Abstract
Most retroviral packaging cell lines were established by a helper virus plasmid cotransfected with a separate plasmid encoding a selection marker. Since this selection marker coexisted in trans with the helper virus sequence, helper virus gene expression could be inactivated by host DNA methylation despite selection for the cotransfected selection marker. We have reported that DNA methylation could occur in the long terminal repeat (LTR) region of helper virus in vector producer cells (VPC) in up to 2% of the population per day (W. B. Young, G. L. Lindberg, and C. J. Link, Jr., J. Virol. 74:3177–3187, 2000). To overcome host cell DNA methylation that suppresses viral gene expression, we constructed a chimeric retroviral helper virus, pAM3-IRES-Zeo, that contains Moloney murine leukemia virus as a helper virus and a picornavirus internal ribosome entry site (IRES) sequence followed by a Zeocin selection marker at the 3′ end of the env sequence. This pAM3-IRES-Zeo permitted selection for intact and functional helper virus in transfected cells without subcloning. By selection with Zeocin, a mixed population of pAM3-IRES-Zeo-transfected NIH3T3 cells (AMIZ cells) was maintained with little or no DNA methylation of the helper virus 5′ LTR. The high level of pAM3-IRES-Zeo gene expression resulted in no detectable vector superinfection and in high vector titers (2 × 106 to 1.5 × 107 CFU/ml) after introduction of a retroviral vector. When Zeocin selection was withdrawn from AMIZ cells, methylation of the 5′ LTR increased from 17 to 36% of the population during 67 days of continuous culture and the cells became susceptible to superinfection. During this period, gene expression of pAM3-IRES-Zeo decreased and vector titer production was reduced to 2 × 104 CFU/ml. These data demonstrate an important role of DNA methylation in the genetic instability of VPC. The chimeric helper virus allows the establishment of a mixed population of packaging cells capable of high-level and sustained vector production without cloning procedures.
Our laboratory is interested in the genetic instability of retroviral vector producer cells (VPC) caused by host cell DNA methylation. We have observed that extensive DNA methylation can occur in murine LTKOSN.2 VPC of retroviral helper virus sequences at a rate of 2% of the cell population per day. The DNA methylation of the helper virus 5′ long terminal repeat (LTR) in LTKOSN.2 VPC correlated with reduced helper virus gene expression. These cells had significantly reduced Env-receptor interference and became target cells for vector reentry (superinfection). The VPC developed increasing genetic instability manifested by increasing vector copy numbers. The decreased helper virus gene expression, secondary to DNA methylation, dramatically reduced the vector titer of VPC (54). To overcome these limitations caused by host DNA methylation, we redesigned a retroviral helper virus to improve vector packaging efficiency and used this helper virus to study the interaction between host cells and retroviral sequences, especially host DNA methylation.
Mammalian DNA methyltransferase catalyzes the transfer of a methyl group to cytosines located 5′ to guanosine (CpG dinucleotide) and causes epigenetic effects which usually involve gene silencing. Methylated CpG dinucleotides inactivate gene expression by altering the DNA conformation (8, 22, 36) or attracting the binding of methylated CpG-binding proteins (13, 23, 38, 39) to impede transcription. The majority of DNA methylation patterns in mammalian genomes are found in retrovirus-related sequences, such as retrotransposons and endogenous or exogenous retroviruses (52). Evidence suggests that DNA methylation may act as a host defense system against retroviral invasion of the cellular genome (3, 51, 52). DNA methylation can be triggered by insertion of viral DNA sequence into chromosomes regardless of whether DNA transfection (2) or viral infection (16, 29, 45) was used to introduce the viral DNA sequences.
In several experimental systems, host cell methylation of retroviral provirus or retrotransposons has been evaluated. In a transgenic-mouse model, a retroviral provirus altered the methylation pattern within 1 kb of the retroviral integration site. The provirus was methylated, leading to an inactivation of transcription (17, 18). Sequences of small interspersed repetitive elements contained in the rat α-fetoprotein promoter region were associated with increased DNA methylation and decreased downstream reporter gene expression (12). Reduction of host DNA methylation leads to amplification and retrotransposition of kangaroo endogenous retroviral element 1 and xenologous recombination of chromosomes in interspecific mammalian hybrids of the Australian wallaby (41). Interestingly, retroviruses may benefit from host DNA methylation as well. Human immunodeficiency virus type 1 (HIV-1) infection may induce host DNA methylation activity, and as a consequence, the promoter region of gamma interferon was downregulated by DNA methylation (29). This may alter the balance of cytokines and reduce immune surveillance (28, 29). The inactivation of HIV-1 or human T-cell leukemia virus type 1 gene expression by host DNA methylation of viral LTR regions may also induce latency of HIV-1 or human T-cell leukemia virus type 1 infection (1, 28, 44, 45).
These prior experiments did not evaluate DNA methylation of helper virus 5′ LTR in VPC. Many commonly used retroviral vector packaging cell lines were established by cotransfection of two plasmids, one containing a helper virus genome and the other encoding a drug selection marker (26, 31–33). In this cotransfection system, selection for drug resistance does not require active helper virus gene expression, and so the 5′ LTR promoter region can be silenced by DNA methylation (54). Prior studies have demonstrated the concept of including an antibiotic selection marker (7) or a cell surface fluorescence-activated cell sorter marker (human Phoenix cell line; http://www.stanford.edu/group/nolan/NL-phoenix.html) downstream of gag-pol to monitor the gene expression. As reported here, a chimeric helper virus, pAM3-IRES-Zeo, was designed containing an internal ribosome entry site (IRES) sequence of the encephalomyocarditis virus (19), a member of the picornaviruses (43), and a Zeocin resistance gene (Zeo) (10) to allow selection against DNA methylation that might occur in the helper virus 5′ LTR region.
During translation of most eukaryotic mRNAs, ribosomes scan mRNA from the 5′ cap sequence until an initiation codon is reached. In contrast, in picornavirus mRNA, ribosomes initiate translation by an alternative mechanism that involves internal initiation rather than scanning. The IRES sequences of picornavirus enable ribosomes to bind in a cap-independent fashion and start translation at the next AUG codon downstream (20). Ligation of the IRES sequence followed by Zeo at the 3′ end of the env gene permits the translation of helper virus open reading frames and a selection marker from this mRNA (Fig. 1). Selection with Zeocin eliminates cells with methylated helper virus 5′ LTR from the population. This design should ensure sustained helper virus gene expression, which would increase virion production and create sufficient Env receptor interference to prevent superinfection. The prevention of superinfection may in turn reduce replication-competent retrovirus (RCR) formation (34, 35). One additional advantage is that pAM3-IRES-Zeo allows the establishment of packaging cell lines within a shorter time. This advantage might be critical when making human VPC from a primary cell culture or stem cells to avoid immune rejection (48, 49), while transplantation of VPC into patients is necessary for continuous gene transfer (42).
