Abstract
The nonpolymorphic major histocompatibility complex E (MHC-E) molecule is up-regulated on many cancer cells, thus contributing to immune evasion by engaging inhibitory NKG2A/CD94 receptors on NK cells and tumor-infiltrating T cells. To investigate whether MHC-E expression by cancer cells can be targeted for MHC-E–restricted T cell control, we immunized rhesus macaques (RM) with rhesus cytomegalovirus (RhCMV) vectors genetically programmed to elicit MHC-E–restricted CD8+ T cells and to express established tumor-associated antigens (TAAs) including prostatic acidic phosphatase (PAP), Wilms tumor-1 protein, or Mesothelin. T cell responses to all three tumor antigens were comparable to viral antigen-specific responses with respect to frequency, duration, phenotype, epitope density, and MHC restriction. Thus, CMV-vectored cancer vaccines can bypass central tolerance by eliciting T cells to noncanonical epitopes. We further demonstrate that PAP-specific, MHC-E–restricted CD8+ T cells from RhCMV/PAP-immunized RM respond to PAP-expressing HLA-E+ prostate cancer cells, suggesting that the HLA-E/NKG2A immune checkpoint can be exploited for CD8+ T cell–based immunotherapies.
Cytomegalovirus-based vaccines induce T cells recognizing cancer antigens via conserved MHC-E molecules.
INTRODUCTION
The human leukocyte antigen-E (HLA-E) is a highly conserved major histocompatibility class Ib (MHC-Ib) molecule that shows very limited polymorphism in the human population, unlike the highly polymorphic MHC-Ia molecules HLA-A, HLA-B, and HLA-C (1). Similar to other MHC-I molecules, HLA-E associates with β2-microglobulin (β2m), but HLA-E predominantly binds the peptides VMAPRTL(L,I,V,F)L (VL9) encoded in the signal sequence of HLA-A, HLA-B, HLA-C, and HLA-G heavy chains (2). VL9 is cotranslationally released by signal peptide peptidase and loaded onto the HLA-E/β2m dimer in a process that involves the peptide transporter associated with antigen processing (TAP) and the chaperone Tapasin (3, 4). The resulting trimeric complex of HLA-E/β2m/VL9 is transported to the cell surface upon assembly. In contrast to other MHC-I/peptide complexes, however, this trimeric complex generally does not engage T cell receptors (TCRs) but instead serves as ligand for inhibitory NKG2A/CD94 and activating NKG2C/CD94 heterodimeric C-type lectin receptors expressed on natural killer (NK) cells and a subset of CD8+ T cells (5, 6). This regulation of NK and T cell activity via presentation of MHC-Ia signal peptides by MHC-Ib molecules is conserved across mammals (7). This system thus serves as an indirect sensor for TAP function and MHC-I expression, which is often down-regulated by intracellular pathogens, particularly viruses, or lost during cancer progression.
While expression of HLA-E is ubiquitous, it is generally low on the surface of most cell types in part due to limited VL9 supply (8). On cells that express high levels of HLA-E, such as monocyte/macrophages, HLA-E is rapidly internalized and targeted to intracellular vesicles suggesting continuous turnover (8, 9). HLA-E is up-regulated by some intracellular pathogens, as well as on some tumor and senescent cells which prevents immune clearance by engaging the inhibitory NKG2A/CD94 receptor on NK cells and T cells (10–13). A well-known example of this strategy is human cytomegalovirus (HCMV) which eliminates endogenous VL9 by degrading nascent MHC-Ia and blocking TAP while encoding a viral VL9 peptide in the gene UL40 which is loaded onto HLA-E independent of TAP, thus ensuring high levels of HLA-E expression despite inhibition of MHC-Ia antigen presentation (14, 15). Up-regulation of HLA-E has also been reported for gynecological, breast, non–small cell lung, renal cell, and colorectal cancers, and this up-regulation often correlates with NKG2A expression on a large percentage of tumor-infiltrating CD8+ T cells (TILs) [reviewed in (11, 16)]. A particularly notable example was reported for human papilloma virus (HPV)–positive head and neck cancers where 50% of the CD8+ tumor–infiltrating lymphocytes expressed NKG2A/CD94, whereas CD8+ T cells in the blood rarely expressed this inhibitory receptor (17). In the same study, it was shown that T cells elicited by vaccination against HPV up-regulated NKG2A which limited their effectiveness in controlling experimental tumors in animal models. The HLA-E/NKG2A ligand receptor pair thus seems to represent an immune checkpoint that limits both the effectiveness of tumor-infiltrating NK cells and tumor antigen–targeting T cells (11, 18). For this reason, NKG2A-targeting antibodies are now in clinical testing and have shown some promise, particularly in combination with immunotherapies (16, 19, 20). In addition, these antibodies might improve the activity of active vaccination against tumor antigens (17, 21).
Although engaging the NKG2A/C receptors seems to be its main function, HLA-E can also present peptides to the TCR of CD8+ T cells (22, 23). Such HLA-E–restricted CD8+ T cells have been reported for Mycobacterium tuberculosis and other intracellular bacteria (24–26). Moreover, polymorphism in the VL9 sequence of HCMV UL40 results in the elicitation of HLA-E/VL9–specific CD8+ T cells, particularly when there is a mismatch with the VL9 sequence of the host (27–29). Furthermore, using HLA-E tetramers containing HLA-E binding viral peptides, it was possible to identify and expand human HLA-E–restricted CD8+ T cells that recognized HIV-infected and severe acute respiratory syndrome coronavirus 2–infected cells (30, 31). While such HLA-E–restricted CD8+ T cells have not yet been reported for cancer, self-peptides have been shown to be presented by HLA-E (32–36). Moreover, it has been shown that HLA-E in TAP-deficient cancer cells contains many non-VL9 peptides derived from cellular proteins (37).
On the basis of these observations, it has been hypothesized that HLA-E–positive cancer cells could be targeted by HLA-E–restricted CD8+ T cells recognizing non-VL9 peptides (35), but testing this hypothesis has been limited by the inability to elicit HLA-E–restricted CD8+ T cells to specific antigens in vivo. However, in the context of studying rhesus cytomegalovirus (RhCMV) as a vaccine platform, we observed that the RhCMV strain 68-1 elicits CD8+ T cells that are restricted by Mamu-E, the rhesus macaque (RM) homolog of HLA-E (we use MHC-E as a general term for both the human and rhesus molecules) as well as MHC-II instead of MHC-Ia (38, 39). One of the underlying reasons for the ability of RhCMV to elicit MHC-E–restricted CD8+ T cells is the up-regulation of MHC-E in infected cells by Rh67, the RhCMV homolog of HCMV UL40 (40). However, wild-type RhCMV counteracts this mechanism by expressing a series of small, chemokine-like proteins (homologs of HCMV UL128, UL130, UL146, and UL147) that each prevent the induction of MHC-E–restricted CD8+ T cells (41). Wild-type RhCMV thus elicits conventional, MHC-Ia–restricted CD8+ T cells.
Coincidentally, in the course of in vitro passaging, strain 68-1 lost expression of these inhibitory gene products, thus enabling the induction of MHC-E–restricted CD8+ T cells (41, 42). Previous work showed that strain 68-1 RhCMV vectors expressing simian immunodeficiency virus (SIV) antigens protected against challenge with highly pathogenic strains of SIV by a unique “control and clear” mechanism, i.e., the durable, high-frequency effector memory T cell responses elicited by RhCMV/SIV vaccines intercepted SIV early during infection, arrested its replication, and ultimately cleared the virus (43–46). Similar results were obtained in cynomolgus macaques using CyCMV/SIV-vectors (47). Notably, this protection was lost when RhCMV/SIV vectors that lacked the ability to elicit MHC-E–restricted CD8+ T cells were used as vaccines (40, 41, 48). In contrast, tropism-modified RhCMV/SIV vectors that exclusively elicited MHC-E–restricted CD8+ T cells maintained protection against SIV (48). On the basis of these studies, genetically modified HCMV/HIV vectors were designed that are now undergoing clinical testing (49). In addition to uncovering a previously unknown mechanism of control, these studies also demonstrated that MHC-E–restricted CD8+ T cells recognizing SIV-derived peptides completely unrelated in sequence to VL9 were effective controllers of SIV replication (38, 47, 50).
The ability of RhCMV vectors to elicit MHC-E–restricted CD8+ cells is not limited to SIV but has also been demonstrated for M. tuberculosis, malaria and hepatitis B virus (HBV) antigens (51–53). However, to date, all antigens inserted into RhCMV vectors previously were foreign, i.e., derived from viruses, bacteria, or parasites, to elicit antipathogen MHC-E–restricted CD8+ T cells, leaving open the question of whether such T cells can also be elicited to tumor-associated antigens (TAAs), i.e., self-antigens overexpressed in tumor cells (54). As proof of concept, we focus on prostate cancer (PCa), a cancer that has so far resisted immunotherapy notwithstanding the fact that the first antigen-specific immunotherapy, Sipuleucel-T, was approved for this cancer (55). Sipuleucel-T consists of autologous dendritic cells loaded ex vivo with a recombinant fusion protein of granulocyte-macrophage colony-stimulating factor and prostatic acidic phosphatase (PAP) and reintroduced into the patient to elicit PAP-specific T cells (56). Since PCa is particularly attractive for T cell–mediated immunotherapy due to the nonessential nature of prostate tissue and the presence of multiple organ-specific TAAs, many TAA-targeting vaccine approaches are now in clinical trials, either as monotherapies or in combination with checkpoint inhibitors (57). Here, we explore the possibility of targeting HLA-E for the immunotherapy of PCa. We demonstrate HLA-E expression in primary tumors and show that RhCMV vectors are capable of eliciting broadly targeted, MHC-E–restricted T cells to autologous PAP as well as other well-established other tumor antigens. We observed that such MHC-E–restricted, PAP-specific T cells recognize HLA-E– and PAP-expressing PCa cells and cell lines. Our data thus suggest that targeting HLA-E with HLA-E–restricted, TAA-specific CD8+ T cells could be a new approach to immunotherapy of PCa and possibly other cancers.
RESULTS
HLA-E is up-regulated in PCa
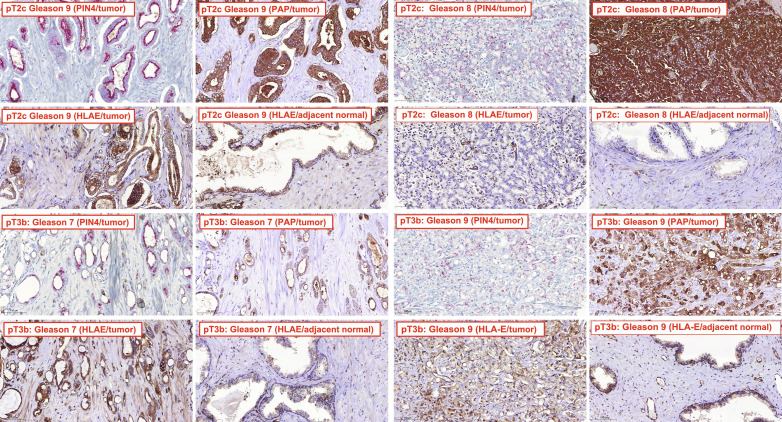
To determine HLA-E expression in PCa, we used immunohistochemistry (IHC) to analyze in-house generated or commercially available tissue microarrays (TMAs). In-house TMAs 1 to 4 contained a total of 258 tumor cores and 130 matched normal prostate sections from 86 radical prostatectomy cases. One hundred sixty-three cores showed tumor purity, and 93% of these were determined to be positive for HLA-E as detected by polyclonal antiserum (data file S1). TMA1 was additionally stained for PAP and the prostatic intraepithelial neoplasia-4 (PIN-4) marker cocktail. As expected, expression of PIN4 and PAP was found in 98% of tumor cores but not in normal tissue (data file S1). Moreover, 80% of PAP-positive tumors were also positive for HLA-E. Representative images demonstrate that high-grade tumors (Gleason 7 and above) were positive for HLA-E, whereas adjacent normal tissue was negative (Fig. 1). Low-grade tumors (Gleason 6 or less) were generally negative for HLA-E (data file S1). Similar results were obtained using the commercially available HLA-E–specific monoclonal antibody (mAb) 4D12 (fig. S1). Fourteen of 18 cores with tumor purity stained positive for HLA-E (data file S2). High HLA-E expression was also observed in metastatic PCa (fig. S2). These data suggest that HLA-E is widely expressed in advanced PCa, and HLA-E is coexpressed with PAP.
Fig. 1. HLA-E expression in PCa.
IHC analysis of PCa or adjacent normal tissue for expression of HLA-E, PAP, and PIN4. TMAs were prepared and processed as described in Materials and Methods. Representative IHC of PCa tumor tissue and adjacent normal tissue are shown for Gleason 7, 8, and 9 tumors stained for HLA-E using a polyclonal antiserum. Pathological tumor (pT) stages of PCa are indicated with pT2c representing localized bilateral PCa, whereas pT3b represents locally advanced PCa. Staining for PAP and PIN4 is additionally shown for the tumor tissue. The results for all TMAs are summarized in data file S1.
