Abstract
Genome-wide association studies (GWASs) have identified specific genetic variants associated with complex human traits and behaviors, such as educational attainment, mental disorders, and personality. However, small effect sizes for individual variants, uncertainty regarding the biological function of discovered genotypes, and potential “outside-the-skin” environmental mechanisms leave a translational gulf between GWAS results and scientific understanding that will improve human health and well-being. We propose a set of social, behavioral, and brain-science research activities that map discovered genotypes to neural, developmental, and social mechanisms and call this research program phenotypic annotation. Phenotypic annotation involves (a) elaborating the nomological network surrounding discovered genotypes, (b) shifting focus from individual genes to whole genomes, and (c) testing how discovered genotypes affect life-span development. Phenotypic-annotation research is already advancing the understanding of GWAS discoveries for educational attainment and schizophrenia. We review examples and discuss methodological considerations for psychologists taking up the phenotypic-annotation approach.
Keywords: genetics, GWAS, gene–environment correlation, polygenic scores, development
Differences in DNA sequences between people are an important source of individual differences in their psychology (Turkheimer, 2000). More closely related relatives more strongly resemble one another in factors such as memory, intelligence, personality, self-esteem, physical health, and mental health; everything is heritable (Polderman et al., 2015). Yet these family-based estimates of heritability are “black boxes” regarding mechanisms: Heritability estimates reveal neither which genetic variants are important nor how differences in DNA sequences result in, for example, a person being smarter, more prone to depression, or more likely to have a psychotic break.
Still a Black Box? From Twin Studies to Genome-Wide Association Studies (GWASs)
Now, GWASs (see the Appendix for a glossary of key terms used in this article) are peering inside the black box of heritability. The GWAS method is hypothesis free, meaning that it does not focus on specific genetic variants selected on the basis of prior knowledge about biological function. Rather, a GWAS surveys genetic variation across the genome. Each one of millions of common genetic variants known as single-nucleotide polymorphisms (SNPs) is tested, with a rigorous statistical correction to control the Type I error rate. Despite initial skepticism, GWAS discoveries for human diseases, traits, and behaviors now number in the thousands (Visscher et al., 2017). Specific SNPs have been identified in GWASs of psychology-relevant phenotypes, including neuroticism, schizophrenia, reproductive behavior, intelligence, and educational attainment (https://www.ebi.ac.uk/gwas/).
Despite the accelerating pace of GWAS discovery, heritabilities of human traits and behaviors largely remain black boxes. A key finding from GWASs has been confirmation that genetic influence on variation in most human phenotypes reflects the combined effects of very large numbers of individual genetic variants, each of which has a tiny effect size (R2 < .01; Chabris, Lee, Cesarini, Benjamin, & Laibson, 2015). However, discovered SNPs have turned up only rarely in or near the genes that researchers had hypothesized would be important, and, as we discuss below, the biology linking discovered SNPs to phenotypes is often unclear (Boyle, Li, & Pritchard, 2017). Thus, GWASs have opened the black box of heritability only to find thousands on thousands of smaller black boxes—genotypes of uncertain function that are correlated with phenotypes via unknown mechanisms.
The dominant approach to making sense of GWAS results is bioinformatics annotation. To annotate is to add notes that explain and interpret a text. Bioinformatics annotation takes the minimal text rendered by a GWAS, a list of associations between individual SNPs and a phenotype, and attempts to explain that text using insights from biology. For example, bioinformatics annotation might draw on research about which tissues and in what types of cells genes are expressed, how genes have changed over the course of human evolution, or whether gene products are targeted by known pharmacological agents (e.g., Wray et al., 2018). This approach is particularly powerful when (a) links between GWAS-identified variants and genes are clear (e.g., the SNP rs6265, a genome-wide significant “hit” in GWASs of obesity and smoking, changes the protein encoded by the gene BDNF from valine to methionine); (b) the biology of the phenotype is well known, as in GWASs of well-characterized blood molecules such as lipids; and (c) knowledge of genes relevant to that biology is available, as in GWASs of blood proteins, in which genes encoding the protein or its regulators are known.
Often, however, these three conditions are not met, and this may be especially true for GWASs of phenotypes relevant to behavioral and brain scientists. In the first case, links between GWAS-identified variants and the genes whose function they affect are not straightforward. For example, an early GWAS discovery for obesity, a variant in the gene FTO, was recently revealed to influence obesity primarily through the regulation not of FTO but of the gene IRX3, nearly 1 million nucleotides away (Claussnitzer et al., 2015). This in-trans mechanism, in which a variant affects a phenotype by regulating a spatially distal gene, is likely to be common. For example, in a recent GWAS of blood proteins, 10% of replicated genome-wide significant associations were in trans, with some variants even on different chromosomes from DNA sequences known to encode those proteins (Suhre et al., 2017). In addition to the difficulty of annotating variants to genes, knowledge of the biology that influences phenotypic variation is frequently incomplete (Johnson et al., 2017). In fact, it is precisely this challenge that motivated the GWAS approach in the first place.
