Abstract
The human immunodeficiency virus type 1 (HIV-1) Gag precursor, Pr55Gag, is necessary and sufficient for the assembly and release of viruslike particles. Binding of Gag to membrane and Gag multimerization are both essential steps in virus assembly, yet the domains responsible for these events have not been fully defined. In addition, the relationship between membrane binding and Gag-Gag interaction remains to be elucidated. To investigate these issues, we analyzed, in vivo, the membrane-binding and assembly properties of a series of C-terminally truncated Gag mutants. Pr55Gag was truncated at the C terminus of matrix (MAstop), between the N- and C-terminal domains of capsid (CA146stop), at the C terminus of capsid (p41stop), at the C terminus of p2 (p43stop), and after the N-terminal 35 amino acids of nucleocapsid (NC35stop). The ability of these truncated Gag molecules to assemble and release viruslike particles and their capacity to copackage into particles when coexpressed with full-length Gag were determined. We demonstrate that the amount of truncated Gag incorporated into particles is incrementally increased by extension from CA146 to NC35, suggesting that multiple sites in this region are involved in Gag multimerization. Using membrane flotation centrifugation, we observe that MA shows significantly reduced membrane binding relative to full-length Gag but that CA146 displays steady-state membrane-binding properties comparable to those of Pr55Gag. The finding that the CA146 mutant, which contains only matrix and the N-terminal domain of capsid, exhibits levels of steady-state membrane binding equivalent to those of full-length Gag indicates that strong Gag-Gag interaction domains are not required for the efficient binding of HIV-1 Gag to membrane.
The production of lentivirus particles from infected cells involves multiple steps, including viral protein transport to and association with the plasma membrane, assembly of the structural proteins, encapsidation of the genomic RNA, and particle release (for reviews, see references 11 and 49). The Gag polyprotein precursor plays a central role in these events and, in fact, is both necessary and sufficient for the assembly and release of immature viruslike particles (VLPs). The human immunodeficiency virus type 1 (HIV-1) Gag precursor, Pr55Gag, is composed of four major domains: matrix (MA or p17), capsid (CA or p24), nucleocapsid (NC or p7), and p6. Two spacer peptides are also present within Pr55Gag: p2, located between CA and NC, and p1, located between NC and p6. Pr55Gag is cleaved by the viral protease (PR) during or immediately after virus budding from the cell to produce the mature Gag proteins.
The primary determinants of Gag membrane binding appear to lie within MA. This domain contains a signal for N-terminal myristylation, and it is well established that the covalent modification of Gag with myristic acid is essential for efficient membrane binding of Pr55Gag (11). Accumulating evidence suggests that not only the presence but also the degree of exposure of this fatty acid is important for Gag membrane binding and that the latter may be controlled by the conformation of MA. This hypothesis, referred to as the myristyl switch model, originally derived from studies involving cellular proteins (e.g., recoverin) that bind membrane in a reversible manner (3, 34). The myristyl switch model has been applied to HIV-1 Gag to explain the observation that membrane binding of mature p17(MA) is much weaker than that of Pr55Gag. According to this model, the myristate moiety is highly exposed in the context of Pr55Gag but is sequestered within p17(MA) following proteolytic cleavage of the precursor by PR (21, 48, 63). We have shown that single amino acid substitutions in MA can increase or decrease membrane binding of Pr55Gag in a manner dependent on myristylation, perhaps by changing the degree of myristate exposure (37).
Although the myristic acid moiety attached to the N terminus of the MA domain is clearly critical for Gag membrane binding, thermodynamic considerations suggest that the myristate alone cannot provide sufficient binding energy to anchor Pr55Gag at the lipid bilayer (41). It therefore seems likely that domains elsewhere in the protein contribute to Gag membrane binding. Indeed, structural (22, 32) and biochemical (62) data have implicated a highly basic domain spanning MA amino acids 17 to 31 in membrane binding. However, seemingly contradictory results have been reported from deletion mutagenesis studies in which Gag proteins lacking most or all of MA are able to efficiently produce virus particles (29, 46, 53, 55). We recently reported that mutations in the MA basic domain influence both membrane binding and the targeting of Gag to the plasma membrane and that the roles of MA in membrane binding and Gag targeting are genetically separable (38).
Multiple domains in Pr55Gag have been proposed to mediate Gag multimerization. Based on X-ray crystallography studies, both HIV-1 and simian immunodeficiency virus (SIV) MA were reported to form trimers (22, 44), and biochemical assays utilizing MA expressed in bacteria or in a baculovirus system also suggested the ability of MA to trimerize (35, 36). Although mutations that disrupted MA trimerization impaired virus particle assembly in a baculovirus overexpression system (36), the biological relevance of MA trimerization in virus replication remains in question since, as mentioned above, deletion mutants lacking most or all of MA are able to efficiently assemble and release virus particles. A number of studies utilizing various expression systems have suggested a role for Gag domains spanning CA, p2, and NC in virus assembly (for a review, see reference 11). However, the relative contribution of each of these domains in promoting Gag-Gag interactions is unclear. Bacterially expressed CA has been shown to assemble in vitro (10, 20, 51), and the C-terminal domain of CA dimerizes in the crystal structure in the absence of other domains (16). Mutations affecting the C-terminal domain of HIV-1 CA have also been shown to disrupt virus assembly (8, 26, 45, 50, 54, 59). In contrast, mutations in the N-terminal domain of CA generally do not disrupt virus assembly but rather perturb proper virion maturation (45, 52). In some reports, the p2 spacer peptide was observed to play a major role in VLP assembly (1, 25, 28, 35, 53), although another study observed a role for p2 in virion morphogenesis but not in Gag assembly itself (42). It has been proposed that basic domains in NC are essential for retroviral Gag-Gag interaction (4–6), and data from several groups have suggested that HIV-1 NC promotes efficient virus particle assembly (7, 9, 17, 23, 53, 60, 61). In some cases, however, NC appears to be dispensable for efficient VLP assembly and release (25, 35). The latter studies were performed using a high-level baculovirus overexpression system in which Gag concentrations are likely to substantially exceed physiological levels.
In addition to its proposed role in promoting Gag-Gag interactions, it has also been reported that NC is critical for high-level binding of Gag to membrane (43, 47). Together with the reports implicating NC in Gag multimerization, these findings suggested the possibility that Gag multimerization is a prerequisite for efficient membrane binding.
