Summary
Lynch syndrome (LS) is the most common hereditary cancer syndrome. Heterozygous loss-of-function variants in PMS2 are linked to LS. While these variants are not directly cancer causing, reduced PMS2 function results in the accumulation of somatic variants and increased cancer risk over time due to DNA mismatch repair dysfunction. It is reasonable that other types of genetic variation that impact the expression of PMS2 may also contribute to cancer risk. The Kozak sequence is a highly conserved translation initiation motif among higher eukaryotes and is defined as the nine base pairs upstream of the translation start codon through the first four bases of the translated sequence (5’-[GTT]GCATCCATGG-3’; human PMS2: NM_000535.7). While Kozak sequence variants in PMS2 have been reported in ClinVar in patients with suspected hereditary cancer, all variants upstream of the translation start site are currently classified as variants of undetermined significance (VUSs). We hypothesized that variants significantly disrupting the Kozak sequence of PMS2 would decrease PMS2 protein expression, contributing to increased cancer risk over time. Using a dual-luciferase reporter plasmid and site-directed mutagenesis, we generated the wild-type human PMS2 and the ClinVar VUSs within the PMS2 Kozak sequence. Besides the c.1A>C variant, which is already known to be pathogenic, we implicate six additional variants as American College of Medical Genetics and Genomics (ACMG)/Association for Molecular Pathology (AMP) pathogenic supporting (PP) variants and classify ten as benign supporting (BP). In summary, we present a method developed for the classification of human PMS2 Kozak sequence variants that can contribute to the re-classification of VUSs identified in patients.
Keywords: Lynch syndrome, hereditary cancer, cancer risk, non-coding variation, Kozak sequence, PMS2
Graphical abstract

This study investigates the role of 5′ UTR PMS2 Kozak sequence genetic variation in cancer risk. It accomplishes this through the development of a mid-throughput reporter assay where variants can be tested for protein translation efficiency. The results highlight the importance of continued study of the Kozak sequence related to human disease.
Main text
Lynch syndrome (LS; MIM: 120435, 609310, 613244, 614337, and 614350) affects 1 in 279 people, making it the most common hereditary cancer syndrome.1 LS is caused by germline loss-of-function mutations in the DNA mismatch repair (MMR) genes MLH1 (MIM: 120436), MSH2 (MIM: 609309), MSH6 (MIM: 600678), and PMS2 (MIM: 600259).1 While these variants do not directly initiate cancer, reduced MMR activity results in the accumulation of somatic variants and thus increases cancer risk over time. Heterozygous variants in PMS2 that reduce the expression of the PMS2 protein or decrease its function are linked to LS type 4 (MIM: 614337). LS type 4 indicates LS caused by mutations in PMS2 as opposed to the other three LS genes mentioned above. However, the vast majority of PMS2 clinical sequencing efforts focus only on the coding sequence. As more is learned about the structure and function of the human genome, additional non-coding regulatory mechanisms (e.g., enhancer elements) that contribute to gene/protein expression are being discovered.2 This includes the “Kozak sequence,” a nucleotide motif lying immediately upstream of translation start sites that was first described by Marilyn Kozak in 1984.3,4 The Kozak sequence consists of 10–13 nucleotides spanning the gene translation initiation AUG codon (5’-[GTT]GCATCCATGG-3’; human PMS2 [hPMS2] NCBI: NM_000535.7) and is a highly conserved protein translation initiation motif among eukaryotes. The importance surrounding the Kozak bases −7 to −9 (in brackets above) is currently unclear. It is thought that the Kozak sequence contributes to slowing scanning 40S ribosomes at the appropriate mRNA start codon (generally the closest to the 5′ cap) allowing for pairing of the Met-tRNAi with the AUG start sequence. This then allows for recruitment of the 60S ribosome to initiate translation. However, the exact mechanism by which the Kozak sequence promotes translation initiation remains elusive. It is possible that the 40S ribosomal complex directly interacts with the sequence slowing down scanning. Additionally, it has been proposed that the Kozak sequence contributes to secondary mRNA hairpin structures that pause 40S ribosome scanning and promote translation initiation.5
