Abstract
The P143 protein of Autographa californica nuclear polyhedrosis virus is essential for replication of viral DNA. To determine the function of P143, the protein was purified to near homogeneity from recombinant baculovirus-infected cells that overexpress P143. ATPase activity copurified with P143 protein during purification and also during gel filtration at a high salt concentration. The ATPase activity did not require the presence of single-stranded DNA, but was stimulated fourfold by the addition of single-stranded DNA. The ATPase activity of P143 had a Km of 60 μM and a turnover of 4.5 molecules of ATP hydrolyzed/s/molecule of enzyme, indicating moderate affinity for ATP and high catalytic efficiency. P143 unwound a 40-nucleotide primer in an ATP-dependent manner, indicating that the enzyme possesses in vitro DNA helicase activity. Based on this result, it seems likely that P143 functions as a helicase in viral DNA replication.
Autographa californica nuclear polyhedrosis virus (AcNPV) is the best-understood member of the Baculoviridae, a family of large double-stranded DNA (dsDNA) viruses that infect arthropod insects. Baculoviruses provide an interesting system in which to study eukaryotic DNA replication. As large DNA viruses that encode all the factors essential for viral DNA replication (20), they have the necessary complexity to supply information applicable to nonviral systems. Because it is a viral system, AcNPV is simpler to study than its host because it encodes only one DNA polymerase and one DNA helicase, rather than the multiple specialized polymerases and helicases found in eukaryotic cells. The study of herpes simplex virus and vaccinia virus DNA replication has added to our understanding of eukaryotic DNA replication. We believe that investigation of baculovirus DNA replication will also make significant contributions to the field of DNA replication.
Transient expression assays have shown that six genes are necessary and sufficient for DNA replication: pol, p143, lef-1, lef-2, lef-3, and ie-1 (13). The six essential genes were predicted to encode an origin binding protein, a priming factor to provide a free 3′ hydroxyl, a DNA polymerase, a helicase to unwind the DNA duplex, and a single-stranded DNA (ssDNA) binding protein (SSB) to maintain the ssDNA exposed by the helicase. The AcNPV proteins DNApol and LEF-3 were shown to exhibit DNA polymerase and ssDNA binding activity, respectively (8, 23). IE-1 may function as the origin binding protein, as it is the only essential replication protein shown to bind specifically to a putative origin of replication (2). Proteins with helicase and primase activities have yet to be identified biochemically.
P143 was classified as an essential DNA replication protein by sequencing a temperature-sensitive mutant, ts8, which was unable to replicate viral DNA at a nonpermissive temperature (19). The mutation was found to be in the open reading frame that encodes P143. Analysis of the P143 amino acid sequence revealed a nucleoside triphosphate (NTP) binding domain and six motifs associated with helicase activity, and therefore P143 was termed a putative DNA helicase. P143 homologues from other sequenced baculoviruses contain these recognized helicase motifs and the NTP binding domain, suggesting that these sequences are important for enzyme function (1, 10, 11). However, mutations in three of the six helicase motifs failed to alter the ability of P143 to complement a temperature-sensitive mutation in the p143 gene (17). Because previous mutagenesis studies on known helicases had found that all six motifs were essential for activity in vivo, this result appeared to contradict the hypothesis that P143 was a viral DNA helicase.
Additional experiments with P143 revealed that the purified protein bound dsDNA, but not ssDNA, in a sequence-nonspecific manner (15). P143 was also shown to coimmunoprecipitate with LEF-3 (5). Furthermore, expression of LEF-3 was shown to be required for the transport of P143 into the nucleus (30).
DNA helicases are necessary for the replication of double-stranded genomes, as the DNA duplex must be unwound in order to form a replication fork. All helicases bind and hydrolyze the γ-phosphates of NTPs. ATP is usually the preferred energy source for helicases. The ATPase activity of helicases is almost always DNA dependent or DNA stimulated (22). The energy released by ATP hydrolysis is coupled to the breaking of hydrogen bonds in duplex DNA or to translocation of helicase along DNA (18).
To determine whether P143 was a DNA helicase, it was purified to near homogeneity from Spodoptera frugiperda (Sf9) cells infected with a recombinant baculovirus designed to overexpress P143. Purified recombinant protein bound to both dsDNA and ssDNA in a sequence-nonspecific manner. DNA-stimulated ATPase activity copurified with P143 through the final purification step and a subsequently used gel filtration column. P143 was found to have moderate affinity for ATP, with a Km of 60 μM and a fairly high catalytic efficiency. In an in vitro DNA helicase assay, P143 unwound a 40-nucleotide primer annealed to single-stranded M13 (ssM13) in an ATP-dependent manner. We therefore conclude that p143 encodes a DNA helicase.
MATERIALS AND METHODS
Cells and virus.
S. frugiperda (Sf9) cells were cultured in TNM-FH medium supplemented with 10% fetal calf serum. AcNPV strain E2 was propagated and maintained as previously described (29).
Construction of a recombinant baculovirus expressing P143.
Site-directed mutagenesis (4) was used to insert a BamHI site upstream of the P143 open reading frame by using the primer sequence 5′-GTCAATCATGTTGGATTCCGTG TTTGTACTTTT-3′. The resulting plasmid was digested with BamHI and XbaI, which cuts downstream of P143 in the vector. The fragment containing p143 was ligated into pVL1393 which was previously digested with BamHI and XbaI. The resulting transfer vector and RP6-S/C DNA, previously digested with Bsu36I, were transfected into Sf9 cells. Polyhedron-deficient progeny virus was selected by plaque assay. Viral DNAs were screened by EcoRI digestion to verify that double-crossover recombination occurred. A plaque isolate with the correct insertion was termed vAc-P143.
Purification of P143.