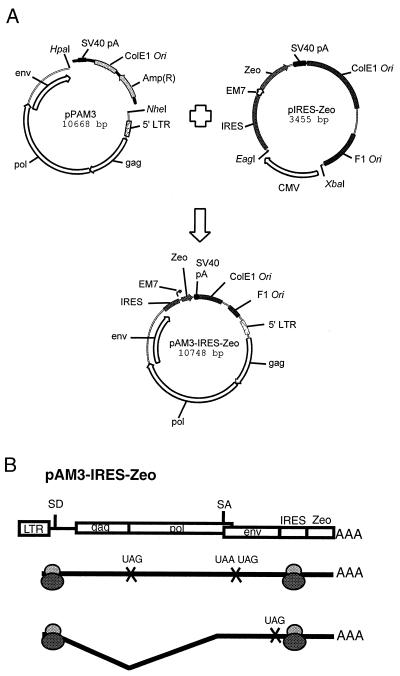
FIG. 1.
Construction and cap-independent translation mechanism of the chimeric pAM3-IRES-Zeo helper virus. (A) Plasmids used in this study. For details of their construction, see Materials and Methods. Briefly, a 2.8-kb fragment including the IRES-Zeo expression cassette, an SV40 polyadenylation signal sequence, the bacterial replication origin (ColE1 Ori), and phage replication origin (F1 Ori) was excised from pIRES-Zeo. The ColE1 Ori and ampicillin resistance gene (Ampr) of pPAM3 were replaced with the above 2.8-kb IRES-Zeo-containing fragment from pIRES-Zeo. The EM7 prokaryotic promoter located at the 5′ end of the Zeo gene permits selection for pAM3-IRES-Zeo in bacteria. (B) Genomic RNA of MoMLV contains two internal stop codons at the 3′ ends of the gag and pol genes that terminate cap-dependent translation and allow appropriate ratios of viral structural proteins. In pAM3-IRES-Zeo-derived transcripts, ribosomes also recognize the IRES sequence and initiate translation from the first AUG codon of Zeo downstream of the IRES sequence. A portion of genomic RNA is spliced into env transcripts that are translated in a cap-dependent mechanism. SD, splicing donor; SA, splicing acceptor.
MATERIALS AND METHODS
Construction of helper virus pAM3-IRES-Zeo and LEIN vector.
An IRES sequence of encephalomyocarditis virus was isolated from the LXIN retroviral vector (Clontech, Palo Alto, Calif.) by NsiI and PstI digestions and inserted into a PstI-linearized pZeoSV mammalian expression vector (Invitrogen, Carlsbad, Calif.) immediately 5′ of the EM-7 prokaryotic promoter/Zeocin resistance gene (Zeo) to create an IRES-Zeo expression cassette in plasmid pIRES-Zeo-SV40. SalI digestion of pIRES-Zeo-SV40 deleted the simian virus 40 (SV40) promoter and downstream polyadenylation signal to generate pIRES-Zeo. A 2.8-kb fragment consisting of the IRES-Zeo expression cassette, SV40 poly(A) signal, bacterial replication origin (ColE1 Ori), and phage replication origin (F1 Ori) was excised from pIRES-Zeo by EagI digestion, Klenow fill-in (GIBCO BRL, Life Technology Co., Gaithersburg, Md.), and, finally, XbaI digestion. To construct pAM3-IRES-Zeo, an amphotropic helper virus, pPAM3 (31) (kindly provided by A. Dusty Miller, Fred Hutchinson Cancer Research Center, Seattle, Wash.), was digested by HpaI at the 3′ end of the env gene and NheI at the 5′ end of the LTR to delete the ColE1 Ori and ampicillin resistance gene (Ampr). This deleted region was replaced with the 2.8-kb IRES-Zeo fragment described above (Fig. 1). The resulting chimeric helper virus plasmid, pAM3-IRES-Zeo, allows selection with Zeocin in bacterial culture and mammalian cells.
The LEIN retroviral vector carrying an enhanced green fluorescent protein (6, 11, 37) reporter gene was constructed by replacing the SV40 promoter-neomycin phosphotransferase gene (Neor) cassette of pLESN (27) with a 1.4-kb IRES-Neo cassette, excised from pIRES-Neo by NaeI and NsiI digestions.
Cell culture and transfection.
Cell cultures were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL, Life Technology Co.) plus 10% fetal calf serum under 5% CO2 at 37°C. The subclones of LTKOSN.2 VPC were obtained by limiting dilution of parental LTKOSN.2 VPC onto two 96-well plates (54). Helper virus and vector gene expression, DNA methylation status, and vector production in these subclones have been previously characterized (54). To rescue LTKOSN and ΔLTKOSN vectors from preexisting LTKOSN VPC subclones with methylated and silenced helper virus DNA, the subclones were transfected with pAM3-IRES-Zeo using Fugene 6 transfection reagent (Roche Molecular Biochemicals, Indianapolis, Ind.). To study the effects of host DNA methylation on retroviral helper virus without interference from chromosomal copies of pPAM3 present in LTKOSN VPC, pAM3-IRES-Zeo plasmid was transfected into NIH 3T3 tk− cells (ATCC CRL1658) utilizing Fugene 6 transfection reagent. A mixed population of pAM3-IRES-Zeo-transfected NIH 3T3 tk− cells, termed AMIZ cells, was established. Prior to transfection, pAM3-IRES-Zeo plasmid was linearized by BspHI digestion and 6 to 10 μg of pAM3-IRES-Zeo was then transfected to each well in six-well plates. Selection with Zeocin (350 μg/ml; Invitrogen) began 48 h after transfection and continued for at least 2 weeks. Transfection of the LEIN vector into the AMIZ cell pool and GP+E86 packaging cells (26) (kindly provided by Arthur Bank, Columbia University, New York, N.Y.) was completed by using DOTAP liposomal transfection reagent (Roche Molecular Biochemicals) with 5 μg of LEIN plasmid for each well in six-well plates. Selection with G418 (1 mg/ml; GIBCO) was started 48 h after transfection and continued for 2 weeks.
Retroviral infection, superinfection, and titer assays.
Supernatants collected from pAM3-IRES-Zeo-transfected LTKOSN.2 VPC subclones were diluted in 10-fold serial dilutions to transduce NIH3T3 tk− cells, A375 cells (ATCC CRL1619) (human melanoma), and IGROV cells (human ovarian carcinoma) (50), which were plated at 105 cells/well in six-well plates with 10 μg of protamine sulfate per ml. At 24 h after transduction, cells were selected for 10 to 14 days in medium containing G418 (1 mg/ml). Titers were calculated by multiplying the number of resistant colonies by the dilution factor.
To perform superinfection assays on AMIZ cells, supernatants containing LEIN vector collected from LEIN-transfected AMIZ cells were passed through a 0.4-μm-pore-size syringe filter and diluted 10-fold and 100-fold before being used in superinfection assays. Along with AMIZ cells, NIH 3T3 tk− and PA317 cells were transduced as Env receptor interference-negative and -positive controls, respectively. Selection with G418 (1 mg/ml) on these transduced cells started 24 h after a single exposure to LEIN vector and continued for 10 to 14 days. The number of G418-resistant colonies was used as the index for superinfection on PA317 and AMIZ cells. To investigate the vector production capability of AMIZ cells, a LEIN vector from the ecotropic Moloney murine leukemia virus (MoMLV) packaging cell line, GP+E86, was transduced into AMIZ cells without further subcloning.
