Summary
Background
Invasive pneumococcal disease due to serotype 3 (S3-IPD) is associated with high mortality rates and long-term adverse effects. The introduction of the 13-valent pneumococcal conjugate vaccine (PCV13) into the Spanish paediatric immunisation programme has not led to a decrease in the adult S3-IPD. We aimed to analyse the incidence, clinical characteristics and genomics of S3-IPD in adults in Spain.
Methods
Adult IPD episodes hospitalized in a Southern Barcelona hospital were prospectively collected (1994–2020). For genomic comparison, S3-IPD isolates from six Spanish hospitals (2008–2020) and historical isolates (1989–1993) were analysed by WGS (Illumina and/or MinION).
Findings
From 1994 to 2020, 270 S3-IPD episodes were detected. When comparing pre-PCV (1994–2001) and late-PCV13 (2016–2020) periods, only modest changes in S3-IPD were observed (from 1.58 to 1.28 episodes per 100,000 inhabitants year). In this period, the incidence of the two main lineages shifted from 0.38 to 0.67 (CC180-GPSC12) and from 1.18 to 0.55 (CC260-GPSC83). The overall 30-day mortality remained high (24.1%), though a decrease was observed between the pre-PCV (32.4%; 95.0% CI, 22.0–45.0) and the late-PCV13 period (16.7%; 95.0% CI, 7.5–32.0) (p = 0.06). At the same time, comorbidities increased from 77.3% (95.0% CI, 65.0–86.0) to 85.7% (95.0% CI, 71.0–94.0) (p = 0.69). There were no differences in clinical characteristics or 30-day mortality between the two S3 lineages. Although both lineages were genetically homogeneous, the CC180-GPSC12 lineage presented a higher SNP density, a more open pan-genome, and a major presence of prophages and mobile genetic elements carrying resistance genes.
Interpretation
Adult S3-IPD remained stable in our area over the study period despite PCV13 introduction in children. However, a clonal shift was observed. The decrease in mortality rates and the increase in comorbidities suggest a change in clinical management and overall population characteristics. The low genetic variability and absence of clinical differences between lineages highlight the role of the S3 capsule in the disease severity.
Funding
This study has been funded by Instituto de Salud Carlos III (ISCIII) “PI18/00339”, “PI21/01000”, “INT22/00096”, “FI22/00279”, CIBER “CIBERES-CB06/06/0037”, “CIBERINFEC-CB21/13/00009” and MSD grant “IISP 60168”.
Keywords: Streptococcus pneumoniae, Serotype 3, Genomics
Research in context.
Evidence before this study
Streptococcus pneumoniae has been extensively studied over the decades, particularly after the introduction of vaccines. However, despite the large number of articles on it, only a few are specific to serotypes and provide a contextual analysis of the clinical data and genomic features. To address this gap, we conducted a literature search on PubMed for articles published up to 1st September 2023, using the search terms “Streptococcus pneumoniae”, “Serotype 3″, “IPD” (invasive pneumococcal disease), “Dynamics”, “Genomics”, and “Capsule”. There is evidence respecting the stability of the incidence of serotype 3 in some countries, despite the introduction of the PCV13. Regarding genetic analysis, the assessment of the globally disseminated lineage CC180 revealed a differentiation into clades following the introduction of the PCV13. Additionally, a genetic analysis conducted in the UK linked the increase in serotype 3 cases to a specific subclade within CC180. Notably, there were no analyses available covering an extended time period in the same geographical area, including on the strains collected prior to the introduction of PCVs.
Added value of this study
This study offers a comprehensive perspective on serotype 3, shedding light on its clinical significance and providing valuable insights into its genomic evolution over more than 30 years, including isolates predating the introduction of PCVs. By analysing a collection of 344 isolates, we have not only contextualised serotype 3 within clinical data, but have also detailed the evolutionary trajectories of various lineages in the context of PCVs. Our disease analysis over an extended period has revealed changes in the population affected by S3-IPD and in the clinical presentation. Additionally, we have examined the potential impact of serotype 3 lineages on IPD outcomes. Furthermore, this extensive dataset has allowed us to identify differences not only in clinical characteristics, but also in genomic features among the major clonal complexes, thereby enhancing our understanding of this important pathogen.
Implications of all the available evidence
Despite high levels of childhood vaccination with the PCV13 in Spain, no herd protection has been observed in adults for serotype 3. Understanding its evolution could reveal new strategies to prevent the prevalence of this serotype in a public health context. Furthermore, the analysis of clinical characteristics of patients over the study period could contribute to our understanding of the epidemiology of this serotype. The holistic and multidisciplinary approach of this work sheds new light on the persistence of serotype 3 in the era of PCV13.
Introduction
Pneumococcal infections, such as pneumonia, meningitis and bacteraemia, are one of the main causes of morbidity and mortality worldwide.1 The introduction of the pneumococcal conjugate vaccines (PCVs) has reduced rates of disease and mortality in both children and adults.2 Nevertheless, the burden of disease and associated mortality are still a global health problem, mainly in adults.2 Among them, invasive pneumococcal disease (IPD) due to serotype 3 (S3-IPD), which is one of the targets of the PCV13, remains predominant. Moreover, S3-IPD has been associated with high mortality rates, particularly in the elderly,3 as well as the severity of pneumonia presentation4 and long-term effects such as major adverse cardiovascular events.5
The polysaccharide capsule is the main virulence factor in pneumococci and forms the basis of vaccine development.6 To date, more than 100 serotypes have been identified, with this diversity associated with differences in invasiveness and mortality among the serotypes. In Spain, the PCV7 was licensed in 2001, while the PCV13, targeting serotype 3, was licensed in 2011. Children were vaccinated on a voluntary basis until 2016, when the PCV13 was introduced into the official immunisation programme for children, leading to an uptake of 97.8% in 2020 (https://www.sanidad.gob.es/areas/promocionPrevencion/vacunaciones/calendario-y-coberturas/coberturas/home.htm). In contrast to the incidence of IPD caused by other PCV13 serotypes (such as 7F, 1 and 23F), which decreased after vaccine introduction in all age groups, the prevalence of IPD due to S3 has increased among adults over 60, suggesting poor herd protection.7,8 The PCV13 effectiveness against serotype 3 has been reported to be as low as 25.9% (95.0% CI, −65.3 to 66.8) in children.9 Thus, a study suggested that antibody titres above 2.83 μg/mL were needed to reach protection, which is higher than those achieved for serotype 3 after PCV13 vaccination.10 In the present study, we analysed changes in clinical characteristics among the adult S3-IPD cases and compared genomes of the responsible pneumococcal isolates before and after the introduction of the PCVs in children.
