Abstract
The T-cell-mediated immune response plays a central role in the defense against intracellular pathogens. To avoid this immune response, viruses have evolved elaborate mechanisms that target and modulate many different aspects of the host's immune system. A target common to many of these viruses is the major histocompatibility complex (MHC) class I molecules. Kaposi's sarcoma-associated herpesvirus (KSHV) encodes K3 and K5 zinc finger membrane proteins which remove MHC class I molecules from the cell surface. K3 and K5 exhibit 40% amino acid identity to each other and localize primarily near the plasma membrane. While K3 and K5 dramatically downregulated class I molecules, they displayed different specificities in downregulation of HLA allotypes. K5 significantly downregulated HLA-A and -B and downregulated HLA-C only weakly, but not HLA-E, whereas K3 downregulated all four HLA allotypes. This selective downregulation of HLA allotypes by K5 was partly due to differences in amino acid sequences in their transmembrane regions. Biochemical analyses demonstrated that while K3 and K5 did not affect expression and intracellular transport of class I molecules, their expression induced rapid endocytosis of the molecules. These results demonstrate that KSHV has evolved a novel immune evasion mechanism by harboring similar but distinct genes, K3 and K5, which target MHC class I molecules in different ways.
A major immune defense against viral infection is mediated by cytotoxic T lymphocytes (CTLs), which recognize and lyse infected cells upon engagement of the T-cell receptor with major histocompatibility complex (MHC) class I molecules presenting viral peptides (10, 26). Viral proteins are degraded to peptides by the proteasomes in the cytosol. These peptides are translocated by the transporter associated with antigen processing (TAP) to the endoplasmic reticulum (ER), where they assemble with the MHC class I heavy chain and β2 microglobulin to form a trimeric complex. This mature complex is then transported from the ER to the plasma membrane, where the MHC class I complexes present the peptide to CTLs (20, 35).
Herpesviruses establish lifelong infections despite the presence of an active immune system. To achieve this, herpesviruses encode gene products that affect MHC class I expression, either at the level of transcription or of expression at the cell surface (26). For example, the herpes simplex virus type 1 US12 gene product, called ICP47, binds to TAP and prevents the delivery of cytosolic antigen peptides to assembling class I molecules in the ER (7, 11, 37). Epstein-Barr virus-encoded EBNA-1 protein inhibits antigen processing, thus affecting presentation of certain peptides by MHC class I complexes (18). Human cytomegalovirus (HCMV) and murine CMV contain numerous lytic glycoproteins, each of which is sufficient to cause increased turnover of MHC class I molecules. Two ER-resident glycoproteins, US2 and US11, of HCMV are sufficient to induce the rapid dislocation of newly synthesized MHC class I proteins from the ER to the cytosol, where the class I heavy chains are degraded by the proteasome (26, 34). HCMV US6 prevents peptide loading of MHC class I molecules by inhibiting TAP-mediated peptide translocation into the ER (1). In addition, the HCMV US3 and the murine CMV m152 proteins prevent the transport of MHC class I molecules to the cell surface by retaining the molecules within the ER (12, 38).
DNA sequences of a member of the herpesvirus group, Kaposi's sarcoma-associated herpesvirus (KSHV) or human herpesvirus 8, have been consistently identified in Kaposi's sarcoma tumors from human immunodeficiency virus (HIV)-positive and HIV-negative patients (2, 3, 19). KSHV has also been identified in body cavity-based lymphoma and some forms of Castleman's disease (2, 3, 27). The genomic sequence indicates KSHV to be a gamma-2 herpesvirus that is closely related to herpesvirus saimiri (HVS) (21a, 29) and the recently isolated rhesus monkey rhadinovirus (5, 32) and retroperitoneal fibromatosis herpesvirus (28). Despite the colinearity of genomic structures and the similarity of genomic sequences among these gamma-2 herpesviruses, each virus contains nonconserved, unique regions (29). DNA sequence analysis of a 13-kb nonconserved region of the KSHV genome revealed a number of cellular homologs (21a, 24, 29). These include a virus-encoded interleukin 6 (21, 22, 25), MIP1-α and -β chemokines (13, 21, 25), Bcl-2 (30), dihydrofolic reductase, and thymidylate synthetase. In addition, this region contains several unique open reading frames, called K3, K4.2, K5, and K7, which do not have apparent homology with any known cellular genes (24, 29). Interestingly, K3 and K5 exhibit 40% amino acid identity with each other (24, 29) and are expressed during the early lytic cycle of viral replication (24, 33). Both the K3 and K5 gene products are related to the immediate-early gene (IE1) product of bovine herpesvirus 4 and ORF12 of HVS (24). All of these open reading frames contain two C4HC3 zinc finger motifs at the amino terminus and hydrophobic transmembrane regions in the center but are of various sizes in the carboxyl-terminal tail (24).
In this report, we demonstrate that K3 and K5 downregulate the surface expression of MHC class I molecules with different activities and specificities. Biochemical analyses demonstrate that downregulation of class I molecules induced by K3 and K5 is likely due to their rapid endocytosis. Thus, KSHV encodes similar but distinct genes, K3 and K5, which may protect the infected cells from destruction by immune effector cells.
MATERIALS AND METHODS
Cell culture and transfection.
BJAB and 221 cells were grown in RPMI medium supplemented with 10% fetal calf serum. COS-1 cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum. 221 cells harboring defined HLA allotypes have been described previously (4). A Fusin lipofection (Boehringer Mannheim) transfection procedure was used for transient expression in COS-1 cells. The pEF1-derived expression vector (20 μg) was introduced into BJAB cells by electroporation at 250 V and 960 μF in serum-free Dulbecco's modified Eagle's medium. After a 48-h incubation, the cells were cultured with selection medium containing 2 mg of neomycin/ml for 5 weeks.
Plasmid construction.
Based on the KSHV genomic sequence (29), DNA containing the KSHV K3, K4.2, or K5 open reading frame was amplified from BCBL-1 genomic DNA by PCR using a 5′ primer which corresponds to the amino-terminal sequence of each gene and a 3′ primer which corresponds to the carboxyl-terminal sequence of each gene (15). The primers used for PCR contain EcoRI and XbaI recognition sequences for subsequent cloning. Amplified DNA was ligated into the EcoRI and XbaI cloning sites of the pEpiTag vector (Invitrogen, Carlsbad, Calif.) for six-His tagging at the carboxyl terminus of each gene. To construct green fluorescent protein (GFP) expression vectors, each gene was subcloned into the pQBIGFP vector. PCR-amplified DNA fragments were completely sequenced to verify the presence of expected sequences and the absence of aberrant alterations.