In this study, we examine the relationship between Gag multimerization and membrane binding by analyzing C-terminally truncated Gag proteins for their ability to form VLPs when singly expressed, to copackage into VLPs when coexpressed with full-length Gag, and to bind membrane. Since some of the discrepancies in previous studies regarding the mapping of Gag multimerization domains likely resulted from the use of different expression systems, we expressed the Gag mutants under relatively physiological conditions, i.e., in human (HeLa) cells in the context of HIV-1 proviral clones. We demonstrate, in vivo, that strong interactions between Gag proteins are promoted by multiple determinants within the C-terminal region of CA, p2, and the N-terminal region of NC. Significantly, we observe that a truncated Gag protein lacking these sequences, and containing only MA and the N-terminal domain of CA, binds membrane at levels comparable to those of full-length Pr55Gag. These results indicate that neither NC nor other strong Gag multimerization domains are required for efficient binding of HIV-1 Gag to membrane.
MATERIALS AND METHODS
Cell culture and transfection.
HeLa cells were maintained as previously described (12). Transfection of HeLa cells was performed by the calcium phosphate precipitation method as reported before (13).
Plasmids, mutagenesis, and DNA cloning.
Construction of the derivative of the HIV-1 proviral molecular clone pNL4-3 (2) containing a Gly→Ala change at MA amino acid 1 (the 1GA mutant) has been reported previously (13). The pNL4-3/PR− molecular clone, which contains a PR active-site mutation (Asp→Asn change at PR amino acid 25), has been described previously (24). The pNL4-3 derivatives pNL4-3/p41stop and pNL4-3/MAstop, which contain stop codons in the sequences encoding p2 amino acid 1 and CA amino acids 1 and 2, respectively, have been described previously (37). Construction of the double mutants pNL4-3/1GA/MAstop, pNL4-3/1GA/p41stop, and pNL4-3/1GA/PR− has been detailed previously (37).
The pNL4-3 derivatives pNL4-3/CA146stop, pNL4-3/p43stop, and pNL4-3/NC35stop, which contain stop codons in the sequences encoding CA amino acids 146 and 147, NC amino acids 1 and 2, and NC amino acids 35 and 36, respectively, were constructed by oligonucleotide-directed mutagenesis using an M13mp18 subclone harboring the 1.4-kbp SphI-PstI fragment from pNL4-3 (nucleotides 1443 to 2839) as a template. The following oligonucleotides were used in the mutagenesis reactions: for pNL4-3/CA146, 5′-AGAATGTATTAATAAACCAGCATT-3′; for pNL4-3/p43stop; 5′-GCTACCATAATGTAATAGAAAGG-3′; and for pNL4-3/NC35stop, 5′-AGGAAAAAGTGATGATGGAAATGT-3′. Nonmyristylated (1GA) versions of molecular clones containing the CA146stop, p43stop, and NC35stop mutations were constructed by introducing the SphI-EcoRI fragments (nucleotides 1443 to 5743) from pNL4-3/CA146stop, pNL4-3/p43stop, and pNL4-3/NC35stop, respectively, into pNL4-3/1GA.
VLP incorporation assay and Western blotting.
HeLa cells were cotransfected with pNL4-3 derivatives expressing truncated Gag proteins and pNL4-3/PR− (expressing full-length Pr55Gag) at a DNA ratio of 1:1. Two days posttransfection, virions were pelleted from culture supernatants; VLP and cell lysates were prepared as detailed previously (12, 57). Cell and VLP lysates were analyzed by Western blotting essentially as previously described (27). Gag proteins were detected with a mixture of three mouse monoclonal antibodies against MA (from Advanced Biotechnologies, Columbia, Md.; Cellular Products, Buffalo, N.Y.; and Capricorn, Scarborough, Maine) as primary antibodies and horseradish peroxidase-conjugated anti-mouse immunoglobulin (Ig; Amersham) as a secondary antibody. Detection was performed with enhanced chemiluminescence Western blotting reagents (Amersham). To avoid overexposure of blots of cell-associated material, eightfold less lysate was loaded for cell- than for VLP-associated material. For quantification of Gag proteins, alkaline phosphatase-conjugated anti-mouse Ig (Amersham) was used as a secondary antibody; detection with enhanced chemifluorescence Western blotting reagents (Amersham) was performed in accordance with the manufacturer's instructions using a Fuji FLA-2000 image analyzer.
Confocal microscopy.
The methods used to examine transfected HeLa cells by confocal microscopy have been described recently (38). Briefly, HeLa cells were cultured in chamber slides (Nunc) and transfected by the calcium phosphate precipitation method without glycerol shock. Two days posttransfection, cells were fixed with 3.7% formaldehyde, permeabilized with phosphate-buffered saline (PBS) containing 0.1% Triton X-100, and incubated with anti-p17 monoclonal antibody (Cellular Products, Inc.) and Texas red-conjugated anti-mouse IgG. Cells were mounted with Fluoromount G (Virotech International) and examined with a Zeiss LSM410 laser scanning microscope.
Analysis of VLP density.
VLPs were pelleted from the culture supernatant of transfected HeLa cells and resuspended in 0.4 ml of PBS. Resuspended VLPs were placed onto a sucrose gradient composed of 20, 30, 40, 50, and 60% sucrose layers in PBS (0.95 ml each). The gradients were centrifuged at 100,000 × g for 16 h at 4°C in a Beckman SW55Ti rotor. Eleven fractions were collected from the top of the centrifuge tubes. Fractionated samples were analyzed as described above.
Membrane-binding assay.
Membrane flotation centrifugation was performed as detailed previously (37, 38). Briefly, HeLa cells were collected in PBS, washed once with 10 mM Tris-HCl (pH 7.5) containing 1 mM EDTA and 1 mM EGTA, and resuspended in 10 mM Tris-HCl containing 1 mM EDTA, 6% (wt/vol) sucrose, and Complete protease inhibitor cocktail (Boehringer Mannheim). Postnuclear supernatants obtained after sonication of cell suspensions were mixed with 85.5% (wt/vol) sucrose and placed on the bottom of a centrifuge tube. On top of this postnuclear supernatant-containing 73% (wt/vol) sucrose mixture was layered 65 and 10% (wt/vol) sucrose. The gradients were centrifuged at 100,000 × g for 18 h. Ten fractions were collected from the top of the centrifuge tube. Fractionated samples were analyzed by Western blotting as previously described (27). Quantitation of Western blotting data was performed by densitometry scanning. Similar results were obtained when Pr55Gag-expressing cells were disrupted by homogenization rather than sonication (unpublished results).
RESULTS
Construction of HIV-1 Gag truncation mutants.