Methods
Hospital setting and study design
We conducted two analyses of S3-IPD in adults using three collections of isolates (Supplementary Figure S1). The first analysis was a retrospective analysis of a prospective laboratory-based study that included all adult (≥18 years old) S3-IPD episodes (n = 270) from 1994 to 2020 detected in Hospital Universitari de Bellvitge (HUB), a tertiary hospital located in the Southern Barcelona area. For this analysis, clinical data on patients, including demographics, early (≤7 days) and 30-day mortality, underlying diseases and previous antibiotic therapy, were prospectively collected along with available isolates (n = 252, 1st collection). The second analysis included the whole genome sequencing (WGS) of 344 S3-IPD isolates: 252 genomes from the first analysis (1st collection), 7 isolates (2nd collection) recovered from the HUB historical collection (1989–1993), and 85 isolates (3rd collection) from a multicentre study.11 The last collection includes strains from Catalonia (Hospital Universitari Vall d’Hebron [VH, n = 22], Hospital Universitari Germans Trias i Pujol [HUGTiP, n = 17] and Hospital Universitari Parc Taulí [Taulí, n = 12], Madrid (Hospital General Universitario Gregorio Marañón [HGUGM, n = 19]) and the Basque Country (Hospital Universitario Donostia [HUD, n = 15]).
Identification, antibiotic susceptibility testing and serotyping
Pneumococci were identified using standard microbiological procedures, including optochin susceptibility and/or bile solubility. Serotyping was performed at the Spanish Pneumococcal Reference Laboratory for Pneumococci by the quellung reaction, dot blot and/or capsular sequence typing. The antimicrobial susceptibility was assessed by microdilution, following the guidelines of the European Committee on Antimicrobial Susceptibility Testing (EUCAST) (www.eucast.org/clinical_breakpoints/). The antibiotics tested were penicillin, ampicillin, amoxicillin, tetracycline, chloramphenicol, cefotaxime, clindamycin, erythromycin, ciprofloxacin and trimethoprim/sulfamethoxazole.
Statistical analysis
The incidence of S3-IPD was calculated as the number of episodes per 100,000 inhabitants per year. The population was obtained from IDESCAT (http://www.idescat.cat/). To analyse temporal trends, five different periods were defined based on the introduction of the PCVs in Spain: pre-PCV (1994–2001), early-PCV7 (2002–2005), late-PCV7 (2006–2010), early-PCV13 (2011–2015) and late-PCV13 (2016–2020). The Mantel-Haenszel trend test for categorical and the Pearson test for numerical variables were used to examine trends across periods. A complete case analysis was performed and the number of missingness are reported in Table 1.
Table 1.
Differences in clinical characteristics of patients with S3-IPD over the five study periods.
| pre-PCV n (%) | early-PCV7 n (%) | late-PCV7 n (%) | early-PCV13 n (%) | late-PCV13 n (%) | p-trenda | |
|---|---|---|---|---|---|---|
| Available cases | 76 | 48 | 54 | 48 | 44 | |
| Clinical characteristics | ||||||
| Age (years), Mean (±SD) | 67.6 (±17.5) | 66.1 (±11.9) | 66.8 (±15.6) | 62.5 (±16.2) | 70.8 (±12.9) | 0.872 |
| Male | 46 (60.5) | 34 (70.8) | 36 (66.7) | 30 (62.5) | 30 (68.2) | 0.613 |
| Clinical presentation | 0.642 | |||||
| Pneumonia | 73 (96.1) | 43 (89.6) | 38 (70.4) | 42 (87.5) | 35 (79.5) | |
| Meningitis | 2 (2.6) | 2 (4.2) | 10 (18.5) | 2 (4.2) | 6 (13.6) | |
| Others | 1 (1.3) | 3 (6.3) | 6 (11.1) | 4 (8.3) | 3 (6.8) | |
| Bacteraemia | 62 (81.6) | 45 (93.8) | 48 (88.9) | 44 (91.7) | 43 (97.7) | 0.011 |
| Available cases | 65 | 46 | 54 | 48 | 42 | |
| Nosocomial acquisitionb | 4 (6.2) | 2 (4.4) | 3 (5.6) | 2 (4.2) | 1 (2.4) | 0.414 |
| Risk factors | ||||||
| Smoking | 21 (32.3) | 15 (32.6) | 17 (31.5) | 18 (37.5) | 13 (31.0) | 0.841 |
| Alcohol abuse | 12 (18.5) | 8 (17.4) | 6 (11.3) | 8 (16.7) | 10 (23.8) | 0.756 |
| Prior antibiotic therapy | 8 (12.3) | 3 (6.5) | 13 (24.1) | 10 (20.8) | 7 (16.7) | 0.201 |
| Comorbidities | ||||||
| One or more | 51 (77.3) | 31 (67.4) | 34 (63.0) | 32 (66.7) | 36 (85.7) | 0.692 |
| Chronic pulmonary disease | 14 (21.2) | 13 (28.3) | 14 (25.9) | 16 (33.3) | 6 (14.3) | 0.864 |
| Chronic heart disease | 20 (30.3) | 12 (26.1) | 9 (16.7) | 8 (16.7) | 15 (35.7) | 0.800 |
| Diabetes mellitus | 12 (18.2) | 10 (21.7) | 9 (16.7) | 10 (20.8) | 21 (50.0) | 0.003 |
| Malignancies | 10 (15.2) | 9 (19.6) | 10 (18.5) | 13 (27.1) | 10 (23.8) | 0.146 |
| Liver cirrhosis | 5 (7.6) | 2 (4.3) | 3 (5.6) | 1 (2.1) | 4 (9.5) | 0.955 |
| HIV infection | 2 (3.0) | 1 (2.2) | 2 (3.7) | 2 (4.2) | 0 (0.0) | 0.639 |
