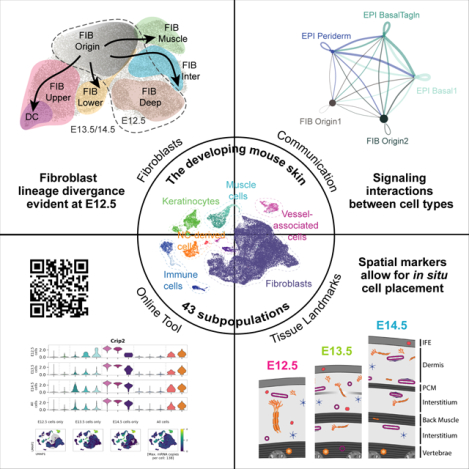
SUMMARY
A wealth of specialized cell populations within the skin facilitates its hair producing, protective, sensory and thermoregulatory functions. How the vast cell-type diversity and tissue architecture develops is largely unexplored. Here, with single-cell transcriptomics, spatial cell-type assignment and cell-lineage tracing, we deconstruct early embryonic mouse skin during the key transitions from seemingly uniform developmental precursor states to a multilayered, multilineage epithelium and complex dermal identity. We identify the spatiotemporal emergence of hair-follicle-inducing, muscle-supportive, and fascia-forming fibroblasts. We also demonstrate the formation of the panniculus carnosus muscle, sprouting blood vessels without pericyte coverage, and the earliest residence of mast and dendritic immune cells in skin. Finally, we identify an unexpected epithelial heterogeneity within the early single-layered epidermis and a signaling-rich periderm layer. Overall, this cellular and molecular blueprint of early skin development – which can be explored at http://kasperlab.org/tools – establishes histological landmarks and highlights unprecedented dynamical interactions among skin cells.
Graphical Abstract
eTOC blurb
Jacob et al. establish a cellular and molecular blueprint of early mouse skin development using single-cell transcriptomics, lineage tracing, and multiplex RNA in situ staining. Considering all cell types, they determine molecular and histological key transitions, cross-cell type communications, as well as the onset of lineage-diversification during skin development.
INTRODUCTION
During skin development, one of the most remarkable changes occurs when the epidermis transforms from a single-layered epithelium to a multi-layered and appendage-producing epithelium. Mouse epidermis develops from the surface ectoderm at embryonic day (E) 9.5, starting as a single layer of basal keratinocytes that is subsequently covered by a transient protective layer of squamous cells called ‘periderm’. Within the following days until birth at approximately E19.5 a fully stratified epidermis is formed, which acts as a reliable barrier keeping pathogens outside and water inside1. During these 10 days, also the epidermal appendages form. In dorsal skin, hair follicles develop in three waves, with the first epithelial thickenings – so-called hair placodes – being morphologically visible at E14.5. Hair placodes maintain a tight dialogue with the underlying dermal condensate (DC), a mesenchymal signaling center that stays in close contact with the hair follicle throughout its lifetime2–5. The vast majority of studies on embryonic skin to date have focused on the skin’s epidermis6–8. Nevertheless, important aspects of epidermal development remain unresolved, such as the maturation of the periderm and its signaling potential, basal-cell heterogeneity prior to placode formation, and the involvement of mature placode cells in shaping the skin’s dermal architecture.
In the dermis, fibroblasts are the most abundant cell type, yet little is known about their heterogeneity and contributions to early skin development. The few studies that focused on the developing dermis were mostly centered around the molecular and cellular establishment of hair follicles (e.g.,9–12) leaving a major gap in knowledge about the non-hair-follicle-related mesenchymal cell types during early skin development. It has been proposed that dermal fibroblasts derive from a single fibroblast lineage that diverges at E16.5 forming the upper (papillary) dermis including the hair follicle-associated dermal papillae, dermal sheath and arrector pili muscles, and the lower (reticular) dermis and adipocytes of the hypodermis13. Although the existence of fibroblast heterogeneity and potential fate-specification prior to the lineage divergence at E16.5 has been proposed14, major questions remain. How heterogeneous are fibroblasts during early skin development? When does fibroblast heterogeneity emerge? By which means do early fibroblasts support tissue specification and maturation?
A major challenge to answer any of these questions is the complete lack of histological or molecular tissue landmarks in early developing skin. In adult mouse skin, the only certain landmark to date that defines ‘skin-associated’ cells is the panniculus carnosus muscle (PCM). Only the cells above the PCM (epidermis/dermis), the PCM itself, and a thin layer of connective tissue cells (called fascia) just below the PCM are considered skin-associated. When the PCM is formed has not been reported. At E12.5, the future skin dermis and fascia, as well as non-skin-associated cells, are part of a seemingly homogenous tissue space spanning from the vertebrae to the epidermis. Similarly, at E13.5 and 14.5, little is known about dermal tissue architecture and cell type diversity, placing more questions. When does the PCM form? What is the spatiotemporal diversity of all other cell types, such as neural crest-derived, vessel-associated, or immune cells, during early skin development?
Here, through comprehensive computational analysis of all cell types sampled at E12.5, E13.5, and E14.5 combined with cell-type localization in situ and in vivo cell-fate mapping, we i) determined fibroblast heterogeneity and onset of lineage commitment, ii) showed when the PCM forms, iii) resolved the periderm-transcriptome and epidermal cell heterogeneity prior to placode formation, iv) characterized all other major cell types, v) portrayed the comprehensive interplay between skin cell types, and v) provided histological landmarks which are essential to place cells in their spatial tissue context.
RESULTS
Single-cell profiling-assisted generation of histological landmarks in E12.5, E13.5 and E14.5 skin
To unveil the cellular diversity and decisive signaling events driving early skin maturation, we profiled E12.5, E13.5 and E14.5 mouse back skin. We isolated full-thickness dorsal skin and generated single-cell transcriptome (10x v2) libraries of epithelial and stromal cells. To assure true biological replicates, 5 embryos per embryonic time point were processed, sequenced, and quality-controlled individually (Figure S1A–B). Moreover, histological analysis of the remaining body of each sequenced embryo as well as intact littermates ensured correct embryonic age (Figure S1C). After quality control, all three timepoints were analyzed together (Figure S1D–F; Methods) to better capture developmental trends and the dynamics of cell populations.
The complete dataset contains 32,194 single-cell transcriptomes with 11,280 cells coming from E12.5, 9,964 cells from E13.5 and 10,950 cells from E14.5 (Figure 1A). Based on cluster-specific gene expression we identified keratinocytes, fibroblasts, immune cells, vessel-associated mural (pericytes and vascular smooth muscle cells) and endothelial cells, neural crest-derived melanocytes and Schwann cells, and muscle cells (Figure 1B–C; Table S1). Through fluorescent in situ hybridization (FISH for mRNA) and immunofluorescence staining (IF for protein) of cell-type specific marker genes, we mapped all major cell types within E12.5, E13.5, and E14.5 skin tissue (Figure 1D–L). We also used these cell-type-specific markers together with wheat germ agglutinin (WGA) cell-membrane staining to establish histological landmarks of early skin development, which the commonly used H&E staining cannot resolve (Figure S1G–I). Notably, staining for ACTC1 revealed the stepwise development of multiple muscle layers including the PCM (Figure 1D–F), PTPRC highlights the exclusively dermal location of immune cells (Figure 1J), Sox10 shows large nerve trunks growing towards the epidermis at E12.5 and more spread-out nerves at E14.5 (Figure 1K–L), and Rgs5/Pecam1 co-staining depicts the dense vascular network (Figure 1G–I). For this work, these landmarks (summarized in Figure 1M–O) were instrumental for the mapping and placement of numerous cell populations within the rapidly developing full-thickness skin.
Figure 1. Anatomy of embryonic skin from E12.5, E13.5, and E14.5.
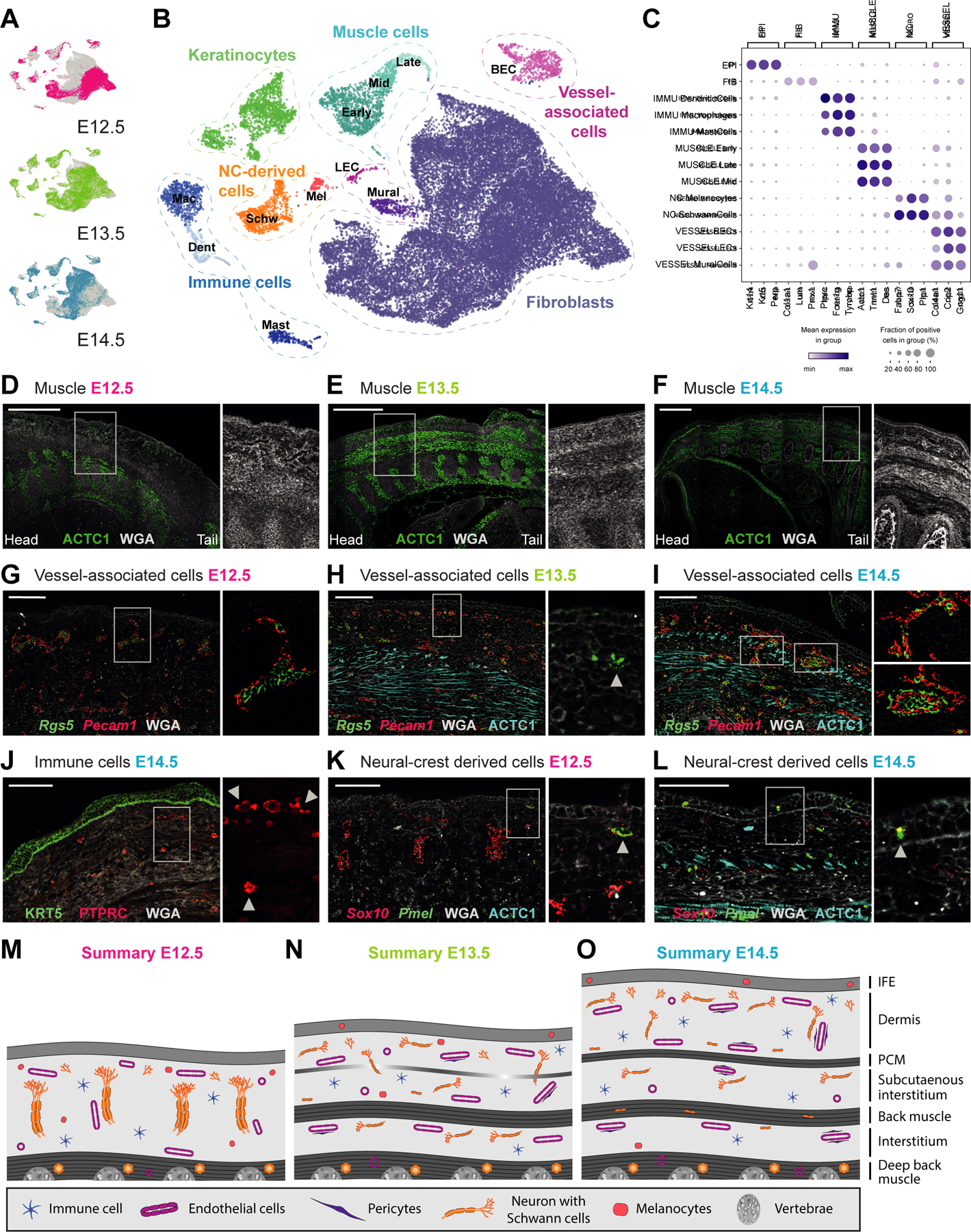
(A) UMAP visualization of all cells from the different embryonic ages (n = 11 280 cells of E12.5, 9 964 of E13.5, and 10 950 of E14.5).
(B) UMAP visualization of first-level clustering of all cells (n = 32 194 cells).
(C) Marker gene expression dot plot for main cell classes.
(D-L) mRNA (italics) and protein (capital letters) stainings revealing the major anatomical landmarks of dorsal embryonic skin (sagital sections). Microscope images originate from larger tile scans (n = 3 mice). Scale bars, 500μm (D-F) and 100μm (G-L). * denotes erythrocyte autofluoresence
(D-F) Muscle layers (ACTC1) with zoom ins to visualize anatomical layers using membraneous counterstain with WGA. M marks developing muscle layers.
(G-I) Endothelial (Pecam1) and mural (Rgs5) cells. Arrowhead in (H) marks earliest evidence of mural cells at E13.5. Upper zoom in in (I) shows smaller vessel with discontinuous mural cell lining while lower zoom in in (I) shows larger vessel with continuous mural cell lining.
(J) Immune cells (PTPRC) and epidermis (KRT5). Arrowheads highlight immune cells with dendritic phenotype.
(K-L) Melanocytes (Sox10 + Pmel) and Schwann cells (Sox10). Arrowhead shows the arrival of melanocytes in the epidermis.
(M-O) Schemes summarizing anatomical landmarks at E12.5, E13.5 and E14.5.
See also Figure S1.
Fibroblast heterogeneity exists long before the reported establishment of papillary and reticular dermis
Our dataset contains in total 25,944 fibroblasts out of 32,194 randomly sampled cells, from 15 individual embryos, allowing for robust identification of 22 fibroblast subpopulations (Figure S2A; Table S2). Overall, fibroblasts isolated from the skin and underlying (non-skin) tissue (Methods and Discussion) separated into seven major cell groups. We named these groups FIB Origin, FIB Deep, FIB Upper & DC, FIB Lower, FIB Muscle, FIB Inter and CHOND (Figure 2B, 3B, and S2A) based on several criteria such as their appearance in development, their gene expression profiles, RNAvelocity analysis, and tissue location.
