Abstract
Equine arteritis virus (EAV), the prototype Arterivirus, is a positive-stranded RNA virus that expresses its replicase in the form of two large polyproteins of 1,727 and 3,175 amino acids. The functional replicase subunits (nonstructural proteins), which drive EAV genome replication and subgenomic mRNA transcription, are generated by extensive proteolytic processing. Subgenomic mRNA transcription involves an unusual discontinuous step and generates the mRNAs for structural protein expression. Previously, the phenotype of mutant EAV030F, which carries a single replicase point mutation (Ser-2429→Pro), had implicated the nsp10 replicase subunit (51 kDa) in viral RNA synthesis, and in particular in subgenomic mRNA transcription. nsp10 contains an N-terminal (putative) metal-binding domain (MBD), located just upstream of the Ser-2429→Pro mutation, and a helicase activity in its C-terminal part. We have now analyzed the N-terminal domain of nsp10 in considerable detail. A total of 38 mutants, most of them carrying specific single point mutations, were tested in the context of an EAV infectious cDNA clone. Variable effects on viral genome replication and subgenomic mRNA transcription were observed. In general, our results indicated that the MBD region, and in particular a set of 13 conserved Cys and His residues that are assumed to be involved in zinc binding, is essential for viral RNA synthesis. On the basis of these data and comparative sequence analyses, we postulate that the MBD may employ a rather unusual mode of zinc binding that could result in the association of up to four zinc cations with this domain. The region containing residue Ser-2429 may play the role of “hinge spacer,” which connects the MBD to the rest of nsp10. Several mutations in this region specifically affected subgenomic mRNA synthesis. Furthermore, one of the MBD mutants was replication and transcription competent but did not produce infectious progeny virus. This suggests that nsp10 is involved in an as yet unidentified step of virion biogenesis.
Equine arteritis virus (EAV) (17) is the prototype of the Arteriviridae, a recently established family of positive-stranded, enveloped RNA viruses (for reviews see references 47 and 53). The other three members of the Arteriviridae are Lactate dehydrogenase-elevating virus (LDV), Porcine reproductive and respiratory syndrome virus (PRRSV), and Simian hemorrhagic fever virus (SHFV). Based on their presumed common ancestry (7), the arteriviruses have been united with the coronaviruses in the order of the Nidovirales. Important common properties of nidoviruses (Fig. 1) are the presence of a unique array of conserved domains in the viral replicase, the use of ribosomal frameshifting and proteolytic processing to regulate replicase gene expression, and the use of discontinuous transcription to generate a nested set of subgenomic (sg) mRNAs for structural protein expression (for reviews see references 15, 35, and 53).
FIG. 1.
EAV genome organization and expression. (Top) EAV genome organization and the nested set of sg mRNAs (RNAs 2 to 7). Black boxes, common leader sequences; gray boxes, ORFs expressed from each sg mRNA. (Bottom) ORF1a and ORF1ab replicase polyproteins. The ORF1a-encoded protease domains and their corresponding cleavage sites are indicated with white or shaded boxes and arrowheads, respectively. Black boxes, hydrophobic domains and conserved ORF1b-encoded domains. Abbreviations: P, papainlike cysteine protease; C, cysteine protease; H, hydrophobic domain; S, serine protease; RdRp, putative RNA-dependent RNA polymerase; M, predicted MBD; Hel, putative helicase domain; N, conserved nidovirus-specific domain.
The EAV nonstructural proteins are generated by proteolytic processing of two large replicase polyproteins. These precursors, the open reading frame 1a (ORF1a) and ORF1ab proteins, are expressed from the viral genome as polypeptides of 1,727 (187 kDa) and 3,175 (345 kDa) amino acids (aa), respectively. The ORF1ab protein is only produced upon ORF1a/ORF1b ribosomal frameshifting (10). The two replicase polyproteins are proteolytically processed by three ORF1a-encoded proteinases (55, 57, 58). This results in the production of 12 nonstructural proteins (nsp1 to nsp12; Fig. 1) and multiple processing intermediates. Eight of the processing end products are derived from the ORF1a polyprotein (56, 63, 64, 68). Of the four cleavage products that are derived from the ORF1b polypeptide, three proteins contain highly conserved domains that are assumed to have important functions in arterivirus (nidovirus) replication. nsp9 harbors the predicted RNA-dependent RNA polymerase function (48), nsp10 comprises a putative metal-binding domain (MBD) (10, 23) and the nucleoside triphosphate-binding/helicase (Hel) motif (22), and nsp11 contains a conserved domain of unknown function which is only found in nidoviruses (10, 52). With the exception of nsp1, all EAV nonstructural proteins localize exclusively to membranes in the perinuclear region of the infected cell (61, 64). Viral RNA synthesis colocalizes with these replicase subunits, indicating that a membrane-bound replication complex is formed. Most likely, this complex is anchored to vesicular double-membrane structures via hydrophobic domains, which are present in nsp2, nsp3, and nsp5 (46, 61).
EAV produces a nested set of six sg mRNAs (10, 13) to express the seven genes that are located downstream of the replicase gene. At least six of these genes encode structural proteins (14, 54). The sg mRNAs are not only 3′-coterminal but also share a common 5′ leader sequence of 211 nucleotides (nt), which is derived from the 5′ end of the genome (9, 13). Nidovirus discontinuous transcription is still only partially understood, but it has become clear that a conserved transcription-regulating sequence (TRS) plays an essential role (65). In the genome, TRSs are present at the 3′ end of the leader sequence and at the 5′ end of each of the mRNA body sequences. In the sg mRNAs, the TRS is found at the site where leader- and body-derived sequences have been fused. These features, which were first described for coronaviruses (34, 59), suggested that discontinuous transcription involves base pairing between the genomic leader TRS and the TRS complements in the viral negative strand. Recently, we obtained direct evidence for such a key role of TRS base pairing in leader-to-body joining (65) following site-directed mutagenesis of TRSs in an EAV infectious cDNA clone (62). Our data were most compatible with a model in which discontinuous transcription yields sg minus strands, which subsequently function as templates for sg mRNA synthesis (51, 65).
In addition to exploring the function of specific RNA sequences, we took advantage of the availability of a reverse genetics system for EAV to address the role of proteins or protein domains which have been implicated in nidovirus genome replication and/or sg mRNA transcription. Among these, EAV nsp10 (51 kDa) can be considered a primary target. With its conserved MBD and Hel domains, this protein represents the most conserved nidovirus replicase subunit. Furthermore, it is also the first nonstructural protein for which direct experimental evidence for a role in EAV RNA synthesis was obtained. The phenotype of a mutant (EAV030F) carrying a single point mutation in nsp10 (Ser-2429 to Pro; amino acid numbering is based on the full-length ORF1ab protein sequence) strongly suggested a specific role for this protein in sg mRNA synthesis. Although EAV030F replicated its genomic RNA with wild-type (wt) efficiency, it was defective in sg RNA synthesis (62). The latter was reduced by at least 100-fold at the levels of both sg minus strand and sg mRNA synthesis (66).
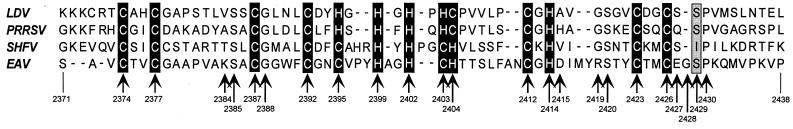
The putative MBD of the arterivirus nsp10 contains 13 conserved Cys and His residues (Fig. 2) (10, 23). For the purpose of this paper, we have chosen to define the most N-terminal and most C-terminal of these residues (Cys-2374 and Cys-2426, respectively) as the borders of the MBD. The Ser-2429→Pro mutation of mutant EAV030F is located in a region which appears to connect the MBD and Hel domains of nsp10. However, because residue Ser-2429 is just downstream of the last conserved Cys residue (Cys-2426), it is clear that we cannot exclude the possibility that Ser-2429 is actually part of the functional MBD or influences its function in a more indirect manner. In this study, we have analyzed the function of the MBD (residues 2374 to 2426, as defined above) and that of the downstream Ser-2429 region by site-directed mutagenesis. A large panel of mutants was characterized at the level of genome replication and sg mRNA transcription, protein synthesis, and virus production. We concluded that the nsp10 MBD region plays a crucial role in viral RNA synthesis. Furthermore, the results obtained with one of our mutants suggested that the MBD genetic locus controls an additional function which is essential for the production of infectious progeny virus.
FIG. 2.
Alignment of four arterivirus nsp10 MBD sequences: LDV-C (21), PRRSV Lelystad (43), SHFV LVR (T. R. Maines, et al. personal communication), and EAV Bucyrus (10). The conserved His and Cys residues and the position of EAV Ser-2429 (and its equivalents) are indicated with black and grey boxes, respectively. Arrows, EAV residues that were targeted by site-directed mutagenesis (Table 1). EAV amino acid numbers are based on the ORF1ab replicase polyprotein sequence.
MATERIALS AND METHODS
Cells, virus, and antisera.