Metabolic labeling, immunoprecipitation, and endo H digestion.
For metabolic labeling, cells were rinsed three times with phosphate-buffered saline (PBS), washed once with labeling medium (RPMI minus methionine and cysteine plus 10% dialyzed fetal calf serum), and then incubated with 5 ml of the same medium containing 200 μCi of [35S]methionine and [35S]cysteine (New England Nuclear, Boston, Mass.) for 16 h. For pulse-chase analysis, cells were labeled for 15 min and chased for 20 and 40 min. For immunoprecipitation, cells were harvested and lysed with lysis buffer (0.15 M NaCl, 1% Nonidet P-40, and 50 mM HEPES buffer [pH 8.0]) containing 1 mM Na2VO3, 1 mM NaF, and protease inhibitors (leupeptin, aprotinin, phenylmethylsulfonyl fluoride, and bestatin). Immunoprecipitation was performed with a 1:500-diluted anti-six-His antibody (Santa Cruz Biotech, Santa Cruz, Calif.) together with 30 μl of protein A- and protein G-agarose beads. Washed immunoprecipitates were resuspended in 20 μl of 50 mM sodium citrate (pH 5.5)–0.2% sodium dodecyl sulfate, heated for 5 min at 95°C, and incubated for 6 h at 37°C with endo-β-N-acetylglucosaminidase H (endo H).
Immunofluorescence.
Cells were fixed with 4% paraformaldehyde for 15 min, permeabilized with 70% ethanol for 15 min, blocked with 10% goat serum in PBS for 30 min, and reacted with 1:100-diluted primary anti-six-His antibody (Santa Cruz Biotech) for K3, K4.2, and K5 staining in PBS for 30 min at room temperature. After incubation, the cells were washed extensively with PBS, incubated with 1:100-diluted secondary antibody (Vector, Burlingame, Calif.) in PBS for 30 min at room temperature, and washed three times with PBS. For Golgi staining, cells were metabolically labeled with 5 μg of BODIPY-FL C5-ceramide (Molecular Probes, Eugene, Oreg.)/ml for 30 min as recommended by the manufacturer's protocol. Finally, the cells were mounted in mounting medium (Vector). Confocal microscopy was performed using a TCS SP laser scanning microscope (Leica Microsystems, Exton, Pa.) fitted with a 100× Leica objective (PL APO; 1.4 NA) and using the Leica image software. Images were collected at 512- by 512-pixel resolution. The stained cells were optically sectioned in the z axis, and the images in the different channels (photomultiplier tubes) were collected simultaneously. The step size in the z axis varied from 0.2 to 0.5 μm to obtain 30 to 50 slides/imaged field. The images were transferred to a Macintosh G3 computer (Apple Computer, Cupertino, Calif.), and NIH Image version 1.61 software was used to render the images.
Immunofluorescence-based endocytosis.
Cells were stained with a fluorescein isothiocyanate (FITC)-conjugated class I antibody and a Texas red-conjugated transferrin receptor antibody at 4°C for 30 min, washed twice with cold RPMI culture medium, and incubated at 37°C to allow for the internalization of antibody-bound class I molecules and transferrin receptor. At various time points, the localization of class I molecules and transferrin receptor was determined with a Leica confocal immunofluorescence microscope.
Flow cytometry analysis.
Cells (5 × 105) were washed with RPMI medium containing 10% fetal calf serum and incubated with FITC- or phycoerythrin (PE)-conjugated monoclonal antibodies for 30 min at 4°C. After being washed, each sample was fixed with 1% paraformaldehyde solution and flow cytometry analysis was performed with a FACScan (Becton Dickinson Co., Mountainview, Calif.). W6/32 antibody for MHC class I, B43 antibody for CD19, RPA-T8 antibody for CD8, M-A712 antibody for transferrin receptor, and TU39 antibody for MHC class II used for fluorescence-activated cell sorter and confocal immunofluorescence analyses were obtained from PharMingen Becton Dickinson Company.
RESULTS
Downregulation of surface expression of MHC class I molecules by KSHV K3 and K5.
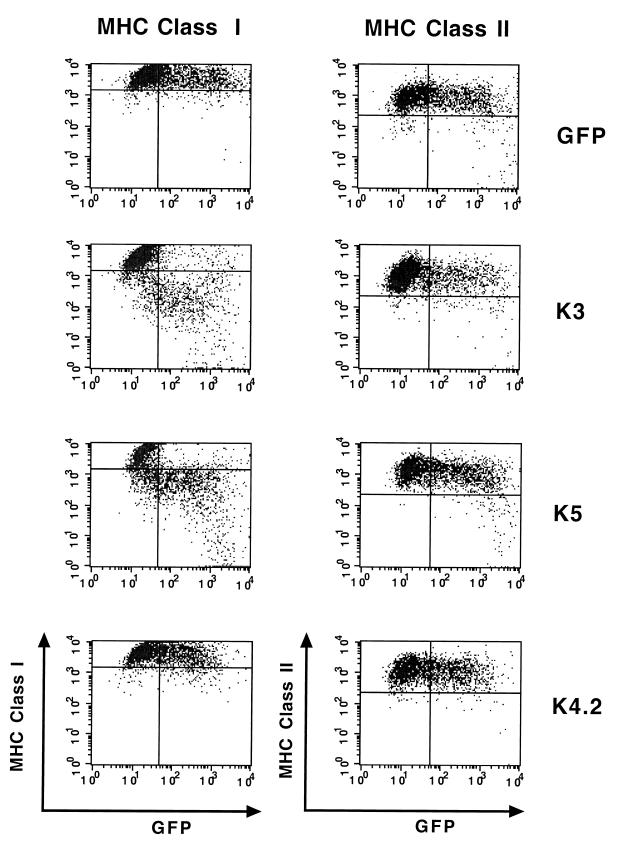
HVS viral superantigen is expressed in the immediate-early lytic phase and powerfully induces the proliferation of CD4-positive cells (6, 14, 36). At a position equivalent to that of the HVS viral superantigen, KSHV contains unique membrane proteins called K3, K4.2, and K5 (25). While none of these membrane proteins demonstrated a detectable level of superantigenic activity (data not shown), we found that either K3 or K5 expression significantly downregulated surface expression of MHC class I molecules (Fig. 1). However, the level of class I downregulation by K3 was much higher than that by K5 (Fig. 1). In contrast, K4.2 did not affect class I surface expression under the same conditions (Fig. 1). Furthermore, the downregulation of MHC class I molecules by K3 and K5 was specific; their expression did not affect the surface expression of other lymphocyte antigens, including MHC class II and immunoglobulin M under the same conditions (Fig. 1 and data not shown).
