Abstract
Viral peptides are recognized by cytotoxic T lymphocytes (CTL) as a complex with major histocompatibility complex (MHC) class I molecules, but the extent to which a single HLA allele can accommodate epitope peptides of different length and sequence is not well characterized. Here we report the identification of clonal CTL responses from the same donor that independently recognize one of two HLA-B57-restricted epitopes, KAFSPEVIPMF (KF11; p24Gag residues 30 to 40) and KAFSPEVI (KF8; p24Gag residues 30 to 37). Although lysis studies indicated that the KF11 peptide stabilized the HLA-B57-peptide complex more efficiently than the KI8 peptide, strong clonal responses were directed at each epitope. In samples from a second donor, the same phenomenon was observed, in which distinct CTL clones recognized peptide epitopes presented by the same HLA class I allele (in this case, HLA-A3) which were entirely overlapping. These data are relevant to the accurate characterization of CTL responses, which is fundamental to a detailed understanding of MHC class I-restricted immunity. In addition, these studies demonstrate marked differences in the length of peptides presented by HLA-B57, an allele which is associated with nonprogressive human immunodeficiency virus infection.
Cytotoxic T lymphocytes (CTL) play a central role in the control of chronic viral infections such as AIDS virus infection (30). However, the generation of high numbers of CTL alone does not necessarily confer protection against disease. Studies in lymphochoriomeningitis virus infection of mice have shown very persuasively that there are differences in the effectiveness with which particular CTL clear virus and, indeed, that immunodominant CTL are not necessarily the most effective (11). Strikingly, substantial numbers of CTL which can be stained by peptide-major histocompatibility complex (MHC) tetramers may be unable to elaborate the normal effector functions of cytotoxicity or cytokine release in response to the appropriate antigenic stimulus (12, 36). Likewise, in human viral infections, ample evidence has accumulated which demonstrates that CTL differ in their effectiveness. Numerous studies have identified differences in outcome which are associated with the presence or absence of particular HLA molecules (7, 14, 20, 22, 24, 25). The possible mechanism by which HLA class I molecules might influence speed to progression is unclear. Potential explanations include that the particular human immunodeficiency virus (HIV)-specific epitopes presented by individual class I molecules may be of significance either by virtue of sequence conservation or due to the binding affinity of the peptide to the MHC molecule.
Thus, descriptions of CTL numbers which collectively recognize unspecified epitopes within given proteins may inadequately represent the magnitude of the clonal CTL response. Understanding the role played by CTL in these different circumstances depends first on the precise definition of the HLA class I restriction of the principal responses and of the optimal epitopes which are recognized. In this way, these CTL responses may be studied using the most sensitive reagents available, enzyme-linked immunospot (Elispot) and peptide-MHC tetramer assays (2, 27). A second major advantage in precise epitope definition is in the induction of CTL responses using peptides. The therapeutic option offered by infusions of autologous dendritic cells pulsed with optimal epitope peptides is fast becoming a reality (10), and use of the optimal peptide, as opposed to a suboptimal, longer peptide, is likely to be more effective in inducing strong CTL responses, since suboptimal peptides generally bind with lower affinity (8).
A complication in the precise definition of CTL activity is that epitopes are clustered together into regions within proteins of high immunogenicity, and very similar peptides which differ by only a few amino acids are recognized by distinct CTL clones (5, 13a). The phenomenon of different MHC class I alleles competing for presentation of overlapping viral epitopes has been suggested to play a role in determining the immunodominance of the CTL response (31). For example, an HLA-B8-restricted influenza virus nucleoprotein-specific epitope ELRSRYWAI entirely overlaps the HLA-B*2702-restricted epitope LRSRYWAI. In one person studied, who expressed both B8 and B27, it appeared that B8 and B27 competed for a common peptide fragment in the endoplasmic reticulum. This led to suboptimal loading of HLA-B8 and consequently to failed presentation of the B8 epitope peptide.
In the studies described below, two distinct HLA-B57-restricted HIV-specific CTL epitopes, which also completely overlap, are defined by using CTL clones from the same donor. One is the 8-mer KAFSPEVI, p24Gag residues 30 to 37, a novel epitope, and the second is the previously described 11-mer epitope KAFSPEVIPMF, p24Gag residues 30 to 40 (14). Comparisons of the ability of these two peptides to stabilize B57-peptide complexes show that the 11-mer, which fits the B57 motif (4), binds considerably more stably than the 8-mer, which does not closely conform to the peptide-binding motif. However, strong in vivo CTL responses were generated to both peptides. Thus, even in a situation in which the same HLA molecule is supplied in the endoplasmic reticulum with alternative peptides which overlap entirely in sequence and which differ greatly in their ability to stabilize the peptide-MHC complex, these B57 molecules can be adequately loaded with the weakly binding epitope to induce strong CTL responses.
MATERIALS AND METHODS
Subjects studied.
The two donors described were both infected by HIV-1 via mother-to-child transmission. Donor 026-BMC is a healthy, 9-year-old Hispanic child. At the time of study, he was receiving antiretroviral therapy of d4T, 3TC. His most recent viral load was 1,261 RNA copies per ml of plasma, and his most recent absolute CD4 count was 681 × 109/liter. Donor 021-BMC is a healthy, 14-year-old African-American child. His antiretroviral therapy at the time of study was also d4T and 3TC. His most recent viral load was 11,850/μl, and the absolute CD4 count was 650 × 109/liter. Fresh peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque (Sigma, St. Louis, Mo.) density gradient centrifugation. HLA typing was performed by SSP-PCR (6). The class I HLA type of 026-BMC was HLA A3/—B42/57 Cw7/17 and that of 021-BMC was A3/3001 B42/— Cw17/—.
Peptides.
Twenty-two 20-mer peptides spanning the p24Gag protein, each overlapping by 10 amino acids the previous peptide in the panel (NIBSC Centralized Facility for AIDS Reagents, supported by EU Program EVA and the United Kingdom Medical Research Council), were used to screen for p24Gag-specific CTL responses in Elispot assays. A similar panel of 24 15-mer peptides, each overlapping by 10 amino acids, were used to screen for CTL responses within p17Gag. Shorter peptides were synthesized on a Synergy peptide synthesizer (432A; Applied Biosystems, Foster City, Calif.) using conventional F-moc chemistry. All peptides were analyzed for purity by reverse-phase high-pressure liquid chromatography. Peptides in all cases were >80% pure.
