Summary
Hypertrophic cardiomyopathy (HCM) is a heritable cardiovascular disorder that affects 1 in 500 people. A significant percentage of HCM is attributed to mutations in β-cardiac myosin, the motor protein that powers ventricular contraction. This study reports how two early onset HCM mutations, D239N and H251N, affect the molecular biomechanics of human β-cardiac myosin. We observe significant increases (20% - 90%) in actin gliding velocity, intrinsic force and ATPase activity compared to wild type myosin. Moreover, for H251N, we find significantly lower binding affinity between the S1 and S2 domains of myosin, suggesting that this mutation may further increase hyper-contractility by releasing active motors. Unlike previous HCM mutations studied at the molecular level using human β-cardiac myosin, early onset HCM mutations lead to significantly larger changes in the fundamental biomechanical parameters and show clear hyper-contractility.
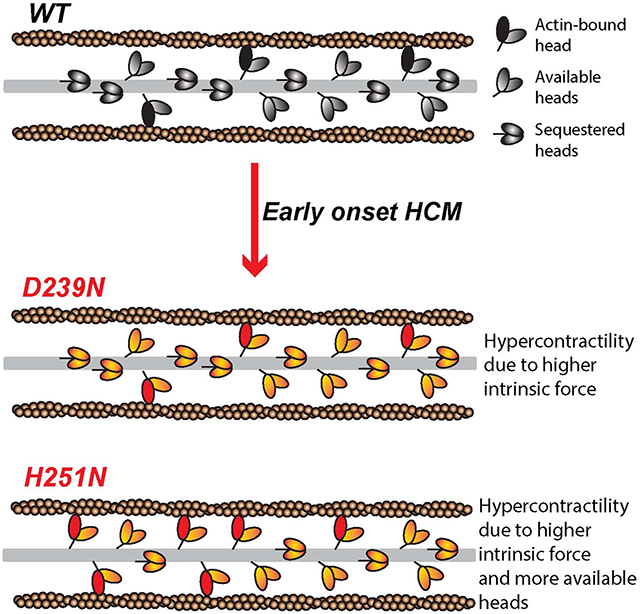
Graphical Abstract
Introduction
Hypertrophic cardiomyopathy (HCM) is a heritable cardiovascular disorder characterized by abnormal thickening of the left ventricular walls (Nishimura et al., 2003), preserved or increased systolic function and reduced diastolic function (Maron, 2002). HCM is typically diagnosed in late adolescence or adulthood, and is the leading cause of sudden cardiac death in those under the age of 35 (Maron, 2003). Disease presentation in infancy or childhood is associated with poorer outcomes; rates of death/cardiac transplantation approach 20% (Colan et al., 2007).
HCM is most commonly caused by mutations in genes encoding sarcomeric proteins, principally those encoding β-cardiac myosin (MYH7) and cardiac myosin binding protein-C (MYBP3) (Ho et al., 2015; Walsh et al., 2016). Despite the identification of the genetic basis of this disease, relatively little is known about the molecular mechanisms by which these mutations result in the HCM disease phenotype. It has been hypothesized that mutations in β-cardiac myosin, the mechanoenzyme that drives ventricular contraction, cause HCM by affecting the power output of the myosin motor (Spudich, 2014). Power output is the product of force and velocity, and so the effects of HCM-causing mutations on myosin power output at the molecular level can be determined by assaying the velocity of actin filament gliding on ensembles of mutant myosin motors and the force produced by those ensembles. Ensemble force can be estimated by determining the key parameters that contribute: Fensemble = Fintrinsic (ts/tc) Na, where Fintrinsic is the intrinsic force of the motor, ts is the time of the ATPase cycle that myosin is tightly bound to actin, tc is the total cycle time of the actin-activated myosin ATPase, ts/tc is the duty ratio, and Na is the number of myosin heads that are functionally accessible for interaction with actin in the sarcomere.
Previous studies of the effects of HCM mutations on these biomechanical parameters using reconstituted systems have shown variable results (reviewed in Table S1 and (Moore et al., 2012)). Studies using protein derived from mouse model systems showed considerable increases in either ATPase activity, velocity of actin filament gliding in in vitro motility assays, or intrinsic force (Debold et al., 2007; Tyska et al., 2000). However, these mutations were expressed in a mouse α-cardiac myosin backbone that has greater than 80 amino acid differences from the human β-cardiac myosin motor domain, and subsequent studies have shown that the same HCM mutation (R403Q) caused significantly different biomechanical effects on mouse α vs β-cardiac myosin (Lowey et al., 2008). Thus it is important to analyze mutations in the context of the human β-cardiac myosin backbone. However, studies using protein derived from human biopsy samples have also provided conflicting results, likely due to the fact that these samples contain only small amounts of variably preserved mixtures of WT and mutant β-cardiac myosin, as well as some α-cardiac myosin (Alpert et al., 2005; Moore et al., 2012; Palmiter et al., 2000). Patient derived cardiomyocytes or myofibrils contain secondary modifications to multiple sarcomeric proteins due to the end stage nature of the biopsy tissue, further complicating analysis of the underlying effects of the myosin mutations (Kirschner et al., 2005; Kraft et al., 2013; Lankford et al., 1995).
