Abstract
It has been suggested that nonstructural protein 5A (NS5A) of hepatitis C virus (HCV) plays a role in the incapacitation of interferon by inactivation of RNA-dependent protein kinase PKR. In order to further investigate the role of NS5A, we tried to identify cellular proteins interacting with NS5A by using the yeast two-hybrid system. The karyopherin β3 gene was isolated from a human liver cell library as a protein interacting with NS5A. The protein-protein interaction between NS5A and karyopherin β3 was confirmed by in vitro binding assay and an in vivo coimmunoprecipitation method. The effect of NS5A on the karyopherin β3 activity was investigated using a yeast cell line containing mutations in both PSE1 and KAP123, genes that are homologous to the human karyopherin β3 gene. Human karyopherin β3 complemented the loss of the PSE1 and KAP123 functions, supporting growth of the double mutant cells. However, expression of NS5A hampered the growth of the double mutant cells supplemented with human karyopherin β3. On the other hand, expression of NS5A by itself had no effect on the growth of the double mutant expressing wild-type yeast PSE1. This indicates that NS5A may inhibit karyopherin β3 function via protein-protein interaction. The role of NS5A in HCV replication is discussed.
Hepatitis C virus (HCV) is the major etiologic agent of non-A, non-B hepatitis (1, 8, 38). Chronic infection with HCV results in liver cirrhosis and hepatocellular carcinoma (7, 45). HCV belongs to the family Flaviviridae, having a positive-sense RNA genome (32, 42, 47). The RNA encodes a polyprotein (∼3,010 amino acids) with the following gene order: 5′-C-E1-E2-p7-NS2-NS3-NS4A-NS4B-NS5A-NS5B-3′. During and/or after translation, the polyprotein is processed into functional proteins by host- and virus-encoded proteases. Core (C) and envelope (E1 and E2) proteins are believed to compose the structural elements of the virion particle. Nonstructural protein 2 (NS2), NS3, and NS4A are involved in the proteolytic processing of the HCV polyprotein (4, 5, 15, 18, 25, 26, 27, 28, 30, 40, 50, 52). RNA-dependent RNA polymerase and RNA helicase activities are assigned to NS5B and the C-terminal two-thirds of NS3, respectively (6, 36). No function has yet been assigned to NS4B.
NS5A exists in two different forms (p56 and p58) in cells. The proteins differ in their phosphorylation status (2, 34, 51). NS4A or NS3-4A-4B augments hyperphosphorylation of NS5A (43, 51). The sequence around the middle part of NS5A (amino acids 2209 to 2248) is termed the interferon sensitivity-determining region, since it correlates with interferon sensitivity of the HCV genotype 1b (16, 17, 39). The sequence in the interferon sensitivity-determining region was shown to play a key role in the inhibition of the protein kinase PKR, a mediator of interferon-induced resistance, through protein-protein interaction (20, 21). NS5A was also shown to interact with a SNARE-like protein (53). The C-terminal region of NS5A contains a potential transcriptional activation domain, but the role of its activity in viral replication is not known (10, 35, 49). Recently, NS5A was shown to perturb Grb2-mediated signaling pathways by selectively targeting the growth factor receptor-bound protein 2 (Grb2) adapter protein (48), and the introduction of NS5A into murine fibroblasts (NIH 3T3) promoted anchorage-independent growth and tumor formation in nude mice (22). This suggests that NS5A may also have a role in cell growth regulation. However, no biochemical function has yet been assigned to the N terminus of HCV NS5A.
To investigate the various roles of HCV NS5A in viral replication, we searched for cellular proteins interacting with the NS5A protein by yeast two-hybrid screening of a human hepatocyte cDNA library. We identified karyopherin β3, a member of the karyopherin β family also known as RanBP5, as the cellular counterpart of HCV NS5A. Karyopherins are a group of proteins mediating transport of proteins and possibly RNAs (reference 44 and references therein). For instance, karyopherin β1 (importin β), in association with karyopherin α (importin α), facilitates nuclear import of proteins containing classical nuclear localization signals (24). Karyopherin β3, a 124-kDa protein, exhibits a significant level of similarity to karyopherin β1 (44.4% similarity and 17.6% identity). The similarity of karyopherin β3 to other members of the karyopherin β family, its localization in the cytoplasm and nuclear rim, and its binding to repeat-containing nucleoporins and to Ran-GTP strongly suggest that karyopherin β3 may play a role in nucleoplasmic transport (13, 55). Karyopherin β3 is highly homologous to Saccharomyces cerevisiae protein PSE1 (KAP121) (65.2% similarity and 28.3% identity) and to KAP123 (58.9% similarity and 23% identity). Functional relationships among these proteins are yet to be elucidated. Several potential activities of karyopherin β3 and PSE1 have been proposed. Karyopherin β3 facilitated nuclear import of ribosomal proteins in an in vitro transportation assay system (33). Overexpression of yeast PSE1 resulted in an increase in protein secretion and stimulated mitochondrial import of hydrophobic proteins in yeast cells (9, 12). In addition, the conditional loss of PSE1 in a strain lacking KAP123 resulted in a specific blockage of mRNA export from the nucleus (46). The molecular bases of these phenomena remain obscure.
Here we show protein-protein interaction between NS5A and karyopherin β3 by an in vitro binding assay and an in vivo coimmunoprecipitation method. The effect of NS5A on the karyopherin β3 activity was investigated using a yeast cell line with mutations in both PSE1 (pse1-1) and KAP123 (Δkap123), genes that are homologous to the human karyopherin β3 gene. Human karyopherin β3 complemented the loss of both PSE1 and KAP123 functions and supported growth of the double mutant cells at a nonpermissive temperature, but expression of NS5A hampered the growth of the double mutant cells supplemented with human karyopherin β3. On the other hand, expression of NS5A by itself had no effect on the growth of the double mutant expressing introduced wild-type yeast PSE1. This indicates that NS5A may inhibit karyopherin β3 function via protein-protein interaction. Therefore, it is likely that HCV NS5A modulates cellular activities by inhibiting the activity of karyopherin β3.
MATERIALS AND METHODS
Plasmid construction.