Baby hamster kidney (BHK-21) cells were used for propagation of the EAV Bucyrus strain (17) and RNA transfections. The EAV ORF1a and ORF1b protein-specific antisera have been described previously (46, 56, 64). Indirect double immunofluorescence assays (IFAs) were carried out as described before (61) using an anti-nsp3 rabbit serum and a mouse monoclonal antibody (MAb) recognizing the ORF5-encoded glycoprotein GL (20). Alternatively, the anti-nsp3 serum was used in combination with a mouse MAb (3E2) directed against the ORF7-encoded nucleocapsid protein (N), referred to as anti-N (39). As secondary antibodies, Cy3-coupled donkey anti-rabbit immunoglobulin G (IgG) and fluorescein isothiocyanate-coupled goat anti-mouse IgG conjugates were used (both from Jackson ImmunoResearch Laboratories).
Mutagenesis of the EAV full-length cDNA clone and construction of EAV replicons.
Standard recombinant DNA techniques were carried out as described by Sambrook et al. (50). Site-directed PCR mutagenesis was performed as described by Landt et al. (36). Mutations were introduced into a shuttle vector containing a HindIII-KpnI fragment (nt 6973 to 7526) of pEAV030H, a wt EAV full-length cDNA clone containing an engineered HindIII site at position 6973 (62). Following complete sequence analysis of the cloned PCR fragments, restriction fragments specifying an MBD mutation(s) (Table 1) were introduced into pEAV030H. In full-length construct pEP the sequence encoding the EAV MBD (nt 7340 to 7526; replicase aa 2374 to 2433) was exchanged for the corresponding sequence of the Lelystad strain of porcine arterivirus PRRSV (42) (nt 9336 to 9502; replicase aa 3044 to 3097).
TABLE 1.
EAV nsp10 mutants and their phenotypes
| Construct | Substitution | Codon
|
Result ofa:
|
Description of phenotypeb | |||
|---|---|---|---|---|---|---|---|
| Wild type | Mutant | nsp3 IFA | N protein IFA | Infectivity | |||
| EAV030H | None (wild-type) | + | + | + | Wild type | ||
| C2374H | Cys-2374→His | UGC | CAC | − | − | − | Nonviable |
| C2377H | Cys-2377→His | UGU | CAU | − | − | − | Nonviable |
| K2384A | Lys-2384→Ala | AAG | GCG | − | − | − | Nonviable |
| S2385A | Ser-2385→Ala | UCU | GCU | + | + | + | Wild type |
| S2385T | Ser-2385→Thr | UCU | ACU | + | + | + | Wild type |
| C2387H | Cys-2387→His | UGU | CAU | − | − | − | Nonviable |
| G2388A | Gly-2388→Ala | GGA | GCA | + | + | + | Viable, delayed |
| G2388V | Gly-2388→Val | GGA | GUA | + | + | + | Viable, delayed |
| C2392H | Cys-2392→His | UGU | CAU | − | − | − | Nonviable |
| C2395H | Cys-2395→His | UGU | CAC | + | + | + | Viable, delayed |
| H2399C | His-2399→Cys | CAC | UGC | − | − | − | Nonviable |
| H2402C | His-2402→Cys | CAU | UGU | − | − | − | Nonviable |
| C2403H | Cys-2403→His | UGU | CAU | − | − | − | Nonviable |
| H2404C | His-2404→Cys | CAC | UGU | − | − | − | Nonviable |
| CCH→HHC | Cys-2395→His | UGU | CAC | − | − | − | Nonviable |
| Cys-2403→His | UGU | CAU | |||||
| His-2404→Cys | CAC | UGC | |||||
| C2412H | Cys-2412→His | UGC | CAU | − | − | − | Nonviable |
| H2414C | His-2414→Cys | CAC | UGC | + | + | − | RNA synthesis, but not viable |
| H2414Y | His-2414→Tyr | CAC | UAC | − | − | − | Nonviable |
| D2415A | Asp-2415→Ala | GAC | GCC | − | − | − | Nonviable |
| R2419A | Arg-2419→Ala | CGC | GCG | + | + | + | Viable, delayed |
| S2420A | Ser-2420→Ala | UCC | GCU | + | + | + | Wild type |
| S2420T | Ser-2420→Thr | UCC | ACU | + | + | + | Viable, cold sensitive |
| C2423H | Cys-2423→His | UGC | CAC | − | − | − | Nonviable |
| C2426H | Cys-2426→His | UGU | CAU | − | − | − | Nonviable |
| C2426N | Cys-2426→Asn | UGU | AAU | − | − | − | Nonviable |
| E2427A | Glu-2427→Ala | GAG | GCG | + | + | + | Viable, delayed |
| ΔE2427 | Glu-2427 deletion | GAG | − | − | − | Nonviable | |
| ΔG2428 | Gly-2428 deletion | GGU | − | − | − | Nonviable | |
| EAV030F | Ser-2429→Pro | UCC | CCC | + | − | − | Defect in sg mRNA synthesis |
| S2429A | Ser-2429→Ala | UCC | GCC | + | + | + | Wild type |
| S2429C | Ser-2429→Cys | UCC | UGC | + | + | + | Wild type |
| S2429G | Ser-2429→Gly | UCC | GGC | + | + | + | Wild type |
| S2429H | Ser-2429→His | UCC | CAC | + | + | + | Wild type |
| S2429L | Ser-2429→Leu | UCC | CUC | + | + | + | Wild type |
| S2429T | Ser-2429→Thr | UCC | ACC | + | + | + | Wild type |
| S2429G/P2430G | Ser-2429→Gly | UCC | GGC | + | ± | − | Defect in sg mRNA synthesis |
| Pro-2430→Gly | CCA | GGA | |||||
| S2429P/P2430S | Ser-2429→Pro | UCC | CCC | + | − | − | Defect in sg mRNA synthesis |
| Pro-2430→Ser | CCA | UCA | Delayed genome replication | ||||
+, positive; −, negative; ±, intermediate.
Delayed, replication delayed compared to that of wt virus at the same temperature, as judged by IFA (onset of nonstructural and structural protein synthesis) and virus spread to untransfected cells
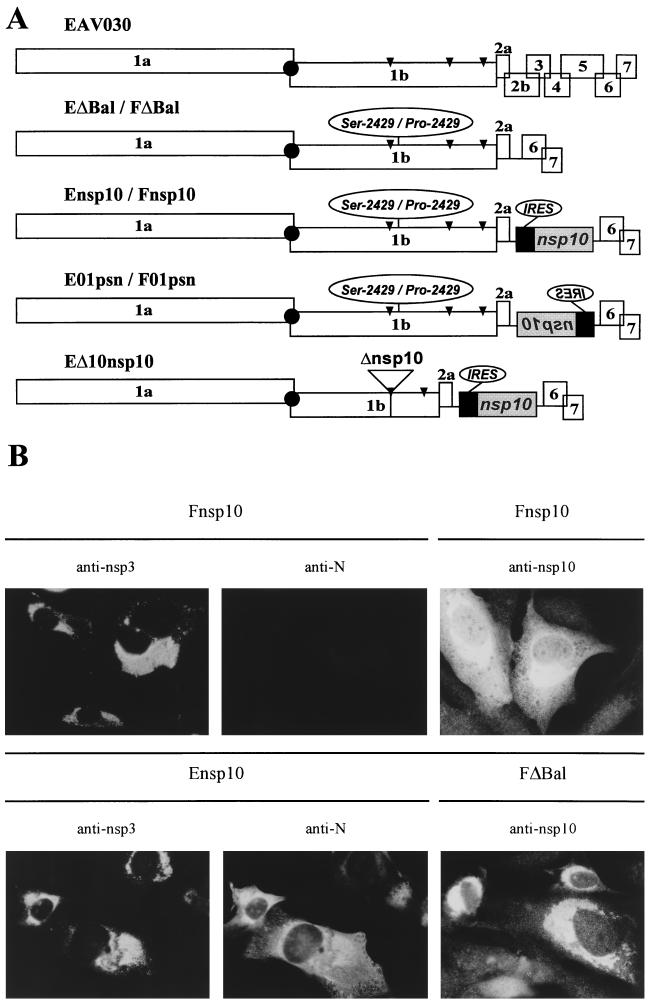
To obtain autonomous EAV replicons, different parts of the 3′ genomic region containing ORFs 2a to 7 were deleted from wt full-length clone pEAV030 (Fig. 3). Replicon EΔBal contained a deletion from nt 10023 to 11638 (between two BalI restriction sites in pEAV030), which did not affect the TRSs for sg mRNA2, mRNA6, and mRNA7 transcription or ORFs 2a, 6, and 7 (Fig. 3A). In construct pEnsp10, a cassette was inserted at the site of the BalI deletion (Fig. 3A); the cassette consisted of the encephalomyocarditis virus (EMCV) internal ribosomal entry site (IRES) (29) followed by the coding sequence for wt EAV nsp10 (nt 7334 to 8734 of the EAV genome; replicase residues 2371 to 2837 [63]). The IRES-nsp10 gene cassette was taken from the previously described nsp10 expression vector pL(2371-2837) (63). In control construct pE01psn, the IRES-nsp10 gene cassette was inserted in the antisense orientation (Fig. 3A). The same set of 3′ deletions and insertions was made in the pEAV030F clone containing the Ser-2429→Pro mutation (generating constructs pFΔBal, pFnsp10, and pF01psn). In addition, we constructed pΔ10Ensp10, a pEnsp10 derivative in which the complete nsp10-coding region was deleted from the replicase gene (Fig. 3A). To this end, the nsp9- and nsp11-coding regions were fused and an artificial Glu-2370/Ser-2838 cleavage site was engineered (nt 7333 was fused to nt 8735).