RhCMV elicits high-frequency T cell responses to tumor antigens
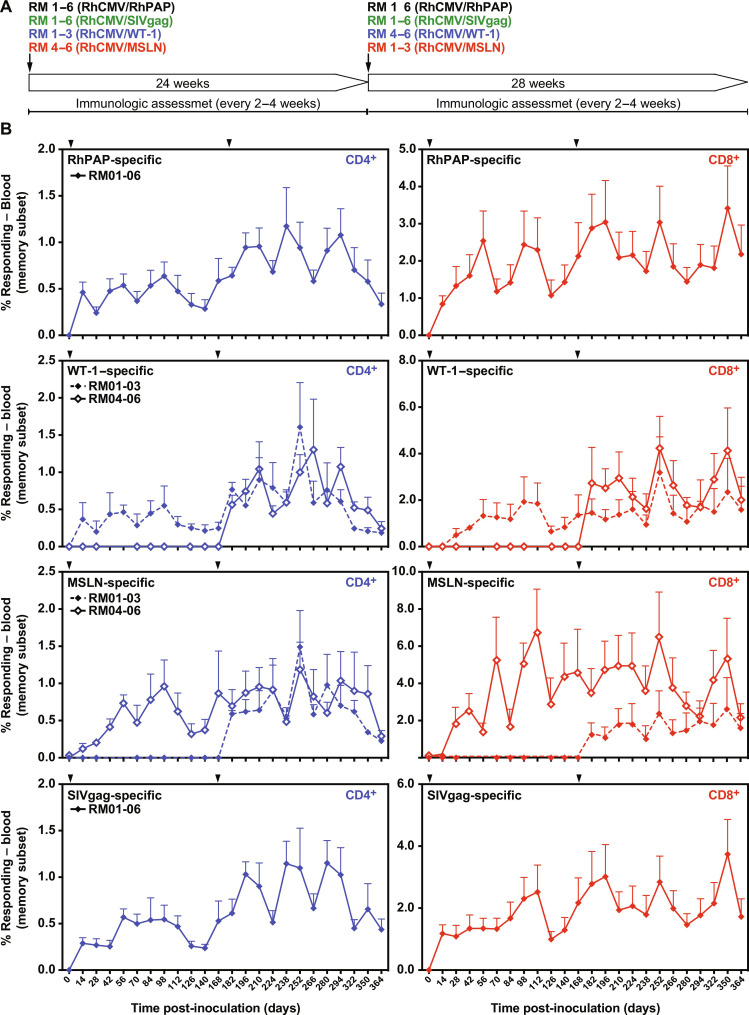
To determine whether CMV-based vectors could be used to elicit MHC-E–restricted CD8+ T cells to TAAs, we generated three RhCMV recombinants by inserting synthetic full-length or partial genes encoding rhesus PAP (RhPAP), the human transcription factor Wilms tumor 1 (WT-1) and the human cell adhesion protein Mesothelin (MSLN) into 68-1 RhCMV (fig. S3). The TAAs chosen in addition to PAP have been previously targeted for T cell–focused immunotherapy approaches that were either TCR-T cell based (WT-1) (58) or chimeric antigen receptor (CAR)–T cell based (MSLN) (59). RhPAP was expressed by replacing the nonessential RhCMV gene Rh107 (ortholog of HCMV UL78), whereas WT-1 and MSLN were expressed by replacing Rh110 (ortholog of HCMV UL82) which renders the resulting vectors deficient for spreading and shedding (60). We inoculated six male, RhCMV-seropositive RMs with 5 × 106 plaque-forming units (PFU) of 68-1 RhCMV/RhPAP. Three of these RMs were additionally inoculated with the same dose of RhCMV/WT-1, whereas the remaining three RMs were inoculated with RhCMV/MSLN. To compare the TAA-specific T cell responses to those observed to typical pathogen antigen-specific responses, we also inoculated all six RM with previously described 68-1 RhCMV expressing the SIV Gag protein (61). The CD4+ and CD8+ T cell response to each of the four antigens was monitored by intracellular cytokine staining (ICS) for interferon-γ (IFNγ) and tumor necrosis factor–α (TNFα) using pools of 15–amino acids oligomer peptides overlapping by 11 amino acids corresponding to each of the vaccine insert antigens. Each of the immunized RMs developed both CD4+ and CD8+ T cell responses to RhPAP, WT-1, and MSLN that were comparable to those elicited to SIV Gag with respect to average frequency, durability (Fig. 2), and phenotype (fig. S4). Furthermore, when we inoculated 68-1 RhCMV/WT-1 and 68-1 RhCMV/MSLN into the three animals of the cohort that had not received these recombinants initially, each of the RM rapidly developed a WT-1 or MSLN-specific T cells (Fig. 2). This observation is consistent with our previous demonstration that vector-specific immunity does not affect the ability of these vectors to elicit insert-specific T cell responses upon subsequent reinoculations (61, 62). Moreover, these results also demonstrated that both spreading-competent and spreading-impaired vectors were able to elicit TAA-specific responses. PAP, WT-1, and MSLN are nonmutated self-antigens that are expressed not only in cancerous tissue but also in some healthy tissues, albeit at a lower level (63–65). Since self-antigens are generally subject to central and peripheral tolerance (66), the strong T cell responses observed in all animals against each of the tumor antigens thus warranted further in depth characterization.
Fig. 2. RhCMV elicits robust and durable T cell responses to TAAs.
(A) Six male RMs were inoculated with 68-1 RhCMV/RhPAP and with 68-1 RhCMV/SIVgag. RMs 1 to 3 were additionally coinoculated with RhCMV/WT-1, whereas RMs 4 to 6 were coinoculated with 68-1 RhCMV/MSLN. At day 140, RMs 1 to 3 received 68-1 RhCMV/MSLN, whereas RMs 4 to 6 received 68-1 RhCMV/WT-1. (B) CD4+ and CD8+ T cell responses were measured in peripheral blood mononuclear cells (PBMCs) using overlapping peptide pools for each of the antigens by ICS for CD69 and IFNγ and/or TNFα at each of the indicated time points. The background subtracted mean response frequencies are shown (+SEM). Days of inoculation are indicated by arrowheads. Individual response frequencies are shown in data file S3.
RhCMV elicits broadly targeted MHC-II and MHC-E–restricted CD8+ T cell responses to TAAs
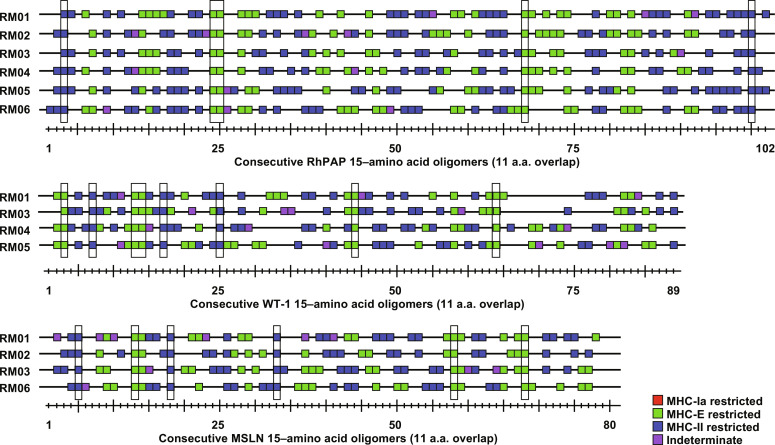
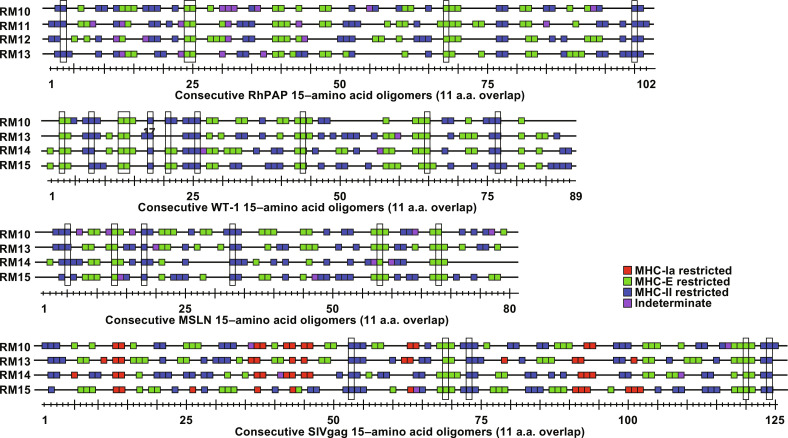
We previously demonstrated that strain 68-1 RhCMV expressing antigens of various pathogens (SIV, HBV, M. tuberculosis, and the malaria parasite Plasmodium knowlesi) elicits MHC-II and MHC-E–restricted but not MHC-Ia–restricted, CD8+ T cells to these antigens (38, 39, 51–53). In addition, we reported that the average number of epitopes per antigen recognized by CD8+ T cells of 68-1 RhCMV/SIV-immunized RM was substantially higher than observed for SIV-infected RM or RM immunized with other SIV vaccine platforms such as DNA, pox-, or adeno-vectors (39). To determine whether TAA-specific CD8+ T cells recognized a similarly high density of epitopes, we measured the response to each individual 15–amino acid oligomer peptide of RhPAP, WT-1, and MSLN by ICS in peripheral blood mononuclear cells (PBMCs) of immunized RM. As shown in Fig. 3, on average, 51% (range: 43 to 58% in individual RM) of the overlapping 15–amino acid oligomer peptides were recognized by CD8+ T cells from immunized RMs (note that the number of minimal 8– to 9–amino acid epitopes is expected to be smaller due to overlap). Because this result is comparable to the average number of peptides reported for SIV Gag (39), we conclude that 68-1 RhCMV is able to elicit CD8+ T cells to a very high number of epitopes within a given TAA. Given the fact that PAP was derived from RM and the high conservation of WT-1 and MSLN (93 and 88% identity, respectively, between human and rhesus), this result also rules out the possibility that the observed T cell responses were directed against a small number of epitopes mismatched between the immunogen and the self-antigen. This further supports the conclusion that 68-1 RhCMV vector–elicited CD8+ T cell responses are not impeded by immune tolerance to self-proteins.
Fig. 3. Epitope specificity and MHC restriction of TAA-targeting CD8+ T cells.
CD8+ T cell responses in PBMC of RMs 1 to 6 (Fig. 2) were measured by ICS for IFNγ and TNFα to individual overlapping consecutive 15–amino acid oligomer peptides that span the indicated proteins. Assay limit of detection was determined as described previously (51), with 0.05% after background subtraction being the minimum threshold. Above background responses (IFNγ and/or TNFα) are shown by a square along the length of each protein with the peptide numbers shown below. Results of blocking experiments are indicated by the color of the square: Peptides restricted by MHC-II (blue) were blocked with class II-associated invariant chain peptide (CLIP) and anti–human leukocyte antigen-DR (HLA-DR) antibody, whereas MHC-E–restricted peptides (green) were blocked with VL9 peptide and pan–MHC-I antibody. Indeterminate responses are shown in purple. Supertopes, i.e., epitopes recognized in all RMs (including Fig. 5), are boxed in. Overall analysis of 15–amino acid oligomer peptide responses is shown in data file S3. a.a., amino acid.
Next, we determined the MHC restriction of CD8+ T cell–stimulating peptides in RMs of the cohort by adding MHC-Ia, MHC-E, or MHC-II blocking antibodies or peptides to the ICS assay (38, 39). Similar to previously reported responses to heterologous antigens, we observed that peptides were either presented by MHC-II or by MHC-E but not by MHC-Ia, with only a small number of peptides failing to be clearly assigned (Fig. 3). A further unusual characteristic of the CD8+ T cell epitope specificity observed in 68-1 RhCMV-immunized RM is that some peptides are always recognized in each RM irrespective of their MHC allotypes. Such universal shared epitopes, we refer to them as “supertopes,” were also observed for TAA-immunized RM in this cohort (highlighted in Fig. 3) and confirmed in additional cohorts. Thus, TAA-specific responses elicited by 68-1 RhCMV are not different from pathogen-specific responses with respect to magnitude, durability, epitope density, MHC restriction, and supertope recognition.
RhCMV can elicit MHC-Ia–restricted CD8+ T cell responses to tumor antigens
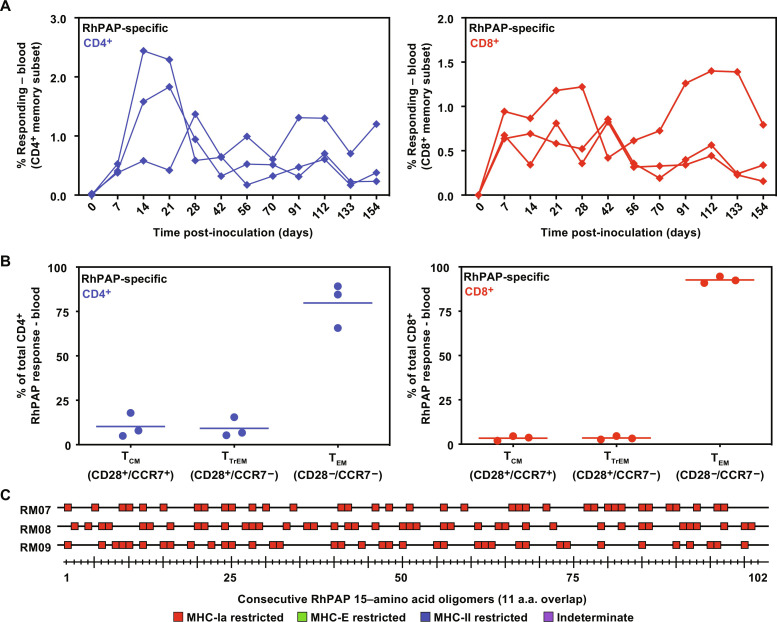
The ability of 68-1 RhCMV vectors to elicit CD8+ T cell responses to self-antigens could be due to MHC-II– and MHC-E–restricted CD8+ T cells not being subject to the same immunological selection against self-reactivity as MHC-Ia–restricted T cells. We previously showed that clone 68-1.2 RhCMV vectors, in which the RhCMV homologs of the HCMV pentameric complex subunits UL128 and UL130 were repaired, lost the ability to elicit MHC-II– and MHC-E–restricted CD8+ T cells; instead, these vectors elicit MHC-Ia–restricted CD8+ T cell responses similar to wild-type RhCMV (38, 39, 41). To explore whether 68-1.2 RhCMV would elicit MHC-Ia–restricted T cell responses to TAAs, we inserted RhPAP by replacing the UL78 ortholog Rh107 (fig. S3). When the resulting vector was inoculated into a new cohort of three RMs, we observed robust CD4+ and CD8+ T cell responses to RhPAP (Fig. 4A) that displayed the typical effector memory phenotype (Fig. 4B). Notably, CD8+ T cell responses were now exclusively restricted by MHC-Ia but still broadly targeted (Fig. 4C). These results demonstrate that wild-type–like RhCMV vectors are capable of eliciting CD8+ T cell responses to TAAs that recognize peptides in the context of MHC-Ia, suggesting that CMV vectors can overcome tolerance to self-antigens irrespective of their MHC restriction.
Fig. 4. The 68-1.2 RhCMV vectors elicit MHC-Ia–restricted CD8+ T cells to RhPAP.
(A) Three male RMs were inoculated with 68-1.2 RhCMV/RhPAP. CD8+ and CD4+ T cell responses were determined by ICS in PBMC using an overlapping peptide mix for RhPAP at the indicated time points. (B) Memory differentiation phenotypes of RhPAP-specific T cells responding to peptide pools in each RM were determined on the basis of CD28 and CCR7 expression, delineating central memory (TCM, CD28+, CCR7+), transitional effector-memory (TTrEM, CD28+, CCR7−), and effector-memory (TEM, CD28−, CCR7−). (C) CD8+ T cell responses and MHC restriction to individual 15–amino acid oligomer peptides from the RhPAP mix were determined as in Fig. 3. All peptide-specific responses were MHC-Ia restricted (shown in red color), i.e., they were blocked by pan–MHC-I antibody but not by VL9 peptide or anti–MHC-II antibodies. Results for each RM in (A) and (B) and overall analysis of 15–amino acid oligomer peptide responses in (C) are listed in data file S3.
