Abstract
Chronic infection with hepatitis B virus (HBV) is one of the major etiological factors in the development of human hepatocellular carcinoma. Transgenic mice that express the HBV X protein (HBx) have previously been shown to be more sensitive to the effects of hepatocarcinogens, although the mechanism for this cofactor role remains unknown. The ability of HBx to inhibit DNA repair in transiently transfected cell lines suggests one possible pathway. In the present study, primary hepatocytes isolated from transgenic mice that possess the HBV X gene under the control of the human α-1-antitrypsin regulatory region (ATX mice) were found to be deficient in their ability to conduct unscheduled DNA synthesis in response to UV-induced DNA damage. In order to measure the impact of HBx expression on DNA repair in vivo, double-transgenic mice that express HBx and possess a bacteriophage lambda transgene were sacrificed at 30, 90, and 240 days of age. Mutation frequency was determined for high-molecular-weight liver DNA of ATX and control mice by functional analysis of the lambda transgene. Expression of HBx did not significantly increase the accumulation of spontaneous mutations. These results are consistent with previous studies of HBx transgenic mice in which no effect of HBx on liver histology was apparent. This new animal model provides a powerful system in which to investigate the in vivo cooperation between HBx expression and environmental carcinogens.
The hepatitis B virus (HBV) X protein (HBx) is a 17-kDa regulatory protein necessary for the establishment of hepadnaviral infection in woodchucks and, presumably, in all mammals (9, 65). Detectable in both the cytoplasm and nucleus of infected cells (13, 38, 48, 56), the essential role(s) for HBx in the life cycle of the virus remains to be established. Although HBx does not bind DNA directly, it is capable of transactivating cellular genes (reviewed in references 6, 8, 45, and 51) and can induce protein kinase signaling cascades (3, 13, 29, 37). In addition to interacting with numerous cellular proteins (reviewed in reference 16), HBx demonstrates a deoxy-ATPase activity (12).
Chronic infection with HBV is considered a major risk factor for the development of human hepatocellular carcinoma (HCC) (1, 49), but the exact mechanism(s) by which HBV infection leads to the development of liver cancer remains to be elucidated. An increased rate of hepatocyte cell death and regeneration caused by the host immune response to viral antigens undoubtedly contributes to the pathology of chronic HBV infection (reviewed in references 11 and 49). Several lines of evidence indicate that HBx may also contribute to the development of HCC. Although the majority of HBx transgenic mouse lines do not demonstrate an increased incidence of liver cancer under normal conditions (5, 14, 21, 34, 39, 43), such mice are more susceptible to the tumorigenic effects of hepatocarcinogens (14, 50) or the oncogene c-myc (54). Based on these observations, HBx is believed to be an important cofactor in HBV-associated liver cancer.
HBx may influence the development of liver cancer by multiple mechanisms. Its transactivation of cellular genes (6, 8, 45, 51) as well as the induction of one or more signaling pathways (3, 13, 26, 29, 35, 37) may lead to the up-regulation of cellular oncogenes (54, 63) or to changes in cell cycle progression (4) and/or regulation. Indeed, HBx was shown to induce cell cycle progression in quiescent skin fibroblasts (31). HBx has also been shown to interact with at least 3 proteins or protein complexes that are directly involved in DNA repair: p53 (17, 57), TFIIH (42), and DDB1 (previously known as UVDDB1 or XAP1) (33, 48). The latter data suggests that HBx expression could cause an accumulation of DNA mutations by indirectly compromising the repair ability of cells. This is particularly intriguing given the strong correlation between chronic HBV infection, aflatoxin B1 exposure, and the development of HCC (18, 53, 62).
Experimental evidence that HBx inhibits DNA repair comes from studies using transiently transfected immortalized tissue culture cell lines and primary mouse hepatocytes. These studies concluded that there was significant inhibition (25 to 60%) of global nucleotide excision repair (NER) in response to either UV or aflatoxin B1 exposure (2, 20, 25, 41). Although the studies clearly demonstrate that HBx compromises NER in eukaryotic cells, the actual mechanism by which it inhibits repair remains unknown. In addition, the impact of HBx-mediated inhibition of repair in vivo remains to be established.
The purpose of the present study was to extend the cell culture DNA repair studies into an in vivo setting. The unscheduled DNA synthesis assay was first used to demonstrate that primary hepatocyte cultures derived from HBx transgenic mice are deficient in NER relative to primary hepatocytes derived from a wild-type mouse. In order to measure the impact of HBx on the accumulation of mutations in vivo, we generated a double-transgenic mouse line that expresses HBx and possesses an integrated lambda transgene, allowing measurement of mutation frequency (MF) (30). Histological examination of mouse liver tissue did not reveal any consistent liver abnormalities in these double-transgenic mice. The expression of HBx did not significantly increase the accumulation of spontaneous mutations in high-molecular-weight (HMW) liver DNA, nor did it alter the spectrum of mutations that occurred. Importantly, these studies establish and characterize a new animal model in which the effects of HBx on MF can be measured upon exposure to mutagenic agents, a situation that more closely resembles the synergism between chronic HBV infection and environmental carcinogens in the etiology of human liver cancer.
MATERIALS AND METHODS
Transgenic mice.
Transgenic mice harboring the X gene (nucleotides 1376 to 1840 of subtype adw2) under the control of the human α-1-antitrypsin inhibitor regulatory region (ATX mice) (34, 50) were maintained by breeding of hemizygous ATX males (ICR × B6C3) with wild-type females (ICR). Hemizygous ATX females (ICR × B6C3) were then mated with homozygous λ males (C57BL/6 Big Blue) (30) obtained from Stratagene Corporation, and the male F1 progeny were used for this study. At appropriate time points (30, 90, and 240 days of age), mice were sacrificed and portions of three liver lobes were fixed in 10% neutral buffered formalin (16 h) and 70% ethanol. Tissues were paraffin embedded, and coded hematoxylin-and-eosin-stained sections submitted for histological analysis by M.J.F. Remaining tissue was frozen in liquid nitrogen and stored for later experiments.
Southern hybridization.