To examine the relationship between Gag-Gag interaction and membrane binding, we constructed a series of C-terminal Gag truncation mutants (Fig. 1). To minimize the disruption of native Gag conformation, stop codons were introduced either immediately following Pr55Gag cleavage sites or adjacent to known structural domains. The NC35stop mutant is truncated just before the second NC zinc finger and thus includes the two highly basic domains of NC (located at the NC N terminus and between the two zinc finger motifs). The p43stop mutant is truncated immediately after the p2/NC cleavage site to produce an MA-CA-p2 Gag protein. p41stop contains all of MA and CA, while CA146stop is truncated at CA amino acid 146 (in the linker region between the N- and C-terminal domains of CA [15, 58]) and therefore contains MA and the N-terminal domain of CA. The MAstop mutant expresses only p17(MA). These mutations were introduced into the HIV-1 molecular clone pNL4-3 (2). All Gag proteins were synthesized in the absence of PR; to express full-length Pr55Gag, we used a PR− version of pNL4-3, pNL4-3/PR− (24). The truncated clones were also PR−, since the stop codons in Gag prevented the synthesis of Pr160Gag-Pol.
FIG. 1.
Schematic representation of HIV-1 Gag C-terminal truncation mutants. The domain structure of the Gag precursor Pr55Gag is shown at the top. The regions expressed in the truncation mutants analyzed in this study are indicated. The two heavy lines over CA represent the two structural domains of this protein. The zinc finger motifs (Zn) of NC are represented by stippled boxes.
Sequences within the NC domain of HIV-1 Gag are essential for efficient VLP assembly.
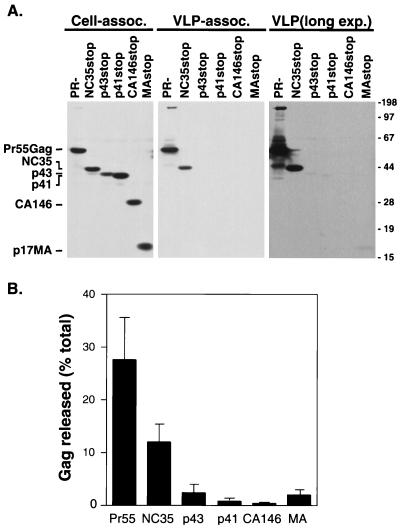
We first tested the ability of the Gag truncation mutants to form VLPs. HeLa cells were transfected with pNL4-3/PR− or pNL4-3 derivatives expressing the Gag truncation mutants. Cell and VLP lysates prepared 2 days posttransfection were analyzed by Western blotting with a mixture of monoclonal antibodies against MA (Fig. 2). Approximately 28% of the total amount of full-length Gag expressed was VLP associated. Relative to full-length Gag, pNL4-3/NC35stop-transfected cells produced a reduced but readily detectable amount (12%) of VLP-associated Gag protein. Approximately 2% of p43 Gag was recovered in the medium in a pelletable form. Less than 1% of p41 and CA146 Gag proteins were found in the pelleted medium. We observed a low level (approximately 2%) of pelletable material released into the medium of cells expressing MA alone (MAstop). These results are in agreement with studies (7, 9, 60) suggesting that domains within HIV-1 NC play an important role in VLP assembly and release.
FIG. 2.
VLP production of singly expressed truncation mutants. (A) HeLa cells were transfected with pNL4-3/PR− or pNL4-3 derivatives expressing the indicated truncated Gag proteins. VLPs were pelleted from the supernatant of transfected cells by ultracentrifugation. Cell-associated (assoc., left panel) and VLP-associated (middle and right panels) material was analyzed by Western blotting with a mixture of three monoclonal antibodies against MA. As a percentage of total material recovered, eightfold more VLP-associated than cell-associated material was loaded onto the gels. The rightmost panel shows a longer exposure (exp.) (12-fold) of the middle panel. The positions of the full-length Gag precursor Pr55Gag, NC35, p43, p41, CA146, and MA proteins are shown. The positions of molecular size markers are indicated at the right (in kilodaltons). (B) Percentage of total Gag expressed that is released from the cell in a pelletable form. Data represent averages of three experiments; standard deviations are indicated by error bars.
Gag multimerization is promoted by the C-terminal domain of CA and by p2 and NC.
To analyze Gag-Gag interaction further, we next assessed the ability of the truncated Gag mutants to be assembled into VLPs when coexpressed with full-length Gag. HeLa cells were transfected with a 1:1 ratio of pNL4-3/PR− and pNL4-3 derivatives expressing the truncated Gags. Cell and VLP lysates were analyzed by Western blotting using a mixture of monoclonal antibodies against MA (Fig. 3). Release of VLP-associated Pr55Gag was not affected by coexpression of any of the truncated Gag proteins, indicating that, at least at a 1:1 DNA ratio, the truncated Gag molecules did not transdominantly interfere with wild-type assembly and release. When NC35, p43, and p41 were coexpressed with full-length Gag, approximately 27, 21, and 12%, respectively, of the total amount of truncated Gag expressed was incorporated into VLPs. In contrast, only a very small amount of CA146 and MA were VLP associated (approximately 3 and 4%, respectively). Consistent with data obtained using similar constructs in an in vitro binding assay (6), these results suggest that the CA146 and MA proteins lack domains involved in promoting strong Gag-Gag interactions in vivo.
FIG. 3.
Coexpression of full-length Gag with the myristylated (WT) forms of the truncated Gag proteins. (A) HeLa cells were singly transfected with pNL4-3/PR− (lanes -) or cotransfected with pNL4-3/PR− and pNL4-3 derivatives expressing the indicated truncated Gag proteins. Cell- (left panel) and VLP- (right panel) associated material was analyzed as described in the legend to Fig. 2. The positions of the full-length Gag precursor Pr55Gag, NC35, p43, p41, CA146, and MA proteins are shown. (B) Percentage of total truncated Gag expressed that is released from the cell in a pelletable form. Data represent averages of three experiments; standard deviations are indicated by error bars.
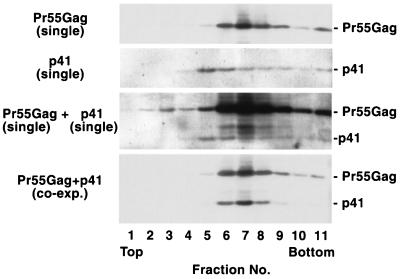
The finding that p43 and p41 do not efficiently form VLPs when expressed alone (Fig. 2) yet are readily detected in VLP preparations produced upon coexpression with full-length Gag (Fig. 3) suggests that these truncated proteins coassemble with full-length Gag via Gag-Gag interactions. However, it is formally possible that expression of full-length Gag in some manner stimulates assembly and release of VLPs composed solely of truncated molecules. To test this possibility, and to demonstrate that p41 Gag is copackaged into VLPs with full-length Pr55Gag, we analyzed the density of p41-containing VLPs released in the presence and absence of coexpressed Pr55Gag (Fig. 4). HeLa cells were either singly transfected with pNL4-3/PR− or pNL4-3/p41stop or cotransfected with these two molecular clones. VLP preparations recovered from the culture supernatants were analyzed by sucrose density gradient centrifugation. Eleven fractions were recovered, and the amount of Gag in each fraction was determined by Western blotting. VLPs derived from cells expressing full-length Pr55Gag displayed a density characteristic of retroviral particles; Gag was found primarily in fractions 6 to 9, with the peak in fraction 7 (corresponding to 1.17 g/cm3) (Fig. 4, top panel). In contrast, VLP-associated Gag derived from cells singly transfected with the pNL4-3/p41stop clone peaked in fraction 5 (1.13 g/cm3) (Fig. 4, second panel), consistent with previous reports (47, 53). The distinct density of Pr55Gag and p41 VLPs was also observed when VLPs produced from cells singly transfected with pNL4-3/PR− or pNL4-3/p41stop were mixed (Fig. 4, third panel). However, when Pr55Gag and p41 were coexpressed, the distribution of p41 in the sucrose gradient matched that of singly expressed Pr55Gag (Fig. 4, bottom panel). These results demonstrate that the presence of p41 Gag in VLP preparations derived from cells cotransfected with pNL4-3/PR− and pNL4-3/p41stop is due to the coassembly of p41 and full-length Gag molecules.