RNA analysis of helper virus and vector gene expression.
Total cellular RNA was isolated from transfected cells and VPC by using the RNAeasy kit (Qiagen Inc., Valencia, Calif.) and Northern blotted from a 1% agarose–0.4 M formaldehyde gel. Vector transcripts were detected with a Neo probe. Helper virus transcripts were detected by a 1.4-kb env probe, which was isolated from pPAM3 after XhoI digestion. Human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) cDNA was used to demonstrate similar RNA loading and to standardize the helper virus gene expression to allow comparisons between selected and unselected cells. For analysis, the band intensities of both unspliced and spliced helper virus transcripts were divided by the intensity of GAPDH to determine relative expression levels.
DNA methylation analysis.
In AMIZ cells transfected with LEIN vector, the methylation status of provirus and vectors was determined by evaluating the resistance to digestion with a DNA methylation-sensitive restriction endonuclease, SmaI, in the 5′ LTR region. Genomic DNA was digested with DraI and EcoRV to reduce the DNA fragment size, precipitated with ethanol, and then redissolved in sterile water. This DNA digest was divided into two equal portions, one of which was subjected to SmaI digestion. The Southern blot membrane was hybridized with a 428-bp fragment of the gag sequence (PvuII-DraI) from pPAM3 to detect helper virus and a GFP probe to detect the LEIN vector. Densitometric analyses were performed with a GS300 densitometer (Hoefer Scientific Instruments) to measure the relative densities of the SmaI-sensitive band and the DraI-EcoRV band. Due to interference from endogenous retroviral elements, the fraction of SmaI methylation in 5′ LTR was calculated as 1 − (intensity ratio of the SmaI-sensitive band at 1.5 kb to the DraI-EcoRV band at 1.8 kb) (see Fig. 6).
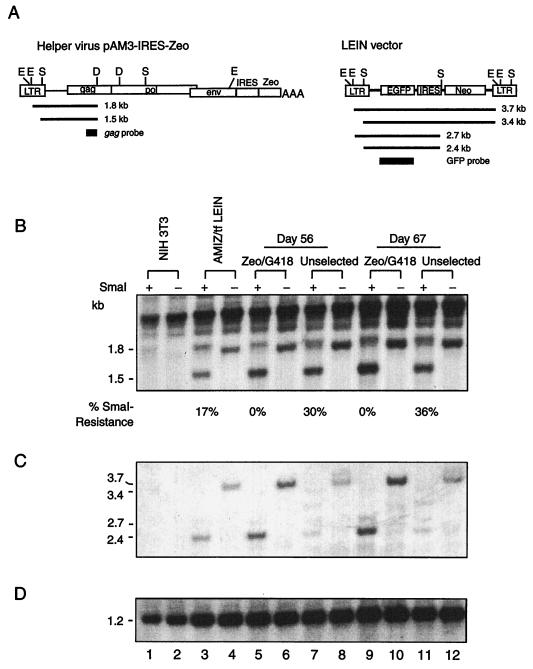
FIG. 6.
Drug selection eliminates DNA methylation of helper virus and vector from the VPC population. Genomic DNA extracted from AMIZ cells transfected with the LEIN vector with Zeocin and G418 selection or without drug selection on days 0 (lanes 3 and 4), 56 (lanes 5 to 8), and 67 (lanes 9 to 12) was first digested with DraI and EcoRV and divided into two equal portions. One portion was subjected to methylation-sensitive SmaI restriction endonuclease digestion, and the other was not. (A) Schema of the helper virus and vectors showing the locations of restriction enzyme sites and probes used for methylation analysis. D, DraI; E, EcoRV; S, SmaI; AAA, SV40 polyadenylation signal. Drawings are not to scale. (B) Hybridization of the gag DNA probe to detect helper virus 5′ LTR. SmaI digestion reduced the 1.8-kb band (even-numbered lanes from 4 to 12) to 1.5 kb (odd-numbered lanes from 3 to 11). DNA from NIH 3T3 cells was used to show the presence of endogenous retroviral elements (lanes 1 and 2). Since a 1.8-kb band was generated from the endogenous retroviral element after SmaI digestion (lane 1), the values for SmaI resistance were measured by densitometry as the relative intensities of the 1.8-kb bands without SmaI digestion and the 1.5-kb bands after SmaI digestion. (C) A green fluorescent protein DNA probe was used to detect the 5′ LTR of the LEIN vector. SmaI digestion reduced the 3.7-kb fragment to 3.4-, 2.7-, and 2.4-kb fragments, depending on the methylation status of the SmaI site in LEIN vector. (D) A 0.68-kb DNA fragment digested from the gag gene of pPAM3 by BstXI was used as a probe to detect a 1.2-kb band of the endogenous retroviral element to demonstrate relative loading in paired digestions with and without SmaI.
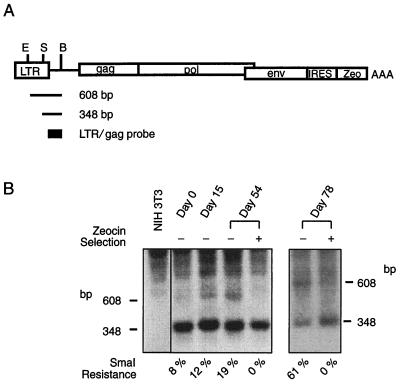
Without the interference of vector and endogenous retroviral sequences mentioned above, the DNA methylation status of the 5′ LTR region of pAM3-IRES-Zeo in AMIZ cells was determined by digesting genomic DNA with EcoRV, BstEII, and SmaI. If methylation occurred at the SmaI site, a 608-bp fragment would be excised instead of a 348-bp fragment when the DNA was probed with a 261-bp fragment excised from pAM3-IRES-Zeo by KpnI and AflII digestions. The degree of DNA methylation was calculated as the intensity of the SmaI-insensitive band (608 bp) divided by the sum of the intensities of this 608-bp fragment and the SmaI-sensitive fragment (348 bp) (see Fig. 3).
FIG. 3.
DNA methylation of helper virus 5′ LTR over time with and without Zeocin selection. (A) Schema of the pAM3-IRES-Zeo helper virus, showing the restriction enzyme sites and the probe used for the methylation analysis. B, BstEII; E, EcoRV; S, SmaI; AAA, SV40 polyadenylation signal. The drawing is not to scale. (B) A genomic DNA Southern blot membrane was probed with a 261-bp fragment excised from pAM3-IRES-Zeo with KpnI and AflII digestion. If methylation was present at the SmaI site, a 608-bp fragment would result instead of a 348-bp fragment. The degree of DNA methylation was calculated as the intensity of the SmaI-insensitive band (608 bp) divided by the sum of the intensities of this 608-bp band and the SmaI-sensitive fragment (348 bp).