Mutations in the Kozak sequence can cause alterations in the translation efficiency of a gene; this has been largely attributed to the sequence’s ability to slow down the scanning ribosome.5 Some studies have shown that even modest changes to translation efficiency can lead to severe consequences.6 Specific Kozak sequence variants have been shown to reduce translation in genes necessary for heart development, resulting in cases of congenital heart disease.7 Recent studies have shown that the CD40 (MIM: 190535) Kozak sequence variant c.–1C>T is associated with acute coronary syndrome8 and Graves’ disease,9 while the c.–5C>T variant in WWOX (MIM: 605131) has been implicated in oral cancer risk.10 Additionally, in one study, a mutation at the c.–6G position found in the β-globin (MIM: 141900) Kozak sequence resulted in a 30% decrease in translation and contributed to an overall increased risk for β-thalassemia.11 Another study reported this same mutation in the Kozak sequence of GATA4 (MIM: 600576), which contributed to the pathogenesis of atrial septal defects.7
All non-coding Kozak sequence variants upstream of the c.+1A in PMS2 in ClinVar are currently classified as variants of undetermined significance (VUSs) or likely benign.12 This is likely due to the paucity of clinical research on this non-coding region. The declining costs and increasing availability of whole-genome sequencing continue to highlight our significant lack of understanding in how non-coding regions contribute to gene expression and disease biology. Current limitations include (1) the lack of functional and/or computational data for classification rubrics, (2) sequencing challenges in C-G-rich regions (e.g., promoters), and (3) limited annotation of non-coding regions.13,14 Indeed, most of the recent Kozak sequence studies have centered around “optimizing” the sequence as a tool to increase translation efficiency, with little focus on mutation classification or investigations of translation deficiency.15,16,17,18 We hypothesized that variants significantly disrupting the Kozak sequence of PMS2 would decrease PMS2 protein expression, contributing to increased cancer risk over time. To test this hypothesis, we developed a reporter assay for the classification of hPMS2 Kozak sequence variants identified in patients.
We used the psiCHECK-2 dual-luciferase reporter vector (Promega, Madison, WI, USA) as the backbone for all experiments. This vector was chosen because it contains both a Renilla luciferase and an independently transcribed firefly luciferase gene. Using both allows one locus to be genetically modified while the other one remains genetically fixed, resulting in the ability to normalize transfection efficiency within each independent experiment. In this plasmid, both the Renilla and firefly luciferase Kozak sequences are defined as 5′-GCCACCATGG-3′. The Q5 Site-Directed Mutagenesis (SDM) Kit (New England Biolabs, Ipswich, MA, USA) was used to modify the psiCHECK-2 Renilla Kozak sequence to the hPMS2 (NCBI: NM_000535.7) sequence: 5′-[GTT]GCATCCATGG-3′ (Tables 1 and 2) per the manufacturer’s protocol. It is notable that the PMS2 Kozak sequence differs from the accepted eukaryotic consensus sequence 5′-GCC(A/G)CCATGG-3′ at the −4 and −3 positions. The −3 position has been shown to be highly conserved,19 and a T at this position in other gene contexts has been shown to significantly decrease translation efficiency.20 However, we found no significant difference (p = 0.6; two-tailed Student’s t test; data not shown) between the translation efficiency of the hPMS2 Kozak sequence and the endogenous plasmid sequence (i.e., the conserved eukaryotic sequence). The same SDM protocol was used to generate each of the 5′ UTR Kozak sequence variants that have been reported in NCBI ClinVar12 (Tables 1 and 2). Further, we generated a positive control mutant, c.1A>C. The c.1A>C mutation has multiple pathogenic submissions in ClinVar12 and is predicted to result in loss of the translation start codon, causing reduced protein expression.21 This specific mutation is linked to LS presentations including, but not limited to, hereditary nonpolyposis colorectal neoplasms and hereditary breast and ovarian cancer.12 Finally, we generated a c.−6G>C mutation that has not been reported in the context of PMS2 in ClinVar, but the −6 position is known to be important in translation initiation. We hypothesized that this variant is disruptive based on its association with other gene/disease contexts as mentioned previously.7,11
Table 1.
