Significance
Skeletal muscle secretes important biomolecules and vesicles, known as exosomes, that can affect how brain cells function. However, how nerve signals control muscle secretion, through a connection called the neuromuscular junction, is not completely understood. Our research bridges this gap using a tissue model representing the neuron–muscle interface. Stimulated by nerve impulses, skeletal muscles secrete higher levels of substances such as irisin, a hormone, and exosomes that contain more diverse microRNA cargos. These muscle-derived factors foster the growth of neuronal connections, enhance signal transmission along the axons, and improve communication between brain cells. This study highlights the role of nerve stimulation in muscle secretions and offers a method for producing substances that support brain development and function.
Keywords: neuromuscular junction, myokine, exosome, skeletal muscle, innervation
Abstract
Myokines and exosomes, originating from skeletal muscle, are shown to play a significant role in maintaining brain homeostasis. While exercise has been reported to promote muscle secretion, little is known about the effects of neuronal innervation and activity on the yield and molecular composition of biologically active molecules from muscle. As neuromuscular diseases and disabilities associated with denervation impact muscle metabolism, we hypothesize that neuronal innervation and firing may play a pivotal role in regulating secretion activities of skeletal muscles. We examined this hypothesis using an engineered neuromuscular tissue model consisting of skeletal muscles innervated by motor neurons. The innervated muscles displayed elevated expression of mRNAs encoding neurotrophic myokines, such as interleukin-6, brain-derived neurotrophic factor, and FDNC5, as well as the mRNA of peroxisome-proliferator-activated receptor γ coactivator 1α, a key regulator of muscle metabolism. Upon glutamate stimulation, the innervated muscles secreted higher levels of irisin and exosomes containing more diverse neurotrophic microRNAs than neuron-free muscles. Consequently, biological factors secreted by innervated muscles enhanced branching, axonal transport, and, ultimately, spontaneous network activities of primary hippocampal neurons in vitro. Overall, these results reveal the importance of neuronal innervation in modulating muscle-derived factors that promote neuronal function and suggest that the engineered neuromuscular tissue model holds significant promise as a platform for producing neurotrophic molecules.
Skeletal muscles not only are responsible for voluntary body movement but also play a crucial role in maintaining homeostasis and regulating body-wide organ functions. Specifically, skeletal muscle secretes various peptides and cytokines, named myokines, which exert autocrine, paracrine, and endocrine effects (1). For instance, interleukin-6 (IL-6) and interleukin-15 (IL-15) secreted by muscles can move into the surrounding tissue and bloodstream (2) and regulate the metabolic activity of multiple organs, such as the liver, adipose tissue, and the muscle itself (3). Besides, muscles produce brain-derived neurotrophic factor (BDNF), a neurotrophic factor that supports growth, survival, and differentiation of neurons (4). Moreover, irisin, another myokine secreted by muscles, can cross the blood–brain barrier, supporting brain activity (5) and promoting neurogenesis (6). Additionally, muscle releases extracellular vesicles (EVs), including exosomes (7). Muscle-secreted exosomes act as intercellular messengers by delivering proteins and nucleic acids, including microRNA (miRNA) cargoes (7). In vivo studies have shown that levels of myokines and exosomes in plasma and muscle increase with sustained exercise (2, 8). Intriguingly, myokines and exosomes collected from plasma have thus the potential to reduce and ameliorate neurodegenerative diseases, including Alzheimer’s disease (9, 10).
There is increasing evidence that patients with muscular dystrophy and neuromuscular disorders often experience multiorgan dysfunction and immune system failure (11–13). For instance, patients with myotonic dystrophy frequently encounter pulmonary and cardiac impairments (14). Additionally, aged individuals with sarcopenia present a higher risk of developing infections, possibly due to disruptions in normal muscle-derived soluble factors (15). Cognitive dysfunction has also been observed in patients with amyotrophic lateral sclerosis (ALS) (16) and Duchenne muscular dystrophy (17). These complications are likely attributed to abnormal secretion of myokines and muscle-derived exosomes following muscle damage. However, the underlying mechanisms behind these observations are not yet well understood. Considering that these diseases often involve neuronal denervation, the question naturally arises: How do motor neuron innervation and firing regulate secretion and bioactivity of muscle-derived factors?
Two-dimensional (2D) skeletal muscles engineered in vitro have been employed for studying muscle secretion. In particular, electrical stimulation (18) was applied to induce contractions, thereby probing the effect of muscle activity on secretion activities (19). However, these platforms overlook the crucial role of neuromuscular junctions (NMJs), which are fundamental to the physiological function of muscles. The NMJ is a chemical synapse formed between a motor neuron and a muscle fiber. At this junction, the nerve impulse triggers the release of neurotransmitters from the motor neurons. These neurotransmitters bind to the postsynaptic receptors on the muscles, leading to muscle contraction while also supporting maintenance of muscle homeostasis (20). At the same time, the muscle sends retrograde signals to motor neurons to maintain the NMJ plasticity (21). Therefore, neural denervation, observed in aging, chronic inflammation, and various muscular diseases, inevitably leads to muscle weakening (22).
To address these overlooked aspects of NMJ, there has been a growing focus on engineering neuron-innervated skeletal muscle in vitro (23). For instance, researchers have assembled microfluidic platforms to induce innervation of optogenetic motor neurons to myoblasts-derived muscle, where each tissue was initially compartmentalized (24). Additionally, both 2D and three-dimensional neuromuscular platforms have been developed to study intercellular cross talk during innervation and neuron-fired actuation (25–27). However, these models have not yet been utilized to investigate how neuronal innervation and firing affect secretion yield, composition, and biological consequences of muscle-derived myokines and exosomes.
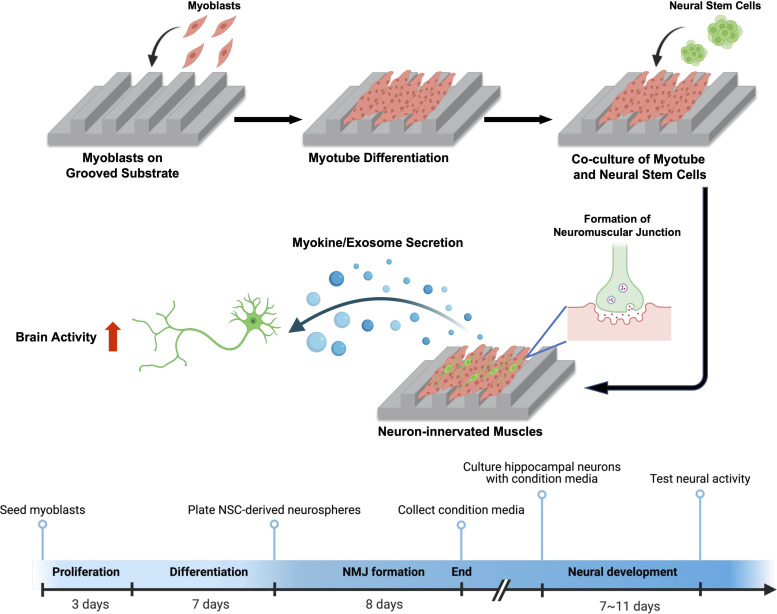
In this study, we hypothesize that neuronal innervation and firing regulate secretion, composition, and bioactivity of muscle-secreting myokines and exosomes. This hypothesis was examined by using a neuromuscular engineered tissue model we recently developed (26). We recreated a 2D skeletal muscle sheet on a flat or microgrooved poly(urethane acrylate) (PUA) substrate to modulate neuronal innervation and muscle function (Fig. 1). The groove width of the patterned substrate was maintained at 1.6 μm to promote the active clustering of acetylcholine receptors (AChRs) in muscle, thereby facilitating neural innervation. Neural stem cells (NSCs) were differentiated into motor neurons on top of the muscle sheet to form NMJs. We evaluated the impact of neural innervation on muscle contraction and metabolism, myokine-encoding messenger RNA (mRNA) expression, myokine synthesis, and exosome secretion. In addition, we profiled exosomal miRNAs from the neuron-free and neuron-innervated muscle and investigated their effects on neural development. Furthermore, we assessed the neurotrophic effects of these muscle-derived factors on branching, intracellular transport, and spontaneous activities of primary hippocampal neuronal cultured in vitro. Overall, this study highlights the significance of neural innervation in regulating the secretion of skeletal muscle to produce neurotrophic myokines and exosomes, which can have profound implications for in vitro nervous system cultures.
Fig. 1.
Schematic of creating a neuron-innervated muscle model using a grooved pattern substrate and subsequent analysis of muscle secretory activity. Created with BioRender.com.
Results
Substrate Topology Regulates Neural Innervation of Skeletal Muscles.