| Chronic renal failure | 4 (6.1) | 0 (0.0) | 2 (3.7) | 6 (12.5) | 4 (9.5) | 0.108 |
| Cerebrovascular disease/dementia | 11 (16.7) | 2 (4.3) | 5 (9.3) | 0 (0.0) | 6 (14.3) | 0.247 |
| Immunosuppressive therapy | 5 (7.7) | 5 (10.9) | 3 (5.6) | 7 (14.6) | 6 (14.3) | 0.217 |
| Septic shock | 19 (29.2) | 13 (28.3) | 11 (20.8) | 17 (35.4) | 11 (26.2) | 0.977 |
| Available cases | 71 | 46 | 54 | 48 | 42 | |
| 7-day mortality | 18 (25.4) | 7 (15.2) | 10 (18.5) | 7 (14.6) | 6 (14.3) | 0.127 |
| 30-day mortality | 23 (32.4) | 11 (23.9) | 11 (20.4) | 11 (22.9) | 7 (16.7) | 0.062 |
| Available cases | 18 | 29 | 53 | 48 | 44 | |
| PPV23c | 3 (16.7) | 6 (20.7) | 14 (26.4) | 21 (43.8) | 29 (65.9) | 0.519 |
Statistically significant results are highlighted in bold; SD (Standard deviation), n (number of cases), % (percentage).
p-value of first (pre-PCV) and last (late-PCV13) period comparison.
Acquisition can be nosocomial or extrahospitalary.
Includes any vaccination with the pneumococcal polysaccharide vaccine 23-valent (PPV23).
Whole genome sequencing and bioinformatics analysis
Genomic DNA was extracted (QIAamp-DNA-MiniKit, Qiagen) and quantified (Qubit, Thermo Fisher). All isolates were sequenced using Nextera XT followed by paired-end sequencing (2 × 300 bp) on a MiSeq platform (Illumina). Sequences were assembled using the INNUca v4.2 pipeline (github.com/B-UMMI/INNUca), with default parameters. The oldest genome of each major lineage (HUB-01001 for CC180-GPSC12 and HUB-01990 for CC260-GPSC83) and isolates presenting resistance genes associated with mobile genetic elements (MGEs) were long-read sequenced (MinION, ONT) and subjected to further hybrid assembly using the Unicycler pipeline.12 Reads were deposited in the European Nucleotide Archive and the metadata from all the genomes are summarised in Supplementary Table S1.
Using the Global pneumococcal Sequencing project (GPS) resources (pathogen.watch/), we grouped each genotype into one global pneumococcal sequencing cluster (GPSC). Furthermore, to contextualize our collection (n = 344), the metadata of all adult S3-IPD cases from 1994 to 2020 (n = 383) available at www.pneumogen.net/gps/gps-database-overview/(accessed December 20th 2022) were downloaded and classified according to the GPSC, ST, period and country (Supplementary Table S2). Regarding the CC180-GSPC12 lineage, a correlation between our clades and those described worldwide13,14 was performed. A phylogenetic tree including 400 genomes from Groves et al.14 (clade Ia [n = 212], clade Ib [n = 3], clade II [n = 102] and an external clade [n = 83] plus our collection [n = 344]) was constructed, as described above (Supplementary Table S3).
MLST was performed in silico using the MLST v4.2 software (github.com/tseemann/mlst) with the PubMLST database (pubmlst.org/organisms/streptococcus-pneumoniae/). Prokka v1.13.715 was used for the genome annotations and Roary16 was applied for the pan-genome analysis. A whole genome SNP-based alignment was constructed by Snippy v4.4.0 (github.com/tseemann/snippy) using the closed genomes of HUB-01001 (CC180-GPSC12) and HUB-01990 (CC260-GPSC83) as references. A phylogenetic tree was constructed by filtering recombination using Gubbins v3.017 and the data were visualised with ggtree from ggplot2 (ggplot2.tidyverse.org/) or iTol (itol.embl.de/). Genomes were considered to belong to the same clade when the number of SNPs was lower than 200. In order to assess the temporal structure of CC180 and CC260, the Bactdating (github.com/xavierdidelot/BactDating) was used.
The capsular operon of S3 was analysed using Geneious R9 (Biomatters) from wzg-wze and ugd-pgm in order to exclude transposases since short-reads sequencing is not appropriated to study these genes. The 524/62 (CR931634) capsular operon was used as the reference.
The chromosomal genes involved with antibiotic resistance (parC, gyrA, folA, folP, pbp1a, pbp2x and pbp2b) were studied using Geneious R9. The PBP alleles (pbp1a, pbp2x and pbp2b) were analysed on the Pathogenwatch website (pathogen.watch/). Acquired resistance genes were screened using the ResFinder database.18 To characterise MGEs, a BLASTn search between the MGE candidates containing resistance genes and their flanking regions was performed to compare them with previously described MGEs in public databases. The genes involved in integration, excision, mobilisation and replication were identified using a BLASTx search to compare the predicted ORFs with the NCBI NR database and to classify them following the recommendations of Ambroset et al.19 Finally, the Easyfig programme20 was used to illustrate the nucleotide sequence alignments.