Primer list
| Primer name | Sequence (5′→3′) |
|---|---|
| Univ_F | CCATGGCTTCCAAGGTGTACGACC |
| WT_R | ATGCAACCCTATAGTGAGTCGTATTAAG |
| psiCHECK2_seq_F | TTCTCTCCACAGGTGTCCAC |
| Kozak_Var01_F | GGTTGCATCCCTGGCTTCCAA |
| Kozak_Var02_F | GGGTTGCATCGATGGCTTCCA |
| Kozak_Var03_F | GGGTTGCATCTATGGCTTCCA |
| Kozak_Var04_F | GGGTTGCATCAATGGCTTCCA |
| Kozak_Var05_F | AGGGTTGCATGCATGGCTTCC |
| Kozak_Var06_F | AGGGTTGCATACATGGCTTCC |
| Kozak_Var07_F | AGGGTTGCATTCATGGCTTCC |
| Kozak_Var08_F | TAGGGTTGCAGCCATGGCTTC |
| Kozak_Var09_F | ATAGGGTTGCGTCCATGGCTT |
| Kozak_Var10_F | TATAGGGTTGGATCCATGGCT |
| Kozak_Var11_F | TATAGGGTTGAATCCATGGCTTC |
| Kozak_Var12_F | CTATAGGGTTACATCCATGGC |
| Kozak_Var13_F | TCACTATAGGCTTGCATCCATG |
| Kozak_Var14_F | ATCCATGGCTTCCAAGGT |
| Kozak_Var15_F | CATCCATGGCTTCCAAGG |
| Kozak_Var16_F | CTATAGGGTTCCATCCATGGC |
| Kozak_Var17_F | ACTATAGGGTCGCATCCATGG |
| Kozak_Var01_R | CTATAGTGAGTCGTATTAAGTAC |
| Kozak_Univ01_R | TATAGTGAGTCGTATTAAGTACTCTAG |
| Kozak_Var05_R | ATAGTGAGTCGTATTAAGTACTCTAGC |
| Kozak_Univ02_R | ATAGTGAGTCGTATTAAGTACTCTAG |
| Kozak_Var08_R | TAGTGAGTCGTATTAAGTACTCTAGC |
| Kozak_Var09_R | AGTGAGTCGTATTAAGTACTCTAG |
| Kozak_Univ03_R | GTGAGTCGTATTAAGTACTCTAG |
| Kozak_Univ04_R | TGAGTCGTATTAAGTACTC |
| Kozak_Var13_R | GTCGTATTAAGTACTCTAGC |
| Kozak_Var14_R | ACCCTATAGTGAGTCGTATTAAG |
| Kozak_Var15_R | CCCTATAGTGAGTCGTATTAAG |
| Kozak_Var17_R | GAGTCGTATTAAGTACTCTAG |
Table 2.
Site-directed mutagenesis specifications
| Variant name | Sequence changea | Fwd. primer | Rev. primer | Annealing temperature (°C) |
|---|---|---|---|---|
| Kozak_hPMS2 | N/A | Univ_F | WT_R | 59 |
| Kozak_Var01 | c.1A>C | Kozak_Var01_F | Kozak_Var01_R | 60 |
| Kozak_Var02 | c.−1C>G | Kozak_Var02_F | Kozak_Univ01_R | 63 |
| Kozak_Var03 | c.−1C>T | Kozak_Var03_F | Kozak_Univ01_R | 63 |
| Kozak_Var04 | c.−1C>A | Kozak_Var04_F | Kozak_Univ01_R | 63 |
| Kozak_Var05 | c.−2C>G | Kozak_Var05_F | Kozak_Var05_R | 65 |
| Kozak_Var06 | c.−2C>A | Kozak_Var06_F | Kozak_Univ02_R | 63 |
| Kozak_Var07 | c.−2C>T | Kozak_Var07_F | Kozak_Univ02_R | 63 |
| Kozak_Var08 | c.−3T>G | Kozak_Var08_F | Kozak_Var08_R | 65 |
| Kozak_Var09 | c.−4A>G | Kozak_Var09_F | Kozak_Var09_R | 63 |
| Kozak_Var10 | c.−5C>G | Kozak_Var10_F | Kozak_Univ03_R | 61 |
| Kozak_Var11 | c.−5C>A | Kozak_Var11_F | Kozak_Univ03_R | 61 |
| Kozak_Var12 | c.−6G>A | Kozak_Var12_F | Kozak_Univ04_R | 57 |
| Kozak_Var13 | c.−9G>C | Kozak_Var13_F | Kozak_Var13_R | 57 |
| Kozak_Var14 | c.−7_−5del | Kozak_Var14_F | Kozak_Var14_R | 63 |
| Kozak_Var15 | c.−8_−6del | Kozak_Var15_F | Kozak_Var15_R | 61 |
| Kozak_Var16 | c.−6G>C | Kozak_Var16_F | Kozak_Univ04_R | 57 |
| Kozak_Var17 | c.−7T>C | Kozak_Var17_F | Kozak_Var17_R | 58 |
Fwd., forward; Rev., reverse.