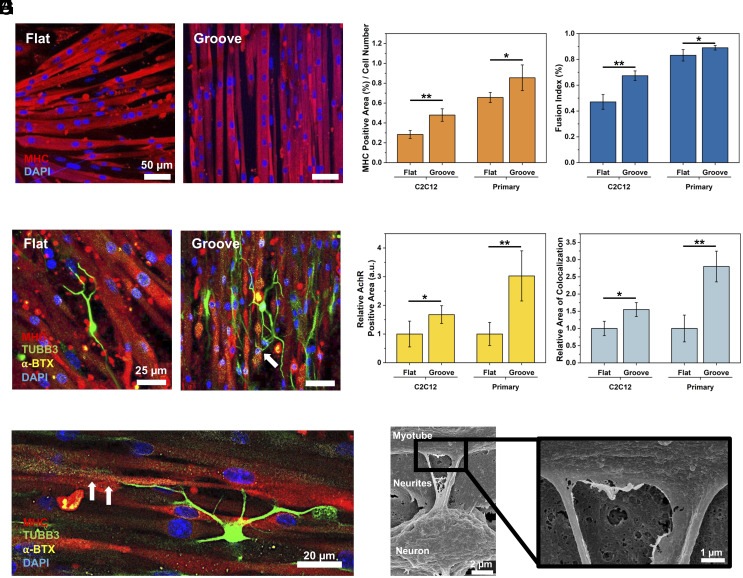
We scaled up our previous neuromuscular platform (26) to achieve a larger dimension of 1 × 1 cm2. This enlarged muscle sheet allowed us to obtain sufficient myokines and EVs for analysis. First, C2C12 and primary myoblasts were differentiated on either flat or microgrooved PUA substrates prepared using capillary force lithography (28) (SI Appendix, Fig. S1A). The microgrooved substrate presented patterned grooves of 1.6 µm width, similar to the cross-sectional diameter of a myofibril (SI Appendix, Fig. S1B) (29). The resulting myotubes aligned anisotropically on the microgrooved substrate, as confirmed by the low dispersion values (SI Appendix, Fig. S2). In contrast, myotubes formed on the flat substrate were oriented randomly. The aligned myotubes displayed a larger area positive for myosin heavy chain (MHC) and a higher fusion index than randomly oriented myotubes formed on flat substrates (Fig. 2 A–C and SI Appendix, Fig. S3A), confirming that the substrate with myofibril-like topology is advantageous for myogenesis.
Fig. 2.
Effect of substrate topology on recreation of neuron-innervated muscle. (A) Immunofluorescence images of myotubes (MHC, red) and nucleus (DAPI, blue) of primary myoblasts differentiated on flat and microgrooved (Groove) substrates. (B) MHC-positive area divided by total cell number and (C) fusion index of C2C12-derived or primary myoblast-derived myotubes. (n = 5, *P < 0.05). (D) Immunofluorescence images of the neuron-innervated muscle made with primary myoblasts. [myotubes (MHC, red), neurons (TUBB3, green), AChRs (α-BTX, yellow), and nuclei (DAPI, blue)] (E) Innervation of neuron on primary myoblast-derived myotube on a grooved substrate. The white arrows indicate the innervation spot of the axonal terminal toward AChR clusters. (F) Relative AChR expression area on myotubes. (G) Relative colocalization area of axon and AChR on innervated muscles. (n ≥ 3, *P < 0.05, **P < 0.01). (H) Scanning electron microscopy (SEM) images of the innervated muscle.
Next, neurospheres derived from NSCs were plated on the muscle sheet to create neuron-innervated muscles (Fig. 2D and SI Appendix, Fig. S3B). Within 3 d, cells migrated from the neurospheres and differentiated into motor neurons. After 8 d of coculture, the formations of NMJ were confirmed with the colocalization of AChRs labeled with α-Bungarotoxin (α-BTX) and axonal terminals immunostained with an antibody to tubulin beta-3 (TUBB3) (Fig. 2 D and E and SI Appendix, Fig. S3B). Neuron-innervated muscles formed on the microgrooved substrate expressed higher levels of AChRs and exhibited a greater number of axon terminals colocalized with AChR clusters than those on the flat substrate (Fig. 2 F and G and SI Appendix, Fig. S3C). Intriguingly, the increases in AChR expression and colocalization of axon and AChR were more evident in the primary myoblast-derived myotubes. The innervation between neurons and muscles was further confirmed with scanning electron microscopy (SEM) (Fig. 2H). Notably, the neuron-innervated muscle on the microgrooved substrate had a 1.2-fold larger MHC-positive area and thicker myotubes than the neuron-free muscle (SI Appendix, Fig. S4).
Substrate Topology Modulates the Contraction of Engineered Neuromuscular Tissues.
To evaluate the impact of substrate topology on NMJ functionality, we examined the ability of motor neurons to control the contraction of the primary myoblasts-derived muscle using three conditions: 1) neuron-free muscle (Neuron-free Muscle), 2) neuron-innervated muscle (Innervated Muscle), and 3) neuron-innervated muscle incubated with 500 µM glutamate [Innervated Muscle(G)] starting from day 4 of coculture. Primary myoblasts were used to create the Innervated Muscle because of their capability for more active neuronal innervation than C2C12. Glutamate was used to stimulate motor neurons. The quadrupole time-of-flight electrospray ionisation mass spectrometry (Q-TOFESI-MS) analysis showed that the glutamate added to the culture media of the Innervated Muscle was consumed within 24 h, whereas the glutamate level in the culture media of Neuron-free Muscle was barely reduced (SI Appendix, Fig. S5).
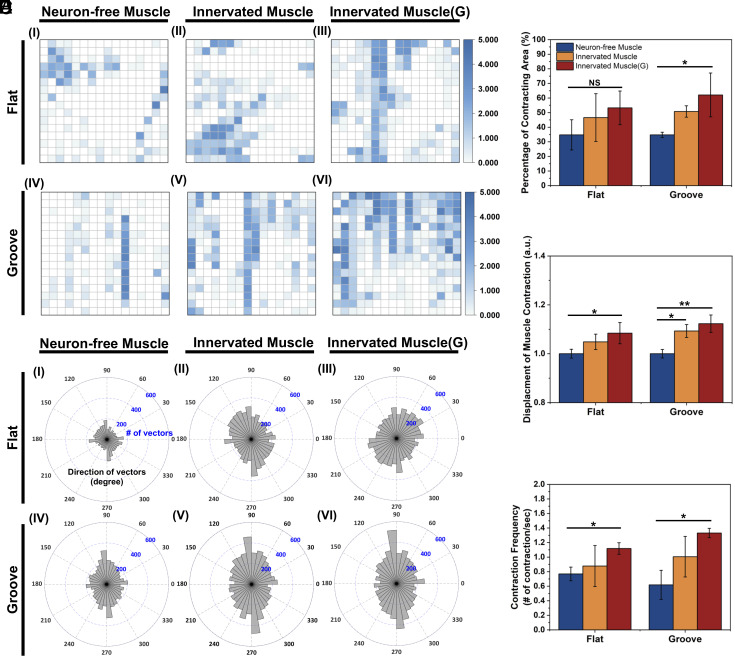
After 8 d of coculture, the Innervated Muscle showed increased twitching compared to the Neuron-free Muscle (Fig. 3A and Movies S1 and S2). Quantitative analysis showed that 30% of muscle sheets twitched locally and feebly without neuronal innervation, as indicated by the scattered blocks marking the faint contracting area (Fig. 3 A, I and IV and Fig. 3B). On the microgrooved substrate, neuronal innervation (Fig. 3 A, V) and dosage of glutamate (Fig. 3 A, VI) drove 60% of the muscle sheet to contract (Fig. 3B) and increased displacement per stroke by 10% (Fig. 3C). In contrast, the muscle sheet formed on the flat substrate exhibited minimal changes in contraction, regardless of coculture with NSCs (Fig. 3 A, II) and glutamate exposure (Fig. 3 A, III). In addition, the muscle sheet formed on the flat substrate exhibited a more considerable variance in the contracting area than the microgrooved substrate when cocultured with NSCs (Fig. 3B).
Fig. 3.
Analysis of muscle contraction of Neuron-free Muscle, Innervated Muscle, and Innervated Muscle(G), created on a flat and a microgrooved (Groove) substrate. (A) Visualization of the muscle contraction on the flat and the grooved substrate. Size of the view: 299.5 μm × 224.6 μm. The z value represented the number of contractions in each block per second. (B) Percentage of contracting myotubes. (C) Relative contraction displacement of myotubes. The “y” value was normalized to the value of Neuron-free Muscle on the flat substrate. (D) Direction of muscle contraction 90 and 270 degrees were aligned with the grooved pattern direction. (E) Average contraction frequency of the muscles (n= 3, *P < 0.05, **P < 0.01).
More interestingly, muscle sheets on the microgrooved substrate contracted anisotropically along with the groove direction, while muscle sheets on the flat substrate contracted in random directions (Fig. 3D). Neuronal innervation and glutamate stimulation further enhanced the synchronized anisotropic contraction of myotubes on the microgrooved substrate, as displayed by the continuity of contracting blocks. Additionally, neuronal innervation and glutamate stimulation increased the average contraction frequency from 0.6 to 1.3/s on the microgrooved substrate (Fig. 3E). Moreover, the variation in the contraction frequency between samples decreased when Innervated Muscles were exposed to glutamate.
Neuronal Innervation Increases Myokine-Encoding MRNA Expression and Secretory Activity of Skeletal Muscles.
We evaluated the extent to which neuronal innervation regulates myokine synthesis and secretory activity of the skeletal muscles using the three conditions noted above. We focused on analyzing biomolecules that have an impact on muscle growth and neural development, including IL-6, IL-15, BDNF, and fibronectin type III domain-containing protein 5 (FNDC5) (the precursor of irisin) (30) (Fig. 4A).
Fig. 4.