Elispot assays.
Fresh PBMC were plated in 96-well polyvinylidene plates (Millipore, Bedford, Mass.) which had been precoated with 0.5 μg of anti-gamma interferon (IFN-γ) monoclonal antibody (MAb) 1-DIK (Mabtech, Stockholm, Sweden). PBMC were added at 50,000 cells per well in a volume of 180 μl, and the peptides were added in a volume of 20 μl. The end concentration of the peptides was 10 μM. The plates were incubated overnight at 37°C in 5% CO2 and washed with phosphate-buffered saline (PBS) before addition of the second, biotinylated anti-IFN-γ MAb, 7-B6-1 biotin (Mabtech) at 0.5 μg/ml and incubated at room temperature for 100 min. Following six washes, streptavidin-conjugated alkaline phosphatase (Mabtech) was added at room temperature for 40 min. Individual cytokine-producing cells were detected as dark spots after a 20-min reaction with 5-bromo-4-chloro-3-indolyl phosphate and nitroblue tetrazolium using an alkaline phosphatase conjugate substrate (Bio-Rad, Richmond, Calif.). The number of specific T cells was calculated by substracting the negative control values. The background was less than 40 T cells per 106 PBMC (2 spots/well at 50,000 PBMC/well). Assays were repeated using lower input numbers of cells and in quadruplicate in order to quantitate responses to individual peptides more accurately.
Generation of CTL clones.
CTL clones were generated using methods previously described (33). In brief, PBMC were plated out in 96-well plates at limiting dilution (100 to 10 cells/well) and cultured with irradiated allogeneic feeder PBMC at 50,000 cells/well in a final volume per well of 200 μl of R10 medium (RPMI 1640 [Sigma] with 10% fetal calf serum [Sigma], 10 mM HEPES buffer [Sigma], antibiotics, 2 mM l-glutamine, 50 U of penicillin-streptomycin per ml). The anti-CD3 MAb 12F6 was added at 100 μg/ml. On day 5 and once weekly thereafter, the medium was changed with R10 medium containing 50 U of recombinant interleukin 2 (IL-2) per ml. Clones were screened for specific recognition of HLA-matched, peptide-pulsed, chromium 51 (New England Nuclear, North Billerica, Mass.)-labeled Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell line (B-LCL) target cells after 21 to 28 days in culture. The contents of wells showing high specific recognition of the relevant peptide were then transferred to 24-well plates and restimulated as above, except that 106 feeders and recombinant IL-2 were added to each well. Expanded wells were then retested for lytic activity from 14 days of culture onwards and maintained in culture by monthly restimulations as described above.
Chromium release assays and peptide-MHC stabilization assay.
Effector cells were tested for recognition of peptide-pulsed EBV-transformed B-LCL in standard 51Cr release assays (34). In these standard assays, target cells were incubated with peptide for 1 h prior to being washed thrice and then utilized immediately in the assay. In the peptide-MHC stabilization assays, as previously described (13), target cells were initially prepared in the same way, by pulsing with 10−5 M peptide. However, the thrice-washed targets were not used immediately but remained incubating at 37°C in 5% CO2 for between 1 and 78 h before the chromium release assay began.
RESULTS
Identification of two distinct CTL responses within the same p24Gag 20-mer, NAWVKVIEEKAFSPEVIPMF.
Screening of CTL responses to p24Gag using PBMC from donor 026-BMC (HLA class I type A3/— B42/57 Cw7/17) showed that two 20-mer peptides out of the panel of 22 tested were recognized in an Elispot assay. One of the two major responses was directed towards the 20-mer p24.3 (NAWVKVIEEKAFSPEVIPMF; p24Gag residues 21 to 40). Repeating the assay using two 15- to 16-mer overlapping peptides contained within the 20-mer p24.3, the major response was observed towards the 15-mer KVIEEKAFSPEVIPMF (>1,000/106 PBMC). An additional response which was not characterized further was detected towards the 16-mer NAWVKVIEEKAFSPEV (760/106 PBMC).
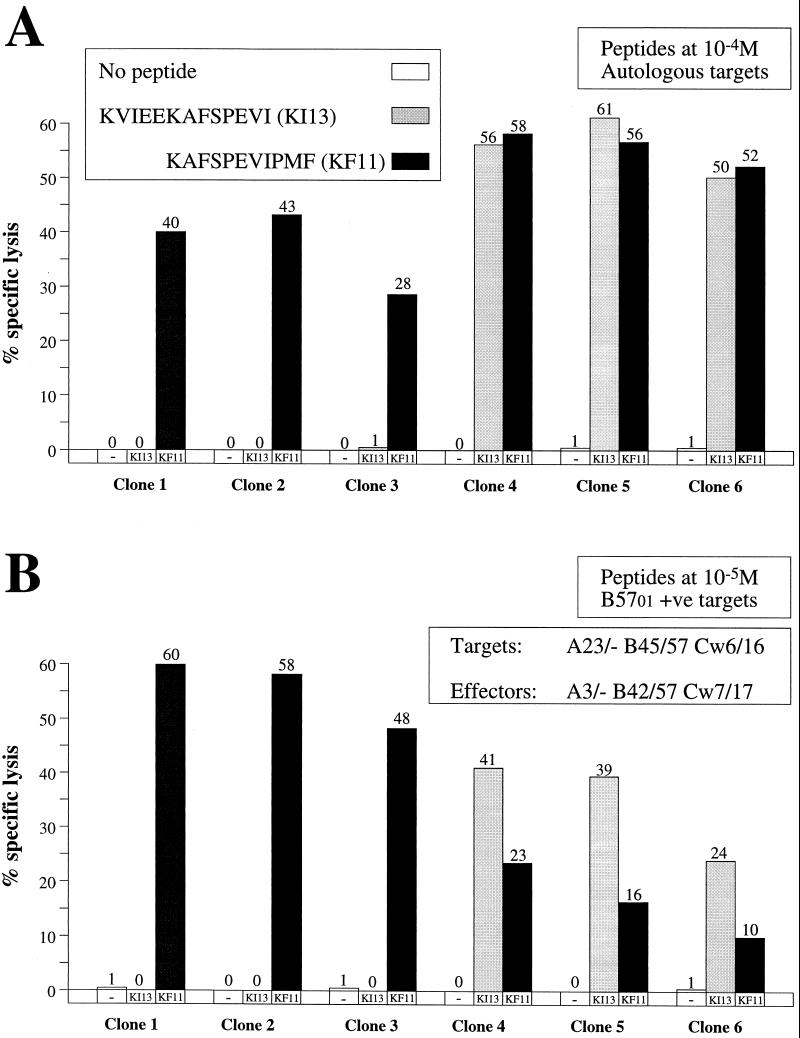
Clones were generated from the PBMC, and 10 clones showed recognition of the 15-mer KVIEEKAFSPEVIPMF. However, comparing recognition by these clones of the 13-mer VKVIEEKAFSPEVI (KI13) and the 11-mer KAFSPEVIPMF (KF11), a previously defined HLA-restricted B57-restricted optimal CTL epitope (14), 5 of 10 recognized only the 11-mer and 5 of 10 clones recognized both the 13-mer and 11-mer peptides (six representative clones are shown in Fig. 1A). These assays were repeated using six EBV-transformed B-LCL target cell lines matched through only one HLA class I molecule, and both sets of clones recognized the peptides only when presented by two different sets of target cells matched only through HLA-B57 (HLA restriction is demonstrated in Fig. 1B). Since the effector cells did not express HLA-Cw6, which is often found in linkage disequilibrium with B57 (9), these responses were clearly defined as B57 restricted. Peptide-pulsed HLA-mismatched targets were not lysed by these clones (not shown).