RhCMV does not elicit MHC-Ia–restricted CD8+ T cell responses to canonical, MHC-Ia–restricted epitopes in TAAs
We previously reported that MHC-Ia–restricted epitopes recognized by CD8+ T cells from 68-1.2 RhCMV/SIV-immunized RM did not include recognition of “canonical” epitopes that dominate the responses of RM immunized with conventional vaccines or infected with SIV (39). Thus, 68-1.2 RhCMV vectors elicit CD8+ T cells to “noncanonical” MHC-Ia–restricted epitopes, i.e., epitopes that are subdominant in the context of conventional vaccines or other viruses (39). In contrast, CD8+ T cells to well-established, canonical MHC-Ia–restricted epitopes can be elicited by RhCMV vectors when the gene Rh189 is deleted (39). This is observed regardless of the vector backbone, i.e., 68-1.2–derived, Rh189-deleted vectors elicit CD8+ T cells to both noncanonical and canonical MHC-Ia epitopes whereas 68-1–derived, Rh189-deleted vectors elicit CD8+ T cells to MHC-Ia–restricted canonical epitopes alongside unconventional responses restricted by MHC-II and MHC-E (39). Since the Rh189 protein is the homolog of HCMV US11 targeting newly synthesized MHC-Ia heavy chains for proteasomal destruction (67, 68) we hypothesize that deletion of Rh189 enables direct priming of MHC-Ia-restricted CD8+ T cells similar to other vaccines or virus infections (69). To examine whether RhCMV vectors would be capable of eliciting CD8+ T cell responses to canonical MHC-Ia–restricted epitopes, we deleted Rh189 from the TAA-expressing 68-1 RhCMV vectors (fig. S3). As previously demonstrated for P. knowlesi antigens (52), this approach obviates the need for a priori knowledge of canonical epitopes since deletion of Rh189 from 68-1 RhCMV results in CD8+ T cells that recognize canonical epitopes presented by MHC-Ia in a background of MHC-II– and MHC-E–restricted, such that any MHC-Ia–restricted response elicited by such vectors can be considered to canonical epitopes (39). We coinoculated six male RMs with 68-1 RhCMVΔRh189/RhPAP together with the previously described 68-1 RhCMVΔRh189/SIVgag (39). Three RMs were additionally coinoculated with 68-1 RhCMVΔRh189/WT-1 or 68-1 RhCMVΔRh189/MSLN with the remaining three RMs inoculated with these recombinants approximately 6 months later (fig. S5A). As observed for Rh189-intact RhCMV, we observed robust and durable T cell responses to each of the TAAs that were comparable to SIV Gag with respect to overall frequency (fig. S5B) and phenotype (fig. S4). Moreover, the percentage of epitopes recognized was similar to that observed for Rh189-intact vectors (Fig. 5). However, when we determined the MHC restriction of these peptides, we did not observe any MHC-Ia–restricted epitopes for any of the three TAAs (Fig. 5). In contrast, SIV Gag-specific T cells targeted MHC-Ia epitopes in addition to MHC-II and MHC-E consistent with our previous report (39). These results suggest that Rh189-deleted RhCMV elicits CD8+ T cell responses by direct priming to canonical epitopes presented by MHC-Ia to foreign antigens but not to self-antigens. The most likely explanation for this lack of canonical responses is that self-reactive T cells to these epitopes are subject to central tolerance, i.e., they are eliminated during thymic selection (66). In contrast, MHC-Ia–restricted responses elicited by 68-1.2 RhCMV, most likely via cross-presentation, do not seem to be subject to thymic elimination consistent with these epitopes being noncanonical or subdominant in the context of other vaccines. Similarly, CD8+ T cells to MHC-II or MHC-E–restricted epitopes elicited by 68-1 RhCMV avoid central tolerance. CMV-based vaccines can thus circumvent immunological tolerance to self-antigens by activating T cells that recognize subdominant or unconventionally restricted epitopes.
Fig. 5. Rh189-deleted 68-1 RhCMV elicits canonical MHC-Ia–restricted T cells to SIV Gag but not to TAAs.
Six RMs were coinoculated with 68-1 RhCMVΔRh189 expressing the indicated antigens as shown in fig. S3. MHC restriction of individual 15–amino acid oligomer peptides from each antigen for CD8+ T cells from four RMs of this cohort was determined as described in Fig. 3. Above background responses are shown as squares at the position along the antigen with the colors indicating blocking by reagents specific for MHC-II (blue), MHC-E (green), or MHC-Ia (red). Supertope peptides are shown by boxes. Indeterminate responses are shown in purple. Overall analysis of 15–amino acid oligomer peptide responses is shown in data file S3.
MHC-E–restricted, TAA-specific CD8+ T cells recognize endogenously processed cancer antigens
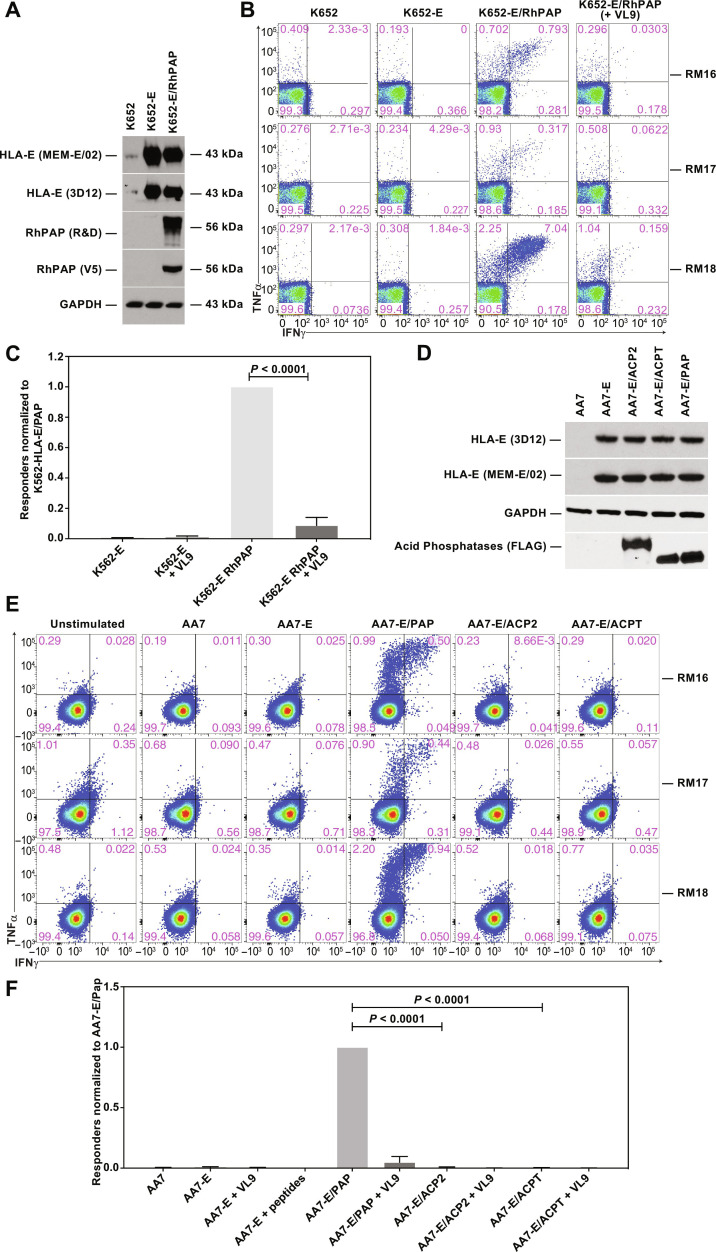
Our results suggest that CMV-based vectors could be used to elicit effector memory CD8+ T cell responses that target TAA-derived, noncanonical epitopes presented in the context of MHC-Ia, MHC-II, or MHC-E. In contrast, other cancer vaccination or immunotherapy strategies, as well as immune checkpoint inhibitors, generally aim to elicit MHC-Ia–restricted CD8+ T cells to canonical epitopes. The ability to elicit MHC-E–restricted CD8+ T cells to TAA is particularly unique to the CMV-based vector platform. However, it is not known whether HLA-E expressed on cancer cells can present TAA-derived peptides and thus can be targeted by MHC-E–restricted, TAA-specific CD8+ T cells. To determine whether endogenously expressed TAA would be recognized by MHC-E–restricted CD8+ T cells, we transfected RhPAP into previously described MHC-Ia–negative K562 cells expressing HLA-E*01:03 (K562-E) (50). Expression of HLA-E and RhPAP in the respective cell lines was confirmed by immunoblot (Fig. 6A). To generate MHC-E–restricted CD8+ T cells specific for RhPAP, we inoculated a new cohort of three RMs with 68-1 RhCMV/RhPAP and demonstrated the development of RhPAP-specific CD4+ and CD8+ T cells over time (fig. S6). Next, we coincubated K562-E and K562-E/RhPAP cells with CD8+ T cells isolated from each of the three RMs and monitored their cytokine response by ICS in the presence or absence of pre- and coincubation with the MHC-E binding (and blocking) peptide VL9. CD8+ T cells from each RM were stimulated when exposed to K562-E/RhPAP but not by K562-E cells (Fig. 6B). Moreover, stimulation by K562-E/RhPAP was prevented by exogenous VL9 peptide blocking. Similar results were obtained in multiple independent experiments (Fig. 6C). These results strongly suggest that endogenously expressed PAP-derived peptides can be loaded onto HLA-E and presented to MHC-E–restricted CD8+ T cells. Moreover, these data also confirm earlier data that MHC-E–restricted T cells obtained from 68-1 RhCMV-immunized animals recognize HLA-E across species (38, 50).
Fig. 6. Stimulation of MHC-E–restricted, PAP-specific CD8+ T cells by PAP-expressing K562 cells.
(A) Immunoblot of cell lysates of indicated K562-derived cell lines. Expression of endogenous and transfected HLA-E in K562 cells was demonstrated with two HLA-E–specific antibodies MEM-E/02 and 3D12 (5, 90, 91). Note that nontransfected K562 cells express low amounts of HLA-E (88). V5 epitope–tagged RhPAP was detected with PAP-specific (MAB6240, R&D Systems) or V5-specific mouse monoclonal antibodies. A GAPDH-specific mAb was used as loading control. (B) ICS for IFNγ and TNFα of CD8+ T cells from three 68-1 RhCMV/RhPAP-immunized RM after coincubation with the indicated cells in the presence or absence of VL9 peptide. (C) Results from five independent experiments showing the average relative frequency (background-subtracted and normalized to K562-E/PAP) of CD69 and IFNγ and/or TNFα-positive CD8+ T cells from 68-1 RhCMV/RhPAP-immunized RM responding to the indicated cell lines in the presence or absence of VL9. Individual results are shown in data file S3. Statistical significance was determined by paired t test (not significant, P > 0.05). (D) Immunoblot for HLA-E and FLAG-tagged acid phosphatases. Lysates of stable cell lines of AA7 cells (= K562 cells deleted for HLA-E; fig. S7) transfected with HLA-E alone or together with human PAP, ACP2, or ACPT were electrophoretically separated and probed with the indicated antibodies by immunoblot. (E) ICS for IFNγ and TNFα of CD8+ T cells from three 68-1 RhCMV/RhPAP-immunized RM after coincubation with the indicated cell lines. (F) Average frequencies (+SEM) of CD69 and IFNγ and/or TNFα-positive CD8+ T cells responding to the indicated cell lines were background subtracted and normalized to AA7-E/PAP. Individual results are shown in data file S3. Statistical significance was determined using paired t test.
As member of the acid phosphatase family, PAP displays sequence similarity with other phosphatases, with the highest similarity to lysosomal acid phosphatase 2 (ACP2) (50% identity) (70) and testicular acid phosphatase (ACPT) (46% identity) (71), including stretches of identical 8– to 9–amino acid sequences representing potential T cell epitopes. The high epitope density of the T cell response elicited to RhPAP in RM (Fig. 3) thus raised the possibility that MHC-E–restricted T cells would cross-react with other acid phosphatases. To examine this possibility, we generated a new series of cell lines expressing human PAP, ACP2, or ACPT. While K562 cells are negative for MHC-Ia, they express residual amounts of HLA-E (Fig. 6A and fig. S7). Therefore, we introduced V5 epitope–tagged versions of these genes into a K562 derivative, termed AA7 cells, in which HLA-E was deleted using CRISPR-Cas9 followed by reintroduction of HLA-E*01:03 (AA7-E) (fig. S7). Expression of HLA-E as well as the human acid phosphatases was confirmed by immunoblot (Fig. 6D). Expression of human PAP in these cells stimulated CD8+ T cells isolated from the three RhPAP-immunized RMs as shown by ICS (Fig. 6E). In contrast, T cell simulation was not observed with AA7-E cells expressing either ACP2 or ACPT in three independent experiments (Fig. 6, E and F). These results are consistent with MHC-E–restricted RM T cells recognizing peptides derived from both human and RhPAP. In contrast, peptides presented from endogenously expressed ACP2 or ACPT do not appear to include cross-reactive epitopes.