For the yeast two-hybrid system, the plasmids pAS2 and pACT2 (Clontech, Inc.) were used as sources of the GAL4 DNA-binding domain (BD) and GAL4 transcriptional activation domain (AD), respectively. The plasmids pYBD-5A(1973-2419), pYBD-5A(1973-2302), pYBD-5A(1973-2204), pYBD-5A(1973-2119), and pYBD-5A(2120-2204) were constructed as described previously (10). For the construction of pYBD-5A(1973-2172), HCV cDNA corresponding to amino acids 1973 to 2172 was amplified by PCR using the DNA of pTHE1964-3011 as a template (27). Oligonucleotides 5′-TACCCATACCCGGGTACCATGTCCGGCTCGTGGCTAAG-3′ and 5′-AGGTTACCCGGGTCAAGTGAGCACTGCTACATC-3′ were used as plus- and minus-strand DNA primers, respectively. The PCR product was digested with XmaI and then inserted into the XmaI site of pAS2 (Clontech, Inc). The construct pYAD-5A(1973-2419), which contains the GAL4 AD and the full-length HCV NS5A protein, was constructed by inserting the DNA insert of pYBD-5A(1973-2419) excised with XmaI into the XmaI site of pACT2. pYBD-karyopherin β3(1007-1097) was constructed by inserting the NcoI fragment of pYAD-karyopherin β3(1007-1097) into the NcoI site of pACT2. pTM-NS5A, used for in vitro translation of the HCV NS5A protein, was constructed by inserting the blunt-ended XmaI fragment from pYBD-NS5A(1973-2419) into the blunt-ended XmaI site of pTM-1. The plasmid pGEX-karyopherin β3, expressing a glutathione S-transferase (GST)–karyopherin β3 fusion protein in Escherichia coli, was constructed by ligating the blunt-ended ApaI-XhoI fragment from pSK-karyopherin β3 to the blunt-ended XmaI-XhoI fragment of pGEX-KG. To generate a Myc epitope-tagged full-length protein of karyopherin β3 (pCMV/myc-karyopherin β3), PCR was performed to amplify cDNA of karyopherin β3 using the primers: 5′-ACCCATACCCGGGACCATGGAACAAAAACTCATCTCAGAAGAGG ATCTGATGGCGGCGGCCGCGGCGGAG-3′ and 5′-CCTCCAGAAGTCTGTACTTGGCG-3′ (GSP2). SmaI- and NotI-Klenow-treated pEGEP-N1 (Clontech, Inc), a SmaI-BamHI-treated fragment of the PCR product, and the BamHI-EcoRV fragment of pSK-karyopherin β3 were ligated to generate plasmid pCMV/myc-karyopherin β3. The plasmid pCMV/HA-NS5A, encoding HCV NS5A and a hemagglutinin (HA) epitope tag at the N terminus, was constructed by inserting a SmaI-digested PCR product generated with the primers 5′-TAC CCATACCCGGGACCATGTACCCATACGATGTTCCAGATTACGCTTC CGGCTCGTGGCTAAGGG-3′ and 5′-AGGTTACCCGGGTCAGCAGCAGACGACGTCCTC-3′ into the blunt-ended AgeI-NotI site of pEGEP-N1 (Clontech, Inc.). The yeast expression vector pRS316/ADH-AD was constructed by inserting a blunt-ended SphI fragment of pGAD424 (Clontech, Inc.) into the blunt-ended XbaI-XhoI site of pPS1066 (46). pKM84, a URA CEN plasmid containing an SphI fragment of the ADH1 promoter, was obtained by self-ligation of HindIII-digested pRS316/ADH-AD. The yeast expression plasmids pKM84-karyopherin β3 and pKM84/myc-karyopherin β3 were constructed by inserting the blunt-ended ApaI-XhoI fragment of pSK-karyopherin β3 or the blunt-ended SalI-StuI fragment of pCMV/myc-karyopherin β3 into pKM84 digested with HindIII. The plasmids pGal, a TRP1 2μ-ori plasmid containing the GAL1 promoter, was generated by inserting the PvuII-SmaI fragment of pGBT9 (Clontech, Inc.) into the blunt-ended NheI-NcoI site of pYES2 (Invitrogen). The plasmids pGal-NS5A(1973-2419) and pGal-NS5A(1973-2172), galactose-inducible vectors encoding full-length NS5A and the N-terminal region of NS5A, respectively, were constructed by inserting the blunt-ended XmaI fragment of pYBD-5A(1973-2419) or pYBD-5A(1973-2172), respectively, into the blunt-ended EcoRI site of pGal. pGal-NS5A(2173-2419), a galactose-dependent vector encoding the C-terminal region of NS5A, was constructed by PCR amplification using the primers 5′-TACCCATACCCGGGTACCATGTCCATGCTCACCGACCC-3′ and 5′-AGGTTACCCGGGTCAGCAGCAGACGACGTCCTC-3′. The SmaI-digested PCR fragment was then inserted into the blunt-ended EcoRI site of pGal.
Yeast cell culture, transformation, and β-galactosidase assay.
Yeast cells were grown on YPD (1% yeast extract, 2% peptone, 2% dextrose, 1.5% agar [for plates]) or on synthetic minimal medium (0.67% yeast nitrogen base, the appropriate auxotrophic supplements, 1.5% agar [for plates]) containing 2% dextrose (SD) or 2% galactose and 2% raffinose (SGAL). Yeast was transformed with appropriate plasmids by the lithium acetate method (23), and the transformants were selected on the appropriate synthetic minimal medium. For the β-galactosidase assay, yeast cells grown on synthetic minimal plates were transferred to a filter (Whatman no. 1). The filter was placed in liquid nitrogen for 30 s and then incubated in Z buffer (60 mM Na2HPO4, 40 mM NaH2PO4, 10 mM KCl, 1 mM MgSO4) containing 0.82 mM 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal). The filters were kept at 30°C and monitored for color change indicating β-galactosidase activity.
Two-hybrid screening.
The yeast strain HF7c [MATa ura3-52 his3-200 lys2-801 ade2-101 trp1-901 leu2-3,112 gal4-542 gal80-538 LYS2::GAL1UAS-GAL1TATA-HIS3 URA3::GAL417mer(x3)-CyClTATA-lacZ] was used for the two-hybrid selection (19). The plasmid pYBD-5A(1973-2302) was used as bait. The pACT cDNA library (Clontech) from human liver was used as a source of prey genes. The bait plasmid and the pACT2 cDNA library were introduced into the yeast strain HF7c by the lithium acetate method. Transformants were selected for tryptophan, leucine, and histidine prototrophy. Isolated colonies were tested for β-galactosidase activity. The prey plasmids were selected from yeast colonies giving a positive signal according to the manufacturer's protocol. False positives were eliminated by retransforming the host HF7c strain containing pYBD-5A(1973-2302) or other nonspecific baits with the isolated pACT plasmids.
cDNA cloning of human karyopherin β3.
cDNA corresponding to the 5′ end of karyopherin β3 mRNA was obtained by a rapid amplification of cDNA ends (5′-RACE) procedure using a 5′-RACE kit (Marathon-Ready cDNA; Clontech). The B1 (5′-GCATTTGCCCAGTCCTCATCTTC-3′; corresponding to positions 949 to 971 of karyopherin β3 cDNA) and B2 (5′-ATGAATTCTGAGGAATAGTCTGTGC-3′; positions 904 to 922) primers were used as the gene-specific reverse primers, and the forward primers were provided in the 5′-RACE kit (Clontech). The middle region of the karyopherin β3 cDNA was obtained by reverse transcription-nested PCR (RT-PCR). A1 (5′-CAGGCGGTAAATGACTCGTGC-3′; positions 667 to 687), A2 (5′-ATGAATTCCAGAATGATGATTCTGTCC-3′; positions 690 to 709), GSP1 (5′-GCTGCTCAGGACTGAGCTGTGC-3′; positions 3238 to 3259), and GSP2 (5′-CCTCCAGAAGTCTGTACTTGGCG-3′; positions 3196 to 3218) were used as the primers. The first round of reverse transcription-nested PCR was performed with A1 and GSP1 as primers, and the second round of PCR was performed with A2 and GSP2. The 5′ end, middle region, and 3′ end of karyopherin β3 cDNA obtained from the interactive trap library plasmid [pYAD-karyopherin β3(1007-1097)] were ligated into pBluescript SK(−) (Invitrogen) to generate pSK-karyopherin β3. The karyopherin β3 cDNA was sequenced by the standard dideoxy method.