FIG. 1.
(A) Patterns of recognition of peptides KI13 (KVIEEKAFSPEVI) and KF11 (KAFSPEVIPMF) by six representative CTL clones derived from donor 026-BMC. The peptide concentration was 100 μM, the effector-target cell (E:T) ratio in each case was 5:1, and the target cells were autologous EBV-transformed B cells (B-LCL). (B) Pattern of recognition of the same peptides by the same CTL clones as in panel A but using HLA-B57-matched targets from donor 003-BMC (HLA class I type of target cells, A23/—B45/57 Cw6/16; HLA class I type of 026-BMC CTL clones, A3/—B42/57 Cw7/17). The peptide concentration used was 10 μM, and the E:T ratio was 5:1. There was no recognition of target cells matched through class I molecules other than B57 or of HLA-mismatched targets (data not shown).
Fine mapping of a novel HLA-B57-restricted CTL epitope.
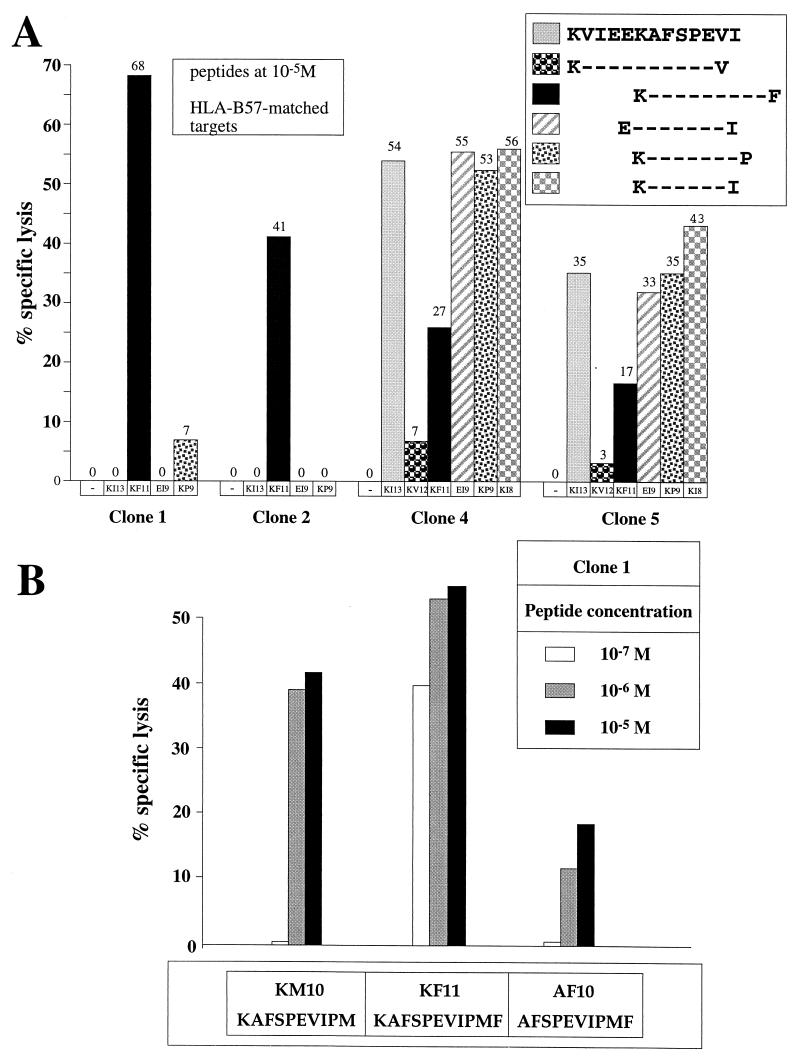
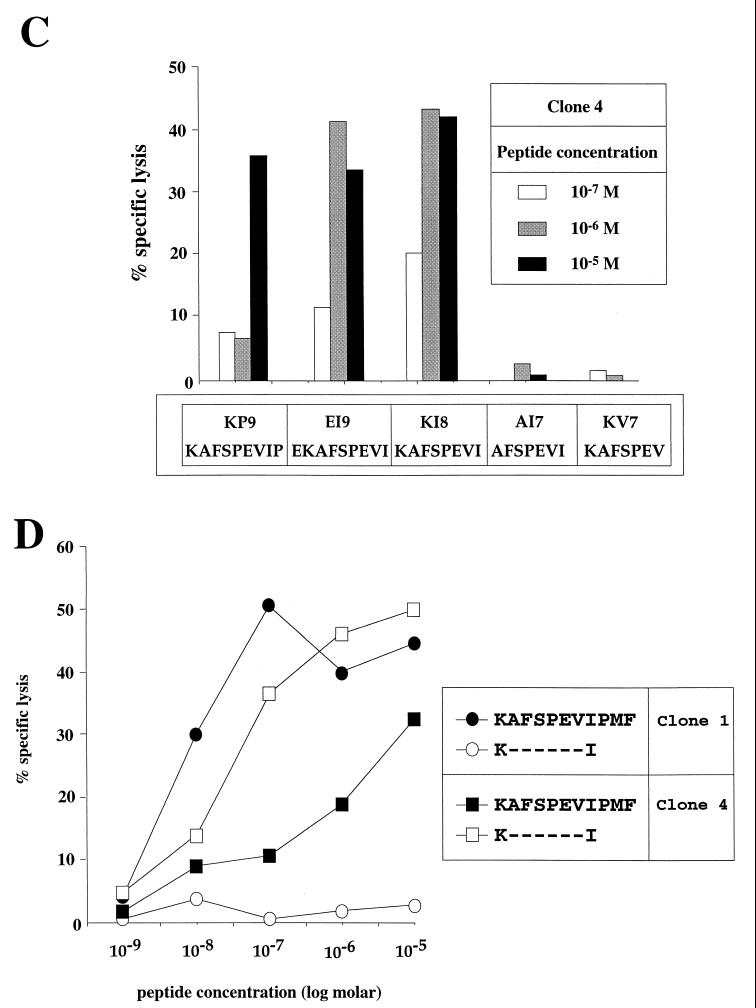
The five clones which recognized KF11 but not KI13 did not see peptide truncations of KF11 except at concentrations of 10−6 M or above, whereas similar levels of lysis were observed using 1/1,000 the concentration of KF11 (Fig. 2A and B). Thus, these five clones were specific for the previously optimized epitope KF11 (p24Gag residues 30 to 40). The other five clones were specific for the 8-mer KAFSPEVI (KI8; p24Gag residues 30 to 37), since longer peptides did not improve sensitization of target cells for lysis and peptides such as AFSPEVIPMF and KVIEEKAFSPEV, which did not contain the critical residues at the N- or C-terminal positions of the 8-mer, were recognized very poorly (Fig. 2A and C). Comparison of the affinity of the KF11- and KI8-specific clones for the respective cognate peptide-MHC complex as described by peptide titration curves consistently showed that the KF11-specific clones needed approximately 1/10 the peptide concentrations required by the KI8-specific clones for equivalent lysis (Fig. 2D; values are representative of three similar assays).
FIG. 2.
(A) Recognition of truncations of IK13 (KVIEEKAFSPEVI) and KF11 (KAFSPEVIPMF) by four representative CTL clones. The peptide concentration was 10 μM, the targets were B57-matched B-LCL from donor 003-BMC, and the E:T ratio was 5:1. (B) Recognition of truncations of KF11 at concentrations of peptide of 1, 10, and 100 μM by CTL clone 1 (similar pattern of recognition was observed for clone 2; not shown). Target cells were the same as in panel A. (C) Recognition of peptides differing by one amino acid at the N-terminal and C-terminal residues