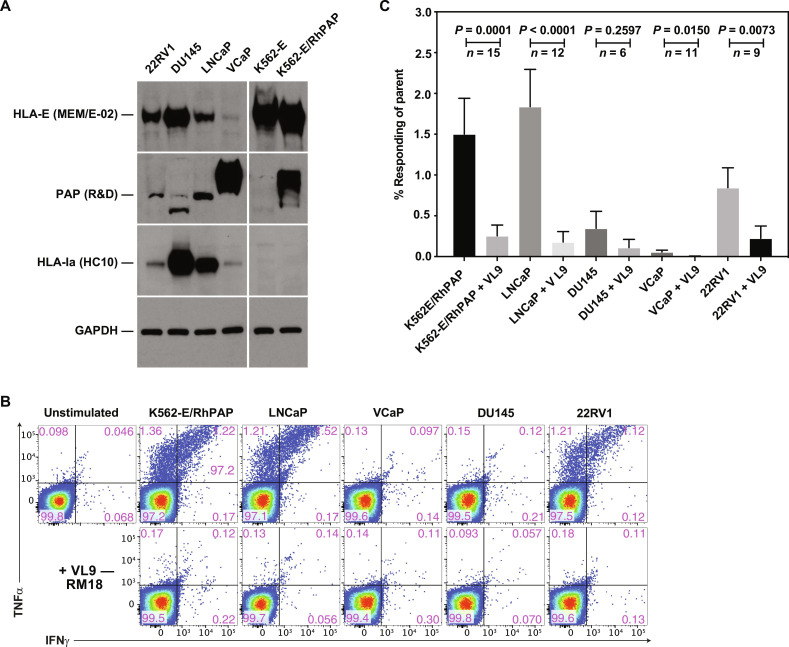
Next, we wanted to determine whether MHC-E–restricted, PAP-specific RM CD8+ T cells would be stimulated in the presence of human PCa cell lines. We selected the four cell lines 22RV1, LNCaP, VCaP, and DU145 representing a range of HLA-I, HLA-E, and PAP expression based on published mRNA expression profiles (72). We confirmed by immunoblot that 22RV1 and LNCaP expressed both PAP and HLA-E, whereas DU145 was strongly positive for HLA-E and HLA-Ia but negative for PAP expression (Fig. 7A) VCaP cells were highly positive for PAP but expressed very low levels of both HLA-I and HLA-E. Upon coincubations with MHC-E–restricted, PAP-specific CD8+ T cells, we observed stimulation of T cells by cell lines that were both positive for HLA-E and PAP, including the positive control K562-E/PAP; and this simulation was blocked by exogenous VL9 consistent with HLA-E–dependent antigen presentation. A representative experiment is shown in Fig. 7B, and a summary of multiple experiments is shown in Fig. 7C and extended data file S3. These results are consistent with PCa cells being able to be targeted by MHC-E–restricted CD8+ T cells.
Fig. 7. Recognition of PCa cell lines by MHC-E–restricted, PAP-specific CD8+ T cells.
(A) Immunoblot of cell lysates of the indicated cell lines for HLA-E, PAP, HLA-Ia, and GAPDH. Antibodies used were MEM-E/02 for HLA-E (90), HC10 for HLA-Ia heavy chains (92), and MA5-15738 (Invitrogen) for GAPDH. The anti-PAP antibody (MAB6240 R&D Systems) detected a lower molecular weight band in DU145 cells that is likely nonspecific since DU145 cells are known to be PAP negative (93, 94). (B) ICS for IFNγ and TNFα production of CD8+ T cells from a 68-1 RhCMV/RhPAP-immunized RM after coincubation with the indicated cell lines in the presence or absence of VL9 peptide. In addition, we inhibited potential MHC-II presentation by adding anti-DR and CLIP peptide to all stimulations. (C) Average frequencies (+SEM) of CD69 and IFNγ and/or TNFα-positive CD8+ T cells from 68-1 RhCMV/RhPAP-immunized RM after background subtraction. The number of repeat experiments is indicated. Individual results are shown in data file S3. Statistical significance was calculated using paired Mann-Whitney U test (not significant, P > 0.05).
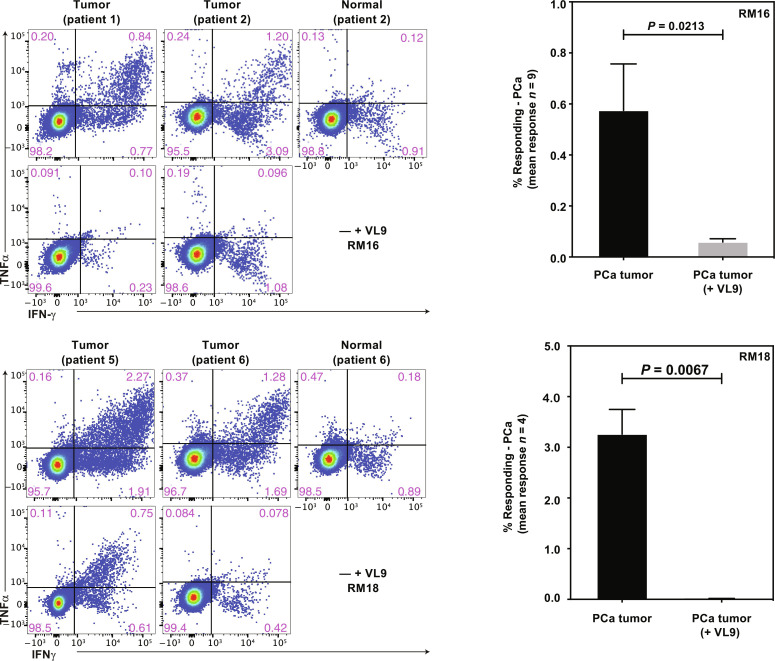
To further examine whether MHC-E–restricted, PAP-specific CD8+ T cells would be able to respond to primary PCa cells, we generated cell suspensions from patients with PCa who underwent radical prostatectomy. When these cell suspensions were coincubated with CD8+ T cells isolated from PBMC of 68-1 RhCMV/RhPAP-immunized RMs, we observed HLA-E–dependent stimulation as demonstrated by VL9 inhibition in about half of the samples tested (15 of 32) (Fig. 8). These observations thus suggest that PCa tumors can be targeted by MHC-E–restricted CD8+ T cells.
Fig. 8. Recognition of primary PCa cells by MHC-E–restricted, PAP-specific CD8+ T cells.
Left: ICS for IFNγ and TNFα production by CD8+ T cells from two 68-1 RhCMV/RhPAP-immunized RM after coincubation with PCa cell suspensions in the presence or absence of VL9 peptide. In addition, we inhibited MHC-II presentation by adding anti-DR antibody and CLIP peptide in all stimulations. Right: Summary of results from the indicated number of primary PCa samples showing the average frequency of CD69 and IFNγ and/or TNFα-positive CD8+ T cells from 68-1 RhCMV/RhPAP-immunized RM in the absence or presence of VL9 peptide after background subtraction. Individual results are shown in data file S3. Statistical significance was calculated using paired t test.
DISCUSSION
Our data suggest that CMV-based vectors can be used to generate MHC-E–restricted CD8+ T cells to common cancer antigens and that cancer cells can present tumor antigens to these T cells via HLA-E. Targeting HLA-E is therefore a new modality for cancer immunotherapy. The vectors used in our study elicited both MHC-E–restricted CD8+ T cells and MHC-II–restricted CD8+ T cells to TAAs. Recently, we reported that insertion of targeting sites for the endothelial cell–specific microRNA 126 into essential viral genes of strain 68-1 RhCMV prevented the induction of MHC-II–restricted CD8+ T cells, resulting in a vector backbone that exclusively elicited MHC-E–restricted CD8+ T cells (48). Similar “HLA-E–only” HCMV vectors could be used to specifically target HLA-E expressing cancer cells with TAA-specific CD8+ T cells.
In contrast to RhCMV vectors lacking two gene regions encoding either the homologs of HCMV UL128 and UL130 or homologs of UL146 and UL147, RhCMV with wild-type configuration or partially deleted RhCMV elicit CD8+ T cells that are exclusively restricted by MHC-Ia (39, 41). Using the UL128/130-repaired RhCMV clone 68-1.2 as a representative of the latter type of vector configuration, we were able to elicit MHC-Ia–restricted CD8+ T cell responses to RhPAP. Except for their MHC restriction, these responses appeared to be similar to the MHC-II– and MHC-E–restricted responses with respect to breadth, magnitude, duration, and phenotype. This result indicates that the potent and lasting T cell responses elicited by CMV vectors are not limited to unconventionally restricted CD8+ T cells but can also include conventional, MHC-Ia–restricted CD8+ T cells. However, we failed to elicit MHC-Ia–restricted CD8+ T cell responses to TAAs when Rh189, the RhCMV homolog of HCMV US11, was deleted. In the same RM, deletion of Rh189 enabled 68-1 RhCMV to elicit CD8+ T cells to canonical, MHC-Ia–restricted SIV Gag epitopes in addition to MHC-E– and MHC-II–restricted epitopes, consistent with our previous report (39). The lack of such canonical responses upon immunization with Rh189-deleted 68-1–based vectors expressing tumor antigens is likely attributable to the fact that the TAAs used in our studies represent a class of cancer antigens that is also found in healthy tissue, and thus, any canonical MHC-Ia–restricted epitope in these proteins is recognized as “self” by the immune system. In general, T cell responses to self-antigens are limited by central immunological tolerance, i.e., negative selection in the thymus, as well as peripheral tolerance, e.g., by up-regulation of inhibitory coreceptors (66).
While great progress has been made to overcome peripheral tolerance, e.g., by using checkpoint inhibitors, central tolerance is more difficult to overcome because the majority of such T cells have been eliminated from the peripheral pool (66). We therefore interpret our finding that Rh189-deleted RhCMV is unable to elicit MHC-Ia–restricted CD8+ T cells as evidence for the elimination of such MHC-Ia–restricted, canonical T cells in the thymus due to negative selection. In contrast, MHC-Ia–restricted CD8+ T cells elicited by pentamer-repaired 68-1.2 RhCMV likely represent lower avidity T cells recognizing subdominant epitopes that escape negative selection. Immunization with RhCMV thus revealed two different classes of MHC-Ia–restricted TAA-specific T cells, with T cells to canonical epitopes likely generated via direct priming by RhCMV-infected cells upon Rh189-deletion, whereas T cells to noncanonical epitopes likely result from cross-priming of infected cells by professional antigen presenting cells (APC). Since a correlation between cross-priming and T cell responses to subdominant epitopes has been established previously (73), it is conceivable that efficient cross-priming was the reason why pentamer-intact 68-1.2 RhCMV was able to elicit broadly targeted MHC-Ia–restricted T cells to noncanonical epitopes. In contrast, we previously concluded that MHC-E– and MHC-II–restricted CD8+ T cells are elicited by direct priming since the former require expression of Rh67 and can be inhibited by abrogation of infection in miR142-expressing myeloid-derived cells, whereas the latter can be inhibited by abrogation of infection in miR126-expressing endothelial cells (40, 48). Thus, MHC-II– and MHC-E–restricted CD8+ T cells to TAAs likely escape thymic elimination for a different reason, e.g., as a result of the low affinity of peptides targeted by RhCMV-elicited unconventional T cells. CMV-based cancer vaccines thus seem to bypass immunological tolerance mechanisms by either cross-priming MHC-Ia–restricted T cells to noncanonical, subdominant epitopes or direct priming of MHC-II or MHC-E to CD8+ T cells to low-affinity epitopes that are not subject to thymic negative selection. These mechanisms of bypassing immunological tolerance seem to be another unique feature of CMV vectors.
The high conservation of MHC-E between human and nonhuman primates allowed us to use MHC-E–restricted RM T cells to detect PAP-derived epitope presentation by HLA-E on human cancer cell lines and primary cancer cells. The observation that HLA-E expressing cancer cells are able to present a given TAA via HLA-E is consistent with our previous observations that MHC-E expressing T cells or hepatocytes infected with SIV or HBV, respectively, are recognized by MHC-E–restricted T cells (50, 53). While the exact nature of antigen-derived peptides presented on MHC-E still needs to be determined in all of these cases, these results taken together strongly suggest that the presentation of peptides other than VL9 is not an exception but a hitherto underappreciated function of MHC-E. How such non-VL9 peptides are loaded onto MHC-E still needs to be elucidated, but some of the characteristics of the MHC-E molecule render it likely that such peptides are picked up in a post–endoplasmic reticulum compartment upon loss of VL9 and recycling of empty HLA-E molecules (8, 40). Although VL9 is the strongest HLA-E binding peptide, its affinity for HLA-E is low compared to MHC-Ia binding peptide (74). Moreover, unlike MHC-Ia, peptide-free HLA-E is unusually stable in vitro (75, 76). Therefore, it is possible that, upon loss of VL9, endocytosed MHC-E acquires other, non-optimal peptides in recycling endosomes in a process that is more similar to MHC-II loading than MHC-Ia (8, 9). Such non-VL9 peptides are likely bound with lower affinity than VL9 itself, resulting in a transient antigen presentation that is sufficient to be recognized by MHC-E/peptide-specific T cells but insufficient to elicit a de novo T cell response. Such peptides are also difficult to detect by peptide elution from immune-precipitated MHC-I molecules or direct binding assays due to their low affinity and transient interaction (74). However, despite these characteristics, the complexes of non-VL9 peptides and MHC-E can be detected in MHC-E–restricted CD8+ T cells. We thus conclude that cancer cells that express both a given TAA and HLA-E can be targeted by MHC-E–restricted CD8+ T cells.
Our demonstration that HLA-E is up-regulated in PCa is consistent with reports from other cancer cell types (11, 17, 21, 77–80). This up-regulation of HLA-E in cancer correlates with the expression of the inhibitory HLA-E receptor NKG2A/CD94 on a high percentage of TILs. In one report, on average, 50% of tumor-infiltrating CD8+ (but not CD4+) T cells in head and neck cancer biopsies expressed NKG2A, whereas NKG2A was only found on NK cells in PBMC of the same patients (17). The up-regulation of HLA-E in solid tumors thus likely represents an immune evasion mechanism that dampens the antitumor response of tumor-specific or cancer vaccine–induced T cells. Targeting MHC-E/TAA peptide complexes with MHC-E–restricted CD8+ T cells thus potentially turns an immune evasion mechanism into an immunological liability. Moreover, any expression of NKG2A on vaccine-induced MHC-E–restricted CD8+ T cells could be neutralized with NKG2A-targeting antibodies. Presently, modified RhCMV vectors are the only platform capable of eliciting MHC-E–restricted CD8+ T cells to selected antigens. Ongoing clinical trials are asking the question whether this capability can be translated into HCMV-based vectors in humans. Regardless of the outcome of these trials, however, it has already been demonstrated that HLA-E–restricted T cells to non-VL9 peptides derived from HIV can be generated from HIV-naïve individuals using peptide stimulation and tetramer staining (30). Conceivably, a similar approach could be used to elicit TAA peptide–specific, HLA-E–restricted T cells. In turn, the TCRs of such T cells could then be used to generate TCR-transgenic T cells as TAA-targeting immunotherapy. Unlike all other TCR-T cells, these reagents could be used regardless of the recipient’s HLA allotype offering an off-the-shelf approach to TCR T cell therapy. The supertopes identified in these studies could potentially be used to select such TCRs since these epitopes seem to be the common denominator in the CMV-induced T cell response and as such almost certainly involved in generating the cancer cell–recognizing T cells. Moreover, it is possible that TCRs from RhCMV-induced T cells in RMs could be isolated and inserted into human T cells to convey MHC-E/peptide specificity. Thus, both CMV vaccine–based and TCR-T cell–based approaches will be available to target HLA-E on cancer cells.