Expression and purification of recombinant proteins.
From the plasmid pGEX-karyopherin β3, a GST-fused karyopherin β3 was expressed in E. coli BL21(DE3)/pLys S. Recovery of the GST fusion protein was carried out as previously described (13, 55). Briefly, a 2-liter culture induced with isopropyl-β-d-thiogalactopyranoside (IPTG) at a final concentration of 1 mM was lysed in buffer L (20 mM Na-phosphate [pH 7.6], 300 mM NaCl, 10% glycerol, 0.2% Tween 20, 1 mM β-mercaptoethanol) with protease inhibitors. After centrifugation, the supernatant was applied to a glutathione-Sepharose 4B column (Pharmacia) and eluted with 20 mM glutathione. The eluted protein was pooled and dialyzed into buffer D (20 mM Na-phosphate [pH 7.6], 50 mM NaCl, 10% glycerol, 5 mM β-mercaptoethanol).
In vitro binding assay.
The GST fusion proteins were adsorbed onto glutathione beads prewashed three times with 10 volumes of buffer D by incubation at 4°C for 1 h on a rotating mixer. The beads were then washed three times with 1 ml of buffer D and stored at 4°C as a 50% slurry in buffer D. Radiolabeled NS5A was generated using an in vitro transcription-translation system (Promega) and [35S]methionine (DuPont NEN). Equal amounts of 35S-labeled translation products (as determined by sodium dodecyl sulfate-polyacrylamide gel electrophoresis [SDS-PAGE] and a BAS Radioanalytic Imaging System) were incubated with 80 μl of GST beads (50% slurry) in 1 ml of GB buffer (final concentrations of 20 mM Tris-HCl [pH 8.0], 0.25% NP-40, 50 mM NaCl, and 1 mM EDTA). After 2 h of incubation at 4°C on a rotating mixer, the beads were washed five times with 1 ml of GB buffer and boiled for 3 min in 30 μl of 2× SDS sample buffer before analysis by SDS-PAGE. Gels were dried and exposed to X-ray films.
Coimmunoprecipitation.
Cos-7 cells were transiently transfected with the indicated plasmids using an electroporation method described previously (37). After 48 h of cultivation, the cells were washed and resuspended in lysis buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.1% NP-40, 1 mM phenylmethylsulfonyl fluoride). Equal amounts of cleared cell lysates were subjected to immunoprecipitation with monoclonal anti-HA antibody (F-7; Santa Cruz), followed by adsorption to protein G-agarose (Boehringer Mannheim). The beads were washed three times with washing buffer (50 mM Tris-HCl [pH 7.5], 150 mM NaCl, 1 mM EDTA, 0.01% NP-40). The antibody-protein complexes were then resolved by SDS-PAGE, and the Myc-tagged protein was identified by Western blotting with a monoclonal anti-Myc antibody (9E10; Santa Cruz) probe using an enhanced chemiluminescence system.
Complementation of a yeast PSE1 and KAP123 double mutation with human karyopherin β3.
Yeast strain PSY1042 (MATa ura3-52 leuΔ1 trpΔ63 GAL+ pse1 Δkap123), which contains the PSE1 temperature-sensitive allele (pse1-1) and the KAP123 null mutation (Δkap123), was used for the complementation test (46). These cells grow well at 25°C but do not grow at 36°C. Yeast strain PSY1042 was transformed with plasmid pKM84-karyopherin β3 or pKM84/myc-karyopherin β3 and then cultivated on a uracil-deficient SD plate at 25°C. The resulting transformants were streaked onto uracil-deficient SD plates and then cultivated at either 25 or 36°C. In order to investigate the effect of HCV NS5A on the human karyopherin β3 in the transformed yeast, yeast cells containing pKM84/myc-karyopherin β3 were subsequently transformed with plasmid pGal-NS5A(1973-2419), pGal-NS5A(1973-2172), or pGal-NS5A(2173-2419). The resulting transformants were selected on uracil- and tryptophan-deficient SD. The effect of NS5A was determined by cultivating the transformants on an SGAL plate at 36°C.
Western blot analysis of NS5A in yeast cells.
The yeast transformants with galactose-inducible expression plasmids were grown at 25°C in SGAL. Cells were harvested at an optical density at 600 nm of 0.7 and then lysed by vigorous sonication and vortexing together with glass beads. The yeast lysate was centrifuged at 12,000 rpm as previously described (46), and the supernatant was used for Western blot analysis. Equal amounts of proteins were resolved by SDS-PAGE, and NS5A and its derivatives were identified by Western blotting using a polyclonal antibody against NS5A that was kindly provided by R. Bartenschlager.
RESULTS
Identification of cellular proteins interacting with HCV NS5A in the yeast two-hybrid system.
To identify cellular proteins interacting with HCV NS5A, a yeast two-hybrid system was employed to screen a human liver cDNA library (MATCHMAKER cDNA library from Clontech) using the C-terminally truncated HCV NS5A (amino acids 1973 to 2302) as bait. Nine positive clones were obtained from the screening of 2 × 106 independent yeast colonies. DNA sequence analysis showed that four of the nine positive clones encoded the C-terminal portion of karyopherin β3 encompassing amino acid residues 1007 to 1097 (Fig. 1). A full-length cDNA clone of karyopherin β3 was obtained from mRNA of HeLa cells as described in Materials and Methods. The entire cDNA clone of the karyopherin β3 gene was then sequenced to confirm the identity. Alignment of its deduced amino acid sequence with the yeast PSE1 (or KAP121) amino acid sequence revealed 65.2% similarity and 28.3% identity over the entire length of the protein (13, 55), which suggested that karyopherin β3 may have functions equivalent to those of the yeast PSE1 product. Furthermore, the human karyopherin β3 exhibited 58.9% homology and 23% identity to the yeast KAP123 product (55). The mammalian homologue of yeast KAP123 has not yet been identified.
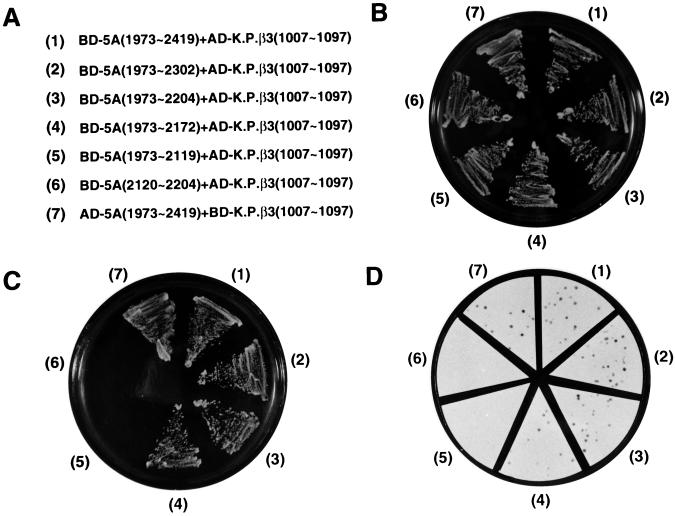
FIG. 1.
