Abstract
Nef is a myristoylated protein of 27 to 35 kDa that is conserved in primate lentiviruses. In vivo, Nef is required for high viral load and full pathological effects. In vitro, Nef has at least four activities: induction of CD4 and major histocompatibility complex (MHC) class I downregulation, enhancement of viral infectivity, and alteration of T-cell activation pathways. We previously reported that the Nef protein from human immunodeficiency virus type 1 interacts with a novel human thioesterase (hTE). In the present study, by mutational analysis, we identified a region of the Nef core, extending from the residues D108 to W124, that is involved both in Nef-hTE interaction and in Nef-induced CD4 downregulation. This region of Nef is located on the oligomer interface and is in close proximity to the putative CD4 binding site. One of the mutants carrying a mutation in this region, targeted to the conserved residue D123, was also found to be defective in two other functions of Nef, MHC class I downmodulation and enhancement of viral infectivity. Furthermore, mutation of this residue affected the ability of Nef to form dimers, suggesting that the oligomerization of Nef may be critical for its multiple functions.
The nef gene of the primate lentiviruses encodes a 27-kDa protein anchored at the cytoplasmic face of the plasma membrane through a myristate at its amino terminus (for reviews, see references 13 and 37). Experiments in rhesus monkeys infected with simian immunodeficiency virus (SIV) strain SIVmac with a disrupted nef gene have demonstrated the importance of the Nef protein for maintenance of high virus loads and disease progression (30).
In vitro studies revealed that Nef downregulates both CD4 and major histocompatibility complex class I (MHC-I) cell surface expression, which may contribute to the preservation of viral infectivity and immune evasion, respectively (for a review, see reference 46). These two activities mainly result from Nef-induced CD4- and MHC-I-accelerated endocytosis, involving a dileucine sequence and a Tyr-based motif located in the cytoplasmic domains of CD4 and MHC-1, respectively. However, the mechanisms by which Nef downregulates CD4 and MHC-I have not yet been fully elucidated. Whereas Nef could act as a physical connector between CD4 and the cellular endocytotic pathway, it may rather reveal a cryptic endocytosis motif in MHC-I (34, 38; for a review, see reference 47).
In addition to the induction of CD4 and MHC-I downregulation, Nef enhances virus infectivity. At least two mechanisms could account for this activity. First, Nef stimulates viral infectivity in a CD4-independent manner. This effect could be the consequence of stimulation of proviral DNA synthesis. Nef might also facilitate phosphorylation of the matrix protein by a cellular protein kinase required for the maintenance of optimal virion infectivity (58, 60). Second, Nef may counteract, by downregulating CD4, the inhibitory effect of CD4 on envelope virion incorporation and function (48, 50; for reviews, see references 26 and 32). Conflicting results concerning the effects of Nef on the T-cell activation pathways have been reported, depending on its intracellular localization (5, 10, 15, 57).
In order to elucidate the mechanisms of Nef actions, attempts have been made to identify the cellular mediators and the regions required for its functions. Nef has been shown to interact with various cellular proteins, including CD4 (26, 51), β-COP (6, 46), human thioesterase (hTE) (35), the μ1 and μ2 chains of clathrin adaptor protein (AP) complexes (34, 58), Hck (42, 52), Lck protein tyrosine kinase (10, 21), and serine threonine kinases (4, 43, 53). More recently, it has been shown that Nef proteins from SIV and human immunodeficiency virus (HIV) interact with the T-cell receptor ζ chain (28, 61). This interaction is involved in the HIV-1 Nef-induced upregulation of Fas ligand (61). Nef also interacts with a guanine nucleotide exchange factor, Vav, and leads to increased activity of Vav and its downstream effectors, such as cytoskeletal changes and the activation of c-Jun N-terminal kinase (17).
Although membrane localization through myristoylation is required for all Nef functions (1, 19, 54), different regions of Nef have been implicated in CD4 and MHC-I downregulation and in enhancement of viral infectivity. It has been reported that the proline-rich SH3-binding region of HIV-1 Nef (PxxP, residues 72 to 77) is implicated in the enhancement of virion infectivity and in MHC-I downregulation but not in CD4 downregulation (21, 39, 52). However, a recent study showed that this motif also contributed to CD4 downregulation and that this effect could be observed only when Nef was expressed at low levels (12). In addition, a dileucine-based protein sorting motif in Nef has been shown to be required for CD4 downregulation and optimal viral infectivity but not for downregulation of MHC-I (11, 12). Structural and biochemical studies indicated that Nef might form oligomers (2, 16, 31). However, the functional significance of Nef oligomerization remains unknown.
We reported that Nef proteins from various HIV-1 isolates interact with a novel hTE homologous to the thioesterase II of Escherichia coli (35). We found that a Nef mutant generated by random mutagenesis and mutated in several amino acid residues, which had lost the ability to interact with hTE, was also defective for CD4 downregulation activity (35). In the present work, using site-directed mutagenesis, we demonstrate that mutations in a region of the Nef core extending from residues D108 to W124 abolish both Nef-hTE interaction and CD4 downregulation. One of these mutants carrying a mutation targeted to the conserved residue D123 was also defective for MHC-I downmodulation and enhancement of viral infectivity. Furthermore, mutation of this residue resulted in the impairment of Nef oligomerization.
MATERIALS AND METHODS
Plasmid constructions and generation of Nef mutants.
A PCR-generated BamHI-SalI fragment corresponding to the coding sequence of the Nef protein from isolate HIV-1Lai was cloned into the pcDNA3 mammalian expression vector (Invitrogen) by insertion at the BamHI and XhoI sites, resulting in construct pcDNA3-NefLai. NefLai mutants were generated by PCR with pfu polymerase (Stratagene) and appropriate primers as described by Chowers et al. (8). The resulting amplified products were then purified and inserted in various expression vectors. All inserts were entirely sequenced using an automated ABI sequencer to check that the desired mutants had been obtained. The XhoI-PmlI fragments of the Nef4 and NefD123G mutants, which contain corresponding mutations, were inserted into a proviral infectious HIV-1 NL4-3-derived clone containing a unique PmlI site (52).
Two-hybrid assays.
The PCR fragments corresponding to different Nef mutants, generated as described above, were inserted into the yeast expression vector pGBT10 by using the restriction sites BamHI at the 5′ end and SalI at the 3′ end to fuse these Nef mutants in frame with the GAL4 binding domain (Gal4BD). The resulting constructs were cotransformed into yeast strain HF7c with vector pGAD1318, expressing hTE in fusion with the Gal4 activation domain (Gal4AD), as previously described (6, 34, 35). β-Galactosidase quantitative assays were performed with strain SFY526 as described previously (7). Assays were performed in duplicate.
