Abstract
The structure-activity analysis (SAR) and machine learning were used to investigate potential anti-S. aureus agents in a faster method. In this study, 24 oxygenated benzene ring components with S. aureus inhibition capacity were confirmed by literature exploring and in-house experiments, and the SAR analysis suggested that the hydroxyl group position may affect the anti-S. aureus activity. The 2D-MLR-QSAR model with 9 descriptors was further evaluated as the best model among the 21 models. After that, hesperetic acid and 2-HTPA were further explored and evaluated as the potential anti-S. aureus agents screening in the natural product clustering library through the best QSAR model calculation. The antibacterial capacities of hesperetic acid and 2-HTPA had been investigated and proved the similar predictive pMIC value resulting from the QSAR model. Besides, the two novel components were able to inhibit the growth of S. aureus by disrupting the cell membrane through the molecular dynamics simulation (MD), which further evidenced by scanning electron microscopy (SEM) test and PI dye results. Overall, these results are highly suggested that QSAR can be used to predict the antibacterial agents targeting S. aureus, which provides a new paradigm to research the molecular structure-antibacterial capacity relationship.
Keywords: Anti-S. aureus, QSAR, Molecular dynamic simulation, Hesperetic acid, 2-HTPA
Highlights
-
•
24 oxygenated benzene ring components with anti-S. aureus capacity were collected.
-
•
2D-MLR-QSAR model with 9 parameters was the best model.
-
•
Hesperetic acid and 2-HTPA had been explored as potential anti-S. aureus agents.
-
•
Hesperetic acid and 2-HTPA showed significant anti-S. aureus capacity by breaking the membrane.
1. Introduction
Staphylococcus aureus (S. aureus) is a food-borne pathogen widely in variable food products. It can produce Staphylococcal enterotoxins and further cause serious public health problems, for instance, gastroenteritis [1]. In the United States, there were 1019 illnesses and 127 hospitalizations caused by S. aureus, which represented 12.8 % of all foodborne outbreaks [2]. Besides, the European Food Safety Authority highlighted Staphylococcal enterotoxins as one-fourth bacterial toxins which led to foodborne outbreaks in the European Union in 2020 [3]. It has been reported that S. aureus can contaminate many different types of foods, such as raw meat, milk and milk products, and even the ready-to-eat foods, as well as frozen foods [4]. S. aureus has been considered a majority threat to public health. Therefore, it is urgent to limit the growth of S. aureus and reduce economic costs.
Natural products with nontoxic properties and significant antimicrobial capacities have been considered as the substitutions for artificial bacteriostatic based on the recent demand by consumers. The antimicrobial properties of the secondary metabolites from plants have been proved by the FDA and applied in food, cosmetic, as well as pharmaceutical industries [5]. Among these, oxygenated benzene ring components formed with the aromatic structure showed significant bacteriostatic effects. For instance, eugenol, thymol, and carvacrol, which were structured as an aromatic ring with a hydroxyl group binding, were reported with a broad spectrum of antibacterial capacity [6]. Cinnamaldehyde was also proven with significant antimicrobial capacity and has already been applied in food preservation. It was suggested that the aldehyde structure containing may strengthen the antimicrobial capacity compared with carbonyl groups containing [7]. Besides, aromatic acids, such as cinnamic acid, gallic acid, and vanillic acid, also exhibited broad antibacterial capacity [8,9]. However, with the emergence of resistance of S. aureus, traditional methods of antibacterial agent screening required higher cost and longer time. Exploring a data-driven, sustainable, and expeditious approach toward discovering natural inhibitors against S. aureus represents a promising avenue for future research [10,11].
Quantitative structure-activity relationship (QSAR) is an alternative approach to reduce the cost and improve the efficiency of bacteriostatic findings [12]. QSAR model can be used for interpreting the relationships between the structure and the biological properties [13], and for predicting biological, chemical, as well as physical properties of specific structured components by machine learning [14]. In addition, natural products have been investigated and identified variable biological activities in food applications by the QSAR model [15]. The predicted model was applied to the inhibitory activity of oxygenated aromatic compounds against Geotrichum citri-aurantii [16]. In addition, the inhibitory effect of oxygenated aromatic compounds was suggested to be related to the number and position of hydroxyl, and aldehyde groups, as well as the length of ester chain in the structure-activity relationship (SAR) analysis. It further provided theoretical guidance for the design of anti-Geotrichum citri-aurantii drugs for the effective controlling of citrus sour rot. However, to the best of our knowledge, the SAR study has not been used to investigate the relationships between oxygenated benzene ring components and their anti-S. aureus activities.
In this research article, there first aimed to set up a reliable QSAR model among the selected data, and then predict the potential bacteriostatic agents featured with oxygenated benzene ring structures through the QSAR model. The novel natural components were evidenced the investigated the anti-S. aureus activities and mechanisms, which also confirmed the efficiency of the QSAR model. Finally, there could be a new paradigm for researching the molecular structure−antibacterial capacity relationship.
2. Materials and methods
2.1. Materials
Hesperetic acid, 2-hydroxybenzene-1,4-dicarboxylic acid (2-HTPA), thymol, carvacrol, eugenol, cinnamaldehyde, salicylic acid, trans-cinnamic acid, and trans-ferulic acid, as well as ethanol were purchased from Shanghai Macklin Biochemical Co., Ltd. Tween 80 was supplied by Beijing Solarbio Science & Technology Co., Ltd. LB broth was purchased from Beijing Aoboxing Bio-tech Co., Ltd. S. aureus 186335 was obtained from Beijing Beina Chuanglian Biotechnology Institute. Bacillus subtilis CMCC63501 (B. subtilis), Pseudomonas aeruginosa ATCC27853 (P. aeruginosa), and Escherichia coli ATCC25922 (E. coli) were purchased from Beijing Solarbio Science & Technology Co., Ltd.
2.2. MIC test
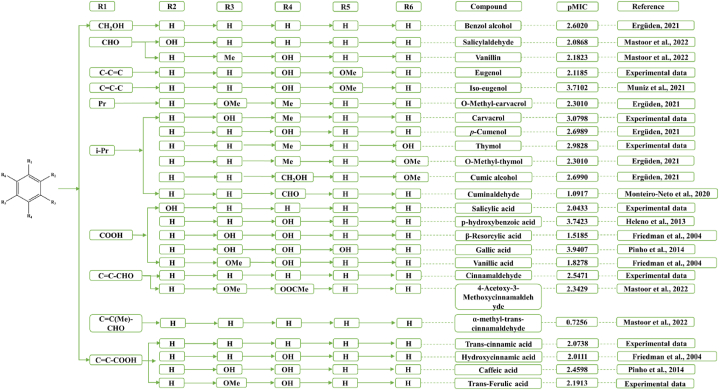
MIC test of thymol, carvacrol, eugenol, cinnamaldehyde, salicylic acid, trans-cinnamic acid, and trans-ferulic acid was processed in the 96-well plate method. The concentration of these components was diluted following the double dilution method. The antibacterial agents were completely mixed with S. aureus (1:1, v: v) and inoculated for 24 h at 37 °C [17]. The first clear well was recorded as the MIC value respectively. The results were further concluded with the data from online literature and converted to pMIC value [16], which was calculated for further model building, and sorted in Fig. 1.