Finally, even a perfect understanding of biology would be an imperfect understanding of mechanism, because genetic effects can involve “outside-the-skin” processes—mechanisms that are not located entirely inside a person’s body or brain but rather operate through exposure to physical or social environments. For example, among the first GWAS discoveries for lung cancer were nicotine-receptor gene polymorphisms, which exert their effect via smoking behavior causing exposure to carcinogens in cigarette smoke (Wassenaar et al., 2011). Other, less-easy-to-annotate GWAS discoveries may similarly have biology → behavior → environment → phenotype mechanisms of action on disease risk. In sum, small effect sizes, uncertain biology, and outside-the-skin processes leave a translational gulf between bioinformatics-annotated GWAS results and scientific understanding that can improve human health and well-being. We propose phenotypic annotation as a research agenda that can help bridge that gulf.
Phenotypic Annotation: DNA Variants as Building Blocks of Life Courses
Phenotypic annotation comprises a set of social, behavioral, and brain-science research activities that map connections between GWAS discoveries and the neural, developmental, and social processes that give rise to psychological experiences and behavior. Whereas current biologically focused approaches to GWAS translation are bottom-up research strategies intended to answer the question, “How do the genetic variants associated with this phenotype change genome biology?” phenotypic annotation is a top-down strategy designed to investigate the question, “How do the genetic variants associated with this phenotype change the development and behavior of an organism?” (Belsky, Moffitt, & Caspi, 2013). Phenotypic annotation involves three important shifts in thinking about genotype–phenotype relationships: (a) from genotypes to genomes, (b) from discovery phenotypes to nomological networks, and (c) from proximate biology to life-course development.
From genotypes to genomes
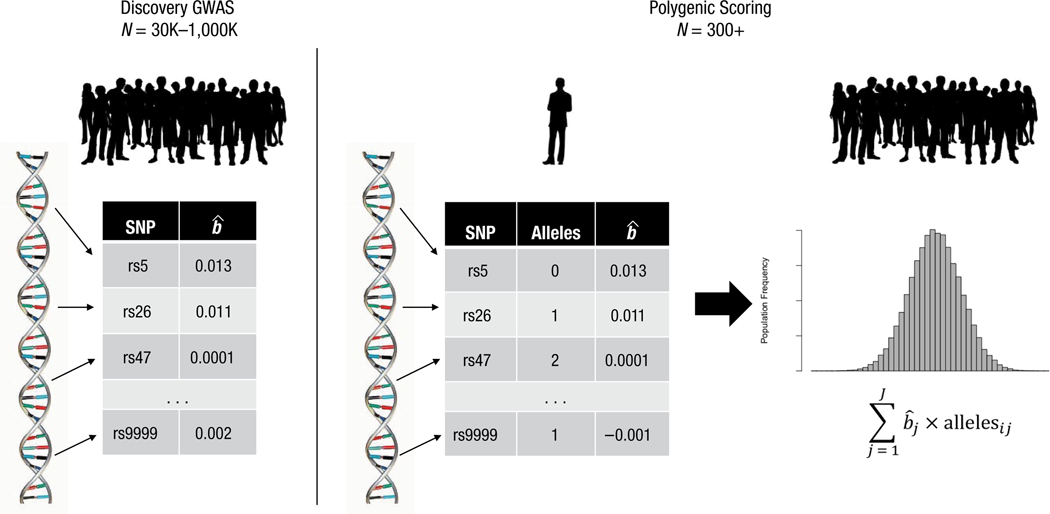
Following up the tiny individual effects identified in a GWAS presents a challenge in terms of statistical power, and studies that include the measurements needed to investigate mechanisms of genetic effects rarely have sufficient sample sizes. A solution to this challenge is suggested by evidence that genetic effects tend to combine additively, resulting in a quantitative polygenic distribution of genetic influence (Plomin, Haworth, & Davis, 2009). This polygenic distribution can be measured by applying GWAS results as a scoring algorithm to genetic data from an independent sample of participants. Specifically, a participant’s polygenic score is calculated as the genome-wide weighted average of phenotype-associated alleles, where weights are typically effect sizes from an independent-discovery GWAS (Dudbridge, 2013; Fig. 1). Polygenic scores solve the small-effects problem by aggregating signals from SNPs across the genome into a single measure with a larger effect. For example, polygenic scores based on GWASs of education and intelligence can explain as much as 10% of phenotypic variance (Plomin & von Stumm, 2018).
Fig. 1.
