Abstract
Background
Hepatocellular carcinoma (HCC) continues to endanger human health worldwide. Regulatory networks of competing endogenous RNAs (ceRNAs) play important roles in HCC. TP53 is the second most often altered gene in HCC and has a significant role in regulating target genes such as miRNAs and lncRNAs.
Methods
Data from patients with TP53 mutation were collected through the cBioPortal database and differential analysis was performed to screen RNAs related to TP53 mutation. The lncRNA-miRNA–mRNA relationship was predicted by the miRcode, miRDB, and TargetScan databases. The ceRNA networks were screened and visualized by Cytoscape. Core ceRNA networks were generated by differential analysis, coexpression analysis, prognostic analysis and subcellular localization. Finally, methylation, mutation, PPI, GSEA, immunity and drug sensitivity analyses of MEX3A were performed to determine the role of MEX3A in HCC.
Results
We identified 1508 DEmRNAs, 85 DEmiRNAs and 931 DElncRNAs and obtained a ceRNA network including 28 lncRNAs, 4 miRNAs and 31 mRNAs. Twenty hub DERNAs in the TP53-altered-related ceRNA network were screened out by Cytoscape and the core ceRNA network (LINC00491/TCL6-hsa-miR-139-5p-MEX3A) was obtained by multiple analyses. In addition, we discovered that the methylation level of MEX3A was decreased and the mutation frequency was raised in HCC. Furthermore, elevated MEX3A expression was associated with alterations in the HCC immunological microenvironment.
Conclusion
We successfully constructed a reciprocal ceRNA network, which could provide new ideas for exploring HCC mechanisms and therapeutic approaches.
Keywords: Hepatocellular carcinoma, TP53, Competing endogenous RNA, LINC00491, TCL6, miR-139-5p, MEX3A
Abbreviation
Abbreviation Full name
- AFP
Alpha fetoprotein
- BP
Biological process
- CC
Cellular component
- ceRNA
Competing endogenous RNA
- ceRNETs
Competing endogenous RNA networks
- cfDNA
Cell-free DNA
- CNV
Copy number variation
- CTCs
Circulating tumor cells
- ctDNA
Circulating tumor DNA
- CTLs
Cytotoxic T lymphocytes
- CTRP
Cancer therapeutics response portal
- DE
Differentially expressed
- DFS
Disease-free survival
- DSS
Disease-specific survival
- GSEA
Gene Set Enrichment Analysis
- HCC
Hepatocellular carcinoma
- ICIs
Immune checkpoint inhibitors
- IPS
Immunophenoscore
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- MEX3A
Mex-3 RNA binding family member A
- MF
Molecular function
- MREs
MiRNA response elements
- OS
Overall survival
- PFS
Progression-free survival
- PPI
Protein–protein interaction
- TACE
Transarterial chemoembolization
- TCGA
The Cancer Genome Atlas
- TCL6
T cell leukemia/lymphoma 6
- TIDE
Tumour immune dysfunction and exclusion
- TIMER
Tumour immune estimation resource
- TKIs
Tyrosine kinase inhibitors
- TP53
Tumour protein p53
1. Introduction
Primary liver cancer is a significant global health concern, ranking as the sixth most prevalent cancer and the third leading cause of cancer mortality, with its incidence increasing annually. In 2020, there were 905,677 new cases of liver cancer and 830,180 liver cancer-related deaths worldwide [1]. Hepatocellular carcinoma (HCC) is the most common type of primary liver cancer, accounting for approximately 75%–85 % of all liver malignancies. The primary risk factors for liver cancer include viral hepatitis, alcohol consumption, obesity, type 2 diabetes, and smoking [1]. The 5-year survival rate for HCC is a mere 18 % [2], highlighting the urgent need for comprehensive understanding of the molecular mechanisms underlying HCC to improve liver cancer treatment. By 2030, HCC is predicted to become the third leading cause of cancer-related mortality [3].
With the advancement of high-throughput sequencing technology, research on the pathogenic genes responsible for the development of HCC has become increasingly sophisticated. Among the various driver gene mutations implicated in HCC, TP53 mutations are the second most common, present in 25 %–30 % of HCC patients [4]. Strikingly, the prevalence of TP53 mutations increases to almost 60 % in Hepatitis B virus-induced HCC [5]. Wild-type TP53 protein is essential for DNA damage repair, cell cycle regulation, apoptosis, and cellular senescence [6]. Patients with TP53 mutations exhibit poorer prognoses than those without such mutations, with lower overall survival (OS) and recurrence-free survival (RFS) [7]. Moreover, TP53 mutation impacts serum alpha fetoprotein (AFP) levels [8], promotes HCC microvascular invasion [9], and contributes to a poorly differentiated tumor [10]. Hence, it is imperative to understand the precise role of TP53 in the development of HCC.
Noncoding RNAs (ncRNAs) are a class of RNAs that do not encode proteins, with lncRNAs, miRNAs, pseudogenes, and circRNAs being primarily involved in posttranscriptional regulation [11]. LncRNAs, circRNAs, mRNAs, and pseudogenes can modulate miRNA function by binding to miRNA response elements (MREs) in miRNA coding regions [12]. When multiple ncRNAs have the same MRE, they compete for miRNA binding, resulting in the formation of competing endogenous RNAs (ceRNAs) [13]. These RNAs can engage in crosstalk and create ceRNA networks (ceRNETs), which regulate various cellular processes. Dysregulation of ceRNETs in normal cells can cause malignant changes [14]. Although the composition and function of most ceRNETs remain unknown, exploring ceRNA-related pathways in HCC can aid in the diagnosis and treatment of the disease.
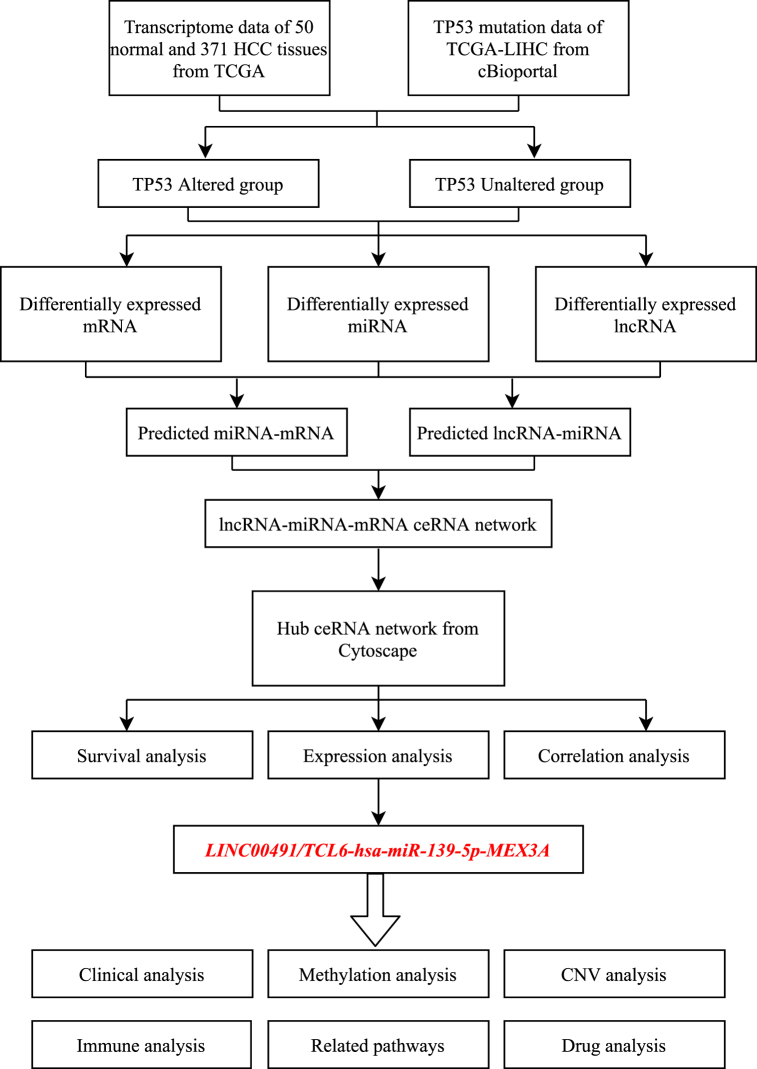
In this study, we aimed to establish a comprehensive ceRNA network for HCC based on the TP53 mutation status. The workflow of the study is depicted in Fig. 1. Firstly, we segregated 347 HCC samples into two groups, the TP53-mutated group and the TP53-wild type group. We performed differential expression analysis to identify differentially expressed (DE) RNAs (lncRNAs, miRNAs, mRNAs), and then constructed a lncRNA-miRNA–mRNA triple regulatory network. Furthermore, we conducted functional enrichment analysis to understand the functional roles and underlying mechanisms of the ceRNA network. Key ceRNA networks were identified based on the results of expression analysis, prognostic analysis, and subcellular localization prediction. We also performed correlation analysis between the predicted genes and clinical parameters to determine their clinical significance. Our findings revealed that the LINC00491/TCL6-hsa-miR-139-5p-MEX3A axis plays a pivotal role in HCC. We further explored the changes in MEX3A in HCC through methylation and mutation analyses, ascertained the role of MEX3A in liver cancer through protein–protein interaction (PPI) and Gene Set Enrichment Analysis (GSEA), and conducted immune infiltration analysis and related drug screening to investigate the potential biological function of MEX3A in HCC.
