Significance
Toll-like receptor 7 (TLR7) is vital for innate immunity, recognizing single-stranded RNAs (ssRNAs) as ligands. While microRNAs have been the primary focus of endogenous TLR7 ligand research, recent advances in short noncoding RNA biology have expanded the range of candidate ssRNA molecules. This study comprehensively explored extracellular (ex-) transfer RNA (tRNA)-derived RNAs in extracellular vesicles (EVs) secreted from human macrophages and identified the 5′-tRNAValCAC/AAC half as a potent activator of TLR7. The terminal GUUU sequence of the 5′-tRNAValCAC/AAC half was identified as a universal signature for TLR7 activation. In patients infected with Mycobacterium tuberculosis, the levels of the extracellular 5′-tRNAValCAC/AAC half are drastically upregulated. These findings provide insights into a variety of TLR7-related pathologies and potential therapeutic strategies.
Keywords: tRNA half, TLR7, short non-coding RNA, macrophage, Valine
Abstract
Toll-like receptors (TLRs) are crucial components of the innate immune system. Endosomal TLR7 recognizes single-stranded RNAs, yet its endogenous ssRNA ligands are not fully understood. We previously showed that extracellular (ex-) 5′-half molecules of tRNAHisGUG (the 5′-tRNAHisGUG half) in extracellular vesicles (EVs) of human macrophages activate TLR7 when delivered into endosomes of recipient macrophages. Here, we fully explored immunostimulatory ex-5′-tRNA half molecules and identified the 5′-tRNAValCAC/AAC half, the most abundant tRNA-derived RNA in macrophage EVs, as another 5′-tRNA half molecule with strong TLR7 activation capacity. Levels of the ex-5′-tRNAValCAC/AAC half were highly up-regulated in macrophage EVs upon exposure to lipopolysaccharide and in the plasma of patients infected with Mycobacterium tuberculosis. The 5′-tRNAValCAC/AAC half-mediated activation of TLR7 effectively eradicated bacteria infected in macrophages. Mutation analyses of the 5′-tRNAValCAC/AAC half identified the terminal GUUU sequence as a determinant for TLR7 activation. We confirmed that GUUU is the optimal ratio of guanosine and uridine for TLR7 activation; microRNAs or other RNAs with the terminal GUUU motif can indeed stimulate TLR7, establishing the motif as a universal signature for TLR7 activation. These results advance our understanding of endogenous ssRNA ligands of TLR7 and offer insights into diverse TLR7-involved pathologies and their therapeutic strategies.
One of the fundamental properties of the immune system is to initiate immune responses against invasive pathogens by recognizing conserved pathogen-associated molecular patterns (PAMPs) (1, 2). The immune system can also recognize intracellular molecules of host cells released by stressed or injured tissues, so-called damage-associated molecular patterns (DAMPs), such as histones, self-DNAs, or -RNAs (3–5). Toll-like receptors (TLRs) and other pathogen-recognition receptors in the innate immune system detect PAMPs and DAMPs, initiating protective responses (6, 7). Among the 10 TLRs characterized in humans, TLR1, -2, -4, -5, -6, and -10 localize to the cell surface (surface TLRs), while TLR3, -7, -8, and -9 localize to intracellular compartments such as endosomes (endosomal TLRs). While the surface TLRs mainly recognize bacterial and fungal cell wall components, viral envelope proteins, and protozoal components, endosomal TLRs are nucleic acid-sensing receptors; TLR7 and -8 are specialized in detecting single-stranded RNAs (ssRNAs), while TLR3 and -9 sense double-stranded RNAs (dsRNAs) and single-stranded DNAs, respectively (8). ssRNA recognition by TLR7 or -8 results in the recruitment of the adaptor protein MyD88, leading to IRF7- and NF-κB-mediated transcription and downstream induction of interferon and cytokine production (6, 8).
While “foreign” ssRNAs from bacteria or viruses have been well studied as the ligands of TLR7 and -8 (9–14), “endogenous” ssRNA ligands from host cells have not been fully characterized yet. To be recognized by TLR7 and -8, ssRNAs should be present inside the endosomes where TLR7 and -8 reside, which can be achieved via extracellular vesicle (EV)-mediated delivery of ssRNAs to endosomes. Indeed, certain EV-contained extracellular (ex-) microRNAs (miRNAs) encapsulated within EVs are transported to the endosomes of recipient cells where they activate TLR7 and -8 (15–18). This miRNA-mediated TLR7/8 activation is relevant not only to immune response (19, 20) but also to neurodegeneration (15), cancers (21, 22), autoimmunity (23, 24), and various other diseases (25).
Although miRNAs have dominated current research on both ex-short noncoding RNAs (sncRNAs) and endogenous ssRNA ligands of TLRs, recent advances in our understanding of “previously hidden” sncRNAs have widened the pool of the candidate ssRNA molecules of TLR7/8 ligands. sncRNAs generally possess either a hydroxyl group (OH), a phosphate (P), or a 2′,3′-cyclic phosphate (cP) at their termini (26). To date, most sncRNA sequencing studies have relied on a standard small RNA-seq method in which 5′- and 3′-adaptors (AD) can be ligated only to the 5′-P and 3′-OH ends of RNAs; thus, current sncRNA analyses significantly underrepresent non-miRNA-sncRNAs lacking the 5′-P or 3′-OH ends (26–28). This notion is especially important when it comes to the sequencing analyses of ex-sncRNAs because many ex-sncRNAs lack 5′-P or 3′-OH (29–31). Pretreatment of RNAs with T4 polynucleotide kinase (T4 PNK) to convert the RNA termini to 5′-P/3′-OH-ends (thus rendering them available for 5′-/3′-AD ligation) is required to capture whole ex-sncRNAs (29–31). In our sequencing study of sncRNAs in EVs secreted from human monocyte-derived macrophages (HMDMs), T4 PNK treatment was required to amplify the majority of the cDNAs (32). Treating with a mutant form of T4 PNK, which is deficient in 3′-dephosphorylation activity, resulted in a dramatic reduction of cDNA yield (32), indicating that the majority of sncRNAs within HMDM EVs contain 3′-P or cP, while miRNAs and other 3′-OH-containing RNAs constitute only a minor proportion. Among the EV-sncRNAs, 5′-tRNA halves were found to be one of the major species, comprising over 93% of tRNA-derived sncRNAs in HMDM-secreted EVs (32).
tRNA halves are the most abundant class of tRNA-derived sncRNAs. In mammalian cells, they are generated from angiogenin (ANG)-mediated anticodon cleavage of tRNAs, which can be induced by various biological factors/phenomena such as stress stimuli (33–35) and sex-hormone signaling pathways (36). tRNA halves are expressed as functional sncRNAs that promote stress granule formation (37), regulate translation (33, 38), and promote cell proliferation (36). Our previous study found that mycobacterial infection and accompanying surface TLR activation lead to upregulation of NF-κB-mediated ANG gene transcription, inducing the accumulation of tRNA halves in HMDMs (32). Then, the 5′-tRNA halves are selectively and abundantly packaged into EVs of HMDMs. Among ex-5′-tRNA halves in HMDM EVs, the ex-5′-tRNAHisGUG half is abundantly accumulated (>210 times more than miR-150, the most abundant ex-miRNA) and delivered to endosomes of recipient HMDMs, where it strongly activates TLR7 (32). The strength of its activity was equal to or even more than that of HIV-derived ssRNA40, a widely used positive control ssRNA known as a strong activator of endosomal TLRs (9), suggesting that the 5′-tRNAHisGUG half has the capacity to produce an immune response (32). While these findings suggest that 5′-tRNA halves serve as immune activators (39), only the 5′-tRNAHisGUG half had been demonstrated to activate endosomal TLR.
In this study, by widely exploring immunostimulatory ex-5′-tRNA half molecules, we identified the 5′-tRNAValCAC/AAC half as another strong TLR7 activator. We further investigated sequence determinants within these 5′-tRNA halves for TLR7 activation to shed light on why only the 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half exhibit strong activity. Our findings provide a fuller understanding of the role of endogenously produced 5′-tRNA half molecules in activating the endosomal TLR pathway and suggest the wide existence of other endogenous ssRNA molecules that function as ligands for endosomal TLRs.
Results
The 5′-tRNAValCAC/AAC Half Activates TLR7.
Our previous sequencing of T4 PNK-treated sncRNAs from HMDM-secreted EVs revealed 5′-tRNA half as the most major class of ex-tRNA-derived sncRNAs (32). To explore active 5′-tRNA halves in endosomal TLR stimulation, we focused on abundant 5′-tRNA half species whose read numbers in the sequencing data of HMDM EV samples were >1% of total 5′-tRNA half reads (SI Appendix, Table S1). Given that the human genome encodes 55 cytoplasmic (cyto) tRNA isoacceptors, each with distinct anticodon sequences (40), the 17 species of 5′-tRNA halves that were extracted originate from a specific subset of cyto tRNA isoacceptors: cyto tRNAValCAC/AAC, tRNAHisGUG, tRNAGlyGCC/CCC, and tRNAGluCUC (the 5′-tRNA half-derived sequences of some isodecoders of tRNAValCAC and tRNAValAAC, or tRNAGlyGCC and tRNAGlyCCC, are identical and thus indistinguishable). The four cyto tRNAs were in aggregate the sources of over 90% of the identified EV-5′-tRNA halves (SI Appendix, Table S2). Among 5′-tRNA halves from these tRNAs, the 5′-tRNAHisGUG half has already been shown to activate TLR7, while the 5′-tRNAGluCUC half was inactive (32). We further explored the activity of the rest of the major 5′-tRNA halves, 5′-tRNAValCAC/AAC half, and 5′-tRNAGlyGCC/CCC half, whose most abundant sequences are derived from nucleotide position [np: as per the nucleotide numbering system specific to tRNAs (41)] 1 to 33 and 1 to 32, respectively (SI Appendix, Tables S1 and S3).
