Abstract
Three previously unidentified dihydrostilbene glycosides, named oleiferaside A (1), oleiferaside B (2), and oleiferaside C (3), were discovered through a phytochemical exploration on Camellia oleifera Abel. leaves. Additionally, nine known secondary metabolites (4–12) were also identified. The undescribed secondary metabolites 1–3 were elucidated as 3,5-dimethoxydihydrostilbene 4′-O-α-l-arabinofuranosyl-(1 → 6)-β-d- glucopyranoside, 3,5-dimethoxydihydrostilbene 4′-O-α-l-arabinopyranosyl-(1 → 6)-β-d- glucopyranoside and 3,5-dimethoxydihydrostilbene 4′-O-β-d-apiofuranosyl-(1 → 6)-β-d- glucopyranoside, respectively. HR-MS and NMR spectroscopy were utilized for determining the structures of the isolates. The natural products were assessed for their anti-inflammatory effect using RAW264.7 macrophage stimulated by LPS. The findings demonstrated that compounds 1–4 exhibited inhibitory activities on NO and PGE2 production without causing cytotoxicity. These observations suggest that these compounds may have potential anti-inflammatory properties.
Keywords: Camellia oleifera Abel., Dihydrostilbene glycosides, Anti-inflammatory, Oleiferaside A, Oleiferaside B, Oleiferaside C
Highlights
-
•
The phytochemical investigation on the leaves of C. oleifera resulted in the discovery of twelve compounds.
-
•
Three previously undescribed compounds, oleiferaside A-C (1-3), were isolated from C. oleifera leaves.
-
•
The tested dihydrostilbene glycosides (1-4) showed potential anti-inflammatory activities with no cytotoxicity.
1. Introduction
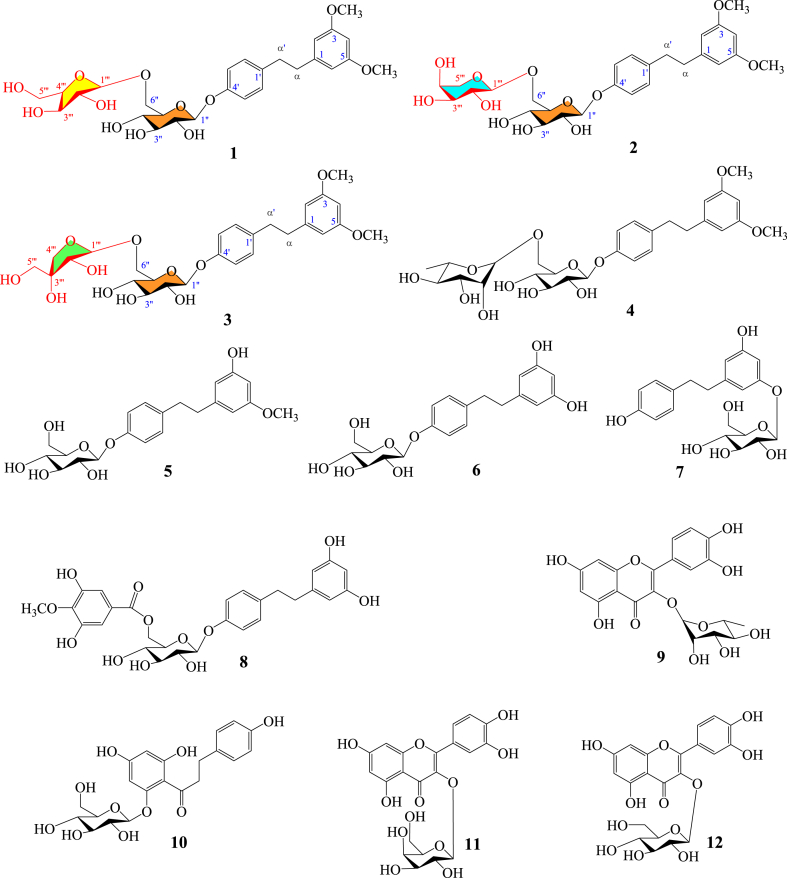
Camellia oleifera Abel., a member of the Camellia genus (Theaceae family), has a wide distribution across several provinces in China, including Jiangxi, Hunan, Fujian, Guangdong, and Guangxi [1]. Recognized as a traditional Chinese herb-medicine, the dried leaves of C. oleifera are officially listed in the National Compilation of Chinese Herbal Medicine and Chinese Materia Medica. The plant leaves are known for their various therapeutic properties, including clearing heat and detoxifying, astringent hemostasis, refreshing the brain, promoting blood circulation and dispersing stasis, relieving pain. They have been traditionally employed as a remedy for conditions such as epistaxis, skin ulceration and itching, ulcerative gangrene, acute pharyngitis, stomachache, as well as sprain and contusion [2,3]. The identification of triterpenoids and their saponins, flavonoids and their glycosides, dihydrostilbenes and their glycosides, anthraquinones, lignans, phenols, organic acids, steroids, and fatty acids have been significant outcomes of previous research conducted on this plant [[4], [5], [6], [7], [8], [9], [10]]. These compounds have exhibited diverse pharmacological actions, such as anti-tumor effects [4], anti-inflammatory properties [5], anti-neuroinflammatory and neuroprotective effects [11,12], antioxidant activity [8,13,14], antibacterial effects [5], hypoglycemic effects [15,16], and anti-thrombotic effects [17]. In our current study, we focused on the methanol extracts of C. oleifera leaves and identified three new bioactive dihydrostilbene glycosides (1–3) and nine known secondary metabolites (4–12) (Fig. 1). This article presents the isolation, structure determination, and assessment of the cytotoxicity and anti-inflammatory behaviour of these natural products.