HMW DNA was purified from transgenic mouse tail samples by using the Wizard Genomic DNA Purification Kit (Promega). DNA was digested with BamHI, was resolved on a 1% agarose gel, and was transferred to a nylon membrane (Boehringer Mannheim). X-gene-specific probe DNA was prepared by PCR amplification of HBV plasmid DNA by using the primer set 5′-ATGGCTGCTAGGCTGTACTG-3′ and 5′-CTACAAGAGATGATTAGGCAGA-3′. Probe DNA for the cII gene was amplified from 3 μg of homozygous Big Blue mouse DNA by using the cII-specific primer set 5′-ACCACACCTATGGTGTATGCA-3′ and 5′-GTCATAATGACTCCTGTTGATAG-3′. DNA was radiolabeled with [32P]dCTP (3,000 Ci/mmole) (ICN) by using the Rediprime II random priming labeling kit (Amersham Pharmacia). Standard conditions for hybridizations and X-ray film exposures were used (46). All mice (ATX and wild type) used for this study possessed approximately 40 copies of the λ transgene. ATX mice also harbored 10 to 15 copies of the ATX transgene.
Immunoprecipitation and Western blot verification of HBx expression.
Tissue extracts were prepared by homogenizing liver tissue in Extraction buffer (50 mM Tris-HCl, pH 8.0; 100 mM NaCl; and 1% NP-40). Immunoprecipitation and Western blot analysis were used to verify expression of HBx, as described previously (50). Briefly, following electrophoresis on sodium dodecyl sulfate–15% polyacrylamide gels, separated proteins were transferred to nitrocellulose filters. The presence or absence of HBx was verified by using rabbit anti-HBx polyclonal serum, an avidin-biotin detection kit (Vector Laboratories), and chemiluminescence (Amersham Pharmacia).
Isolation of primary hepatocytes.
Primary mouse hepatocytes from ATX and wild-type mice were obtained by liver perfusion (59). In brief, mice were anesthetized and a V incision was made to expose the internal organs. The flushed liver was perfused with Earle's balanced salt solution plus collegenase (0.3 mg/ml) and ovomucoid trypsin inhibitor (0.04 mg/ml). After several minutes of perfusion, the liver was removed, and loose hepatocytes were dispersed in Williams Media E (WME) plus 1% fetal calf serum. Following filtration to remove nonhepatocyte debris, cells were washed and plated on 60-mm-diameter tissue culture dishes. After 2 h of incubation at 37°C, the adherent hepatocytes were refed with serum-free WME (WME plus 30 nM selenium, 10 μg of insulin per ml, 10 μg transferrin, 100 nM somatotropin, 1 μM thyroid hormone T3, 1 μM dexamethasone, 50 ng of epidermal growth factor per ml, and 25 μg of gentamycin per ml [15]) and were incubated at 37°C overnight.
UDS assay.
The induction of unscheduled DNA synthesis (UDS) by UV and the subsequent incorporation of [3H]thymidine into repaired DNA was performed as previously described (2). Approximately 24 h after attachment, hepatocyte cultures were washed with buffered saline and were exposed to 0 or 20 J of UV (254 nm) per m2 in a Stratalinker (Stratagene Biocrest). Cultures were immediately refed with serum-free WME plus 5 μCi [3H]thymidine per ml and were incubated for 1 h at 37°C. Cells were harvested by incubation with phosphate-buffered saline plus trypsin and were rinsed with phosphate-buffered saline. HMW DNA was isolated from the cell suspensions by using the QIAamp tissue kit (QIAGEN). The amount of [3H]thymidine incorporation was measured by standard liquid scintillation counting techniques. DNA concentration was determined spectrophotometrically and used to normalize values obtained by liquid scintillation counting. Results reported for each animal were based on counts per minute per microgram of DNA calculated from at least three plate replicates (three UV-exposed and three unexposed plates). To calculate fold induction, average values for UV-exposed plates were divided by the average value for unexposed hepatocytes of the same genotype. To facilitate comparison among different matched sets of hepatocyte cultures, fold induction values for wild-type cultures were set to 100%, and the values for ATX cultures were then converted to percentage relative to wild type in that experiment.
Packaging of lambda phage DNA and plaque assays.
Liver HMW DNA for use in the Big Blue mutagenesis assay was isolated by using the RecoverEase system (Stratagene Biocrest) under conditions recommended by the supplier. Importantly, the RecoverEase system uses no phenol or chemical extractions that may further damage the DNA. Liver HMW DNA was subsequently incubated with Transpack (Stratagene Biocrest) to excise and package the lambda phage genome. Packaged DNA was then incubated with a suspension (optical density at 600 nm = 0.5) of Escherichia coli G1250 (G1217hflA::Tn5 hflB29 Tn10). Determination of relative mutation frequency was accomplished by assaying for inactivating mutations in the bacteriophage lambda cII gene (24). A portion of diluted culture (1/100) was then plated and incubated at 37°C for 24 h to determine total (cII− plus cII+) PFU. The remaining culture was plated and incubated at 24°C for 48 h, conditions allowing growth of only mutant plaques. The relative MF was calculated as the ratio of mutant PFU to total PFU. All mutant plaques were subsequently isolated and replated under the same conditions to verify the mutant phenotype.
DNA sequencing.
DNA obtained from cored mutant plaques (heated to 95°C for 5 min) was amplified by a standard PCR procedure using cII-specific primers (listed above). Amplified cII DNA was then sequenced by using a Thermosequenase cycle sequencing kit (Amersham Pharmacia), and the products were resolved by electrophoresis on 6% polyacrylamide-urea gels.
TUNEL assays.
Sections cut from paraffin-embedded tissues were analyzed for apoptosis by using a commercially available kit (Trevigen) and the manufacturer's protocol. Following the terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) reaction, cells were counterstained, and the percentage of apoptotic cells was determined by counting 10 consecutive fields, each containing approximately 300 hepatocytes.
Statistical analysis.
Probability calculations for the UDS, mutation frequency, and apoptosis assays were performed by using a two-tailed Student's t test (Microsoft Excel software package). Standard deviation (SD) and mean values were calculated by using the same software. The MF value of one ATX mouse lay more than two SDs from the mean and, on that basis, was excluded from the final reported results.
RESULTS
Inhibition of UDS in primary ATX mouse hepatocytes.