Polygenic scoring. Polygenic scores aggregate information from up to millions of single-nucleotide polymorphisms (SNPs) across the genome into a single composite index summarizing genome-wide genetic influence on a target phenotype. The procedure of computing a polygenic score is conducted using whole-genome SNP data on a research participant and scoring information from some external database, typically a published genome-wide association study (GWAS). In the first step, the participant’s SNP genotypes are assigned weights that indicate the direction and magnitude of that SNP’s association with a target phenotype. Most often these weights are the effect sizes, , estimated in a GWAS that did not include the research participant. Next, the number of phenotype-associated alleles (0, 1, or 2) at each SNP () is counted, and the count is multiplied by the weight, . Finally, the weighted count is summed across SNPs to compute the participant’s polygenic score. The resulting distribution of polygenic scores across participants is normal. Polygenic-score analysis often involves two additional features. First, because polygenic scores draw information from many sites across the genome, they are sensitive to bias arising from allele-frequency differences between populations of different ancestry, called population stratification (Martin et al., 2017). Polygenic-score analysis is therefore typically conducted within populations that share genetic ancestry (e.g., Europeans), and the analysis usually includes covariate adjustment for principal components estimated from genome-wide SNP data to account for any residual population stratification (Price et al., 2006). Second, polygenic-score analysis is sometimes restricted to a subset of SNPs. This includes procedures to select SNPs that are statistically independent of one another and procedures to select SNPs that meet other criteria, such as having p values in the discovery GWAS that fall below a certain threshold. To date, evidence suggests that polygenic scores are best constructed using data from all available SNPs (Dudbridge & Newcombe, 2016; Ware et al., 2017).
As a result, samples numbering only in the hundreds to thousands can be well powered to test genetic effects, opening the door to a breadth of behavioral and brain-science research designs, including randomized trials of behavioral interventions, longitudinal cohort studies, and neuroimaging studies. This gain in statistical power comes with a loss of granularity: Associations with polygenic scores cannot be attributed to specific genes. However, much of biology is itself polygenic (Iacono, Vaidyanathan, Vrieze, & Malone, 2014). Polygenic scores, then, are useful tools for testing whether biological intermediaries, such as brain structure or function, might mediate GWAS-discovered genetic associations with more complex traits and behaviors.
From GWAS-discovery phenotypes to nomological networks
It would be a mistake to conceptualize the SNPs discovered in a GWAS as narrowly measuring genetic risk for the precise phenotype studied in the GWAS itself. For example, genes discovered in a GWAS of educational attainment are not “education genes” per se (Belsky et al., 2016). Discovered SNP associations may arise from genetic influence on any correlate of the discovery phenotype (Belsky & Israel, 2014). In this way, results from hypothesis-free GWAS discoveries pose an interpretive challenge similar to the classic construct-validity problem. Newly discovered SNPs have been selected on the basis of their criterion validity (i.e., their ability to predict the phenotype used in the original GWAS), but the constructs measured by the resulting polygenic score remains largely unexplored. As Cronbach and Meehl described more than 60 years ago, “‘learning more about’ a theoretical construct is a matter of elaborating the nomological network in which it occurs, or of increasing the definiteness of its components” (Cronbach & Meehl, 1955, p. 290). We suggest that learning more about the theoretical construct of genetic influence, as measured by GWAS discoveries, can be advanced by elaborating the nomological network in which SNP–phenotype associations are embedded.
As an example, studies focused on genetics discovered in GWASs of educational attainment have revealed a surprisingly consistent nomological network, including behavioral patterns of achievement leading up to and extending beyond the completion of formal schooling, cognitive and personological characteristics known to influence educational success, early realization of developmental milestones in language and reading, and environments conducive to educational success, including family socioeconomic status, neighborhood conditions, and peer characteristics (Barth, Papageorge, & Thom, 2017; Belsky, Domingue, et al., 2018; Belsky et al., 2016; Conley et al., 2015; Domingue, Belsky, Conley, Harris, & Boardman, 2015; Krapohl et al., 2017).
Phenotypic-annotation analyses of genetic loci discovered in GWASs of schizophrenia have yielded a more complicated picture. Polygenic scores illustrate expected associations with childhood mental health problems (Nivard et al., 2017), early neurocognitive deficits (Riglin et al., 2017), and life-course cognitive decline (McIntosh et al., 2013). Moreover, schizophrenia-associated genetic variants appear to be more common among individuals in creative professions (Power et al., 2015) and are associated with putative environmental risk factors for the disorder, including neighborhood disadvantage and illicit drug use (Power et al., 2015; Sariaslan et al., 2016). However, puzzlingly, polygenic scores from schizophrenia GWASs are not consistently associated with symptom severity or frequency of psychotic episodes (Jones et al., 2016; Stepniak et al., 2014), and there is a surprising positive genetic correlation between schizophrenia and educational attainment (Bansal et al., 2018). A further possible research challenge is that schizophrenia genetics may be related to nonparticipation or loss to follow up (Taylor et al., 2018). Overall, more work, particularly focused on early development, is needed to elucidate what neural, cognitive, and behavioral constructs are being tapped by genes discovered in GWASs of schizophrenia.