Fig. 1.
Chemical structures of compounds 1–12.
2. Materials and methods
2.1. Instrument and reagent
An AUTOPOL I automatic polarimeter from Rudolph Research Analytical (U.S.A) was used for optical rotation test. A SPECORD 50 PLUS UV spectrophotometer purchased from Analytik Jena AG (Germany) was employed for UV spectra. A NICOLETIS50 FT-IR spectrometer from Thermo Fisher (Germany) was applied for IR spectra. An AVANCE III Bruker-400 spectrometer (Switzerland) was utilized for NMR spectra. Residual solvent peaks were used as references for the chemical shifts of NMR. A Xevo G2-XS TOF instrument (Waters) was operated for obtaining MS data. The instruments and reagents used below, including silica gel, TLC, C18 column, semi-preparative HPLC system, LPS, MTT, FBS, DMEM, and ELISA kits for NO and PGE2, are consistent with our previous literature reports [18].
2.2. Plant materials
The leaves of Camellia oleifera Abel. were gathered from their natural habitat in Xingguo, Jiangxi Province, China, in May 2020. Then, the medicinal materials were verified by Xiao-Jun Li from the Department of Pharmacy at Gannan Medical University. The voucher specimens have been stored at the Key Laboratory of Natural Product Research and Development in Jiangxi Province Universities, Gannan Medical University, under the reference number CO202005.
2.3. Extraction and purification
2.8 kg of dried leaves of C. oleifera were pulverized into powder and subjected to three extractions using 70 % methanol (3 × 10 L) at 60 °C with reflux for 2 h on each occasion. Afterwards, the solvent was evaporated, resulting in a MeOH extract that was subsequently dispersed in distilled water. Sequential partitioning was carried out using organic solvents with different polarities for giving petroleum ether part (36.7 g), EtOAc part (201 g), and n-BuOH part (357.4 g), respectively.
The 201 g of EtOAc part was first isolated by silica gel column chromatography (CC). The elution process involved a gradient of CH2Cl2–CH3OH with varying ratios of 100:1, 50:1, 30:1, 20:1, 10:1, 5:1, 3:1, 1:1, and 1:3 (v/v). Each fraction was collected, yielding seven major fractions (E1-E7) by TLC analysis. 84.55 g of E5 was performed on a silica gel CC applying a mobile phase of dichloromethane/methanol/water (10:2:0.1–1:3:0.2, v/v/v). This process yielded six sub-fractions (E5.1-E5.6). A C18 semi-preparative HPLC was operated for the further purification of E5.4 (350 mg), using a gradient elution of methanol/water (flow rate: 3 mL/min, 30:70–70:30, v/v). As a result, compounds 4 (132.2 mg), 9 (18.4 mg), 5 (7.1 mg), 1 (12.9 mg), 2 (8.3 mg), 3 (7.3 mg), and 10 (12.6 mg) were obtained consecutively. E5.6 (126 mg) was chromatographed by C18 semi-preparative HPLC using a mobile phase of methanol/water (flow rate: 3 mL/min, 30:70–70:30, v/v). This process resulted in the isolation of compounds 6 (7.1 mg) and 7 (8.6 mg), along with sub-fractions E5.6.1-E5.6.3. Subsequently, in order to obtain compounds 11 (5.9 mg) and 12 (7.6 mg), a C18 semi-preparative HPLC method was established for the further purification of E5.6.1 (30.2 mg), with the gradient elution of methanol/water (flow rate: 3 mL/min, 20:70–50:50, v/v). Simultaneously, compounds 8 (6.8 mg) and 10 (8.7 mg) were yield from E5.6.2 (33.7 mg) via purification of C18 semi-preparative HPLC using a elution of methanol/water (flow rate: 3 mL/min, 50:50, v/v).
2.4. Spectroscopic data
Oleiferaside A (1): white amorphous powder; HR-ESI-MS m/z: 575.2092 [M + Na]+ (calcd. for C27H36O12Na, 575.2099); molecular formula: C27H36O12; UV (MeOH) λmax (log ε) 203 (0.94), 218 (0.41); [α]20 D −69.97 (c 0.23, MeOH); IR (microscope) νmax 3650, 1635, 1540, 1507, 1150, 1074, 831, 685 cm−1; NMR data, refer to Table 1.
Table 1.
NMR data of undescribed compounds 1–3. 1H/400 MHz,13C/100 MHz, δ in ppm, J in Hz.
