Abstract
The cytosolic enzymes N-Acetyl Transferases 1 and 2 (NATs) transfer an acetyl group from acetyl-CoA to a xenobiotic substrate. NATs are regulated at the genetic and epigenetic levels by deacetylase enzymes such as sirtuins. The enzymatic expression of NAT1, NAT2, and SIRT1 was evaluated by flow cytometry, as well as the enzymatic activity of NATs by cell culture and HPLC analysis. Six SNPs were determined through genotyping. T2D patients (n = 29) and healthy subjects (n = 25) with a median age of 57 and 50, respectively, were recruited. An increased enzyme expression and a diminished NAT2 enzymatic activity were found in cells of T2D patients compared to the control group, while NAT1 was negatively correlated with body fat percentage and BMI. In contrast, Sirtuin inhibition increased NAT2 activity, while Sirtuin agonism decreased its activity in both groups. The analysis of NAT2 SNPs showed a higher frequency of rapid acetylation haplotypes in T2D patients compared to the control group, possibly associated as a risk factor for diabetes. The enzymatic expression of CD3+NAT2+ cells was higher in the rapid acetylators group compared to the slow acetylators group. The levels and activity of NAT1 were associated with total cholesterol and triglycerides. Meanwhile, CD3+NAT2+ cells and NAT2 activity levels were associated with HbA1c and glucose levels. The results indicate that NAT2 could be involved in metabolic processes related to the development of T2D, due to its association with glucose levels, HbA1c, and the altered SIRT-NAT axis. NAT1 may be involved with dyslipidaemias in people who are overweight or obese.
Keywords: N-acetyltransferases, Diabetes, Obesity, Sirtuins, Metabolism
Highlights
-
•
The expression and regulation of NAT1 and NAT2 enzymes in T lymphocytes indicate a role in the immune system of Type 2 Diabetes and obese patients.
-
•
T lymphocytes from patients with type 2 diabetes have high levels of NAT2 protein but decreased enzymatic activity.
-
•
NAT1 protein negatively correlates with fat percentage, BMI, and serum lipid levels in T lymphocytes of obese patients.
-
•
Sirtuin 1 (SIRT1) regulates the enzymatic activity of NAT2 in Type 2 diabetes.
Abbreviations
- NAT
arylamine N-acetyltranferase
- SIRT1
sirtuin 1
- T2D
Type 2 Diabetes
- BMI
Body Mass Index
- SNP
Single Nucleotide Polymorphism
- PDH
pyruvate dehydrogenase
- Ac-INH
acetyl-Isoniazid
- Ac-PABA
acetyl-p-aminobenzoic acid
- APC
allophycocyanin
- FITC
fluorescein IsoTioCyanate
- HPLC
high performance liquid chromatography
- INH
isoniazid
- NAM
nicotinamide
- PABA
p-aminobenzoic acid
- PBMC
peripheral blood mononuclear cells
- PBS
phosphate-buffered saline
- MFI
Mean Fluorescence Intensity
- RPMI
Roswell Park Memorial Institute medium
- ACN
acetonitrile
- NAM
Nicotinamide
- SIRT1720
SIRT agonist
- HbA1c
glycated haemoglobin
- HDL
high-density lipoprotein
- LDL
low-density lipoprotein
1. Introduction
Diabetes Mellitus (DM) is a group of metabolic disorders characterised by chronic hyperglycaemia and is due to defects in the secretion or action of insulin, or both. Type 2 diabetes (T2D), the most common form of DM, is a combination of an alteration of the beta cells of the pancreas and insulin resistance [1]. T2D affects around 463 million adults between 20 and 79 years of age (9.3 % of the world population in this age group) and has been associated with obesity [2].
Chronic low-grade inflammation due to lymphocyte infiltration, cytokine synthesis, and decreased regulatory T cells (Treg) is characteristic of metabolic diseases such as T2D and obesity. Insulin resistance is a condition of the metabolic syndrome present in these conditions, generated by different factors, whether genetic, epigenetic, or cellular. Furthermore, there is an increase in proinflammatory subpopulations such as Th1, Th17, and Th2 in adipose tissue and the blood of patients with T2D [3]. The differentiation of T cells depends on different metabolic pathways: for the generation of effector T cells, pyruvate generated from glycolysis is required to be converted into lactate by the enzyme lactate dehydrogenase, while for Treg cells, pyruvate is required to enter the Krebs cycle in the mitochondria to generate Acetyl CoA by the enzyme pyruvate dehydrogenase (PDH). Enzymes that participate in the regulatory pathways of PDH are the Arylamine N-acetyltransferase 1 and 2 (NAT) enzymes [4].
The cytosolic phase II enzymes, NAT1 and NAT2, transfer an acetyl group from Acetyl-CoA to a xenobiotic substrate. These enzymes share three domains: the domains I and II are more conserved between NAT enzymes than domain III and have a conserved catalytic triad composed of three residues Cys68-His107-Asp122 in their functional structure, which forms part of the active site. NAT1 show wide tissue distribution and has an affinity for p-aminobenzoic acid (PABA), p-aminosalicylic acid, and p-aminobenzoyl glutamic acid. NAT2 presents a more restricted distribution in tissues and has an affinity for sulfamethazine (SMZ), isoniazid (INH), procainamide, and dapsone. This difference in substrate recognition between the two enzymes is a consequence of a smaller substrate binding pocket in NAT1 (162 Å3) than in NAT2 (257 Å3) as a consequence of two key residue substitutions, R127 and Y129 in NAT1. In NAT2, serine residues occupy these positions [5,6]. Studies in NAT1-knockout cell lines (NAT2 in humans) show low levels of insulin-mediated glucose uptake, suggesting that NAT2 in humans could be involved in insulin sensitivity [7]. Furthermore, NATs are expressed in immune system cells, with a high expression of NAT2 having been reported in lymphocytes from patients with tuberculosis compared to healthy subjects [8]. On the other hand, the enzymatic activity of NATs can be regulated at the genetic and epigenetic levels by deacetylase enzymes such as sirtuins that can promote their degradation.
The sirtuin family includes seven protein deacetylases (SIRT1–SIRT7) in the nucleus, cytoplasm, and mitochondria. SIRT3, SIRT4, and SIRT5 are mitochondrial proteins, while SIRT1, SIRT6, and SIRT7 are nuclear enzymes that may take part in the epigenetic regulation of the cellular phenotype [9]. SIRT1 has a crucial role in the immune and metabolic response by regulating the deacetylation of transcription factors relevant to inflammatory processes, carbohydrate and lipid metabolism, apoptosis, cellular senescence, and oxidative stress [10].