CD4 downregulation assay.
A 10-μg amount of pcDNA3 vector carrying wild-type or mutated Nef sequences was cotransfected into P4-2 cells (HeLa CD4+ cells expressing β-galactosidase under the control of the HIV-1 LTR) (9) with 2 μg of pcDNA3-GFP (expressing green fluorescent protein [GFP]) by electroporation as previously described (34). Briefly, P4-2 cells (8 × 106 cells per point) resuspended in 200 μl of Dulbecco's modified Eagle's medium with 10% fetal calf serum and 10 mM HEPES were mixed with 50 μl of 200 mM NaCl containing the appropriate plasmids. Electroporation was performed at 200 V and 960 μF in 4-mm-wide cuvettes in a Bio-Rad Gene Pulser. At 24 h after transfection, cells were labeled with phycoerythrin-conjugated anti-CD4 monoclonal antibodies (Leu3A; Becton Dickinson) as described previously (55). The surface expression of CD4 was analyzed on GFP-positive cells with a FACScan cytofluorimeter (Becton Dickinson).
MHC-I downregulation assay.
The Nef-FT vector carrying the NefLai coding sequence is a gift from F. Bachelerie (Institut Pasteur, Paris, France) (3). The Nef-FT-D123G vector was constructed by replacing the BlpI-BspEI fragment in the NefLai coding sequence of the Nef-FT vector with the corresponding BlpI-BspEI fragment of the PCR product containing the mutation D123G. The HLA-A2 vector was a kind gift from F. Lemonnier (Institut Pasteur, Paris, France). The HLA-A2 gene was subcloned in a pcDNA plasmid (Biolabs), yielding pCA2. pCA2 (4 μg) and pCDNA3-GFP (2 μg) were cotransfected into HeLa cells together with either 12 μg of Nef-FT, Nef-FT-D123G, or Nef-mock plasmid. The cellular surface expression level of the HLA-A2 allele on GFP-positive cells was analyzed with anti-HLA-A2 BB7.2 antibodies by flow cytometry as previously described (55).
Virus production and infectivity assay.
Infectious viruses were prepared from the supernatant of HeLa cells transfected with proviral DNA. Cells transfected by the calcium phosphate method were washed 24 h posttransfection, and infectious supernatant was collected after a further 24 h, filtered through a 0.45-μm-pore-size filter, and used immediately for infection assays. The p24 content in supernatant aliquots was simultaneously quantified using a commercial test kit (p24-ELISA; Innogenetics, Ghent, Belgium). Viral infectivity was determined by incubating different dilutions of supernatants on semiconfluent HeLa-P4 indicator cells in six-well plates in a final volume of 2 ml. After 24 h, the cells were fixed in 0.5% glutaraldehyde, and infected cells were stained blue with X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) substrate as described before (14). Blue cells were scored from randomly selected fields under 40× magnification, and the total number of infected cells per well was calculated and correlated with the amount of input p24.
Assay for oligomerization of Nef.
A plasmid encoding hybrid Nef protein fused to the extracellular and transmembrane domain of CD8 (CD8Nefwt) was constructed as previously described (17). The mutation D123V was introduced into the CD8Nef construct by PCR-mediated mutagenesis, resulting in CD8NefD123V. The truncated protein CD8T, which contained only the extracellular and transmembrane domains of CD8 with no intracellular residues (17), was used as a control. 293T cells were transfected with CD8T, CD8Nefwt, or CD8NefD123V by using lipofectamine (Gibco) according to the manufacturer's instructions. Cells were harvested at 48 h posttransfection, lysed in kinase extraction buffer (17), and analyzed by Western blotting. For the visualization of Nef homodimers, membranes were probed with the anti-Nef monoclonal antibody 158 (16).
Immunoblot analyses.
Transfected or infected cells were lysed in 25 mM HEPES (pH 7.4)–150 mM KCl–5 mM EDTA–1% Triton X-100 at 4°C for 30 min. After centrifugation at 13,000 × g for 5 min, the clarified supernatant containing cellular lysates from about 600,000 transfected or infected cells was electrophoresed on sodium dodecyl sulfate–12% polyacrylamide gel electrophoresis (SDS-PAGE) followed by electrophoretic transfer onto nitrocellulose membranes as previously described (35). Immunoblot analyses of Nef were performed with either an anti-HIV-1BH10 Nef antiserum (1:2,000) (National Institutes of Health AIDS repository, catalog no. 2121) (see Fig. 2E) or an anti-HIV-1Lai antiserum (1:1,000) (see Fig. 5B), raised in rabbits by immunization with the fused protein glutathione-S-transferase (GST)-NefLai (N. Heveker, unpublished results), and then developed by the enhanced chemiluminescence system (Amersham Pharmacia Biotech).
FIG. 2.
Two-hybrid assays of hTE binding to wild-type and mutant Nef. The amino acid substitution in each Nef mutant is indicated at the corresponding position of the delineated sequence between residues D108 and N126. hTE binding of wild-type and mutant Nef proteins was determined as described in the legend to Fig. 1A. The percentage of wild-type activity is shown in parentheses.
FIG. 5.
Mutation D123G in NefLai abolishes the ability of the protein to enhance viral infectivity. (A) Comparison of infectivity. HeLa cells were transiently transfected with pNL4-3wt, pNL4-3ΔNef, pNL4-3Nef4, or pNL4-3NefD123G proviral DNA. Infectious virions were collected 48 h later and used immediately to infect HeLa P4-2 indicator cells. Infectivity was evaluated by the number of infected blue cells assayed, with different quantities of viruses titrated by the p24 antigen content. These results are representative of experiments performed with three different virus stocks. (B) Nef protein expression in virus producer cells, estimated by Western blotting of cell lysates with anti-Nef antibodies.
RESULTS
D123G mutation in HIV-1 Nef abolishes the ability of the protein to interact with hTE and to downregulate CD4 cell surface expression.