| Position | 1a,b |
2a,b |
3a,b |
|||
|---|---|---|---|---|---|---|
| δH | δC | δH | δC | δH | δC | |
| 1 | – | 145.3 | – | 145.3 | – | 145.3 |
| 2 | 6.30 (d, J = 2.2) | 107.6 | 6.30 (d, J = 2.2) | 107.6 | 6.30 (d, J = 2.2) | 107.6 |
| 3 | – | 162.2 | – | 162.2 | – | 162.2 |
| 4 | 6.27 (d, J = 2.2) | 98.9 | 6.27 (d, J = 2.2) | 98.9 | 6.27 (d, J = 2.2) | 98.9 |
| 5 | – | 162.2 | – | 162.2 | – | 162.2 |
| 6 | 6.30 (d, J = 2.2) | 107.6 | 6.30 (d, J = 2.2) | 107.6 | 6.30 (d, J = 2.2) | 107.6 |
| α | 2.80 (m) | 39.5 | 2.80 (m) | 39.5 | 2.80 (m) | 39.5 |
| α′ | 2.81 (m) | 38.1 | 2.81 (m) | 38.1 | 2.81 (m) | 38.1 |
| 1′ | – | 137.1 | – | 137.1 | – | 137.1 |
| 2′ | 7.10 (d, J = 8.6) | 130.5 | 7.10 (d, J = 8.6) | 130.5 | 7.10 (d, J = 8.6) | 130.5 |
| 3′ | 7.00 (d, J = 8.6) | 117.6 | 7.00 (d, J = 8.6) | 117.6 | 7.00 (d, J = 8.6) | 117.6 |
| 4′ | – | 157.4 | – | 157.4 | – | 157.4 |
| 5′ | 7.00 (d, J = 8.6) | 117.6 | 7.00 (d, J = 8.6) | 117.6 | 7.00 (d, J = 8.6) | 117.6 |
| 6′ | 7.10 (d, J = 8.6) | 130.5 | 7.10 (d, J = 8.6) | 130.5 | 7.10 (d, J = 8.6) | 130.5 |
| 3,5-OCH3 | 3.71 (s) | 55.6 | 3.71 (s) | 55.6 | 3.71 (s) | 55.6 |
| Sugar moiety | Glc | Glc | Glc | |||
| 1″ | 4.83 (d, J = 7.3) | 102.5 | 4.85 (d, J = 7.2) | 102.3 | 4.81 (d, J = 7.5) | 102.6 |
| 2″ | 3.43 (m) | 74.9 | 3.43 (m) | 74.9 | 3.43 (m) | 74.9 |
| 3″ | 3.43 (overlapped) | 77.9 | 3.43 (overlapped) | 77.8 | 3.43 (overlapped) | 77.9 |
| 4″ | 3.36 (m) | 71.8 | 3.37 (m) | 71.5 | 3.35 (m) | 71.6 |
| 5″ | 3.59 (m) | 76.8 | 3.62 (m) | 77.3 | 3.59 (m) | 77.0 |
| 6a′′ | 3.60 (m) | 68.2 | 3.62 (overlapped) | 69.3 | 3.61 (m) | 68.8 |
| 6b′′ | 4.05 (dd, J = 5.4, 14.3) | 4.10 (dd, J = 2.1, 11.5) | 4.01 (dd, J = 1.5, 10.6) | |||
| Sugar moiety | Araf | Arap | Apif | |||
| 1‴ | 4.93 (d, J = 1.5) | 110.0 | 4.30 (d, J = 6.8) | 104.9 | 4.98 (d, J = 2.4) | 111.1 |
| 2‴ | 4.00 (dd, J = 3.4, 1.5) | 83.3 | 3.57 (m) | 72.5 | 3.91 (d, J = 2.4) | 78.0 |
| 3‴ | 3.82 (dd, J = 6.0, 3.4) | 78.9 | 3.49 (dd, J = 3.3, 8.9) | 74.1 | – | 80.5 |
| 4‴ | 3.96 (m) | 85.8 | 3.75 (m) | 69.5 | 3.76 (d, J = 9.6) | 75.0 |
| 3.98 (d, J = 9.6) | ||||||
| 5‴ | 3.62 (m) | 63.0 | 3.42 (m) | 66.7 | 3.58 (s) | 65.5 |
| 3.70 (m, overlapped) | 3.82 (dd, J = 3.1, 12.4) | |||||
a) Recorded in CD3OD. b) Multiplicities inferred from DEPT and HSQC experiments. Glc: glucopyranose; Araf: arabinofuranose; Arap: arabinopyranose; Apif: apiofuranose.
Oleiferaside B (2): white amorphous powder; HR-ESI-MS m/z 575.2091 [M + Na]+ (calcd. for C27H36O12Na, 575.2099); molecular formula: C27H36O12; UV (MeOH) λmax (log ε) 205 (2.25), 218 (1.04); [α]20 D −68.84 (c 0.05, MeOH); IR (microscope) νmax 3628, 1593, 1540, 1508, 1455, 1204, 1066, 829, 683 cm−1; NMR data, refer to Table 1.
Oleiferaside C (3): white amorphous powder; HR-ESI-MS m/z 575.2091 [M + Na]+ (calcd. for C27H36O12Na, 575.2099); molecular formula: C27H36O12; UV (MeOH) λmax (log ε) 205 (2.33), 218 (1.24); [α]20 D −25.31 (c 0.13, MeOH); IR (microscope) νmax 3490, 1593, 1508, 1457, 1226, 1203, 1149, 1056, 823, 691 cm−1; NMR data, refer to Table 1.