FIG. 3.
Expression of wt nsp10 from an IRES element inserted in the 3′ end of a EAV030F-derived replicon. (A) Schematic representation and nomenclature of the replicons constructed for this approach. The complete EAV genome and the ΔBal replicons are depicted at the top. Black box, EMCV IRES (29), which is followed by a wt nsp10 coding sequence. Arrowheads, cleavage sites in the ORF 1b protein (63). (B) IFAs of BHK-21 cells transfected with Fnsp10, Ensp10, or FΔBal (24 h after electroporation). Cells were stained for nsp10 or double labeled for nsp3 and the ORF7-encoded N protein.
RNA transcription, RNA transfection, and RNA analysis.
The methods for in vitro transcription of infectious RNA from EAV full-length cDNA clones and for the transfection of BHK-21 cells by electroporation have been described previously (62). For double transfections, equal amounts of the two transcripts were mixed prior to addition of the RNA to the cells. Unless stated otherwise, RNA for a reverse transcription-PCR (RT-PCR) analysis for the MBD-coding region of the nsp10 gene was isolated at 24 h posttransfection. The RT primer was complementary to nt 7534 to 7550, and the PCR was performed using the RT primer and a sense primer corresponding to nt 6336 to 6356. The PCR products were subsequently cloned and used for sequence analysis. RNA synthesis was monitored from 8 to 10 h posttransfection by metabolic labeling (10 μg of dactinomycin and 200 μCi of [3H]uridine/ml), as described previously (11). Cell lysates were prepared as described by Spaan et al. (60), and intracellular RNA was analyzed by gel electrophoresis in 1.5% agarose gels (11).
RESULTS
Analysis of EAV030F revertants.
We have previously described mutant EAV030F, which displays a severe defect in sg mRNA synthesis due to a single point mutation (Ser-2429→Pro) in nsp10 (62, 66). Residue 2429 is located just 3 aa downstream of the last conserved Cys residue (Cys-2426) of the predicted nsp10 MBD (10, 23), which contains a total of 13 Cys and His residues that are conserved in arteriviruses (Fig. 2). To characterize the EAV030F mutant and its phenotype in more detail, we first analyzed reversion of the Ser-2429→Pro mutant. Thus far, the error rate of the arterivirus replicase has not been studied in any detail, but, as for other positive-stranded RNA viruses, it was expected to be sufficiently high to allow the rapid reversion of the single EAV030F point mutation. This was expected to result in the onset of sg mRNA synthesis, and thus in the subsequent generation of structural proteins and infectious progeny virus.
In the course of our studies, we monitored reversion during more than 20 experiments with EAV030F or one of its derivatives (see below). By using IFAs, EAV030F-transfected cells were routinely tested for structural protein synthesis and virus spread at 48 to 96 h postinfection (p.i.) (which equals 4 to 8 cycles of EAV replication). Indeed, reversion was observed in a number of these experiments, although it was always detected at late(r) time points posttransfection (24 h p.i. at the earliest). Once reversion had occurred, complete cytopathic effect developed within the next 24 h due to the spread of revertant virus. In an infectious center assay, transfection of 1 μg of EAV030 RNA yielded approximately 1,200 plaques after 72 h. No plaques could be detected using the same amount of EAV030F transcript, indicating that in this particular experiment reversion occurred with a frequency of less than 10−3.
To study reversion in more detail, medium was harvested at 96 h p.i. from one of the EAV030F transfections in which reversion was detected. The medium was used to infect fresh BHK-21 cells, and intracellular RNA was isolated (at 22 h p.i.) and used for an RT-PCR analysis of the nsp10-coding region of the replicase gene (see Materials and Methods). The PCR fragment was cloned, and the complete sequences of two subclones were analyzed. Surprisingly, these clones did not reveal reversion from the Pro-2429 codon back to the wt Ser codon (CCC to UCC). Instead, the second nucleotide of the triplet had been mutated: one clone carried a His codon (CAC), while the other had a Leu codon (CUC). To confirm that either of these mutations is indeed sufficient for reversion to the wt phenotype, small restriction fragments (HpaI-KpnI; nt 7305 to 7526) containing the two mutations were cloned back to the wt pEAV030 clone. Upon transfection of RNA derived from both mutant full-length clones (S2429H and S2429L), efficient sg mRNA synthesis and virus spread were observed. Thus, replacement of the Pro-2429 residue of mutant EAV030F by either His or Leu yielded a phenotype which could not be distinguished from that of the wt EAV030 virus.
The region immediately downstream of the MBD is important for both genomic and sg RNA synthesis.
Following the unexpected results of our revertant analysis, four other substitutions (Ala, Cys, Gly, and Thr) at replicase position 2429 were tested (Fig. 2B). These novel mutations were also tolerated, and the mutants behaved like wt EAV030. A metabolic ([3H]uridine) labeling of viral RNA synthesis revealed that the levels of sg mRNA produced by the four new mutants, the two revertants described above, and wt EAV030 were comparable (data not shown).
The above results demonstrated that the position of Ser-2429 can be occupied by at least six other residues without significantly compromising sg mRNA transcription or genome replication. In terms of side chain properties, some of these substitutions (e.g., His and Leu) are radically different from the wt Ser residue. Furthermore, despite the conservation of a Ser-2429 equivalent in LDV and PRRSV, the SHFV sequence contains Ile at the corresponding position (Fig. 2; T. R. Maines, H. H. Starling, S. L. Methven, L. Chen, S. N. Kumar, and M. A. Brinton, personal communication). Taken together, these observations suggested that Ser-2429 does not play a specific role in EAV sg mRNA synthesis and that, in principle, its replacement does not result in an sg mRNA transcription defect as displayed by EAV030F. Instead, our data supported the view that the EAV030F phenotype results from the unique structural properties of the Pro residue introduced at position 2429, possibly in conjunction with the presence of an adjacent Pro residue at position 2430 (Fig. 2). To study this region in more detail, two additional mutants were generated: the Ser-2429–Pro-2430 dipeptide was replaced by either a Pro-Ser dipeptide (S2429P/P2430S), thereby inverting the sequence of these two residues in the protein, or by a Gly-Gly dipeptide (S2429G/P2430G). Remarkably, both these mutants displayed phenotypes resembling that of EAV030F. Based on our standard analysis by IFA (Table 1), they were both capable of efficient genome replication, but their sg mRNA synthesis was clearly affected. Since a very low level of structural protein expression could be detected for mutant S2429G/P2430G, this mutant appeared to be less defective in sg RNA synthesis than EAV030F. This was not the case for inversion mutant S2429P/P2430S, which also displayed a delayed onset of genome replication. For both mutants, temperature sensitivity was tested by growing the transfected cells at 34 and 39.5°C, but no phenotypic differences were observed.
The final two mutations in the region immediately downstream of the MBD concerned Glu-2427 and Gly-2428, the two residues which separate the last conserved Cys (Cys-2426) of the MBD and Ser-2429. In the other arterivirus sequences (Fig. 2) only one residue separates these two amino acids. We investigated the effect of the deletion of either Glu-2427 or Gly-2428 from EAV nsp10 (Table 1), thereby generating two mutants with increased sequence similarity compared to the situation in other arteriviruses. However, both these deletions turned out to be lethal since no sign of viral RNA synthesis could be detected (see Discussion).
EAV genome replication is extremely sensitive to a variety of mutations in the nsp10 MBD.
We previously hypothesized (62) that the nsp10 MBD might have (partially) different functions in arterivirus genome replication and sg RNA synthesis and could thereby play a key role in the regulation of the latter process. The functions of the EAV nsp10 MBD were now analyzed in more detail by generating a large panel of mutants. Depending on the substituted amino acid(s), the mutants could be placed in one of three groups.
The first group (16 mutants) was characterized by substitutions of the 13 conserved Cys and His residues, which may be involved in the coordination of Zn2+ (10, 23). Most of these Cys and His replacements were designed to maximize the chance of preserving the (predicted) metal-binding properties (Cys for His, His for Cys; Table 1 and Fig. 2). One mutant (CCH→HHC) carried three substitutions (Cys-2395, Cys-2403, and His-2404 to His, His, and Cys, respectively), since the latter order of Cys and His residues was observed in LDV and PRRSV (Fig. 2). In the second set of mutants, charged residues were replaced by Ala, a method which was previously shown to increase the chance of inducing temperature-sensitive mutations (2, 16, 26, 69). The third set of MBD mutants was obtained by replacing one of three conserved residues not expected to be directly involved in metal binding (Ser-2385, Gly-2388, and Ser-2420; Fig. 2). Finally, we replaced the entire MBD of EAV nsp10 (residues 2374 to 2433) with that of the porcine arterivirus PRRSV (residues 3044 to 3097), resulting in 31 replacements and deletion of six residues not predicted to be involved in metal binding (construct pEP).