Schematic diagram of HCV NS5A and karyopherin β3 used in the yeast two-hybrid system. The top panel represents NS5A and its derivatives fused to the GAL4 BD or GAL4 AD, and the bottom panel depicts the C-terminal end of karyopherin β3 fused to the GAL4 BD or GAL4 AD. Solid, dotted, hatched, and wave-lined boxes represent the GAL4 BD, HCV NS5A (5A), GAL4 AD, and karyopherin β3 (K.P. β3), respectively.
The N-terminal region of NS5A is essential for interaction with karyopherin β3.
In order to determine the region in HCV NS5A required for interaction with karyopherin β3, two-hybrid analyses were carried out using NS5A, NS5A derivatives, and truncated karyopherin β3 genes (Fig. 1). The yeast plasmids used for the two-hybrid system (Fig. 2A) were cointroduced into yeast strain HF7c, and the transformants were grown on a medium lacking tryptophan and leucine (Fig. 2B). In the two-hybrid system, a protein-protein interaction is indicated by viability of yeast cells on histidine-deficient plates (Fig. 2C) and by β-galactosidase activity in the yeast cells (Fig. 2D). As shown in Fig. 2C and D, yeast cells containing plasmid BD-NS5A(1973-2172) and plasmids encoding larger NS5A constructs [BD-NS5A(1973-2204), BD-NS5A(1973-2302), and BD-NS5A(1973-2419)] grew on histidine-deficient plates (Fig. 2C, sectors 1, 2, 3, and 4) and exhibited β-galactosidase activity (Fig. 2D, sectors 1, 2, 3, and 4). A further C-terminal deletion of NS5A [BD-NS5A(1973-2119)] and an N-terminal deletion of NS5A [BD-5A(2120-2204)] abolished the protein-protein interaction (Fig. 2C and D, sectors 5 and 6). This indicates that the amino acid residues 2120 to 2172 of HCV NS5A include an essential part for the interaction with karyopherin β3 and that residues 1973 to 2172 are sufficient for the interaction. The protein-protein interaction in the two-hybrid system was also detected when the bait and the prey plasmids were exchanged reciprocally (Fig. 2C and D, sector 7). On the other hand, the full-length karyopherin β3 did not give a positive signal in the yeast two-hybrid system (data not shown). Misfolding of the fusion protein and/or exclusion of the protein from the nucleus is a possible reason for this phenomenon. Nevertheless, the full-length karyopherin β3 did bind to HCV NS5A in in vitro and in vivo assay systems (see below).
FIG. 2.
Determination of the domain in HCV NS5A responsible for the interaction with karyopherin β3. (A) Plasmid pairs used in the two-hybrid analysis shown in panels B, C, and D. The numbers in panels B, C, and D refer to these plasmid pairs. (B) Yeast cells transformed with the plasmid pairs in panel A were cultured on an SD plate lacking tryptophan and leucine. (C) Viability of the yeast transformants shown in panel B on an SD plate lacking tryptophan, leucine, and histidine and containing 2 mM 3-amino-1,2,4-triazole. Interaction between the two hybrid proteins is indicated by the growth of the yeast cells on this medium. (D) β-Galactosidase activities of the transformants. The dots indicate yeast colonies with β-galactosidase activity.
HCV NS5A binds to karyopherin β3 in vitro.
In vitro binding assays were performed to confirm the interaction between HCV NS5A and human karyopherin β3. The full-length karyopherin β3 cDNA was connected in frame to the C-terminal end of the GST gene in a bacterial expression vector to produce a GST-karyopherin β3 fusion protein. The protein was expressed in E. coli and then partially purified. Direct in vitro binding assays were carried out using the purified GST-karyopherin β3 and 35S-labeled NS5A generated by in vitro translation. The radiolabeled NS5A efficiently coprecipitated with the GST-karyopherin β3 but not with the GST negative control protein (Fig. 3, lanes 4 and 6). Luciferase, another negative control protein, did not bind to either GST or GST-karyopherin β3 (Fig. 3, lanes 3 and 5). This indicates that NS5A directly interacted with karyopherin β3.
FIG. 3.
In vitro analysis of HCV NS5A-karyopherin β3 interaction. The in vitro translation products of luciferase and NS5A are shown in lanes 1 and 2, respectively. These 35S-labeled proteins were incubated with resin-bound GST (lanes 3 and 4) or GST-karyopherin β3 (lanes 5 and 6). After the samples were washed with GB buffer (20 mM Tris-HCl [pH 8.0], 0.25% NP-40, 50 mM NaCl, 1 mM EDTA), the resin-bound proteins were resolved in an SDS–12.5% polyacrylamide gel.
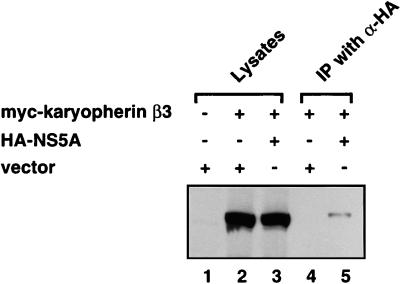
HCV NS5A interacts with karyopherin β3 in mammalian cells.
To determine whether HCV NS5A is capable of binding to karyopherin β3 inside cells, coimmunoprecipitation was performed using cells that expressed both of the proteins. Cos-7 cells were transfected with vectors encoding both the HA-tagged HCV NS5A (HA-NS5A) and the Myc-tagged karyopherin β3 (myc-karyopherin β3). The cell lysates were then subjected to immunoprecipitation using an anti-HA monoclonal antibody (Fig. 4, lanes 2 and 3). Immunoprecipitates were resolved by SDS-PAGE and transferred to a nitrocellulose membrane for Western blot analysis using the anti-Myc monoclonal antibody. The myc-karyopherin β3 was coimmunoprecipitated with HA-NS5A by the anti-HA antibody (Fig. 4, lane 5). In contrast, myc-karyopherin β3 was not precipitated by the anti-HA antibody when NS5A was absent from the cell (Fig. 4, lane 4). This indicates that NS5A interacted with karyopherin β3 in vivo.
FIG. 4.
In vivo coimmunoprecipitation of HCV NS5A and karyopherin β3. Cos-7 cells were transfected with plasmids expressing Myc-tagged karyopherin β3 and HA-tagged NS5A (lanes 3 and 5) or with a plasmid expressing Myc-tagged karyopherin β3 and the vector (lanes 2 and 4). Forty-eight hours after transfection, the Cos-7 cells were lysed and subjected to immunoprecipitation (IP) using an anti-HA monoclonal antibody (α-HA). The immunoprecipitated proteins were resolved by SDS-PAGE and then analyzed by Western blot assay using an anti-Myc antibody.
Human karyopherin β3 complements the double mutation of yeast PSE1 and KAP123.
Since the sequence of human karyopherin β3 is highly homologous to the sequences of yeast PSE1 and KAP123, we investigated whether human karyopherin β3 might be functionally homologous to yeast PSE1 and KAP123 products. The null mutation of KAP123 (Δkap123) does not exhibit any phenotypic difference from the wild type yeast (46). The temperature-sensitive mutation of PSE1 (pse1-1) caused delayed cell growth at the nonpermissive temperature (46). On the other hand, the PSE1 and KAP123 double mutant (PSY1042 [pse1-1 Δkap123]) grew at the permissive temperature (25°C) but not at the nonpermissive temperature (36°C) (Fig. 5, vector) (46). However, the double mutant could grow at the nonpermissive temperature (36°C) when the yeast cell contained plasmids encoding either yeast PSE1, human karyopherin β3, or Myc-tagged karyopherin β3 (Fig. 5, PSE1, karyopherin β3, and myc-karyopherin β3, respectively). This indicates that human karyopherin β3 can complement the function of the yeast PSE1 and KAP123 products.