A Nef mutant (Nef4) was previously isolated from a random Nef mutant library generated by PCR mutagenesis on the basis of its lack of interaction with hTE (35). This mutant contained five substitutions, W57R, F68S, D123G, H166R, and L170Q, and was found to be defective for CD4 downregulation. In order to determine which of these mutations was responsible for the loss of Nef-hTE interaction and CD4 downregulation, we generated the corresponding Nef single mutants, each containing one of these five mutations. These mutants were then fused to the Gal4BD for hTE-binding analysis in a two-hybrid quantitative assay and were expressed in HeLa CD4+ cells to determine their ability to downregulate CD4 surface expression in a transient-transfection assay. As shown in Fig. 1A, the two-hybrid assays revealed that the single mutation D123G was sufficient to abolish Nef association with hTE. Another substitution of the D123 residue, the D123V mutation, gave identical results (data not shown), further demonstrating that the D123 residue is required for interaction of Nef with hTE. The binding of hTE to NefW57R was reduced to about 58% of that of wild-type Nef, whereas the three other mutations (NefF68S, NefH166R, and NefL170Q) had no effect on hTE binding. Since the D123 residue is located in a loop exposed on the surface of the protein (2, 23, 32), mutation of this residue is not predicted to significantly alter the three-dimensional structure of the Nef core domain. This is consistent with our observation that mutation of the D123 residue did not affect the interaction of Nef with other cellular partners, such as the Hck SH3 domain, the μ1 chain of the AP1 adaptor complex, and β-COP (data not shown).
FIG. 1.
D123G mutation in NefLai abolishes interaction of Nef with hTE and Nef-induced CD4 downregulation. Five single mutants containing each of the five mutations found in the Nef4 multiple mutant were analyzed for hTE binding and for CD4 downregulation. (A) Quantitative β-galactosidase (β-gal) assays of wild-type Nef and Nef mutants binding to hTE. Strain SFY526 expressing both hTE fused to Gal4AD and wild-type Nef or Nef mutants fused to Gal4BD was assayed by a β-galactosidase assay. The results are expressed as β-galactosidase activity units determined for each wild-type or mutated Nef protein (indicated in parentheses). The background level is approximately 2 U and corresponds to SFY526 expressing both Gal4BD-Nefwt and Gal4-AD-SNF4 hybrids (not shown). (B and C) CD4 downregulation activity of Nef mutants. Wild-type and mutant Nef proteins were transiently expressed in HeLa CD4+ P4-2 cells in combination with GFP as described in the text. Surface levels of CD4 antigen were evaluated on GFP-positive cells by flow cytometry and are represented by different colored areas as indicated. The control was cells transfected with an empty pcDNA3 vector. Data are representative of three independent experiments. (D) Expression levels of Nef mutants in transfected cells used in the CD4 downregulation assays. Immunoblotting of a total cell lysate with anti-Nef antibodies. Cells were transfected with empty or recombinant expression vectors as follows: pcDNA3 vector (lane 1), pcDNA3-Nefwt (lane 2), pcDNA3-Nef4 (lane 3), pcDNA3-NefW57R (lane 4), pcDNA3-NefF68S (lane 5), pcDNA3-NefD123G (lane 6), pcDNA3-NefH166R (lane 7), and pcDNA3-NefL170Q (lane 8).
Flow cytometry analysis performed in P4-2 cells transiently transfected with these Nef mutants showed that NefW57R and NefD123G (Fig. 1B) were unable, like the Nef4 mutant, to downmodulate CD4 expression at the cell surface, whereas the three other mutants, NefF68S, NefH166R, and NefL170Q, were still fully or partially active (Fig. 1C, pink, light blue, and yellow areas, respectively). As judged from immunoblotting, the expression of wild-type and mutated Nef proteins in transfected cells was comparable except for the mutant NefW57R which displayed a lower expression level than the wild-type Nef protein (Fig. 1D). These results indicate that the residue D123 in HIV-1Lai Nef is critical for both Nef-hTE binding and Nef-induced CD4 downregulation, whereas the inability of the NefW57R mutant to downregulate CD4 could be due to its instability.
Region extending from D108 to W124 of HIV-1 Nef is critical for both hTE binding and CD4 downregulation.
The D123 residue, together with most of the contiguous residues from D108 to N126, is highly conserved in Nef proteins from many HIV-1 isolates (56). Furthermore, several Nef multiple mutants that we selected for lack of hTE binding in the two-hybrid assay contain mutations in this region (L. X. Liu and R. Benarous, unpublished results), suggesting that this region might be important for Nef functions. To test this hypothesis, different Nef point mutants containing various substitutions in this region have been generated (Fig. 2). The NefD108A, NefD111G, NefW124R, and NefN126S mutants were generated based on the mutations found in different Nef multiple mutants selected for the loss of hTE binding in a two-hybrid assay (unpublished data). The NefL112D, NefF121G, and NefP122R mutants (kind gift from George Cohen, Massachusetts Institute of Technology) were selected from a library of Nef single mutants randomly generated at defined residues. These Nef mutants were analyzed first in a two-hybrid assay for interaction with hTE and then in a transient-transfection assay for CD4 downregulation. As shown in Fig. 2, all of these mutants except N126S did not interact with hTE, as indicated by the absence of β-galactosidase activity in two-hybrid quantitative assays. Accordingly, all hTE-binding-deficient mutants were also deficient for CD4 downregulation (Fig. 3A), although they were expressed at a level comparable to the wild-type Nef protein (Fig. 3B). In contrast, mutant N126S was nearly as efficient as Nef wild-type protein in inducing CD4 downregulation (Fig. 3A). These results clearly demonstrate that there is a tight correlation between the ability of Nef to interact with hTE and to mediate CD4 downregulation.
FIG. 3.
CD4 downregulation activity of Nef mutants. (A) Analysis of CD4 downregulation activity. HeLa P4-2 cells were transfected with wild-type Nef, mutant Nef, or empty pcDNA3 (control) vector, along with the pcDNA3-GFP vector. Surface expression of CD4 was analyzed on GFP-positive cells by flow cytometry. Data are representative of three independent experiments. (B) Expression levels of Nef mutants in the transfected cells used in the CD4 downregulation assay, estimated by immunoblotting of total cell lysates with anti-Nef antibodies.
NefD123G mutant fails to induce cell surface MHC-I downregulation.
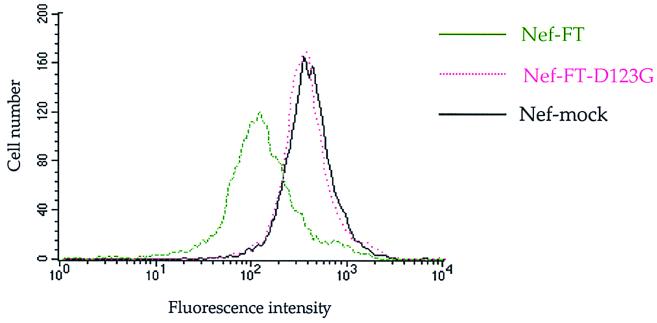
We then analyzed whether the D123G mutation also affected Nef-dependent MHC-I downregulation. The NefD123G mutant, wild-type NefLai, and an antisense Nef sequence were cloned into the pCMV expression vector, resulting in the Nef-FT-D123G, Nef-FT, and Nef-mock constructs, respectively. Each of these constructs was then cotransfected into P4-2 cells with the pCA2 construct, which allowed expression of the HLA-A2 allele of MHC-I. HLA-A2 surface expression was analyzed by flow cytometry with the anti-HLA-A2 monoclonal antibody BB7.2 (34, 55). As shown in Fig. 4, HLA-A2 surface expression was reduced in cells transfected with Nef-FT compared with cells transfected with the Nef-mock construct. In the cells transfected with the Nef-FT-D123G construct, no downregulation of HLA-A2 was observed, although the expression levels of wild-type Nef and the NefD123G mutant were comparable (data not shown). Therefore, the D123 residue is important for both CD4 and MHC-I downregulation. In contrast, some other mutations in this region (D108A and L112D), although causing deficient CD4 downregulation (Fig. 3A), conserved the ability to downregulate HLA-A2 (G. Cohen, personal communication), confirming that different residues of the protein are involved in these two activities of Nef.
