Abstract
The Epstein-Barr virus (EBV) nuclear antigen 3C (EBNA-3C) protein is a transcriptional regulator of viral and cellular genes that is essential for EBV-mediated immortalization of B lymphocytes in vitro. EBNA-3C can inhibit transcription through an association with the cellular DNA-binding protein Jκ, a function shared by EBNA-3A and EBNA-3B. Here, we report a mechanism by which EBNA-3C can activate transcription from the EBV latent membrane protein 1 (LMP-1) promoter in conjunction with EBNA-2. Jκ DNA-binding sites were not required for this activation, and a mutant EBNA-3C protein unable to bind Jκ activated transcription as efficiently as wild-type EBNA-3C, indicating that EBNA-3C can regulate transcription through a mechanism that is independent of Jκ. Furthermore, activation of the LMP-1 promoter is a unique function of EBNA-3C, not shared by EBNA-3A and EBNA-3B. The DNA element through which EBNA-3C activates the LMP-1 promoter includes a Spi-1/Spi-B binding site, previously characterized as an important EBNA-2 response element. Although this element has considerable homology to mouse immunoglobulin light chain promoter sequences to which the mouse homologue of Spi-1 binds with its dimerization partner IRF4, we demonstrate that the IRF4-like binding sites in the LMP-1 promoter do not play a role in EBNA-3C-mediated activation. Both EBNA-2 and EBNA-3C were required for transcription mediated through a 41-bp region of the LMP-1 promoter encompassing the Spi binding site. However, EBNA-3C had no effect on transcription mediated in conjunction with the EBNA-2 activation domain fused to the GAL4 DNA-binding domain, suggesting that it does not function as an adapter between EBNA-2 and the cellular transcriptional machinery. Like EBNA-2, EBNA-3C bound directly to both Spi-1 and Spi-B in vitro. This interaction was mediated by a region of EBNA-3C encompassing a likely basic leucine zipper (bZIP) domain and the ets domain of Spi-1 or Spi-B, reminiscent of interactions between bZIP and ets domains of other transcription factors that result in their targeting to DNA. There are many examples of regulation of the hematopoietic-specific Spi transcription factors through protein-protein interactions, and a similar regulation by EBNA-3C, in conjunction with EBNA-2, is likely to be an important and unique contribution of EBNA-3C to EBV-mediated immortalization.
The human herpesvirus Epstein-Barr virus (EBV) establishes a latent infection within B lymphocytes that is maintained for the lifetime of the host. Since most EBV-related diseases occur years to decades after primary infection, the establishment of a latent infection is an essential step in the development of EBV-associated malignancies. Following EBV infection in vitro, primary B lymphocytes are immortalized and able to proliferate indefinitely in culture. Of the 12 viral genes expressed during latency in these cells, 6 encode proteins considered essential for efficient EBV-mediated immortalization in vitro: EBV nuclear antigen 1 (EBNA-1), EBNA-2, EBNA-3A, EBNA-3C, EBNA-LP, and latent membrane protein 1 (LMP-1) (10, 18, 23, 28, 29, 50, 52).
The molecular basis for the role of the EBV oncoprotein LMP-1 in transformation is its ability to constitutively activate the tumor necrosis factor receptor signal transduction pathway (36). While LMP-1 is capable of transforming immortal rodent cell lines (11), overexpression of LMP-1 in B cells results in cytotoxicity or cytostasis (15, 31). The expression of LMP-1 in EBV-transformed B lymphocytes is regulated by the concerted actions of viral and cellular proteins through promoter elements targeted by ubiquitous as well as B-cell-specific proteins. One key regulator of LMP-1 expression, EBNA-2, activates transcription through interactions with cellular proteins, including Jκ (for which there are two binding sites in the LMP-1 promoter, located in the regions from bp −298 to −290 and from bp −223 to −213) and Spi-1/Spi-B, related proteins of the ets family of transcription factors that bind to a single site in the LMP-1 promoter (bp −169 to −158) (17, 20, 22, 24, 25, 27, 48). Not only is Jκ the downstream signaling protein of the Notch pathway, but it directly interacts with the intracellular domain of the Notch protein. Following activation of Notch, the intracellular domain is released by proteolysis and migrates to the nucleus to bind to DNA through its interaction with Jκ (16). The presence of Notch provides a signal that activates transcription (21). In this respect, EBNA-2 is functionally analogous to activated Notch: EBNA-2 binds to the promoter through Jκ (17, 20, 27) and provides a strong activation domain that contacts various proteins of the basal transcription machinery (54, 55). Since activation of Notch is associated with several types of cancer (13, 59), the interaction of EBNA-2 with Jκ is likely to play an important role in EBV-mediated immortalization. The mechanism by which EBNA-2 activates transcription through Spi proteins is less clearly defined. Since the Spi binding site is critical for EBNA-2-mediated activation of the LMP-1 promoter and EBNA-2 binds to Spi-1 in vitro (22, 24), it is assumed that EBNA-2 binds to the promoter in a complex with Spi proteins; indeed, Spi-1 is regulated by interactions with a variety of transcription factors (12, 37). The facts that Spi-1 was originally identified as an oncoprotein (35) and that Spi proteins play important roles in the differentiation and proliferation of B lymphocytes (33, 49) suggest that the interaction of EBNA-2 with Spi proteins is important in the immortalization of B lymphocytes by EBV. In addition to interaction with Jκ and Spi proteins, EBNA-2-mediated activation of the LMP-1 promoter can be potentiated by EBNA-LP; this activity appears to involve the transactivation domain of EBNA-2, suggesting a possible role in facilitating contact with the cellular transcription machinery (19, 38), although a direct interaction between EBNA-2 and EBNA-LP has not been demonstrated.
EBNA-3A, -3B, and -3C are also important regulators of LMP-1 expression. The EBNA-3 proteins are encoded by three distinct genes similar in structure and positioned in tandem within the viral genome (45). The EBNA-3 proteins share limited homology in a region near the amino terminus, and this conserved domain mediates binding to Jκ (60). In contrast to the interaction of EBNA-2 with Jκ, the EBNA-3 proteins prevent Jκ from binding to its cognate DNA element, thereby suppressing transcription mediated through Jκ-responsive elements (26, 42, 43, 57, 60).
EBNA-3C, in addition to its well-defined role as a repressor of EBNA-2-mediated activation of transcription (via interaction with Jκ), can also activate gene expression. EBNA-3C contains a potential basic leucine zipper (bZIP) motif close to its amino terminus and a glutamine-proline-rich domain in its carboxyl terminus that functions as a transactivation domain in gene fusion assays (30). The presence of these motifs, therefore, suggests that EBNA-3C might activate gene expression through a direct association with DNA. Indeed, we have reported that EBNA-3C binds to nonspecific DNA in the presence of cellular proteins (44), though a direct or indirect interaction with specific DNA sequences has not been demonstrated. The first experimental evidence implicating EBNA-3C as a transcriptional activator was provided by gene transfer experiments that demonstrated the ability of EBNA-3C to activate expression of both cellular and viral genes. Specifically, in an EBV-negative Burkitt lymphoma (BL) cell line, EBNA-3C induces expression of the complement receptor CD21, also a B-cell activation marker, that functions as the EBV receptor (58). Furthermore, in the EBV-positive BL cell line Raji, which harbors a virus lacking the majority of the EBNA-3C gene, restoration of EBNA-3C expression results in increased LMP-1 protein (1, 2). Our laboratory demonstrated that EBNA-3C activates the LMP-1 promoter in the presence of EBNA-2, suggesting that activation by EBNA-3C occurs at the level of transcription (30).
Here, to ascertain the mechanism(s) by which EBNA-3C activates transcription, we first identified the DNA element(s) in the LMP-1 promoter that is responsive to EBNA-3C. Our data indicate that sequences between positions −181 and −141 of the LMP-1 promoter, encompassing the Spi-1/Spi-B binding site (−169 to −158), mediate activation in trans with EBNA-3C. This activation is clearly distinct from the interaction of EBNA-3C with Jκ that we and others have reported, and is a unique property of EBNA-3C relative to EBNA-3A and EBNA-3B. EBNA-3C-mediated activation requires an intact Spi binding site as well as a fully functional EBNA-2 protein. Furthermore, we have demonstrated a specific interaction between EBNA-3C and both Spi-1 and Spi-B that is mediated by a domain of EBNA-3C encompassing a likely leucine zipper and the ets domain of both Spi-1 and Spi-B. Since interactions between the bZIP and ets domains of several transcription factors mediate binding of both proteins to DNA (4, 47), this suggests the possibility that EBNA-3C may be targeted to DNA in a similar manner.
MATERIALS AND METHODS
Cell culture.
The EBV-negative human BL cell line BL2, the EBV-positive BL cell line Raji, and the EBV-transformed lymphoblastoid cell line IB4 were maintained in RPMI 1640 medium supplemented with 10% fetal bovine serum (Hyclone) and 2 mM l-glutamine.