From proximate biology to life-course development
Individual differences in human psychology do not spring forth like Athena from Zeus’s head but are rather shaped over time through developmental processes in which early emerging differences structure trajectories and shape future outcomes (Belsky, Moffitt, & Caspi, 2013). Thus, a necessary step in understanding mechanisms through which GWAS-discovered genetics influence psychology and behavior is to address the question of when in human development genetic influences manifest. For example, SNPs discovered in GWASs of adult body mass index are associated with accelerations in weight gain during early and middle childhood (Belsky et al., 2012). SNPs discovered in GWASs of adult smoking behavior are associated with accelerated progression from smoking initiation to dependence during adolescence (Belsky, Moffitt, Baker, et al., 2013). SNPs discovered in GWASs of educational attainment are associated with an accelerated pace of cognitive development and the acquisition of self-control and interpersonal skills from infancy through middle childhood (Belsky et al., 2016). By interrogating how polygenic scores constructed from GWASs of adult samples are related to phenotypes measured in early life, these studies illuminate the developmental intermediaries between genotypes that are established at conception and adult phenotypes that are canalized later in development.
The developmental processes linking GWAS discoveries with mature phenotypes might involve gene–environment correlations. Childhood social and physical environments that predict individual differences in health and achievement across life are themselves heritable (Plomin & Bergeman, 1991). One implication of gene–environment correlations is that genetic differences potentially confound putative environmental effects. A second, less appreciated implication is that environments might mediate genetic effects (Scarr & McCartney, 1983). Specifically, genetic differences between people might cause them to select into different environments, a process known as active/evocative gene–environment correlation. These environments could then reinforce or magnify differences in traits or behaviors. For example, a child’s genes might influence his or her tendencies toward antisocial behavior in ways that lead to social assortment with delinquent peers (Mann et al., 2016). This peer environment might, in turn, incentivize or facilitate opportunities for more antisocial behavior. Genetic differences between people might also become correlated with their environments when those environments are shaped by genetic relatives. For example, genes influence sexual behavior in ways that make some adolescents more likely to experience early, out-of-wedlock parenthood. Children of such unions will inherit their parents’ genes along with a single-parent environment that may affect sexual behavior (Mendle et al., 2009). Such “passive” gene–environment correlations (so-called because they arise without any active niche picking on the part of the child) are a known potential confound of associations between family environments and child outcomes. However, they can also confound or contaminate GWAS discoveries of genetic effects. Because genotypes are shared between relatives, an association between a child’s genotype and his or her phenotype could reflect the effect of a parental genotype that is mediated through an environmental pathway (Koellinger & Harden, 2018; Kong et al., 2018).
Research that integrates environmental measures and polygenic scores can now be used to test specific hypotheses about environmental processes that mediate and moderate genetic effects. For example, polygenic scores for educational attainment, age at first birth, and schizophrenia show correlations with a range of measured environments, including family social class (Belsky, Domingue, et al., 2018), growing up without a father in the home (Gaydosh, Belsky, Domingue, Boardman, & Harris, 2018), peer delinquency (Krapohl et al., 2017), and neighborhood conditions (Belsky, Caspi, et al., 2018; Sariaslan et al., 2016). Polygenic scores will be particularly useful in longitudinal studies, which can trace reciprocal associations between people and their environments, and in multigenerational family designs, which provide opportunities to test for indirect genetic effects, that is, genetic effects that are mediated via the family environment provided by siblings, parents, and grandparents (Bates et al., 2018; Kong et al., 2018; Liu, 2018). Finally, studies capitalizing on shifts in macroenvironmental contexts, such as policy reforms or government changes, can test the environmental conditions under which genotype–phenotype relationships are preserved or disrupted (Barcellos, Carvalho, & Turley, 2018; Rimfeld et al., 2018).
What’s Next?
Previous efforts to integrate genetics into psychological science have been strained by an enduring fear that studying genetics will reinvigorate the eugenics movement, by the practical difficulties of addressing certain research questions within twin studies, and, more recently, by the poor reproducibility of candidate gene findings. But the ethical and reproducible integration of psychology and genetics is not only possible but also essential to the success of both fields.
Now, large-scale GWASs and polygenic-score analysis offer new opportunities to bring genetics and psychological science together (Table 1). Phenotypic annotation is an approach to understanding GWAS discoveries that leverages the expertise of psychological scientists in how to measure traits, behaviors, and environments and the strength of psychological theories for understanding how individuals and their environments interact. By shifting focus from the proximate biology of genomes to the life-course development of humans, including the environments that individuals grow up in and that they build for themselves, phenotypic annotation provides an opportunity for psychological scientists to help unpack the many black boxes the GWAS era has delivered.
Table 1.
