Abstract
Bitter substances in functional foods and beverages can act as nutraceuticals, offering potential health benefits. However, their unpleasant sensory impact reduces the consumption of these foods. Consequently, the discovery of bitter masking compounds is crucial for enhancing the intake of bioactive compounds in functional foods and beverages. Bitter taste is mediated by TAS2Rs, a sub-family of G-protein-coupled receptors. TAS2R14 is especially pivotal in the perception of bitterness, as it is one of the most broadly tuned bitter receptors. In this study, allspice was extracted and purified to yield five single compounds based on sensory guided fractionation. The structures of each compound were determined based on nuclear magnetic resonance (NMR) and high-resolution mass spectrometry (HR-MS). In a sensory evaluation, compound 1 exhibited bitter masking activity against quinine. Molecular docking analysis revealed that compound 1 could act as an antagonist of the TAS2R14 bitter receptor.
Keywords: Food flavor, Taste modulator, Bitter taste receptor, TAS2R, Bitterness
Chemical compounds studied in this article: (2R)-3-(4-hydroxy-3-methoxy-phenyl) propane-1,2-diol (PubChem CID: 101154580); Sphalleroside A (PubChem CID: 101702475); Pimentol (PubChem CID: 9917512), (4S)-R-terpineol 8-O-β-D-(6-O-galloyl) glucopyranoside (PubChem CID: 10552230); (4R)-R-terpineol 8-O-β-D-(6-O-galloyl) glucopyranoside (PubChem CID: 10695593)
Graphical abstract
Highlights
-
•
Bitter masking compound was identified from allspice.
-
•
Compound was isolated based on sensory guided fractionation.
-
•
Sensory test revealed its bitter masking effect.
-
•
Compound 1 could behave as a TAS2R14 antagonist in molecular docking analysis.
1. Introduction
People avoid bitter tastes due to their negative hedonic impact and an instinctual avoidance of toxic substances. This aversion to bitterness remains high throughout the life and reduces the intake of bitter substances (Mennella, Pepino, & Beauchamp, 2003; Mennella, Reed, Mathew, Roberts, & Mansfield, 2015). Newborns rejected bitter solutions, displaying negative facial expressions (Ganchrow, Steiner, & Daher, 1983). Adult tasters with a large number of papillae tend to consume fewer vegetables than adults with a smaller number of papillae. This is because they perceive bitterness more intensely due to their heightened sense of taste (Duffy et al., 2010). However, bitter functional foods or beverages contain numerous bioactive compounds, including flavonoids, terpenes, and polyphenolic acids beneficial for our health (Günther-Jordanland, Dawid, Dietz, & Hofmann, 2016; Mancuso, Borgonovo, Scaglioni, & Bassoli, 2015). Thus, there have been many trials to decrease the bitter taste. For example, resin absorption method has been used to remove naringin, a bitter compound in grapefruit juice (Muñoz, Holtheuer, Wilson, & Urrutia, 2022; Wilson, Wagner Jr, & Shaw, 1989). However, this resin absorption process also removes other phytochemicals from grapefruit juice (Gordon et al., 2021). Similarly, several methods have been developed to decrease the caffeine content in coffee and tea. During decaffeination, several organic solvents, including dichloromethane, ethyl acetate, or hexane are used to extract caffeine (Sharif, Ahmad, Anjum, Ramzan, & Malik, 2014; Welton, 2015). The issue is that these organic solvents are toxic and harmful to our health. Therefore, the discovery of bitter masking substances from natural sources is essential for the food industry.
Sweet and umami tastes are mediated by TAS1R, whereas bitter taste is mediated by TAS2R, both of which are expressed in the oral tissues. Both TAS1R and TAS2R are G-protein-coupled receptors (GPCRs), each belonging to a distinct group. TAS1R genes are composed of 3 isoforms such as TAS1R1, TAS1R2, and TAS1R3 to assemble the sweet or umami taste receptor (Carey, Kim, Cohen, Lee, & Nead, 2022). On the other hand, TAS2R genes consist of 26 isoforms to form the bitter taste receptor. TAS1R is categorized within the class C GPCR group, whereas TAS2R is classified within the frizzled/taste2 receptor group, mainly associated with the detection of bitter taste compounds. Notably, the sequence of TAS2R bear little resemblance to that of other GPCR families, including TAS1R, suggesting a considerable degree of structural diversity (Behrens & Ziegler, 2020; Di Pizio & Niv, 2015; Fredriksson, Lagerström, Lundin, & Schiöth, 2003; Lang, Di Pizio, Risso, Drayna, & Behrens, 2023; Lipchock, Mennella, Spielman, & Reed, 2013; Maehashi & Huang, 2009; Melis, Errigo, Crnjar, Pes, & Tomassini Barbarossa, 2019).
The process of bitter taste perception relies on two key signaling pathways, namely, the Gαgust and Gβγ cascades (Chandrashekar et al., 2000; De Giorgio et al., 2016). Prior to the engagement with bitter tastants, Gαgust and the Gβγ heterodimer are colocalized at the taste receptor, ready to be triggered into action. In the Gαgust pathway, once a bitter substance binds to the receptor, Gαgust undergoes a nucleotide exchange, swapping guanosine diphosphate (GDP) for guanosine triphosphate (GTP), and dissociates from both the taste receptor and the Gβγ heterodimer. Following this separation, Gαgust stimulates phosphodiesterase (PDE), leading to a reduction in the cellular concentration of cyclic adenosine monophosphate (cAMP). This interaction deactivates protein kinase (PKA), known as an inhibitor of Ca2+ release. Consequently, the activation of the Gαgust pathway aids the release of intracellular Ca2+, which plays an important role in signal transmission in the nervous system (Ueda, Ugawa, Yamamura, Imaizumi, & Shimada, 2003; Boto, Gomez-Diaz, & Alcorta, 2010; McLaughlin, McKinnon, & Margolskee, 1992a). Concurrently, the binding of bitter compound also activates the Gβγ pathway. Once the bitter compound binds to receptor, Gβγ separates from the taste receptor. This event activates phospholipase C β2 (PLCβ2) to cleave phosphatidylinositol 4,5-bisphosphate (PIP2) into two molecules, namely, inositol triphosphate (IP3) and diacylglycerol (DAG). IP3 subsequently binds to its receptor IP3R3, prompting the release of intracellular Ca2+. The released Ca2+ activates TRPM5, a monovalent selective cation channel, leading to Na+ influx. The resultant increase in Na+ levels stimulates voltage-gated Na+ channels, causing membrane depolarization and neurotransmitter release, thus conveying the signal to the nervous system. (Behrens & Meyerhof, 2016; McLaughlin, McKinnon, & Margolskee, 1992b; Vilardaga, Agnati, Fuxe, & Ciruela, 2010).
However, if an antagonist of the bitter receptor is present, it would prevent the activation of the transduction cascade, thus resulting in the attenuation of the bitter taste. Therefore, the identification of bitter receptor antagonist can be a potential method to inhibit bitterness. As mentioned above, 26 human bitter receptors have been reported, and among them, TAS2R14 is one of the most broadly tuned bitterness receptors (Meyerhof et al., 2010; Nowak et al., 2018). Thus, finding an antagonist to TAS2R14 is critical to reducing the perception of bitterness, thereby potentially improving the acceptance of functional foods and beverages.
To date, only a small number of bitter reducing compounds have been reported from nature (Ley, Blings, Paetz, Krammer, & Bertram, 2006; Li et al., 2014; Suess, Brockhoff, Meyerhof, & Hofmann, 2016). However, some bitter-masking substances may pose health concerns. Several fractions from cheese have been identified as bitter substances, which include high levels of fatty acids (Homma et al., 2012). The intake of large quantities of fatty acids can be harmful to our health. Sweetness or saltiness are also known to reduce bitterness (Breslin & Beauchamp, 1997; Walters, 1996). However, many sweet tasting foods have high levels of sugars that are associated with metabolic diseases (Herman & Birnbaum, 2021; Malik et al., 2010). Also, salt consumption for the use of masking bitterness can cause hypertension (Frisoli, Schmieder, Grodzicki, & Messerli, 2012). Therefore, finding small molecule compounds that mask bitterness would be an ideal solution, as they are likely to have fewer side effects.
