Significance
Immunotherapy by immune checkpoint inhibitors, e.g., PD-1/PD-L1 inhibitors, is now recognized as one of the most promising treatment modalities for solid tumors. However, clinical efficacy prediction based on PD-L1 expression in tumor tissues is currently imprecise and lacks the capability for real-time monitoring. Here, we describe a unique nanoplatform for in situ intracellular detection of expression of PD-L1 RNA, which is combined with in vitro cellular behavior analysis, to enable better prediction of efficacy of treatment of cancer in patients. By using a 5 mL blood sample, the system generates analytical outputs that, when combined, provide an index that enables quantitative and more accurate prediction of the outcomes of patients under immunotherapeutic treatment, when compared to traditional biopsy.
Keywords: microfluidics, single cell, circulating tumor cells, immunotherapy, prediction
Abstract
Accurate prediction of the efficacy of immunotherapy for cancer patients through the characterization of both genetic and phenotypic heterogeneity in individual patient cells holds great promise in informing targeted treatments, and ultimately in improving care pathways and clinical outcomes. Here, we describe the nanoplatform for interrogating living cell host-gene and (micro-)environment (NICHE) relationships, that integrates micro- and nanofluidics to enable highly efficient capture of circulating tumor cells (CTCs) from blood samples. The platform uses a unique nanopore-enhanced electrodelivery system that efficiently and rapidly integrates stable multichannel fluorescence probes into living CTCs for in situ quantification of target gene expression, while on-chip coculturing of CTCs with immune cells allows for the real-time correlative quantification of their phenotypic heterogeneities in response to immune checkpoint inhibitors (ICI). The NICHE microfluidic device provides a unique ability to perform both gene expression and phenotypic analysis on the same single cells in situ, allowing us to generate a predictive index for screening patients who could benefit from ICI. This index, which simultaneously integrates the heterogeneity of single cellular responses for both gene expression and phenotype, was validated by clinically tracing 80 non–small cell lung cancer patients, demonstrating significantly higher AUC (area under the curve) (0.906) than current clinical reference for immunotherapy prediction.
Immunotherapy by immune checkpoint inhibitors (ICI), including PD-1/PD-L1 inhibitors, is recognized as one of the most promising treatment modalities for solid tumors (1). However, only a minority of patients benefit from ICI treatment, and an increasing body of evidence has suggested that using a single biomarker in tumor tissue, such as PD-L1 expression, cannot accurately predict the response to PD-1/PD-L1 inhibitors, limiting the effectiveness rate to 40 to 60% (2, 3). In addition, current clinical practice, using localized sampling and an endpoint analysis strategy, often falls short of providing sufficiently granular spatiotemporal information to enable the accurate evaluation of underlying intratumor heterogeneity, resulting in a less well-understood knowledge of the cellular responses in tumorigenesis and immunotherapy (4).
Liquid biopsy based on circulating tumor cells (CTCs) has emerged as an alternative approach, enabling dynamic monitoring throughout the course of treatment (5). Due to the limited number of CTCs in blood (1 to 20 CTCs/mL) and their high heterogeneities, clinical success in CTC-based diagnosis has a heavy dependence on the technologies for both cell capture and detection (6). For CTC capture, microfluidic-based techniques have demonstrated the advantages of easy operation, high throughput, and automation (7).
Current techniques can also be classified into two categories depending on the strategy for identifying CTCs, either involving those based upon membrane protein markers [where CTCs or leukocytes are specifically separated by magnetic nanoparticles modified with recognition tools such as antibodies (8)]; or those based upon the differences in physical characteristics between CTCs and leukocytes [using differences in size and deformability to enable label-free separation in the microfluidic chips (9, 10)]. In both cases, due to the variability of CTCs, the enrichment efficiency in cell capture is reduced (11). By using a combination of membrane markers and physical characteristics, it is proposed that it is possible to significantly increase capture efficiency by reducing the loss of heterogeneous CTCs (12).
For genetic analysis of CTCs, single-cell sequencing technologies enable understanding in-depth genetic features such as specific gene expressions and gene mutations (13). Analysis of key proteins using immunofluorescence staining is also commonly used (14). In one example, CTCs isolated by a size-based vortex chip were stained with fluorescent antibodies to quantify the PD-L1 protein expression, enabling the identification of patients that are highly responsive to immunotherapy (15).
Notwithstanding these examples, current methods of genetic or protein analysis are typically based on dead CTCs, which results in challenges to comprehensively analyze cellular heterogeneities around both gene expression and cancer cell behaviors (6, 11). Clinical practice has yet to validate evidence for the impact of such techniques on improving its accuracy in prediction in cancer immunotherapy (16).
In this study, we introduce a Nanoplatform for Interrogating living Cell Host-gene and (micro-) Environment (NICHE) as a microfluidics approach, based upon magnetic cell removal and “bagatelle traps,” for the highly efficient capture of CTCs from blood samples. The platform also integrates a unique nanopore-enhanced electrodelivery technique that generates safe and efficient cell perforation, and rapidly transports a DNA tetrahedron (DNAT) fluorescence probe for real-time in situ quantification of mRNA in the living CTCs. On-chip coculture of CTCs and T cells further allows for real-time analysis of cellular proliferation and phenotype alteration in response to treatment, while correlating responses to gene expression heterogeneity.
We demonstrate the platform’s capabilities of capturing CTCs and characterizing both their genotype and phenotype, with the technique validated in a study involving 80 non–small cell lung cancer (NSCLC) patients. Based upon PD-L1 expression and CTC phenotype, we defined a NICHE index for predicting immunotherapy efficacy in cancer patients. Analysis of outcomes for the cancer patients was used to validate the effectiveness of the NICHE index, demonstrating significantly higher AUC (0.906) than the current clinical standard reference method of using the tumor proportion score (TPS) calculated based on PD-L1 expression of tumor tissue (AUC=0.578) in predicting immunotherapy efficacy.
Results
Design and Functionalities of NICHE.
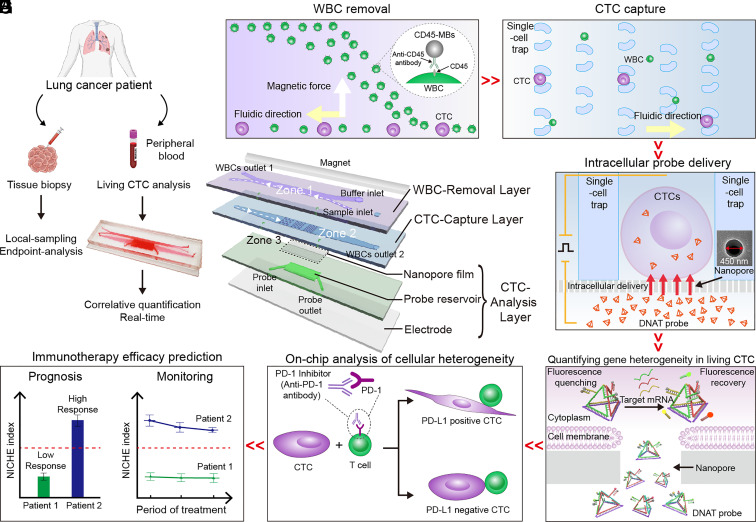
We have previously designed microfluidic single-cell traps and developed understanding of the flow and geometric conditions to realize high trapping efficiencies as well as optimizing optical signal collection for understanding the treatment of cancers in blood (17–19). Building upon this experience, the NICHE platform was designed for on-chip real-time quantification of PD-L1 expression in living single CTCs from patients’ liquid biopsies, as well as for correlating phenotypic behaviors against immune cells, to provide precise, granular spatiotemporal information for the prediction of immunotherapy in NSCLC patients, with the clinical workflow illustrated in Fig. 1A.
Fig. 1.
Design and functionality of NICHE. (A) NICHE is designed to interrogate CTCs from peripheral blood as a liquid biopsy, adopting an alternative route to the traditional biopsy. (B) NICHE enables removal of WBCs, capturing CTCs, delivery of DNAT probes into living CTCs through the nanopore-enhanced electrodelivery (NEED) technique for intracellular mRNA relative quantification, and analysis of CTC behaviors in response to immune cells for immunotherapy response prediction. (C) In the WBC-Removal Layer, WBCs are tagged with CD45-MBs and sorted out of the cell-flow channel under an external magnetic field. (D) CTCs are subsequently captured on three groups of “bagatelle”-shaped traps in an array, while remaining WBCs pass through the microtraps and flow to WBCs outlet 2. (E) Captured CTCs settle on the nanopores where intracellular probe delivery is implemented by NEED under a pulsed electric field. (F) DNAT probes are used to quantify PD-L1 expression in living CTCs. (G) CTCs that settled on the nanopores are cocultured with immune T cells under the effect of ICI to observe their behavior and response to immune environments. (H) Finally, the efficacy of immunotherapy is evaluated and monitored by detecting PD-L1 expression and analyzing the behavior of single living CTCs.