FIG. 1.
Downregulation of surface expression of MHC class I molecules by K3 and K5. BJAB cells were electroporated with GFP reporter, GFP-K3, GFP-K4.2, or GFP-K5 vector. The cell surface levels of MHC class I molecules were assessed 48 h posttransfection by staining the cells with a W6/32 antibody for MHC class I or TU39 antibody for MHC class II (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. Three populations of cells were distinguished by the applied gates; in the upper left quadrant are the untransfected cells, in the upper right quadrant are the GFP-positive transfected cells which have not downregulated class I or class II, and in the lower right quadrant are the GFP-positive transfected cells that have downregulated class I or class II. The data were reproduced in three independent experiments.
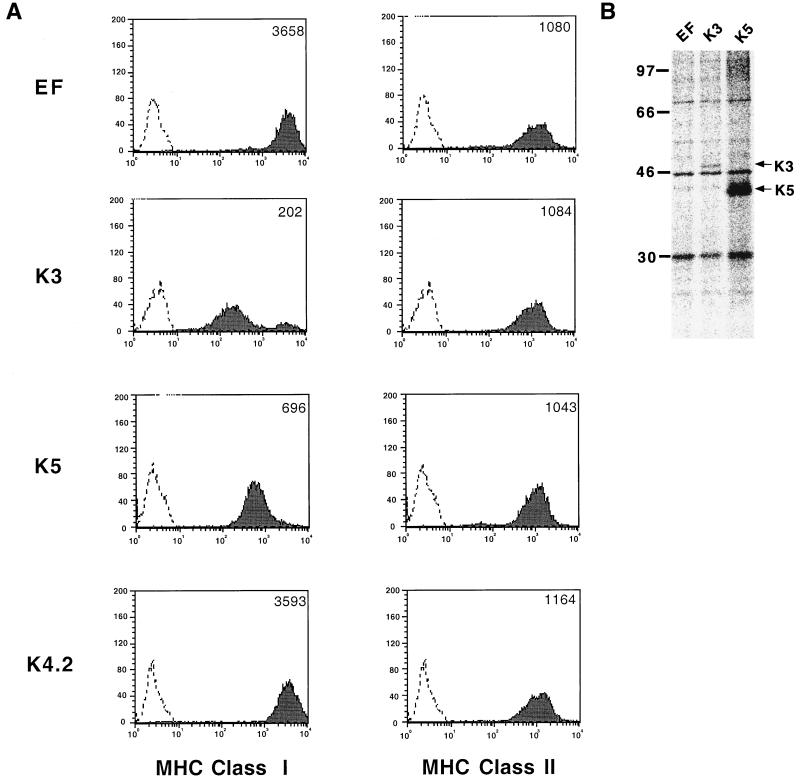
To further demonstrate the specific downregulation of class I molecules by K3 and K5, BJAB cells were used to establish cell lines stably expressing the K3, K4.2, or K5 gene. Full-length K3, K4.2, and K5 genes were modified to encode a six-histidine epitope tag at their carboxyl termini and cloned into the expression vector pEpiTag (EF), which contains the elongation factor 1 promoter for gene expression. After the electroporation of the K3, K4.2, or K5 expression vector, cells were selected by growth in medium containing 2 mg of G418/ml and examined for MHC class I surface expression by flow cytometry. The stable expression of K3 and K5 in BJAB cells induced a dramatic downregulation of MHC class I molecules on their surfaces, whereas K4.2 expression did not affect MHC class I surface expression (Fig. 2A). Consistent with results from a transient-expression assay (Fig. 1), K3 downregulated class I molecules to a greater extent than K5 (Fig. 2A). The downregulation of MHC class I by K3 and K5 was specific; their expression did not affect the surface levels of MHC class II, immunoglobulin M, and CD19 under the same conditions (Fig. 2A and data not shown). When G418-resistant cells were examined for K3 or K5 expression by radioactive immunoprecipitation with an anti-six-His antibody, a 50-kDa protein for K3 and a 38-kDa protein for K5 were detected (Fig. 2B, lanes 2 and 3). In contrast, these proteins were not detected in control BJAB cells transfected with an empty vector (Fig. 2B, lane 1).
FIG. 2.
Downregulation of MHC class I molecules on cells stably expressing K3 or K5. (A) Downregulation of MHC class I molecules by K3 or K5. G418-resistant cells were stained with a PE-conjugated W6/32 antibody or a PE-conjugated pan-class I antibody and analyzed by flow cytometry. Two hundred thousand events were collected on a FACScan flow cytometer. As a control, a histogram of each cell line (shaded) is overlaid with a dotted-line histogram of an isotype antibody control. The mean value of the relative level of class I and II surface expression is presented inside each graph. (B) Expression of K3 and K5. BJAB cells were labeled with [35S]methionine and [35S]cysteine overnight. Radioactive cell lysates were used for immunoprecipitation with an anti-six-His antibody. Lane EF, BJAB/EF; lane K3, BJAB/K3; lane K5, BJAB/K5. The arrows indicate the K3 and K5 proteins.
Subcellular localization of K3 and K5.