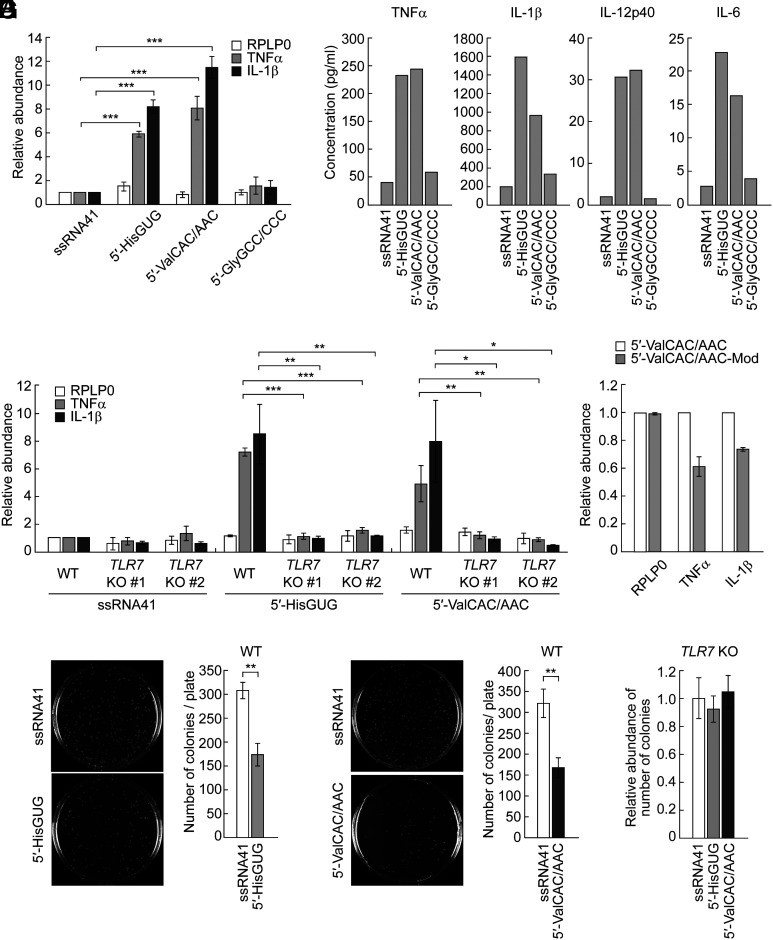
To deliver the 5′-tRNA halves into the endosomes of HMDMs, the cells were transfected with the RNAs using the cationic liposome 1,2-dioleoyloxy-3-trimethylammonium-propane (DOTAP). DOTAP, serving as an EV mimic, actively facilitates the delivery of encapsulated RNAs into the endosomes of recipient cells and has been widely adopted for the endosomal delivery of RNAs in previous studies (32, 42–44). As a negative control, an inactive mutant of 20-nucleotide (nt) HIV-1-derived ssRNA termed ssRNA41 (9, 32) was also transfected. As shown in Fig. 1A, transfections of the 5′-tRNAValCAC/AAC half, as well as the 5′-tRNAHisGUG half, enhanced the levels of TNFα and IL-1β mRNAs, whereas transfections of the 5′-tRNAGlyGCC/CCC half and ssRNA41 did not. We further observed the induction of the secretion of cytokines, such as TNFα, IL-1β, IL-12p40, and IL-6, into the culture medium upon the transfection of the 5′-tRNAValCAC/AAC half and the 5′-tRNAHisGUG half (Fig. 1B). The upregulation of cytokine expression upon DOTAP-mediated transfection of the two 5′-tRNA halves was not observed in both of the two clones of TLR7 knockout (KO) HMDMs whose TLR7 expression was completely depleted (32) (Fig. 1C). Collectively, these results identified the 5′-tRNAValCAC/AAC half, in addition to the 5′-tRNAHisGUG half, as a TLR7-stimulating molecule, while the 5′-tRNAGlyGCC/CCC half has been confirmed to be inactive in endosomal TLR stimulation.
Fig. 1.
The 5′-tRNAValCAC/AAC half activates TLR7. (A) Using DOTAP, 5′-tRNA halves and ssRNA41 (negative control) were transfected into HMDMs. Total RNAs from the cells were subjected to RT-qPCR for the indicated mRNAs. The quantified mRNA levels were normalized to the levels of GAPDH mRNA. For all graphs in the present study, error bars indicate mean ± SD of triplicate measurements (*P < 0.05, **P < 0.01, and ***P < 0.001; two-tailed t test). (B) After RNA transfection into HMDMs using DOTAP, the culture medium was subjected to measurement of concentration of the indicated cytokines. Means of two independent experiments are shown. The experimental results from three replicates for both the 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half are presented in Fig. 3B, while those for the 5′-tRNAGlyGCC/CCC half are shown in SI Appendix, Fig. S1. (C and D) The experiments in (A) were performed by using two different TLR7 KO THP-1 cell clones (C) or the 5′-tRNAValCAC/AAC half with modifications (D). (E and F) HMDMs transfected with the 5′-tRNAHisGUG half (F) or the 5′-tRNAValCAC/AAC half (G) were subjected to bacterial infection and invasion assay. Transfection of ssRNA41 was used as a negative control. Representative pictures of the plates with E. coli colonies and bar graphs of the counted numbers of colonies are shown. (G) The experiments in (F) and (G) were performed by using TLR7 KO THP-1 cell #1.
Earlier studies have shown that posttranscriptional modifications in tRNAs or other RNAs can affect their activity in stimulating endosomal TLRs (45–47). Among the two active 5′-tRNA halves, our previous study has demonstrated that the 5′-tRNAHisGUG half containing endogenous modifications [dihydrouridine (D) at np 16, 19, and 20, and pseudouridine (Ψ) at np 32] maintains a similar potency to its unmodified version in stimulating TLR7. Regarding the 5′-tRNAValCAC/AAC half, based on the information from mature tRNAValCAC/AAC (48), it is expected to contain the following three species of modifications: Ψ at np 13 and 27; D at np 16, 19, and 20; and 2-methylguanosine (m2G) at np 26. As shown in Fig. 1D, the 5′-tRNAValCAC/AAC half containing all these modifications exhibited approximately 30 to 40% reduced activity compared to its unmodified version. These results suggest that the modified endogenous 5′-tRNAValCAC/AAC half is still a potent TLR7 activator, although the modifications negatively influence the activity.
We further examined whether the 5′-tRNA half-mediated pathways contribute to bacterial elimination. After DOTAP-mediated transfection of the 5′-tRNAHisGUG half or 5′-tRNAValCAC/AAC half, HMDMs were infected with Escherichia coli. Subsequent analysis revealed a significant reduction in E. coli colony numbers in the plates from HMDMs transfected with the 5′-tRNAHisGUG half or 5′-tRNAValCAC/AAC half compared to those transfected with negative control ssRNA41 (Fig. 1 E and F). In contrast, E. coli colony numbers were unchanged even upon the transfection of the 5′-tRNA halves when TLR7 KO HMDMs were used (Fig. 1G). Taken together, TLR7 activation by these 5′-tRNA half molecules enhances the antibacterial properties of HMDMs, effectively eradicating the infected bacteria.
The Levels of the 5′-tRNAValCAC/AAC Half Are Up-Regulated in HMDMs Exposed to Lipopolysaccharide (LPS) and in Patients Infected with Mycobacterium tuberculosis (Mtb).
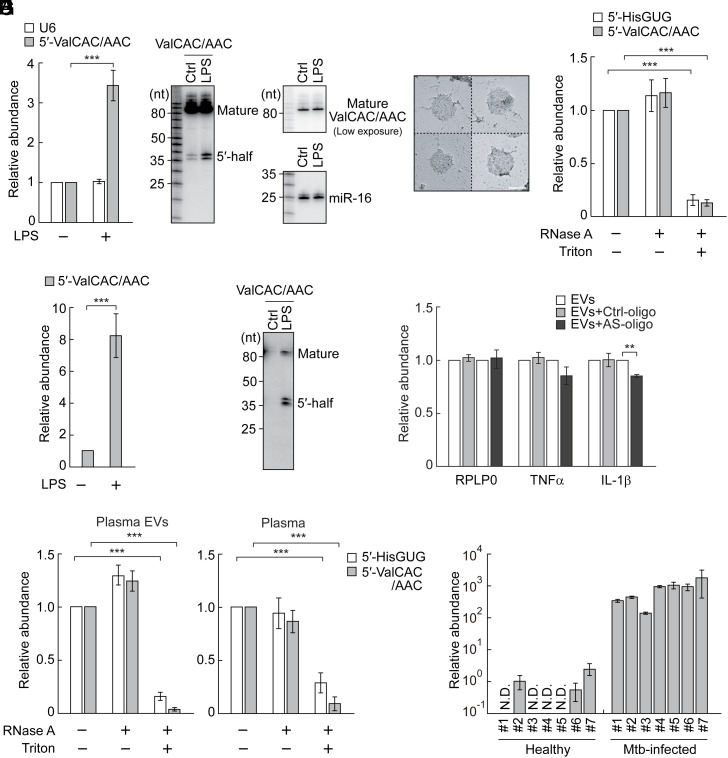
tRNAValCAC/AAC are the most major sources of ex-5′-tRNA halves (SI Appendix, Table S1), and the 5′-tRNAValCAC/AAC half is the most abundant ex-tRNA-derived sncRNA in HMDM EVs (SI Appendix, Table S2). Given its activity in TLR7 stimulation and bacterial elimination, we further elucidated its expressional regulation. Our TaqMan RT-qPCR quantification indicated upregulation of the levels of the 5′-tRNAValCAC/AAC half in HMDMs upon stimulation of TLR4 by LPS treatment (Fig. 2A). Northern blot confirmed the LPS-induced upregulation of the 5′-tRNAValCAC/AAC half, while the levels of mature tRNAValCAC/AAC were not affected by LPS treatment (Fig. 2B). To confirm whether EVs secreted from HMDMs carry the 5′-tRNAValCAC/AAC half, we obtained EVs from the culture medium of LPS-treated HMDMs using an ultracentrifugation-based method. Transmission electron microscopy (Fig. 2C), as well as nanoparticle tracking analysis and western blot, verified the successful isolation of HMDM EVs as in our previous study (32). EVs treated solely with RNase A and those left untreated showed comparable amplification signals in TaqMan RT-qPCR for the 5′-tRNAValCAC/AAC half. In contrast, EVs treated with both RNase A and a detergent exhibited significantly diminished signals (Fig. 2D), confirming that the 5′-tRNAValCAC/AAC half is located inside the EVs, rather than present as non-EV contaminants. LPS treatment up-regulated the levels of the 5′-tRNAValCAC/AAC half not only in HMDMs (Fig. 2 A and B) but also in their EVs (Fig. 2 E and F); the fold change of the upregulation was more evident in EVs than in HMDMs. In our northern blot analyses, the lower 33 nt band of the 5′-tRNAValCAC/AAC half is more abundant than its longer 34 nt band in EVs (Fig. 2F), while the 34 nt band is more prominent than the 33 nt band in HMDMs (Fig. 2B). These results are consistent with our sequencing data (32), and suggest selective packaging of the 33 nt version of the 5′-tRNAValCAC/AAC half into EVs. Collectively, these results suggest that the 5′-tRNAValCAC/AAC half is an immune-responsive molecule whose accumulation and packaging to EVs are induced by surface TLR activation.