Fig. 1.
Flow chart of this study.
2. Materials and methods
2.1. Data source
We acquired the lncRNA and mRNA expression data of 50 normal and 371 HCC patients, and miRNA expression data of 50 normal and 375 HCC patients, from the Cancer Genome Atlas (TCGA) database. Clinical data was also obtained from TCGA. TP53 mutation status was identified using cBioPortal (www.cbioportal.org/), and after merging the mutation data and transcriptome data, we selected 347 patients (111 patients with TP53 mutation and 236 patients without TP53 mutation) for subsequent analysis. We also obtained the copy number variation (CNV) data for TP53 and MEX3A from cBioPortal.
2.2. DE genes and functional enrichment analysis
To investigate the DE RNAs associated with TP53 mutation status in HCC, we performed a differential expression analysis [15,16]. Specifically, we applied a statistical threshold of p < 0.05 and log2|fold change| > 0.7 to identify DElncRNAs, DEmRNAs, and DEmiRNAs. The volcano plots of DE genes were constructed to visualize the expression differences, and the top 15 DE genes (including lncRNAs, mRNAs, and miRNAs) were further displayed in heatmaps. To gain insights into the potential functions of these DE genes, Kyoto Encyclopedia of Genes and Genomes (KEGG), biological process (BP), cellular component (CC), and molecular function (MF) enrichment analyses were performed by Metascape for DEmRNAs and FunRich for DEmiRNAs’ targeted genes.
2.3. Construction of a ceRNA network in HCC
To investigate the interactions between lncRNAs and miRNAs, we utilized the miRcode database (http://www.mircode.org/) and integrated DElncRNAs and DEmiRNAs to identify potential lncRNA-miRNA pairs. We then utilized the miRDB (http://mirdb.org/) and TargetScan (http://www.targetscan.org/) databases to predict the targeted mRNAs of these miRNAs and intersected them with DEmRNAs to generate miRNA-mRNA pairs. Using these predicted pairs, we constructed a ceRNA network for HCC utilizing Cytoscape. The cytoHubba plug-in was used to identify a hub ceRNA network composed of 20 genes based on the betweenness among all nodes.
2.4. Prediction of the subcellular locations of lncRNAs
As a ceRNA, lncRNA can act as a miRNA sponge by binding to miRNA in the cytoplasm, which results in the reduced activity of miRNA and indirectly upregulates the expression of miRNA target genes. To determine the subcellular location of the four lncRNAs, we obtained their sequences from lncBook (https://ngdc.cncb.ac.cn/lncbook/) and submitted them to lncLocator (http://www.csbio.sjtu.edu.cn/bioinf/lncLocator/) for prediction.
2.5. Co-expression analysis
We performed Pearson's correlation analysis to investigate the potential regulatory relationships between lncRNA-miRNA, lncRNA-mRNA, and miRNA-mRNA pairs in HCC. P-value <0.05 and |R| > 0.1 were used as the criteria to identify pairs with potential regulatory effects.
2.6. Methylation analysis of MEX3A
We utilized the Tumor Immune Estimation Resource (TIMER) database to explore the correlation between MEX3A expression and three DNA methyltransferases. To examine the methylation status of MEX3A in normal and HCC tissues, we performed an analysis using UALCAN (http://ualcan.path.uab.edu/). We also investigated the association between MEX3A expression and its methylation level through cBioPortal. Moreover, we assessed the relationships between various methylation sites and their impact on expression and prognosis using MethSurv (https://biit.cs.ut.ee/methsurv/) and MEXPRESS (https://mexpress.be/).
2.7. PPI and GSEA
In order to investigate the potential mechanism of MEX3A in HCC, we conducted a thorough analysis of its PPI using GeneMANIA (http://genemania.org/). Moreover, to identify the key signaling pathways and immune-related pathways that might be implicated in this process, we performed GSEA based on the expression of MEX3A. By doing so, we aimed to shed light on the underlying molecular mechanisms of MEX3A in HCC.
2.8. Immune cell infiltration, immune checkpoints and immune therapy analysis
We utilized the ESTIMATE algorithm to calculate immune, stromal, and ESTIMATE scores in our analysis. To determine the correlation between MEX3A expression and immune cell infiltration, we employed several algorithms including TIMER, CIBERSORT, CIBERSORT-ABS, QUANTISEQ, MCPCOUNTER, XCELL, and EPIC. Furthermore, we used ten immune checkpoints, namely ADORA2A, CSF1R, CTLA4, IL10, IL10RB, LGALS9, PDCD1, TGFB1, TGFBR1, and VTCN1, to assess their association with MEX3A expression using the TIMER algorithm. To evaluate the immune dysfunction and exclusion in HCC patients, we utilized the Tumor Immune Dysfunction and Exclusion (TIDE) algorithm. By uploading the transcriptome data to the TIDE website (http://tide.dfci.harvard.edu), we obtained the TIDE score and the responsiveness of HCC patients to immunotherapy. The immunophenoscore (IPS), including MHC molecules, effector cells, immune checkpoints and immunosuppressive cells, can determine immunogenicity. We obtained IPS, IPS-CTLA4 blocker, IPS-PD1/PDL1/PDL2 blocker and IPS-CTLA4-and PD1/PDL1/PDL2 blocker from The Cancer Immunome Atlas (TCIA, https://tcia.at/).
2.9. Clinical correlation analysis
We conducted a comparative analysis of the expression levels of LINC00491, TCL6, hsa-miR-139-5p, MEX3A, and TP53 across different tumor stages and degrees of tumor differentiation. Additionally, we utilized the prognostic data available on cBioPortal to calculate OS, disease-free survival (DFS), disease-specific survival (DSS), and progression-free survival (PFS) to gain insights into the potential clinical significance of these genes.
2.10. Drug sensitivity analysis
We performed drug sensitivity analysis of MEX3A using GSCALite (http://bioinfo.life.hust.edu.cn/GSCA/) by analyzing the small molecules and drugs obtained from the cancer therapeutics response portal (CTRP). The top six drugs that showed the highest correlation with MEX3A expression levels were selected for further investigation. To gain a better understanding of the chemical structures of these drugs, we generated 3D figures of the six drugs using PubChem (https://pubchem.ncbi.nlm.nih.gov/).
2.11. Dual luciferase reporter gene assay
The dual-luciferase reporter assay was performed in accordance with the manual provided with the Dual Luciferase Reporter Assay Kit (Vazyme, China). First, the amount of plasmid needed for transfection was calculated. Cell lysis and fuorescence intensity assessments were performed 48h after transfection.
3. Results
3.1. Functional analysis of TP53 in HCC
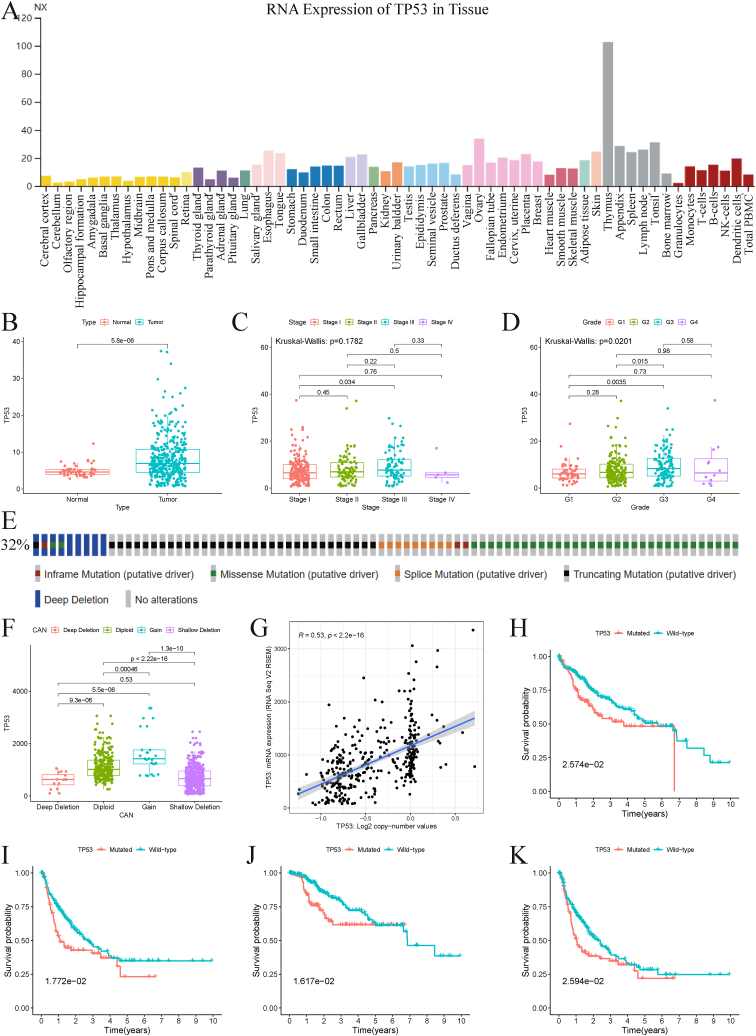
TP53 is a well-known tumor-associated gene that encodes a tumor suppressor protein and plays a crucial role in various human cancers, including HCC. In this study, we first investigated the role of TP53 in HCC. Using the HPA database, we analyzed the RNA expression levels of TP53 in various tissues (Fig. 2A) and found that TP53 was upregulated in HCC compared to normal tissues (Fig. 2B). Further analysis of the relationship between TP53 and clinicopathological features revealed that TP53 expression showed no significant difference in different tumor stages (Fig. 2C) but increased with increasing tumor grade (Fig. 2D).