MATERIALS AND METHODS
Study design
The first objective of this study was to determine whether RhCMV-based vectors can elicit T cell responses to TAAs in RMs despite immunological tolerance. The second objective was to compare the MHC restriction of the TAA-specific CD8+ T cell responses to those observed to heterologous antigens. The third objective was to determine whether MHC-E–restricted, PAP-specific CD8+ T cells elicited in RM would be stimulated by PAP- and HLA-E–expressing PCa cells. The fourth objective was to monitor HLA-E expression in PCa tumor tissue. Objective 2 was dependent on completion of objective 1, specifically the demonstration of T cell responses to cancer antigens. Objectives 3 and 4 were dependent on achieving objective 2, specifically the demonstration of MHC-E–restricted, TAA-specific T cell responses.
Animal studies were approved by the Institutional Animal Care and Use Committee (IACUC). To minimize the number of animals used in these experiments, we coinoculated animals with multiple RhCMV vectors expressing TAAs and control antigens using three to six animals per group. These low numbers were sufficient to map the MHC restriction, including defining shared epitopes (supertopes), of CD8+ T cells. All immunological results were independently observed in multiple experiments by monitoring the same animal over time and in at least three different animals. PAP-specific responses were additionally confirmed in independent cohorts. The ability of RM T cells to recognize PAP-expressing PCa cells in vitro was assessed using multiple cell lines that expressed PAP and HLA-E naturally or that were transfected with PAP and HLA-E. The number of independent experiments and the number of replicates per experiment are indicated in the figure legends.
PCa samples used for IHC and for the generation of PCa cell suspensions were obtained under a protocol that was reviewed and approved by the institutional review board (IRB). All samples were obtained with consent from patients with PCa undergoing prostatectomy. Primary data are reported in data files S1 and S2.
RhCMV strains and recombinants
All RhCMV recombinants were derived from strain 68-1 RhCMV cloned as bacterial artificial chromosome (BAC) (81, 82) (GenBank accession MT157325). Clone 68-1.2 was generated from 68-1 by repair of the antiapoptotic UL36 homolog Rh61/Rh60 and insertion of the UL128, UL130-homologous open reading frames (ORFs) Rh175.4 and Rh157.5 from RhCMV strain 190.82 as described previously (83) (GenBank accession MT157326). RhCMV recombinants were propagated on primary rhesus fibroblasts, telomerized rhesus fibroblasts (TRF), or pp71-expressing TRF, and virus stocks were generated and titered as described previously (42, 60, 81).
To generate 68-1 RhCMV/WT-1 and 68-1 RhCMV/MSLN vectors, we inserted synthetic genes for human WT-1 (GenBank accession NP_001393977.1) or human MSLN (GenBank accession AAH03512.1). The inserts were modified as follows: The N-terminal fragment (amino acids 7 to 385) of WT-1 was codon-optimized without two proline-rich regions (amino acids 127 to 141 and amino acids 188 to 190); the MSLN insert was not codon-optimized and contained the C-terminal fragment of the isoform 1 preprotein starting at amino acid 296. The synthetic genes also included a fusion of the influenza hemagglutinin (HA) epitope tag (YPYDVPDYA) at the C terminus of the encoded proteins. The synthetic genes were inserted into 68-1 RhCMV by replacing the pp71-encoding ORF Rh110 using galactokinase (galK)/aminoglycoside 3′-phosphotransferase (KanR)–mediated positive selection after homologous BAC recombination in the galK-negative Escherichia coli strain SW105 (84). In this two-step process, an expression cassette containing the galK and KanR genes flanked by 80-bp homology arms corresponding to the Rh110 gene borders was introduced into the SW105 E. coli strain carrying the 68-1 RhCMV BAC. Recombinants were selected for kanamycin and chloramphenicol resistance and validated by Sanger sequencing of the inserted region. Next, we replaced the galK/KanR cassette by homologous recombination with polymerase chain reaction (PCR) fragments of the MSNL or WT-1 genes using primers that contained 50-bp homology to the upstream and downstream regions of Rh110 followed by negative selection against the presence of the galK gene on M63 2-deoxy-galactose–containing chloramphenicol plates. The resulting BAC constructs were analyzed by restriction digest and Sanger sequencing of the inserted region and by next-generation sequencing (NGS) of the complete genome using an Illumina MiSeq or iSeq sequencing platform.
To generate the Rh189-deleted versions of 68-1 RhCMV/WT-1 and MSLN, we deleted the Rh189 gene by insertion of galK/KanR using a second round of galK/KanR recombination and selection as described above, followed by replacement of the galK/KanR cassette by homologous recombination with a 100-bp oligonucleotide (containing 50-bp sequences flanking the Rh189 ORF) and selection as described above. The resulting BACs were characterized and sequence verified as described above.
The 68-1 and 68-1.2 RhCMV/RhPAP recombinants were generated using Lambda Red recombination to replace the RhCMV ORF Rh107 with the full-length synthetic gene encoding RhPAP (GenBank accession EHH16763.1) carrying a C-terminal paramyxovirus V5 epitope tag (GKPIPNPLLGLDST). Briefly, the synthetic gene fragments were cloned into pORI and amplified by PCR together with a KanR marker flanked by modified flippase recognition target (FRT-5) sites (85). PCR primers included 50 bp homologous to the Rh107 flanking region so that bacteriophage Lambda recombination enzymes Red α,β,γ-dependent homologous recombination resulted in exchange of Rh107 in either 68-1 or 68-1.2 BAC with RhPAP. The KanR resistance cassette was subsequently removed by induction of the yeast derived flippase (FLP) recombinase leaving an FRT-5 “scar” in the untranslated region. To generate the Rh189-deleted version of 68-1 RhCMV/RhPAP, we additionally deleted Rh189 using Lambda Red recombination. To avoid recombination with the previously retained FRT-5 site, we used wild-type FRT sites flanking the KanR cassette. The final BACs were analyzed as above by restriction digest, Sanger sequencing of inserted regions, and NGS of the entire genome on an Illumina MiSeq sequencer.
Recombinant viruses were reconstituted by electroporation of BAC DNA into primary rhesus fibroblasts. Rh110 (pp71)-deleted vectors were recovered on pp71-expressing fibroblasts as described previously (60). The loxP-flanked BAC cassette encodes a Cre recombinase under control of a mammalian promoter resulting in spontaneous excision of the BAC cassette (86). Viral stocks for in vivo and in vitro experiments were generated, and PFU were determined by 50% tissue culture infective dose on primary rhesus fibroblasts or pp71-expressing TRF. Expression of TAAs was confirmed by immunoblot using HA or V5 epitope tag specific antibodies (fig. S3).
Cell lines
PCa cell lines were obtained from American Type Culture Collection (ATCC) and grown as recommended: LNCaP (ATCC catalog no. CRL-1740) and 22Rv1 (ATCC catalog no. CRL-2505, RRID:CVCL-1045) were maintained in RPMI 1640 medium containing 10% fetal bovine serum (FBS) (Gibco catalog no. 26140-079). VCaP (ATCC catalog no. CRL-2876) and DU145 (ATCC catalog no. HTB-81) cells were maintained in Dulbecco’s modified Eagle/F12 medium (DMEM/F12) (ATCC catalog no. 30-2006) containing 10% FBS.
The chronic myeloid leukemia cell line K562 was obtained from ATCC (ATCC catalog no. CFRL-3344). This cell line lacks MHC-I expression (87) but maintains low levels of MHC-E (88). K562-E cells stably transfected with HLA-E*01:03 (K562-E cells) were generated by transfection with pCEP4-HLA-E*01:03 followed by fluorescence-activated cell sorting as described previously (38). The K562-E/RhPAP cell line was generated by lentivirus transduction after inserting a synthetic gene encoding RhPAP (GenBank accession EHH16763.1) with a C-terminal V5 epitope tag (GKPIPNPLLGLDST) into pLVX EF1α IRES-Puromycin (Clontech). K562-E/RhPAP cells were selected using puromycin (4 μg/ml; Invivogen) and cultured in DMEM supplemented with 10% FBS, penicillin (100 U/ml), and streptomycin (100 μg/ml). Expression of RhPAP was confirmed by immunoblotting using anti-V5 antibody (Invitrogen #37-7500).
To generate the MHC-E–negative K562 derivative cell line AA7, single guide RNA targeting HLA-E was designed and constructed in the CRISPR-Cas9 two part guide RNA (crRNA and tracrRNA, ATT0 488 format by Integrated DNA Technologies). K562 cells were transfected with Cas9 ribonucleoprotein using the AMAXA 4D nucleofector electroporation apparatus (Lonza). At 2 days posttransfection, ATT0 488+ K562 cells were isolated by FACS and incubated at 37°C for 2 days to recover. Transfected cells were then cultured at 27°C overnight to increase HLA-E cell surface expression. The VL9 peptide was added to the transfected cells before staining with anti–HLA-E antibody 3D12. HLA-E–negative cells were isolated by FACS. Cell sorting was performed on a Sony MA900 multi-application cell sorter. Targeted sequencing at the indel site of HLA-E knockout (KO) cells was performed by The Genome Engineering and Stem Cell Center (Washington University School of Medicine in St. Louis), and Cas9 KO efficiency was determined to be >99%. AA7 cells expressing HLA-E (AA7-E) were generated by lentivirus transduction. To generate HLA-E expressing lentivirus, HLA-E 01:03 was cloned into the lentiGuide-Puro vector and transfected into 293T cells along with packaging vector psPAX2 and pseudotyping vector pMD2.G using Lipofectamine 2000 (Thermo Fisher Scientific). At 48 hours posttransduction, HLA-E–positive cells were isolated by FACS as described above.
Acid phosphatases were expressed in AA7-E cells by transfection with pCM6-Entry expression vectors (OriGene) containing human PAP (GenBank accession NM_001134194, OriGene catalog no. RC225669), human ACPT (GenBank accession NM_033068, OriGene catalog no. RC220923), and human ACP2 (GenBank accession NM_001610, OriGene catalog no. RC210562). Cells were selected by including G418 (0.5 mg/ml) in media and maintained in Iscove’s Modified Dulbecco’s Medium (IMDM; Gibco catalog no. 12440-053) containing 10% FBS. Expression of HLA-E and acid phosphatases was verified by flow cytometry and immunoblot.
Immunoblots
Cells were harvested with trypsin/EDTA (Corning 25-051-CI) and washed with PBS. The cell pellets were then resuspended at 1 × 107 cells/ml in water with 1% NP-40 and HALT protease inhibitors (Thermo Fisher Scientific) and incubated at 4°C for 2 hours to lyse the cells. The resulting lysates were centrifuged at 15,000 rpm at 4°C for 45 min. The supernatants were harvested and frozen at −20°C until use. Lysates were separated on 10% polyacrylamide gels and transferred to polyvinylidene difluoride membranes (Millipore). Following blocking with 5% milk in PBS–0.1% Tween-20 (Thermo Fisher Scientific) (PBS-T), membranes were probed with the following antibodies in 5% milk in PBS-T: anti–MHC-E 3D12 (1:1000; Invitrogen 14-9953-82), anti–MHC-E MEM-E/02 (1:5000; Invitrogen MA1-19304), anti-FLAG epitope (1:5000; Sigma-Aldrich #F3165), anti-V5 epitope (1:500; Invitrogen #37-7500), anti–glyceraldehyde phosphate dehydrogenase (GAPDH; 1:5000; Invitrogen #MA5-15738), anti-HA epitope (1:2000; clone HA-7, Sigma-Aldrich H9658), anti-PAP (1:500; MAB6240, R&D Systems) anti–MHC-I heavy chain (1:5000; HC10, Thermo Fisher Scientific), or anti-RhCMV IE2 (15 μg/ml; clone IIA5.2) (53). Membranes were washed three times with PBS-T and then incubated with goat anti-mouse horseradish peroxidase (HRP; 1:5000; Invitrogen #A28177) secondary antibody in 5% milk in PBS-T. Membranes were incubated with Supersignal West Pico chemiluminescent substrate (Thermo Fisher Scientific) and exposed to chemiluminescent film (GE Healthcare) or scanned on a Bio-Rad ChemiDoc MP imager.
Generation of primary PCa cell suspensions
Samples were obtained under a protocol approved by the IRB of Mount Sinai School of Medicine from individuals who consented to the study and underwent prostatectomy (study ID no. 14-00024; GC0 #14-0318). The procedure to isolate primary PCa cells is compliant with federal regulations for use of clinical biospecimens [45 CFR 46.102(f)]. At tissue grossing, ~4-mm3 tissues corresponding to adjacent normal and/or tumor area were freshly dissected by a resident pathologist, and de-identified tissue specimens were transported in magnetic-activated cell sorting (MACS) tissue storage solution (Miltenyi Biotech, catalog no. 130-100-008) supplemented with 10 μM Rock Inhibitor (Y-27632; Selleck Chemicals, catalog no. S1049), to the laboratory. Tissues were washed twice in DMEM and minced into smaller 1-mm3 pieces. The cell suspension was prepared using a tumor dissociation kit (Miltenyi Biotech, catalog no. 130-096-334) and gentle MACS dissociator (program 37C_h_TDK_2) for 60 min. The cell suspension was filtered using a 70-μm strainer and collected by centrifugation at 430g for 10 min. Cryopreserved cells were stored in liquid N2 and transported on dry ice.
Immunohistochemistry
IHC was used to analyze HLA-E expression in PCa TMAs. PCa TMAs (1 to 4) containing a total of 258 tumor cores and 130 matched normal from 86 radical prostatectomy cases were generated in-house (study ID no. 14-00024; MT Sinai Cohort). TMA slides were processed using the Ventana RUO discovery protocol. Antigen retrieval was performed using EDTA (pH 9.0), and tissue cores were incubated with primary anti–HLA-E rabbit polyclonal antibody (1:100 dilution; HPA031454, Sigma Prestige Ab), followed by incubation with HRP-coupled secondary antibody and detection using the Ventana-DAB system. A summary of the results is shown in data file S1.
TMA1 was additionally stained with PIN4 cocktail (CK5 + CK14 + p63 + P504S; prediluted double stain antibody; catalog no. PPM 225 DS AA, H,L; Biocare Medical, CA, USA) or anti-PAP antibody (1:1000; dilution, MO79201-2, clone PASE/4LJ, Dako Laboratories, USA) and processed using the Ventanna RUO discovery protocol. A summary of the results is shown in data file S2.
HLA-E expression was additionally analyzed by IHC in PCa TMA T195c US (Biomax) consisting of 24 cores from 12 cases (pT1-T3N0M0) and 4 cores from normal prostates. TMA slides were processed using the Ventana RUO discovery protocol. Antigen retrieval was performed using EDTA (pH 9.0), and tissue cores were incubated with primary anti–HLA-E mouse mAb 4D12 (1:1000; LSBio, LS-C179742) for 60 min followed by incubation with HRP-coupled secondary antibody and detection using the Ventana-DAB system.