Fig. 2.
Enhanced accumulation of the ex-5′-tRNAValCAC/AAC half in EVs from LPS-treated HMDMs and in plasma from Mtb-infected patients. (A) Total RNAs from HMDMs treated with LPS were subjected to TaqMan RT-qPCR for the 5′-tRNAValCAC/AAC half. U6 snRNA was quantified as a control. The quantified RNA levels were normalized to the levels of 5S rRNA. (B) Total RNAs from HMDMs treated with LPS were subjected to northern blot for the 5′-tRNAValCAC/AAC half and its corresponding mature tRNA. miR-16 was analyzed as a control. (C) Transmission electron microscopic evaluation of the isolated EVs. Four representative EV images are shown. (Scale bar, 100 nm.) (D) HMDM-secreted EVs were treated with RNase A and/or Triton X-100 and then subjected to TaqMan RT-qPCR for quantification of 5′-tRNA halves. (E and F) Total RNAs from HMDM-secreted EVs were subjected to TaqMan RT-qPCR (E) and northern blot (F) for the 5′-tRNAValCAC/AAC half. (G) EVs from LPS-treated HMDMs were mixed with DOTAP-fused AS-oligo of the 5′-tRNAValCAC/AAC half or Ctrl-oligo with scrambled sequences and applied to recipient HMDMs, followed by quantification of the indicated mRNAs. (H) Human plasma and its isolated EVs were treated with RNase A and/or Triton X-100 and then subjected to TaqMan RT-qPCR for quantification of 5′-tRNA halves. (I) RNAs isolated from plasma samples of healthy individuals or Mtb-infected patients were subjected to TaqMan RT-qPCR for the 5′-tRNAValCAC/AAC half. The quantified 5′-tRNA half levels were normalized to spike-in RNA levels. The relative abundances to the healthy sample #2 (set as 1) are shown.
Considering the abundant accumulation of the 5′-tRNAValCAC/AAC half in HMDM EVs, we further analyzed the activity of the EV-contained, endogenous 5′-tRNAValCAC/AAC half in stimulating TLR7 by utilizing antisense oligonucleotide (AS-oligo) of the 5′-tRNAValCAC/AAC half and control oligonucleotide (Ctrl-oligo) with scrambled sequences. Application of the isolated HMDM EVs to recipient HMDMs up-regulated the levels of TNFα and IL-1β mRNA as described in our previous study (32). The AS-oligo reduced the EV-induced upregulation of cytokine mRNAs, while Ctrl-oligo did not show such activity (Fig. 2G). The reduction by the AS-oligo was not as pronounced as that observed for the 5′-tRNAHisGUG half in our previous study (32). This difference might be due to the inherently lower TLR7 stimulation activity of the endogenous 5′-tRNAValCAC/AAC half compared to the 5′-tRNAHisGUG half. While posttranscriptional modifications do not adversely affect the TLR7 stimulation activity of the 5′-tRNAHisGUG half (32), they reduce the activity of the 5′-tRNAValCAC/AAC half (Fig. 1D). Despite this, our findings suggest that the endogenous 5′-tRNAValCAC/AAC half, incorporated in EVs, still possesses significant potential to induce cytokine production by stimulating TLR7.
We further examined the accumulation of the 5′-tRNAValCAC/AAC half in human plasma samples. Our upgraded version of TaqMan RT-qPCR for limited amounts of RNA materials (49) successfully detected the 5′-tRNAValCAC/AAC half in the EVs isolated from human plasma (Fig. 2H). As in the case of HMDM EVs, drastic reduction of amplification signals upon treatments with both RNase A and detergent, but not with RNase A treatment alone, confirmed the presence of the 5′-tRNAValCAC/AAC half inside the plasma EVs. As the quantification of the 5′-tRNA half using plasma (not the isolated plasma EVs) exhibited similar amplification patterns (Fig. 2H), the detected 5′-tRNAValCAC/AAC half molecules were anticipated to predominantly reside within plasma EVs. Subsequently, we quantified the abundance of the 5′-tRNAValCAC/AAC half in the plasma samples from healthy individuals or patients infected with Mtb. Considering the potential effects of sex hormones (36) and aging (50) on tRNA half expression, we restricted our analysis to males aged 30 to 35 y, aligning with the parameters of our prior research (32, 49). During the RNA extraction process, we included a synthetic control RNA as a spike-in to serve as a reference for normalization. As shown in Fig. 2I, the levels of the plasma 5′-tRNAValCAC/AAC half were drastically enhanced by approximately 1,000-fold in Mtb-infected patients compared to healthy individuals. These findings demonstrate that the upregulation and secretion of the 5′-tRNAValCAC/AAC half during the immune response occur not only in cell culture conditions but also in the plasma of patients infected with microbes, indicating its relevance in actual pathological contexts.
The Presence of Successive Uridine Sequences in Single-Stranded Regions of 5′-tRNA Halves Is Not Sufficient for TLR7 Activation.
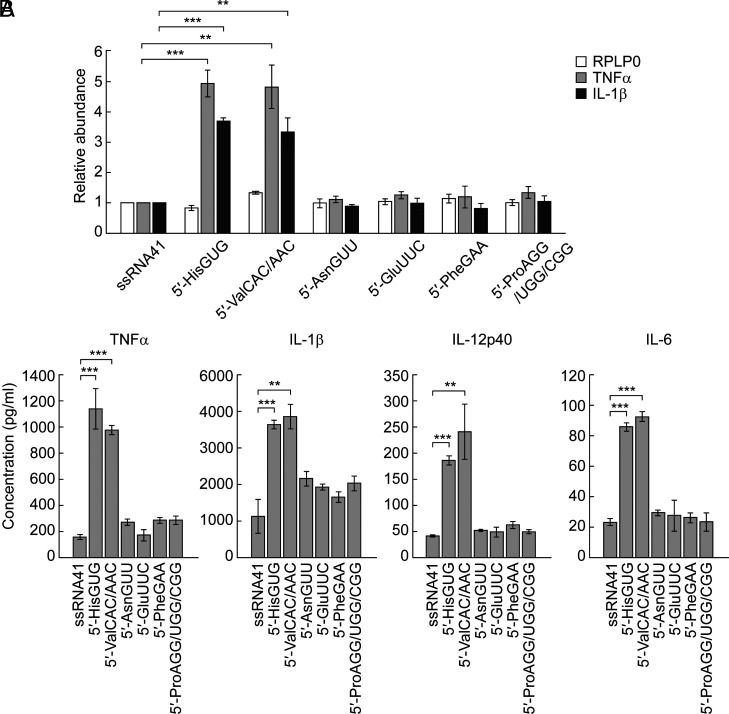
Considering that the 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half are active but the 5′-tRNAGluCUC half and the 5′-tRNAGlyGCC/CCC half are inactive in TLR7 stimulation, we attempted to characterize sequence determinants required for TLR7 stimulation. TLR7 possesses two ligand-binding sites for guanosine and ssRNA, respectively (10). Successive uridine sequences (such as UU and UUU) in ligand ssRNAs have been shown to be important for TLR7-binding (51), and indeed, both of the two active 5′-tRNA halves contain successive Us in single-stranded regions in their secondary structures; the 5′-tRNAHisGUG half contains one UU in a loop and one UU at the 3′-end, while the 5′-tRNAValCAC/AAC half contains two UUs in loops and one UUU at the 5′-end (SI Appendix, Fig. S2 A and B). To explore other active 5′-tRNA halves identified in HMDM-secreted EVs, we screened the most abundant sequences of the ex-5′-tRNA halves for the presence of successive Us in single-stranded regions (SI Appendix, Table S4), leading us to select four 5′-tRNA halves (5′-tRNAAsnGUU half, 5′-tRNAGluUUC half, 5′-tRNAPheGAA half, and 5′-tRNAProAGG/UGG/CGG half) with more than two UUs in single-stranded regions (SI Appendix, Fig. S2C) for examination of their activity in endosomal TLR stimulation. As shown in Fig. 3 A and B, none of the four 5′-tRNA halves showed the strong activity of cytokine induction upon their DOTAP-mediated transfection, while the two active 5′-tRNA halves served as positive controls, suggesting that the presence of successive Us in the single-stranded region is not sufficient for TLR7 activation.
Fig. 3.
The 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half are the only 5′-tRNA halves that strongly activate endosomal TLR. (A) The indicated 5′-tRNA halves and ssRNA41 (negative control) were subjected to DOTAP-mediated transfection into HMDMs. Total RNAs from the cells were subjected to RT-qPCR for the indicated mRNAs. (B) After the RNA transfection, the culture medium was subjected to measurement of the concentration of the indicated cytokines.
The Presence of Terminal GUUU Sequences in 5′-tRNA Halves Is Necessary for TLR7 Activation.