The integrated platform comprises three zones formed by the assembly of associated functional layers, including those for WBC removal, CTC capture, and CTC analysis with a nanopore film, a probe reservoir, and a bottom electrode (Fig. 1B and SI Appendix, Fig. S1A). Collected from the blood of NSCLC patients, the peripheral blood mononuclear cells (PBMCs) were preincubated with anti-CD45 antibody modified with magnetic beads (CD45-MBs) and were injected through the sample inlet, into Zone 1 (Fig. 1C), located on the upper WBC-Removal Layer. A cross-shaped microfluidic channel was designed for separating white blood cells (WBCs) from the CTCs. The WBCs specifically labeled with CD45-MBs were deflected across the cell-flow channel where a magnet was placed to remove the WBCs in WBCs outlet 1.
These CTCs, together with any WBCs missed in Zone 1, keep their original flow trajectory until they reach Zone 2, fabricated on the CTC-Capture Layer, where CTCs are retained in a bagatelle array (Fig. 1D), comprising three tiers of arch-shaped microfluidic cell traps. To guarantee high capture efficiency, the sizes of entrance (EN) and exit (EX) of the traps were designed with consideration for the size heterogeneity of CTCs. The remaining WBCs whose sizes are smaller than the exit of the traps pass through and flow to WBCs outlet 2. The captured CTCs settle in situ on a nanopore film of the CTC-Analysis Layer (Zone 3) aligned with the traps, that form a “cell-nanopore” juxtaposed configuration. This was used to deliver probes intracellularly for measuring target mRNAs in living CTCs, assisted with a pulsed electric field (Fig. 1E) (20, 21).
The unique “nanopore-enhanced electrodelivery” (NEED) technique enabled high-efficiency transport of fluorescence probes across the cell membrane, which has been experimentally and numerically validated in this work. We designed a tetrahedron (DNAT) probe with a molecular structure tailored to maintain its stability against DNA-degrading enzymes in the cytosol (Fig. 1F). The DNAT probes used three fluorescence detection channels, providing stable internal reference for precise quantification of target mRNA (e.g., PD-L1) expression and CTC identification at the single-cell level. Subsequently, NICHE was used for coculturing the captured CTCs with immune cells (e.g., T cells), for analysis of cellular responses (e.g., phenotype alteration, proliferation) in response to ICI (Fig. 1G), combined into a specific NICHE index that stems from PD-L1 and phenotype heterogeneities within the CTCs (Fig. 1H).
NICHE for Efficient Capture and Quantifying PD-L1 mRNAs in Living Cells.
To quantify the ability of NICHE to eliminate WBCs from CTCs, we designed a model experiment using Jurkat T cells (lymphocytic leukemia cell line) as a proxy for WBCs, and two human lung cancer cell lines (A549 and HCC827) that have been widely used as CTC-mimic in lung cancer research (22, 23). On the WBC-Removal Layer, the WBCs (labeled with CD-45 MBs) were magnetically deflected from the original flow direction, while CTCs maintained their original trajectory toward Zone 2 (CTC-Capture Layer), enabling separation (SI Appendix, Fig. S1 B–D and Movies S1–S3). The cell flow rate was also optimized at 80 μL/min, to shorten the removal time without impacting the efficiency of separation, for which >93% of T cells, labeled with >10 CD45-MBs per cell, were removed (SI Appendix, Fig. S1 E and F). Considering the diameters of CTCs, which vary between 8 μm and 18 μm (8), we fabricated three tiers of bagatelle cell trap arrays in Zone 2 along the flow direction, with a gradually reduced ratio of EN/EX, i.e., 28 μm/18 μm; 20 μm/10 μm; 14 μm/8 μm (SI Appendix, Fig. S2A).
Numerical analysis indicated that the arch-shaped trap modified the flow velocity and shape, enabling cell capture (SI Appendix, Fig. S2 B–H). Each tier was interleaved row-by-row, significantly reducing the possibility of cells clogging in front of the trap, and further facilitating the removal of remaining WBCs missed in Zone 1 (SI Appendix, Fig. S2 I and J and Movie S4). Statistically, the platform displayed highly efficient (~95%) on-chip CTC capture (SI Appendix, Fig. S2 K and L), as well as a high recovery rate (>70%) in the blood-mimetic samples, for various staring CTC numbers, from 500 down to 10 cells/mL (SI Appendix, Fig. S2M) (24, 25).
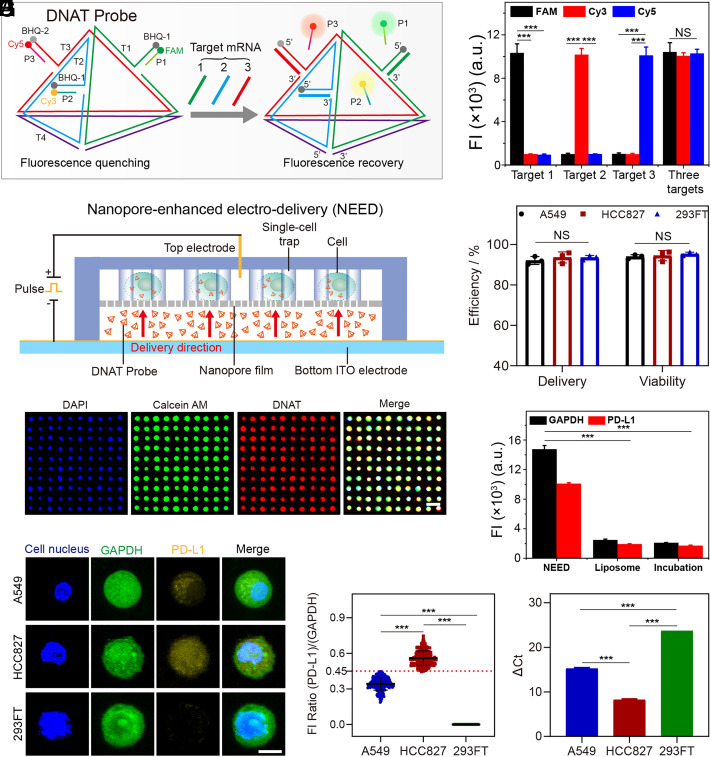
On the CTC-Analysis Layer, the living CTCs captured by the microfluidic cell trap arrays were interrogated in situ with a single-cell resolution. For precise quantification of PD-L1 expression, we designed a DNAT probe that possessed three detection channels, simultaneously identifying PD-L1 mRNA, GAPDH mRNA, as a housekeeper control and a reference for normalizing the fluorescence intensities among different cells (Fig. 2A and SI Appendix, Fig. S3 A–D), and CK (cytokeratin) mRNA that was used to distinguish CTCs (24, 25). The DNAT probe was formed through self-assembly with seven DNA sequences (i.e., T1, T2, T3, T4, P1, P2, and P3; SI Appendix, Table S1). The sequence T1, T2, T3, and T4 constitute a tetrahedral structure, which demonstrated a significantly higher resistance against DNA-degrading enzymes than conventional probes due to the steric hindrance for binding of endonucleases (SI Appendix, Fig. S4 A and B) (26). The sequence P1, P2, and P3 were prelabeled with fluorophores FAM (green), Cy3 (yellow), and Cy5 (red), with each being quenched by a corresponding fluorescence quenching group on the 5′ end of sequence T1, T2, and T3. After probe hybridization, the fluorescence intensities of the three channels (FAM, Cy3, and Cy5) were significantly decreased, indicating successful pairing (P1-T1, P2-T2, and P3-T3) (SI Appendix, Fig. S4 C–E). However, in the presence of target RNAs, we show that the DNATs sequentially demonstrate a rapid response to strand displacement (<10 min; SI Appendix, Fig. S4 F–H), high sensitivity with good linearity of response in the range between 100 pM and 100 nM, and a low limit of detection (LOD) of 100 pM (SI Appendix, Fig. S4 I–K). The probes also showed high specificity when challenged simultaneously by three RNA targets (Fig. 2B).
Fig. 2.
NICHE for quantifying target mRNA in living cells. (A) A schematic diagram of DNAT probes for detecting three target mRNA. T1, T2, T3, and T4 constitute the structural skeleton of the DNAT probe. The 5′ end of T1, T2, and T3 are partially complementary to P1, P2, and P3, respectively. Target 1, GAPDH mRNA. Target 2, PD-L1 mRNA. Target 3, CK mRNA. (B) The specificity of DNAT probes for detection of three target mRNAs. FAM, Cy3, and Cy5 represent the fluorescence signals from P1, P2, and P3, respectively. FI, fluorescent intensity. (C) A cross-sectional view of NEED for delivering DNAT probes into living CTCs. (D) The delivery efficiency of NEED and the viability of cells after probe delivery. DNAT probes labeled with FAM dye (without quencher) are delivered into cells for calculating the delivery efficiency. Propidium iodide (PI) dye is used to evaluate cell viability. The statistical data are obtained by counting 1,000 cells/chip from three independent assays. (E) Fluorescent characterization of probe delivery and cell viability on a compact single-cell trap array (the distance between the two cells is 10 μm). DAPI, blue dye for cell nucleus. Calcein AM, green dye, is used to evaluate cell viability. DNAT probe is labeled with Cy5 (without quencher). (Scale bar, 50 μm.) (F) Delivery dose of DNAT probes in HCC827 cells (10,000 cells) by NEED, liposome-based delivery, and simple incubation with free probe, using fluorescence intensity obtained via flow cytometry. (G) Fluorescent images of cells detected by DNAT probes. (Scale bar, 5 μm.) (H) Ratios of fluorescent intensity from 300 cells detected in three separated experiments. Error bars represent the SD. (I) Relative quantification of RNAs detected by qPCR. ΔCt is the difference between the Ct value of PD-L1 RNA and the Ct value of GAPDH RNA. The value of ΔCt is negatively correlated with target RNA concentration. PD-L1 is not expressed in 293FT cells, and the Ct value of PD-L1 is set at 40. ***P-value < 0.001. NS, no significant difference. Error bars in (B), (D), (F), and (I) represent the SD of three independent experiments.