from the optimal peptide KI8 (KAFSPEVI) recognized by CTL clone 4 (a similar pattern of recognition was observed for clone 5; not shown) using concentrations of peptide of 1, 10, and 100 μM. Target cells were as in panel A. (D) Peptide titration curves showing specific recognition of KF11 by clone 1 and specific recognition of KI8 by clone 4. Target cells were as in panel A.
Instability of the KI8-B57 complex compared with KF11-B57 complex.
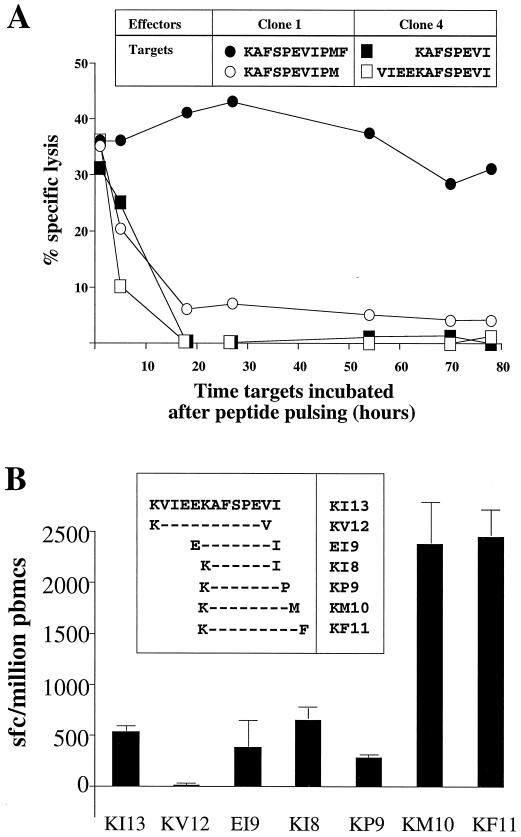
Six previously defined HLA-B57-restricted CTL optimal epitopes (14, 26) and 14 peptides eluted from B57 molecules (4) were peptides 9 to 11 amino acids in length, each containing a large hydrophobic residue, either tryptophan or phenylalanine, at the C terminus. Thus, it was surprising that an 8-mer containing a medium-sized hydrophobic residue (isoleucine) in the C-terminal position should also represent an HLA-B57-restricted epitope. In order to test whether the KI8 peptide pulsed onto target cells remained bound for CTL recognition for days on the target cells, as would be expected for typical optimal epitope peptides (13, 15, 18), or only for a matter of hours, target cells were pulsed with saturating concentrations of peptide (10−5 M) for 1 h, washed thrice, and incubated for between 1 and 78 h prior to the chromium release assay. Lysis of targets was reduced to 0% within 18 h of the cells' being pulsed with 10−5 M KI8, whereas little diminution in lysis by KF11-specific clones was observed even following 78 h of pulsing target cells with KF11 (Fig. 3A).
FIG. 3.
(A) Stabilization of peptide-B57 complexes by the peptides KF11 and KI8. Clone 1 (specific for KF11) shows recognition of KF11-pulsed targets >75 h after peptide pulsing. Clone 4 (specific for KI8) showed recognition of KI8-pulsed target cells for <18 h after peptide pulsing. The peptide concentration used was 10 μM. Targets and E:T ratios were as in Fig. 2. (B) Frequency of peptide-specific responses by Elispot assay following epitope optimization using fresh PBMC. The peptide concentration was 10 μM. Error bars show SD from the mean of four replicates for each peptide shown.
Frequency of CTL specific for KF11 and KI8 determined by Elispot assay.
The optimal and suboptimal peptides were utilized in further Elispot assays to determine the frequency of CTL within PBMC specific for each (Fig. 3B). The frequency of KI8-specific cells was 692/106 PBMC (standard deviation [SD], 63/106 PBMC), and that of KF11-specific cells was 2,442/106 PBMC (SD, 266/106 PBMC).