As the conflicting results described above illustrate, studies to define the functional consequences of these and other β-cardiac myosin mutations have been hampered by the lack of available expressed and purified human cardiac myosin; however, this has been solved by the establishment of a mouse myoblast expression system (Liu et al., 2008; Resnicow et al., 2010; Srikakulam and Winkelmann, 2004). Thus, highly purified recombinant human β-cardiac myosin short Subfragment 1 (sS1) motor domains have been examined for the effects of several different HCM mutations. In contrast to the studies using mouse alpha-cardiac myosin, the results using these recombinant human motors have shown smaller changes in myosin’s fundamental biomechanical parameters, and typically a hyper-contractile change in one parameter is accompanied by a hypo-contractile change in another (Nag et al., 2015; Sommese et al., 2013). Recently, Kawana et al. (Kawana et al., 2016, doi: http://dx.doi.org/10.1101/065649) studied three converter domain HCM mutations, and found only very subtle changes. It is unclear whether these small changes reflect the fact that mutations that give rise to HCM typically cause relatively small perturbations in myosin biomechanics or rather that current assays do not accurately measure the effects of HCM mutations on β-cardiac myosin’s force-velocity relationship.
In order to address this question, we studied the effects of mutations that cause severe, early onset HCM. Although the causative mutation is present at birth, most individuals with HCM do not manifest evidence of the disease until late adolescence or adulthood. However, in a subset of individuals, symptomatic HCM can present in infancy or childhood (pediatric HCM incidence ~5 per million) (Colan et al., 2007). Disease progression can be swift, with significant morbidity and mortality, especially in infants. (Colan et al., 2007). Three quarters of all pediatric-onset HCM is classified as idiopathic (Colan et al., 2007). It is known that up to 50-60% of idiopathic HCM is associated with mutations primarily in genes encoding sarcomeric proteins, and roughly 30-50% of those are in β-cardiac myosin (Ho et al., 2015; Walsh et al., 2016).
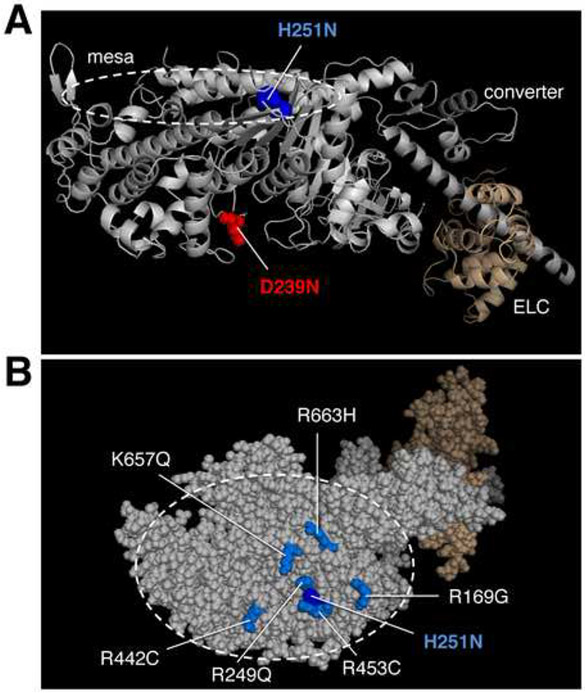
Recently, novel β-cardiac mutations have been found that cause severe disease in infants and children (Kaski et al., 2009; Morita et al., 2008). Some of the HCM mutations identified in pediatric patients have not been reported, or reported only rarely, in adult patient populations (Kaski et al., 2009; Morita et al., 2008). While the number of patients with these mutations is small, statistical analysis suggests that these mutations may be associated with an HCM subtype characterized by more severe, early onset disease (Knight and Leinwand, personal communication). We chose two early onset HCM mutations, H251N on the myosin mesa, a relatively flat surface of the catalytic domain that has been proposed to be a binding site for the myosin coiled-coil tail and myosin binding protein-C (Spudich, 2015) and D239N within the Switch-1 (nucleotide binding) domain (Figure 1), to test whether mutations associated with such a severe clinical phenotype would exhibit more significant, and consistently hyper-contractile, changes in myosin biomechanical parameters. We utilized assays for actin-activated myosin ATPase activity (De La Cruz and Ostap, 2009), in vitro motility (Kron and Spudich, 1986), loaded in vitro motility (Aksel et al., 2015), and single molecule intrinsic force (Fintrinsic) by optical tweezers force spectroscopy (Rice et al., 2003) to elucidate the effect of each mutation on biochemical cross-bridge cycling parameters. We also analyzed binding between myosin sS1 and Subfragment 2 (S2) domains (Nag et al. 2016, doi: http://dx.doi.org/10.1101/065508) to examine the hypothesis that HCM mutations in the myosin mesa disrupt a sequestered state of myosin heads in the sarcomere that limit the number of functionally accessible heads for interaction with actin (Spudich, 2015). Together, we show that these mutations exhibit clear, uniform and significant gain of function, unlike many recently studied HCM-causing mutations in human β-cardiac myosin. Furthermore, we find that the H251N mutation, located on the myosin mesa, disrupts binding of the motor to the proximal part of the myosin tail and may further contribute to hyper-contractilty by increasing the number of functionally accessible heads for interaction with actin.
Figure 1.
Structure of a homology-modeled human β-cardiac sS1 domain showing the positions of two mutations that cause early onset HCM. (a) Structure of homology-modeled human β-cardiac sS1, containing residues 1-808 of the MHC (grey) and the ELC (brown). The positions of the HCM mutations H251N (blue) and D239N (red) are shown. (b) Sphere model of sS1 shown in (a) rotated ~90° about the horizontal axis toward the reader, viewing the mesa from the top. The position of the mutation H251N on the mesa is seen in the middle of a cluster of other positively charged residues, all of which cause HCM when mutated (Spudich, 2015).