Figure 2. Deconstruction of fibroblast heterogeneity at E12.5 (expression and location).
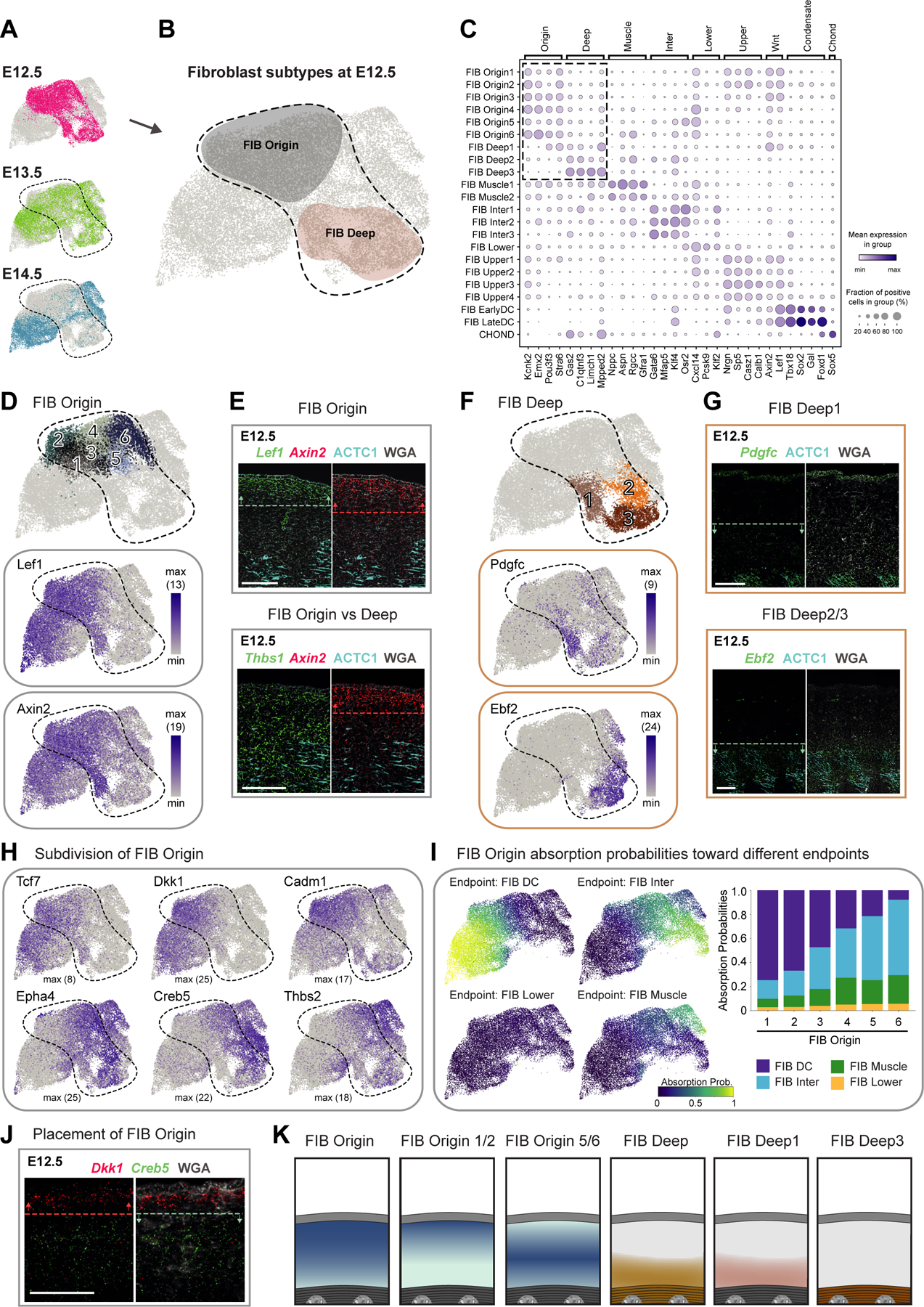
(A) UMAP visualization of fibroblasts from the different embryonic ages (n = 10 008 cells of E12.5, 8 016 of E13.5, and 7 920 of E14.5).
(B) Major fibroblast subtypes at E12.5 highlighted on UMAP.
(C) Marker gene expression dot plot for major fibroblast groups. Highlighted are the clusters mostly present at E12.5.
(D,F) Subclustering of early fibroblast groups (upper panels). Expression pattern of characteristic marker genes (lower panels). Number in brackets shows max number of RNA copies detected per cell (absolute abundance).
(E,G,J) mRNA (italics) and protein (capitalized) stainings of fibroblast subpopulations. Dashed lines and arrows highlight the region with highest expression. Microscope images originate from larger tile scans (n = 3 mice). Scale bars, 100μm.
(H) Marker gene expression of FIB Origin subpopulations on UMAP.
(I) Absorption probabilities towards the differentiation endpoints projected onto UMAP (left panels) and quantified for each FIB Origin subpopulation (right panel).
(K) Schemes summarizing major fibroblast groups at E12.5 and their location.
Figure 3. Deconstruction of fibroblast heterogeneity at E13.5 and E14.5 (expression and location).
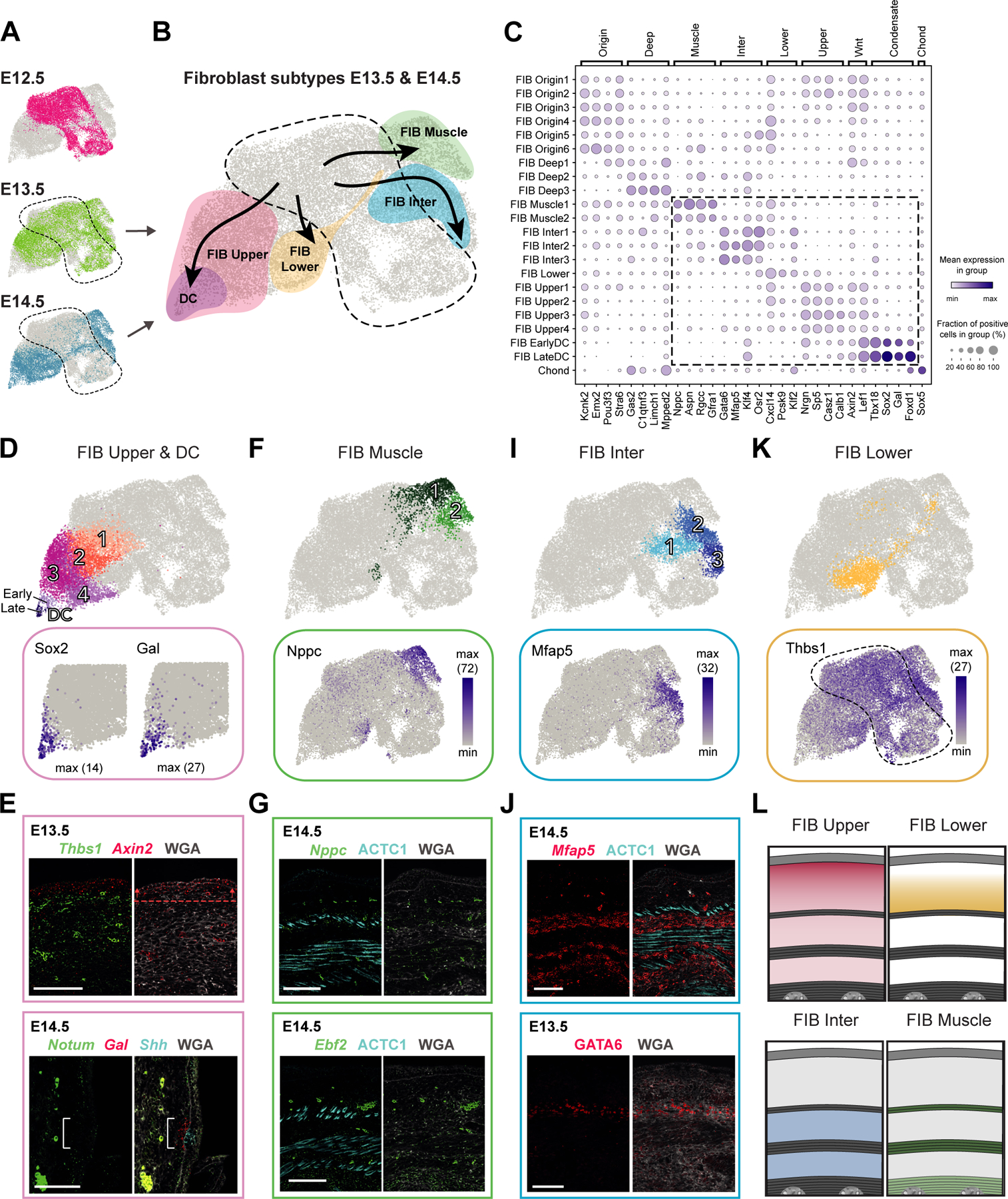
(A) UMAP with fibroblasts from different embryonic ages (as shown in Fig 2A).
(B) UMAP showing major fibroblast subtypes at E13.5 and E14.5 together with their differentiation trajectories (velocity trends).
(C) Marker gene expression dot plot for major fibroblast groups. Highlighted are clusters mostly present at E13.5 and E14.5.
(D,F,I,K) Subclustering of major fibroblast groups (upper panels). Expression pattern of characteristic marker genes (lower panels).
(E,G,J) mRNA (italics) and protein (capitalized) stainings highlighting fibroblast subpopulations. Dashed line with arrows highlights the region of highest expression. Bracket shows reduced Notum expression. M marks developing muscle layers. Microscope images originate from larger tile scans (n = 3 mice). Scale bars, 100μm.
(L) Schemes summarizing major fibroblast groups at E14.5 and their location. Similar at E13.5, but PCM is not fully developed yet.
FIB Origin and FIB Deep – Notable fibroblast heterogeneity exists already at E12.5
At E12.5, dermis contains the two major fibroblast subsets FIB Origin and FIB Deep, characterized by expression of unique gene sets (Figure 2A–C and S2C) and different tissue locations (summarized in Figure 2K). The FIB Origin cells constitute a Wnt-pathway-activated Lef1+Axin2+ fibroblast subset that maps to the upper dermis (Figure 2D–E) and expresses remarkably few receptors and ligands (Figure S2E). According to RNAvelocity analysis, which can predict differentiation paths based on the expression of unspliced and spliced mRNA15, FIB Origin fibroblasts may serve as the source for almost all other fibroblast clusters emerging at E13.5 and E14.5 (Figure S2B). Lef1 and Axin2 is also expressed in one of the FIB Deep subpopulations at E12.5 (FIB Deep1), which is further characterized by co-expression of Pdgfc (Figure 2F) and Hoxb8 (Figure S2F) and maps to the lower half of the sub-epidermal space (Figure 2G). FIB Origin and FIB Deep1 also share the expression of the myofibroblast markers Acta2 (also known as α-SMA) and Tagln (also known as SM22-α) (Figure S2G). FIB Deep2 and FIB Deep3 constitute Ebf2+ Postn+ cell populations (Figure 2F and S2H), with FIB Deep3 additionally expressing Limch1 (Figure S2F). FIB Deep2/3 were mapped within the deep back muscle at E12.5 (Figure 2G and S2H).
As the sub-epidermal/dermal tissue at E12.5 is still a single compartment, which is not yet separated by any muscle layers (see Figure 1M–O), we considered that not all sampled fibroblasts (and/or their respective lineages) will be part of the skin-associated tissue compartment (i.e. fibroblasts above PCM, within PCM, and in fascia). Based on histological landmarks and tissue placement of FIB Origin/Deep subpopulations, FIB Origin cells were the most likely source for skin-associated fibroblasts. Thus, we probed whether FIB Origin cells are at E12.5 transcriptionally still uniform or show already heterogeneity that may point towards future fibroblast lineages. Unbiased clustering assigned FIB Origin cells into 6 subgroups (Figure 2D), which arranged in dimensionality-reduced space (UMAP) into ‘left’ (FIB Origin1/2), ‘middle’ (FIB Origin3/4) and ‘right’ (FIB Origin5/6) subpopulations. The FIB Origin1/2 cells are enriched for Wnt-pathway components such as Lef1, Tcf7, Dkk1, Sp5 and the adhesion molecule Cadm1 (Figure 2C,D,H), while the FIB Origin5/6 cells are marked by e.g. Epha4, Creb5, and Thbs2 (Figure 2H). FIB Origin3/4 cells are in the UMAP placed between FIB Origin1/2 and 5/6 cells and express genes of both. Co-staining of markers characteristic for FIB Origin1/2 (Dkk1-enriched) or FIB Origin5/6 (Creb5-enriched) fibroblast subsets revealed a clearly distinct spatial placement, with FIB Origin1/2 mapping closer to epidermis than FIB Origin5/6 (Figure 2J). Notably, when overlaying the marker gene expression for FIB Origin1/2 and Origin5/6 subsets on the fibroblast UMAP, the subsets seem to extend into FIB Upper/DC and FIB Inter/Muscle, respectively (Figure 2H; see Figure 3B), suggesting that the subdivision of FIB Origin cells may already reflect early commitment towards future fibroblast fates (presented in Figure 3). Indeed, fate simulation of the FIB Origin subpopulations confirmed this observation at a global gene expression level (Figure 2I and S2I; Methods). In summary, we found that skin-associated fibroblast heterogeneity already exists at E12.5 (FIB Origin subsets), representing the earliest reported fibroblast heterogeneity – transcriptionally and spatially – during the development of mouse skin.