FIG. 5.
Functional complementation of S. cerevisiae containing mutations in both the PSE1 and KAP123 genes by the human karyopherin β3. The temperature-sensitive pse1-1 Δkap123 strain (PSY1042) was transformed with the URA CEN plasmid expressing no protein (vector), karyopherin β3, Myc-tagged karyopherin β3, or PSE1. Each transformant was streaked on selective medium (SD lacking uracil) and incubated at 25°C or 36°C for 7 days. The proteins expressed in the transformants are noted.
HCV NS5A inhibits the function of karyopherin β3.
In order to evaluate the biological importance of the interaction between HCV NS5A and karyopherin β3, we investigated the effect of HCV NS5A on the function of karyopherin β3. We constructed yeast plasmids encoding HCV NS5A and its derivatives under the control of the GAL1 promoter and tested the effect of the proteins on yeast cells complemented with human karyopherin β3 (yeast strain PSY1042 containing plasmid pKM84/myc-karyopherin β3). Without the induction of NS5A and its derivatives, yeast cells complemented with human karyopherin β3 grew well at the nonpermissive temperature (Fig. 6A). On the other hand, upon the induction of the full-length HCV NS5A and the N-terminal domain of NS5A that binds to karyopherin β3, yeast cells complemented with human karyopherin β3 could not grow at the nonpermissive temperature [Fig. 6B, NS5A(1973-2419) and NS5A(1973-2172)]. Under the same conditions, the C-terminal domain of NS5A, which does not bind to karyopherin β3, did not affect the growth of the yeast cells [Fig. 6B, NS5A(2173-2419)]. We also tested the effect of full-length HCV NS5A on yeast cells supplemented with yeast PSE1 instead of human karyopherin β3. The yeast cells producing yeast PSE1 grew well with or without expression of HCV NS5A [Fig. 6C and D, NS5A(1973-2419)]. The expression of the full-length HCV NS5A and of the deletion mutants of NS5A was confirmed by Western blot analysis using anti-NS5A antibody, which was kindly provided by R. Bartenschlager (Fig. 6E). Almost the same amounts of full-length NS5A were detected in yeast cells transformed with either the yeast PSE1- or the human karyopherin β3-expressing vector (Fig. 6E, lanes 3 and 4). Apparently, the band intensity of the C-terminal region of NS5A, which does not inhibit human karyopherin β3 (Fig. 6E, lane 6), was stronger than that of the N-terminal region of NS5A, which inhibits human karyopherin β3 (Fig. 6E, lane 5). This strongly suggests that the growth inhibition by NS5A (shown in Fig. 6B) is not due to the toxic effect of NS5A per se but that it is related to the human karyopherin β3 activity. Taken together, our observations indicate that HCV NS5A inhibits the function of human karyopherin β3 most likely through direct protein-protein interaction.
FIG. 6.
HCV NS5A inhibits the function of human karyopherin β3. (A and B) HCV NS5A inhibits human karyopherin β3 in vivo. The yeast double mutant (pse1-1 Δkap123) complemented with plasmid pKM84/myc-karyopherin β3 was transformed with galactose-inducible expression construct pGal-NS5A(1973-2419) [NS5A(1973-2419)], pGal-NS5A(1973-2172) [NS5A(1973-2172)], or pGal-NS5A(2173-2419) [NS5A(2173-2419)] or negative control vector pGal (vector). Each transformant was streaked onto SD (A) or SGAL (B) and observed for growth at 36°C for 7 days. (C and D) HCV NS5A does not inhibit yeast PSE1 in vivo. The yeast double mutant (pse1-1 Δkap123) complemented with yeast PSE1 (pPS1066) was transformed with galactose-inducible expression construct pGal-NS5A(1973-2419) [NS5A(1973-2419)] or the negative control vector pGal (vector). Each transformant was streaked on SD (C) or SGAL (D), and growth at 36°C was scored after 2 days. (E) Western blot analysis. The yeast transformants shown in panels A and C were grown at 25°C in SGAL. Extracts prepared from the transformants were subjected to Western blot analysis using anti-NS5A antibody. Either PSE1- or human karyopherin β3-expressing plasmid was cotransformed with vector (lanes 1 and 2), full-length NS5A (lanes 3 and 4), N-terminal NS5A (lane 5), or C-terminal NS5A (lane 6). The positions of marker proteins are indicated. Bands of NS5A and its derivatives are indicated by arrowheads.
DISCUSSION
We found that HCV NS5A specifically interacted with karyopherin β3, blocking its activity in vivo. The N-terminal part of HCV NS5A (amino acids 1973 to 2172) was required for the protein-protein interaction and the inhibition of karyopherin β3. On the other side, the C-terminal end of karyopherin β3 was sufficient for the interaction with HCV NS5A. It has been seen before that the C-terminal regions of proteins of the karyopherin β family are required for direct interaction with target molecules or with adapter molecules binding to substrates (24). Thus, it is possible that HCV NS5A may compete with substrates that naturally bind to karyopherin β3. The N-terminal parts of karyopherin βs, which are the most conserved regions among the karyopherin β family genes, compose interfaces that interact with components of the translocation apparatus. In the case of karyopherin β1, this region is required for binding to the nuclear pore complex and to Ran (54). Likewise, the N-terminal portion of karyopherin β3 has been shown to bind to Ran (55).
How would NS5A contribute to viral proliferation by interacting with karyopherin β3? It is hard to formulate a conclusive hypothesis with such limited studies of the functions of NS5A and karyopherin β3. Nevertheless, we can speculate about possible physiological roles of the protein-protein interaction by referring to previous reports about functions of karyopherin β3. Karyopherin β3 is a member of the karyopherin β family, which facilitates transportation of proteins and/or RNAs between different compartments of the cell. The cytoplasmic and nuclear rim localization of karyopherin β3 and its binding to the repeat sequence of nucleoporins and to Ran-GTP strongly suggest that karyopherin β3 is involved in nucleocytoplasmic transport (13, 55). The high homology of karyopherin β3 to the yeast PSE1 and KAP123 products and the results of our complementation experiments using the yeast double mutant strain (PSY1042 [pse1-1 Δkap123]) lead us to conclude that the human karyopherin β3 can replace the function(s) of yeast PSE1 and KAP123.
Several functions of karyopherin β3, PSE1, and/or KAP123 have been reported. First, karyopherin β3 may function in the nuclear import of macromolecules. In vitro nuclear import experiments have shown that karyopherin β3, importin beta, transportin, and RanBP7 facilitated ribosomal protein transportation into the nucleus (33). Second, yeast PSE1 and/or KAP123 may be involved in mRNA export (46). Third, yeast PSE1 may enhance secretion of proteins (9). Fourth, yeast PSE1 may augment mitochondrial import of hydrophobic mitochondrial proteins (12). It is, however, not clear whether these effects of karyopherin β3, PSE1, and/or KAP123 are direct or indirect ones. We should consider all of these possible functions of karyopherin β3 when we think about a biological role(s) for the interaction between NS5A and karyopherin β3.