RESULTS
Construction of a chimeric retroviral helper virus with IRES and selection marker to allow direct selection of helper virus gene expression.
We previously determined that DNA methylation occurred in 2% of the cell population per day within the 5′ LTR region of helper virus to inactivate helper virus gene expression in VPC (54). To eliminate methylated helper virus 5′ LTR from the packaging-cell population, a chimeric retroviral helper virus, pAM3-IRES-Zeo (Fig. 1), was constructed. The pAM3-IRES-Zeo construction allows Zeocin selection of cells with 5′ LTR promoter function, since helper virus and Zeor gene expression are transcribed from the 5′ LTR promoter (Fig. 1B). The selection with Zeocin maintains cells that also express helper virus and therefore counteract DNA methylation effects. Packaging cells based on this pAM3-IRES-Zeo helper virus should maintain high-titer production. The evaluation of these pAM3-IRES-Zeo-transfected cells with or without Zeocin selection provide a methylation profile of helper virus 5′ LTR and helper virus gene expression.
Analysis of chimeric pAM3-IRES-Zeo vector packaging ability in preexisting LTKOSN.2 VPC subclones.
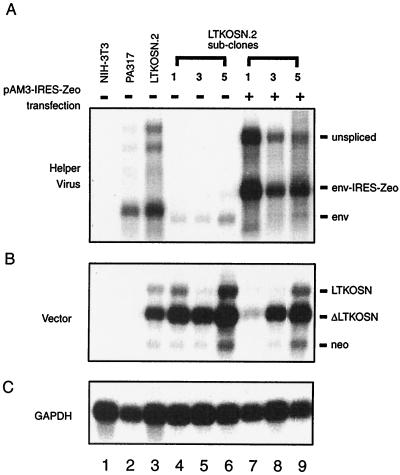
To test the packaging ability of pAM3-IRES-Zeo helper virus, pAM3-IRES-Zeo was transfected into three individual subclones of LTKOSN.2 VPC. LTKOSN.2 VPC contain one LTKOSN vector and an additional ΔLTKOSN, which is derived from the LTKOSN vector with a herpes simplex virus tk deletion mutation (53). The pPAM3 helper virus gene expression in these three LTKOSN.2 VPC subclones, 1, 3, and 5 (Fig. 2A, lanes 4 to 6), was inactivated by DNA methylation with impeded vector production ability (Table 1) (54). However, the LTKOSN (4.0-kb) and ΔLTKOSN (2.8-kb) vectors in these subclones were still transcribed (Fig. 2B, lanes 4 to 6) and no significant DNA methylation of these vectors was observed (54). This indicated that a key limitation of vector production in LTKOSN.2 VPC subclones is the lack of helper virus gene expression. Rescue of LTKOSN and ΔLTKOSN vectors from these three subclones was performed by transfection of pAM3-IRES-Zeo followed by 2 weeks of Zeocin selection. This restored high level of vector production was shown by titer determination on human IGROV ovarian carcinoma, human A375 melanoma, and murine NIH 3T3 tk− target cells. The titers ranged from 4 × 105 to 1.6 × 107 CFU/ml (Table 1). In addition, this increased packaging activity with pAM3-IRES-Zeo resulted in a reduction of retained LTKOSN and ΔLTKOSN vectors inside VPC when analyzed by Northern blot analysis (Fig. 2B, lanes 7 to 9).
FIG. 2.
Gene expression of pAM3-IRES-Zeo and vectors in LTKOSN.2 VPC subclones. (A) Northern blot analysis of cellular RNA extracted from pAM3-IRES-Zeo-transfected LTKOSN.2 subclones hybridized with the env probe. Expression of unspliced MoMLV transcript (gag-pol-env-IRES-Zeo) and spliced RNA (env-IRES-Zeo) were significantly greater than pPAM3 gene expression, which exhibited only spliced env transcripts. (B) Gene expression of LTKOSN and ΔLTKOSN vectors. Shown is hybridization of the same Northern blot membrane with the Neor probe to detect full-length LTKOSN (4.0 kb) and ΔLTKOSN (2.5 kb) RNA transcripts and a Neor transcript (1.2 kb) expressed from the internal SV40 promoter. Fewer vector transcripts were retained in pAM3-IRES-Zeo-transfected cells, since transcripts were packaged into virions. (C) Hybridization of the same Northern blot membrane with a human GAPDH cDNA probe demonstrates fairly equivalent RNA loading.
TABLE 1.
Titer of LTKOSN.2 VPC subclones before and after transfection of pAM3-IRES-Zeo helper virus
| Subclone | pAM3-IRES-Zeo | Titer of subclone on target cells (CFU/ml)
|
||
|---|---|---|---|---|
| NIH 3T3 | A375 | IGROV | ||
| 1 | + | 1.1 × 107 | 1 × 107 | 1 × 107 |
| − | 1 × 101 | 2 × 100 | 0 | |
| 3 | + | 5 × 106 | 5.5 × 105 | 4 × 105 |
| − | 0 | 0 | 0 | |
| 5 | + | 1.6 × 107 | 5 × 106 | 4 × 106 |
| − | 0 | 0 | 0 | |
Analysis of gene expression in pAM3-IRES-Zeo-transfected LTKOSN.2 VPC subclones demonstrated significantly greater helper virus gene expression compared to that for pPAM3 in PA317 packaging cells and parental LTKOSN.2 VPC (Fig. 2A). In addition to env transcripts, only one population of unspliced helper virus (gag-pol-env-IRES-Zeo) was detected in pAM3-IRES-Zeo-transfected subclones, which indicates that the integration of pAM3-IRES-Zeo should be intact in transfected cells after selection. In contrast, cotransfection of pPAM3 without direct selection for pPAM3 gene expression but other selection markers in trans could result in randomly interrupted pPAM3 for integration. This was shown by two additional transcripts of lower molecular weight detected in PA317 and LTKOSN.2 VPC (Fig. 2, lanes 2 and 3) (54). These results demonstrate that enhanced and sustained helper virus gene expression can be obtained in polyclonal packaging cells when pAM3-IRES-Zeo is used to allow Zeocin selection without the need to perform time-consuming cell subcloning. This implies a potential use of pAM3-IRES-Zeo to establish new packaging cells from other cells such as human primary cells.
Cells transfected with pAM3-IRES-Zeo provide a model to study DNA methylation of retroviral sequences.