2.5. Acidic hydrolysis of 1–3
Acid hydrolysis and derivatization methods referred to previous studies [19,20]. In short, each of compounds 1–3 (1.5 mg each) was added to 2 M HCl (500 μL) and subjected to heating for 2 h at 90 °C. After the hydrolysis process, the reaction mixture was neutralized with 500 μL of 2 M NH4OH and subsequently dried using an evaporator. Hydrolyzed samples 1–3, along with standard sugars (d-glucopyranose, l-arabinofuranose, l-arabinopyranose, and d-apiofuranose, 5 mg each), and 5 mg of l-cysteine methyl ester hydrochloride were dissolved in 1 mL of pyridine and reacted for 1 h at 60 °C. Subsequently, 5 μL of 2-methylphenyl isothiocyanate was introduced into the mixture, followed by an additional hour of heating. The resulting reaction mixture (20 μL) was then analyzed using RP-HPLC and detected at 250 nm to determine the types of sugars present. This was done by comparing with authentic samples, utilizing the elution system CH3CN–H2O in 0.1 % HCOOH (v/v, 15:85–35:65, flow rate: 0.8 mL/min). The tR values obtained were 35.087 min for d-glucopyranose, 37.319 min for l-arabinofuranose, 37.011 min for l-arabinopyranose, and 44.219 min for d-apiofuranose (Table S1).
2.6. Cell culture
The mouse RAW264.7 macrophages (RRID: CVCL_0493) were purchased from the American Type Culture Collection (ATCC, Manassas, VA). The cell cultures employed in our investigation underwent examination and were validated to be free from mycoplasma contamination. Each RAW264.7 cell was cultured separately in DMEM culture medium with 100 U/mL of penicillin G, 100 mg/L of streptomycin, and 10 % heat-inactivated FBS. Following that, the RAW264.7 macrophage was placed in a controlled environment with 5 % CO2 at a temperature of 37 °C for incubation. Every 2 days, the culture medium was refreshed. When the cell bottle reached 80 % confluency, the previous medium was discarded, and the cells underwent 2–3 washes with PBS. Subsequently, the addition of 0.25 % trypsin facilitated cell digestion, and repeated pipetting with a pipette ensured even dispersion of the cells. The resulting single-cell suspension was seeded at a 1:5 ratio in the cell culture medium.
2.7. MTT assay for cell viability
The step-by-step protocols for MTT assay were documented in our prior studies [21,22]. The analysis was performed in triplicate, independently.
2.8. Nitrite assay
As stated in our previous research, we employed the ELISA kit to accurately quantify the levels of NO [21,22].
2.9. Prostaglandin E2 assay
The measurement of Prostaglandin E2 (PGE2) levels for selected compounds were conducted utilizing commercially obtainable kits. Three separate assays were carried out on the basis of operation instructions. Briefly, RAW264.7 macrophages were seeded in 24-well culture plates at a density of 5 × 104 cells/well. The test compounds were exposed to varying concentrations of treatment, and subsequently stimulated with 1 μg/mL of LPS for 24 h. Following the incubation, the supernatant was collected and utilized for PGE2 concentration measurement via the application of a PGE2 ELISA kit.
2.10. Statistical analysis
One-way ANOVA was employed for statistical analysis of normally distributed data to assess variations among mean values. The findings are shown as the mean ± standard deviation (S.D.). The statistical significance was considered at p < 0.05.
3. Results and discussion
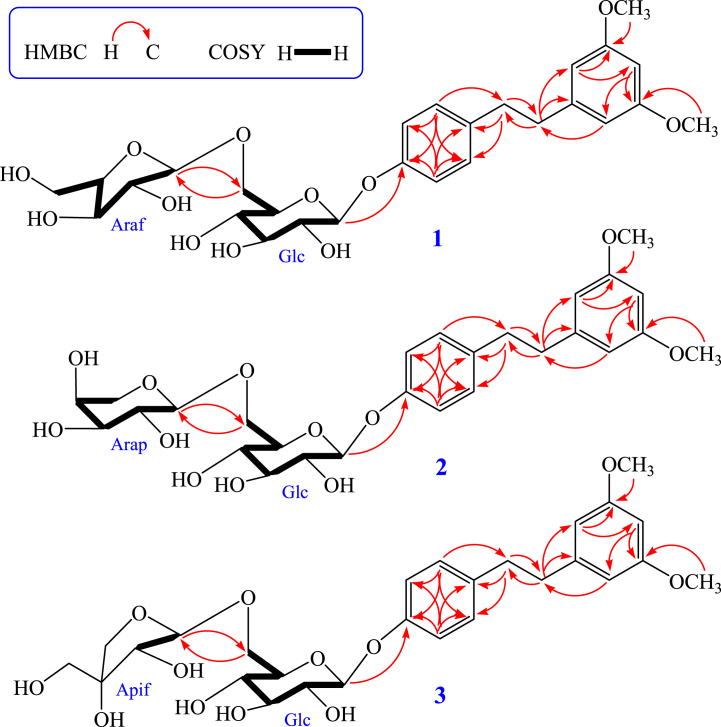
Compound 1 was acquired in the form of a white amorphous powder. HR-ESI-MS data which revealed a m/z peak at 575.2092 [M+Na]+ (calculated for C27H36O12Na, 575.2099) demonstrated that the molecular formula was C27H36O12. The 1H NMR spectrum of compound 1 (recorded in CD3OD) exhibited the following signals: seven aromatic protons at δ 7.10 (2H, d, J = 8.6 Hz), 7.00 (2H, d, J = 8.6 Hz), 6.30 (2H, d, J = 2.2 Hz) and 6.27 (1H, d, J = 2.2 Hz); two methoxy groups at δ 3.71 (6H, s); two methylenes at δ 2.80 (2H, m) and 2.81 (2H, m). In addition, the analysis of the spectra indicated the presence of two sugars, as evidenced by two anomeric protons at δ 4.83 (1H, d, J = 7.3 Hz) and 4.93 (1H, d, J = 1.5 Hz). Confirmation of the presence of 27 carbon signals was achieved through additional