All MBD mutants were tested in the context of an infectious cDNA clone, using wt EAV030 and mutant EAV030F as controls. Transfected cells were plated at 34 and 39.5°C and were subsequently assayed for genomic and sg RNA synthesis by means of the previously described double IFA for nonstructural and structural protein synthesis (62). Cells grown at 39.5°C were analyzed at 12, 24, and 36 h posttransfection. Cells kept at 34°C were analyzed at 24, 48, and 72 h posttransfection, since the replication of EAV is delayed by about twofold at this lower temperature. The medium of transfected-cell cultures was harvested at these time points and passaged to test for the production of infectious progeny virus. The results obtained during this extensive analysis are presented in Table 1.
In the first group of mutants, replacement of 11 of the 13 conserved Cys and His residues completely abolished viral RNA synthesis (Fig. 2 and Table 1). Replicase gene expression and virus production could not be detected, and (in multiple experiments) none of these mutants yielded revertants, suggesting that viral RNA synthesis was fully blocked by these mutations. Distinct phenotypes were observed for the mutants carrying the Cys-2395→His or the His-2414→Cys substitutions. The C2395H mutant was viable, but its reproduction was clearly delayed compared to that of wt EAV030. This conclusion was based on the later onset of nonstructural and structural protein synthesis and the slower spread of progeny virus. The H2414C mutant appeared to replicate and generate sg mRNAs with wt efficiency. Surprisingly, however, despite the abundant expression of both replicase and structural proteins, this mutant turned out to be defective in the production of infectious progeny virus (see Discussion), since fresh BHK-21 cells could not be infected with the medium from H2414C transfections. So far, reversion of this mutation, which would require a double nucleotide substitution (Table 1), has not been observed.
None of the charged residue-to-Ala mutants (Table 1) displayed a conditionally lethal phenotype, although certain phenotypic differences were observed. The Arg-2419→Ala and Glu-2427→Ala mutants showed the delayed phenotype described above for C2395H. The Lys-2384→Ala and Asp-2415→Ala mutants did not replicate at all. Likewise, the mutants from the third group exhibited different phenotypes. Replacement of Ser-2385 with Ala or Thr produced mutants with a wt phenotype. The two Gly-2388 mutants (G2388A and G2388V) were viable, but their replication was delayed compared to that of wt EAV030 (i.e., comparable to the C2395H phenotype). One of the two Ser-2420 mutants (S2420T) displayed a cold-sensitive phenotype: at 39.5°C it behaved like wt EAV030, but at 34°C its RNA synthesis and spread were at least three times slower than those of the wt virus. Finally, the mutant (pEP) in which the EAV nsp10 MBD was replaced with that of PRRSV was not replication competent (see Discussion).
In summary, our mutagenesis study of the nsp10 MBD and sequences immediately downstream produced a variety of phenotypes (Table 1). It became clear that the MBD is essential for arterivirus genome replication and that it may also play a (direct or indirect) role in virion biogenesis. Although none of the mutations within the MBD itself induced the intriguing EAV030F phenotype, two novel double mutations in the Ser-2429 region (S2429P and P2430S and S2429G and P2430G) yielded phenotypes which strongly resembled that of EAV030F. This suggests that the structural properties of the residues in this region can affect the functionality of nsp10 in a specific step in sg mRNA synthesis, although it may be too early to unequivocally attribute this function to the MBD (see also Discussion).
Development of replicons for nsp10 trans complementation experiments.
To characterize the properties of nsp10 in more detail, an alternative approach to site-directed mutagenesis was chosen. We attempted to complement the EAV030F defect by expression of the wt nsp10 from the 3′-terminal region of the viral RNA, which normally encodes the structural proteins. To this end, we first developed autonomous EAV replicons containing the full-length replicase gene but lacking a substantial part of the downstream structural protein-coding region. By generating a variety of truncations in this part of the genome, it was established that the region covering ORFs 2b to 5 could be deleted without affecting genome replication or sg mRNA transcription from the remaining TRSs. The properties of the full set of replicons are described elsewhere (R. Molenkamp, H. van Tol, B. C. D. Rozier, Y. van der Meer, and E. J. Snijder, unpublished data). For the studies described below, we used deletion mutant EΔBal (Fig. 3), which lacked functional ORFs 2b to 5 and was therefore unable to generate progeny virus. Replicon EΔBal replicated with wt efficiency and generated sg mRNAs from the remaining TRSs (mRNAs 2, 6, and 7; data not shown). As expected, this replicon did not produce significant levels of sg mRNAs when the Ser-2429→Pro mutation was introduced via a mutation of its replicase gene (replicon FΔBal). Like the original EAV030F mutant, FΔBal was negative for structural protein expression in our standard IFA.
The EAV030F defect cannot be complemented by coexpression of wt nsp10.
To analyze whether expression of wt nsp10 in EAV030F-transfected cells would suffice to trans complement the sg mRNA transcription defect, we constructed an EAV030F-derived replicon which expressed wt nsp10 from an IRES element in its 3′-terminal part (Fig. 3A). The EMCV IRES (29) was inserted at about 275 nt downstream of the replicase gene, followed by sequences encoding the wt nsp10. Internal initiation of translation from this IRES element was expected to result in wt nsp10 expression without the need for sg mRNA synthesis. Complementation of the nsp10 defect should result in the transcription of sg mRNAs 2, 6, and 7 and the expression of a subset of structural proteins. We constructed wt and Ser-2429→Pro variants carrying the IRES-nsp10 gene cassette in the sense (pEnsp10 and pFnsp10, respectively) and antisense (pE01psn and pF01psn, respectively) orientations (Fig. 3A).
Following transfection of RNA transcripts, cells were fixed for IFAs after 12, 24, 36, and 48 h. First, we established that the IRES-nsp10 gene cassette did not interfere with genome replication or sg mRNA transcription. Replicon Ensp10 replicated efficiently and sg mRNAs were generated, as evidenced by the double staining for nsp3 and the mRNA7-encoded N protein (Fig. 3B). Surprisingly, the two RNAs containing the IRES-nsp10 gene cassette in the antisense orientation (E01psn and F01psn) did not show any sign of replication (data not shown), an outcome that resembles results obtained with similar constructs for the flavivirus Kunjin virus (31). Unfortunately, upon transfection of replicon Fnsp10, complementation of the nsp10 Ser-2429→Pro mutation by coexpression of wt nsp10 was not observed. Fnsp10 RNA replicated efficiently, but sg mRNA synthesis was not detected, as evidenced by the nsp3/N double staining shown in Fig. 3B.
It should be noted that our EAV nsp10-specific antiserum does not discriminate between wt and mutant (Ser-2429→Pro) nsp10. Hence, we could not establish that the IRES element of replicon Fnsp10 (or Ensp10) was functional and that wt nsp10 was indeed expressed from this locus. However, recent experiments using similar constructs with non-EAV reporter genes downstream of the IRES have firmly established that efficient internal initiation of translation from this site does indeed occur (M. A. Tijms, L. C. van Dinten, A. E. Gorbalenya, and E. J. Snijder, unpublished data). Furthermore, a comparison of the nsp10 staining in cells transfected with FΔBal (which does not contain the IRES-nsp10 gene cassette) or Fnsp10 strongly suggested increased expression of nsp10 by the latter (Fig. 3B). Instead of the usual membrane-associated nsp10 pattern, Fnsp10-transfected cells showed a clearly enhanced staining that was spread throughout the cytoplasm. The same observation was made when replicons EΔBal and Ensp10 were compared (data not shown). In contrast, the other subunits of the Fnsp10 replicase (e.g., nsp3; Fig. 3B) localized to the perinuclear region, as usual (46, 61). Taken together, our observations suggested that the IRES-nsp10 gene cassette in replicons Ensp10 and Fnsp10 was indeed functional but that wt nsp10 expressed from this site was incapable of complementing the defect caused by the Ser-2429→Pro mutation. This result might be explained by the failure of the wt nsp10 to associate with the rest of the replication complex, as suggested by the apparently cytoplasmic localization of IRES-expressed wt nsp10 (Fig. 3B).
Finally, to test whether deletion of nsp10 from the replicase polyprotein might promote the incorporation of IRES-expressed nsp10 in a functional replication complex, we deleted the nsp10-coding sequence from the replicase gene of Ensp10 (construct pΔ10Ensp10; Fig. 3A). To this end, the sequences encoding nsp9 and nsp11 were fused to generate a Glu-2370/Ser-2838 nsp9/nsp11 cleavage site for the nsp4 serine protease. The functionality of this cleavage site was confirmed by detection of fully cleaved nsp9 (data not shown) upon coexpression of the mutant ORF1b protein and nsp4 in the recombinant vaccinia virus/T7 expression system, as described previously (63). Unfortunately, pΔ10Ensp10-derived RNA was not replication competent, suggesting that it is impossible to establish nsp10 function when this subunit is not produced as an integral part of the EAV replicase polyprotein (see also Discussion).