Results
H251N and D239N have significantly increased actin-activated ATPase activity compared to WT
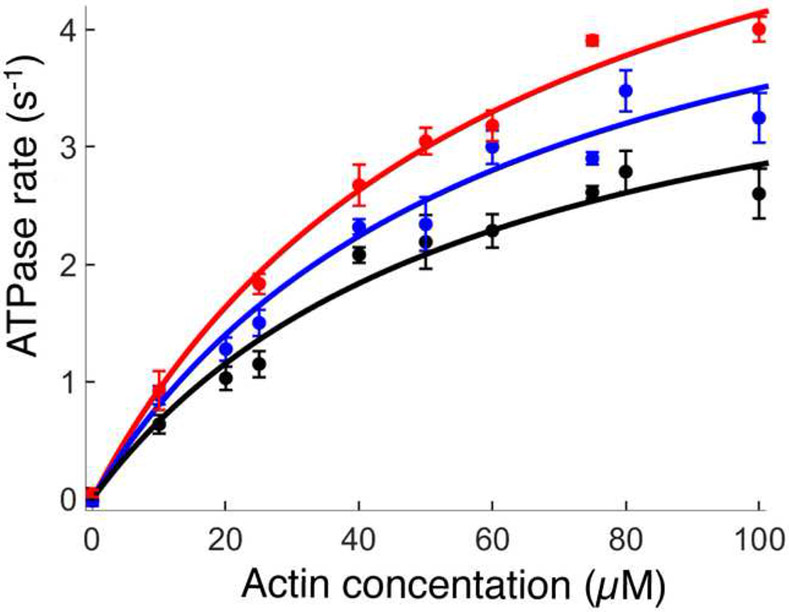
To assess the effects of the mutations on total ATPase cycle time (tc), we measured the maximal ATPase activity of the actin-myosin complex (kcat) for WT and each mutant (tc =1/kcat). We found that the kcat for H251N (5.6 ± 0.4 s−1, 8 replicates from 3 sS1 preparations) and for D239N (6.7 ± 0.5 s−1, 6 replicates from 3 sS1 preparations) were significantly increased compared to WT sS1 (4.5 ± 0.4 s−1, 12 replicates, 5 sS1 preparations) (Figure 2 and Supplemental Table S2). These increases in catalytic rates (24% for H251N and 50% for D239N) are the largest observed for any mutation assayed in the human β-cardiac myosin backbone, and thus the largest decreases in tc. There were no changes in the values of KM for either of the human β-cardiac mutant myosins as compared to WT (Supplemental Table S2).
Figure 2.
Actin-activated ATPase activities of WT, H251N and D239N human β-cardiac sS1s. The actin-activated ATPase activities of WT (black) (12 replicates from 5 sS1 preparations), H251N (blue) (8 replicates from 3 sS1 preparations), and D239N (red) (6 replicates from 3 sS1 preparations) sS1s are fitted to a Michaelis-Menten curve. The error bars are the standard error of the mean (S.E.M.) from multiples myosin preparations.
Single molecule optical tweezer force spectroscopy shows an increase in the intrinsic force of both H251N and D239N
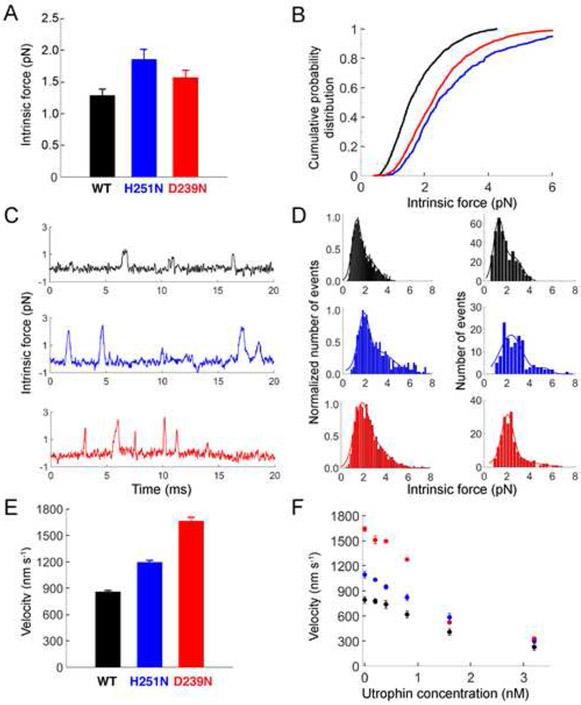
To measure the effect of these mutations on the intrinsic force of human β-cardiac sS1, we used a dual beam optical trap, as described previously (Rice et al., 2003). Force histograms of individual molecules were used to calculate the mean force for each of the proteins (Nag et al., 2015) (Figure 3A-D). The WT sS1 single molecule mean force was measured as 1.3 ± 0.1 pN (2 preparations, 6 molecules, ~350 events). Compared to the WT sS1, both H251N (1.9 ± 0.2 pN; 2 preparations, 6 molecules, ~350 events) and D239N (1.6 ± 0.1 pN; 2 preparations, 6 molecules, >1000 events) showed appreciable increases in force (H251N = 46% and D239N = 23% increase, respectively) (Figure 3A). Additionally, we used a cumulative probability distribution function to independently assess the increase in contractile force at the single molecule level (Nag et al., 2015). For all the events measured with all the molecules, we calculated the cumulative probability distribution (Nag et al., 2015). From Figure 3B it is clear that at all cumulative probability values, the single molecule force of WT < D239N < H251N.
Figure 3.