We explored different methods for intracellular delivery of DNAT probes into CTCs, including passive diffusion or endocytosis, such as simple incubation with a free probe or liposome-based delivery, although in all cases they showed limited uptake efficiency (SI Appendix, Fig. S5A). To address this limitation, as well as for in situ monitoring of CTCs, we designed the NEED technique to be carried out on the CTC-Analysis Layer (Fig. 2C). In this methodology, the cells were captured by the trap array and settled on the nanopore film (pore-size distribution CV = 1.1%; pore density 125 ± 5/100 μm2) (SI Appendix, Fig. S5 B and C). Under a low voltage (15 V) applied between a top electrode (a silver needle) and a bottom indium tin oxide (ITO) electrode, the NEED generated a consistent transmembrane potential (1 V to 6 V) for different cells at the nanopore-cell interface, which enabled efficient electroporation of the cell membrane (SI Appendix, Table S2 and Fig. S5 D–I) (27, 28).
As the equivalent resistance of the nanopore is several orders of magnitudes greater than the equivalent resistance of the cell membrane (29), the majority of the potential drop is applied over the two terminals of the nanopores, which drives electrophoresis of the surface-charged molecules (i.e., DNAT probes) within the nanopore. The DNAT probes are accelerated and transported into cells from the probe reservoir within several seconds (SI Appendix, Fig. S5 J and K and Movie S5). By optimizing the electrical conditions (15 V, 1 ms pulse duration, 150 pulses) and the probe conditions (60 min incubation, 1 μM), we achieved a high and uniform delivery efficiency (95%) among each cell while keeping high cell viability (90%) (Fig. 2 D and E and SI Appendix, Fig. S6 A–F). We also verified that NEED does not cause changes in endogenous PD-L1 expression (SI Appendix, Fig. S6 G–I). These results, when combined, demonstrate that the intracellular signal of DNAT probes was increased at least fivefold compared to that delivered by either simple incubation or liposome-based delivery (Fig. 2F and SI Appendix, Fig. S6 J–L).
To evidence the performance of NICHE in quantifying PD-L1 expression in living cells, we selected two PD-L1-positive cell lines, i.e., human lung cancer cells A549 and HCC827, and one PD-L1-negative cell line, i.e., human embryonic kidney cells 293FT. The PD-L1 fluorescence signals are emitted intracellularly (Fig. 2 G and H and SI Appendix, Fig. S7 A–C), with its intensity directly proportional to the PD-L1 expression level, as confirmed by qRT-PCR (Fig. 2I and SI Appendix, Fig. S7D).
For further confirmation of these outcomes, we used a A549 cell that expressed low level of PD-L1 as the cell model to demonstrate our ability to detect the up- and downregulation of the PD-L1 expression using the NICHE platform. We found increased fluorescence intensity of PD-L1 in PD-L1-overexpressed A549 cells (A549-PD-L1ꜛ), while a decreased signal in PD-L1-knockdown A549 cells (A549-PD-L1ꜜ), thus validating the specificity of the DNAT probe for quantifying PD-L1 expression in living cells (SI Appendix, Fig. S8 A–E).
Further by using normalization based upon GAPDH as a house-keeping gene in conventional platforms, we were able to define the fluorescent intensity ratio of PD-L1/GAPDH = 0.45 as the threshold to categorize the subtype of PD-L1high and PD-L1low in cancer cells (see details in Materials and Methods). As stated, the shape of the DNATs is not only essential for their stability in real samples, but also provides the opportunity to multiplex the analysis. We demonstrated this capability by detecting CK mRNA in HCC827 cells (SI Appendix, Fig. S9 A–C), providing the ability of identifying CTCs from WBCs and increasing the robustness of the analysis.
NICHE for Correlative Analysis of PD-L1 Expression and Immune-Response.
Within tumor microenvironments, cancer cells (including CTCs) inhibit the immune response through specific interaction with the ligand expressed by the cancer cells and the immune checkpoint on immune cells (2). PD-1 and PD-L1 proteins expressed on T cells and cancer cells act as surface receptors to trigger inhibitory signaling to attenuate the T cell response. ICI-based therapy exploits these interactions and uses antibodies against either PD-L1 or PD-1 to block the specific interaction for reinvigorating T cell response (Fig. 3A). However, the exact role of PD-L1 expression and its impact upon the response of CTCs against immunotherapy, particularly at single-cell resolution, has not been fully elucidated (11, 30). The success of NICHE in real-time in situ quantification of PD-L1 expression in living CTCs allows for on-chip observation of the behaviors of CTCs in response to immune environments (e.g., T cell attack) while correlating these responses to the heterogeneity in PD-L1 expression.
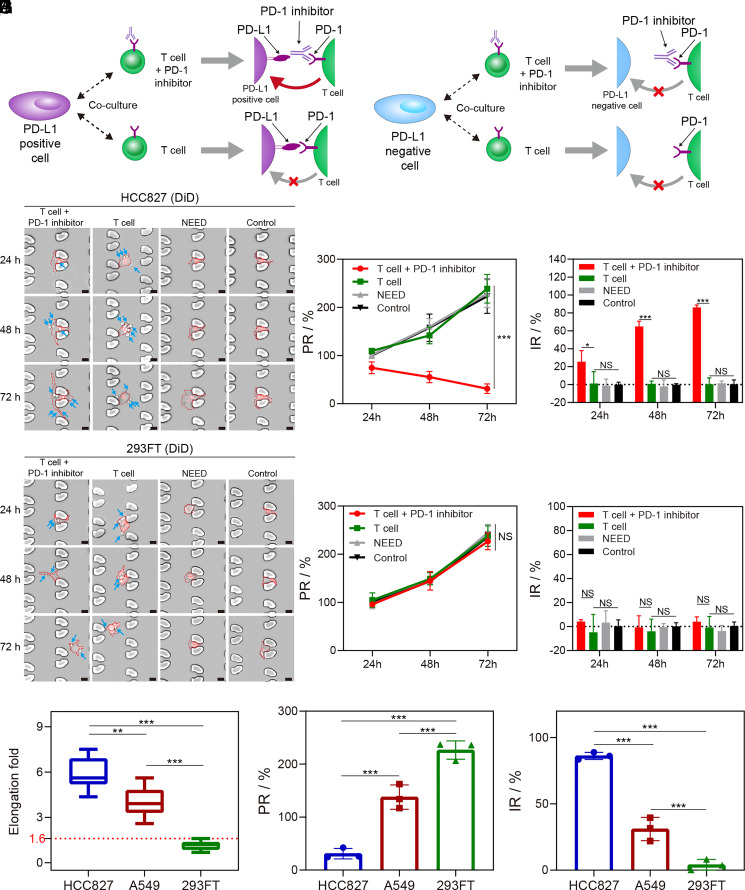
Fig. 3.
NICHE pairs tumor cells and immune T cells to quantify immune-response to PD-1 inhibitor. (A) Schematic of the interaction between tumor cells and T cells in coculture. The immune response to the cancer cell expressing PD-L1 is suppressed and can be restored using a PD-1 inhibitor, while it has no impact on a PD-L1-negative cell. (B) and (C) Time-lapse images of cocultured cells in a single Regions of Interest (ROI). T cell + PD-1 inhibitor, cells were cocultured with Jurkat T cells and PD-1 inhibitor. T cell, cells were cocultured with Jurkat T cells. Control, the cells without coculture with Jurkat T cells. HCC827 cells (B) and 293FT cells (C) are stained with DiD dye (Red) for long-term tracking. The blue arrows indicate Jurkat T cells. (Scale bar, 20 μm.) (D) and (E) The proliferation rate (PR) (D) and inhibition rate (IR) (E) of HCC827 cells in coculture with Jurkat T cells. (F) and (G) PR (F) and IR (G) of 293FT cells. (H) Amplitude of morphological elongation after coculture with Jurkat T cells (blocked with anti-PD-1 antibodies) for 72 h. Three hundred cells are counted in each experiment. (I) and (J) PR (I) and IR (J) of cancer cells after coculture with Jurkat T cells (block with anti-PD-1 antibodies) for 72 h. *P-value < 0.05. **P-value < 0.01. ***P-value < 0.001. NS, no significant difference. Error bars in (D–G), (I), and (J) are SD of three independent experiments.
