Abstract
Previous research has found that living in a disadvantaged neighborhood is associated with poor health outcomes. Living in disadvantaged neighborhoods may alter inflammation and immune response in the body, which could be reflected in epigenetic mechanisms such as DNA methylation (DNAm). We used robust linear regression models to conduct an epigenome-wide association study examining the association between neighborhood deprivation (Area Deprivation Index; ADI), and DNAm in brain tissue from 159 donors enrolled in the Emory Goizueta Alzheimer’s Disease Research Center (Georgia, USA). We found one CpG site (cg26514961, gene PLXNC1) significantly associated with ADI after controlling for covariates and multiple testing (p-value=5.0e-8). Effect modification by APOE ε4 was statistically significant for the top ten CpG sites from the EWAS of ADI, indicating that the observed associations between ADI and DNAm were mainly driven by donors who carried at least one APOE ε4 allele. Four of the top ten CpG sites showed a significant concordance between brain tissue and tissues that are easily accessible in living individuals (blood, buccal cells, saliva), including DNAm in cg26514961 (PLXNC1). Our study identified one CpG site (cg26514961, PLXNC1 gene) that was significantly associated with neighborhood deprivation in brain tissue. PLXNC1 is related to immune response, which may be one biological pathway how neighborhood conditions affect health. The concordance between brain and other tissues for our top CpG sites could make them potential candidates for biomarkers in living individuals.
Keywords: DNA methylation, brain tissue, neighborhood deprivation, neuropathology, epigenetics
INTRODUCTION
Neighborhood socioeconomic status (SES) is complex and has unique social, cultural, physical, and economic attributes that can impact human health [1]. Residing in a deprived neighborhood has been associated with increased incidence of mental health conditions such as depression [2], increased risk of chronic conditions such as cardiovascular disease [2], and increased risk of brain-health diseases including Alzheimer’s disease [3, 4]. Research has demonstrated that living in a disadvantaged neighborhood can lead to chronic stress in the body, mainly through the immune and inflammatory response system [5]. The specific biological mechanisms that link neighborhood conditions to health outcomes are not fully understood.
A growing body of evidence suggests that epigenetics may help explain how neighborhood conditions impact health [6, 7]. DNA methylation (DNAm) is a well-studied epigenetic mechanism that involves the addition of a methyl group to DNA, typically at the 5-carbon of cytosine at cytosine-phosphate-guanine (CpG) dinucleotides, which can influence gene expression [8]. While the link between individual-level socioeconomic factors and differential DNAm has been well established [9–11], the effect of neighborhood-level socioeconomic factors on DNAm is less well known.
Existing studies on the relationship between neighborhood deprivation and DNAm are limited due to the novelty of the field of social epigenomics. Additionally, the implications of this relationship in the context of neuropsychiatric disorders are not well characterized. One study using blood samples and one study using saliva samples both found increased global DNAm among those living in more disadvantaged neighborhoods [12, 13]. Another study using blood samples identified three CpG sites that were associated with neighborhood deprivation, with one being linked to a gene (MAOB) that is related to Parkinson’s Disease [14]. Two other studies using blood samples found increased DNAm in nine genes related to stress and inflammation in the body [6, 15]. However, none of these identified CpG sites or genes were replicated across different studies. It is also important to note that none of these existing studies have examined the association between neighborhood deprivation and DNAm in brain tissue. DNAm changes in the brain specifically are important to study because they can provide indications of neuropathology outcomes such as Alzheimer’s disease (AD) [16–22] and depression [23, 24]. Many of these brain health outcomes have themselves been associated with neighborhood deprivation [2, 25, 26].
Given this gap in knowledge of how neighborhood deprivation impacts differential DNAm in the brain, we evaluated the association between the most established measure of neighborhood deprivation (Area Deprivation Index; ADI) and DNAm measured from brain tissue samples in a sample of mainly cognitively impaired, deceased donors from Georgia, USA, and analyzed whether those associations were independent of the observed AD neuropathology. DNAm at any CpG sites showing an association with ADI was further investigated in terms of their concordance across other (more accessible) tissues to explore their potential for serving as biomarkers in living individuals.
RESULTS
Description of study population
Our study included 159 donors. In the total study population, 89 (56.0%) were male, 142 (89.3%) were white, and the mean age at death was 76.6 years (SD 10.0) (Table 1). Of the total population, 56% had at least one APOE ε4 allele and 95.7% were clinically diagnosed with AD or some other form of dementia before death. Overall, 45.9% were classified as having the highest Braak Stage of 6, 69.2% were classified as having frequent CERAD, and 58.5% were classified as having a high ABC score. The mean ADI was 36.7 (SD 25.6), which is less deprived than the national average of ADI=50. Overall, 116 (73.0%) were classified into the lower ADI group (ADI<50; less deprived). Compared to those in the high ADI group (ADI≥50; more deprived), those in the low ADI group were more likely to be white (95.7% vs. 72.1% in the high ADI group) and have at least a college degree (79.3% vs. 72.1% in the high ADI group), with the two groups being similar in other demographic categories. Additionally, those in the low ADI group were more likely to be diagnosed with AD or some other form of dementia (97.4.1% vs. 90.7% in the high ADI group) but were similar on other clinical categories including Braak Stage, CERAD, ABC score, and APOE ε4 alleles. The study characteristics of our analysis sample did not significantly differ from the full cohort (Supplementary Table 8).
Table 1. Characteristics of individuals from the ADRC cohort, stratified by Area Deprivation Index group (low=ADI<50 corresponding to less deprived than the national average), (high=ADI ≥50 corresponding to more deprived than the national average).