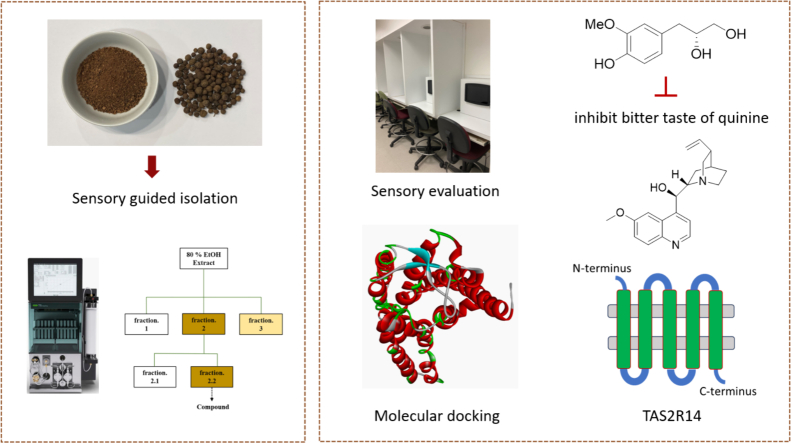
In this study, allspice was extracted with 80% ethanol and separated by food grade SP-70 resin using ethanol and water. Among them, the 30% ethanol-eluted allspice fraction showed the highest bitter masking activity, and the 50% ethanol-eluted allspice fraction presented moderate activity. Therefore, sensory guided isolation was performed to obtain a bitter masking compound from allspice. And its binding mechanism with TAS2R14 was analyzed via molecular docking model.
2. Materials and methods
2.1. General procedures
LC – MS grade acetonitrile, high-performance liquid chromatography (HPLC) grade methanol, and ACS grade methanol were obtained from Fisher Scientific (Fair Lawn, NJ). Food grade ethanol was purchased from Decon Labs (King of Prussia, PA). SP-70 resin was obtained from Fisher Scientific (Fair Lawn, NJ). HPLC reverse phase column was purchased from Waters (Milford, MA). Flash column chromatography was performed using a Buchi Pure C-810 (Flawil, Switzerland) equipped with reverse phase column (EcoFlex C18, 220 g). Sephadex LH-20 gel was purchased from GE Healthcare (Piscataway, NJ). Quinine monohydrochloride dihydrate and caffeine were purchased from Sigma-Aldrich (St. Louis, MO). 1H and 13C NMR data were collected on a Bruker Avance II 600 MHz spectrometer at the University of Florida. A HPLC system from Jasco (Tokyo, Japan) was utilized at a flow rate of 3 mL/min with UV detection at 210 and 254 nm using an X-select C18 column (10 × 250 mm, 5 μm particle size; Waters, Milford, MA). The Lyovapor L-300 from Buchi (Flawil, Switzerland) was used as a high-vacuum system to remove the solvents.
2.2. Precautions taken for sensory analysis
The fractions and test compounds were freeze-dried by a high vacuum system (0.1 mbar) to remove the solvents and determined to contain no remaining solvents based on analytical method. In a sensory test, the “sip and spit” method was employed to limit the consumption of sensory evaluation samples. A sensory evaluation was carried out in a sensory room at room temperature. Sensory testing was conducted in the morning, and panelists were advised not to drink coffee or tea to avoid developing a tolerance to bitter tastes and caffeine (Dawid et al., 2012).
2.3. Screening and training the sensory panel
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and national research committees and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The University of Florida's Institutional Review Board (IRB) granted ethical approval for the study protocol and the process for obtaining consent (Feng, Gmitter Jr, Grosser, & Wang, 2021). Every individual participant involved in the study provided their informed consent. Eight subjects (three women and five men, ages 27 – 40 years) were screened to test their sensitivity to bitter taste. In triangle and scale test, three participants (two men and one woman, ages 28 – 39) demonstrated consistent and reliable scoring patterns and were thus selected for the bitter-masking sensory panel. The selected three subjects were further trained to assess the bitter intensity of quinine and caffeine solution.
2.4. Measurement of bitter masking effect of single compounds against quinine and caffeine
Two individual standard solutions were prepared using quinine and caffeine. Distilled water was used to dissolve the reagents. To establish the optimal concentration for the standard solutions, the panel tasted and rated six different concentrations each of quinine (2, 4, 6, 8, 10, 12 ppm) and caffeine (300, 400, 500, 600, 700, 800 ppm). Six ppm quinine and 500 ppm caffeine solutions were scored as having medium bitterness intensity in 0 (no sensation) to 15 (strongest sensation) scale evaluation. Thus, solutions of 6 ppm of quinine and 500 ppm of caffeine were used as standards. The concentrations of both the quinine and caffeine in our standard solutions are the same or very similar to those reported in previous literature (Ley et al., 2006; Ley, Krammer, Reinders, Gatfield, & Bertram, 2005; Riemer, 1994). For the preparation of test solutions, each compound was added to the standard solution at three different concentrations (25, 50, and 100 ppm). The bitterness intensity of test solutions was compared with that of standard solution and rated using a scale from 0 (no sensation) to 15 (strongest sensation). Compound 1, being an oil, was first dissolved in food grade ethanol at a concentration of 10.0% (w/v) before it was added to the solution. For the statistical analysis, One-way analysis of variance (ANOVA) (p < 0.05) was carried out using SPSS v.28.0 (IBM Corp., Armonk, NY).
2.5. Sensory guided fractionation
Total extraction of allspice was separated using various columns including SP-70, reverse phase C18, and Sephadex LH-20. In each separation step, fractions were tested for their bitter masking activity. A 10-ppm quinine solution in distilled water was used as a standard solution. To prepare the test solutions, each fraction was added to the standard solution to reach a final concentration of 2 mg/mL. The bitterness intensity of the standard solution was set to a score of 9.0. The bitterness intensity of test solutions was then compared with that of standard solution and rated using a scale from 0 (no sensation) to 15 (strongest sensation). Fractions that reduced bitterness underwent further separation to obtain single compounds. Detailed isolation procedures for compounds 1 – 5 are as follows.