Biomechanical parameters of WT, H251N and D239N human β-cardiac sS1s. (a) Intrinsic force measurements from a dual beam single molecule laser trap. Force histograms of individual molecules were used to calculate the mean force for WT (2 preparations, 6 molecules, ~350 events) and mutant motors ( H251N = 2 preparations, 6 molecules, ~350 events; D239N = 2 preparations, 6 molecules, >1000 events). (b) Cumulative frequency distribution calculated from all single molecule force events. (c) Representative force traces for WT, H251N, and D239N sS1 motors show multiple binding events for each motor. Note the increased amplitude (force) of the H251N and D239N sS1 binding events as compared to WT sS1. (d) Intrinsic force measurements of WT, H251N and D239N sS1 motors. Histograms of force measurements of all molecules (left panels) and representative individual molecules (right panels) are shown with double Gaussian fits (see Supplemental Methods for details). (e) Unloaded average actin gliding velocities (WT=25 replicates from 5 sS1 preparations; H251N=10 replicates from 3 sS1 preparations; D239N=6 replicates from 3 sS1 preparations). (f) Loaded in vitro motility results, using utrophin as the load molecule (all motors =6 replicates from 3 sS1 preparations). All error bars represent the S.E.M. In all panels, WT is black, H251N blue, and D239N red.
H251N and D239N generate higher actin gliding velocities than WT in an in vitro motility assay
We next assessed the effect of the mutations on the velocity at which human β-cardiac sS1 propels actin filaments in an in vitro motility assay (Kron and Spudich, 1986).The average unloaded velocities of H251N (1200 ± 20 nm/s,10 replicates from 3 sS1 preparations) and D239N (1670 ± 40 nm/s, 6 replicates from 3 sS1 preparations) were significantly greater than that of WT sS1 (860 ± 10 nm/s, 25 replicates from 5 sS1 preparations) (Figure 3E, Supplemental Table S2, Supplemental videos 1-3). As with the catalytic rate constants, these extremely large increases in actin gliding velocities compared to the WT (D239N = 94% and H251N = 40%) are the largest changes observed for human β-cardiac myosin containing HCM-causing mutations.
H251N and D239N both generate higher ensemble force than WT in a loaded in vitro motility assay
To gain further insight into how the biomechanical properties of myosin are altered due to these mutations, we utilized a loaded in vitro motility assay. Our loaded in vitro motility assay is an adaptation of the actin gliding assay, where the actin binding protein utrophin is introduced on the surface to act as a load molecule against which myosin works (Aksel et al., 2015). Determining the average gliding velocities at various utrophin concentrations gives a load-velocity relationship for the ensemble of myosin motors on the surface (Figure 3F) (Aksel et al., 2015). Previous loaded motility studies have used full length myosin as the motor and alpha-actinin as the load molecule (Greenberg and Moore, 2010; Karabina et al., 2015) (See Supplemental information for discussion). Utrophin contains the same actin binding site as alpha-actinin but is a smaller molecule, and thus better suited for use with the smaller sS1 motor (Aksel et al., 2015). It is currently unclear how best to model such loaded velocity data (e.g., see Greenberg and Moore, 2010 versus Aksel et al. 2015), but examination of the curves for the human β-cardiac WT and mutant sS1s clearly shows that at each applied load (utrophin concentration) the velocities of the mutant motors were higher than that of WT, suggesting a higher relative ensemble force and power output (power = force x velocity) for the two mutants.
H251N sS1 binds to human cardiac proximal S2 with significantly decreased affinity compared to WT
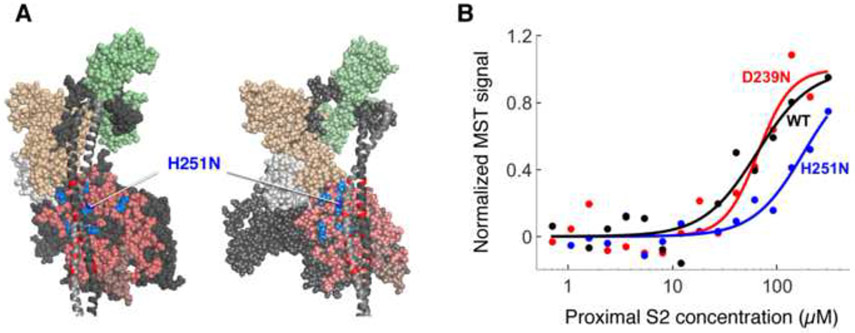
Structural studies of several types of myosins, including cardiac myosin, have shown that the myosin S1 head domains fold back onto the proximal portion of the α-helical coiled-coil tail of myosin (Al-Khayat et al., 2013; Alamo et al., 2008). It has been hypothesized that such folding back could sequester heads and thus alter the number of functionally accessible myosin heads for exerting force on actin (Spudich, 2015). It has also been suggested that HCM-causing mutations may disrupt this sequestered state, releasing more heads to interact with actin, thus resulting in the hyper-contractility phenotype observed clinically (Spudich, 2015). Nag et al. (Nag et al. 2016, doi: http://dx.doi.org/10.1101/065508) have shown that the myosin mesa mutation R453C, which is located at an interface between the putative S1-S2 interaction region, significantly weakens the binding of sS1 to proximal S2. H251N is next to R453C on the surface of the myosin mesa, at the interface between the putative S1-S2 interaction region, whereas D239N is remote from this region.
To test if H251N also disrupts the S1-S2 interaction, we used microscale thermophoresis (MST) to assess the relative binding affinities of WT and mutant sS1s with human proximal S2, as described by Nag et al. (Nag et al. 2016, doi: http://dx.doi.org/10.1101/065508). MST measurements revealed that, as for the R453C mutation (Nag et al. 2016, doi: http://dx.doi.org/10.1101/065508), there was a significant decrease in sS1-proximal S2 binding affinity due to the H251N mutation (200 ± 30 μM; 8 runs and 3 myosin preparations, p-value = 0.04) in comparison to WT (45 ± 10 μM; 10 runs and 5 myosin preparations), but statistically significant change was not observed for D239N (60 ± 8 μM; 5 runs and 2 myosin preparations, p-value = 0.69) (Figure 4B, Supplemental Table S2), which is located in a region where there is no proposed S1-S2 interaction.