Sf9 cells (109) were infected with vAc-P143 at a multiplicity of infection of 10 and harvested 48 h postinfection. The cells were washed three times in cold phosphate-buffered saline and resuspended in one packed cell volume of hypotonic buffer (20 mM HEPES [pH 7.5], 5 mM KCl, 1.5 mM MgCl2, 1 mM dithiothreitol [DTT], 1 μg of leupeptin/ml, 1% aprotinin). After 10 min on ice, cells were Dounce homogenized and centrifuged at 2,000 × g. The pellet, containing the nuclear fraction, was resuspended in 1 packed cell volume of hypotonic buffer. An equal volume of hypotonic buffer plus 3.4 M NaCl was added, and the cell suspension was shaken gently for 1 h on ice. The nuclear extract was centrifuged for 1 h at 100,000 × g.
The salt concentration of the supernatant was reduced to 0.33 M KCl by addition of hypotonic buffer, and protein-DNA complexes were precipitated by the dropwise addition of 5% Polymin-P (pH 8.0 in water) to a final concentration of 0.2%. The precipitate was collected by centrifugation at 100,000 × g for 30 min. Proteins were eluted from the Polymin-P pellet by suspension in 10 ml of buffer A (20 mM KH2PO4 [pH 7.2], 1 M KCl, 1 mM EDTA, 1 mM DTT). The DNA–Polymin-P complex was removed by centrifugation at 100,000 × g for 30 min. Proteins in the high-salt supernatant were precipitated by the addition of 0.53 g of ammonium sulfate/ml. The pellet was suspended in 10 ml of buffer B (20 mM KH2PO4 [pH 7.2], 50 mM KCl, 1 mM EDTA, 1 mM DTT) and dialyzed against five 1-liter volumes of the same buffer.
The protein was then loaded onto a 5-ml heparin column connected to a Pharmacia fast protein liquid chromatography system. The column was washed in buffer B and eluted in a 20-ml gradient from 0.05 to 1 M KCl. Peak fractions were analyzed by sodium dodecyl sulfate-polyacrylamide electrophoresis (SDS-PAGE). Fractions containing the most P143 were pooled, dialyzed against buffer B, and loaded onto a Mono S column. P143 was eluted with a linear salt gradient of 0.05 to 1 M KCl. Peak fractions were dialyzed against buffer B and loaded onto a 1.2-ml ssDNA agarose column (Bethesda Research Laboratories) and eluted with a step gradient in 0.2 M increments from 0.1 to 1 M KCl in buffer B. P143 was shown to be purified to near homogeneity by SDS-PAGE and then dialyzed against buffer B plus 50% glycerol.
Electrophoretic mobility shift assays.
A DNA probe of 252 bp containing the homologous repeat 5 (hr5) sequence was made by restriction enzyme digestion by BamHI of pUC-hr5, a plasmid that contains a 252-bp fragment of hr5. Treatment with calf intestinal alkaline phosphatase was followed by T4 kinase labeling with [γ-32P]ATP, EcoRV digestion, and low-melting-point agarose gel purification. An ssDNA probe was generated from the double-stranded probe by dilution in water followed by boiling and rapid cooling on ice. DNA binding reaction mixtures (30 μl) contained 5 fmol of radioactively labeled DNA, 10 mM Tris-HCl (pH 8.0), 0.15 M NaCl, 1 mM DTT, 10% glycerol, and the indicated concentration of purified P143. Reaction mixtures were incubated on ice for 20 min and then loaded onto a 3.5% nondenaturing polyacrylamide gel and fractionated at 4°C by using a running buffer of 50 mM Tris-borate, pH 8.3, and 1 mM EDTA at 200 V for 3 h. Gels were dried and exposed to film overnight.
ATPase assays.
Reaction mixtures (20 μl) contained the indicated amounts of purified P143 and DNA, 20 mM Tris-acetate (pH 7.3), 15 mM NaCl, 8 mM Mg-acetate, 0.1 mM ATP, 1 μCi of [γ-32P]ATP/mmol. Samples were incubated at 30°C for 30 min and then were terminated by the addition of 25 mM EDTA. Polyethyleneimene thin-layer plates were spotted with 1 μl of each reaction mixture and the radiolabeled, hydrolyzed phosphate separated from the di- and triphosphates by elution with 1 M formic acid and 0.5 M LiCl. Thin-layer chromatography plates were dried and exposed to PhosphorImager screens. Results were quantitated by using a Storm PhosphorImager. Free γ-phosphate detected in the absence of enzyme was subtracted as background.
Helicase assays.
Reaction mixtures (20 μl) contained 20 mM Tris-acetate (pH 7.3), 15 mM NaCl, 3 mM MgCl, 10 mM NTP (as labeled in the figure legends), 6.75 fmol of template, and the indicated amount of purified P143. The template was constructed by labeling a 40-nucleotide oligonucleotide (5′TTAAATGCAATGCCTGAGTAATGTAGGTAAAGATT3′) with T4 kinase and [γ-32P]ATP. The radiolabeled primer was then annealed with ssM13mp18 in a 2:1 ratio. Unannealed primer was removed by using a 1-ml Bio-Gel A-5m column (Bio-Rad). Reaction mixtures were incubated at 37°C for 30 min, then stopped by the addition of 2 μl of 10× stop buffer (25 mM EDTA, 50 mM NaCl, 10% glycerol, and 0.2% bromophenol blue) plus 1% SDS. Samples were fractionated on a 12% Tris-borate-EDTA (TBE) gel, fixed, dried, and exposed to PhosphorImager screens.
RESULTS
Overexpression and purification of P143.
The p143 gene was placed under control of the polyhedrin promoter by cloning it into transfer vector pVL1393. Recombinant virus was made by cotransfecting pVL1393-p143 and RP6-S/C viral DNA into Sf9 cells. A resulting polyhedrin-negative plaque was isolated, purified, and shown to contain the P143 gene in the expected location by restriction enzyme digestion of extracted viral DNA (data not shown). The virus was named vAc-P143.