Fig. 2.
Overview of the tumor suppressor TP53 in HCC. (A) RNA expression of TP53 in various normal tissues. (B) Relative TP53 expression in normal and tumor tissues of HCC patients. (C) TP53 expression in different tumor stages. (D) TP53 expression in different degrees of tumor differentiation. (E) Frequency of genetic alterations with TP53 in HCC from cBioPortal. (F) TP53 expression in different copy-number alterations of TP53. (G) Correlation between expression and copy-number values of TP53. Prognostic analysis between TP53 mutated and wild type groups, including OS (H), DFS (I), DSS (J), and PFS (K). HCC, Hepatocellular carcinoma; OS, Overall survival; DFS, Disease Free survival; DSS, Disease-specific survival; PFS, Progression Free survival.
We also analyzed the alterations of TP53 in 114 (32 %) of 353 HCC patients and found various types of mutations, including in-frame mutations, missense mutations, splice mutations, truncating mutations, and deep deletions (Fig. 2E). HCC samples with deep and shallow deletions had lower TP53 expression than those with diploid and gain (Fig. 2F). Additionally, we observed a positive correlation between TP53 copy number values and mRNA expression (Fig. 2G). Moreover, we found that patients in the TP53-mutated group had worse survival outcomes than those in the TP53-wild type group, regardless of OS, DFS, DSS, or PFS (Fig. 2H–K).
3.2. DE RNA analysis based on TP53 mutations in HCC
To gain further insight into the interplay between TP53 mutations and the RNA network involved in HCC tumorigenesis, we retrieved expression data for mRNA, lncRNA, and miRNA from both normal and HCC samples in TCGA. We divided the patients into TP53-mutated and TP53-wild type groups. Based on the criteria for identifying DE RNAs, we identified 1508 DEmRNAs, 85 DEmiRNAs, and 931 DElncRNAs. Of these, 1314 mRNAs, 82 miRNAs, and 884 lncRNAs were upregulated, while 194 mRNAs, 3 miRNAs, and 47 lncRNAs were downregulated (Figs. S1A–C). We generated a heatmap of the top 15 RNAs with the most significant expression differences between the two groups (Figs. S1D–F).
3.3. Enrichment analysis of the DEmRNAs and DEmiRNAs’ targeted genes
To further investigate potential mechanisms underlying HCC, we conducted enrichment analyses using the Metascape database and FunRich. KEGG analysis identified several pathways, including the cell cycle, drug metabolism, DNA replication, and p53 signaling pathways (Fig. S2A), as well as different signaling events such as the ErbB receptor, IFN-gamma, IGF1, and LKB1 pathways (Fig. S2E). BP analysis showed that most of the pathways were related to cell cycle processes (Fig. S2B), cell communication, and signal transduction (Fig. S2F). The most significant pathways in CC were the chromosomal region (Fig. S2C), the nucleus, and the cytoplasm (Fig. S2G). For MF items, the top 5 significant pathways involved in mRNA were gated channel activity, tubulin binding, ATPase acting on DNA, signaling receptor regulator activity, and calcium ion binding (Fig. S2D). The most significant MF pathway was transcription factor activity (Fig. S2H).
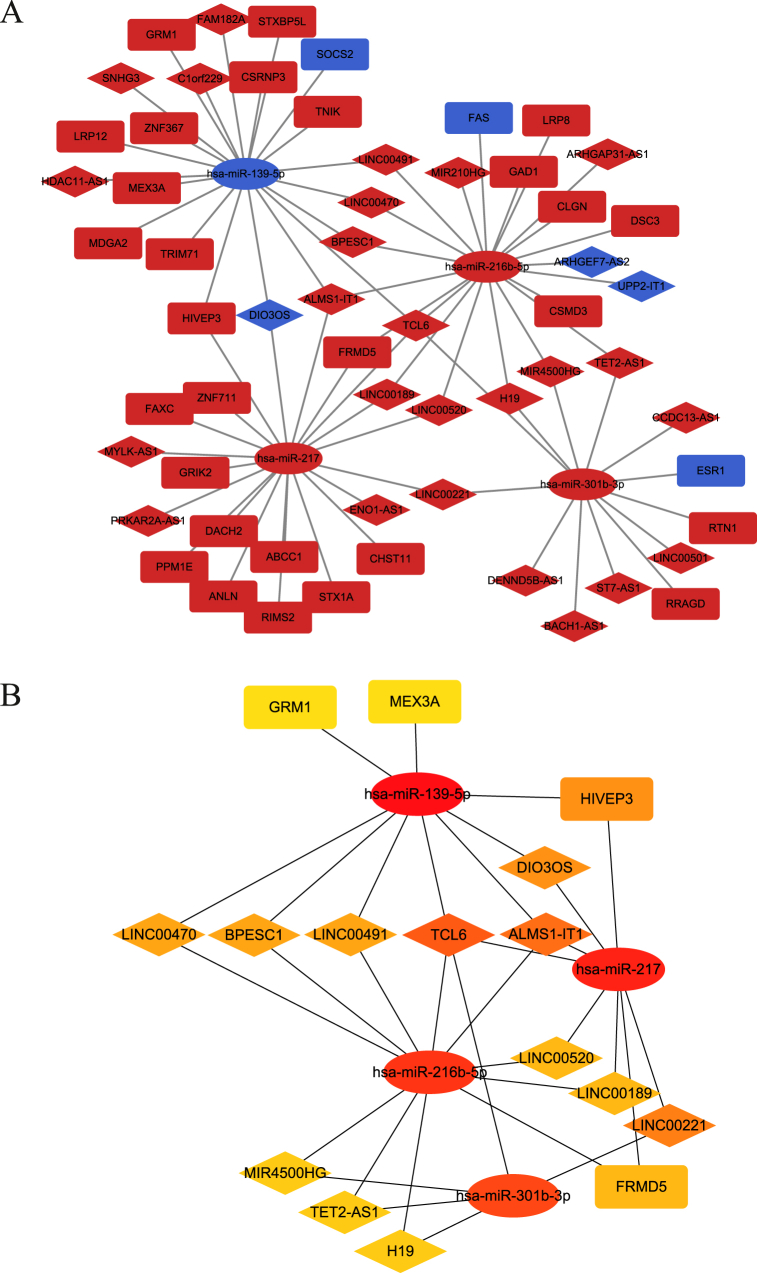
3.4. Construction of the ceRNA network and identification of the hub network in HCC
Using the miRcode, miRDB and TargetScan databases, we predicted lncRNA-miRNA pairs and miRNA-mRNA pairs to construct a ceRNA network. The network, consisting of 28 lncRNAs (25 upregulated and 3 downregulated), 4 miRNAs (3 upregulated and 1 downregulated), and 31 mRNAs (28 upregulated and 3 downregulated), was visualized using Cytoscape (Fig. 3A). To identify the most important nodes in the network, we calculated the betweenness of all nodes and selected the top 20 RNAs with the highest betweenness scores. The hub ceRNA network, consisting of 12 lncRNAs, 4 miRNAs and 4 mRNAs, was constructed and identified as the hub ceRNA ntwork (Fig. 3B).
Fig. 3.
Construction of the lncRNA-miRNA-mRNA regulatory network and identification of core network. (A) Construction of TP53-mutated-related ceRNA network in HCC. The diamond represents lncRNAs, the ellipse represents miRNAs and the round rectangle represents mRNAs. Red represents up-regulated genes and blue represents down-regulated genes. (B) The hub ceRNA network of 20 genes identified by Cytoscape.