Dos and Don’ts of Integrating Genetics and Psychological Science
| Do … genotype your participants. Even if your study is too small for a primary genetic-discovery analysis such as a GWAS, there are lots of opportunities for high-impact research. Genetic data are special. DNA sequences do not change, so they have to be measured only once. And they are all purpose. The same genetic data can be used to follow-up results from any genetic-association study, including studies that will be conducted in the future. Collecting genetic data is already inexpensive, and costs are falling. For example, companies such as Gencove now offer services that include sample collection, DNA analysis, and data cleaning and processing for less than $100 per subject. |
| Do … focus on whole genomes rather than individual genes. Unless you have many thousands of participants, your research is probably underpowered to study single genetic variants. For smaller studies, polygenic-score analysis is more likely to yield reproducible results. |
| Don’t … forget about statistical power. GWAS is a data-mining method. Samples greater than 10,000 are a minimum threshold for successful discovery analysis of most phenotypes, and samples an order of magnitude larger are much more valuable. Polygenic scores developed from an underpowered GWAS are less likely to yield reproducible results. While psychologists are trained to value measurement precision, a coarsely measured phenotype in a very large sample can result in more replicable GWAS discoveries than a precisely measured phenotype in a smaller sample. For example a polygenic score derived from a GWAS of years of education in 1 million people predicts performance on an IQ test better than a polygenic score derived from GWAS of cognitive test scores in a sample one third that size (Lee et al., 2018). |
| Don’t … wait for a GWAS of your exact phenotype of interest. Avoid thinking of GWAS results as measuring genes “for” the GWAS target phenotype. Instead, think about how your phenotype of interest might be related to an existing, well-powered GWAS. New methods and analytic approaches are expanding opportunities. For example, methods are now available to combine results from multiple GWASs of related traits to refine polygenic prediction (Grotzinger et al., 2018; Turley et al., 2018). And multiple polygenic scores could be combined into a single analysis (Krapohl et al., 2018). |
|
Do … pick the low-hanging fruit. GWAS results are as much questions as answers. The ever-growing crop of GWAS discoveries for social and behavioral phenotypes creates opportunities for psychological scientists. Several examples of low-hanging fruit ripe for picking by psychological scientists include the following: • Genetic analysis of brain-imaging studies to test genetic associations with structural and functional differences in the brain • Genetic analysis of randomized trials to test genetic heterogeneity in response to interventions, ranging from quality preschool to psychotherapy, as well as to evaluate genetic differences in participation and adherence |
| Don’t … ignore development. Most GWASs are conducted in samples of adults. But most GWAS phenotypes have early-developmental origins. Phenotypic-annotation analysis should consider developmental antecedents that fall within the nomological network of a target GWAS phenotype. For example, the onset of schizophrenia typically occurs in adulthood, but it has a neurodevelopmental etiology that could influence brain development and behavior from infancy. |
|
Do … think about how genotypes can help understand the environment. Testing Gene × Environment interactions is one way that genetic data can help us understand environments, but there are many others. • Running an intervention study? Including polygenic scores as control variables can boost statistical power in analyses testing treatment effects (Rietveld et al., 2013). • Studying school, peer, or family environments? Integrating polygenic scores offers an opportunity to test environmental processes as mechanisms for genetic effects. • Studying families? Genetic analysis can test environmental effects, called “indirect genetic effects” or “genetic nurture.” • Running longitudinal studies with high-density measurements? Genetic analysis can test hypotheses about reciprocal transactions between people and their environments. |
Note: GWAS = genome-wide association study.
Acknowledgments
D. W. Belsky and K. P. Harden contributed equally to this manuscript.
Funding
D. W. Belsky and K. P. Harden are supported by Early Career Research Fellowships from the Jacobs Foundation.
Appendices
Appendix
Glossary of key terms
Genome-wide association study (GWAS): data-mining analysis in which variants scattered across the genome, typically single-nucleotide polymorphisms, are individually regressed on a phenotype. Because of the large number of statistical tests, which can number in the millions, a GWAS adopts stringent p-value thresholds (e.g., p < 5 × 10−8) to avoid false discoveries.
Heritability: the proportion of variance in a measured phenotype that can be statistically accounted for by genetic differences between people.
Missing heritability: the difference in heritability estimate from twin studies and the heritability that can be explained by measured DNA differences between unrelated people. Typically, twin-study estimates of heritability are about twice as large as the variance explained by measured single-nucleotide polymorphisms. Causes of missing heritability remain much debated in genetics. Potential causes include interactions between genes (epistasis), unmeasured Gene × Environment interactions, and rare DNA variants that have not been measured.
Phenotype: anything that is not a genotype—including physical and personality characteristics, behaviors, disease diagnoses, brain structure and function, gene-expression levels, and DNA methylation states.
Single-nucleotide polymorphism (SNP): a single-nucleotide change in the human DNA sequence prevalent in more than 1% of a population. A strand of DNA is composed of a unique sequence of four nucleotides: guanine (G), cytosine (C), thiamine (T), and adenine (A). For example, one person might have an A at a particular location on his or her genome, whereas another person has a C.
Footnotes
Declaration of Conflicting Interests
The author(s) declared that there were no conflicts of interest with respect to the authorship or the publication of this article.
Action Editor
Randall W. Engle served as action editor for this article.