DNA methylation in mammalian cells is site dependent within the genome (14). Therefore, a mixed population of pAM3-IRES-Zeo-transfected cells would be required to study the DNA methylation of helper virus 5′ LTR in order to minimize the effects of positional interference. To establish a pooled population of packaging cells without chromosomal pPAM3 effects, pAM3-IRES-Zeo was transfected into NIH 3T3 tk− cells, and this was followed by selection with Zeocin without further subcloning. This pool of newly established packaging cells was named AMIZ packaging cells (pAM-IRES-Zeo). To allow DNA methylation to occur, AMIZ cells were released from Zeocin selection for 1 month and then placed in continuous culture with or without Zeocin selection for 78 days (10 passages). DNA methylation and gene expression of pAM3-IRES-Zeo were examined at 15, 54, and 78 days after being released from selection. Over the first 54 days of the cell culture period, DNA methylation of the 5′ LTR increased from 8 to 19%, and by day 78 it reached 61% (Fig. 3). The DNA methylation rate of helper virus 5′ LTR averaged 0.7% of the population per day during a 78-day period. AMIZ cells with continued Zeocin selection did not exhibit any detectable DNA methylation (Fig. 3). This drug selection effectively eliminated methylated pAM3-IRES-Zeo from the pooled AMIZ population.
Retroviral superinfection is blocked by enhanced helper virus gene expression.
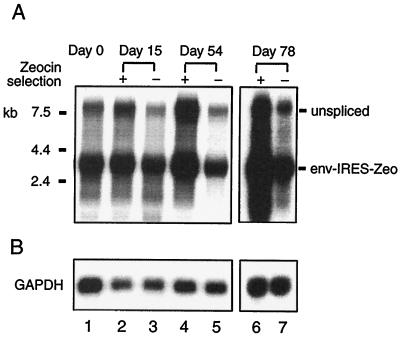
The effect of Zeocin selection on AMIZ cells was analyzed by gene expression of pAM3-IRES-Zeo in AMIZ cells. Gene expression of pAM3-IRES-Zeo in AMIZ cells with constant Zeocin selection showed a twofold increase compared to AMIZ cells without selection on day 15 and at least a fourfold increase on days 54 and 78 (Fig. 4). In contrast, pAM3-IRES-Zeo gene expression in AMIZ cells without Zeocin selection declined over time (Fig. 4, lanes 3, 5, and 7). Continuous Zeocin selection may have selected integration sites that are highly transcriptionally active and have less DNA methylation activity (5, 22).
FIG. 4.
The effectiveness of Zeocin selection on helper virus gene expression in AMIZ cells over time. (A) Unspliced MoMLV transcript (gag-pol-env-IRES-Zeo) and spliced RNA (env-IRES-Zeo) were detected by an env probe in cellular RNA extracted from AMIZ cells with and without continuous Zeocin selection on days 0, 15, 54, and 78. (B) Hybridization with the GAPDH cDNA probe demonstrates the relative RNA loading.
We directly determined whether decreased pAM3-IRES-Zeo gene expression reduced Env receptor interference and increased vector superinfection. The susceptibility to superinfection was measured by exposing AMIZ cells from the above experiment to amphotropic LEIN vector supernatants and subjecting them to G418 selection. The number of G418-resistant colonies obtained from AMIZ cells with continued Zeocin selection was reduced from 23 on day 15 to no superinfection observed on days 54 and 78 (Table 2). In contrast, G418-resistant colonies obtained from AMIZ cells without Zeocin selection ranged from 1.2 × 103 to 5.6 × 103. These results demonstrate that increased gene expression of helper virus correlates with reduced susceptibility to superinfection.
TABLE 2.
Increased resistance of superinfection by Zeocin selection
| Daya | No. of G418-resistant colonies on target cells
|
|||
|---|---|---|---|---|
| NIH 3T3 | PA317 | AMIZ
|
||
| No selection | Selection | |||
| 15 | 7 × 105 | 3.2 × 104 | 4.3 × 103 | 2.3 × 101 |
| 54 | 6 × 104 | 1 × 104 | 1.2 × 103 | 0 |
| 78 | 5 × 105 | 1.9 × 104 | 5.6 × 103 | 0 |
Number of days after AMIZ cells were cultured in parallel either with or without Zeocin.
A high level of vector production is maintained by Zeocin selection.
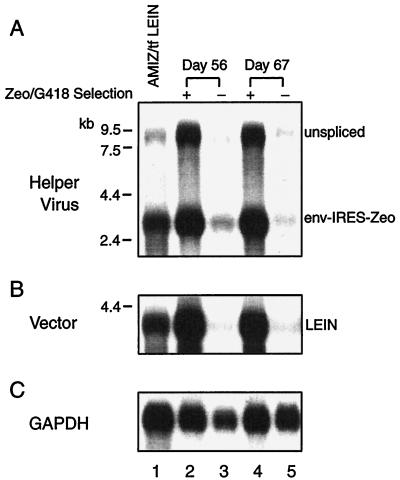
Vector production was analyzed in this AMIZ cell pool by transfecting LEIN vector into AMIZ cells and performing G418 selection to establish a VPC for titer assay. Zeocin selection was withdrawn from the AMIZ cell culture during the first 3 weeks of G418 selection after transfection with the LEIN vector. The titer obtained from this newly established uncloned population of AMIZ cells was 3.5 × 106 CFU/ml, which is 100-fold higher than the titer observed from a mixed population of PA317 cells transfected with LEIN vector (4 × 104 CFU/ml). In addition, AMIZ cells were transduced with LEIN vector collected from LEIN-transfected GP+E86 cells, and an improved titer of 9 × 106 CFU/ml was obtained from a mixed cell population. To investigate whether selection with both Zeocin and G418 would adversely affect vector production, LEIN-transfected AMIZ cells were evaluated 56 (8 passages) and 67 (10 passages) days after transfection. Titers obtained from AMIZ cells transfected with LEIN (3.5 × 106 CFU/ml on day 0) and placed under continuous selection with Zeocin and G418 were 2 × 106 CFU/ml (day 56) and 1.5 × 107 CFU/ml (day 67). In contrast, titers obtained from the same AMIZ cells transfected with LEIN but not subjected to G418 and Zeocin selection were only 2 × 104 and 4 × 104 CFU/ml on days 56 and 67, respectively. The reduced titer correlated with a significant decrease of both helper virus and vector gene expression when time points with and without selection were compared (Fig. 5). No significant increase of titer or helper virus gene expression was observed when the 17% DNA methylation present on day 0 was further reduced to 0% by day 56 after selection. This suggests a threshold effect, as we previously observed in cloned VPC (54). Substantial decreases of vector production, helper virus gene expression, and Env receptor interference were observed only when at least 60% methylation of the helper virus 5′ LTR occurred.
FIG. 5.
Gene expression of the pAM3-IRES-Zeo helper virus and LEIN vector in AMIZ cells transfected with the LEIN vector. (A) Northern blot analysis of cellular RNA extracted from AMIZ cells transfected with the LEIN vector evaluated at passage 3 (day 0, lane 1) and on days 56 and 67 (lanes 2 to 5) by hybridization with the env probe. Zeocin and G418 selection resulted in higher levels of unspliced MoMLV RNA transcript (gag-pol-env-IRES-Zeo) and spliced RNA transcripts (env-IRES-Zeo). (B) The level of LEIN vector was also higher in AMIZ cells under Zeocin and G418 selection. (C) Rehybridization with the GAPDH cDNA probe was used to demonstrate fairly equivalent RNA loading.