To validate this, we cocultured the cells, i.e., PD-L1-positive cells (HCC827, A549), or PD-L1-negative cells (293FT), with immune cells (Jurkat T and HuT-78) using NICHE (31). The results showed that the phenotypes of cells do not change after the delivery of DNAT probes by NEED (Fig. 3 B and C and SI Appendix, Fig. S10A). In the presence of Jurkat T cells, PD-L1-positive cells with the addition of PD-1 inhibitors (anti-PD-1 antibodies) significantly change the phenotype to elongated morphologies (above 100 μm in length) from 24 h to 72 h (Fig. 3B and SI Appendix, Fig. S10A and Movies S6–S8), while PD-L1-negative cells (293FT) retain their native phenotype when cocultured with Jurkat T cells, irrespective of the PD-1 inhibitor treatment (Fig. 3C). We noted that the number of immune cells bound to the tumor cells changes in real time, demonstrating a dynamic interaction between these two types of cells.
We also evaluated both proliferation rate (PR) and inhibition rate (IR), as defined in Materials and Methods, of HCC827 cells, A549 cells, and 293FT cells when cocultured with Jurkat T cells. The PR and IR between control cells and cells subjected to NEED analysis were comparable, demonstrating the negligible effect of NEED on cell behaviors (Fig. 3 D–G and SI Appendix, Fig. S10 B and C). HCC827 cells display the lowest PR and highest IR among all groups under treatment with PD-1 inhibitors. However, there is no significant difference in cell proliferation among the 293FT groups, with significantly lower IR than the PD-L1-positive cells treated with the antibodies (<5% vs. >20%). The Jurkat T cells have negligible effects on the PD-L1-negative cells (293FT) in inhibiting cell growth. Results based on HuT-78 cell line remain consistent with the results based on the Jurkat T cell line, in terms of phenotype morphologies, PR and IR in the coculture experiments (SI Appendix, Fig. S11 A–G), validating the robustness of our findings.
Because the proliferation of PD-L1-positive cells was compromised, to validate the correlation between PD-L1 expression and cellular behaviors in response to immune cells, we modified 293FT cells to that with low expression of PD-L1 (PD-L1low) and with high expression of PD-L1 (PD-L1high), respectively (SI Appendix, Fig. S12 A–C). The results indicate neither phenotypic morphology change nor significant increased length in the PD-L1-positive 293 FT cells (SI Appendix, Fig. S12 D and E). Both PR and IR of these cells further indicated no significant differences between wild-type 293FT cells (PD-L1−) and PD-L1-positive 293 FT cells (SI Appendix, Fig. S12 F and G). These results imply that 293FT cells did not reverse their phenotype morphologies by forced expression of PD-L1 only, which indicated that PD-L1 was not a deterministic factor to the morphological change of the cells.
To further confirm this finding, we detected the protein expression of cells that had undergone phenotypic morphology change (HCC827 and A549) by label-free protein mass spectrometry and compared them with those of control groups (without immune cell coculture), for the specific purpose of screening determinants of morphological changes in cells. The results show 26 up-regulated proteins and 14 down-regulated proteins within the cells that underwent phenotypic morphology change (SI Appendix, Fig. S13 A–D). However, PD-L1, was not among those directly involving regulation, a finding that is consistent with other literature reporting that PD-L1 is not directly involved in the regulation of cellular morphological changes (32, 33). We noted that up-regulated proteins that contribute to the control of cell migration and cytoskeleton activity included VIM, PLOD2, and MACF1 (34–36). In contrast, down-regulated proteins primarily controlled cell proliferation, for example, SLC3A2, ABCC3, and ITGA3 (37–39).
Based on the phenomenon that the cells showed a lengthened phenotype, we hypothesized that the change in their phenotype was to evade the killing effect of immune cells. In relation to their elongated morphology, this coculture microenvironment would trigger epithelial–mesenchymal transition (EMT) that increases cellular deformability to facilitate their travel through the organ (32, 40). To explore this hypothesis, we measured the expression of both epithelial cell markers (E-cadherin and cytokeratin) and mesenchymal cell markers (vimentin and N-cadherin) (SI Appendix, Fig. S14 A and B). Both western blot and immuno-fluorescent results show upregulation of vimentin and N-cadherin and downregulation of E-cadherin and cytokeratin in the elongated cells under the attack from T cells, as compared to the control. These results indicated that the elongated cancer cells may undergo EMT-mediated phenotype alteration in response to immune attack.
Furthermore, by comparing the morphology and proliferation results among the three types of cells, we found a significant inhibition in the proliferation of the cells that underwent morphological elongation (Fig. 3 H–J), which enables the establishment of a quantitative threshold to identify the subgroup highly responsive to ICI. We therefore used the maximum elongation that could be achieved by 293FT cells (a cell line without phenotypic morphological elongation), as the cutoff value for determining phenotype alteration positive cells (PA+, cell elongation fold >1.6).
NICHE for Predicting Efficacy of Immunotherapy.
To assess the clinical performance of NICHE in predicting the efficacy of immunotherapy, we analyzed blood samples from 80 NSCLC patients before operative or drug treatment (SI Appendix, Table S3). First of all, we validated the capability of the microfluidic cell traps to capture living CTCs from clinical blood samples, and NICHE’s specificity for in situ probing of PD-L1 expression and CTC identification, which was confirmed by CD45−/CK+ immunostaining (Fig. 4A and SI Appendix, Fig. S15A) (25). The results also indicate that the cell trap array with different sizes enabled efficient capture of the CTCs of different sizes. We observed that the CTC number of locally advanced or advanced treatment-naïve NSCLC patients clinically defined as stage III to IV were significantly higher than that of patients in clinical stage I to II (Fig. 4B), confirming previous studies which identified CTCs as a marker of aggressiveness (6, 41).
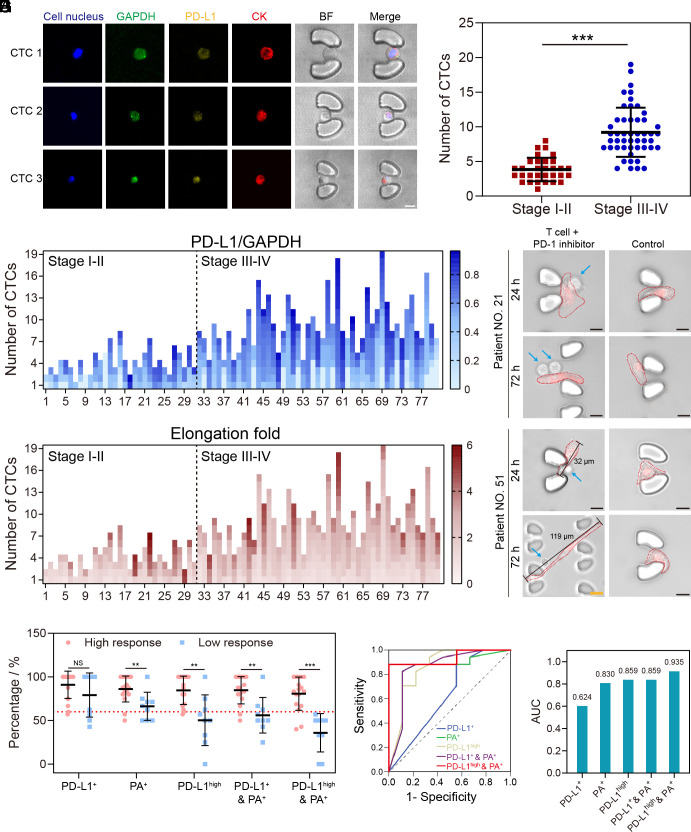
Fig. 4.
Evaluation of clinical samples using NICHE. (A) Images of CTCs of different sizes analyzed by NICHE. (Scale bar, 10 μm.) CTC 1, CTC 2, and CTC 3 are isolated from the blood sample of patient No. 27. (B) A scatter plot of CTC numbers isolated from 80 NSCLC patients. (C) Ratios of the fluorescent intensity of PD-L1 (Cy3 channel) and GAPDH (FAM channel) from individual CTCs obtained from 80 NSCLC patients. (D) Confocal images of cocultured CTCs with Jurkat T cells (supplemented with anti-PD-1 antibodies). CTCs are stained with DiD dye (Red) for long-term tracking. The blue arrows indicate Jurkat T cells. (Scale bar (Black), 10 μm.) (Scale bar (Yellow), 20 μm.) (E) Elongation fold of individual CTCs obtained from 80 NSCLC patients. Each square represents a CTC. (F) Scatterplots of different indexes for immunotherapy prediction. High response, patients with the response evaluation of pCR and PR. Low response, patients with the response evaluation of MPR, MPR-, SD, and PD. (G) The receiver operating characteristic (ROC) curves comparing NICHE with different indexes. (H) Comparison of AUC (area under the curve) among different indexes, including PD-L1+, PD-L1high, PA+, PD-L1+ & PA+, and PD-L1high & PA+. **P-value < 0.01. ***P-value < 0.001. NS, no significant difference. Error bars in (B) and (F) are SD of each group’s patient samples.