CHOND – Embryonic chondrocytes transcriptionally map with skin fibroblasts
Among the 22 fibroblast subsets we identified a small cluster of chondrocytes or their precursors (CHOND) that located next to the FIB Deep populations in the UMAP (Figure S2A,J). Chondrocytes share their developmental origin (paraxial mesoderm-derived somites) with skeletal muscle and the dermis16. They express Pdgfra, as well as the transcription factors Sox5 and Sox6, which activate the cartilage-promoting factor Sox9 resulting in the expression of chondrocyte-specific genes such as Col2a1, Col9a3, Acan, and Matn417–19 (Figure S2K). mRNA staining for the chondrocyte differentiation marker Mia20 showed strong expression in the developing vertebrae (Figure S2L–M). Future studies may benefit from the realization that chondrocytes can cluster with fibroblasts.
FIB Upper and FIB DC – Acute loss of Wnt inhibitors marks dermal condensate formation
Starting from E13.5, as expected for the time just prior to hair follicle induction, we identified a fibroblast subset (FIB Upper) that shows high Wnt-pathway activity (e.g. Lef1 and Axin2) and can mature into the hair follicle-inductive dermal condensate (FIB DC) (Figure 2D and 3A–D). This is in line with the current view that Wnt-signaling activated fibroblasts are a prerequisite for embryonic hair follicle development9,10,21–27. Axin2 mRNA staining revealed that these Wnt-signaling activated fibroblasts become confined to only a few layers in the uppermost dermis at E13.5 (compare Figure 2E with 3E), a pattern that has been noted before21. In line with previous reports9,10,12, we also detect that cells exit the cell cycle just prior to DC commitment (upregulation of G0/G1 genes in FIB DC, e.g. Cdkn1a and Btg1; Figure S2D and S3A) and start expressing Sox2 when the morphologically recognizable DC is forming (E14.5) (Figure 3D and S2C). These cells also express pre-DC marker Fst, early DC markers Sema6a and Fgf10, and later DC markers Dll1 and Bmp3 (Figure S3B)9.
Additionally, our data revealed a most striking and abrupt gene-expression change at both initial and final DC-lineage commitment, each signified by a sharp downregulation of Wnt-pathway inhibitors. At the FIB Origin to FIB Upper border (E12.5 to E13.5; initial commitment) we noted acute and permanent downregulation of Dkk2 (Figure S3A), which is expressed in non-hairy, but absent in hair-bearing skin28. Together with our data, this suggests that the absence of Dkk2 may be a key determinator for fibroblasts becoming competent to enter a DC fate.
As FIB Upper cells become more Wnt-pathway activated, they also upregulate Dkk1 (Figure 2D,H). This parallel upregulation continues until DC-fated cells acquire Sox2 (final commitment), when Dkk1 expression drops acutely9,10. Strikingly, in our data we observe the same switch-like pattern with sharp downregulation at the border between FIB Upper3/4 and FIB EarlyDC also for Notum and Cav1 (Figure S3A), both acting as Wnt-signaling inhibitors29,30. The loss of Notum expression in mature DC cells was confirmed by co-staining of Notum and the DC-marker Gal in E14.5 skin (Figure 3E). The fact that several prominent Wnt-signaling inhibitors are first up-regulated in FIB Upper and then abruptly down-regulated in EarlyDC cells suggests that this is a functional feature of DC formation and/or DC commitment, which remains an exciting route to be explored.
FIB Lower – Dermal fibroblasts without unique marker gene expression
At E13.5 the FIB Lower subset emerges. As this cell cluster lacked unique marker genes we used exclusion criteria that placed this population. At E13.5, Thbs1 expression becomes confined to the lower dermis as well as the subcutaneous interstitial layer, illustrated in Figure 1N–O, that starts below the PCM and reaches until the spine31,32 (Figure 2E, 3E,K,L, and S3I). In addition to Thbs1, the interstitium expresses Mfap5 (see below), which places Thbs1+/Mfap5- FIB Lower cells to the lower dermis (Figure 3I–J).
FIB Muscle – Perimuscular Nppc+ fibroblasts possess the ability to support the developing muscle
Also at E13.5, a group of Nppc+ fibroblasts (FIB Muscle) was first observed (Figure 3B and S2C). This cell population is characterized by expression of Nppc, Rgcc and Gfra1 (Figure 3C,F) and is exclusively located within the developing muscle layers (Figure 3G, upper image). Sub-clustering further separated FIB Muscle cells into two subgroups, characterized by expression of Aspn and Wnt4 (FIB Muscle1), and Ebf2 and Igfbp3 (FIB Muscle2), respectively (Figure 2F, 3F, and S3C).
As Wnt4 has been reported to maintain satellite cell quiescence33, while Igfbp3 is known to support myoblast differentiation34, it is conceivable that the two FIB Muscle subpopulations are involved in balancing activation and quiescence of satellite cells, as has been found in adult settings35. Moreover, FIB Muscle cells express high levels of collagen isoforms I, III, IV, and VI (Figure S3D), which are the primary components of extracellular matrix (ECM) within the skeletal muscles and help to mediate force transmission36–46.
FIB Inter – Fibroblasts constituting fascia fibroblasts and serving as a cellular source for adipose stem cells
From E13.5, a distinct group of Mfap5+/Gata6+ fibroblasts (FIB Inter) can be detected (Figure 3B,C,I), which mapped to the interstitial layers via ECM component Mfap547 staining (Figure 3J).
The FIB Inter1/2 subcluster expressed genes characteristic for the fascia underlying the PCM, such as Nov, Dpp4 and Plac814,48, as well as additional fascia-associated genes Mfap5, Wnt2, Creb5, Col14a1, and Tmeff2 (Figure 3I and S3E–F)48. On the other hand, FIB Inter3 cells express the key adipogenic transcription factors Pparg and Cebpa (Figure S3G–H; Table S1)49,50. Given that bundles of fascial fibres are often found mixed with fat to form pressure-tolerant fibro-adipose tissue associated with skin (e.g. soles, palms, or finger tips in humans)51, we followed up on the intriguing possibility that FIB Inter cells might include adipogenic cells. FIB Inter2 expresses early regulators of adipogenesis Junb, Fos, Atf3 and Klf452, while FIB Inter3 expresses later adipogenesis regulators Cebpa49 and Pparg50 (Figure S3G), suggesting that FIB Inter2 cells mature to FIB Inter3 during early adipogenesis. However, these clusters lack the terminal differentiation markers such Fabp4 and Cd3653 (Figure S3G), which appear at approximately E16 and are followed by the characteristic lipid droplets of mature adipocytes appearing at E18.554. In sum, our data suggests that a fibroblast subset may already be fated for the adipose lineage as early as E13.5.
Lineage tracing confirms FIB Muscle and FIB Inter populations
Because little is known about functionally specialized fibroblasts such as muscle-associated or interstitial fibroblasts, we performed lineage tracing to determine if FIB Muscle cells remain muscle restricted and if FIB Inter cells indeed contribute primarily to lower skin layers such as the fascia and adipose tissue as the scRNA-seq data suggested.
For tracing the FIB Inter cells, we identified Gata6 as one of the most specific cell markers among all skin populations (Figure 4A–B). Thus, we used Gata6-EGFP-CreERT2/R26-tdTomato mice (hereafter Gata6-Tom) and traced Gata6+ cells from E13.5 to E15.5 (initial tracing), or to postnatal days P5 and P35 when hair follicles are in active growth (anagen), which is accomponied with an enlarged and mature dermal white adipose tissue (DWAT) compartment (Figure 4C). At E15.5 adipocytes are not yet formed, however we found Gata6-Tom traced cells abundantly present in the fascia and subcutaneous interstitium (Figure 4D). Moreover, tracing to P5 and P35 revealed the persistence of the FIB Inter lineage in the fascia from postnatal stage to early adulthood (Figure 4E–F). Unexpectedly however, we did not detect Gata6-Tom traced DWAT adipocytes (marked by PLP1 staining) (Figure 4E–F), and due to technical limitations we could not determine if the subcutanous white adipose tissue (SWAT) was traced. This leaves open two possibilities; FIB Inter cells either do not represent adipocyte precursers or FIB Inter cells only contribute to SWAT formation and thus DWAT and SWAT originate from independent precursors (see Discussion).
Figure 4. Tracing the fate of Ebf2+ and Gata6+ fibroblasts.
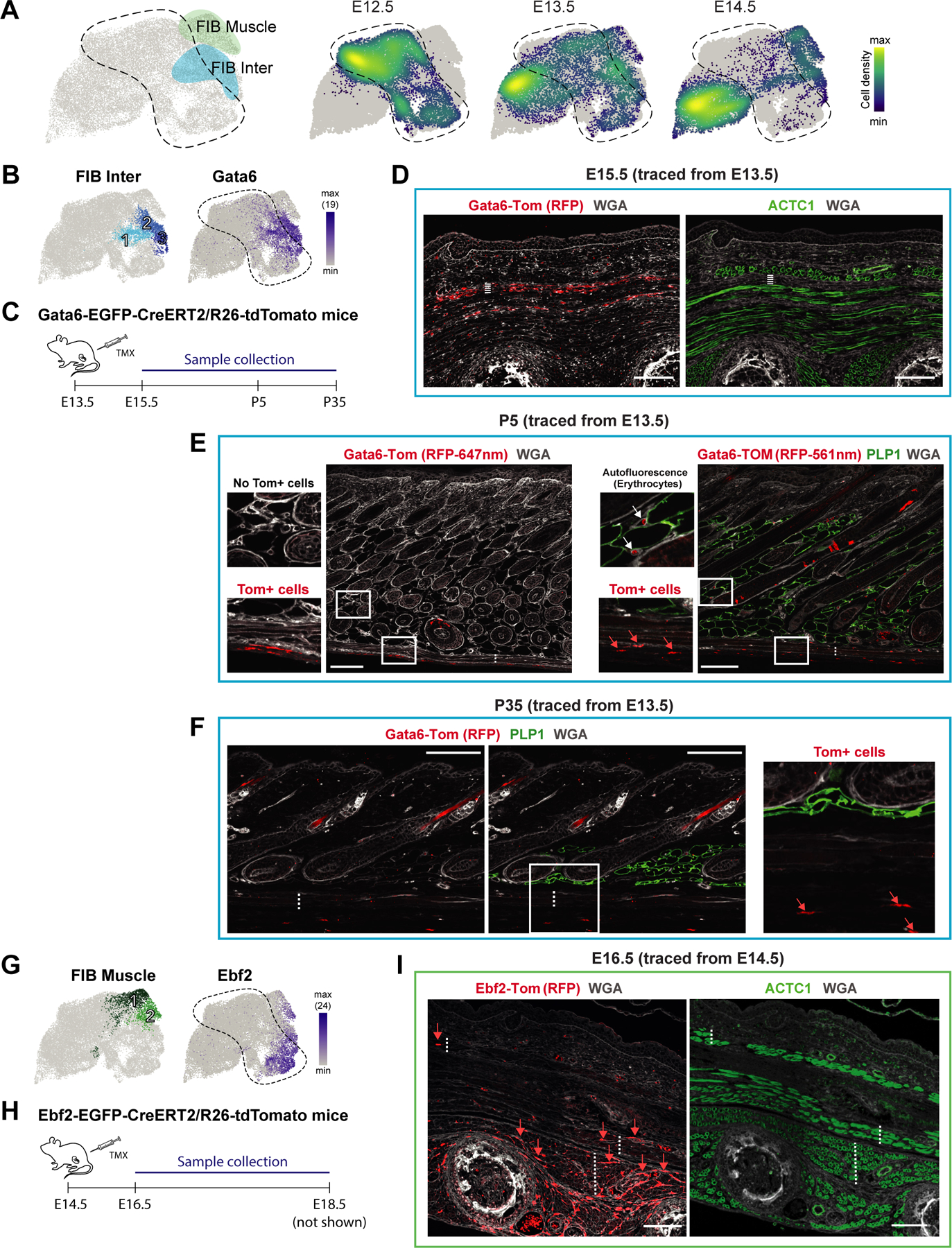
(A) FIB Muscle and FIB Inter fibroblasts highlighted on UMAP (left panel). Density plot showing the distribution of fibroblasts from the different embryonic ages on the UMAP (right panel).
(B) UMAPs with FIB Inter subpopulations (left panel) and Gata6 expression (right panel).
(C) Experimental setup for lineage tracing of Gata6+ cells.
(D) Initial 2-day-tracing pattern of Gata6+ cells (left panel) and staining of the developing muscle layers (right panel). Dashed line marks Fascia/SWAT layer on the two consecutive sections. M marks developing muscle layers. Scale bars, 100μm.
(E, F) Tracing pattern of Gata6+ cells at postnatal day 5 (P5; E) or P35 (F) and PLP1 protein staining of lipid droplets. Note the strong erythrocyte autofluorescence within DWAT at 561nm (E). Dotted lines indicate the PCM. Scale bars, 100μm.
(G) UMAPs with FIB Muscle subpopulations (left panel) and Ebf2 expression (right panel).
(H) Experimental setup for lineage tracing of Ebf2+ cells.