DISCUSSION
The arterivirus nsp10 MBD plays an essential role in genomic and sg RNA synthesis.
The previously described transcription defect of mutant EAV030F (62) prompted us to study both the Ser-2429 region of nsp10 and the predicted upstream MBD (defined above as the sequence between Cys-2374 and Cys-2426). The latter was previously implicated in viral RNA synthesis (10, 23) and was postulated to coordinate Zn2+ through a number of conserved Cys and His residues. Obviously, the functional domain of which the MBD is part may be larger than the sequence between residues 2374 and 2426. Its logical N-terminal border is the nsp9/nsp10 cleavage site (Glu-2370/Ser-2371), but a C-terminal border is less easy to predict. Sequence comparisons (Fig. 2) suggest that it may be close to Ser-2429, downstream of which the arterivirus sequences are clearly less conserved.
The 36 mutants analyzed in this study displayed a variety of phenotypes (Table 1). Of the 22 mutants that were nonviable, the large majority were completely defective in RNA synthesis. One mutant (H2414C) was apparently defective in virion biogenesis, and two double mutations in the Ser-2429 region (S2429G/P2430G and S2429P/P2430S) severely compromised sg mRNA synthesis. These observations provide genetic evidence for a crucial role of the MBD in EAV RNA synthesis, and possibly also in the generation of progeny virus (see below). Genome replication was abolished when 9 of the 10 absolutely conserved Cys or His residues (Fig. 2) were replaced by His or Cys, respectively. We had hoped that (some of) these replacements would have been tolerated, since they were designed to maximize the chance of preserving Zn2+ binding, and thereby the folding of the MBD. Also a triple mutant (C2395H/C2403H/H2404C) and the mutant with the EAV MBD exchanged for that of PRRSV were nonviable. Since these replacements have been accepted by other arteriviruses in the course of natural evolution (Fig. 2), we believed that they might have been compatible with the folding of the EAV MBD. However, two deletion mutants (ΔE2427 and ΔG2428) with increased sequence similarity to other arteriviruses were also deficient in RNA synthesis. In these mutants, the conserved residues Cys-2426 and Pro-2430 were separated by two instead of three residues, a situation encountered in the three other arteriviruses (Fig. 2). Taken together, these results suggest that the nsp10 MBD and the components of the EAV transcription machinery with which it interacts have coevolved in a species-specific manner.
The EAV030F-like phenotype of two novel mutants in the Ser-2429 region (S2429G/P2430G and S2429P/P2430S) can be interpreted as additional evidence for a specific role of the MBD in sg mRNA synthesis (62). Unfortunately, none of the MBD Cys and His mutants displayed the EAV030F phenotype, a finding which would have strongly supported this hypothesis. However, it should be noted that in our biological assay sg RNA transcription completely depends on genome replication. Thus, the results presented in this study cannot be used to draw any conclusions on the ability of the many replication-negative mutants to generate sg mRNAs.
More-convincing data were obtained for the Ser-2429 region, in particular for residues Ser-2429 and Pro-2430. Of the nine single or double mutations tested at these positions (Table 1) none yielded a replication-deficient phenotype, but three mutants were severely impaired in sg mRNA transcription. Remarkably, each of the mutants from the latter group carries replacements that may strongly affect protein structure. Our results suggest that the presence of Pro at position 2429 or Gly at position 2430 (the latter in combination with additional Gly residues at positions 2428 and 2429) interferes with efficient sg mRNA synthesis. Interestingly, Pro and Gly are known for imposing the most- and least-severe structural constraints, respectively, on a protein backbone. Thus, the presence of these residues might strongly influence the flexibility of the nsp10 structure in this particular region. We therefore speculate that residues 2429 and 2430 are part of a “hinge spacer” region that connects the N-terminal MBD with the rest of nsp10. Apparently, the flexibility of the Ser-2429 region is most crucial for sg mRNA transcription, possibly because it controls specific interactions between the MBD and other (RNA or protein) components of the viral transcription complex during discontinuous RNA transcription (65). Interestingly, a similarly positioned 14-aa region containing a Ser-Pro dipeptide is adjacent to the C-terminal Zn2+ binding Cys of the proven Zn2+ fingers in the GAL4 and LAC9 proteins (49, 70–72).
The arterivirus MBD may adopt a unique multinuclear organization and bind four Zn2+s.
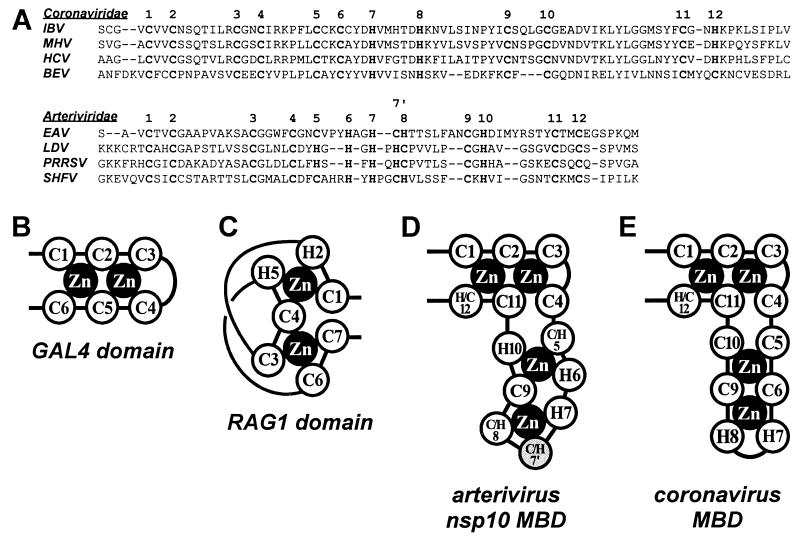
The combination of a putative MBD and a Hel domain in one protein, as in EAV nsp10, is found in a number of viral and cellular proteins (18, 27, 30, 33, 40, 45, 67), including the replicases of related nidoviruses (12, 23, 28, 52). At the primary structure level, the MBDs of all these proteins are dissimilar from the arterivirus one. However, unlike the other proteins, the coronavirus MBDs could be arbitrarily aligned with their arterivirus counterparts on the basis of similar patterns of conserved Cys and His residues in the two sets of proteins (Fig. 4A). In coronaviruses, 12 Cys and His residues are clearly conserved, while in arteriviruses, between 11 and 13 conserved Cys and His residues could be identified by using different alignments (data not shown). Because of our genetic data (see below) and the absolute conservation of the Cys and His residues in the replicases of LDV, PRRSV, and SHFV (data not shown), we have chosen to present an alignment in which 13 Cys and His residues are conserved in the arterivirus MBD (Fig. 4A).
FIG. 4.
Comparison of nidovirus MBD sequences. (A) Alignment of the MBD sequences of the coronaviruses human coronavirus (HCV), mouse hepatitis virus (MHV), and infectious bronchitis virus (IBV); the torovirus Berne virus (BEV); and the arteriviruses EAV, PRRSV, LDV, and SHFV. The conserved Cys and His residues are in boldface, with putative equivalent residues of coronaviruses and arteriviruses being indicated with the same numbers. The residues indicated with 7 and 7′ represent alternatives in the arterivirus alignment. Note that alternative alignments are feasible for the central part of the arterivirus MBD but that these changes do not (necessarily) affect the zinc binding model presented in panel D. (B to E) Schematic representation of the Zn2Cys6 binuclear cluster found in GAL4-like proteins (B) (41), the Zn2Cys5His2 binuclear cluster of the RAG1 dimerization domain (C) (19), the postulated Zn4Cys9His4/Zn4Cys8His5 conformation of the arterivirus nsp10 MBD (D) (see Discussion), and a similar conformation which can be envisioned for the related MBDs of coronaviruses.
The apparent selective pressure to conserve Cys and His residues is compatible with the original suggestion that they coordinate several structural Zn2+s (23). Our genetic data proved that all 13 Cys and His residues are essential for EAV viability, with two residues (Cys-2395 and His-2414) partly tolerating replacement by another residue with Zn2+-coordinating potential (Fig. 2B). The nonuniform susceptibility to His→Cys and Cys→His replacements (see above) is not surprising since similar observations have been reported for a number of other Zn2+ fingers (37, 70). Therefore, we believe that our data indirectly support the Zn2+ binding properties of the MBD, although we acknowledge that the Zn2+ binding capacity of this nidovirus domain remains to be proven experimentally.