analysis of the 13C NMR and DEPT spectra. Among these, 16 signals were attributed to a dihydrostilbene aglycone moiety, while the remaining 11 signals were assigned to a disaccharide residue. The dihydrostilbene skeleton of compound 1 was identified through the analysis of its 1H and 13C NMR spectra (refer to Table 1), with the presence of characteristic aromatic carbons at δ 145.3 (C-1), 107.6 (C-2, C-6), 162.2 (C-3, C-5), 98.9 (C-4) and 137.1 (C-1′), 130.5 (C-2′, C-6′), 117.6 (C-3′, C-5′), 157.4 (C-4′), two methylene carbons at δ 39.5 (C-α) and 38.1 (C-α′), respectively. The NMR spectral data of compound 1 closely resembled those of compound 4, which had been previously documented from C. oleifera [7], except the presence of different sugar moiety (arabinofuranose) resonances [δH 4.93 (1H, d, J = 1.5 Hz, araf-H-1‴)/δC 110.0, 83.3, 78.9, 85.8, 63.0 (araf-C-1‴-5‴)] in 1, which was located at glc-C-6′′ (δ 68.2), instead of rhamnopyranose in 4. The coupling constants of anomeric protons glc-H-1′′ (δ 4.83, 1H, d, J = 7.3 Hz) and araf-H-1‴ (δ 4.93, 1H, d, J = 1.5 Hz) showed that the glucopyranose and arabinofuranose were β-positioned and α-positioned, respectively. Additionally, assignment of the sugar chain (6-O-α-l-arabinofuranosyl-β-d- glucopyranoside) was conducted through comparing its NMR spectral data with known compound 2-phenylethyl 6-O-α-l-arabinofuranosyl-β-d-glucopyranoside and literature values [23,24]. The HMBC correlations from protons H6 (2 × OMe) at δ 3.71 (6H, s) to carbons C-3 and C-5 (δ 162.2); proton H-1″ at δ 4.83 (1H, d, J = 7.3 Hz) correlated with C-4′ (δ 157.4); the correlations from H-1‴ at δ 4.93 (1H, d, J = 1.5 Hz) to C-6′′ (δ 68.2) and from H-6a′′ at δ 3.60 (1H, m) and H-6b′′ at δ 4.05 (1H, m) to C-1‴ (δ 110.0) were further confirmed that the methoxyl moiety, β-d-glucopyranose and α-l-arabinofuranose were located at C-3/C-5, C-4′ and glc-C-6″, respectively (Fig. 2). In addition, the monosaccharides of 1 were further confirmed to be d-glucopyranose and l-arabinofuranose via the RP-HPLC analysis after acid hydrolysis and derivatization of 1, with the retention time (tR) were 35.087 min and 37.319 min for d-glucopyranose and l-arabinofuranose (Table S1). As a result, compound 1 was conclusively identified as 3,5-dimethoxydihydrostilbene 4′-O-α-l-arabinofuranosyl-(1 → 6)-β-d-glucopyranoside, a previously unreported compound that has been designated as oleiferaside A.
Fig. 2.
Important 1H–1H COSY (highlighted) and HMBC (indicated by arrows) correlations of undescribed compounds 1–3.
Compound 2 was achieved in the form of a white amorphous powder. The analysis of HR-ESI-MS data revealed the molecular formula of C27H36O12 for compound 2, indicating the presence of ten degrees of unsaturation. Comparing the 1H and 13C NMR spectra of compounds 1 and 2, it was evident that they shared a high degree of similarity. The main difference observed was the presence of a α-l-arabinopyranose [δH 4.30 (1H, d, J = 6.8 Hz, arap-H-1‴)/δC 104.9, 72.5, 74.1, 69.5, 66.7 (arap-C-1‴-5‴)] in 2, instead of a α-l-arabinofuranose [δH 4.93 (1H, d, J = 1.5 Hz, araf-H-1‴)/δC 110.0, 83.3, 78.9, 85.8, 63.0 (araf-C-1‴-5‴)] in 1 (see Table 1). In addition, the assignment of the sugar chain (6-O-α-l-arabinopyranosyl-β-d-glucopyranoside) was accomplished by comparing its spectral data with those of the known compound 2-phenylethyl 6-O-α-l-arabinopyranosyl- β-d-glucopyranoside and relevant literature values [23]. These observations suggested that the relative configurations of glucopyranose and arabinopyranose in 2 were β-positioned and α-positioned. Also, the β-glucopyranose and α-arabinopyranose located at C-4′ and glc-C-6″ were further confirmed by HMBC spectrum (Fig. 2). Moreover, RP-HPLC analysis further confirmed that the monosaccharides of 2 are d-glucopyranose and l-arabinopyranose, which were obtained through acid hydrolysis of 2 with 2 M HCl, with the tR were 35.087 min and 37.011 min for d-glucopyranose and l-arabinopyranose (Table S1). As a result, compound 2 was conclusively identified as 3,5-dimethoxydihydrostilbene 4′-O-α-l-arabinopyranosyl- (1 → 6)-β-d-glucopyranoside, a previously unreported compound that has been designated as oleiferaside B.