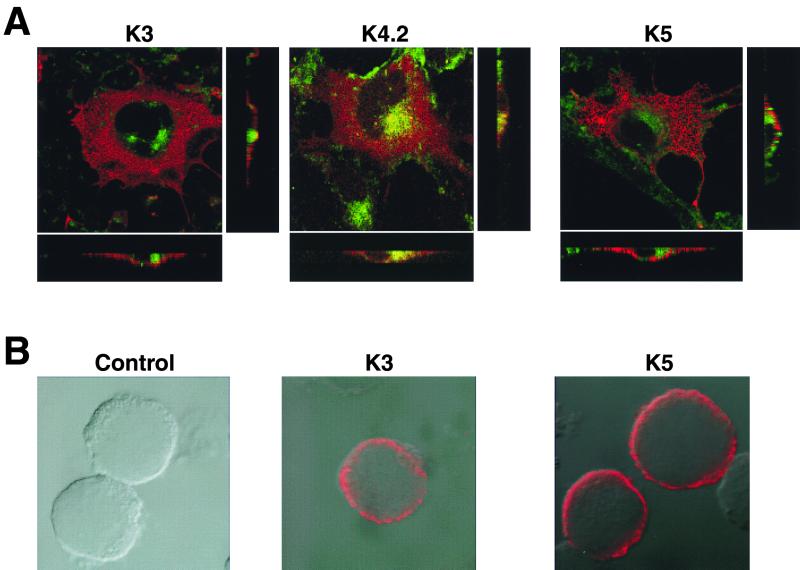
To investigate the roles of K3 and K5 in the downregulation of MHC class I surface expression, we examined their subcellular localization by indirect-immunofluorescence tests. COS-1 cells were transfected with an expression vector containing a six-His epitope-tagged K3, K4.2, or K5 gene. Two days posttransfection, the cells were costained with a Texas red-conjugated anti-six-His antibody and a FITC-conjugated Golgi-specific dye and examined under a confocal immunofluorescence microscope. The K3, K4.2, and K5 proteins were all primarily present in the cytoplasm. While a high degree of overlapping staining between K4.2 and the Golgi complex was detected, no overlapping staining or a minimal level of overlapping staining between K3 or K5 and the Golgi complex was observed (Fig. 3A). Furthermore, cross sections of transfected cells demonstrated that K3 and K5 were localized near the plasma membrane and that a large portion of K4.2 was localized at the Golgi complex (Fig. 3A). Immunofluorescence tests with BJAB cells stably expressing K3 and K5 also indicated their presence in the cytoplasm and near the plasma membrane (Fig. 3B).
FIG. 3.
Localization of K3 and K5 near the plasma membrane. (A) Localization in COS-1 cells. COS-1 cells were transfected with pEF-K3-6xHis (K3), pEF-K4.2-6xHis (K4.2), or pEF-K5-6xHis (K5) DNA. The cells were permeabilized with ethanol and reacted with a Texas red-conjugated anti-six-His antibody (red) and FITC-conjugated Golgi-specific dye (green). Immunofluorescence was examined with a Leica confocal immunofluorescence microscope. To determine their specific locations, stained cells were cross-sectioned and viewed under the Leica confocal immunofluorescence microscope and are presented in the side boxes. The overlapping staining was visualized as yellow color. (B) Localization in BJAB cells. BJAB/EF, BJAB/K3, and BJAB/K5 cells were permeabilized with ethanol and reacted with a Texas red-conjugated anti-six-His antibody. Immunofluorescence was examined with a Leica confocal immunofluorescence microscope.
Endo H digestion.
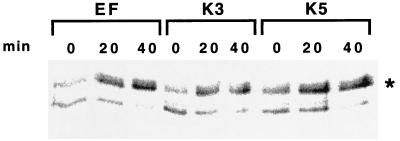
Carbohydrate moieties of MHC class I complexes contain high-mannose-type oligosaccharides in the ER which are susceptible to endo H digestion. After transport to the medial Golgi, however, their carbohydrate moieties acquire resistance to cleavage by endo H (23). To investigate the effect of K3 and K5 on the intracellular transport of MHC class I complexes, BJAB/EF, BJAB/K3, and BJAB/K5 cells were metabolically labeled with [35S]methionine and [35S]cysteine for 15 min and chased for 20 or 40 min. MHC class I complexes were then precipitated with a monoclonal W6/32 antibody and subjected to endo H digestion. The heavy chain of MHC class I in BJAB/K3 and BJAB/K5 cells acquired resistance to endo H cleavage equivalent to that in control BJAB/EF cells (Fig. 4). In addition, a similar level of class I molecules was expressed in BJAB/EF, BJAB/K3, and BJAB/K5 cells (Fig. 4 and data not shown). These results indicate that K3 and K5 do not affect the expression and acquisition of endo H resistance by MHC class I molecules at a detectable level.
FIG. 4.
Endo H sensitivity of class I molecules. BJAB/EF, BJAB/K3, and BJAB/K5 cells were metabolically labeled with [35S]methionine and [35S]cysteine for 15 min and chased for 0, 20, or 40 min. Radioactively labeled lysates were used for immunoprecipitation with an anti-class I antibody. Immunoprecipitates were treated with endo H prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis. The portion of the gel displaying the heavy chain of class I molecules is shown. ∗, the endo H-resistant heavy chain of class I molecules.
Acceleration of endocytosis of class I molecules by K3 and K5.