When it plays a structural role, Zn2+ is typically coordinated by using tetrahedral geometry (4, 5). Accordingly, the original model of the nidovirus MBD predicted the coordination of three Zn2+s by 12 Cys and His residues, forming three mononuclear finger-like modules (23). If the arterivirus nsp10 MBD actually employs 13 rather than 12 Zn2+-coordinating residues, the original tetrahedron-based mononuclear model must be revised. The MBD might use a more complex mode of Zn2+ binding, e.g., a variant which is known as a binuclear cluster, in which a Cys residue coordinates two closely spaced Zn2+s. Consequently, these clusters bind the same number of Zn2+s as the mononuclear ones but require fewer Cys and His residues. To date, two types of binuclear clusters have been described: the Zn2Cys6 variant found in GAL4-like proteins (Fig. 4B) [41] and the modified mononuclear RING site, known as the Zn2Cys5His2 cluster, of the RAG1 dimerization domain (Fig. 4C) [3]. In these clusters two Zn2+s are bound by six and seven Cys and His residues, respectively. When combined into one protein, these two clusters form a unique structure in which four Zn2+s could be coordinated by 13 Cys and His residues, the exact number found in the arterivirus nsp10 MBD (Fig. 4D). The RAG1 protein, which contains two extra mononuclear Zn2+-binding sites in addition to the binuclear cluster, shows that a complex combination of Zn2+-binding structural components, including a binuclear cluster, is not unprecedented (3). We therefore speculate that the arterivirus MBD has the formula Zn4Cys9His4 or Zn4Cys8His5 and binds four rather than three Zn2+s as originally suggested. A similar Zn4 organization could be maintained in the coronavirus MBD by using 12 conserved Cys and His residues (Fig. 4E) and replacing 2 His residues (H6 and H7 in the model in Fig. 4D) with 1 Cys residue (C6 in Fig. 4E). Interestingly, the four N-terminal and two C-terminal residues, which may form the conserved GAL4-like cluster, are in the most conserved part of the nidovirus alignment (Fig. 4A) and their replacement always abolished EAV RNA synthesis (Table 1). In contrast, mutagenesis of the Cys and His residues in the less conserved central region of the MBD was partly tolerated in two instances (C5 and H10 in Fig. 4D).
Finally, the variety of phenotypes observed for the mutants with MBDs lacking Cys or His mutations indicated that additional residues are important for MBD function. This phenomenon has been observed in many other mutagenesis studies in which residues other than Cys or His were probed (1, 6, 8, 32, 38, 70, 71, 73). These residues might be present on the surface of the nsp10 structure and might interact with other proteins or nucleic acid sequences, a function that could be impaired or abolished upon their replacement.
A link between nsp10 and virion biogenesis?
To our surprise, one of the MBD mutants (H2414C) was competent in genome replication and sg RNA synthesis but defective in the production of infectious progeny virus. Spread of this mutant in transfected cell cultures was not observed, and fresh cells could not be infected by passaging the culture supernatant. We recently described a similar phenotype for another EAV nsp10 mutant (Q2837P), in which the processing of the nsp10/nsp11 cleavage site was abolished (63). For the latter mutant, we showed that this phenotype was probably not due to changes at the level of RNA structure, which may have been induced by the introduction of mutations into ORF1b (63). Furthermore, the absence of the MBD-coding region from a recently isolated natural defective interfering RNA did not interfere with the packaging of this RNA into EAV virions (44).
Remarkably, unlike the His-2414→Cys mutation, the introduction of Tyr at position 2414 completely abolished viral RNA synthesis. The phenotypic differences between the H2414C and H2414Y mutants can be reconciled when the MBD (or the entire nsp10) is assumed to be multifunctional and involved in both RNA synthesis and virus production. According to this model, Cys but not Tyr, which lacks the Zn2+-coordinating properties of Cys, might be able to replace His-2414 without affecting its role in one of these processes, namely, RNA synthesis. At the same time, this His-2414→Cys substitution might still induce subtle changes in the zinc binding or other properties of the MBD, since His and Cys are not completely equivalent. In this manner, an unknown function of nsp10 in virion biogenesis (e.g., nucleocapsid formation, virus assembly, or virus maturation) could be affected.
Zn2+ finger domains of retrovirus nucleocapsid proteins have also been reported to have multiple roles. There, His and Cys, unlike other residues, could be interchanged at some positions of a Zn2+ finger without impairing RNA binding or RNA encapsidation. However, virus infectivity was affected in these mutants (8, 24, 25). Although a more detailed analysis of the EAV H2414C mutant is clearly required, the available data are compatible with the involvement of an arterivirus nonstructural protein (cleaved nsp10 or an nsp10-containing processing intermediate) in virion biogenesis, as previously proposed by our group (63).
nsp10 function requires replicase polyprotein expression.
As outlined in Results, our attempts to complement the Ser-2429→Pro mutation by coexpression of wt nsp10 were not successful (Fig. 3). Although we achieved expression of wt nsp10 from an IRES element in the 3′-terminal part of the viral genome, efficient sg RNA transcription was not restored. Possibly, the exchange of mutant nsp10 in the replication complex for wt nsp10 was hampered by the fact that the mutant protein remains fully functional in genome replication. Consequently, the molecule is probably tightly associated with other components of the RNA-dependent RNA polymerase complex by interactions that may arise immediately after or even prior to proteolytic cleavage of the replicase polyprotein. Alternatively, we cannot exclude the possibility that sg RNA transcription requires an nsp10-containing processing intermediate. In this manner, the intrinsic properties of nsp10 may have prevented the successful use of this complementation assay to study the functions of this protein in more detail.
Recently, we obtained evidence that sg mRNA transcription from an EAV030F RNA template is indeed feasible. Upon cotransfection of autonomous replicons expressing either a wt or a Ser-2429→Pro mutant replicase, the transcription of a specific sg mRNA from the EAV030F-derived replicon could be demonstrated (data not shown). It is possible that, in contrast to the IRES-directed complementation attempt, the double-transfection approach resulted in true trans complementation at the level of nsp10 or an nsp10-containing processing intermediate. However, we consider it more likely that the full-length wt replicase (or replication complex) was able to use the EAV030F-derived replicon as the template for genome replication and/or sg mRNA synthesis.
ACKNOWLEDGMENTS
We are indebted to Janneke Meulenberg for PRRSV cDNA clones and to Udeni Balasuriya, James MacLachlan, and Amy Glaser for supplying EAV-specific MAbs. We are grateful to Margo Brinton for sharing the SHFV replicase sequence with us prior to publication. We thank Willy Spaan for helpful comments and suggestions and Yvonne van der Meer for all photographic material.
A. E. Gorbalenya was funded in part by the National Cancer Institute, National Institutes of Health, under contract no. NO1-CO-56000.