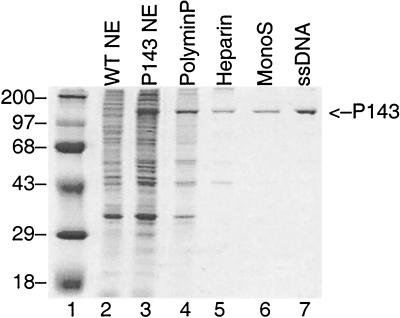
Nuclear extracts were prepared from Sf9 cells 48 h after infection with vAc-P143. Analysis of total cell proteins by SDS-PAGE showed overexpression of a protein with the expected size of P143 (Fig. 1, compare lanes 2 and 3). The nuclear extract from the recombinant virus-infected preparation was precipitated with Polymin-P. P143 precipitated with Polymin-P at a relatively high salt concentration (0.33 M) by binding to Polymin-P directly or through a DNA intermediate. The Polymin-P pellet was then resuspended in a high salt concentration to release bound proteins. The high-salt supernatant was then precipitated by 0.53 g of ammonium sulfate/ml to remove the Polymin-P (lane 4). The dialyzed pellet was loaded onto a heparin affinity column, from which P143 eluted between 298 and 384 mM KCl. Peak fractions were pooled (lane 5) and loaded onto a Mono S column. A cation-exchange column was chosen because P143 has an isoelectric point of 8.3, so the protein would be positively charged in pH 7.2 buffer. P143 eluted in a fairly broad peak between 128 and 287 mM KCl. Peak fractions were pooled (lane 6), diluted to reduce the salt concentration to 50 mM KCl, and then fractionated on an ssDNA agarose column. P143 eluted from ssDNA agarose at 200 to 400 mM KCl. SDS-polyacrylamide gel analysis of the peak fraction (Fig. 1, lane 7) followed by Coomassie blue staining showed that P143 appeared to be purified to homogeneity.
FIG. 1.
Purification of P143. Nuclear extracts (NE) from vAc-P143 infected Sf9 cells (lane 3) were precipitated with Polymin-P, the pellet was resuspended in high-salt buffer to release proteins, and the supernatant was precipitated by ammonium sulfate. The resuspended pellet (lane 4) was subjected to chromatography on heparin. Peak fractions from the heparin column were pooled (lane 5) and fractionated on a Mono S column. Mono S peak fractions (lane 6) were diluted and loaded onto an ssDNA agarose. Lane 7, 4 μg of protein of the peak fraction from ssDNA agarose; lane 2, crude nuclear extract prepared from cells infected with RP6-S/C, the parental virus; lane 1, positions of protein molecular markers are (in kilodaltons) on the left. The arrow on the right indicates the position of P143. Samples were separated on an SDS–8% polyacrylamide gel and stained with Coomassie blue.
P143 binds dsDNA and ssDNA.
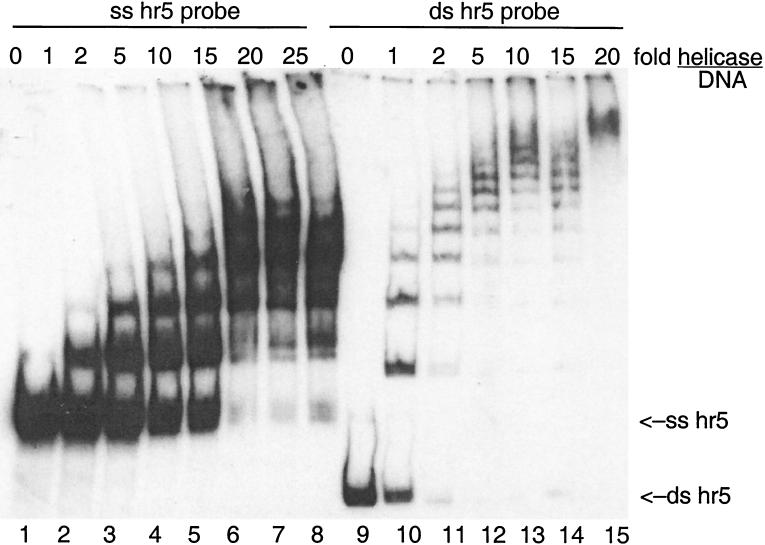
The DNA binding activity of purified P143 was examined by electrophoretic mobility shift assay (Fig. 2). For these experiments, we used a 252-bp fragment of hr5 that had been 5′-end labeled with T4 kinase. P143 bound to the dsDNA probe, as shown previously by Laufs et al. (15). The purified recombinant P143 shifted dsDNA with the same distinctive step-wise pattern previously observed for P143 purified from insect cells infected with wild-type AcNPV. Six retarded bands can be detected in lane 11, suggesting that six P143 monomers bound to the probe in the slowest migrating band and one monomer bound to the probe in the fastest migrating band. Increasing amounts of enzyme produced even slower migrating bands that finally compressed into one diffuse band in lane 15.
FIG. 2.
Binding of purified P143 to dsDNA and ssDNA. Lanes 1 to 8, ssDNA probe; lanes 9 to 15, dsDNA probe. The molar ratio of purified P143 to DNA is indicated at the top. Reaction mixtures were separated on a native 3.5% acrylamide–TBE gel, dried, and exposed to film.
Our results differed from those of Laufs et al. in that we also observed a shift of the corresponding ssDNA by P143, while their enzyme preparation shifted only dsDNA. Interestingly, the pattern of bands seen with the single-stranded probe differs from that of the dsDNA probe. Fewer retarded bands were seen with ssDNA than with dsDNA at the same relative ratios of enzyme to DNA. At the highest amount of protein to DNA, the ssDNA was not retarded as much as was the dsDNA, and also fewer retarded bands could be distinguished.