(I) 2-day-tracing pattern of Ebf2+ cells (left panel) and staining of the developing muscle layers (right panel). Dotted lines indicate the PCM, underlying back muscle layers (M), as well as deep-tissue interstitial space on the two consecutive sections. Scale bars, 100μm.
(D-F, I) Microscope images originate from larger tile scans (n = 3 mice).
To trace the FIB Muscle population, Ebf2 was one of the most suitable markers (at E14.5 expressed in FIB Muscle2 and FIB Inter3; Figure 4A,G). Thus, we utilized Ebf2-EGFP-CreERT2/R26-tdTomato mice (hereafter Ebf2-Tom) to trace Ebf2+ cells from E14.5 to E16.5 or E18.5 (Figure 4H). Both 2- and 4-day tracing gave rise to Ebf2-Tom cells within the superficial and deep back muscles and more rarely in the PCM, suggesting that the FIB Muscle1/2 cell group identified by scRNA-seq indeed constitutes a muscle-associated fibroblast subtype (Figure 4I). As expected from the scRNA-expression pattern (Figure 4A,G), Ebf2-Tom tracing also gave rise to some interstitial cells (Figure 4I). Having identified muscle-associated (FIB Muscle) and muscle-adjacent (FIB Inter) fibroblast populations, we were curious about their potential role in muscle development and its maintenance.
The PCM layer forms de novo at E13.5 likely in direct signaling dialogue with FIB Muscle and FIB Inter cells
Our scRNA-seq data analysis captured the full process of early skeletal myogenesis (Figure 5A). The MUSCLE Early population, which encompass the skeletal muscle stem cells, or Pax7+ satellite cells55,56, constitute the majority (~70%) of our detected muscle cells (Figure 5A). In comparison, satellite cells in early postnatal life account for 30–35% and in adulthood 1–4%57. The MUSCLE Mid and MUSCLE Late populations recapitulate the stepwise expression of myogenic regulatory factors that govern muscle cell differentiation, with Myod1 and Myf5 being early markers for satellite cells that have committed to differentiation and Myog and Myf6 driving terminal differentiation (Figure 5A)58,59. MUSCLE Late cells further express markers of mature muscle fiber subtypes e.g., Tnnc1 for Type I and Tnnc2 for Type II fibers (Figure 5A)60,61. Notably, Syndecans (Sdc1, Sdc2, Sdc3), which allow MUSCLE Early cells to sense a wide array of signaling supporting muscle formation and maintenance62, are downregulated in MUSCLE Mid and MUSCLE Late cells (Figure S4A). Interestingly, we captured each of the muscle subpopulations (MUSCLE Early, Mid, Late) at all sampled time points (Figure 5A), suggesting that there is no major transcriptional difference between the PCM, superficial and deep back muscle layers at E12.5, E13.5 and E14.5.
Figure 5. Developing muscle layers in embryonic skin.
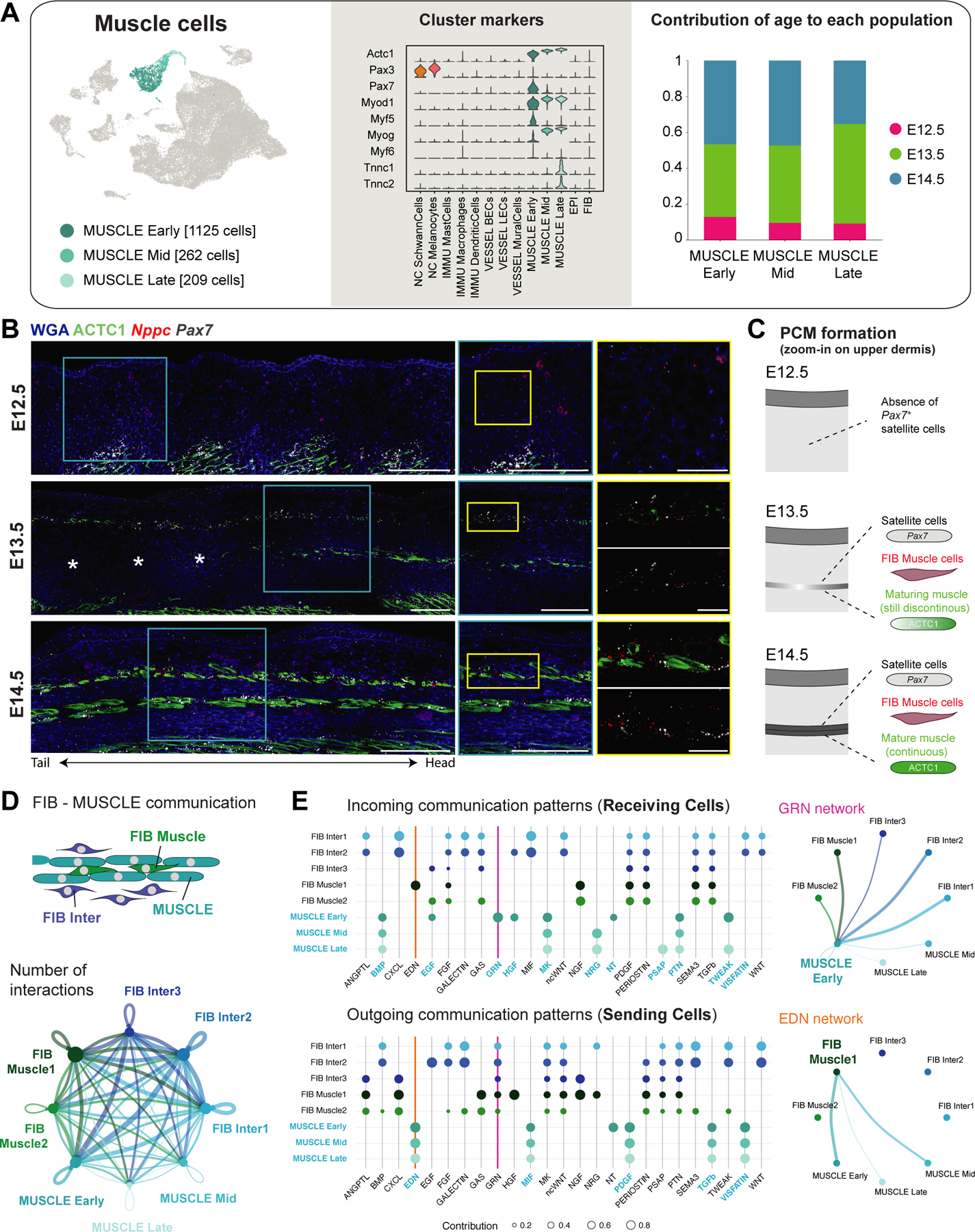
(A) UMAP (from Figure 1B) with subpopulations of muscle cells (left panel), violin plot of marker gene expression (center panel), and contribution of each embryonic time point to subpopulations (right panel).
(B) mRNA (italics) and protein (capitalized) stainings highlighting developing muscle layers. Microscope images originate from larger tile scans (n = 3 mice). Scale bars, 100μm (panorama, blue zoom-in) and 25μm (yellow zoom-in). Asterisks: indicate a discontiunous upper back muscle layer (dependent on histological cut and distance to dorsal midline).
(C) Scheme of panniculus carnosus muscle (PCM) formation.
(D) Circle plot visualizing number of interactions between FIB Inter, FIB Muscle, and MUSCLE subpopulations. Edge width proportional to the number of interactions. Edges colored according to sending cell population.
(E) Dot plot showing outgoing and incoming signaling patterns between FIB Inter, FIB Muscle, and MUSCLE subpopulations (left panels). Dot size proportional to enrichment of signaling pathway in the cell population. Circle plots for selected signaling pathways with significant interactions (right panels).
As the PCM is entirely absent at E12.5, we next asked whether the PCM is established through migration of back muscle cells or via de novo differentiation within the dermis. To this end, we stained all three developmental timepoints for Pax7 mRNA (satellite cells), Nppc mRNA (FIB Muscle) and ACTC1 protein (actin alpha cardiac muscle 1; the predominant actin isoform in early muscle development63) (Figure 5B). At E13.5, we detected the appearance of a non-continuous PCM layer, with intermingling Pax7+ satellite cells, Nppc+ fibroblasts, and some ACTC1+ cells. As Pax7+ cells were entirely absent between muscle layers (i.e., no migrating muscle precursor cells), we conclude that the PCM may form via direct differentiation at the destined location (Figure 5B–C). Moreover, CellChat analysis, which can probe for signaling communication patterns between cell types, suggests strong interactions between muscle cells and fibroblasts (FIB Muscle/Inter) strengthening our hypothesis that muscle-surrounding fibroblasts likely support muscle formation and its maintenance (Figure 5D–E).
Finally, the PCM and back muscles are (like other skeletal muscles in the body) innervated by motor neurons which connect to the muscle via neuromuscular junctions scattered along the myofibers64–66. We indeed find evidence of those neuromuscular junctions in our data (Table S3 and Figure S4B), such as MUSCLE Late cells expressing Musk, that upon binding of ARGN (expressed e.g., by motor neurons) induces clustering of acetylcholine receptors to form neuromuscular junctions67,68. Subunits of those acetylcholine receptors (e.g. Chrna1, Chrna4, Chrnd, Chrng) are expressed in MUSCLE Mid and MUSCLE Late cells (Figure S4B).
Cellular heterogeneity of vascular, immune, and neural crest-derived cells in the developing dermis
To reveal the full complexity of dermal cell types, we molecularly characterized the subtypes of vessel-associated cells, immune cells, and neural crest-derived cells (Figure 6A–C and S4C–F). We then probed for their autocrine and paracrine signaling communication potentials across cell types using CellChat69 (Figure 6D) as well as an alternative receptor-ligand (R-L) interaction analysis as used in Joost et al.70 (Figure S5) with scoring based on a comprehensive hand-curated list of known annotations (Figure S5; Table S3; Methods).
Figure 6. Vessel, immune and neural-crest (NC)-derived subtypes and signaling interactions for their respective establishment in embryonic skin.
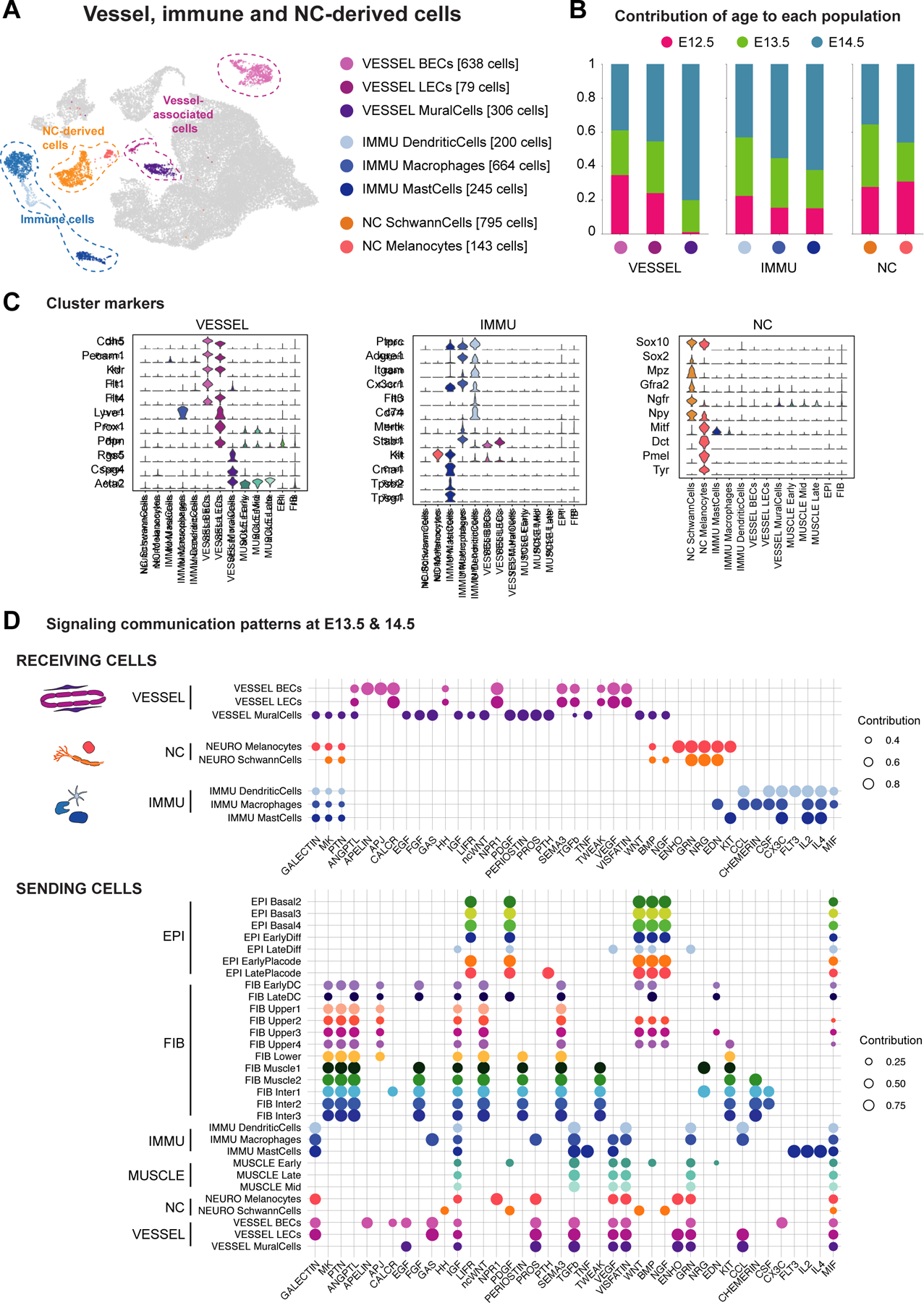
(A) UMAP (from Figure 1B) with subpopulations of vessel-associated cells, immune cells, and neural crestderived cells.