Identification of two distinct CTL responses within the same p17Gag 15-mer, WEKIRLRPGGKKKYK.
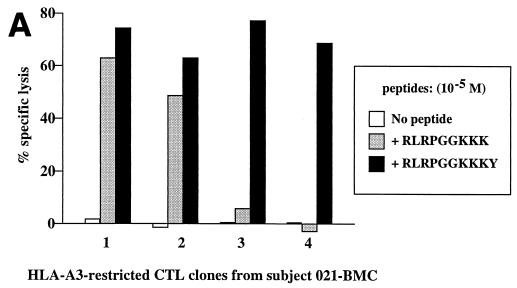
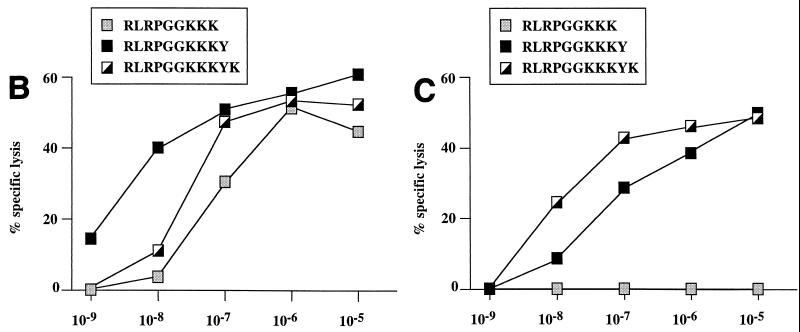
Screening for CTL responses to p17Gag overlapping peptides using PBMC from donor 021-BMC (HLA class I type A3/3001 B42/—Cw17/—) revealed a dominant response within this protein to the 15-mer WEKIRLRPGGKKKYK (WK15; p17Gag residues 16 to 30), at a frequency of >1,000/106 PBMC. Four CTL clones were generated which recognized this peptide, WK15, all through HLA-A3-restricted responses (Fig. 4). Peptide-pulsed targets not expressing HLA-A3 were not lysed by these clones (not shown). The fine specificity of these clones was analyzed further initially using previously defined HLA-A3-restricted CTL epitope peptides within this region (5), namely, KIRLRPGGK (KK9; residues 18 to 26), RLRPGGKKK (RK9; residues 20 to 28), and RLRPGGKKKKY (RY10; residues 20 to 29). None of the four CTL clones showed recognition of the KK9 epitope. All four clones recognized the RY10 epitope. However, whereas clones 1 and 2 showed strong recognition of the RK9 epitope, clones 3 and 4 did not recognize RK9 even at high peptide concentrations (Fig. 1A). The optimal epitopes recognized were, respectively, RY10 and a previously undescribed HLA-A3 epitope, RLRPGGKKKYK (RK11; p17Gag residues 20 to 30) (Fig. 1B and C). Thus, in a second subject, clonal characterization of the CTL response revealed the presence of distinct responses to two epitopes presented by the same HLA allele, one of which is entirely contained within the other.
FIG. 4.
(A) Recognition of previously defined HLA-A3-restricted CTL epitopes RLRPGGKKK (RK9) and RLRPGGKKKY (RY10) by four clones derived from PBMC from donor 021-BMC (HLA A3/3001 B42/—Cw17/—). Targets were B-LCL from donor KS (HLA class I type A3/— B7/— Cw7/—) at an E:T ratio of 5:1. Specific lysis by the clones of targets pulsed with the peptide KIRLRPGGK (KK9) was between −1 and 0% (not shown). (B) Fine specificity of CTL clone 1. The best-recognized peptide was RY10. Similar data using CTL from clone 2 are not shown. Targets were the same as in panel A. (C) Fine specificity of CTL clone 3. The best-recognized peptide was RLRPGGKKKYK (RK11). Similar data using CTL from clone 4 are not shown. Targets were the same as in panel A.
DISCUSSION
These results demonstrate that HLA-B57 can present efficiently two Gag peptides for CTL recognition, one of which is entirely contained within the other. Although one of these peptides, KI8, bound much less effectively to the B57 molecule, it was still apparently able to compete in vivo with the longer peptide for binding and presentation. The detectable magnitude of CTL response to KI8 was lower than to KF11 (692 versus [2,442 − 692 =] 1,750 per 106 PBMC), but both were persistently present in vivo in substantial numbers. That this is not an isolated phenomenon is supported by the finding of a second set of HLA-A3-restricted epitopes, one of which is entirely contained within the other. These studies point to differences between CTL targeting the same epitopic region and restricted by the same HLA class I allele, which may have important consequences for their effectiveness.
Detailed studies were performed for the HLA-B57-restricted responses to further define this phenomenon. These data illustrate that CTL responses may be induced by an epitope, KI8, which not only shows a rapid off-rate from the restriction element, but which also might be expected to compete unfavorably with a longer peptide, KF11, which can stabilize the B57-peptide complex for long periods of time. Optimized epitopes which are 11 amino acids in length are relatively rare; 98 of 110 (89%) optimized HIV-specific CTL epitope peptides listed in the HIV Immunology Database (5) were shorter than 11 amino acids in length. One may speculate that proteasomal cleavage and peptide transport by the transporter associated with antigen presentation (TAP) might favor delivery of KI8 into the endoplasmic reticulum, but there are no data to support this. It appears that 8- to 12-mers are all efficiently transported (3), nor are there data that would suggest preferential cleavage of KI8 by the proteasome (29) or transport by TAP (3). Once it has reached the surface of the infected cell, it is evident that the duration of time in which the KI8 peptide is able to stabilize the B57-peptide complex is, although short, sufficient to induce specific CTL responses.
An additional possible explanation for the presence of KI8-specific CTL would be that the KF11-B57 complexes not only stimulate the generation of KF11-specific CTL but are also responsible for the generation of the KI8-specific CTL. CTL which can be induced by particular peptides but which cannot lyse infected cells expressing those same peptides have been described previously (19, 28).