The presence of single nucleotide polymorphisms (SNPs) can also regulate NATs: NAT1 and NAT2 are highly polymorphic, especially NAT2. These polymorphisms modify enzyme function and expression, giving rise to rapid, intermediate, and slow acetylation phenotypes. The slow acetylation phenotype is the most common in the European population and is highly heterogeneous in Africa and America [11]. In the Mexican population, the intermediate acetylator phenotype is the most common [8].
Our group reported higher levels of SIRT1 compared to SIRT6 in lymphocytes from healthy subjects, and in addition, an increase in the enzymatic activity of NAT2 was detected when sirtuins were inhibited in mononuclear cell cultures [12]. SIRT1 may regulate energy metabolism and insulin sensitivity through its ability to activate peroxisome proliferator-activated receptor alpha (PPAR-α) by deacetylation of peroxisome proliferator-activated receptor-gamma coactivator (PGC-1α), leading to an increased expression of transcription factors, increased fatty acid oxidation, and protection against metabolic damage induced by T2D and a high-fat diet [13]. Together, these data indicate that sirtuins regulate activity of NATs to induce their degradation in cells from patients with T2D; however, it is unknown whether this mechanism of modulation of NATs by sirtuins or by the presence of SNPs is altered in patients with T2D, and in those who are overweight or obese. This study therefore aimed to analyse the enzymatic activity, expression, and modulation of NAT1 and NAT2 in T lymphocytes from patients with T2D and obesity.
2. Materials and methods
2.1. Population
In the present study, 29 patients diagnosed with T2D and 25 healthy volunteers were recruited from the hospital "Dr. Ignacio Morones Prieto" in San Luis Potosí, Mexico. Blood samples were collected in 8 mL EDTA vacutainer tubes (BD VACUTAINER®) for enzyme expression and activity analyses. The laboratory results and clinical history of the patients were recorded. For the healthy group, we included subjects with normal biochemical parameters. Subjects with infectious diseases, autoimmune diseases, or other disorders such as high blood pressure, cancer, kidney failure, or any pharmacological treatments, alcohol consumption, or use of tobacco or illicit drugs were excluded. The study adhered to the Declaration of Helsinki and was approved by the Ethics and Research Committee of the hospital "Dr. Ignacio Morones Prieto” with registration number 63-19. A written informed consent was obtained from each participant.
2.2. Isolation of peripheral blood mononuclear cells
Peripheral blood mononuclear cells (PBMC) were isolated by density gradient centrifugation as described below: blood was diluted 1:1 with phosphate-buffered saline (PBS) at a pH of 7.4, was slowly transferred to a 15 mL conical tube with Ficoll-Hypaque, and centrifuged at 2,500 rpm for 20 min at 25 °C. The PBMC was washed twice with PBS at room temperature, then resuspended in RPMI medium supplemented with 50 U/mL penicillin and 50 μg/mL streptomycin. The trypan blue exclusion assay was used to assess cell viability.
2.3. Expression of intracellular proteins by flow cytometry
The percentage of cells expressing the intracellular proteins of interest was evaluated using different monoclonal antibodies. First, the cells were incubated with 10 μL of 1:10 autologous serum for 15 min at 4 °C to block the Fc receptors. Subsequently, the cells were incubated with an anti-CD3-PE antibody (eBioscience catalog #12-0037-42), then fixed and permeabilised with the commercial Buffer Fix/Perm Kit (eBioscience catalog #00-5523-00). The cells were then incubated with rabbit anti-NAT1 (abcam catalog #ab109114) or rabbit anti-NAT2 (abcam catalog #ab194114) primary antibody in separate tubes for 1.5 h at 4 °C in the dark. Next, the cells were incubated with an anti-rabbit APC secondary antibody (eBioscience catalog #31984) in the dark for 30 min at 4 °C. To measure SIRT1 levels in lymphocytes, a rabbit anti-SIRT1-Alexa Fluor 488 antibody (abcam catalog #ab196368) was added to the cells for 30 min at 4 °C in the dark. Finally, cells were treated with 1 % paraformaldehyde for analysis on the FACSCanto II cytometer using the FlowJo V10.8.1 software (LLC, BD®). The results included the percentage of cells positive for NAT1, NAT2, or SIRT1. In addition, the Mean Fluorescence Intensity (MFI) was analysed.
2.4. In situ arylamine N-acetyltransferase assay
PBMC (2x105 cells) were cultured in RPMI medium supplemented with 50 U/mL penicillin and 50 μg/mL streptomycin at 37 °C in a humid atmosphere with 5 % CO2. A SIRT antagonist, Nicotinamide (NAM) at 0 and 30 μmol/L, or the SIRT agonist, SIRT1720, at 0, 0.5, and 1 μmol/L, was added to the cells and incubated for 3 h at 37 °C. After this time, the medium was replaced with fresh medium with the specific substrate for each enzyme: 100 μM PABA or INH. The cells were then incubated for 24 h. The supernatant was isolated and stored at −80 °C until high-performance liquid chromatography (HPLC) analysis in the Biopharmacy Laboratory of the Faculty of Chemical Sciences of the Autonomous University of San Luis Potosi (UASLP).
2.5. In situ determination of the enzymatic activity of arylamine N-acetyltransferases
The enzymatic activity of NAT1 and NAT2 was determined by HPLC analysis of PBMC culture supernatants, quantifying the metabolites of each enzyme: p-acetyl aminobenzoic acid (Ac-PABA) for NAT1 and acetyl-isoniazid (Ac-INH) for NAT2. To extract the metabolites, we evaporated 100 μL of supernatant in 1.5 mL Eppendorf tubes, and the pellet was subsequently resuspended in 80 % 50 mM acetic acid and 20 % acetonitrile (ACN) for NAT1; and 80 % 20 mM 1-heptanesulfonic acid, 2.5 mM phosphate buffer (pH = 2.8), 10 % ACN, and 10 % trichloroacetic acid w/v for NAT2. Once resuspended, 20 μL was injected into the HPLC equipment, which was composed of a Waters model 1525 continuous flow binary pump, Autosampler 717 Plus water loop injection from 10 to 100 μL, Waters model 2487 UV–Vis detector, and Breeze software version 3.20. The PABA/Ac-PABA and INH/Ac-INH standards were USP grade, and the solvents were HPLC grade. A Waters X-terra RP18 125 A, 5 μm, 3 × 20 mm column was used to separate the analytes. The mobile phase for NAT1 consisted of 80 % 50 mM acetic acid and 20 % ACN, with a wavelength of 270 nm. In the case of NAT2, the mobile phase was 97 % 20 mM 1-heptanesulfonic acid and phosphate buffer (pH = 2.8). In both cases, 2.5 mM and 3 % ACN with a wavelength of 266 nm at a constant flow of 0.4 mL/min was used. NOM-177-SSA1-2013 validated the described method. The concentration of each analyte in the sample was calculated using a calibration curve in a concentration range of 0.57 μg/mL to 18 μg/mL. All conditions were performed in duplicate. The enzymatic activity of NATs is expressed as nmol of acetylated substrate per million cells over 24 h incubation.