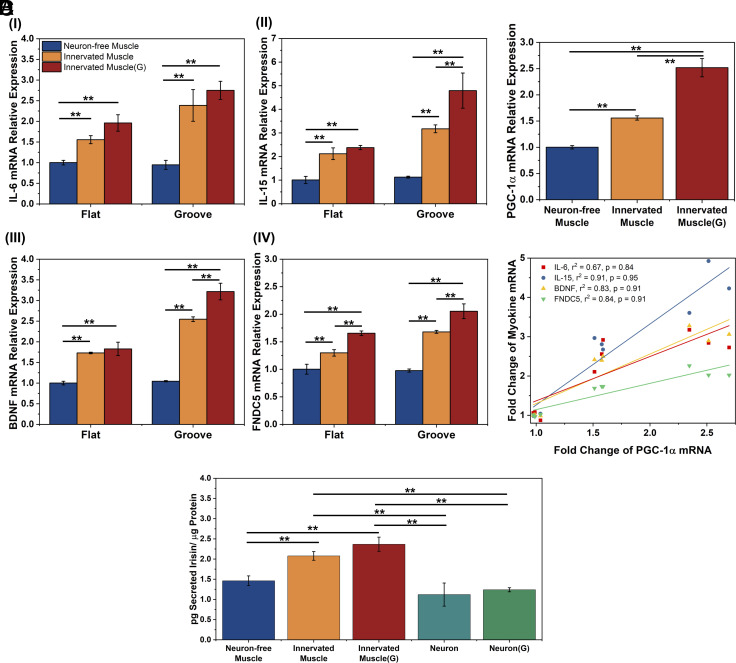
Effect of neuronal innervation on myokine-encoding mRNA expression and irisin secretion of skeletal muscles. Primary myoblasts were used in this analysis. (A) IL-6 (I), IL-15 (II), BDNF (III), and FNDC5 (IV)-encoding mRNA levels of Neuron-free Muscle, Innervated Muscle, and Innervated Muscle(G) on the flat and the microgrooved substrates after 8 d of coculture. mRNA levels were quantified with a fold change to the expression of Neuron-free Muscle formed on the flat substrate. (B) PGC-1α-encoding mRNA expression of muscles on the grooved substrate. (C) Correlation of IL-6, IL-15, BDNF, and FNDC5-encoding mRNA expressions to PGC-1α-encoding mRNA expression of the muscles on the grooved substrates. (r2: R-squared value, p: correlation coefficient) (D) Extracellular irisin concentration normalized to the total intracellular protein level of muscles or NSC-derived neurons (Neuron) and NSC-derived neurons incubated with 500 µM glutamate [Neuron(G)] on the microgrooved substrate. The protein concentration was measured after 8 d of coculture (n = 3, *P < 0.05, **P < 0.01).
The Innervated Muscle expressed higher levels of IL-6, IL-15, and BDNF-encoding mRNA than Neuron-free Muscle (Fig. 4 A, I–III). The mRNA expression levels were further increased when the Innervated Muscle was exposed to glutamate. These increases were more significant with muscle formed on the microgrooved substrate. Particularly on the microgrooved substrate, stimulating Innervated Muscle with glutamate increased IL-6, IL-15, and BDNF-encoding mRNA expression levels by 2.7-fold, 4.8-fold, and 3.2-fold, respectively, compared to the Neuron-free Muscle on the flat substrate. The FNDC5-encoding mRNA expression level in muscle was also increased with neuronal innervation and glutamate-triggered stimulation by 1.7-fold and 2.0-fold, respectively. (Fig. 4 A, IV). To confirm that the changes in mRNA expression were primarily attributed to muscle cells, the same number of NSCs used for innervating muscle was plated and differentiated on the PUA substrate. Protein and RNA quantities in NSC-derived neural cells were measured relative to monocultured muscle cells. Protein levels in NSC-derived neural cells were 7.5-fold lower, and RNA levels were 13.7-fold lower compared to those in muscle cells (SI Appendix, Fig. S6). The extracted mRNA quantity from neural cells was insufficient to ascertain the cycle threshold (Ct) for IL-6, IL-15, BDNF, and FNDC5-encoding mRNAs, even after 40 amplification cycles, while the housekeeping gene GAPDH only reaching a Ct value of 27.
We further examined whether the myokine gene expression is related to muscle energy metabolism by measuring the gene expression of peroxisome-proliferator-activated receptor γ coactivator 1α (PGC-1α) in the muscles (Fig. 4B). On the microgrooved substrate, neuronal innervation to the muscle increased the PGC-1α-encoding mRNA expression level by 1.5-fold compared to the neuron-free muscle. Additionally, glutamate stimulation increased the PGC-1α-encoding mRNA expression level in the Innervated Muscle by 2.5-fold. Consequently, the expression levels of IL-6, IL-15, BDNF, and FNDC5-encoding mRNA in the muscles showed a strong positive correlation to the PGC-1α mRNA level (Fig. 4C). However, correlations between the mRNA expression levels of IL-6, IL-15, and BDNF and that of PGC-1α were markedly weaker in muscles cultured on flat substrates (SI Appendix, Fig. S7).
Next, we examined the intracellular IL-6, BDNF, and irisin levels of the cells cultured on the grooved substrate due to their potential roles in regulating neurogenesis and neural function (4, 31). Innervated Muscle(G) exhibited increased levels of IL-6 and irisin, but not the intracellular BDNF, compared to Neuron-free Muscle (SI Appendix, Fig. S8). We further evaluated the secretion levels of these myokines in the cell culture media and normalizing them to the total intracellular protein concentration. Only irisin was detectable in the culture media, while IL-6 and BDNF concentrations were below the detection limit (Fig. 4D). The Innervated Muscle exhibited a 45% higher irisin secretion than the Neuron-free Muscle. Furthermore, glutamate stimulation elevated the irisin secretion level of the Innervated Muscle by 62%. In parallel, we measured the secreted irisin concentration in the culture media of NSC-derived neurons. Neurons secreted a lower level of irisin than the Innervated Muscles, regardless of the exposure to glutamate. Given that the cell number and intracellular protein of NSCs in the Innervated Muscle were significantly lower than muscle cells, we concluded that the increases in intracellular proteins and secreted protein of the muscles were mainly influenced by neuronal signals.
Neuronal Innervation Modulates Exosome Secretion of Skeletal Muscles.
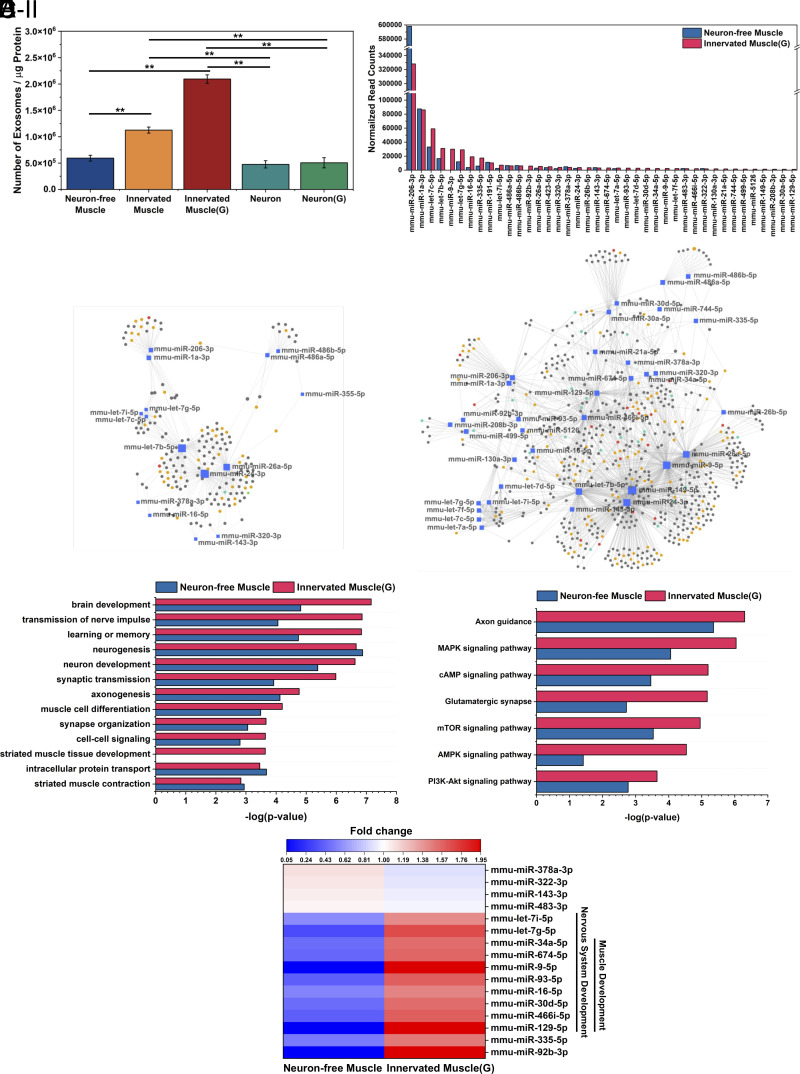
We investigated the effects of neuronal innervation and glutamate on the secretion of exosomes from the skeletal muscles. Exosomes secreted from muscles cultured on the microgrooved substrate were confirmed by transmission electron microscopy (TEM) (SI Appendix, Fig. S9A). The average hydrodynamic diameter of the exosomes was determined to be 79 nm, ranging from 30 to 280 nm, using nanoparticle tracking analysis (SI Appendix, Fig. S9B). Intriguingly, neuronal innervation resulted in a 1.8-fold increase in exosome secretion compared to the Neuron-free Muscle (Fig. 5A). Furthermore, glutamate stimulation doubled the exosome secretion from the Innervated Muscle. In addition, media from NSC-derived neurons showed a comparable exosome concentration to the Neuron-free Muscle, regardless of glutamate addition. We assessed the cellular origin of exosomes from the Innervated Muscle by analyzing desmin expression, a muscle-specific protein, using liquid chromatography-mass spectrometry. Exosomes from both Neuron-free Muscle and Innervated Muscle contained identical amounts of desmin, while exosomes from NSC-derived neuron cultures had negligible amounts of desmin (SI Appendix, Fig. S9C). This result indicated that the exosomes secreted from the Innervated Muscle were predominantly derived from muscle cells.
Fig. 5.