3.5. Identification of the LINC00491/TCL6-hsa-miR-139-5p-MEX3A axis in HCC
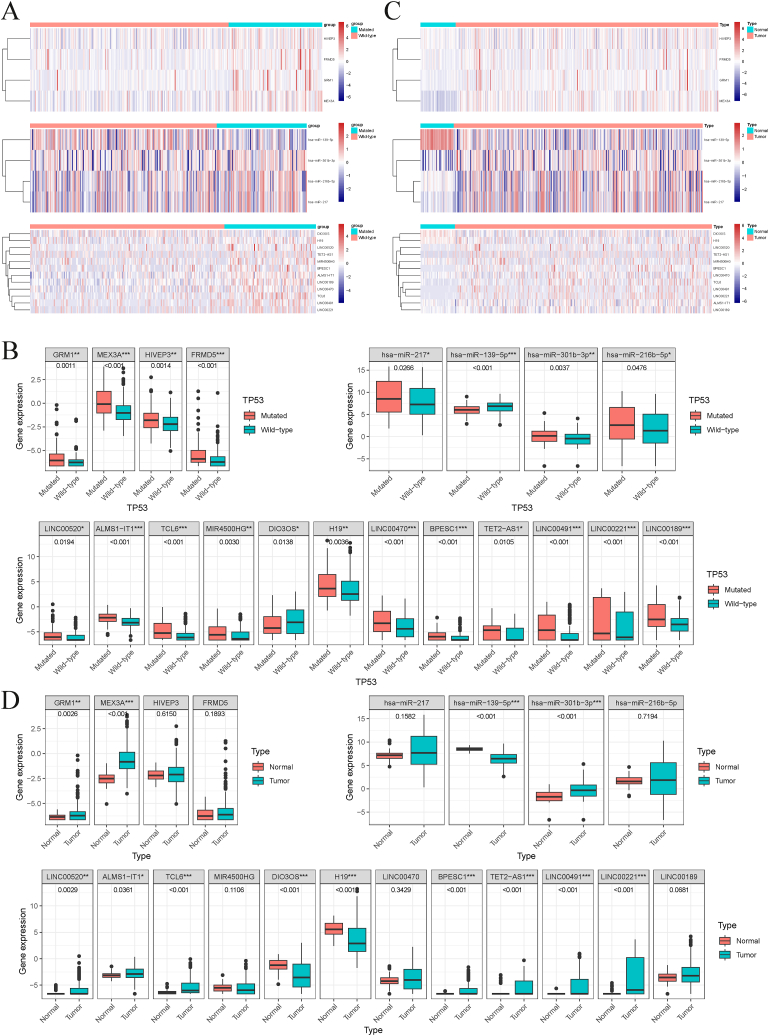
To investigate the ceRNA network underlying the pathogenesis of HCC, we first examined the expression differences of 20 RNAs between TP53-mutated and TP53-wild type groups as well as normal liver and HCC tissues using the Mann-Whitney test. We found 4 upregulated mRNAs (HIVEP3, FRMD5, GRM1, MEX3A), 3 upregulated miRNAs (hsa-miR-301b-3p, hsa-miR-216b-5p, hsa-miR-217), 1 downregulated miRNA (hsa-miR-139-5p), 11 upregulated lncRNAs (H19, LINC00520, TET2-AS1, MIR4500HG, BPESC1, ALMS1-IT1, LINC00189, LINC00470, TCL6, LINC00491, LINC00221) and 1 downregulated lncRNA (DIO3OS) in the TP53-mutated group compared with the TP53-wild type group (Fig. 4A and B). Further analysis of the expression levels of the 20 RNAs in normal and HCC tissues showed 2 upregulated mRNAs (GRM1, MEX3A), 1 upregulated miRNA (hsa-miR-301b-3p), 1 downregulated miRNA (hsa-miR-139-5p), 8 upregulated lncRNAs (H19, LINC00520, TET2-AS1, BPESC1, ALMS1-IT1, TCL6, LINC00491, LINC00221) and 1 downregulated lncRNA (DIO3OS) in HCC (Fig. 4C and D). Combined with the differential analysis of 20 RNAs between TP53 mutated and wild type groups as well as normal and HCC tissues, we believe that 2 mRNAs (GRM1, MEX3A), 2 miRNA (hsa-miR-301b-3p, hsa-miR-139-5p), and 9 lncRNAs (H19, LINC00520, TET2-AS1, BPESC1, ALMS1-IT1, TCL6, LINC00491, LINC00221, DIO3OS) were identified as critical in HCC progression.
Fig. 4.
Differential expression of 20 RNAs in HCC. Heatmaps (A) and scatter plots (B) of the selected 20 genes between TP53 mutated and wild type groups. Heatmaps (C) and scatter plots (D) of the selected 20 genes between normal and tumor tissues in HCC. *P < 0.05; **P < 0.01; ***P < 0.001.
Next, we conducted Kaplan-Meier (KM) survival analysis among the 20 RNAs to identify those that were significantly associated with HCC prognosis. Three mRNAs (HIVEP3, FRMD5, MEX3A) showed higher expression levels, indicating a shorter survival time (Fig. S3A). The higher the hsa-miR-301b-5p expression is, the worse the prognosis. The effect of hsa-miR-139-5p showed the opposite result (Fig. S3B). The results of the effects of lncRNAs on prognosis showed that the expression levels of LINC00520, BPESC1, ALMS1-IT1, LINC00470, TCL6, LINC00491, and LINC00221 were negatively correlated with prognosis, while DIO3OS expression was positively correlated with prognosis (Fig. S3C).
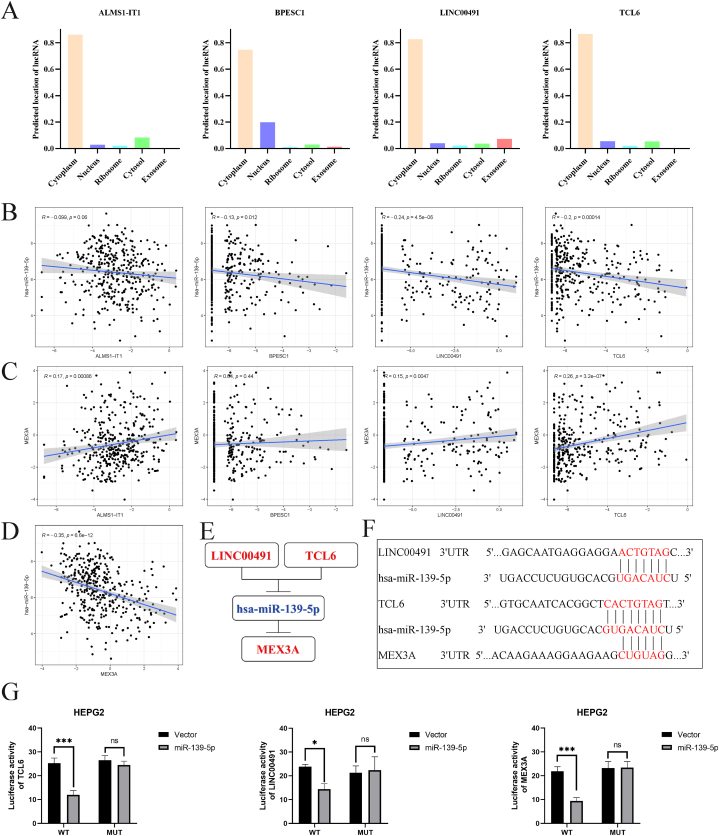
In summary, the mRNA with intersection is MEX3A, the miRNAs with intersection are hsa-miR-301b-3p, hsa-miR-139-5p, and the lncRNAs with intersection are LINC00520, BPESC1, ALMS1-IT1, TCL6, LINC00491, LINC00221, DIO3OS. Based on the molecular sponge mechanism of ceRNA network, we believe that miRNA expression is negatively correlated with mRNA. Since MEX3A is highly expressed in tumors, we selected hsa-miR-139-5p, which is lowly expressed in tumors, and excluded hsa-miR-301b-3p. Similarly, we selected lncRNAs (LINC00520, BPESC1, ALMS1-IT1, TCL6, LINC00491, LINC00221) that were highly expressed in tumors. LINC00520 and LINC00221 did not have a potential predictive relationship with hsa-miR-139-5p, so we excluded these two lncRNAs. Finally, 4 lncRNAs (BPESC1, ALMS1-IT1, TCL6, LINC00491), hsa-miR-139-5p and MEX3A were included for further analysis.
We predicted the subcellular localization of the 4 lncRNAs and found that they were mainly located in the cytoplasm (Fig. 5A). Correlation analysis indicated that hsa-miR-139-5p was negatively related to BPESC1, LINC00491, and TCL6 (Fig. 5B), while MEX3A was positively related to ALMS1-IT1, LINC00491, and TCL6 (Fig. 5C). Notably, MEX3A and hsa-miR-139-5p showed a significant negative correlation (Fig. 5D). These findings suggested that LINC00491/TCL6 may act as a ceRNA to regulate MEX3A expression by sponging hsa-miR-139-5p (Fig. 5E). The binding sites between LINC00491/TCL6/MEX3A and hsa-miR-139-5p shown in Fig. 5F. In addition, Fig. 5G shows that luciferase activities of wild-type LINC00491, TCL6 and MEX3A in HEPG2 cells was obviously reduced after co-transfection of miR-106a-3p mimic, but did not change the activities of mutant form, which suggest that miR-139-5p is a direct target of LINC00491, TCL6 and MEX3A.
Fig. 5.
Construction and correlation analysis of the ceRNA network. (A) The subcellular localization of the 4 lncRNAs (ALMS1-IT1, BPESC1, LINC00491 and TCL6) predicted by lncLocator. (B) Correlation between the expression of hsa-miR-139-5p and the 4 lncRNAs. (C) Correlation between the expression of MEX3A and the 4 lncRNAs. (D) Correlation between the expression of hsa-miR-139-5p and MEX3A. (E) Schematic model of the ceRNA network. Red represents up-regulated and blue represents down-regulated. (F) Base pairing between LINC00491, TCL6 and hsa-miR-139-5p predicted by miRcode, hsa-miR-139-5p and MEX3A predicted by Targetscan. (G) Luciferase activities of wild type (WT) and mutated (Mut) LINC00491, TCL6 and MEX3A reporter plasmid in HEPG2 cells co-transfected with miR-139-5p mimic.