Allspice was extracted with 80% ethanol (three times) at room temperature in a sonicator. The extract was dried in vacuo using a rotary evaporator. The extraction was separated by SP-70 open column with a step-gradient system of ethanol/water (0/10 → 3/7 → 5/5 → 7/3 → 10/0 [v:v]). In a sensory test, the 30% EtOH eluted fraction (Frac.30) and 50% EtOH eluted fraction (Frac.50) showed bitter masking activities and these two fractions were further separated (Table S1). First, the 30% EtOH eluted fraction (Frac.30) was loaded on flash column chromatography (C18, 50 μm spherical) with a gradient system of methanol/water (1/9 → 10/0 [v:v]) to obtain four sub-fractions (Frac.30.1 – Frac.30.4). In the following sensory test, two fractions (Frac.30.2 and Frac.30.3) reduced bitterness intensity. Thus, Frac.30.2 was further separated via Sephadex LH-20 with a gradient system of ethanol/water (7/3 → 10/0 [v:v]) to yield three sub-fractions (Frac.30.2.1 – Frac.30.2.3). Frac.30.2.3 was once again separated using Sephadex LH-20 with a gradient system of ethanol/water (8.5/1.5 → 10/0 [v:v]) to yield two sub-fractions (Frac.30.2.3.1 – Frac.30.2.3.2). Frac.30.2.3.2 was purified by semi-preparative HPLC column (Xselect CSH OBD 10 × 250 mm, 5 μm particle size, solvent A: water containing 0.1% formic acid, solvent B: methanol containing 0.1% formic acid / 0 – 3 min: 15% B, 3 – 19 min 20% B, 20 – 34 min 23% B, flow rate 3 mL/min) to obtain compound 2 (tR = 18.6 min) and 3 (tR = 26.2 min). Frac.30.3 was separated using Sephadex LH-20 with a gradient system of ethanol/water (7/3 → 10/0 [v:v]) to obtain Frac.30.3.1 and Frac.30.3.2. Then, Frac.30.3.2 was purified using semi-preparative HPLC (21% aqueous MeOH, flow rate 3 mL/min) to isolate compound 1 (tR = 16.7 min). Similar to Frac.30, 50% EtOH eluted fraction (Frac.50) was also loaded on flash column chromatography (C18, 50 μm spherical) with a gradient system of methanol/water (1/9 → 10/0 [v:v]) to obtain three sub-fractions (Frac.50.1 – Frac.50.3). Frac.50.2 was separated using Sephadex LH-20 with a gradient system of ethanol/water (7/3 → 10/0 [v:v]) to obtain three sub-fractions (Frac.50.2.1 – Frac.50.2.3). Frac.50.2.2 was purified using semi-preparative HPLC (solvent A: water containing 0.1% formic acid, solvent B: methanol containing 0.1% formic acid / 0–3 min: 15% B, 3–19 min 20% B, 20–34 min 23% aqueous methanol, flow rate 3 mL/min) to yield compound 4 (tR = 25.0 min) and 5 (tR = 27.5 min).
2.6. Sweet enhancement test
Sucrose was dissolved in distilled water at 6 ppm and used as a standard solution (Li, Servant, & Tachdjia, 2011). Compound 1 was added to the standard solution at three different concentrations (25, 50, and 100 ppm) for the preparation of the test solution. The sweetness intensity of test solutions was compared with that of standard solution. The sweetness was rated using a scale from 0 (no sensation) to 15 (strongest sensation).
2.7. LC – MS quantification
Quantification analysis was conducted utilizing an Ultimate 3000 LC system linked to a Quantiva triple quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA). Compound 1 was separated using a Phenomenex Gemini C18 (3.0 × 150 mm, particle size 3.0 μm) column at 30 °C with a gradient elution system (solvent A: water containing 0.1% formic acid, solvent B: acetonitrile containing 0.1% formic acid). The gradient profile was as follows: 0 – 3 min 5% B, 3 – 20 min 5 – 50% B, 20 – 21.5 min 50 – 100%, and 21.5 – 28.5 min 100% B. The re-equilibration was maintained for 6 min using the initial gradient of the mobile phase. The system operated at a flow rate of 0.3 mL/min, with an injection volume set to 5 μL. The mass spectrometer equipped with an electrospray ionization (ESI) interface operated in negative ionization mode. The settings for the ESI were: spray voltage, 2500 V; vaporizer temperature, 300 °C; ion transfer tube temperature, 325 °C; aux gas, 12 Arb; sheath gas, 40 Arb; and sweep gas, 1 Arb. Selective reaction monitoring (SRM) mode was applied for the MS2 detection. For each analyte, MS/MS parameters were optimized using flow injection analysis (Table S2). Data processing and instrument management were conducted using Xcalibur software (Version 3.0). The high-resolution electrospray ionization mass spectrometry (HR – ESI – MS) values were recorded with UPLC (Thermo Vanquish Flex Binary RSLC platform) coupled to a Q Exactive Plus mass spectrometer (Thermo Fisher Scientific, Waltham, MA).
2.8. Molecular docking analysis
Docking analysis was performed using GOLD software version 5.3 (Genetic Optimisation for Ligand Docking; the Cambridge Crystallographic Data Centre, Cambridge, UK) with default parameters. The bitter receptor TAS2R14 was downloaded from the BitterDB Protein Data Bank (http://bitterdb.agri.huji.ac.il/dbbitter.php) (BitterDB ID: 14, Uniprot: Q9NYV8, accessed December 21, 2022). The binding pocket for the GOLD molecular docking calculations was defined based on previous literature and PrankWeb prediction (https://prankweb.cz/) (Yu et al., 2022). The best GOLD PLP CHEM score was chosen and visualization of 2D models of complexes was depicted by BIOVIA Discovery Studio 2021.5.
2.9. Spectroscopic data
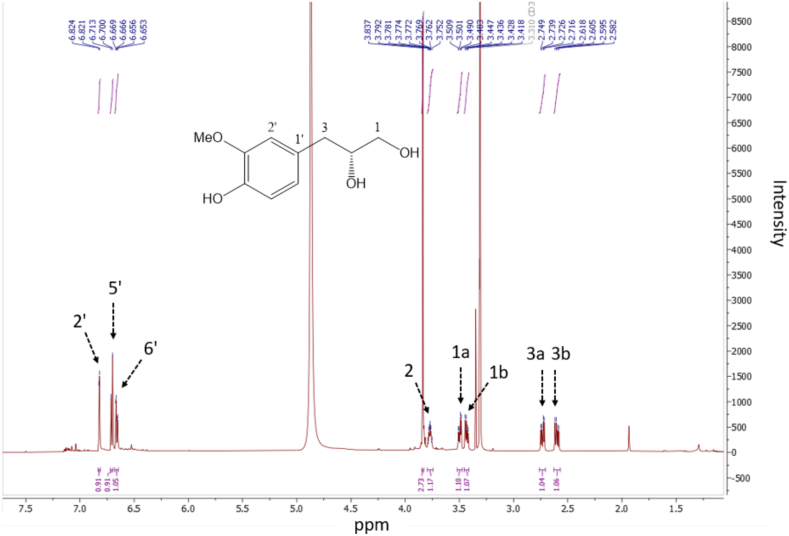
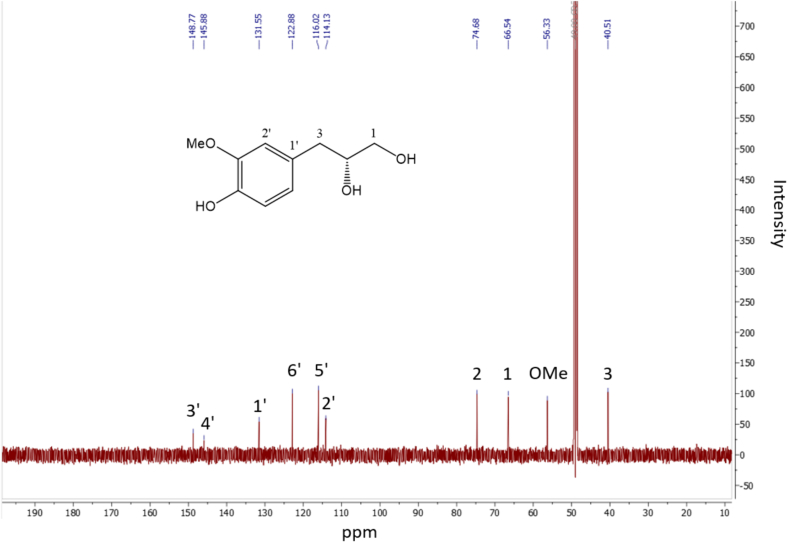
1H and 13C NMR spectra of compound 1 are presented in Fig. 1, Fig. 2. 1H and 13C NMR spectra of compounds 2 – 5 are shown in Figs. S1 – S8. Tables S3 – S4 depict the NMR information of compounds 2 – 5. The HR – ESI – MS data for 2 – 5 are as follows. Compound 2: HR – ESI – MS peak at m/z 341.1241 [M – H] – (calcd for C16H21O8, 341.1236); Compound 3: HR – ESI – MS peak at m/z 493.1352 [M – H] – (calcd for C23H25O12, 493.1346); Compound 4: HR – ESI – MS peak at m/z 467.1918 [M – H] – (calcd for C23H31O10, 467.1917); Compound 5: HR – ESI – MS peak at m/z 467.1915 [M – H] – (calcd for C23H31O10, 467.1917).