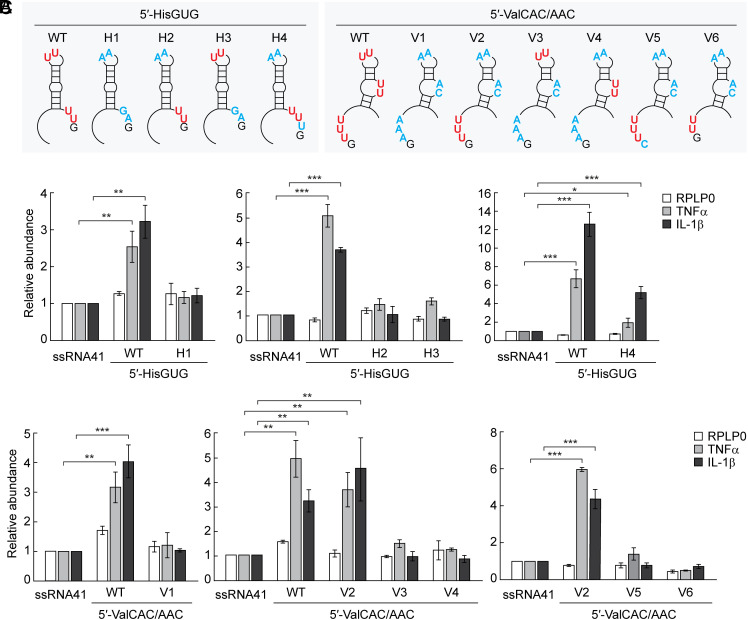
To further explore sequence determinants of active 5′-tRNA halves, we made various mutants of the 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half whose successive Us were replaced with As (Fig. 4A and SI Appendix, Fig. S2 A and B). To keep the secondary structures of the mutants identical to those of corresponding wild-type molecules, Us were replaced with G or C when replacement with A affected the secondary structure. The mutant 5′-tRNAHisGUG half lacking both of the two UUs (H1) did not activate TLR7 (Fig. 4B), indicating the necessity of successive Us. The mutant 5′-tRNAHisGUG half lacking either one of the two UUs (H2 and H3) showed significantly reduced activity (Fig. 4B), suggesting that two UUs are required to strongly activate TLR7. As in the case of the 5′-tRNAHisGUG half, the mutant 5′-tRNAValCAC/AAC half lacking three successive Us (V1) did not show TLR7 stimulation activity (Fig. 4C). However, the mutant retaining UUU at the 5′-end but lacking two UUs in loop regions (V2) activated TLR7 as strongly as the wild-type molecule, while the mutants containing only one UU in loop regions (V3 and V4) did not strongly activate TLR7 (Fig. 4C). The replacement of the 5′-terminal G (next to the UUU sequence) with C (V5) abolished the activity (Fig. 4C), suggesting that 5′-terminal GUUU sequences are necessary for TLR7 stimulation activity. Replacement of the UUU with UU (V6) also abolished the activity (Fig. 4C), indicating that three Us are required and two Us are not enough to stimulate TLR7. Collectively, these results suggest that the 5′-terminal GUUU sequences in the 5′-tRNAValCAC/AAC half are important determinants for TLR7 stimulation activity.
Fig. 4.
Determining sequence elements of 5′-tRNA halves required for TLR7 activation. (A) Positions of mutations in respective mutants in the 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half (the mutated nucleotides are shown in blue). (B and C) The mutants of 5′-tRNAHisGUG half (B) or 5′-tRNAValCAC/AAC half (C) were subjected to DOTAP-mediated transaction into HMDMs. Total RNAs from the cells were subjected to RT-qPCR for the indicated mRNAs.
To test whether 3′-terminal UUUG sequences also act as the determinant, we added U to 3′-terminal UU sequences in the mutant H2 to make a 5′-tRNAHisGUG half containing 3′-terminal UUUG sequences but lacking any other successive U sequences (mutant H4, Fig. 4A and SI Appendix, Fig. S2A). As shown in Fig. 4B, the changes partially restored the TLR7 stimulation activity. This suggests that not only the 5′-terminal GUUU but also the 3′-terminal UUUG could be important determinants for TLR7 activation, although changing 3′-terminal UUG sequences of the wild-type 5′-tRNAHisGUG half to UUUG (mutant H5, SI Appendix, Fig. S2A) resulted in TLR7 stimulation activity that was comparable to, but not greater than, that of the wild-type molecule (SI Appendix, Fig. S3).
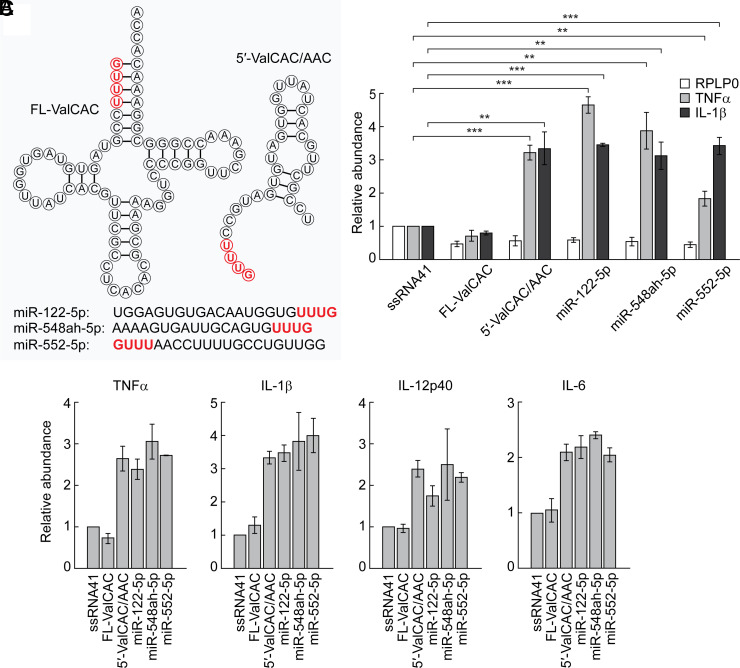
The Presence of Terminal GUUU Sequences Could Be Sufficient for Endosomal TLR Stimulation.
Although it has been reported that mature, full-length tRNAs accumulate in EVs (52), it is noteworthy that the full-length tRNAValCAC/AAC cannot activate TLR7 (Fig. 5 A–C). Unlike the 5′-tRNAValCAC/AAC whose 5′-terminal GUUU is located in the single-stranded region, the 5′-terminal GUUU sequences in the full-length tRNA form base-pairs in the acceptor stem, suggesting that the terminal GUUU sequences need to exist as ssRNAs to act as TLR7 ligands. Given the terminal GUUU sequences are important for TLR7 activation, we further explored endogenous RNA molecules containing terminal GUUU sequences; our screening of human miRNA sequences extracted three miRNAs (miR-122-5p, miR-548ah-5p, and miR-552-5p) as those that contain 5′-terminal GUUU or 3′-termiunal UUUG sequences. As shown in Fig. 5 A–C, DOTAP-mediated transfection of all three miRNAs induced cytokine secretions, suggesting that all of the examined miRNAs are active in endosomal TLR stimulation. These results suggested the universality of the single-stranded terminal GUUU sequences to act as a TLR7 stimulator.
Fig. 5.
Requirements of terminal GUUU sequences for TLR7 activation. (A) Sequences of the indicated sncRNAs. (B and C) The indicated RNAs were subjected to DOTAP-mediated transfection into HMDMs. Total RNAs from the cells were subjected to RT-qPCR for the indicated mRNAs (B) or culture medium was subjected to measurement of concentration of the indicated cytokines (C).
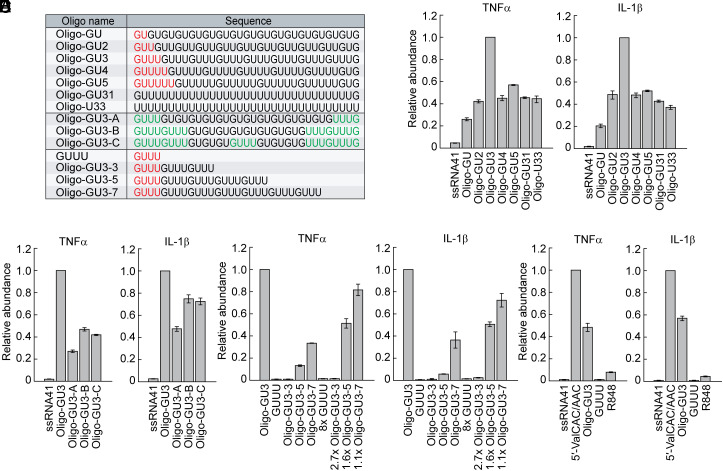
GUUU Is the Best Ratio of Guanosine and Uridine for Endosomal TLR Activation.
The ligands for the first and second binding pockets of TLR7 are G nucleotide and ssRNAs with successive Us, respectively (10, 51). To determine the optimal ratio of G and U nucleotides, we investigated the activity of 33-nt ssRNAs with repeat sequences of GU, GUU (GU2), GUUU (GU3), GUUUU (GU4), GUUUUU (GU5), or poly U with terminal G residues (GU31), as well as poly U without any G residues (U33; Fig. 6A). The GUUU repeats exhibited the highest activity in endosomal TLR stimulation following DOTAP transfection (Fig. 6B). Replacement of some GUUU sequences in the GUUU repeats to GU repeats (GU3-A-C; Fig. 6A) led to the reduction of the activity (Fig. 6C), confirming that the GUUU repeats are the sequences with the optimal G/U sequence signature for endosomal TLR activation. The activity of poly U (U33) was significantly weaker than that of the GUUU repeats (Fig. 6B), suggesting that the presence of G nucleotides in ssRNA, which would become the ligand of the first binding pocket of TLR7, is important. To explore the role of the length of GUUU repeats in TLR7 activation, we analyzed the activity of ssRNAs with varying numbers of GUUU repeats: GUUU (a single repeat), Oligo-GU3-3 (3 repeats), Oligo-GU3-5 (5 repeats), Oligo-GU3-7 (7 repeats), and Oigo-GU3 (8 repeats). As shown in Fig. 6D, an increase in GUUU repeat numbers corresponded to enhanced TLR7 stimulation activity. To determine whether a greater quantity of shorter RNAs could offset their lower activity, we further performed the experiments with the adjusted amounts to match the molar quantity of GUUU sequences in Oligo-GU3. Specifically, we used 8, 2.7, 1.6, and 1.1 times the molar amounts of GUUU, Oligo-GU3-3, Oligo-GU3-5, and Oligo-GU3-7, respectively, compared to Oligo-GU3. Despite the equivalent amounts of GUUU molecules, the longer RNAs consistently demonstrated higher activity (Fig. 6D), underscoring the significant impact of RNA length on TLR7 stimulation activity. Interestingly, the activity of the 5′-tRNAValCAC/AAC half was found to be stronger than that of the GUUU repeats with the same length (Oligo-GU3, Fig. 6E), likely due to unidentified molecular factors. The activity of the 5′-tRNAValCAC/AAC half in our experiments at a specific time point was further shown to be stronger than that of R848, an agonist of TLR7 and -8 which have been utilized for therapies of cancer and infectious diseases (53–56) (Fig. 6E).