3.6. Correlation between clinical parameters and the ceRNA network
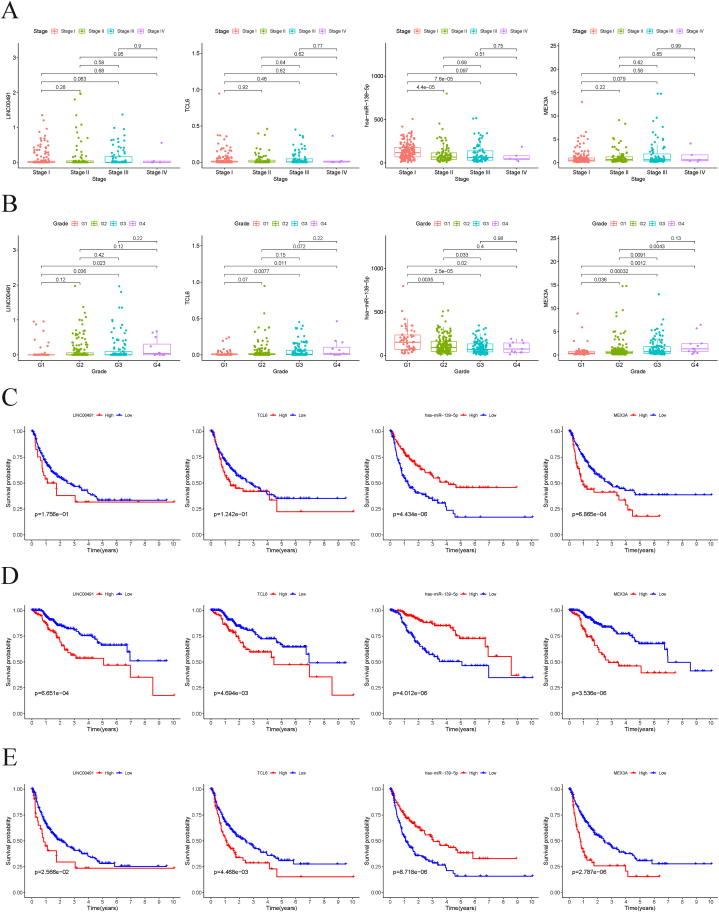
To highlight the significance of the ceRNA network we identified, we examined its correlation with several clinical parameters, such as tumor stage, grade, DFS, DSS, and PFS. Our findings revealed that the expression of hsa-miR-139-5p was inversely associated with tumor stage, indicating that its downregulation could be a contributing factor in HCC progression (Fig. 6A). Furthermore, we observed that TCL6, hsa-miR-139-5p, and MEX3A expression levels were significantly associated with tumor grade (Fig. 6B). Notably, patients with low hsa-miR-139-5p and high MEX3A expression had a shorter DFS (Fig. 6C). Regarding DSS and PFS, our results demonstrated that two lncRNAs (LINC00491 and TCL6) and MEX3A were negatively correlated with patient prognosis, while the expression of hsa-miR-139-5p was positively correlated with prognosis (Fig. 6D and E).
Fig. 6.
The distribution of expression of LINC00491, TCL6, hsa-miR-139-5p and MEX3A in different tumor stages (A) and degrees of tumor differentiation (B). DFS (C), DSS (D) and PFS (E) analysis of LINC00491, TCL6, hsa-miR-139-5p and MEX3A in HCC.
3.7. Analysis of the methylation, mutation status and function of MEX3A in HCC
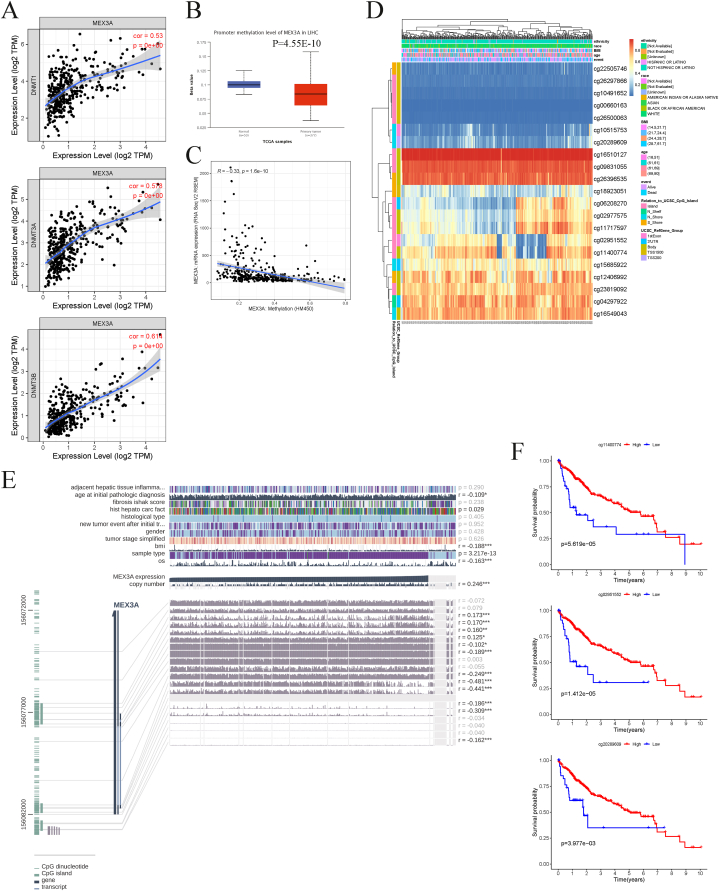
We analyzed the methylation status of MEX3A in HCC using various databases. We investigated the correlations between MEX3A and three DNA methyltransferases, which showed significant positive correlations (Fig. 7A). The promoter methylation of MEX3A was found to be decreased in HCC compared to normal tissues (Fig. 7B), and we observed a negative correlation between the methylation level of MEX3A and its mRNA level (Fig. 7C). A heatmap summarizing the methylation extents at various sites in MEX3A was also generated (Fig. 7D). We further found that 12 of 19 methylation sites were significantly correlated with MEX3A expression (Fig. 7E). Three methylation sites (cg11400774, cg02951552, and cg20289609) of MEX3A showed markedly negative correlations with its expression, and we explored the correlation between each methylation site and prognosis, demonstrating that high methylation of the three sites indicated a better prognosis (Fig. 7F). Seven of the other nine sites were also significantly correlated with the prognosis of HCC (Fig. S4).
Fig. 7.
Methylation analysis of MEX3A. (A) Correlation between MEX3A and three DNA methyltransferases (DNMT1, DNMT3A, and DNMT3B). (B) Promoter methylation level of MEX3A in normal and HCC tissues using UALCAN. (C) Correlation between expression and methylation level of MEX3A using cBioPortal. (D) Methylation site of MEX3A in HCC using MethSurv. (E) Correlation between MEX3A expression and various methylation site using MEXPRESS. (F) Overall survival analysis of three methylation sites (cg11400774, cg02951552 and cg20289609) of MEX3A in HCC patients.
We also found that HCC samples with amplification and shallow deletion had higher MEX3A expression than those with diploid and gain (Fig. S5A). Furthermore, we found a positive correlation between MEX3A copy number values and expression (Fig. S5B). The alteration frequency of MEX3A in HCC was 11 %, dominated by amplification (Fig. S5C). To further elucidate the functions of MEX3A in HCC, we performed PPI and GSEA analyses. The PPI network showed that MEX3A may be involved in the regulation of mRNA processing and RNA splicing (Fig. S5D). GSEA analysis indicated that MEX3A was involved in various classical signaling pathways associated with tumor occurrence and development, such as the JAK-STAT, MAPK, NOTCH, and p53 signaling pathways (Fig. S5E).
3.8. Immune landscape, immunotherapy analysis and drug screening related to MEX3A in HCC
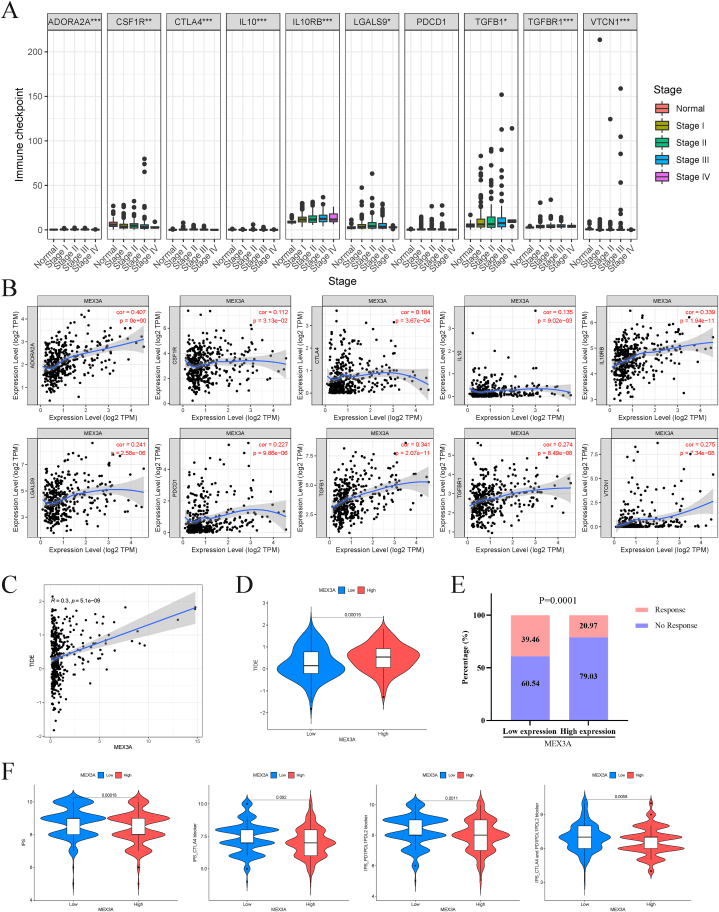
Regarding immune-related pathways, GSEA analysis revealed that the high-MEX3A expression group had enriched chemokine signaling and B- and T-cell receptor signaling pathways (Fig. S6A). The ESTIMATE algorithm revealed that the high-MEX3A expression group had lower immune and stromal scores (Fig. S6B). We also assessed immune cell infiltration using seven algorithms, which showed significant differences in a variety of immune cells, such as B cells, macrophages, monocytes, and Tregs, between the high- and low-MEX3A expression groups (figs. S6C–D). We conducted an analysis of immune checkpoint expression in normal and different tumor stages, with a focus on ADORA2A, CSF1R, CTLA4, IL10, IL10RB, LGALS9, TGFB1, TGFBR1, and VTCN1 (Fig. 8A). We also performed a correlation analysis and observed positive associations between MEX3A expression and these immune checkpoints (Fig. 8B). This prompted us to investigate the potential of MEX3A as a therapeutic target for immunotherapy. Using the TIDE website, we predicted the responsiveness to immunotherapy based on TIDE scores and found a positive correlation between MEX3A expression and TIDE score (Fig. 8C). Furthermore, we discovered that the high-MEX3A expression group had a higher TIDE score (Fig. 8D). Notably, our immunotherapy predictions revealed that the low-MEX3A expression group had a better likelihood of responding to immunotherapy (Fig. 8E). To evaluate the benefits of immunotherapy (anti-CTLA4 and anti-PD1) for patients, we utilized the TCIA website and compared the potential benefits between the high- and low-MEX3A expression groups (Fig. 8F). Our analysis demonstrated that the low-MEX3A expression group had a higher IPS (immunophenoscore) than the high-MEX3A expression group across all subgroups (CTLA4 blocker and/or PD1/PDL1/PDL2 blocker).