Plasmids.
The mammalian expression vector pSG5 (Stratagene) was used to express EBNA proteins in transient transfection assays as well as to generate mRNA for in vitro translation. The full length Spi-1 cDNA was provided by F. Moreau-Gachelin (Institut Curie, Paris, France). Part of the 5′ untranslated region of Spi-1 which inhibits in vitro translation was removed by restriction endonuclease digestion with Bsu36I, and the truncated cDNA, containing the full-length open reading frame, was subcloned into pSG5 to generate pSG5-Spi-1. Full-length Spi-B and IRF4 cDNAs, provided by D. G. Tenen (Harvard Medical School) and T. W. Mak (University of Toronto), respectively, were subcloned into pSG5 to generate pSG5-Spi-B and pSG5-IRF-4.
To identify sequences of the LMP-1 promoter mediating activation by EBNA-3C, the following reporter constructs were generated. The mutations in the Jκ binding sites have been previously reported in the context of a promoter extending to bp −512 (30); a fragment extending from bp −512 to −54 was removed from this construct and used to replace the wild-type sequences in a fragment of the LMP-1 promoter extending to bp −2350. The BamHI-MluI (−215/−54 [the numbers given represent the first and last nucleotides of a fragment, relative to the transcription start site]) LMP-1 promoter fragment was subcloned into pBLCAT2, which contains a herpes simplex virus (HSV) thymidine kinase (TK) TATA box, to generate LMP−215/−54BLCAT2. LMP−215/−144BLCAT2 was generated by deletion of the NlaIII-MluI (−144/−54) fragment. The BspHI-BamHI fragment was deleted from the LMP-1 promoter to generate −2350LMPCATd−548/−210. −2350LMPCATd−548/−54 was generated by deletion of BspHI-MluI (−548/−54) fragment. All plasmids used for transfections were purified using anion-exchange resin (Qiagen), followed by cesium chloride density gradient purification.
In vitro transcription and translation.
pSG5-derived expression vectors were linearized at the end of the coding region by restriction endonuclease digestion for use as DNA templates. Spi-1, Spi-B, and IRF4 were generated by using the coupled TNT reticulocyte lysate system (Promega) in the presence of either [35S]methionine (Du Pont) or unlabeled methionine.
Preparation of nuclear extracts.
Nuclear extracts were prepared by a modification of the method of Dignam as previously described. Cells were harvested by centrifugation, washed with ice-cold phosphate-buffered saline, and repelleted by centrifugation. The cells were resuspended in lysis buffer (1× phosphate-buffered saline, 20% glycerol, 0.2% NP-40, 0.5 mM phenylmethylsulfonyl fluoride [PMSF]) and lysed by 10 strokes of a Dounce homogenizer. The nuclei were pelleted by centrifugation and resuspended in buffer C (20 mM HEPES-KOH [pH 7.9], 0.42 M NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM dithiothreitol [DTT]). Nuclei were lysed by 10 strokes of a Dounce homogenizer and incubated at 4°C for 30 min on a rotator. The debris was removed by centrifugation. The supernatant fraction was dialyzed against buffer D (20 mM HEPES-KOH [pH 7.9], 0.1 M KCl, 20% glycerol, 0.2 mM EDTA, 0.5 mM PMSF, 0.5 mM DTT) at 4°C for at least 6 h. The supernatant fraction was clarified by centrifugation and frozen at −70°C. The protein concentration was determined by the Bradford method with a protein assay kit (Bio-Rad).
GST fusion chromatography.
Glutathione S-transferase (GST) fusion proteins were expressed in Escherichia coli and purified on glutathione-Sepharose beads as described previously. All fusion proteins were subjected to sodium dodecyl sulfate (SDS)-polyacrylamide gel electrophoresis (PAGE) to verify migration at the expected size. Purified fusion proteins, bound to glutathione-Sepharose beads, were incubated with 35S-labeled in vitro-translated proteins at 4°C for 30 min in 400 μl of NET-N (120 mM NaCl, 20 mM Tris-HCl, pH 8.0, 1 mM EDTA, and 0.5% Nonidet P-40). Proteins bound to beads were collected by centrifugation and washed five times in 1 ml of NET-N. Bound proteins were eluted by boiling in SDS-PAGE sample buffer, separated by SDS-PAGE, and detected by autoradiography of the dried gel.
Electrophoretic mobility shift assay (EMSA).
The BamHI-NlaIII fragment (−215/−144) of the LMP-1 promoter containing the Spi-1/Spi-B binding site was labeled with [α-32P]dCTP, using the Klenow fragment of DNA polymerase I. Proteins were incubated for 30 min in a 20-μl volume containing 4 μg of poly (dI-dC) (Pharmacia), 10 mM HEPES-KOH (pH 7.9), 60 mM KCl, 4% glycerol, 1 mM EDTA and 1 mM DTT, together with 32P-labeled oligonucleotide (10,000 cpm). DNA-protein complexes were resolved by electrophoresis on a 5% nondenaturing acrylamide gel in 90 mM Tris-HCl, 88 mM boric acid and 2 mM EDTA. For competition, a 100-fold excess of unlabeled double-stranded oligonucleotide was included. Wild-type LMP-1 promoter (−181/−141) or oligonucleotides with mutations in either the Spi-1/Spi-B binding sites or the potential interferon responsive elements (IREs) were used as the competitor.
Transfection and CAT assay.
EBV-negative BL2 cells were transfected at a concentration of 8 × 106 cells per 250 μl of RPMI 1640. Reporter plasmids and expression vectors were introduced into cells by electroporation at 250 V and 960 μF. The total amount of DNA used in each transfection was maintained at a constant level of 30 μg by adding appropriate amounts of empty expression vector. Cells were harvested 36 to 48 h posttransfection and lysed by freeze-thaw cycles. The chloramphenicol acetyltransferase (CAT) activity was determined by standard two-phase partition method and quantitated using a phosphorimager. CAT activity was calculated as the ratio of acetylated [14C]chloramphenicol to the total acetylated and unacetylated [14C]chloramphenicol. Activity was then presented relative to that obtained with the expression vector containing no insert. A pCMV-hGH plasmid in which the human growth hormone gene is under the control of cytomegalovirus immediate-early gene promoter was used as an internal control for differences in the transfection efficiency between samples. The level of growth hormone was determined by radioimmunoassay with a commercial kit (Nichols Institute). All transfections were performed in duplicate and repeated at least three times.
Site-directed mutagenesis.
To determine the roles of the Spi-1/Spi-B binding site and the potential IRE sequences in EBNA-3C-mediated activation, site-directed mutagenesis of the LMP-1 promoter was performed by PCR with the Quikchange site-directed mutagenesis kit (Stratagene). The AAGG core of the PU.1 binding site was mutated to CCTG with primers 5′-CACACGCTTTCTACGGACCCTTTCTACGCTTAC-3′ and 5′-GTAAGCGTAGAAAGGGTCCGTAGAAAGCGTGTG-3′. The IRE-like sequence CCTTTC proximal to the TATA box was mutated to AAGGGA with primers 5′-CGCTTTCTACTTCCAAGGGATACGCTTACATGC-3′ and 5′-GCATGTAAGCGTATCCCTTGGAAGTAGAAAGCG-3′.
An IRE-like sequence GCTTTC distal to the TATA box was mutated to TAGGGA with primers 5′-CACAAACACACTAGGGATACTTCCCCTTTC-3′ and 5′-GAAAGGGGAAGTATCCCTAGTGTGTTTGTG-3′. The mutations were confirmed by sequence analysis, and fragments containing the desired mutations were cloned into LMP-1 reporter gene constructs.
RESULTS
Localization of EBNA-3C-responsive DNA element in the LMP-1 promoter.
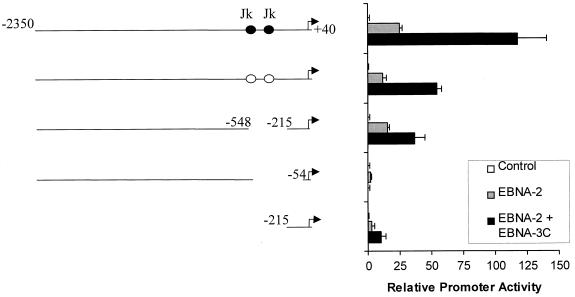
We previously demonstrated that EBNA-3C can increase expression of reporter genes controlled by the LMP-1 promoter in the presence of EBNA-2 (30). Since both EBNA-2 and EBNA-3C can regulate the LMP-1 promoter through interactions with Jκ, we first addressed whether the Jκ binding sites in the LMP-1 promoter contribute to EBNA-3C-mediated activation by examining the ability of EBNA-3C to activate transcription from a fragment of the LMP-1 promoter extending from bp −2350 to +40 in which both Jκ sites were mutated. As shown in Fig. 1, EBNA-3C increased expression from this LMP-1 promoter fragment 4.5-fold relative to that obtained with EBNA-2 alone. Mutation of both Jκ sites (located at bp −298 to −290 and bp −223 to −213) lowered expression due to the loss of these important EBNA-2-responsive elements. However, mutation of these sites did not affect the ability of EBNA-3C to activate expression in the presence of EBNA-2, suggesting that binding of Jκ to the LMP-1 promoter is not required for EBNA-3C-mediated activation.