Finally, the PHASTER website (phaster.ca/) was used for the first screening for bacterial prophages. Then, a classification was performed using Geneious R9 and the bacterial prophages described in Brueggemann et al.21 and Martín-Galiano et al.22
Ethics statement
This work has been approved by The Clinical Research Ethics Committee of Hospital Universitari de Bellvitge (PR065/21). This study was in accordance with the Declaration of Helsinki from the World Medical Association. Written informed consent was waived as this is an observational study with isolates obtained as part of the normal microbiological routine. According to national normative data, all the data were anonymized, and patient confidentiality was always protected following the current legal normative in Spain (LOPD 3/2018 and RD 1720/2007).
Role of the funding source
The funders of the study had no role in the study design, data collection, data analysis, data interpretation, or the writing of the report.
Results
Trends and clinical characteristics of S3-IPD
Among the 2570 IPD episodes detected over the study period (1994–2020), 270 (10.5%) were caused by serotype 3 isolates. As previously reported, we observed an increase in the overall IPD incidence from 15.2/100,000 inhabitants year in the pre-PCV period to 21.0/100,000 in the late-PCV7 period (Fig. 1). After the introduction of the PCV13, the IPD rate decreased to 12.9/100,000 (late-PCV13). When focusing on S3-IPD, its incidence remained stable after the introduction of the PCV7 (from 1.58/100,000 inhabitants year in the pre-PCV period to 1.61/100,000 in the late-PCV7 period) and showed a non-significant decreased in the late-PCV13 period (1.28/100,000).
Fig. 1.
Evolution of serotype 3 lineages that cause IPD in adults. The black line (right axis) represents the overall incidence (episodes/100,000) of IPD in adults. The bars represent the incidence of S3-IPD by periods (left axis) caused by different STs. Other-CC180 includes ST8561, ST9420 and ST18057. Other-CC260 includes ST1377, ST18059 and ST18060. Other Lineages represents isolates belonging to ST113 (GPSC50), ST458 (GPSC458), ST717 (GPSC3), ST6193 (GPSC309) and ST18058.
The analysis of the clinical characteristics of patients with S3-IPD through the whole period showed differences regarding to the clinical presentation and comorbidities, between the first (pre-PCV) and last (late-PCV13) period. However, due to the low number of cases only the increase in the proportion of patients with diabetes mellitus reach statistical significance (Table 1). In addition, the 30-day mortality (32.4%–16.7%; p = 0.062) and 7-day mortality (25.4%–14.3%; p = 0.127), usually related to the severity of the disease at the onset of infection, showed a decrease over the whole period. Over the study period, the rate of S3-IPD patients vaccinated with the 23-valent pneumococcal polysaccharide vaccine (PPV23) increased from 16.7% (pre-PCV) to 65.9% (late-PCV13) (p = 0.519). However, in many cases, the vaccine was administered at an interval greater than 5 years. Over the study period, 33 vaccine failures were detected (Supplementary Table S4) among 31 who had received the PPV23 alone, one who had received a sequential schedule with the PCV7 and PPSV23 and one who had received the PCV13 in the four years before to the onset of infection.
Changes in the lineage composition of serotype 3 pneumococci and clinical data
The analysis of 252 serotype 3 isolates obtained between 1994 and 2020 (Fig. 1) revealed the presence of two main lineages, CC180-GPSC12 (n = 107) and CC260-GPSC83 (n = 135), in addition to other minor ones grouped as other lineages: ST113 (GPSC50), ST458 (GPSC51), ST717 (GPSC3), ST6193 (GPSC309) and ST18058 (not assigned). A lineage shift was observed during the study period. The incidence of disease due to CC260-GPSC83 decreased from 1.18/100,000 in the pre-PCV period to 0.55/100,000 in the late-PCV13 period (p < 0.05), whereas disease due to CC180-GPSC12 increased from 0.38/100,000 to 0.67/100,000 (p = 0.06). Within CC260-GPSC83, disease caused by ST1220 progressively increased, accounting for nearly half of the cases of this lineage in the late-PCV13 period. Globally, the GPS metadata (Supplementary Table S2) showed that 54.8% of the available serotype 3 genomes belonged to CC180-GPSC12, while 8.6% belonged to CC260-GPSC83. GPSC12 was highly prevalent in North America (95.8%), Oceania (85.7%), Europe (75.5%) and South America (69.8%), whereas Asia and Africa presented more genetic variability (GPSC12; 46.8% and 12.5%, respectively). GPSC83 was present in most continents, but the frequency was higher in Europe (17.0%) and Africa (13.5%).
There were no significant differences in the clinical characteristics of the episodes between the CC180-GPSC12 and CC260-GPSC83 lineages (Supplementary Table S5). Nevertheless, pneumonia was more frequent among the episodes caused by CC260-GPSC83 (90.5% vs 82.3%; p = 0.062), while meningitis was more common in those caused by CC180-GPSC12 (5.8% vs 10.6%; p = 0.241). Early mortality (19.7% vs 18.2%; p = 0.869) and 30-day mortality (27.3% vs 22.7%; p = 0.459) were also similar between the groups. Among the vaccine failures, 54.5% were episodes caused by CC260-GPSC83 (n = 18), and 36.4% were due to CC180-GPSC12 (n = 12) (Supplementary Table S4).
Phenotypic and genetic features of the serotype 3 lineages
The capsular operon of serotype 3 showed a low number of polymorphic sites, a total of 28 in the wgz-wze region and 45 in the ugd-pgm region. Most of them (n = 35) were sporadic variations present in a few genomes, 13 were polymorphisms present in all the genomes compared to the references, and 21 differentiated the capsular operon of the two major lineages. No differences were found between S3-ST260 and S3-ST1220 (Supplementary Table S6A). The N57K and N375H amino acid replacements in Ugd, A341T and H367N in WchE and E58G in GalU were present in all the isolates, whereas other minor substitutions were present in a single isolate (Supplementary Table S6B).