For proliferation, differentiation, and changes in metabolism, cells respond to intra- and extracellular signals, including virus infection. The transmission of cellular signals is often executed by signal-transducing molecules shuttling between different subcellular compartments. It is therefore not surprising that some viruses use strategies that block the transport of cellular signaling molecules in order to incapacitate a host antiviral defense system and/or to perturb cellular homeostasis. For instance, the matrix protein (M protein) of vesicular stomatitis virus blocks transportation of RNAs and proteins between the nucleus and the cytoplasm by inhibiting Ran guanosine triphosphatase-dependent nuclear transport (29). HCV NS5A might function in a similar way as the M protein of vesicular stomatitis virus. The subcellular localization patterns of NS5A and karyopherin β3 support this possibility. HCV NS5A is localized in the cytoplasm and enriched in the perinuclear space region (31, 37, 51), which is similar to the distribution pattern of karyopherin β3 (13, 55). Therefore, it is plausible to consider that karyopherin β3 might be sequestered from its normal active sites by binding to NS5A.
It is also possible that the export of RNAs from the nucleus could be impeded by HCV NS5A, as suggested by the phenotype of the yeast PSE1 and KAP123 double mutant (46). The blockage of RNA export in turn may inhibit expression of genes exerting antiviral activities that normally would be induced by viral infection. Alternatively, NS5A may inhibit the protein secretion-enhancing activity of karyopherin β3, as was shown by overexpression of PSE1 in yeast cells (9). In these respects, NS5A might block production and/or secretion of cytokines from HCV-infected cells. For instance, NS5A may inhibit secretion of alpha interferon, one of the first cytokines produced in response to virus infection, from HCV-infected cells, thus preventing the initiation of antiviral activities of neighboring cells. Since alpha interferon activates NK cell cytotoxicity and induces lysis of virus-infected cells, the inhibition of the protein secretion apparatus in the virus-infected cells would be advantageous to virus proliferation. In addition, the blockage of the protein secretion pathway may also hamper presentation of viral antigens in association with major histocompatibility complex class I molecules, which is required to make cytotoxic T lymphocytes recognize the virus-infected cell. In fact, suppression of antiviral activities through blockage of protein secretion has been discovered for several viruses. The poliovirus proteins 2B and 3A and the Epstein-Barr virus BARF1 inhibit secretion of cellular proteins (14) and alpha interferon (11), respectively, which modulates innate host responses to the viruses. In this respect, it is worth noting that the release of tumor necrosis factor alpha and interleukin-1 beta by phorbol myristate acetate was reduced for peripheral blood monocytes that had been collected from patients chronically infected with HCV (41). The inhibition of protein secretion by HCV NS5A, therefore, may be related to the correlation that exists between the nucleotide sequence of NS5A and the sensitivity of HCV to interferon treatment, even though the interferon sensitivity-determining region identified by Enomoto et al. (17) lies outside of the segment required for the interaction with karyopherin β3.
Analysis of yeast overexpressing PSE1 led to the conclusion that karyopherin β3 may also play a role in mitochondrial protein import (12). It is tempting to speculate that NS5A binding to karyopherin β3 could prevent normal mitochondrial protein import, resulting in alterations of mitochondrial functions. Intriguingly, frequent ultrastructural alterations of the mitochondria have been observed in patients' hepatocytes infected with HCV genotype 1b (3). The increased production of free radicals in the damaged mitochondria might contribute to the development of the severe liver diseases caused by HCV.
Due to the lack of a reliable in vitro cultivation system for HCV, it is difficult to confirm the interaction between NS5A and karyopherin β3 in the presence of all other viral proteins and to investigate all of these possible roles for HCV NS5A in HCV-infected cells. Instead, we used yeast cells to investigate the roles of karyopherin β3 and HCV NS5A in vivo. A study of the effect of HCV NS5A on the transport of macromolecules in mammalian cells, utilizing an NS5A-expressing cell line, is in progress.
ACKNOWLEDGMENTS
We are grateful to R. Bartenschlager for the gift of the NS5A-specific antiserum.
This study was supported in part by grants from the G7 program and the Molecular Medicine Research Group Program of MOST and by HMP-98-B-3-0020.
REFERENCES
- 1.Alter H J, Purcell R H, Shih J W, Melpolder J C, Houghton M, Choo Q L, Kuo G. Detection of antibody to hepatitis C virus in prospectively followed transfusion recipients with acute and chronic non-A, non-B hepatitis. N Engl J Med. 1989;321:1494–1500. doi: 10.1056/NEJM198911303212202. [DOI] [PubMed] [Google Scholar]
- 2.Asabe S I, Tanji Y, Satoh S, Kaneko T, Kimura K, Shimotohno K. The N-terminal region of hepatitis C virus-encoded NS5A is important for NS4A-dependent phosphorylation. J Virol. 1997;71:790–796. doi: 10.1128/jvi.71.1.790-796.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barbaro G, DiLorenzo G, Asti A, Ribersani M, Belloni G, Grisorio B, Filice G, Barbarini G. Hepatocellular mitochondrial alterations in patients with chronic hepatitis C: ultrastructural and biochemical findings. Am J Gastroenterol. 1999;94:2198–2205. doi: 10.1111/j.1572-0241.1999.01294.x. [DOI] [PubMed] [Google Scholar]
- 4.Bartenschlager, Ahlborn-Laake R L, Mous J, Jacobsen H. Kinetic and structural analyses of hepatitis C virus polyprotein processing. J Virol. 1994;68:5045–5055. doi: 10.1128/jvi.68.8.5045-5055.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bartenschlager R, Ahlborn-Laake L, Mous J, Jacobsen H. Nonstructural protein 3 of the hepatitis C virus encodes a serine-type proteinase required for cleavage at the NS3/4 and NS4/5 junctions. J Virol. 1993;67:3835–3844. doi: 10.1128/jvi.67.7.3835-3844.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Behrens S E, Tomei L, De Francesco R. Identification and properties of the RNA-dependent RNA polymerase of hepatitis C virus. EMBO J. 1996;15:12–22. [PMC free article] [PubMed] [Google Scholar]
- 7.Bruix J, Barrera J M, Calvet X, Ercilla G, Costa J, Sanchez-Tapias J M, Ventura M, Vall M, Bruguera M, Bru C, Rodes J. Prevalence of antibodies to hepatitis C virus in Spanish patients with hepatocellular carcinoma and hepatic cirrhosis. Lancet. 1989;ii:1004–1006. doi: 10.1016/s0140-6736(89)91015-5. [DOI] [PubMed] [Google Scholar]