Fig. 1.
1H NMR spectrum (methanol‑d4, 600 MHz) of compound 1.
Fig. 2.
13C NMR spectrum (methanol‑d4, 150 MHz) of compound 1.
2.10. Spectroscopic data of compound 1
1H NMR (600 MHz, methanol‑d4): δH 3.50 (1H, dd, J = 11.2, 4.4 Hz, H – 1a), 3.43 (1H, dd, J = 11.2, 6.2 Hz, H – 1b), 3.77 (1H, m, H – 2), 2.73 (1H, dd, J = 13.8, 5.8 Hz, H – 3a), 2.60 (1H, dd, J = 13.8, 7.4 Hz, H – 3b), 6.82 (1H, d, J = 1.9 Hz, H – 2′), 6.71 (1H, d, J = 8.0 Hz, H – 5′), 6.66 (1H, dd, J = 8.0, 1.9 Hz, H – 6′), 3.84 (3H, s, OMe). 13C NMR (methanol‑d4, 150 MHz): δC 66.5 (C – 1), 74.7 (C – 2), 40.5 (C – 3), 131.6 (C – 1′), 114.1 (C – 2′), 148.8C – 3′), 145.9 (C – 4′), 116.0 (C – 5′), 122.9 (C – 6′), 56.3 (OMe). HR – ESI – MS peak at m/z 197.0818 [M – H] – (calcd for C10H13O4, 197.0813).
3. Results
3.1. Sensory guided isolation
Sensory guided isolation strategies have been increasingly utilized to identify taste modulators from natural sources (Duggan, Gilch, Stark, Dawid, & Hofmann, 2022; Zhang, Wang, & Du, 2022; Zhao, Chen, Li, & Xu, 2018). A search on Google Scholar using the term “sensory guided fractionation” revealed only 1 paper published before the year 2000, while 21 papers appeared between 2001 and 2010, and 179 papers were reported from 2011 to 2023 (assessed November 6, 2023). The advantage of sensory guided fractionation is that it quickly addresses the sensory modifying compounds, thereby saving time and labor.
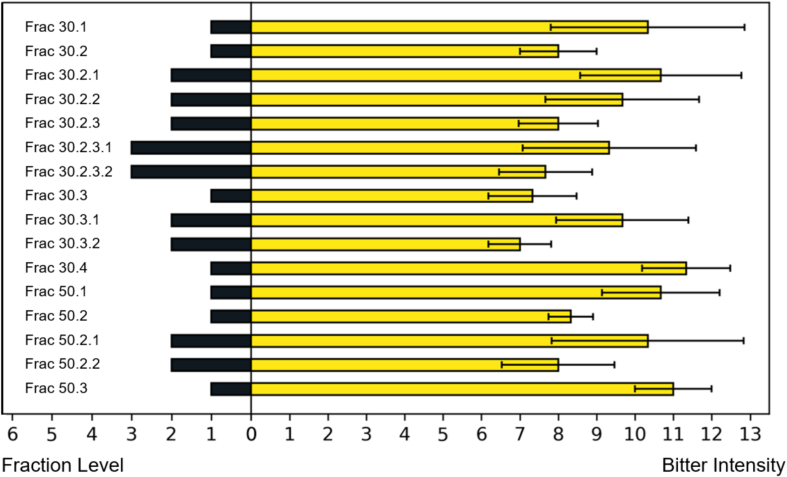
In this study, the total extract of allspice was divided using various columns, including SP-70, RP-C18, and Sephadex LH-20. At each stage of separation, fractions were assessed for their ability to mask bitter taste. Fractions that reduced bitterness intensity underwent further separation. Fig. 3 depicts the bitter masking effect of each fraction. Finally, as described in section 2.5 (sensory guided fractionation), five single compounds (1 – 5) were isolated. Their chemical structures were determined based on spectroscopic data, such NMR and HR – MS – ESI.
Fig. 3.
Bitterness intensity of each fraction. On the left side, bar lengths correspond to three distinct values, representing different separation stages: 1. SP-70/C-18; 2. SP-70/C-18/LH-20; 3. SP-70/C-18/LH-20/LH-20. The right side displays the bitterness intensity of test solutions (Each fraction was added to standard solution and used as test solutions). Values are mean ± SD (n = 3).
3.2. Sensory test
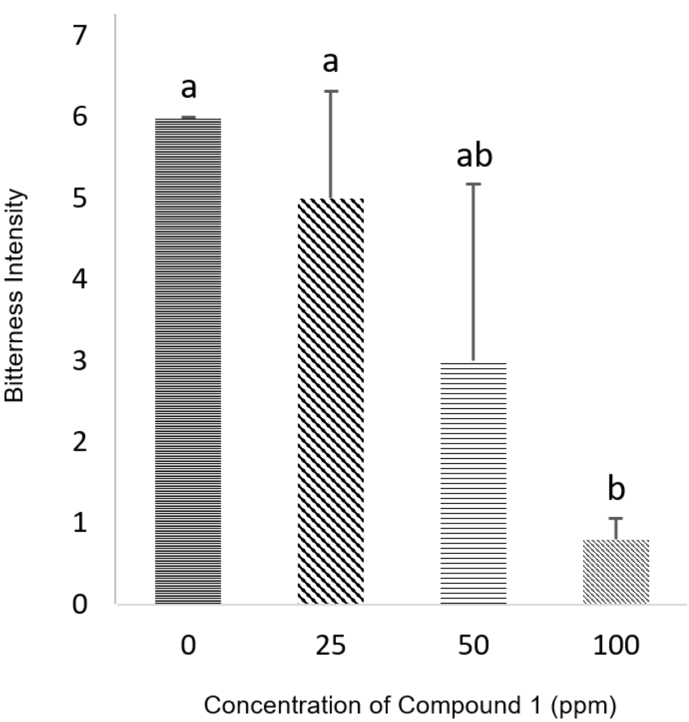
In the sensory test, compound 1 inhibited the bitterness of quinine according to its concentration in the range of 25 – 100 ppm (Fig. 4). A consistent reduction in perceived bitterness with an increased concentration of compound 1 revealed that compound 1 was responsible for the bitter masking activity. To assess if the bitterness inhibition was due to sweetness enhancement, a sweet enhancer sensory test was also conducted. Compound 1 did not present any sweet enhancing activity, indicating its bitter masking effect was not attributable to sweetness enhancement (Table S5). The bitter inhibitory effect of compound 1 was also tested against caffeine, however, it did not show any reducing effect (Table S6). Compounds 2 – 5 did not reduce the bitterness of either quinine or caffeine.
Fig. 4.
Bitterness intensity of standard and test solutions (6 ppm quinine was used as standard solution; compound 1 was added to standard solution at three different concentrations and used as test solutions). Statistical analyses were performed using ANOVA followed by Scheffe's post hoc test, and the criterion for statistical significance was set to p < 0.05. Values are mean ± SD (n = 3).