| Sample characteristic n (%) or mean [SD] | Total (n=159) | Low ADI (n=116) | High ADI (n=43) | |||
| ADI1 | 36.7 | [25.6] | 24.1 | [15.5] | 70.5 | [14.1] |
| Demographics | ||||||
| Race | ||||||
| White | 142 | (89.3) | 111 | (95.7) | 31 | (72.1) |
| Black | 17 | (10.7) | 5 | (4.3) | 12 | (27.9) |
| Sex | ||||||
| Male | 89 | (56.0) | 63 | (54.3) | 26 | (60.5) |
| Female | 70 | (44.0) | 53 | (45.7) | 17 | (39.5) |
| Age at death | 76.6 | [10.0] | 76.6 | [9.6] | 76.6 | [11.1] |
| Education attainment | ||||||
| High school or less | 36 | (22.6) | 24 | (20.7) | 12 | (27.9) |
| College degree | 76 | (47.8) | 56 | (48.3) | 20 | (46.5) |
| Graduate degree | 47 | (29.6) | 36 | (31.0) | 11 | (25.6) |
| Clinical variables | ||||||
| Braak Stage | ||||||
| Stage 1 | 16 | (10.1) | 11 | (9.5) | 5 | (11.6) |
| Stage 2 | 11 | (6.9) | 6 | (5.2) | 5 | (11.6) |
| Stage 3 | 20 | (12.6) | 17 | (14.7) | 3 | (7.0) |
| Stage 4 | 17 | (10.7) | 12 | (10.3) | 5 | (11.6) |
| Stage 5 | 22 | (13.8) | 18 | (15.5) | 4 | (9.3) |
| Stage 6 | 73 | (45.9) | 52 | (44.8) | 21 | (48.8) |
| CERAD2 | ||||||
| No | 35 | (22.0) | 26 | (22.4) | 9 | (20.9) |
| Sparse | 4 | (2.5) | 4 | (3.4) | 0 | (0.0) |
| Moderate | 10 | (6.3) | 7 | (6.0) | 3 | (7.0) |
| Frequent | 110 | (69.2) | 79 | (68.1) | 31 | (72.1) |
| ABC3 | ||||||
| Not | 15 | (9.4) | 10 | (8.6) | 5 | (11.6) |
| Low | 29 | (18.2) | 21 | (18.1) | 8 | (18.6) |
| Intermediate | 22 | (13.8) | 17 | (14.7) | 5 | (11.6) |
| High | 93 | (58.5) | 68 | (58.6) | 25 | (58.1) |
| APOE4 ε4 Allele(s) | ||||||
| 0 | 70 | (44.0) | 51 | (44.0) | 19 | (44.2) |
| 1 | 68 | (42.8) | 53 | (45.7) | 15 | (34.9) |
| 2 | 21 | (13.2) | 12 | (10.3) | 9 | (20.9) |
| Cognitive classification | ||||||
| No dementia | 7 | (4.4) | 3 | (2.6) | 4 | (9.3) |
| Other dementia | 66 | (41.5) | 51 | (44.0) | 15 | (34.9) |
| AD5 | 86 | (54.2) | 62 | (53.4) | 24 | (55.8) |
1Area Deprivation Index; 2Consortium to establish a register for AD; 3Amyloid, Braak and CERAD; 4Apolipoprotein E; 5Alzheimer’s disease.
Association between neighborhood deprivation and DNA methylation in the brain
One CpG site (cg26514961, gene PLXNC1) was significantly associated with ADI when controlling for self-reported race, sex, APOE ε4, education, age at death, cell type proportions, and post-mortem interval (p-value=5.0e-8) (Figures 1, 2 and Table 2). A 20-unit increase in ADI was associated with a -0.0052 decrease in DNAm beta value (Table 2). No other CpG sites were significantly associated with ADI (Figure 1). The other top nine CpG sites and their associated genes were cg08087060 (KLHDC4), cg01291468 (UGT1A10, UGT1A7, UGT1A9, and UGT1A8), cg16241648 (ARPC1A), cg20912923 (a CSMD1), cg09431774 (KIAA1671) and the intergenic CpG sites cg05419854, cg15953452, cg06787422, and cg13521319 (Table 2 and Supplementary Figure 2). The epigenome-wide summary statistics are available online (Supplementary EWAS Output.xlsx). Similar results were found after additional adjustment for neuropathology markers of AD (CERAD, Braak Stage, and ABC); thus indicating that these results were independent of the degree of neuropathology (Supplementary Table 2). Results were also similar after excluding the 2.5% cognitively normal donors from the EWAS (Supplementary Table 7). Our regional analysis using DMRs did not find any regions to be statistically significant. The top ten regions are summarized in Supplementary Table 10.
Figure 1.
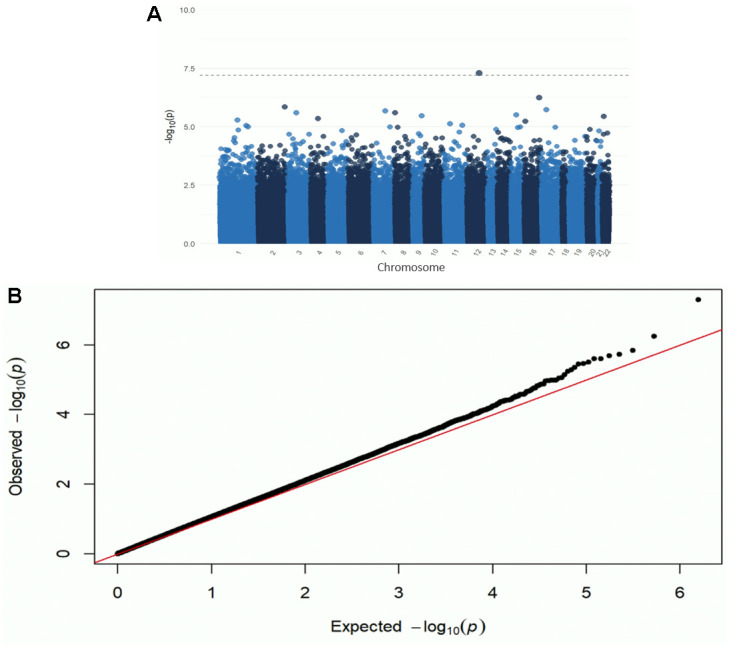
Manhattan (A) and QQ plot (B) from the EWAS of DNAm with the ADI. Adjusted for race, sex, educational attainment, age at death, APOE genotype, cell type, and post-mortem interval. Bonferroni-threshold: 0.05/789889 = 6.33e-8 (λ=0.94).
Figure 2.
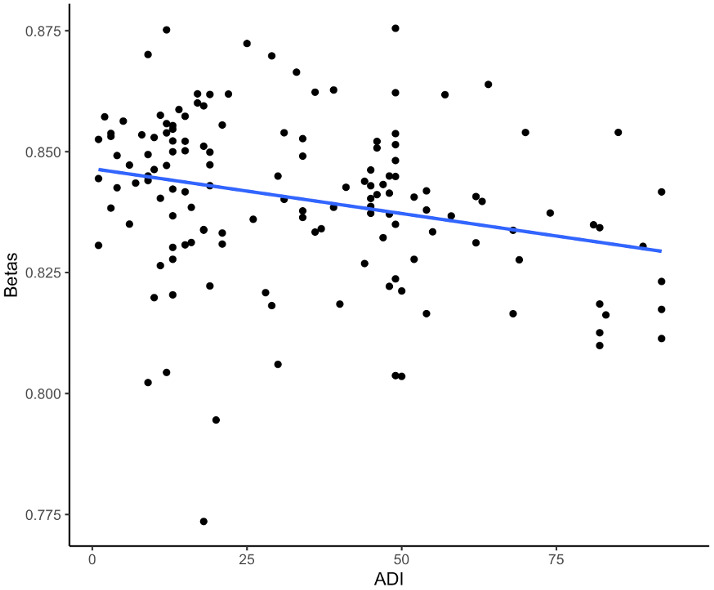
Scatterplot of DNAm beta values and the ADI from the EWAS of DNAm with the ADI for the CpG site cg26514961 (PLXNC1). The dots represent the DNAm beta and ADI values for a participant, and the blue line represents the (unadjusted) linear relationship between the DNAm beta values and the ADI.