References
- Bansal V, Mitjans M, Burik CAP, Linnér RK, Okbay A, Rietveld CA, … Koellinger PD (2018). Genome-wide association study results for educational attainment aid in identifying genetic heterogeneity of schizophrenia. Nature Communications, 9(1), Article 3078. doi: 10.1038/s41467-018-05510-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barcellos SH, Carvalho LS, & Turley P. (2018). Education can reduce health disparities related to genetic risk of obesity: Evidence from a British reform. BioRxiv. doi: 10.1101/260463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth D, Papageorge NW, & Thom K. (2017). Genetic ability, wealth, and financial decision-making (IZA Discussion Paper No. 10567). Rochester, NY: Social Science Research Network. Retrieved from SSRN website: https://papers.ssrn.com/abstract=2923653 [Google Scholar]
- Bates TC, Maher BS, Medland SE, McAloney K, Wright MJ, Hansell NK, … Gillespie NA (2018). The nature of nurture: Using a virtual-parent design to test parenting effects on children’s educational attainment in genotyped families. Twin Research and Human Genetics, 21, 73–83. doi: 10.1017/thg.2018.11 [DOI] [PubMed] [Google Scholar]
- Belsky DW, Caspi A, Arseneault L, Corcoran D, Domingue BW, Harris KM, … Odgers C. (2018). Genetics and the geography of health, behavior, and attainment. BioRxiv. doi: 10.1101/376897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Domingue BW, Wedow R, Arseneault L, Boardman JD, Caspi A, … Harris KM (2018). Genetic analysis of social-class mobility in five longitudinal studies. Proceedings of the National Academy of Sciences, USA, 115, E7275–E7284. doi: 10.1073/pnas.1801238115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, & Israel S. (2014). Integrating genetics and social science: Genetic risk scores. Biodemography and Social Biology, 60, 137–155. doi: 10.1080/19485565.2014.946591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Baker TB, Biddle AK, Evans JP, Harrington H, … Caspi A. (2013). Polygenic risk and the developmental progression to heavy, persistent smoking and nicotine dependence: Evidence from a 4-decade longitudinal study. JAMA Psychiatry, 70, 534–542. doi: 10.1001/jamapsychiatry.2013.736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, & Caspi A. (2013). Genetics in population health science: Strategies and opportunities. American Journal of Public Health, 103(Suppl. 1), S73–S83. doi: 10.2105/AJPH.2012.301139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington H, Hogan S, … Caspi A. (2016). The genetics of success: How single-nucleotide polymorphisms associated with educational attainment relate to life-course development. Psychological Science, 27, 957–972. doi: 10.1177/0956797616643070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belsky DW, Moffitt TE, Houts R, Bennett GG, Biddle AK, Blumenthal JA, … Caspi A. (2012). Polygenic risk, rapid childhood growth, and the development of obesity: Evidence from a 4-decade longitudinal study. Archives of Pediatrics and Adolescent Medicine, 166, 515–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyle EA, Li YI, & Pritchard JK (2017). An expanded view of complex traits: From polygenic to omnigenic. Cell, 169, 1177–1186. doi: 10.1016/j.cell.2017.05.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabris CF, Lee JJ, Cesarini D, Benjamin DJ, & Laibson DI (2015). The fourth law of behavior genetics. Current Directions in Psychological Science, 24, 304–312. doi: 10.1177/0963721415580430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claussnitzer M, Dankel SN, Kim K-H, Quon G, Meuleman W, Haugen C, … Kellis M. (2015). FTO obesity variant circuitry and adipocyte browning in humans. New England Journal of Medicine, 373, 895–907. doi: 10.1056/NEJMoa1502214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conley D, Domingue B, Cesarini D, Dawes CT, Rietveld CA, & Boardman J. (2015). Is the effect of parental education on offspring biased or moderated by genotype? Sociological Science, 2(6), 82–105. doi: 10.15195/v2.a6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cronbach LJ, & Meehl PE (1955). Construct validity in psychological tests. Psychological Bulletin, 52, 281–302. [DOI] [PubMed] [Google Scholar]
- Domingue BW, Belsky DW, Conley D, Harris KM, & Boardman JD (2015). Polygenic influence on educational attainment: New evidence from the National Longitudinal Study of Adolescent to Adult Health. AERA Open, 1(3). doi: 10.1177/2332858415599972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F. (2013). Power and predictive accuracy of polygenic risk scores. PLOS Genetics, 9(3), Article e1003348. doi: 10.1371/journal.pgen.1003348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudbridge F, & Newcombe PJ (2016). Accuracy of gene scores when pruning markers by linkage disequilibrium. Human Heredity, 80, 178–186. [DOI] [PubMed] [Google Scholar]