Compound 3 was gained in the form of a white amorphous powder. The analysis of HR-ESI-MS data revealed the molecular formula of C27H36O12 for compound 3, indicating the presence of ten degrees of unsaturation. The 1H and 13C NMR spectra showed a high degree of similarity between compounds 1 (or 2) and 3, with the main distinguishing feature being the presence of a terminal connected sugar moiety β-d-apiofuranose [δH 4.98 (1H, d, J = 2.4 Hz, apif-H-1‴)/δC 111.1, 78.0, 80.5, 75.0, 65.5 (apif-C-1‴-5‴)] in 3, instead of a α-l-arabinofuranose or α-l-arabinopyranose in 1 or 2 (see Table 1). Furthermore, the sugar chain assignment (6-O-β-d-apiofuranosyl-β-d-glucopyranoside) was determined by comparing its NMR spectral data with a known compound, 2-phenylethyl 6-O-β-d- apiofuranosyl-β-d-glucopyranoside, using δ values for glc and apif, as well as references from literature [23,24]. Based on these observations, it can be inferred that the relative configurations of glucopyranose and apiofuranose in compound 3 are both located in the β position. Additionally, the HMBC spectrum further confirmed the presence of β-glucopyranose at C-4′ and β-apiofuranose at glc-C-6''. (Fig. 2). Moreover, the monosaccharides in compound 3 were further identified as d-glucopyranose and d-apiofuranose through the RP-HPLC analysis following acid hydrolysis of 3 using 2 M HCl and derivatization of sugars, with the tR were 35.087 min and 44.219 min for d-glucopyranose and d-apiofuranose (Table S1). As a result, compound 3 was conclusively identified as 3,5-dimethoxydihydrostilbene 4′-O-β-d-apiofuranosyl-(1 → 6)-β-d- glucopyranoside, a previously unreported compound that has been designated as oleiferaside C.
Nine previously described compounds 4–12 were determined to be 1-(3′,5′-dimethoxy) phenyl-2-[4″-O-β-d-glucopyranosyl(6 → 1)-O-α-l-rhamnopyranosyl]phenylethane (4) [7], sasastilboside A (5) [25], 3,5-dihydroxydihydrostilbene 4′-O-β-d-glucopyranoside (6) [7], 5,4′-dihydroxy-dihydrostilbene 3-O-β-d-glucopyranoside (7) [26], 3,5-dihydroxyl- dihydrostilbene 4′-O-[6″-O-(4‴-methoxylgalloyl)]-β-d-glucopyranoside (8) [12], quercitrin (9) [27], phlorizin (10) [28], quercetin-3-O-β-d-galactopyranoside (11) [9], and quercetin-3-O-β-d-glucopyranoside (12) [9] via comparing the NMR and mass data of the compounds with the values reported in the literature.
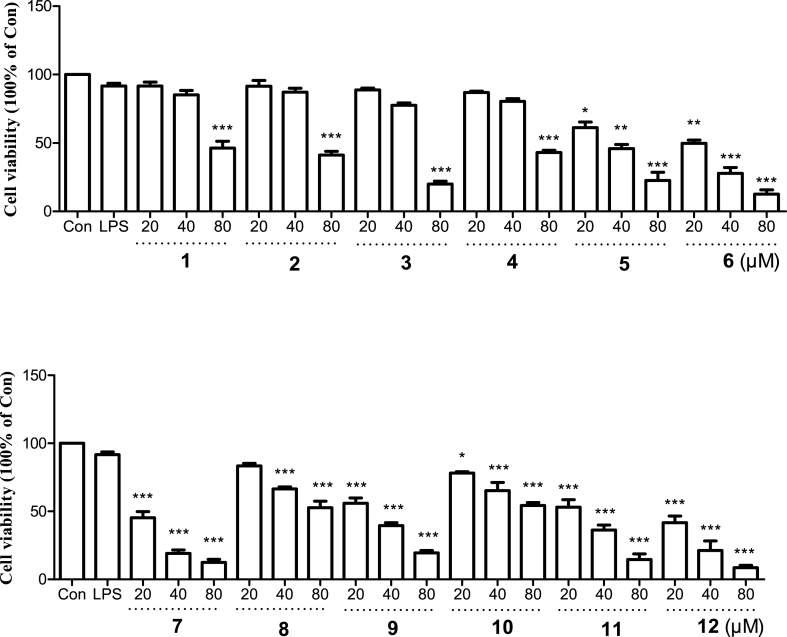
To assess the anti-inflammatory activities of the isolates, LPS-induced RAW264.7 cells were utilized. Prior to conducting the experiments, the cytotoxicity of all compounds was tested up to a concentration of 80 μM using the MTT assay. We found that 20 μM and 40 μM of tested compounds 1–4 showed very weak impact on cell viability of RAW264.7 macrophages, while 80 μM of all compounds exhibited significant cytotoxicity (Fig. 3). As shown in Table 2, the results of nitrite and PGE2 assay demonstrated that the IC50 values of NO and PGE2 for compounds 1–4 were 28.56 ± 0.16 and 15.14 ± 0.11 (1), 42.21 ± 0.21 and 20.22 ± 0.18 (2), 37.88 ± 0.32 and 12.25 ± 0.24 (3), 23.47 ± 0.15 and 13.31 ± 0.22 (4), respectively. These findings uncovered the potential anti-inflammatory activity of those secondary metabolites. Additional investigations are necessary to assess the mechanism of anti-inflammation for active phytochemical constituents.
Fig. 3.
Cell viability of compounds 1–12 on LPS-induced RAW264.7 cells. The values are expressed as mean of three experiments ±S.D. *p < 0.05, **p < 0.01 and ***p < 0.001 compared with Con group.