REFERENCES
- 1.Barlow P N, Luisi B, Milner A, Elliot M, Everett R. Structure of the C3HC4 domain by 1H-nuclear magnetic resonance spectroscopy: a new structural class of zinc-finger. J Mol Biol. 1994;237:201–211. doi: 10.1006/jmbi.1994.1222. [DOI] [PubMed] [Google Scholar]
- 2.Bass S H, Mulkerrin M G, Wells J A. A systematic mutational analysis of hormone-binding determinants in the human growth hormone receptor. Proc Natl Acad Sci USA. 1991;88:4498–4502. doi: 10.1073/pnas.88.10.4498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bellon S F, Rodgers K K, Schatz D G, Coleman J E, Steitz T A. Crystal structure of the RAG1 dimerization domain reveals multiple zinc-binding motifs including a novel zinc binuclear cluster. Nat Struct Biol. 1997;4:586–591. doi: 10.1038/nsb0797-586. [DOI] [PubMed] [Google Scholar]
- 4.Berg J M, Godwin H A. Lessons from zinc-binding peptides. Annu Rev Biophys Biomol Struct. 1997;26:357–371. doi: 10.1146/annurev.biophys.26.1.357. [DOI] [PubMed] [Google Scholar]
- 5.Berg J M, Shi Y. The galvanization of biology: a growing appreciation for the roles of zinc. Science. 1996;271:1081–1085. doi: 10.1126/science.271.5252.1081. [DOI] [PubMed] [Google Scholar]
- 6.Borden K L B, Boddy M N, Lally J, O'Reilly N J, Martin S, Howe K, Solomon E, Freemont P S. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cavanagh D. Nidovirales: a new order comprising Coronaviridae and Arteriviridae. Arch Virol. 1997;142:629–633. [PubMed] [Google Scholar]
- 8.Dannull J, Surovoy A, Jung G, Moelling K. Specific binding of HIV-1 nucleocapsid protein to PSI RNA in vitro requires N-terminal zinc finger and flanking basic amino acid residues. EMBO J. 1994;13:1525–1533. doi: 10.1002/j.1460-2075.1994.tb06414.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.den Boon J A, Kleijnen M F, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic mRNA synthesis: analysis of leader-body junctions and replicative-form RNAs. J Virol. 1996;70:4291–4298. doi: 10.1128/jvi.70.7.4291-4298.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.den Boon J A, Snijder E J, Chirnside E D, de Vries A A F, Horzinek M C, Spaan W J M. Equine arteritis virus is not a togavirus but belongs to the coronaviruslike superfamily. J Virol. 1991;65:2910–2920. doi: 10.1128/jvi.65.6.2910-2920.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.den Boon J A, Spaan W J M, Snijder E J. Equine arteritis virus subgenomic RNA transcription: UV inactivation and translation inhibition studies. Virology. 1995;213:364–372. doi: 10.1006/viro.1995.0009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Denison M R, Spaan W J M, van der Meer Y, Gibson C A, Sims A C, Prentice E, Lu X T. The putative helicase of the coronavirus mouse hepatitis virus is processed from the replicase gene polyprotein and localizes in complexes that are active in viral RNA synthesis. J Virol. 1999;73:6862–6871. doi: 10.1128/jvi.73.8.6862-6871.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.de Vries A A F, Chirnside E D, Bredenbeek P J, Gravestein L A, Horzinek M C, Spaan W J M. All subgenomic mRNAs of equine arteritis virus contain a common leader sequence. Nucleic Acids Res. 1990;18:3241–3247. doi: 10.1093/nar/18.11.3241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.de Vries A A F, Chirnside E D, Horzinek M C, Rottier P J M. Structural proteins of equine arteritis virus. J Virol. 1992;66:6294–6303. doi: 10.1128/jvi.66.11.6294-6303.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Vries A A F, Horzinek M C, Rottier P J M, de Groot R J. The genome organization of the Nidovirales: similarities and differences between arteri-, toro-, and coronaviruses. Semin Virol. 1997;8:33–47. doi: 10.1006/smvy.1997.0104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Diamond S E, Kirkegaard K. Clustered charged-to-alanine mutagenesis of poliovirus RNA-dependent RNA polymerase yields multiple temperature-sensitive mutants defective in RNA synthesis. J Virol. 1994;68:863–876. doi: 10.1128/jvi.68.2.863-876.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Doll E R, Bryans J T, McCollum W H, Crowe M E W. Isolation of a filterable agent causing arteritis of horses and abortion by mares. Its differentiation from the equine abortion (influenza) virus. Cornell Vet. 1957;47:3–41. [PubMed] [Google Scholar]
- 18.Dracheva S, Koonin E V, Crute J J. Identification of the primase active site of the herpes simplex virus type 1 helicase-primase. J Biol Chem. 1995;270:14148–14153. doi: 10.1074/jbc.270.23.14148. [DOI] [PubMed] [Google Scholar]
- 19.Freemont P S. The RING finger: a novel protein sequence motif related to the zinc finger. Ann N Y Acad Sci. 1993;684:174–192. doi: 10.1111/j.1749-6632.1993.tb32280.x. [DOI] [PubMed] [Google Scholar]
- 20.Glaser A L, de Vries A A F, Dubovi E J. Comparison of equine arteritis virus isolates using neutralizing monoclonal antibodies and identification of sequence changes in GL associated with neutralization resistance. J Gen Virol. 1995;76:2223–2233. doi: 10.1099/0022-1317-76-9-2223. [DOI] [PubMed] [Google Scholar]
- 21.Godeny E K, Chen L, Kumar S N, Methven S L, Koonin E V, Brinton M A. Complete genomic sequence and phylogenetic analysis of the lactate dehydrogenase-elevating virus (LDV) Virology. 1993;194:585–596. doi: 10.1006/viro.1993.1298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gorbalenya A E, Koonin E V. Comparative analysis of the amino acid sequences of the key enzymes of the replication and expression of positive-strand RNA viruses. Validity of the approach and functional and evolutionary implications. Sov Sci Rev Sect D. 1993;11:1–84. [Google Scholar]
- 23.Gorbalenya A E, Koonin E V, Donchenko A P, Blinov V M. Coronavirus genome: prediction of putative functional domains in the non-structural polyprotein by comparative amino acid sequence analysis. Nucleic Acids Res. 1989;17:4847–4861. doi: 10.1093/nar/17.12.4847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorelick R J, Chabot D J, Ott D E, Gagliardi T D, Rein A, Henderson L E, Arthur L O. Genetic analysis of the zinc finger in the Moloney murine leukemia virus nucleocapsid domain: replacement of zinc-coordinating residues with other zinc-coordinating residues yields noninfectious particles containing genomic RNA. J Virol. 1996;70:2593–2597. doi: 10.1128/jvi.70.4.2593-2597.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gorelick R J, Fu W, Gagliardi T D, Bosche W J, Rein A, Henderson L E, Arthur L O. Characterization of the block in replication of nucleocapsid protein zinc finger mutants from Moloney murine leukemia virus. J Virol. 1999;73:8185–8195. doi: 10.1128/jvi.73.10.8185-8195.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hassett D E, Condit R C. Targeted construction of temperature-sensitive mutations in vaccinia virus by replacing clustered charged residues with alanine. Proc Natl Acad Sci USA. 1994;91:4554–4558. doi: 10.1073/pnas.91.10.4554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hayward-Lester A, Hewetson A, Beale E G, Oefner P J, Doris P A, Chilton B S. Cloning, characterization, and steroid-dependent posttranscriptional processing of RUSH-1 alpha and beta, two uteroglobin promoter-binding proteins. Mol Endocrinol. 1996;10:1335–1349. doi: 10.1210/mend.10.11.8923460. [DOI] [PubMed] [Google Scholar]
- 28.Heusipp G, Harms U, Siddell S G, Ziebuhr J. Identification of an ATPase activity associated with a 71-kilodalton polypeptide encoded in gene 1 of the human coronavirus 229E. J Virol. 1997;71:5631–5634. doi: 10.1128/jvi.71.7.5631-5634.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Johnson R E, Henderson S T, Petes T D, Prakash S, Bankmann M, Prakash L. Saccharomyces cerevisiae RAD5-encoded DNA repair protein contains DNA helicase and zinc-binding sequence motifs and affects the stability of simple repetitive sequences in the genome. Mol Cell Biol. 1992;12:3807–3818. doi: 10.1128/mcb.12.9.3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Khromykh A A, Westaway E G. Subgenomic replicons of the flavivirus Kunjin: construction and applications. J Virol. 1997;71:1497–1505. doi: 10.1128/jvi.71.2.1497-1505.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kuger P, Godecke A, Breunig K D. A mutation in the Zn-finger of the GAL4 homolog LAC9 results in glucose repression of its target genes. Nucleic Acids Res. 1990;18:745–751. doi: 10.1093/nar/18.4.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kusakabe T, Richardson C C. The role of the zinc motif in sequence recognition by DNA primases. J Biol Chem. 1996;271:19563–19570. doi: 10.1074/jbc.271.32.19563. [DOI] [PubMed] [Google Scholar]
- 34.Lai M M C, Baric R S, Brayton P R, Stohlman S A. Characterization of leader RNA sequences on the virion and mRNAs of mouse hepatitis virus, a cytoplasmic RNA virus. Proc Natl Acad Sci USA. 1984;81:3626–3630. doi: 10.1073/pnas.81.12.3626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lai M M C, Cavanagh D. The molecular biology of coronaviruses. Adv Virus Res. 1997;48:1–100. doi: 10.1016/S0065-3527(08)60286-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Landt O, Grunert H-P, Hahn U. A general method for rapid site-directed mutagenesis using the polymerase chain reaction. Gene. 1990;96:125–128. doi: 10.1016/0378-1119(90)90351-q. [DOI] [PubMed] [Google Scholar]
- 37.Lium E K, Silverstein S. Mutational analysis of the herpes simplex virus type 1 ICP0 C3HC4 zinc ring finger reveals a requirement for ICP0 in the expression of the essential α27 gene. J Virol. 1997;71:8602–8614. doi: 10.1128/jvi.71.11.8602-8614.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Loeber G, Stenger J E, Ray S, Parsons R E, Anderson M E, Tegtmeyer P. The zinc finger region of simian virus 40 large T antigen is needed for hexamer assembly and origin melting. J Virol. 1991;65:3167–3174. doi: 10.1128/jvi.65.6.3167-3174.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.MacLachlan N J, Balasuriya U B, Hedges J F, Schweidler T M, McCollum W H, Timoney P J, Hullinger P J, Patton J F. Serologic response of horses to the structural proteins of equine arteritis virus. J Vet Diagn Investig. 1998;10:229–236. doi: 10.1177/104063879801000302. [DOI] [PubMed] [Google Scholar]