The DNA methylation status of 5′ LTRs of helper virus and vector were significantly increased in AMIZ cells transfected with LEIN vector and cultured without either G418 or Zeocin selection (Fig. 6). This increased methylation corresponded to the above-mentioned decreased vector titer and significantly reduced the gene expression of the helper virus and vector (Fig. 5). The DNA methylation of the helper virus 5′ LTR increased from 17% on day 0 to 30 and 36% on days 56 and 67, respectively. The average DNA methylation rate of helper virus 5′ LTR in AMIZ cells transfected with LEIN was estimated to be only 0.3% of the cell population per day during 67 days of continuous cell culture. In contrast, DNA methylation was not detected in AMIZ cells transfected with LEIN vector and placed under continuous G418 and Zeocin selection. No detectable DNA methylation occurred in the LEIN vector on day 0 (Fig. 6C, lanes 3 and 4), while the 5′ LTR helper virus showed 17% DNA methylation (Fig. 6B, lanes 3 and 4). This may be secondary to the timing of G418 and Zeocin selection. AMIZ cells transfected with LEIN vector were placed under G418 selection for 3 weeks to select for a LEIN-positive population, and Zeocin selection was not applied until day 0 in the experiment.
DISCUSSION
The experimental model described has demonstrated an approach using a retroviral helper virus combining a picornavirus IRES sequence and a selection marker gene that allows efficient elimination of methylated helper virus from packaging-cell populations. This strategy of using drug selection maintained high levels of helper virus gene expression and high-titer vector production (1.5 × 107 CFU/ml) from a nonsubcloned population of VPC. The presence of greater Env receptor interference blocks vector superinfection and may reduce other potential problems with retroviral vectors, including replication-competent retrovirus formation and multiple copies of vectors. A new packaging cell pool, AMIZ cells, established by transfection of pAM3-IRES-Zeo chimeric helper virus into NIH 3T3 tk− cells without any subcloning procedure, has proved a useful system to study the effect of host DNA methylation on retroviral sequences.
The selection of transfected cells (AMIZ cells) with Zeocin to maintain pAM3-IRES-Zeo gene expression eliminated DNA methylation from AMIZ cells and may also select cells with pAM3-IRES-Zeo helper virus integrated in optimal and active chromosomal regions. Ratios of pAM3-IRES-Zeo gene expression in selected AMIZ cells compared to nonselected AMIZ cells were about 2:1 on day 15 and at least 4:1 on days 54 and 78 (Fig. 4), while helper virus showed only 12, 19, and 61% DNA methylation, respectively (Fig. 3). Similar results were also observed in AMIZ cells transfected with LEIN vector. Cells under continuous selection showed no detectable DNA methylation of the 5′ LTR, but 30% (day 56) and 36% (day 67) DNA methylation was detected in cells without selection (Fig. 6). LEIN-transfected AMIZ cells under continuous selection had a vector titer of 1.5 × 107 CFU/ml on day 67, compared to 4 × 104 CFU/ml on day 67, in cells without selection. This 1,000-fold difference in titer probably reflects the fact that structural proteins of viruses function as multimers (15). The formation of multimers occurs in a sigmoid rather than a linear dose-response fashion with respect to protein concentration that correlates more directly with helper virus gene expression and DNA methylation. The effect of host DNA methylation on the helper virus 5′ LTR is therefore amplified by transcription, viral assembly, and then vector production.
To maintain efficient Env receptor interference and active viral production, a threshold level of helper virus gene expression is required. In retrovirus infection, this threshold level of gene expression is established by the accumulation of a sufficient copy number of virus through superinfection until efficient Env receptor interference is achieved and maintained (40). In our study, the threshold level of helper virus gene expression was achieved by Zeocin selection rather than by increasing the copy number of helper virus. Superinfection was observed when selection pressure was released and helper virus gene expression declined. These results support the conclusion that continuous selection of helper virus in VPC might enhance Env receptor interference and reduce the possibility of RCR formation.
For continuous virus production, retroviral gene expression has to be regulated at a sufficient level without interfering with host cell growth and differentiation. Increased levels of viral RNAs and proteins in infected cells can cause cytopathic effects, usually at the cost of cell death, by interrupting the production or translation of host mRNA (47). Although we observed that AMIZ cells under continuous selection did proliferate more slowly than AMIZ cells without selection, AMIZ cells under continuous G418 and Zeocin selection for high gene expression for 67 days (Fig. 5) still proliferated (data not shown). We did not attempt to select for pPAM3 gene expression by drug selection against the herpes simplex virus tk selection marker plasmid cotransfected into PA317 cells. This approach is unlikely to be successful, since the selection marker plasmid is separate from pPAM3. An alternative approach to reverse methylation is treatment with 5′-azacytidine (5-aza-C) to reverse DNA methylation (21, 24). In previous experiments, we found that only a minor portion of pPAM3 helper virus expression could be restored by 5-aza-C (54). Treatment with 5-aza-C does not specifically reverse helper virus DNA methylation; it also inhibits cellular DNA methyltransferase and causes cytotoxicity to treated cells (21). The data from this study suggest that a combination of helper virus and IRES sequences with selectable markers is a viable option to eliminate host DNA methylation of helper virus from VPC.
One potential application of this chimeric helper virus to gene therapy would be to allow packaging cells to be established from primary cell culture without subcloning. This might be useful for transplanting VPC into patients (42) without the immune elimination of murine VPC and virions (48, 49). Several studies have aimed at establishing a retroviral packaging cell line by using either adenovirus (4, 9, 25) or herpes simplex virus (46) to import retroviral helper virus genome into target cells in vivo or ex vivo. In this study, the chimeric retroviral helper virus pAM3-IRES-Zeo was used to generate a pooled population of pAM3-IRES-Zeo-transfected cells, AMIZ cells. AMIZ cells transfected with a retroviral vector maintained titers between 3.5 × 106 and 1.5 × 107 CFU/ml. These titers are comparable to reported titers from individually cloned VPC, which generally ranged from 104 to 107 CFU/ml (30). Transfection of pAM3-IRES-Zeo into cells followed by selection for positive populations can take only 2 weeks or less, depending on the transfection efficiency. Since some primary cell cultures are too sensitive to allow effective antibiotic selection, replacing the Zeocin selection marker with a cellular surface marker or GFP gene might be required to overcome obstacles to making VPC from primary cell lines.
ACKNOWLEDGMENTS
We thank John P. Levy for providing the LESN vector and George Cook for editorial correction.
This work was supported in part by a grant from Iowa Methodist Medical Center, Des Moines, and Research Project Grant RPG-98-091-01-MBC from the American Cancer Society (C.J.L.). W.-B. Young is a recipient of a predoctoral fellowship grant from the Molecular, Cellular and Developmental Biology Program, Iowa State University, Ames.