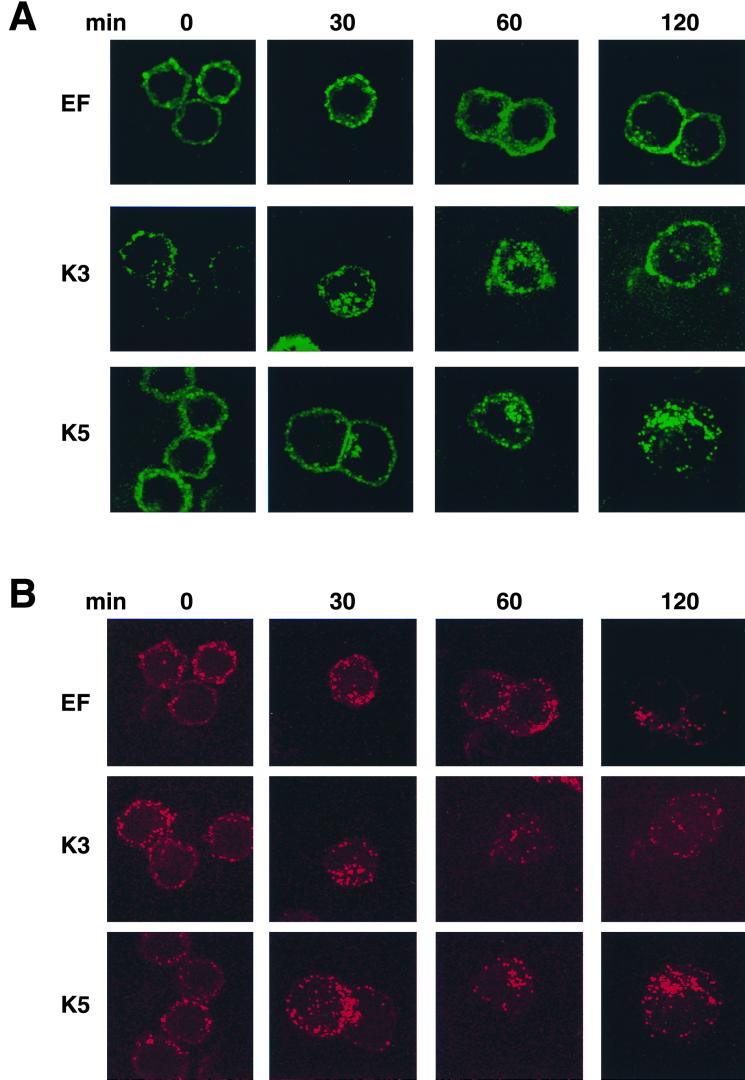
HIV Nef has been shown to downregulate MHC class I molecules by accelerating their endocytosis (9, 16). To examine whether K3 and K5 affected the internalization of class I molecules, we used an immunofluorescence-based endocytosis assay. After being stained with anti-class I and anti-transferrin receptor antibodies at 4°C for 30 min, BJAB/EF, BJAB/K3, and BJAB K5 cells were washed twice with cold culture medium and incubated at 37°C to allow for the internalization of antibody-bound class I molecules and transferrin receptor. At appropriate time points, the localization of class I and transferrin receptor was determined by confocal immunofluorescence microscope analysis. Unlike control cells, in which an apparent endocytosis of class I molecules was detected after approximately 120 min of incubation, K3-expressing cells exhibited a rapid endocytosis of class I molecules; the internalization of class I molecules was evident within 30 min and was almost completed after 60 min of incubation (Fig. 5A). K5-expressing cells also showed an accelerated endocytosis of class I molecules compared to control cells: however, the rate of endocytosis in K3-expressing cells was much higher than that in K5-expressing cells (Fig. 5A). This is consistent with our observation that the effect of K3 on downregulation of class I molecules is more pronounced than that of K5, as shown in Fig. 1 and 2. In contrast, similar rates of endocytosis of the transferrin receptor were detected in all three cell lines under the same conditions (Fig. 5B). Furthermore, class I molecules were predominantly organized into punctate patterns on the surfaces of K3- and K5-expressing cells, whereas they were evenly distributed on the surfaces of control BJAB/EF cells (Fig. 5A). These results suggest that the downregulation of class I molecules induced by K3 and K5 is likely due to their rapid endocytosis.
FIG. 5.
Rapid endocytosis of class I molecules by K3 and K5. BJAB/EF, BJAB/K3, and BJAB/K5 cells were stained with a FITC-conjugated class I antibody (A) and a Texas red-conjugated transferrin receptor antibody (B) at 4°C for 30 min, washed twice with cold culture medium, and incubated at 37°C to allow for the internalization of antibody-bound class I molecules and transferrin receptor. At the indicated time points (in minutes), the localization of class I and transferrin receptor was determined with a Leica confocal immunofluorescence microscope.
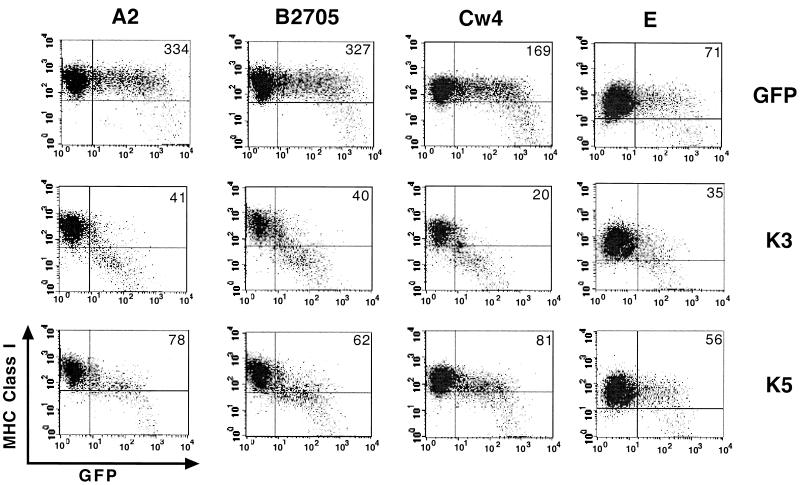
Selective downregulation of HLA allotypes by K5.
HLA-A and HLA-B proteins are primarily responsible for presenting processed antigen peptides to CTLs, whereas HLA-C and HLA-E are the predominant class I alleles responsible for preventing natural killer (NK) cells from killing lymphoid cells (17). To investigate potential selectivity of downregulation of HLA allotypes by K3 and K5, 221 cells stably expressing defined HLA allotypes (4) were electroporated with GFP, GFP-K3, or GFP-K5 expression vector. The effect of K3 and K5 expression on the cell surface levels of HLA allotypes was assessed 48 h posttransfection by staining for the MHC class I protein and gating of the GFP-positive cell population by flow cytometry. A dramatic downregulation of HLA-A2, HLA-B2705, HLA-Cw4, and HLA-E was observed in the K3-expressing GFP-positive cell population (Fig. 6). The K5-expressing GFP-positive cell population exhibited approximately four- to fivefold downregulation of HLA-A2 and HLA-B2705 (Fig. 6). However, K5 induced only twofold downregulation of HLA-Cw4 and it caused almost no downregulation of HLA-E (Fig. 6). Transfection with the GFP reporter vector resulted in no effect on surface expression of HLA allotypes under the same conditions (Fig. 6).
FIG. 6.
Selective downregulation of HLA allotypes by K3 and K5. 221 cells stably expressing defined HLA allotypes were electroporated with GFP, GFP-K3, or GFP-K5 vector. Cell surface levels of MHC class I molecules were assessed 48 h posttransfection by staining the cells with PE-conjugated pan-class I W6/32 antibody (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. The numbers inside the boxes indicate the average mean values of class I surface expression of the GFP-positive cell population. The data were reproduced in three independent experiments.