The established peptide-binding motif for HLA-B57 (4) is consistent with these data showing impaired stabilization of the B57-peptide complex by KI8 compared with that by KF11. The large F pocket in B57 made available by the presence of the small serine residue at MHC position 116 can accommodate large hydrophobic residues such as tryptophan, phenylalanine, and tyrosine at the C-terminal position of binding peptides. All 20 peptides which were either previously identified as optimal B57-restricted epitopes (14, 26) or eluted from B57 class I molecules (4) carry one of these residues at the C-terminal anchor position. Apart from being the only B57-restricted 8-mer described, KI8 is therefore the only B57-restricted epitope not to carry a large hydrophobic residue at the C-terminal position. It is theoretically possible that the C-terminal Ile of the KI8 peptide would not reach the F pocket but would fit, for example, in the D pocket of the B57 peptide-binding groove, but there is no evidence for this. Relevant crystallographic data examining the secondary structure of 9-mer and 10-mer peptides bound to HLA-Aw68 showed that both peptides used the same primary anchor positions tethering the peptide to the MHC at each end of the peptide, but that the 10-mer simply bowed out from the groove compared to the more extended 9-mer (16). However, recent studies showed that an 8-mer epitope peptide could be extended by 4 residues at the C-terminal end and still continue to form productive peptide-MHC complexes, whereas extensions of the 8-mer by even 1 residue at the N-terminal end of the peptide rendered it unable to bind (21).
Having identified CTL which are specific for a peptide which has a very rapid off-rate from the presenting MHC class I molecule, we faced the question of whether these CTL are indeed effective in vivo in killing virus-infected cells. Other CTL epitopes have been defined which, when subjected to scrutiny, appear to operate only at very high concentrations of peptide (17, 23, 32). Clearly, from the point of view of vaccine design, it is vital to be aware of which CTL specificities are effective in terms of controlling viremia. Assays comparing inhibition of viral replication (35) by different CTL clones are currently being undertaken in this laboratory (Goulder et al., unpublished data).
The second point which is illustrated by these studies relates to CTL enumeration by peptide-MHC tetramers and Elispot assays. In this instance, use of the longer peptide KF11 in the high concentrations used in the Elispot assays (10−5 M), or within a B57-KF11 tetramer complex, would be likely to detect KI8-specific cells in addition to KF11-specific cells (Fig. 2D). Thus, in the instance illustrated (Fig. 3B), the calculation of number of KF11-specific CTL per 106 PBMC is likely to represent an overestimate in this instance by approximately 40%. This is clearly of less importance than if the peptide being used to test for the dominant response is the shorter peptide KI8, which is not recognized even at high concentrations by the KF11-specific CTL. In this instance, the error would underestimate the true frequency of KF11-specific CTL by >70%.
This situation, in which dominant CTL responses are missed altogether by use of an epitope peptide which is shorter than the actual epitope recognized, is less of a problem when overlapping peptides are used in conjunction with “optimized” peptides in screening assays. Use of the inappropriate peptide-MHC tetramer similarly may potentially altogether miss a response which dominates the antiviral CTL activity. Recent optimization of the Mamu-A*01-restricted p27Gag-specific epitope (1) illustrated the point in which the majority of CTL clones recognized the peptide CTPYDINQM (CM9), while a minority of clones were specific for the 10-mer peptide CTPYDINQML (CL10). The CL10-specific clones showed no recognition of the 9-mer, and thus it is very unlikely that these clones would have been detected by CM9-Mamu-A*01 tetramers.
In relation to vaccine design, it is probable that processing of the longer epitope (that is, CL10, KF11, and RK11 in the examples described above) would enable CTL responses to be generated to both the longer and the shorter epitopes (that is, CM9, KI8, and RY10). However, the converse clearly would be unlikely to apply. In instances in which CTL specific for the longer peptide were particularly effective in killing virus-infected cells, it would be important to include the longer epitope within the vaccine construct.
A final consideration relates to the possible mechanism by which particular HLA molecules may owe their association with rapid or slow progression to disease (7, 14, 20, 22, 24, 25). In the case of HLA-B57, these studies raise the possibility that the wide diversity of peptides may be accommodated as B57-binding epitopes may partially explain the association of B57 with slow progression in HIV infection which has been described previously (14, 24, 25). It is also noteworthy that the epitope described (KAFSPEVIPMF) shows remarkable sequence conservation (HIV web site http://www.hiv-lanl.gov). Of 160 published B clade sequences, 158 are conserved in this region. This implies that purifying selection pressure exists against amino acid change in this epitope, presumably to maintain adequate viral fitness.
In conclusion, precise epitope definition is critical to detailed understanding of CTL responses and the variability of effectiveness which can be evident between CTL of different specificities. These data challenge the frequently made assumption that the optimal epitope for one CTL clone tested necessarily represents the optimal epitope peptide for other HLA-matched CTL clones targeting the same region, either in the same person studied or in other HLA-matched persons studied. Approaches to studying CTL using only published optimized epitopes corresponding to the HLA class I type of the subject being studied or using peptide-MHC tetramers designed using previously described epitopes entail a significant risk of missing important CTL responses which are present. Further studies are needed to determine which CTL responses are effective in the control of virus infection in order that the appropriate CTL response be induced by vaccine constructs.
ACKNOWLEDGMENTS
Assistance from Eileen Macnamara in the collection of blood samples to study the CTL responses described above is gratefully acknowledged.
This work was supported by grants to P.J.R.G. from the Elizabeth Glaser Pediatric AIDS Foundation and the Medical Research Foundation (United Kingdom) (grant G108/274) and to B.D.W. through the National Institutes of Health (AI28568 and AI30914) and the Doris Duke Charitable Foundation. P.J.R.G. is an Elizabeth Glaser Scientist of the Elizabeth Glaser Pediatric AIDS Foundation. B.D.W. is a Doris Duke Distinguished Clinical Science Professor.