3.3. Chemical structure of compound 1
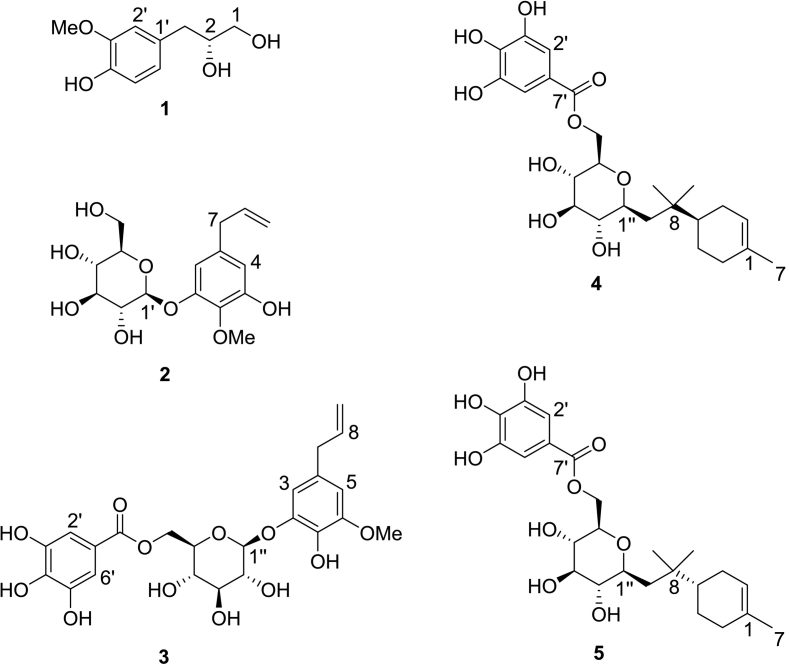
Compound 1 was isolated as a colorless oil. The HR – ESI – MS data for compound 1 displayed a peak at m/z 197.0818 [M – H] – (calcd for 197.0813, C10H13O4), indicating a molecular formula of C10H14O4. Together with mass spectrometry, NMR data established the chemical structure of compound 1. Compound 1 mainly consists of two parts, namely, a benzene ring and a chain structure. In the benzene ring part, three aromatic protons were shown at [δH 6.82 (1H, d, J = 1.9 Hz, H – 2′), 6.71 (1H, d, J = 8.0 Hz, H – 5′), 6.66 (1H, dd, J = 8.0, 1.9 Hz, H – 6′)], suggesting a typical ABX system. In this ABX system, two ortho protons were presented at [δH 6.71 (1H, d, J = 8.0 Hz, H – 5′), 6.66 (1H, dd, J = 8.0, 1.9 Hz, H – 6′)] with a coupling constant of 8.0 Hz. Additionally, two para protons were shown at [δH 6.82 (1H, d, J = 1.9 Hz, H – 2′), 6.66 (1H, dd, J = 8.0, 1.9 Hz, H – 6′)] with a coupling constant of 1.9 Hz. Their corresponding carbons were confirmed based on heteronuclear single quantum coherence (HSQC) spectrum (δH 6.82 / δC 114.1, δH 6.71/ δC 116.0, δH 6.66 / δC 122.9). In 13C NMR spectrum, the other two aromatic carbons in the benzene ring showed relatively deshielded chemical shift at δC 145.9 and 148.8, indicating that either a hydroxy or a methoxy group is attached to them. Among these two carbons, one carbon (δC 148.8) showed heteronuclear multiple bond correlation (HMBC) correlation with the methoxy group (δH 3.84, s, 3H), indicating the methoxy group is attached to this carbon (δC 148.8). The position of the methoxy group is also confirmed based on previous literature (Luyen et al., 2014). The rest part of compound 1 was determined as a chain segment with a chemical formula of CH2CH(OH)CH2OH. The HSQC spectrum showed correlation from two methylene protons (H-1a/1b) to their corresponding carbon (δC 66.5, C-1). The relatively deshielded carbon chemical shift of C-1 (δC 66.5) suggested that C-1 is substituted with a hydroxy group. Two methylene protons at C-1 showed HMBC correlation with methine carbon (C-2, δC 74.7), suggesting that C-2 is also substituted by hydroxy group. The HSQC spectrum confirmed that proton at δH 3.77 (m, H-2) is coupled with C-2 (δC 74.7). C-2 (δC 74.7) showed HMBC correlation with another two methylene protons [δH 2.73 (1H, dd, J = 13.8, 5.8 Hz, H – 3), 2.60 (1H, dd, J = 13.8, 7.4 Hz, H – 3)] those positioned on C-3. Consequently, 1H and 13C NMR data of compound 1 were in good agreement with those of previous literature (Luyen et al., 2014). Therefore, compound 1 was characterized as (2R)-3-(4-hydroxy-3-methoxy-phenyl) propane-1,2-diol. The chemical structure of compounds 2 – 5 were determined based on HR – ESI – MS and NMR information as well (Fig. 5). Compounds 2 – 5 were characterized as sphalleroside A, pimentol, (4S)-R-terpineol 8-O-β-D-(6-O-galloyl) glucopyranoside, and (4R)-R-terpineol 8-O-β-D-(6-O-galloyl) glucopyranoside, respectively (Chen, Chen, Shi, Sun, & Ji, 1997; Kikuzaki, Sato, Mayahara, & Nakatani, 2000; Oya, Osawa, & Kawakishi, 1997).
Fig. 5.
Chemical structures of five compounds isolated from allspice based on sensory guided fractionation.
LC – MS quantification analysis showed that the contents of compound 1 in allspice was 1.248 ± 0.071 mg/kg of total extract. Linearity was defined with calibration curves calculated using least-squares linear regression (Table S7).
3.4. Molecular docking analysis
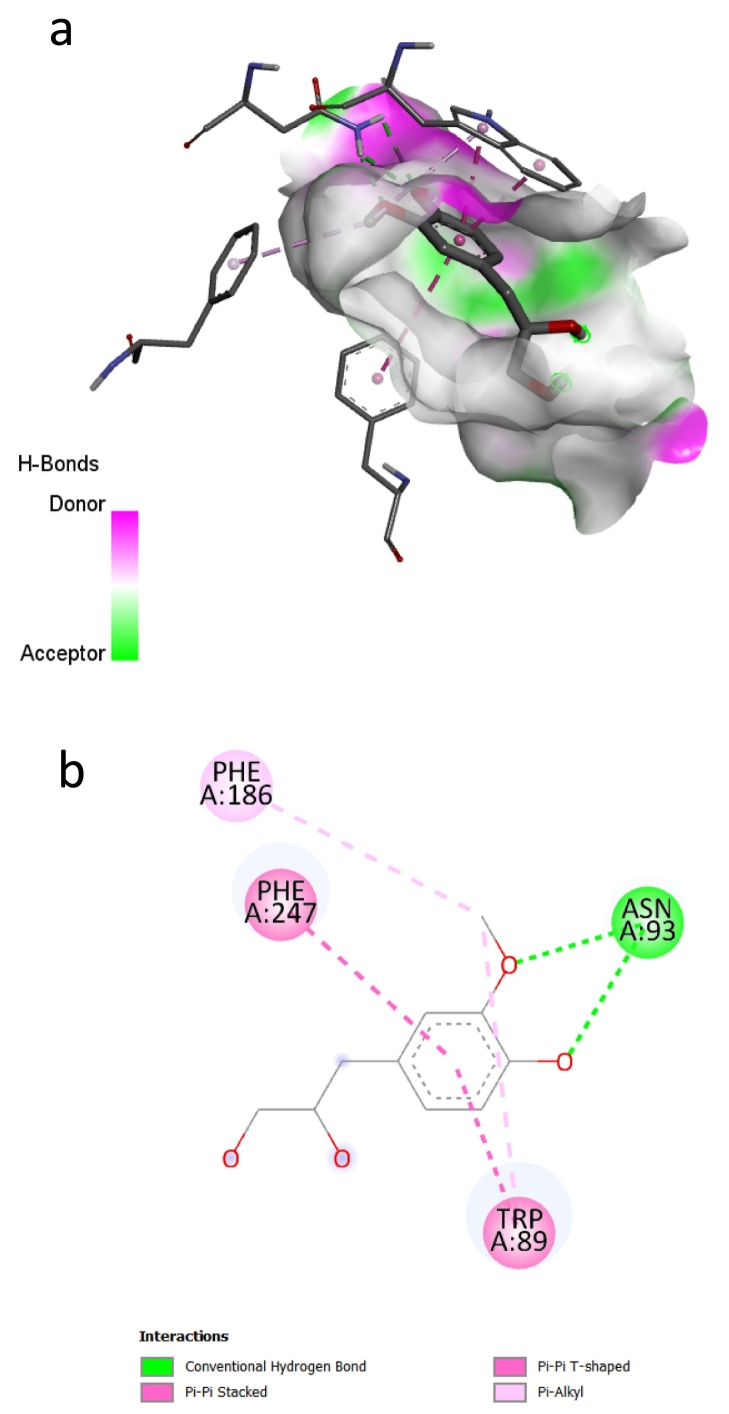
To investigate the cellular mechanism of the bitter inhibitory effect, we performed a molecular docking analysis. Compound 1 was docked into the binding site of TAS2R14 using the GOLD 3.01 docking program, achieving a fitness value of 64.72. Leveraging recent advancements in our understanding of human bitter taste receptors, we have utilized a homology-based three-dimensional model of TAS2R14, enhancing the accuracy of predictions concerning the receptor's structure and function. The interaction of compound 1 with the active site of hTAS2R31 is visually represented in Fig. 6. A docking study revealed compound 1 interacting with Phe 186, Phe 247, Asn 93, and Trp 89. Compound 1 predominantly consists of a benzene ring and a chain segment, with an adjacent methoxy and a hydroxy group on the benzene ring. Thus, the electron donating properties of the benzene ring, along with methoxy and hydroxy groups contributed to the compound's binding affinity with the amino residues of the receptor.
