G, Argos P. Primary structural comparison of RNA-dependent polymerase from plant, animal and bacterial viruses. Nucleic Acids Res. 1984;12:7269–7282. doi: 10.1093/nar/12.18.7269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Koonin E V, Dolja V V. Evolution and taxonomy of positive-strand RNA viruses: implications of comparative analysis of amino acid sequences. Crit Rev Biochem Mol Biol. 1993;28:375–430. doi: 10.3109/10409239309078440. [DOI] [PubMed] [Google Scholar]
- 15.Lemm J A, Rice C M. Assembly of functional Sindbis virus RNA replication complexes: requirement for coexpression of P123 and P34. J Virol. 1993;67:1905–1915. doi: 10.1128/jvi.67.4.1905-1915.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lemm J A, Rice C M. Roles of nonstructural polyproteins and cleavage products in regulating Sindbis virus RNA replication and transcription. J Virol. 1993;67:1916–1926. doi: 10.1128/jvi.67.4.1916-1926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemm J A, Rümenapf T, Strauss E G, Strauss J H, Rice C M. Polypeptide requirements for assembly of functional Sindbis virus replication complexes: a model for the temporal regulation of minus-strand and plus-strand RNA synthesis. EMBO J. 1994;13:2925–2934. doi: 10.1002/j.1460-2075.1994.tb06587.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lemm J A, Bergqvist A, Read C M, Rice C M. Template-dependent initiation of Sindbis virus RNA replication in vitro. J Virol. 1998;72:6546–6553. doi: 10.1128/jvi.72.8.6546-6553.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18a.Liang, Y., J. Yao, and S. Gillam. Rubella virus nonstructural protein protease domains involved in trans- and cis-cleavage activities. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 19.Maniatis T, Fritsch E F, Sambrook J. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1982. [Google Scholar]
- 20.Marr L D, Wang C-Y, Frey T K. Expression of the rubella virus nonstructural protein ORF and demonstration of proteolytic processing. Virology. 1994;198:1–7. doi: 10.1006/viro.1994.1070. [DOI] [PubMed] [Google Scholar]
- 21.Nestorowicz A, Chambers T S, Rice C M. Mutagenesis of the yellow fever virus NS2A/2B cleavage site: effects on proteolytic processing, viral replication and evidence for alternative processing of the NS2A protein. Virology. 1994;199:114–123. doi: 10.1006/viro.1994.1103. [DOI] [PubMed] [Google Scholar]
- 22.Novak J E, Kirkegaard K. Improved method for detecting poliovirus negative strands used to demonstrate specificity of positive-strand encapsidation and the ratio of positive to negative strands in infected cells. J Virol. 1991;65:3384–3387. doi: 10.1128/jvi.65.6.3384-3387.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pugachev K V, Abernathy E S, Frey T K. Genomic sequence of the RA27/3 vaccine strain of rubella virus. Arch Virol. 1997;142:1165–1180. doi: 10.1007/s007050050150. [DOI] [PubMed] [Google Scholar]
- 24.Rozanov M N, Koonin E V, Gorbalenya A E. Conservation of the putative methyltransferase domain: a hallmark of the “Sindbis-like” supergroup of positive-strand RNA virus. J Gen Virol. 1992;73:2129–2134. doi: 10.1099/0022-1317-73-8-2129. [DOI] [PubMed] [Google Scholar]
- 25.Shirako Y, Strauss J H. Regulation of Sindbis virus RNA replication: uncleaved P123 and nsP4 function in minus-strand RNA synthesis, whereas cleaved products from P123 are required for efficient plus-strand RNA synthesis. J Virol. 1994;68:1874–1885. doi: 10.1128/jvi.68.3.1874-1885.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Strauss J H, Strauss E G. The alphaviruses: gene expression, replication, evolution. Microbiol Rev. 1994;58:491–562. doi: 10.1128/mr.58.3.491-562.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wolinsky J S. Rubella. In: Fields B N, Knipe D M, Howley P M, et al., editors. Fields virology, third ed. Philadelphia, Pa: Lipincott-Raven Publishers; 1996. pp. 899–921. [Google Scholar]
- 28.Xu J, Mendez E, Caron P R, Lin C, Murcko M A, Collett M S, Rice C M. Bovine viral diarrhea virus NS3 serine proteinase: polyprotein cleavage sites, cofactor requirements, and molecular model of an enzyme essential for pestivirus replication. J Virol. 1997;71:5312–5322. doi: 10.1128/jvi.71.7.5312-5322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yao J, Yang D, Chong P, Hwang D, Liang Y, Gillam S. Proteolytic processing of rubella virus nonstructural proteins. Virology. 1998;246:74–82. doi: 10.1006/viro.1998.9179. [DOI] [PubMed] [Google Scholar]
- 30.Yao J, Gillam S. Mutational analysis, using a full-length rubella virus cDNA clone, of rubella virus E1 transmembrane and cytoplasmic domains required for virus release. J Virol. 1999;73:4622–4630. doi: 10.1128/jvi.73.6.4622-4630.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]