CTCs can also cluster, as assemblies which are composed of several cells with a larger size and some specific properties, including high metastatic potential (42). Despite their existence, we did not observe any in our study of clinical samples from patients. This may be a consequence of the design of trap structures, which could not easily immobilize CTC clusters. We also note that the level of PD-L1 expression in CTCs has only a weak correlation to cancer stages (Fig. 4C and SI Appendix, Fig. S15B), and that the percentage of PD-L1-positive cells in CTCs is higher than the percentage of PD-L1-positive cells in tissues in our study. These results are consistent with other literature that describes that the percentage of PD-L1-positive cells in CTCs was significantly more than that in tissue cells (14, 43), as CTCs are able to survive in immune cell-rich peripheral blood, which relies on their specific ability to resist the attack of immune cells (2, 16). High PD-L1 expression is a pathway known to enable tumor cells to escape the immune response (3, 44).
We next measured the phenotype alteration of the captured CTCs under immune environments and observed that PD-L1-positive CTCs from 18 of the clinical samples all underwent distinct phenotype alteration with the introduction of T cells, while in other samples, it was only a proportion of PD-L1-positive CTCs (Fig. 4 D and E and SI Appendix, Fig. S15 C–E). These reveal high genetic and phenotypic heterogeneities of the CTCs in clinical blood samples. We therefore established an indicator, combining genetic and phenotypic heterogeneities within CTCs, for predicting a patient’s immunotherapy response. As a measure of genetic heterogeneity, we chose to use the proportion of the PD-L1-positive CTCs in a blood sample, and based upon the results of cell lines (Fig. 3H), we established a threshold (PD-L1high vs. PD-L1low, see Materials and Methods for definition) as a precise parameter in compartmenting the heterogeneity. For phenotypic heterogeneity, we defined phenotype alteration (measured as PA+/PA−, see Materials and Methods for definition). When using these measures together, we adopted the proportion of PD-L1high & PA+ CTCs in blood sample as an indicator (NICHE index), then validated its performance in predicting the outcomes of the NSCLC patients. For comparison, four other indicators, i.e., the proportion of PD-L1+ CTCs, the proportion of PD-L1high CTCs, the proportion of PA+ CTCs, and the proportion of PD-L1+ & PA+ CTCs, were also measured in parallel.
Out of the 80 diagnosed patients, 35 underwent neoadjuvant immunotherapy based on PD-1 inhibitor, and 34 out of 35 successfully underwent surgical resection and their therapeutic outcomes were recorded by pathological evaluation (pathologic complete response, pCR; major pathological remission, MPR; major pathological remission was not achieved, MPR-) (SI Appendix, Table S4). In addition, 26 only underwent standard PD-1 inhibitor immunotherapy and the therapeutic outcomes of 18 out of 26 were recorded by imaging (partial response, PR; stable disease, SD; progressive disease, PD) (SI Appendix, Table S5). These two groups of patients (comprising 52 patients in total) allowed us to further validate the effective prediction of the NICHE index.
In order to predict the efficacy based on the NICHE index, we randomly split the patient into a training cohort (n = 26, SI Appendix, Table S6) and a validation cohort (n = 26, SI Appendix, Table S7). All patients were divided into a High response group and a Low response group. For patients receiving resection, according to pathological examination, pCR was treated as High response and non-pCR (MPR and MPR-) was treated as Low response. For those without resection, PR was treated as High response and non-PR (SD and PD) were treated as Low response.
In the training cohort, based on the actual therapeutic response, the samples were divided into two groups (High response and Low response), and then the above five parameters were compared separately. The results show that PA+, PD-L1high, PD-L1+ & PA+, and NICHE index were able to differentiate the effects of immunotherapy, except for PD-L1+ (Fig. 4F). Among all parameters, the NICHE index resulted in the most pronounced distinction between High response and Low response. Further, we calculated the ROC curves for each parameter (Fig. 4G), and found that the NICHE index provided the highest Jordon index (0.882) and the highest AUC (area under the curve) value (0.935) (Fig. 4H and SI Appendix, Table S8).
We next determined the efficacy of each sample in the validation cohort based on the optimal thresholds established with the training cohort to test its validity for clinical prediction. The sensitivity and specificity of the NICHE index achieved 87.5 % and 100%, respectively (PD-L1+, 100%/10%; PA+, 43.8%/100%; PD-L1high, 93.8%/70%; PD-L1+ & PA+, 50%/90%) (SI Appendix, Table S9). In addition, the NICHE index shows a significantly higher AUC (0.906) than TPS (0.578) for the analysis of a subset of 36 patients with tissue biopsies from 80 diagnosed patients (SI Appendix, Fig. S15F). These results demonstrate the impact of combining the genetic heterogeneity (PD-L1high/PD-L1low) and the cellular heterogeneity (PA+/PA−) in a single index increased the prediction accuracy of cancer immunotherapy.
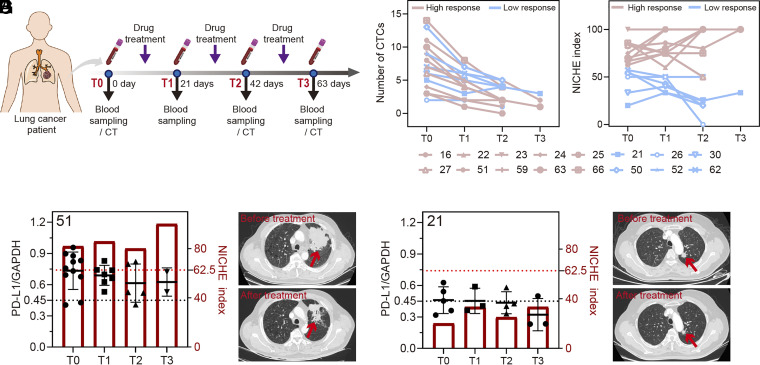
Clinically, patients undergoing immunotherapy may exhibit changes that lead to drug resistance, disease recurrence, or development of progressive disease (45). Building upon using the NICHE index as a prediction before any treatment, we explored the dynamic change of the NICHE index along with the trends of immunotherapy outcome over time. Of the 61 patients who underwent immunotherapy, we randomly selected 16 patients and tracked their outcomes throughout their drug treatment periods as a longitudinal evaluation of the index over time (Fig. 5A and SI Appendix, Table S10). The number of CTCs in all 16 patients shows a clear reduction after treatment (Fig. 5B), as for other studies (6, 46). However, the NICHE index provides more nuanced information (Fig. 5C). As discussed previously, patients with a high initial NICHE index obtained high response (Fig. 5 D and E and SI Appendix, Fig. S16 A–C and Table S10), relative to the patient with a low NICHE index (No. 21) whose final treatment outcome was clinically evaluated as MPR- (Fig. 5 F and G and SI Appendix, Fig. S16D). These results indicate a correlation between the initial NICHE index and the therapeutic efficacy over time.
Fig. 5.
NICHE for monitoring NSCLC patients throughout drug treatment periods. (A) A schematic diagram of treatment process and sampling for monitoring cancer progression. (B) The CTC numbers isolated from blood samples of NSCLC patients at different drug treatment periods. T0, T1, T2, and T3 represent the period of treatment. High response, patients with the response evaluation of pCR and PR. Low response, patients with the response evaluation of MPR, MPR-, SD, and PD. (C) NICHE index determined based on blood samples of NSCLC patients at different drug treatment periods. (D) NICHE indexes and ratios of the fluorescent intensity of PD-L1 (Cy3 channel) and GAPDH (FAM channel) for each CTCs isolated from the blood samples of patient No. 51 (D). (E) The CT images of patient No. 51 before and after drug treatment. (F) presents the same data as (D) for patient No. 21, whilst (G) provides the CT images before and after drug treatment also for patient No. 21. The red arrows indicate tumor location.
Notably, for this latter patient (No. 21) with a poor outcome, the NICHE index stayed constant throughout treatment (Fig. 5F), while for patient No. 51, the index increased upon treatment. This highlights the complexity of cellular responses and Fig. 5D may indicate that the NICHE index may be a better parameter for monitoring treatment, after the initial diagnostic decision (and further emphasizes the importance of monitoring cell heterogeneity at both a genetic and phenotypic level, in therapy prediction and personalized medicine).
Discussion
Microfluidic technologies have been widely used for the enrichment of low-abundance CTCs, and once captured, this has afforded the opportunity to explore cell heterogeneity, either through observing the cells’ phenotypic behavior or analyzing their transcriptome by direct lysis of captured CTCs (47). The design strategy and functionality of NICHE are different from many microfluidics methods for CTC detection, enabling us to simultaneously capture CTCs, quantify gene expression, and monitor the phenotype of the cells in situ in coculture with immune cells (with real-time observation of the cell response to immune attack). Using this platform, we demonstrate sensitive, specific, and accurate prediction of response to ICI-based therapy in NSCLC patients. In contrast to previous methods, there is no need to release cells from the device (all analysis is in situ), nor are there complicated analytical workflows during the genetic analysis (e.g., cell lysis for gene extraction and purification), both of which may lead to CTC being lost.