Fig. 6.
Molecular docking analysis of the ligand-binding pocket of TAS2R14. a: 3D hydrogen bond surface plot at binding site. b: 2D representation showing interactions at binding site: hydrogen bond (green dashed line), π-alkyl hydrophobic bond (pink dashed line), and π-π bond (red dashed line). (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article.)
The methoxy group in the benzene ring interacted with Phe 186 and Trp 89 through a π – alkyl hydrophobic bond. The π – alkyl hydrophobic bond is a non-covalent interaction between the π – electrons of the aromatic ring and the alkyl group of the methoxy group. The π-electrons of the aromatic ring are attracted to the electron-deficient carbon atoms of the methoxy group, influenced by the oxygen atom's electron-withdrawing properties (McMurray, 2011a). This interaction contributed to the stability of the protein-ligand complexes. Besides the methoxy group, the benzene ring itself also formed π – π interactions with Phe 247 and Trp 89 of the receptor. The π – π interaction is a non-covalent interaction occurring between the π – electrons of the aromatic ring in compound 1 and those in the amino acid residues Phe247 and Trp89. The π – electrons of the aromatic rings are attracted to each other due to the electron rich π system around the benzene ring, which generates a partial negative charge (McMurray, 2011b). This interaction plays a critical role in stabilizing the complexes between compound 1 and the receptor. As mentioned above, in compound 1, one methoxy and one hydroxy group are attached to the benzene ring in adjacent position. Each oxygen atom from these methoxy and hydroxy groups formed a hydrogen bond with Asn 93. The hydroxy group is a common functional group that is capable of forming hydrogen bonds due to the presence of the polar O – H bond. Additionally, the oxygen in a methoxy group can also act as a hydrogen bond acceptor.
Previous research also showed that a benzene ring interacted with Phe247 and Trp89 via π – π interactions. Several peptides derived from Oncorhynchus mykiss nebulin exhibited high affinity for TAS2R14 in molecular docking analysis, with their inhibitory effects against quinine assessed using an electronic tongue (Yu et al., 2022). Zhao et al. (2021) reported another peptide as a potential TAS2R14 blocker. They hydrolyzed Mizuhopecten yessoensis myosin in silico using gastrointestinal proteases to produce peptides. From the 405 hydrolyzed peptides, the 10 most promising candidates were selected based on physical and chemical properties, including toxicity, solubility, and bioactivity predictions. Six peptides, including QRPR, GFPSR, NPPK, DPDF, LEGSLE, and KPM, presented docking interaction energies over 60.0 (kcal/mol). Among them, LEGSLE formed interactions with Asn 93 (via a conventional hydrogen bond), Trp 89 (via an electrostatic bond), and Phe 247 (via a hydrophobic bond). These three amino acid residues correspond to those identified in our study. In 2019, a subfraction of hen protein hydrolysate (protex 50FP) was found to inhibit TAS2R14, thereby reducing bitterness (Greene et al., 2011). The protex 50FP hydrolysate was fractionated by a reverse phase column, and one of its subfractions inhibited calcium mobilization in HEK293T cells and reduced the bitter intensity of a quinine solution in electronic tongue analysis.
4. Conclusion
Many bioactive compounds, including alkaloids, polyphenolics, or terpenes, are bitter, discouraging the consumption of phytochemicals. The US Department of health and human services reported that only 26.3% of adults in the US consumed dark green vegetables from 2015 to 2018 (Ansai & Wambogo, 2021). A lack of vegetable consumption is associated with adult diseases such as cancer or cardiovascular disease (Aune et al., 2017; Law & Morris, 1998). Significant progress has been made in elucidating the structure of bitter taste receptors to understand the cellular mechanism of bitterness. However, a challenge arises because the number of bitter taste receptors, around 26, greatly outnumbers the sweet receptors, of which there are only two. Moreover, the low similarity in sequence between bitter and sweet taste receptors complicates the situation, making it difficult to predict sequencing outcomes.
In this study, sensory guided isolation yielded five candidate single compounds from allspice. Among them, compound 1 showed a bitter masking effect against quinine. In the concentration range of 25 to 100 ppm, compound 1 reduced the bitter taste of 6 ppm quinine. Based on the spectroscopic data, including NMR and HR – ESI – MS, the chemical structure of compound 1 was characterized as (2R)-3-(4-hydroxy-3-methoxy-phenyl) propane-1,2-diol. The cellular mechanism was determined using molecular docking software analysis. Compound 1 showed significant binding affinity with TAS2R14, interacting with ASN93, Phe 186, Phe 247, and Trp 89, suggesting its potential as an antagonist.
CRediT authorship contribution statement
Jin-Pyo An: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation. Xin Liu: Writing – review & editing, Validation. Yu Wang: Writing – review & editing, Supervision, Conceptualization.
Declaration of competing interest
The authors declare no known competing interests that could influence the work reported in this article.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.fochx.2024.101426.
Contributor Information
Jin-Pyo An, Email: anjinpyo@ufl.edu.
Xin Liu, Email: xin.liu@ufl.edu.
Yu Wang, Email: yu.wang@ufl.edu.
Appendix A. Supplementary data
Supplementary material
Data availability
Data will be made available on request.