Fig. 6.
GUUU is the best ratio of G and U for TLR7 activation. (A) Sequences of the ssRNAs used. (B–E) The indicated RNAs were subjected to DOTAP-mediated transfection into HMDMs. Total RNAs from the cells were subjected to RT-qPCR for the indicated mRNAs.
Discussion
Although miRNAs have been the best-explored endogenous ssRNA ligands for endosomal TLRs thus far (16–18), 3′-OH-containing RNAs, such as miRNAs, are the minor species of ex-sncRNAs (29, 32, 57). On the other hand, it is increasingly acknowledged that tRNA-derived sncRNAs are a plentiful type of ex-RNAs (39, 58), and cP-containing 5′-tRNA half molecules are the most abundant class of ex-tRNA-derived sncRNAs (32). Although recent studies have provided evidence that certain tRNA halves exist in the form of nicked mature tRNAs (59, 60), the scarcity of 3′-tRNA halves in HMDM EVs (32) and in the plasma of Mtb-infected patients (57) suggests that 5′- and 3′-tRNA halves can be regulated through distinct and independent pathways. This aligns well with multiple studies that have reported a much higher accumulation of 5′-tRNA halves compared to 3′-tRNA halves in various cells and tissues (61–65). The much greater abundance of ex-5′-tRNA halves compared to that of miRNAs (32) suggests ex-5′-tRNA halves form a more significant, biologically relevant class of endogenous ligands of immune receptors. Upon conducting a thorough investigation of ex-5′-tRNA halves in EVs secreted by HMDMs, we have identified the 5′-tRNAValCAC/AAC half as the most abundant and as an immunostimulatory molecule activating TLR7. This finding represents a second example of an immune-active 5′-tRNA half, with the 5′-tRNAHisGUG half being the first identified (32). We propose that, like cytokines, these 5′-tRNA halves may function as “immune activators” by stimulating TLR7 upon delivery to endosomes in neighboring immune cells and other types of cells via EV-mediated cell–cell communication.
Surface TLR activation induces the accumulation of the 5′-tRNAValCAC/AAC half in HMDMs and their secreted EVs. The expressional induction and secretion were not limited to cell culture settings but further occurred in plasma samples of Mtb-infected patients which showed dramatic enrichment of the 5′-tRNAValCAC/AAC half by an approximately 1,000-fold. The enhanced levels of the 5′-tRNAValCAC/AAC half, as well as the 5′-tRNAHisGUG half (49), and their activity in TLR7 stimulation could be involved in the immune response against Mtb infection. In fact, the genetic polymorphisms in the TLR7 gene, which reduce its activity, increase susceptibility to Mtb infection (66), and upregulation or stimulation of TLR7 in macrophages suppress Mtb growth (67, 68). In addition to Mtb infection, a dramatic increase in the levels of serum ex-5′-tRNA halves has also been observed in patients infected with hepatitis B virus infection (69), and the infections with respiratory syncytial virus (RSV) and Rickettsia have been reported to induce the expression of 5′-tRNA halves (70–72). Therefore, the induction of 5′-tRNA halves and their secretion as ex-5′-tRNA halves could potentially be a universal phenomenon across various infectious diseases. Further investigations may establish 5′-tRNA half molecules as potential circulating biomarkers for the noninvasive evaluation of infection severity and immune response status.
Full-length tRNAValCAC/AAC was incapable of stimulating TLR7. Shortening it by anticodon cleavage into less-rigid 5′-half molecules to expose the 5′-terminal GUUU sequence as the single-stranded region is crucial to producing immunostimulatory sncRNAs. We identified ANG as the enzyme which produces immune-responsive tRNA halves in HMDMs (32). An increase in ANG levels has been observed upon RSV and Rickettsia infections (72, 73). Further research is warranted on immune-responsive ribonucleases, which cleave tRNAs and other RNAs to produce ex-sncRNAs. ANG-mediated cleavage leaves a cP in 3′-cleavage products (74), and thus 5′-tRNA halves contain a cP (36). Other unidentified immune-responsive ribonucleases, if any, may also produce cP-containing RNAs, because cP-containing RNAs (and possibly 3′-P-containing RNAs) are the majority of ex-sncRNAs (29, 32). The formation of cP by such enzymes could contribute to TLR7 activation, because cP-containing guanosine (G-cP) exhibits significantly higher affinity compared to guanosine without cP, and thus G-cP is proposed as an endogenous ligand for the first binding site of TLR7 (51). The cP-formation is also implicated in the activation of TLR8 (75). Given that this study employed synthetic RNAs lacking cP in the analysis of TLR7 stimulation activity, further research is necessary to elucidate the potential impact of cP in tRNA halves on the activation of TLR7.
Posttranscriptional modifications of RNA molecules, such as 2′-O-methylation, m6A, m5C, m5U, s2U, and Ψ, have been recognized for their inhibitory effects on the activation of TLR7 and -8 (45, 46, 76–78). The 5′-tRNAValCAC/AAC half containing modifications showed a reduced TLR7 stimulation activity compared to its unmodified version. Although this experiment was performed under the premise that tRNA half molecules possess modifications identical to those of their corresponding mature tRNAs, and that their modification status remains consistent both within cells and in EVs, further research is required to confirm these assumptions. Despite this, the modified version still retained some level of TLR7 stimulation activity, and it is worth noting that the 5′-tRNAValCAC/AAC half is the most abundantly accumulated tRNA-derived sncRNAs in HMDM EVs. Further, our experiments using EVs and AS-oligo have indeed demonstrated the activity of endogenous 5′-tRNAValCAC/AAC half molecules within EVs.
While ssRNA sequence determinants for TLR7 stimulation have not yet been fully defined, it is known that successive U sequences and/or GU-rich sequences are necessary to activate TLR7. In this study, we have identified the terminal GUUU sequence as a universal signature for strong TLR7 activation. Structural characterization of TLR7 showed G nucleotide (G-cP, preferably) and ssRNAs with successive Us as the ligands for the first and second binding pockets of TLR7, respectively (10, 51). Considering that endosomal RNase T2 has been reported to preferentially cleave between G and U to produce G-cP (75), it is plausible that the 5′-terminal GUUU might be cleaved by such an enzyme, yielding both G-cP and the remaining UUU-containing ssRNA. These products would perfectly fit to the first and second binding pockets of TLR7, respectively, although TLR7 activation via the 3′-terminal UUUG requires different explanation. The replacement of the 5′-terminal G with C abolished TLR7 simulation activity, supporting the necessity of the presence of G in the sequence signature. Given this notion, it is not surprising that the activity of poly-U was significantly weaker than that of the GUUU repeats, which is possibly due to not enough concentration of endogenous G nucleotide in the endosomes. TLR7 activation by the GUUU repeats was stronger than that by GUUU alone, further suggesting the involvement of RNA cleavages inside the endosome in the production of effective TLR7 ligands. Our finding of GUUU as a universal sequence signature suggests the possibility of various endogenous RNAs with such sequences, as well as exogenous RNAs from pathogens, to be recognized by TLR7 for promotion of immune response, some examples of which were shown for GUUU-containing miRNAs in this study. The activity of the 5′-tRNAValCAC/AAC half was stronger than that of GUUU repeats of the same length, suggesting that beyond the GUUU motif, there are other, yet unidentified molecular factors within the 5′-tRNAValCAC/AAC half, such as secondary or tertiary structures, that facilitate endosomal enzymatic cleavage and/or TLR7 binding. In light of this, the roles of secondary and tertiary structures of tRNA-derived sncRNAs across various biological processes have been discussed in recent reviews (79, 80).
In our experiment at a specific time point, the activity of the 5′-tRNAValCAC/AAC half in stimulating TLR7 was stronger than that of R848, a synthetic agonist for TLR7 and -8. It is conceivable that the 5′-tRNAValCAC/AAC half may undergo cleavages by RNase T2 or other enzymes, generating G-cP and ssRNAs containing successive Us in higher molar abundance. R848 has been shown to have potent antiviral (81) and antitumor effects (82, 83) and has been used in cancer immunotherapy (53), vaccine adjuvant development (54), and the treatment of infectious diseases such as hepatitis C virus (55) and HIV (56). However, the systemic use of R848 is associated with adverse effects such as strong immune-related toxicities (84), liver toxicity (85), brain swelling, and central nervous system edema (86). As the 5′-tRNAValCAC/AAC half, as well as the 5′-tRNAHisGUG half, are endogenous molecules and possess a greater TLR7 activation potential than R848, they may be promising candidate molecules for therapeutic applications with reduced or no toxicity. Further characterization of this line could make 5′-tRNA halves useful as adjuvants in vaccine development and as immune boosters in treatments of cancer and infectious diseases for which R848 is currently in use.