ATPase activity copurifies with P143.
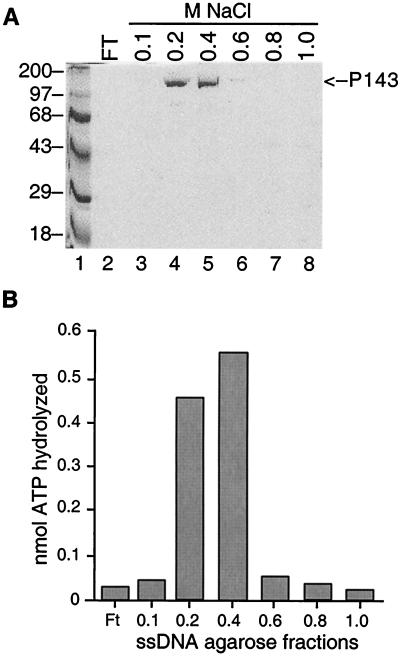
The amino acid sequence of P143 suggests that it may function as a helicase in DNA replication. All known DNA helicases hydrolyze one or more NTPs, most commonly ATP. Therefore, we assayed the ssDNA agarose fractions for ATPase activity to determine if P143 could be a DNA helicase. ATPase activity was measured by the inclusion of [γ-32P]ATP in the reaction mixture. Cleavage of the γ-phosphate was detected by separation of free phosphate from unutilized substrate on a thin-layer chromatography plate due to its greater mobility. Hydrolysis was quantitated by measuring the ratio of the faster migrating radioactive species (the cleaved γ-32P) to the total radioactivity. We found that the ssDNA agarose column fractions containing the most P143 also had the highest level of ATPase activity (Fig. 3).
FIG. 3.
Test of ssDNA agarose column fractions for ATPase activity. (A) Purification of P143 by affinity chromatography on ssDNA agarose. Peak fractions from the Mono S column were loaded onto an ssDNA agarose gravity column and eluted with an NaCl step gradient. Proteins in each fraction were separated on an SDS–8% polyacrylamide gel and stained with Coomassie blue. Lanes 4 and 5 contain P143 purified to near homogeneity. The positions of protein molecular size markers in lane 1 are shown (in kilodaltons) on the left. The arrow on the right indicates the position of P143. (B) ATPase activity. Each fraction was separately dialyzed to 50 mM KCl, and 1 μl of each was assayed for ATPase activity.
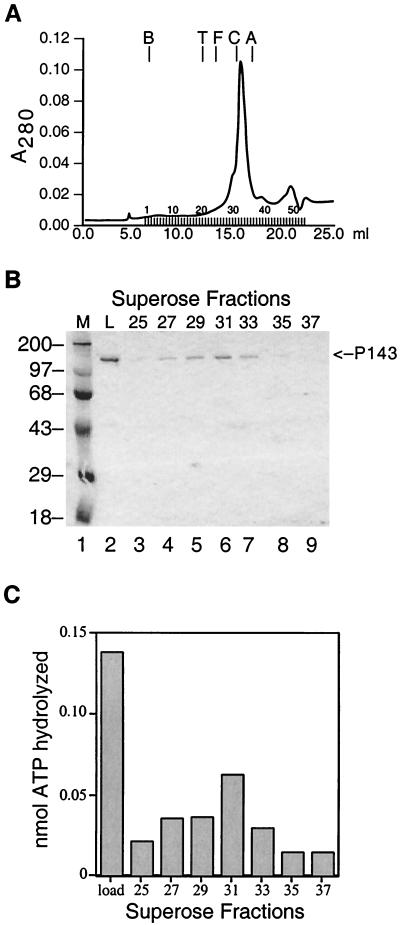
To further confirm that the ATPase activity was due to P143 and not to minor contaminants that coeluted from the ssDNA agarose column, the peak fractions of P143 were then filtered through a Superose 6 column, which separates proteins by size (Fig. 4). It was necessary to use a 1 M KCl column buffer in order to prevent interaction between P143 and the column matrix, a phenomenon observed with several other helicases filtered through sizing columns (22). Analysis of alternate fractions on SDS-polyacrylamide gels confirmed that the UV peak corresponded to the P143 protein peak (Fig. 4b).
FIG. 4.
Gel filtration of P143. (A) P143 peak fractions from ssDNA agarose chromatography were adjusted to 1 M KCl and filtered through Superose 6. Fractions (0.4 ml) were collected from 6 to 22 ml. The positions of elution of blue dextran 2000, thyroglobulin (T; 669 kDa), ferritin (F; 443 kDa), catalase (C; 232 kDa), and aldolase (A; 158 kDa) were determined by elution of protein standards. (B) The indicated fractions across the peak of UV absorbance were separated by electrophoresis on an SDS–8% polyacrylamide gel and visualized by staining with Coomassie blue. Lane 2, the load; lanes 3 to 9, fractions 25 to 37, odd numbers only. The position of P143 is indicated on the left. The migration of protein molecular weight markers is indicated on the left. (C) Aliquots of the corresponding fractions were dialyzed to 50 mM KCl, and 2 μl of each was assayed for ATPase activity. Background activity from a no-enzyme control was subtracted.
Fractions were assayed for ATPase activity across the peak of protein, and once again, ATPase activity was found in fractions containing P143, but not in other fractions (Fig. 4c). The amount of P143 was found to increase and decrease concomitantly with the peak of enzymatic activity, as well as with the peak in the UV trace.
P143 ATPase activity is ssDNA stimulated.