Analysis of exosomes released from neuron-free muscle and innervated muscle. (A) Number of exosomes in the conditioned media normalized to intracellular proteins after 24-h incubation (n = 3, *P < 0.05, **P < 0.01). (B) Normalized read counts of the top 95% abundant exosomal miRNAs. (C) miRNA-target gene network of the top 95% miRNAs of (C, I) Neuron-free Muscle and (C, II) Innervated Muscle(G). Genes targeted by at least two miRNAs were selected for the network analysis. Genes related to nervous system development were marked with a yellow circle, and muscle development-related genes were marked with red circles. Genes regulating both nerve and muscle development were marked with green circles. (D) Enriched nervous system and muscle development-related biological processes and (E) Enriched KEGG pathways regulated by the top 95% miRNA in the exosomes secreted by Neuron-free Muscle or Innervated Muscle(G). (F) Heat map showing the fold change of top 95% exosomal miRNA expression between the Neuron-free Muscle and Innervated Muscle(G) (P-value < 0.05, n = 3). The color key indicates the fold change of miRNA expression; red indicates upregulation, and blue indicates downregulation.
To examine the effect of neuronal innervation on miRNA cargoes in the exosomes, we also conducted whole miRNA sequencing. Among a total of 467 identified miRNAs (SI Appendix, Fig. S9D and Dataset S1), we focused on the miRNAs accounting for 95% of the total read (Fig. 5B). The sequencing results revealed that miR-1 and miR-206 were predominantly expressed in the exosome. This aligned with a previous report which referred to these miRNA as “myomiRs”, noting their high expression levels in skeletal muscle (32). Notably, exosomes secreted by the Innervated Muscle(G) presented a 1.8-fold larger number of miRNA types in the top 95% read (40 miRNA) than those secreted by the Neuron-free Muscle (22 miRNA). We further estimated the potential effects of these miRNAs on gene expression using the miRTarBase database. Collectively, 2,643 genes were predicted to be affected by the 40 miRNAs present in the exosomes released by the Innervated Muscle(G), while 1,228 genes were affected by the 22 miRNAs found in the exosomes from the Neuron-free Muscle (Dataset S1).
To gain insights into the biological processes regulated by genes that are affected by these miRNAs, we performed miRNA functional analysis using miRNet (33) and GO enrichment analysis. The resulting miRNA-target gene networks revealed connections to nervous system development (represented by yellow and green circles) and muscle development (represented by red and green circles) (Fig. 5C). The target network associated with exosomes secreted by the Innervated Muscle(G) (Fig. 5 C, II) was more extensive than that of the Neuron-free Muscle (Fig. 5 C, I) due to the diversity of miRNA content. The proximity visualization highlighted miRNAs targeting similar gene clusters, such as the let-7 family, miR-1a-3p, and miR-206-3p. Among these miRNAs, mmu-let-7b-5p, mmu-miR-149-5p, mmu-miR-24-3p, mmu-miR-26a-5p, mmu-miR-466i-5p, mmu-miR-9-5p showed significant effects on targets related to nervous system development (43 targets), while mmu-miR-24-3p, mmu-miR-466i-5p, and mmu-miR-9-5p had a notable impact on targets involved in muscle development (16 targets). Detailed information on the affected biological processes pertaining to nervous system development and muscle development is shown in Fig. 5D. The miRNAs in the exosomes derived from the Innervated Muscle(G) showed higher significance in regulating these biological processes, such as brain development, transmission of nerve impulse, and striated muscle tissue development. Additionally, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was applied to examine the biologic effect of these miRNAs. Notably, miRNAs in exosomes from the Innervated Muscle(G) demonstrated more significant enrichment in specific pathways, including the PI3K-Akt, mTOR, and AMPK signaling pathways that govern cellular metabolism, cell growth, and proliferation (34) (Fig. 5E). Other enriched pathways include the glutamatergic synapse and axon guidance pathways, which are crucial in modulating the formation of neural network connections, synaptic transmission, and plasticity (35). Throughout this analysis, we compared the expression level of the top 95% read miRNA in exosomes between the Neuron-free Muscle and the Innervated Muscle(G) (Fig. 5F). Interestingly, 10 miRNAs involved in nervous system and muscle development processes were up-regulated by neuronal innervation and glutamate stimulation.
Effects of Innervated Muscle-Conditioned Media on Neural Phenotype and Activity.
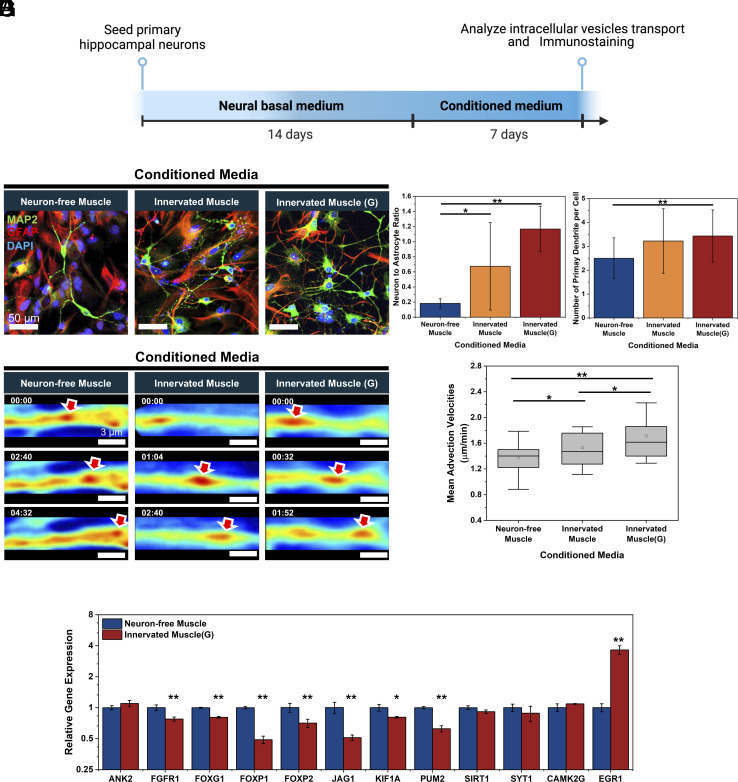
We evaluated the paracrine effects of muscle-secreted myokines and exosomes on neurogenesis, intracellular transport, and neuron-firing activities of primary rat hippocampal neurons. Conditioned media of the muscle models created on the microgrooved substrates were added separately to primary rat hippocampal neurons, which had been precultured in the basal neural media for 14 d (Fig. 6A). The concentrations of irisin and exosome in the conditioned media were quantified (SI Appendix, Fig. S10). After 7 d, primary cells cultured with the Innervated Muscle and Innervated Muscle(G) media increased the number of microtubule-associated protein 2 (MAP2)-positive neurons compared to those cultured with Neuron-free Muscle medium (Fig. 6B). Consequently, the Innervated Muscle and Innervated Muscle(G) media increased the ratio between neuron and glial fibrillary acidic protein (GFAP)-positive astrocyte by threefold and sixfold, respectively, compared with Neuron-free Muscle medium (Fig. 6C). In addition, neurons incubated in the Innervated Muscle(G) medium displayed more primary dendrites than those incubated in the Neuron-free Muscle medium (Fig. 6D).
Fig. 6.
Effects of muscle-derived conditioned media on neural development and intracellular axon transport. (A) Timeline of testing effects of muscle-derived conditioned media on hippocampal neurogenesis. Three different conditioned media were harvested from Neuron-free Muscle, Innervated Muscle, and Innervated Muscle(G) cultured on the microgrooved substrate. (B) Immunofluorescence images of hippocampal neurons cultured with conditioned media for 7 d. [neurons (MAP2, green), astrocytes (GFAP, red), and nuclei (DAPI, blue)] (C) Neuron-to-astrocyte ratio and (D) number of primary dendrites per cell altered with three different conditioned media (n > 5 per group, *P < 0.05, **P < 0.01). (E) Representative SLIM-based images of vesicular transport in neurites incubated with conditioned media (minutes:seconds). Red arrows indicated the tracked vesicles. (F) Mean advection velocities of vesicles transported in neurites quantified from SLIM images (n > 40 neurites, *P < 0.05, **P < 0.01). (G) Comparative gene expression analysis in hippocampal neurons after 7 d culture with conditioned media from Innervated Muscle(G) versus Neuron-free Muscle (n = 3, *P < 0.05, **P < 0.01).
The conditioned media influenced the axonal transport of intracellular substances, as measured using a label-free quantitative phase imaging module called spatial light interference microscopy (SLIM) (36) (Fig. 6E and Movie S3). The intracellular transport of substances including vesicles plays a critical role in the growth of neurites and synaptogenesis (37). When neurons were cultured with the medium from the Innervated Muscle, the mean advection velocity of intracellular substances transported in neurites increased by 10% compared to neurons cultured with the Neuron-free Muscle medium (Fig. 6F). The mean advection velocities of substances further increased by 24% when neurons were cultured in the Innervated Muscle(G) medium (Fig. 6F).
Further analysis was conducted to investigate the potential mechanisms affected by the muscle-conditioned media in regulating neural development. The expression levels of genes targeted by specific miRNAs and genes related to neuronal activity in hippocampal neurons were analyzed. The RT-PCR analysis revealed that seven neurogenesis-suppressing genes were significantly down-regulated in neurons cultured with the Innervated Muscle(G) medium, compared to the Neuron-free Muscle medium (Fig. 6G). Downregulation of mRNAs encoding FGFR1, FOXG1, FOXP1, and FOXP2, known to be inhibited by miR-9, promotes neural differentiation, migration, and outgrowth (38–40). Furthermore, the observed inhibition of JAG1, a target of miR-124, is associated with enhanced neurogenesis (41). In addition, the downregulation of KIF1A, inhibited by miR-101or miR-466i-5p, is beneficial for synapse function, and the downregulation of PUM2, inhibited by miR-9 or miR-129-5p, is advantageous for dendritic growth (42). Notably, miR-9-5p, miR-466i-5p, and miR-129-5p were more abundant in exosomes derived from the Innervated muscle(G) than those from the Neuron-free Muscle (Fig. 5F). Separately, EGR1, a critical mediator of synaptic plasticity and neuronal activity (43), was up-regulated by 3.6-fold when neurons were cultured with conditioned media from the Innervated Muscle(G) compared to the Neuron-free Muscle.