Fig. 8.
Immune checkpoints and immunotherapy related to MEX3A in HCC. (A) The distribution of expression of 10 immune checkpoint genes in different tumor stages. (B) Correlation between MEX3A expression and 10 immune checkpoint genes. (C) Correlation between MEX3A expression and TIDE score. (D) Difference of TIDE score between the low- and high-MEX3A expression groups. (E) The comparison of beneficiaries from immunotherapy between the low and high-MEX3A expression groups. (F) Differences of IPS, IPS-CTLA4 blocker, IPS-PD1/PDL1/PDL2 blocker and IPS-CTLA4 and PD1/PDL1/PDL2 blocker between the low and high-MEX3A expression groups. TIDE, Tumor Immune Dysfunction and Exclusion. *P < 0.05; **P < 0.01; ***P < 0.001.
Finally, we explored the correlations between drug sensitivity and MEX3A expression using the GSCALite website. Our analysis revealed significant negative correlations between MEX3A expression and the IC50 of various drugs (Fig. S7A), with six drugs (CAY10618, GSK-J4, STF-31, belinostat, daporinad, and tivantinib) exhibiting the strongest relevance. We further obtained the 3D structures of these six drugs through PubChem (Fig. S7B).
4. Discussion
Primary liver cancer, specifically HCC, is a significant public health concern as it is the sixth most prevalent cancer worldwide and the third leading cause of cancer mortality [1]. However, the symptoms of HCC are often subtle and can go unnoticed in the early stages, leading to a delayed diagnosis and a limited range of treatment options. Consequently, most HCC patients are diagnosed at an advanced stage, where the therapeutic efficacy of standard treatment modalities such as surgical resection, liver transplantation, ablation, and transarterial chemoembolization (TACE) is poor. This is reflected in the low five-year survival rate for liver cancer. To address this issue, complementary systemic therapies such as immune checkpoint inhibitors (ICIs), tyrosine kinase inhibitors (TKIs), and monoclonal antibodies are being developed and used in advanced cancer patients [17]. Therefore, it is essential to investigate the novel pathogenesis of HCC to enhance our ability to identify, prevent, and treat the disease, with the ultimate goal of improving patient outcomes.
Currently, the advancements in omics liquid biopsy have developed rapidly as a diagnostic and monitoring tool. The term “liquid biopsy” encompasses circulating tumor DNA (ctDNA)/cell-free DNA (cfDNA), circulating tumor cells (CTCs), circulating miRNAs, and exosomes [18,19]. The use of liquid biopsy may make it possible to improve HCC early diagnosis. According to a study, the frequency of detected mutations of ctDNA responded with the physical mutations in the pathocellular carcinoma were 44 % (RAS), 63.0 % (TERT), 48.1 % (TP53), and 37.0 % (CTNNB1) respectively [20]. Additionally, in Chinese cohorts with high HBV infection frequency, 38.6 % of HCC patients contained a mutation gene and TP53 had the highest mutation rate (60.0 %) [21]. HCC has a high incidence of ctDNA TP53 mutation identification. Despite this, because the TP53 mutation is common in many malignant tumors, there is little evidence that it is tissue-specific in HCC ctDNA [22]. TP53 acts as a transcription factor and participates in cancer-related biological processes by regulating target genes such as miRNAs and lncRNAs [23,24]. However, the downstream changes in the ceRNA network mediated by TP53 mutations are not yet fully understood. Therefore, exploring the mutation of TP53 in HCC patients and identifying characteristic TP53 mutations is particularly important for improving the early diagnosis of HCC. In this study, we aimed to shed light on this topic by analyzing TP53-related transcriptomic and mutational data in HCC patients obtained from the TCGA and cBioPortal databases. We compared data from TP53-mutated patients with TP53-wild type patients to identify downstream DE lncRNAs, miRNAs, and mRNAs. Next, we used the miRcode database to predict lncRNA-miRNA pairs and the miRDB and TargetScan databases to predict miRNA-mRNA pairs. By combining this information, we constructed a ceRNA network (LINC00491/TCL6-has-miR-139-5p-MEX3A), which we visualized using Cytoscape. Our findings provide novel insights into the role of TP53 mutations in HCC and may contribute to the development of new therapeutic strategies for this disease.
Studies have shown that LINC00491 is highly expressed in HCC tumor tissues and cells. Interference with LINC00491 expression in MHCC-97H and LF cell lines, which are associated with HCC, significantly reduced tumor cell proliferation, migration, and invasion, as well as tumor cell cycle progression. In BALB/c mice, knockout of LINC00491 resulted in a significant reduction in HCC tumor volume and weight [25]. Overexpression of LINC00491 promoted proliferation and distant lung metastases in mice in another study [26]. TCL6 was initially linked to T-cell leukemia and clear cell renal cell carcinoma [27,28]. However, recent research has found that Lnc-TCL6 shows a substantial increase in whole blood at an early clinical stage and is strongly predictive of liver cirrhosis. Furthermore, TCL6 expression is significantly linked to HBV infection status and projected risk-metastasis features [29,30]. The KM analysis in this research also revealed that TCL6 and LINC00491 expression levels were negatively correlated with the prognosis of HCC patients.
MicroRNAs (miRNAs) play a critical role in ceRNA networks by connecting lncRNAs and mRNAs through shared MRE structures. Among the DEmiRNAs driven by TP53 mutation, miR-139-5p exhibited the most significant differential expression, and its expression was notably downregulated in HCC tumor tissues. Moreover, low levels of miR-139-5p were associated with poor prognosis. Previous research has demonstrated that silencing miR-139-5p promoted cell viability, colony formation, migration, and invasion of HCC cells, whereas overexpressing miR-139-5p had the opposite effect. In a xenograft mouse model, miR-139 also inhibited HCC growth [31]. Interestingly, TP53 could directly bind to the TP53 responsive site in the 5′ flanking region of miR-139-5p in the colon cancer cell line HCT116, which activated miR-139-5p transcription and suppressed phosphodiesterase 4D protein production, thereby inhibiting colon cancer progression [32]. These findings are consistent with the data in our study, which also showed lower expression of miR-139-5p in the TP53 mutated group.
MEX3A is a member of the MEX3 family and has been implicated in cancer progression. Elevated expression of MEX3A has been observed in several human cancers, including liver, colon, gastric, bladder, and breast cancer. Studies have shown that MEX3A promotes tumor cell proliferation, invasion, and migration in gastric and breast cancer [[33], [34], [35], [36], [37]]. Notably, a dual-luciferase assay found that miR-139-5p can bind to and inhibit MEX3A expression in breast cancer cells, confirming the binding site predicted in our research [36]. We also analyzed the methylation status and mutation of MEX3A in HCC. Methylation of MEX3A was decreased and mRNA expression was elevated in HCC. There was a positive association between the three DNA methyltransferases and MEX3A. We identified 12 methylation sites linked to MEX3A expression and investigated the relationships between these sites. MEX3A mutations were also identified in HCC, with a frequency of 11 % mainly due to amplifications. We found a positive correlation between MEX3A copy number values and expression. In addition, we analyzed the immune landscape, immunotherapy, and drug screening related to MEX3A in HCC. We found that the high MEX3A expression group had lower immune and stromal scores, and significant differences in immune cells and immune checkpoints compared to the low expression group. Immunotherapy predictions suggested better responsiveness of the low MEX3A expression group to immunotherapy. We also screened six MEX3A-related drugs (CAY10618, GSK-J4, STF-31, belinostat, daporinad, and tivantinib) to explore the feasibility of treating HCC by targeting MEX3A.