2.6. DNA extraction
Genomic DNA was extracted using the commercial Wizard® Genomic DNA Purification Kit (Promega Corporation, WI, USA). gDNA was quantified spectrophotometrically with a SynergyTM HT Multi-Mode Microplate Reader (BioTek Instruments, VT, USA) and stored at −20 °C.
2.7. NAT2 genotyping
The SNPs in the coding region of NAT2, as well as their alleles and haplotypes, were determined by analysing a panel of six SNPs: rs1041983 (282C > T), rs1801280 (341T > C), rs1799929 (481C > T), rs1799930 (590G > A), rs1208 (803A > G), and rs1799931 (857G > A). The SNPs were evaluated by allelic discrimination real-time polymerase chain reaction PCR using pre-designed TaqMan probes from Applied Bio-Systems/Thermofisher Scientific. TaqMan® probes labelled with a reporter fluorochrome (VIC or FAM) are specific for one of the two possible bases reported for the SNPs analysed. A 2x TaqMan® Genotyping Master Mix (Applied Biosystem™), a 20x probe, and a primer mix were used for each assay. The gDNA was adjusted to a concentration of 10 ng/μL. The thermocycler conditions for the experiment were 95 °C for 10 min, followed by 50 cycles at 92 °C for 15 s, and 60 °C for 90 s. PCR amplification and real-time reading were performed on a CFX96 Touch™ real-time PCR detection system (Bio-Rad© Laboratories, Inc.). Control samples were run without DNA in each experiment to ensure that there was no amplification by contaminating DNA.
2.8. NAT2 phenotype
Subjects with two of the alleles associated with rapid acetylation activity (NAT2*4, NAT2*11, NAT2*12, and NAT2*13 families) were classified as rapid acetylators; subjects with two alleles associated with slow acetylation activity (NAT2*5, NAT2*6, NAT2*7, NAT2*10, and NAT2*14 families) were classified as slow acetylators; and subjects with one rapid allele and one slow allele were classified as intermediate acetylators, as described in the consensus nomenclature (http://nat.mbg.duth.gr/).
2.9. Statistical analysis
A univariate analysis was conducted for each variable to determine its distribution using the normality and log normality tests (Anderson-Darling, D'Agostino & Pearson; Shapiro-Wilk, Kolmogorov-Smirnov test). Continuous variables were described using the mean and standard deviation or median and interquartile ranges according to the distribution of the data. Categorical variables were described by frequency and percentage, and categorical and dichotomous variables were analysed in contingency tables using Fisher's exact test. Differences between groups were estimated with a Student's T test or an ANOVA in parametric data, and a Mann-Whitney U or Kruskal-Wallis test in cases of non-parametric data. A 95 % confidence interval and a p-value <0.05 were considered to represent statistical significance. All statistical analyses were performed in R version 4.3.2.
3. Results
3.1. Clinical and anthropometric characteristics of the study subjects
A total of 29 T2D patients and 25 healthy subjects were recruited. The median age of the subjects was 57 years (±12.3) and 50 years (±9.0), respectively. The population was classified according to body mass index (BMI) by the following criteria: normal weight (NW) = 18.5–24.9 kg/m2, overweight (OW) = 25–29.9 kg/m2, and obese (OB) = ≥ 30 kg/m2. Additionally, the subjects were classified according to their percentage of body fat, classifying obesity as men with a body fat percentage greater than or equal to 25 % and women with a body fat percentage greater than or equal to 35 %, according to a body adiposity estimator (BAE) [14,15]. The anthropometric, clinical, and biochemical characteristics of the study population are described in Table 1. As expected, subjects with T2D had a significantly higher BMI, glucose levels, and glycosylated haemoglobin. Regarding the biochemical variables, a decrease in high-density lipoprotein (HDL), low-density lipoprotein (LDL), and total cholesterol levels was observed in T2D patients compared to the control group.
Table 1.
Anthropometric and clinical characteristics.
| T2D patients | Healthy group | p | |
|---|---|---|---|
| n | 29 | 25 | |
| Female, n (%) | 18 (42) | 16 (44) | >0.9999χ |
| Male, n (%) | 11 (38) | 9 (36) | |
| Age, n | 57 (±12.3) | 50 (±9.0) | 0.0299‡ |
| Weight, kg | 80.0 (±15.2) | 70.3 (±14.3) | 0.0274‡ |
| BMI, kg/m2 | 30.1 (±5.4) | 27.1 (±4.7) | 0.0057‡ |
| Glucose, mg/dL | 161.0 (±60.01) | 86.86 (±8.71) | <0.0001‡ |
| HbA1c, % | 8.68 (±1.77) | 5.75 (±0.31) | <0.0001‡ |
| Controlled HbA1c < 7, n (%) | 6 (17.6) | – | - |
| Non-controlled HbA1c > 7, n (%) | 23 (82.3) | – | - |
| Normoweight, n (%) | 3 (10) | 11 (44) | 0.0182χ |
| Overweight, n (%) | 8 (28) | 5 (20) | |
| Obesity, n (%) | 18 (62) | 9 (36) | |
| Non-obese (% body fat), n (%) | 8 (28) | 7 (28) | 0.4501χ |
| Obese (% body fat), n (%) | 21 (72) | 18 (72) | |
| Metabolic syndrome, n (%) | 24 (83) | 0 (0) | - |
| Total Cholesterol, mg/dL | 166.2 (±44.33) | 190.0 (±36.05) | 0.0168† |
| HDL Cholesterol, mg/dL | 39.20 (±13.88) | 49.26 (±17.27) | 0.0111† |
| LDL Cholesterol, mg/dL | 105.0 (±96.19) | 112.4 (±26.93) | 0.0077† |
| Triglycerides, mg/dL | 247.3 (±135.5) | 155.9 (±83.85) | 0.0137‡ |
| Atherogenic index, Total-Chol/HDL-Chol | 4.44 (±1.19) | 4.29 (±1.24) | 0.7147‡ |
| Serum urea, mg/dL | 38.93 (±23.89) | 55.05 (±48.65) | 0.5237† |
| Serum creatinine, mg/dL | 0.975 (±0.85) | 0.829 (±0.20) | 0.5654‡ |
Values are represented as mean (standard deviation). T2D, Type 2 Diabetes; BMI, Body Mass Index; HbA1c, Glycosylated hemoglobin. †U Mann-Whitney, ‡ Unpaired t-test. Χ Chi-Square (Fisher's exact) test.