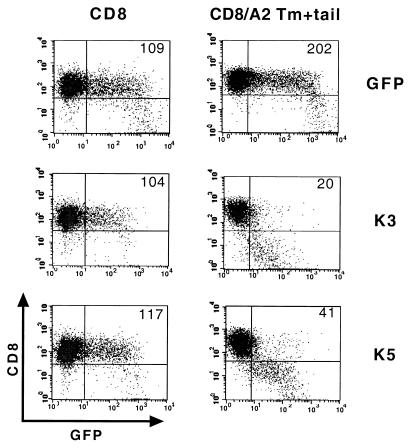
Specific regions of HLA required for K3- and K5-mediated downregulation.
To identify the region of HLA-A2 that accounted for K3- and K5-dependent downregulation, 221 cells containing a chimeric protein between human CD8α and HLA-A2 were electroporated with GFP, GFP-K3, and GFP-K5 expression vectors. This chimera contained the extracellular region of CD8α joined to the transmembrane (Tm) and cytoplasmic tail region of HLA-A2 (CD8/A2-Tm+tail) (4). Upon transfection, wild-type CD8α did not undergo appreciable downregulation by K3 and K5, whereas CD8α/A2-Tm+tail was drastically downregulated in a K3- and K5-dependent manner (Fig. 7). This indicates that the transmembrane and cytoplasmic regions of HLA-A2 are responsible for the downregulation by K3 and K5.
FIG. 7.
Downregulation of CD8 chimera containing transmembrane and cytoplasmic regions of HLA-A2 by K3 and K5. 221 cells stably expressing CD8α or the CD8/A2-Tm+tail chimera were electroporated with GFP reporter, GFP-K3, or GFP-K5 vector. The cell surface level of CD8 molecules was assessed 48 h posttransfection by staining the cells with an anti-CD8 antibody (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. The numbers inside the boxes indicate the average mean values of CD8 surface expression of the GFP-positive cell population. The data were reproduced in three independent experiments.
Since K5 induced a minimal level of downregulation of HLA-Cw4, we further investigated whether a transfer of the transmembrane and cytoplasmic tail (A2-Tm+tail) or the cytoplasmic tail (A2-tail) of HLA-A2 to HLA-Cw4 conferred K5-dependent downregulation. 221 cells expressing the HLA-Cw4/A2-Tm+tail or HLA-Cw4/A2-tail chimera were electroporated with GFP, GFP-K3, or GFP-K5 expression vector, and the levels of surface expression of these chimeras were examined by flow cytometry. The replacement of the cytoplasmic region of HLA-Cw4 with that of HLA-A2 did not affect the level of K5-mediated downregulation; K5 induced approximately twofold downregulation of HLA-Cw4 and HLA-Cw4/A2-tail (Fig. 8). In addition, the replacement of the cytoplasmic region of HLA-Cw4 with that of HLA-B27 did not affect the level of K5-mediated downregulation (data not shown). However, a transfer of both transmembrane and cytoplasmic regions of HLA-A2 moderately augmented the K5-mediated downregulation of HLA-Cw4/A2 chimeras (Fig. 8). The level of K5-mediated downregulation of Cw4/A2-Tm+tail was comparable to that of HLA-A2 (Fig. 6 and 8). In contrast, K3 was able to drastically downregulate all forms of HLA proteins under the same conditions (Fig. 8). These studies indicate that K5 downregulates class I molecules in a way that is similar to but distinct from that of K3. In addition, the transmembrane regions of HLA-A and HLA-B potentially contribute to their susceptibilities to K5-mediated downregulation.
FIG. 8.
Regions of class I molecules required for downregulation by K3 and K5. 221 cells stably expressing class I chimeric molecules were electroporated with GFP reporter, GFP-K3, or GFP-K5 vector. The level of MHC class I surface expression was assessed 48 h posttransfection by staining the cells with a PE-conjugated W6/32 antibody for class I (y axis) and gating the GFP-positive cell population (x axis) by flow cytometry. The numbers inside the boxes indicate the average mean values of MHC class I surface expression of the GFP-positive cell population. The data were reproduced in three independent experiments.
DISCUSSION
To establish lifelong infection in the host, herpesviruses have evolved mechanisms for avoiding the host immune response. Among these mechanisms is the downregulation of the surface expression of MHC class I molecules (26). In this report, we add two KSHV proteins to an increasing list of viral proteins which apparently interfere with cellular immunity. KSHV is closely related to other gamma-2 herpesviruses by virtue of the colinearity of their genomic structures and their common possession of cellular homologs. However, the K3 and K5 genes are unique to KSHV. These genes are adjacent in the viral genome and are expressed during the early lytic cycle of viral replication, and their gene products exhibit 40% amino acid identity (24). These properties suggest that one of the genes has arisen via a gene duplication event. The advantage of a virus simultaneously expressing two proteins with similar functions is not immediately clear. However, HCMV uses an even more complex strategy to prevent class I presentation. HCMV has four glycoproteins, US2, US3, US6, and US11, all of which downregulate class I-mediated antigen presentation (20, 26). One hypothesis for the overlapping kinetics and functions of these proteins is that they may assure complete prevention of class I-mediated antigen presentation and thus T-cell recognition of virus-infected cells.
Despite their sequence identity and functional similarity, K3 and K5 exhibit differences in the activity and specificity of class I downregulation. For example, K3 drastically downregulates HLA-A, -B, -C, and -E, whereas K5 exclusively downregulates HLA-A and -B. This suggests that K3 and K5 downregulate class I molecules in similar but distinct ways. In addition, studies of HLA-Cw4/A2 and HLA-Cw4/B27 chimeras indicate that the transmembrane regions of HLA-A2 and HLA-B27 potentially contribute to their susceptibilities to K5-mediated downregulation.
While the dramatic downregulation of HLA allotypes by K3 likely confers a protection from killing by CTLs, its indiscriminate downregulation of HLA-C and -E may invite NK cells to attack infected cells. To protect against this, a virus may employ additional tactics: a dominant selective activity of K5 over K3 in HLA downregulation, differential expression between K3 and K5 in the viral lytic cycle, or a yet-to-be-discovered, novel mechanism. Further study will be directed to providing evidence for or against this hypothesis. Nevertheless, this study suggests that KSHV has evolved a novel immune evasion mechanism by harboring similar but distinct genes, K3 and K5, which target MHC class I molecules with different activities and specificities.