FIG. 1.
Jκ DNA-binding sites are not required for EBNA-3C-mediated activation of the LMP-1 promoter. The ability of EBNA-3C to activate the LMP-1 promoter was evaluated using a CAT reporter gene assay following transfection of EBV-negative BL2 cells. The truncated or internally deleted fragments of the LMP-1 promoter controlling expression of the CAT reporter gene are depicted schematically at the left; numbers indicate base pair positions relative to the transcriptional start site. Black circles indicate Jκ binding sites. Specific mutations of the Jκ binding sites are indicated by open circles. A 10-μg quantity of each of the reporters indicated was transfected with either empty pSG5 expression vector (control) or pSG5-EBNA-2 in the presence or absence of pSG5-EBNA-3C. CAT activity was measured 36 h after transfection and is quantified relative to the activity obtained with empty expression vector. Error bars represent standard deviations.
Consistent with our mutational analysis described above, deletion of nucleotides between bp −548 and −215 (containing both Jκ sites) from the promoter fragment extending to bp −2350 did not preclude EBNA-3C-mediated activation. However, extension of this deletion from bp −548 to −54 abolished the ability of EBNA-3C to activate this promoter, suggesting that the EBNA-3C-responsive DNA element might lie between bp −215 and −54. Indeed, the region from bp −215 to +40 alone was responsive to EBNA-3C (Fig. 1), although overall promoter activity was reduced, perhaps due to the loss of binding sites for transcription factors that cooperate with EBNA-2 and EBNA-3C to increase promoter activity. However, no effect of EBNA-2 or EBNA-3C was seen when these upstream elements alone were linked to a heterologous promoter and used in reporter gene assays (data not shown). Thus, while other transcription factors probably contribute to total promoter activity, they are not capable of activating transcription themselves, perhaps due to a lack of proximity to the TATA box. By contrast, DNA elements between bp −215 and +40 were sufficient to mediate EBNA-3C responsiveness.
Although the levels of EBNA-2 and EBNA-3C expressed following transfections appeared similar to those present within latently infected cell lines, as monitered by immunofluorescence, it was possible that EBNA-3C-mediated activation was due to overexpression of EBNA-3C. To address this concern, we repeated the transfections using increasing amounts of the EBNA-3C expression vector ranging from 1 to 10 μg. As little as 1 μg of pSG5-EBNA-3C was sufficient to activate expression from the LMP-1 promoter (data not shown). Additionally, immunoblot analysis of transfected cells revealed no change in EBNA-2 protein levels in the presence of EBNA-3C under conditions where a twofold change in EBNA-2 could readily be detected (data not shown), demonstrating that EBNA-3C does not function simply by increasing EBNA-2 levels. To determine whether EBNA-3C could also augment transcription in the presence of levels of EBNA-2 that occur within a latently infected cell line where EBNA-2 is expressed from the EBV genome, we transfected EBNA-3C into the latently infected EBV-positive BL cell line Raji that expresses EBNA-2 but not EBNA-3C and that has been used previously to demonstrate that EBNA-3C activates expression of LMP-1 protein (1, 2). As shown in Fig. 2, EBNA-3C did activate transcription in the presence of physiological levels of EBNA-2.
FIG. 2.
EBNA-3C activated the LMP-1 promoter in the presence of endogenous levels of EBNA-2 in an EBV-positive BL cell line. The ability of EBNA-3C to activate the LMP-1 promoter in the presence of levels of EBNA-2 that occur within EBV latently infected cells was examined by transfection of EBV-positive Raji cells, which express EBNA-2 but not EBNA-3C, with a reporter gene controlled by the LMP-1 promoter and an EBNA-3C expression vector or empty vector (control). Error bars represent standard deviations.
Activation of the LMP-1 promoter is a unique property of EBNA-3C unrelated to interaction with Jκ.
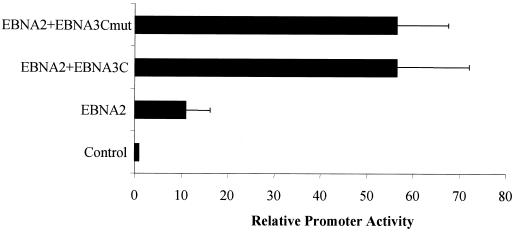
Although the results shown in Fig. 1 demonstrated that activation did not require the Jκ-responsive DNA elements, it was possible that EBNA-3C could bring Jκ to the promoter through an interaction with other DNA elements. As a final test of whether the EBNA-3C–Jκ interaction plays any role in EBNA-3C-mediated activation of the LMP-1 promoter, we used a mutant EBNA-3C protein that, as we have previously demonstrated, does not interact with Jκ and is incapable of regulating transcription through Jκ response elements (60). As shown in Fig. 3, this mutation had no effect on the ability of EBNA-3C to activate expression in the presence of EBNA-2; similar levels of wild-type and mutant EBNA-3C protein were detected by immunoblotting (data not shown).
FIG. 3.
An EBNA-3C mutant that does not interact with Jκ activates the LMP-1 promoter as efficiently as wild-type EBNA-3C. The involvement of the Jκ binding domain in EBNA-3C-mediated activation was investigated by transfection of BL2 cells with a reporter gene controlled by the LMP-1 promoter together with empty expression vector (control) or expressing EBNA-2 alone or in the presence of either wild-type EBNA-3C or a mutant EBNA-3C protein specifically mutated within the Jκ binding domain (EBNA-3Cmut). Error bars represent standard deviations.
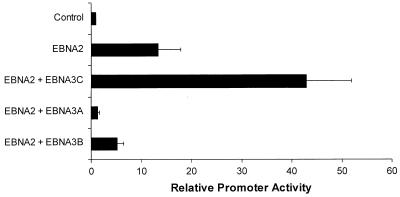
The EBNA-3A and EBNA-3B proteins are distantly related to EBNA-3C, and though they have little actual sequence homology, they share some common properties. These include the ability to bind to Jκ as well as the presence of C-terminal activation domains (D. R. Marshall and C. E. Sample, unpublished data), suggesting that this is a family of transcriptional regulators. Because of these similarities, we investigated whether the ability to activate the LMP-1 promoter was also a conserved property of the EBNA-3 proteins. However, unlike EBNA-3C, neither EBNA-3A nor EBNA-3B was able to activate the LMP-1 promoter (Fig. 4). Together, these data indicate that the ability of EBNA-3C to activate expression from the LMP-1 promoter is clearly distinct from its interaction with Jκ and is a function that is unique to EBNA-3C.
FIG. 4.
Activation of the LMP 1 promoter is a unique property of EBNA-3C not shared by other EBNA-3 proteins. Expression vectors for either EBNA-3A, EBNA-3B, or EBNA-3C were transfected into BL2 cells together with an EBNA-2 expression vector and a reporter plasmid controlled by the LMP-1 promoter. Error bars represent standard deviations.
Sequences between bp −215 and −144 are sufficient for EBNA-3C-mediated activation.
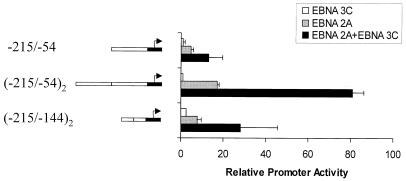
The 255-bp fragment of the LMP-1 promoter (bp −215 to +40) that we determined contains the EBNA-3C response element (Fig. 1) encompasses a TATA box that has the atypical sequence TACATAA, compared with the conventional TATAA. To determine whether the atypical TATA box played a role in EBNA-3C-mediated activation, a fragment of the LMP-1 promoter extending from bp −215 to −54 was cloned into pBLCAT2, which contains a conventional TATA box from the HSV TK promoter. EBNA-3C was able to activate expression through this sequence in the presence of EBNA-2 (Fig. 5), indicating that the LMP-1 TATA box is not required. The total activity from this small fragment was low, however, possibly due to the fact that only a limited number of transcription factors can assemble on this small piece of DNA. When two copies of this 160-bp LMP-1 promoter fragment were cloned into the reporter plasmid, EBNA-3C strongly activated expression in the presence of EBNA-2. Further deletional analysis identified a 71-bp fragment, located between bp −215 and −144, that was activated in trans by EBNA-3C when two copies were placed in pBLCAT2. Activation achieved in the presence of EBNA-3C was greater than that obtained by a 10-fold-greater amount of EBNA-2 expression vector. This 71-bp region of the promoter contains the Spi site (bp −169 to −158) previously demonstrated to be an EBNA-2 responsive element (22, 24). Therefore, one possibility raised by this result is that EBNA-3C-mediated potentiation of EBNA-2's transactivation of the LMP-1 promoter requires sequences adjacent to or coincident with the Spi binding site.