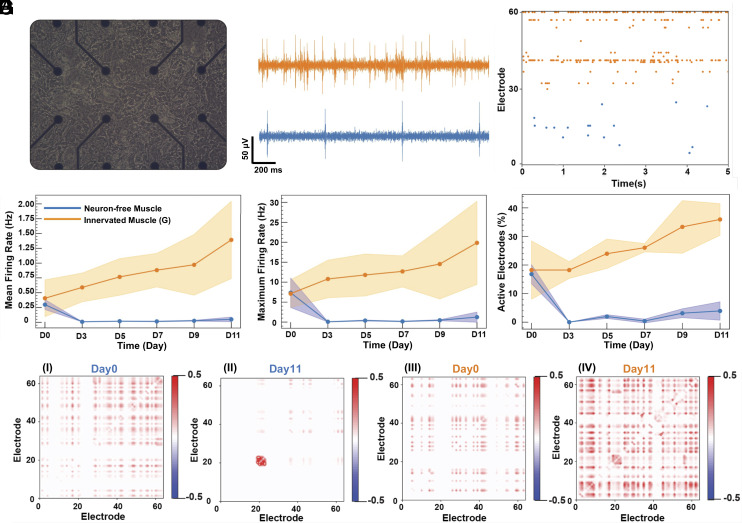
We further examined the extent to which muscle-derived conditioned media modulate spontaneous network activities of primary rat hippocampal neurons using multielectrode arrays (MEA) recording (Fig. 7A). After 11 d in the conditioned media, neurons incubated with the Innervated Muscle(G) medium showed a higher number of spontaneous firings than those incubated with the Neuron-free Muscle medium (Fig. 7B). The raw signals were further filtered to confirm they originated from neural impulses (SI Appendix, Fig. S11A). Neurons cultured in the Innervated Muscle(G) medium showed active signals across electrodes within a chosen time window, whereas neurons cultured with the Neuron-free Muscle medium rarely exhibited spikes (Fig. 7C).
Fig. 7.
Effects of muscle-derived conditioned media on neural electrophysiology. (A) Bright-field image of hippocampal neurons on MEA after 14 d of culture in the neurobasal medium. (B) Firing signals from one electrode of MEA. These signals were captured after incubating the neurons with conditioned media derived from Innervated Muscle(G) (orange) and Neuron-free Muscle (blue) for 11 d. (C) The raster plots of recording from each MEA with neurons incubated with the Innervated Muscle(G) medium (orange, electrode number 31 to 60) and Neuron-free Muscle medium (blue, electrode number 0 to 30) for 11 d. (D) Mean firing rate, (E) maximum firing rate, and (F) percentage of active electrodes from the neurons cultured with Neuron-free Muscle medium (blue) and Innervated Muscle(G) medium (orange) (n = 3). The data were analyzed from day 0 to day 11 after starting the culture of neurons with conditioned media. (G) Correlation between electrodes with neurons incubated in Neuron-free Muscle medium (G, I at day 0 and G, II at day 11) and Innervated Muscle(G) medium (G, III at day 0 and G, IV at day 11).
We further characterized the neural activity by measuring the mean firing rate (Fig. 7D), maximum firing rate (Fig. 7E), and number of active electrodes (Fig. 7F). Neurons cultured with the Innervated Muscle(G) medium displayed continuous increases in all three variables over 11 d. In contrast, neurons cultured with Neuron-free Muscle medium underwent decreases in firing activities and number of active electrodes after 3 d. We also quantified the correlation among the electrodes of MEAs to estimate neural networks’ underlying connectivity. A high correlation value between pairs of electrodes signifies dependence and synchronicity. Neurons cultured with the Neuron-free Muscle medium showed poor correlations due to sporadic and asynchronous firings (Fig. 7 G, I and II). A small signal cluster observed in Fig. 7 G, II suggests a strong correlation among adjacent electrodes, potentially due to the signal from the same neuron or a local cluster of neurons. Additionally, the single cluster might reflect the formation of confined neuronal networks within hippocampal neurons cultured in the Neuron-free Muscle medium. In contrast, the correlation of neurons cultured with the Innervated Muscle(G) medium became more pronounced after 11 d (Fig. 7 G, III and IV). Interestingly, the neurons cultured with the conditioned media exhibited delta (1 to 4 Hz), beta (11 to 30 Hz), and gamma (30 to 140 Hz) oscillations, associated with the signal processing for memory storage and retrieval (SI Appendix, Fig. S11 B and C) (44, 45).
Furthermore, we evaluated the impacts of purified exosomes and proteins from the conditioned media derived from Innervated Muscle(G) on neural functionality by dosing these factors to primary hippocampal neurons cultured on MEAs. Similar to conditioned media, both exosomes and proteins significantly boosted the spontaneous neuronal network activity, as exhibited by increases in average and maximum firing rates, along with a greater number of active electrodes, as shown in SI Appendix, Fig. S11D. Enhanced synchronicity of the neuronal network was also observed through increased correlations between electrodes (SI Appendix, Fig. S11E).
Discussion
This study demonstrates that neuronal innervation plays a key role in stimulating the secretion activity of skeletal muscle using a neuromuscular tissue model. This tissue model was created by differentiating skeletal myoblasts on a microgrooved substrate in which the width of individual microgroove closely resembled the diameter of myofibrils. In our previous study, we examined the influence of substrate topography on NMJ formation by coculturing skeletal muscle with NSCs on substrates featuring groove widths of 200 nm, 800 nm, and 1,600 nm, as well as on flat substrates. Notably, skeletal muscle cultured on substrates with patterned grooves exhibited an increased expression of AChRs, particularly on substrates with 1,600 nm grooves (26). A subsequent study supported these findings by demonstrating that substrates with grooved patterns enhanced the expression of integrin α7 and the AChR ε-subunit, thereby promoting AChR clustering and facilitating the assembly of human NMJs (46). Consequently, the grooved-patterned substrate serves as a pivotal component to investigate the effects of neuronal innervation on the skeletal muscle function. Our results showed that the Innervated Muscle cultured using anisotropically aligned myofibers formed on the microgrooved substrate responded more effectively to glutamate compared to the Innervated Muscle recreated with randomly oriented myofibers formed on the flat substrate. We propose that this is attributed to the increased number of postsynaptic AChR clusters that guide neural innervation and subsequently increase the formation of NMJs.
By comparing the Neuron-free Muscle and the Innervated Muscle, we could analyze the impact of neuronal innervation on the neurotrophic myokines gene expression. Neuronal innervation up-regulated the expression of mRNAs encoding myokines such as IL-6, IL-15, BDNF, and FNDC5, resulting in increased protein synthesis. These mRNA expressions were further up-regulated when the Innervated Muscle was stimulated with glutamate. Glutamate stimulation of the Innervated Muscle also up-regulated the expression of PGC-1α, a transcriptional coactivator that regulates the genes involved in mitochondrial bioenergetics, metabolic function, and myokine synthesis (47, 48). PGC-1α also plays a role in maintaining and remodeling NMJs (49). Therefore, we propose that neuronal firing induces muscle contraction and modulates the bioenergetic metabolism within the innervated muscles, thus up-regulating cellular pathways governing the transcription of myokine genes such as PGC-1α. The up-regulated PGC-1α expression and increased irisin secretion in glutamate-triggered neuromuscular contraction is in accordance with previous studies showing the involvement of PGC-1α in regulating FNDC5, which is subsequently cleaved and secreted as irisin (5). Interestingly, these up-regulated expressions of myokine and PGC-1α-encoding mRNAs and increased secretion of myokines and exosomes were more evident in the Innervated Muscle cultured on the grooved substrate than the flat substrate. Therefore, engineering of substrate topology is critical for creating the neuron-innervated muscle to produce neurotrophic factors.
Our study also revealed that Innervated Muscle(G) increased secretion of exosomes carrying miRNA cargoes involved in the nervous system development. Exosomes are cell-derived EVs containing various RNA species, proteins, peptides, and metabolites that regulate physiological and pathological processes in biosystems (50). Previous studies have shown that muscle contraction and physical activity elevated exosome concentration in circulation after exercise (51, 52) and altered miRNA content in circulating exosomes, thereby influencing the development and homeostasis of skeletal muscle and neurons (42). Several in vivo experiments reported that up to 5% of the exosomes in plasma were derived from skeletal muscle (53). These exosomes included muscle-enriched miRNAs, such as miR-1, miR-133, miR-206, and miR-208, which contribute to NMJ formation and maintenance (54). However, underlying mechanisms by which neuron–muscle interface plays a role in regulating exosome secretion require further investigation.
Our analysis, using the Neuron-free Muscle and the Innervated Muscle(G), demonstrated that glutamate-triggered neuronal signals to skeletal muscle significantly contribute to the enrichment of diverse miRNA cargos in muscle-derived exosomes. Notably, the most abundant miRNA, miR-206, is associated with neuroprotective effects in seizure-induced brain injury, ALS, and various neuromuscular disorders (55–57). Additionally, miRNAs such as miR-206 and miR-1 are crucial in skeletal muscle development and related pathologies (58, 59). The up-regulated expression of miRNAs in the Innervated Muscle(G)-derived exosomes such as miR-9-5p and let-7 have been reported to modulate the biological process of neurogenesis and synaptic plasticity (60, 61). Let-7 also regulates NSC proliferation and differentiation (62) and central nervous system development (63). Although PGC-1α overexpression in skeletal muscle is reported to alter exosomal cargos (64), the mechanism by which neuronal innervation and myokines modulate exosome secretion yield should be investigated further. In this regard, the Innervated Muscle will serve as a valuable model to investigate how muscles regulate exosome packaging and release in response to neuronal signaling.