All serotype 3 isolates were uniformly susceptible to penicillin (MIC ≤0.03 mg/L), amoxicillin (MIC ≤0.03 mg/L) and cefotaxime (MIC ≤0.06 mg/L) regardless of genotype or PBP pattern. The activity of the remaining antibiotics tested was above 95%. The exception was tetracycline which showed a 92.9% of susceptibility rate among isolates of GPSC12 (Supplementary Table S8). Among the isolates harboring resistance determinants, 20 tetracycline-resistant isolates carried the tet(M) gene, 9 presented resistance to chloramphenicol through the expression of cat and 8 exhibited resistance to macrolides via the expression of erm(B). Three different transposon families and eight structures (Supplementary Figure S2) were found: the Tn916 family (Acc. num. U09422.1) (structures 1 and 2), the Tn1549 family (structure 3) and the Tn5252 family (Acc. num. EU351020.1) (structures 4, 5, 6, 7 and 8). One isolate presented resistance to trimethoprim/sulfamethoxazole and another three were not susceptible to quinolones (Supplementary Table S7).
An in-depth analysis of 170 CC180-GPSC12 genomes showed six major clades (named 180–1 to 180–6), with a maximum of 5468 SNPs when comparing with the oldest genome of our series (HUB-01001) (Fig. 2). Temporal structure analysis using Bactdating did not find a good temporal signal. Nevertheless, clade 180–6 was dominant throughout the whole study period (n = 140), followed by the clades 180–5 (n = 8), 180–3 (n = 7), 180–1 (n = 6), 180–4 (n = 5) and 180–2 (n = 4). The phylogenetic tree presented in Supplementary Figure S3 reveals that the previously described13,14 clade Ia includes clade 180–6, clade Ib contains 180–1, 180–2 and 180–3, and clade II comprises 180–4 and 180–5. Ninety-four CC180-GPSC12 isolates carried phages and their prevalence was clade-specific. A total of 83 genomes from clade 180–6 (59.2%) had the IPP57 phage (Acc. num. KY065494) (Brueggemann et al.)21 also known as ΦOXC141, while one isolate had the IPP58 phage (Acc. num. KY065495). Five genomes from clade 180 to 5 had the IPP19 prophage (Acc. num. KY065460), 2 genomes from clade 180 to 4 had the PPH020_2 prophage (Acc. num. NZ_LR216031.1) and 3 genomes from 180 to 2 had the PPH115_1 phage (Acc. num. NZ_LR216034.1).22 Finally, we did not find phages in the isolates from clades 180–1 to 180–3. In all cases, phages of the same type were inserted into the same location in the genome. In general, the presence of MGEs in this lineage was rare (n = 12). Six isolates from clade 180 to 5 had the Tn916 transposon (structure 2), carrying the tet(M) gene, while 2 had the integrative and conjugative element belonging to the Tn5252 family (structure 7) that carried both, tet(M) and erm(B). The remaining 4 harbored Tn5252, with 2 isolates from 180 to 6 containing structure 4, and 2 isolates from clade 180 to 4 containing structure 8 (Supplementary Figure S2).
Fig. 2.
Genetic diversity of CC180-GPSC12. Heatmap displaying differences in the SNPs, using the oldest isolate HUB-01001 as a reference. The phylogenetic tree groups genomes into different clades based on the SNPs, with their origins shown as black triangles in the tree. Circles at the end of the branches represent hospitals. The other information displayed includes the ST (shown as squares) and the period, as well as the presence of transposons and phages (shown as dots).
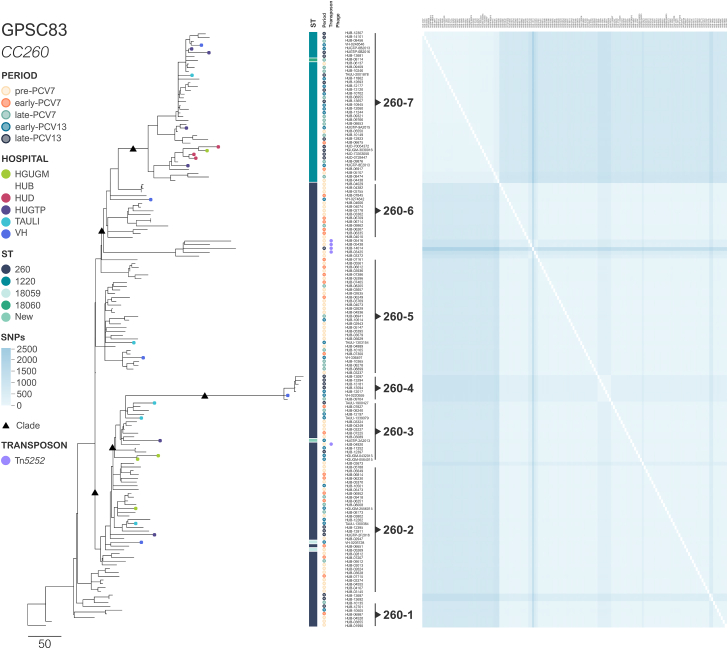
Isolates from the second lineage, CC260-GPSC83, had a lower number of SNPs (2126 SNPs) than CC180-GPSC12 ones (Fig. 3). As in CC180-GPSC12 a good temporal signal was not obtained. However, seven different clusters were detected among the 164 genomes, most being ST260 and sporadic ST isolates (termed here clades 260–1 to 260–6) while the remaining one was related to ST1220 isolates (260–7). No phages were found in this lineage and the presence of MGEs was also sporadic and associated with Tn5252. Three genomes from the clades 260–5 (n = 1) to 260–6 (n = 2) had structure 5, whereas one isolate from clade 260 to 3 had structure 3 (Supplementary Figure S2).