Currently, a gold standard is still lacking as a predictor guiding ICI treatment. The clinical reference TPS is currently based upon PD-L1 expression from a conventional tissue biopsy, with localized sampling and endpoint analysis, which do not readily afford the opportunity either for continuous sampling and/or for monitoring (4). In contrast to such conventional biopsy endpoint-analysis, studying CTCs in blood offers the possibility of monitoring therapeutic impacts or prediction, including using multiple markers such as PD-L1 and Ki-67 for example (48). Expanding beyond molecular markers only, our proposed NICHE index is calculated from both phenotypic and genetic data collected using a versatile micro- and nanofluidic system which generates real-time, correlative analysis from single living CTCs. This significantly improves spatiotemporal precision in understanding the specific behavior of individual cancer cells, providing the opportunity to understand important clinical phenomena, critical to in-patient care, including immune escape and drug resistance.
Systematic analysis highlighted the versatility of the NICHE platform, combining the multilayered functional microfluidics with magnetic actuation and microfluidic cell trap arrays to achieve a high efficiency of WBC removal (>93%) and on-chip CTC capture (~95%) in the blood-mimetic samples. These samples, representative of advanced disease, provide strong confidence that the technology will also be able to access the important information carried by the very few CTCs present in samples from patient at earlier stages (often 1 to 5 CTCs/samples). The stable, multichannel fluorescence probes (DNAT) provide real-time, precise quantification of mRNAs in living cells, demonstrating the capacity to distinguish gene heterogeneity between individual cells, while the NEED technique ensures safe, efficient, and rapid intracellular delivery of probes for in situ interrogation within living cells. The platform allowed us to explore in detail the role of ICI and the interplay of gene expression and cell phenotypic behaviors within immune environments in a specific lung cancer scenario, with important preliminary findings, supported by patient data, indicating that the PD-L1 mRNA expression in CTCs is not a strong indicator of the cancer stage. We also show that the phenotype alteration of CTCs collected from patient blood samples behaves in a manner consistent with CTC-mimic (cell lines) in response to immune cells, albeit with higher heterogeneity. Although we established the correlation between phenotype alteration of CTCs within immune environments with patients’ response to ICI-based therapy, the specific mechanism at play will require further exploration.
We also demonstrate that the NICHE index changes during the course of treatment, thereby providing a dynamic assessment of the cancer progression and the efficacy of treatment, with the future potential to be used as a predictive indicator to tailor the individual treatment. While we emphasize that early detection remains the key predictive indicator for survival and treatment efficacy, our results demonstrate the great potential for the NICHE platform and the associated index as a route to enable precision medicine for treating a range of cancers. The NICHE system could also in future be used for a wider range of cells as a tool to study the effect of the immune microenvironment and reveal further regulation mechanisms.
As CTC populations, even from the same patient sample, are well known to be highly heterogenous, causing significant variations in gene expressions and cellular behaviors (11, 45), the NICHE index [which considers two important factors (PD-L1 expression and cell elongation)] could be advantageously combined with other parameters or biomarkers to provide more information. For example, other liquid biopsy biomarkers such as ctDNA (49) and exosomal PD-L1 (50) have recently shown promise in following lung cancer patients treated with immunotherapy (2).
In addition, in the scenario where there are a limited number of CTCs, such as in early stage of the disease or following effective treatment, variations in the characteristics of CTCs may cause fluctuations in the efficacy of the NICHE index. SI Appendix, Table S3 allows to appraise the accuracy of the index for different numbers of CTCs, for the 52 samples that had clinical efficacy assessment results (SI Appendix, Tables S6 and S7). A valid NICHE index could be obtained for the sample with as few as two CTCs present. The accuracy of the index was strong for samples with more than eight CTCs (>80%), although more samples will be required to form a robust conclusion. To achieve a high confidence in the NICHE index in predicting patients, taking the blood sample more than one more time may increase the numbers of CTCs for analysis.
Materials and Methods
Materials.
The cell culture medium Dulbecco’s modified Eagle medium (DMEM, C11995500BT), RPMI 1640 medium (C11875500BT), streptavidin-modified magnetic beads (1 μm), cell membrane dye DiD (V-22887), DiO (V-22886), cell nuclear dye DAPI (62247), and fetal bovine serum (FBS, 10099141C) were purchased from Thermo Fisher Scientific. Calcein AM (C2012) was purchased from Beyotime. Penicillin-streptomycin (15140-122) and biotin-labeled anti-CD45 antibody (SAB4700477) were obtained from Sigma-Aldrich. Liposomes (029) were purchased from Fluorescence. Density gradient centrifugal fluid (Ficoll-Paque PREMIUM) was purchased from Cytiva. The DNA sequences and RNA sequences were synthesized by Sangon. The polycarbonate nanopore membrane (PET117745) was purchased from Wuweikejinxinfa Technology Co., Ltd. Indium tin oxide (ITO)–coated glass (thickness, 2 mm. ITO layer, 185 nm.) was purchased from Xiangcheng Technology Co., Ltd. The SYLGARD 184 Silicone Elastomer Kit (01673921) was purchased from Corning.
DNAT Probe Synthesis.
The components of DNAT probes including the sequences of T1, T2, T3, T4, P1, P2, and P3 (SI Appendix, Table S1) were respectively dissolved in 10 mM Tris-HCl solution containing 5 mM MgCl2 (pH 8), with a final concentration of 10 μM for each sequence. The obtained mixture was incubated at 95 °C for 5 min and then rapidly cooled down on ice. The structure of DNAT probe was verified by 1% agarose gel electrophoresis. For the target detection in buffer, the DNAT probes (1 μM) were mixed with the target sequence at 37 °C. The fluorescence intensity of the mixture was detected using a microplate reader (Spark, Tecan). In the enzymatic digestion experiments, fluorescence intensity was detected every 5 min after DNase I (1 U/mL, GD201-01, TransGen Biotech) was mixed with the probes.
Fabrication of NICHE Platform.
The microfluidic chip was assembled with the WBC-Removal Layer, CTC-Capture Layer, nanopore film, probe reservoir, and electrodes. The WBC-Removal Layer and CTC-Capture Layer were fabricated with standard soft lithography (see SI Appendix for details). The nanopore film (10 mm × 20 mm) was prepared with a commercial nanomembrane (nanopore diameter, 450 nm). For probe reservoir preparation, channels and chamber were punched in a PDMS layer (thickness, 1 mm).
The WBC-Removal Layer, CTC-Capture Layer, probe reservoir, and bottom electrodes were assembled by oxygen bonding, and the nanopore film and the CTC-Capture Layer were bonded by treating the CTC-Capture layer with an aminosilane, [aminoethylaminopropyl(trimethoxysilane), AEAPS], and then bonded to the nanopore film. The bottom ITO-coated glass and silver needle were used as electrodes to respectively connect the negative pole and positive pole of the electroporation instrument (ECM830, BTX) during probe delivery.
Isolation of CTCs and Gene Relative Quantification.
The cell samples (cancer cells within PBMCs) were mixed with CD45-MBs (10 μL CD45-MBs/mL cell sample) and cultured at 37 °C for 1 h. The samples were injected into the chip from the inlet by an injection pump. At the same time, PBS was injected from the buffer inlet. After separation with a magnet, the tumor cells and the remaining few WBCs were flown into the CTC-Capture Layer. After the tumor cells were trapped in the microfluidic cell traps, DNAT probes (1 μM) were injected into the probe reservoir. The needle-like electrode was inserted into the chip. The bottom electrode (negative pole) and the needle electrode (positive pole) were connected to an electroporation instrument for probe delivery. The cells were cultured in culture medium for 1 h. For probe delivery by liposome, 1 μM of DNAT probe was mixed with a liposome solution and then added into the cell culture medium with the cells for 1 h. For probe delivery by incubation, 1 μM of DNAT probe was added into the cell culture medium and incubated with the cells for 1 h. The cells were observed by a confocal microscope (CISM 100, Sunny). Flow cytometry (NovoCyte, Agilent) was used to determine the fluorescence intensity distribution of cells. To evaluate the delivery efficiency, DNAT probes that were only labeled with a FAM dye were used. The delivery efficiency was calculated based on the following equation:
To evaluate the cell viability, propidium iodide (PI, Solarbio) that can only enter porated cells was used to stain the cells for 10 min. The cell viability was calculated based on the following equation:
Coculture of Tumor Cells and Immune Cells.