FIG. 4.
VLP density analysis. HeLa cells were singly transfected with pNL4-3/PR− (top panel) or pNL4-3/p41stop (second panel) or cotransfected with both pNL4-3/PR− and pNL4-3/p41stop (bottom panel). VLPs recovered from the transfected cell supernatants were loaded onto 20 to 60% sucrose gradients. In the third panel, VLPs derived from pNL4-3/PR−-transfected cells were mixed with those from a fourfold-greater number of pNL4-3/p41stop-transfected cells before sucrose gradient centrifugation. The second and third panels were intentionally overexposed to allow visualization of the low-level p41 VLP-associated material. Gag proteins were detected as described in the legend to Fig. 2. Pr55Gag and p41 are shown.
In the experiment presented in Fig. 3, the truncated Gag proteins could be incorporated into VLPs via direct interaction with full-length Gag or by a more passive mechanism resulting from their association with the plasma membrane. In addition, as indicated in Fig. 2, NC35 Gag produces significant amounts of VLP-associated material when singly expressed. To examine Gag-Gag interactions more directly, we coexpressed pNL4-3/PR− with nonmyristylated (1GA) versions of truncated Gag proteins. The 1GA mutation largely abolishes the ability of full-length or truncated Gag proteins to bind membrane and produce VLPs (37) (data not shown). We then examined the incorporation of the nonmyristylated, truncated Gag proteins into VLPs (Fig. 5). Nonmyristylated NC35, p43, and p41 were incorporated into VLPs when coexpressed with full-length Gag; approximately 12, 6, and 2% of these proteins, respectively, were VLP associated. These results suggest that NC35, p43, and p41 are incorporated into VLPs in this assay by their interaction with full-length Gag. In contrast, CA146 and MA were largely unable to become incorporated into VLPs, confirming that these truncated Gag proteins lack the ability to interact tightly with Pr55Gag. Taken together, these results suggest that the C-terminal domains of CA, p2, and NC function cooperatively to promote strong Gag-Gag interactions. The ability of the nonmyristylated Gag proteins to become incorporated into VLPs also confirms that membrane binding is not a prerequisite for HIV-1 Gag multimerization (6, 31, 40). However, the increased incorporation of myristylated versus nonmyristylated Gags (compare Fig. 3 and 5) suggests that membrane binding may enhance the formation of Gag-Gag contacts (see Discussion).
FIG. 5.
Coexpression of full-length Gag with nonmyristylated (1GA) forms of the truncated Gag proteins. (A) HeLa cells were singly transfected with pNL4-3/PR− (lanes -) or cotransfected with pNL4-3/PR− and pNL4-3 derivatives expressing the indicated truncated Gag proteins. Cell- (left panel) and VLP- (right panel) associated material was analyzed as described in the legend to Fig. 2. The positions of the full-length Gag precursor Pr55Gag, NC35, p43, p41, CA146, and MA proteins are shown. (B) Percentage of total truncated Gag expressed that is released from the cell in a pelletable form. Data represent averages of three experiments; standard deviations are indicated by error bars.
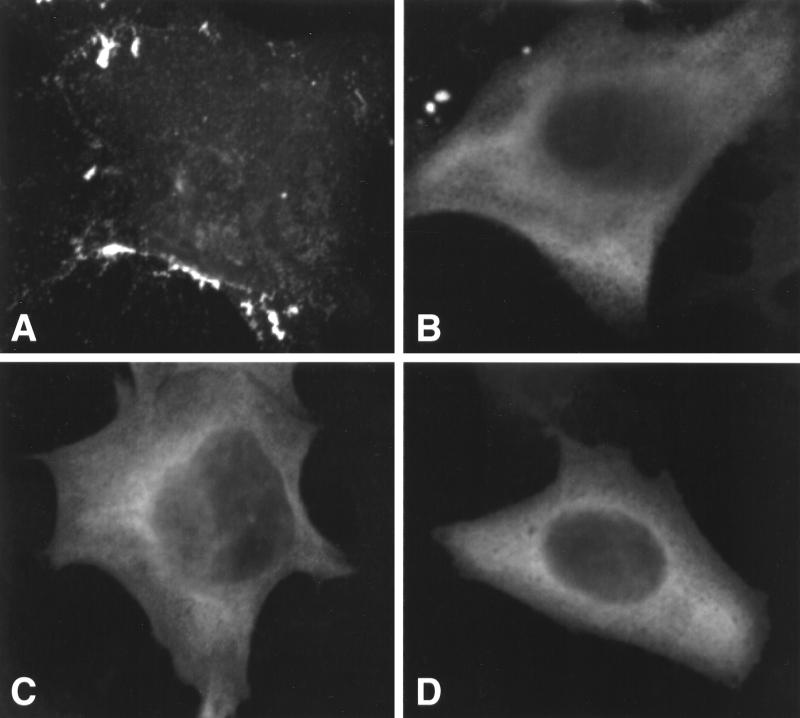
A possible explanation for the lack of efficient VLP incorporation of the truncated Gag mutants (e.g., CA146 and MA) is that they might be targeted to a location in the cell distinct from the site to which full-length Gag is localized. We recently demonstrated that a mutant Gag that is targeted to the Golgi apparatus rather than the plasma membrane still interacts efficiently with wild-type Gag and is rescued into VLPs when coexpressed with wild-type (38). This observation suggests that Gag multimerization takes place early after synthesis, before targeting differences are imposed. Nevertheless, we wished to examine directly whether differences in Gag localization might account for the low efficiency of nonmyristylated (1GA) MA and CA146 Gag incorporation into VLPs upon coexpression with full-length Gag. To this end, we used confocal microscopy to examine cells expressing full-length, myristylated Gag or nonmyristylated full-length, NC35, or CA146 Gag proteins. The results demonstrated that cells transfected with pNL4-3/PR− showed the punctate staining pattern characteristic of HIV-1 Gag (Fig. 6A), whereas cells expressing nonmyristylated versions of full-length (Fig. 6B) or truncated (Fig. 6C and D) Gag displayed the hazy, diffuse localization pattern typical of nonmyristylated Gag mutants (47, 48, 54). No differences in localization pattern were observed between cells expressing a mutant that is readily incorporated into VLPs (1GA/NC35, Fig. 6C) and cells expressing a truncated Gag that is not efficiently incorporated (1GA/CA146, Fig. 6D). Thus, we conclude that differential Gag localization does not explain the inability, evident in Fig. 5, of 1GA/MA or 1GA/CA146 Gag to interact with full-length Gag and become incorporated into VLPs.