Fig. 1.
Structure and pMIC value of 24 oxygenated benzene ring compounds (Notation: Me, methyl; Pr, propyl; i-Pr, isopropyl. Experimental data: in-house activity data).
2.3. Literature data collection
The online data of oxygenated benzene ring components with anti-S. aureus capacity was indexed from “Web of Science” and “PubMed” with the keywords “against S. aureus”, “antibacterial”, “antibacterial agent”, “against Staphylococcus aureus”, “Natural product antibacterial Staphylococcus aureus”, which was more than 100 papers in total. The certain MIC value of the target components against S. aureus was selected artificially and presented in Fig. 1. The expression of pMIC as a decimal logarithm was aimed to arrive a better mathematical value of biological activities in the model setting [18].
2.4. Machine learning
2.4.1. Data set processing
MIC of natural products structured with a benzene ring and substituted with oxygenated groups was collected and further converted to pMIC value which was calculated by the following formula (1) [16]:
| (1) |
where M referred to the molecular weight of the compound (g/mol), MIC was calculated to mg/mL. Higher pMIC values correspond to stronger anti-S. aureus capacity.
2.4.2. Molecular descriptor processing
Three descriptors: 2D, 3D, and RDKit were used for characterizing the oxygenated benzene ring components. The descriptors 2D and 3D were generated by the PaDEL tool, where the 2D descriptor contained 1444 descriptors (1D and 2D descriptors), and the 3D descriptor contained 431 descriptors. The supplementary materials explained the species and meanings of these descriptors. The open-source chemical information tool, RDKit, is used to read SMILES information and generate descriptors with 208 structural properties.
2.4.3. Feature processing and QSAR model construction
Feature processing and QSAR model construction were processed by Molaical software [19]. Genetic algorithm (GA) and elitism way were the multiple regression models to feature optimization and build leave-one cross-validation (Q2-LOO) [16]. The relative parameters in detail: mutation ratio is 0.5, population size is 200, cross-validation Q2-LOO is selected as the cross-validation index, number of evolutions is 150000, interval steps for repeating population generation are 100. In addition, to select the best model, the number of variables in the model ranges from 3 to 9, where 21 QSAR models were set up.
2.4.4. Model evaluation and screening
Q2-LOO, R2 fitting, MSE, and MAE were used for calculating and screening the QSAR model. Q2 coefficient values between 0.5 and 1.0 indicate that an acceptable model has been established; the closer the R2fitting to 1, the better the model fitting and prediction ability. The smaller the MSE, and MAE values, the smaller the error of model prediction.
2.5. Anti-S. aureus oxygenated benzene ring component screening
The MolNatSim tool [20] was used to match the similarity of each oxygenated benzene ring component in this study to establish a clustering library of the target natural product compounds. To rapidly locate the most similar (MFS score >0.8) natural product molecules and perform conditional screening. After that, the best QSAR model was used to predict the natural products based on the selected components. Besides, the activity of anti-S. aureus was provided according to the average value of pMIC within the 24 structured components against S. aureus.
2.6. The activity study of potential oxygenated benzene ring components against S. aureus
The MIC test of hesperetic acid and 2-hydroxyterephthalic acid (2-HTPA) against S. aureus was the same as the method described in Section 2.2. The clear wells were further placed on plate account ager, where the concentration without colony growth on the plate was marked as the minimum bactericidal concentration (MBC) [21,22].
The inhibition zone of hesperetic acid and 2-HTPA was determined following the referenced method with some modification [23]. Briefly, S. aureus was cultured and diluted by sterile saline to 108 CFU/mL 100 μL of each suspension was spread on the culture plate, and holes were placed on the Petri dishes. After that, samples were added to the wells. The dishes were incubated at 37 °C for 24 h. The inhibition zones were further measured and taken photographs. Triplication was processed.
2.7. Effects of hesperetic acid and 2-HTPA on bacterial morphology
Hesperetic acid and 2-HTPA were added to bacterial suspension with 0.5 × MIC, 1 × MIC, 2 × MIC and inoculated at 37 °C for 2 h. The suspension was later centrifuged and washed with PBS, and fixed with glutaraldehyde (2.5 %) at 4 °C overnight [4]. The morphologies of S. aureus followed processing with some modification [24]. Target bacteria were treated by each bacteriostatic agent at 0.5 × MIC, 1 × MIC, and 2 × MIC for 4 h respectively. After that, the suspensions were centrifuged and rinsed with PBS twice. Glutaraldehyde (2.5 %) was added to fix bacterial cells at 4 °C overnight. The samples were further dehydrated in ethanol at different concentrations and evaporated for observation [25].
2.8. Electrostatic potential and molecular orbital calculation of molecular surface
The ORCA program with the B3LYP(D3) def2-SVP def2/J functional was applied in the geometric optimization of Hesperetic acid and 2-HTPA, and the single point energies were calculated with B3LYP(D3) def2-TZVP def2/J. The electrostatic potential and molecular orbital (HOMO and LUMO) of the two antibacterial agents were calculated by the Multiwfn program, and visualized plot analysis was performed with VMD [26].
2.9. Molecular dynamics (MD) simulation
The MD simulation was processed with the reference of Drew Bennett et al. (2016) [27]. The bilayer containing 170 lipid layers both above and below was constructed using CHARMM-GUI server and Membrane Builder, where DOPC and DOPG in the ratio of 1:1 served as the simple cell membrane model [[28], [29], [30]]. In which, the topological parameters of DOPC and DOPG were obtained by using Amber14sb force fields. ORCA program with the r2 r2SCAN-3c functional was employed in the geometric optimization of hesperetic acid and 2-HTPA. Each single point energy was calculated by B3LYP/G D3 def2-TZVP def2/J RIJCOSX. Multiwfn was carried out to match the RESP charges and give GAFF topological parameters [31]. Then hesperetic acid and 2-HTPA molecules were placed on one side of the membrane as the model system, while the cell membrane was set up as the blank system. There consumed 300 ns in the MD simulation. All MD simulations were executed employing the GROMACS 21.6 package with the TIP3P explicit water model [32]. The electrical neutrality of each system was maintained through the incorporation of counterions, followed by 0.15 M sodium chloride. Energy minimization (1000.0 kJ/mol/nm) for each system was performed in 5000 steps which using the steepest descent method. Following this, the membrane structure underwent complete relaxation through six 500 ps restricted MD simulations in each system [29], which ensured the system maintained a constant temperature (310.5 K) and pressure (1.0 bar). Subsequently, 300 ns production simulations were conducted. The Nose-Hoover and the Parrinello-Rahman protocols were set up as the temperature and pressure control in the system [33,34]. Hydrogen bonds (H-bonds) were constrained using the default linear constraint (LINCS) solver algorithm [35]. The Particle-Mesh Ewald (PME) method was applied in dealing with long-range interactions, as well as a 1.2 nm cutoff for van der Waals interactions [36]. After the MD simulation, the GROMACS package (version 21.6) was performed to analyze the trajectories.