HLA-E expression was further analyzed using IHC in radical prostatectomy specimens (Gleason 9) with perineural invasion or in tumor positive lymph nodes. Five-micrometer formalin-fixed paraffin-embedded (FFPE) sections were processed using the Ventana RUO discovery protocol. Antigen retrieval was performed using EDTA (pH 9.0), and sections were incubated with primary anti–HLA-E rabbit polyclonal antibody (1:100; Sigma-Aldrich) for 60 min followed by incubation with HRP-coupled secondary antibody and detection using the Ventana-DAB system.
Rhesus macaques
A total of 18 purpose-bred male RMs (Macaca mulatta) of Indian genetic background were used in this study. All RMs were classified as specific pathogen free defined by being free of cercopithecine herpesvirus 1, D-type simian retrovirus, simian T-lymphotropic virus type 1, SIV, and M. tuberculosis but naturally infected with RhCMV. All RM used in this study were housed at the Oregon National Primate Research Center (ONPRC) in Animal Biosafety level 2 rooms. RM care and all experimental protocols and procedures were approved by the ONPRC IACUC. The ONPRC is a category I facility. The Laboratory Animal Care and Use Program at the ONPRC is fully accredited by the American Association for Accreditation of Laboratory Animal Care and has an approved assurance (#A3304-01) for the care and use of animals on file with the National Institutes of Health Office for Protection from Research Risks. The IACUC adheres to national guidelines established in the Animal Welfare Act (7 U.S.C. Sections 2131 to 2159) and the Guide for the Care and Use of Laboratory Animals (eighth edition) as mandated by the US Public Health Service Policy. RhCMV vectors were dosed at 1 × 106 to 1 × 107 PFU per vector for immunogenicity analysis, all via subcutaneous administration. Mononuclear cell preparations for immunologic assays were obtained at indicated time points from blood or from frozen splenocytes obtained by necropsy, as previously described (43).
T cell assays
MHC-E–restricted CD8+ T cell responses to cancer cell lines and PCa cells were determined by flow cytometric ICS as described previously for RhCMV-infected or HBV-infected cells (40, 53). Briefly, adherent cells were harvested by trypsinization, and non-adherent cell were harvested and resuspended at 4 × 106 cells/ml in DMEM plus 10% FBS. MHC-blocking reagents were added 2 hours before 50-μl aliquots of these infected cell suspensions (2 × 105 cells) were added to 5 × 105 CD8β+ T cells in 50 μl. The T cells were isolated from PBMC samples of indicated RM using a nonhuman primate CD8+ T cell isolation kit (Miltenyi Biotec) and LS columns (Miltenyi Biotec). The ICS assay for these analyses was performed as described below for PBMC.
TAA and SIV-specific CD4+ and CD8+ T cell responses were measured in PBMC by flow cytometric ICS as described in detail previously (38, 39, 41, 45, 46, 89). T cell responses to total antigens were measured by ICS using mixes of sequential 15–amino acid oligomer peptides (11–amino acid overlap) spanning RhPAP, WT-1, MSLN, or SIVmac239 Gag proteins. T cell responses to individual peptides were measured similarly by ICS, and the MHC restriction type (MHC-Ia, MHC-E, and MHC-II) of a peptide response was determined by preincubating isolated mononuclear cell aliquots with either the pan anti–MHC-I mAb W6/32 (10 μg/ml), the MHC-II–blocking mAb G46.6 (10 μg/ml), or the MHC-E–blocking peptide VMAPRTLLL (VL9; 20 μM). To be considered MHC-E–restricted by blocking, the individual peptide response must have been blocked by both W6/32 and VL9 and not blocked by G46.6. MHC-II–restricted responses were blocked by G46.6 but not W6/32 or VL9, and MHC-Ia–restricted responses were blocked by W6/32 only (38, 39). Responses that did not meet these inhibition criteria were considered indeterminate.
Stained samples were analyzed on an LSR-II or FACSymphony A5 flow cytometer (BD Biosciences). Data analysis was performed using FlowJo software (Tree Star). In all analyses, gating on the lymphocyte population was followed by the separation of the CD3+ T cell subset and progressive gating on CD4+ and CD8+ T cell subsets. Antigen-responding cells in both CD4+ and CD8+ T cell populations were determined by their intracellular expression of CD69 and either or both of the cytokines IFNγ and TNFα [or in polycytokine analyses, expression of CD69 and any combination of the cytokines: IFNγ, TNFα, interleukin-2 (IL-2), and macrophage inflammatory protein-1β (MIP-1β)]. Assay limit of detection was determined as previously described (51), with 0.05% after background subtraction being the minimum threshold used in this study. After background subtraction, the raw response frequencies above the assay limit of detection were “memory-corrected” (i.e., percent responding out of the memory population), as previously described (38, 39, 41, 45, 46, 89). For memory phenotype analysis of antigen-specific T cells, all CD4+ or CD8+ T cells expressing CD69 plus IFNγ and/or TNFα were first Boolean “OR” gated, and then this overall Ag-responding population was subdivided into the memory subsets of interest on the basis of surface phenotype (CCR7 versus CD28). Similarly, for polycytokine analysis of SIV or TAA-specific T cells, all CD4+ or CD8+ T cells expressing CD69 plus cytokines were Boolean “OR” gated, and polyfunctionality was delineated with any combination of the four cytokines tested (IFNγ, TNFα, IL-2, and MIP-1β) using the Boolean “AND” function.
Statistical analysis
Statistical comparison of in vitro T cell responses between groups of assays was performed using paired t test, Student’s t test, or Mann Whitney U test.
Acknowledgments
We thank the ONPRC virology core for generating viral stocks for in vivo experiments. We also thank M. McAllaster, Vir Biotechnology for CRISPR design assistance and H. Tucker for technical assistance with generating modified K562 cells. We thank A. Townsend for help with finalizing the figures and J. Ford for assembling supplemental data files.
Funding: This work was funded by US Army Medical Research and Development Command grant W81XWH1910358 to K.F., sponsored research agreement 84517 by Vir Biotechnology to K.F., the National Institute of Allergy and Infectious Diseases (NIAID) (R01 AI095113, P01 AI094417, and R37 AI054292 to L.J.P.; R01 AI059457 to K.F.), and the National Institutes of Health Office of the Director (P51OD011092) to K.F. and L.J.P.
Author contributions: K.F. and L.J.P. conceived the overall experimental strategy, supervised experiments, acquired funding, analyzed and interpreted data, and wrote the paper. S.H. and M.K.A. planned and performed animal experiments and immunologic assays, assisted by A.S. S.H. additionally contributed to conceptualization, method development, data curation and validation, as well as data visualization together with A.S. A.K.T., N.B., D.C., and S.S.N. contributed to experimental design and interpretation, provided PCa tissues, and supervised or performed IHC. T.H.T. and S.D. performed pathology analysis and scoring of tissue samples and TMA. R.F.I., M.C.V., and D.Mo. designed, performed, and analyzed the T cell experiments with tumor cells. M.M., L.U., and D.Ma. designed, generated, and characterized RhCMV/PAP constructs. M.M. and T.B. characterized antigen expression by cancer cells and RhCMV-infected fibroblasts and helped with data analysis. J.D. and C.M. generated and characterized the WT-1 and MSLN constructs. E.J.L. generated and characterized modified K562 (AA7) cells under supervision by A.K. and C.H.-D. N.J. characterized AA7-derived cell lines.
Competing interests: OHSU, L.J.P., S.H., and K.F. have a substantial financial interest in Vir Biotechnology Inc., a company that may have a commercial interest in the results of this research and technology. L.J.P., S.H., D.Ma., and K.F. are coinventors of OHSU patents licensed to Vir (US2011/036657, US2014/020690, US2016/017373, and US2020/03648). L.J.P., S.H., and K.F. have received compensation for consulting for Vir. The potential individual and institutional conflicts of interest have been reviewed and managed by OHSU. C.M., E.J.L., C.H.-D., J.D., and A.K. performed these experiments as employees of Vir. The authors declare that they have no other competing interests.
Data and materials availability: All data needed to evaluate the conclusions in the paper are present in the paper and/or the Supplementary Materials. All reagents can be obtained from OHSU. The AA7 and AA7-derived cells can be provided by OHSU pending scientific review and a completed material transfer agreement from ATCC for obtaining the parental K562 cells. Requests for the AA7 and AA7-derived cells should be submitted to the senior authors.
Supplementary Materials
This PDF file includes:
Figs. S1 to S7
Legends for data files S1 to S3
Other Supplementary Material for this manuscript includes the following:
Data files S1 to S3
REFERENCES AND NOTES
- 1.Geraghty D. E., Stockschleader M., Ishitani A., Hansen J. A., Polymorphism at the HLA-E locus predates most HLA-A and -B polymorphism. Hum. Immunol. 33, 174–184 (1992). [DOI] [PubMed] [Google Scholar]
- 2.Braud V., Jones E. Y., McMichael A., The human major histocompatibility complex class Ib molecule HLA-E binds signal sequence-derived peptides with primary anchor residues at positions 2 and 9. Eur. J. Immunol. 27, 1164–1169 (1997). [DOI] [PubMed] [Google Scholar]
- 3.Lee N., Goodlett D. R., Ishitani A., Marquardt H., Geraghty D. E., HLA-E surface expression depends on binding of TAP-dependent peptides derived from certain HLA class I signal sequences. J. Immunol. 160, 4951–4960 (1998). [PubMed] [Google Scholar]
- 4.Braud V. M., Allan D. S., Wilson D., McMichael A. J., TAP- and tapasin-dependent HLA-E surface expression correlates with the binding of an MHC class I leader peptide. Curr. Biol. 8, 1–10 (1998). [DOI] [PubMed] [Google Scholar]
- 5.Lee N., Llano M., Carretero M., Ishitani A., Navarro F., Lopez-Botet M., Geraghty D. E., HLA-E is a major ligand for the natural killer inhibitory receptor CD94/NKG2A. Proc. Natl. Acad. Sci. U.S.A. 95, 5199–5204 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Braud V. M., Allan D. S., O’Callaghan C. A., Söderström K., D’Andrea A., Ogg G. S., Lazetic S., Young N. T., Bell J. I., Phillips J. H., Lanier L. L., McMichael A. J., HLA-E binds to natural killer cell receptors CD94/NKG2A, B and C. Nature 391, 795–799 (1998). [DOI] [PubMed] [Google Scholar]
- 7.Ruibal P., Franken K. L. M. C., van Meijgaarden K. E., van Loon J. J. F., van der Steen D., Heemskerk M. H. M., Ottenhoff T. H. M., Joosten S. A., Peptide binding to HLA-E molecules in humans, nonhuman primates, and mice reveals unique binding peptides but remarkably conserved anchor residues. J. Immunol. 205, 2861–2872 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He W., Gea-Mallorqui E., Colin-York H., Fritzsche M., Gillespie G. M., Brackenridge S., Borrow P., McMichael A. J., Intracellular trafficking of HLA-E and its regulation. J. Exp. Med. 220, e20221941 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Camilli G., Cassotta A., Battella S., Palmieri G., Santoni A., Paladini F., Fiorillo M. T., Sorrentino R., Regulation and trafficking of the HLA-E molecules during monocyte-macrophage differentiation. J. Leukoc. Biol. 99, 121–130 (2016). [DOI] [PubMed] [Google Scholar]
- 10.Mancini M., Vidal S. M., Mechanisms of natural killer cell evasion through viral adaptation. Annu. Rev. Immunol. 38, 511–539 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Borst L., van der Burg S. H., van Hall T., The NKG2A-HLA-E axis as a novel checkpoint in the tumor microenvironment. Clin. Cancer Res. 26, 5549–5556 (2020). [DOI] [PubMed] [Google Scholar]
- 12.Pereira B. I., Devine O. P., Vukmanovic-Stejic M., Chambers E. S., Subramanian P., Patel N., Virasami A., Sebire N. J., Kinsler V., Valdovinos A., LeSaux C. J., Passos J. F., Antoniou A., Rustin M. H. A., Campisi J., Akbar A. N., Senescent cells evade immune clearance via HLA-E-mediated NK and CD8+ T cell inhibition. Nat. Commun. 10, 2387 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu X., Song J., Zhang H., Liu X., Zuo F., Zhao Y., Zhao Y., Yin X., Guo X., Wu X., Zhang H., Xu J., Hu J., Jing J., Ma X., Shi H., Immune checkpoint HLA-E:CD94-NKG2A mediates evasion of circulating tumor cells from NK cell surveillance. Cancer Cell 41, 272–287.e9 (2023). [DOI] [PubMed] [Google Scholar]
- 14.Ulbrecht M., Martinozzi S., Grzeschik M., Hengel H., Ellwart J. W., Pla M., Weiss E. H., Cutting edge: The human cytomegalovirus UL40 gene product contains a ligand for HLA-E and prevents NK cell-mediated lysis. J. Immunol. 164, 5019–5022 (2000). [DOI] [PubMed] [Google Scholar]
- 15.Tomasec P., Braud V. M., Rickards C., Powell M. B., McSharry B. P., Gadola S., Cerundolo V., Borysiewicz L. K., McMichael A. J., Wilkinson G. W., Surface expression of HLA-E, an inhibitor of natural killer cells, enhanced by human cytomegalovirus gpUL40. Science 287, 1031 (2000). [DOI] [PubMed] [Google Scholar]