Table 2. Top ten CpG sites from the epigenome-wide association study of DNA methylation with the Area Deprivation Index, stratified by the presence or absence of the APOE ε4 allele.
| Total (n=159) | ε4 present (n=89) | ε4 absent (n=70) | Effect modification | |||||||
| CpG | Chromosome | Position | Gene(s) | Effect estimate | P-value | Effect estimate | P-value | Effect estimate | P-value | P-value |
| cg26514961 | 12 | 94566784 | PLXNC1 | -0.0052 | 5.0e-8 | -0.0050 | 0.0033 | -0.0008 | 0.0002 | 8.7e-6 |
| cg08087060 | 16 | 87795808 | KLHDC4 | -0.0040 | 5.7e-7 | -0.0036 | 0.0030 | -0.0007 | 0.0009 | 4.6e-5 |
| cg01291468 | 2 | 234589374 |
UGT1A10;UGT1A7;
UGT1A9;UGT1A8 |
0.0034 | 1.4e-6 | 0.0027 | 0.0098 | 0.0004 | 0.0019 | 0.0003 |
| cg05419854 | 17 | 19398395 | - | -0.0058 | 1.8e-6 | -0.0042 | 0.0190 | -0.0008 | 0.0004 | 1.0e-5 |
| cg16241648 | 7 | 98923114 | ARPC1A | 0.0016 | 2.1e-6 | 0.0018 | 0.0046 | 0.0003 | 0.0020 | 0.0002 |
| cg20912923 | 8 | 2885516 | CSMD1 | -0.0026 | 2.5e-6 | -0.0021 | 0.0083 | -0.0002 | 0.0008 | 3.1e-5 |
| cg15953452 | 3 | 63053400 | - | -0.0050 | 2.5e-6 | -0.0042 | 0.0105 | 0.0008 | 0.0001 | 6.2e-6 |
| cg06787422 | 15 | 63331851 | - | -0.0024 | 3.1e-6 | -0.0015 | 0.0411 | -0.0002 | 0.0012 | 0.0001 |
| cg13521319 | 9 | 133423844 | - | -0.0018 | 3.4e-6 | -0.0020 | 0.0019 | -0.0001 | 0.0046 | 0.0007 |
| cg09431774 | 22 | 25465561 | KIAA1671 | -0.0028 | 3.6e-6 | -0.0020 | 0.0249 | -0.0002 | 0.0002 | 8.6e-6 |
Bold: statistically significant at the Bonferroni threshold of 6.33e-8.
Effect estimates can be interpreted per a 20-unit increase in ADI. All models were adjusted for the following covariates: race, sex, educational attainment, age at death, apolipoprotein E (APOE) genotype, cell type, and post-mortem interval.
Next, we investigated whether the associations with the top ten CpG sites from the EWAS of ADI were modified by APOE ε4 allele. We found nominally significant (p-value < 0.05) effect modifications by presence versus absence of the APOE ε4 allele for all our top ten CpG sites. Effect estimates for associations between the ADI and DNAm observed in the whole study population were similar as the estimates observed among donors with at least one APOE ε4 allele. Effect estimates were alleviated toward the null among donors without any APOE ε4 alleles. No CpG sites were found to be significantly associated with ADI in either APOE ε4 group (Supplementary Figure 1A, 1B).
We then examined whether any of the top ten CpG sites from the EWAS of ADI were associated with AD pathology (CERAD, Braak Stage, and ABC). None of the ten CpG sites were significantly associated with any of the three neuropathology outcomes (Table 3).
Table 3. Association between the top ten CpG sites from the epigenome-wide association study of Area Deprivation Index (compare Table 2), and their association with neuropathology markers (CERAD, ABC and Braak stage).
| CERAD1 | ABC2 | Braak stage | ||||
| CpG | Chromosome | Position | Gene(s) | Effect estimate (95%-CI) | Effect estimate (95%-CI) | Effect estimate (95%-CI) |
| cg26514961 | 12 | 94566784 | PLXNC1 | 0.41 (-0.85, 1.67) | 0.67 (-0.35, 1.70) | 0.41 (-0.78, 2.56) |
| cg08087060 | 16 | 87795808 | KLHDC4 | 0.57 (-1.17, 2.32) | 0.08 (-1.47, 1.63) | 0.57 (-2.73, 1.92) |
| cg01291468 | 2 | 234589374 |
UGT1A10;UGT1A7;
UGT1A9;UGT1A8 |
-1.20 (-3.50, 1.09) | -1.14 (-3.13, 0.82) | -1.20 (-4.67, 1.49) |
| cg05419854 | 17 | 19398395 | - | -0.89 (-2.17, 0.37) | -0.39 (1.50, 0.71) | -0.89 (-1.05, 2.52) |
| cg16241648 | 7 | 98923114 | ARPC1A | 0.93 (-3.42, 5.30) | -1.08 (-4.96, 2.77) | 0.93 (-6.89, 5.12) |
| cg20912923 | 8 | 2885516 | CSMD1 | 0.46 (-2.25, 3.18) | 1.54 (-0.66, 3.77) | 0.46 (-1.82, 5.46) |
| cg15953452 | 3 | 63053400 | - | 0.11 (-1.33, 1.55) | 0.56 (-0.62, 1.75) | 0.11 (-0.66, 2.93) |
| cg06787422 | 15 | 63331851 | - | -0.13 (-3.33, 3.06) | 0.50 (-2.22, 3.22) | -0.13 (-3.20, 4.94) |
| cg13521319 | 9 | 133423844 | - | -1.37 (-5.24, 2.46) | -0.69 (-4.24, 2.84) | -1.37 (-6.76, 4.03) |
| cg09431774 | 22 | 25465561 | KIAA1671 | 0.37 (-2.17, 2.91) | -0.10 (-2.41, 2.21) | 0.37 (-4.31, 2.78) |
1Consortium to Establish a Register for AD; 2Amyloid, Braak, and CERAD.
Effect estimates can be interpreted per a 0.1-unit increase in DNAm. All models were adjusted for the following covariates: race, sex, educational attainment, age at death, apolipoprotein E (APOE) genotype, cell type, and post-mortem interval.