2.10. Statistical analysis
The data were analyzed using OriginPro 2021. One-way analysis of variance (ANOVA) and Duncan's new multiple-range tests were utilized to evaluate the significant differences among the data from different groups by SPSS Statistics 23. The differences were considered significant when p < 0.05 [37,38]. All experiments were conducted in triplicate.
3. Results and discussion
3.1. SAR analysis of the oxygenated benzene ring compounds
The structure of 24 oxygenated benzene ring compounds and their pMIC values were listed in Fig. 1. The pMIC value was here to suggest the antibacterial capacity and increased with the strength of the bacteriostatic effect. Seven oxygenated benzene ring compounds have been evidenced with anti-S. aureus capacity in this research, where the pMIC value was between 2.0433 and 3.0798. Within these compounds, the pMIC value of carvacrol was 3.0798, it presented a much better inhibition activity on S. aureus, and only a little less than iso-eugenol, p-hydroxybenzoic acid, or gallic acid reported before [9,39,40]. The pMIC values of the other six oxygenated benzene ring compounds were higher than 2, which showed higher anti-S. aureus capacity than vanillic acid, α-methyl-trans-cinnamaldehyde, hydroxycinnamic acid, and cuminaldehyde [8,41,42].
The SAR analysis of the oxygenated benzene ring compounds was further conducted in detail. In the oxygenated benzene ring compounds with isopropyl benzene as the parent nucleus, the position of the methoxy linkage had almost no effect on the anti-S. aureus activity, with the pMIC values of o-methyl-carvacrol and o-methyl-thymol were both 2.301, which may be contributed by the stronger hydrophobic property of the methoxy group. Notably, the anti-S. aureus activity was strengthened by the substitution of hydrophobic Me at R4 with the hydrophilic CH2OH group. It could be told that cumic alcohol presented a higher inhibition capacity on S. aureus than O-Methyl-thymol, with the pMIC of cumic alcohol was 2.699. Unlike the methoxy group, the connecting position of the hydroxyl group may affect the anti-S. aureus capacity significantly. The phenolic hydroxyl group can lose hydrogen electrons easily, and it therefore can exert the antibacterial effect as the electron donor [16,43]. In terms of phenolic hydroxyl positions, the hydroxyl groups close to the methyl position (carvacrol, pMIC = 3.0798) were more favorable to enhance the antibacterial activity of the oxygenated benzene ring components than those close to the isopropyl position (thymol, pMIC = 2.9828). This situation may be due to the isopropyl group at R2 producing higher hydrophobicity which attenuates the contribution of the phenolic hydroxyl group to antibacterial capacity. In addition, in the oxygenated benzene ring compounds with styrenic acid as the parent nucleus, the anti-S. aureus activity of caffeic acid with hydroxyl groups attached at both the R3 and R4 positions was superior to that of hydroxycinnamic acid with hydroxyl groups attached at the R4 hydroxyl group and trans-cinnamic acid, which does not contain hydroxyl groups. While, the pMIC value decreased when the hydroxyl group at R3 of caffeic acid was replaced by the methoxy group, which is similar to the SAR results of thymol and carvacrol. However, in the oxygenated benzene ring components with benzoic acid as the parent nucleus, the different positions of the hydroxyl group showed a significant effect on the anti-S. aureus activity. Compared to o-hydroxybenzoic acid, p-hydroxybenzoic acid showed more the anti-S. aureus activity (pMIC = 3.7423) with an increased pMIC of 1.699. However, the number of the hydroxyl groups did not show a direct correlation with anti-S. aureus activity. For instance, the anti-S. aureus capacity of β-Resorcylic acid with a hydroxyl group at R3 was lower, while further increased when substituted by OMe. Compared with β-Resorcylic acid, gallic acid with a hydroxyl group attached to both R3, R4, and R5 positions had the strongest anti-S. aureus effect and the pMIC value reached 3.9407. The difference may be correlated to the different antibacterial mechanisms of the different compounds, such as disrupting the integrity of bacterial cell membranes and interacting with different targets of bacteria [16,44,45].
3.2. MLR-QSAR analysis
The three molecular descriptors characterized the structure of oxygenated benzene ring components, and 21 MLR-QSAR model with S. aureus inhibition activity was set up based on the optimized descriptors and the pMIC values by genetic algorithm. The validation parameters such as Q2-LOO, R2fitting, MAE, MSE, etc. were used to evaluate the robustness and accuracy of the models and placed in Table .1. With the increase of equation variables, the R2fitting of MLR-QSAR model constructed based on the three molecular descriptors increased, while MAE and MSE decreased gradually. In the consideration of smaller datasets, the overfitting of the model may be contributed by too many variables [20]. Therefore, in this study, there trained 3 to 9 variables of the MLR-QSAR model. In the MLR-QSAR model constructed by 9 variables, the 2D-MLR-QSAR model was superior to the 3D-MLR-QSAR and RDKit-MLR-QSAR models. Where the R2fitting of 2D-MLR-QSAR was 0.8988, and Q2-LOO was 0.7776, indicating that the constructed model was acceptable [16]. The MAE and MSE of 2D-MLR-QSAR were 0.3091 and 0.1256 respectively, which were lower than 3D-MLR-QSAR (MAE = 0.3245, MSE = 0.1705) and RDKit-MLR-QSAR (MAE = 0.3347, MSE = 0.1870), which further explained the prediction error of 2D-MLR-QSAR was smaller (Fig. 2). Besides, the reasonability of the equation was evaluated by External Q2, which was calculated following equation (2) below:
| (2) |
Where yi, and fi, are experimental and predicted values in the test set, respectively. ӯ is the average value of train set data. When external Q2 is a negative value, it indicates that there are outliers in the data set [19]. The result indicated that the external Q2 of 2D-MLR-QSAR, 3D-MLR-QSAR, and RDKit-MLR-QSAR constructed by 9 variables were greater than 0, and the external Q2 of 2D-MLR-QSAR was 0.9404, which indicating that the constructed model achieved the examination and can be used for further prediction [46]. Similar results were also obtained in Potdar's study, where the new synthesis components were evaluated and the 3D-MLR-QSAR model was selected and provided the binding sites with their characteristics [47].
Table 1.