References
- Ansai N., Wambogo E.A. 2021. Fruit and vegetable consumption among adults in the United States, 2015–2018. NCHS Data Brief, Article 397. [DOI] [PubMed] [Google Scholar]
- Aune D., Giovannucci E., Boffetta P., Fadnes L.T., Keum N., Norat T.…Tonstad S. Fruit and vegetable intake and the risk of cardiovascular disease, total cancer and all-cause mortality-a systematic review and dose-response meta-analysis of prospective studies. International Journal of Epidemiology. 2017;46(3):1029–1056. doi: 10.1093/ije/dyw319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens M., Meyerhof W.G. In: Chemosensory transduction. Zufall F., Munger S.D., editors. Academic Press; 2016. Protein–coupled taste receptors; pp. 227–244. [Google Scholar]
- Behrens M., Ziegler F. Structure-function analyses of human bitter taste receptors—Where do we stand? Molecules. 2020;25(19) doi: 10.3390/molecules25194423. Article 4423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boto T., Gomez-Diaz C., Alcorta E. Expression analysis of the 3 G-protein subunits, Gα, Gβ, and Gγ, in the olfactory receptor organs of adult Drosophila melanogaster. Chemical Senses. 2010;35(3):183–193. doi: 10.1093/chemse/bjp095. [DOI] [PubMed] [Google Scholar]
- Breslin P.A.S., Beauchamp G.K. Salt enhances flavour by suppressing bitterness. Nature. 1997;387(6633):563. doi: 10.1038/42388. [DOI] [PubMed] [Google Scholar]
- Carey R.M., Kim T., Cohen N.A., Lee R.J., Nead K.T. Impact of sweet, umami, and bitter taste receptor (TAS1R and TAS2R) genomic and expression alterations in solid tumors on survival. Scientific Reports. 2022;12 doi: 10.1038/s41598-022-12788-z. Article 8937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandrashekar J., Mueller K.L., Hoon M.A., Adler E., Feng L., Guo W.…Ryba N.J. T2Rs function as bitter taste receptors. Cell. 2000;100(6):703–711. doi: 10.1016/s0092-8674(00)80706-0. [DOI] [PubMed] [Google Scholar]
- Chen N.Y., Chen T., Shi J., Sun H., Ji L. Two new phenolic glucosides from Sphallerocarpus gracilis. Indian Journal of Chemistry. 1997;36B:107–109. [Google Scholar]
- Dawid C., Henze A., Frank O., Glabasnia A., Rupp M., Büning K., Orlikowski D., Bader M., Hofmann T. Structural and sensory characterization of key pungent and tingling compounds from black pepper (Piper nigrum L.) Journal of Agricultural and Food Chemistry. 2012;60(11):2884–2895. doi: 10.1021/jf300036a. [DOI] [PubMed] [Google Scholar]
- De Giorgio R., Mazzoni M., Vallorani C., Latorre R., Bombardi C., Bacci M.L.…Clavenzani P. Regulation of α-Transducin and α-Gustducin expression by a high protein diet in the pig gastrointestinal tract. PLoS One. 2016;11(2) doi: 10.1371/journal.pone.0148954. Article e0148954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pizio A., Niv M.Y. Promiscuity and selectivity of bitter molecules and their receptors. Bioorganic & Medicinal Chemistry. 2015;23(14):4082–4091. doi: 10.1016/j.bmc.2015.04.025. [DOI] [PubMed] [Google Scholar]
- Duffy V.B., Hayes J.E., Davidson A.C., Kidd J.R., Kidd K.K., Bartoshuk L.M. Vegetable intake in college-aged adults is explained by oral sensory phenotypes and TAS2R38 genotype. Chemosensory Perception. 2010;3:137–148. doi: 10.1007/s12078-010-9079-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duggan T., Gilch J., Stark T.D., Dawid C., Hofmann T. Identification, quantitation and sensory elucidation of off-taste compounds in wheat bran. Journal of Food Bioactives. 2022;20:29–39. doi: 10.31665/JFB.2022.18326. [DOI] [Google Scholar]
- Feng S., Gmitter F.G., Jr., Grosser J.W., Wang Y. Identification of key flavor compounds in citrus fruits: A flavoromics approach. Food Science and Technology. 2021;1(11):2076–2085. doi: 10.1021/acsfoodscitech.1c00304. [DOI] [Google Scholar]
- Fredriksson R., Lagerström M.C., Lundin L.G., Schiöth H.B. The G-protein-coupled receptors in the human genome form five main families. Phylogenetic analysis, paralogon groups, and fingerprints. Molecular Pharmacology. 2003;63(6):1256–1272. doi: 10.1124/mol.63.6.1256. [DOI] [PubMed] [Google Scholar]
- Frisoli T.M., Schmieder R.E., Grodzicki T., Messerli F.H. Salt and hypertension: Is salt dietary reduction worth the effort? The American Journal of Medicine. 2012;125(5):433–439. doi: 10.1016/j.amjmed.2011.10.023. [DOI] [PubMed] [Google Scholar]
- Ganchrow J.R., Steiner J.E., Daher M. Neonatal facial expressions in response to different qualities and intensities of gustatory stimuli. Infant Behavior and Development. 1983;6(4):473–484. doi: 10.1016/S0163-6383(83)90301-6. [DOI] [Google Scholar]
- Gordon R.M., Washington T.L., Sims C.A., Goodrich-Schneider R., Baker S.M., Yagiz Y., Gu L. Performance of macroporous resins for debittering HLB-affected grapefruit juice and its impacts on furanocoumarin and consumer sensory acceptability. Food Chemistry. 2021;352 doi: 10.1016/j.foodchem.2021.129367. Article 129367. [DOI] [PubMed] [Google Scholar]
- Greene T.A., Alarcon S., Thomas A., Berdougo E., Doranz B.J., Breslin P.A., Rucker J.B. Probenecid inhibits the human bitter taste receptor TAS2R16 and suppresses bitter perception of salicin. PLoS One. 2011;6(5) doi: 10.1371/journal.pone.0020123. Article e20123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Günther-Jordanland K., Dawid C., Dietz M., Hofmann T. Key phytochemicals contributing to the bitter off-taste of oat (Avena sativa L.) Journal of Agricultural and Food Chemistry. 2016;64(51):9639–9652. doi: 10.1021/acs.jafc.6b04995. [DOI] [PubMed] [Google Scholar]
- Herman M.A., Birnbaum M.J. Molecular aspects of fructose metabolism and metabolic disease. Cell Metabolism. 2021;33(12):2329–2354. doi: 10.1016/j.cmet.2021.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Homma R., Yamashita H., Funaki J., Ueda R., Sakurai T., Ishimaru Y., Abe K., Asakura T. Identification of bitterness-masking compounds from cheese. Journal of Agricultural and Food Chemistry. 2012;60(18):4492–4499. doi: 10.1021/jf300563n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kikuzaki H., Sato A., Mayahara Y., Nakatani N. Galloylglucosides from berries of Pimenta dioica. Journal of Natural Products. 2000;63(6):749–752. doi: 10.1021/np9906121. [DOI] [PubMed] [Google Scholar]
- Lang T., Di Pizio A., Risso D., Drayna D., Behrens M. Activation profile of Tas2r2, the 26th human bitter taste receptor. Molecular Nutrition & Food Research. 2023;67(11) doi: 10.1002/mnfr.202200775. Article 2200775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law M.R., Morris J.K. By how much does fruit and vegetable consumption reduce the risk of ischaemic heart disease? European Journal of Clinical Nutrition. 1998;52(8):549–556. doi: 10.1038/sj.ejcn.1600603. [DOI] [PubMed] [Google Scholar]
- Ley J.P., Blings M., Paetz S., Krammer G.E., Bertram H.J. New bitter-masking compounds: Hydroxylated benzoic acid amides of aromatic amines as structural analogues of homoeriodictyol. Journal of Agricultural and Food Chemistry. 2006;54(22):8574–8579. doi: 10.1021/jf0617061. [DOI] [PubMed] [Google Scholar]
- Ley J.P., Krammer G., Reinders G., Gatfield I.L., Bertram H.J. Evaluation of bitter masking flavanones from Herba Santa (Eriodictyon californicum (H. & A.) Torr., Hydrophyllaceae) Journal of Agricultural and Food Chemistry. 2005;53(15):6061–6066. doi: 10.1021/jf0505170. [DOI] [PubMed] [Google Scholar]
- Li J., Pan L., Fletcher J.N., Lv W., Deng Y., Vincent M.A.…Kinghorn A.D. In vitro evaluation of potential bitterness-masking terpenoids from the Canada goldenrod (Solidago canadensis) Journal of Natural Products. 2014;77(7):1739–1743. doi: 10.1021/np5001413. [DOI] [PubMed] [Google Scholar]