Annotations on NM_000535.7(PMS2).
Subsequently, all SDM plasmids were individually transformed into 5-alpha chemically competent E. coli cells (Invitrogen, Waltham, MA, USA) and Sanger verified following miniprep extraction (Invitrogen) using the psiCHECK2_seq_F primer (Table 1) at Genewiz/Azenta Life Sciences (South Plainfield, NJ, USA). Plasmids used for the in vitro assay were maxiprepped (Invitrogen) and quantified using the Qubit dsDNA BR Assay Kit (Thermo Fisher Scientific, Waltham, MA, USA). All SDM primers were designed using NEBaseChanger v.1.3.3, and oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA, USA).
HEK293T cells were maintained in media containing DMEM (4.5 g/L glucose, L-glutamine, and sodium pyruvate; NY/Corning, Corning, NY, USA) with 10% fetal bovine serum (Gibco, Grand Island, NY, USA/Corning) and 1× penicillin-streptomycin (Gibco). Cells were seeded at 4 × 104 cells per well in a 24-well plate with 1 mL of culture media 24 h before transfection. Experiments were plated in three biological replicates. Following a medium change, cells were transfected with plasmid complexed with PolyJet In Vitro DNA Transfection Reagent (SignaGen Laboratories, Rockville, MD, USA) per the manufacturer’s protocol. Media were changed again 24 h after transfection. Forty-eight hours after transfection, cells were mechanically dissociated from the well bottom, 50 μL of cells were removed from each well in triplicate to an opaque, white 96-well plate (PerkinElmer, Waltham, MA, USA), and luciferase expression data were collected using the Dual-Glo Luciferase Assay (Promega) per the manufacturer’s protocol on a BioTek Synergy LX (Agilent, Santa Clara, CA, USA). Kozak sequence translation efficiency was calculated as the proportion of Renilla luciferase/firefly luciferase expression per well.16 The data were normalized to the hPMS2 wild-type (WT) plasmid. In vitro assay results were compared across genotypes using a one-way ANOVA test followed by post hoc Fisher’s least significant difference test compared to the hPMS2 sequence on GraphPad Prism v.9.4.0.
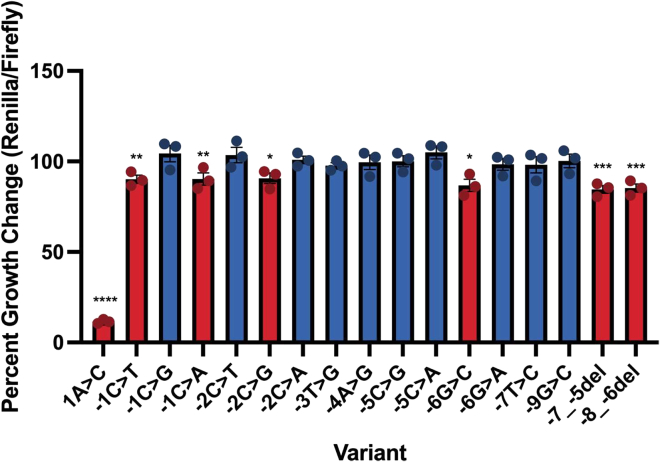
Translation efficiency was measured for each PMS2 Kozak sequence mutant. As expected, the positive control variant showed significantly reduced protein translation compared to the WT sequence (Figure 1): c.+1A>C (p < 0.0001; 88% translation reduction). Remaining Renilla luciferase expression in this mutant is hypothesized to derive from alternative AUG usage, as there are eight additional in-frame AUG codons in this gene, several of which are predicted to maintain the amino acids required for the protein active site.22 In total, we tested 18 PMS2 UTR Kozak sequence variants: the WT sequence, a positive control, 15 VUS(s) identified from ClinVar, and the c.−6G>C variant. We identified six variants with significantly decreased in vitro translation efficiency indicating a potentially pathogenic classification (Figure 1). These included c.−1C>A (p = 0.0081), c.−1C>T (p = 0.0072), c.−2C>G (p = 0.0106), c.−6G>C (p = 0.0135), c.−7_−5del (p = 0.0001), and c.−8_−6del (p = 0.0002). The remaining ten variants had translation efficiencies that did not significantly differ from the hPMS2 WT sequence, supporting a benign classification: c.−1C>G (p = 0.20), c.−2C>A (p = 0.84), c.−2C>T (p = 0.43), c.−3T>G (p = 0.61), c.−4A>G (p = 0.90), c.−5C>G (p = 0.98), c.−5C>A (p = 0.15), c.−6G>A (p = 0.64), c.−7T>C (p = 0.59), and c.−9G>C (p = 0.93; Figure 1).