Before coculture, HCC827 cells, A549 cells, or 293FT cells were stained with DiD dye (1:100) for 15 min. After washing three times, ca. 2,000 cells were dispensed into 96-well plates. To stimulate immune cells, phorbol 12-myristate 13-acetate (PMA, 50 ng/mL) and ionomycin (1 μM) were added into the culture medium and incubated for 48 h (31). After three PBS washes, 5,000 immune cells were added in the wells containing HCC827 cells, A549 cells, or 293FT cells. Then, 0.5 μL PD-1 inhibitor (0.6 mg/mL, Toripalimab, AtaGenix) was added into the corresponding wells. The morphology of HCC827 cells, A549 cells, or 293FT cells were recorded after coculture for 24 h, 48 h, and 72 h. In clinical samples, after imaging of CTCs, they were stained with DiD membrane dye and washed using culture medium. CTCs were then cultured on the chip until adherence. The steps for CTC coculture with immune cells were the same as for cell lines. The proliferation rate and inhibition rate were calculated by the absorbance of HCC827 cells, A549 cells, or 293FT cells (removal of immune cells) which were detected by a CCK8 kit (CK04, DOJINDO).
where As is the absorbance of experimental groups; Ac is the absorbance of each group for the first day; Ab is absorbance of blank groups.
where As is the absorbance of experimental groups; Ac is the absorbance of the control groups for that day (the cells without any treatment in the control groups); Ab is absorbance of blank groups.
NICHE Index.
The NICHE index is defined as the percentage of the CTC subgroup showing both PD-L1 high expression (PD-L1high) and clear phenotype alteration (PA+) in response to immune cells stimulation, as analyzed on the NICHE platform. The threshold for categorizing the subtype of PD-L1high and PD-L1low in cancer cells, including CTCs, is defined as the fluorescence intensity ratio of PD-L1/GAPDH (internal reference) emitted from DNAT probes within each single cell. Based upon the statistical analysis in Fig. 2H, we adopted the mean of fluorescent intensity ratio of PD-L1/GAPDH of 300 A549 cells and 300 HCC827 cells to be the threshold, which is given as 0.45. Within the coculture experiment with 293FT, we used 1.6, the maximum elongation that could be achieved by 293FT cells (a cell line without phenotypic morphological elongation), as the cutoff value for determining PA positive CTCs (PA+, cell elongation fold > 1.6) and PA negative CTCs (PA−, cell elongation fold ≤ 1.6). The change of cell elongation was calculated by dividing the length of the cell at 72 h by the length at 24 h. The cell length was defined as the longest path between two points on the cell membrane.
Clinical Samples.
Peripheral venous blood samples from each NSCLC patient were collected at Peking University Cancer Hospital & Institute. This study was approved by the Ethics Committee of Peking University Cancer Hospital & Institute (Institutional Review Board reference 2022KT45). Before sampling, each patient provided written informed consent (SI Appendix, Supplementary Methods).
Data Analysis.
The flow rate distribution and electric field distribution were simulated by COMSOL software (COMSOL Multiphysics 5.5), with the model and parameters given in SI Appendix, Fig. S5 D and E and Table S2. The SDs were calculated from the data of three independent experiments. For statistical analysis, two-sided Student t tests were used to calculate the P-value, with the CI labeled in each figure. Sensitivity = TP/(TP+FN). Specificity = TN/(TN+FP) (TP, number of true positive samples. TN, number of true negative samples. FP, number of false positive samples. FN, number of false negative samples). The raw data are displayed in SI Appendix, Table S9. All numerical results were analyzed by Origin 2017 and GraphPad Prism 8. Images were analyzed using ImageJ (21). The detection limit is defined as a signal from the test group greater than the blank value plus three times the blank error value.
Supplementary Material
Appendix 01 (PDF)
WBC removal inlet. The inlet area of the microfluidic chip, at the entrance of the WBC removal layer. The cells are flowing into the layer at the bottom of the video pushed by the buffer inlet on the top.
WBC removal through magnetic force. shows the WBC removal functionality (further from the area of Movie S1). Cells flow at the bottom of the channel when the magnet is off. When it is turned on, the labeled WBCs are attracted to the top of the channel.
WBC removal outlet. shows the outcome of the WBC removal (futher from the area of Movie S2). Cells that were deflected are exiting the channel on the top outlet, whilst unaffected cells can be seen leaving the channel by the bottom inlet.
CTC-capture. Demonstrates the CTC-capture functionality, with a single cell being trapped in the highlighted “micro-fence“.
NEED function. shows a fluorescent observation of a cell trapped in the capture layer (artificially recolored in red for ease of referencing), undergoing the NEED treatment to incorporate a cy5-DNAT probe.
Cell morphology – elongated. shows a single cell tagged with DiO, overlayed with a brightfield image showing the elongation after stimulation over 72h.
Cell morphology – no elongation. Same condition as Movie S6 but with an unstimulated cell, which does not change in morphology.
Cell morphology – control. Same condition as Movies S6 and S7 as a control (no activation).
Acknowledgments
This work was supported by the following funding: National Natural Science Foundation of China (22304006), National Key Research and Development Program of China (2022YFB3205600), National Key Research and Development Program of China (2021YFC2302100), Beijing Natural Science Foundation (7212204), National Natural Science Foundation of China (32071407), National Natural Science Foundation of China (62003023), Fundamental Research Funds for the Central Universities (JKF-YG-22-B001), Science Foundation of Peking University Cancer Hospital 2022-5 (A002476), and United Kingdom Research and Innovation, Engineering and Physical Sciences Research Council (EP/S02347X/1).
Author contributions
N.W., M.Y., and L.C. designed research; Z.D., Yusen Wang, Yang Wang, P.M., F.L., Y.Z., Z.J., and M.Y. performed research; B.L., S.Y., M.Z., B.J., J.F., and P.Z. contributed new reagents/analytic tools; Z.D., Yusen Wang, G.X., Yang Wang, J.R., P.J., Y.Z., K.Y., Z.H., N.W., M.Y., J.M.C., and L.C. analyzed data; and Z.D., G.X., J.R., P.J., M.Y., J.M.C., and L.C. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission. C.T.L. is a guest editor invited by the Editorial Board.
Contributor Information
Nan Wu, Email: nanwu@bjmu.edu.cn.
Mingzhu Yang, Email: yixin051@163.com.
Jonathan M. Cooper, Email: Jon.Cooper@glasgow.ac.uk.
Lingqian Chang, Email: lingqianchang@buaa.edu.cn.
Data, Materials, and Software Availability
Raw data have been deposited in research data/Enlighten (51).