- 16.Fisher J. G., Doyle A. D. P., Graham L. V., Khakoo S. I., Blunt M. D., Disruption of the NKG2A:HLA-E immune checkpoint axis to enhance NK cell activation against cancer. Vaccine 10, 1993 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Montfoort N., Borst L., Korrer M. J., Sluijter M., Marijt K. A., Santegoets S. J., van Ham V. J., Ehsan I., Charoentong P., André P., Wagtmann N., Welters M. J. P., Kim Y. J., Piersma S. J., van der Burg S. H., van Hall T., NKG2A blockade potentiates CD8 T cell immunity induced by cancer vaccines. Cell 175, 1744–1755.e15 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Abd Hamid M., Wang R.-Z., Yao X., Fan P., Li X., Chang X.-M., Feng Y., Jones S., Maldonado-Perez D., Waugh C., Verrill C., Simmons A., Cerundolo V., McMichael A., Conlon C., Wang X., Peng Y., Dong T., Enriched HLA-E and CD94/NKG2A interaction limits antitumor CD8+ tumor-infiltrating T lymphocyte responses. Cancer Immunol. Res. 7, 1293–1306 (2019). [DOI] [PubMed] [Google Scholar]
- 19.Tinker A. V., Hirte H. W., Provencher D., Butler M., Ritter H., Tu D., Azim H. A. Jr., Paralejas P., Grenier N., Hahn S. A., Ramsahai J., Seymour L., Dose-ranging and cohort-expansion study of monalizumab (IPH2201) in patients with advanced gynecologic malignancies: A trial of the canadian cancer trials group (CCTG): IND221. Clin. Cancer Res. 25, 6052–6060 (2019). [DOI] [PubMed] [Google Scholar]
- 20.André P., Denis C., Soulas C., Bourbon-Caillet C., Lopez J., Arnoux T., Bléry M., Bonnafous C., Gauthier L., Morel A., Rossi B., Remark R., Breso V., Bonnet E., Habif G., Guia S., Lalanne A. I., Hoffmann C., Lantz O., Fayette J., Boyer-Chammard A., Zerbib R., Dodion P., Ghadially H., Jure-Kunkel M., Morel Y., Herbst R., Narni-Mancinelli E., Cohen R. B., Vivier E., Anti-NKG2A mAb is a checkpoint inhibitor that promotes anti-tumor immunity by unleashing both T and NK cells. Cell 175, 1731–1743.e13 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.van Hall T., Andre P., Horowitz A., Ruan D. F., Borst L., Zerbib R., Narni-Mancinelli E., van der Burg S. H., Vivier E., Monalizumab: Inhibiting the novel immune checkpoint NKG2A. J. Immunother. Cancer 7, 263 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García P., Llano M., de Heredia A. B., Willberg C. B., Caparrós E., Aparicio P., Braud V. M., López-Botet M., Human T cell receptor-mediated recognition of HLA-E. Eur. J. Immunol. 32, 936–944 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Grant E. J., Nguyen A. T., Lobos C. A., Szeto C., Chatzileontiadou D. S. M., Gras S., The unconventional role of HLA-E: The road less traveled. Mol. Immunol. 120, 101–112 (2020). [DOI] [PubMed] [Google Scholar]
- 24.McMurtrey C., Harriff M. J., Swarbrick G. M., Duncan A., Cansler M., Null M., Bardet W., Jackson K. W., Lewinsohn D. A., Hildebrand W., Lewinsohn D. M., T cell recognition of Mycobacterium tuberculosis peptides presented by HLA-E derived from infected human cells. PLOS ONE 12, e0188288 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salerno-Gonçalves R., Fernandez-Viña M., Lewinsohn D. M., Sztein M. B., Identification of a human HLA-E-restricted CD8+ T cell subset in volunteers immunized with Salmonella enterica serovar Typhi strain Ty21a typhoid vaccine. J. Immunol. 173, 5852–5862 (2004). [DOI] [PubMed] [Google Scholar]
- 26.Heinzel A. S., Grotzke J. E., Lines R. A., Lewinsohn D. A., McNabb A. L., Streblow D. N., Braud V. M., Grieser H. J., Belisle J. T., Lewinsohn D. M., HLA-E-dependent presentation of Mtb-derived antigen to human CD8+ T cells. J. Exp. Med. 196, 1473–1481 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sullivan L. C., Nguyen T. H. O., Harpur C. M., Stankovic S., Kanagarajah A. R., Koutsakos M., Saunders P. M., Cai Z., Gray J. A., Widjaja J. M. L., Lin J., Pietra G., Mingari M. C., Moretta L., Samir J., Luciani F., Westall G. P., Malmberg K. J., Kedzierska K., Brooks A. G., Natural killer cell receptors regulate responses of HLA-E-restricted T cells. Sci. Immunol. 6, eabe9057 (2021). [DOI] [PubMed] [Google Scholar]
- 28.Jouand N., Bressollette-Bodin C., Gerard N., Giral M., Guerif P., Rodallec A., Oger R., Parrot T., Allard M., Cesbron-Gautier A., Gervois N., Charreau B., HCMV triggers frequent and persistent UL40-specific unconventional HLA-E-restricted CD8 T-cell responses with potential autologous and allogeneic peptide recognition. PLOS Pathog. 14, e1007041 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pietra G., Romagnani C., Mazzarino P., Falco M., Millo E., Moretta A., Moretta L., Mingari M. C., HLA-E-restricted recognition of cytomegalovirus-derived peptides by human CD8+ cytolytic T lymphocytes. Proc. Natl. Acad. Sci. U.S.A. 100, 10896–10901 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang H., Rei M., Brackenridge S., Brenna E., Sun H., Abdulhaqq S., Liu M. K. P., Ma W., Kurupati P., Xu X., Cerundolo V., Jenkins E., Davis S. J., Sacha J. B., Früh K., Picker L. J., Borrow P., Gillespie G. M., McMichael A. J., HLA-E-restricted, Gag-specific CD8+ T cells can suppress HIV-1 infection, offering vaccine opportunities. Sci. Immunol. 6, eabg1703 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yang H., Sun H., Brackenridge S., Zhuang X., Wing P. A. C., Quastel M., Walters L., Garner L., Wang B., Yao X., Felce S. L., Peng Y., Moore S., Peeters B. W. A., Rei M., Gomes J. C., Tomas A., Davidson A., Semple M. G., Turtle L. C. W., Openshaw P. J. M., Baillie J. K., Mentzer A. J., Klenerman P., Investigators I. C., Borrow P., Dong T., McKeating J. A., Gillespie G. M., McMichael A. J., HLA-E-restricted SARS-CoV-2-specific T cells from convalescent COVID-19 patients suppress virus replication despite HLA class Ia down-regulation. Sci. Immunol. 8, eabl8881 (2023). [DOI] [PubMed] [Google Scholar]
- 32.Terrazzano G., Sica M., Gianfrani C., Mazzarella G., Maurano F., De Giulio B., de Saint-Mezard S., Zanzi D., Maiuri L., Londei M., Jabri B., Troncone R., Auricchio S., Zappacosta S., Carbone E., Gliadin regulates the NK-dendritic cell cross-talk by HLA-E surface stabilization. J. Immunol. 179, 372–381 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Wooden S. L., Kalb S. R., Cotter R. J., Soloski M. J., Cutting edge: HLA-E binds a peptide derived from the ATP-binding cassette transporter multidrug resistance-associated protein 7 and inhibits NK cell-mediated lysis. J. Immunol. 175, 1383–1387 (2005). [DOI] [PubMed] [Google Scholar]
- 34.Michaelsson J., Teixeira de Matos C., Achour A., Lanier L. L., Karre K., Soderstrom K., A signal peptide derived from hsp60 binds HLA-E and interferes with CD94/NKG2A recognition. J. Exp. Med. 196, 1403–1414 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pietra G., Romagnani C., Manzini C., Moretta L., Mingari M. C., The emerging role of HLA-E-restricted CD8+ T lymphocytes in the adaptive immune response to pathogens and tumors. J. Biomed. Biotechnol. 2010, 907092 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Celik A. A., Kraemer T., Huyton T., Blasczyk R., Bade-Doding C., The diversity of the HLA-E-restricted peptide repertoire explains the immunological impact of the Arg107Gly mismatch. Immunogenetics 68, 29–41 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lampen M. H., Hassan C., Sluijter M., Geluk A., Dijkman K., Tjon J. M., de Ru A. H., van der Burg S. H., van Veelen P. A., van Hall T., Alternative peptide repertoire of HLA-E reveals a binding motif that is strikingly similar to HLA-A2. Mol. Immunol. 53, 126–131 (2013). [DOI] [PubMed] [Google Scholar]
- 38.Hansen S. G., Wu H. L., Burwitz B. J., Hughes C. M., Hammond K. B., Ventura A. B., Reed J. S., Gilbride R. M., Ainslie E., Morrow D. W., Ford J. C., Selseth A. N., Pathak R., Malouli D., Legasse A. W., Axthelm M. K., Nelson J. A., Gillespie G. M., Walters L. C., Brackenridge S., Sharpe H. R., López C. A., Früh K., Korber B. T., McMichael A. J., Gnanakaran S., Sacha J. B., Picker L. J., Broadly targeted CD8+ T cell responses restricted by major histocompatibility complex E. Science 351, 714–720 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hansen S. G., Sacha J. B., Hughes C. M., Ford J. C., Burwitz B. J., Scholz I., Gilbride R. M., Lewis M. S., Gilliam A. N., Ventura A. B., Malouli D., Xu G., Richards R., Whizin N., Reed J. S., Hammond K. B., Fischer M., Turner J. M., Legasse A. W., Axthelm M. K., Edlefsen P. T., Nelson J. A., Lifson J. D., Früh K., Picker L. J., Cytomegalovirus vectors violate CD8+ T cell epitope recognition paradigms. Science 340, 1237874 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verweij M. C., Hansen S. G., Iyer R., John N., Malouli D., Morrow D., Scholz I., Womack J., Abdulhaqq S., Gilbride R. M., Hughes C. M., Ventura A. B., Ford J. C., Selseth A. N., Oswald K., Shoemaker R., Berkemeier B., Bosche W. J., Hull M., Shao J., Sacha J. B., Axthelm M. K., Edlefsen P. T., Lifson J. D., Picker L. J., Früh K., Modulation of MHC-E transport by viral decoy ligands is required for RhCMV/SIV vaccine efficacy. Science 372, eabe9233 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Malouli D., Hansen S. G., Hancock M. H., Hughes C. M., Ford J. C., Gilbride R. M., Ventura A. B., Morrow D., Randall K. T., Taher H., Uebelhoer L. S., McArdle M. R., Papen C. R., Trethewy R. E., Oswald K., Shoemaker R., Berkemeier B., Bosche W. J., Hull M., Greene J. M., Axthelm M. K., Shao J., Edlefsen P. T., Grey F., Nelson J. A., Lifson J. D., Streblow D., Sacha J. B., Früh K., Picker L. J., Cytomegaloviral determinants of CD8+ T cell programming and RhCMV/SIV vaccine efficacy. Sci. Immunol. 6, eabg5413 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taher H., Mahyari E., Kreklywich C., Uebelhoer L. S., McArdle M. R., Moström M. J., Bhusari A., Nekorchuk M., E X., Whitmer T., Scheef E. A., Sprehe L. M., Roberts D. L., Hughes C. M., Jackson K. A., Selseth A. N., Ventura A. B., Cleveland-Rubeor H. C., Yue Y., Schmidt K. A., Shao J., Edlefsen P. T., Smedley J., Kowalik T. F., Stanton R. J., Axthelm M. K., Estes J. D., Hansen S. G., Kaur A., Barry P. A., Bimber B. N., Picker L. J., Streblow D. N., Früh K., Malouli D., In vitro and in vivo characterization of a recombinant rhesus cytomegalovirus containing a complete genome. PLOS Pathog. 16, e1008666 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hansen S. G., Piatak M. Jr., Ventura A. B., Hughes C. M., Gilbride R. M., Ford J. C., Oswald K., Shoemaker R., Li Y., Lewis M. S., Gilliam A. N., Xu G., Whizin N., Burwitz B. J., Planer S. L., Turner J. M., Legasse A. W., Axthelm M. K., Nelson J. A., Früh K., Sacha J. B., Estes J. D., Keele B. F., Edlefsen P. T., Lifson J. D., Picker L. J., Immune clearance of highly pathogenic SIV infection. Nature 502, 100–104 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Picker L. J., Hansen S. G., Lifson J. D., New paradigms for HIV/AIDS vaccine development. Annu. Rev. Med. 63, 95–111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hansen S. G., Ford J. C., Lewis M. S., Ventura A. B., Hughes C. M., Coyne-Johnson L., Whizin N., Oswald K., Shoemaker R., Swanson T., Legasse A. W., Chiuchiolo M. J., Parks C. L., Axthelm M. K., Nelson J. A., Jarvis M. A., Piatak M. Jr., Lifson J. D., Picker L. J., Profound early control of highly pathogenic SIV by an effector memory T-cell vaccine. Nature 473, 523–527 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hansen S. G., Vieville C., Whizin N., Coyne-Johnson L., Siess D. C., Drummond D. D., Legasse A. W., Axthelm M. K., Oswald K., Trubey C. M., Piatak M. Jr., Lifson J. D., Nelson J. A., Jarvis M. A., Picker L. J., Addendum: Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 17, 1692 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Malouli D., Gilbride R. M., Wu H. L., Hwang J. M., Maier N., Hughes C. M., Newhouse D., Morrow D., Ventura A. B., Law L., Tisoncik-Go J., Whitmore L., Smith E., Golez I., Chang J., Reed J. S., Waytashek C., Weber W., Taher H., Uebelhoer L. S., Womack J. L., McArdle M. R., Gao J., Papen C. R., Lifson J. D., Burwitz B. J., Axthelm M. K., Smedley J., Früh K., Gale M. Jr., Picker L. J., Hansen S. G., Sacha J. B., Cytomegalovirus-vaccine-induced unconventional T cell priming and control of SIV replication is conserved between primate species. Cell Host Microbe 30, 1207–1218.e7 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hansen S. G., Hancock M. H., Malouli D., Marshall E. E., Hughes C. M., Randall K. T., Morrow D., Ford J. C., Gilbride R. M., Selseth A. N., Trethewy R. E., Bishop L. M., Oswald K., Shoemaker R., Berkemeier B., Bosche W. J., Hull M., Silipino L., Nekorchuk M., Busman-Sahay K., Estes J. D., Axthelm M. K., Smedley J., Shao D., Edlefsen P. T., Lifson J. D., Früh K., Nelson J. A., Picker L. J., Myeloid cell tropism enables MHC-E-restricted CD8+ T cell priming and vaccine efficacy by the RhCMV/SIV vaccine. Sci. Immunol. 7, eabn9301 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Caposio P., Van Den Worm S., Crawford L., Perez W., Kreklywich C., Gilbride R. M., Hughes C. M., Ventura A. B., Ratts R., Marshall E. E., Malouli D., Axthelm M. K., Streblow D., Nelson J. A., Picker L. J., Hansen S. G., Früh K., Characterization of a live-attenuated HCMV-based vaccine platform. Sci. Rep. 9, 19236 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wu H. L., Wiseman R. W., Hughes C. M., Webb G. M., Abdulhaqq S. A., Bimber B. N., Hammond K. B., Reed J. S., Gao L., Burwitz B. J., Greene J. M., Ferrer F., Legasse A. W., Axthelm M. K., Park B. S., Brackenridge S., Maness N. J., McMichael A. J., Picker L. J., O’Connor D. H., Hansen S. G., Sacha J. B., The role of MHC-E in T cell immunity is conserved among humans, rhesus macaques, and cynomolgus macaques. J. Immunol. 