FIG. 5.
Delineation of the LMP 1 promoter DNA element required for EBNA-3C-mediated activation. Reporter plasmids (5 μg) containing one or two copies of small fragments of the LMP-1 promoter (indicated on the left by open bars; numbers given are coordinates of the LMP-1 promoter relative to the start site of transcription) and the HSV TK TATA box (black bar) were transfected into BL2 cells, together with expression vectors for EBNA-2, EBNA-3C, or both. Error bars represent standard deviations.
ets family member Spi-1/Spi-B binding site is required for EBNA-3C-mediated activation.
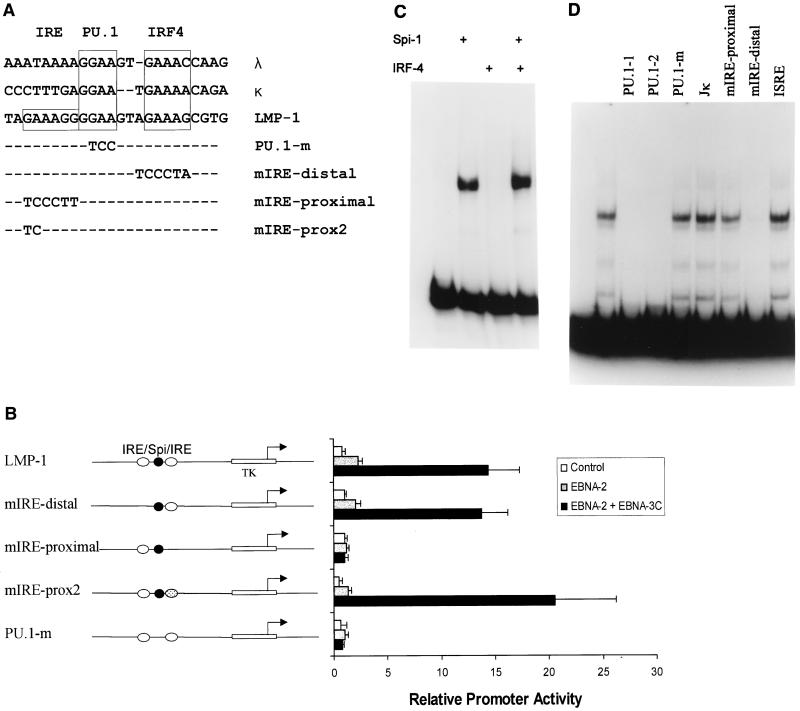
Since ets proteins, including PU.1, the mouse homologue of Spi-1, often bind DNA as heterodimers with other cellular DNA-binding proteins (3, 4, 6, 7, 14, 37, 40, 47, 51), we examined the sequences adjacent to the Spi binding site to determine whether there were any potential binding sites for known transcription factors. Juxtaposed to the Spi site is a potential IRE; together, these sequences are highly homologous to sequences within the mouse immunoglobulin light chain enhancers (Fig. 6A), through which the mouse homologue of Spi-1 recruits IRF4 to activate transcription (12, 40). In the LMP-1 promoter, a second potential IRE site lies on the other side of the Spi site. To determine whether either of these IRE-like sequences are required for EBNA-3C-mediated activation, site-directed mutagenesis was used to introduce mutations into each of these potential IREs as well as within the Spi site itself (Fig. 6A). Mutation of the IRE-like sequences upstream of, or distal to, the TATA box had no effect on EBNA-3C-mediated activation (Fig. 6B), while our initial mutation of the IRE-like sequences downstream of, or proximal to, the TATA box abolished the ability of EBNA-3C to activate this reporter. However, further investigation demonstrated that the mutation proximal to the TATA box affected the ability of Spi proteins to bind to the LMP-1 promoter (see below). Therefore, a smaller mutation that would affect only the downstream IRE-like core sequence and not the Spi site was generated; this mutation had no effect on activation by EBNA-3C (Fig. 6B). By contrast, mutation of the Spi binding site abolished activation by EBNA-2 alone as well as by EBNA-2 in the presence of EBNA-3C.
FIG. 6.
The Spi-1/Spi-B binding site on the LMP-1 promoter is required for EBNA-3C-mediated activation. (A) The EBNA-3C-responsive promoter element has homology to the immunoglobulin light chain λ and κ enhancers as indicated by the boxes. These enhancers contain a PU.1/Spi-1 site and an IRE recognized by the mouse homologue of IRF4. A second potential IRE sequence lies towards the TATA box on the LMP-1 promoter as indicated. Mutations were generated in each of these sites, and the altered bases are indicated below. To demonstrate homology, the lower strand of the LMP-1 DNA is shown; all DNAs are given in a 5′ to 3′ orientation such that the LMP-1 TATA box lies toward the left. (B) The potential IREs (open ovals) in a reporter plasmid containing an HSV TK TATA box and a 71-bp LMP-1 promoter fragment extending from bp −215 to −144 (indicated on the left) were mutated entirely (mIRE-distal and mIRE-proximal, absence of oval) or partially (mIRE-prox2, dotted oval). The Spi binding site (black circle) was also mutated (PU.1m, absence of circle). The ability of EBNA-2 and EBNA-3C to activate these reporters was tested by transfection of BL2 cells with expression vectors for EBNA-2, EBNA-3C, or both. (C) This 71-bp fragment of the LMP-1 promoter (−215/−144) was incubated with either Spi-1 or IRF4, generated by in vitro translation, or with both together, as indicated by the plus signs above the lanes. Protein-DNA complexes were analyzed by EMSA. (D) EMSA was performed with proteins from EBV-positive Raji cells and the EBNA-3C response element from the LMP-1 promoter (−215/−144). Unlabeled double-stranded competitor oligodeoxynucleotides, added to binding reactions in 100-fold excess of probe, included two smaller oligonucleotides containing the PU.1 site (PU.1-1, bp −175 to −154, and PU.1-2, bp −181 to −141) or a mutated PU.1 site (PU.1-m), oligonucleotides containing the mutations shown in panel A (mIRE-proximal and mIRE-distal), the Jκ site from the EBV BamHIC promoter, or the response element from the interferon-stimulated gene 15 promoter.
To determine whether IRF4 could bind to the potential IREs in the LMP-1 promoter, we performed EMSA using the bp −215 to −144 fragment of the LMP-1 promoter. Spi-1 bound to this fragment, as demonstrated by the formation of a single DNA-protein complex (Fig. 6C). Despite the striking sequence homology to the mouse immunoglobulin (Ig) enhancer elements on which Spi-1 and IRF4 form a complex, IRF4 did not bind to the LMP-1 promoter either alone or in the presence of Spi-1. Similar results were obtained with Spi-B (data not shown). These data suggest that, for the LMP-1 promoter, Spi-1 does not function in conjunction with its known dimerization partner, IRF4.
To identify other cellular proteins that form a complex on this minimal EBNA-3C response element and to investigate the effect of EBNA-3C on these complexes, we performed EMSA with nuclear extracts from Raji cells (Fig. 6D), although similar results were obtained with a variety of EBV-positive and -negative B-cell extracts. Multiple protein-DNA complexes could be detected, and to determine the DNA element required to generate these complexes, various oligonucleotides were included as competitors. The majority of complexes could be competed by two separate oligonucleotides containing a Spi-1 site, but neither a mutated Spi-1 site nor a Jκ binding site (included as a negative control) had any effect. These data are similar to the results obtained by others (22, 24), and suggest that the majority of the protein-DNA complexes generated with this fragment are due to binding at or close to the Spi site. To test the effects of the mutations in the IRE-like elements described above on protein binding, we used oligonucleotides containing these mutations as the competitors in EMSAs (Fig. 6D). Only an oligonucleotide containing the largest mutation of the proximal IRE failed to compete for binding of the cellular proteins, likely due to disruption of Spi-1/Spi-B binding. An oligonucleotide containing an interferon-stimulated response element, which IRF4 binds (32) and which has homology to the LMP-1 promoter, did not compete for binding. Collectively, these data suggest that the IRE-like sequences do not play a role in EBNA-3C-mediated activation.