Look-up of top hits in mQTL or cross-tissue databases
Genetic variants influence DNAm patterns, so we investigated whether the identified DNAm associations were likely driven by genetic variant effects (mQTLs). According to the GoDMC database [27], of the top ten CpG sites from the EWAS of ADI, three were associated with at least one mQTL, namely cg26514961 (PLXNC1), cg01291468 (UGT1A10, UGT1A7, UGT1A9, and UGT1A8), and cg06787422 (intergenic) (Supplementary Table 3). To evaluate the correlation of DNAm at our top ten CpG sites across different tissues, we used the BECon tool and Gene Expression Omnibus Database [Accession code GSE111165]. Two CpG sites (cg20912923 (CSMD1) and cg06787422 (intergenic)) exhibited blood–brain concordance using the BECon tool (Supplementary Table 4A). Both of these sites exhibited 75-90% percentile mean correlations between blood and brain samples. Using the Gene Expression Omnibus Database, only cg15953452 (intergenic) exhibited significant blood-brain concordance (Supplementary Table 4B). Two CpG sites, cg26514961 (PLXNC1) and cg16241648 (ARPC1A), exhibited significant buccal cell-brain concordance. Four CpG sites (cg26514961 (PLXNC1), cg16241648 (ARPC1A), cg15953452 (intergenic), and cg06787422 (intergenic)) exhibited significant saliva-brain concordance.
Pathway enrichment analysis
To further aid the interpretation of our top associations, we performed a gene ontology (GO) and KEGG pathway enrichment analysis based on the top 1000 CpG sites with lowest raw p-values. After correction for multiple testing (FDR <0.05), we did not identify any GO terms or KEGG pathways with an overrepresentation of genes containing significantly, differentially methylated CpGs that would indicate an enriched biological pathway. GO terms and KEGG pathways that were nominally significant (raw p<0.05) are included in the supplement (Supplementary Table 5A, 5B).
DISCUSSION
In the ADRC autopsy cohort of 159 donors, we found one CpG site (cg26514961, gene PLXNC1) that was significantly associated with the ADI in brain tissue after controlling for covariates and multiple testing. Effect modification by APOE ε4 was found to be statistically significant for the top ten CpG sites from the EWAS, indicating that the observed associations between ADI and DNAm were mainly driven by donors who carried at least one APOE ε4 allele. Four of the top ten CpG sites showed a significant concordance between brain tissue and tissues that are easily accessible in living individuals (blood, buccal cells, saliva), including DNAm in cg26514961 (PLXNC1). This suggests that differential DNAm in these CpG sites could potentially be detected prior to death. None of the top ten CpG sites from the EWAS of ADI were associated with AD pathology in this autopsy cohort and the EWAS results were robust to additional adjustment for neuropathology markers. This indicates that the identified associations between ADI and differential DNAm in the brain were independent of the degree of AD-related neuropathology.
The EWAS identified cg26514961 as being significantly associated with the ADI, which is associated with the PLXNC1 gene. This gene is believed to be related to the immune response in the body [28]. Additionally, the corresponding RNA and protein levels are altered in the brains of people with AD [29]. The protein that this gene encodes regulates melanocyte adhesion, and viral semaphorins are thought to modulate the immune response through binding to this receptor [29]. Previous research has suggested the immune response as a potential biological pathway of how neighborhood deprivation affects the body [5]. This hypothesis is further supported by three additional genes that were among the top three CpG sites (cg01291468 [UGT1A7, UGT1A8, and UGT1A9]) and which have all been linked to immunosuppression [30–32]; thus providing further evidence that neighborhood deprivation impacts health through the immune response. Two additional genes among our top ten CpG sites have been associated with brain-related health outcomes and aging. KLHDC4 (cg08087060) is associated with Huntington’s disease [33], and CSMD1 (cg20912923) is related to learning and memory [34]. In a meta-analysis of brain tissue-based EWAS in Alzheimer’s disease (n=1453), our top ten CpG sites were not found in any of the studies the authors examined, and none of the top 25 CpG sites they found to be statistically significant for Alzheimer’s disease were associated with ADI in our analysis (Supplementary Table 9) [35].
Our study found concordance between brain and other tissues in four of our top ten CpG sites. It is important to examine the concordance between brain tissue and other tissues (such as blood, saliva, and buccal) because brain tissue samples are not accessible from living donors, whereas these three other tissues are. Differential DNAm in tissues that are easily accessible in living individuals can serve as biomarkers of exposures or to predict related health outcomes. Thus, if DNAm profiles in brain tissue are correlated with other tissues, those profiles can potentially be used to identify individuals at heightened risk, and may lead to earlier access to preventative care.
None of the top ten CpG sites have been identified in prior studies as being related to DNAm and ADI, most likely due to the different tissues that were used. Two prior studies found increased global DNAm among those living in more disadvantaged neighborhoods [12, 13]. These studies did not examine particular CpG sites or genes, so it is unclear which locations experienced increased or decreased DNAm levels. Another study found three CpG sites that were associated with neighborhood deprivation, with one being linked to a gene that is related to Parkinson’s Disease [14]. None of these three CpG sites were identified in our EWAS (Supplementary Table 6), but it is of note that their study also identified genes associated with an aging-related disease. Two other studies found increased DNAm in genes related to stress and inflammation in the body [6, 15], which is closely linked to the immune response pathway that two of our top ten CpG sites were linked to [36]. Overall, our findings related to stress and inflammation align with pathways identified in previous research, but more studies are needed to replicate our findings and to identify other CpG sites and genes which are related to neighborhood deprivation.
We found evidence of effect modification by APOE ε4 in the EWAS of ADI, indicating that the observed associations between ADI and DNAm were mainly driven by donors who carried at least one APOE ε4 allele. This aligns with previous research, which suggests that there are differences in epigenome-wide methylation among APOE ε4 carriers and non-carriers in blood samples in many genetic positions and loci [37]. Further research is needed to investigate how DNAm differs by APOE ε4 being present or absent, especially in brain tissue.
None of the top ten CpG sites from the EWAS of ADI were associated with AD pathology in this autopsy cohort. This finding could be due to most participants in our sample being cognitively impaired, which limits the statistical power to detect differences between impaired and non-impaired individuals. More research on this association with a larger sample of non-impaired individuals is needed to better understand the relationship between these CpG sites and AD.
Lastly, three of our top ten CpG sites were associated with at least one known mQTL, which is an indicator of the genetic influence on DNAm levels [27]. While we are unable to disambiguate the effects of the environment and genes on DNAm levels, only a proportion of the variation in DNAm levels is explained by genetic effects. In fact, the joint effects of environmental factors and single nucleotide polymorphisms (SNP) have been found to be larger contributors to DNAm variation than SNPs alone [38].