Figure 1.
Translation efficiency of hPMS2 Kozak sequence variants
Bar graph shows the percentage of Renilla/firefly expression normalized to the hPMS2 wild-type (WT) control sequence. Data shown represent three independent transfections measured in technical triplicate (n = 3; ±SEM). ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001. One-way ANOVA with post hoc Fisher’s least significant difference test compared to the hPMS2 WT control in each experiment. Blue bars indicate the variant is likely benign, and red bars indicate potentially pathogenic variants.
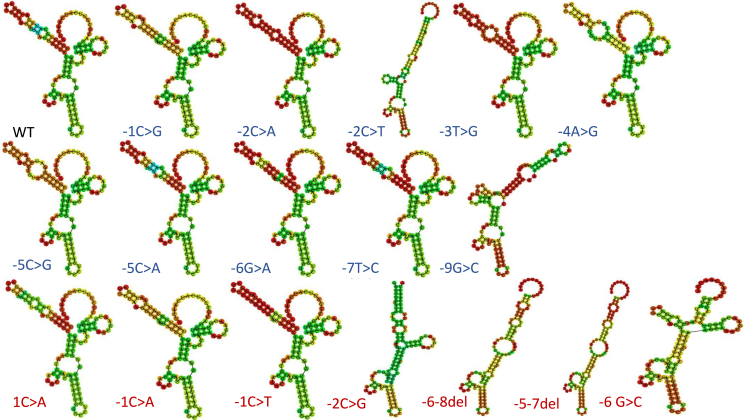
The current theory of eukaryotic translation is that a 40S ribosomal subunit and the initiator tRNA bind to the 5′ cap of mRNA. This complex then scans until it reaches the appropriate start codon (AUG), where it slows down, due to interactions with the Kozak sequence, and initiates translation. One hypothesis regarding this mechanism is that the Kozak sequence contributes to a secondary mRNA structure that physically slows ribosomal scanning. To investigate whether secondary structural changes could distinguish significant versus non-significant variants, we used an in silico tool called RNA fold23 on each of the 18 Kozak sequence variants tested. This experiment, however, did not reveal a conserved structural mechanism for potentially pathogenic versus benign variants at this locus (Figure 2). In fact, quite diverse structures were predicted among our functionally benign variants, particularly the c.−9G>C and c.−2C>T variants, which shared some structural features with predicted pathogenic variants (Figure 2). While we do not know whether these in silico predictions reflect the in vivo state, these data suggest that mutations in the hPMS2 Kozak sequence do not change translation efficiency by altering the conserved secondary mRNA structure.
Figure 2.
Predicted secondary RNA structures for hPMS2 Kozak sequence variants
In silico RNAfold predictions are shown for hPMS2 Kozak sequence variants within the context of the Renilla luciferase plasmid sequence. Blue text indicates likely benign, and red text indicates potentially pathogenic variants.
Our study highlights the potential role for PMS2 5′ UTR Kozak sequence variation in cancer risk. This was accomplished through the development of a mid-throughput reporter assay to measure translation efficiency linked to mutations in this region. Importantly, our data implicate five potentially pathogenic variants that warrant additional study in humans. Further, our data support a pathogenic classification for c.–6G>C in the context of PMS2, as this variant has been observed and associated with diseases at other loci.7,11 These data, coupled with the existing translation initiation theory, emphasize the importance of the Kozak sequence for proper translation initiation of PMS2 transcripts and the need for further genetic research in this region.