3.2. Intracellular expression of NAT1, NAT2, and SIRT1 in T2D patients
The detection strategy for NAT enzymes by flow cytometry is described in Fig. 1A. The results showed high levels of NAT2 in total lymphocytes in T2D patients compared to control subjects (p = 0.0088). Protein expression in T lymphocytes (CD3+) was evaluated, and T2D patients had higher levels of double positive cells for CD3+NAT2+ compared to the control group (p = 0.0086) (Fig. 1B). The analysis of median fluorescence intensity (MFI) in T lymphocytes showed higher levels of NAT2 in T2D patients compared to the control group (p = 0.0110) (Fig. 1C). NAT1 tended to increase in T2D patients, while SIRT1 showed similar levels in both groups. Therefore, no significant differences were found in the expression of NAT1 and SIRT1 between T2D patients and the control group. Furthermore, it was observed that patients who had higher levels of CD3+NAT2+ cells also had the highest levels of CD3+SIRT1+ cells (r = 0.6314, p = 0.0066, data not shown).
Fig. 1.
Expression of NAT1 or NAT2 in CD3+ T lymphocytes in the control group and T2D patients by flow cytometry. (A) Experimental strategy used to determine the expression of double positive cells. (B) NAT1, NAT2, and SIRT1 expression in CD3+ T lymphocytes (p = 0.0086 by the Wilcoxon Test). (C) Mean Fluorescence Intensity of NAT1, NAT2, and SIRT1 in CD3+ T lymphocytes (p = 0.0110 by the Wilcoxon Test). *p < 0.05, **p < 0.01. Boxes show interquartile ranges, and bars represent the highest and lowest values.
3.3. In situ enzymatic activity of NAT1 and NAT2 in mononuclear cell cultures
A substrate-specific metabolic activity assay was performed by HPLC analysis to determine the enzymatic activity of NAT1 and NAT2 (Supplementary Fig. 1). The NAT1 enzymatic activity in PBMC showed a wide variability in both groups, which did not allow for finding significant differences. On the other hand, the enzymatic activity of NAT2 was lower in T2D patients compared to the control group (p = 0.0054) (Fig. 2A).
Fig. 2.
Enzymatic activity of NAT1 and NAT2. (A) NAT1 and NAT2 (p = 0.0054) (Wilcoxon test). (B) Effect of SIRT1 on NAT1 enzymatic activity. (C) Effect of SIRT1 on NAT2 enzymatic activity (Kruskal-Wallis test). NAT enzymatic activity is reported as nmol Ac PABA/24h/million cells (NAT1) and nmol Ac INH/24h/million cells (NAT2). The enzymatic activity was normalised by nanomoles of acetylated substrate per million cells over 24 h incubation. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Boxes show interquartile ranges, and bars represent the highest and lowest values.
3.4. SIRT1 and its possible modulation of arylamine N-acetyltransferase 2 activity
The behaviour of the NAT1 enzymatic activity was not affected in T2D patients or the control group (Fig. 2B), while NAT2 showed a decrease in enzymatic activity in the presence of SRT1720 at 1 μM (p = 0.0052) and an increase of enzymatic activity in the presence of NAM at 30 μM (p < 0.0001) in the control group. A similar behaviour was observed in T2D patients (Fig. 2C), which was significant under NAM at 30 μM (p = 0.0003); however, NAM's effect on NAT2 activity was smaller in T2D patients than in control subjects (p = 0.0004).
3.5. Association of NAT1+ and NAT2+ cell levels with overweight and obesity
Overweight and obesity are metabolic conditions associated with T2D. When classifying the study population into subgroups according to BMI (normal weight, overweight, and obesity) no differences were identified in the expression levels of NATs (data not shown). In contrast, the correlation analysis showed a negative correlation between NAT1 expression and the percentage of body fat (r = −0.4128, p = 0.0361) and BMI (r = −0.5389, p = 0.0045) in T2D patients (Table 2).
Table 2.
Association of NAT1+ and NAT2+ cell levels with BMI and Body fat percentage.
|
Lymphocytes CD3+NAT1+ |
Lymphocytes CD3+NAT2+ |
|||
|---|---|---|---|---|
| r Spearman | P value | r Spearman | P value | |
| T2D | ||||
| BMI (kg/m2) | −0.5389 | 0.0045** | 0.03997 | 0.8400 |
| Body fat (%) | −0.4128 | 0.0361* | 0.1289 | 0.5050 |
| Healthy group | ||||
| BMI (kg/m2) | 0.2105 | 0.3598 | 0.1417 | 0.5401 |
| Body fat (%) | 0.5747 | 0.0126* | −0.2430 | 0.9237 |
T2D, Type 2 Diabetes; BMI, Body Mass Index. P value < 0.05 was taken as significant.
3.6. Correlation analysis of NAT1, NAT2, and SIRT1 with biochemical and anthropometric variables
The association of NATs and SIRT1 with the clinical and anthropometric characteristics of the subjects' multiparametric analyses was performed. NAT1 showed a negative correlation between CD3+NAT1+ T lymphocytes and the total cholesterol levels (r = −0.5399, p = 0.0140) and triglyceride levels (r = multiparametric analyses 0.5049, p = 0.0326) of control subjects. There was a positive association of CD3+NAT1+ T lymphocytes with the atherogenic index (r = 0.5232, p = 0.0259) in T2D patients, while the enzymatic activity of NAT1 showed a negative relationship with triglyceride levels in both study groups. The statistics are shown in Fig. 3. On the other hand, NAT2 showed a positive association between CD3+NAT2+ T cells and the percentage of muscle mass of control subjects (r = 0.6311, p = 0.0207) (Supplementary Fig. 2A). Interestingly, in T2D patients, this association was inversely proportional (r = −0.5729, p = 0.0204).
Fig. 3.
Association of NAT1 with serum cholesterol and triglyceride levels. (A) Association of NAT1+ T lymphocytes with the atherogenic index in healthy subjects and T2D patients. (B) Association of NAT1+ T lymphocytes with total cholesterol levels in healthy subjects and T2D patients. (C) Association of NAT1+ T lymphocytes with triglyceride levels in healthy subjects and T2D patients. (D) Association of NAT1 enzymatic activity with triglyceride levels in healthy subjects and T2D patients.
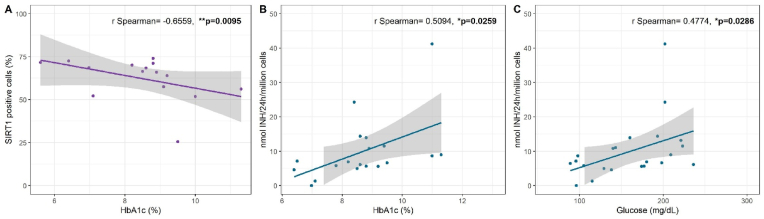
Another clinical parameter that determines glycaemic control and supports defining treatment is the glycated haemoglobin (HbA1c) concentration. The study variables of NATs and SIRT1 and glycaemic control were therefore analysed in T2D patients. The expression of CD3+SIRT1+ T cells showed a negative correlation with HbA1c levels (r = −0.6559, p = 0.0095) (Fig. 4A). On the other hand, the NAT2 enzymatic activity of T2D patients showed a positive correlation with HbA1c (r = 0.5020, p = 0.0259) (Fig. 4B), as well as with glucose levels (r = 0.4774, p = 0.0286) (Fig. 4C). No association of NAT1 activity and enzymatic expression with HbA1c was found (data not shown).