Additionally, primary rat hippocampal neurons cultured with the conditioned medium of the Innervated Muscle(G) exhibited an increased neuron-to-astrocyte ratio, branching of neurites, intracellular mass transport in the axons, and neuronal firing. These enhancements in neurogenesis and spontaneous network activity can be attributed to the increased secretion of neurotrophic factors. We validated this hypothesis by adding the purified proteins and exosomes from the conditioned media to the hippocampal neuron culture and demonstrating the promoted neuronal network activity. Previously, peripheral delivery of FNDC5-encoding viral vectors to the liver was shown to elevate the plasma irisin level, inducing higher expression of neuroactive genes in the hippocampus (5). Further, FNDC5-facilitated neurogenesis was confirmed by transduction-induced overexpression of FNDC5 in mouse embryonic stem cells, enhancing neural precursor and neuron markers at a differentiation state (6). Another study demonstrated that exosomes from induced pluripotent stem cell-derived neurons increased the proliferation of neuronal cells in the hippocampal dentate gyrus (65). These findings collectively highlight the potential role of muscle-derived irisin and exosomes as intercellular communicators for regulating neuron development.
In summary, this study demonstrated, through an in vitro neuromuscular tissue model, that neuronal innervation and firing play an important role in regulating muscle metabolism, promoting the expression of neurotrophic myokine-encoding mRNAs, and increasing the secretion yield and neurotrophic activity of myokines and exosomes. In response to glutamate, the Innervated Muscle produced a higher level of irisin and a wider array of neurotrophic miRNAs enclosed in exosomes than muscle without neurons. These biological factors promoted axonal transport, firing activity, and synchrony of primary hippocampal neurons cultured in vitro, illuminating the crucial role of neuron–muscle interface in cross talk between skeletal muscle and brain cells. We propose that this finding provides invaluable perspectives on how neuronal innervation or its absence may influence the interplay between exercise and muscle secretion, although further research will be needed to investigate the combined effects of neuron–muscle interface and exercise on secretion. We further envisage that this neuromuscular model will be instrumental in investigating changes in muscle secretion associated with neuromuscular diseases using patient-derived stem cells. This platform is poised to produce neurotrophic factors critical to developing and maintaining nervous systems in vitro.
Materials and Methods
Culture of Primary and C2C12 Myoblasts for Myogenic Differentiation.
The PUA substrates were fabricated using capillary force lithography (28). The detailed fabrication procedures were described in Supporting Information. PUA substrates were cut into 1 cm × 1 cm, sterilized with 70% ethanol, and washed with phosphate buffer saline (PBS). The substrates were coated with 0.2 mg/mL Matrigel (Corning) and 0.2 mg/mL collagen in PBS for 1 h at room temperature. C2C12 myoblasts (American Type Culture Collection) were plated on the PUA substrates at the density of 4 × 104 cells cm−2 and incubated in growth medium [10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (PS) in DMEM (Dulbecco's Modified Eagle Medium)]. Separately, primary myoblasts (detailed information of primary myoblast isolation was provided in SI Appendix) were plated on the substrates at the density of 10 × 104 cells cm−2 and cultured in primary myoblast medium (20% FBS and 10% v/v AmnioMAX C-100 supplement in DMEM/F12). After 3 d of proliferation, both C2C12 myoblasts and primary myoblasts were incubated with differential medium [10% horse serum (HS), 1% PS, 1% L-glutamine] for another 7 d to differentiate into myotubes. The cells were incubated at the condition of 5% CO2 and 37 °C. All cells were used below passage 5.
Culture of NSCs for Neurosphere Formation.
Following the manufacturer’s protocol, mouse NSCs (R&D Systems) were expanded in the form of neurospheres. Briefly, NSCs were incubated in DMEM/F12 medium supplemented with 1% N-2 MAX Media supplement (R&D Systems), 1% PS, 1× basic recombinant human fibroblast growth factor (bFGF) (R&D Systems), and 1× epidermal growth factor (EGF) (R&D Systems) for 5 d. Fresh EGF and bFGF were added to the medium daily. After 5 d of culturing, the NSCs-derived neurospheres were cultured in neural differential medium (1% N-2 MAX Media supplement and 1% PS in DMEM/F12) for another 3 d.
Coculture of Myotubes and Neurospheres to Make Neuromuscular Tissue.
To create the neuron-innervated muscle, C2C12 myoblasts or primary myoblasts were plated on the PUA substrates and differentiated to form a myotube layer, as described above. Then, the differentiated NSC-derived neurospheres were collected and seeded on top of the muscle layer. The ratio of muscle cells and NSCs used for coculture was 6 to 1. The cells were incubated with the neural differential medium overnight. The next day, the medium was replaced with the coculture medium (neural differential medium supplemented with 2% HS) for another 8 d.
Immunofluorescence Analysis.
Immunofluorescence staining of myotubes.
After 7 d differentiation, myotubes derived from C2C12 myoblasts or primary myoblasts were fixed with 4% w/v paraformaldehyde (Sigma) in PBS for 15 min, permeabilized with 0.1% v/v Triton X-100 (Sigma) in PBS for 5 min, and blocked with 2% bovine serum albumin (Sigma) in PBS for 45 min at room temperature. After blocking, myotubes were incubated with MF-20 anti-MHC (1:300) (iT FX, Developmental Studies Hybridoma Bank) at 4 °C overnight. The next day, the cells were rinsed with PBS three times, and MHC was labeled with Alexa Fluor-594 goat anti-mouse IgG (Invitrogen) (1:500) at room temperature for 1 h. Last, the nuclei were stained with DAPI (Sigma) (1:500) at room temperature for 5 min. The samples were imaged with a confocal microscope, LSM 700 (Carl Zeiss). The MHC-positive area was quantified with ImageJ by counting the area of Alexa Fluor 594 signals. The fusion index was quantified by calculating the ratio of the number of nuclei in myotubes to the total number of nuclei.
Immunofluorescence staining of neuron-innervated muscles.
After an 8 d coculture, the neuron-innervated muscles were fixed, permeabilized, and blocked as previously described. After blocking, the samples were primarily stained with MF-20 anti-MHC (1:300) and rabbit monoclonal anti-TUBB3 (Abcam) (1:500) at 4 °C overnight. The next day, MHC and TUBB3 were labeled with Alexa Fluor-594 goat anti-mouse IgG (1:500) and Alexa Fluor-488 donkey anti-rabbit IgG (Invitrogen) (1:500), respectively, at room temperature for 1 h. AChR were labeled with α-BTX, Alexa Fluor 647 conjugate (Invitrogen) (1:1,000) at room temperature for 1 h. Finally, DAPI (1:500) was used to stain the nuclei. The samples were imaged under a LSM 710 confocal microscope (Carl Zeiss). The AChR-positive area was quantified with ImageJ. The colocalized area between the axon and AChR was quantified with the BIOP plugin in ImageJ.
Immunofluorescence staining of primary hippocampal neurons.
Primary rat hippocampal neurons were harvested from Long-Evans BluGill rats on postnatal day 2. The detailed isolation procedure was provided in Supporting Information. Neurons were seeded at a density of 3.5 × 105 cell/cm2 on the glass-bottom dish. The glass-bottom dish was coated with 50 μg/mL poly-D-lysine and poly-L-lysin at room temperature for 1 h and 20 μg/mL laminin at 4 °C overnight. The neurons were cultured with the neurobasal growth medium, consisting of Neurobasal-A (Gibco), B27 Plus (Gibco, 2% v/v), GlutaMAX (Gibco, 1% v/v), and penicillin–streptomycin (Gibco, 1% v/v), for 14 d and incubated with respective conditioned media for another 7 d. After 7 d incubation, the cells were fixed, permeabilized, and blocked. The samples were stained with primary antibodies, rabbit polyclonal anti-MAP2 (Abcam) (1:200), and chicken polyclonal anti-GFAP (Abcam) (1:1,000) at 4 °C overnight. The next day, MAP2 and GFAP were labeled with secondary antibodies of Alexa Fluor-488 donkey anti-rabbit IgG (Invitrogen) (1:500) and Alexa Fluor-594 goat anti-chicken IgY (Abcam) (1:1,000), respectively, for 1 h at room temperature. The nuclei of cells were separately stained with DAPI (1:500) for 5 min. The samples were imaged by a LSM 700 confocal microscope (Carl Zeiss).
SEM for Innervation Imaging.
After an 8 d coculture, the neuron-innervated muscles were fixed with 4% paraformaldehyde in 0.1 M potassium phosphate buffer (PPB) for 1 h at room temperature. After the fixation, the samples were washed with 0.1 M PPB 3 times. Then, the samples were dehydrated with ascending ethanol concentration (50%, 70%, 90%, 95%, and 100%) for 10 min twice in each concentration and dried in a vacuum overnight. The samples were sputter-coated with gold. SEM imaging was performed using the FEI Helios 600i dual-beam system.
Analysis of Muscle Contraction.
The contraction of skeletal muscle was quantified through custom MATLAB scripts. Frames were divided into subframes, each containing a single boundary of myotubes. A tracking function detected the movement of the boundary. Each deflection was counted as a contraction in the form of the peak for downstream analysis. Detailed information regarding the analysis of contraction frequency, contracting visualization, direction of contraction, and contraction displacement was provided in SI Appendix.
mRNA Expression Analysis.
RNA from muscles, NSC-derived neural cells, or primary hippocampal neurons was extracted and purified using the RNeasy Plus Mini Kit (Qiagen) following the manufacturer’s protocol. The RNA concentration of each sample was quantified using a NanoDrop™ OneC Spectrophotometer (Thermo Scientific). cDNA was synthesized up to 1 μg of RNA using the iScript cDNA Synthesis Kit (Bio-Rad). The mixture was reacted in a thermal cycler (Bio-Rad C1000) following the manufacturer’s protocol. Two microliters of cDNA was used for each quantitative polymerase chain reaction (qPCR) using the iTaq Universal SYBR Green Supermix. The qPCR primers for each target gene are listed in SI Appendix, Table S1. qPCR was run with three replicates for each condition on Light Cycler 480 (Roche) with 40 amplification cycles following the manufacturer’s protocol. Relative gene expression levels were calculated by the ΔΔCt method. GAPDH was chosen as the housekeeping gene.