QSAR models based on three molecular descriptors (2D, 3D, and RDKit).
| Descriptor type | Equation variable | Q2-LOO | External Q2 | R2fitting | MAE | MSE |
|---|---|---|---|---|---|---|
| 2D | 3 | 0.2788 | 0.1577 | 0.4195 | 0.4848 | 0.4075 |
| 4 | 0.3317 | 0.7915 | 0.5315 | 0.4801 | 0.3776 | |
| 5 | 0.4183 | 0.5703 | 0.6125 | 0.4754 | 0.3287 | |
| 6 | 0.4927 | 0.9018 | 0.6988 | 0.4560 | 0.2866 | |
| 7 | 0.4974 | 0.4689 | 0.6431 | 0.3855 | 0.2840 | |
| 8 | 0.5582 | 0.8010 | 0.7945 | 0.3735 | 0.2496 | |
| 9 | 0.7776 | 0.9404 | 0.8988 | 0.3091 | 0.1256 | |
| MLR | y = 218.98704 + (0.50735) * AATS7e + (−122.97401) * SCH-6 + (−0.63369) * MDEC-23 + (−0.11651) * AATS3i + (−49.62828) * SpMax1_Bhm + (−3.73674) * PetitjeanNumber + (9.20161) * SpMin8_Bhm + (−2.18225) * nHdCH2 + (−0.00136) * ATSC8v | |||||
| 3D | 3 | 0.1661 | −1.6794 | 0.3687 | 0.5474 | 0.4712 |
| 4 | 0.3112 | 0.3249 | 0.5270 | 0.4527 | 0.3892 | |
| 5 | 0.4161 | 0.5715 | 0.5971 | 0.4446 | 0.3300 | |
| 6 | 0.5056 | 0.1541 | 0.6721 | 0.4031 | 0.2793 | |
| 7 | 0.5171 | 0.5071 | 0.7000 | 0.4107 | 0.2728 | |
| 8 | 0.5290 | 0.2649 | 0.7851 | 0.3788 | 0.2661 | |
| 9 | 0.6983 | 0.5009 | 0.8682 | 0.3245 | 0.1705 | |
| MLR | y = −112.00837 + (2.39451) *L2u + (−0.00206) * TDB5v + (0.00683) * TDB6m + (−0.24531) * RDF60u + (−0.00316) * TDB7m + (−0.53723) * RDF60v + (−0.25830) * TDB1m + (61.22847) * TDB2u + (0.01484) * DPSA-2 | |||||
| RDKit | 3 | 0.1429 | 0.7147 | 0.4046 | 0.5515 | 0.4843 |
| 4 | 0.2862 | 0.8292 | 0.4921 | 0.4641 | 0.4033 | |
| 5 | 0.5138 | 0.5799 | 0.6792 | 0.3711 | 0.2747 | |
| 6 | 0.3897 | 0.8544 | 0.5731 | 0.3882 | 0.3449 | |
| 7 | 0.5974 | 0.8337 | 0.7774 | 0.3653 | 0.2275 | |
| 8 | 0.5915 | 0.7053 | 0.8146 | 0.3683 | 0.2308 | |
| 9 | 0.6690 | 0.8763 | 0.8776 | 0.3347 | 0.1870 | |
| MLR | y = 5.85428 + (0.09940) * EState_VSA10 + (−0.15843) * EState_VSA4 + (−0.82728) * fr_para_hydroxylation + (0.11647) * SlogP_VSA4 + (0.05932) * VSA_EState3 + (3.50872) * HallKierAlpha + (0.16995) * EState_VSA1 + (0.72483) * VSA_EState7 + (−0.32154) * fr_Al_COO | |||||
Note: Equations referred to MLR equations with different molecular descriptors under nine parameter calculation.
Fig. 2.
The QSAR models with Leave-One-Out (LOO). (A) 2D. descriptor (B) 3D descriptor. (C) RDKit descriptor.
3.3. Key descriptor analysis
For the best MLR-QSAR equation, there were 9 key descriptors relating to the antibacterial effect on S. aureus in the structure of oxygenated benzene ring components: AATS7e, SCH-6, MDEC-23, AATS3i, SpMax1_Bhm, PetitjeanNumber, SpMin8_Bhm, nHdCH2, and ATSC8v (Table 1). AATS7e and SpMin8_Bhm were correlated with the model positively, and the pMIC value increased with increasing of AATS7e and SpMin8_Bhm. Among, the coefficient value of SpMin8_Bhm is the maximum (9.20161). AATS7e is corresponded to average Broto-Moreau autocorrelation-lag7/weighted by Sanderson electronegativities, while SpMin8_Bhm refers to the smallest absolute eigenvalue of Burden modified matrix-n8/weighted by relative mass [48]. Other descriptors were negatively correlated to the model. SCH-6 is simple chain, order 6 [49], MDEC-23 is molecular distance edge between all secondary and tertiary carbons [50], AATS3i is average Broto-Moreau autocorrelation-lag3/weighted by first ionization potential, SpMax1_Bhm is largest absolute eigenvalue of Burden modified matrix-n1/weighted by relative mass, nHdCH2 [51] and ATSC8v are Count of atom-type H E-State:=CH2 and Centered Broto-Moreau autocorrelation-lag8/weighted by van der Waals volumes separately [48]. The PetitjeanNumber refers to the shape of the cation [52]. These descriptors with negative coefficients were negatively correlated to the antibacterial activity of oxygenated benzene ring components. In this case, the smaller the value of the descriptors, the stronger the inhibitory effect against S. aureus.
3.4. Discovery and prediction of potential oxygenated benzene ring components
The selected best model (2D-MLR-QSAR) was further used to discover and predict the potential oxygenated benzene ring components with anti-S. aureus capacity, which was also able to confirm the efficacy of the model. A total of 50 highly similar oxygenated benzene ring components (MFS >0.8) were explored using the MolNatSim tool based on the structural similarity strategy of extended connectivity fingerprints (Supplementary materials sheet 8) [20]. Among these components, there were 22 components enriched with higher predicted pMIC value (pMICpre >2.5) calculated by 2D-MLR-QSAR equation, and better drug-like properties (QED >0.5) [53]. After that, we tried to purchase these components for experimental screening. However, most of them were still in the identified stage, which was not produced commercially. In this condition, two components were purchased, namely hesperetic acid (pMICpre = 2.881) and 2-HTPA (pMICpre = 2.501).
The ESP map presented the visualized distribution of charges and electrostatic potentials of the molecule (Fig. 3 a and b) [54,55]. The blue region represented the positive charges of 2-HTPA and hesperetic acid, and appeared on the hydrogen atoms of carboxyl groups structurally, which suggests more preference for electrophilic activity. On the other hand, the red region represented the negative charges of hydroxyl groups and alkyl groups, which presented a potential positively related to nucleophilic reactivity [55,56]. The frontier molecular orbitals, which reflected the electronic density distribution and also helped to predict the active sites of molecules [57], were placed in Fig. 3. EHOMO represented the highest occupied molecular orbital and suggested the susceptibility of electron-donating capacity [58], while ELUMO stood for the lowest unoccupied molecular orbitals and reflected the ability of electron-accepting of molecules [59]. The electron density of HOMO on 2-HTPA and hesperetic acid was well distributed on the phenolic benzene ring and hydroxyl group. This implemented these parts may have more tendency to donate electrons and the ability against S. aureus may be stronger [16]. The electron density of LUMO was clouded on the carboxyl group and the carboxyl group with the alkene of 2-HTPA and hesperetic acid respectively. The functional groups may accept electrons easily [60]. Comparing the EHOMO and ELUMO of each molecule, the ability to donate electrons of these components was stronger than accepting. The energy gap (ΔEL-H) between HOMO and LUMO is used to determine the chemical reactivity of the molecule [55]. The energy gaps of 2-HTPA and hesperetic acid were 4.789 eV and 4.268 eV respectively, where a higher energy gap of 2-HTPA suggesting higher kinetic stability than hesperetic acid, and this result was consistent with the pMICpre value resulting from the 2D-QSAR model [59].