REFERENCES
- 1.Bednarik D P, Cook J A, Pitha P M. Inactivation of the HIV LTR by DNA CpG methylation: evidence for a role in latency. EMBO J. 1990;9:1157–1164. doi: 10.1002/j.1460-2075.1990.tb08222.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bednarik D P, Mosca J D, Raj N B. Methylation as a modulator of expression of human immunodeficiency virus. J Virol. 1987;61:1253–1257. doi: 10.1128/jvi.61.4.1253-1257.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bestor T H, Tycko B. Creation of genomic methylation patterns. Nat Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- 4.Caplen N J, Higginbotham J N, Scheel J R, Vahanian N, Yoshida Y, Hamada H. Adeno-retroviral chimeric viruses as in vivo transducing agents. Gene Ther. 1999;6:454–459. doi: 10.1038/sj.gt.3300835. [DOI] [PubMed] [Google Scholar]
- 5.Cedar H. DNA methylation and gene activity. Cell. 1988;53:3–4. doi: 10.1016/0092-8674(88)90479-5. [DOI] [PubMed] [Google Scholar]
- 6.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of the green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 7.Cosset F L, Takeuchi Y, Battini J L, Weiss R A, Collins M K. High-titer packaging cells producing recombinant retroviruses resistant to human serum. J Virol. 1995;69:7430–7436. doi: 10.1128/jvi.69.12.7430-7436.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Feil R, Boyano M D, Allen N D, Kelsey G. Parental chromosome-specific chromatin conformation in the imprinted U2af1-rs1 gene in the mouse. J Biol Chem. 1997;272:20893–20900. doi: 10.1074/jbc.272.33.20893. [DOI] [PubMed] [Google Scholar]
- 9.Feng M, Jackson W H, Jr, Goldman C K, Rancourt C, Wang M, Dusing S K, Siegal G, Curiel D T. Stable in vivo gene transduction via a novel adenoviral/retroviral chimeric vector. Nat Biotechnol. 1997;15:866–870. doi: 10.1038/nbt0997-866. [DOI] [PubMed] [Google Scholar]
- 10.Gatignol A, Durand H, Tiraby G. Bleomycin resistance conferred by a drug-binding protein. FEBS Lett. 1988;230:171–175. doi: 10.1016/0014-5793(88)80665-3. [DOI] [PubMed] [Google Scholar]
- 11.Haas J, Park E C, Seed B. Codon usage limitation in the expression of HIV-1 envelope glycoprotein. Curr Biol. 1996;6:315–324. doi: 10.1016/s0960-9822(02)00482-7. [DOI] [PubMed] [Google Scholar]
- 12.Hasse A, Schulz W A. Enhancement of reporter gene de novo methylation by DNA fragments from the alpha-fetoprotein control region. J Biol Chem. 1994;269:1821–1826. [PubMed] [Google Scholar]
- 13.Hendrich B, Bird A. Identification and characterization of a family of mammalian methyl-CpG binding proteins. Mol Cell Biol. 1998;18:6538–6547. doi: 10.1128/mcb.18.11.6538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoeben R C, Migchielsen A A, van der Jagt R C, van Ormondt H, van der Eb A J. Inactivation of the Moloney murine leukemia virus long terminal repeat in murine fibroblast cell lines is associated with methylation and dependent on its chromosomal position. J Virol. 1991;65:904–912. doi: 10.1128/jvi.65.2.904-912.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hunter E. Macromolecular interactions in the assembly of HIV and other retroviruses. Semin Virol. 1994;5:71–83. [Google Scholar]
- 16.Jähner D, Jaenisch R. Retrovirus-induced de novo methylation of flanking host sequences correlates with gene inactivity. Nature. 1985;315:594–597. doi: 10.1038/315594a0. [DOI] [PubMed] [Google Scholar]
- 17.Jähner D, Jaenisch R. Chromosomal position and specific demethylation in enhancer sequences of germ line-transmitted retroviral genomes during mouse development. Mol Cell Biol. 1985;5:2212–2220. doi: 10.1128/mcb.5.9.2212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jähner D, Stuhlmann H, Stewart C L, Harbers K, Lohler J, Simon I, Jaenisch R. De novo methylation and expression of retroviral genomes during mouse embryogenesis. Nature. 1982;298:623–628. doi: 10.1038/298623a0. [DOI] [PubMed] [Google Scholar]
- 19.Jang S K, Krausslich H G, Nicklin M J, Duke G M, Palmenberg A C, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jang S K, Wimmer E. Cap-independent translation of encephalomyocarditis virus RNA: structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes Dev. 1990;4:1560–1572. doi: 10.1101/gad.4.9.1560. [DOI] [PubMed] [Google Scholar]
- 21.Juttermann R, Li E, Jaenisch R. Toxicity of 5-aza-2′-deoxycytidine to mammalian cells is mediated primarily by covalent trapping of DNA methyltransferase rather than DNA demethylation. Proc Natl Acad Sci USA. 1994;91:11797–11801. doi: 10.1073/pnas.91.25.11797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keshet I, Lieman-Hurwitz J, Cedar H. DNA methylation affects the formation of active chromatin. Cell. 1986;44:535–543. doi: 10.1016/0092-8674(86)90263-1. [DOI] [PubMed] [Google Scholar]
- 23.Lamb B T, Satyamoorthy K, Li L, Solter D, Howe C C. CpG methylation of an endogenous retroviral enhancer inhibits transcription factor binding and activity. Gene Expr. 1991;1:185–196. [PMC free article] [PubMed] [Google Scholar]
- 24.Lengauer C, Kinzler K W, Vogelstein B. DNA methylation and genetic instability in colorectal cancer cells. Proc Natl Acad Sci USA. 1997;94:2545–2550. doi: 10.1073/pnas.94.6.2545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin X. Construction of new retroviral producer cells from adenoviral and retroviral vectors. Gene Ther. 1998;5:1251–1258. doi: 10.1038/sj.gt.3300720. [DOI] [PubMed] [Google Scholar]
- 26.Markowitz D, Goff S, Bank A. A safe packaging line for gene transfer: separating viral genes on two different plasmids. J Virol. 1988;62:1120–1124. doi: 10.1128/jvi.62.4.1120-1124.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mazo I A, Levy J P, Muldoon R R, Link C J, Jr, Kain S R. Retroviral expression of green fluorescent protein. Methods Enzymol. 1999;302:329–341. doi: 10.1016/s0076-6879(99)02030-3. [DOI] [PubMed] [Google Scholar]