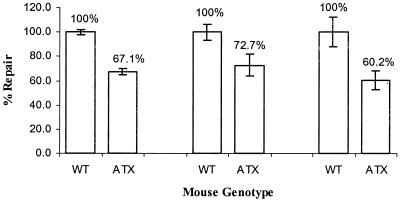
The expression of HBx is known to inhibit NER in transiently transfected cell culture, and we hypothesized that HBx might exert a similar effect in primary mouse hepatocytes. To demonstrate this, primary hepatocyte cultures from ATX and wild-type littermates were obtained by collagenase perfusion, and the ability of these cells to repair UV-induced DNA damage was then measured in the UDS assay as previously described (2). Compared to hepatocytes from wild-type mice, hepatocytes derived from ATX mice demonstrated a significant reduction in their ability to repair UV damage (Fig. 1) (P < 0.012). The 27 to 40% reduction measured in murine ATX hepatocytes is similar to the level of inhibition observed in transiently transfected human HepG2 cell cultures (2, 20). The expression of HBx in hepatocyte cultures obtained from ATX mice was verified by immunoprecipitation and Western blot analyses (data not shown). These results establish that the murine NER pathway is inhibited in hepatocytes that constitutively express HBx and validate the use of ATX mice for further studies on inhibition of DNA repair.
FIG. 1.
Induction of UDS in primary ATX and wild-type mouse hepatocytes. The incorporation of [3H]thymidine was measured in cultures harvested 60 min after exposure to 0 or 20 J of UV per m2. Results shown were obtained from three separate experiments using sex-matched ATX and wild-type mouse littermates. Mean and SD values were calculated from three unexposed- and three exposed-plate replicates for each genotype. To facilitate comparison among different matched sets of hepatocyte cultures, fold induction values for wild-type cultures were set to 100%, and the values for ATX cultures were then converted to a percentage relative to the wild type.
Generation of double-transgenic mice.
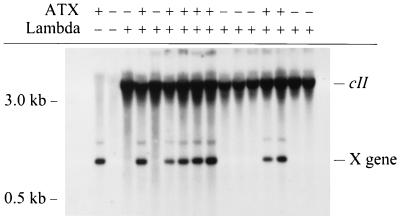
After demonstrating that NER was inhibited in ATX hepatocytes cultured in vitro, we wanted to examine the effect of HBx expression on the accumulation of endogenous mutations in vivo. To accomplish this goal, hemizygous ATX females were crossed with homozygous Big Blue males (Stratagene Biocrest) (30) which contain 80 copies of an intact bacteriophage λ transgene. Progeny mice were genotyped by Southern blot hybridization (Fig. 2). For the purposes of clarity, single-transgenic mice harboring only λ are referred to as wild-type mice, while double-transgenic mice harboring both λ and ATX transgenes are referred to as ATX mice. Male mice were subsequently sacrificed (at 30, 90, and 240 days of age), and portions of their livers were used for purification of HMW DNA.
FIG. 2.
Detection of ATX and λ transgenes by Southern blot hybridization. Transgenic male mice used for this study were generated as described in Materials and Methods. The migration of cII and X transgenes is indicated at the right.
Determination of in vivo mutation frequency.
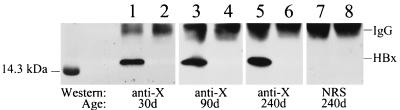
Purified HMW DNA was packaged into λ phage and used to infect E. coli. The relative MF of each individual liver was then determined as described in Materials and Methods. Comparison of the relative mutation frequencies in 30-day-old mice did not reveal any significant differences between ATX and wild-type mice (Table 1), (P > 0.29). Similarly, animals sacrificed at 90 and 240 days did not show any genotype-specific differences in the accumulation of mutations (all P values > 0.34). As a positive control, treatment of a mouse with the liver carcinogen diethylnitrosamine resulted in a sixfold increase in MF, verifying that this assay reliably detects changes in mutation accumulation (data not shown). Expression of HBx was confirmed for all ATX mice in these experiments by using a combination immunoprecipitation-Western blot procedure (Fig. 3). These results demonstrate that the expression of HBx in vivo does not influence the frequency of spontaneous mutations in liver DNA.
TABLE 1.
Determination of MF in transgenic mice
| Age (days) | Genotype | Animal no. | Total PFU (105)a | Mutant PFUb | MF (10−5) | Mean ± SD (10−5) |
|---|---|---|---|---|---|---|
| 30 | ATX | 2087 | 2.60 | 9 | 3.46 | |
| 2089 | 3.98 | 17 | 4.27 | |||
| 2141 | 4.07 | 14 | 3.44 | |||
| 2142 | 4.18 | 17 | 4.06 | 3.10 ± 1.02 | ||
| 2194 | 8.25 | 11 | 1.33 | |||
| 2195 | 3.94 | 10 | 2.54 | |||
| 2200 | 2.68 | 7 | 2.61 | |||
| Wild type | 2086 | 6.37 | 16 | 2.51 | ||
| 2088 | 3.92 | 19 | 4.85 | |||
| 2161 | 5.52 | 19 | 3.44 | |||
| 2165 | 5.94 | 25 | 4.21 | 3.71 ± 1.08 | ||
| 2193 | 4.33 | 18 | 4.16 | |||
| 2196 | 4.23 | 20 | 4.73 | |||
| 2197 | 3.41 | 7 | 2.05 | |||
| 90 | ATX | 2046 | 4.10 | 13 | 3.17 | |
| 2048 | 4.10 | 15 | 3.66 | 3.18 ± 0.47 | ||
| 2071 | 10.1 | 29 | 2.72 | |||
| Wild type | 2053 | 3.18 | 12 | 3.77 | ||
| 2073 | 3.45 | 11 | 3.18 | 3.59 ± 0.35 | ||
| 2074 | 4.72 | 18 | 3.81 | |||
| 240 | ATX | 2016 | 6.75 | 27 | 4.00 | |
| 2020 | 5.77 | 17 | 2.95 | 3.43 ± 0.53 | ||
| 2029 | 3.60 | 12 | 3.33 | |||
| Wild type | 2017 | 3.06 | 10 | 3.26 | ||
| 2023 | 3.78 | 12 | 3.18 | 3.06 ± 0.28 | ||
| 2033 | 9.12 | 25 | 2.74 |
Calculated from dilutions incubated at 37°C.
Isolated mutants replated and incubated at 24°C to verify phenotype.
FIG. 3.
Detection of HBx in transgenic mouse liver tissue. Shown is a representative result using immunoprecipitation and Western blot hybridization to detect HBx in 30-day-old (lanes 1 and 2), 90-day-old (lanes 3 and 4), and 240-day-old (lanes 5 to 8) mice. HBx expression was clearly demonstrated in all ATX mice (lanes 1, 3, and 5) and absent in all wild-type mice (lanes 2, 4, and 6). No protein bands were detected for 240-day-old ATX and wild-type mice (lanes 7 and 8, respectively) when nonspecific rabbit serum was substituted for rabbit anti-HBx polyclonal serum during the Western blot hybridization procedure. All mice used in this study were similarly screened for HBx expression by this method (data not shown). IgG, immunoglobulin G.