Figure 4.
Binding of H251N and D239N human β-cardiac sS1s to human cardiac proximal S2. (a) Structural model of homology-modeled sequestered heads of human β-cardiac S1 based on the 3D-reconstructed structure of tarantula skeletal muscle myosin thick filaments (Alamo et al., 2008). A short version of myosin HMM, showing only 126 residues of the coiled-coil S2 domain, is illustrated in two projections. The heavy chain residues of the S1 head on the left are colored pink (mesa residues), dark blue (H251N), light blue (arginine HCM mutations), white (the converter domain), and dark grey (all remaining residues). The ELC is colored light brown and the RLC is light green. HCM-causing mutations of glutamate and aspartate residues in the proximal S2 tail are shown in red. H251N is at the interaction site of S1 and S2, while D239N is remote from this site and not visible in this projection (b) Representative MST binding curves (fit to the Hill Equation) of WT and mutant human β-cardiac sS1s tagged with a C-terminal eGFP to human proximal S2 give the affinity of the sS1 to S2 (KD)
Discussion
Elucidating the effect of HCM mutations on human β-cardiac myosin function and contractility at the molecular level has been challenging due to the small differences seen in molecular parameters such as intrinsic force and duty ratio, some of which contribute to hyper-contractility and others to hypo-contractility (Supplemental Table S1). Thus, how these mutations cause the typical hyper-contractile phenotype of clinical HCM is unclear. Two distinct but not mutually exclusive hypotheses are (1) these small changes reflect the fact that mutations that give rise to HCM typically cause relatively small perturbations in myosin biomechanics, and the net effect of the combination of such small effects is difficult to assess; and/or (2) current assays do not measure all of the important effects of HCM mutations on β-cardiac myosin’s ability to produce power. In this study we examine both hypotheses.
To address whether HCM causing mutations can result in only relatively small changes in β-cardiac myosin biomechanics, we studied the effects of mutations that cause phenotypically severe disease, reasoning that mutations linked predominantly to an earlier onset and more severe clinical phenotype would show greater changes at the molecular level in comparison to previously studied HCM mutations that most often cause adult-onset disease. In this paper, we present the molecular characterization of two early-onset mutations, H251N and D239N, which show very large increases in function for all the human β-cardiac myosin biomechanical parameters measured. D239N is in the switch I region of the nucleotide binding pocket. While it is difficult to accurately predict the functional effects of specific mutations given the highly allosteric nature of the myosin motor domain, mutagenesis studies of residues in this region of myosin from a variety of sources have revealed their importance in coupling the energy of ATP hydrolysis to lever arm movement (Coureux et al., 2004; Forgacs et al., 2009) while structural studies suggest their importance in nucleotide release (Holmes et al., 2003; Reubold et al., 2003). The velocity and ATPase rates of D239N are the highest of any of the human β-cardiac S1 mutations measured thus far. H251N is in the central β-sheet that undergoes twisting upon binding of ATP and communicates to the 50K domains to release actin. This allows switch II to move into the closed position and ATP to be hydrolyzed (Reubold et al., 2003). While these two mutations are located in structurally different regions of the head, they both could alter how nucleotide binding translates to actin binding and force production. This idea is supported by the large changes observed in their biomechanical properties, and we hypothesize that these large changes alone may account for the more severe, early onset clinical phenotype observed for patients carrying these mutations.
The question remains as to what is causing hyper-contractility in patients with mutations that cause relatively small changes in myosin biomechanical parameters, as seen in the adult onset mutations shown in supplemental Table S1. We hypothesize that many HCM mutations that cause only minor changes in myosin motor function may exert a major effect by increasing the number of myosin heads that are functionally accessible (Na) for interaction with actin in the sarcomere (Spudich, 2015). Specifically, we hypothesize that HCM mutations located on a flat surface of the motor domain termed the myosin mesa disrupt a sequestered state in which myosin heads are folded back onto their own coiled-coil tail (S2) (Spudich, 2015). EM reconstructions provide structural evidence that such a sequestered, or closed, state occurs in smooth, skeletal, and cardiac muscle (Al-Khayat et al., 2013; Alamo et al., 2008; Wendt et al., 2001). Nag et al. (Nag et al. 2016, doi: http://dx.doi.org/10.1101/065508) used MST to show that the sS1-proximal S2 interaction is weakened as a result of myosin HCM mutations on the myosin mesa, in a manner consistent with the structural model shown in Fig. 4. We tested this hypothesis using the two early onset mutations, and found that H251N, which is on the mesa, significantly decreased the affinity of the sS1-proximal S2 interaction while D239N, which is remote from the mesa, had no effect on the interaction (Figure 4). Thus, in the case of H251N human β-cardiac myosin, an additional parameter that may be increased by the mutation is Na. Of all the human β-cardiac myosin HCM mutations investigated, H251N is the only HCM mutation to show increases in function for all measured parameters, and D239N shows increases for all parameters, except the affinity of the sS1-promixal S2 interaction. Moreover, ensemble force studies carried out on these motors using the loaded in vitro motility assay show elevated velocities for both D239N and H251N human β-cardiac sS1 at all utrophin concentrations, thereby suggesting a larger power output (P = force x velocity) for the mutant myosins. Taken together, these data show increased function above what has been previously published for human β-cardiac myosin carrying HCM mutations (Nag et al., 2015; Sommese et al., 2013), supporting the idea that, in concert with the more severe clinical phenotype, these early-onset mutations show more severe effects at the molecular level.