FIG. 4.
Mutation D123G in NefLai abolishes the ability of the protein to downregulate MHC-I. HeLa P4-2 cells were transfected with wild-type Nef-FT (green curve), mutant Nef-FT-D123G (pink dotted curve), or Nef-mock (black curve) vector, along with the pCA (HLA-A2) and pcDNA3-GFP vectors. Surface expression of HLA-A2 was analyzed on GFP-positive cells with anti-HLA-A2 BB7.2 antibodies by flow cytometry. Data are representative of three independent experiments.
NefD123G mutant also defective for Nef-induced enhancement of viral infectivity.
Previous studies suggested that Nef-induced CD4 downregulation and enhancement of viral infectivity are independent activities which rely on different structural domains of Nef (19, 52, 54, 60). To determine whether the D123G mutation affects the ability of Nef to enhance viral infectivity, the Nef4 and NefD123G mutants were cloned into the infectious NL4-3 HIV-1 strain. As a negative control, we used an HIV-1 NL4-3 proviral clone carrying a frameshift in the nef coding region (41, 52). These proviral clones were transfected into HeLa cells to produce infectious viruses. At 48 h after transfection, viral supernatants were used to infect P4-2 (HeLa CD4+) cells. The infectivity of these viral preparations was analyzed by counting the number of infected blue cells with respect to the amounts of p24 antigen found in the viral supernatant. As shown in Fig. 5A, the infectivity obtained with the NL4-3 virus harboring wild-type Nef was about three times higher than that obtained with the Nef-minus virus. The infectivity of the viruses harboring the Nef4 or NefD123G mutant was equivalent to that obtained with the Nef-minus virus (Fig. 5A), indicating that the single point mutant NefD123G was also deficient for Nef-induced enhancement of infectivity. The expression level of the wild-type and mutated Nef proteins in cells producing virions was comparable, as shown by immunoblotting with anti-Nef antibodies (Fig. 5B). By contrast, the mutant D108A, deficient in hTE binding (Fig. 2) and CD4 downregulation (Fig. 3), conserved a significant ability to enhance virion infectivity (G. Cohen, personal communication). Altogether, these results indicate that the conserved D123 residue is important not only for Nef-induced CD4 and MHC-I downregulation, but also for the enhancement of viral infectivity.
D123 residue required for formation of Nef dimers.
The importance of the D123 residue for the biological activity of Nef could be explained by its involvement in the interaction of the viral protein with hTE. Alternatively, the crystal structure of Nef (2) suggests that this residue may be critical for the formation of Nef dimers (Fig. 6). We therefore investigated whether mutation of the D123 residue in Nef affects its ability to dimerize. Homodimers of NefLai in cellular lysates that were denatured under reducing conditions in the presence of SDS have been described before for nonfusion as well as CD8-Nef fusion proteins (16, 30; O. T. Fackler, unpublished results). Thus, hybrid CD8-Nef proteins were expressed in 293T cells, and the cellular lysates were assayed for the presence of Nef dimers by Western blotting. As shown in Fig. 7, higher-molecular-weight species with an apparent molecular mass that corresponds to that expected for homodimers were detected for the wild-type Nef fusion protein (lane 3). In sharp contrast, higher-molecular-weight species were detected neither for the CD8NefD123V mutant fusion protein (lane 2) nor for the CD8T truncated construct lacking intracellular residues and used as a control (lane 1). Given the similar expression levels of these Nef proteins, we conclude that residue D123 is critical for the formation of Nef homodimers.
FIG. 6.
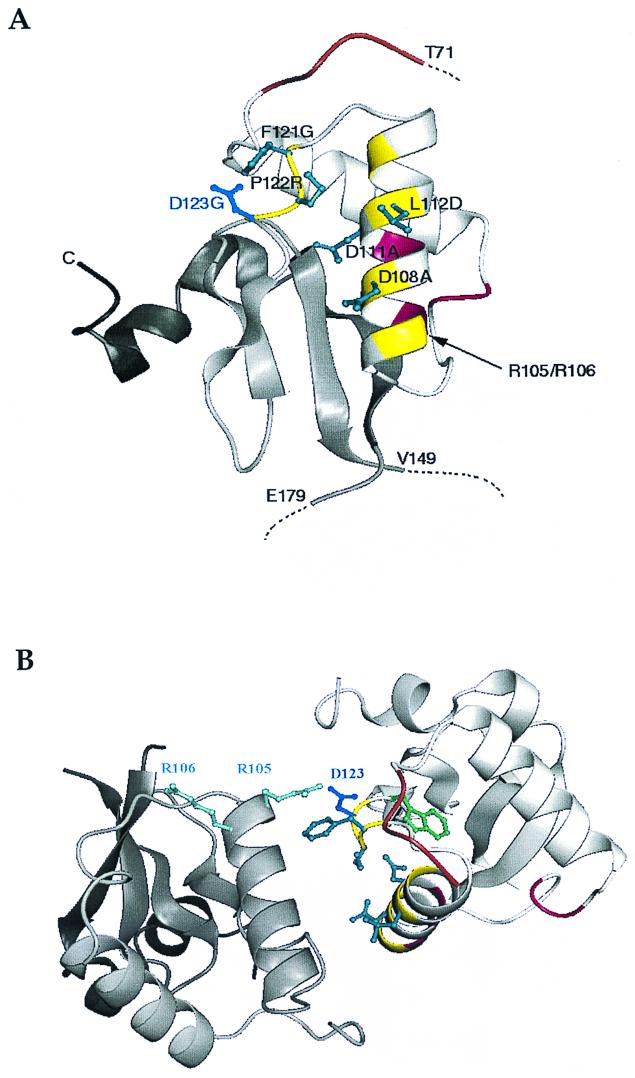
Localization of mutated residues within the Nef core domain. (A) Secondary-structure representation of the conserved core domain of HIV-1 Lai Nef, showing side chains of mutated residues. The residues of which mutation disrupts interaction with hTE and downregulation of CD4 are shown in blue. D123 is shown in dark blue. All these residues (D108, D111, L112, F121, P122, and D123) are solvent exposed in the monomer. Displayed on the secondary structure are the SH3 binding PxxPxR motif (brown), the region involved in homomeric contacts (yellow), and the putative CD4-binding surface (magenta). Note that all mutated residues localize within the oligomer interface. Dotted lines indicate where the unstructured N terminus (residues 1 to 70) and central loop region (residues 149 to 177) project out of the Nef core domain. (B) Ribbon diagram of the Nef dimer. Colors are the same as for panel A. Residue D123 is located at the bottom of the interface and makes a salt bridge with residue R105 of the opposite subunit.