Supporting Information
References
- 1.Sangro B., Sarobe P., Hervás-Stubbs S., Melero I., Advances in immunotherapy for hepatocellular carcinoma. Nat. Rev. Gastroenterol. Hepatol. 18, 525–543 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ribas A., Wolchok J. D., Cancer immunotherapy using checkpoint blockade. Science 359, 1350–1355 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Doroshow D. B., et al. , PD-L1 as a biomarker of response to immune-checkpoint inhibitors. Nat. Rev. Clin. Oncol. 18, 345–362 (2021). [DOI] [PubMed] [Google Scholar]
- 4.Li W., et al. , Liquid biopsy in lung cancer: Significance in diagnostics, prediction, and treatment monitoring. Mol. Cancer 21, 25 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ignatiadis M., Sledge G. W., Jeffrey S. S., Liquid biopsy enters the clinic—implementation issues and future challenges. Nat. Rev. Clin. Oncol. 18, 297–312 (2021). [DOI] [PubMed] [Google Scholar]
- 6.Deng Z., Wu S., Wang Y., Shi D., Circulating tumor cell isolation for cancer diagnosis and prognosis. EBioMedicine 83, 104237 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Guo L., et al. , Recent progress of nanostructure-based enrichment of circulating tumor cells and downstream analysis. Lab Chip 23, 1493–1523 (2023). [DOI] [PubMed] [Google Scholar]
- 8.Mishra A., et al. , Ultrahigh-throughput magnetic sorting of large blood volumes for epitope-agnostic isolation of circulating tumor cells. Proc. Natl. Acad. Sci. U.S.A. 117, 16839–16847 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Boya M., et al. , High throughput, label-free isolation of circulating tumor cell clusters in meshed microwells. Nat. Commun. 13, 3385 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki T., Kaji N., Yasaki H., Yasui T., Baba Y., Mechanical low-pass filtering of cells for detection of circulating tumor cells in whole blood. Anal. Chem. 92, 2483–2491 (2020). [DOI] [PubMed] [Google Scholar]
- 11.Keller L., Pantel K., Unravelling tumour heterogeneity by single-cell profiling of circulating tumour cells. Nat. Rev. Cancer 19, 553–567 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Bu J., et al. , Surface engineering for efficient capture of circulating tumor cells in renal cell carcinoma: From nanoscale analysis to clinical application. Biosens. Bioelectron. 162, 112250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng Y. H., et al. , Hydro-Seq enables contamination-free high-throughput single-cell RNA-sequencing for circulating tumor cells. Nat. Commun. 10, 2163 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Guibert N., et al. , PD-L1 expression in circulating tumor cells of advanced non-small cell lung cancer patients treated with nivolumab. Lung Cancer 120, 108–112 (2018). [DOI] [PubMed] [Google Scholar]
- 15.Dhar M., et al. , Evaluation of PD-L1 expression on vortex-isolated circulating tumor cells in metastatic lung cancer. Sci. Rep. 8, 2592 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Topalian S. L., Taube J. M., Pardoll D. M., Neoadjuvant checkpoint blockade for cancer immunotherapy. Science 367, eaax0182 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wlodkowic D., Faley S., Zagnoni M., Wikswo J. P., Cooper J. M., Microfluidic single-cell array cytometry for the analysis of tumor apoptosis. Anal. Chem. 81, 5517–5523 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Faley S. L., et al. , Microfluidic single cell arrays to interrogate signalling dynamics of individual, patient-derived hematopoietic stem cells. Lab Chip 9, 2659–2664 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Faley S., Copland M., Reboud J., Cooper J. M., Intracellular protein trafficking kinetics in chronic myeloid leukemia stem cells using a microfluidic platform. Integr. Biol. (Camb) 4, 368–373 (2012). [DOI] [PubMed] [Google Scholar]
- 20.Dong Z., et al. , Single living cell analysis nanoplatform for high-throughput interrogation of gene mutation and cellular behavior. Nano Lett. 21, 4878–4886 (2021). [DOI] [PubMed] [Google Scholar]
- 21.Sun H., et al. , Companion-probe & race platform for interrogating nuclear protein and migration of living cells. Biosens. Bioelectron. 210, 114281 (2022). [DOI] [PubMed] [Google Scholar]
- 22.Dhar M., et al. , Functional profiling of circulating tumor cells with an integrated vortex capture and single-cell protease activity assay. Proc. Natl. Acad. Sci. U.S.A. 115, 9986–9991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hücker S. M., et al. , Single-cell microRNA sequencing method comparison and application to cell lines and circulating lung tumor cells. Nat. Commun. 12, 4316 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poudineh M., et al. , Tracking the dynamics of circulating tumour cell phenotypes using nanoparticle-mediated magnetic ranking. Nat. Nanotechnol. 12, 274–281 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Dong Z., et al. , A microwell-assisted multiaptamer immunomagnetic platform for capture and genetic analysis of circulating tumor cells. Adv. Healthcare Mater. 7, 1801231 (2018). [DOI] [PubMed] [Google Scholar]
- 26.Dong Z., Xue X., Liang H., Guan J., Chang L., DNA nanomachines for identifying cancer biomarkers in body fluids and cells. Anal. Chem. 93, 1855–1865 (2021). [DOI] [PubMed] [Google Scholar]
- 27.Gallego-Perez D., et al. , Topical tissue nano-transfection mediates non-viral stroma reprogramming and rescue. Nat. Nanotechnol. 12, 974–979 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sheng Y., et al. , Living cell nanoporation and exosomal RNA analysis platform for real-time assessment of cellular therapies. J. Am. Chem. Soc. 144, 9443–9450 (2022). [DOI] [PubMed] [Google Scholar]
- 29.Chang L., et al. , 3D nanochannel electroporation for high-throughput cell transfection with high uniformity and dosage control. Nanoscale 8, 243–252 (2016). [DOI] [PubMed] [Google Scholar]
- 30.Hong X., et al. , Molecular signatures of circulating melanoma cells for monitoring early response to immune checkpoint therapy. Proc. Natl. Acad. Sci. U.S.A. 115, 2467–2472 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jiang X., et al. , Cancer-on-a-chip for modeling immune checkpoint inhibitor and tumor interactions. Small 17, e2004282 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amorim S., et al. , Hyaluronic acid of low molecular weight triggers the invasive “Hummingbird” phenotype on gastric cancer cells. Adv. Biosyst. 4, e2000122 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Bessède E., et al. , Helicobacter pylori generates cells with cancer stem cell properties via epithelial-mesenchymal transition-like changes. Oncogene 33, 4123–4131 (2014). [DOI] [PubMed] [Google Scholar]
- 34.Costello A. M., et al. , Selection and characterization of vimentin-binding aptamer motifs for ovarian cancer. Molecules (Basel, Switzerland) 26, 6525 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wan J., et al. , Hypoxia-induced PLOD2 regulates invasion and epithelial-mesenchymal transition in endometrial carcinoma cells. Genes Genomics 42, 317–324 (2020). [DOI] [PubMed] [Google Scholar]
- 36.Zhao W., Qian H., Zhang R., Gao X., Gou X., MicroRNA targeting microtubule cross-linked protein (MACF1) would suppress the invasion and metastasis of malignant tumor. Med. Hypotheses 104, 25–29 (2017). [DOI] [PubMed] [Google Scholar]
- 37.Liang J., Sun Z., Overexpression of membranal SLC3A2 regulates the proliferation of oral squamous cancer cells and affects the prognosis of oral cancer patients. J. Oral Pathol. Med. 50, 371–377 (2021). [DOI] [PubMed] [Google Scholar]
- 38.Adamska A., et al. , ABCC3 is a novel target for the treatment of pancreatic cancer. Adv. Biol. Regul. 73, 100634 (2019). [DOI] [PubMed] [Google Scholar]
- 39.Zhang H., Cui X., Cao A., Li X., Li L., ITGA3 interacts with VASP to regulate stemness and epithelial-mesenchymal transition of breast cancer cells. Gene 734, 144396 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Dongre A., Weinberg R. A., New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 20, 69–84 (2019). [DOI] [PubMed] [Google Scholar]
- 41.de Bono J. S., et al. , Circulating tumor cells predict survival benefit from treatment in metastatic castration-resistant prostate cancer. Clin. Cancer Res. 14, 6302–6309 (2008). [DOI] [PubMed] [Google Scholar]
- 42.Fabisiewicz A., Grzybowska E., CTC clusters in cancer progression and metastasis. Med. Oncol. 34, 12 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Sinoquet L., et al. , Programmed cell death ligand 1-expressing circulating tumor cells: A new prognostic biomarker in non-small cell lung cancer. Clin Chem. 67, 1503–1512 (2021). [DOI] [PubMed] [Google Scholar]
- 44.Jiang X., et al. , Role of the tumor microenvironment in PD-L1/PD-1-mediated tumor immune escape. Mol. Cancer 18, 10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Dagogo-Jack I., Shaw A. T., Tumour heterogeneity and resistance to cancer therapies. Nat. Rev. Clin. Oncol. 15, 81–94 (2018). [DOI] [PubMed] [Google Scholar]
- 46.Strati A., et al. , Prognostic significance of PD-L1 expression on circulating tumor cells in patients with head and neck squamous cell carcinoma. Ann. Oncol. 28, 1923–1933 (2017). [DOI] [PubMed] [Google Scholar]
- 47.Lim S. B., et al. , Addressing cellular heterogeneity in tumor and circulation for refined prognostication. Proc. Natl. Acad. Sci. U.S.A. 116, 17957–17962 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Spiliotaki M., et al. , Dynamic monitoring of PD-L1 and Ki67 in circulating tumor cells of metastatic non-small cell lung cancer patients treated with pembrolizumab. Mol Oncol. 17, 792–809 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Assaf Z. J. F., et al. , A longitudinal circulating tumor DNA-based model associated with survival in metastatic non-small-cell lung cancer. Nat. Med. 29, 859–868 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.de Miguel-Perez D., et al. , Extracellular vesicle PD-L1 dynamics predict durable response to immune-checkpoint inhibitors and survival in patients with non-small cell lung cancer. J. Exp. Clin. Cancer Res. 41, 186 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dong Z., et al. , Data from “Genetic and phenotypic profiling of single living circulating tumor cells from patients with microfluidics”. Figshare. 10.6084/m9.figshare.3178534.v2. Deposited 15 April 2024. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
WBC removal inlet. The inlet area of the microfluidic chip, at the entrance of the WBC removal layer. The cells are flowing into the layer at the bottom of the video pushed by the buffer inlet on the top.
WBC removal through magnetic force. shows the WBC removal functionality (further from the area of Movie S1). Cells flow at the bottom of the channel when the magnet is off. When it is turned on, the labeled WBCs are attracted to the top of the channel.
WBC removal outlet. shows the outcome of the WBC removal (futher from the area of Movie S2). Cells that were deflected are exiting the channel on the top outlet, whilst unaffected cells can be seen leaving the channel by the bottom inlet.
CTC-capture. Demonstrates the CTC-capture functionality, with a single cell being trapped in the highlighted “micro-fence“.
NEED function. shows a fluorescent observation of a cell trapped in the capture layer (artificially recolored in red for ease of referencing), undergoing the NEED treatment to incorporate a cy5-DNAT probe.
Cell morphology – elongated. shows a single cell tagged with DiO, overlayed with a brightfield image showing the elongation after stimulation over 72h.
Cell morphology – no elongation. Same condition as Movie S6 but with an unstimulated cell, which does not change in morphology.
Cell morphology – control. Same condition as Movies S6 and S7 as a control (no activation).
Data Availability Statement
Raw data have been deposited in research data/Enlighten (51).