Table 2.
Effects of 4 selected natural products on production of LPS-stimulated NO and PGE2 in RAW264.7 cells.
| Compounds | NO (IC50, μM) | PGE2 (IC50, μM) |
|---|---|---|
| 1 | 28.56 ± 0.16 | 15.14 ± 0.11 |
| 2 | 42.21 ± 0.21 | 20.22 ± 0.18 |
| 3 | 37.88 ± 0.32 | 12.25 ± 0.24 |
| 4 | 23.47 ± 0.15 | 13.31 ± 0.22 |
| Butein | 5.56 ± 0.13 | 8.79 ± 0.17 |
Butein: positive control. The data is presented as mean ± SD based on triple separate experiments.
4. Conclusions
To sum up, the research on the chemical constituents of C. oleifera leaves resulted in identifying twelve different compounds. Among them, three undescribed compounds named oleiferaside A (1), oleiferaside B (2), and oleiferaside C (3) were identified, along with nine previously reported natural products (4–12). Additionally, the toxicity of the separated substances was evaluated, along with their ability to inhibit the generation of NO and PGE2 caused by LPS in RAW264.7 macrophages. The tested dihydrostilbene glycosides (1–4) demonstrated moderate anti-inflammatory activities without causing any cytotoxic effects.
Data availability statement
Data included in article/supp. material/referenced in article.
CRediT authorship contribution statement
Yi Xu: Writing – original draft, Investigation. Si-Qi Tang: Writing – original draft, Methodology, Investigation. Zong-Wu Suo: Formal analysis. Kai-Xin Wei: Formal analysis. Walter Luyten: Validation. Hao Huang: Validation, Supervision, Resources. Xiao-Jun Li: Writing – review & editing, Supervision, Project administration, Funding acquisition, Conceptualization.
Declaration of competing interest
The authors declare no conflict of interest.
Acknowledgments
This project were supported financially by the National Natural Science Foundation of China (Grant 82111530101 and 81803493), Jiangxi Provincial Key R & D project (Grant 20212BBG73041), Ganzhou Science and Technology Innovation Youth Talent Plan (Grant 202101094465), the Key R & D project of Ganzhou (Grant 2023PCG16850) and Gannan Medical University (Grant QD201905, ZD201911 and YB201910).
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.heliyon.2024.e30507.
Contributor Information
Hao Huang, Email: huanghao_26@163.com.
Xiao-Jun Li, Email: xjli@gmu.edu.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article.
References
- 1.Delectis Florae Reipublicae Popularis Sinicae. Academiae Sinicae Edita Agendae. vol. 49. Science Press; Beijing: 1998. p. 13. (Flora Reipublicae Popularis Sinicae). [Google Scholar]
- 2.Editorial Committee of the Chinese Materia Medica of the State Administration of Traditional Chinese Medicine, Chinese Materia Medica (Zhonghua Bencao) Shanghai Science and Technology Press; Shanghai: 1999. p. 585. [Google Scholar]
- 3.Compilation Group of the National Compilation of Chinese Herbal Medicine . People's Medical Publishing House; Beijing: 1975. National Compilation of Chinese Herbal Medicine (Quanguo Zhongcaoyao Huibian) p. 313. [Google Scholar]
- 4.Xu Y., Tang S.Q., Huang H., Liu X.Q., Li X.J. Research progress on triterpenoids from Camellia oleifera and their pharmacological activities. Chin. Tradit. Herb. Drugs. 2022;53:1210–1219. [Google Scholar]
- 5.Duan Y., Zhou Y.H., Li S.X., Li J. Advances in phytochemical constituents and antibacterial and anti-inflammatory activity of Camellia oleifera Abel. Nat. Prod. Res. Dev. 2021;33:1603–1615. [Google Scholar]
- 6.Quan W., Wang A., Gao C., Li C. Applications of Chinese Camellia oleifera and its by-products: a review. Front. Chem. 2022;10 doi: 10.3389/fchem.2022.921246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen Y.L., Tang L., Feng B.M., Shi L.Y., Wang H.G., Wang Y.Q. New bibenzyl glycosides from leaves of Camellia oleifera Abel. with cytotoxic activities. Fitoterapia. 2011;82:481–484. doi: 10.1016/j.fitote.2010.12.009. [DOI] [PubMed] [Google Scholar]
- 8.Zhang J.L., Qiu A.Y. Studies on the antioxidants in the leaves of Camellia oleifera Abel. for edible oils. J. Food Sci. Biotechnol. 1990;9:37–42. [Google Scholar]
- 9.Chen Y.L., Feng B.M., Tang L., Li H.B., Shi L.Y., Wang Y.Q. Chemical constituents from leaves of Camellia oleifera Abel. J. Shenyang Pharm. Univ. 2010;27:292–294. [Google Scholar]
- 10.Cao Q.M., Zhong H.Y., Wu J.Y., Bao L.Y., Sun Y.J. Compounds and structure identification in extracts from Camellia oleifera leaves by HPLC preparation. Nonwood Forest Res. 2015;33:68–74. [Google Scholar]