FIG. 7.
D123 is critical for dimerization of Nef. Immunoblot analysis with anti-Nef monoclonal antibody 158 of total cell lysates of 293T cells expressing the CD8T (lane 1), CD8NefD123V (lane 2), and CD8Nefwt (lane 3) fusion proteins.
DISCUSSION
In this study, by combining mutagenesis with protein interaction and function studies, we identify a region of HIV-1 NefLai from residues D108 to W124 which is required for Nef-mediated CD4 downregulation and for Nef-hTE interaction. In addition, we have shown that the D123 residue in NefLai is critical not only for CD4 downregulation but also for two other essential functions of Nef, MHC-I downregulation and enhancement of viral infectivity.
Site-directed mutagenesis has been used by several groups to define the structural determinants required for Nef functions, and in general, these different functions were linked to distinct structural determinants in the core domain of the protein. The only motif unequivocally known to be required for all functions of Nef was the N-terminal myristoylated glycine, which is essential for the targeting of this protein to the membrane. Our results suggest that besides this N-terminal anchor domain, the conserved residue D123 in the core domain of Nef is also involved in multiple functions of the protein.
The three-dimensional structure of the Nef protein, both alone and in complex with Src family SH3 domains, has been solved (2, 24, 33). This structure consists of four antiparallel β-strands flanked by two outer layers composed of a polyproline type II helix and four α-helixes. All mutated residues reported herein localize on one side of the core domain formed by α-helix B (residues Q104 to T117) and the coil (residues Q118 to Q125), leading into β-strand A (Fig. 6A). The side chains of these mutated residues (except for W124) are exposed to aqueous solvents on the tertiary structure of Nef. They may therefore be available for interactions with cellular ligands. Furthermore, the Nef D123 mutants, which have lost several Nef functions and interaction with hTE, are still capable of binding to several other Nef-binding proteins such as the μ1 chain (p47) of the clathrin adaptor AP1 complex, the SH3 domain of Hck, and β-COP, ruling out the possibility that the abrogation of different activities of Nef by this mutation is due to alteration of the global structure of Nef.
The hypothesis has been raised that Nef could act as a connector between the cellular endocytic machinery and the receptors whose internalization and cellular trafficking are modified by Nef. As recently reported, several cellular cofactors which could be required for Nef-mediated CD4 and MHC-I downregulation are the medium chains of clathrin-associated AP complexes and the β-COP subunit of the COP-I complex (6, 20, 34, 36, 45, 46). Surprisingly, the mutants carrying a mutation at position D123 described in this study, in spite of complete deficiency for CD4 and MHC-I downregulation, still bind to the μ1 chain and β-COP as efficiently as wild-type Nef. A reasonable interpretation of these results is that binding of Nef to the μ chains of AP complexes, as well as binding to β-COP, although necessary, is not sufficient for Nef-induced receptor downregulation. Thus, the D123 residue could be required for the interaction of Nef with its target receptors rather than with the endocytic machinery. From this point of view, it is of interest to note that this residue is in the proximity of the putative CD4-binding site (residues 95 to 97, 106, and 109 to 110) mapped by Grzesiek et al. (23) (magenta regions in Fig. 6).
The D123 residue is located close to a hydrophobic “spot” of the surface of Nef, formed by residues L112, Y115, F121, and P122, which has been reported to constitute the interface for oligomer formation by Nef (2; S. Arold et al., unpublished results) (Fig. 6). Monomeric and oligomeric Nef molecules coexist in a concentration-dependent equilibrium in solution (25; Arold et al., unpublished results). L112, F121, and P122 form the hydrophobic core of this interface, and D108 and D123 are important for charge complementarity between Nef monomers. Nonconservative mutation of the D123 residue could destabilize or alter the oligomeric state of Nef. Since this residue is also important for the interaction between Nef and hTE, alteration of Nef oligomerization may explain why the D123 mutations affect both receptor downmodulation and binding to hTE. Interestingly, in Nef oligomers but not in the Nef monomer, D123 is interacting with the conserved diarginine motif R105-R106 of the opposite protomer. Substitution of this RR motif leads to abrogation of Nef-induced CD4 downregulation and enhancement of viral infectivity (29, 60). R106 is also required for the interaction of Nef with the Nef-associated kinase (40, 54, 60). A unifying explanation for the numerous effects of the D123 substitution could therefore be that this mutation affects both the oligomer interface of Nef and the conformation of the RR motif. Indeed, the experiment shown in Fig. 7 is in favor of a disruption of the oligomeric state of Nef provoked by such a mutation. Nef oligomers have been reported by several groups in vivo and in vitro (4, 16, 22, 31, 37), but their importance in Nef activities has not yet been elucidated. Our results support the idea that oligomerization plays a critical role in the different activities of the Nef protein. Since most receptors are internalized as dimers, it is conceivable that homomeric interactions between Nef molecules reinforce the interaction of Nef with an oligomeric target, such as the CD4 dimer. It has also been reported that the recognition of tyrosine-based endocytotic signals clearly involves a dimeric μ2 chain of the clathrin adapter AP2 complex (49).
The functional relevance of Nef-hTE interaction remains an open question. While all the Nef mutations targeted in the region D108 to W124 caused deficiency in both hTE binding and CD4 downregulation, suggesting a possible role of hTE binding in CD4 downregulation, the importance of this interaction for Nef-mediated MHC-I downregulation and enhancement of virion infectivity is less clear, since some Nef mutants (D108A and L112D) deficient in hTE binding are still able to downregulate MHC-I and/or to enhance virion infectivity (G. Cohen, personal communication). Thus, the loss of the three functions of Nef observed with mutant D123 seems to be more specifically associated with the loss of Nef oligomerization than with the lack of hTE interaction resulting from this substitution.