Based on the GSEA results, we found that MEX3A was associated with a variety of classical signaling pathways, such as JAK-STAT, MAPK, P53, WNT and so on. Fang et al. found that MEX3A can promote HCC progression and inhibit sorafenib chemotherapy sensitivity through Hippo signaling pathway [38]. In addition, MEX3A can regulate the malignant progression of breast cancer and lung cancer by activating the PI3K/AKT pathway [39,40]. MEX3A promoted tumor progression by activating EMT and regulating the Wnt/β-catenin pathway via DVL3 in endometrial cancer [41]. Xiang et al. found that MEX3A may promote nasopharyngeal carcinoma development and progression via the miR-3163/SCIN axis by regulating NF-κB signaling [42]. MEX3A was also found to be involved in the p53 signaling pathway, and knockdown of MEX3A in ovarian cancer did not alter p53 mRNA levels but increased p53 protein stability. MEX3A-mediated p53 protein degradation was the key to inhibit ferroptosis and promote tumorigenesis [43]. In our core ceRNA network, TCL6 and LINC00491 were identified as the key lncRNAs. Immune-related pathways such as JAK-STAT signaling pathway were regulated by TCL6 in breast cancer [44]. In addition, TCL6 promoted the development of lung cancer through AKT signaling pathway by interacting with PDK1 [44]. Kulkarni et al. found that TCL6 recruited STAU1 and mediated the attenuation of Src mRNA in renal clear cell carcinoma, which subsequently significantly downregulated the AKT and EMT pathways [45]. In addition, LINC00491 directly interacted with MTSS1 to promote lung cancer proliferation, migration and invasion by affecting the ubiquitination of MTSS1, thereby activating Wnt/β-catenin signaling pathway [45]. KEGG analysis identified TP53 mutation had a major effect on the cell cycle pathway. TP53 can protect cells from growth and division, thus mediating cell-cycle arrest, DNA repair and apoptosis After being activated by various cellular stressors. The cell cycle pathway is altered in at least half of HCC patients with frequent TP53 mutations (12%–48 %), and especially the TP53 R249S mutation related to AFB1 exposure may provide a special growth and survival advantage in human liver cancer cells [[46], [47], [48]]. In the protein regulatory aspect, cytoplasmic TP53 R249S mutant was reported to form complex with TANK-binding protein kinase 1 or viral hepatitis B protein X to evade innate immune surveillance or promote HCC proliferation [49,50]. Transcriptionally, TP53 R249S could interact with other transcriptional factors, such as c-Myc to enhance ribosomal DNA transcription [51], or direct promoter occupancy and activation of chromatin regulators [52].
There are some limitations that should be addressed. Firstly, experimental validation is required to confirm the predicted binding affinities of the lncRNAs, miRNAs, and mRNAs in cellular experiments. Secondly, it is essential to further explore the role and mechanism of the ceRNA network (LINC00491/TCL6-has-miR-139-5p-MEX3A) in HCC through in vivo and in vitro experiments. These experiments will provide more conclusive evidence regarding the functionality of the ceRNA network and its potential as a therapeutic target in HCC.
5. Conclusion
In this study, we utilized a variety of bioinformatic techniques to establish a ceRNA network linked with HCC, based on TP53 mutations. The network provides innovative insights into targeting the pathogenic processes and treatment options for HCC.
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Fundings
Not applicable.
Data availability
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database (https://portal.gdc.cancer.gov/) and cBioPortal database (www.cbioportal.org/).
CRediT authorship contribution statement
Dong Wang: Validation, Software, Investigation. Wenxiang Shi: Writing – review & editing, Writing – original draft, Supervision, Methodology, Formal analysis, Data curation, Conceptualization. Chenjie Qiu: Writing – review & editing, Writing – original draft, Supervision, Project administration, Methodology, Investigation, Formal analysis, Data curation, Conceptualization.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
Not applicable.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30066.
Contributor Information
Wenxiang Shi, Email: swx1995@sjtu.edu.cn.
Chenjie Qiu, Email: qiuchenjie2020@163.com.
Appendix A. Supplementary data
The following are the Supplementary data to this article.
References
- 1.Sung H., Ferlay J., Siegel R.L., et al. Global cancer Statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–249. doi: 10.3322/caac.21660. [DOI] [PubMed] [Google Scholar]
- 2.Lee I.C., Lei H.J., Chau G.Y., et al. Predictors of long-term recurrence and survival after resection of HBV-related hepatocellular carcinoma: the role of HBsAg. Am. J. Cancer Res. 2021;11:3711–3725. [PMC free article] [PubMed] [Google Scholar]
- 3.Rahib L., Smith B.D., Aizenberg R., Rosenzweig A.B., Fleshman J.M., Matrisian L.M. Projecting cancer incidence and deaths to 2030: the unexpected burden of thyroid, liver, and pancreas cancers in the United States. Cancer Res. 2014;74:2913–2921. doi: 10.1158/0008-5472.CAN-14-0155. [DOI] [PubMed] [Google Scholar]
- 4.Long J., Wang A., Bai Y., et al. Development and validation of a TP53-associated immune prognostic model for hepatocellular carcinoma. EBioMedicine. 2019;42:363–374. doi: 10.1016/j.ebiom.2019.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gao Q., Zhu H., Dong L., et al. Integrated proteogenomic characterization of HBV-related hepatocellular carcinoma. Cell. 2019;179:1240. doi: 10.1016/j.cell.2019.10.038. [DOI] [PubMed] [Google Scholar]
- 6.Shi W., Lu J., Li J., et al. Piperlongumine attenuates high calcium/phosphate-induced arterial calcification by preserving P53/PTEN signaling. Front Cardiovasc Med. 2020;7 doi: 10.3389/fcvm.2020.625215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liu J., Ma Q., Zhang M., et al. Alterations of TP53 are associated with a poor outcome for patients with hepatocellular carcinoma: evidence from a systematic review and meta-analysis. Eur. J. Cancer. 2012;48:2328–2338. doi: 10.1016/j.ejca.2012.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Atta M.M., el-Masry S.A., Abdel-Hameed M., Baiomy H.A., Ramadan N.E. Value of serum anti-p53 antibodies as a prognostic factor in Egyptian patients with hepatocellular carcinoma. Clin. Biochem. 2008;41:1131–1139. doi: 10.1016/j.clinbiochem.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 9.Park N.H., Chung Y.H., Youn K.H., et al. Close correlation of p53 mutation to microvascular invasion in hepatocellular carcinoma. J. Clin. Gastroenterol. 2001;33:397–401. doi: 10.1097/00004836-200111000-00011. [DOI] [PubMed] [Google Scholar]
- 10.Nogueira J.A., Ono-Nita S.K., Nita M.E., et al. 249 TP53 mutation has high prevalence and is correlated with larger and poorly differentiated HCC in Brazilian patients. BMC Cancer. 2009;9:204. doi: 10.1186/1471-2407-9-204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wong C.M., Tsang F.H., Ng I.O. Non-coding RNAs in hepatocellular carcinoma: molecular functions and pathological implications. Nat. Rev. Gastroenterol. Hepatol. 2018;15:137–151. doi: 10.1038/nrgastro.2017.169. [DOI] [PubMed] [Google Scholar]
- 12.Sen R., Ghosal S., Das S., Balti S., Chakrabarti J. Competing endogenous RNA: the key to posttranscriptional regulation. Sci. World J. 2014;2014 doi: 10.1155/2014/896206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sanchez-Mejias A., Tay Y. Competing endogenous RNA networks: tying the essential knots for cancer biology and therapeutics. J. Hematol. Oncol. 2015;8:30. doi: 10.1186/s13045-015-0129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Afify A.Y., Ibrahim S.A., Aldamsisi M.H., Zaghloul M.S., El-Ekiaby N., Abdelaziz A.I. Competing endogenous RNAs in hepatocellular carcinoma-the pinnacle of rivalry. Semin. Liver Dis. 2019;39:463–475. doi: 10.1055/s-0039-1688442. [DOI] [PubMed] [Google Scholar]
- 15.Han Q., Zhang X., Ren X., et al. Biological characteristics and predictive model of biopsy-proven acute rejection (BPAR) after kidney transplantation: evidences of multi-omics analysis. Front. Genet. 2022;13 doi: 10.3389/fgene.2022.844709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y., Wang J., Li L., et al. AC010973.2 promotes cell proliferation and is one of six stemness-related genes that predict overall survival of renal clear cell carcinoma. Sci. Rep. 2022;12:4272. doi: 10.1038/s41598-022-07070-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Llovet J.M., Montal R., Sia D., Finn R.S. Molecular therapies and precision medicine for hepatocellular carcinoma. Nat. Rev. Clin. Oncol. 2018;15:599–616. doi: 10.1038/s41571-018-0073-4. [DOI] [PubMed] [Google Scholar]