Besides the c.1A>C mutation, most of the variants in this study are currently classified as VUS(s), which are not clinically actionable. Of note, the c.−7T>C variant was found to be likely benign in our assay (Figure 1). This variant has the highest population frequency compared to the other variants tested in our assay (NM_000535.7:c.−7T>C; minor-allele frequency: 0.00032 across all genetic ancestry groups). We recently identified a family with suspected hereditary cancer carrying the c.−7T>C variant in PMS2 (data not shown). While there are conflicting interpretations of pathogenicity for this variant in ClinVar,12 segregation analysis within this family, population frequency data, and the functional data presented here collectively support a benign classification for this variant.
One of the most intriguing results of this study involved the −1 position. Two out of the three base changes tested in this location resulted in decreased translation efficiency (C>A and C>T but not C>G; Figure 1). This is consistent with the original publications from Marilyn Kozak highlighting the –1C and −2 bases as key residues within the 5′ UTR Kozak sequence.3,6,24 However, we did not see this same pattern at the –2C base; only one variant, c.–2C>G, resulted in a significant decrease in translation efficiency (Figure 1), indicating that variants may have different phenotypes and outcomes depending on the context of the Kozak sequence. This deviation from the Kozak literature may be due to the hPMS2 Kozak sequence itself. The hPMS2 Kozak sequence differs from the eukaryotic consensus at two highly conserved positions, c.−4C>A and c.−3A/G>T. The c.−3A/G base has been reported as a crucial site for translation initiation at other loci.20 The utilization of a −3T in the hPMS2 context is very rare according to original publications from Marilyn Kozak.19 Indeed, our own analysis of the human genome identified only three other genes sharing the same Kozak sequence with the hPMS2 locus (ASB1 [MIM: 605758], ETV2 [MIM: 609358], and LENG9 [MIM: N/A]; data not shown). hPMS2 bases −1, −2, −3, −4, and −6 are highly conserved across species, and PMS2 protein expression is important for human survival, suggesting that this is not a “weak” Kozak sequence. Importantly, our reversion of c.−3T>G (i.e., one of the consensus −3 bases) did not increase translation efficiency, supporting the idea that key bases are likely context specific.
Marilyn Kozak also posed the idea that varying gene-to-gene effects can be impacted by downstream factors and/or secondary structure from the mutated sequence.25 Our RNA folding data did not show a consistent deviation in secondary structure for all potentially pathogenic variants in this study (Figure 2). However, our data do show that the hPMS2 Kozak sequence −1 base must be a C or a G for WT gene expression (Figures 1 and 2) that could support either a ribosomal interaction or the presence of local secondary structures surrounding the start codon that we could not model. A potential caveat of our functional assay is that several other in-frame start codons exist within the Renilla luciferase sequence that would likely still result in a functional protein. From Marilyn Kozak’s early work,19 we know that 40S ribosomes can have “leaky scanning” such that an AUG codon may be skipped and another utilized when in a more favorable context. Indeed, it is possible that some of our likely benign hPMS2 Kozak sequence variants simply utilized a downstream, in-frame start codon that could not be distinguished by our assay.
Functional genomics studies such as this are required for variant re-classification and will remove diagnostic barriers in clinical settings for better and earlier cancer prevention and treatment. When the cancer risk of mutations is not clearly defined, prophylactic measures are often overlooked or ignored and rarely re-evaluated.26,27 That is not to say this study was without limitations. One limitation is that these assays were performed in the haploid state driving the expression of a reporter gene rather than in their native context. Indeed, LS mutations are most often in the heterozygous state, and we do not know if or how the cell compensates for reduced translation efficiency. Official ACMG re-classification of these variants will require that patient-derived tissues/cells recapitulate these effects.14 However, this does not alter the conclusions nor the importance of this study. Rather, our data strongly support continued research into the Kozak sequence to better understand how mutations in this region impact cancer risk.
Acknowledgments
This study was funded by Creighton University Kicks for a Cure and LB595 research funds to H.A.F.S.