Although we found no differences in protein-DNA complexes formed on the LMP-1 promoter in EBV-positive and EBV-negative cell extracts, it is possible that the amount of EBNA-3C present in the cell is insufficient to form a stable protein-DNA complex detectable by EMSA analysis. Indeed, large amounts of exogenous EBNA-2 are required to detect EBNA-2-Jκ complexes (17, 27, 46, 56). To test this possibility, we added exogenous EBNA-3C, generated by either in vitro translation or baculovirus expression (44), to the binding reactions. Despite a variety of experimental conditions tested, no changes were detected in the binding of any of the complexes in the presence of EBNA-3C. However, we were also unable to detect any changes in complexes in the presence of EBNA-2 included as a control. Although EBNA-2 has been demonstrated to bind Spi-1 (22) and the Spi-1 binding site is critical for EBNA-2 responsiveness of the LMP-1 promoter (22, 24, 25, 48), a protein complex containing both EBNA-2 and Spi-1 has not been demonstrated by EMSA analysis. Furthermore, it has been difficult to demonstrate EBNA-2 targeting through the association with Jκ. Thus, even though no complex formation was detected in the presence of EBNA-3C by EMSA, it remains possible that the effects of EBNA-3C are mediated through direct or indirect interactions with elements coincident with or immediately adjacent to the Spi site.
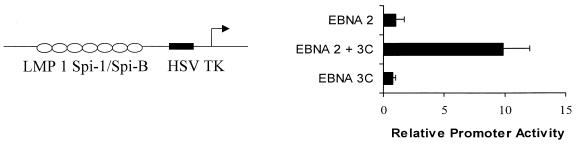
EBNA-3C activates a heterologous promoter controlled by multiple copies of the Spi-1/Spi-B binding site.
Since the protein-DNA complexes generated with the LMP-1 promoter required the Spi site and EBNA-3C-mediated activation of the LMP-1 promoter occurs only in the presence of EBNA-2 and an intact Spi-1/Spi-B binding site, we questioned whether the Spi binding site and the immediately adjacent sequences were sufficient for transactivation by EBNA-3C and EBNA-2. To answer this question, we generated a reporter gene plasmid controlled solely by multiple copies of an oligonucleotide corresponding to bp −181 to −141 that contain the Spi-1/Spi-B binding site located between bp −169 and −158. Interestingly, although EBNA-2 requires the Spi-1/Spi-B binding site for transactivation of the LMP-1 promoter and binds to Spi-1 in vitro, EBNA-2 alone did not activate transcription through the Spi-1 site in this context (Fig. 7), even in the presence of 10 μg of the EBNA-2 expression vector, reinforcing our conclusion that EBNA-3C does not function simply by increasing the levels of EBNA-2. Similarly, EBNA-3C alone did not activate this promoter. However, coexpression of both EBNA-3C and EBNA-2 resulted in activation of reporter gene expression (Fig. 7). The observation that EBNA-2 did not activate transcription of this construct in the absence of EBNA-3C suggests the possibility that EBNA-3C may specifically affect transcription mediated through Spi family members and/or their associated proteins.
FIG. 7.
Multiple copies of a Spi-1/Spi-B oligonucleotide are sufficient for EBNA-3C-mediated activation. Seven copies of the 40-bp LMP-1 promoter fragment (−181/−141) containing the Spi binding site (open ovals) were cloned into a reporter gene plasmid containing the HSV TK TATA box (black bar). The ability of EBNA-2 and EBNA-3C to activate these reporters was determined by transfection of BL2 cells with this reporter gene plasmid in the presence of an expression vector for EBNA-2, EBNA-3C, or both. Error bars represent standard deviations.
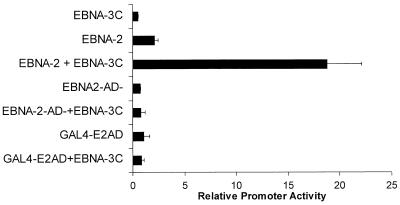
Transcriptionally active EBNA-2 is required for EBNA-3C-mediated activation.
One possible mechanism suggested by our findings is that EBNA-3C participates in a protein-DNA complex that is stabilized in the presence of EBNA-2. We reasoned that if this were true an EBNA-2 protein deleted for the activation domain might be able to participate in the formation of the complex and that the glutamine-proline-rich transactivation domain of EBNA-3C might be sufficient to activate transcription. To test this, we used a mutant EBNA-2 protein from which the transactivation domain had been deleted (9). Although EBNA-3C strongly activated the LMP-1 promoter in the presence of wild-type EBNA-2, no activation was detected in the presence of this mutant EBNA-2 protein (Fig. 8). Similarly, EBNA-3C did not activate the LMP-1 promoter in the presence of the EBNA-2 activation domain alone, furnished as a fusion protein of the GAL4 DNA-binding domain and the EBNA-2 transactivation domain. Thus, neither the EBNA-2 domain that interacts with Spi-1 nor the transactivation domain alone is sufficient to participate in EBNA-3C-mediated activation.
FIG. 8.
Transcriptionally active EBNA-2 is required for EBNA-3C-mediated activation of the LMP-1 promoter. Expression vectors for EBNA-2, a transcriptionally inactive EBNA-2 with a deletion in the transactivation domain (EBNA2-AD-), or a fusion between the EBNA-2 transactivation domain (amino acids 426 to 462) and the GAL4 DNA-binding domain (Gal4-E2AD) were transfected into BL2 cells with a reporter gene controlled by the EBNA-3C-responsive element from the LMP-1 promoter in the presence or absence of EBNA-3C.
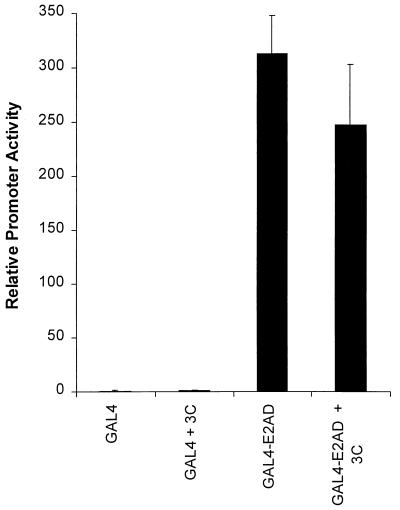
A second possible mechanism that would require the activation domain of EBNA-2 is that EBNA-3C functions as an adapter between EBNA-2 and the transcription machinery. To test this hypothesis, we examined the ability of a protein containing the EBNA-2 transactivation domain and the GAL4 DNA-binding domain (8) to activate a GAL4-responsive reporter in the presence and absence of EBNA-3C. As illustrated in Fig. 9, EBNA-3C had no effect on the ability of the GAL4 EBNA-2 fusion protein to activate expression from a GAL4-responsive promoter. This data suggests that EBNA-3C does not simply function as an adapter between the EBNA-2 activation domain and the transcription machinery.
FIG. 9.
EBNA-3C has no effect on transcription mediated by the activation domain of EBNA-2. To test the effect of EBNA-3C on transcription activated by the activation domain of EBNA-2, a GAL4-responsive reporter gene plus expression vectors for the GAL4 DNA-binding domain or GAL4-E2AD were transfected into BL2 cells in the presence or absence of EBNA-3C.
EBNA-3C interacts with Spi-1 and Spi-B in vitro.
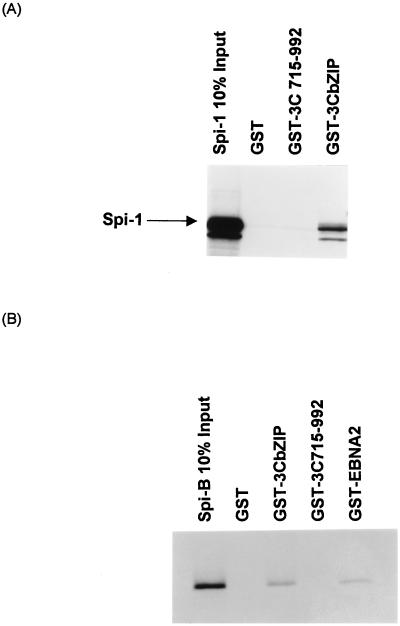
Since a 40-bp oligonucleotide encompassing the Spi binding site functions as an EBNA-3C response element (Fig. 7) and the basic regions of several bZIP family transcription factors interact with ets family members, including Spi-1 (4, 47), it is possible that a similar direct physical interaction might occur between EBNA-3C and the Spi transcription factors. To examine this possibility, Spi-1 generated by in vitro translation was incubated with a series of GST–EBNA-3C fusion proteins. Spi-1 did not bind to GST or GST fused to the C terminus of EBNA-3C (amino acids 751 to 952). However, Spi-1 did associate with a domain of EBNA-3C (amino acids 181 to 365) encompassing the potential bZIP motif (amino acids 255 to 290) fused to GST (Fig. 10A). Identical results were obtained with Spi-B (Fig. 10B). Furthermore, the association between Spi-B and the bZIP motif of EBNA-3C appeared equivalent to that obtained with GST–EBNA-2.
FIG. 10.