The possible role of hTE in Nef-mediated CD4 downregulation could be related to its TE enzymatic activity. As previously reported, the preferred substrates of hTE are myristoyl-coenzyme A and palmitoyl-coenzyme A (35). By releasing free myristate or palmitate, TE activity could be indirectly involved in the control of the lipid modifications of proteins, which are important for membrane anchoring of proteins and receptor internalization. The observation that Nef enhances in vitro hTE activity to cleave the TE bond of palmitoyl-coenzyme A supports this hypothesis (59). Further investigations are required to verify this possibility.
In this study, our attempt to map the hTE-binding site of Nef led to identification of a conserved D123 residue which is critical for CD4 and MHC-I downregulation and enhancement of viral infectivity. This residue is also important for Nef oligomerization. This suggests that the oligomerization of Nef may be important for its functions.
ACKNOWLEDGMENTS
This work was supported by grants from ANRS, Sidaction, ARC, and Comité de Paris of the Ligue Nationale Contre le Cancer. L.X.L. is supported by ANRS.
We thank George Cohen for his generous gift of Nef mutants, Isabelle Bouchaert for expertise in FACS analysis, and Franck Letourneur for DNA sequencing.
REFERENCES
- 1.Aiken C, Konner J, Landau N R, Lenburg M E, Trono D. Nef induces CD4 endocytosis: requirement for a critical dileucine motif in the membrane-proximal CD4 cytoplasmic domain. Cell. 1994;76:853–864. doi: 10.1016/0092-8674(94)90360-3. [DOI] [PubMed] [Google Scholar]
- 2.Arold S, Franken P, Strub M P, Hoh F, Bénichou S, Benarous R, Dumas C. The crystal structures of HIV-1 Nef protein bound to the Fyn kinase SH3 domain suggests a role for this complexe in altered T cell receptor signaling. Structure. 1997;15:1361–1372. doi: 10.1016/s0969-2126(97)00286-4. [DOI] [PubMed] [Google Scholar]
- 3.Bachelerie F, Alcami J, Hazan U, Israël N, Goud B, Arenzana-Seisdedos F, Virelizier J L. Constitutive expression of human immunodeficiency virus (HIV) Nef protein in human astrocytes does not influence basal or induced HIV long terminal repeat activity. J Virol. 1990;64:3059–3062. doi: 10.1128/jvi.64.6.3059-3062.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barber S A, Flaherty M T, Plafker S M, Clements J E. A novel kinase activity associated with Nef derived from neurovirulent simian immunodeficiency virus. Virology. 1998;251:165–175. doi: 10.1006/viro.1998.9408. [DOI] [PubMed] [Google Scholar]
- 5.Baur A S, Sawai E T, Dazin P, Fantl W J, Cheng-Mayer C, Peterlin B M. HIV-1 Nef leads to inhibition or activation of T cells depending on its intracellular localization. Immunity. 1994;1:373–384. doi: 10.1016/1074-7613(94)90068-x. [DOI] [PubMed] [Google Scholar]
- 6.Bénichou S, Bomsel M, Bodeus M, Durand H, Douté M, Letourneur F, Camonis J, Benarous R. Physical interaction of the HIV-1 Nef protein with beta-COP, a component of non-clathrin-coated vesicle for membrane traffic. J Biol Chem. 1994;269:30073–30076. [PubMed] [Google Scholar]
- 7.Bouhamdan M, Bénichou S, Rey F, Navarro J M, Agostini I, Spire B, Camonis J, Slupphaug G, Vigne R, Benarous R, Sire J. Human immunodeficiency virus type 1 Vpr protein binds to the uracil DNA glycosylase DNA repair enzyme. J Virol. 1996;70:697–704. doi: 10.1128/jvi.70.2.697-704.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chowers M Y, Spina C A, Kwoh T J, Fitch N J S, Richman D D, Guatelli J C. Optimal infectivity in vitro of human immunodeficiency virus type 1 requires an intact nef gene. J Virol. 1994;68:2906–2914. doi: 10.1128/jvi.68.5.2906-2914.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clavel F, Charneau P. Fusion from without directed by human immunodeficiency virus particles. J Virol. 1994;68:1179–1185. doi: 10.1128/jvi.68.2.1179-1185.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collette Y, Dutartre H, Benziane A, Ramos-Morales F, Benarous R, Harris M, Olive D. Physical and functional interaction of Nef with Lck. J Biol Chem. 1996;271:6333–6341. doi: 10.1074/jbc.271.11.6333. [DOI] [PubMed] [Google Scholar]
- 11.Craig H M, Pandori M W, Guatelli J C. Interaction of HIV-1 Nef with the cellular dileucine-based sorting pathway is required for CD4 downregulation and optimal viral infectivity. Proc Natl Acad Sci USA. 1998;95:11229–11234. doi: 10.1073/pnas.95.19.11229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig H M, Pandori M W, Riggs N L, Richman D D, Guatelli J C. Analysis of the SH3-binding region of HIV-1 nef: partial functional defects introduced by mutations in the polyproline helix and the hydrophobic pocket. Virology. 1999;262:55–63. doi: 10.1006/viro.1999.9897. [DOI] [PubMed] [Google Scholar]
- 13.Cullen B R. HIV-1 auxiliary proteins: making connections in a dying cell. Cell. 1998;93:685–692. doi: 10.1016/s0092-8674(00)81431-2. [DOI] [PubMed] [Google Scholar]
- 14.Drajic T, Hazan U, Alizon M. Detection of cell fusion mediated by the envelopes of human retroviruses by transactivation of a reporter gene. Methods Mol Genet. 1995;7:218–236. [Google Scholar]
- 15.Du Z, Lang S M, Sasseville V G, Lackner A A, Ilyinskii P O, Daniel M D, Jung J U, Desrosiers R C. Identification of a nef allele that causes lymphocyte activation and acute disease in macaque monkeys. Cell. 1995;82:665–674. doi: 10.1016/0092-8674(95)90038-1. [DOI] [PubMed] [Google Scholar]
- 16.Fackler O T, Kienzle N, Kremmer E, Boese A, Schramm B, Klimkait T, Kucherer C, Mueller-Lantzsch N. Association of human immunodeficiency virus Nef protein with actin is myristoylation dependent and influences its subcellular localization. Eur J Biochem. 1997;247:843–851. doi: 10.1111/j.1432-1033.1997.00843.x. [DOI] [PubMed] [Google Scholar]