- 28.Mikovits J A, Raziuddin, Gonda M, Ruta M, Lohrey N C, Kung H F, Ruscetti F W. Negative regulation of human immune deficiency virus replication in monocytes. Distinctions between restricted and latent expression in THP-1 cells. J Exp Med. 1990;171:1705–1720. doi: 10.1084/jem.171.5.1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mikovits J A, Young H A, Vertino P, Issa J P, Pitha P M, Turcoski-Corrales S, Taub D D, Petrow C L, Baylin S B, Ruscetti F W. Infection with human immunodeficiency virus type 1 upregulates DNA methyltransferase, resulting in de novo methylation of the gamma interferon (IFN-gamma) promoter and subsequent downregulation of IFN-gamma production. Mol Cell Biol. 1998;18:5166–5177. doi: 10.1128/mcb.18.9.5166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miller A D. Retrovirus packaging cells. Hum Gene Ther. 1990;1:5–14. doi: 10.1089/hum.1990.1.1-5. [DOI] [PubMed] [Google Scholar]
- 31.Miller A D, Buttimore C. Redesign of retrovirus packaging cell lines to avoid recombination leading to helper virus production. Mol Cell Biol. 1986;6:2895–2902. doi: 10.1128/mcb.6.8.2895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miller A D, Chen F. Retrovirus packaging cells based on 10A1 murine leukemia virus for production of vectors that use multiple receptors for cell entry. J Virol. 1996;70:5564–5571. doi: 10.1128/jvi.70.8.5564-5571.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Miller A D, Garcia J V, Von Shur N, Lynch C M, Wilson C, Eiden M V. Construction and properties of retrovirus packaging cells based on gibbon ape leukemia virus. J Virol. 1991;65:2220–2224. doi: 10.1128/jvi.65.5.2220-2224.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller A D, Trauber D R, Buttimore C. Factors involved in production of helper virus-free retrovirus vectors. Somat Cell Mol Genet. 1986;12:175–183. doi: 10.1007/BF01560664. [DOI] [PubMed] [Google Scholar]
- 35.Muenchau D D, Freeman S M, Cornetta K, Zwiebel J A, Anderson W F. Analysis of retroviral packaging lines for generation of replication-competent virus. Virology. 1990;176:262–265. doi: 10.1016/0042-6822(90)90251-l. [DOI] [PubMed] [Google Scholar]
- 36.Muiznieks I, Doerfler W. The topology of the promoter of RNA polymerase II-and III-transcribed genes is modified by the methylation of 5′-CG-3′ dinucleotides. Nucleic Acids Res. 1994;22:2568–2575. doi: 10.1093/nar/22.13.2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Muldoon R R, Levy J P, Kain S R, Kitts P A, Link C J., Jr Tracking and quantitation of retroviral-mediated transfer using a completely humanized, red-shifted green fluorescent protein gene. BioTechniques. 1997;22:162–167. doi: 10.2144/97221rr03. [DOI] [PubMed] [Google Scholar]
- 38.Nan X, Campoy F J, Bird A. MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin. Cell. 1997;88:471–481. doi: 10.1016/s0092-8674(00)81887-5. [DOI] [PubMed] [Google Scholar]
- 39.Nan X, Ng H H, Johnson C A, Laherty C D, Turner B M, Eisenman R N, Bird A. Transcriptional repression by the methyl-CpG-binding protein MeCP2 involves a histone deacetylase complex. Nature. 1998;393:386–389. doi: 10.1038/30764. [DOI] [PubMed] [Google Scholar]
- 40.Odawara T, Oshima M, Doi K, Iwamoto A, Yoshikura H. Threshold number of provirus copies required per cell for efficient virus production and interference in Moloney murine leukemia virus-infected NIH 3T3 cells. J Virol. 1998;72:5414–5424. doi: 10.1128/jvi.72.7.5414-5424.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.O'Neill R, O'Neill M, Graves J A. Undermethylation associated with retroelement activation and chromosome remodelling in an interspecific mammalian hybrid. Nature. 1998;393:68–72. doi: 10.1038/29985. [DOI] [PubMed] [Google Scholar]
- 42.Ram Z, Culver K W, Oshiro E M, Viola J J, DeVroom H L, Otto E, Long Z, Chiang Y, McGarrity G J, Muul L M, Katz D, Blaese R M, Oldfield E H. Therapy of malignant brain tumors by intratumoral implantation of retroviral vector-producing cells. Nat Med. 1997;3:1354–1361. doi: 10.1038/nm1297-1354. [DOI] [PubMed] [Google Scholar]
- 43.Rueckert R R. Picornaviridae: the viruses and their replication. In: Fields B N, et al., editors. Fields virology. New York, N.Y: Raven Press; 1996. pp. 609–654. [Google Scholar]
- 44.Saggioro D, Forino M, Chieco-Bianchi L. Transcriptional block of HTLV-I LTR by sequence-specific methylation. Virology. 1991;182:68–75. doi: 10.1016/0042-6822(91)90649-v. [DOI] [PubMed] [Google Scholar]
- 45.Saggioro D, Panozzo M, Chieco-Bianchi L. Human T-lymphotropic virus type I transcriptional regulation by methylation. Cancer Res. 1990;50:4968–4973. [PubMed] [Google Scholar]
- 46.Savard N, Cosset F L, Epstein A L. Defective herpes simplex virus type 1 vectors harboring gag, pol, and env genes can be used to rescue defective retrovirus vectors. J Virol. 1997;71:4111–4117. doi: 10.1128/jvi.71.5.4111-4117.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Somasundaran M, Robinson H L. Unexpectedly high levels of HIV-1 RNA and protein synthesis in a cytocidal infection. Science. 1988;242:1554–1557. doi: 10.1126/science.3201245. [DOI] [PubMed] [Google Scholar]
- 48.Takeuchi Y, Cosset F L, Lachmann P J, Okada H, Weiss R A, Collins M K. Type C retrovirus inactivation by human complement is determined by both the viral genome and the producer cell. J Virol. 1994;68:8001–8007. doi: 10.1128/jvi.68.12.8001-8007.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Takeuchi Y, Porter C D, Strahan K M, Preece A F, Gustafsson K, Cosset F L, Weiss R A, Collins M K. Sensitization of cells and retroviruses to human serum by (alpha 1-3) galactosyltransferase. Nature. 1996;379:85–88. doi: 10.1038/379085a0. [DOI] [PubMed] [Google Scholar]
- 50.Teyssier J R, Benard J, Ferre D, Da Silva J, Renaud L. Drug-related chromosomal changes in chemoresistant human ovarian carcinoma cells. Cancer Genet Cytogenet. 1989;39:35–43. doi: 10.1016/0165-4608(89)90227-6. [DOI] [PubMed] [Google Scholar]
- 51.Yoder J A, Bestor T H. Genetic analysis of genomic methylation patterns in plants and mammals. Biol Chem. 1996;377:605–610. [PubMed] [Google Scholar]
- 52.Yoder J A, Walsh C P, Bestor T H. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]
- 53.Young W-B, Beecham E J, Lindberg G L, Link C J., Jr Restriction mapping of retroviral vector episomal DNA. BioTechniques. 2000;28:562–565. doi: 10.2144/00283cr01. [DOI] [PubMed] [Google Scholar]
- 54.Young W-B, Lindberg G L, Link C J., Jr DNA methylation of helper virus increases genetic instability of retroviral vector producer cells. J Virol. 2000;74:3177–3187. doi: 10.1128/jvi.74.7.3177-3187.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]