TLR7 is involved in various biological processes and diseases (87–92). For example, TLR7 mutations are associated with the symptom severity of COVID-19 (93–95). TLR7 has also been implicated in the progression of Parkinson’s disease (96), Alzheimer’s disease (15, 97, 98), and autoimmune diseases such as systemic lupus erythematosus (99) and atherosclerosis (100, 101). Although the growing evidence of TLR7 involvement in the pathogenesis of noninfectious diseases highlights the need for a better understanding of endogenous ssRNA ligands of immune receptors, their exploration and characterization are still at an initial stage, and we are just beginning to appreciate the previously hidden classes of sncRNAs not detected by standard RNA-seq methods. We expect that the 5′-tRNAHisGUG half and the 5′-tRNAValCAC/AAC half are just the early examples of immune receptor-stimulating endogenous ssRNAs, and other ex-sncRNAs would be found to be similarly immunostimulatory through the TLR7 or -8 axis. Beyond tRNA-derived sncRNAs, our first genome-wide identification of cP-containing sncRNAs revealed abundant expression of rRNA- and mRNA-derived sncRNAs in various tissues (50), and, indeed, rRNA- and mRNA-derived sncRNAs have been shown to be abundantly secreted as ex-RNAs (30, 57, 102, 103). Updated sncRNA sequencing methods to address the issue of reverse transcription impairment by posttranscriptional modifications, such as PANDORA-seq (104) and TGIRT-seq (105), present viable options for future exploration of immunostimulatory ex-sncRNAs, especially those focusing on heavily modified sncRNAs such as tRNA- and rRNA-derived sncRNAs. The exploration should review studies looking into the characteristics of ssRNA ligands of TLR7 and -8, including terminal GUUU sequence signatures for TLR7 activation that this study revealed. Further research into endogenous ssRNA ligands of TLR7 and -8 would help our fuller understanding of the immune system in the body’s response to diverse pathologies.
Materials and Methods
In Vitro RNA Synthesis.
The synthetic RNAs used in this study are shown in SI Appendix, Table S3 [the information on sncRNA “license plates” (106) and names via tDRnamer (107) are included]. These RNAs were synthesized by an in vitro transcription reaction as described previously (32). dsDNA templates were synthesized using PrimeSTAR GXL DNA Polymerase (Takara Bio) and the primers shown in SI Appendix, Table S5. The templates were then subjected to an in vitro transcription reaction with T7 RNA polymerase (New England Biolabs) at 37 °C for 4 h. For the RNAs whose 5′-terminal nucleotide is not G, the in vitro synthesized RNAs contained the ribozyme sequence to generate a mature 5′-end as described previously (108). After incubation for in vitro RNA synthesis, the reaction mixture was further incubated for three cycles at 90 °C for 2.5 min and 37 °C for 15 min, allowing the ribozyme reaction. The synthesized RNAs were then gel-purified using denaturing PAGE with single-nucleotide resolution. For full-length tRNAValCAC, we performed annealing by incubating it in the annealing buffer, consisting of 50 mM Tris-HCl (pH 8.0) and 100 mM MgCl2 at 70 °C for 3 min, followed by incubation at 37 °C for 20 min. The 5′-tRNAValCAC/AAC half containing posttranscriptional modifications (the 5′-ValCAC/AAC-Mod in SI Appendix, Table S3) was synthesized by Integrated DNA Technologies.
Cell Culture, TLR4 Activation, RNA and R848 Delivery, and Measurement of Cytokine Concentration.
THP-1 human acute monocytic leukemia cells (American Type Culture Collection) were cultured in RPMI 1640 medium (Corning) with 10% FBS and differentiated into HMDMs using phorbol 12-myristate 13-acetate (PMA; Sigma-Aldrich), as described previously (32, 109). For activation of TLR4, HMDMs were cultured with a medium containing 100 ng/mL of LPS from E. coli O111:B4 (Sigma-Aldrich) for 12 h as described previously (32). TLR7 KO THP-1 cell lines, whose TLR7 expression is completely depleted, were previously generated using a CRISPR/Cas9 approach (32). Before transfection, the cells were primed with 100 units/mL of interferon γ (Thermo Fisher Scientific) for 18 to 24 h (42). To deliver RNAs to endosomes, we used the cationic liposome DOTAP (Sigma-Aldrich) as previously described (32, 110, 111). In brief, 230 pmol of synthetic RNAs or Resiquimod (R848, InvivoGen) were mixed with 60 µL of HBS buffer and 15 µL of DOTAP reagent and incubated for 15 min. The RNA-DOTAP solution was then added to 1 mL RPMI 1640 medium with 2% FBS, followed by incubation of the cells for 16 h. Cytokine concentrations of the cultured medium of HMDMs were measured by Multiplexing LASER Bead Technology (Eve Technologies) or ELISA kit (R&D Systems), as previously described (32).
EVs and Their Treatments.
EVs were isolated from the culture medium of LPS-treated HMDMs according to an ultracentrifugation-based method. As described previously (32), the abundant presence of EVs in the EV solution was confirmed by NTA and transmission electron microscopy (JEM-2100, JEOL) at Materials Characterization Core at Drexel University. To confirm the presence of EV-RNAs, the isolated EVs were incubated with PureLink RNase A (200 ng/μL, Thermo Fisher Scientific) with or without 0.1% Triton X-100 at 37 °C for 30 min.
EV-Mediated RNA Delivery to Endosomes.
The experiments using EVs and antisense oligonucleotides were performed as described previously (32). AS-oligo or Ctrl-oligo with scrambled sequences (SI Appendix, Table S3) was first infused with DOTAP as described above. EVs isolated from LPS-treated HMDMs were mixed with the DOTAP-oligonucleotide solution and then were applied to recipient HMDMs, followed by incubation for 16 h. To eliminate possible effects of potential endotoxin (LPS) contamination, EVs isolated from LPS-treated HMDMs were incubated with 10 mg/mL polymyxin B (PMB) (Sigma-Aldrich) at 4 °C for 1 h prior to adding to HMDMs.
Quantification of RNAs by TaqMan RT-qPCR and Standard RT-qPCR.
Total RNA was isolated from the cells and EVs using TRIsure (Bioline). TaqMan RT-qPCR for specific quantification of 5′-tRNA halves was performed as described previously (32, 35, 36, 50, 112). Briefly, to remove cP from 5′-tRNA halves, total RNA was treated with T4 polynucleotide kinase (T4 PNK; New England Biolabs), followed by ligation to a 3′-RNA AD by T4 RNA ligase (T4 Rnl; Thermo Fisher Scientific). Ligated RNA was then subjected to TaqMan RT-qPCR using the One Step PrimeScript RT-PCR Kit (Takara Bio), 200 nM of a TaqMan probe targeting the boundary of the target RNA and the 3′-AD, and forward and reverse primers. The TaqMan probe and primer sequences are shown in SI Appendix, Table S6. Although both 33-nt and 34-nt versions of the tRNAValCAC/AAC half are present in HMDMs and their EVs, the TaqMan probe designed for this study specifically targets the 33-nt version. Due to limited amounts of starting RNA materials, for quantification of plasma 5′-tRNA halves, we utilized an upgraded, more efficient version of TaqMan RT-qPCR in which T4 PNK treatment and 3′-AD ligation are simultaneously performed in a one-step reaction (49). For quantification of mRNAs by standard RT-qPCR, total RNA was treated with DNase I (Promega) and subjected to reverse transcription using RevertAid Reverse Transcriptase (Thermo Fisher Scientific) and a reverse primer. The synthesized cDNAs were then subjected to PCR using 2×qPCR Master Mix (Bioland Scientific) and forward and reverse primers. Sequences of the primers used are shown in SI Appendix, Table S7.
Northern Blot.
Northern blot was performed as previously described (36) with the following antisense probes: 5′-tRNAValCAC/AAC half, 5′-GCGAACGTGATAACCACTAC-3′; and miR-16, 5′-GCCAATATTTACGTGCTGCTA-3′.
Bacterial Infection and Invasion Assay.
After 16 h of DOTAP transfection of RNAs, HMDMs (1 × 106 cells) were plated on 6-well plates and were incubated with E. coli [100 multiplicities of infection (MOI)] in RPMI 1640 (no antibiotics) for 60 min at 37 °C. HMDMs were then washed with PBS three times and incubated with RPMI 1640 containing high concentration (3×) of penicillin–streptomycin (Thermo Fisher Scientific) at 37 °C for 60 min. Subsequently, the medium was replaced with RPMI 1640 containing normal concentration (1×) of penicillin-streptomycin, followed by further incubation at 37 °C for 24 h. HMDMs were then washed and lysed with 0.5% Triton X-100. Intracellular bacteria were enumerated by plating on LB agar plates.
Ethical Approval, Human Plasma Samples, and RNA Isolation.
The Office of Human Research of Thomas Jefferson University approved our use of patient samples without private information in accordance with all federal, institutional, and ethical guidelines. We obtained the plasma samples from a biological specimen company, BioIVT, without receiving patients’ information. The plasma samples were derived from healthy or Mtb-infected males aged 30 to 35 y. RNA isolation from the plasma samples was performed as described previously (32, 49). First, 500 µL plasma was centrifuged at 16,060 g for 5 min, and then 400 µL supernatant was mixed with 1 fmol of the spike-in control RNA (SI Appendix, Table S3) and subjected to RNA extraction using TRIzol LS Reagent (Thermo Fisher Scientific). The extracted RNAs were further subjected to purification using the miRNeasy Mini Kit (Qiagen).
Supplementary Material
Appendix 01 (PDF)
Acknowledgments
We are grateful to the members of Kirino lab for helpful discussions and to Dr. Craig Johnson for his technical assistance in handling transmission electron microscope at the Materials Characterization Core at Drexel University. This study was supported in part by NIH Grant (GM106047, HL150560, AI151641, AI168975, and AI171366 to Y.K.) and American Cancer Society Research Scholar Grant (RSG-17-059-01-RMC, to Y.K.).
Author contributions
K.P., T.K., and Y.K. designed research; K.P. and T.K. performed research; K.P. and T.K. analyzed data; and K.P., T.K., and Y.K. wrote the paper.
Competing interests
The authors declare no competing interest.
Footnotes
This article is a PNAS Direct Submission.
Data, Materials, and Software Availability
There are no data underlying this work.