Fig. 4.
Correlation of NAT2 and SIRT1 with the average glucose level in T2D patients. (A) SIRT1+ T cell expression. (B) Association of NAT2 enzymatic activity with HbA1c. (C) Association of NAT2 enzymatic activity with glucose levels.
Regarding anthropometric characteristics, the age and sex of the subjects imply different inflammatory and hormonal states that can lead to alterations in the expression of NATs. In healthy subjects, CD3+NAT2+ T cells showed a negative correlation with the age of the subjects (r = −0.4106, p = 0.0463) (Supplementary Fig. 2B). Furthermore, lower expression levels were found in females compared to males for both CD3+NAT1+ T lymphocytes (p = 0.0153) and CD3+NAT2+ T lymphocytes (p = 0.0014) (Supplementary Fig. 3). T2D patients did not demonstrate any associations.
3.7. NAT2 genotyping
A panel of six SNPs were evaluated by allelic discrimination real-time polymerase chain reaction PCR analysis of 32 samples (15 healthy subjects and 17 patients with T2D). The evaluation of the panel of six SNPs allowed haplotypes to be assigned according to the nomenclature consensus (http://nat.mbg.duth.gr). The haplotypes that presented fast acetylating capacity were NAT2*4, NAT2*12, and NAT2*13, while those that presented slow acetylating capacity were NAT2*5, NAT2*6, and NAT2*7. The distribution of allele frequencies is represented in Table 3. Haplotypes with rapid acetylation were more frequent in T2D patients (p = 0.0463; OR 2.956, 95 % CI 1.03–8.56) compared to the control group. In the control group, the rapid acetylation phenotypes were presented in 13.3 % of the population made up of the NAT2*4/*4 diplotype. The intermediate acetylation phenotype was present in 33.3 % of the population as the NAT2*5A/*12A, NAT2*6A/*13A, NAT2*4/*7A, and NAT2*4/*6A diplotypes (Supplementary Fig. 4A). The slow acetylation phenotype was presented in 53.3 % of the population made up of the diplotypes NAT2*5B/*5B, NAT2*5A/*7B, NAT2*7C/*6B, NAT2*5B/*6A, and NAT2*5B /*7A. In the group of T2D patients, rapid acetylation phenotypes were present in 35.2 % of the population made up of the NAT2*4/*4 and NAT2*4/*12A diplotypes (Supplementary Fig. 4B). The intermediate acetylation phenotype occurred in 41.1 % of the population due to the presence of the diplotypes NAT2*5A/*12A, NAT2*7A/*13A, NAT2*4/*6B, NAT2*4/*7A, and NAT2*6A/*13A. The slow acetylation phenotype occurred in 23.5 % of the population made up of the NAT2*5B/*7B, NAT2*5KA/*7B, NAT2*7B/*7B, and NAT2*7B/*6A diplotypes. Hardy-Weinberg equilibrium was present in both groups (Supplementary Table 1).
Table 3.
NAT2 haplotype distribution.
| Haplotype | Phenotype |
Allele frequency |
P value | |
|---|---|---|---|---|
|
T2D patients n (%) |
Healthy group n (%) |
|||
| 34 | 30 | |||
| NAT2*4 | Rapid | 11 (32.3) | 6 (20) | 0.8995 |
| NAT2*12 | 5 (14.7) | 2 (6.7) | ||
| NAT2*13 | 3 (8.8) | 1 (3.3) | ||
| NAT2*5 | Slow | 4 (11.8) | 10 (33.3) | 0.2975 |
| NAT2*6 | 3 (8.8) | 5 (16.7) | ||
| NAT2*7 | 8 (23.6) | 6 (20) | ||
| Haplotype | ||||
| Rapid | 19 (55.9) | 9 (30) | 0.0463* | |
| Slow | 15 (44.1) | 21 (70) | ||
| Phenotype | Phenotype frequency | |||
| 17 | 15 | |||
| Rapid | 6 (35.3) | 2 (13.3) | 0.1690 | |
| Intermediate | 7 (41.2) | 5 (33.3) | ||
| Slow | 4 (23.5) | 8 (53.3) | ||
All the values are represented as n (%); d.f. = 1 for all tests. P value < 0.05 was taken as significant.
3.8. Association of the NAT2 acetylator phenotype with enzyme activity and expression
An association analysis was performed considering the expression levels and NAT2 enzymatic activity with the acetylator phenotype. A positive association of enzyme expression with the acetylating capacity of NAT2 was observed (Fig. 5A). The enzymatic expression of CD3+NAT2+ cells was higher in the fast acetylator group compared to the slow acetylator group (p = 0.0358). The enzymatic activity tended to be higher in subjects with a rapid acetylation phenotype (Fig. 5B). In the control group, a non-significant decrease in the enzymatic activity of NAT2 was observed in individuals who had one or two mutated alleles of the SNPs 282C > T, 341T > C, 481C > T, 590G > A, and 803>G (Fig. 5C). In contrast, in T2D patients, the presence of a mutated allele increased the enzymatic activity in individuals who had the 341T > C (p = 0.0407) and 481C > T (p = 0.0098) polymorphisms.
Fig. 5.
(A) Association of the NAT2 acetylator phenotype with enzymatic expression in CD3+NAT2+ T lymphocytes (p = 0.0358 by the ANOVA test). (B) Association of the NAT2 acetylator phenotype with enzymatic activity. (C) Association of the genotype with NAT2 enzymatic activity. *p < 0.05, **p < 0.01.
4. Discussion
The aetiology of T2D is multifactorial, with the primary factor being insulin resistance. Furthermore, the insulin receptor activation associated with increased levels of tumour necrosis factor (TNF-α) participates in the development of the inflammatory process with obesity [16,17]. The enzymes NAT1 and NAT2 transform xenobiotics and, recently, cellular function and homeostasis. In order to determine the levels of NAT1 and NAT2 in cells of the immune system of T2D patients, we evaluated their expression at the protein level. We observed an increase in the expression of NAT2 in patients with T2D, with percentages similar to those reported in healthy subjects [12].