Our study has several strengths. One major strength is that our study is the first known study on the association between neighborhood deprivation and DNAm in brain tissue, which is difficult to obtain and the most relevant tissue to study brain-related health outcomes. Studying DNAm changes in brain tissue is especially important because it can provide insight into neuropathology outcomes such as Alzheimer’s disease [16–22] and depression [23, 24]. These brain health outcomes have themselves been associated with neighborhood deprivation [2, 25, 26], which is the reason more research on neighborhood deprivation using brain tissue is needed. Another strength of our study is that we had diversity of neighborhood deprivation. In our study, the ADI ranged from 1 to 95, thus including very deprived and very privileged neighborhoods. Another strength of our study is that we used the Infinium methylation EPIC array as opposed to the Illumina Infinium HumanMethylation450 (450K) BeadChip array. The EPIC array covers more than 850,000 methylation sites whereas the 450K array only covers 450,000 methylation sites. Only one of the five previous studies on the association between neighborhood deprivation and DNAm used the EPIC array [13]. Of the top ten CpG sites associated with ADI in our cohort, four CpG sites were only available on the EPIC array.
Our study has a few limitations. Our sample size was relatively small (n=159), which limited the statistical power to detect associations. Additionally, our sample was not racially diverse and only contained self-reported White and Black donors. Only 10.7% of participants in our sample were Black, limiting our ability to detect racial differences. Thus, we are unable to generalize our results to other racial or ethnic groups. Another limitation of our study is that we only had information on the donors’ last known address. It is possible that the donors moved around a lot during their life, or only moved to their last address at the end of their life. In these cases, the long-term or even life-term exposure to neighborhood deprivation would not be captured in the data. It is possible that the neighborhood conditions of where someone grew up or lived during most of their life are more relevant to studying the association with DNA methylation as opposed to where they lived at the end of their life, but further research is needed to elucidate these effects throughout the lifespan. Another limitation of our study is that the 2020 ADI measure we used does not correspond with the donors’ years of death. This could lead to measurement error in our study, which may result in biased estimates. A final limitation of our study is that very few participants were not cognitively impaired (2.5%). Because the majority of participants had some form of cognitive impairment, the statistical power to detect differences between impaired and non-impaired participants was rather limited. Furthermore, most participants exhibited Braak Stage 6 (45.9%), had frequent CERAD (69.2%), and had a high ABC score (58.5%). These are extreme values as compared to the general US population, demonstrating that our population was not representative of the larger US or Georgia population.
Overall, our study identified one CpG site (cg26514961, PLXNC1 gene) that was significantly associated with neighborhood deprivation in brain tissue. We also found evidence of effect modification by APOE ε4, suggesting that the observed associations between ADI and DNAm were mainly driven by donors who carried at least one APOE ε4 allele. Our study provides motivation to conduct larger studies on the association between neighborhood deprivation and DNAm in the brain to replicate and expand upon our findings. The identification of significant CpG sites could provide novel insights into the etiology of health disparities, and the concordance between brain and other tissues for our top CpG sites could make them potential candidates for biomarkers in living individuals.
MATERIALS AND METHODS
Study population
The study population was derived from brain tissue donors recruited by the Emory Goizueta Alzheimer’s Disease Research Center (ADRC). Most of the donors in this study were patients diagnosed as having Alzheimer’s Disease and were treated at the Emory Clinic or Emory University Hospital. In total, 1011 donors enrolled in the study until the third quarter of 2020 (Supplementary Table 1). The inclusion criteria for our study were the following: 1) residential addresses within Georgia; 2) age at death of at least 55; 3) died after 1999; 4) no missing values in outcomes and key covariates which include race, sex, educational attainment, APOE genotype; 5) DNAm data was available. Based on these criteria, 159 donors remained in the analysis. Written consent for brain donation was obtained from next of kin as required under Georgia law. Emory University’s Institutional Review Board approved this study.
Assessment of neighborhood deprivation
Neighborhood deprivation was defined using the Area Deprivation Index (ADI), a census-based socioeconomic index developed by Kind et al. [39]. The ADI is calculated using socioeconomic status domains of income, education, employment, and housing quality indicators obtained from the American Community Survey. Using these domains, the ADI is calculated from 17 census indicators that are multiplied by previously published factor weights and summed for each census block group and then transformed into a standardized index [20]. The ADI assigns ranked percentiles that range from 1 to 100, where 100 represents the most deprived neighborhood. A neighborhood is defined as a census block group, which is the smallest geographic unit used by the United States Census Bureau to tabulate 100-percent data. A census block group comprises a set of blocks that generally contain 600 to 3000 people and is the smallest unit with detailed demographic-economic characteristics [40]. We linked the 2020 ADI to each participant’s geocoded residential address at the time of their death using Federal Information Processing Standards codes [41].
Assessment of neuropathologic markers
The ADRC conducted neuropathologic evaluations on every donor’s brain using diagnostic criteria and established research evaluations. The neuropathologic assessments evaluated the severity of AD-related neuropathology changes, which included a variety of stains and immunohistochemical preparations as well as semi-quantitative scoring of multiple neuropathologic changes in brain regions by experienced neuropathologists using published criteria. AD neuropathology was assessed using the Consortium to Establish a Register for AD (CERAD) score, Braak stage, and a combination of Amyloid, Braak, and CERAD (ABC) score. CERAD score represents the prevalence of neuritic plaques with four levels from zero neuritic plaques to frequent. Braak stage is a staging scheme which represents neurofibrillary tangles (NFTs) and has six stages (Stage I-VI), with higher stages indicating a wider distribution of NFTs in the brain. ABC score combines CERAD and Braak Stage with the prevalence of Amyloid plaques and is converted to one of four levels of AD neuropathologic changes: not, low, intermediate, or high.
Assessment of DNA methylation
Fresh, frozen prefrontal cortex samples were collected from donors at autopsy, and DNA was isolated from these samples using the QIAGEN GenePure kit. Illumina Infinium HumanMethylationEPIC BeadChips arrays were used to assess DNAm in the 159 samples and 6 replicates for quality control to assess the background technical variation (root mean square error (RMSE) ranged from 0.022-0.028). We followed a validated quality control and normalization pipeline as previously published [42]. Pre-processing and statistics were completed using R (v4.2.0). All DNAm data were preprocessed to identify low-quality samples, exclude specific probes, and reduce the impact of batch effects. Raw intensity files were converted to methylation beta values ranging on a continuous scale from 0 to 1 for each of the CpG sites measured on the array. The Illumina’s 636 control probes were used via the R package ewastools to assess technique parameters including array staining, extension, hybridization, target removal, specificity, and bisulfite conversion [43]. Additional sample outlier detection was implemented based on detection p-value, beadcount, and distance from the group average in principal components. The Funnorm function and Combat function were used to normalize the distributions to reduce technical variation and correct for differences between type I and type II probe signals. The following probes were further removed: XY probes, low-quality probes with missing in more than 5% of samples, probes with poor detection p-values, probes predicted to cross-hybridize, probes that bind to the sex chromosomes, polymorphic probes, and probes with infinite values. In total, after all preprocessing steps, 159 samples and 789,286 CpG sites remained for the down-stream analysis. We used the estimateCellCounts function in the R package minfi to obtain the cell-type proportions (neuronal vs. non-neuronal cells) for each sample using the most recent prefrontal cortex database [44, 45].