Fig. 3.
Genetic diversity of CC260-GPSC83. Heatmap displaying differences in the SNPs, using the oldest isolate HUB-01990 (1991) as a reference. The phylogenetic tree groups genomes into different clades based on the SNPs, with their origins shown as black triangles in the tree. Circles at the end of the branches represent hospitals. The other information displayed includes the ST (shown as squares) and the period, as well as the presence of transposons and phages (shown as dots).
Regarding the minor lineages, four CC458-GPSC51 genomes (n = 6) contained the PPH060 prophage (Acc. num. NZ_LR216035.1) and one of them also had a Tn1549 transposon (structure 3). CC717-GPSC3, which is frequently associated with serotype 33F, consisted of a single isolate harbouring Tn6002 [erm(B)] (Acc. num. AY898750.1), with the IPP28 phage (Acc. num. KY065469) located upstream of the transposon.
Genomes of the serotype 3 isolates from different parts of the country were spread along the phylogenetic trees of both lineages, without the presence of any clusters that would suggest clonal differences between the Spanish regions.
The pan-genome analysis highlighted the differences between the two lineages (Fig. 4). Regarding the relationship between the total and conserved genes (Fig. 4a), CC180-GPSC12 had a higher total gene count (2286 vs 2061 genes) and a more open genome compared to CC260-GPSC83, resulting in fewer conserved genes content (76.2% vs 88.5%). These differences also correlated with those observed in Fig. 4c, where the line representing the non-conserved genes was higher for CC180-GPSC12. Furthermore, when this information was stratified per lineage (Fig. 4b), CC180-GPSC12 had more accessory genomes than CC260-GPSC83 (22.8% vs 11.4%), representing almost a quarter of the total compared to the 10.0% for CC260-GPSC83.
Fig. 4.
Analysis of the overall serotype 3 pangenome and major lineages. (a) The X-axis shows the number of genomes included in the analysis, while the Y-axis presents the number of genes in the pan-genome analysis for each added genome. The squares display the conserved genes of the genome, whereas the dots represent the accessory genes. (b) Representation of the pan-genome according to the core genome (in dark green), the soft-core genome (in light green), the shell genome (in orange) and the cloud genome (in beige). The grey line represents the core genome (core plus soft-core genome) and the dark purple line represents the accessory genome (shell plus cloud genome). (c) Differences in the genes between the conserved genome and the accessory genome of GPSC12 (red line) and GPSC83 (blue line) are shown.
Discussion
In this study, we analysed S3-IPD cases over a 30-year period in which different PCVs were licensed for use in Spain. Our results show that the incidence of S3-IPD remained stable after PCV13 introduction, with no observable herd protection. This has also been described in other countries, suggesting that the vaccination of children is not enough to achieve protection in adults. This is probably due to the low impact of the PCV13 in carriage that is the basis for herd protection.14,23,24 The pneumococcal diseases caused by S3 are associated with a high risk of death,2 severe disease,4 and long-term adverse effects.5 In our study, a non-significant decline in the 30-day mortality rates over the study period was observed, probably reflecting the progressive improvement in the management of pneumococcal diseases such as meningitis and pneumonia.25 In addition, we observed similar mortality rates between the two major lineages, CC180-GPSC12 and CC260-GPSC83, highlighting the fact that the type 3 capsular polysaccharide is the main factor associated with disease fatality. A change in the patient population over the study period was observed, with progressive increases in the mean age of patients and the presence of comorbidities (especially diabetes), as observed previously, and a decrease in the prevalence of pneumonia.2 This could be linked to an overall increase in life expectancy and underscores the need to protect these populations that are at risk of IPD. However, this comparison could yield significant results with a larger population sample, thereby increasing the statistical power, or applying a better alternative that could be analysing within a multiple imputation framework.
Antibiotic resistance has been always low among S3 isolates, despite the presence of MGEs.13 We did not find resistance to beta-lactam antibiotics, while resistance to other antibiotics was low. Furthermore, acquired resistance through MGEs was linked to the Tn916 family of transposons that conferred resistance to a single or multiple antibiotics.13 Interestingly, despite the high macrolide resistance rates in Spain and the high antibiotic consumption rates (especially long-acting ones),26 these MGEs had not spread throughout these S3 lineages, highlighting the stability of their genomes.
Most studies analysing the genomic composition of invasive serotype 3 pneumococci have highlighted its clonal homogeneity, with a worldwide dominance of CC180-GPSC12. In our setting, the CC260-GPSC83 lineage was gradually replaced by CC180-GPSC12, especially in the early-PCV13 period. This replacement has also been observed in Portugal,27 a country where CC260-GPSC83 was also an important lineage. Likewise in Mexico,28 after PCV13 introduction, CC180-GPSC12 progressively replaced CC260-GPSC83. In addition to the capsule, other factors could enhance the vaccine resistance of CC180-GPSC12 pneumococci. Some studies have highlighted the progressive increase in the prevalence of clade II within CC180-GPSC12 lineage. This clade is associated with higher antibiotic non-susceptibility rates and a lower prophage content compared to clade Ia.13,14 In our study, clade Ia was dominant in all the periods, while an increase in clade II was not observed. Nevertheless, in agreement with previous studies,13,29 our clade Ia genomes (180–6) had higher rates of integrated ΦOXC141 phages when compared to clade II (clade 180–4 and clade 180–5), which was more likely to have antibiotic resistance, and clade Ib (180–1 to 180–3), which has few phages and little resistance. The strict conservation of gene vicinity indicates that the integration of this prophage occurred just once in the history of the lineage. The presence of the ΦOXC141 prophage in GPSC12-CC180 was associated with the inhibition of transformation, which could explain the higher diversity of the CPSC12-CC180 genomes from clade 180–1 to 180–4 (without prophages) that was observed in our study when compared to the clades 180–5 and 180–6 (mainly with a prophage).13 The lower prevalence of this lineage before PCVs introduction in our setting and the existence of other dominant lineages could explain these differences. On the other hand, little is known about the genomics of the CC260-GPSC83 lineage. This lineage showed greater genetic homogeneity with few differences from the oldest isolate, a reduced pan-genome and an absence of phages. We hypothesise that this could be explained by the narrower spread of this lineage compared to that of CC180-GPSC12.