(B) Contribution of each embryonic time point to subpopulations.
(C) Violin plots of marker gene expression.
(D) Dot plot showing enrichment of signaling pathways received by vessel-associated, immune, and/or NC-derived cells.
See also Figures S4 and S5 and Tables S1, S2 and S3.
Vascular remodeling starts around E13, and sprouting vessels form without pericyte coverage in dorsal skin
Vascular endothelial cells (VESSEL BECs) and lymphatic endothelial cells (VESSEL LECs) were present in all analyzed timepoints (Figure 6B), which we confirmed in tissue via pan-endothelial Pecam1 (CD31) staining (Figure 1G–I)71. Cells of the VESSEL BEC population express characteristic genes like Efnb2, Ephb4, Apln, and Aplnr (Figure 6C and S4C), encoding for proteins that regulate arterial-venous alignment through repulsion (via arterial EFNB2 and venous EPHB4) and attraction (via arterial APLN and venous APLNR)72,73. VESSEL LECs express genes, such as Lyve1, Prox1, and Pdpn (Figure 6C), which are involved in the formation of lymphatic endothelium from venous endothelium as early as E12.574–77.
Ongoing angiogenesis78 is reflected by VESSEL BECs and VESSEL LECs from all analyzed timepoints expressing Mmp2 and Dll4 (Figure S4C). Tip cells of sprouting vessels use MMP2 to degrade the vascular basement membrane and DLL4 to prevent neighboring Notch1+/Notch4+ cells from responding to the key angiogenic factor Vegfa (Figure S4C)79–81. VEGF receptor expression allows to distinguish between VESSEL BECs and VESSEL LECs, with Flt1 (VEGFR1) expressed in vascular angiogenesis, Flt4 (VEGFR3) in lymph angiogenesis, and Kdr (VEGFR2) during both processes82 (Figure 6C). A variety of cells provide typical angiogenic factors (i.e., VEGF family, PDGF family, BMPs, Tgfb1, Pgf, ECM components such as Pcolce, Col1a1, Sparc), and molecules implicated in angiogenesis (Semaphorins, Netrins, Neuropillins, Slit proteins) (Figure 6D and S5A)82–88. Notably, keratinocytes (EPI populations) also express high levels of factors such as Pdgfa, Bmp2, or Bmp7 (Figure 6D and S5A), fitting the earlier notion that avascular epidermis can stimulate dermal blood supply89.
Angiogenesis is followed by vascular remodeling, i.e., the recruitment of mural cells90,91 encompassing vascular Smooth Muscle Cells (vSMCs) and pericytes, which presented as one Rgs5+/Cpsg4+/Acta2+ population (Figure 6C)92,93. Endothelial cells (VESSEL BECs) attract the PDGFRB+ EGFR+ mural cells94 by providing respective ligands PDGFB and HBEGF (Figure S4C; Pdgfb and Hbegf). Interestingly, we found that mural cells first appear at E13.5, represented by rare Rgs5+ cells (Figure 1H and 6B). By E14.5 Rgs5+ cells abundantly line small and large vessels (likely representing pericytes and vSMCs, respectively) (Figure 1I). The absence of pericytes at E12.5 is significant because of a longstanding controversy whether sprouting vessels initially form without pericyte coverage95, or if pericytes are present from the beginning and actively assist vessel sprouting96,97. Our data supports the first model for embryonic mouse back skin.
Immature skin is already primed with mast cells, dermal dendritic cells and immature macrophages
Between E12.5 and E14.5, dorsal skin is exclusively populated by myeloid cells, i.e. dermal dendritic cells (IMMU DendriticCells), macrophages (IMMU Macrophages) and mast cells (IMMU MastCells) (Figure 1B–C and 6A–B). Each of these populations express Ptprc (CD45), they are present at all analyzed embryonic time points (Figure 6C), and exclusively locate to the dermis (Figure 1J).
While dendritic cells and macrophages share the expression of many markers such as Adgre1 (F4/80), Itgam, and Cx3cr198,99, they clearly differ in their development and function. Dendritic cell development is critically linked to Flt3 (Figure 6C and 6D)100, they specialize on antigen presenting via MHC-II complexes (e.g. H2-Aa, H2-Ab1, H2-Eb1, Cd74 and Ciita) (Figure 6C and S4D)101,102, and express Ccr2 and Ccr7 enabling their migration to the skin-draining lymph nodes to activate T cells (Figure S4D)103,104. Our dendritic cell population is constituted by classical dendritic cells with the majority being marked by Itgam (CD11b) and a smaller fraction expressing Itgae (CD103) (Figure 6C and S4D)98,105,106. To our knowledge, it has not been reported when dendritic cells start seeding the mouse dermis; our data shows that they are already present at E12.5 (Figure 6B).
Dermal macrophages are tissue-resident cells that are specialized to scavenge damaged cells or invading bacteria107 through expressing receptors like Mertk and Stab1108 (Figure 6C). Macrophage-like cells seed the dermis as early as E10.5109,110. As expected, we detect them at every time point, however they lack MHC-II expression and thus mostly represent immature macrophages (Figure S4D). There is a possibility that our macrophage population also contains precursors of Langerhans Cells as those can derive from yolk sac-derived macrophages and share a number of molecular features (Adgre1+, Ptprc+, Itgam+, Cx3cr1+, Flt3−) (Figure 6C and S4D)111,112.
Mast cells (MCs) are characterized by expression of Kit and serine proteases (e.g. Cma1, Tpsb2, Tpsg1), which are typical for their secretory granules (Figure 6C)113. We find MCs already at E12.5 (Figure 6B), displaying the signature of yolk sac-derived MCs (Grm6+, Cma1+, Prss34+, Smpx+) but still lacking the adult MC signature (Adrb2−, Il1rap−, C2−, Lyz1−) (Figure 6C and S4D). Earlier reports observed sparse dermal mast cells only at around E14.5/E15114,115
Leukocyte recruitment to peripheral tissues is directed by chemokines. Via CellChat and R-L analysis we provide a comprehensive expression pattern overview including the plethora of chemokines involved in leukocyte recruitment (Table S3 Figure 6D and S5C). Strikingly, mostly skin-resident immune cells (and partly mural cells) express those chemokines, suggesting their active involvement in skin homing of more immune cells.
Melanocytes and Schwann cells in early embryonic skin
Our dataset contains two neural crest-derived populations: Schwann cells (NC SchwannCells) and melanocytes (NC Melanocytes), which are sampled at all three time points and marked e.g., by Sox10 expression (Figure 1B–C and 6A–B)116.
Peripheral neurons have entered embryonic skin at the studied time points117,118, which is reflected by the presence of Sox2+, Mpz+, Gfra2+ Schwann cells119 in our dataset (Figure 6C). Likely due to the distant location of neuronal-cell bodies (spinal cord for motor neurons, paravertebral sympathetic ganglia for sympathetic neurons, dorsal root ganglia for sensory neurons)120, we did not detect transcriptomes of neuronal cells. Yet, visualizing the nerve-encasing Schwann cells with Sox10 mRNA and PPARG protein staining121 revealed thick sensory nerve trunks traversing the dermis towards the epidermis at E12.5 (Figure 1K and S4E), seemingly splitting up and spreading out as the embryo continues to grow (Figure 1L). At E14.5, nerves are mainly located underneath the epidermis (sensory neurons) and under the PCM (sensory and motor neurons) (Figure 1L and S4E).
Skin innervation is facilitated by neuronal and non-neuronal cells (e.g., fibroblasts and keratinocytes) expressing neurotrophins which are crucial for neuron growth and maintenance (e.g. Ntf3, Ntf5, Bdnf and Ngf) and a variety of genes that direct the growth cones of developing axons (e.g. Ephrins, Netrins, Slit proteins and Semaphorins) (Figure 6D, S4F, and S5B)122–127. In turn, cutaneous nerves also release a variety of neuropeptides, such as Npy (Neuropeptide Y) (Figure 6C), to increase vascular permeability, support immune cell recruitment, and induce angiogenesis128,129.
The melanocytes in our dataset express the master melanocyte transcription factor Mitf, as well as Dct, Pmel, and Tyr (Figure 6C)130–132. Melanocytes migrate through the dermis around E12.5 and enter the epidermis around E13.5133,134, supporting our finding of gradual migration of Sox10+/Pmel+ cells from the dermis at E12.5 (Figure 1K) to spread throughout the epidermis by E14.5 (Figure 1L), which will eventually persist in adult hair-follicles135. This melanocyte recruitment is supported by fibroblast and epidermal keratinocytes, expressing factors like Edn1, Edn3, Kitl and a (agouti) (Figure 6D, S4F, and S5B)130,136.
Basal epidermal keratinocyte heterogeneity starts already at E12.5
At E12.5, the epidermis consists of keratinocytes that form a morphologically uniform basal layer that is covered by the periderm. Surprisingly, the E12.5 basal cells separated into two transcriptionally distinct populations, which we termed EPI BasalTagln and EPI Basal1 (Figure 7A–C and S6A,D). EPI Basal1 cells have no unique molecular signature within keratinocytes. Their characteristic genes are shared with either EPI BasalTagln from E12.5 skin (e.g., Olfm1, Bfsp2, Acvr2a, Podxl), with EPI Basal2–4 populations from E13.5 and E14.5 skin (e.g., Lmo1, Dcn, Ifitm3) or with all basal populations (e.g., Krt5, Krt14, Krt15, Sostdc1, Vcan) (Figure S6B). In contrast, EPI BasalTagln cells express a unique set of genes including C1qtnf3, Hapln1, Tagln, Cldn11, Amtn, Myl9, Bambi, and many more (Figure 7D–E; Table S1). Remarkably, EPI BasalTagln cells also express smooth muscle genes such as Tagln (SM22a) and Myl9 (Figure 7D), which is highly unexpected for epithelial cells under physiological conditions. Compared to all basal cell clusters, EPI BasalTagln cells express a high number of receptors and ligands, including higher levels of Wnt3a, Wnt4 and Wnt and exclusive Wnt2 expression (Figure 7D and S6E; Table S1), which were commonly believed to be uniformly expressed throughout embryonic epidermis22,137.
Figure 7. Epidermal development from a single basal layer towards a HF-inducing and stratified epithelium.
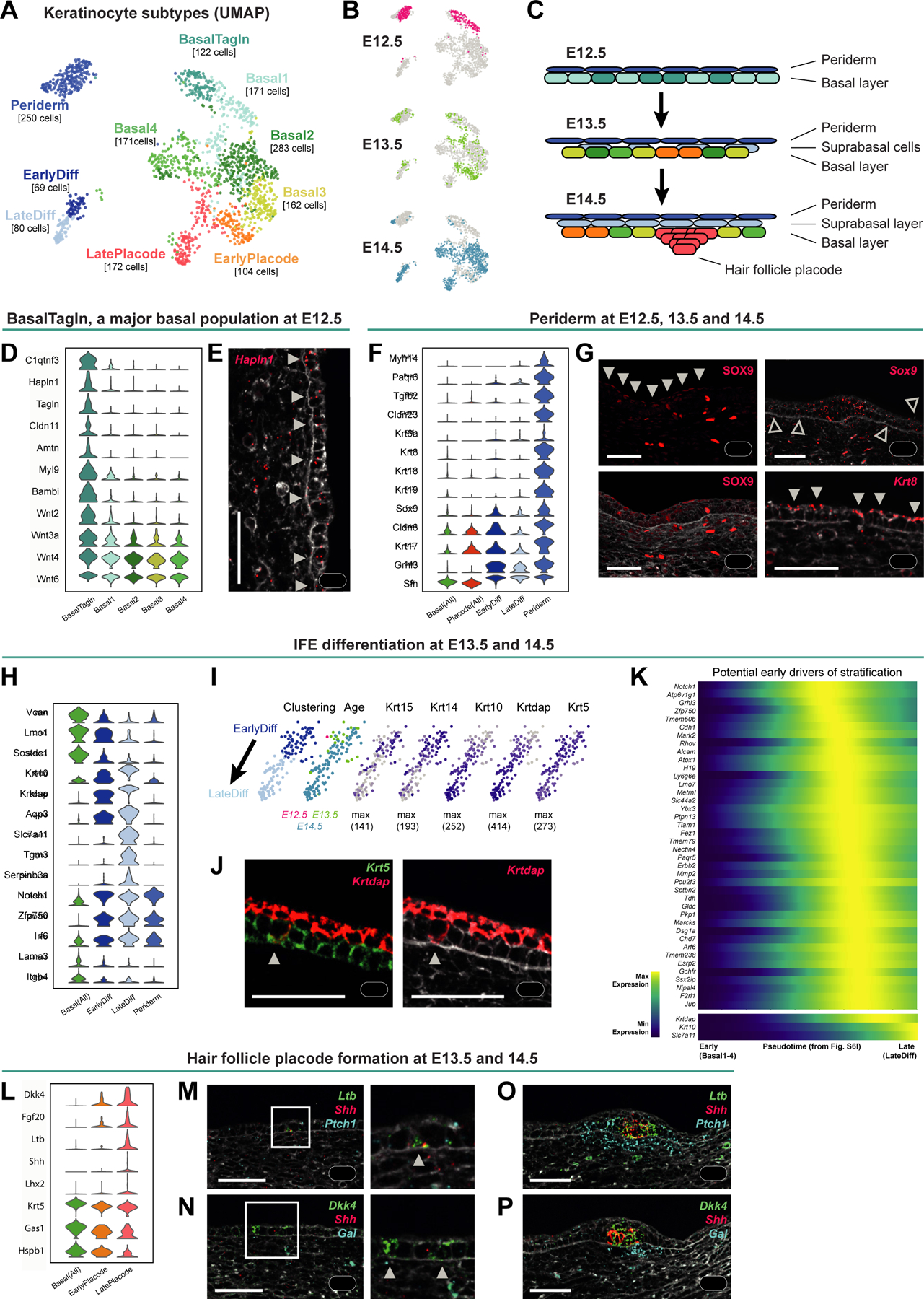
(A-B) UMAP visualization of all keratinocyte subclusters (A) and their embryonic ages (B; n = 360 cells of E12.5, 347 of E13.5, and 877 of E14.5).