- 40.Mannhaupt G, Stucka R, Ehnle S, Vetter I, Feldmann H. Molecular analysis of yeast chromosome II between CMD1 and LYS2: the excision repair gene RAD16 located in this region belongs to a novel group of double-finger proteins. Yeast. 1992;8:397–408. doi: 10.1002/yea.320080507. [DOI] [PubMed] [Google Scholar]
- 41.Marmorstein R, Carey M, Ptashne M, Harrison S C. DNA recognition by GAL4: structure of a protein-DNA complex. Nature. 1992;356:379–380. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- 42.Meulenberg J J M, de Meijer E J, Moormann R J M. Subgenomic RNAs of Lelystad virus contain a conserved leader-body junction sequence. J Gen Virol. 1993;74:1697–1701. doi: 10.1099/0022-1317-74-8-1697. [DOI] [PubMed] [Google Scholar]
- 43.Meulenberg J J M, Hulst M M, de Meijer E J, Moonen P L J M, den Besten A, de Kluyver E P, Wensvoort G, Moormann R J M. Lelystad virus, the causative agent of porcine epidemic abortion and respiratory syndrome (PEARS), is related to LDV and EAV. Virology. 1993;192:62–72. doi: 10.1006/viro.1993.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Molenkamp R, Rozier B C D, Greve S, Spaan W J M, Snijder E J. Isolation and characterization of an arterivirus defective interfering RNA genome. J Virol. 2000;74:3156–3165. doi: 10.1128/jvi.74.7.3156-3165.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ouzounis C A, Blencowe B J. Bacterial DNA replication initiation factor priA is related to proteins belonging to the ‘DEAD-box’ family. Nucleic Acids Res. 1991;19:6953. doi: 10.1093/nar/19.24.6953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedersen K W, van der Meer Y, Roos N, Snijder E J. Open reading frame 1a-encoded subunits of the arterivirus replicase induce endoplasmic reticulum-derived double-membrane vesicles which carry the viral replication complex. J Virol. 1999;73:2016–2026. doi: 10.1128/jvi.73.3.2016-2026.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Plagemann P G W. Lactate dehydrogenase-elevating virus and related viruses. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. 3rd ed. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. pp. 1105–1120. [Google Scholar]
- 48.Poch O, Sauvaget I, Delarue M, Tordo N. Identification of four conserved motifs among the RNA dependent polymerase encoding elements. EMBO J. 1989;8:3867–3874. doi: 10.1002/j.1460-2075.1989.tb08565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Salmeron J M, Jr, Johnston S A. Analysis of the Kluyveromyces lactis positive regulatory gene LAC9 reveals functional homology to, but sequence divergence from, the Saccharomyces cerevisiae GAL4 gene. Nucleic Acids Res. 1986;14:7767–7781. doi: 10.1093/nar/14.19.7767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 51.Sawicki S G, Sawicki D L. Coronaviruses use discontinuous extension for synthesis of subgenome-length negative strands. Adv Exp Biol Med. 1995;380:499–506. doi: 10.1007/978-1-4615-1899-0_79. [DOI] [PubMed] [Google Scholar]
- 52.Snijder E J, den Boon J A, Bredenbeek P J, Horzinek M C, Rijnbrand R, Spaan W J M. The carboxyl-terminal part of the putative Berne virus polymerase is expressed by ribosomal frameshifting and contains sequence motifs which indicate that toro- and coronaviruses are evolutionarily related. Nucleic Acids Res. 1990;18:4535–4542. doi: 10.1093/nar/18.15.4535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Snijder E J, Meulenberg J J M. The molecular biology of arteriviruses. J Gen Virol. 1998;79:961–979. doi: 10.1099/0022-1317-79-5-961. [DOI] [PubMed] [Google Scholar]
- 54.Snijder E J, van Tol H, Pedersen K W, Raamsman M J B, de Vries A A F. Identification of a novel structural protein of arteriviruses. J Virol. 1999;73:6335–6345. doi: 10.1128/jvi.73.8.6335-6345.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Snijder E J, Wassenaar A L M, Spaan W J M. The 5′ end of the equine arteritis virus replicase gene encodes a papainlike cysteine protease. J Virol. 1992;66:7040–7048. doi: 10.1128/jvi.66.12.7040-7048.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Snijder E J, Wassenaar A L M, Spaan W J M. Proteolytic processing of the replicase ORF1a protein of equine arteritis virus. J Virol. 1994;68:5755–5764. doi: 10.1128/jvi.68.9.5755-5764.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Snijder E J, Wassenaar A L M, Spaan W J M, Gorbalenya A E. The arterivirus nsp2 protease: an unusual cysteine protease with primary structure similarities to both papain-like and chymotrypsin-like proteases. J Biol Chem. 1995;270:16671–16676. doi: 10.1074/jbc.270.28.16671. [DOI] [PubMed] [Google Scholar]
- 58.Snijder E J, Wassenaar A L M, van Dinten L C, Spaan W J M, Gorbalenya A E. The arterivirus nsp4 protease is the prototype of a novel group of chymotrypsin-like enzymes, the 3C-like serine proteases. J Biol Chem. 1996;271:4864–4871. doi: 10.1074/jbc.271.9.4864. [DOI] [PubMed] [Google Scholar]
- 59.Spaan W J M, Delius H, Skinner M, Armstrong J, Rottier P J M, Smeekens S, van der Zeijst B A M, Siddell S G. Coronavirus mRNA synthesis involves fusion of non-contiguous sequences. EMBO J. 1983;2:1839–1844. doi: 10.1002/j.1460-2075.1983.tb01667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Spaan W J M, Rottier P J M, Horzinek M C, van der Zeijst B A M. Isolation and identification of virus-specific mRNAs in cells infected with mouse hepatitis virus (MHV-A59) Virology. 1981;108:424–434. doi: 10.1016/0042-6822(81)90449-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van der Meer Y, van Tol H, Krijnse Locker J, Snijder E J. ORF1a-encoded replicase subunits are involved in the membrane association of the arterivirus replication complex. J Virol. 1998;72:6689–6698. doi: 10.1128/jvi.72.8.6689-6698.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.van Dinten L C, den Boon J A, Wassenaar A L M, Spaan W J M, Snijder E J. An infectious arterivirus cDNA clone: identification of a replicase point mutation which abolishes discontinuous mRNA transcription. Proc Natl Acad Sci USA. 1997;94:991–996. doi: 10.1073/pnas.94.3.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.van Dinten L C, Rensen S, Gorbalenya A E, Snijder E J. Proteolytic processing of the open reading frame 1b-encoded part of arterivirus replicase is mediated by nsp4 serine protease and is essential for virus replication. J Virol. 1999;73:2027–2037. doi: 10.1128/jvi.73.3.2027-2037.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.van Dinten L C, Wassenaar A L M, Gorbalenya A E, Spaan W J M, Snijder E J. Processing of the equine arteritis virus replicase ORF1b protein: identification of cleavage products containing the putative viral polymerase and helicase domains. J Virol. 1996;70:6625–6633. doi: 10.1128/jvi.70.10.6625-6633.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.van Marle G, Dobbe J C, Gultyaev A P, Luytjes W, Spaan W J M, Snijder E J. Arterivirus discontinuous mRNA transcription is guided by base-pairing between sense and antisense transcription-regulating sequences. Proc Natl Acad Sci USA. 1999;96:12056–12061. doi: 10.1073/pnas.96.21.12056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.van Marle G, van Dinten L C, Luytjes W, Spaan W J M, Snijder E J. Characterization of an equine arteritis virus replicase mutant defective in subgenomic mRNA synthesis. J Virol. 1999;73:5274–5281. doi: 10.1128/jvi.73.7.5274-5281.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Visse R, de Ruijter M, Ubbink M, Brandsma J A, van de Putte P. The first zinc-binding domain of UvrA is not essential for UvrABC-mediated DNA excision repair. Mutat Res. 1993;294:263–274. doi: 10.1016/0921-8777(93)90009-6. [DOI] [PubMed] [Google Scholar]
- 68.Wassenaar A L M, Spaan W J M, Gorbalenya A E, Snijder E J. Alternative proteolytic processing of the arterivirus replicase ORF1a polyprotein: evidence that NSP2 acts as a cofactor for the NSP4 serine protease. J Virol. 1997;71:9313–9322. doi: 10.1128/jvi.71.12.9313-9322.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Wertman K F, Drubin D G, Botstein D. Systematic mutational analysis of the yeast ACT1 gene. Genetics. 1992;132:337–350. doi: 10.1093/genetics/132.2.337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Witte M M, Dickson R C. Cysteine residues in the zinc finger and amino acids adjacent to the finger are necessary for DNA binding by the LAC9 regulatory protein of Kluyveromyces lactis. Mol Cell Biol. 1988;8:3726–3733. doi: 10.1128/mcb.8.9.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Witte M M, Dickson R C. The C6 zinc finger and adjacent amino acids determine DNA-binding specificity and affinity in the yeast activator proteins LAC9 and PPR1. Mol Cell Biol. 1990;10:5128–5137. doi: 10.1128/mcb.10.10.5128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Wray L V, Jr, Witte M M, Dickson R C, Riley M I. Characterization of a positive regulatory gene, LAC9, that controls induction of the lactose-galactose regulon of Kluyveromyces lactis: structural and functional relationship to GAL4 of Saccharomyces cerevisiae. Mol Cell Biol. 1987;7:1111–1121. doi: 10.1128/mcb.7.3.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Zang W Q, Veldhoen N, Romaniuk P J. Effects of zinc finger mutations on the nucleic acid binding activities of Xenopus transcription factor IIIA. Biochemistry. 1995;34:15545–15552. doi: 10.1021/bi00047a021. [DOI] [PubMed] [Google Scholar]