- Li X., Servant G., Tachdjia C. The discovery and mechanism of sweet taste enhancers. Biomolecular Concepts. 2011;2(4):327–332. doi: 10.1515/bmc.2011.021. [DOI] [PubMed] [Google Scholar]
- Lipchock S.V., Mennella J.A., Spielman A.I., Reed D.R. Human bitter perception correlates with bitter receptor messenger RNA expression in taste cells. The American Journal of Clinical Nutrition. 2013;98(4):1136–1143. doi: 10.3945/ajcn.113.066688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luyen B.T.T., Tai B.H., Thao N.P., Yang S.Y., Cuong N.M., Kwon Y.I.…Kim Y.H. A new phenylpropanoid and an alkylglycoside from Piper retrofractum leaves with their antioxidant and α-glucosidase inhibitory activity. Bioorganic & Medicinal Chemistry Letters. 2014;24(17):4120–4124. doi: 10.1016/j.bmcl.2014.07.057. [DOI] [PubMed] [Google Scholar]
- Maehashi K., Huang L. Bitter peptides and bitter taste receptors. Cellular and Molecular Life Sciencies. 2009;66(10):1661–1671. doi: 10.1007/s00018-009-8755-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik V.S., Popkin B.M., Bray G.A., Després J.P., Willett W.C., Hu F.B. Sugar-sweetened beverages and risk of metabolic syndrome and type 2 diabetes: A meta-analysis. Diabetes Care. 2010;33(11):2477–2483. doi: 10.2337/dc10-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso G., Borgonovo G., Scaglioni L., Bassoli A. Phytochemicals from Ruta graveolens activate TAS2R bitter taste receptors and TRP channels involved in gustation and nociception. Molecules. 2015;20(10):18907–18922. doi: 10.3390/molecules201018907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin S.K., McKinnon P.J., Margolskee R.F. In: Chemical signals in vertebrates 6. Doty R.L., Müller-Schwarze D., editors. Springer; 1992. α Gustducin: A taste cell specific G protein subunit closely related to the α transducins; pp. 9–11. [Google Scholar]
- McLaughlin S.K., McKinnon P.J., Margolskee R.F. Gustducin is a taste-cell-specific G protein closely related to the transducins. Nature. 1992;357(6379):563–569. doi: 10.1038/357563a0. [DOI] [PubMed] [Google Scholar]
- McMurray J.E. In: Organic chemistry. 8th ed. McMurray J.E., editor. Cengage Learning; 2011. Benzene and aromaticity; pp. 451–477. [Google Scholar]
- McMurray J.E. In: Organic chemistry. 8th ed. McMurray J.E., editor. Cengage Learning; 2011. Chemistry of benzene; pp. 478–524. [Google Scholar]
- Melis M., Errigo A., Crnjar R., Pes G.M., Tomassini Barbarossa I. TAS2R38 bitter taste receptor and attainment of exceptional longevity. Scientific Reports. 2019;9(1) doi: 10.1038/s41598-019-54604-1. Article 18047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella J.A., Pepino M.Y., Beauchamp G.K. Modification of bitter taste in children. Developmental Psychobiology. 2003;43(2):120–127. doi: 10.1002/dev.10127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella J.A., Reed D.R., Mathew P.S., Roberts K.M., Mansfield C.J. “A spoonful of sugar helps the medicine go down”: Bitter masking by sucrose among children and adults. Chemical Senses. 2015;40(1):17–25. doi: 10.1093/chemse/bju053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyerhof W., Batram C., Kuhn C., Brockhoff A., Chudoba E., Bufe B., Appendino G., Behrens M. The molecular receptive ranges of human TAS2R bitter taste receptors. Chemical Senses. 2010;35(2):157–170. doi: 10.1093/chemse/bjp092. [DOI] [PubMed] [Google Scholar]
- Muñoz M., Holtheuer J., Wilson L., Urrutia P. Grapefruit debittering by simultaneous naringin hydrolysis and limonin adsorption using naringinase immobilized in agarose supports. Molecules. 2022;27(9):2867–2879. doi: 10.3390/molecules27092867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowak S., Di Pizio A., Levit A., Niv M.Y., Meyerhof W., Behrens M. Reengineering the ligand sensitivity of the broadly tuned human bitter taste receptor TAS2R14. Biochimica et Biophysica Acta, General Subjects. 2018;1862(10):2162–2173. doi: 10.1016/j.bbagen.2018.07.009. [DOI] [PubMed] [Google Scholar]
- Oya T., Osawa T., Kawakishi S. Spice constituents scavenging free radicals and inhibiting pentosidine formation in a model system. Bioscience, Biotechnology, and Biochemistry. 1997;61(2):263–266. doi: 10.1271/bbb.61.263. [DOI] [PubMed] [Google Scholar]
- Riemer J. Bitterness inhibitors. (U.S. Patent No. 5,336,513) U.S. Patent and Trademark Office. 1994. https://patentcenter.uspto.gov/applications/08163494
- Sharif R., Ahmad S.W., Anjum H., Ramzan N., Malik S.R. Effect of infusion time and temperature on decaffeination of tea using liquid–liquid extraction technique. Journal of Food Process Engineering. 2014;37(1):46–52. doi: 10.1111/jfpe.12058. [DOI] [Google Scholar]
- Suess B., Brockhoff A., Meyerhof W., Hofmann T. The odorant (R)-citronellal attenuates caffeine bitterness by inhibiting the bitter receptors TAS2R43 and TAS2R46. Journal of Agricultural and Food Chemistry. 2016;66(10):2301–2311. doi: 10.1021/acs.jafc.6b03554. [DOI] [PubMed] [Google Scholar]
- Ueda T., Ugawa S., Yamamura H., Imaizumi Y., Shimada S. Functional interaction between T2R taste receptors and G-protein α subunits expressed in taste receptor cells. Journal of Neuroscience. 2003;23(19):7376–7380. doi: 10.1523/JNEUROSCI.23-19-07376.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vilardaga J.P., Agnati L.F., Fuxe K., Ciruela F. G-protein-coupled receptor heteromer dynamics. Journal of Cell Science. 2010;123:4215–4220. doi: 10.1242/jcs.063354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters D.E. How are bitter and sweet tastes related? Trends in Food Science & Technology. 1996;7(12):399–403. doi: 10.1016/S0924-2244(96)10040-6. [DOI] [Google Scholar]
- Welton T. Solvents and sustainable chemistry. Proceedings of the Royal Society A: Mathematical, Physical and Engineering Sciences. 2015;471 doi: 10.1098/rspa.2015.0502. Article 20150502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson C.W., Wagner C.J., Jr., Shaw P.E. Reduction of bitter components in grapefruit and navel orange juices with. beta-cyclodextrin polymers or XAD resins in a fluidized bed process. Journal of Agricultural and Food Chemistry. 1989;37(1):14–18. doi: 10.1021/jf00085a004. [DOI] [Google Scholar]
- Yu Z., Wang Y., Zhao W., Li J., Shuian D., Liu J. Identification of Oncorhynchus mykiss nebulin-derived peptides as bitter taste receptor TAS2R14 blockers by in silico screening and molecular docking. Food Chemistry. 2022;368 doi: 10.1016/j.foodchem.2021.130839. Article 130839. [DOI] [PubMed] [Google Scholar]
- Zhang H.X., Wang Z.Z., Du Z.Z. Sensory-guided isolation and identification of new sweet-tasting dammarane-type saponins from Jiaogulan (Gynostemma pentaphyllum) herbal tea. Food Chemistry. 2022;388 doi: 10.1016/j.foodchem.2022.132981. Article 132981. [DOI] [PubMed] [Google Scholar]
- Zhao T., Chen S., Li H., Xu Y. Identification of 2-hydroxymethyl-3, 6-diethyl-5-methylpyrazine as a key retronasal burnt flavor compound in soy sauce aroma type baijiu using sensory-guided isolation assisted by multivariate data analysis. Journal of Agricultural and Food Chemistry. 2018;66(40):10496–10505. doi: 10.1021/acs.jafc.8b03980. [DOI] [PubMed] [Google Scholar]
- Zhao W., Li D., Wang Y., Kan R., Ji H., Su L.…Li J. Identification and molecular docking of peptides from Mizuhopecten yessoensis myosin as human bitter taste receptor T2R14 blockers. Food & Function. 2021;12(23):11966–11973. doi: 10.1039/d1fo02447g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Data Availability Statement
Data will be made available on request.