Confounder assessment
Confounders were identified based on existing literature. All models were adjusted for the following covariates: race, sex, educational attainment, age at death, apolipoprotein E (APOE) genotype, cell type, and post-mortem interval. Due to the sample only containing White and Black participants, the race variable was binary. Educational attainment was defined as the highest level of education completed by the participant and classified into high school or less, college degree, and graduate degree. APOE genotype had three levels in the analysis: no ε4 allele, single ε4 allele, and double ε4 allele. The APOE ε4 allele is a well-known risk factor of developing Alzheimer’s disease, and the current analysis considered: 0, 1, and 2 ε4 alleles. Also, a binary APOE genotype (ε4 absent vs. present) was used for testing the effect modification by the genotype. Binary APOE genotype was used for effect modification analyses to conserve statistical power in analyses (see Table 1 for a distribution of APOE ε4 genotypes).
Statistical analysis
To identify DNAm patterns in brain tissue that are associated with ADI, we conducted an epigenome-wide association study (EWAS) of single CpG sites and an analysis of differentially methylated regions (DMRs). For the EWAS, we ran a robust linear regression model using the RLM function within the MASS package with ADI as the independent variable and DNAm beta values at each CpG site as a dependent variable, adjusting for self-reported race, sex, APOE genotype, education, age at death, cell type, and post-mortem interval. We applied a Bonferroni threshold to correct for multiple testing based on the number of tested CpG sites (threshold: 0.05/789889 = 6.33e-8). Associations between ADI and DMRs were analyzed using the R package dmrff.
We conducted several sensitivity analyses to evaluate the robustness of our EWAS findings. First, we adjusted for neuropathology markers (CERAD, Braak Stage, and ABC) to investigate whether the identified associations were independent of the degree of neuropathology. Second, we conducted an EWAS of ADI after excluding the 2.5% cognitively normal donors. Third, since APOE ε4 is a well-known risk factor for developing AD, we included a multiplicative interaction term between ADI and APOE genotype (presence or absence of ε4 allele) in our EWAS to test for effect modification and presented the stratified effect estimates derived from that interaction model.
Next, we investigated whether DNAm patterns in brain tissue that are associated with ADI are also linked with neuropathology markers. We ran linear regression models using each of the top ten CpGs as the independent variables, and three neuropathology outcomes (CERAD, Braak Stage, and ABC) as dependent variables in separate models, adjusting for ADI, self-reported race, sex, APOE genotype, education, age at death, cell type, and post-mortem interval.
We conducted additional analyses for the top ten CpG sites in the EWAS analysis to evaluate their correlation across different tissues and how methylation at those sites is affected by genotypic variation. This included blood–brain concordance analysis using the Blood–Brain Epigenetic Concordance (BECon) tool [46], blood-brain, buccal-brain, and saliva-brain concordance using the data from Braun et al. (2019) on the Gene Expression Omnibus Database [Accession code GSE111165] [47, 48] and methylation quantitative trait loci (mQTL) mapping using the GoDMC database [27]. To further aid the interpretation of our top associations, we conducted a gene ontology (GO) and KEGG pathway enrichment analysis which was a look-up of top hits in mQTL and cross-tissue databases using the R package missMethyl based on the top 1000 CpG sites with lowest raw p-values [49].
Supplementary Material
Abbreviations
- ABC
Amuloid, Braak, and CERAD
- AD
Alzheimer’s disease
- ADI
Area Deprivation Index
- ADRC
Alzheimer’s Disease Research Center
- APOE
Apolipoprotein E
- BECon
Blood-Brain Epigenetic Concordance
- CERAD
Consortium to Establish a Register for AD
- CpG
Cytosine-phosphate-guanine
- DNAm
DNA methylation
- EWAS
Epigenome-wide association study
- GO
Gene ontology
- mQTL
Methylation quantitative trait loci
- NFTs
Neurofibrillary tangles
- RMSE
Root mean square error
- SES
Socioeconomic status
- SNP
Single nucleotide polymorphism
Footnotes
AUTHOR CONTRIBUTIONS: LP wrote the original draft and carried out the statistical analyses. ZL assisted with the main statistical analyses. SA assisted with secondary analyses. AH conceived and supervised the project and helped with the preparation of the original draft. The remaining authors contributed data and participated in editing the manuscript. All authors read and approved the final manuscript.
CONFLICTS OF INTEREST: We have no conflicts of interest to declare.
Ethical Statement and Consent: Written informed consent for brain donation was obtained from next of kin as required under Georgia state law. Emory University’s Institutional Review Board approved this study. This study was determined to be non-human subjects research. Emory does not require IRB review of studies that do not meet the definitions of "human subjects research".
FUNDING: This work was supported by the HERCULES Pilot Project via NIEHS P30ES019776 (Huels), the Goizueta Alzheimer’s Disease Research Center: Pilot Grant via NIA P50AG025688 (Huels/Liang), the Rollins School of Public Health Dean’s Pilot and Innovation Grant (Huels) and NIA R01AG079170 (Huels/Wingo).