FIG. 6.
Confocal microscopy of Gag-expressing cells. HeLa cells were transfected with pNL4-3/PR− (A), pNL4-3/1GA/PR− (B), pNL4-3/1GA/NC35stop (C), or pNL4-3/1GA/CA146stop (D) and examined by confocal microscopy as described in Materials and Methods.
Strong Gag-Gag interaction domains are not necessary for efficient binding of HIV-1 Gag to membrane.
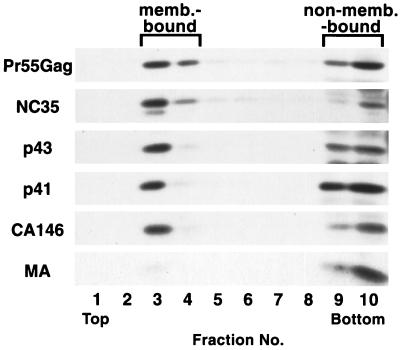
To examine the relationship between Gag-Gag interaction and membrane binding, we next measured the ability of the truncated Gag proteins to bind membrane. We performed membrane flotation centrifugation (Fig. 7), which we and others have previously used to analyze steady-state HIV-1 Gag membrane binding (37, 39, 48). This method can distinguish oligomeric, non-membrane-bound Gag complexes from membrane-bound Gag, a separation that cannot be achieved by conventional subcellular fractionation techniques. In a previous study, we used this approach to confirm that MA binds membrane much less efficiently than does full-length Gag and to demonstrate that mutations near the N terminus of MA impair Gag membrane binding without affecting the addition of myristate (37). HeLa cells were transfected with pNL4-3/PR− or pNL4-3 derivatives expressing the C-terminal Gag truncation mutants. Postnuclear supernatants of cell homogenates were subjected to membrane flotation centrifugation, followed by Western blotting analysis of the fractions derived from the centrifugation (Materials and Methods). Fractions 3 and 4 contain membrane-bound material; non-membrane-bound proteins are recovered in fractions 9 and 10 (37) (see Materials and Methods). Consistent with previous data obtained in HeLa cells (37, 39) approximately 40% of full-length Pr55Gag was recovered in the membrane-containing fractions (Fig. 7), whereas approximately 3% of MA floated to fractions 3 and 4. NC35, p43, and p41 showed approximately 75, 50, and 25% membrane binding, respectively. Interestingly, CA146 was recovered in membrane fractions to the same extent as full-length Pr55Gag (approximately 40%). These results, together with the data presented in Fig. 2, 3, and 5, indicate that strong Gag-Gag interaction domains are not required for HIV-1 Gag to achieve a high level of membrane binding.
FIG. 7.
Effects of C-terminal truncation on Gag membrane (memb.) binding. HeLa cells were transfected with pNL4-3/PR− (Pr55Gag) or its derivatives expressing the indicated truncated Gag proteins. Postnuclear supernatants were prepared and subjected to equilibrium flotation centrifugation (Materials and Methods), during which membrane-bound material floated to the interface between 10 and 65% sucrose (fractions 3 and 4). Gag proteins were detected as described in the legend to Fig. 2.
The data presented in Fig. 3 and 5 indicate that truncation mutants lacking NC and the C terminus of CA are very inefficiently incorporated into VLPs upon coexpression with Pr55Gag. These results suggest that these mutants are unable to interact strongly with full-length Gag. To examine whether MA and CA146 might interact with Pr55Gag in the cell but not be incorporated into VLPs, we performed the following experiment. pNL4-3PR− was cotransfected with 1GA (nonmyristylated) derivatives of pNL4-3/MAstop, CA146stop, or NC35stop. Postnuclear supernatants were subjected to membrane flotation centrifugation, and the distribution of Gag was determined by Western blotting. The results indicated that 1GA/NC35 Gag was recruited by Pr55Gag into the membrane fraction with an efficiency that was markedly higher than the efficiency with which either 1GA/MA or 1GA/CA146 was recruited to the membrane (data not shown). These observations support the conclusion, drawn from the VLP incorporation data, that MA and CA146 interact only weakly with full-length Gag.
DISCUSSION
In this study, we demonstrate that the region of HIV-1 Gag spanning the C-terminal domain of CA, p2, and the N-terminal 35 amino acids of NC promotes strong Gag-Gag interaction (Fig. 2, 3, and 5). Extension of Gag from amino acid 146 of CA (CA146) to residue 35 of NC (NC35) incrementally increases truncated Gag incorporation into VLPs through interaction with full-length Pr55Gag. It is therefore likely that multiple sequences act cooperatively to promote Gag multimerization during assembly, either by contributing directly to Gag-Gag interaction or by influencing the conformation of interacting sequences. Although truncations that remove the C-terminal domain of CA and downstream sequences block the ability of truncated molecules to interact with full-length Pr55Gag and become packaged into VLPs, a truncated version of Gag containing only MA and the N-terminal domain of CA (CA146) is capable of high-level membrane binding (Fig. 7). These results indicate that strong Gag-Gag interaction domains are not required for efficient binding of Gag to membrane.
It is well established that MA binds membrane quite weakly relative to full-length Pr55Gag (21, 37, 48, 63). It has been proposed that after Pr55Gag cleavage, MA undergoes a conformational transition that results in the sequestration of the N-terminal myristate moiety and a reduction in membrane-binding potential (see Introduction). The difference between MA and CA146 in membrane-binding ability can also be explained by this conformational switch model: the addition of the N-terminal domain of CA to MA may induce a conformational change in the MA domain that exposes membrane-binding determinants, including the N-terminal myristate. We consider it unlikely that the N-terminal domain of CA itself functions directly in promoting membrane binding, since cryoelectron microscopy of immature HIV-1 particles suggests that Gag sequences C-terminal to MA are oriented away from the membrane in a rod-like fashion (14). The hypothesis that efficient membrane binding of CA146 is highly dependent on its conformation is supported by our observation that the addition of a short polypeptide sequence (the FLAG epitope tag) to the C terminus of CA146 significantly reduces its membrane-binding ability (A. Ono and E. O. Freed, unpublished data). In an attempt to define further the amount of CA sequence necessary to obtain high-level membrane binding, we constructed Gag truncation mutants which terminated at CA residue 14 (CA14) or 93 (CA93). However, poor detection or instability of these truncated molecules hampered our efforts to definitively determine their membrane-binding properties (unpublished data).