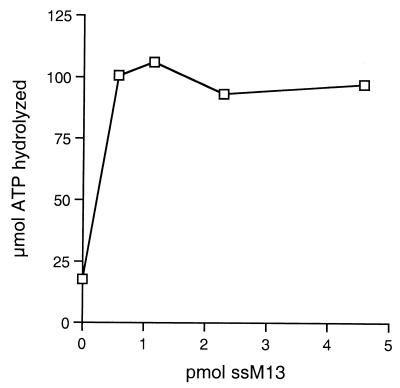
Addition of ssDNA increased the amount of ATP hydrolyzed more than fourfold (Fig. 5). dsDNA and RNA did not significantly increase ATP hydrolysis (data not shown). The degree of stimulation by ssDNA may be underestimated due to the presence of ssDNA from the ssDNA agarose column, the final column in the purification of P143 used in this assay. However, we have determined that the ATPase activity of P143 is DNA independent. This was verified by fractionating P143 from ssDNA agarose on a MonoQ anion-exchange column to remove any DNA contamination. The resulting fractions were assayed for ATPase activity. Fractions containing P143 hydrolyzed ATP in the absence of DNA (data not shown).
FIG. 5.
Effect of DNA on P143 ATPase activity. ATPase assay contained 140 fmol of P143 and the indicated amounts of ssM13mp18 per reaction. Each point represents the average of three experiments.
Determination of the Km of P143 ATPase activity.
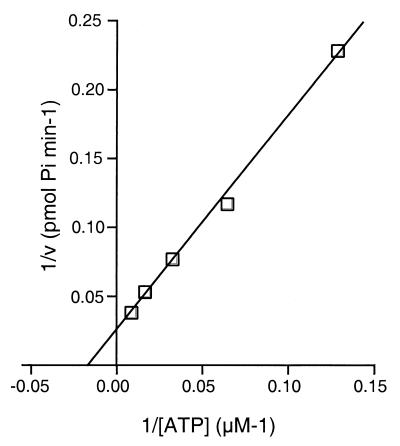
We further characterized the ATPase activity of P143 by determining the Km of the reaction (Fig. 6). Km is a measure of affinity between enzyme and substrate; specifically, it is the concentration of substrate at half of the maximum reaction rate. We measured the initial rate of formation of free phosphate over a range of ATP concentrations after a 5-min incubation. A Lineweaver-Burk plot was used to calculate a Km of 60 μM ATP. This is well within the range calculated for known helicases, which range from 5 μM for simian virus 40 large T antigen (3) to 750 μM for bovine papillomavirus type 1 E1 (28). The kcat, or turnover number, was calculated to be 4.5 molecules of ATP hydrolyzed/s/molecule of P143. The kcats of known helicases range from 0.5 to 0.8 molecules/s/molecule of Rep68 of adeno-associated virus (32) to 30 to 50 molecules/s/molecule of DNA helicase I of Schizosaccharomyces pombe (24).
FIG. 6.
Km of P143 ATP hydrolysis. The rate of ATP hydrolysis was determined for 7.8125, 15.625, 31.25, 62.5, and 125 mM ATP concentrations in the presence of 1 pmol of ssM13 DNA. Reaction mixtures were incubated with 140 fmol of purified P143 for 5 min at 37°C. A double-reciprocal plot of the rate of 32Pi formation versus the ATP concentration is shown.
P143 unwinds DNA in an in vitro helicase assay.
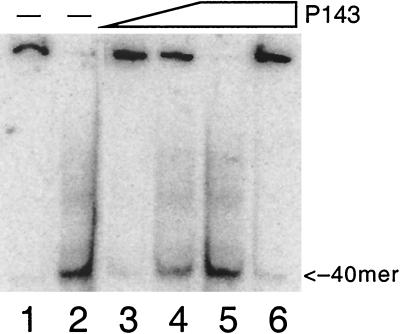
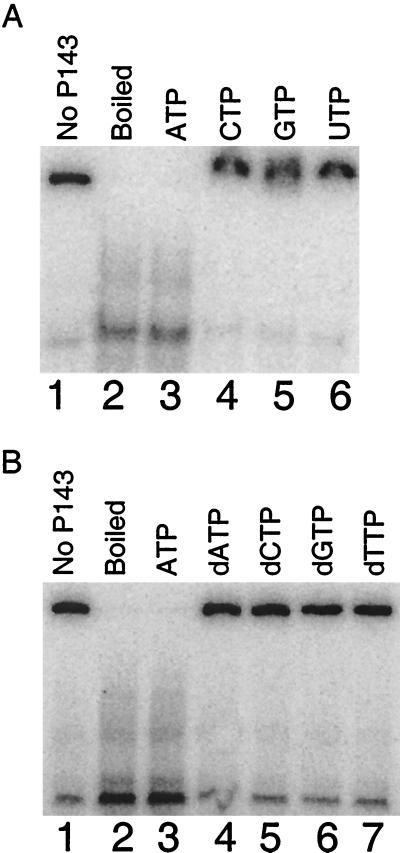
Helicase activity was measured by the displacement of a 32P-labeled 40-mer from ssM13 DNA. Increasing amounts of P143 resulted in increased unwinding (Fig. 7). Displacement of primer was dependent on ATP hydrolysis, as no unwinding occurred in the absence of ATP (compare lanes 5 and 6). There was also no displacement when ATP was replaced with ATPγS (data not shown). No other ribonucleotides or deoxyribonucleotides could replace ATP as a cofactor (Fig. 8a and b). These data indicate that P143 functions as a DNA helicase in vitro, and that it uses only the hydrolysis energy of ATP to fuel DNA unwinding.
FIG. 7.
DNA helicase activity of P143. Purified P143 was tested for helicase activity by using the oligonucleotide displacement assay. The substrate consists of an ssM13 molecule with a radiolabeled 40-nucleotide primer annealed; there was 6.75 fmol of substrate per lane. Lane 1, substrate in the absence of enzyme; lane 2, substrate after boiling for 3 min; lanes 3, 4, and 5, 70, 140, and 280 fmol of P143, respectively. The reaction mixture shown in lane 6 was incubated with 280 fmol of P143 in the absence of ATP. Samples were fractionated on a 12% acrylamide–TBE gel. The positions of the free 40-mer are indicated on the right.