200, 49–60 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen S. G., Zak D. E., Xu G., Ford J. C., Marshall E. E., Malouli D., Gilbride R. M., Hughes C. M., Ventura A. B., Ainslie E., Randall K. T., Selseth A. N., Rundstrom P., Herlache L., Lewis M. S., Park H., Planer S. L., Turner J. M., Fischer M., Armstrong C., Zweig R. C., Valvo J., Braun J. M., Shankar S., Lu L., Sylwester A. W., Legasse A. W., Messerle M., Jarvis M. A., Amon L. M., Aderem A., Alter G., Laddy D. J., Stone M., Bonavia A., Evans T. G., Axthelm M. K., Früh K., Edlefsen P. T., Picker L. J., Prevention of tuberculosis in rhesus macaques by a cytomegalovirus-based vaccine. Nat. Med. 24, 130–143 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hansen S. G., Womack J., Scholz I., Renner A., Edgel K. A., Xu G., Ford J. C., Grey M., Laurent B. S., Turner J. M., Planer S., Legasse A. W., Richie T. L., Aguiar J. C., Axthelm M. K., Villasante E. D., Weiss W., Edlefsen P. T., Picker L. J., Früh K., Cytomegalovirus vectors expressing Plasmodium knowlesi antigens induce immune responses that delay parasitemia upon sporozoite challenge. PLOS ONE 14, e0210252 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Burwitz B. J., Hashiguchi P. K., Mansouri M., Meyer C., Gilbride R. M., Biswas S., Womack J. L., Reed J. S., Wu H. L., Axthelm M. K., Hansen S. G., Picker L. J., Früh K., Sacha J. B., MHC-E-restricted CD8+ T cells target hepatitis B virus-infected human hepatocytes. J. Immunol. 204, 2169–2176 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Buonaguro L., Tagliamonte M., Selecting target antigens for cancer vaccine development. Vaccine 8, 615 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cheever M. A., Higano C. S., PROVENGE (Sipuleucel-T) in prostate cancer: The first FDA-approved therapeutic cancer vaccine. Clin. Cancer Res. 17, 3520–3526 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Sims R. B., Development of sipuleucel-T: Autologous cellular immunotherapy for the treatment of metastatic castrate resistant prostate cancer. Vaccine 30, 4394–4397 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Bansal D., Reimers M. A., Knoche E. M., Pachynski R. K., Immunotherapy and immunotherapy combinations in metastatic castration-resistant prostate cancer. Cancer 13, 334 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chapuis A. G., Egan D. N., Bar M., Schmitt T. M., McAfee M. S., Paulson K. G., Voillet V., Gottardo R., Ragnarsson G. B., Bleakley M., Yeung C. C., Muhlhauser P., Nguyen H. N., Kropp L. A., Castelli L., Wagener F., Hunter D., Lindberg M., Cohen K., Seese A., McElrath M. J., Duerkopp N., Gooley T. A., Greenberg P. D., T cell receptor gene therapy targeting WT1 prevents acute myeloid leukemia relapse post-transplant. Nat. Med. 25, 1064–1072 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hassan R., Thomas A., Alewine C., Le D. T., Jaffee E. M., Pastan I., Mesothelin immunotherapy for cancer: Ready for prime time? J. Clin. Oncol. 34, 4171–4179 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Marshall E. E., Malouli D., Hansen S. G., Gilbride R. M., Hughes C. M., Ventura A. B., Ainslie E., Selseth A. N., Ford J. C., Burke D., Kreklywich C. N., Womack J., Legasse A. W., Axthelm M. K., Kahl C., Streblow D., Edlefsen P. T., Picker L. J., Früh K., Enhancing safety of cytomegalovirus-based vaccine vectors by engaging host intrinsic immunity. Sci. Transl. Med. 11, eaaw2603 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hansen S. G., Vieville C., Whizin N., Coyne-Johnson L., Siess D. C., Drummond D. D., Legasse A. W., Axthelm M. K., Oswald K., Trubey C. M., Piatak M. Jr., Lifson J. D., Nelson J. A., Jarvis M. A., Picker L. J., Effector memory T cell responses are associated with protection of rhesus monkeys from mucosal simian immunodeficiency virus challenge. Nat. Med. 15, 293–299 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hansen S. G., Powers C. J., Richards R., Ventura A. B., Ford J. C., Siess D., Axthelm M. K., Nelson J. A., Jarvis M. A., Picker L. J., Früh K., Evasion of CD8+ T cells is critical for superinfection by cytomegalovirus. Science 328, 102–106 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Quintero I. B., Araujo C. L., Pulkka A. E., Wirkkala R. S., Herrala A. M., Eskelinen E.-L., Jokitalo E., Hellstrom P. A., Tuominen H. J., Hirvikoski P. P., Vihko P. T., Prostatic acid phosphatase is not a prostate specific target. Cancer Res. 67, 6549–6554 (2007). [DOI] [PubMed] [Google Scholar]
- 64.Pritchard-Jones K., Fleming S., Davidson D., Bickmore W., Porteous D., Gosden C., Bard J., Buckler A., Pelletier J., Housman D., van Heyningen V., Hastie N., The candidate Wilms’ tumour gene is involved in genitourinary development. Nature 346, 194–197 (1990). [DOI] [PubMed] [Google Scholar]
- 65.Ordóñez N. G., Application of mesothelin immunostaining in tumor diagnosis. Am. J. Surg. Pathol. 27, 1418–1428 (2003). [DOI] [PubMed] [Google Scholar]
- 66.Makkouk A., Weiner G. J., Cancer immunotherapy and breaking immune tolerance: New approaches to an old challenge. Cancer Res. 75, 5–10 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wiertz E. J., Jones T. R., Sun L., Bogyo M., Geuze H. J., Ploegh H. L., The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell 84, 769–779 (1996). [DOI] [PubMed] [Google Scholar]
- 68.Pande N. T., Powers C., Ahn K., Früh K., Rhesus cytomegalovirus contains functional homologues of US2, US3, US6, and US11. J. Virol. 79, 5786–5798 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Früh K., Picker L., CD8+ T cell programming by cytomegalovirus vectors: Applications in prophylactic and therapeutic vaccination. Curr. Opin. Immunol. 47, 52–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sharief F. S., Lee H., Leuderman M. M., Lundwall A., Deaven L. L., Lee C. L., Li S. S., Human prostatic acid phosphatase: cDNA cloning, gene mapping and protein sequence homology with lysosomal acid phosphatase. Biochem. Biophys. Res. Commun. 160, 79–86 (1989). [DOI] [PubMed] [Google Scholar]
- 71.Yousef G. M., Diamandis M., Jung K., Diamandis E. P., Molecular cloning of a novel human acid phosphatase gene (ACPT) that is highly expressed in the testis. Genomics 74, 385–395 (2001). [DOI] [PubMed] [Google Scholar]
- 72.Ghandi M., Huang F. W., Jané-Valbuena J., Kryukov G. V., Lo C. C., McDonald E. R. III, Barretina J., Gelfand E. T., Bielski C. M., Li H., Hu K., Andreev-Drakhlin A. Y., Kim J., Hess J. M., Haas B. J., Aguet F., Weir B. A., Rothberg M. V., Paolella B. R., Lawrence M. S., Akbani R., Lu Y., Tiv H. L., Gokhale P. C., de Weck A., Mansour A. A., Oh C., Shih J., Hadi K., Rosen Y., Bistline J., Venkatesan K., Reddy A., Sonkin D., Liu M., Lehar J., Korn J. M., Porter D. A., Jones M. D., Golji J., Caponigro G., Taylor J. E., Dunning C. M., Creech A. L., Warren A. C., McFarland J. M., Zamanighomi M., Kauffmann A., Stransky N., Imielinski M., Maruvka Y. E., Cherniack A. D., Tsherniak A., Vazquez F., Jaffe J. D., Lane A. A., Weinstock D. M., Johannessen C. M., Morrissey M. P., Stegmeier F., Schlegel R., Hahn W. C., Getz G., Mills G. B., Boehm J. S., Golub T. R., Garraway L. A., Sellers W. R., Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 569, 503–508 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Otahal P., Hutchinson S. C., Mylin L. M., Tevethia M. J., Tevethia S. S., Schell T. D., Inefficient cross-presentation limits the CD8+ T cell response to a subdominant tumor antigen epitope. J. Immunol. 175, 700–712 (2005). [DOI] [PubMed] [Google Scholar]
- 74.Walters L. C., McMichael A. J., Gillespie G. M., Detailed and atypical HLA-E peptide binding motifs revealed by a novel peptide exchange binding assay. Eur. J. Immunol. 50, 2075–2091 (2020). [DOI] [PubMed] [Google Scholar]
- 75.Walters L. C., Harlos K., Brackenridge S., Rozbesky D., Barrett J. R., Jain V., Walter T. S., O’Callaghan C. A., Borrow P., Toebes M., Hansen S. G., Sacha J., Abdulhaqq S., Greene J. M., Früh K., Marshall E., Picker L. J., Jones E. Y., McMichael A. J., Gillespie G. M., Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat. Commun. 9, 3137 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Walters L. C., Harlos K., Brackenridge S., Rozbesky D., Barrett J. R., Jain V., Walter T. S., O’Callaghan C. A., Borrow P., Toebes M., Hansen S. G., Sacha J. B., Abdulhaqq S., Greene J. M., Fruh K., Marshall E., Picker L. J., Jones E. Y., McMichael A. J., Gillespie G. M., Pathogen-derived HLA-E bound epitopes reveal broad primary anchor pocket tolerability and conformationally malleable peptide binding. Nat. Commun. 9, 4833 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kamiya T., Seow S. V., Wong D., Robinson M., Campana D., Blocking expression of inhibitory receptor NKG2A overcomes tumor resistance to NK cells. J. Clin. Invest. 129, 2094–2106 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Seliger B., Jasinski-Bergner S., Quandt D., Stoehr C., Bukur J., Wach S., Legal W., Taubert H., Wullich B., Hartmann A., HLA-E expression and its clinical relevance in human renal cell carcinoma. Oncotarget 7, 67360–67372 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Derre L., Corvaisier M., Charreau B., Moreau A., Godefroy E., Moreau-Aubry A., Jotereau F., Gervois N., Expression and release of HLA-E by melanoma cells and melanocytes: Potential impact on the response of cytotoxic effector cells. J. Immunol. 177, 3100–3107 (2006). [DOI] [PubMed] [Google Scholar]
- 80.Marin R., Ruiz-Cabello F., Pedrinaci S., Mendez R., Jimenez P., Geraghty D. E., Garrido F., Analysis of HLA-E expression in human tumors. Immunogenetics 54, 767–775 (2003). [DOI] [PubMed] [Google Scholar]
- 81.Chang W. L., Tarantal A. F., Zhou S. S., Borowsky A. D., Barry P. A., A recombinant rhesus cytomegalovirus expressing enhanced green fluorescent protein retains the wild-type phenotype and pathogenicity in fetal macaques. J. Virol. 76, 9493–9504 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Malouli D., Nakayasu E. S., Viswanathan K., Camp D. G. II, Chang W. L., Barry P. A., Smith R. D., Fruh K., Reevaluation of the coding potential and proteomic analysis of the BAC-derived rhesus cytomegalovirus strain 68-1. J. Virol. 86, 8959–8973 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lilja A. E., Shenk T., Efficient replication of rhesus cytomegalovirus variants in multiple rhesus and human cell types. Proc. Natl. Acad. Sci. U.S.A. 105, 19950–19955 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G., Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33, e36 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Borst E. M., Benkartek C., Messerle M., Use of bacterial artificial chromosomes in generating targeted mutations in human and mouse cytomegaloviruses. Curr. Protoc. Immunol. Chapter 10, 10.32.1-10.32.30 (2007). [DOI] [PubMed] [Google Scholar]
- 86.Yu D., Smith G. A., Enquist L. W., Shenk T., Construction of a self-excisable bacterial artificial chromosome containing the human cytomegalovirus genome and mutagenesis of the diploid TRL/IRL13 gene. J. Virol. 76, 2316–2328 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Drew S. I., Terasaki P. I., Billing R. J., Bergh O. J., Minowada J., Klein E., Group-specific human granulocyte antigens on a chronic myelogenous leukemia cell line with a Philadelphia chromosome marker. Blood 49, 715–718 (1977). [PubMed] [Google Scholar]
- 88.Lo Monaco E., Tremante E., Cerboni C., Melucci E., Sibilio L., Zingoni A., Nicotra M. R., Natali P. G., Giacomini P., Human leukocyte antigen E contributes to protect tumor cells from lysis by natural killer cells. Neoplasia 13, 822–830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hansen S. G., Marshall E. E., Malouli D., Ventura A. B., Hughes C. M., Ainslie E., Ford J. C., Morrow D., Gilbride R. M., Bae J. Y., Legasse A. W., Oswald K., Shoemaker R., Berkemeier B., Bosche W. J., Hull M., Womack J., Shao J., Edlefsen P. T., Reed J. S., Burwitz B. J., Sacha J. B., Axthelm M. K., Früh K., Lifson J. D., Picker L. J., A live-attenuated RhCMV/SIV vaccine shows long-term efficacy against heterologous SIV challenge. Sci. Transl. Med. 11, eaaw2607 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tremante E., Lo Monaco E., Ingegnere T., Sampaoli C., Fraioli R., Giacomini P., Monoclonal antibodies to HLA-E bind epitopes carried by unfolded β2 m-free heavy chains. Eur. J. Immunol. 45, 2356–2364 (2015). [DOI] [PubMed] [Google Scholar]
- 91.Lo Monaco E., Sibilio L., Melucci E., Tremante E., Suchanek M., Horejsi V., Martayan A., Giacomini P., HLA-E: Strong association with β2-microglobulin and surface expression in the absence of HLA class I signal sequence-derived peptides. J. Immunol. 181, 5442–5450 (2008). [DOI] [PubMed] [Google Scholar]
- 92.Stam N. J., Spits H., Ploegh H. L., Monoclonal antibodies raised against denatured HLA-B locus heavy chains permit biochemical characterization of certain HLA-C locus products. J. Immunol. 137, 2299–2306 (1986). [PubMed] [Google Scholar]
- 93.Lin M. F., Lee C. L., Clinton G. M., Tyrosyl kinase activity is inversely related to prostatic acid phosphatase activity in two human prostate carcinoma cell lines. Mol. Cell. Biol. 6, 4753–4757 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Namekawa T., Ikeda K., Horie-Inoue K., Inoue S., Application of prostate cancer models for preclinical study: Advantages and limitations of cell lines, patient-derived xenografts, and three-dimensional culture of patient-derived cells. Cells 8, 74 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figs. S1 to S7
Legends for data files S1 to S3
Data files S1 to S3