Supporting Information
References
- 1.Medzhitov R., Origin and physiological roles of inflammation. Nature 454, 428–435 (2008). [DOI] [PubMed] [Google Scholar]
- 2.Takeuchi O., Akira S., Pattern recognition receptors and inflammation. Cell 140, 805–820 (2010). [DOI] [PubMed] [Google Scholar]
- 3.Kono H., Rock K. L., How dying cells alert the immune system to danger. Nat. Rev. Immunol. 8, 279–289 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gong T., Liu L., Jiang W., Zhou R., DAMP-sensing receptors in sterile inflammation and inflammatory diseases. Nat. Rev. Immunol. 20, 95–112 (2020). [DOI] [PubMed] [Google Scholar]
- 5.Remick B. C., Gaidt M. M., Vance R. E., Effector-triggered immunity. Annu. Rev. Immunol. 41, 453–481 (2023). [DOI] [PubMed] [Google Scholar]
- 6.Kawasaki T., Kawai T., Toll-like receptor signaling pathways. Front. Immunol. 5, 461 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zindel J., Kubes P., DAMPs, PAMPs, and LAMPs in immunity and sterile inflammation. Annu. Rev. Pathol. 15, 493–518 (2020). [DOI] [PubMed] [Google Scholar]
- 8.Tan X., Sun L., Chen J., Chen Z. J., Detection of microbial infections through innate immune sensing of nucleic acids. Annu. Rev. Microbiol. 72, 447–478 (2018). [DOI] [PubMed] [Google Scholar]
- 9.Heil F., et al. , Species-specific recognition of single-stranded RNA via Toll-like receptor 7 and 8. Science 303, 1526–1529 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Zhang Z., et al. , Structural analysis reveals that Toll-like receptor 7 is a dual receptor for guanosine and single-stranded RNA. Immunity 45, 737–748 (2016). [DOI] [PubMed] [Google Scholar]
- 11.Lund J. M., et al. , Recognition of single-stranded RNA viruses by Toll-like receptor 7. Proc. Natl. Acad. Sci. U.S.A. 101, 5598–5603 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melchjorsen J., et al. , Activation of innate defense against a paramyxovirus is mediated by RIG-I and TLR7 and TLR8 in a cell-type-specific manner. J. Virol. 79, 12944–12951 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Triantafilou K., et al. , Human cardiac inflammatory responses triggered by Coxsackie B viruses are mainly Toll-like receptor (TLR) 8-dependent. Cell Microbiol. 7, 1117–1126 (2005). [DOI] [PubMed] [Google Scholar]
- 14.Gantier M. P., et al. , Genetic modulation of TLR8 response following bacterial phagocytosis. Hum. Mutat. 31, 1069–1079 (2010). [DOI] [PubMed] [Google Scholar]
- 15.Lehmann S. M., et al. , An unconventional role for miRNA: Let-7 activates Toll-like receptor 7 and causes neurodegeneration. Nat. Neurosci. 15, 827–835 (2012). [DOI] [PubMed] [Google Scholar]
- 16.Fabbri M., Paone A., Calore F., Galli R., Croce C. M., A new role for microRNAs, as ligands of Toll-like receptors. RNA Biol. 10, 169–174 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chen X., Liang H., Zhang J., Zen K., Zhang C. Y., microRNAs are ligands of Toll-like receptors. RNA 19, 737–739 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bayraktar R., Bertilaccio M. T. S., Calin G. A., The interaction between two worlds: MicroRNAs and Toll-like receptors. Front. Immunol. 10, 1053 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Feng Y., et al. , Extracellular microRNAs induce potent innate immune responses via TLR7/MyD88-dependent mechanisms. J. Immunol. 199, 2106–2117 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zou L., et al. , Brain innate immune response via miRNA-TLR7 sensing in polymicrobial sepsis. Brain Behav. Immun. 100, 10–24 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fabbri M., TLRs as miRNA receptors. Cancer Res. 72, 6333–6337 (2012). [DOI] [PubMed] [Google Scholar]
- 22.Casadei L., et al. , Exosome-derived miR-25-3p and miR-92a-3p stimulate liposarcoma progression. Cancer Res. 77, 3846–3856 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu W. C., Song S. J., Zhang Y., Li X., Role of extracellular vesicles in autoimmune pathogenesis. Front. Immunol. 11, 579043 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Salvi V., et al. , Exosome-delivered microRNAs promote IFN-alpha secretion by human plasmacytoid DCs via TLR7. JCI Insight 3, e98204 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Groot M., Lee H., Sorting mechanisms for microRNAs into extracellular vesicles and their associated diseases. Cells 9, 1044 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shigematsu M., Kawamura T., Kirino Y., Generation of 2’,3’-cyclic phosphate-containing RNAs as a hidden layer of the transcriptome. Front. Genet. 9, 562 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shigematsu M., Kirino Y., Making invisible RNA visible: Discriminative sequencing methods for RNA molecules with specific terminal formations. Biomolecules 12, 611 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gumas J., Kirino Y., Challenges inherent to the sequencing and quantification of short non-coding RNAs and how to address them. Clin. Transl. Discovery 2, e93 (2022). [Google Scholar]
- 29.Giraldez M. D., et al. , Phospho-RNA-seq: A modified small RNA-seq method that reveals circulating mRNA and lncRNA fragments as potential biomarkers in human plasma. EMBO J. 38, e101695 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Akat K. M., et al. , Detection of circulating extracellular mRNAs by modified small-RNA-sequencing analysis. JCI Insight 5, e127317 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Qin Y., et al. , High-throughput sequencing of human plasma RNA by using thermostable group II intron reverse transcriptases. RNA 22, 111–128 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pawar K., Shigematsu M., Sharbati S., Kirino Y., Infection-induced 5’-half molecules of tRNAHisGUG activate Toll-like receptor 7. PLoS Biol. 18, e3000982 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yamasaki S., Ivanov P., Hu G. F., Anderson P., Angiogenin cleaves tRNA and promotes stress-induced translational repression. J. Cell Biol. 185, 35–42 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fu H., et al. , Stress induces tRNA cleavage by angiogenin in mammalian cells. FEBS Lett. 583, 437–442 (2009). [DOI] [PubMed] [Google Scholar]
- 35.Shigematsu M., Kirino Y., Oxidative stress enhances the expression of 2’,3’-cyclic phosphate-containing RNAs. RNA Biol. 17, 1060–1069 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Honda S., et al. , Sex hormone-dependent tRNA halves enhance cell proliferation in breast and prostate cancers. Proc. Natl. Acad. Sci. U.S.A. 112, E3816–E3825 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Emara M. M., et al. , Angiogenin-induced tRNA-derived stress-induced RNAs promote stress-induced stress granule assembly. J. Biol. Chem. 285, 10959–10968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ivanov P., Emara M. M., Villen J., Gygi S. P., Anderson P., Angiogenin-induced tRNA fragments inhibit translation initiation. Mol. Cell 43, 613–623 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gumas J., Kirino Y., Extracellular tRNA-derived RNAs as emerging activators of endosomal Toll-like receptors: A narrative review. ExRNA 5, 0002 (2023). [Google Scholar]
- 40.Chan P. P., Lowe T. M., GtRNAdb: A database of transfer RNA genes detected in genomic sequence. Nucleic Acids Res. 37, D93–D97 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sprinzl M., Horn C., Brown M., Ioudovitch A., Steinberg S., Compilation of tRNA sequences and sequences of tRNA genes. Nucleic Acids Res. 26, 148–153 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gantier M. P., et al. , TLR7 is involved in sequence-specific sensing of single-stranded RNAs in human macrophages. J. Immunol. 180, 2117–2124 (2008). [DOI] [PubMed] [Google Scholar]
- 43.Sioud M., Single-stranded small interfering RNA are more immunostimulatory than their double-stranded counterparts: A central role for 2’-hydroxyl uridines in immune responses. Eur. J. Immunol. 36, 1222–1230 (2006). [DOI] [PubMed] [Google Scholar]
- 44.Forsbach A., et al. , Identification of RNA sequence motifs stimulating sequence-specific TLR8-dependent immune responses. J. Immunol. 180, 3729–3738 (2008). [DOI] [PubMed] [Google Scholar]
- 45.Kariko K., Buckstein M., Ni H., Weissman D., Suppression of RNA recognition by Toll-like receptors: The impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23, 165–175 (2005). [DOI] [PubMed] [Google Scholar]
- 46.Jockel S., et al. , The 2’-O-methylation status of a single guanosine controls transfer RNA-mediated Toll-like receptor 7 activation or inhibition. J. Exp. Med. 209, 235–241 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gehrig S., et al. , Identification of modifications in microbial, native tRNA that suppress immunostimulatory activity. J. Exp. Med. 209, 225–233 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen E. Y., Roe B. A., Sequence studies on human placenta tRNAVal: Comparison with the mouse myeloma tRNAVal. Biochem. Biophys. Res. Commun. 78, 631–640 (1977). [DOI] [PubMed] [Google Scholar]
- 49.Kawamura T., Shigematsu M., Kirino Y., In vitro production and multiplex quantification of 2’,3’-cyclic phosphate-containing 5’-tRNA half molecules. Methods 203, 335–341 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shigematsu M., Morichika K., Kawamura T., Honda S., Kirino Y., Genome-wide identification of short 2’,3’-cyclic phosphate-containing RNAs and their regulation in aging. PLoS Genet. 15, e1008469 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang Z., et al. , Structural analyses of Toll-like receptor 7 reveal detailed RNA sequence specificity and recognition mechanism of agonistic ligands. Cell Rep. 25, 3371–3381.e5 (2018). [DOI] [PubMed] [Google Scholar]
- 52.Shurtleff M. J., et al. , Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. U.S.A. 114, E8987–E8995 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kim S. Y., et al. , Lyophilizable and multifaceted Toll-like receptor 7/8 agonist-loaded nanoemulsion for the reprogramming of tumor microenvironments and enhanced cancer immunotherapy. ACS Nano 13, 12671–12686 (2019). [DOI] [PubMed] [Google Scholar]