The alteration in the expression of these enzymes has been reported in different pathologies, such as infectious diseases, i.e., tuberculosis, and in some types of neoplasms, such as Acute Lymphoblastic Leukaemia, Chronic Myeloid Leukaemia, and breast cancer [8,18,19]. However, in patients with non-communicable diseases such as diabetes and obesity, it is not yet known. In NAT2-knockout animal models and cell lines, an increase in insulin resistance, glucose, lipid levels, and mitochondrial dysfunction has been reported [7,20]. This study found an increase in NAT2 levels in T2D patients at the protein level. Similar changes in expression have been described at the protein level in patients with tuberculosis or in primary breast tumour tissue [8,21]. Immune alterations in T2D reflect the overproduction of TNF-α and adipokine leptin, which increases the regulation of the Th17 transcription factor RORyt and the cytokine IL-17 [16]. NATs could have a relevant role in the phenotype of CD4+ T cells in inflammatory microenvironments, such as those in adipose tissue.
It has been reported that the differentiation of Treg cells requires an increase in acetyl-CoA concentrations through the enzyme pyruvate dehydrogenase (PDH), which is regulated by, among other enzymes, NATs [4,22]. Therefore, the increase in CD3+NAT2+ T cells in T2D patients may indicate a possible modulatory mechanism of NAT2 to favour the differentiation of the anti-inflammatory phenotype. Additionally, the lower expression of both NATs in healthy women compared to men in the same group can be attributed to the protective effects of oestrogens, which have been shown to decrease the proportion of Th17 cells and increase the proportion of Treg cells, improving insulin sensitivity in women with T2D and obesity [23,24]. To our knowledge, this is the first study to show an association between NATs and the sex of subjects; however, more studies are necessary in subpopulations of T lymphocytes to determine the modulatory function of NATs. Thus, it would be essential to evaluate the expression of the NAT1 and NAT2 proteins in the Th1 and Th17 subpopulations that directly participate in the inflammatory process and that are involved in the pathogenesis of T2D.
While NAT2 is associated with T2D, NAT1 was shown to be associated with comorbidities such as obesity. Similar levels of NAT1+ cells between T2D patients and controls showed a negative correlation with BMI, percentage of body fat, total cholesterol, and triglycerides in healthy subjects, as well as a negative association of NAT1 enzymatic activity with levels of triglycerides in both groups. Previous studies have shown an association between total cholesterol, HDL, LDL, and triglyceride levels with the NAT2 acetylator phenotype in rats (NAT1 in humans) [25]. A proteomic analysis in NAT1-knockout cell lines revealed that its deletion causes mitochondrial alteration through inhibition of ATP synthase and damage to the mitochondrial membrane [26]. Therefore, metabolic pathways such as fatty acid oxidation and ketogenesis are affected, which could partly explain why obese subjects had lower NAT1 levels. Our NAT1 data could point to a possible role in obesity and metabolic syndrome; however, to date, there has been a lack of studies in overweight and obese populations with the characteristics of metabolic syndrome to demonstrate the preliminary data of our study.
On the other hand, studies on mouse 3T3-L1 adipocytes have classified NAT2 as an insulin sensitivity gene [7], which could explain why T2D patients had lower enzymatic activity than healthy subjects in this study. The alteration of its expression in T cells could indicate that NAT2 is involved in some metabolic pathway in T2D. Our results with sirtuins could support the above, as NAT2 was the only enzyme that was shown to be regulated by sirtuins, which is consistent with studies previously carried out by our group in healthy subjects [12,27]. The mechanism by which sirtuins regulate NAT2 is not well described; acetylation of NATs, like many other cytosolic proteins, occurs at different amino acids throughout its structure. This prevents the protein from being degraded; however, acetyl groups must be removed to maintain balance in the cell. Sirtuins and deacetylase enzymes can participate in this regulation mechanism in the specific case of lysine residues [6,28]. In contrast, there was no change when promoting SIRT1 with SRT1720 in T2D patients; however, NAM's increase in NAT2 enzymatic activity was lower in patients with DM2 than in healthy subjects, which could indicate that the SIRT-NAT axis is affected in this metabolic condition. To our knowledge, these data could be the first study that reports the alteration of NAT2 in both expression and function in immune cells from patients with T2D.
The acetylator phenotype of NATs is mainly associated with polymorphisms in the coding region, and NAT2 is the most polymorphic of both enzymes [29]. Therefore, the principal SNPs of NAT2 were additionally analysed. The linkage disequilibrium between the polymorphisms gives rise to six main haplotypes [30,31]. Studies carried out in the Mexican population reported a predominance of the wild-type allele [8], consistent with this study's findings; however, in the Mexican population, the allelic frequency of these NAT2 polymorphisms in T2D patients is unknown. Likewise, the expression of CD3+NAT2+ T cells was shown to be higher in individuals who had a rapid acetylation phenotype. The above would indicate that the polymorphism favours NAT2 expression at the protein level, possibly preventing its degradation at the post-transcriptional level.
In contrast, when the association of each SNP with the enzymatic function of NAT2 was evaluated, the presence of one or more mutated alleles decreased the enzymatic activity in the control group, as has been observed in genotyping studies in hepatocytes from the Caucasian, African-American, and Hispanic populations [30]. However, these differences were not significant in the control group. In contrast, T2D patients who had a mutated allele for rs1801280 (341T > C) and rs1799929 (481C > T) showed an increase in the enzymatic activity of NAT2. The 341T > C polymorphism involves a change from isoleucine to threonine at position 114, changing this hydrophobic residue to a polar hydrophilic one. The threonine residue can reduce hydrophobic interactions at the interface between domain II and domain I, altering the protein folding in that region [32]. However, many subjects presented both SNP 341T > C and 481C > T, the latter being associated with a rapid acetylation phenotype, explaining the increase in enzymatic activity in these subjects. The association of NATs SNPs in diabetes have been inconsistent in different populations: some studies have found that there is an excess of genotypes encoding intermediate acetylation in T2D and that slow haplotypes such as the rs1799931(G > A) genotype was present in the control population but not in the T2D population and had a protective effect for T2D [33,34]. However, it is still necessary to increase the number of study subjects to determine the relationship of these SNPs with NAT2 and the development and progression of T2D.
In T2D and obesity with increased TNF-α, free fatty acids and reactive oxygen species (ROS) activate IKKβ and JNK1 in adipose tissue and the liver, which induces inhibition of the insulin receptor substrate (IRS- 1) and, therefore, insulin resistance [17]. As mentioned, because diabetic complications are directly related to the average blood glucose value, measured by the HbA1c concentration [35], we evaluated the association of NATs with the concentration of HbA1c and glucose and, then, the progression of T2D in this study. Interestingly, NAT2 showed higher levels of expression in patients with uncontrolled T2D. Inflammation and insulin resistance could induce null or decreased activity of NAT2. At the same time, the cell increases the expression of the NAT2 protein as a compensatory mechanism of the T cell attributable to the positive effect of NAT2 on mitochondrial function to seek to reduce ROS [20], increase the anti-inflammatory phenotype and, therefore, modulate the state of insulin resistance and inflammation that occurs; however, studies are still necessary to better describe the mechanism of these NAT2 alterations. A negative correlation of CD3+SIRT1+ T cells with HbA1c levels was found. Studies carried out on insulin-resistant adipocyte cell lines found that SIRT1 improves glucose consumption in the presence or absence of insulin through the synthesis of GLUT4 [36], which would explain why the subjects who had higher levels of CD3+SIRT1+ T cells were those who had better glycaemic control. SIRT1 could be a potential therapeutic target for insulin-resistant patients; however, this would be the first study to evaluate its relationship with NATs in T2D patients.