- Gaydosh L, Belsky DW, Domingue BW, Boardman JD, & Harris KM (2018). Father absence and accelerated reproductive development in non-Hispanic White women in the United States. Demography, 55, 1245–1267. doi: 10.1007/s13524-018-0696-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grotzinger AD, Rhemtulla M, de Vlaming R, Ritchie SJ, Mallard TT, Hill WD, … Tucker-Drob EM (2018). Genomic SEM provides insights into the multivariate genetic architecture of complex traits. BioRxiv, Article 305029. doi: 10.1101/305029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iacono WG, Vaidyanathan U, Vrieze SI, & Malone SM (2014). Knowns and unknowns for psychophysiological endophenotypes: Integration and response to commentaries. Psychophysiology, 51, 1339–1347. doi: 10.1111/psyp.12358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson EC, Border R, Melroy-Greif WE, de Leeuw CA, Ehringer MA, & Keller MC (2017). No evidence that schizophrenia candidate genes are more associated with schizophrenia than noncandidate genes. Biological Psychiatry, 82, 702–708. doi: 10.1016/j.biopsych.2017.06.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones HJ, Stergiakouli E, Tansey KE, Hubbard L, Heron J, Cannon M, … Zammit S. (2016). Phenotypic manifestation of genetic risk for schizophrenia during adolescence in the general population. JAMA Psychiatry, 73, 221–228. doi: 10.1001/jamapsychiatry.2015.3058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koellinger PD, & Harden KP (2018). Using nature to understand nurture. Science, 359, 386–387. doi: 10.1126/science.aar6429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Frigge ML, Vilhjalmsson BJ, Young AI, Thorgeirsson TE, … Stefansson K. (2018). The nature of nurture: Effects of parental genotypes. Science, 359, 424–428. doi: 10.1126/science.aan6877 [DOI] [PubMed] [Google Scholar]
- Krapohl E, Hannigan LJ, Pingault J-B, Patel H, Kadeva N, Curtis C, … Plomin R. (2017). Widespread covariation of early environmental exposures and trait-associated polygenic variation. Proceedings of the National Academy of Sciences, USA, 114, 11727–11732. doi: 10.1073/pnas.1707178114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krapohl E, Patel H, Newhouse S, Curtis CJ, von Stumm S, Dale PS, … Plomin R. (2018). Multi-polygenic score approach to trait prediction. Molecular Psychiatry, 23, 1368–1374. doi: 10.1038/mp.2017.163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee JJ, Wedow R, Okbay A, Kong E, Maghzian O, Zacher M, … Cesarini D. (2018). Gene discovery and polygenic prediction from a genome-wide association study of educational attainment in 1.1 million individuals. Nature Genetics, 50, 1112–1121. doi: 10.1038/s41588-018-0147-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H. (2018). Social and genetic pathways in multigenerational transmission of educational attainment. American Sociological Review, 83, 278–304. doi: 10.1177/0003122418759651 [DOI] [Google Scholar]
- Mann FD, Patterson MW, Grotzinger AD, Kretsch N, Tackett JL, Tucker-Drob EM, & Harden KP (2016). Sensation seeking, peer deviance, and genetic influences on adolescent delinquency: Evidence for person-environment correlation and interaction. Journal of Abnormal Psychology, 125, 679–691. doi: 10.1037/abn0000160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, … Kenny EE (2017). Human demographic history impacts genetic risk prediction across diverse populations. The American Journal of Human Genetics, 100, 635–649. doi: 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AM, Gow A, Luciano M, Davies G, Liewald DC, Harris SE, … Deary IJ (2013). Polygenic risk for schizophrenia is associated with cognitive change between childhood and old age. Biological Psychiatry, 73, 938–943. doi: 10.1016/j.biopsych.2013.01.011 [DOI] [PubMed] [Google Scholar]
- Mendle J, Harden KP, Turkheimer E, Van Hulle CA, D’Onofrio BM, Brooks-Gunn J, … Lahey Benjamin B. (2009). Associations between father absence and age of first sexual intercourse. Child Development, 80, 1463–1480. doi: 10.1111/j.1467-8624.2009.01345.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nivard MG, Gage SH, Hottenga JJ, van Beijsterveldt CEM, Abdellaoui A, Bartels M, … Middeldorp CM (2017). Genetic overlap between schizophrenia and developmental psychopathology: Longitudinal and multivariate polygenic risk prediction of common psychiatric traits during development. Schizophrenia Bulletin, 43, 1197–1207. doi: 10.1093/schbul/sbx031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plomin R, & Bergeman CS (1991). The nature of nurture: Genetic influence on “environmental” measures. Behavioral & Brain Sciences, 14, 414–427. doi: 10.1017/S0140525X00070588 [DOI] [Google Scholar]
- Plomin R, Haworth CMA, & Davis OSP (2009). Common disorders are quantitative traits. Nature Reviews Genetics, 10, 872–878. doi: 10.1038/nrg2670 [DOI] [PubMed] [Google Scholar]
- Plomin R, & von Stumm S. (2018). The new genetics of intelligence. Nature Reviews Genetics, 19, 148–159. doi: 10.1038/nrg.2017.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polderman TJC, Benyamin B, de Leeuw CA, Sullivan PF, van Bochoven A, Visscher PM, & Posthuma D. (2015). Meta-analysis of the heritability of human traits based on fifty years of twin studies. Nature Genetics, 47, 702–709. doi: 10.1038/ng.3285 [DOI] [PubMed] [Google Scholar]