- 54.Dowling D. J., et al. , Development of a TLR7/8 agonist adjuvant formulation to overcome early life hyporesponsiveness to DTaP vaccination. Sci. Rep. 12, 16860 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee J., et al. , Activation of anti-hepatitis C virus responses via Toll-like receptor 7. Proc. Natl. Acad. Sci. U.S.A. 103, 1828–1833 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nian H., et al. , R-848 triggers the expression of TLR7/8 and suppresses HIV replication in monocytes. BMC Infect. Dis. 12, 5 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Gumas J., Kawamura T., Shigematsu M., Kirino Y., Immunostimulatory short non-coding RNAs in the circulation of patients with tuberculosis infection. Mol. Ther. Nucleic Acids 35, 1–15 (2024). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tosar J. P., Cayota A., Extracellular tRNAs and tRNA-derived fragments. RNA Biol. 17, 1149–1167 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Costa B., et al. , Nicked tRNAs are stable reservoirs of tRNA halves in cells and biofluids. Proc. Natl. Acad. Sci. U.S.A. 120, e2216330120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen X., Wolin S. L., Transfer RNA halves are found as nicked tRNAs in cells: Evidence that nicked tRNAs regulate expression of an RNA repair operon. RNA 29, 620–629 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dhahbi J. M., et al. , 5’ tRNA halves are present as abundant complexes in serum, concentrated in blood cells, and modulated by aging and calorie restriction. BMC Genomics 14, 298 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Su Z., Kuscu C., Malik A., Shibata E., Dutta A., Angiogenin generates specific stress-induced tRNA halves and is not involved in tRF-3-mediated gene silencing. J. Biol. Chem. 294, 16930–16941 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mo X., et al. , Lactate induces production of the tRNA(His) half to promote B-lymphoblastic cell proliferation. Mol. Ther. 28, 2442–2457 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Krishna S., et al. , Dynamic expression of tRNA-derived small RNAs define cellular states. EMBO Rep. 20, e47789 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L., et al. , 5’ Half of specific tRNAs feeds back to promote corresponding tRNA gene transcription in vertebrate embryos. Sci. Adv. 7, eabh0494 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Lai Y. F., et al. , Functional polymorphisms of the TLR7 and TLR8 genes contribute to Mycobacterium tuberculosis infection. Tuberculosis (Edinb) 98, 125–131 (2016). [DOI] [PubMed] [Google Scholar]
- 67.Bao M., Yi Z., Fu Y., Activation of TLR7 inhibition of Mycobacterium tuberculosis survival by autophagy in RAW 264.7 macrophages. J. Cell Biochem. 118, 4222–4229 (2017). [DOI] [PubMed] [Google Scholar]
- 68.Lee H. J., et al. , TLR7 stimulation with imiquimod induces selective autophagy and controls Mycobacterium tuberculosis growth in mouse macrophages. Front. Microbiol. 11, 1684 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zhang Y., et al. , Identification and characterization of an ancient class of small RNAs enriched in serum associating with active infection. J. Mol. Cell Biol. 6, 172–174 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Zhou J., et al. , Identification of two novel functional tRNA-derived fragments induced in response to respiratory syncytial virus infection. J. Gen. Virol. 98, 1600–1610 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Deng J., et al. , Respiratory syncytial virus utilizes a tRNA fragment to suppress antiviral responses through a novel targeting mechanism. Mol. Ther. 23, 1622–1629 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gong B., et al. , Compartmentalized, functional role of angiogenin during spotted fever group rickettsia-induced endothelial barrier dysfunction: Evidence of possible mediation by host tRNA-derived small noncoding RNAs. BMC Infect. Dis. 13, 285 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wang Q., et al. , Identification and functional characterization of tRNA-derived RNA fragments (tRFs) in respiratory syncytial virus infection. Mol. Ther. 21, 368–379 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Shapiro R., Riordan J. F., Vallee B. L., Characteristic ribonucleolytic activity of human angiogenin. Biochemistry 25, 3527–3532 (1986). [DOI] [PubMed] [Google Scholar]
- 75.Greulich W., et al. , TLR8 is a sensor of RNase T2 degradation products. Cell 179, 1264–1275.e13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rimbach K., Kaiser S., Helm M., Dalpke A. H., Eigenbrod T., 2’-O-methylation within bacterial RNA acts as suppressor of TLR7/TLR8 activation in human innate immune cells. J. Innate. Immun. 7, 482–493 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Keller P., et al. , Double methylation of tRNA-U54 to 2’-O-methylthymidine (Tm) synergistically decreases immune response by Toll-like receptor 7. Nucleic Acids Res. 46, 9764–9775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Freund I., Eigenbrod T., Helm M., Dalpke A. H., RNA modifications modulate activation of innate Toll-like receptors. Genes (Basel) 10, 92 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kuhle B., Chen Q., Schimmel P., tRNA renovatio: Rebirth through fragmentation. Mol. Cell 83, 3953–3971 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen Q., Zhou T., Emerging functional principles of tRNA-derived small RNAs and other regulatory small RNAs. J. Biol. Chem. 299, 105225 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Vanwalscappel B., Tada T., Landau N. R., Toll-like receptor agonist R848 blocks Zika virus replication by inducing the antiviral protein viperin. Virology 522, 199–208 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bahmani B., et al. , Intratumoral immunotherapy using platelet-cloaked nanoparticles enhances antitumor immunity in solid tumors. Nat. Commun. 12, 1999 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Michaelis K. A., et al. , The TLR7/8 agonist R848 remodels tumor and host responses to promote survival in pancreatic cancer. Nat. Commun. 10, 4682 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Lu R., et al. , Formulation and preclinical evaluation of a Toll-like receptor 7/8 agonist as an anti-tumoral immunomodulator. J. Control Release 306, 165–176 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Seki R., Nishizawa K., Use of TLR9 and TLR7/8 agonists in combination with d-galactosamine in exploring models for distinct severities of systemic inflammation relative to liver injury. Physiol. Res. 69, 1125–1129 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zahr N. M., Zhao Q., Goodcase R., Pfefferbaum A., Systemic administration of the TLR7/8 agonist resiquimod (R848) to mice is associated with transient, in vivo-detectable brain swelling. Biology (Basel) 11, 274 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lind N. A., Rael V. E., Pestal K., Liu B., Barton G. M., Regulation of the nucleic acid-sensing Toll-like receptors. Nat. Rev. Immunol. 22, 224–235 (2022), 10.1038/s41577-021-00577-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Miyake K., et al. , Nucleic acid sensing by Toll-like receptors in the endosomal compartment. Front. Immunol. 13, 941931 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Trivedi S., Greidinger E. L., Endosomal Toll-like receptors in autoimmunity: Mechanisms for clinical diversity. Therapy 6, 433–442 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kano N., Ong G. H., Ori D., Kawai T., Pathophysiological role of nucleic acid-sensing pattern recognition receptors in inflammatory diseases. Front. Cell Infect. Microbiol. 12, 910654 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Crump K. E., Sahingur S. E., Microbial nucleic acid sensing in oral and systemic diseases. J. Dent. Res. 95, 17–25 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.He L., et al. , Nucleic acid sensing pattern recognition receptors in the development of colorectal cancer and colitis. Cell Mol. Life Sci. 74, 2395–2411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Asano T., et al. , X-linked recessive TLR7 deficiency in ~1% of men under 60 years old with life-threatening COVID-19. Sci. Immunol. 6, eabl4348 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Solanich X., et al. , Genetic screening for TLR7 variants in young and previously healthy men with severe COVID-19. Front. Immunol. 12, 719115 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Fallerini C., et al. , Association of Toll-like receptor 7 variants with life-threatening COVID-19 disease in males: Findings from a nested case-control study. Elife 10, e67569 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Campolo M., et al. , TLR7/8 in the pathogenesis of Parkinson’s disease. Int. J. Mol. Sci. 21, 9384 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Frank S., Copanaki E., Burbach G. J., Muller U. C., Deller T., Differential regulation of toll-like receptor mRNAs in amyloid plaque-associated brain tissue of aged APP23 transgenic mice. Neurosci. Lett. 453, 41–44 (2009). [DOI] [PubMed] [Google Scholar]
- 98.Dembny P., et al. , Human endogenous retrovirus HERV-K(HML-2) RNA causes neurodegeneration through Toll-like receptors. JCI Insight 5, e131093 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Brown G. J., et al. , TLR7 gain-of-function genetic variation causes human lupus. Nature 605, 349–356 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Karadimou G., et al. , Low TLR7 gene expression in atherosclerotic plaques is associated with major adverse cardio- and cerebrovascular events. Cardiovasc. Res. 113, 30–39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Salagianni M., et al. , Toll-like receptor 7 protects from atherosclerosis by constraining "inflammatory" macrophage activation. Circulation 126, 952–962 (2012). [DOI] [PubMed] [Google Scholar]
- 102.Li M., et al. , Analysis of the RNA content of the exosomes derived from blood serum and urine and its potential as biomarkers. Philos. Trans. R. Soc. Lond. B Biol. Sci. 369, 20130502 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Jia J., et al. , Distinct extracellular RNA profiles in different plasma components. Front. Genet. 12, 564780 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Shi J., et al. , PANDORA-seq expands the repertoire of regulatory small RNAs by overcoming RNA modifications. Nat. Cell Biol. 23, 424–436 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Xu H., Yao J., Wu D. C., Lambowitz A. M., Improved TGIRT-seq methods for comprehensive transcriptome profiling with decreased adapter dimer formation and bias correction. Sci. Rep. 9, 7953 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Pliatsika V., Loher P., Telonis A. G., Rigoutsos I., MINTbase: A framework for the interactive exploration of mitochondrial and nuclear tRNA fragments. Bioinformatics 32, 2481–2489 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Holmes A. D., et al. , A standardized ontology for naming tRNA-derived RNAs based on molecular origin. Nat. Methods 20, 627–628 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Fechter P., Rudinger J., Giege R., Theobald-Dietrich A., Ribozyme processed tRNA transcripts with unfriendly internal promoter for T7 RNA polymerase: Production and activity. FEBS Lett. 436, 99–103 (1998). [DOI] [PubMed] [Google Scholar]
- 109.Pawar K., Hanisch C., Palma Vera S. E., Einspanier R., Sharbati S., Down regulated lncRNA MEG3 eliminates mycobacteria in macrophages via autophagy. Sci. Rep. 6, 19416 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fabbri M., et al. , MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. U.S.A. 109, E2110–E2116 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Honda K., et al. , Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature 434, 1035–1040 (2005). [DOI] [PubMed] [Google Scholar]
- 112.Honda S., et al. , The biogenesis pathway of tRNA-derived piRNAs in Bombyx germ cells. Nucleic Acids Res. 45, 9108–9120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix 01 (PDF)
Data Availability Statement
There are no data underlying this work.