EBNA-3C interacts with Spi-1 and Spi-B in vitro. A fragment of the EBNA-3C cDNA encoding the Jκ binding domain and the bZIP motif (GST-3CbZIP, containing amino acids 181 to 365 of EBNA-3C), a C-terminal domain of EBNA-3C (GST-3C715-992), or the C-terminal two-thirds of EBNA-2, between amino acids 117 and 484 (GST-EBNA2), were used to generate GST fusion proteins. These fusion proteins bound to glutathione beads were incubated with 35S-labeled Spi-1 (A) or Spi-B (B) generated by in vitro translation. Proteins bound to the fusion proteins were separated by SDS-PAGE and detected by autoradiography.
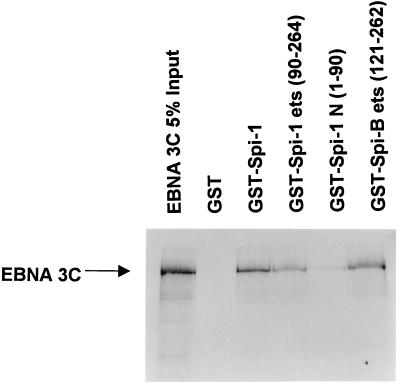
Other leucine zipper proteins that have been reported to bind to ets proteins interact specifically with the ets domain. To identify the domain of Spi-1 and Spi-B that interacts with EBNA-3C, various GST-Spi fusion proteins were generated. Full-length Spi-1 (as well as the ets domain of both Spi-1 and Spi-B) bound to EBNA-3C, whereas no binding by the amino-terminal activation domain of Spi-1 was observed (Fig. 11). These data suggest a possible model whereby EBNA-3C is targeted to DNA through an interaction between its leucine zipper domain and the ets domain of the Spi proteins.
FIG. 11.
EBNA-3C binds to the ets domain of Spi-1 and Spi-B. GST fusion proteins that contained full-length Spi-1 (GST-Spi-1), the amino-terminal activation domain [GST-Spi-1 N (1-90)], or the C-terminal ets domain of either Spi-1 [GST-Spi-1 ets (90-264)] or Spi-B [GST-Spi-B ets (121-262)] were generated. These proteins were incubated with 35S-labeled EBNA-3C generated by in vitro translation.
DISCUSSION
Genetic experiments have demonstrated that both EBNA-3A and EBNA-3C are essential for EBV-mediated immortalization of B lymphocytes (52), suggesting that each protein plays a unique role in this process. Here, we have shown that a unique function of EBNA-3C is its ability to activate expression from the LMP-1 promoter in the presence of EBNA-2. Since all three EBNA-3 proteins bind Jκ, one would predict that the ability of EBNA-3C to activate the LMP-1 promoter is distinct from its interaction with Jκ. Indeed, we have demonstrated that a mutant EBNA-3C protein incapable of binding to Jκ is able to activate expression from the LMP-1 promoter and that a 41-bp LMP-1 promoter fragment, containing a Spi-1/Spi-B but not a Jκ site, was sufficient for EBNA-3C-mediated activation.
The Spi binding site in the LMP-1 promoter is also essential for EBNA-2-mediated activation of this promoter (22, 24, 48), and EBNA-2, like EBNA-3C, binds to Spi-1 in vitro (22). Since EBNA-2 has no demonstrable DNA-binding capability itself, it has been postulated that EBNA-2 is targeted to DNA through its interaction with Spi-1 (22), though a protein-DNA complex containing EBNA-2 and Spi-1 has yet to be demonstrated. Since ets proteins typically bind to DNA as heterodimers (3, 4, 6, 14, 47, 51), it is possible that other cellular proteins participate in this complex. For example, the murine homologue of Spi-1, PU.1, recruits IRF4 (also known as PIP or LSIRF) to the Ig κ and λ 3′ enhancers to facilitate the formation of a transcriptionally active complex that also includes AP-1 (12, 40). However, despite its striking sequence homology to the Spi/IRF4 binding sites in the Ig κ enhancer, the Spi site in the LMP-1 promoter did not support the assembly of a similar complex between Spi proteins and IRF4. Instead, our data demonstrate that the ets domains of both Spi-1 and Spi-B proteins interact with a region of EBNA-3C containing a likely bZIP domain. Although we have thus far been unable to detect a protein-DNA complex containing EBNA-3C and Spi proteins by EMSA, there is ample precedent for interactions between a bZIP domain and an ets domain mediating the formation of a protein-DNA complex (4, 47). Thus, although EBNA-3C has no known DNA-binding capability, by providing a strong activation domain, it may provide a function analogous to that of IRF4 in its interaction with Spi-1. The simplest model that incorporates both the previous findings with EBNA-2 and the data presented here, therefore, is that EBNA-2, EBNA-3C, and Spi proteins are each needed to form a transcriptionally active complex on DNA.
The targeting of a Spi protein by EBNA-2 and EBNA-3C is reminiscent of the interaction of both EBV proteins with Jκ. Clearly then, one function of the EBNA-3 proteins is to regulate the activity of EBNA-2. EBNA-3C is thus far unique among the EBNA-3 proteins in that it both positively and negatively regulates EBNA-2 activity on the same promoter, albeit through distinct DNA elements. Although the significance of this is not yet clear, certainly levels of LMP-1 must be tightly regulated because, while essential for immortalization, high levels of LMP-1 lead to cytostasis (15, 31). Thus, one possibility is that EBNA-3C may mediate each of these effects at a distinct point in the cell cycle in order to fine-tune levels of LMP-1. In support of such a hypothesis, interaction of Spi-1 with at least one cellular protein is regulated by phosphorylation (39); we have previously demonstrated that EBNA-3C itself is a phosphoprotein, though it is not known whether this contributes to any regulatory role it might play (44). What advantage might be provided by the interaction of both EBNA-2 and EBNA-3C with Spi proteins? One possibility is that, individually, EBNA-3C and EBNA-2 are only weakly tethered to the LMP-1 promoter in conjunction with Spi-1 or Spi-B but that a more stable complex is formed in the presence of both viral proteins. Indeed, neither EBNA-2 nor EBNA-3C alone is able to activate transcription through the 41-bp promoter fragment containing the Spi binding site. Transcription initiation likely requires the acidic domain of EBNA-2 that binds to TFIIB, TAF40, and TFIIH (53, 55). Since the EBNA-2 acidic domain does not bind to TBP, whereas glutamine-proline-rich transactivation domains in other transcription factors have been shown to bind to TBP, the glutamine-proline-rich activation domain of EBNA-3C may therefore be required to facilitate the formation of an active transcriptional complex. Previous results have indicated that EBNA-2 is unable to activate expression of the LMP-1 promoter through Spi-1 alone, and the data suggested that an unidentified factor, termed LMP-1 binding factor 7 (LBF7), that binds to bp −215 to −205 of the LMP-1 promoter might also be required to mediate EBNA-2 responsiveness (22). Although we also find that EBNA-2 cannot activate expression through Spi binding sites alone, the addition of EBNA-3C is sufficient to restore EBNA-2 responsiveness, demonstrating that a factor binding to bp −215 to −205 is not absolutely required, though it may contribute to maximal promoter activity. Interestingly, the mutation generated in the upstream IRE-like sequence overlaps the binding site for a second factor, LBF4 (22). Mutation of the LBF4 binding site has no effect on the activation of LMP-1 by EBNA-2 (22). Our finding that a similar mutation has no effect on the ability of EBNA-3C to activate the LMP-1 promoter suggests that LBF4 does not play a role in EBNA-3C-mediated activation.
Although the detailed mechanism(s) through which activation of LMP-1 expression occurs is not yet known, EBNA-3C can clearly activate gene expression, at least of LMP-1, via a cooperative mechanism involving both viral (EBNA-2) and cellular (Spi-1/Spi-B) transcription factors. Spi-1 and Spi-B, which have more divergent ets domains than other ets family members (34, 41), play important roles in the development and differentiation of B lymphocytes (33, 49), and regulate the expression of a variety of genes critical for the functions of hematopoietic-lineage cells (5). Clearly, EBNA-3C is likely to affect cellular genes that contribute to the immortalization of B lymphocytes through its interaction with Spi proteins. Future experiments, therefore, will explore the mechanism(s) through which EBNA-3C regulates transcription and attempt to identify those cellular genes regulated by EBNA-3C to clarify its role(s) in EBV-mediated immortalization and viral latency.
ACKNOWLEDGMENTS
This research was supported by U.S. Public Health Research grants CA-56645 and CA73561 from the National Cancer Institute, Cancer Center Support (CORE) grant CA-21765, and the American Lebanese Syrian Associated Charities (ALSAC).
We thank Jennifer Moore, Evelyn Stigger-Rosser, and Mary Manspeaker for excellent technical support and Jeff Sample and Rozenn Dalbiès for critical reading of the manuscript.