Of the arylamine N-acetyltransferases, NAT2 is the main one to be implicated in T2D due to its association with glucose levels and HbA1c, and as a gene involved in insulin sensitivity [7]. However, its role as a modulator in cellular metabolism has not yet been described. Studies carried out in animal models have found that its deletion induces mitochondrial dysfunction, causing an increase in ROS and a 19 % decrease in energy capacity at the physiological level [20]. In this study, the control group showed a positive correlation of NAT2 levels with the subjects' muscle mass percentage. If NAT2 is associated with mitochondrial function, an increase in mitochondria due to greater muscle mass would explain the increase in NAT2 expression.
Furthermore, healthy subjects had a decrease in the expression of NAT2 with advancing age; this could be related to the findings mentioned above, as it is known that muscle loss is related to age [37] and, therefore, the loss of NAT2. Interestingly, in T2D patients, the association of NAT2 and muscle mass was inverse, indicating an alteration in the possible modulatory mechanism that NAT2 may have on this metabolic pathway. However, more studies are required to address this type of NAT2 involvement.
5. Conclusion
In summary, the results indicate that NAT2 could be involved in metabolic processes related to the development of T2D, while NAT1 could be involved in the dyslipidaemia of people who are overweight or obese. These findings advance our understanding human N-acetyltransferases and their involvement in cellular metabolism.
Funding information
Paz-Rodríguez Víctor Alejandro received a scholarship (1145960) from CONACYT, Mexico.
CRediT authorship contribution statement
Víctor Alejandro Paz-Rodríguez: Writing – original draft, Visualization, Software, Methodology, Investigation, Formal analysis. Diana Judith Herrera-Vargas: Methodology, Investigation. Eneida Turiján-Espinoza: Validation, Supervision. Miguel Ernesto Martínez-Leija: Supervision, Methodology, Investigation, Data curation. Emmanuel Rivera-López: Supervision, Methodology, Conceptualization. Oswaldo Hernández-González: Methodology. Daniel Zavala-Reyes: Methodology. Mariana Haydee García-Hernández: Supervision, Resources, Project administration, Funding acquisition. Juan Manuel Vargas-Morales: Supervision, Methodology. Rosa del Carmen Milán-Segovia: Validation, Supervision. Diana Patricia Portales-Pérez: Writing – review & editing, Validation, Supervision, Resources, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.bbrep.2024.101716.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Data availability
Data will be made available on request.
References
- 1.Blair M. Diabetes mellitus review. Urol. Nurs. 2016;36(1):27–36. [PubMed] [Google Scholar]
- 2.Russo M.P., Grande-Ratti M.F., Burgos M.A., Molaro A.A., Mb B. Prevalence of diabetes, epidemiological characteristics and vascular complications. Arch. Cardiol. Mex. 2023;93(1):30–36. doi: 10.24875/ACM.21000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Touch S., Clement K., Andre S. T cell populations and functions are altered in human obesity and type 2 diabetes. Curr Diab Rep. 2017;17(9):81. doi: 10.1007/s11892-017-0900-5. [DOI] [PubMed] [Google Scholar]
- 4.Kurniawan H., Soriano-Baguet L., Brenner D. Regulatory T cell metabolism at the intersection between autoimmune diseases and cancer. Eur. J. Immunol. 2020;50(11):1626–1642. doi: 10.1002/eji.201948470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sim E., Walters K., Boukouvala S. Arylamine N-acetyltransferases: from structure to function. Drug Metab. Rev. 2008;40(3):479–510. doi: 10.1080/03602530802186603. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X., Ma Z., Dong D., Wu B. Arylamine N-acetyltransferases: a structural perspective. Br. J. Pharmacol. 2013;169(4):748–760. doi: 10.1111/bph.12182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Knowles J.W., Xie W., Zhang Z., Chennamsetty I., Assimes T.L., Paananen J., et al. Identification and validation of N-acetyltransferase 2 as an insulin sensitivity gene. J. Clin. Invest. 2015;125(4):1739–1751. doi: 10.1172/JCI74692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salazar-Gonzalez R., Milán-Segovia R.C., Portales-Pérez D.P. Expression of NAT2 in immune system cells and the relation of NAT2 gene polymorphisms in the anti-tuberculosis therapy in Mexican mestizo population. Mol. Biol. Rep. 2014;41(12):7833–7843. doi: 10.1007/s11033-014-3677-5. 12. [DOI] [PubMed] [Google Scholar]
- 9.Tanno M., Kuno A., Horio Y., Miura T. Emerging beneficial roles of sirtuins in heart failure. Basic Res. Cardiol. 2012;107(4):273. doi: 10.1007/s00395-012-0273-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Howitz K.T., et al. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305(5682):390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 11.Taja-Chayeb L., Agúndez J.A., Miguez-Muñoz C., Chavez-Blanco A., Dueñas-Gonzalez A. Arylamine N-acetyltransferase 2 genotypes in a Mexican population. Genet. Mol. Res. 2012;11(2):1082–1092. doi: 10.4238/2012.April.27.7. [DOI] [PubMed] [Google Scholar]
- 12.Turijan-Espinoza E., Salazar-Gonzalez R.A., Uresti-Rivera E.E., Hernandez-Hernandez G.E., Ortega-Juarez M., Milan R., et al. A pilot study of the modulation of sirtuins on arylamine N-acetyltransferase 1 and 2 enzymatic activity. Acta Pharm. Sin. B. 2018;8(2):188–199. doi: 10.1016/j.apsb.2017.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pfluger P.T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M.H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc Natl Acad Sci U S A. 2008;105(28):9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dávila-Batista V., Gómez-Ambrosi J., Fernández-Villa T., Molina A.J., Frühbeck G., Martín V. Colour scale percent body fat by CUN-BAE adiposity estimator. Atención Primaria. 2016;48(6):422–423. doi: 10.1016/j.aprim.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Castro-Porras L.V., Rojas-Russell M.E., Villanueva-Sánchez J., López-Cervantes M. An anthropometry-based equation of fat mass percentage as a valid discriminator of obesity. Publ. Health Nutr. 2019;22(7):1250–1258. doi: 10.1017/S1368980018004044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.SantaCruz-Calvo S., Bharath L., Pugh G., SantaCruz-Calvo L., Lenin R.R., Lutshumba J., et al. Adaptive immune cells shape obesity-associated type 2 diabetes mellitus and less prominent comorbidities. Nat. Rev. Endocrinol. 