(C) Scheme summarizing epidermis development in the analyzed time window, with cell colored according to the cluster colors in (A).
(D) Violin plot of EPI BasalTagln marker gene expression.
(E) Hapln1 mRNA staining revealing expression in basal IFE (arrowheads).
(F) Violin plots of periderm marker gene expression.
(G) SOX9 protein staining (left panels); expression in periderm (filled arrowheads) and hair placode cells (asterisks). Sox9 mRNA staining (upper right); expression in cells within and outside of hair placode (asterisk and empty arrowheads, respectively). Krt8 mRNA staining; expression in periderm cells (filled arrowheads).
(H) Violin plot of differentiation marker gene expression.
(I) UMAPs of differentiating keratinocytes (EPI EarlyDiff and EPI LateDiff from A) colored according to subclustering and embryonic age (left two panels) or expression of basal and suprabasal marker genes (right panels).
(J) Krt5 and Krtdap mRNA staining reveals a representative basal cell with a differentiation signature (arrowhead).
(K) Heatmap of potential early drivers of stratification along the pseudotime from EPI Basal1–4 to EPI EarlyDiff and EPI LateDiff cells from E14.5 (Figure S6I). Krtdap, Krt10, and Slc7a11 are plotted for comparison.
(L) Violin plot of hair placode marker gene expression.
(M-P) Ltb, Shh and Ptch1 (M, O) or Dkk4, Shh, and Gal (N, P) mRNA staining at E13.5 (M, N) and E14.5 (O, P), showing early placode cells (M-N; arrowheads) and mature hair placodes (marked by Ltb, Shh, and Dkk4) as well as dermal condensates (marked by Ptch1 and Gal).
(E, G, J, M-P) Images originate from larger tile scans (n = 3 mice). Scale bars: 50μm.
See also Figures S6 and S7 and Tables S1 and S2.
Notably, CellChat analysis predicts that both basal populations are equally engaged in auto- and paracrine cell-type interactions (Figure S6F–J), leaving the role of these two transcriptionally distinct populations entirely open. Based on gene expression and their position in dimensionality-reduced space it seems more likely that EPI Basal1 cells are the precursors for the general interfollicular epidermis (IFE) at E13.5 and E14.5 (Figure 7A and S6A). In comparison to E12.5, basal IFE at E13.5 and E14.5 appears transcriptionally rather uniform. The EPI Basal2–4 populations do not display unique marker genes and overall only show minor expression differences (Figure S6B; Table S1) and fewer specific receptors or ligands (Figure S6E).
The periderm matures and exhibits a signaling-rich molecular signature
Periderm is a curious specialization of embryonic epidermis. It is a layer composed of squamous cells that cover the epidermis and its presence is crucial for preventing pathological epithelial adhesions within the embryo138. Periderm cells start covering dorsal epidermis around E10 (delaminating from the basement membrane to cover basal keratinocytes) and are shed around E17/18 when the cornified layer forms139–141. Although the existence of this layer has long been known, its molecular characterization has remained incomplete even in the era of scRNA-seq (see Discussion). In our data, we robustly identified periderm cells (‘EPI Periderm’) at all three time points (Figure 7A and S6A), likely due to the large cell proportion relative to all epithelial cells at E12.5 (Figure 7B and S6D). We identified a number of periderm markers including well-known (e.g. Cldn6/23, Krt6/8/17/18/19, Grhl3, Sfn) and additional ones (e.g. Myh14, Paqr6, Tgfb2, Sox9)138,141–144, of which Cldn6, Krt17, Grhl3, Sfn and Sox9 are not uniquely expressed in EPI Periderm among EPI populations (Figure 7F; Table S1). Interestingly, periderm cells are the unique receivers of GRN signaling and constitute a source for TGFb signalling which can be received by epidermis-near E12.5 fibroblasts (Figure 7G). Interestingly, Sox9 and Krt8, known for their role in hair follicle formation and as a marker of the immature epidermis, respectively, show their highest mRNA expression in periderm cells (Figure 7F–G). Additionally, we observed a basal (placode periphery) and suprabasal (IFE associated) Sox9 expression pattern as reported recently (Figure 7G)145.
Cells within the periderm still divide (Figure S6C)138. In addition, we found that the periderm undergoes a molecular maturation characterized by increased expression of known IFE differentiation markers (e.g., Krtdap, Lgals3, Dkkl1), and multiple genes that have not been linked to epidermal differentiation, such as Foxq1, Krt4, Lingo2, Mal, Pllp, Prss27, and Tchh (Figure S7A). Surprisingly, within keratinocyte clusters, Periderm cells express the highest number of receptors and ligands (Figure S6E), including those facilitating e.g., Ephrin signaling, Notch signaling and Tgfb signaling (Table S1).
Deconstructing early epidermal stratification
Differentiating keratinocytes separate into an early differentiating group (EPI EarlyDiff) marked by co-expression of basal (e.g., Krt15, Krt14, Vcan, Lmo1, Sostdc1) as well as suprabasal genes (e.g., Krt10, Krtdap, Aqp3), and a more mature differentiating population (EPI LateDiff) which has gradually lost basal gene expression and further increased differentiation-related gene expression (e.g., Slc7a11, Tgm3, Serpinb3a, Lor) (Figure 7H–I). In line with previous reports of differentiation initiating at E13146,147, we detect the first differentiating EPI EarlyDiff cells at E13.5, and EPI LateDiff at E14.5 (Figure S6D). In situ staining further revealed that some rare Krt5+ basal layer cells already start upregulating Krt10 and Krtdap even though they have not delaminated yet (Figure 7J)148. Interestingly, even though Keratin 5 and 14 usually co-polymerize, in suprabasal cells of embryonic skin Krt14 is strictly downregulated while Krt5 expression persists, and in the hair placode it’s vice versa (Figure 7I and S7B–C).
As the early signals that make a basal cell commit to differentiation are not fully resolved in embryonic skin, we utilized our dataset to identify potential drivers. After performing rigorous cell cycle corrections (this was the dominating factor in initial velocity analysis) we obtained a clear differentiation pseudotime trajectory from EPI Basal2/4 cells toward EPI EarlyDiff/LateDiff cells (Figure S7D) and early pseudotime-dependent genes (Figure 7K). Notch1, a known commitment switch in epidermal differentiation149,150, Cdh1 (E-CAD), which is responsible for altered adhesion properties that allow keratinocytes to differentiate151, and Grhl3, which facilitates epidermal stem cell differentiation152 were among the top hits in our list suggesting that our gene list likely contains additional (not yet functionally tested) differentiation drivers. The upregulation of these genes appears to be transient or at least most pronounced in early differentiation supporting a potential switch-like function (Figure 7K and S7D).
Hair placodes engage in the establishment of blood vessels, nerves, and immune environment
The epithelial counterpart to the dermal condensate, which is necessary for hair follicle formation, is the hair follicle placode. While placodes became morphologically first visible at E14.5 (Figure S1H–I), cells with transcriptional signs of a placode fate were already detected at E13.5 (EPI EarlyPlacode and EPI LatePlacode) (Figure 7A–C and S6D). EPI EarlyPlacode cells express the placode markers Fgf20 and Dkk4, but still lack Shh and Lhx2153,154 (Figure 7L). In situ, they can be captured at E13.5 by Dkk4 and Ltb staining (Figure 7M–N). Shh was detected centrally within the more mature placode (as shown in145), while other markers such as Ltb showed a slightly broader expression pattern (Figure 7O–P and S7E). Interestingly, placode cells downregulated only a handful of genes (e.g., Gas1, Krt5, and Hspb1) but upregulated numerous, suggesting that placode commitment is rather determined by the gain than the loss of expression (Figure 7L). While reporter mice and in situ mRNA stainings have long revealed that placode patterning begins prior to E14.527,155–157, previous scRNA-seq studies of embryonic skin did not reveal those E13.5 cells with early placode markers likely due to the choice of a different analysis strategy10.
Finally, our receptor-ligand analysis (Figure 6D and S5A–C) suggests that the nascent placode and dermal condensate cells immediately engage in reciprocal interactions with crucial cell types for blood supply (Bmp2, Bmp7, and Mfge8), innervation (Bdnf, Nrtn, Ntf3, and Edn3), and immune support (Tnf and Ltb)158–161(Figure S7F).
DISCUSSION
The most insightful transcriptional investigations of early embryonic skin to date have relied on known markers and averaged transcriptomes (bulk RNA-seq of FACS-sorted populations)162 or focused their scRNA-seq analysis on specific cell types/processes such as the molecular origin of IFE cells163, the cellular origin of hair follicle stem cells145, or placode- or dermal condensate-fate specification9,10,164.
The work at hand leveraged scRNA-seq analysis of randomly sampled cells at E12.5, E13.5, and E14.5 to paint a holistic picture of early skin development. Through comprehensive computational analysis of all sampled cells, cell-type localization in situ, and in vivo cell-fate mapping, we answered major outstanding questions in mouse skin development and made unexpected discoveries. When and where does skin begin? How heterogeneous are fibroblasts prior to dermal condensate formation? Is the periderm merely a signaling-inert protective layer to be shed at birth?
When and where does skin begin – setting detailed anatomical and molecular landmarks
Until now, E12.5 dermis and underlying non-skin-associated cells were perceived as a seemingly homogenous tissue covering the area between the vertebrae and epidermis. Similarly, little was known about dermal tissue architecture and cell type diversity at E13.5 and E14.5. Thus, it was critical to unbiasedly sample the skin and the underlying tissue at full thickness from E12.5–14.5 (see Methods) to establish anatomical and molecular tissue landmarks of the skin and the underlying tissue.
One of the most important landmarks to define cell populations as skin-associated in mouse is the PCM. While it is well-known that the PCM originates from the E9.5 Pax7+ dermomyotome165–167, its histological emergence had not been documented. By combining RNA- and protein-staining for early and mature muscle cells, and muscle-associated fibroblasts defined in this study, we revealed the emergence of the PCM (Figure 5B–C). The PCM as well as other developing muscle layers were used as landmarks to place our scRNA-seq subpopulations. Altogether, this extensive back-mapping effort generated a detailed molecular tissue guide that complements previous findings and accelerates the interpretation of future findings.
Fibroblast heterogeneity and the emergence of papillary and reticular dermis
It has been the accepted view that dermal fibroblasts constitute a ‘uniform cell type’ until E16.5, when they finally commit to two different lineages generating upper papillary and lower reticular dermis13. Our data reveal that molecular and functional diversity of fibroblasts is already established at E13.5, with further specification at E14.5, when the dermal condensate forms (Figure 3). We also find clear transcriptional heterogeneity in E12.5 dermis (Figure 2), pointing at a fate bias towards distinct fibroblast subtypes. To what degree these early lineages remain plastic or are already fate-restricted under homeostatic conditions remains to be determined.
Driskell et al. also established a molecular distinction of fibroblasts into papillary dermis (DPP4+/DLK1−/LY6A−), reticular dermis (DPP4−/DLK1+/LY6A+) and hypodermis (DPP4−/DLK1−/LY6A+) starting from late embryogenesis. They detected DLK1 protein expression throughout the dermis until E16.5, while lineage-specific DPP4 (CD26) and LY6A emerged around E16.513. We observed Dlk1 expression throughout the dermis (see online tool) and detected cells with a Ly6a+/Dpp4+ double signature in the FIB Inter population starting from E13.5 (Figure S3E), raising the question if there is a relationship between our Dpp4+/Ly6a+ FIB Inter cells and the papillary and reticular dermis. As we observe FIB Inter cells at a time when FIB Upper and FIB Lower cells (the tentative precursors of papillary and reticular dermis; Dpp4−/Ly6a+) have already been established, it is likely that FIB Upper and FIB Lower cells develop in parallel to FIB Inter cells.
Remarkably, the early existence of spatially defined fibroblast layers resembles the dermal structure of healing wounds. Similar to the developing skin, where spatial layering of fibroblasts is one of the earliest morphogenic events and intriguingly precedes morphologically definable development events, such as hair follicle morphogenesis and dermal adipose morphogenesis, also the healing wound shows spatial segregation of transcriptionally distinct fibroblasts even days before de novo hair follicles are established in regenerating epidermis13,168–172.