REFERENCES
- 1.Olden K, Olden HA, Lin YS. The Role of the Epigenome in Translating Neighborhood Disadvantage Into Health Disparities. Curr Environ Health Rep. 2015; 2:163–70. 10.1007/s40572-015-0048-x [DOI] [PubMed] [Google Scholar]
- 2.Diez Roux AV, Mair C. Neighborhoods and health. Ann N Y Acad Sci. 2010; 1186:125–45 10.1111/j.1749-6632.2009.05333.x [DOI] [PubMed] [Google Scholar]
- 3.Pase MP, Rowsthorn E, Cavuoto MG, Lavale A, Yassi N, Maruff P, Buckley RF, Lim YY. Association of Neighborhood-Level Socioeconomic Measures With Cognition and Dementia Risk in Australian Adults. JAMA Netw Open. 2022; 5:e224071. 10.1001/jamanetworkopen.2022.4071 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Christensen GM, Li Z, Pearce J, Marcus M, Lah JJ, Waller LA, Ebelt S, Hüls A. The complex relationship of air pollution and neighborhood socioeconomic status and their association with cognitive decline. Environ Int. 2022; 167:107416. 10.1016/j.envint.2022.107416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Iyer HS, Hart JE, James P, Elliott EG, DeVille NV, Holmes MD, De Vivo I, Mucci LA, Laden F, Rebbeck TR. Impact of neighborhood socioeconomic status, income segregation, and greenness on blood biomarkers of inflammation. Environ Int. 2022; 162:107164. 10.1016/j.envint.2022.107164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JA, Zhao W, Wang X, Ratliff SM, Mukherjee B, Kardia SLR, Liu Y, Roux AV, Needham BL. Neighborhood characteristics influence DNA methylation of genes involved in stress response and inflammation: The Multi-Ethnic Study of Atherosclerosis. Epigenetics. 2017; 12:662–73. 10.1080/15592294.2017.1341026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Giurgescu C, Nowak AL, Gillespie S, Nolan TS, Anderson CM, Ford JL, Hood DB, Williams KP. Neighborhood Environment and DNA Methylation: Implications for Cardiovascular Disease Risk. J Urban Health. 2019. (Suppl 1); 96:23–34. 10.1007/s11524-018-00341-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Moore LD, Le T, Fan G. DNA methylation and its basic function. Neuropsychopharmacology. 2013; 38:23–38. 10.1038/npp.2012.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Laubach ZM, Perng W, Cardenas A, Rifas-Shiman SL, Oken E, DeMeo D, Litonjua AA, Duca RC, Godderis L, Baccarelli A, Hivert MF. Socioeconomic status and DNA methylation from birth through mid-childhood: a prospective study in Project Viva. Epigenomics. 2019; 11:1413–27. 10.2217/epi-2019-0040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Needham BL, Smith JA, Zhao W, Wang X, Mukherjee B, Kardia SLR, Shively CA, Seeman TE, Liu Y, Diez Roux AV. Life course socioeconomic status and DNA methylation in genes related to stress reactivity and inflammation: The multi-ethnic study of atherosclerosis. Epigenetics. 2015; 10:958–69. 10.1080/15592294.2015.1085139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stringhini S, Polidoro S, Sacerdote C, Kelly RS, van Veldhoven K, Agnoli C, Grioni S, Tumino R, Giurdanella MC, Panico S, Mattiello A, Palli D, Masala G, et al. Life-course socioeconomic status and DNA methylation of genes regulating inflammation. Int J Epidemiol. 2015; 44:1320–30. 10.1093/ije/dyv060 [DOI] [PubMed] [Google Scholar]
- 12.Shen J, Fuemmeler BF, Sheppard VB, Bear HD, Song R, Chow WH, Zhao H. Neighborhood disadvantage and biological aging biomarkers among breast cancer patients. Sci Rep. 2022; 12:11006. 10.1038/s41598-022-15260-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsu PC, Kadlubar S, Su LJ, Acheampong D, Rogers LJ, Runnells G, McElfish PA, Schootman M. County poverty levels influence genome-wide DNA methylation profiles in African American and European American women. Transl Cancer Res. 2019; 8:683–92. 10.21037/tcr.2019.02.07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawrence KG, Kresovich JK, O’Brien KM, Hoang TT, Xu Z, Taylor JA, Sandler DP. Association of Neighborhood Deprivation With Epigenetic Aging Using 4 Clock Metrics. JAMA Netw Open. 2020; 3:e2024329. 10.1001/jamanetworkopen.2020.24329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reuben A, Sugden K, Arseneault L, Corcoran DL, Danese A, Fisher HL, Moffitt TE, Newbury JB, Odgers C, Prinz J, Rasmussen LJ, Williams B, Mill J, Caspi A. Association of Neighborhood Disadvantage in Childhood With DNA Methylation in Young Adulthood. JAMA Netw Open. 2020; 3:e206095. 10.1001/jamanetworkopen.2020.6095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Horvath S, Zhang Y, Langfelder P, Kahn RS, Boks MP, van Eijk K, van den Berg LH, Ophoff RA. Aging effects on DNA methylation modules in human brain and blood tissue. Genome Biol. 2012; 13:R97. 10.1186/gb-2012-13-10-r97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yokoyama AS, Rutledge JC, Medici V. DNA methylation alterations in Alzheimer’s disease. Environ Epigenet. 2017; 3:dvx008. 10.1093/eep/dvx008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nicolia V, Cavallaro RA, López-González I, Maccarrone M, Scarpa S, Ferrer I, Fuso A. DNA Methylation Profiles of Selected Pro-Inflammatory Cytokines in Alzheimer Disease. J Neuropathol Exp Neurol. 2017; 76:27–31. 10.1093/jnen/nlw099 [DOI] [PubMed] [Google Scholar]
- 19.Wang SC, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer’s disease. PLoS One. 2008; 3:e2698. 10.1371/journal.pone.0002698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Iwata A, Nagata K, Hatsuta H, Takuma H, Bundo M, Iwamoto K, Tamaoka A, Murayama S, Saido T, Tsuji S. Altered CpG methylation in sporadic Alzheimer’s disease is associated with APP and MAPT dysregulation. Hum Mol Genet. 2014; 23:648–56. 10.1093/hmg/ddt451 [DOI] [PubMed] [Google Scholar]