- 8.Choo Q L, Kuo G, Weiner A J, Overby L R, Bradley D W, Houghton M. Isolation of a cDNA clone derived from a blood-borne non-A, non-B viral hepatitis genome. Science. 1989;244:359–362. doi: 10.1126/science.2523562. [DOI] [PubMed] [Google Scholar]
- 9.Chow T Y, Ash J J, Dignard D, Thomas D Y. Screening and identification of a gene, PSE-1, that affects protein secretion in Saccharomyces cerevisiae. J Cell Sci. 1992;101:709–719. doi: 10.1242/jcs.101.3.709. [DOI] [PubMed] [Google Scholar]
- 10.Chung K M, Song O K, Jang S K. Hepatitis C virus nonstructural protein 5A contains potential transcriptional activator domains. Mol Cell. 1997;7:661–667. [PubMed] [Google Scholar]
- 11.Cohen J I, Lekstrom K. Epstein-Barr virus BARF1 protein is dispensable for B-cell transformation and inhibits alpha interferon secretion from mononuclear cells. J Virol. 1999;73:7627–7632. doi: 10.1128/jvi.73.9.7627-7632.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Corral-Debrinski M, Belgareh N, Blugeon C, Claros M G, Doye V, Jacq C. Overexpression of yeast karyopherin Pse1p/Kap121p stimulates the mitochondrial import of hydrophobic proteins in vivo. Mol Microbiol. 1999;31:1499–1511. doi: 10.1046/j.1365-2958.1999.01295.x. [DOI] [PubMed] [Google Scholar]
- 13.Deane R, Schafer W, Zimmermann H P, Mueller L, Gorlich D, Prehn S, Ponstingl H, Bischoff F R. Ran-binding protein 5 (RanBP5) is related to the nuclear transport factor importin-beta but interacts differently with RanBP1. Mol Cell Biol. 1997;17:5087–5096. doi: 10.1128/mcb.17.9.5087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Doedens J R, Kirkegaard K. Inhibition of cellular protein secretion by poliovirus proteins 2B and 3A. EMBO J. 1995;14:894–907. doi: 10.1002/j.1460-2075.1995.tb07071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eckart M R, Selby M, Masiarz F, Lee C, Berger K, Crawford K, Kuo C, Kuo G, Houghton M, Choo Q L. The hepatitis C virus encodes a serine protease involved in processing of the putative nonstructural proteins from the viral polyprotein precursor. Biochem Biophys Res Commun. 1993;192:399–406. doi: 10.1006/bbrc.1993.1429. [DOI] [PubMed] [Google Scholar]
- 16.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Izumi N, Marumo F, Sato C. Comparison of full-length sequences of interferon-sensitive and resistant hepatitis C virus 1b. Sensitivity to interferon is conferred by amino acid substitutions in the NS5A region. J Clin Invest. 1995;96:224–230. doi: 10.1172/JCI118025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Enomoto N, Sakuma I, Asahina Y, Kurosaki M, Murakami T, Yamamoto C, Ogura Y, Izumi N, Marumo F, Sato C. Mutations in the nonstructural protein 5A gene and response to interferon in patients with chronic hepatitis C virus 1b infection. N Engl J Med. 1996;334:77–81. doi: 10.1056/NEJM199601113340203. [DOI] [PubMed] [Google Scholar]
- 18.Failla C, Tomei L, De Francesco R. Both NS3 and NS4A are required for proteolytic processing of hepatitis C virus nonstructural proteins. J Virol. 1994;68:3753–3760. doi: 10.1128/jvi.68.6.3753-3760.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feilotter H E, Hannon G J, Ruddell C J, Beach D. Construction of an improved host strain for two hybrid screening. Nucleic Acids Res. 1994;22:1502–1503. doi: 10.1093/nar/22.8.1502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gale M, Jr, Blakely C M, Kwieciszewski B, Tan S L, Dossett M, Tang N M, Korth M J, Polyak S J, Gretch D R, Katze M G. Control of PKR protein kinase by hepatitis C virus nonstructural 5A protein: molecular mechanisms of kinase regulation. Mol Cell Biol. 1998;18:5208–5218. doi: 10.1128/mcb.18.9.5208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gale M J, Jr, Korth M J, Tang N M, Tan S L, Hopkins D A, Dever T E, Polyak S J, Gretch D R, Katze M G. Evidence that hepatitis C virus resistance to interferon is mediated through repression of the PKR protein kinase by the nonstructural 5A protein. Virology. 1997;230:217–227. doi: 10.1006/viro.1997.8493. [DOI] [PubMed] [Google Scholar]
- 22.Ghosh A K, Steele R, Meyer K, Ray R, Ray R B. Hepatitis C virus NS5A protein modulates cell cycle regulatory genes and promotes cell growth. J Gen Virol. 1999;80:1179–1183. doi: 10.1099/0022-1317-80-5-1179. [DOI] [PubMed] [Google Scholar]
- 23.Gietz D, St. Jean A, Woods R A, Schiestl R H. Improved method for high efficiency transformation of intact yeast cells. Nucleic Acids Res. 1992;20:1425. doi: 10.1093/nar/20.6.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gorlich D. Nuclear protein import. Curr Opin Cell Biol. 1997;9:412–419. doi: 10.1016/s0955-0674(97)80015-4. [DOI] [PubMed] [Google Scholar]
- 25.Grakoui A, McCourt D W, Wychowcki C, Feinstone S M, Rice C M. A second hepatitis C virus-encoded proteinase. Proc Natl Acad Sci USA. 1993;90:10583–10587. doi: 10.1073/pnas.90.22.10583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Grakoui A, McCourt D W, Wychowski C, Feinstone S M, Rice C M. Characterization of the hepatitis C virus-encoded serine proteinase: determination of proteinase-dependent polyprotein cleavage sites. J Virol. 1993;67:2832–2843. doi: 10.1128/jvi.67.5.2832-2843.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hahm B, Han D S, Back S H, Song O K, Cho M J, Kim C J, Shimotohno K, Jang S K. NS3-4A of hepatitis C virus is a chymotrypsin-like protease. J Virol. 1995;69:2534–2539. doi: 10.1128/jvi.69.4.2534-2539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Han D S, Hahm B, Rho H M, Jang S K. Identification of the protease domain in NS3 of hepatitis C virus. J Gen Virol. 1995;76:985–993. doi: 10.1099/0022-1317-76-4-985. [DOI] [PubMed] [Google Scholar]
- 29.Her L S, Lund E, Dahlberg J E. Inhibition of Ran guanosine triphosphatase-dependent nuclear transport by the matrix protein of vesicular stomatitis virus. Science. 1997;276:1845–1848. doi: 10.1126/science.276.5320.1845. [DOI] [PubMed] [Google Scholar]
- 30.Hijikata M, Mizushima H, Akagi T, Mori S, Kakiuchi N, Kato N, Tanaka T, Kimura K, Shimotohno K. Two distinct proteinase activities required for the processing of a putative nonstructural precursor protein of hepatitis C virus. J Virol. 1993;67:4665–4675. doi: 10.1128/jvi.67.8.4665-4675.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ide Y, Zhang L, Chen M, Inchauspe G, Bahl C, Sasaguri Y, Padmanabhan R. Characterization of the nuclear localization signal and subcellular distribution of hepatitis C virus nonstructural protein NS5A. Gene. 1996;182:203–211. doi: 10.1016/s0378-1119(96)00555-0. [DOI] [PubMed] [Google Scholar]