Our genomic analysis is a significant strength in the study as it contributes to enhance our understanding of the evolution and diversification of pneumococcal serotype 3 lineages using a large collection of isolates from across the country. Moreover, we have a large collection of isolates from before the introduction of the PCVs. The main limitation of our study is that the overall analysis of both clinical and genetic data could only be conducted at a single centre. Nevertheless, the analysis of both the clinical and genetic characteristics of this serotype over an extended period compensates for this limitation, thereby adding strength to our study.
In conclusion, the number of adult S3-IPD cases barely decreased in Spain, despite the introduction of the PCV13 in children. The progressive decline in mortality rates could be rather linked to an improvement in clinical patient management. Despite the clonal shift towards a higher prevalence of CC180-GPSC12, the two main lineages presented high internal genetic homology without any noticeable clinical difference. Nevertheless, these results should be confirmed with other studies. But, this observation, together with the similarity of the capsular operon, highlights the role of the serotype 3 capsule in IPD. Continuous surveillance of this serotype will be needed to support this hypothesis, along with the introduction of new PCVs that aim to prevent pneumococcal pneumonia in adults.
Contributors
SCS, AGD, IG, JMM, EC, MDQ, AC, NL, JY, DB, MA, FT, ICJ, AJMG, MAD, SM, JL, RP, JC, CA.
CA, AGD conceptualised, designed, and supervised the study. IG, JMM, EC, MDQ, AC, NL, JY, MA, FT, MAD, JL, RP contributed to collect IPD cases and clinical characteristics. SCS, AGD, ICJ, SM, SB designed and performed the whole genome sequencing approaches. SCS, AGD, JC, DB, CA conducted the final analysis of data. SCS, AGD, CA verified the data of the study and wrote the original draft of the manuscript and had full access to all the data in the study. All authors participated in manuscript review and editing, and had final responsibility for the decision to submit for publication.
Data sharing statement
The data supporting the findings of this study are available within the paper and its Supplementary materials. Genomes are available for downloading through the accession number provided in the dataset (see Supplementary materials). Additionally, the de-identified raw data are available upon reasonable request from the corresponding author c.ardanuy@bellvitgehospital.cat.
Declaration of interests
C.A. has been a scientific adviser for, and/or has received research funding from, Merck Sharp & Dohme Corp and Pfizer. J.Y. has been a scientific adviser for, and/or has received research funding from, Merck Sharp & Dohme Corp, Pfizer and GSK. All other authors declare that they have no conflicts of interest regarding this research.
Acknowledgements
This study has been funded by Instituto de Salud Carlos III (ISCIII) through the grant “PI18/00339” and “PI21/01000”, “INT22/00096” (co-funded by European Social Fund), CIBER de Enfermedades Respiratorias (CIBERES-CB06/06/0037) and CIBER de Enfermedades Infecciosas (CIBERINFEC-CB21/13/00009). SCS was supported by a PFIS predoctoral grant number FI22/00279. The study is partially supported by an Investigator Study Program of MSD grant (IISP 60168). The opinions expressed in this manuscript are from the authors who did not represent those of MSD.
We would like to thank to the Microbiology Departments of Hospital Donostia, Hospital General Universitario Gregorio Marañón, Hospital Universitari Germans Trias i Pujol, Hospital Universitari Parc Taulí and Hospital Universitari Vall d’Hebrón, and the staff of the Microbiology Laboratory of Hospital Universitari de Bellvitge who with their daily work contributed to this project. We thank also the CERCA Programme/Generalitat de Catalunya for institutional support.
Footnotes
Translation: For the Spanish translation of the abstract see Supplementary Materials section.
Supplementary data related to this article can be found at https://doi.org/10.1016/j.lanepe.2024.100913.
Contributor Information
Aida González-Díaz, Email: agonzalezd@bellvitgehospital.cat.
Carmen Ardanuy, Email: c.ardanuy@bellvitgehospital.cat.