Fig. 3.
ESP maps encoded onto the total electron density surface of 2-HTPA (a) and hesperetic acid (b). Molecular orbital surfaces for the HOMO and LUMO of 2-HTPA (c) and hesperetic acid (d).
3.5. Anti- S. aureus activities of hesperetic acid and 2-HTPA
To evaluate the antibacterial activity of hesperetic acid and 2-HTPA, four respective bacteria including S.aureus, B.subtilis, E.coli, and P.aeruginosa were selected to test. The MIC values of hesperetic acid and 2-HTPA on S. aureus showed the same value which was 0.5 mg/mL. While the MIC values on B.subtilis, E.coli, and P.aeruginosa were around 1.0 mg/mL (Table S1). This result suggested that the screened bacteriostatic agents presented obvious inhibition against S. aureus. while the similar MIC values between hesperetic acid and 2-HTPA may be affected by the different solubility. Besides, the pMIC values of hesperetic acid and 2-HTPA were 2.589 and 2.561 respectively, smaller than the MAE value (0.3091). Moreover, the inhibition zone of hesperetic acid was 5.75 ± 0.96 cm, which was a little higher than 2-HTPA (5.00 ± 0.82 cm). The inhibition zone of these two components showed no significant difference (Fig. 4A and B). Furthermore, the morphological changes of S. aureus treated with different concentrations of hesperetic acid and 2-HTPA were observed by SEM to analyze the potential inhibition mechanisms (Fig. 4a–g). Comparing with the control group, the addition of hesperetic acid at different concentrations were able to cause obvious breakages on the membrane, and the degree of the membrane damage was positively correlated with the concentration of the new antibacterial agent. Similarly, the same trend was obtained in the addition of 2-HTPA, and the severe damage on the membrane was more obvious. Propidium Iodide (PI) staining method can be used to evaluated the integrity of the cell membrane. The red fluorescence intensity was observed in the treatment groups (H-MIC and 2H-MIC), while the cell in control group without antibacterial agent addition was no red fluorescence observed. After addition of hesperetic acid and 2-HTPA, the red fluorescence was detected (Fig. S1). PI dye is able to penetrate through a damaged cell membrane, and bound to DNA, finally resulted in red fluoresces [61]. Therefore, this suggested that hesperetic acid and 2-HTPA could damage the membrane integrity of S. aureus [62].
Fig. 4.
Antibacterial activity of inhibition zone of S. aureus treated by hesperetic acid and 2-HTPA (p<0.05) (A and B). SEM of S. aureus treated by different concentration of bacteriostatic agents. (a) S. aureus without treatment, (b) Hesperetic acid at 0.5MIC, (c) Hesperetic acid at MIC, (d) Hesperetic acid at 2MIC, (e) 2-HTPA at 0.5MIC, (f) 2-HTPA at MIC, (g) 2-HTPA at 2MIC.
These results suggested that both hesperetic acid and 2-HTPA were able to inhibit the growth of S. aureus. and the antibacterial mechanisms may be associated with membrane destruction [63].
3.6. Interaction between 2-HTPA and hesperetic acid with the membrane of S. aureus
The MD simulation trajectories of 2-HTPA and hesperetic acid with cell membrane were recorded in Fig. 5A and B. There were lasted 300 ns, and the molecules landed and attacked the cell membrane quickly, which suggested 2-HTPA and hesperetic acid may directly affect the cell membrane visually. On the other hand, the mean square displacement (MSD) of the cell membrane (Fig. 5a) showed an increasing trend with the development of time, which suggested the mobility of the cell membrane increased with the addition of antibacterial agents [64]. It could be the interaction between the cell membrane and the agents, which destroyed the stability of the cell membrane, and accelerated the MSD value with water phase breaking in. This reaction may be related to the hydrogen binding between the cell membrane and the antibacterial agents in the simulation system (Fig. 5b). The differences in membrane number density treated with blank, 2-HTPA, and hesperetic acid (Fig. 5 c to e.) also proved the effective targets of these two molecules were placed on the membrane. Furthermore, the influence of different antibacterial agents (2-HTPA, hesperetic acid) in the lipid bilayer qualified by mass density along the axis perpendicular was shown in Fig. 5e–h. Within 300 ns, the density range of aqueous phase increased and introduced more water molecules into the hydrophobic region of the cell membrane, which meant that the fluidity of the cell membrane was enhanced and loosened, and the stability of the cell membrane could also be disrupted. These figures provided the possible locations of antibacterial agent molecules in the hydrophobic region of the simulation systems [65].
Fig. 5.
MD simulation trajectory of 2-HTPA and cell membrane (A), hesperetic acid and cell membrane (B) (From 0 to 300 ns). Interaction between 2-HTPA and Hesperetic acid with the membrane of S. aureus. MSD value (a), Hydrogen bond number (b), Membrane number densities (c-e represented Blank, 2-HTPA, and hesperetic acid, respectively), Mass densities (f-h represented Blank, 2-HTPA, and hesperetic acid, respectively).
4. Conclusion
In conclusion, MLR-QSAR models with three descriptors (2D, 3D, RDkit) were set up based on the SAR analysis of 24 oxygenated benzene ring components with anti-S. aureus activity and further evaluated by validation parameters (Q2-LOO, R2fitting, MAE, MSE, and external Q2). The SAR results first indicated the position and number of hydroxyl groups at different parent nuclei can affect the inhibition of S. aureus differently. In 21 QSAR models, the 2D-MLR-QSAR model was the best model with higher R2fitting and Q2-LOO, but lower MAE or MSE value, and the external Q2 was also used to examine the acceptability of this model. MLR equation analysis shows AATS7e and SpMin8_Bhm descriptors were correlated with the model positively, while others (SCH-6, MDEC-23, AATS3i, SpMax1_Bhm, nHdCH2, ATSC8v, and PetitjeanNumber) descriptor were negatively correlated to the antibacterial activity. Further, oxygenated benzene ring components were enriched using QSAR modeling combined with a natural product clustering library tool of over 400,000 natural products, and obtained a total of 50 highly similar new oxygenated benzene ring components, among which 22 compounds were considered to be highly inhibitory to S. aureus. MD simulation results suggested that 2-HTPA and hesperetic acid can attack the cell membrane and change the fluidity of the cell membrane. Besides, under the treatments of 2-HTPA and hesperetic acid, the water molecules were much easier into the hydrophobic region, and further disrupted the stability of cells. Antibacterial test results proved that hesperetic acid and 2-HTPA had a better inhibitory effect on S. aureus (pMIC>2.5), and its antibacterial mechanism may be related to the membrane since the disrupted membrane observed by SEM and PI staining tests (Fig. S1). Overall, the effectiveness of SAR and machine learning applications in screening anti-S. aureus of oxygenated benzene ring components, and the novel components, hesperetic acid and 2-HTPA, have been proved the efficient anti-S. aureus capacities. It therefore provided a new insight for the discovery and design of new anti-S. aureus bacteriostatic agents.