- 11.Vitalini S., Cicek S.S., Granica S., Zidorn C. Dihydroresveratrol type dihydrostilbenoids: chemical diversity, chemosystematics, and bioactivity. Curr. Med. Chem. 2018;25:1194–1240. doi: 10.2174/0929867324666170830112343. [DOI] [PubMed] [Google Scholar]
- 12.Cuc N.T., Yen D.T.H., Yen P.H., Hang D.T.T., Tai B.H., Seo Y., Namkung W., Kim S.H., Cuong P.V., Kiem P.V., Nhiem N.X., Ngoc T.M. Dihydrostilbene glycosides from Camellia sinensis var. assamica and their cytotoxic activity. Nat. Prod. Res. 2022;36:3931–3937. doi: 10.1080/14786419.2021.1900844. [DOI] [PubMed] [Google Scholar]
- 13.Li J.J., Huang K.Y., Gong J.L., Ma W. Research on the scavenging effect of ethanol extracts from Camellia oleifera Abel. leaves on 2,2-diphenyl-1-picrylhydrazyl radical. Chem. Ind. For. Prod. 2008;28:82–86. [Google Scholar]
- 14.Li J.J., Gong J.L., Zhou J.H., Chen C.J., Li S.F. Research on extraction and antioxidant activities of the total flavonoids from Camellia oleifera leaves. Food Res. Dev. 2008;29:93–96. [Google Scholar]
- 15.Peng L., Zhu B.F., Liu Z. Study on blood glucose lowering and antioxidation of extracts from flesh fruit and flesh leaf of Camellia oleifera Abel. Chin. Pharmacol. Bull. 2007;23:1679–1680. [Google Scholar]
- 16.Zhang W.Y., Hong Z.F., Chen Q.C., Xu C.J., Wang X.Y. Effect of ethanol extract from Camellia oleifera Abel. on blood glucose in type 2 diabetic mice. Strait Pharm. J. 2017;29:21–24. [Google Scholar]
- 17.Qiang H.B., Wang X.P. Study on anticoagulability and antithrombotic activity of water extracts from the leaves of Camellia oleifera Abel. J. Anhui Agric. Sci. 2010;38:11136–11137. [Google Scholar]
- 18.Li X.J., Song X.H., Tang S.Q., Wei K.X., Suo Z.W., Xu Y., Luo J., Huang H., Li C.Y., Liu D.X., Liu X.Q. Phytochemical constituents from rhizomes of Dryopteris crassirhizoma and their anti-inflammatory activity. Nat. Prod. Res. 2023;10:1–7. doi: 10.1080/14786419.2023.2256446. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka T., Nakashima T., Ueda T., Tomii K., Kouno I. Facile discrimination of aldose enantiomers by reversed-phase HPLC. Chem. Pharm. Bull. 2007;55:899–901. doi: 10.1248/cpb.55.899. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y.H., Avula B., Fu X., Wang M., Khan I.A. Simultaneous determination of the absolute configuration of twelve monosaccharide enantiomers from natural products in a single injection by a UPLC-UV/MS method. Planta Med. 2012;78:834–837. doi: 10.1055/s-0031-1298432. [DOI] [PubMed] [Google Scholar]
- 21.Li X.J., Kim K.W., Kim D.C., Oh H., Liu X.Q., Kim Y.C. Three novel monoterpenoid glycosides from fruits of Eleutherococcus henryi. Nat. Prod. Res. 2021;35:1299–1306. doi: 10.1080/14786419.2019.1645661. [DOI] [PubMed] [Google Scholar]
- 22.Li X.J., Zou Q.P., Wang X., Kim K.W., Lu M.F., Ko S.K., Yook C.S., Kim Y.C., Liu X.Q. Lupane triterpenes from the leaves of Acanthopanax gracilistylus. Molecules. 2018;23:87. doi: 10.3390/molecules23010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma S.J., Mizutani M., Hiratake J., Hayashi K., Yagi K., Watanabe N., Sakata K. Substrate specificity of beta-primeverosidase, a key enzyme in aroma formation during oolong tea and black tea manufacturing. Biosci. Biotech. Bioch. 2001;65:2719–2729. doi: 10.1271/bbb.65.2719. [DOI] [PubMed] [Google Scholar]
- 24.Yang D., Xie H.H. Aromatics and flavonoids from Strawberry cv. Falandi, J. Trop. Subtrop. Bot. 2017;25:617–624. [Google Scholar]
- 25.Cuc N.T., Cuong N.T., Anh L.T., Yen D.T.H., Tai B.H., Trang D.T., Yen P.H., Kiem P.V., Nam N.H., Minh C.V., Nhiem N.X. Dihydrostilbene glycosides from Camellia sasanqua and their α-glucosidase and α-amylase inhibitory activities. Nat. Prod. Res. 2021;35:4025–4031. doi: 10.1080/14786419.2020.1756802. [DOI] [PubMed] [Google Scholar]
- 26.Feng W.S., Cao X.W., Kuang H.X., Zheng X.K. A new stilbene glycoside from Dryopteris sublaeta. Acta Pharm. Sin. 2005;40:1131–1134. [PubMed] [Google Scholar]
- 27.Li Y.L., Li K.M., Su M.X., Liang J.T., Chen Y.W., Zhang Y.W. Studies on antiviral constituents in stems and leaves of Pithecellibium clypearia, China J. Chin. Mater. Med. 2006;31:397–400. [PubMed] [Google Scholar]
- 28.Cho J.Y., Lee K.D., Park S.Y., Jeong W.C., Moon J.H., Ham K.S. Isolation and identification of α-glucosidase inhibitors from the stem bark of the nutgall tree (Rhus javanica Linné) J. Korean Soc. Appl. Biol. Chem. 2013;56:547–552. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data included in article/supp. material/referenced in article.