REFERENCES
- 1.Allday M J, Crawford D H, Thomas J A. Epstein-Barr virus (EBV) nuclear antigen 6 induces expression of the EBV latent membrane protein and an activated phenotype in Raji cells. J Gen Virol. 1993;74:361–369. doi: 10.1099/0022-1317-74-3-361. [DOI] [PubMed] [Google Scholar]
- 2.Allday M J, Farrell P J. Epstein-Barr virus nuclear antigen EBNA3C/6 expression maintains the level of latent membrane protein 1 in G1-arrested cells. J Virol. 1994;68:3491–3498. doi: 10.1128/jvi.68.6.3491-3498.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bassuk A G, Anandappa R T, Leiden J M. Physical interactions between Ets and NF-κB/NFAT proteins play an important role in their cooperative activation of the human immunodeficiency virus enhancer in T cells. J Virol. 1997;71:3563–3573. doi: 10.1128/jvi.71.5.3563-3573.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bassuk A G, Leiden J M. A direct physical association between ETS and AP-1 transcription factors in normal human T cells. Immunity. 1995;3:223–237. doi: 10.1016/1074-7613(95)90092-6. [DOI] [PubMed] [Google Scholar]
- 5.Bassuk A G, Leiden J M. The role of Ets transcription factors in the development and function of the mammalian immune system. Adv Immunol. 1997;64:65–104. doi: 10.1016/s0065-2776(08)60887-1. [DOI] [PubMed] [Google Scholar]
- 6.Bradford A P, Conrad K E, Wasylyk C, Wasylyk B, Gutierrez-Hartmann A. Functional interaction of c-Ets-1 and GHF-1/Pit-1 mediates Ras activation of pituitary-specific gene expression: mapping of the essential c-Ets-1 domain. Mol Cell Biol. 1995;15:2849–2857. doi: 10.1128/mcb.15.5.2849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brass A L, Kehrli E, Eisenbeis C F, Storb U, Singh H. Pip, a lymphoid-restricted IRF, contains a regulatory domain that is important for autoinhibition and ternary complex formation with the Ets factor PU.1. Genes Dev. 1996;10:2335–2347. doi: 10.1101/gad.10.18.2335. [DOI] [PubMed] [Google Scholar]
- 8.Cohen J I, Kieff E. An Epstein-Barr virus nuclear protein 2 domain essential for transformation is a direct transcriptional activator. J Virol. 1991;65:5880–5885. doi: 10.1128/jvi.65.11.5880-5885.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cohen J I, Wang F, Kieff E. Epstein-Barr virus nuclear protein 2 mutations define essential domains for transformation and transactivation. J Virol. 1991;65:2545–2554. doi: 10.1128/jvi.65.5.2545-2554.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen J I, Wang F, Mannick J, Kieff E. Epstein-Barr virus nuclear protein 2 is a key determinant of lymphocyte transformation. Proc Natl Acad Sci USA. 1989;86:9558–9562. doi: 10.1073/pnas.86.23.9558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dambaugh T, Wang F, Hennessy K, Woodland E, Rickinson A, Kieff E. Expression of the Epstein-Barr virus nuclear protein 2 in rodent cells. J Virol. 1986;59:453–462. doi: 10.1128/jvi.59.2.453-462.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Eisenbeis C F, Singh H, Storb U. Pip, a novel IRF family member, is a lymphoid-specific, PU.1-dependent transcriptional activator. Genes Dev. 1995;9:1377–1387. doi: 10.1101/gad.9.11.1377. [DOI] [PubMed] [Google Scholar]
- 13.Ellisen L W, Bird J, West D C, Soreng A L, Reynolds T C, Smith S D, Sklar J. TAN-1, the human homolog of the Drosophila notch gene, is broken by chromosomal translocations in T lymphoblastic neoplasms. Cell. 1991;66:649–661. doi: 10.1016/0092-8674(91)90111-b. [DOI] [PubMed] [Google Scholar]
- 14.Fitzsimmons D, Hodsdon W, Wheat W, Maira S M, Wasylyk B, Hagman J. Pax-5 (BSAP) recruits Ets proto-oncogene family proteins to form functional ternary complexes on a B-cell-specific promoter. Genes Dev. 1996;10:2198–2211. doi: 10.1101/gad.10.17.2198. [DOI] [PubMed] [Google Scholar]
- 15.Floettmann J E, Ward K, Rickinson A B, Rowe M. Cytostatic effect of Epstein-Barr virus latent membrane protein-1 analyzed using tetracycline-regulated expression in B cell lines. Virology. 1996;223:29–40. doi: 10.1006/viro.1996.0452. [DOI] [PubMed] [Google Scholar]
- 16.Fortini M E, Artavanis-Tsakonas S. The suppressor of hairless protein participates in notch receptor signaling. Cell. 1994;79:273–282. doi: 10.1016/0092-8674(94)90196-1. [DOI] [PubMed] [Google Scholar]
- 17.Grossman S R, Johannsen E, Tong X, Yalamanchili R, Kieff E. The Epstein-Barr virus nuclear antigen 2 transactivator is directed to response elements by the Jκ recombination signal binding protein. Proc Natl Acad Sci USA. 1994;91:7568–7572. doi: 10.1073/pnas.91.16.7568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hammerschmidt W, Sugden B. Genetic analysis of immortalizing functions of Epstein-Barr virus in human B lymphocytes. Nature. 1989;340:393–397. doi: 10.1038/340393a0. [DOI] [PubMed] [Google Scholar]
- 19.Harada S, Kieff E. Epstein-Barr virus nuclear protein LP stimulates EBNA-2 acidic domain-mediated transcriptional activation. J Virol. 1997;71:6611–6618. doi: 10.1128/jvi.71.9.6611-6618.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 21.Jarriault S, Brou C, Logeat F, Schroeter E H, Kopan R, Israel A. Signalling downstream of activated mammalian Notch. Nature. 1995;377:355–358. doi: 10.1038/377355a0. [DOI] [PubMed] [Google Scholar]
- 22.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by Jκ and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kaye K M, Izumi K M, Kieff E. Epstein-Barr virus latent membrane protein 1 is essential for B-lymphocyte growth transformation. Proc Natl Acad Sci USA. 1993;90:9150–9154. doi: 10.1073/pnas.90.19.9150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Laux G, Adam B, Strobl L J, Moreau-Gachelin F. The Spi-1/PU.1 and Spi-B ets family transcription factors and the recombination signal binding protein RBP-J kappa interact with an Epstein-Barr virus nuclear antigen 2 responsive cis-element. EMBO J. 1994;13:5624–5632. doi: 10.1002/j.1460-2075.1994.tb06900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laux G, Dugrillon F, Eckert C, Adam B, Zimber-Strobl U, Bornkamm G W. Identification and characterization of an Epstein-Barr virus nuclear antigen 2-responsive cis element in the bidirectional promoter region of latent membrane protein and terminal protein 2 genes. J Virol. 1994;68:6947–6958. doi: 10.1128/jvi.68.11.6947-6958.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Le Roux A, Kerdiles B, Walls D, Dedieu J F, Perricaudet M. The Epstein-Barr virus determined nuclear antigens EBNA-3A, -3B, and -3C repress EBNA-2-mediated transactivation of the viral terminal protein 1 gene promoter. Virology. 1994;205:596–602. doi: 10.1006/viro.1994.1687. [DOI] [PubMed] [Google Scholar]
- 27.Ling P D, Rawlins D R, Hayward S D. The Epstein-Barr virus immortalizing protein EBNA-2 is targeted to DNA by a cellular enhancer-binding protein. Proc Natl Acad Sci USA. 1993;90:9237–9241. doi: 10.1073/pnas.90.20.9237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Longnecker R, Miller C L, Miao X-Q, Tomkinson B, Kieff E. The last seven transmembrane and carboxy-terminal cytoplasmic domains of Epstein-Barr virus latent membrane protein 2 (LMP2) are dispensable for lymphocyte infection and growth transformation in vitro. J Virol. 1993;67:2006–2013. doi: 10.1128/jvi.67.4.2006-2013.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mannick J B, Cohen J I, Birkenbach M, Marchini A, Kieff E. The Epstein-Barr virus nuclear protein encoded by the leader of the EBNA RNAs is important in B-lymphocyte transformation. J Virol. 1991;65:6826–6837. doi: 10.1128/jvi.65.12.6826-6837.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Marshall D, Sample C. Epstein-Barr virus nuclear antigen 3C is a transcriptional regulator. J Virol. 1995;69:3624–3630. doi: 10.1128/jvi.69.6.3624-3630.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Martin J M, Veis D, Korsmeyer S J, Sugden B. Latent membrane protein of Epstein-Barr virus induces cellular phenotypes independently of expression of Bcl-2. J Virol. 1993;67:5269–5278. doi: 10.1128/jvi.67.9.5269-5278.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Matsuyama T, Grossman A, Mittrucker H W, Siderovski D P, Kiefer F, Kawakami T, Richardson C D, Taniguchi T, Yoshinaga S K, Mak T W. Molecular cloning of LSIRF, a lymphoid-specific member of the interferon regulatory factor family that binds the interferon-stimulated response element (ISRE) Nucleic Acids Res. 1995;23:2127–2136. doi: 10.1093/nar/23.12.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKercher S R, Torbett B E, Anderson K L, Henkel G W, Vestal D J, Baribault H, Klemsz M, Feeney A J, Wu G E, Paige C J, Maki R A. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- 34.Moreau-Gachelin F. Spi-1/PU.1: an oncogene of the Ets family. Biochim Biophys Acta. 1994;1198:149–163. doi: 10.1016/0304-419x(94)90011-6. [DOI] [PubMed] [Google Scholar]