Sequence analysis of cII mutants.
While HBx expression did not result in a measurable increase in MF, it was conceivable that it could cause a change in the spectrum of mutations by inhibiting the repair of only a certain subset of DNA lesions. To examine this possibility, the cII genes of mutant phage recovered from a representative ATX mouse were sequenced. All identified mutations resulted in amino acid changes within the coding region of cII (data not shown) and were either transition or transversion events at G/C base pairs (Table 2). This result is consistent with the mutation spectrum of lambda cII and lacI genes reported by other studies utilizing lambda-transgenic mice (24, 44). As in those studies, the majority of transition events (75%) also occurred at CpG islands, a characteristic unique to eukaryotes (23). Based on this evidence, we conclude that the majority of the mutations recorded in this assay occurred in the mouse and not in the E. coli strain used to propagate the bacteriophage. These results also confirm that HBx expression does not cause gross changes in the spectrum of endogenous DNA mutations in vivo.
TABLE 2.
cII mutation spectrum in ATX versus wild-type
| Mutation type | Percent occurrence in studya:
|
||
|---|---|---|---|
| Presentb | Jakubczak et al.c | Ross and Leavittd | |
| Transitions | |||
| G/C→A/T (% at CpG) | 44 (75) | 74 (86) | 50 (76) |
| A/T→G/C | 0 | 0 | 4 |
| Transversions | |||
| G/C→T/A (% at CpG) | 33 (0) | 11 (0) | 16 (38) |
| G/C→C/G | 11 | 0 | 2 |
| A/T→T/A | 0 | 11 | 2 |
| A/T→C/G | 0 | 0 | 0 |
| Othere | 11 | 5 | 26 |
| Total isolates | 9 | 19 | 50 |
Percentage of total mutants bearing the specific mutation type noted.
Mutant cII isolates from 30-day-old ATX mouse 2087.
Mutant cII isolates from mouse mammary tissue.
Mutation spectrum of mutant lacI isolates from mouse liver tissue.
Includes deletions, additions, and jackpot mutations (mutations that occur early in the development of the mouse and are amplified by cellular replication).
Effect of HBx on apoptosis in vivo.
Several recent studies have demonstrated that HBx can promote the induction of apoptosis under specific conditions (28, 40, 52, 55). As DNA damage may also induce apoptosis (61), we considered the possibility that HBx-mediated inhibition of DNA repair could lead to an increase in mutation frequency that was not observable due to a simultaneous induction of apoptosis in ATX mouse hepatocytes. To test this hypothesis, paraffin sections of 30-day-old ATX and wild-type mice were analyzed for apoptosis in situ by TUNEL assay. The prevalence of TUNEL-positive cells (0.25 to 0.33%) was similar to that previously reported for hepatocytes in adult C57BL/6 mice (55). Importantly, no significant difference in the incidence of TUNEL-positive cells was observed between ATX and wild-type mice (Table 3). These results demonstrate that, in the absence of exogenous DNA damage, HBx does not significantly alter the incidence of apoptosis in our ATX mice.
TABLE 3.
% Apoptosis in mouse liver tissue
| Genotype | Animal no.a | % Apoptosisb | Mean ± SD |
|---|---|---|---|
| ATX | 2087 | 0.3 | |
| 2089 | 0.4 | 0.25 ± 0.13 | |
| 2141 | 0.1 | ||
| 2142 | 0.2 | ||
| Wild type | 2086 | 0.1 | |
| 2088 | 0.6 | 0.33 ± 0.22 | |
| 2161 | 0.4 | ||
| 2165 | 0.2 |
Mice harvested at 30 days of age.
Determined by counting TUNEL-positive cells in 10 fields of approximately 300 hepatocytes per field.
Histological examination of mouse liver tissue.
Liver tissue preparations for both ATX and wild-type transgenic mice were coded and submitted for histological examination to assess the impact of HBx expression on the architecture of the liver. All mice appeared normal at the time of sacrifice and, with one exception, did not display any gross histological abnormalities. A single 240-day-old wild-type mouse displayed diffuse perivenous and sinusoidal accumulation of lymphocytes consistent with the development of leukemia. Most histological observations appeared to be random in distribution, with isolated foci of inflammation being the most frequently cited abnormality (Table 4). Although mild nuclear polymorphism (anisocytosis) was noted in several animals, it did not increase in severity with age. These observations indicate that expression of HBx does not lead to any consistent histopathological changes in the liver tissues of ATX-λ double-transgenic mice.
TABLE 4.
Histological observations of liver tissue from ATX and wild-type transgenic mice
| Age at sacrifice (days) | Genotype | No. examineda | Histopathological changes |
|---|---|---|---|
| 30 | ATX | 9 | Decreased glycogen in three animals; rare inflammatory foci; occasional slight anisocytosis |
| Wild type | 10 | Rare inflammatory foci; minimal increased mitosis in six animals | |
| 90 | ATX | 6 | Minimal anisocytosis; rare inflammatory foci |
| Wild type | 5 | Minimal anisocytosis; rare inflammatory foci | |
| 240 | ATX | 4 | Mostly normal, increased anisocytosis; rare inflammatory foci |
| Wild type | 3 | Rare foci of acute inflammation |
Number of animals sacrificed for histological examination at that time point. Data was collected from three separate lobes of liver tissue per animal.
DISCUSSION
Evidence linking HBx expression to the development of HBV-associated liver cancer comes, in part, from studies of transgenic mouse lines. Although the majority of these mouse lines do not show an increased incidence of liver cancer, they are more susceptible to the effects of the hepatocarcinogen diethylnitrosamine and activated oncogenes (14, 50, 54). These observations led us to hypothesize that HBx may act as a cofactor in HCC development by inhibiting DNA repair.
The ability of HBx to inhibit the NER pathway has been demonstrated in vitro. A study from our laboratory showed that human hepatoblastoma cell cultures (HepG2) transiently transfected with an HBx expression plasmid conducted an average of 45% less UDS in response to UV exposure than control cultures (2). Other research laboratories, using a variety of cell types and both UV and aflatoxin B1 as mutagenic agents, have since reported similar results (20, 25, 41). The physiological importance of this inhibition, however, remains to be explored in vivo. The ATX transgenic mouse line, which constitutively expresses HBx throughout the life span of the animal (50), provided an ideal model system for such studies.