Although the MA and CA146 Gag proteins are unable to coassemble with full-length Gag into VLPs, we cannot exclude the possibility that they may retain some ability to form low-order multimers. It has been reported that p17(MA) as well as p41 (MA-CA) Gag proteins can form trimers in a baculovirus overexpression system (35), and SIV MA is reportedly capable of forming VLPs (18, 19). HIV-1 MA has also been observed by some (56) but not by others (18) to form low-density particles. We find a low level of pelletable material produced by expression of MA. Nevertheless, it is clear that the determinants for strong Gag-Gag interaction (as defined by the ability to promote copackaging with full-length Gag in VLPs) are located downstream of the sequences present in CA146. Consistent with these results, Gag molecules truncated after MA or between the N- and C-terminal domains of CA did not interact with full-length Gag in an in vitro multimerization assay (6), and a truncated Gag molecule (p41) lacking NC was unable to form high-molecular-weight, detergent-resistant complexes in Gag-expressing cells (30). We also observed that 1GA/NC35 was recruited into membrane fractions much more efficiently than 1GA/MA or 1GA/CA146 when coexpressed with full-length Pr55Gag (Ono and Freed, unpublished data). This intracellular interaction assay confirms the data obtained with the VLP incorporation studies and supports the conclusion that the region of Gag spanning the C terminus of CA to the N terminus of NC promotes strong Gag-Gag interactions.
The inability of myristylated or nonmyristylated MA and CA146 to become incorporated into VLPs upon coexpression with full-length Gag does not appear to be due to differential subcellular localization of the truncated and full-length molecules. Examination by confocal microscopy of cells expressing 1GA/NC35 (which is readily incorporated into VLPs) and 1GA/CA146 (which is not efficiently incorporated) revealed no differences in subcellular localization (Fig. 6). In addition, several lines of evidence suggest that Gag multimerization may occur efficiently between two Gag molecules which, when expressed alone, are differentially targeted. (i) As demonstrated here and previously (6), nonmyristylated HIV-1 Gag readily interacts with myristylated Gag and becomes packaged into VLPs despite the fact that the nonmyristylated protein displays a diffuse, cytoplasmic localization pattern, whereas the myristylated protein shows a punctate, largely plasma membrane-associated distribution (Fig. 6) (38). (ii) We recently demonstrated that mutant Gags that are retargeted to the Golgi as a result of substitutions in MA are efficiently rescued into VLPs when coexpressed with the wild type (38). (iii) A study of the incorporation of HIV-1 Gag-β-galactosidase fusion proteins into VLPs indicated that the ability of various truncated molecules to become incorporated did not correlate with their patterns of subcellular localization (54). Together, these results suggest that Gag-Gag interactions may take place early after synthesis before sorting to different subcellular compartments takes place.
It is noteworthy that incorporation of truncated Gag proteins, especially NC35, p43, and p41, into VLPs is higher for myristylated than for nonmyristylated forms of the truncated molecules (Fig. 3 and 5). This observation suggests that membrane binding may facilitate Gag-Gag interaction by concentrating Gag molecules on the cytoplasmic surface of the plasma membrane. Thus, although membrane binding is clearly not required for Gag multimerization to occur (Fig. 5) (6, 31), it may enhance the efficiency of Gag-Gag interaction. The higher levels of VLP-associated Gag observed with myristylated forms may also reflect the passive incorporation of molecules solely due to their presence on the plasma membrane where virus particle formation occurs.
It has been reported that sequences in the HIV-1 NC known to promote Gag-Gag interaction (6, 7; this study) are required for HIV-1 Gag to bind membrane at levels observed for full-length Pr55Gag (43, 47). These results led to the proposal that NC is essential for efficient membrane binding or for the stable retention of Gag at the membrane (43, 47). However, our finding that the CA146 Gag protein, which lacks NC sequences, was recovered in membrane fractions to the same extent as full-length Pr55Gag (Fig. 7) clearly indicates that strong Gag-Gag interactions promoted by NC are not necessary for Gag to achieve a high level of membrane binding. The discrepancy between these results in terms of a requirement for NC in Gag membrane binding can be explained by two major differences in the studies: (i) a Gag truncation mutant analogous to CA146stop was not analyzed in the studies that observed a low membrane-binding ability of Gag molecules lacking NC sequences (43, 47), and (ii) we used membrane flotation centrifugation instead of traditional fractionation techniques to measure membrane binding. As we have demonstrated previously (37), membrane flotation centrifugation can separate membrane-bound material from non-membrane-bound Gag complexes, whereas standard fractionation techniques fail to achieve this separation. Since NC promotes the formation of Gag complexes, it increases the amount of pelletable Gag in fractionation assays. While CA146 binds membrane at levels comparable to those of Pr55Gag, the increased membrane-binding ability of p43 versus p41 and NC35 versus p43 (Fig. 7) suggests that Gag multimerization may enhance membrane-binding potential to some extent.
During the course of HIV-1 replication, Gag membrane binding and virus assembly are driven not by the mature Gag proteins but by the Pr55Gag precursor. Thus, information relating to the structure of the MA domain of Pr55Gag would be highly informative for the understanding of the late stages of the virus replication cycle. At this time, the structure of only the mature, nonmyristylated MA protein is available (22, 32, 33, 44). Because of its large size, attempts to obtain nuclear magnetic resonance spectroscopy or X-ray crystallography data with full-length Pr55Gag would be a technically daunting task. However, since it binds membrane at levels comparable to full-length Pr55Gag, the myristylated CA146 protein may prove useful in solving the structure of the membrane-binding-competent form of HIV-1 MA.
ACKNOWLEDGMENTS
We thank T. Murakami for helpful suggestions and critical review of the manuscript and A. Buckler-White for DNA sequencing.