- 18.Li J., Han X., Yu X., et al. Clinical applications of liquid biopsy as prognostic and predictive biomarkers in hepatocellular carcinoma: circulating tumor cells and circulating tumor DNA. J. Exp. Clin. Cancer Res. 2018;37:213. doi: 10.1186/s13046-018-0893-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.von Felden J., Craig A.J., Villanueva A. Role of circulating tumor DNA to help decision-making in hepatocellular carcinoma. Oncoscience. 2018;5:209–211. doi: 10.18632/oncoscience.446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lim H.Y., Merle P., Weiss K.H., et al. Phase II studies with refametinib or refametinib plus sorafenib in patients with RAS-mutated hepatocellular carcinoma. Clin. Cancer Res. 2018;24:4650–4661. doi: 10.1158/1078-0432.CCR-17-3588. [DOI] [PubMed] [Google Scholar]
- 21.Huang A., Zhao X., Yang X.R., et al. Circumventing intratumoral heterogeneity to identify potential therapeutic targets in hepatocellular carcinoma. J. Hepatol. 2017;67:293–301. doi: 10.1016/j.jhep.2017.03.005. [DOI] [PubMed] [Google Scholar]
- 22.Wu X., Li J., Gassa A., et al. Circulating tumor DNA as an emerging liquid biopsy biomarker for early diagnosis and therapeutic monitoring in hepatocellular carcinoma. Int. J. Biol. Sci. 2020;16:1551–1562. doi: 10.7150/ijbs.44024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Di Agostino S. The impact of mutant p53 in the non-coding RNA world. Biomolecules. 2020;10 doi: 10.3390/biom10030472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen S., Thorne R.F., Zhang X.D., Wu M., Liu L. Non-coding RNAs, guardians of the p53 galaxy. Semin. Cancer Biol. 2021;75:72–83. doi: 10.1016/j.semcancer.2020.09.002. [DOI] [PubMed] [Google Scholar]
- 25.Chen R., Chen Y., Huang W., et al. Comprehensive analysis of an immune-related ceRNA network in identifying a novel lncRNA signature as a prognostic biomarker for hepatocellular carcinoma. Aging (Albany NY) 2021;13:17607–17628. doi: 10.18632/aging.203250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang W., Yang T., Li D., Huang Y., Bai G., Li Q. LINC00491 promotes cell growth and metastasis through miR-324-5p/ROCK1 in liver cancer. J. Transl. Med. 2021;19:504. doi: 10.1186/s12967-021-03139-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou M., Sugimoto J., Hatakeyama T., Russo G., Isobe M. Identification of the TCL6 genes within the breakpoint cluster region on chromosome 14q32 in T-cell leukemia. Oncogene. 2000;19:2796–2802. doi: 10.1038/sj.onc.1203604. [DOI] [PubMed] [Google Scholar]
- 28.Su H., Sun T., Wang H., et al. Decreased TCL6 expression is associated with poor prognosis in patients with clear cell renal cell carcinoma. Oncotarget. 2017;8:5789–5799. doi: 10.18632/oncotarget.11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li L.J., Wu X.Y., Tan S.W., et al. Lnc-TCL6 is a potential biomarker for early diagnosis and grade in liver-cirrhosis patients. Gastroenterol Rep (Oxf) 2019;7:434–443. doi: 10.1093/gastro/goz050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo L.H., Jin M., Wang L.Q., et al. Long noncoding RNA TCL6 binds to miR-106a-5p to regulate hepatocellular carcinoma cells through PI3K/AKT signaling pathway. J. Cell. Physiol. 2020;235:6154–6166. doi: 10.1002/jcp.29544. [DOI] [PubMed] [Google Scholar]
- 31.Zan Y., Wang B., Liang L., et al. MicroRNA-139 inhibits hepatocellular carcinoma cell growth through down-regulating karyopherin alpha 2. J. Exp. Clin. Cancer Res. 2019;38:182. doi: 10.1186/s13046-019-1175-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cao B., Wang K., Liao J.M., et al. Inactivation of oncogenic cAMP-specific phosphodiesterase 4D by miR-139-5p in response to p53 activation. Elife. 2016;5 doi: 10.7554/eLife.15978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yang D., Jiao Y., Li Y., Fang X. Clinical characteristics and prognostic value of MEX3A mRNA in liver cancer. PeerJ. 2020;8 doi: 10.7717/peerj.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H., Zhang X., Luo J., et al. Knockdown of hMex-3A by small RNA interference suppresses cell proliferation and migration in human gastric cancer cells. Mol. Med. Rep. 2012;6:575–580. doi: 10.3892/mmr.2012.943. [DOI] [PubMed] [Google Scholar]
- 35.Chatterji P., Rustgi A.K. RNA binding proteins in intestinal epithelial biology and colorectal cancer. Trends Mol. Med. 2018;24:490–506. doi: 10.1016/j.molmed.2018.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chu J., Li T., Li L., Fan H. MicroRNA-139-5p suppresses cell malignant behaviors in breast cancer through targeting MEX3A. Comput. Math. Methods Med. 2021;2021 doi: 10.1155/2021/6591541. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 37.Huang Y., Fang C., Shi J.W., Wen Y., Liu D. Identification of hMex-3A and its effect on human bladder cancer cell proliferation. Oncotarget. 2017;8:61215–61225. doi: 10.18632/oncotarget.18050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang S., Zheng L., Chen X., et al. MEX3A determines in vivo hepatocellular carcinoma progression and induces resistance to sorafenib in a Hippo-dependent way. Hepatol Int. 2023;17(6):1500–1518. doi: 10.1007/s12072-023-10565-2. Dec. [DOI] [PubMed] [Google Scholar]
- 39.Chen W., Hu L., Lu X., et al. The RNA binding protein MEX3A promotes tumor progression of breast cancer by post-transcriptional regulation of IGFBP4. Breast Cancer Res. Treat. 2023;201(3):353–366. doi: 10.1007/s10549-023-07028-5. Oct. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu X., Wang Y., Zhou G., Zhou J., Tian Z., Xu J. circGRAMD1B contributes to migration, invasion and epithelial-mesenchymal transition of lung adenocarcinoma cells via modulating the expression of SOX4. Funct. Integr. Genomics. 2023;23:75. doi: 10.1007/s10142-023-00972-x. [DOI] [PubMed] [Google Scholar]
- 41.Yang P., Zhang P., Zhang S. RNA-binding protein MEX3A interacting with DVL3 stabilizes wnt/β-catenin signaling in endometrial carcinoma. Int. J. Mol. Sci. 2022;24 doi: 10.3390/ijms24010592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xiang X.X., Liu Y.L., Kang Y.F., Lu X., Xu K. MEX3A promotes nasopharyngeal carcinoma progression via the miR-3163/SCIN axis by regulating NF-κB signaling pathway. Cell Death Dis. 2022;13:420. doi: 10.1038/s41419-022-04871-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang C.K., Chen T.J., Tan G.Y.T., et al. MEX3A mediates p53 degradation to suppress ferroptosis and facilitate ovarian cancer tumorigenesis. Cancer Res. 2023;83:251–263. doi: 10.1158/0008-5472.CAN-22-1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhang Y., Li Z., Chen M., et al. lncRNA TCL6 correlates with immune cell infiltration and indicates worse survival in breast cancer. Breast Cancer. 2020;27:573–585. doi: 10.1007/s12282-020-01048-5. [DOI] [PubMed] [Google Scholar]
- 45.Kulkarni P., Dasgupta P., Hashimoto Y., et al. A lncRNA TCL6-miR-155 interaction regulates the src-akt-EMT network to mediate kidney cancer progression and metastasis. Cancer Res. 2021;81:1500–1512. doi: 10.1158/0008-5472.CAN-20-0832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Puisieux A., Ji J., Guillot C., et al. p53-mediated cellular response to DNA damage in cells with replicative hepatitis B virus. Proc Natl Acad Sci U S A. 1995;92:1342–1346. doi: 10.1073/pnas.92.5.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Ponchel F., Puisieux A., Tabone E., et al. Hepatocarcinoma-specific mutant p53-249ser induces mitotic activity but has no effect on transforming growth factor beta 1-mediated apoptosis. Cancer Res. 1994;54:2064–2068. [PubMed] [Google Scholar]
- 48.Zucman-Rossi J., Villanueva A., Nault J.C., Llovet J.M. Genetic landscape and biomarkers of hepatocellular carcinoma. Gastroenterology. 2015;149:1226–1239.e1224. doi: 10.1053/j.gastro.2015.05.061. [DOI] [PubMed] [Google Scholar]
- 49.Ghosh M., Saha S., Bettke J., et al. Mutant p53 suppresses innate immune signaling to promote tumorigenesis. Cancer Cell. 2021;39:494–508.e495. doi: 10.1016/j.ccell.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Gouas D.A., Shi H., Hautefeuille A.H., et al. Effects of the TP53 p.R249S mutant on proliferation and clonogenic properties in human hepatocellular carcinoma cell lines: interaction with hepatitis B virus X protein. Carcinogenesis. 2010;31:1475–1482. doi: 10.1093/carcin/bgq118. [DOI] [PubMed] [Google Scholar]
- 51.Liao P., Zeng S.X., Zhou X., et al. Mutant p53 gains its function via c-myc activation upon CDK4 phosphorylation at serine 249 and consequent PIN1 binding. Mol Cell. 2017;68:1134–1146.e1136. doi: 10.1016/j.molcel.2017.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhu J., Sammons M.A., Donahue G., et al. Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature. 2015;525:206–211. doi: 10.1038/nature15251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Publicly available datasets were analyzed in this study. This data can be found here: TCGA database (https://portal.gdc.cancer.gov/) and cBioPortal database (www.cbioportal.org/).