- 32.Inchauspe G, Zebedee S, Lee D H, Sugitani M, Nasoff M, Prince A M. Genomic structure of the human prototype strain H of hepatitis C virus: comparison with American and Japanese isolates. Proc Natl Acad Sci USA. 1991;88:10292–10296. doi: 10.1073/pnas.88.22.10292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jakel S, Gorlich D. Importin beta, transportin, RanBP5 and RanBP7 mediate nuclear import of ribosomal proteins in mammalian cells. EMBO J. 1998;17:4491–4502. doi: 10.1093/emboj/17.15.4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kaneko T, Tanji Y, Satoh S, Hijikata M, Asabe S, Kimura K, Shimotohno K. Production of two phosphoproteins from the NS5A region of the hepatitis C viral genome. Biochem Biophys Res Commun. 1994;205:320–326. doi: 10.1006/bbrc.1994.2667. [DOI] [PubMed] [Google Scholar]
- 35.Kato N, Lan K H, Ono-Nita S K, Shiratori Y, Omata M. Hepatitis C virus nonstructural region 5A protein is a potent transcriptional activator. J Virol. 1997;71:8856–8859. doi: 10.1128/jvi.71.11.8856-8859.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim D W, Gwack Y, Han J H, Choe J. C-terminal domain of the hepatitis C virus NS3 protein contains an RNA helicase activity. Biochem Biophys Res Commun. 1995;215:160–166. doi: 10.1006/bbrc.1995.2447. [DOI] [PubMed] [Google Scholar]
- 37.Kim J E, Song W K, Chung K M, Back S H, Jang S K. Subcellular localization of hepatitis C viral proteins in mammalian cells. Arch Virol. 1999;144:329–343. doi: 10.1007/s007050050507. [DOI] [PubMed] [Google Scholar]
- 38.Kuo G, Choo Q L, Alter H J, Gitnick G L, Redeker A G, Purcell R H, Miyamura T, Dienstag J L, Alter M J, Stevens C E, Tegtmeier G E, Bonino F, Colombo M, Lee W S, Kuo C, Berger K, Shuster J R, Overby L R, Bradley D W, Houghton M. An assay for circulating antibodies to a major etiologic virus of human non-A, non-B hepatitis. Science. 1989;244:362–364. doi: 10.1126/science.2496467. [DOI] [PubMed] [Google Scholar]
- 39.Kurosaki M, Enomoto N, Murakami T, Sakuma I, Asahina Y, Yamamoto C, Ikeda T, Tozuka S, Izumi N, Marumo F, Sato C. Analysis of genotypes and amino acid residues 2209 to 2248 of the NS5A region of hepatitis C virus in relation to the response to interferon-beta therapy. Hepatology. 1997;25:750–753. doi: 10.1002/hep.510250343. [DOI] [PubMed] [Google Scholar]
- 40.Lin C, Pragai B M, Grakoui A, Xu J, Rice C M. Hepatitis C virus NS3 serine proteinase: trans-cleavage requirements and processing kinetics. J Virol. 1994;68:8147–8157. doi: 10.1128/jvi.68.12.8147-8157.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mendoza E C, Paglieroni T G, Zeldis J B. Decreased phorbol myristate acetate-induced release of tumor necrosis factor-alpha and interleukin-1 beta from peripheral blood monocytes of patients chronically infected with hepatitis C virus. J Infect Dis. 1996;174:842–844. doi: 10.1093/infdis/174.4.842. [DOI] [PubMed] [Google Scholar]
- 42.Miller R H, Purcell R H. Hepatitis C virus shares amino acid sequence similarity with pestiviruses and flaviviruses as well as members of two plant virus supergroups. Proc Natl Acad Sci USA. 1990;87:2057–2061. doi: 10.1073/pnas.87.6.2057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Neddermann P, Clementi A, De Francesco R. Hyperphosphorylation of the hepatitis C virus NS5A protein requires an active NS3 protease, NS4A, NS4B, and NS5A encoded on the same polyprotein. J Virol. 1999;73:9984–9991. doi: 10.1128/jvi.73.12.9984-9991.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pemberton L F, Blobel G, Rosenblum J S. Transport routes through the nuclear pore complex. Curr Opin Cell Biol. 1998;10:392–399. doi: 10.1016/s0955-0674(98)80016-1. [DOI] [PubMed] [Google Scholar]
- 45.Saito I, Miyamura T, Ohbayashi A, Harada H, Katayama T, Kikuchi S, Watanabe Y, Koi S, Onji M, Ohta Y, Choo Q-L, Houghton M, Kuo G. Hepatitis C virus infection is associated with the development of hepatocellular carcinoma. Proc Natl Acad Sci USA. 1990;87:6547–6549. doi: 10.1073/pnas.87.17.6547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seedorf M, Silver P A. Importin/karyopherin protein family members required for mRNA export from the nucleus. Proc Natl Acad Sci USA. 1997;94:8590–8595. doi: 10.1073/pnas.94.16.8590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takamizawa A, Mori C, Fuke I, Manabe S, Murakami S, Fujita J, Onishi E, Andoh T, Yoshida I, Okayama H. Structure and organization of the hepatitis C virus genome isolated from human carriers. J Virol. 1991;65:1105–1113. doi: 10.1128/jvi.65.3.1105-1113.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tan S L, Nakao H, He Y, Vijaysri S, Neddermann P, Jacobs B L, Mayer B J, Katze M G. NS5A, a nonstructural protein of hepatitis C virus, binds growth factor receptor-bound protein 2 adaptor protein in a Src homology 3 domain/ligand-dependent manner and perturbs mitogenic signaling. Proc Natl Acad Sci USA. 1999;96:5533–5538. doi: 10.1073/pnas.96.10.5533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tanimoto A, Ide Y, Arima N, Sasaguri Y, Padmanabhan R. The amino terminal deletion mutants of hepatitis C virus nonstructural protein NS5A function as transcriptional activators in yeast. Biochem Biophys Res Commun. 1997;236:360–364. doi: 10.1006/bbrc.1997.6967. [DOI] [PubMed] [Google Scholar]
- 50.Tanji Y, Hijikata M, Satoh S, Kaneko T, Shimotohno K. Hepatitis C virus-encoded nonstructural protein NS4A has versatile functions in viral protein processing. J Virol. 1995;69:1575–1581. doi: 10.1128/jvi.69.3.1575-1581.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tanji Y, Kaneko T, Satoh S, Shimotohno K. Phosphorylation of hepatitis C virus-encoded nonstructural protein NS5A. J Virol. 1995;69:3980–3986. doi: 10.1128/jvi.69.7.3980-3986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tomei L, Failla C, Santolini E, De Francesco R, La Monica N. NS3 is a serine protease required for processing of hepatitis C virus polyprotein. J Virol. 1993;67:4017–4026. doi: 10.1128/jvi.67.7.4017-4026.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Tu H, Gao L, Shi S T, Taylor D R, Yang T, Mircheff A K, Wen Y, Gorbalenya A E, Hwang S B, Lai M M. Hepatitis C virus RNA polymerase and NS5A complex with a SNARE-like protein. Virology. 1999;263:30–41. doi: 10.1006/viro.1999.9893. [DOI] [PubMed] [Google Scholar]
- 54.Wozniak R W, Rout M P, Aitchison J D. Karyopherins and kissing cousins. Trends Cell Biol. 1998;8:184–188. doi: 10.1016/s0962-8924(98)01248-3. [DOI] [PubMed] [Google Scholar]
- 55.Yaseen N R, Blobel G. Cloning and characterization of human karyopherin beta3. Proc Natl Acad Sci USA. 1997;94:4451–4456. doi: 10.1073/pnas.94.9.4451. [DOI] [PMC free article] [PubMed] [Google Scholar]