As the expression of HBx is known to inhibit NER in transiently transfected cell cultures, we first determined whether it had a similar effect in transgenic mouse hepatocytes. The ability of primary ATX hepatocytes to conduct UDS in response to DNA damage was found to be 27 to 40% less than that measured in wild-type cultures. This result demonstrates that murine hepatocytes which express HBx are deficient in global NER and that the extent of inhibition is comparable to that seen in human HepG2 cell cultures. These results indicate that the ATX mouse model is valid for further studies on the in vivo effects of this repair-inhibitory property of HBx.
By using a system specifically designed for the measurement of relative mutation frequency in vivo (Big Blue mouse mutagenesis system), we were able to measure the effect of HBx expression on mutation accumulation. Despite the clear expression of HBx in the liver tissue of mice up to 240 days of age, there was no significant difference in MF values between ATX and wild-type mice. In a previous study, ATX mice treated with the carcinogenic agent diethylnitrosamine showed a significant increase in the development of liver foci relative to treated nontransgenic mice by 240 days of age (50). By analogy, the mice used for this study were of sufficient age to reveal a similar effect of HBx on the accumulation of spontaneous mutations. Previous studies using Big Blue mice have established the criteria (number of animals analyzed and number of plaques counted) needed to reliably discern a twofold difference in average MF values (7). Our experimental design met or exceeded these guidelines. Furthermore, the MF values we obtained for both ATX and wild-type mice were comparable to those reported for wild-type mouse liver tissue by other studies (24, 44). Therefore, we conclude that the expression of HBx did not have a significant effect on liver DNA mutation frequency.
The finding that HBx does not alter liver MF in vivo is consistent with previous studies of HBx transgenic mice (5, 14, 21, 39, 43). The contribution of HBx expression to oncogenesis in two transgenic mouse lines that do develop HCC is open to interpretation (27, 64). In one study, all experimental observations were made by using mice generated from a single transgenic founder (64). The ability of HBx to increase the incidence of HCC in a second mouse line, which was prone to the development liver cancer (27), agrees with the hypothesis that HBx contributes to, but is not solely responsible for, oncogenesis.
It is difficult to draw conclusions on the possible effect of HBx on apoptosis. The expression of HBx sensitizes many cell types (primary mouse hepatocytes, Chang liver, and HepG2) to a variety of proapoptotic stimuli such as treatment with tumor necrosis factor alpha, doxirubicin, anti-Fas antibodies, and serum starvation (28, 40, 52) but inhibits apoptosis in other cell types (REV2 and primary human fibroblasts) (19, 58). More relevant to the present study, HBx increased the incidence of apoptosis approximately twofold in 15-day-old transgenic mice (55). Although no increase was seen in adult transgenic mice in that study, neither could HBx be detected. Based on all these studies, we considered the possibility that sustained HBx expression in the ATX mice could lead to an increased rate of apoptosis among cells incurring sufficient DNA damage and that this could minimize our ability to detect HBx-related differences in MF. However, we did not detect any differences in the rate of apoptosis between ATX and wild-type mouse liver tissues at 30 days of age, a time point when HBx expression is readily detectable. Furthermore, the percentage of apoptotic cells determined for both ATX and wild-type mice in the present study correlated with the percentage reported previously for nontransgenic control mice (55). These results indicate that HBx did not have a discernable effect on in vivo liver apoptosis in the double-transgenic mice used for these experiments.
The inability of HBx to influence MF under normal conditions was not an unexpected result. DNA repair in mammalian cells can be classified into four different pathways: direct reversal, mismatch, base excision, and nucleotide-excision-mediated repair. At this time, we have no evidence regarding the possible impact of HBx expression on DNA repair pathways other than NER. Presumably, the low level of mutations incurred by mouse hepatocyte DNA under the conditions used for the present study was the result of oxidative metabolism and replication. These spontaneous lesions are readily removed not only by NER, but by other repair pathways as well (reviewed in references 36, 47, and 60). This redundancy would effectively minimize the impact of HBx-mediated inhibition of repair unless either the DNA damage occurred at a much higher rate (by exposure to mutagenic agents) or the types of lesions induced were repaired exclusively by NER. Consequently, the inhibition of NER by HBx may lead to an increase in MF values only after exposure to exogenous mutagenic agents such as diethylnitrosamine or aflatoxin B1. Experiments using mice treated with hepatocarcinogens are now in progress.
The concept that HBV-infected individuals are more susceptible to the damaging effects of mutagenic agents is very intriguing given what is known about the development of HCC. In certain geographic regions where HBV infection is endemic, a strong correlation exists between HBV status, exposure to aflatoxin B1, and incidence of liver cancer (18, 53, 62). In addition, increased production of reactive oxygen species and oxidative damage to DNA occurs in transgenic mice that develop chronic inflammation in response to the overexpression of the HBV surface protein (22). Eventually, those mice develop cirrhosis and liver cancer. Although the expression of HBx has yet to be demonstrated in HBV surface protein mice, they do possess the X open reading frame and its native promoter region (10).
As the role of HBx itself in virus replication remains to be clearly established, we can only speculate as to whether HBx-mediated inhibition of DNA repair is relevant to the virus life cycle. It is conceivable that inhibition of repair confers a selective advantage to the virus by allowing a higher rate of genetic variability during chronic infection. Alternatively, HBx and cellular repair proteins may be involved in the conversion of partially double-stranded viral DNA intermediates into transcription-competent, covalently closed circular DNA. Finally, HBx may interact with a cellular protein, such as DDB1, to enhance viral replication or gene expression and by doing so, inadvertently inhibit DNA repair. Along this line, studies showing a role for DDB1 in transcription indicate yet another property that could be targeted by HBx (32). The potential for HBx to influence the regulatory functions of cellular proteins that are also involved in DNA repair, such as p53 and DDB1, may play an important role in the life cycle of HBV and is the subject of ongoing research in our laboratory.
In summary, we have generated a novel double-transgenic mouse line in which the impact of HBx expression on the accumulation of DNA mutations in vivo can be measured. Although we found that HBx inhibited NER in mouse hepatocytes cultured in vitro following exposure to UV, it did not have any discernible impact on mutation frequency in vivo in the absence of DNA damage. This evidence, in conjunction with previous studies, implies that exposure to DNA-damaging agents may be necessary before HBx significantly affects mutation frequency. With this animal model, it will now be possible to address such questions in a setting that more closely resembles the development of HCC in humans.