A diversity of mechanisms of viral interference with cellular immunity is becoming increasingly apparent. HIV Nef has been shown to downregulate MHC class I molecules by accelerating their endocytosis (9, 16). Immunofluorescence tests indicated a redistribution of MHC class I molecules on the surfaces of K3- and K5-expressing cells: the molecules were predominantly organized into a punctate pattern. This staining pattern is similar to that obtained when the clathrin-coated regions of the plasma membrane are visualized. This suggests that, similar to HIV Nef, K3 and K5 may redistribute class I molecules to clathrin-coated regions of the plasma membrane, resulting in an accelerated endocytosis. Furthermore, similar to K5, HIV Nef has been shown to downregulate HLA-A and -B but not HLA-C and -E (4). Thus, despite the absence of discernible homology between KSHV K5 and HIV Nef, they share the ability to induce the selective downregulation of HLA allotypes. This also suggests that KSHV K5 and HIV Nef may target similar cellular proteins but with different mechanism. Kaposi's sarcoma is a complex tumor, and its progression is consistently associated with the suppression of various host immune controls (8, 31). Thus, a detailed study of molecular mechanisms of immune evasion by KSHV K3 and K5 will not only lead to a better understanding of viral persistence and disease progression but also provide a novel means for investigating cellular immune regulatory systems.
ACKNOWLEDGMENTS
We especially thank B. Damania, L. Alexander, and R. Means for critical reading of the manuscript, K. Toohey for photography support, X. Alvarez for confocal microscope analysis, M. DeMaria for flow cytometry analysis, and B. Roy for manuscript preparation.
This work was supported by U.S. Public Health Service grants CA31363, CA82057, CA86841, AI38131, ACS99295, and RR00168.
REFERENCES
- 1.Ahn K, Gruhler A, Galocha B, Jones T R, Wiertz E J, Ploegh H L, Peterson P A, Yang Y, Fruh K. The ER-luminal domain of the HCMV glycoprotein US6 inhibits peptide translocation by TAP. Immunity. 1997;6:613–621. doi: 10.1016/s1074-7613(00)80349-0. [DOI] [PubMed] [Google Scholar]
- 2.Cesarman E, Chang Y, Moore P S, Said J W, Knowles D M. Kaposi's sarcoma-associated Herpesvirus-like DNA sequences in AIDS-related body-cavity-based lymphomas. N Engl J Med. 1995;332:1186–1191. doi: 10.1056/NEJM199505043321802. [DOI] [PubMed] [Google Scholar]
- 3.Chang Y, Cesarman E, Pessin M S, Lee F, Culpepper J, Knowles D M, Moore P S. Identification of herpesvirus-like DNA sequences in AIDS-associated Kaposi's sarcoma. Science. 1994;266:1865–1869. doi: 10.1126/science.7997879. [DOI] [PubMed] [Google Scholar]
- 4.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 5.Desrosiers R C, Sasseville V G, Czajak S C, Zhang X, Mansfield K G, Kaur A, Johnson R P, Lackner A A, Jung J U. A herpesvirus of rhesus monkeys related to the human Kaposi's sarcoma-associated herpesvirus. J Virol. 1997;71:9764–9769. doi: 10.1128/jvi.71.12.9764-9769.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Duboise M, Guo J, Czajak S, Lee H, Veazey R, Desrosiers R C, Jung J U. A role for herpesvirus saimiri orf14 in transformation and persistent infection. J Virol. 1998;72:6770–6776. doi: 10.1128/jvi.72.8.6770-6776.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fruh K, Ahn K, Djaballah H, Sempe P, van Endert P M, Tampe R, Peterson P A, Yang Y. A viral inhibitor of peptide transporters for antigen presentation. Nature. 1995;375:415–418. doi: 10.1038/375415a0. [DOI] [PubMed] [Google Scholar]
- 8.Ganem D. KSHV and Kaposi's sarcoma: the end of the beginning? Cell. 1997;91:157–160. doi: 10.1016/s0092-8674(00)80398-0. [DOI] [PubMed] [Google Scholar]
- 9.Greenberg M E, Iafrate A J, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class I MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hengel H, Koszinowski U H. Interference with antigen processing by viruses. Curr Opin Immunol. 1997;9:470–476. doi: 10.1016/s0952-7915(97)80097-0. [DOI] [PubMed] [Google Scholar]
- 11.Hill A, Jugovic P, York I, Russ G, Bennink J, Yewdell J, Ploegh H, Johnson D. Herpes simplex virus turns off the TAP to evade host immunity. Nature. 1995;375:411–415. doi: 10.1038/375411a0. [DOI] [PubMed] [Google Scholar]
- 12.Jones T R, Wiertz E J, Sun L, Fish K N, Nelson J A, Ploegh H L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kledal T N, Rosenkilde M M, Coulin F, Simmons G, Johnsen A H, Alouani S, Power C A, Lüttichau H R, Gerstoft J, Clapham P R, Clark-Lewis I, Wells T N C, Schwartz T W. A broad-spectrum chemokine antagonist encoded by Kaposi's sarcoma-associated herpesvirus. Science. 1997;277:1656–1659. doi: 10.1126/science.277.5332.1656. [DOI] [PubMed] [Google Scholar]
- 14.Knappe A, Hiller C, Thurau M, Wittmann S, Hofmann H, Fleckenstein B, Fickenscher H. The superantigen-homologous viral immediate-early gene ie14/vsag in herpesvirus saimiri-transformed human T cells. J Virol. 1997;71:9124–9133. doi: 10.1128/jvi.71.12.9124-9133.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A A, Desrosiers R C, Jung J U. Deregulation of cell growth by the Kaposi's sarcoma-associated herpesvirus K1 gene. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 16.Le Gall S, Erdtmann L, Benichou S, Berlioz-Torrent C, Liu L, Benarous R, Heard J M, Schwartz O. Nef interacts with the mu subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity. 1998;8:483–495. doi: 10.1016/s1074-7613(00)80553-1. [DOI] [PubMed] [Google Scholar]
- 17.Leibson P J. Cytotoxic lymphocyte recognition of HLA-E: utilizing a nonclassical window to peer into classical MHC. Immunity. 1998;9:289–294. doi: 10.1016/s1074-7613(00)80611-1. [DOI] [PubMed] [Google Scholar]