REFERENCES
- 1.Allen T M, Sidney J, del Guerico M F, Glickman R L, Lensmeye G L, Wiebe D A, DeMars R, Pauza C D, Johnson R P, Sette A, Watkins D I. Characterization of the peptide-binding motif of a rhesus MHC class I molecule (Mamu-A*01) that binds an immunodominant CTL epitope from SIV. J Immunol. 1998;160:6062–6071. [PubMed] [Google Scholar]
- 2.Altman J, Moss P A H, Goulder P J R, Barouch D H, McHeyzer-Williams M G, Bell J I, McMichael A J, Davis M M. Direct visualization and phenotypic analysis of virus-specific T lymphocytes in HIV-infected individuals. Science. 1996;274:94–96. [Google Scholar]
- 3.Androlewicz M J, Cresswell P. How selective is the transporter associated with antigen processing? Immunity. 1996;5:1–5. doi: 10.1016/s1074-7613(00)80304-0. [DOI] [PubMed] [Google Scholar]
- 4.Barber L D, Percival L, Arnett K L, Gumperz J E, Chen L, Parham P. Polymorphism in the a1 helix of the HLA-B heavy chain can have an overriding influence on peptide-binding specificity. J Immunol. 1997;158:1660–1669. [PubMed] [Google Scholar]
- 5.Brander C, Walker B D. The HLA class I restricted CTL response in HIV infection: systematic identification of optimal epitopes. In: Korber B T M, Brander C, Walker B D, Koup R A, Moore J, Haynes B, Meyers G, editors. HIV molecular immunology database. Los Alamos, N. Mex: Los Alamos National Laboratory; 1998. [Google Scholar]
- 6.Bunce M, O'Neill C M, Barnardo M C, Krausa P, Browning M, Morris P, Welsh K. Phototyping: comprehensive DNA typing for HLA-A, B, C, DRB1, DRB3, DRB4, DRB5 & DQB1 by PCR with 144 primer mixes utilizing sequence-specific primers (PCR-SSP) Tissue Antigens. 1995;46:355–67. doi: 10.1111/j.1399-0039.1995.tb03127.x. [DOI] [PubMed] [Google Scholar]
- 7.Carrington M, Nelson G W, Martin M P, Kissner T, Vlahov D, Goedert J J, Kaslow R, Buchbinder S, Hoots K, O'Brien S J. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–1752. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 8.Cerundolo V, Elliott T, Elvin J, Bastin J, Rammensee H-G, Townsend A R M. The binding affinity and dissociation rates of peptides for class I major histocompatibility complex molecules. Eur J Immunol. 1991;21:2069. doi: 10.1002/eji.1830210915. [DOI] [PubMed] [Google Scholar]
- 9.Charron D, editor. Proceedings of the twelfth international histocompatibility workshop and conference. Paris, France: EDK; 1997. [Google Scholar]
- 10.Dhodapkar M V, Steinman R M, Sapp M, Desai H, Fossella C, Krasovsky J, Donahoe S M, Dunbar P R, Cerundolo V, Nixon D F, Bhardwaj N. Rapid generation of broad T-cell immunity in humans after a single injection of mature dendritic cells. J Clin Invest. 1999;104:173–180. doi: 10.1172/JCI6909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gallimore A, Dumrese T, Hengartner H, Zinkernagel R M, Rammensee H-G. Protective immunity does not correlate with the hierarchy of virus-specific CTL responses to naturally processed peptides. J Exp Med. 1998;187:1647–1657. doi: 10.1084/jem.187.10.1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gallimore A, Glithero A, Godkin A, Tissot A C, Pluckthun A, Elliott T, Hengartner H, Zinkernagel R M. Induction and exhaustion of lymphocytic choriomeningitis virus-specific cytotoxic T lymphocytes visualized using soluble tetrameric major histocompatibility complex class I-peptide complexes. J Exp Med. 1998;187:1383–1393. doi: 10.1084/jem.187.9.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goulder P J R, Phillips R E, Colbert R A, McAdam S, Ogg G, Nowak M A, Giangrande P L, Luzzi G, Morgan B, Edwards A, McMichael A J, Rowland-Jones S. Late escape from an immunodominant cytotoxic T-lymphocyte response associated with progression to AIDS. Nat Med. 1997;3:212–217. doi: 10.1038/nm0297-212. [DOI] [PubMed] [Google Scholar]
- 13a.Goulder, P. J. R., C. Brander, K. Annamalai, N. Mngqundaniso, U. Govender, Y. Tang, S. He, K. E. Hartman, C. A. O'Callaghan, G. S. Ogg, M. A. Altfeld, E. S. Rosenberg, H. Cao, S. A. Kalams, M. G. Hammond, M. Bunce, S. I. Pelton, S. A. Burchett, K. McIntosh, H. M. Coovadia, and B. D. Walker. Differential narrow focusing of immunodominant human immunodeficiency virus Gag-specific cytotoxic T-lymphocyte responses in infected African and Caucasoid adults and children. J. Virol., in press. [DOI] [PMC free article] [PubMed]
- 14.Goulder P J R, Crowley S, Krausa P, Morgan B, Edwards A, Giangrande P, McIntyre K, McMichael A J. Novel, cross-restricted, conserved and immunodominant CTL epitopes in long-term slow progressors in HIV-1 infection. AIDS Res Hum Retroviruses. 1996;12:1691–1698. doi: 10.1089/aid.1996.12.1691. [DOI] [PubMed] [Google Scholar]
- 15.Goulder P J R, Reid S W, Price D A, O'Callaghan C A, McMichael A J, Phillips R E, Jones E Y. Combined structural and immunological refinement of HIV-1 HLA-B8-restricted cytotoxic T lymphocyte epitopes. Eur J Immunol. 1997;27:1515–1521. doi: 10.1002/eji.1830270630. [DOI] [PubMed] [Google Scholar]
- 16.Guo H C, Jardetzky T S, Garrett T P, Lane W S, Strominger J L, Wiley D C. Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature. 1992;360:364–366. doi: 10.1038/360364a0. [DOI] [PubMed] [Google Scholar]
- 17.Harrer E, Harrer T, Barbosa P, Feinberg M, Johnson R P, Buchbinder S, Walker B D. Recognition of the highly conserved YMDD region in HIV-1 reverse transcriptase by HLA-A2-restricted cytotoxic T lymphocytes from an asymptomatic long-term non-progressor. J Infect Dis. 1996;173:476–479. doi: 10.1093/infdis/173.2.476. [DOI] [PubMed] [Google Scholar]