Author contributions
H.A.F.S. conceived of the study; M.A.B. provided some resources for the study; and E.J.M. and H.A.F.S. designed and/or conducted the experiments and wrote the paper with intellectual input from J.N.P., C.J.W., M.A.B., E.E.B., and C.D.H.
Declaration of interests
The authors declare no competing interests.
Web resources
NEBChanger, v.1.3.3 https://nebasechanger.neb.com
OMIM, https://www.omim.org/
RNAfold, http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi
References
- 1.Hampel H., Hall M.J. Hereditary Aspects of Colorectal Cancer: Mismatch Repair Genes Drive Lynch Syndrome. J. Adv. Pract. Oncol. 2018;9:311–315. [Google Scholar]
- 2.Te Paske I.B.A.W., Mensenkamp A.R., Neveling K., ERN-GENTURIS Lynch-Like Working Group. Hoogerbrugge N., Ligtenberg M.J.L., De Voer R.M. Noncoding Aberrations in Mismatch Repair Genes Underlie a Substantial Part of the Missing Heritability in Lynch Syndrome. Gastroenterology. 2022;163:1691–1694.e7. doi: 10.1053/j.gastro.2022.08.041. [DOI] [PubMed] [Google Scholar]
- 3.Kozak M. Point mutations close to the AUG initiator codon affect the efficiency of translation of rat preproinsulin in vivo. Nature. 1984;308:241–246. doi: 10.1038/308241a0. [DOI] [PubMed] [Google Scholar]
- 4.Kozak M. Compilation and analysis of sequences upstream from the translational start site in eukaryotic mRNAs. Nucleic Acids Res. 1984;12:857–872. doi: 10.1093/nar/12.2.857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kozak M. Initiation of translation in prokaryotes and eukaryotes. Gene. 1999;234:187–208. doi: 10.1016/s0378-1119(99)00210-3. [DOI] [PubMed] [Google Scholar]
- 6.Kozak M. Emerging links between initiation of translation and human diseases. Mamm. Genome. 2002;13:401–410. doi: 10.1007/s00335-002-4002-5. [DOI] [PubMed] [Google Scholar]
- 7.Mohan R.A., van Engelen K., Stefanovic S., Barnett P., Ilgun A., Baars M.J.H., Bouma B.J., Mulder B.J.M., Christoffels V.M., Postma A.V. A mutation in the Kozak sequence of GATA4 hampers translation in a family with atrial septal defects. Am. J. Med. Genet. 2014;164A:2732–2738. doi: 10.1002/ajmg.a.36703. [DOI] [PubMed] [Google Scholar]
- 8.Tian C., Qin W., Li L., Zheng W., Qiu F. A common polymorphism in CD40 Kozak sequence (-1C/T) is associated with acute coronary syndrome. Biomed. Pharmacother. 2010;64:191–194. doi: 10.1016/j.biopha.2009.09.021. [DOI] [PubMed] [Google Scholar]
- 9.Jacobson E.M., Concepcion E., Oashi T., Tomer Y. A Graves' disease-associated Kozak sequence single-nucleotide polymorphism enhances the efficiency of CD40 gene translation: a case for translational pathophysiology. Endocrinology. 2005;146:2684–2691. doi: 10.1210/en.2004-1617. [DOI] [PubMed] [Google Scholar]
- 10.Cheng H.L., Liu Y.F., Su C.W., Su S.C., Chen M.K., Yang S.F., Lin C.W. Functional genetic variant in the Kozak sequence of WW domain-containing oxidoreductase (WWOX) gene is associated with oral cancer risk. Oncotarget. 2016;7:69384–69396. doi: 10.18632/oncotarget.12082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.De Angioletti M., Lacerra G., Sabato V., Carestia C. Beta+45 G--> C: a novel silent beta-thalassaemia mutation, the first in the Kozak sequence. Br. J. Haematol. 2004;124:224–231. doi: 10.1046/j.1365-2141.2003.04754.x. [DOI] [PubMed] [Google Scholar]