Data availability statement
Data will be made available on request directly to the corresponding author.
CRediT authorship contribution statement
Zinan Wang: Writing – review & editing, Writing – original draft, Methodology, Investigation, Formal analysis, Data curation. Fei Pan: Validation, Software, Methodology, Data curation, Conceptualization. Min Zhang: Validation, Supervision. Shan Liang: Supervision, Formal analysis. Wenli Tian: Supervision, Resources.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30389.
Contributor Information
Min Zhang, Email: xzm7777@sina.com.
Shan Liang, Email: liangshan@btbu.edu.cn.
Appendix A. Supplementary data
The following are the Supplementary data to this article:
References
- 1.Umeda K., Ono H.K., Wada T., Motooka D., Nakamura S., Nakamura H., Hu D. High production of egc2-related staphylococcal enterotoxins caused a food poisoning outbreak. Int. J. Food Microbiol. 2021;357 doi: 10.1016/j.ijfoodmicro.2021.109366. [DOI] [PubMed] [Google Scholar]
- 2.Liao X., Chen X., Sant'Ana A.S., Feng J., Ding T. Pre-exposure of foodborne staphylococcus aureus isolates to organic acids induces cross-adaptation to mild heat. Microbiol. Spectr. 2023;11(2) doi: 10.1128/spectrum.03832-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Efsa The European Union one health 2020 zoonoses report. EFSA J. 2021;19(12) doi: 10.2903/j.efsa.2021.6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fan Q., Yan C., Shi C., Xu Y., Ma Y., Zhang C., Peng X., Xia X. Inhibitory effect of coenzyme q0 on the growth of staphylococcus aureus. Foodb. Pathog. Dis. 2019;16(5):317–324. doi: 10.1089/fpd.2018.2559. [DOI] [PubMed] [Google Scholar]
- 5.Hou T., Sana S.S., Li H., Xing Y., Nanda A., Netala V.R., Zhang Z. Essential oils and its antibacterial, antifungal and anti-oxidant activity applications: a review. Food Biosci. 2022;47 [Google Scholar]
- 6.Rao J., Chen B., Mcclements D.J. Improving the efficacy of essential oils as antimicrobials in foods: mechanisms of action. Annu. Rev. Food Sci. Technol. 2019;10(1):365–387. doi: 10.1146/annurev-food-032818-121727. [DOI] [PubMed] [Google Scholar]
- 7.Dorman H.J.D., Deans S.G. Antimicrobial agents from plants: antibacterial activity of plant volatile oils. J. Appl. Microbiol. 2000;88(2):308–316. doi: 10.1046/j.1365-2672.2000.00969.x. [DOI] [PubMed] [Google Scholar]
- 8.Friedman M., Buick R., Elliott C.T. Antibacterial activities of naturally occurring compounds against antibiotic-resistant bacillus cereus vegetative cells and spores, escherichia coli, and staphylococcus aureus. J. Food Protect. 2004;67(8):1774–1778. doi: 10.4315/0362-028x-67.8.1774. [DOI] [PubMed] [Google Scholar]
- 9.Pinho E., Ferreira I.C.F.R., Barros L., Carvalho A.M., Soares G., Henriques M., Santos A. Antibacterial potential of northeastern Portugal wild plant extracts and respective phenolic compounds. BioMed Res. Int. 2014;2014 doi: 10.1155/2014/814590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kou X., Shi P., Gao C., Ma P., Xing H., Ke Q., Zhang D. Data-driven elucidation of flavor chemistry. J. Agric. Food Chem. 2023;71(18):6789–6802. doi: 10.1021/acs.jafc.3c00909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang D., Jia C., Sun D., Gao C., Fu D., Cai P., Hu Q. Data-driven prediction of molecular biotransformations in food fermentation. J. Agric. Food Chem. 2023;71(22):8488–8496. doi: 10.1021/acs.jafc.3c01172. [DOI] [PubMed] [Google Scholar]
- 12.Dias T., Gaudêncio S.P., Pereira F. vol. 17. 2019. https://10.3390/md17010016 (A Computer-Driven Approach to Discover Natural Product Leads for Methicillin-Resistant staphylococcus Aureus Infection Therapy). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Holton T.A., Vijayakumar V., Khaldi N. Bioinformatics: current perspectives and future directions for food and nutritional research facilitated by a food-wiki database. Trends Food Sci. Technol. 2013;34(1):5–17. [Google Scholar]
- 14.Liu Y., Pan F., Wang O., Zhu Z., Li Q., Yang Z., Tian W., Zhao L., Zhao L. Qsar model of pancreatic lipase inhibition by phenolic acids and their derivatives based on machine learning and multi-descriptor strategy. Journal of Agriculture and Food Research. 2023;14 [Google Scholar]
- 15.Zhao Y., Xia Y., Yu Y., Liang G. Qsar in natural non-peptidic food-related compounds: current status and future perspective. Trends Food Sci. Technol. 2023;140 [Google Scholar]
- 16.Che J., Pan F., Chen X., Zhang Y., Tao N., Fu Y. Screening of oxygenated aromatic compounds for potential antifungal activity against geotrichum citri-aurantii through structure–activity relationship analysis. J. Agric. Food Chem. 2022;70(42):13787–13795. doi: 10.1021/acs.jafc.2c04955. [DOI] [PubMed] [Google Scholar]
- 17.Lin C., Luan F., Su S., Jiang A., Tan W., Guo Z. Water-soluble fluorine-functionalized chitooligosaccharide derivatives: synthesis, characterization and antimicrobial activity. Carbohydr. Res. 2023;533 doi: 10.1016/j.carres.2023.108935. [DOI] [PubMed] [Google Scholar]
- 18.N'Dri J.S., Kablan A.L.C., Ouattara B., Koné M., Ouattara L., Kodjo C., Ziao N. Science and Education Publishing Co. Ltd; 2019. Qsar Studies of the Antifungal Activities of α-diaminophosphonates Derived from Dapsone by Dft Method. [Google Scholar]
- 19.Bai Q., Tan S., Xu T., Liu H., Yao X. Molaical: a soft tool for 3d drug design of protein targets by artificial intelligence and classical algorithm. Brief. Bioinf. 2021;22(161) doi: 10.1093/bib/bbaa161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo T., Pan F., Cui Z., Yang Z., Chen Q., Zhao L., Song H. Fapd: an astringency threshold and astringency type prediction database for flavonoid compounds based on machine learning. J. Agric. Food Chem. 2023;71(9):4172–4183. doi: 10.1021/acs.jafc.2c08822. [DOI] [PubMed] [Google Scholar]
- 21.Jabir M. Biomedical applications of eichhornia crassipes. Research journal of biotechnology. 2019;14 [Google Scholar]