2022;18(1):23–42. doi: 10.1038/s41574-021-00575-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Berbudi A., Rahmadika N., Tjahjadi A.I., Ruslami R. Type 2 diabetes and its Impact on the immune system. Curr. Diabetes Rev. 2020;16(5):442–449. doi: 10.2174/1573399815666191024085838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Endo Y., Yamashita H., Takahashi S., Sato S., Yoshimoto N., Asano T., et al. Immunohistochemical determination of the miR-1290 target arylamine N-acetyltransferase 1 (NAT1) as a prognostic biomarker in breast cancer. BMC Cancer. 2014;14:990. doi: 10.1186/1471-2407-14-990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tebien E.M., Khalil H.B., Mills J., Ay E. Evaluation of genetic polymorphisms of N-acetyltransferase 2 and relation with chronic Myeloid Leukemia. Asian Pac. J. Cancer Prev. APJCP. 2020;21(12):3711–3717. doi: 10.31557/APJCP.2020.21.12.3711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chennamsetty I., Coronado M., Contrepois K., Keller M.P., Carcamo-Orive I., Sandin J., et al. Nat1 deficiency is associated with mitochondrial dysfunction and exercise intolerance in mice. Cell Rep. 2016;17(2):527–540. doi: 10.1016/j.celrep.2016.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carlisle S.M., Hein D.W. Retrospective analysis of estrogen receptor 1 and N-acetyltransferase gene expression in normal breast tissue, primary breast tumors, and established breast cancer cell lines. Int. J. Oncol. 2018;53(2):694–702. doi: 10.3892/ijo.2018.4436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wang L., Minchin R.F., Essebier P.J., Nj B. Loss of human arylamine N-acetyltransferase I regulates mitochondrial function by inhibition of the pyruvate dehydrogenase complex. Int. J. Biochem. Cell Biol. 2019;110:84–90. doi: 10.1016/j.biocel.2019.03.002. [DOI] [PubMed] [Google Scholar]
- 23.Chen R.Y., Fan Y.M., Zhang Q., Liu S., Li Q., Ke G.L., et al. Estradiol inhibits Th17 cell differentiation through inhibition of RORγT transcription by recruiting the ERα/REA complex to estrogen response elements of the RORγT promoter. J. Immunol. 2015;194(8):4019–4028. doi: 10.4049/jimmunol.1400806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bitoska I., Krstevska B., Milenkovic T., Subeska-Stratrova S., Petrovski G., Mishevska S.J., et al. Effects of hormone replacement therapy on insulin resistance in postmenopausal diabetic women. Open Access Maced J Med Sci. 2016;4(1):83–88. doi: 10.3889/oamjms.2016.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hong K.U., Doll M.A., Lykoudi A., Salazar-González R.A., Habil M.R., Walls K.M., et al. Acetylator genotype-dependent dyslipidemia in rats congenic for N-acetyltransferase 2. Toxicol Rep. 2020;7:1319–1330. doi: 10.1016/j.toxrep.2020.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong K.U., Gardner J.Q., Doll M.A., Stepp M.W., Wilkey D.W., Benz F.W., et al. Proteomic analysis of arylamine N-acetyltransferase 1 knockout breast cancer cells: implications in immune evasion and mitochondrial biogenesis. Toxicol Rep. 2022;9:1566–1573. doi: 10.1016/j.toxrep.2022.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salazar-González R.A., Turiján-Espinoza E., Hein D.W., Milán-Segovia R.C., Uresti-Rivera E.E., Portales-Pérez D.P. Expression and genotype-dependent catalytic activity of N-acetyltransferase 2 (NAT2) in human peripheral blood mononuclear cells and its modulation by Sirtuin 1. Biochem. Pharmacol. 2018;156:340–347. doi: 10.1016/j.bcp.2018.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yuan H., Rossetto D., Mellert H., Dang W., Srinivasan M., Johnson J., et al. MYST protein acetyltransferase activity requires active site lysine autoacetylation. EMBO J. 2012;31(1):58–70. doi: 10.1038/emboj.2011.382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Walker K., Ginsberg G., Hattis D., Johns D.O., Guyton K.Z., Sonawane B. Genetic polymorphism in N-Acetyltransferase (NAT): population distribution of NAT1 and NAT2 activity. J. Toxicol. Environ. Health B Crit. Rev. 2009;12(5–6):440–472. doi: 10.1080/10937400903158383. [DOI] [PubMed] [Google Scholar]
- 30.Hein D.W., Doll M.A. Accuracy of various human NAT2 SNP genotyping panels to infer rapid, intermediate and slow acetylator phenotypes. Pharmacogenomics. 2012;13(1):31–41. doi: 10.2217/pgs.11.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cascorbi I., Drakoulis N., J B., Maurer A., Sperling K., Roots I. Arylamine N-acetyltransferase (NAT2) mutations and their allelic linkage in unrelated Caucasian individuals: correlation with phenotypic activity. Am. J. Hum. Genet. 1995;57(3):581–592. [PMC free article] [PubMed] [Google Scholar]
- 32.Walraven J.M., Zang Y., Trent J.O., Hein D.W. Structure/function evaluations of single nucleotide polymorphisms in human N-acetyltransferase 2. Curr. Drug Metabol. 2008;9(6):471–486. doi: 10.2174/138920008784892065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Irshaid Y.M., Abujbara M.A., Ajlouni K.M., El-Khateeb M., Yb J. N-acetyltransferase-2 genotypes among Jordanian patients with diabetes mellitus. Int J Clin Pharmacol Ther. 2013;51(7):593–599. doi: 10.5414/CP201883. [DOI] [PubMed] [Google Scholar]
- 34.Al-Shaqha W.M., Alkharfy K.M., Al-Daghri N.M., Ak M. N-acetyltransferase 1 and 2 polymorphisms and risk of diabetes mellitus type 2 in a Saudi population. Ann. Saudi Med. 2015;35(3):214–221. doi: 10.5144/0256-4947.2015.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weykamp C. HbA1c: a review of analytical and clinical aspects. Ann Lab Med. 2013;33(6):393–400. doi: 10.3343/alm.2013.33.6.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S., Zhao Z., Ke L., Li Z., Li W., Zhang Z., et al. Resveratrol improves glucose uptake in insulin-resistant adipocytes via Sirt1. J. Nutr. Biochem. 2018;55:209–218. doi: 10.1016/j.jnutbio.2018.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Wilkinson D.J., Piasecki M., Atherton P.J. The age-related loss of skeletal muscle mass and function: measurement and physiology of muscle fibre atrophy and muscle fibre loss in humans. Ageing Res. Rev. 2018;47:123–132. doi: 10.1016/j.arr.2018.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request.