- 21.Smith RG, Hannon E, De Jager PL, Chibnik L, Lott SJ, Condliffe D, Smith AR, Haroutunian V, Troakes C, Al-Sarraj S, Bennett DA, Powell J, Lovestone S, et al. Elevated DNA methylation across a 48-kb region spanning the HOXA gene cluster is associated with Alzheimer’s disease neuropathology. Alzheimers Dement. 2018; 14:1580–8. 10.1016/j.jalz.2018.01.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hüls A, Robins C, Conneely KN, Edgar R, De Jager PL, Bennett DA, Wingo AP, Epstein MP, Wingo TS. Brain DNA Methylation Patterns in CLDN5 Associated With Cognitive Decline. Biol Psychiatry. 2022; 91:389–98. 10.1016/j.biopsych.2021.01.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabunciyan S, Aryee MJ, Irizarry RA, Rongione M, Webster MJ, Kaufman WE, Murakami P, Lessard A, Yolken RH, Feinberg AP, Potash JB, and GenRED Consortium. Genome-wide DNA methylation scan in major depressive disorder. PLoS One. 2012; 7:e34451. 10.1371/journal.pone.0034451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hüls A, Robins C, Conneely KN, De Jager PL, Bennett DA, Epstein MP, Wingo TS, Wingo AP. Association between DNA methylation levels in brain tissue and late-life depression in community-based participants. Transl Psychiatry. 2020; 10:262. 10.1038/s41398-020-00948-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vassilaki M, Aakre JA, Castillo A, Chamberlain AM, Wilson PM, Kremers WK, Mielke MM, Geda YE, Machulda MM, Alhurani RE, Graff-Radford J, Vemuri P, Lowe VJ, et al. Association of neighborhood socioeconomic disadvantage and cognitive impairment. Alzheimers Dement. 2022. [Epub ahead of print]. 10.1002/alz.12702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marsh K, Shao Y, Zhang Y, Masurkar AV, Vedvyas A, Chodosh J. Association of neighborhood socioeconomic disadvantage and cognitive decline. Alzheimers Dement. 2021; 17:e056584. 10.1002/alz.056584 [DOI] [Google Scholar]
- 27.Min JL, Hemani G, Hannon E, Dekkers KF, Castillo-Fernandez J, Luijk R, Carnero-Montoro E, Lawson DJ, Burrows K, Suderman M, Bretherick AD, Richardson TG, Klughammer J, et al. , and BIOS Consortium. Genomic and phenotypic insights from an atlas of genetic effects on DNA methylation. Nat Genet. 2021; 53:1311–21. 10.1038/s41588-021-00923-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.PLXNC1 plexin C1 [Homo sapiens (human)] - Gene - NCBI. 2023. https://www.ncbi.nlm.nih.gov/gene/10154
- 29.Agora. 2023. https://agora.adknowledgeportal.org/genes/ENSG00000136040/summary
- 30.UGT1A7 Gene - UDP Glucuronosyltransferase Family 1 Member A7. 2023. https://www.genecards.org/cgi-bin/carddisp.pl?gene=UGT1A7
- 31.UGT1A8 Gene - UDP Glucuronosyltransferase Family 1 Member A8. 2023. https://www.genecards.org/cgi-bin/carddisp.pl?gene=UGT1A8
- 32.UGT1A9 Gene - UDP Glucuronosyltransferase Family 1 Member A9. 2023. https://www.genecards.org/cgi-bin/carddisp.pl?gene=UGT1A9
- 33.KLHDC4 Gene - Kelch Domain Containing 4. 2023. https://www.genecards.org/cgi-bin/carddisp.pl?gene=KLHDC4
- 34.CSMD1 Gene - CUB And Sushi Multiple Domains 1. 2023. https://www.genecards.org/cgi-bin/carddisp.pl?gene=CSMD1 [DOI] [PMC free article] [PubMed]
- 35.Smith RG, Pishva E, Shireby G, Smith AR, Roubroeks JAY, Hannon E, Wheildon G, Mastroeni D, Gasparoni G, Riemenschneider M, Giese A, Sharp AJ, Schalkwyk L, et al. A meta-analysis of epigenome-wide association studies in Alzheimer’s disease highlights novel differentially methylated loci across cortex. Nat Commun. 2021; 12:3517. 10.1038/s41467-021-23243-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu YZ, Wang YX, Jiang CL. Inflammation: The Common Pathway of Stress-Related Diseases. Front Hum Neurosci. 2017; 11:316. 10.3389/fnhum.2017.00316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Walker RM, Vaher K, Bermingham ML, Morris SW, Bretherick AD, Zeng Y, Rawlik K, Amador C, Campbell A, Haley CS, Hayward C, Porteous DJ, McIntosh AM, et al. Identification of epigenome-wide DNA methylation differences between carriers of APOE ε4 and APOE ε2 alleles. Genome Med. 2021; 13:1. 10.1186/s13073-020-00808-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Czamara D, Eraslan G, Page CM, Lahti J, Lahti-Pulkkinen M, Hämäläinen E, Kajantie E, Laivuori H, Villa PM, Reynolds RM, Nystad W, Håberg SE, London SJ, et al. , and Major Depressive Disorder Working Group of the Psychiatric Genomics Consortium. Integrated analysis of environmental and genetic influences on cord blood DNA methylation in new-borns. Nat Commun. 2019; 10:2548. 10.1038/s41467-019-10461-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kind AJH, Jencks S, Brock J, Yu M, Bartels C, Ehlenbach W, Greenberg C, Smith M. Neighborhood socioeconomic disadvantage and 30-day rehospitalization: a retrospective cohort study. Ann Intern Med. 2014; 161:765–74. 10.7326/M13-2946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.US Census Bureau. Glossary: Block group. Census.gov. 2023. https://www.census.gov/programs-surveys/geography/about/glossary.html
- 41.University of Wisconsin School of Medicine and Public Health. 2020 Area Deprivation Index v1.0. 2023. https://www.neighborhoodatlas.medicine.wisc.edu/
- 42.Konwar C, Price EM, Wang LQ, Wilson SL, Terry J, Robinson WP. DNA methylation profiling of acute chorioamnionitis-associated placentas and fetal membranes: insights into epigenetic variation in spontaneous preterm births. Epigenetics Chromatin. 2018; 11:63. 10.1186/s13072-018-0234-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Murat K, Grüning B, Poterlowicz PW, Westgate G, Tobin DJ, Poterlowicz K. Ewastools: Infinium Human Methylation BeadChip pipeline for population epigenetics integrated into Galaxy. Gigascience. 2020; 9:giaa049. 10.1093/gigascience/giaa049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Aryee MJ, Jaffe AE, Corrada-Bravo H, Ladd-Acosta C, Feinberg AP, Hansen KD, Irizarry RA. Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics. 2014; 30:1363–9. 10.1093/bioinformatics/btu049 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guintivano J, Aryee MJ, Kaminsky ZA. A cell epigenotype specific model for the correction of brain cellular heterogeneity bias and its application to age, brain region and major depression. Epigenetics. 2013; 8:290–302. 10.4161/epi.23924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Edgar RD, Jones MJ, Meaney MJ, Turecki G, Kobor MS. BECon: a tool for interpreting DNA methylation findings from blood in the context of brain. Transl Psychiatry. 2017; 7:e1187. 10.1038/tp.2017.171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Braun PR, Han S, Hing B, Nagahama Y, Gaul LN, Heinzman JT, Grossbach AJ, Close L, Dlouhy BJ, Howard MA 3rd, Kawasaki H, Potash JB, Shinozaki G. Genome-wide DNA methylation comparison between live human brain and peripheral tissues within individuals. Transl Psychiatry. 2019; 9:47. 10.1038/s41398-019-0376-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GEO Accession viewer. 2023. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE111165
- 49.Phipson B, Maksimovic J, Oshlack A. missMethyl: an R package for analyzing data from Illumina’s HumanMethylation450 platform. Bioinformatics. 2016; 32:286–8. 10.1093/bioinformatics/btv560 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