- 22.Alhujaily M., Jabir M.S., Nayef U.M., Rashid T.M., Sulaiman G.M., Khalil K.A.A., Rahmah M.I., Najm M.A.A., Jabbar R., Jawad S.F. vol. 13. 2023. https://10.3390/met13040735 (Au/zno Nanocomposites Prepared by Laser Ablation for Enhancement of Antibacterial Activity and Cytotoxic Properties against Cancer Cells). [Google Scholar]
- 23.Geng C., Liu X., Ma J., Ban H., Bian H., Huang G. High strength, controlled release of curcumin-loaded zif-8/chitosan/zein film with excellence gas barrier and antibacterial activity for litchi preservation. Carbohydr. Polym. 2023;306 doi: 10.1016/j.carbpol.2023.120612. [DOI] [PubMed] [Google Scholar]
- 24.Qiao Y., Xu Y., Liu X., Zheng Y., Li B., Han Y., Li Z., Yeung K.W.K., Liang Y., Zhu S., Cui Z., Wu S. Microwave assisted antibacterial action of garcinia nanoparticles on gram-negative bacteria. Nat. Commun. 2022;13(1):2461. doi: 10.1038/s41467-022-30125-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jabir M.S., Rashid T.M., Nayef U.M., Albukhaty S., Almalki F.A., Albaqami J., Alyamani A.A., Taqi Z.J., Sulaiman G.M. Inhibition of staphylococcus aureus α-hemolysin production using nanocurcumin capped au@zno nanocomposite. Bioinorgan. Chem. Appl. 2022;2022 doi: 10.1155/2022/2663812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Humphrey W., Dalke A., Schulten K. Vmd: visual molecular dynamics. J. Mol. Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 27.Bennett W.F.D., Hong C.K., Wang Y., Tieleman D.P. Antimicrobial peptide simulations and the influence of force field on the free energy for pore formation in lipid bilayers. J. Chem. Theor. Comput. 2016;12(9):4524–4533. doi: 10.1021/acs.jctc.6b00265. [DOI] [PubMed] [Google Scholar]
- 28.Jo S., Kim T., Iyer V.G., Im W. Charmm-gui: a web-based graphical user interface for charmm. J. Comput. Chem. 2008;29(11):1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- 29.Lee J., Cheng X., Swails J.M., Yeom M.S., Eastman P.K., Lemkul J.A., Wei S., Buckner J., Jeong J.C., Qi Y., Jo S., Pande V.S., Case D.A., Brooks C.L.I., Mackerell A.D., Jr., Klauda J.B., Im W. Charmm-gui input generator for namd, gromacs, amber, openmm, and charmm/openmm simulations using the charmm36 additive force field. J. Chem. Theor. Comput. 2016;12(1):405–413. doi: 10.1021/acs.jctc.5b00935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu E.L., Cheng X., Jo S., Rui H., Song K.C., Dávila-Contreras E.M., Qi Y., Lee J., Monje-Galvan V., Venable R.M., Klauda J.B., Im W. Charmm-gui membrane builder toward realistic biological membrane simulations. J. Comput. Chem. 2014;35(27):1997–2004. doi: 10.1002/jcc.23702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu T., Chen F. Multiwfn: a multifunctional wavefunction analyzer. J. Comput. Chem. 2012;33(5):580–592. doi: 10.1002/jcc.22885. [DOI] [PubMed] [Google Scholar]
- 32.Van Der Spoel D., Lindahl E., Hess B., Groenhof G., Mark A.E., Berendsen H.J.C. Gromacs: fast, flexible, and free. J. Comput. Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 33.Evans D.J., Holian B.L. The nose–hoover thermostat. J. Chem. Phys. 1985;83(8):4069–4074. [Google Scholar]
- 34.Parrinello M., Rahman A. Polymorphic transitions in single crystals: a new molecular dynamics method. J. Appl. Phys. 1981;52(12):7182–7190. [Google Scholar]
- 35.Hess B. P-lincs: a parallel linear constraint solver for molecular simulation. Journal of Chemical Theory and Computation - J CHEM THEORY COMPUT. 2007;4 doi: 10.1021/ct700200b. [DOI] [PubMed] [Google Scholar]
- 36.Darden T., York D., Pedersen L. Particle mesh ewald: an n⋅log(n) method for ewald sums in large systems. J. Chem. Phys. 1993;98(12):10089–10092. [Google Scholar]
- 37.Kadhim W.K.A., Nayef U.M., Jabir M.S. Polyethylene glycol-functionalized magnetic (fe3o4) nanoparticles: a good method for a successful antibacterial therapeutic agent via damage dna molecule. Surf. Rev. Lett. 2019;26(10) [Google Scholar]
- 38.Ali I.H., Jabir M.S., Al-Shmgani H.S.A., Sulaiman G.M., Sadoon A.H. Pathological and immunological study on infection with escherichia coli in ale balb/c mice. J. Phys. Conf. 2018;1003(1) [Google Scholar]
- 39.Heleno S.A., Ferreira I.C.F.R., Esteves A.P., irić A., Glamočlija J., Martins A., Soković M., Queiroz M.J.R.P. Antimicrobial and demelanizing activity of ganoderma lucidum extract, p-hydroxybenzoic and cinnamic acids and their synthetic acetylated glucuronide methyl esters. Food Chem. Toxicol. 2013;58:95–100. doi: 10.1016/j.fct.2013.04.025. [DOI] [PubMed] [Google Scholar]
- 40.Muniz D.F., Dos Santos Barbosa C.R., de Menezes I.R.A., de Sousa E.O., Pereira R.L.S., Júnior J.T.C., Pereira P.S., de Matos Y.M.L.S., Da Costa R.H.S., de Morais Oliveira-Tintino C.D., Coutinho H.D.M., Filho J.M.B., Ribeiro De Sousa G., Filho J.R., Siqueira-Junior J.P., Tintino S.R. In vitro and in silico inhibitory effects of synthetic and natural eugenol derivatives against the nora efflux pump in staphylococcus aureus. Food Chem. 2021;337 doi: 10.1016/j.foodchem.2020.127776. [DOI] [PubMed] [Google Scholar]
- 41.Monteiro-Neto V., de Souza C.D., Gonzaga L.F., Da Silveira B.C., Sousa N.C.F., Pontes J.P., Santos D.M., Martins W.C., Pessoa J.F.V., Júnior A.R.C., Almeida V.S.S., de Oliveira N.M.T., de Araújo T.S., Maria-Ferreira D., Mendes S.J.F., Ferro T.A.F., Fernandes E.S. Cuminaldehyde potentiates the antimicrobial actions of ciprofloxacin against staphylococcus aureus and escherichia coli. PLoS One. 2020;15(5) doi: 10.1371/journal.pone.0232987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mastoor S., Nazim F., Rizwan-Ul-Hasan S., Ahmed K., Khan S., Ali S.N., Abidi S.H. vol. 27. 2022. https://10.3390/molecules27206874 (Analysis of the Antimicrobial and Anti-biofilm Activity of Natural Compounds and Their Analogues against staphylococcus Aureus Isolates). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramos Nino M.E., Clifford M.N., Adams M.R. Quantitative structure activity relationship for the effect of benzoic acids, cinnamic acids and benzaldehydes on listeria monocytogenes. J. Appl. Bacteriol. 1996;80(3):303–310. doi: 10.1111/j.1365-2672.1996.tb03224.x. [DOI] [PubMed] [Google Scholar]