Appendix A. Supplementary data
References
- 1.Wang H., Naghavi M., Allen C., et al. Global, regional, and national life expectancy, all-cause mortality, and cause-specific mortality for 249 causes of death, 1980–2015: a systematic analysis for the global burden of disease study 2015. Lancet. 2016;388:1459–1544. doi: 10.1016/S0140-6736(16)31012-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Grau I., Ardanuy C., Cubero M., et al. Declining mortality from adult pneumococcal infections linked to children's vaccination. J Infect. 2016;72:439–449. doi: 10.1016/j.jinf.2016.01.011. [DOI] [PubMed] [Google Scholar]
- 3.Luck J.N., Tettelin H., Orihuela C.J. Sugar-coated killer: serotype 3 pneumococcal disease. Front Cell Infect Microbiol. 2020;10:1–11. doi: 10.3389/fcimb.2020.613287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Garcia-Vidal C., Ardanuy C., Tubau F., et al. Pneumococcal pneumonia presenting with septic shock: host- and pathogen-related factors and outcomes. Thorax. 2010;65:77–81. doi: 10.1136/thx.2009.123612. [DOI] [PubMed] [Google Scholar]
- 5.Africano H., Serrano-Mayorga C., Ramirez-Valbuena P., et al. Major adverse cardiovascular events during invasive pneumococcal disease are serotype dependent. Clin Infect Dis. 2021;72:711–719. doi: 10.1093/cid/ciaa1427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nelson A.L., Roche A.M., Gould J.M., et al. Capsule enhances pneumococcal colonization by limiting mucus-mediated clearance. Infect Immun. 2007;75:83–90. doi: 10.1128/IAI.01475-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sings H.L., De Wals P., Gessner B.D., et al. Effectiveness of 13-valent pneumococcal conjugate vaccine against invasive disease caused by serotype 3 in children: a systematic review and meta-analysis of observational studies. Clin Infect Dis. 2019;68:2135–2143. doi: 10.1093/cid/ciy920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.van der Linden M., Imöhl M., Perniciaro S. Limited indirect effects of an infant pneumococcal vaccination program in an aging population. PLoS One. 2019;14:1–19. doi: 10.1371/journal.pone.0220453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Domínguez Á., Ciruela P., Hernández S., et al. Effectiveness of the 13-valent pneumococcal conjugate vaccine in preventing invasive pneumococcal disease in children aged 7-59 months. A matched case-control study. PLoS One. 2017;12:1–15. doi: 10.1371/journal.pone.0183191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andrews N., Waight P., Burbidge P., et al. Serotype-specific effectiveness and correlates of protection for the 13-valent pneumococcal conjugate vaccine: a postlicensure indirect cohort study. Lancet Infect Dis. 2014;14:839–846. doi: 10.1016/S1473-3099(14)70822-9. [DOI] [PubMed] [Google Scholar]
- 11.González-Díaz A., Càmara J., Ercibengoa M., et al. Emerging non-13-valent pneumococcal conjugate vaccine (PCV13) serotypes causing adult invasive pneumococcal disease in the late-PCV13 period in Spain. Clin Microbiol Infect. 2020;26:753–759. doi: 10.1016/j.cmi.2019.10.034. [DOI] [PubMed] [Google Scholar]
- 12.Wick R., Judd L., Gorrie C., et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13:1–22. doi: 10.1371/journal.pcbi.1005595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Azarian T., Mitchell P., Georgieva M., et al. Global emergence and population dynamics of divergent serotype 3 CC180 pneumococci. PLoS Pathog. 2018;14 doi: 10.1371/journal.ppat.1007438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Groves N., Sheppard C.L., Litt D., et al. Evolution of Streptococcus pneumoniae serotype 3 in England and Wales: a major vaccine evader. Genes. 2019;10:845–856. doi: 10.3390/genes10110845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 16.Page A., Cummins C., Hunt M., et al. Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics. 2015;31:3691–3693. doi: 10.1093/bioinformatics/btv421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Croucher N., Page A., Connor T., et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43 doi: 10.1093/nar/gku1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zankari E., Hasman H., Cosentino S., et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67:2640–2644. doi: 10.1093/jac/dks261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ambroset C., Coluzzi C., Guédon G., et al. New insights into the classification and integration specificity of Streptococcus integrative conjugative elements through extensive genome exploration. Front Microbiol. 2016;6:1–21. doi: 10.3389/fmicb.2015.01483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sullivan M., Petty N., Beatson S. Easyfig: a genome comparison visualizer. Bioinformatics. 2011;27:1009–1010. doi: 10.1093/bioinformatics/btr039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brueggemann A., Harrold C., Rezaei R., et al. Pneumococcal prophages are diverse, but not without structure or history. Sci Rep. 2017;7:1–13. doi: 10.1038/srep42976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martín-Galiano A., García E. Streptococcus pneumoniae: a plethora of temperate bacteriophages with a role in host genome rearrangement. Front Cell Infect Microbiol. 2021;11:1–21. doi: 10.3389/fcimb.2021.775402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Horácio A., Silva-Costa C., Lopes J., et al. Serotype 3 remains the leading cause of invasive pneumococcal disease in adults in Portugal (2012-2014) despite continued reductions in other 13-valent conjugate vaccine serotypes. Front Microbiol. 2016;7:1616. doi: 10.3389/fmicb.2016.01616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Subramanian R., Liyanapathirana V., Barua N., et al. Persistence of pneumococcal serotype 3 in adult pneumococcal disease in Hong Kong. Vaccines (Basel) 2021;9:756–767. doi: 10.3390/vaccines9070756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ruiz L., Zalacain R., Capelastegui A., et al. Bacteremic pneumococcal pneumonia in elderly and very elderly patients. J Gerontol A Biol Sci Med Sci. 2014;69:1018–1024. doi: 10.1093/gerona/glt288. [DOI] [PubMed] [Google Scholar]
- 26.Berbel D., González-Díaz A., López de Egea G., Camara J., Ardanuy C. An overview of macrolide resistance in streptococci: prevalence, mobile elements and dynamics. Microorganisms. 2022;10:2316. doi: 10.3390/microorganisms10122316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horácio A., Silva-Costa C., Diamantino-Miranda J., et al. Population structure of Streptococcus pneumoniae causing invasive disease in adults in Portugal before PCV13 availability for adults: 2008-2011. PLoS One. 2016;11:2008–2011. doi: 10.1371/journal.pone.0153602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Echániz-aviles G., Guerreiro S., Silva-costa C., et al. Streptococcus pneumoniae serotype 3 in Mexico (1994 to 2017): decrease of the unusual clonal complex 4909 lineage following PCV13 introduction. Am Soc Microbiol. 2019;57:e01354–e01358. doi: 10.1128/JCM.01354-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kwun M., Ion A., Cheng H., et al. Post-vaccine epidemiology of serotype 3 pneumococci identifies transformation inhibition through prophage-driven alteration of a non-coding RNA. Genome Med. 2022;14:1–23. doi: 10.1186/s13073-022-01147-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.