ACKNOWLEDGMENTS
This work was supported by NIH research grant CA54557. C.R.M. was supported by research training grant T32DK07664.
We thank Christopher Wagner, Kristopher Frese, Stephanie Moses, and Melissa Wentz for technical assistance.
REFERENCES
- 1.Beasley R P. Hepatitis B virus. The major etiology of hepatocellular carcinoma. Cancer. 1988;61:1942–1956. doi: 10.1002/1097-0142(19880515)61:10<1942::aid-cncr2820611003>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 2.Becker S A, Lee T H, Butel J S, Slagle B L. Hepatitis B virus X protein interferes with cellular DNA repair. J Virol. 1998;72:266–272. doi: 10.1128/jvi.72.1.266-272.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benn J, Schneider R J. Hepatitis B virus HBx protein activates Ras-GTP complex formation and establishes a Ras, Raf, MAP kinase signaling cascade. Proc Natl Acad Sci USA. 1994;91:10350–10354. doi: 10.1073/pnas.91.22.10350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benn J, Schneider R J. Hepatitis B virus HBx protein deregulates cell cycle checkpoint controls. Proc Natl Acad Sci USA. 1995;92:11215–11219. doi: 10.1073/pnas.92.24.11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Billet O, Grimber G, Levrero M, Seye K A, Briand P, Joulin V. In vivo activity of the hepatitis B virus core promoter:tissue specificity and temporal regulation. J Virol. 1995;69:5912–5916. doi: 10.1128/jvi.69.9.5912-5916.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buendia M A. Hepatitis B viruses and hepatocellular carcinoma. Adv Cancer Res. 1992;59:167–226. doi: 10.1016/s0065-230x(08)60306-1. [DOI] [PubMed] [Google Scholar]
- 7.Callahan J D, Short J M. Transgenic λ/lac 1 mutagenicity assay: statistical determination of sample size. Mutat Res. 1995;327:201–208. doi: 10.1016/0027-5107(94)00191-7. [DOI] [PubMed] [Google Scholar]
- 8.Caselmann W H. Trans-activation of cellular genes by hepatitis B virus proteins: a possible mechanism of hepatocarcinogenesis. Adv Virus Res. 1996;47:253–302. doi: 10.1016/s0065-3527(08)60737-x. [DOI] [PubMed] [Google Scholar]
- 9.Chen H-S, Kaneko S, Girones R, Anderson R W, Hornbuckle W E, Tennant B C, Cote P J, Gerin J L, Purcell R H, Miller R H. The woodchuck hepatitis virus X gene is important for establishment of virus infection in woodchucks. J Virol. 1993;67:1218–1226. doi: 10.1128/jvi.67.3.1218-1226.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chisari F V, Klopchin K, Moriyama T, Pasquinelli C, Dunsford H A, Sell S, Pinkert C A, Brinster R L, Palmiter R D. Molecular pathogenesis of hepatocellular carcinoma in hepatitis B virus transgenic mice. Cell. 1989;59:1145–1156. doi: 10.1016/0092-8674(89)90770-8. [DOI] [PubMed] [Google Scholar]
- 11.Christen S, Hagen T M, Shigenaga M K, Ames B N. Chronic inflammation, mutation, and cancer. In: Parsonet J, editor. Microbes and malignancy: infection as a cause of human cancers. New York, N.Y: Oxford University Press; 1999. pp. 35–88. [Google Scholar]
- 12.De-Medina T, Haviv I, Noiman S, Shaul Y. The X protein of hepatitis B virus has a ribo-deoxy ATPase activity. Virology. 1994;202:401–407. doi: 10.1006/viro.1994.1356. [DOI] [PubMed] [Google Scholar]
- 13.Doria M, Klein N, Lucito R, Schneider R J. The hepatitis B virus HBx protein is a dual specificity cytoplasmic activator of Ras and nuclear activator of transcription factors. EMBO J. 1995;19:4747–4757. doi: 10.1002/j.1460-2075.1995.tb00156.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dragani T A, Manenti G, Farza H, Della Porta G, Tiollais P, Pourcel C. Transgenic mice containing hepatitis B virus sequences are more susceptible to carcinogen-induced hepatocarcinogenesis. Carcinogenesis. 1989;11:953–956. doi: 10.1093/carcin/11.6.953. [DOI] [PubMed] [Google Scholar]
- 15.Enat R, Jefferson D M, Ruiz-Opazo N, Gatmaitan Z, Leinwand L A, Reid L M. Hepatocyte proliferation in vitro: its dependence on the use of serum-free hormonally defined medium and substrata of extracellular matrix. Proc Natl Acad Sci USA. 1984;81:1411–1415. doi: 10.1073/pnas.81.5.1411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feitelson M A, Duan L X. Hepatitis B virus x antigen in the pathogenesis of chronic infections and the development of hepatocellular carcinoma. Am J Pathol. 1997;150:1141–1157. [PMC free article] [PubMed] [Google Scholar]
- 17.Feitelson M A, Zhu M, Duan L X, London W T. Hepatitis B x antigen and p53 are associated in vitro and in liver tissues from patients with primary hepatocellular carcinoma. Oncogene. 1993;8:1109–1117. [PubMed] [Google Scholar]
- 18.Ghebranious N, Sell S. Hepatitis B injury, male gender, aflatoxin, and p53 expression each contribute to hepatocarcinogenesis in transgenic mice. Hepatology. 1998;27:383–391. doi: 10.1002/hep.510270211. [DOI] [PubMed] [Google Scholar]
- 19.Gottlob K, Fulco M, Levrero M, Graessmann A. The hepatitis B virus HBx protein inhibits caspase 3 activity. J Biol Chem. 1998;273:33347–33353. doi: 10.1074/jbc.273.50.33347. [DOI] [PubMed] [Google Scholar]
- 20.Groisman I J, Koshy R, Henkler F, Groopman J D, Alaoui-Jamali M A. Downregulation of DNA excision repair by the hepatitis B virus-x protein occurs in p53-proficient and p53-deficient cells. Carcinogenesis. 1999;20:479–483. doi: 10.1093/carcin/20.3.479. [DOI] [PubMed] [Google Scholar]