Revisiting early progenitors of dermal and subcutaneous white adipose tissue
In 2013, Wojciechowicz et al. postulated a possible presence of pre-adipocytes already at E14.5, which our scRNA-seq data clearly supports54. Cells within the FIB Inter2/3 populations increasingly express typical adipogenic genes (e.g., Pparg and Cebpa), suggesting the presence of adipocyte precursors (Figure 3I and S3G). In situ staining for Pparg mRNA and protein in E14.5 skin (Figure S3H) revealed Pparg+ cells within the subcutanous interstitium or just above the PCM, future sites for the subcutanous white adipose tissue (SWAT) and the dermal white adipose tissue (DWAT), respectively173. Notably, our FIB Inter2/3 cells match recent descriptions for adipose mesenchymal progenitors (Dpp4+/Anxa3+/Wnt2+) in human as well as murine skin32, and Dpp4+/Ly6a+/Cd55+ adipose stem cells53 (Figure S3E–F).
Given this concurrence with the literature, it was very surprising that we did not observe any Gata6-Tom traced adipocytes. As we started tracing at E13.5, when the SWAT and DWAT are not yet separated by the PCM, we in principle should have found Tom-traced cells in both compartments, or at least in one of them. However, we did not find traced DWAT cells and due to technical limitations, SWAT was lost when harvesting postnatal skin. This leaves us with three open possibilities: i) FIB Inter cells do not represent adipocyte precursors at all, which is unlikely based on their expression of adipogenic genes, ii) tracing at E13.5 is not efficient enough to label the adipose progenitors, or iii) DWAT and SWAT originate from independent precursors, where FIB Inter cells only contribute to SWAT formation. This is supported by their locations on opposite sides of the developing PCM and by the current view that DWAT is morphologically and developmentally distinct from SWAT54,173. As the latter view is derived from experiments performed from E14.5 onward, the earliest determination of a fibroblast subset towards generating DWAT and/or SWAT tissue remains an interesting route to be explored.
Unexpected heterogeneity in E12.5 epidermis and a surprisingly signaling-rich periderm
We identify significant heterogeneity in E12.5 epidermis, that to date had been considered a uniform epithelial sheet. The identified distinct EPI BasalTagln population could have a possible role in transient signaling during skin development (e.g. to activate the upper dermis via Wnts)174–176 or it could be a source for the periderm as indicated by a shared transcriptional signature (Table S1), but its exact function remains elusive. In comparison, IFE basal cells at E13.5 and E14.5 were transcriptionally very similar. Subclustering of Basal2–4 likely represents cell cycle influences, rather than populations with distinct behaviors or functions. Interestingly however, we observed rare differentiating basal cells (Figure 7J), reminiscent of delaminating K10+ cells in adult mouse skin148. Thus, it is tempting to speculate that epidermal stratification in embryonic skin may be fueled by two coexisting basal-cell behaviors: i) delamination triggered by basal cell crowding as the predominant mechanism147,151, and ii) delamination through gradual differentiation, which is the main mechanism in postnatal and adult skin148,152.
The existence of periderm cells in embryonic skin has long been known139. However, due to lower periderm cell numbers in previous scRNA-seq studies10,11,163, these cells were likely hidden within other keratinocyte clusters, such as differentiated IFE cells, due to periderm cells expressing typical IFE differentiation genes like Grhl3 and Zfp750 (Figure 7F,H). Notably, transcriptional characterization of the periderm revealed that it is highly signalingprone (Figure S6E) which raises the possibility of previously unrecognized ‘non-canonical’ periderm functions in embryonic skin development.
Limitations of the study
In this work we present all identified cell populations (cell types, subtypes, and states) among all randomly sampled cells from embryonic skin. At first sight an overwhelming amount of information, the analysis and presentation of all cell populations within one study advances our understanding from studying individual cell types (in isolation) to their communal functions at the tissue level – revealing insights that cell type-focused studies would not be able to uncover. However, given the vast amount of data, we had to focus our analysis on major outstanding questions such as the emergence of fibroblast and keratinocyte heterogeneity or the development of the PCM. Therefore, some cell types lack in-depth analysis and the current data interpretations may represent purely data-driven suggestions rather than final conclusions, and require further exploration in future studies.
STAR Methods
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and data should be directed to and will be fulfilled by the lead contact Maria Kasper (maria.kasper@ki.se).
Materials availability
This study did not generate new unique reagents.
Data and code availability
Single-cell RNA-seq data have been deposited on ArrayExpress under the accession number E-MTAB-11920. The original code has been deposited on GitHub (https://github.com/kasperlab; DOI for code release at publication: https://doi.org/10.5281/zenodo.8152645), and input files for the analysis pipelines and the annotated and analyzed sequencing data have been deposited at Zenodo (https://doi.org/10.5281/zenodo.7805311). The raw microscopy data that support the findings of this study are available from the lead contact upon reasonable request. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.
EXPERIMENTAL MODEL AND STUDY PARTICIPANT DETAILS
Mouse work
The study was performed on wild-type C57BL/6J mouse embryos (mix of males and females - gender was only determined in retrospect from the sequencing data (Figure S1E)). Timed matings to obtain embryos of specific embryonic ages were set up in the evenings and the next morning was defined as E0.5. Pregnancy after timed matings was determined by comparing weight difference between the start of the mating and 10 days after. Pregnant moms were sacrificed by cervical dislocation when embryos reached the embryonic age of 12.5, 13.5, or 14.5 days, respectively, and embryos were processed for cell isolation or paraffin-embedding. Lineage-tracing experiments were performed by crossing previously described Gata6-EGFP-CreERT2 177, Ebf2EGFP-CreERT2178 and R26-tdTomato knock-in strains185 (hereafter Gata6-Tom). Gata6-Tom mice received i.p. injection of 2mg tamoxifen (in corn oil at a concentration of 20mg/ml) at embryonic day 13.5. Uninjected mice were used as leakiness control. Tissues were sampled either 2 days after induction of lineage tracing (i.e., E15.5) or postnatally (postnatal day 5). Ebf2/Tom mice were i.p. injected with 2mg tamoxifen (in corn oil at a concentration of 20mg/ml) at E14.5 and tissues were sampled either 2 days after induction of lineage tracing or at E18.5.
FELASA recommendations for harmonized health monitoring were followed. The mice were fed ad libitum and handled and housed under standard conditions. All mouse work (except Gata6-lineage tracings) was performed in the animal facility of Karolinska University Hospital Huddinge and in accordance with Swedish legislation and approved by the Linköping Animal Ethics Committee. Gata6-lineage tracings were performed in the animal facility of the Molecular Biotechnology Center at the University of Turin and in accordance with Italian legislation and approved by the local Animal Ethics Committee and the Italian Ministry of Health.
METHOD DETAILS
Replicates
Sequencing was performed on five embryos per embryonic time point. These five embryos originated from two litters and were sampled on two different days. All 15 samples were processed and sequenced individually and can thus serve as true biological replicates (Table S2).
Each individual staining was performed on skin samples from at least 3 different embryos per embryonic age.
Tissue embedding
Whole embryos and postnatal skin tissue were formaldehyde-fixed in 4% PFA for 24h at room temperature and subsequently processed for FFPE sections (4um thickness). When sectioning whole embryos, tissue sections were collected close to the dorsal midline.
Fluorescent in situ hybridization (FISH)
For independent validation and mapping of cell populations, single-molecule FISH was performed using the RNAscope Multiplex Fluorescent Detection Kit v2 according to manufacturer’s instructions using TSA with Cy3, Cy5, and/or Fluorescein on FFPE sections of the embryos. The used probes are listed in the Key Resources Table. All sections were counterstained with either WGA-405 (1:50), WGA-488 (1:50), WGA-647 (1:50), DAPI (1:500), TO-PRO3 (1:1000) or combinations of those.
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Rabbit polyclonal anti-ACTC1 | Thermo Fisher Scientific | Cat# PA5–21396; RRID: AB_11152296 |
| Rat monoclonal anti-CD45 (clone 30-F11) | Thermo Fisher Scientific | Cat# 14–0451-82; RRID: AB_467251 |
| Rabbit polyclonal anti-KRT5 | US Biological | Cat# C9097–37A2; RRID: AB_2134158 |
| Goat polyclonal anti-GATA6 | R and D Systems | Cat# AF1700; RRID: AB_2108901 |
| Rabbit monoclonal anti-PPARG | Thermo Fisher Scientific | Cat# MA5–14889; RRID: AB_10985650 |
| Goat polyclonal anti-PLP1 | Abcam | Cat# ab61682; RRID: AB_944751 |
| Goat polyclonal anti-PDGFRA | R and D Systems | Cat# AF1062; RRID: AB_2236897 |
| Chemicals, Peptides, and Recombinant proteins | ||
| DAPI | Invitrogen | Cat# D1306; RRID: AB_2629482 |
| TO-PRO-3 | Invitrogen | Cat# T3605 |
| WGA (CF405M conjugate) | Biotium | Cat# 29028 |
| WGA (AF488 conjugate) | Invitrogen | Cat# W11261 |
| WGA (AF647 conjugate) | Invitrogen | Cat# W32466 |
| HBSS | Sigma | Cat# H9394 |
| PBS | Sigma | Cat# D8537 |
| BSA | Sigma | Cat# A3311 |
| Dispase II | Gibco | Cat# 17105041 |
| DNase I | Roche | Cat# 10104159001 |
| Collagenase IA | Sigma | Cat# C2674 |
| Tamoxifen | Sigma | Cat# T5648 |
| Corn Oil | Sigma | Cat# C8267 |
| DIVA Decloaker | Biocare Medical | Cat# DV2004MX |
| Critical Commercial Assays | ||
| Chromium Single Cell 3’ kit v2 | 10X Genomics | Cat# PN-120237 |
| RNAscope Multiplex Fluorescent Reagent Kit v2 | ACDBio/Bio-Techne | Cat# 323100 |
| TSA Cy3, Cy5, TMR, Fluorescein Evaluation Kit | PerkinElmer | Cat# NEL760001KT |
| Deposited Data | ||
| Single-cell RNA-seq data | ArrayExpress | E-MTAB-11920 |
| Input files for analysis code and critical output files (cluster assignment, umap coordinates, h5ad files) | Zenodo | https://doi.org/10.5281/zenodo.7805311 |
| Online tool for visualization of single-cell data | Kasper Lab Website | http://kasperlab.org/tools/embryonicskin |
| Experimental Models: Organisms/Strains | ||
| Mouse: C57BL/6J | Charles River | JAX: 000664; RRID: IMSR JAX 000664 |
| Mouse: R26-tdTomato | Jackson Laboratory | JAX: 007908; RRID: IMRS JAX 007908 |
| Mouse: Gata6-EGFP-CreERT2 | Donati et al.177 | N/A |
| Mouse: Ebf2-EGFP-CreERT2 | Qian et al.178 | N/A |
| Oligonucleotides | ||
| See Table S4 for all oligonucleotides used in this study. | ||
| Software and Algorithms | ||
| Custom scripts and computational analysis workflow | Kasper Lab GitHub / Zenodo | https://github.com/kasperlab/Jacob_et_al_2023_Developmental_Cell (release v1.0; corresponds to https://doi.org/10.5281/zenodo.8152645) |
| Cell Ranger | 10X Genomics | https://github.com/10XGenomics/cellranger (release v2.0.0) |
| Seurat | Stuart et al.179 | https://github.com/satijalab/seurat (release v3.1.1) |
| Velocyto | La Manno et al.15 | https://github.com/velocyto-team/velocyto.py (release v0.17.17) |
| scVelo | Bergen et al.180 | https://github.com/theislab/scvelo (release v0.2.1) |
| Scanpy | Wolf et al.181 | https://github.com/theislab/scanpy (release v1.6.0) |
| Bbknn | Polánski et al.182 | https://github.com/Teichlab/bbknn (release v1.3.9) |
| CellRank | Lange et al.183 | https://github.com/theislab/cellrank (release v1.1.0 for epidermal cells & release v.1.5.1 for fibroblasts) |
| CellChat | Jin et al.69 | https://github.com/sqjin/CellChat (release v1.5.0) |
| Fiji | Schindelin et al.184 | https://fiji.sc |
Immunofluorescence (IF)
Immunofluorescence was performed either alone or after completed RNAscope staining. Combined with RNAscope, sections were washed in TBST once and then blocked and stained as in regular IF stainings. For IF without RNAscope, antigen retrieval was performed using DIVA Decloaker. The following antibody concentrations were used: ACTC1 (1:500), CD45 (1:200), KRT5 (1:50), GATA6 (1:25), PPARG (1:100), PLP1 (1:1000), and RFP (endogenous tdTomato is lost during FFPE processing, 1:200). All sections were counterstained with either WGA-405nm (1:50), WGA-488nm (1:50), WGA-647nm (1:50), DAPI (1:500), TO-PRO3 (1:1000) or combinations of those.
Imaging and image analysis
Images were acquired on a Nikon A1R spinning disk confocal as tiled images (10%–15% overlap) and stitched by NIS Elements software. Subsequently, all images were processed in a uniform way (maximum intensity projection, background removal with the “subtract background” plug-in, brightness adjustment, pseudo-colouring) using Fiji184.
Cell isolation
Dorsal skin of embryos (Figure S1B) was dissected with the help of fine dissection tools and dissected skins were incubated in Dispase II (2mg/ml), Collagenase IA (0.2%), and DNAse I (20U/ul) in PBS for 40 minutes at 37°C in ultra-low attachment plates (Corning Costar) on an orbital shaker. The obtained cell suspension was passed through a 40 mm cell strainer. The flow-through was spun down, and subsequently resuspended in PBS + 0.04% BSA. Samples were transported to core facility in PBS + 0.04% BSA in Eppendorf tubes that had been coated with PBS + 20% BSA overnight. Viability of the cell suspension was determined using trypan blue on an EVE automatic cell counter.