REFERENCES
- 1.Accola M A, Hoglund S, Gottlinger H G. A putative alpha-helical structure which overlaps the capsid-p2 boundary in the human immunodeficiency virus type 1 Gag precursor is crucial for viral particle assembly. J Virol. 1998;72:2072–2078. doi: 10.1128/jvi.72.3.2072-2078.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Adachi A, Gendelman H E, Koenig S, Folks T, Willey R, Rabson A, Martin M A. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J Virol. 1986;59:284–291. doi: 10.1128/jvi.59.2.284-291.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames J B, Ishima R, Tanaka T, Gordon J I, Stryer L, Ikura M. Molecular mechanics of calcium-myristoyl switches. Nature. 1997;389:198–202. doi: 10.1038/38310. [DOI] [PubMed] [Google Scholar]
- 4.Bennett R P, Nelle T D, Wills J W. Functional chimeras of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1993;67:6487–6498. doi: 10.1128/jvi.67.11.6487-6498.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowzard J B, Bennett R P, Krishna N K, Ernst S M, Rein A, Wills J W. Importance of basic residues in the nucleocapsid sequence for retrovirus Gag assembly and complementation rescue. J Virol. 1998;72:9034–9044. doi: 10.1128/jvi.72.11.9034-9044.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burniston M T, Cimarelli A, Colgan J, Curtis S P, Luban J. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol. 1999;73:8527–8540. doi: 10.1128/jvi.73.10.8527-8540.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dawson L, Yu X F. The role of nucleocapsid of HIV-1 in virus assembly. Virology. 1998;251:141–157. doi: 10.1006/viro.1998.9374. [DOI] [PubMed] [Google Scholar]
- 8.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dorfman T, Luban J, Goff S P, Haseltine W A, Gottlinger H G. Mapping of functionally important residues of a cysteine-histidine box in the human immunodeficiency virus type 1 nucleocapsid protein. J Virol. 1993;67:6159–6169. doi: 10.1128/jvi.67.10.6159-6169.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ehrlich L S, Agresta B E, Carter C A. Assembly of recombinant human immunodeficiency virus type 1 capsid protein in vitro. J Virol. 1992;66:4874–4883. doi: 10.1128/jvi.66.8.4874-4883.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 12.Freed E O, Martin M A. Evidence for a functional interaction between the V1/V2 and C4 domains of human immunodeficiency virus type 1 envelope glycoprotein gp120. J Virol. 1994;68:2503–2512. doi: 10.1128/jvi.68.4.2503-2512.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Freed E O, Orenstein J M, Buckler-White A J, Martin M A. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol. 1994;68:5311–5320. doi: 10.1128/jvi.68.8.5311-5320.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fuller S D, Wilk T, Gowen B E, Krausslich H G, Vogt V M. Cryo-electron microscopy reveals ordered domains in the immature HIV-1 particle. Curr Biol. 1997;7:729–738. doi: 10.1016/s0960-9822(06)00331-9. [DOI] [PubMed] [Google Scholar]
- 15.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 16.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 17.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 18.Giddings A M, Ritter G D, Jr, Mulligan M J. The matrix protein of HIV-1 is not sufficient for assembly and release of virus-like particles. Virology. 1998;248:108–116. doi: 10.1006/viro.1998.9284. [DOI] [PubMed] [Google Scholar]
- 19.Gonzalez S A, Affranchino J L, Gelderblom H R, Burny A. Assembly of the matrix protein of simian immunodeficiency virus into virus-like particles. Virology. 1993;194:548–556. doi: 10.1006/viro.1993.1293. [DOI] [PubMed] [Google Scholar]
- 20.Gross I, Hohenberg H, Huckhagel C, Krausslich H G. N-terminal extension of human immunodeficiency virus capsid protein converts the in vitro assembly phenotype from tubular to spherical particles. J Virol. 1998;72:4798–4810. doi: 10.1128/jvi.72.6.4798-4810.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hermida-Matsumoto L, Resh M D. Human immunodeficiency virus type 1 protease triggers a myristoyl switch that modulates membrane binding of Pr55(gag) and p17MA. J Virol. 1999;73:1902–1908. doi: 10.1128/jvi.73.3.1902-1908.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hill C P, Worthylake D, Bancroft D P, Christensen A M, Sundquist W I. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci USA. 1996;93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hoshikawa N, Kojima A, Yasuda A, Takayashiki E, Masuko S, Chiba J, Sata T, Kurata T. Role of the gag and pol genes of human immunodeficiency virus in the morphogenesis and maturation of retrovirus-like particles expressed by recombinant vaccinia virus: an ultrastructural study. J Gen Virol. 1991;72:2509–2517. doi: 10.1099/0022-1317-72-10-2509. [DOI] [PubMed] [Google Scholar]
- 24.Huang M, Orenstein J M, Martin M A, Freed E O. p6Gag is required for particle production from full-length human immunodeficiency virus type 1 molecular clones expressing protease. J Virol. 1995;69:6810–6818. doi: 10.1128/jvi.69.11.6810-6818.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jowett J B, Hockley D J, Nermut M V, Jones I M. Distinct signals in human immunodeficiency virus type 1 Pr55 necessary for RNA binding and particle formation. J Gen Virol. 1992;73:3079–3086. doi: 10.1099/0022-1317-73-12-3079. [DOI] [PubMed] [Google Scholar]
- 26.Kattenbeck B, von Poblotzki A, Rohrhofer A, Wolf H, Modrow S. Inhibition of human immunodeficiency virus type 1 particle formation by alterations of defined amino acids within the C terminus of the capsid protein. J Gen Virol. 1997;78:2489–2496. doi: 10.1099/0022-1317-78-10-2489. [DOI] [PubMed] [Google Scholar]
- 27.Kiernan R E, Ono A, Englund G, Freed E O. Role of matrix in an early postentry step in the human immunodeficiency virus type 1 life cycle. J Virol. 1998;72:4116–4126. doi: 10.1128/jvi.72.5.4116-4126.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Krausslich H G, Facke M, Heuser A M, Konvalinka J, Zentgraf H. The spacer peptide between human immunodeficiency virus capsid and nucleocapsid proteins is essential for ordered assembly and viral infectivity. J Virol. 1995;69:3407–3419. doi: 10.1128/jvi.69.6.3407-3419.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee P P, Linial M L. Efficient particle formation can occur if the matrix domain of human immunodeficiency virus type 1 Gag is substituted by a myristylation signal. J Virol. 1994;68:6644–6654. doi: 10.1128/jvi.68.10.6644-6654.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee Y M, Liu B, Yu X F. Formation of virus assembly intermediate complexes in the cytoplasm by wild-type and assembly-defective mutant human immunodeficiency virus type 1 and their association with membranes. J Virol. 1999;73:5654–5662. doi: 10.1128/jvi.73.7.5654-5662.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lee Y M, Yu X F. Identification and characterization of virus assembly intermediate complexes in HIV-1-infected CD4+ T cells. Virology. 1998;243:78–93. doi: 10.1006/viro.1998.9064. [DOI] [PubMed] [Google Scholar]