- Power RA, Steinberg S, Bjornsdottir G, Rietveld CA, Abdellaoui A, Nivard MM, … Stefansson K. (2015). Polygenic risk scores for schizophrenia and bipolar disorder predict creativity. Nature Neuroscience, 18, 953–955. doi: 10.1038/nn.4040 [DOI] [PubMed] [Google Scholar]
- Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, & Reich D. (2006). Principal components analysis corrects for stratification in genome-wide association studies. Nature Genetics, 38, 904–909. doi: 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- Rietveld CA, Medland SE, Derringer J, Yang J, Esko T, Martin NW, … Koellinger PD (2013). GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science, 340, 1467–1471. doi: 10.1126/science.1235488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riglin L, Collishaw S, Richards A, Thapar AK, Maughan B, O’Donovan MC, & Thapar A. (2017). Schizophrenia risk alleles and neurodevelopmental outcomes in childhood: A population-based cohort study. The Lancet Psychiatry, 4, 57–62. doi: 10.1016/S2215-0366(16)30406-0 [DOI] [PubMed] [Google Scholar]
- Rimfeld K, Krapohl E, Trzaskowski M, Coleman JRI, Selzam S, Dale PS, … Plomin R. (2018). Genetic influence on social outcomes during and after the Soviet era in Estonia. Nature Human Behaviour, 2, 269–275. doi: 10.1038/s41562-018-0332-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sariaslan A, Fazel S, D’Onofrio BM, Långström N, Larsson H, Bergen SE, … Lichtenstein P. (2016). Schizophrenia and subsequent neighborhood deprivation: Revisiting the social drift hypothesis using population, twin and molecular genetic data. Translational Psychiatry, 6(5), Article e796. doi: 10.1038/tp.2016.62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scarr S, & McCartney K. (1983). How people make their own environments: A theory of genotype → environment effects. Child Development, 54, 424–435. doi: 10.2307/1129703 [DOI] [PubMed] [Google Scholar]
- Stepniak B, Papiol S, Hammer C, Ramin A, Everts S, Hennig L, … Ehrenreich H. (2014). Accumulated environmental risk determining age at schizophrenia onset: A deep phenotyping-based study. The Lancet Psychiatry, 1, 444–453. doi: 10.1016/S2215-0366(14)70379-7 [DOI] [PubMed] [Google Scholar]
- Suhre K, Arnold M, Bhagwat AM, Cotton RJ, Engelke R, Raffler J, … Graumann J. (2017). Connecting genetic risk to disease end points through the human blood plasma proteome. Nature Communications, 8, Article 14357. doi: 10.1038/ncomms14357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor AE, Jones HJ, Sallis H, Euesden J, Stergiakouli E, Davies NM, … Tilling K. (2018). Exploring the association of genetic factors with participation in the Avon Longitudinal Study of Parents and Children. International Journal of Epidemiology, 47, 1207–1216. doi: 10.1093/ije/dyy060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turkheimer E. (2000). Three laws of behavior genetics and what they mean. Current Directions in Psychological Science, 9, 160–164. doi: 10.1111/1467-8721.00084 [DOI] [Google Scholar]
- Turley P, Walters RK, Maghzian O, Okbay A, Lee JJ, Fontana MA, … Social Science Genetic Association Consortium. (2018). Multi-trait analysis of genome-wide association summary statistics using MTAG. Nature Genetics, 50, 229–237. doi: 10.1038/s41588-017-0009-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, & Yang J. (2017). 10 years of GWAS discovery: Biology, function, and translation. The American Journal of Human Genetics, 101, 5–22. doi: 10.1016/j.ajhg.2017.06.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ware EB, Schmitz LL, Faul JD, Gard A, Mitchell C, Smith JA, … Kardia SL (2017). Heterogeneity in polygenic scores for common human traits. BioRxiv. doi: 10.1101/106062 [DOI] [Google Scholar]
- Wassenaar CA, Dong Q, Wei Q, Amos CI, Spitz MR, & Tyndale RF (2011). Relationship between CYP2A6 and CHRNA5-CHRNA3-CHRNB4 variation and smoking behaviors and lung cancer risk. Journal of the National Cancer Institute, 103, 1342–1346. doi: 10.1093/jnci/djr237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray NR, Ripke S, Mattheisen M, Trzaskowski M, Byrne EM, Abdellaoui A, … Sullivan PF (2018). Genome-wide association analyses identify 44 risk variants and refine the genetic architecture of major depression. Nature Genetics, 50, 668–681. doi: 10.1038/s41588-018-0090-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
Recommended Reading
- Belsky DW, Moffitt TE, Corcoran DL, Domingue B, Harrington H, Hogan S, … Caspi A. (2016). (See References). An example of comprehensive phenotypic-annotation analyses applied to developmental data. [Google Scholar]
- Plomin R, Haworth CMA, & Davis OSP (2009). (See References). Contains high-quality figures that illustrate polygenic influence on traits. [Google Scholar]
- Scarr S, & McCartney K. (1983). (See References). A classic theoretical article on the importance of the environment for understanding genetic effects. [Google Scholar]
- Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, & Yang J. (2017). (See References). A general summary of recent discoveries in genome-wide association studies. [Google Scholar]