- 17.Fackler O T, Luo W, Geyer M, Alberts A S, Peterlin B M. Activation of Vav by Nef induces cytoskeletal rearrangements and downstream effector functions. Mol Cell. 1999;3:729–739. doi: 10.1016/s1097-2765(01)80005-8. [DOI] [PubMed] [Google Scholar]
- 18.Fujii Y, Otake K, Tashiro M, Adachi A. Human immunodeficiency virus type 1 nef protein on the cell surface is cytocidal for human CD4+ T cells. FEBS Lett. 1996;393:105–108. doi: 10.1016/0014-5793(96)00862-9. [DOI] [PubMed] [Google Scholar]
- 19.Goldsmith M A, Warmerdam M T, Atchison R E, Miller M D, Greene W C. Dissociation of the CD4 downregulation and viral infectivity enhancement functions of human immunodeficiency virus type 1 Nef. J Virol. 1995;69:4112–4121. doi: 10.1128/jvi.69.7.4112-4121.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Greenberg M E, Bronson S, Lock M, Neumann M, Pavlakis G N, Skowronski J. Co-localization of HIV-1 Nef with the AP-2 adaptor protein complex correlates with Nef-induced CD4 down-regulation. EMBO J. 1997;16:6964–6976. doi: 10.1093/emboj/16.23.6964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Greenberg M E, Iafrate A J, Skowronski J. The SH3 domain-binding surface and an acidic motif in HIV-1 Nef regulate trafficking of class 1 MHC complexes. EMBO J. 1998;17:2777–2789. doi: 10.1093/emboj/17.10.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Greenway A, Azad A, McPhe D. Human immunodeficiency virus type 1 Nef protein inhibits activation pathways in peripheral blood mononuclear cells and T-cell lines. J Virol. 1995;69:1842–1850. doi: 10.1128/jvi.69.3.1842-1850.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grzesiek S, Stahl S J, Wingfield P T, Bax A. The CD4 determinant for downregulation by HIV-1 Nef directly binds to Nef mapping of the Nef binding surface by NMR. Biochemistry. 1996;35:10256–10261. doi: 10.1021/bi9611164. [DOI] [PubMed] [Google Scholar]
- 24.Grzesiek S, Bax A, Clore G M, Gronenborn A M, Hu J S, Kaufman J, Palmer I, Stahl S J, Wingfield P T. The solution structure of HIV-1 Nef reveals an unexpected fold and permits delineation of the binding surface for the SH3 domain of Hck tyrosine protein kinase. Nat Struct Biol. 1996;3:340–345. doi: 10.1038/nsb0496-340. [DOI] [PubMed] [Google Scholar]
- 25.Grzesiek S, Bax A, Hu J S, Kaufman J, Palmer I, Stahl S J, Wingfield P T. Refined solution structure and backbone dynamics of HIV-1 Nef. Protein Sci. 1997;6:1248–1263. doi: 10.1002/pro.5560060613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Harris M. HIV: a new role for Nef in the spread of HIV. Curr Biol. 1999;9:R459–R461. doi: 10.1016/s0960-9822(99)80282-6. [DOI] [PubMed] [Google Scholar]
- 27.Harris M P G, Neil J C. Myristoylation-dependent binding of HIV-1 Nef to CD4. J Mol Biol. 1994;241:136–142. doi: 10.1006/jmbi.1994.1483. [DOI] [PubMed] [Google Scholar]
- 28.Howe A Y, Jung J U, Derosiers R C. Zeta chain of the T-cell receptor interacts with Nef of simian immunodeficiency virus and human immunodeficiency virus type 2. J Virol. 1998;72:9827–9834. doi: 10.1128/jvi.72.12.9827-9834.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Iafrate A J, Bronson S, Skowronski J. Separable functions of Nef disrupt two aspects of T cell receptor machinery: CD4 expression and CD3 signaling. EMBO J. 1997;16:673–684. doi: 10.1093/emboj/16.4.673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kestler H W, Ringler D J, Mori K, Panicali D L, Sehgal P K, Daniel M D, Desrosiers R. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell. 1991;65:651–662. doi: 10.1016/0092-8674(91)90097-i. [DOI] [PubMed] [Google Scholar]
- 31.Kienzle N, Freund J, Kalbitzer H R, Mueller-Lantzsch N. Oligomerization of the Nef protein from human immunodeficiency virus (HIV) type 1. Eur J Biochem. 1993;214:451–457. doi: 10.1111/j.1432-1033.1993.tb17941.x. [DOI] [PubMed] [Google Scholar]
- 32.Lama J, Mangasarian A, Trono D. Cell-surface expression of CD4 reduces HIV-1 infectivity by blocking Env incorporation in a Nef- and Vpu-inhibitable manner. Curr Biol. 1999;9:622–631. doi: 10.1016/s0960-9822(99)80284-x. [DOI] [PubMed] [Google Scholar]
- 33.Lee C H, Saksela K, Mirza U A, Chait B T, Kuriyan J. Crystal structure of the conserved core of HIV-1 Nef complexed with a Src family SH3 domain. Cell. 1996;85:931–942. doi: 10.1016/s0092-8674(00)81276-3. [DOI] [PubMed] [Google Scholar]
- 34.Le Gall S, Erdtmann L, Bénichou S, Berlioz-Torrente C, Liu L X, Benarous R, Heard J M, Schwartz O. Nef interacts with the μ subunit of clathrin adaptor complexes and reveals a cryptic sorting signal in MHC I molecules. Immunity. 1998;8:483–495. doi: 10.1016/s1074-7613(00)80553-1. [DOI] [PubMed] [Google Scholar]
- 35.Liu L X, Margottin F, Le Gall S, Schwartz O, Selig L, Benarous R, Bénichou S. Binding of HIV-1 Nef to a novel thioesterase enzyme correlates with Nef-mediated CD4 down-regulation. J Biol Chem. 1997;272:13779–13785. doi: 10.1074/jbc.272.21.13779. [DOI] [PubMed] [Google Scholar]
- 36.Lock M, Greenberg M E, Iafrate A J, Swigut T, Muench J, Kirchhoff F, Shohdy N, Skowronski J. Two elements target SIV Nef to the AP-2 clathrin adaptor complex, but only one is required for the induction of CD4 endocytosis. EMBO J. 1999;18:2722–2733. doi: 10.1093/emboj/18.10.2722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Luo T, Foster J L, Garcia J V. Molecular determinants of Nef function. J Biomed Sci. 1997;4:132–138. doi: 10.1007/BF02255641. [DOI] [PubMed] [Google Scholar]
- 38.Mangasarian A, Foti M, Aiken C, Chin D, Carpentier J L, Trono D. The HIV-1 Nef protein acts as a connector with sorting pathways in the Golgi and at the plasma membrane. Immunity. 1997;6:67–77. doi: 10.1016/s1074-7613(00)80243-5. [DOI] [PubMed] [Google Scholar]