- 12.Landrum M.J., Lee J.M., Benson M., Brown G.R., Chao C., Chitipiralla S., Gu B., Hart J., Hoffman D., Jang W., et al. ClinVar: improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018;46:D1062–D1067. doi: 10.1093/nar/gkx1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kircher M., Ludwig K.U. Systematic assays and resources for the functional annotation of non-coding variants. Med. Genet. 2022;34:275–286. doi: 10.1515/medgen-2022-2161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ellingford J.M., Ahn J.W., Bagnall R.D., Baralle D., Barton S., Campbell C., Downes K., Ellard S., Duff-Farrier C., FitzPatrick D.R., et al. Recommendations for clinical interpretation of variants found in non-coding regions of the genome. Genome Med. 2022;14:73. doi: 10.1186/s13073-022-01073-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ambrosini C., Destefanis E., Kheir E., Broso F., Alessandrini F., Longhi S., Battisti N., Pesce I., Dassi E., Petris G., et al. Translational enhancement by base editing of the Kozak sequence rescues haploinsufficiency. Nucleic Acids Res. 2022;50:10756–10771. doi: 10.1093/nar/gkac799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McClements M.E., Butt A., Piotter E., Peddle C.F., MacLaren R.E. An analysis of the Kozak consensus in retinal genes and its relevance to gene therapy. Mol. Vis. 2021;27:233–242. [PMC free article] [PubMed] [Google Scholar]
- 17.Xu L., Liu P., Dai Z., Fan F., Zhang X. Fine-tuning the expression of pathway gene in yeast using a regulatory library formed by fusing a synthetic minimal promoter with different Kozak variants. Microb. Cell Factories. 2021;20:148. doi: 10.1186/s12934-021-01641-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Acevedo J.M., Hoermann B., Schlimbach T., Teleman A.A. Changes in global translation elongation or initiation rates shape the proteome via the Kozak sequence. Sci. Rep. 2018;8:4018. doi: 10.1038/s41598-018-22330-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kozak M. An analysis of 5'-noncoding sequences from 699 vertebrate messenger RNAs. Nucleic Acids Res. 1987;15:8125–8148. doi: 10.1093/nar/15.20.8125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kozak M. Point mutations define a sequence flanking the AUG initiator codon that modulates translation by eukaryotic ribosomes. Cell. 1986;44:283–292. doi: 10.1016/0092-8674(86)90762-2. [DOI] [PubMed] [Google Scholar]
- 21.Senter L., Clendenning M., Sotamaa K., Hampel H., Green J., Potter J.D., Lindblom A., Lagerstedt K., Thibodeau S.N., Lindor N.M., et al. The clinical phenotype of Lynch syndrome due to germ-line PMS2 mutations. Gastroenterology. 2008;135:419–428. doi: 10.1053/j.gastro.2008.04.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schenkmayerova A., Toul M., Pluskal D., Baatallah R., Gagnot G., Pinto G.P., Santana V.T., Stuchla M., Neugebauer P., Chaiyen P., et al. Catalytic mechanism for Renilla-type luciferases. Nat. Catal. 2023;6:23–38. [Google Scholar]
- 23.Kerpedjiev P., Hammer S., Hofacker I.L. Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams. Bioinformatics. 2015;31:3377–3379. doi: 10.1093/bioinformatics/btv372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pesole G., Gissi C., Grillo G., Licciulli F., Liuni S., Saccone C. Analysis of oligonucleotide AUG start codon context in eukariotic mRNAs. Gene. 2000;261:85–91. doi: 10.1016/s0378-1119(00)00471-6. [DOI] [PubMed] [Google Scholar]
- 25.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc. Natl. Acad. Sci. USA. 1990;87:8301–8305. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Makhnoon S., Bednar E.M., Krause K.J., Peterson S.K., Lopez-Olivo M.A. Clinical management among individuals with variant of uncertain significance in hereditary cancer: A systematic review and meta-analysis. Clin. Genet. 2021;100:119–131. doi: 10.1111/cge.13966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen E., Facio F.M., Aradhya K.W., Rojahn S., Hatchell K.E., Aguilar S., Ouyang K., Saitta S., Hanson-Kwan A.K., Capurro N.N., et al. Rates and Classification of Variants of Uncertain Significance in Hereditary Disease Genetic Testing. JAMA Netw. Open. 2023;6:e2339571. doi: 10.1001/jamanetworkopen.2023.39571. [DOI] [PMC free article] [PubMed] [Google Scholar]