Experimental Procedures
Expression and purification of proteins:
Recombinant human β-cardiac sS1 containing HCM causing mutations were co-expressed with a FLAG-tagged human essential light chain (ELC) in C2C12 mouse myoblast cells using adenoviral vectors, and purified using FLAG affinity and ion exchange chromatography (Figure S1B) as described (Sommese et al., 2013).
Actin was prepared from bovine cardiac muscle (identical in sequence to human cardiac actin), as described (Pardee and Spudich, 1982). PDZ-18 protein is a chimeric protein (erbin PDZ+fibronectin) engineered to bind the PDZ binding peptide (RGSIDTWV) with picomolar affinity. The His-tagged PDZ-18 and His-tagged human proximal S2 myosin are both expressed using the Rosetta (DE3) bacterial expression system and purified using nickel affinity chromatography. His-tagged utrophin is also expressed and purified similarly, but requires an additional inclusion body purification step (Aksel et al., 2015).
Actin-Activated ATPase assays:
The steady-state actin-activated ATPase activities of the WT and mutant human β-cardiac myosins were determined at 23 °C using a colorimetric assay to measure inorganic phosphate production at various time points from a solution containing sS1, ATP and increasing amounts of actin filaments (Trybus, 2000). Kinetic parameters (i.e. kcat) were extracted from the data by fitting to the Michaelis-Menten equation using the curve fitting toolbox in MatLab (De La Cruz and Ostap, 2009).
Unloaded and loaded in vitro motility assays:
Flow chambers were constructed from nitrocellulose-coated cover slips mounted on glass slides. sS1-AC was attached to the surface using PDZ-18 protein. Actin filaments were detected using a 100x objective on a Nikon TiE microscope, and at least 4 movies of 30 seconds were recorded for each condition. The mean velocity (MVEL, as described in Aksel et al.) was calculated using the FAST software (Aksel et al., 2015). All measurements were made at 23°C.
For the loaded motility assay, utrophin-AC was added with sS1-AC, and the motility was measured for each concentration of utrophin. The velocity data from both unloaded and loaded motility assays were analyzed using the FAST algorithm (Aksel et al., 2015).
Optical Tweezers Assay:
Details of the experimental setup and data analysis are described in the supplemental information. A trap stiffness of ~0.15 pN/nm was used. The experiments were performed at 23°C. Force histograms of individual molecules were fitted to a double Gaussian function (Figure 3D). The major force peak from the Gaussian fit yielded force value. Mean force was calculated by averaging force values of individual molecules. Cumulative probability distribution analysis was also used to compare the intrinsic force of the different sS1s.
MST Assay:
We used freshly prepared sS1-eGFP, and bacterially expressed proximal S2 myosin (amino acids 839-968) to investigate the effect of the HCM mutations on the myosin S1-S2 interaction. The proximal S2 construct includes the first 126 amino acids of S2 and begins 4 residues before the end of S1. Both proteins were dialyzed into MST assay buffer (10 mM Imidazole pH7.5, 100 mM KCl, 1 mM EDTA, 2 mM MgCl2, 1 mM DTT, 500 μM ADP and 0.05% tween). We used 16 serial 3-fold dilutions of S2 starting at > 300 μM. The sS1 was kept constant at 50 nM. The affinity measurements were performed using a Nanotemper thermophoresis at MST power = 60. The data was fit to the Hill equation. There were preparation-to-preparation differences in the binding affinity, and the WT KD ranged between 35-50 μM; however, the relative difference between WT and mutant myosin was constant.
Development of human β-cardiac myosin protein models:
We developed our models based on known human β-cardiac myosin S1 motor domain structural data (Winkelmann et al., 2015) as described (Homburger et al., 2016). The templates used to model the poststroke structure were obtained from the human β-cardiac myosin motor domain solved by Winkelmann et al. (Winkelmann et al., 2015) (PDB ID code 4P7H, no nucleotide in the active site), supplemented with the rigor structure from the squid myosin motor domain (Efron, 1982) (PDB ID code 3I5G, no nucleotide in the active site) to model the converter domain, lever arm, and light chains. The long coiled-coil S2 structure was templated using the Myosinome database. The structural model of homology-modeled sequestered heads of human β-cardiac S1 is based on the 3D-reconstructed structure of tarantula skeletal muscle myosin thick filaments by Alamo et al. (Alamo et al., 2008).
Statistical Analysis:.
Unloaded gliding velocities were measured over several replicates and averaged, and the error was calculated as the standard error of the mean (S.E.M.). For actin activated ATPase assay, each measurement at a given actin concentration was averaged over several replicates, and the error was expressed as the S.E.M. The data was then fit to the Michaelis Menten equation, and the error in the fit was calculated using 100 bootstrap iterations. Intrinsic force was measured by fitting the force histogram to a double Gaussian for each molecule (Supplemental Methods). The average was calculated over multiple molecules, and the error was calculated as the S.E.M. Lastly, for the MST experiments, the KD was calculated by fitting the data to the Hill equation, and student t-test was used to determine if the mutant was different from the WT sS1.