Analysis of Protein Concentration.
To analyze the intracellular protein, cells were lysed with RIPA buffer containing 1% of Protease Inhibitor Cocktail (Sigma), 1% of Phosphatase Inhibitor Cocktail 2 (Sigma), and 1% of Phosphatase Inhibitor Cocktail 3 (Sigma) on ice for 30 min with intermittent vortex. The samples were centrifuged at 14,000 g for 15 min, and the supernatants were collected for protein analysis. The total intracellular protein amount was quantified with the Pierce™ BCA Protein Assay Kit (ThermoFisher). The concentrations of IL-6, BDNF, and Irisin in the supernatant were measured using the DuoSet enzyme-linked immunosorbent assay (ELISA) development systems (R&D Systems). The conditioned media were collected after 24 h of culture to measure the protein concentration using the DuoSet ELISA kits, as described above.
Exosome Analysis.
Following an 8 d coculture period, cells were exposed to serum-free coculture medium for 24 h. Exosomes were then extracted from the supernatant using isolation reagent and centrifugation. Concentrations of exosomes were measured with NanoSight NS300 (Malvern Instruments), and exosome morphology was examined with TEM (JEOL 2100 Cryo TEM). For miRNA analysis, RNA was extracted with the Total Exosome RNA & Protein Isolation Kit (Invitrogen). The RNA libraries were prepared using the QIAseq miRNA Library Kit (Qiagen) and sequenced with the NovaSeq 6000 Sequencing System (Illumina). For desmin protein analysis, proteins were extracted, digested, and analyzed with liquid chromatography–mass spectrometry (Bruker Daltonics). Detailed information was provided in SI Appendix.
Measurement of Glutamic Acid in Conditioned Media.
To analyze glutamic acid levels, conditioned media were mixed with isotopically labeled standards and extraction solution, centrifuged to obtain the supernatant, which was then dried and reconstituted with a serotonin standard solution. Microfluidic capillary electrophoresis analysis used a ZipChip interfaced to a Q-TOF ESI-MS (maXis 4G, Bruker) with a high-speed microfluidic chip (908 Devices), a setup adapted from previous study (66). Detailed information was provided in SI Appendix.
Protein Purification from Conditioned Media.
For protein purification from the conditioned media, the media were centrifuged at 300 g for 5 min, followed by a 3,000 g centrifugation for 20 min, and then centrifugated at 100,000 g for 6 h at 4 °C to remove exosomes. The resulting supernatants were passed through a 3k MWCO ultracentrifugal filter (MilliporeSigma) at 3,166 g for 40 min at 4 °C to eliminate metabolites. Protein concentrations in the concentrated samples were quantified using a BCA assay.
SLIM Imaging and Dispersion-Relation Phase Spectroscopy (DPS) Analysis for Axonal Mass Transport.
A SLIM system was constructed by attaching a SLIM module (CellVista SLIM Pro, Phi Optics, Inc.) to one end of a commercial phase contrast microscope (36). The SLIM module was built as described previously (67). Upon simple reconstruction technique, quantitative phase images are acquired. DPS analysis was performed on acquired time-sequential phase images to extract the mean advection velocity associated with vesicles (68). The primary hippocampal cells were cultured on the glass-bottom dish as previously described. After 7 d incubation in conditioned medium, live cells were imaged under SLIM to record the intracellular mass transport activity. The axon regions were segmented for DPS analysis to acquire the mean advection velocity.
Multielectrode Array (MEA) Analysis.
Dissociated hippocampal neurons were plated on MEA (60MEA200/30iR-Ti, multichannel systems) to measure the electrophysiological activity. The MEAs were coated with 100 μg/mL poly-D-lysine and poly-L-lysin at room temperature for 3 h, and with 20 μg/mL laminin overnight. Cells were seeded on the MEAs at a density of 4,800 cells/mm2 and cultured in the neurobasal medium for 14 d. Starting on the 15th day, the medium was switched to the respective condition medium and maintained for 11 d. For testing the effect of innervated muscle-derived proteins and exosomes, the medium was switched to the neurobasal medium supplemented with purified proteins (0.3 mg/mL) or exosomes (108 exosome/mL) and maintained for 7 d. The electrophysiological activity for each MEA was recorded for 5 min every alternate day using a 64-channel open-source acquisition board (69) with an open-source recording platform (70). Postprocessing of the data was performed using an open-source electrophysiology package, MiV-OS (71). Detailed information for data processing was provided in SI Appendix.
Statistical Analysis.
Averaged data were presented as mean ± 1 SD. The statistical analyses between pairs of experimental population were performed using nonpaired Student's t test or one-way ANOVA with Tukey's post hoc test. Statistical differences were considered significant at a P-value < 0.05 (*P < 0.05 and **P < 0.01).
Supplementary Material
Appendix 01 (PDF)
Dataset S01 (XLSX)
Contractions of muscle models created on flat substrates.
Contractions of muscle models created on micro-grooved substrates.
Axonal transports of primary hippocampal neurons cultured with conditioned medium.
Acknowledgments
This work was supported by the NSF (CBET-1932192, Expeditions-2123781), NIH (R61HL159948), Alzheimer’s Disease Association grant (2019-AARG-NTF-644507), and a Chan Zuckerberg Biohub Chicago Acceleration Research Award. J.L. and Q.C. thank Air Force Office of Scientific Research grant AFOSR FA9550-20-1-0257. A.E.-M. thanks the support by the NSF under Grant No. 1922758. Our gratitude goes to Dr. Alvaro G. Hernández, Dr. Chris L. Wright, and Dr. Christopher J. Fields from the Roy J. Carver Biotechnology Center for their assistance in creating miRNA sequencing libraries. We also thank the Center for Advanced Bioenergy and Bioproducts Innovation for providing the instruments for real-time PCR analysis.
Author contributions
K.-Y.H. and H.K. designed research; K.-Y.H., M.S., Y.C., A.C.W., C.H., J.W.M., J.L., Y.T., Y.-H.D., A.E.-M., Z.D., X.Z., and S.K. performed research; Q.C., J.V.S., S.G.I., R.B., H.J.C., G.P., M.U.G., and M.G. contributed new reagents/analytic tools; K.-Y.H., G.U., Y.A., M.S., G.J.P.-D., and C.H. analyzed data; and K.-Y.H., G.U., H.J.C., M.G., and H.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
All study data are included in the article and/or supporting information.
Supporting Information
References
- 1.Severinsen M. C. K., Pedersen B. K., Muscle-organ crosstalk: The Emerging roles of myokines. Endocrine Rev. 41, 594–609 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pedersen B. K., Febbraio M. A., Muscle as an endocrine organ: Focus on muscle-derived interleukin-6. Physiol. Rev. 88, 1379–1406 (2008). [DOI] [PubMed] [Google Scholar]
- 3.León-Ariza H. H., Mendoza-Navarrete M. P., Maldonado-Arango M. I., Botero-Rosas D. A., A systematic review of “myokines and metabolic regulation”. Apunts. Medicina de l’Esport 53, 155–162 (2018). [Google Scholar]
- 4.Kim S., et al. , Roles of myokines in exercise-induced improvement of neuropsychiatric function. Pflügers Arch. 471, 491–505 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Wrann C. D., et al. , Exercise induces hippocampal BDNF through a PGC-1α/FNDC5 pathway. Cell Metab. 18, 649–659 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Forouzanfar M., et al. , Fndc5 overexpression facilitated neural differentiation of mouse embryonic stem cells. Cell Biol. Int. 39, 629–637 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Watanabe S., et al. , Skeletal muscle releases extracellular vesicles with distinct protein and microRNA signatures that function in the muscle microenvironment. PNAS Nexus 1, pgac173 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nederveen J. P., Warnier G., Di Carlo A., Nilsson M. I., Tarnopolsky M. A., Extracellular vesicles and exosomes: Insights from exercise science. Front. Physiol. 11, 604274 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta R., Khan R., Cortes C. J., Forgot to exercise? Exercise derived circulating myokines in Alzheimer’s disease: A perspective. Front. Neurol. 12, 649452 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lourenco M. V., et al. , Exercise-linked FNDC5/irisin rescues synaptic plasticity and memory defects in Alzheimer’s models. Nat. Med. 25, 165–175 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stevens R. D., et al. , Neuromuscular dysfunction acquired in critical illness: A systematic review. Intensive Care Med. 33, 1876–1891 (2007). [DOI] [PubMed] [Google Scholar]
- 12.Latronico N., et al. , Critical illness myopathy and neuropathy. The Lancet 347, 1579–1582 (1996). [DOI] [PubMed] [Google Scholar]
- 13.Sun X., et al. , Multiple organ dysfunction and systemic inflammation after spinal cord injury: A complex relationship. J. Neuroinflammation 13, 260 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaminsky P., et al. , Organ dysfunction and muscular disability in myotonic dystrophy type 1. Medicine 90, 262–268 (2011). [DOI] [PubMed] [Google Scholar]
- 15.Nelke C., Dziewas R., Minnerup J., Meuth S. G., Ruck T., Skeletal muscle as potential central link between sarcopenia and immune senescence. eBioMedicine 49, 381–388 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abe K., et al. , Cognitive function in amyotrophic lateral sclerosis. J. Neurol. Sci. 148, 95–100 (1997). [DOI] [PubMed] [Google Scholar]
- 17.Rae M. G., O’Malley D., Cognitive dysfunction in Duchenne muscular dystrophy: A possible role for neuromodulatory immune molecules. J. Neurophysiol. 116, 1304–1315 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedachi T., Fujita H., Kanzaki M., Contractile C2C12 myotube model for studying exercise-inducible responses in skeletal muscle. Am. J. Physiol. Endocrinol. Metab. 295, E1191–E1204 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Carter S., Solomon T. P. J., In vitro experimental models for examining the skeletal muscle cell biology of exercise: The possibilities, challenges and future developments. Pflügers Arch. 471, 413–429 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Witzemann V., Development of the neuromuscular junction. Cell Tissue Res. 326, 263–271 (2006). [DOI] [PubMed] [Google Scholar]