FIG. 8.
Nucleotide requirement for DNA unwinding activity of P143. Reaction conditions were the same as described in the legend for Fig. 7. (A) Nucleotide specificity of the helicase activity of P143. Lane 1, substrate alone; lane 2, boiled substrate alone; lanes 3 to 6, 280 fmol of P143 with a 10 mM concentration of the indicated recombinant NTP. (B) Sugar specificity of helicase activity. Lanes 3 to 7 contain 280 fmol of P143 with a 10 mM concentration of the indicated dNTP; the reaction mixture shown in lane 3 was incubated in the presence of 10 mM ATP as a positive control.
DISCUSSION
The purpose of this article is to present evidence showing that P143 serves as a helicase in AcNPV DNA replication. To achieve this goal, we have purified P143 to homogeneity from Sf9 cells infected with a recombinant virus designed to overexpress P143 (Fig. 1). We found P143 to be labile, requiring rapid purification to preserve activity. It was necessary to carry out all the purification steps following ammonium sulfate precipitation in 1 day. After purification, the protein was stored in 50% glycerol at −20°C, where it remained active for several months.
Our preparations of purified recombinant P143 bound to dsDNA with the same characteristic pattern as previously reported (15). P143 did not bind specifically to hr sequences (15), which are believed to function as origins of replication (20). This is not surprising, as most helicases bind DNA nonspecifically. One notable exception is simian virus 40 T antigen, which is a multifunctional protein that is both an origin binding protein and a DNA helicase (3). The fact that P143 binds DNA nonspecifically suggests that another viral protein functions as an origin binding protein and recruits helicase to the origin.
We have also shown that P143 binds ssDNA, as expected for a DNA helicase (Fig. 2). Our finding that P143 bound ssDNA contradicts an earlier report by Laufs et al. (15), who found that P143 did not bind to ssDNA. Our gel retardation assays were performed under the same conditions as theirs, although our methods of purification differed and our recombinant viruses were independently obtained. It is possible that one of us has inadvertently isolated a mutant clone. If so, it seems more likely that a spontaneous mutation would result in a loss of function than in a gain of function.
Liu and Carstens (17) found that mutations in motifs I, Ia, II, and III affected the ability of P143 to rescue the replication defect of a ts mutant; however, mutations in motifs IV, V, and VI individually did not affect the ability of P143 to compensate for the ts mutant (ts8). This result was surprising and raised questions regarding their earlier assumption that p143 encodes a helicase. In a similar mutagenesis experiment with the herpes simplex virus type 1 helicase UL-5, mutations in all six helicase motifs were found to be essential. The residues chosen for mutagenesis of UL-5 are highly conserved among the herpesvirus proteins, and while the corresponding residues were mutated in P143, these residues are not absolutely conserved in other baculovirus P143 proteins. Thus, the baculovirus helicase may be more permissive to amino acid substitutions than herpes simplex virus type 1 UL-5. Motifs IV, V, and VI have less-well-defined functions than I, Ia, II, and III, and they may not be essential in every helicase. For example, a mutation in motif IV of Escherichia coli Rep protein did not eliminate in vitro helicase activity (18).
Our preparations of P143 had DNA-independent ATPase activity, which was stimulated by the addition of ssM13 DNA. Most helicases are ssDNA-dependent ATPases, and in the past, it was believed that all helicases were completely DNA dependent (14). This seemed logical, as it is thought that helicases couple the energy of ATP hydrolysis to hydrogen bond disruption and/or translocation along ssDNA. ATP hydrolysis in the absence of DNA would waste a valuable source of energy. However, recent studies have characterized several DNA helicases that appear to be DNA stimulated rather than DNA dependent, such as Rep68 from adeno-associated virus (32), E1 from bovine papillomavirus (28), and the RNA helicase from hepatitis C virus (25). Although the possibility exists that DNA contaminated our protein preparations, as well as these others, the amount of DNA able to escape detection would likely be enzymatically insignificant.
A possible explanation for this apparent uncoupling of ATP hydrolysis and helicase activity is that other factors prevent the DNA-independent hydrolysis of ATP in vitro. LEF-3, the viral SSB, could serve as such a factor in the baculovirus system, as it coimmunoprecipitates with P143 (5) and is needed for localization of P143 to the nucleus (30).
dsDNA failed to significantly stimulate the ATPase activity of P143. This is not surprising since of all of the helicases studied, only the ATPase activity of the RecBCD complex is stimulated to a greater extent by dsDNA than ssDNA (26). RNA also did not stimulate the ATPase activity. This suggests that P143 functions as a DNA helicase only and is not capable of unwinding RNA-RNA duplexes.
The ATPase activity of P143 has a Km of 60 μM, indicating a moderately high affinity for ATP (Fig. 6). The kcat of 4.5 is high, indicating that the ATPase domain of P143 has a relatively high catalytic efficiency. Both the Km and kcat are well within the range of values calculated for known helicases. It is unlikely that the ATPase activity attributed to P143 is the result of a minor contaminant undetectable after Coomassie staining. The turnover of a contaminating enzyme would have to be very high to account for the observed ATP hydrolysis if it were present in amounts at least 100-fold less than P143.
We calculated the Km and kcat under generally used conditions in order to compare the performance of P143 to that of other helicases. The Km and kcat of P143 may change under different experimental conditions. E. coli Rep protein was found to have a Km of 200 mM with ssDNA and a Km of 25 mM with a replication fork. The kcat of phage T4 gene 41 protein varies with protein concentration. Its oligomeric state varies with concentration, and its activity is higher as a hexamer than as a monomer (7). Further study may yield more optimal conditions resulting in different Km and kcat values.