Supplementary Material
Acknowledgements
The authors would like to thank Dr. Leslie Leinwand and Dr. Robert Knight for personal communication about these mutations, Shirley Sutton for helping with virus preparation and cloning,.Dr. Darshan Trivedi and Dr. Suman Nag for preparation of proximal S2, and for help with data acquisition and analysis of MST experiments. The research was funded by NIH grants GM33289 and HL117138 (J.A.S.), Lucile Packard CHRI Postdoctoral Fellowship (A.S.A, K.B.K) (UL1 TR001085), Stanford ChEM-H Postdocs at the Interface Award (A.S.A., K.B.K.), Stanford CVI Postdoctoral Award (A.S.A.), American Heart Association Postdoctoral Fellowship (A.S.A.) (16POST30890005) NIH T32 Training Grant in Myocardial Biology T32 HL094274 (K.B.K), Stanford Spectrum Translational Medicine training grant TL1RR025742 (C.L.), NIGMS of the National Institutes of Health award number T32GM007276 (C.L.), and Stanford Bio-X fellowship (C.L.). J.A.S. is a founder of Cytokinetics and MyoKardia and a member of their advisory boards. K.M.R. is a member of the MyoKardia scientific advisory board.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aksel T, Choe Yu E, Sutton S, Ruppel KM, and Spudich JA (2015). Ensemble force changes that result from human cardiac myosin mutations and a small-molecule effector. Cell Rep 11, 910–920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Khayat HA, Kensler RW, Squire JM, Marston SB, and Morris EP (2013). Atomic model of the human cardiac muscle myosin filament. Proceedings of the National Academy of Sciences of the United States of America 110, 318–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamo L, Wriggers W, Pinto A, Bartoli F, Salazar L, Zhao FQ, Craig R, and Padron R (2008). Three-dimensional reconstruction of tarantula myosin filaments suggests how phosphorylation may regulate myosin activity. Journal of molecular biology 384, 780–797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alpert NR, Mohiddin SA, Tripodi D, Jacobson-Hatzell J, Vaughn-Whitley K, Brosseau C, Warshaw DM, and Fananapazir L (2005). Molecular and phenotypic effects of heterozygous, homozygous, and compound heterozygote myosin heavy-chain mutations. American journal of physiology Heart and circulatory physiology 288, H1097–1102. [DOI] [PubMed] [Google Scholar]
- Colan SD, Lipshultz SE, Lowe AM, Sleeper LA, Messere J, Cox GF, Lurie PR, Orav EJ, and Towbin JA (2007). Epidemiology and cause-specific outcome of hypertrophic cardiomyopathy in children: findings from the Pediatric Cardiomyopathy Registry. Circulation 115, 773–781. [DOI] [PubMed] [Google Scholar]
- Coureux PD, Sweeney HL, and Houdusse A (2004). Three myosin V structures delineate essential features of chemo-mechanical transduction. EMBO J 23, 4527–4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De La Cruz EM, and Ostap EM (2009). Kinetic and equilibrium analysis of the myosin ATPase. Methods in enzymology 455, 157–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debold EP, Schmitt JP, Patlak JB, Beck SE, Moore JR, Seidman JG, Seidman C, and Warshaw DM (2007). Hypertrophic and dilated cardiomyopathy mutations differentially affect the molecular force generation of mouse alpha-cardiac myosin in the laser trap assay. American journal of physiology Heart and circulatory physiology 293, H284–291. [DOI] [PubMed] [Google Scholar]
- Efron B. (1982). The jackknife, the bootstrap, and other resampling plans (Philadelphia, Pa.: Society for Industrial and Applied Mathematics; ). [Google Scholar]
- Forgacs E, Sakamoto T, Cartwright S, Belknap B, Kovacs M, Toth J, Webb MR, Sellers JR, and White HD (2009). Switch 1 mutation S217A converts myosin V into a low duty ratio motor. The Journal of biological chemistry 284, 2138–2149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg MJ, and Moore JR (2010). The molecular basis of frictional loads in the in vitro motility assay with applications to the study of the loaded mechanochemistry of molecular motors. Cytoskeleton (Hoboken) 67, 273–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho CY, Charron P, Richard P, Girolami F, Van Spaendonck-Zwarts KY, and Pinto Y (2015). Genetic advances in sarcomeric cardiomyopathies: state of the art. Cardiovascular research 105, 397–408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes KC, Angert I, Kull FJ, Jahn W, and Schroder RR (2003). Electron cryo-microscopy shows how strong binding of myosin to actin releases nucleotide. Nature 425, 423–427. [DOI] [PubMed] [Google Scholar]
- Homburger JR, Green EM, Caleshu C, Sunitha MS, Taylor RE, Ruppel KM, Metpally RP, Colan SD, Michels M, Day SM, et al. (2016). Multidimensional structure-function relationships in human beta-cardiac myosin from population-scale genetic variation. Proceedings of the National Academy of Sciences of the United States of America 113, 6701–6706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karabina A, Kazmierczak K, Szczesna-Cordary D, and Moore JR (2015). Myosin regulatory light chain phosphorylation enhances cardiac beta-myosin in vitro motility under load. Archives of biochemistry and biophysics 580, 14–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaski JP, Syrris P, Esteban MT, Jenkins S, Pantazis A, Deanfield JE, McKenna WJ, and Elliott PM (2009). Prevalence of sarcomere protein gene mutations in preadolescent children with hypertrophic cardiomyopathy. Circ Cardiovasc Genet 2, 436–441. [DOI] [PubMed] [Google Scholar]
- Kirschner SE, Becker E, Antognozzi M, Kubis HP, Francino A, Navarro-Lopez F, Bit-Avragim N, Perrot A, Mirrakhimov MM, Osterziel KJ, et al. (2005). Hypertrophic cardiomyopathy-related beta-myosin mutations cause highly variable calcium sensitivity with functional imbalances among individual muscle cells. American journal of physiology Heart and circulatory physiology 288, H1242–1251. [DOI] [PubMed] [Google Scholar]