- 21.Guidotti L G, Matzke B, Schaller H, Chisari F V. High-level hepatitis B virus replication in transgenic mice. J Virol. 1995;69:6158–6169. doi: 10.1128/jvi.69.10.6158-6169.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hagen T M, Huang S, Curnutte J, Fowler P, Martinez V, Wehr C M, Ames B N, Chisari F V. Extensive oxidative DNA damage in hepatocytes of transgenic mice with chronic active hepatitis destined to develop hepatocellular carcinoma. Proc Natl Acad Sci USA. 1994;91:12808–12812. doi: 10.1073/pnas.91.26.12808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hill K A, Buettner V L, Glickman B W, Sommer S S. Spontaneous mutations in the Big Blue transgenic system are primarily mouse derived. Mutat Res. 1999;436:11–19. doi: 10.1016/s1383-5742(98)00024-6. [DOI] [PubMed] [Google Scholar]
- 24.Jakubczak J L, Merlino G, French J E, Muller W J, Paul B, Adhya S, Garges S. Analysis of genetic instability during mammary tumor progression using a novel selection-based assay for in vivo mutations in a bacteriophage lambda transgene target. Proc Natl Acad Sci USA. 1996;93:9073–9078. doi: 10.1073/pnas.93.17.9073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jia L, Wang X W, Harris C C. Hepatitis B virus X protein inhibits nucleotide excision repair. Int J Cancer. 1999;80:875–879. doi: 10.1002/(sici)1097-0215(19990315)80:6<875::aid-ijc13>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 26.Kekule A S, Lauer U, Weiss L, Luber B, Hofschneider P H. Hepatitis B virus transactivator HBx uses a tumour promoter signaling pathway. Nature. 1993;361:742–745. doi: 10.1038/361742a0. [DOI] [PubMed] [Google Scholar]
- 27.Kim C-M, Koike K, Saito I, Miyamura T, Jay G. HBx gene of hepatitis B virus induces liver cancer in transgenic mice. Nature. 1991;351:317–320. doi: 10.1038/351317a0. [DOI] [PubMed] [Google Scholar]
- 28.Kim H, Lee H, Yun Y. X-gene product of hepatitis B virus induces apoptosis in liver cells. J Biol Chem. 1998;273:381–385. doi: 10.1074/jbc.273.1.381. [DOI] [PubMed] [Google Scholar]
- 29.Klein N P, Schneider R J. Activation of Src family kinases by hepatitis B virus HBx protein and coupled signaling to Ras. Mol Cell Biol. 1997;17:6427–6436. doi: 10.1128/mcb.17.11.6427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kohler S W, Provost G S, Kretz P L, Fieck A, Short J M. An in vivo assay using transgenic mice to analyze spontaneous and induced mutations at the nucleic acid level. Strategies. 1990;3:19–21. [Google Scholar]
- 31.Koikie K, Moriya K, Yotsuyanagi H, Iino S, Kurokawa K. Induction of cell cycle progression by hepatitis B virus HBx gene expression in quiescent mouse fibroblasts. J Clin Investig. 1994;94:44–49. doi: 10.1172/JCI117343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Krishnamoorthy R R, Lee T-H, Butel J S, Das H K. Apolipoprotein B gene regulatory factor-2 (BRF-2) is structurally and immunologically highly related to hepatitis B virus associated protein-1 (XAP-1) Biochemistry. 1997;36:960–969. doi: 10.1021/bi961407c. [DOI] [PubMed] [Google Scholar]
- 33.Lee T-H, Elledge S J, Butel J S. Hepatitis B virus X protein interacts with a probable cellular DNA repair protein. J Virol. 1995;69:1107–1114. doi: 10.1128/jvi.69.2.1107-1114.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee T-H, Finegold M F, Shen R-F, DeMayo J L, Woo S L C, Butel J S. Hepatitis B virus transactivator X protein is not tumorigenic in transgenic mice. J Virol. 1990;64:5939–5947. doi: 10.1128/jvi.64.12.5939-5947.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lee Y H, Yun Y. HBx protein of hepatitis B virus activates Jak1-STAT signaling. J Biol Chem. 1998;273:25510–25515. doi: 10.1074/jbc.273.39.25510. [DOI] [PubMed] [Google Scholar]
- 36.Marra G, Schar P. Recognition of DNA alterations by the mismatch repair system. Biochem J. 1999;338:1–13. [PMC free article] [PubMed] [Google Scholar]
- 37.Natoli G, Avantaggiati M L, Chirillo P, Puri P L, Ianni A, Balsano C, Levrero M. Ras- and Raf-dependent activation of c-Jun transcriptional activity by the hepatitis B virus transactivator pX. Oncogene. 1994;9:2837–2843. [PubMed] [Google Scholar]
- 38.Nomura T, Lin Y, Dorjsuren D, Ohno S, Yamashita T, Murakami S. Human hepatitis B virus X protein is detectable in nuclei of transfected cells, and is active for transactivation. Biochim Biophys Acta. 1999;1453:330–340. doi: 10.1016/s0925-4439(99)00004-6. [DOI] [PubMed] [Google Scholar]
- 39.Perfumo S, Amicone L, Colloca S, Giorgo M, Pozzi L, Tripodi M. Recognized efficiency of the hepatitis B virus polyadenylation signal is tissue specific in transgenic mice. J Virol. 1992;66:6819–6823. doi: 10.1128/jvi.66.11.6819-6823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pollicino T, Terradillos O, Lecoeur H, Gougeon M L, Buendia M A. Pro-apoptotic effect of the hepatitis B virus X gene. Biomed Pharmacother. 1998;52:363–368. doi: 10.1016/s0753-3322(99)80003-1. [DOI] [PubMed] [Google Scholar]
- 41.Prost S, Ford J M, Taylor C, Doig J, Harrison D J. Hepatitis B x protein inhibits p53-dependent DNA repair in primary mouse hepatocytes. J Biol Chem. 1998;273:33327–33332. doi: 10.1074/jbc.273.50.33327. [DOI] [PubMed] [Google Scholar]
- 42.Qadri I, Conaway J W, Conaway R C, Schaack J, Siddiqui A. Hepatitis B virus transactivator protein, HBx, associates with the components of TFIIH and stimulates the DNA helicase activity of TFIIH. Proc Natl Acad Sci USA. 1996;93:10578–10583. doi: 10.1073/pnas.93.20.10578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Reifenberg K, Lohler J, Pudollek H-P, Schmitteckert E, Spindler G, Kock J, Schlickt H-J. Long-term expression of the hepatitis B virus core-e- and X-proteins does not cause pathologic changes in transgenic mice. J Hepatol. 1999;26:119–130. doi: 10.1016/s0168-8278(97)80018-9. [DOI] [PubMed] [Google Scholar]