- 44.Khameneh B., Iranshahy M., Soheili V., Fazly B.B. Review on plant antimicrobials: a mechanistic viewpoint. Antimicrob. Resist. Infect. Control. 2019;8:118. doi: 10.1186/s13756-019-0559-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ecevit K., Barros A.A., Silva J.M., Reis R.L. vol. 2. 2022. pp. 460–498.https://10.3390/futurepharmacol2040030 (Preventing Microbial Infections with Natural Phenolic Compounds). [Google Scholar]
- 46.Tropsha A. Best practices for qsar model development, validation, and exploitation. Mol. Inform. 2010;29(6–7):476–488. doi: 10.1002/minf.201000061. [DOI] [PubMed] [Google Scholar]
- 47.Potdar S., Sharma P., Pal N., Kumar A. Synthesis of azepane-acridine heterocyclic adducts assisted by the catalytic influence of γ-fe2o3@tio2-vitamin b1 and evaluation of their medicinal potential through in vitro antimicrobial, qsar, and molecular docking studies. Results in Chemistry. 2024;7 [Google Scholar]
- 48.Todeschini R., Consonni V. 2009. Molecular Descriptors for Chemoinformatics/, Molecular Descriptors for Chemoinformatics. [Google Scholar]
- 49.Kier L.B., Hall L.H. 1976. Molecular Connectivity in Chemistry and Drug Research, Molecular Connectivity in Chemistry and Drug Research. [Google Scholar]
- 50.Liu S., Cao C., Li Z. Approach to estimation and prediction for normal boiling point (nbp) of alkanes based on a novel molecular distance-edge (mde) vector, λ. J. Chem. Inf. Model. 1998;38(3):387–394. [Google Scholar]
- 51.Hall L.H., Kier L.B. Electrotopological state indices for atom types: a novel combination of electronic, topological, and valence state information. J. Chem. Inf. Model. 1995;35(6):1039–1045. [Google Scholar]
- 52.Amin S.A., Gayen S. Modelling the cytotoxic activity of pyrazolo-triazole hybrids using descriptors calculated from the open source tool “padel-descriptor”. J. Taibah Univ. Sci. 2016;10(6):896–905. [Google Scholar]
- 53.Bickerton G.R., Paolini G.V., Besnard J., Muresan S., Hopkins A.L. Quantifying the chemical beauty of drugs. Nat. Chem. 2012;4(2):90–98. doi: 10.1038/nchem.1243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guerrab W., Lgaz H., Kansiz S., Mague J.T., Dege N., Ansar M., Marzouki R., Taoufik J., Ali I.H., Chung I., Ramli Y. Synthesis of a novel phenytoin derivative: crystal structure, hirshfeld surface analysis and dft calculations. J. Mol. Struct. 2020;1205 [Google Scholar]
- 55.Guerrab W., Chung I., Kansiz S., Mague J.T., Dege N., Taoufik J., Salghi R., Ali I.H., Khan M.I., Lgaz H., Ramli Y. Synthesis, structural and molecular characterization of 2,2-diphenyl-2h,3h,5h,6h,7h-imidazo[2,1-b][1,3]thiazin-3-one. J. Mol. Struct. 2019;1197:369–376. [Google Scholar]
- 56.Uzun S., Esen Z., Koç E., Usta N.C., Ceylan M. Experimental and density functional theory (mep, fmo, nlo, fukui functions) and antibacterial activity studies on 2-amino-4- (4-nitrophenyl) -5,6-dihydrobenzo [h] quinoline-3-carbonitrile. J. Mol. Struct. 2019;1178:450–457. [Google Scholar]
- 57.Zhang L., Chen R., Li X., Xu X., Xu Z., Cheng J., Wang Y., Li Y., Shao X., Li Z. Synthesis, insecticidal activities, and 3d-qasr of n-pyridylpyrazole amide derivatives containing a phthalimide as potential ryanodine receptor activators. J. Agric. Food Chem. 2022;70(39):12651–12662. doi: 10.1021/acs.jafc.2c03971. [DOI] [PubMed] [Google Scholar]
- 58.Mol G.P.S., Aruldhas D., Joe I.H. Chemical reactivity, molecular electrostatic potential and in-silico analysis on benzimidazole fungicide benomyl. Heliyon. 2022;8(11) doi: 10.1016/j.heliyon.2022.e11417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Vanasundari K., Balachandran V., Kavimani M., Narayana B. Spectroscopic investigation, vibrational assignments, fukui functions, homo-lumo, mep and molecular docking evaluation of 4 – [(3, 4 – dichlorophenyl) amino] 2 – methylidene 4 – oxo butanoic acid by dft method. J. Mol. Struct. 2017;1147:136–147. [Google Scholar]
- 60.Belayachi M., Serrar H., El Assyry A., Oudda H., Boukhris S., Touhami M.E., Zarrouk A., Hammouti B., Ebenso E.E., El Midaoui A. Electrochemical evaluation and dft studies of 2-(4-chlorophenyl)-3-hydroxy-4,6-dioxo-8-phenyl-4,6 dihydropyrimido[2,1-b][1,3]thiazine-7-carbonitrile of carbon steel corrosion in hydrochloric acid. Int. J. Electrochem. Sci. 2015;10(4):3038–3053. [Google Scholar]
- 61.Rao W., Fang Z., Chen Z., Wu J., Fang X. Antibacterial mechanism of metabolites of leuconostoc mesenteroides against serratia liquefaciens. Lebensm. Wiss. Technol. 2023;187 [Google Scholar]
- 62.Lv J., Da R., Cheng Y., Tuo X., Wei J., Jiang K., Monisayo A.O., Han B., Quinn F.D. Mechanism of antibacterial activity of bacillus amyloliquefaciens c-1 lipopeptide toward anaerobic clostridium difficile. BioMed Res. Int. 2020;2020 doi: 10.1155/2020/3104613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yang H., Wang L., Pan F., Yuan L., Du H., Zhang P., Lu K. Self-assembling anti-gram-negative bacterial peptide derivatives with potent broad-spectrum antimicrobial activity. Lebensm. Wiss. Technol. 2023;185 [Google Scholar]
- 64.He H., Wan Y., Li F., Li T., Ren B. Exploring the solvent effect on crystal morphology of naphazoline hydrochloride: molecular dynamic simulations and experiments. J. Mol. Liq. 2023 [Google Scholar]
- 65.Khajeh A., Modarress H. The influence of cholesterol on interactions and dynamics of ibuprofen in a lipid bilayer. Biochim. Biophys. Acta Biomembr. 2014;1838(10):2431–2438. doi: 10.1016/j.bbamem.2014.05.029. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data will be made available on request directly to the corresponding author.