The Km and kcat calculated here may be useful in distinguishing P143 from host helicases when P143 is expressed in E. coli or other heterologous systems in which the helicase has been characterized. The catalytic efficiency of P143 supports the idea that P143 is the replicative helicase, unwinding DNA preceding the replication fork. AcNPV DNApol has been found to add 40 nucleotides/s (9). Most helicases use two ATP molecules to break one hydrogen bond or expose 1 nucleotide (14). With this estimate, P143 could expose only 2 nucleotides per second. However, the rate of replication fork movement may not be as high as the rate of leading-strand replication measured in vitro. One role of helicases is to coordinate leading-strand replication with lagging-strand replication, which is necessarily slower. It is also possible that additional factors, such as LEF-3/SSB, would increase the rate of unwinding to allow P143 to keep pace with the DNA polymerase.
The strongest biochemical evidence that P143 is a DNA helicase comes from its ability to unwind a 40-mer annealed to ssM13 DNA. This unwinding assay is the standard test for in vitro helicase activity. A common variation on this assay is the use of a substrate with a 5′ or 3′ tail, created by annealing a primer to M13 that has a 3′ or 5′ end that is not complementary. Several helicases require a tailed or even a forked substrate; others show greater unwinding activity on such substrates. In the assays shown in Fig. 7 and 8, the primer was entirely complementary to M13mp18; therefore, P143 requires neither a 3′ tail, as in gene 4 of phage T7 (21), nor a fork, as in E. coli DnaB (16).
Of the eight NTPs tested, only ATP served as a cofactor for DNA unwinding. Most known helicases utilize more than one NTP with varying efficiency, although ATP is the most common energy source. Bacteriophage T7 is a noteworthy exception to the rule of ATP being the preferred cofactor, as this helicase/primase uses dTTP (21). ATPγS, a slowly hydrolyzed ATP analog, could not replace ATP in the unwinding assay and decreased oligonucleotide displacement when combined with ATP (data not shown). The fact that ATPγS was a negative competitor of ATP indicates that the analog bound to the NTP binding site of P143. Therefore, the failure of ATPγS to support DNA unwinding indicates that ATP hydrolysis is required for DNA unwinding, not merely ATP binding.
An interesting feature of P143 is that it produces a distinctive binding pattern in electrophoretic mobility shift assays using a 252-bp probe. It suggests a stepwise mechanism of assembly on DNA that will be investigated further. Determining the kinetics of DNA binding and the effects of nucleotide cofactors on DNA binding and oligomerization of P143 may aid in our understanding of helicase mechanisms.
ACKNOWLEDGMENT
This research was supported by grant MCB-9874532 from the National Science Foundation.
REFERENCES
- 1.Ahrens C H, Rohrmann G F. The DNA polymerase and helicase genes of a baculovirus of Orgyia pseudosugata. J Gen Virol. 1996;77:825–837. doi: 10.1099/0022-1317-77-5-825. [DOI] [PubMed] [Google Scholar]
- 2.Choi J, Guarino L A. The baculovirus transactivator IE1 binds to viral enhancer elements in the absence of insect cell factors. J Virol. 1995;69:4548–4551. doi: 10.1128/jvi.69.7.4548-4551.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark R, Lane D P, Tjian R. Use of monoclonal antibodies as probes of simian virus 40 T antigen ATPase activity. J Biol Chem. 1981;256:11854–11858. [PubMed] [Google Scholar]
- 4.Deng W P, Nickoloff J A. Site-directed mutagenesis of virtually any plasmid by eliminating a unique site. Anal Biochem. 1992;200:81–84. doi: 10.1016/0003-2697(92)90280-k. [DOI] [PubMed] [Google Scholar]
- 5.Evans J T, Rosenblatt G S, Leisy D J, Rohrmann G F. Characterization of the interaction between the baculovirus ssDNA-binding protein (LEF-3) and putative helicase (P143) J Gen Virol. 1999;80:493–500. doi: 10.1099/0022-1317-80-2-493. [DOI] [PubMed] [Google Scholar]
- 6.Guarino L A, Dong W. Functional dissection of the Autographa californica nuclear polyhedrosis virus enhancer element hr5. Virology. 1994;200:328–335. doi: 10.1006/viro.1994.1197. [DOI] [PubMed] [Google Scholar]
- 7.Guo S, Tabor S, Richardson C C. The linker region between the helicase and primase domains of the bacteriophage T7 gene 4 is critical for hexamer formation. J Biol Chem. 1999;274:30303–30309. doi: 10.1074/jbc.274.42.30303. [DOI] [PubMed] [Google Scholar]
- 8.Hang X, Dong W, Guarino L A. The lef-3 gene of Autographa californica nuclear polyhedrosis virus encodes a single-stranded DNA binding protein. J Virol. 1995;69:3924–3928. doi: 10.1128/jvi.69.6.3924-3928.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hang X, Guarino L A. Purification of Autographa californica nucleopolyhedrovirus DNA polymerase from infected insect cells. J Gen Virol. 1999;80:2519–2526. doi: 10.1099/0022-1317-80-9-2519. [DOI] [PubMed] [Google Scholar]
- 10.Heldens J G M, Liu Y, Zuidema D, Goldbach R W, Vlak J M. A highly conserved genomic region in baculoviruses—sequence analysis of an 11.3 kbp DNA fragment (46.55-55.1 mu) of the Spodoptera exigua multicapsid nucleopolyhedro virus. Virus Res. 1998;55:187–198. doi: 10.1016/s0168-1702(98)00040-9. [DOI] [PubMed] [Google Scholar]