- 44.Ross J A, Leavitt S A. Induction of mutations by 2-acetylaminofluorene in lac1 transgenic B6C3F1 mouse liver. Mutagenesis. 1998;13:173–179. doi: 10.1093/mutage/13.2.173. [DOI] [PubMed] [Google Scholar]
- 45.Rossner M T. Hepatitis B virus X-gene product: a promiscuous transcriptional activator. J Med Virol. 1992;36:101–117. doi: 10.1002/jmv.1890360207. [DOI] [PubMed] [Google Scholar]
- 46.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Press; 1989. [Google Scholar]
- 47.Sancar A. DNA excision repair. Annu Rev Biochem. 1996;65:43–81. doi: 10.1146/annurev.bi.65.070196.000355. . (Erratum, 66:VII, 1997.) [DOI] [PubMed] [Google Scholar]
- 48.Sitterlin D, Lee T-H, Prigent S, Tiollais P, Butel J S, Transy C. Interaction of the UV-damaged DNA-binding protein with hepatitis B virus X protein is conserved among mammalian hepadnaviruses and restricted to transactivation-proficient X-insertion mutants. J Virol. 1997;71:6194–6199. doi: 10.1128/jvi.71.8.6194-6199.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Slagle B L, Becker S A, Butel J S. Hepatitis viruses and liver cancer. In: Minson A, Neil J, McCrae M, editors. Viruses and cancer. Vol. 51. Cambridge, England: University of Cambridge; 1994. pp. 149–171. [Google Scholar]
- 50.Slagle B L, Lee T-H, Medina D, Finegold M J, Butel J S. Increased sensitivity to the hepatocarcinogen diethylnitrosamine in transgenic mice carrying the hepatitis B virus X gene. Mol Carcinog. 1996;15:261–269. doi: 10.1002/(SICI)1098-2744(199604)15:4<261::AID-MC3>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 51.Slagle B L, Lee T-H, Butel J S. Hepatitis B virus and hepatocellular carcinoma. Prog Med Virol. 1992;39:167–203. [PubMed] [Google Scholar]
- 52.Su F, Schneider R J. Hepatitis B virus HBx protein sensitizes cells to apoptotic killing by tumor necrosis factor α. Proc Natl Acad Sci USA. 1997;94:8744–8749. doi: 10.1073/pnas.94.16.8744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sun Z, Lu P, Gail M H, Pee D, Zhang Q, Ming L, Wang J, Wu Y, Liu G, Zhu Y. Increased risk of hepatocellular carcinoma in male hepatitis B surface antigen carriers with chronic hepatitis who have detectable urinary aflatoxin metabolite M1. Hepatology. 1999;30:379–383. doi: 10.1002/hep.510300204. [DOI] [PubMed] [Google Scholar]
- 54.Terradillos O, Billet O, Renard C A, Levy R, Molina T, Briand P, Buendia M A. The hepatitis B virus X gene potentiates c-myc-induced liver oncogenesis in transgenic mice. Oncogene. 1997;14:395–404. doi: 10.1038/sj.onc.1200850. [DOI] [PubMed] [Google Scholar]
- 55.Terradillos O, Pollicino T, Lecoeur H, Tripodi M, Gougeon M L, Tiollais P, Buendia M A. p53-independent apoptotic effects of the hepatitis B virus HBx protein in vivo and in vitro. Oncogene. 1998;17:2115–2123. doi: 10.1038/sj.onc.1202432. [DOI] [PubMed] [Google Scholar]
- 56.Wang W L, London W T, Lega L, Feitelson M A. HBxAg in the liver from carrier patients with chronic hepatitis and cirrhosis. Hepatology. 1991;14:29–37. doi: 10.1002/hep.1840140106. [DOI] [PubMed] [Google Scholar]
- 57.Wang X W, Forrester K, Yeh H, Feitelson M A, Gu J R, Harris C C. Hepatitis B virus X protein inhibits p53 sequence-specific DNA binding, transcriptional activity, and association with transcription factor ERCC3. Proc Natl Acad Sci USA. 1994;91:2230–2234. doi: 10.1073/pnas.91.6.2230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang X W, Gibson M K, Vermeulen W, Yeh H, Forrester K, Sturzbecher H-W, Hoeijmakers J H J, Harris C C. Abrogation of p53-induced apoptosis by the hepatitis B virus X gene. Cancer Res. 1995;55:6012–6016. [PubMed] [Google Scholar]
- 59.Williams G M, Laspia M F, Dunkel V C. Reliability of the hepatocyte primary culture/DNA repair test in testing of coded carcinogens and non-carcinogens. Mutat Res. 1982;97:359–370. doi: 10.1016/0165-1161(82)90003-6. [DOI] [PubMed] [Google Scholar]
- 60.Wood R D. DNA repair in eukaryotes. Annu Rev Biochem. 1996;67:135–167. doi: 10.1146/annurev.bi.65.070196.001031. [DOI] [PubMed] [Google Scholar]
- 61.Wyllie A H, Bellamy C O, Bubb V J, Clarke A R, Corbet S, Curtis L, Harrison D J, Hooper M L, Toft N, Webb S, Bird C C. Apoptosis and carcinogenesis. Br J Cancer. 1999;80(Suppl. 1):34–37. [PubMed] [Google Scholar]
- 62.Yeh F-S, Yu M C, Mo C-C, Luo S, Tong M J, Henderson B E. Hepatitis B virus, aflatoxins, and hepatocellular carcinoma in southern Guangxi, China. Cancer Res. 1989;49:2506–2509. [PubMed] [Google Scholar]
- 63.Yoo Y D, Ueda H, Park K, Flanders K C, Lee Y I, Jay G, Kim S J. Regulation of transforming growth factor-β1 expression by the hepatitis B virus (HBV) X transactivator. J Clin Investig. 1996;97:388–395. doi: 10.1172/JCI118427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu D Y, Moon H B, Son J K, Jeong S, Yu S L, Yoon H, Han Y M, Lee C S, Park J S, Lee C H, Hyun B H, Murakami S, Lee K K. Incidence of hepatocellular carcinoma in transgenic mice expressing the hepatitis B virus X-protein. J Hepatol. 1999;31:123–132. doi: 10.1016/s0168-8278(99)80172-x. [DOI] [PubMed] [Google Scholar]
- 65.Zoulim F, Saputelli J, Seeger C. Woodchuck hepatitis virus X protein is required for viral infection in vivo. J Virol. 1994;68:2026–2030. doi: 10.1128/jvi.68.3.2026-2030.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]