- 32.Massiah M A, Starich M R, Paschall C, Summers M F, Christensen A M, Sundquist W I. Three-dimensional structure of the human immunodeficiency virus type 1 matrix protein. J Mol Biol. 1994;244:198–223. doi: 10.1006/jmbi.1994.1719. [DOI] [PubMed] [Google Scholar]
- 33.Matthews S, Barlow P, Boyd J, Barton G, Russell R, Mills H, Cunningham M, Meyers N, Burns N, Clark N, Kingsman S, Kingsman A, Campbell I. Structural similarity between the p17 matrix protein of HIV-1 and interferon-gamma. Nature. 1994;370:666–668. doi: 10.1038/370666a0. [DOI] [PubMed] [Google Scholar]
- 34.McLaughlin S, Aderem A. The myristoyl-electrostatic switch: a modulator of reversible protein-membrane interactions. Trends Biochem Sci. 1995;20:272–276. doi: 10.1016/s0968-0004(00)89042-8. [DOI] [PubMed] [Google Scholar]
- 35.Morikawa Y, Hockley D J, Nermut M V, Jones I M. Roles of matrix, p2, and N-terminal myristoylation in human immunodeficiency virus type 1 Gag assembly. J Virol. 2000;74:16–23. doi: 10.1128/jvi.74.1.16-23.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Morikawa Y, Zhang W H, Hockley D J, Nermut M V, Jones I M. Detection of a trimeric human immunodeficiency virus type 1 Gag intermediate is dependent on sequences in the matrix protein, p17. J Virol. 1998;72:7659–7663. doi: 10.1128/jvi.72.9.7659-7663.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ono A, Freed E O. Binding of human immunodeficiency virus type 1 Gag to membrane: role of the matrix amino terminus. J Virol. 1999;73:4136–4144. doi: 10.1128/jvi.73.5.4136-4144.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ono A, Orenstein J M, Freed E O. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol. 2000;74:2855–2866. doi: 10.1128/jvi.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Paillart J C, Gottlinger H G. Opposing effects of human immunodeficiency virus type 1 matrix mutations support a myristyl switch model of Gag membrane targeting. J Virol. 1999;73:2604–2612. doi: 10.1128/jvi.73.4.2604-2612.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Peitzsch R M, McLaughlin S. Binding of acylated peptides and fatty acids to phospholipid vesicles: pertinence to myristolyated proteins. Biochemistry. 1993;32:10436–10443. doi: 10.1021/bi00090a020. [DOI] [PubMed] [Google Scholar]
- 42.Pettit S C, Moody M D, Wehbie R S, Kaplan A H, Nantermet P V, Klein C A, Swanstrom R. The p2 domain of human immunodeficiency virus type 1 Gag regulates sequential proteolytic processing and is required to produce fully infectious virions. J Virol. 1994;68:8017–8027. doi: 10.1128/jvi.68.12.8017-8027.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Platt E J, Haffar O K. Characterization of human immunodeficiency virus type 1 Pr55gag membrane association in a cell-free system: requirement for a C-terminal domain. Proc Natl Acad Sci USA. 1994;91:4594–4598. doi: 10.1073/pnas.91.10.4594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rao Z, Belyaev A S, Fry E, Roy P, Jones I M, Stuart D I. Crystal structure of SIV matrix antigen and implications for virus assembly. Nature. 1995;378:743–747. doi: 10.1038/378743a0. [DOI] [PubMed] [Google Scholar]
- 45.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Reil H, Bukovsky A A, Gelderblom H R, Gottlinger H G. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 1998;17:2699–2708. doi: 10.1093/emboj/17.9.2699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spearman P, Horton R, Ratner L, Kuli-Zade I. Membrane binding of human immunodeficiency virus type 1 matrix protein in vivo supports a conformational myristyl switch mechanism. J Virol. 1997;71:6582–6592. doi: 10.1128/jvi.71.9.6582-6592.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 50.von Poblotzki A, Wagner R, Niedrig M, Wanner G, Wolf H, Modrow S. Identification of a region in the Pr55gag-polyprotein essential for HIV-1 particle formation. Virology. 1993;193:981–985. doi: 10.1006/viro.1993.1210. [DOI] [PubMed] [Google Scholar]
- 51.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang C T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang C T, Lai H Y, Li J J. Analysis of minimal human immunodeficiency virus type 1 Gag coding sequences capable of virus-like particle assembly and release. J Virol. 1998;72:7950–7959. doi: 10.1128/jvi.72.10.7950-7959.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wang C T, Stegeman-Olsen J, Zhang Y, Barklis E. Assembly of HIV Gag-β-galactosidase fusion proteins into virus particles. Virology. 1994;200:524–534. doi: 10.1006/viro.1994.1215. [DOI] [PubMed] [Google Scholar]
- 55.Wang C T, Zhang Y, McDermott J, Barklis E. Conditional infectivity of a human immunodeficiency virus matrix domain deletion mutant. J Virol. 1993;67:7067–7076. doi: 10.1128/jvi.67.12.7067-7076.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Wang J J, Horton R, Varthakavi V, Spearman P, Ratner L. Formation and release of virus-like particles by HIV-1 matrix protein. AIDS. 1999;13:281–283. doi: 10.1097/00002030-199902040-00018. [DOI] [PubMed] [Google Scholar]
- 57.Willey R L, Bonifacino J S, Potts B J, Martin M A, Klausner R D. Biosynthesis, cleavage, and degradation of the human immunodeficiency virus 1 envelope glycoprotein gp160. Proc Natl Acad Sci USA. 1988;85:9580–9584. doi: 10.1073/pnas.85.24.9580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Worthylake D K, Wang H, Yoo S, Sundquist W I, Hill C P. Structures of the HIV-1 capsid protein dimerization domain at 2.6 Å resolution. Acta Crystallogr Sect D Biol Crystallogr. 1999;55:85–92. doi: 10.1107/S0907444998007689. [DOI] [PubMed] [Google Scholar]
- 59.Zhang W H, Hockley D J, Nermut M V, Morikawa Y, Jones I M. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]
- 60.Zhang Y, Barklis E. Effects of nucleocapsid mutations on human immunodeficiency virus assembly and RNA encapsidation. J Virol. 1997;71:6765–6776. doi: 10.1128/jvi.71.9.6765-6776.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang Y, Qian H, Love Z, Barklis E. Analysis of the assembly function of the human immunodeficiency virus type 1 Gag protein nucleocapsid domain. J Virol. 1998;72:1782–1789. doi: 10.1128/jvi.72.3.1782-1789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhou W, Parent L J, Wills J W, Resh M D. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol. 1994;68:2556–2569. doi: 10.1128/jvi.68.4.2556-2569.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou W, Resh M D. Differential membrane binding of the human immunodeficiency virus type 1 matrix protein. J Virol. 1996;70:8540–8548. doi: 10.1128/jvi.70.12.8540-8548.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]