- 18.Hay M, Ruhl D J, Basgoz N, Wilson C C, Billingsley J M, DePasquale M P, D'Aquila R, Wolinsky S, Crawford J M, Montefiori D, Walker B D. Lack of viral escape and defective in vivo activation of HIV-1-specific CTL in rapidly progressive infection. J Virol. 1999;73:5509–5519. doi: 10.1128/jvi.73.7.5509-5519.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hill A B, Lee S P, Haurum J S, Murray N, Yao Q Y, Rowe M, Signoret N, Rickinson A B, McMichael A J. Class I major histocompatibility complex-restricted cytotoxic T lymphocytes specific for Epstein-Barr virus (EBV)-transformed B lymphoblastoid cell line against which they were raised. J Exp Med. 1995;181:2221–2228. doi: 10.1084/jem.181.6.2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hill A V S. The immunogenetics of human infectious disease. Annu Rev Immunol. 1998;16:593–618. doi: 10.1146/annurev.immunol.16.1.593. [DOI] [PubMed] [Google Scholar]
- 21.Horig H, Young A C M, Papadopoulos N J, DiLorenzo T P, Nathenson S G. Binding of longer peptides to the H-2Kb heterodimer is restricted to peptides extended at their C terminus: refinement of the inherent MHC class I peptide binding criteria. J Immunol. 1999;163:4434–4441. [PubMed] [Google Scholar]
- 22.Jeffrey J M, Usuku K, Hall S E, Matsumoto W, Taylor G M, Procter J, Bunce M, Ogg G S, Welsh K I, Weber J N, Lloyd A L, Nowak M A, Nagai M, Kodama D, Izumo S, Osame M, Bangham C R M. HLA alleles determine human T-lymphotropic virus-I (HTLV-I) proviral load and the risk of HTLV-1-associated myelopathy. Proc Natl Acad Sci USA. 1999;96:3848–3853. doi: 10.1073/pnas.96.7.3848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kalams S A, Johnson R P, Dynan M J, Hartman K E, Harrer T, Harrer E, Trocha A, Blattner W A, Buchbinder S P, Walker B D. T cell receptor usage and fine specificity of human immunodeficiency virus 1-specific cytotoxic T lymphocyte clones: analysis of quasispecies recognition reveals a dominant response directed against a minor in vivo variant. J Exp Med. 1996;183:1669–1679. doi: 10.1084/jem.183.4.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kaslow R A, Carrington M, Apple R, Park L, Munoz A, Saah A J, Goedert J J, Winkler C, O'Brien S J, Rinaldo C R, Detels R, Blattner W, Phair J, Ehrlich H, Mann D. Influence of human MHC genes on the course of HIV infection. Natl Med. 1996;2:405–411. doi: 10.1038/nm0496-405. [DOI] [PubMed] [Google Scholar]
- 25.Keet I P, Tang J, Klein M, LeBlanc S, Enger C, Rivers C, Apple R J, Mann D, Goedert J J, Miedema F, Kaslow R A. Consistent associations of HLA class I and II and transporter gene products with progression of human immunodeficiency virus type 1 infection in homosexual men. J Infect Dis. 1999;180:299–309. doi: 10.1086/314862. [DOI] [PubMed] [Google Scholar]
- 26.Klein M R, van der Burg S H, Hovenkamp E, Holwerda A M, Drijfhout J W, Melief C J, Miedema F. Characterization of HLA-B57-restricted human immunodeficiency virus type 1 Gag- and RT-specific cytotoxic T lymphocyte responses. J Gen Virol. 1998;79:2191–2201. doi: 10.1099/0022-1317-79-9-2191. [DOI] [PubMed] [Google Scholar]
- 27.Lalvani A, Brookes R, Hambleton S, Britton W J, Hill A V S, McMichael A J. Rapid effector function in CD8+ memory T cells. J Exp Med. 1997;186:859–865. doi: 10.1084/jem.186.6.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McAdam S, Klenerman P, Tussey L, Rowland-Jones S, Lalloo D, Phillips R, Edwards A, Giangrande P, Brown A L, Gotch F M, McMichael A J. Immunogenic HIV variant peptides that bind to HLA-B8 can fail to stimulate cytotoxic T lymphocyte responses. J Immunol. 1995;155:2729–2736. [PubMed] [Google Scholar]
- 29.Rock K I, Goldberg A L. Degradation of cell proteins and the generation of MHC class I-presented peptides. Annu Rev Immunol. 1999;17:739–780. doi: 10.1146/annurev.immunol.17.1.739. [DOI] [PubMed] [Google Scholar]
- 30.Schmitz J E, Kuroda M, Santra S, Sasseville V, Simon M, Lifton M, Racz P, Tenner-Racz K, Dalesandro M, Scallon B, Ghrayeb J, Forman M, Montefiori D, Rieber E, Letvin N, Reimann K. Control of viremia in simian immunodeficiency virus infection by CD8+ lymphocytes. Science. 1999;277:333–338. doi: 10.1126/science.283.5403.857. [DOI] [PubMed] [Google Scholar]
- 31.Tussey L G, Rowland-Jones S, Zheng T S, Androlewicz M J, Cresswell P, Frelinger J, McMichael A J. Different MHC class I alleles compete for presentation of overlapping viral epitopes. Immunity. 1995;3:65–77. doi: 10.1016/1074-7613(95)90159-0. [DOI] [PubMed] [Google Scholar]
- 32.Van Baalen C A, Schutten M, Huisman R C, Boers P H, Gruters R A, Osterhaus A D. Kinetics of antiviral activity by human immunodeficiency virus type 1-specific cytotoxic T lymphocytes (CTL) and rapid selection of CTL escape virus in vitro. J Virol. 1998;72:6851–6857. doi: 10.1128/jvi.72.8.6851-6857.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker B D, Flexner C, Birch-Limberger K, Fisher L, Paradis T J, Aldovini A, Young R, Moss B, Schooley R T. Long-term culture and fine specificity of human cytotoxic T-lymphocyte clones reactive with human immunodeficiency virus type 1. Proc Natl Acad Sci USA. 1989;86:9514–9518. doi: 10.1073/pnas.86.23.9514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker B D. HIV-1-specific cytotoxic T lymphocytes. In: Aldovini A, Walker B D, editors. Techniques in HIV research. New York, N.Y: Stockton Press; 1990. p. 201. [Google Scholar]
- 35.Yang O O, Kalams S A, Rosenzweig M, Trocha A, Jones N, Koziel M, Walker B D, Johnson R P. Efficient lysis of human immunodeficiency virus type 1-infected cells by cytotoxic T lymphocytes. J Virol. 1996;70:5799–5806. doi: 10.1128/jvi.70.9.5799-5806.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zajac A J, Blattman J N, Murali-Krishna K, Sourdive D J D, Suresh M, Altman J D, Ahmed R. Viral immune evasion due to persistence of activated T cells without effector function. J Exp Med. 1998;188:2205–2213. doi: 10.1084/jem.188.12.2205. [DOI] [PMC free article] [PubMed] [Google Scholar]