- 39.Mangasarian A, Piguet V, Wang J K, Chen Y L, Trono D. Nef-induced CD4 and major histocompatibility complex class I (MHC-I) downregulation are governed by distinct determinants: N-terminal alpha helix and proline repeat of Nef selectively regulate MHC-I trafficking. J Virol. 1999;73:1964–1973. doi: 10.1128/jvi.73.3.1964-1973.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Manninen A, Hiipakka M, Vihinen M, Lu W, Mayer B J, Saksela K. SH3-domain binding function of HIV-1 Nef is required for association with a PAK-related kinase. Virology. 1998;250:273–282. doi: 10.1006/viro.1998.9381. [DOI] [PubMed] [Google Scholar]
- 41.Miller M D, Warmerdam M T, Gaston I, Greene W C, Feinberg M B. The human immunodeficiency virus-1 nef gene product: a positive factor for viral infection and replication in primary lymphocytes and macrophages. J Exp Med. 1994;179:101–113. doi: 10.1084/jem.179.1.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moarefi I, Lafevre-Bernst M, Sicheri F, Huse M, Lee C H, Kuriyan J, Miller W T. Activation of the Src-family tyrosine kinase Hck by SH3 domain displacement. Nature. 1997;385:650–653. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 43.Nunn M F, Marsh J W. Human immunodeficiency virus type 1 Nef associates with a member of the p21-activated kinase family. J Virol. 1996;70:6157–6161. doi: 10.1128/jvi.70.9.6157-6161.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Owen D J, Evans P R. A structural explanation for the recognition of tyrosine-based endocytic signals. Science. 1998;282:1327–1332. doi: 10.1126/science.282.5392.1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Piguet V, Chen Y L, Mangasarian A, Foti M, Carpentier J L, Trono D. Mechanism of Nef-induced CD4 endocytosis: Nef connects CD4 with the mu chain of adaptor complexes. EMBO J. 1998;17:2472–2481. doi: 10.1093/emboj/17.9.2472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier J L, Trono D. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 47.Piguet V, Le Gall S, Schwartz O, Trono D. The downregulation of CD4 and MHC-I by primate lentiviruses: a paradigm for the modulation of cell surface receptors. Immunol Rev. 1999;168:51–63. doi: 10.1111/j.1600-065x.1999.tb01282.x. [DOI] [PubMed] [Google Scholar]
- 48.Piguet V, Trono D. The Nef protein of primate lentiviruses. Rev Med Virol. 1999;9:111–120. doi: 10.1002/(sici)1099-1654(199904/06)9:2<111::aid-rmv245>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 49.Riggs N L, Craig H M, Pandori M W, Guatelli J C. The dileucine-based sorting motif in HIV-1 Nef is not required for down-regulation of class I MHC. Virology. 1999;258:203–207. doi: 10.1006/viro.1999.9736. [DOI] [PubMed] [Google Scholar]
- 50.Ross T M, Oran A E, Cullen B R. Inhibition of HIV-1 progeny virion release by cell-surface CD4 is relieved by expression of the Nef Protein. Curr Biol. 1999;9:613–621. doi: 10.1016/s0960-9822(99)80283-8. [DOI] [PubMed] [Google Scholar]
- 51.Rossi F, Gallina A, Milanesi G. Nef-CD4 physical interaction sensed with the yeast two-hybrid system. Virology. 1996;217:397–403. doi: 10.1006/viro.1996.0130. [DOI] [PubMed] [Google Scholar]
- 52.Saksela K, Cheng G, Baltimore D. Proline-rich (PxxP) motifs in HIV-1 nef bind to SH3 domains of a subset of Src kinases and are required for the enhanced growth of Nef+ viruses but not for down-regulation of CD4. EMBO J. 1995;14:484–491. doi: 10.1002/j.1460-2075.1995.tb07024.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sawai E T, Baur A, Struble H, Peterlin B M, Levy J A, Cheng-Mayer C. Human immunodeficiency virus type 1 Nef associated with a cellular serine kinase in T lymphocytes. Proc Natl Acad Sci USA. 1994;91:1539–1543. doi: 10.1073/pnas.91.4.1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sawai E T, Baur A S, Peterlin B M, Levy J A, Cheng-Mayer C. A conserved domain and membrane targeting of Nef from HIV and SIV are required for association with a cellular serine kinase activity. J Biol Chem. 1995;270:15307–15314. doi: 10.1074/jbc.270.25.15307. [DOI] [PubMed] [Google Scholar]
- 55.Schwartz O, Marechal V, Le Gall S, Lemonnier F, Heard J M. Endocytosis of major histocompatibility complex class I molecules is induced by the HIV-1 Nef protein. Nat Med. 1996;2:338–342. doi: 10.1038/nm0396-338. [DOI] [PubMed] [Google Scholar]
- 56.Shugars D C, Smith M S, Glueck D H, Nantermet P V, Seillier-Moiseiwitsch F, Swanstrom R. Analysis of human immunodeficiency virus type 1 nef gene sequences present in vivo. J Virol. 1993;67:4639–4650. doi: 10.1128/jvi.67.8.4639-4650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Skowronski J, Parks D, Mariani R. Altered T cell activation and development in transgenic mice expressing the HIV-1 nef gene. EMBO J. 1993;12:703–713. doi: 10.1002/j.1460-2075.1993.tb05704.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Swingler S, Gallay P, Camaur D, Song J, Abo A, Trono D. The Nef protein of human immunodeficiency virus type 1 enhances serine phosphorylation of the viral matrix. J Virol. 1997;71:4372–4377. doi: 10.1128/jvi.71.6.4372-4377.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Watanabe H, Shiratori T, Shoji H, Miyatake S, Okazaki Y, Ikuta K, Sato T, Saito T. A novel acyl-CoA thioesterase enhances its enzymatic activity by direct binding with HIV Nef. Biochem Biophys Res Commun. 1997;238:234–239. doi: 10.1006/bbrc.1997.7217. [DOI] [PubMed] [Google Scholar]
- 60.Wiskerchen M, Cheng-Mayer C. HIV-1 Nef association with cellular serine kinase correlates with enhanced virion infectivity and efficient proviral DNA synthesis. Virology. 1996;224:292–301. doi: 10.1006/viro.1996.0531. [DOI] [PubMed] [Google Scholar]
- 61.Xu X N, Laffert B, Screaton G R, Kraft M, Wolf D, Kolanus W, Mongkolsapay J, McMichael A J, Baur A S. Induction of Fas ligand expression by HIV involves the interaction of Nef with the T cell receptor ζ chain. J Exp Med. 1999;189:1489–1496. doi: 10.1084/jem.189.9.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]