- Kraft T, Witjas-Paalberends ER, Boontje NM, Tripathi S, Brandis A, Montag J, Hodgkinson JL, Francino A, Navarro-Lopez F, Brenner B, et al. (2013). Familial hypertrophic cardiomyopathy: functional effects of myosin mutation R723G in cardiomyocytes. Journal of molecular and cellular cardiology 57, 13–22. [DOI] [PubMed] [Google Scholar]
- Kron SJ, and Spudich JA (1986). Fluorescent actin filaments move on myosin fixed to a glass surface. Proceedings of the National Academy of Sciences of the United States of America 83, 6272–6276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lankford EB, Epstein ND, Fananapazir L, and Sweeney HL (1995). Abnormal contractile properties of muscle fibers expressing beta-myosin heavy chain gene mutations in patients with hypertrophic cardiomyopathy. The Journal of clinical investigation 95, 1409–1414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Srikakulam R, and Winkelmann DA (2008). Unc45 activates Hsp90-dependent folding of the myosin motor domain. The Journal of biological chemistry 283, 13185–13193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowey S, Lesko LM, Rovner AS, Hodges AR, White SL, Low RB, Rincon M, Gulick J, and Robbins J (2008). Functional effects of the hypertrophic cardiomyopathy R403Q mutation are different in an alpha- or beta-myosin heavy chain backbone. The Journal of biological chemistry 283, 20579–20589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maron BJ (2002). Hypertrophic cardiomyopathy: a systematic review. JAMA 287, 1308–1320. [DOI] [PubMed] [Google Scholar]
- Maron BJ (2003). Sudden death in young athletes. N Engl J Med 349, 1064–1075. [DOI] [PubMed] [Google Scholar]
- Moore JR, Leinwand L, and Warshaw DM (2012). Understanding cardiomyopathy phenotypes based on the functional impact of mutations in the myosin motor. Circ Res 111, 375–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morita H, Rehm HL, Menesses A, McDonough B, Roberts AE, Kucherlapati R, Towbin JA, Seidman JG, and Seidman CE (2008). Shared genetic causes of cardiac hypertrophy in children and adults. N Engl J Med 358, 1899–1908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nag S, Sommese RF, Ujfalusi Z, Combs A, Langer S, Sutton S, Leinwand LA, Geeves MA, Ruppel KM, and Spudich JA (2015). Contractility parameters of human beta-cardiac myosin with the hypertrophic cardiomyopathy mutation R403Q show loss of motor function. Science advances 1, e1500511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura RA, Ommen SR, and Tajik AJ (2003). Cardiology patient page. Hypertrophic cardiomyopathy: a patient perspective. Circulation 108, e133–135. [DOI] [PubMed] [Google Scholar]
- Palmiter KA, Tyska MJ, Haeberle JR, Alpert NR, Fananapazir L, and Warshaw DM (2000). R403Q and L908V mutant beta-cardiac myosin from patients with familial hypertrophic cardiomyopathy exhibit enhanced mechanical performance at the single molecule level. Journal of muscle research and cell motility 21, 609–620. [DOI] [PubMed] [Google Scholar]
- Pardee JD, and Spudich JA (1982). Purification of muscle actin. Methods in enzymology 85 Pt B, 164–181. [DOI] [PubMed] [Google Scholar]
- Resnicow DI, Deacon JC, Warrick HM, Spudich JA, and Leinwand LA (2010). Functional diversity among a family of human skeletal muscle myosin motors. Proceedings of the National Academy of Sciences of the United States of America 107, 1053–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reubold TF, Eschenburg S, Becker A, Kull FJ, and Manstein DJ (2003). A structural model for actin-induced nucleotide release in myosin. Nature structural biology 10, 826–830. [DOI] [PubMed] [Google Scholar]
- Rice SE, Purcell TJ, and Spudich JA (2003). Building and using optical traps to study properties of molecular motors. Methods in enzymology 361, 112–133. [DOI] [PubMed] [Google Scholar]
- Sommese RF, Sung J, Nag S, Sutton S, Deacon JC, Choe E, Leinwand LA, Ruppel K, and Spudich JA (2013). Molecular consequences of the R453C hypertrophic cardiomyopathy mutation on human beta-cardiac myosin motor function. Proceedings of the National Academy of Sciences of the United States of America 110, 12607–12612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA (2014). Hypertrophic and dilated cardiomyopathy: four decades of basic research on muscle lead to potential therapeutic approaches to these devastating genetic diseases. Biophysical journal 106, 1236–1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spudich JA (2015). The myosin mesa and a possible unifying hypothesis for the molecular basis of human hypertrophic cardiomyopathy. Biochemical Society transactions 43, 64–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikakulam R, and Winkelmann DA (2004). Chaperone-mediated folding and assembly of myosin in striated muscle. Journal of cell science 117, 641–652. [DOI] [PubMed] [Google Scholar]
- Trybus KM (2000). Biochemical studies of myosin. Methods 22, 327–335. [DOI] [PubMed] [Google Scholar]
- Tyska MJ, Hayes E, Giewat M, Seidman CE, Seidman JG, and Warshaw DM (2000). Single-molecule mechanics of R403Q cardiac myosin isolated from the mouse model of familial hypertrophic cardiomyopathy. Circ Res 86, 737–744. [DOI] [PubMed] [Google Scholar]
- Walsh R, Thomson KL, Ware JS, Funke BH, Woodley J, McGuire KJ, Mazzarotto F, Blair E, Seller A, Taylor JC, et al. (2016). Reassessment of Mendelian gene pathogenicity using 7,855 cardiomyopathy cases and 60,706 reference samples. Genetics in medicine : official journal of the American College of Medical Genetics. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wendt T, Taylor D, Trybus KM, and Taylor K (2001). Three-dimensional image reconstruction of dephosphorylated smooth muscle heavy meromyosin reveals asymmetry in the interaction between myosin heads and placement of subfragment 2. Proceedings of the National Academy of Sciences of the United States of America 98, 4361–4366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkelmann DA, Forgacs E, Miller MT, and Stock AM (2015). Structural basis for drug-induced allosteric changes to human beta-cardiac myosin motor activity. Nature communications 6, 7974. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.