- 11.Kamita S G, Maeda S. Abortive infection of the baculovirus Autographa californica nuclear polyhedrosis virus in Sf-9 cells after mutation of the putative DNA helicase gene. J Virol. 1997;70:6244–6250. doi: 10.1128/jvi.70.9.6244-6250.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaplan D L, Steitz T A. DnaB from Thermus aquaticus unwinds forked duplex DNA with an asymmetric tail length dependence. J Biol Chem. 1999;274:6889–6897. doi: 10.1074/jbc.274.11.6889. [DOI] [PubMed] [Google Scholar]
- 13.Kool M, Ahrens C H, Goldbach R W, Rohrmann G F, Vlak J M. Identification of genes involved in DNA replication of the Autographa californica baculovirus. Proc Natl Acad Sci USA. 1994;97:11212–11216. doi: 10.1073/pnas.91.23.11212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kornberg A, Baker T. DNA replication. 2nd ed. New York, N.Y: W. H. Freeman and Co.; 1992. [Google Scholar]
- 15.Laufs S, Lu A, Arell K, Carstens E B. P143 of Autographa californica nuclear polyhedrosis virus is a DNA binding protein. Virology. 1997;228:98. doi: 10.1006/viro.1996.8361. [DOI] [PubMed] [Google Scholar]
- 16.LeBowitz J H, McMacken R. The Escherichia coli DnaB replication protein is a DNA helicase. J Biol Chem. 1986;261:4738–4748. [PubMed] [Google Scholar]
- 17.Liu G, Carstens E B. Site directed mutagenesis of the AcMNPV p143 gene: effects on baculovirus DNA replication. Virology. 1999;253:125–136. doi: 10.1006/viro.1998.9485. [DOI] [PubMed] [Google Scholar]
- 18.Lohman T M, Bjornson K P. Mechanisms of helicase catalyzed DNA unwinding. Annu Rev Biochem. 1996;65:169–214. doi: 10.1146/annurev.bi.65.070196.001125. [DOI] [PubMed] [Google Scholar]
- 19.Lu A, Carstens E B. Nucleotide sequence of a gene essential for viral DNA replication in the baculovirus Autographa californica nuclear polyhedrosis virus. Virology. 1991;195:710. doi: 10.1016/0042-6822(91)90500-b. [DOI] [PubMed] [Google Scholar]
- 20.Lu A, Krell P J, Vlak J M, Rohrmann G F. Baculovirus DNA replication. In: Miller L K, editor. The baculoviruses. New York, N.Y: Plenum Publishing Corp.; 1999. pp. 171–191. [Google Scholar]
- 21.Matson S W, Tabor S, Richardson C C. The gene 4 protein of bacteriophage T7. Characterization of helicase activity. J Biol Chem. 1983;258:14017–14024. [PubMed] [Google Scholar]
- 22.Matson S W, Bean D W. Purification and biochemical characterization of enzymes with DNA helicase activity. Methods Enzymol. 1995;262:389–405. doi: 10.1016/0076-6879(95)62031-1. [DOI] [PubMed] [Google Scholar]
- 23.McDougal V V, Guarino L A. Autographa californica nuclear polyhedrosis virus DNA polymerase: measurements of processivity and strand displacement. J Virol. 1999;73:4908–4918. doi: 10.1128/jvi.73.6.4908-4918.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Park J S, Choi E, Lee S, Lee C, Seo Y. A DNA helicase from Schizosaccharomyces pombe stimulated by single-stranded DNA-binding protein at low ATP concentration. J Biol Chem. 1997;272:18910–18919. doi: 10.1074/jbc.272.30.18910. [DOI] [PubMed] [Google Scholar]
- 25.Pregschat F, Averett D R, Clarke B E, Porter D J T. A steady-state and pre-steady-state kinetic analysis of the NTPase activity associated with the hepatitis C virus NS3 helicase domain. J Biol Chem. 1996;271:24449–24457. doi: 10.1074/jbc.271.40.24449. [DOI] [PubMed] [Google Scholar]
- 26.Roman L J, Kowalczykowski S C. Characterization of the helicase activity of the Escherichia coli RecBCD enzyme using a novel helicase assay. Biochemistry. 1989;28:2863–2873. doi: 10.1021/bi00433a018. [DOI] [PubMed] [Google Scholar]
- 27.Runyon G T, Wong I, Lohman T M. Overexpression, purification, DNA binding, and dimerization of the Escherichia coli uvrD gene product (helicase II) Biochemistry. 1993;32:602–612. doi: 10.1021/bi00053a028. [DOI] [PubMed] [Google Scholar]
- 28.Santucci S, Bonne-Andrea C, Clertant P. Bovine papilloma virus type E1 ATPase activity does not depend on binding to DNA nor to viral E2 protein. J Gen Virol. 1995;76:1129–1140. doi: 10.1099/0022-1317-76-5-1129. [DOI] [PubMed] [Google Scholar]
- 29.Summers M D, Smith G E. A manual of methods for baculovirus vectors and insect cell culture procedures. Bulletin 1555. College Station: Texas Agricultural Experiment Station; 1987. [Google Scholar]
- 30.Wu Y, Carstens E B. A baculovirus single-stranded DNA binding protein, LEF-3, mediates the nuclear localization of the putative helicase P143. Virology. 1998;247:32–40. doi: 10.1006/viro.1998.9235. [DOI] [PubMed] [Google Scholar]
- 31.Zhou L, Weller S K. The six conserved helicase motifs of the UL5 gene product, a component of the herpes simplex virus type 1 helicase-primase, are essential for its function. J Virol. 1992;66:469–479. doi: 10.1128/jvi.66.1.469-479.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhou X, Zolotukhin I, Im D-S, Muzyczka N. Biochemical characterization of adeno-associated virus Rep68 DNA helicase and ATPase activities. J Virol. 1999;73:1580–1590. doi: 10.1128/jvi.73.2.1580-1590.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]