- 21.Arnold A.-S., et al. , Morphological and functional remodelling of the neuromuscular junction by skeletal muscle PGC-1α. Nat. Commun. 5, 3569 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Iyer S. R., Shah S. B., Lovering R. M., The neuromuscular junction: Roles in aging and neuromuscular disease. Int. J. Mol. Sci. 22, 8058 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Osaki T., Uzel S. G. M., Kamm R. D., Microphysiological 3D model of amyotrophic lateral sclerosis (ALS) from human iPS-derived muscle cells and optogenetic motor neurons. Sci. Adv. 4, eaat5847 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uzel S. G. M., et al. , Microfluidic device for the formation of optically excitable, three-dimensional, compartmentalized motor units. Sci. Adv. 2, e1501429 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aydin O., et al. , Development of 3D neuromuscular bioactuators. APL Bioeng. 4, 016107 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ko E., et al. , Matrix topography regulates synaptic transmission at the neuromuscular junction. Adv. Sci. 6, 1801521 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kaufman C. D., et al. , Emergence of functional neuromuscular junctions in an engineered, multicellular spinal cord-muscle bioactuator. APL Bioeng. 4, 026104 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang H. S., et al. , Electroconductive nanopatterned substrates for enhanced myogenic differentiation and maturation. Adv. Healthcare Mater. 5, 137–145 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.DeAguero A. A., et al. , Regulation of fiber-specific actin expression by the Drosophila SRF ortholog blistered. Development 146, dev164129 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young M. F., Valaris S., Wrann C. D., A role for FNDC5/Irisin in the beneficial effects of exercise on the brain and in neurodegenerative diseases. Progress Cardiovas. Dis. 62, 172–178 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kummer K. K., Zeidler M., Kalpachidou T., Kress M., Role of IL-6 in the regulation of neuronal development, survival and function. Cytokine 144, 155582 (2021). [DOI] [PubMed] [Google Scholar]
- 32.McCarthy J. J., MicroRNA-206: The skeletal muscle-specific myomiR. Biochim. Biophys. Acta 1779, 682–691 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chang L., Zhou G., Soufan O., Xia J., miRNet 2.0: Network-based visual analytics for miRNA functional analysis and systems biology. Nucleic Acids Res. 48, W244–W251 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Manning B. D., Cantley L. C., AKT/PKB signaling: Navigating downstream. Cell 129, 1261–1274 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lin L., Lesnick T. G., Maraganore D. M., Isacson O., Axon guidance and synaptic maintenance: Preclinical markers for neurodegenerative disease and therapeutics. Trends Neurosci. 32, 142–149 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Z., et al. , Spatial light interference microscopy (SLIM). Opt. Express 19, 1016–1026 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Guedes-Dias P., Holzbaur E. L. F., Axonal transport: Driving synaptic function. Science 366, eaaw9997 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fineberg S. K., Kosik K. S., Davidson B. L., MicroRNAs potentiate neural development. Neuron 64, 303–309 (2009). [DOI] [PubMed] [Google Scholar]
- 39.Leucht C., et al. , MicroRNA-9 directs late organizer activity of the midbrain-hindbrain boundary. Nat. Neurosci. 11, 641–648 (2008). [DOI] [PubMed] [Google Scholar]
- 40.Barca-Mayo O., De Pietri Tonelli D., Convergent microRNA actions coordinate neocortical development. Cellular Mol. Life Sci. 71, 2975–2995 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cheng L.-C., Pastrana E., Tavazoie M., Doetsch F., miR-124 regulates adult neurogenesis in the subventricular zone stem cell niche. Nat. Neurosci. 12, 399–408 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rajman M., Schratt G., MicroRNAs in neural development: From master regulators to fine-tuners. Development 144, 2310–2322 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Duclot F., Kabbaj M., The role of early growth response 1 (EGR1) in brain plasticity and neuropsychiatric disorders. Front. Behav. Neurosci. 11, 35 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Colgin L. L., Moser E. I., Gamma oscillations in the hippocampus. Physiology 25, 319–329 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Leondopulos S. S., Boehler M. D., Wheeler B. C., Brewer G. J., Chronic stimulation of cultured neuronal networks boosts low-frequency oscillatory activity at theta and gamma with spikes phase-locked to gamma frequencies. J. Neural Eng. 9, 026015 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shimizu K., et al. , Alignment of skeletal muscle cells facilitates acetylcholine receptor clustering and neuromuscular junction formation with co-cultured human iPSC-derived motor neurons. Cells 11, 3760 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schnyder S., Handschin C., Skeletal muscle as an endocrine organ: PGC-1α, myokines and exercise. Bone 80, 115–125 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mormeneo E., et al. , PGC-1α induces mitochondrial and myokine transcriptional programs and lipid droplet and glycogen accumulation in cultured human skeletal muscle cells. PLoS One 7, e29985 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Correia J. C., Ferreira D. M. S., Ruas J. L., Intercellular: Local and systemic actions of skeletal muscle PGC-1s. Trends Endocrinol. Metab. 26, 305–314 (2015). [DOI] [PubMed] [Google Scholar]
- 50.Rome S., Forterre A., Mizgier M. L., Bouzakri K., Skeletal muscle-released extracellular vesicles: State of the art. Front. Physiol. 10, 929 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whitham M., et al. , Extracellular vesicles provide a means for tissue crosstalk during exercise. Cell Metab. 27, 237–251.e234 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Frühbeis C., Helmig S., Tug S., Simon P., Krämer-Albers E.-M., Physical exercise induces rapid release of small extracellular vesicles into the circulation. J. Extracell. Vesicles 4, 28239 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Guescini M., et al. , Muscle releases alpha-sarcoglycan positive extracellular vesicles carrying miRNAs in the bloodstream. PLoS One 10, e0125094 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maggio S., et al. , Signal exchange through extracellular vesicles in neuromuscular junction establishment and maintenance: From physiology to pathology. Int. J. Mol. Sci. 20, 2804 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wu Z., Liu Y., Huang J., Huang Y., Fan L., MiR-206 inhibits epilepsy and seizure-induced brain injury by targeting CCL2. Cytotechnology 71, 809–818 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Williams A. H., et al. , MicroRNA-206 delays ALS progression and promotes regeneration of neuromuscular synapses in mice. Science 326, 1549–1554 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Valsecchi V., et al. , miR-206 reduces the severity of motor neuron degeneration in the facial nuclei of the brainstem in a mouse model of SMA. Mol. Therapy 28, 1154–1166 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Ma G., et al. , MiR-206, a Key modulator of skeletal muscle development and disease. Int. J. Biol. Sci. 11, 345–352 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Chen J.-F., et al. , The role of microRNA-1 and microRNA-133 in skeletal muscle proliferation and differentiation. Nat. Genet. 38, 228–233 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lang M.-F., Shi Y., Dynamic roles of microRNAs in neurogenesis. Front. Neurosci. 6, 71 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mikihito S., Hiromi N., Hiroshi K., Takaya A., Shinichi A., MicroRNA-9 regulates neurogenesis in mouse telencephalon by targeting multiple transcription factors. J. Neurosci. 31, 3407 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhao C., et al. , MicroRNA let-7b regulates neural stem cell proliferation and differentiation by targeting nuclear receptor TLX signaling. Proc. Natl. Acad. Sci. U.S.A. 107, 1876–1881 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Fairchild C. L. A., et al. , Let-7 regulates cell cycle dynamics in the developing cerebralcortex and retina. Sci. Rep. 9, 15336 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kargl C. K., et al. , Peroxisome proliferator-activated receptor γ coactivator 1-α overexpression improves angiogenic signalling potential of skeletal muscle-derived extracellular vesicles. Exp. Physiol. 108, 240–252 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sharma P., et al. , Exosomes regulate neurogenesis and circuit assembly. Proc. Natl. Acad. Sci. U.S.A. 116, 16086–16094 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lee C. J., et al. , d-Amino acids and classical neurotransmitters in healthy and type 2 diabetes-affected human pancreatic islets of langerhans. Metabolites 12, 799 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen X., Kandel M. E., Popescu G., Spatial light interference microscopy: Principle and applications to biomedicine. Adv. Opt. Photon. 13, 353–425 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang R., et al. , Dispersion-relation phase spectroscopy of intracellular transport. Opt. Express 19, 20571–20579 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Siegle J. H., et al. , Open Ephys: An open-source, plugin-based platform for multichannel electrophysiology. J. Neural Eng. 14, 045003 (2017). [DOI] [PubMed] [Google Scholar]
- 70.Zhang X., et al. , Mind in vitro platforms: Versatile, scalable, robust, and open solutions to interfacing with living neurons. Adv. Sci. 11, e2306826 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Gazzola Lab, MiV-OS: Analysis and computation framework on MiV system and simulator. GitHub Repository (2022). https://github.com/GazzolaLab/MiV-OS. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Dataset S01 (XLSX)
Contractions of muscle models created on flat substrates.
Contractions of muscle models created on micro-grooved substrates.
Axonal transports of primary hippocampal neurons cultured with conditioned medium.
Data Availability Statement
All study data are included in the article and/or supporting information.