- 18.Levitskaya J, Coram M, Levitsky V, Imreh S, Steigerwald-Mullen P M, Klein G, Kurilla M G, Masucci M G. Inhibition of antigen processing by the internal repeat region of the Epstein-Barr virus nuclear antigen-1. Nature. 1995;375:685–688. doi: 10.1038/375685a0. [DOI] [PubMed] [Google Scholar]
- 19.Mesri E A, Cesarman E, Arvanitakis L, Rafii S, Moore M A S, Posnett D N, Knowles D M, Asch A S. Human herpesvirus-8/Kaposi's sarcoma-associated herpesvirus is a new transmissible virus that infects B cells. J Exp Med. 1996;183:2385–2389. doi: 10.1084/jem.183.5.2385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Miller D M, Sedmak D D. Viral effects on antigen processing. Curr Opin Immunol. 1999;11:94–99. doi: 10.1016/s0952-7915(99)80017-x. [DOI] [PubMed] [Google Scholar]
- 21.Moore P S, Boshoff C, Weiss R A, Chang Y. Molecular mimicry of human cytokine and cytokine response pathway genes by KSHV. Science. 1996;274:1739–1744. doi: 10.1126/science.274.5293.1739. [DOI] [PubMed] [Google Scholar]
- 21a.Neiple F, Albrecht J-C, Fleckenstein B. Cell-homologous genes in the Kaposi's sarcoma-associated rhadinovirus human herpesvirus 8: determinants of its pathogenicity? J Virol. 1997;71:4187–4192. doi: 10.1128/jvi.71.6.4187-4192.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Neipel F, Albrecht J-C, Ensser A, Huang Y-Q, Li J J, Friedman-Kien A E, Fleckenstein B. Human herpesvirus 8 encodes a homolog of interleukin-6. J Virol. 1997;71:839–842. doi: 10.1128/jvi.71.1.839-842.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ng D T, Randall R E, Lamb R A. Intracellular maturation and transport of the SV5 type II glycoprotein hemagglutinin-neuraminidase: specific and transient association with GRP78-BiP in the endoplasmic reticulum and extensive internalization from the cell surface. J Cell Biol. 1989;109:3273–3289. doi: 10.1083/jcb.109.6.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nicholas J, Ruvolo V, Zong J, Ciufo D, Guo H-G, Reitz M S, Hayward G S. A single 13-kilobase divergent locus in the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) genome contains nine open reading frames that are homologous to or related to cellular proteins. J Virol. 1997;71:1963–1974. doi: 10.1128/jvi.71.3.1963-1974.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nicholas J, Ruvolo V R, Burns W H, Sandford G, Wan X, Ciufo D, Hendrickson S B, Guo H-G, Hayward G S, Reitz M S. Kaposi's sarcoma-associated human herpesvirus-8 encodes homologues of macrophage inflammatory protein-1 and interleukin-6. Nat Med. 1997;3:287–292. doi: 10.1038/nm0397-287. [DOI] [PubMed] [Google Scholar]
- 26.Ploegh H L. Viral strategies of immune evasion. Science. 1998;280:248–253. doi: 10.1126/science.280.5361.248. [DOI] [PubMed] [Google Scholar]
- 27.Renne R, Zhong W, Herndier B, McGrath M, Abbey N, Ganem D. Lytic growth of Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in culture. Nat Med. 1996;2:342–346. doi: 10.1038/nm0396-342. [DOI] [PubMed] [Google Scholar]
- 28.Rose T M, Strand K B, Schultz E R, Schaefer G, Rankin G W, Jr, Thouless M E, Tsai C C, Bosch M L. Identification of two homologs of the Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) in retroperitoneal fibromatosis of different macaque species. J Virol. 1997;71:4138–4144. doi: 10.1128/jvi.71.5.4138-4144.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Russo J J, Bohenzxy R A, Chien M-C, Chen J, Yan M, Maddalena D, Parry J P, Peruzzi D, Edelman I S, Chang Y, Moore P S. Nucleotide sequence of the Kaposi's sarcoma-associated herpesvirus (HHV8) Proc Natl Acad Sci USA. 1996;93:14862–14867. doi: 10.1073/pnas.93.25.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sarid R, Sato T, Bohenzky R A, Russo J J, Chang Y. Kaposi's sarcoma-associated herpesvirus encodes a functional Bcl-2 homologue. Nat Med. 1997;3:293–298. doi: 10.1038/nm0397-293. [DOI] [PubMed] [Google Scholar]
- 31.Schulz T F, Chang Y, Moore P S. Kaposi's sarcoma-associated herpesvirus (human herpesvirus 8) In: McCane D J, editor. Human tumor viruses. Washington, D.C.: American Society for Microbiology; 1998. pp. 87–134. [Google Scholar]
- 32.Searles R P, Bergquam E P, Axthelm M K, Wong S W. Sequence and genomic analysis of a Rhesus macaque rhadinovirus with similarity to Kaposi's sarcoma-associated herpesvirus/human herpesvirus 8. J Virol. 1999;73:3040–3053. doi: 10.1128/jvi.73.4.3040-3053.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sun R, Lin S F, Staskus K, Gradoville L, Grogan E, Haase A, Miller G. Kinetics of Kaposi's sarcoma-associated herpesvirus gene expression. J Virol. 1999;73:2232–2242. doi: 10.1128/jvi.73.3.2232-2242.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 35.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 36.Yao Z, Maraskovsky E, Spriggs M K, Cohen J I, Armitage R J, Alderson M R. Herpesvirus saimiri open reading frame 14, a protein encoded by a T lymphotropic herpesvirus, binds to MHC class II molecules and stimulates T cell proliferation. J Immunol. 1996;156:3260–3266. [PubMed] [Google Scholar]
- 37.York I A, Roop C, Andrews D W, Riddell S R, Graham F L, Johnson D C. A cytosolic herpes simplex virus protein inhibits antigen presentation to CD8+ T lymphocytes. Cell. 1994;77:525–535. doi: 10.1016/0092-8674(94)90215-1. [DOI] [PubMed] [Google Scholar]
- 38.Ziegler H, Thale R, Lucin P, Muranyi W, Flohr T, Hengel H, Farrell H, Rawlinson W, Koszinowski U H. A mouse cytomegalovirus glycoprotein retains MHC class I complexes in the ERGIC/cis-Golgi compartments. Immunity. 1997;6:57–66. doi: 10.1016/s1074-7613(00)80242-3. [DOI] [PubMed] [Google Scholar]