- 35.Moreau-Gachelin F, Tavitian A, Tambourin P. Spi-1 is a putative oncogene in virally induced murine erythroleukaemias. Nature. 1988;331:277–280. doi: 10.1038/331277a0. [DOI] [PubMed] [Google Scholar]
- 36.Mosialos G, Birkenbach M, Yalamanchili R, VanArsdale T, Ware C, Kieff E. The Epstein-Barr virus transforming protein LMP1 engages signaling proteins for the tumor necrosis factor receptor family. Cell. 1995;80:389–399. doi: 10.1016/0092-8674(95)90489-1. [DOI] [PubMed] [Google Scholar]
- 37.Nagulapalli S, Pongubala J M, Atchison M L. Multiple proteins physically interact with PU.1. Transcriptional synergy with NF-IL6 beta (C/EBP delta, CRP3) J Immunol. 1995;155:4330–4338. [PubMed] [Google Scholar]
- 38.Nitsche F, Bell A, Rickinson A. Epstein-Barr virus leader protein enhances EBNA-2-mediated transactivation of latent membrane protein 1 expression—a role for the W1W2 repeat domain. J Virol. 1997;71:6619–6628. doi: 10.1128/jvi.71.9.6619-6628.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pongubala J M, Van B C, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. Effect of PU.1 phosphorylation on interaction with NF-EM5 and transcriptional activation. Science. 1993;259:1622–1625. doi: 10.1126/science.8456286. [DOI] [PubMed] [Google Scholar]
- 40.Pongubala J M R, Nagulapalli S, Klemsz M J, McKercher S R, Maki R A, Atchison M L. PU.1 recruits a second nuclear factor to a site important for immunoglobulin κ 3′ enhancer activity. Mol Cell Biol. 1992;12:368–378. doi: 10.1128/mcb.12.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ray D, Bosselut R, Ghysdael J, Mattei M G, Tavitian A, Moreau-Gachelin F. Characterization of Spi-B, a transcription factor related to the putative oncoprotein Spi-1/PU.1. Mol Cell Biol. 1992;12:4297–4304. doi: 10.1128/mcb.12.10.4297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Robertson E S, Grossman S, Johannsen E, Miller C, Lin J, Tomkinson B, Kieff E. Epstein-Barr virus nuclear protein 3C modulates transcription through interaction with the sequence-specific DNA-binding protein Jκ. J Virol. 1995;69:3108–3116. doi: 10.1128/jvi.69.5.3108-3116.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Robertson E S, Lin J, Kieff E. The amino-terminal domains of Epstein-Barr virus nuclear proteins 3A, 3B, and 3C interact with RBPJκ. J Virol. 1996;70:3068–3074. doi: 10.1128/jvi.70.5.3068-3074.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sample C E, Parker B D. Biochemical characterization of the Epstein-Barr virus nuclear antigen 3A and 3C proteins. Virology. 1994;205:535–539. doi: 10.1006/viro.1994.1675. [DOI] [PubMed] [Google Scholar]
- 45.Sample J, Young L, Martin B, Chatman T, Kieff E, Rickinson A. Epstein-Barr virus types 1 and 2 differ in their EBNA-3A, EBNA-3B, and EBNA-3C genes. J Virol. 1990;64:4084–4092. doi: 10.1128/jvi.64.9.4084-4092.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sauder C, Haiss P, Grasser F A, Zimber-Strobl U, Mueller-Lantzsch N. DNA-binding studies of the Epstein-Barr virus nuclear antigen 2 (EBNA-2): evidence for complex formation by latent membrane protein gene promoter-binding proteins in EBNA-2-positive cell lines. J Gen Virol. 1994;75:3067–3079. doi: 10.1099/0022-1317-75-11-3067. [DOI] [PubMed] [Google Scholar]
- 47.Sieweke M H, Tekotte H, Frampton J, Graf T. MafB is an interaction partner and repressor of Ets-1 that inhibits erythroid differentiation. Cell. 1996;85:49–60. doi: 10.1016/s0092-8674(00)81081-8. [DOI] [PubMed] [Google Scholar]
- 48.Sjoblom A, Jansson A, Yang W, Lain S, Nilsson T, Rymo L. PU box-binding transcription factors and a POU domain protein cooperate in the Epstein-Barr virus (EBV) nuclear antigen 2-induced transactivation of the EBV latent membrane protein 1 promoter. J Gen Virol. 1995;76:2679–2692. doi: 10.1099/0022-1317-76-11-2679. [DOI] [PubMed] [Google Scholar]
- 49.Su G H, Chen H M, Muthusamy N, Garrett-Sinha L A, Baunoch D, Tenen D G, Simon M C. Defective B cell receptor-mediated responses in mice lacking the Ets protein, Spi-B. EMBO J. 1997;16:7118–7129. doi: 10.1093/emboj/16.23.7118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Swaminathan S, Tomkinson B, Kieff E. Recombinant Epstein-Barr virus with small RNA (EBER) genes deleted transforms lymphocytes and replicates in vitro. Proc Natl Acad Sci USA. 1991;88:1546–1550. doi: 10.1073/pnas.88.4.1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thomas R S, Tymms M J, McKinlay L H, Shannon M F, Seth A, Kola I. ETS1, NFkappaB and AP1 synergistically transactivate the human GM-CSF promoter. Oncogene. 1997;14:2845–2855. doi: 10.1038/sj.onc.1201125. [DOI] [PubMed] [Google Scholar]
- 52.Tomkinson B, Robertson E, Kieff E. Epstein-Barr virus nuclear proteins EBNA-3A and EBNA-3C are essential for B-lymphocyte growth transformation. J Virol. 1993;67:2014–2025. doi: 10.1128/jvi.67.4.2014-2025.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tong X, Drapkin R, Reinberg D, Kieff E. The 62- and 80-kDa subunits of transcription factor IIH mediate the interaction with Epstein-Barr virus nuclear protein 2. Proc Natl Acad Sci USA. 1995;92:3259–3263. doi: 10.1073/pnas.92.8.3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tong X, Drapkin R, Yalamanchili R, Mosialos G, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain forms a complex with a novel cellular coactivator that can interact with TFIIE. Mol Cell Biol. 1995;15:4735–4744. doi: 10.1128/mcb.15.9.4735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tong X, Wang F, Thut C J, Kieff E. The Epstein-Barr virus nuclear protein 2 acidic domain can interact with TFIIB, TAF40, and RPA70 but not with TATA-binding protein. J Virol. 1995;69:585–588. doi: 10.1128/jvi.69.1.585-588.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-J kappa repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Waltzer L, Perricaudet M, Sergeant A, Manet E. Epstein-Barr virus EBNA3A and EBNA3C proteins both repress RBP-Jκ-EBNA-2-activated transcription by inhibiting the binding of RBP-Jκ to DNA. J Virol. 1996;70:5909–5915. doi: 10.1128/jvi.70.9.5909-5915.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang F, Gregory C, Sample C, Rowe M, Liebowitz D, Murray R, Rickinson A, Kieff E. Epstein-Barr virus latent membrane protein (LMP1) and nuclear proteins 2 and 3C are effectors of phenotypic changes in B lymphocytes: EBNA-2 and LMP1 cooperatively induce CD23. J Virol. 1990;64:2309–2318. doi: 10.1128/jvi.64.5.2309-2318.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zagouras P, Stifani S, Blaumueller C M, Carcangiu M L, Artavanis-Tsakonas S. Alterations in Notch signaling in neoplastic lesions of the human cervix. Proc Natl Acad Sci USA. 1995;92:6414–6418. doi: 10.1073/pnas.92.14.6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhao B, Marshall D R, Sample C E. A conserved domain of the Epstein-Barr virus nuclear antigens 3A and 3C binds to a discrete domain of Jκ. J Virol. 1996;70:4228–4236. doi: 10.1128/jvi.70.7.4228-4236.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]