Of note, when peeling off dorsal skin tissue there were no means to technically prevent co-isolation of cells from the tissue layers underlying skin (cells from the deeper muscle layers, interstitial cells, and/or chondrocytes) Biologically, sampling the entire embryonic outer layer (skin and underlying tissue) was important as those embryonic timepoints are/were ill-defined in terms of what can be considered skin tissue.
Library preparation, sequencing and processing of sequencing data
Single-cell cDNA libraries were prepared using the 10X Genomics Chromium Single Cell 3’ kit v2 according to the manufacturer’s instruction. Libraries were sequenced on the HiSeq2500 system (Illumina). Raw sequencing data was processed using the 10X Genomics Cell Ranger package and the mm10 reference genome.
QUANTIFICATION AND STATISTICAL ANALYSIS
Data analysis
Analysis workflow
All downstream data analysis was performed using a mix of custom scripts and published analysis packages as described below and in Figure S1F, utilizing a mix of R packages (most importantly Seurat) as well as Python packages (most importantly Scanpy and scVelo)179–181.
Major decisions on analytical approaches will be presented below, while we refer to the pipelines that will be deposited on GitHub (https://github.com/kasperlab) for any questions regarding details such as chosen parameters.
Quality control and pre-processing
Cell-filtering was performed by sample and was based on the following criteria: (a) remove cells with <200 genes/cell, (b) remove cells with low diversity index, i.e. Shannon and inverse Simpson index (this removes red blood cells, that are naturally expressing only a small variety of genes), and (c) remove cells that are simultaneously in the lowest 0.05% quantile for genes/cell (nFeature) and for reads/cell (nUMI) and that have a contribution of mitochondrial genes of <1% or >10%. By using these combinatorial criteria, it was ensured that cells would not be excluded just because they have e.g. a lower respiratory rate (i.e., low mitochondrial percentage only).
Subsequently, all 15 samples were combined into one full dataset and filtered once more on genes being expressed in at least 5 cells. Ribosomal genes (Rps and Rpl gene families), haemoglobin genes (Hba and Hbb gene families), as well as mitochondrial genes (mt gene family) were removed, as they interfered with the identification of meaningful marker genes. Log-normalization was performed using Seurat’s NormalizeData function.
Determining sex of embryos
As it was very challenging to determine the sex of the embryos during sampling (due to early developmental stage and the need to process samples quickly for sequencing), litter mates were randomly chosen for sequencing and their sex was determined in retrospect from the scRNA-seq data based on the percentage of reads coming from the X chromosome and the Y chromosome (Figure S1E). This data revealed the gender identity for each of the embryos (3 females/2 males for E12.5; 1 female/4 males for E13.5; 1 female/4 males for E14.5).
Removal of cell doublets and low-quality cells
During analysis, two small groups of doublets (keratinocyte-fibroblast doublets that clustered with keratinocytes and pericyte-fibroblast doublets that clustered with fibroblasts) were encountered as well as some low-quality keratinocytes that survived global quality control (low nFeature, low nUMI, low perc.mito). Those cells were removed, and analysis was re-run without them.
Furthermore, one cluster was identified during first-level clustering which very likely corresponds to neuronal cells (sensory neurons defined by e.g. Neurod1 and Pou4f1 as well as sympathetic neurons defined by e.g. Stmn2 and Nefm). While the signature was rather clean, the cell population originated only from a single E13.5 embryo and thus was not reproducible and most likely the result of some tissue sampling issue. Hence, the cluster was removed. Please refer to the results section, for a discussion of why neuronal transcriptomes would not be expected in this dataset.
Removing effect of confounding factors
To counteract a slight batch effect (i.e., slightly differing characteristics such as higher percentage of histone reads and pseudogene reads) linked to one of the sampling days, linear regression was performed using Seurat’s ScaleData function – a rather mild measure for data integration. Regression was performed for sampling date, as well as gender, percentage of mitochondrial genes, total read counts, and cell cycle scores (S.Score and G2M.Score) as those could also potentially influence dimensionality reduction and clustering while not representing the biological variables of interest.
Prediction of cell cycle stage
Cell cycle stage was predicted using Seurat’s CellCycleScoring function.
Generation of loom files
To allow for running RNA velocity analysis on spliced and unspliced mRNAs, we generated loom files using Velocyto’s run10x function with default parameters and using the mm10 reference genome.
Feature selection
Feature selection was performed using the mean-dropout-method originally suggested by Andrews and Hemberg186 in our own implementation. The 3000 genes with the highest dropout rate given their mean expression level (across non-zero counts) were chosen to be included in further analysis.
Feature selection was performed separately for the full dataset, fibroblasts, or keratinocytes, respectively, to allow for the detection of more subtle differences within fibroblasts and keratinocytes, respectively, that were hidden in the full dataset where distinct signatures of major cell types dominate the highly variable genes.
Clustering, spatial embedding, and trajectory analysis
Clustering, spatial embedding and trajectory analysis were separately adjusted for each of the three analysed groups (full dataset, fibroblasts, and keratinocytes) as they possessed very dissimilar features. The full dataset contained very distinct cell types, while fibroblasts and keratinocytes constituted a much more homogenous cell population with more gradual expression changes. Also, the biological questions that were of interest differed strongly, so different aspects had to be emphasized and analysis was adjusted accordingly.
Full dataset
Dimensionality reduction was performed in Seurat using PCA with the most highly variable genes as input after initial scaling with Seurat’s NormalizeData function (a scaling factor of 10 000 was chosen as this roughly reflects the median reads/cell among the filtered cells). Subsequently, hierarchical clustering (hclust function) was performed based on PCA-reduced data. While this clustering worked well without any further need for data integration, the downstream dimensionality reduction (UMAP) still showed signs of sampling date-derived batch effects. Thus, a batch-corrected neighbourhood graph from BBKNN182 was used to prevent batch-derived separation in UMAP space. To this aim, the regressed dataset was transferred to Python, principal components were recalculated, BBKNN was run, and dimensionality reduction was performed using Scanpy’s UMAP function, which was then used to display the results of the hierarchical clustering (Figure 1B).
Fibroblasts
Fibroblast batch correction was similarly done in Scanpy using BBKNN for UMAP representation. Next, cell clustering was performed using the Leiden algorithm187, revealing the fibroblast subpopulations for subsequent analysis (Figure S2A). As the dermal condensate (DC) is a structure of great importance to skin development, we decided to further subcluster the DC into FIB EarlyDC and FIB LateDC using the Leiden algorithm.
Next, we imported spliced and unspliced mRNA information (from loom files) for RNA-velocity analysis using scVelo’s velocity function in the stochastic mode on the highly variable genes. The predicted dynamics were then plotted on top of the pre-computed UMAP (Figure S2B).
To further analyse cellular dynamics towards the endpoints, we used CellRank (v1.5.1) pseudotime kernel. As input to the model, we calculated velocity pseudotime with identified root cells in the FIB origin populations and end cells as extreme points based on diffusion maps. Finally, we calculated absorption probabilities for each cell to become any of the identified end points.
Keratinocytes
Dimensionality reduction was performed in Seurat using PCA with the most highly variable genes as input after initial scaling. Subsequently, hierarchical clustering (hclust function) was performed based on UMAP-reduced data. The regressed keratinocyte dataset was then transferred to Python and combined with the loom-file-derived information on spliced and unspliced mRNAs.
To better understand epidermal stratification, a subset of E14.5 cells was studied, as E14.5 is the first embryonic age to capture the full differentiation trajectory. Cells from the EPI Basal1, EPI Basal2, EPI Basal3, EPI Basal4, EPI EarlyDiff, and EPI LateDiff clusters were included in the analysis. Cells related to placode, periderm, or the EPI BasalTagln cluster were excluded as they could potentially interfere with the differentiation trajectory. The subset was processed as described before (PCA, BBKNN, UMAP) and velocity analysis was performed, which revealed striking dominance of the cell cycle in RNA-velocity predictions (Figure S7D). Thus, cell cycle effects were regressed out in spliced and unspliced mRNA using Scanpy’s regress_out function, which resulted in a striking velocity pattern reflecting epidermal stratification (Figure S7D). Finally, CellRank in combination with diffusion maps and RNA-velocity based pseudotimes was used to find macrostates and generate a refined pseudotime, to calculate lineage drivers, and to fit a GAM model for gene expression analysis183. The top 200 pseudotime-dependent genes were obtained and the 40 genes among them with the earliest peak in pseudotime were displayed (Figure 7K).
Using a similar approach, the differentiation trajectory within the EPI Periderm cluster was analysed. Periderm cells from E14.5 were identified as a terminal state and a periderm maturation trajectory could be modelled. The top 200 pseudotime-dependent genes were obtained and the 13 genes among them with the latest peak in pseudotime were displayed (Figure S7A).
Test for differential expression of genes in cell populations
Marker genes overexpressed in certain cell populations were determined using the Wilcoxon rank sum test in the Seurat implementation. Marker genes were required to be detected in at least 20% of cells in the respective population and to have a (natural) log fold change ≥ 0.25 compared to all other cells. Correction for multiple testing was performed using the Bonferroni method and the threshold for the FDR (false discovery rate) adjusted p-value was set to 0.05 (Table S1).
Receptor-ligand interactions
Receptor-ligand pairing was based on the approach presented by70 and CellChat, respectively. In brief, for the Joost et al. approach receptors and ligands contained in the marker gene list were considered for potential receptor-ligand pairs. For each cluster pair, receptor-ligand interactions were identified by querying a receptor-ligand database. In contrast to70, the curated receptor-ligand databases from188 and189 were combined to obtain an even more complete set of potential interactions. Both databases are based on human data, but we assume that the majority of registered interaction pairs are also valid for homologous genes/proteins in mice. The code was furthermore optimized for run-time and parallel computing.
To test for the enrichment of receptor-ligand pairs between two populations, the observed number of receptor-ligand pairs was compared to the number of pairs obtained from an equally sized randomly sampled pool of receptors and ligands. For each cluster pair, this simulation was repeated 10 000 times and significantly enriched interactions (p ≤ 0.05 for Benjamini-Hochberg-corrected p-values) were combined into Table S3. Each ligand and receptor in this table was manually annotated with reported functions and each pair was manually scored for likely involvement in the development of vessels, nerves and the immune system (Figure S5; Table S3).
Receptor-ligand interactions were identified within and between major cell populations, fibroblast subpopulations, and epidermal subpopulations (any possible combination of them with any possible signalling directionality).
As an alternative approach, CellChat was used to identify communication patterns within and between cell groups. Using CellChat’s identifyOverExpressedGenes, identifyOverExpressedInteractions, computeCommunProb, computeCommunProbPathway and aggregateNet functions, a cell-cell communication network was inferred for cell populations present at early (E12.5) or late (E13.5 and E14.5) time points, respectively. Separate analyses were run for signaling between muscle cells and muscle-associated fibroblast populations (in late populations; Figure 5D), signaling from all cell types to vessel, immune, or neural crest-derived cells respectively (in late populations; Figure 6D–E), and signaling between epidermal and fibroblast populations (in early populations; Fig. Figure S6F–J). Circle plots were generated using CellChat’s netVisual_aggregate or netVisual_circle function, while the netAnalysis_dot function was used to generate the dotplots of incoming and outgoing communication patterns (all detected pathways were included).
Supplementary Material
Table S4. Oligonucleotides, Related to STAR Methods and Key Resources Table.
Table S1. Marker genes for populations and subpopulations, Related to Figures 2, 3, 5, 6 and 7. (A) Marker genes for the major populations. Marker genes had to fulfil the following criteria: (i) Upregulated in the cluster of interest (log fold change ≥ 0.25), (ii) Expressed in at least 20% of cells in the cluster of interest, and (iii) False discovery rate (FDR)-adjusted p-value ≤ 0.05. (B) Marker gene details for the major populations incl. adjusted p-value and log-fold change compared to all other cells. Cutting thresholds are the same as in Table S1A. (C) Marker genes for the fibroblast subpopulations. Cutting thresholds are the same as in Table S1A. (D) Marker gene details for the fibroblast subpopulations incl. adjusted p-value and log-fold change compared to all other fibroblasts. Cutting thresholds are the same as in Table S1A. (E) Marker genes for the epidermis subpopulations. Cutting thresholds are the same as in Table S1A. (F) Marker gene details for the epidermis subpopulations incl. adjusted p-value and log-fold change compared to all other epidermal cells. Cutting thresholds are the same as in Table S1A. (G) All considered transcription factors.
Table S2. Contribution of metadata categories to populations and subpopulations, Related to Figures 2, 3, 5, 6 and 7. (A) Contribution of metadata categories to the major populations. (B) Contribution of metadata categories to the fibroblast subpopulations. (C) Contribution of metadata categories to the epidermal subpopulations. (D) Contribution of metadata categories to the cells excluded from the final dataset.
Table S3. Receptor-Ligand-Pairs between all major, fibroblast and epidermis subclusters, Related to Figures 5, 6, S5, S7 and STAR Methods. (A) Identified receptor-ligand pairs incl. functional annotation. Receptor-ligand pairs with FDR-corrected p-value < 0.05 were included. (B) Scoring of identified receptor-ligand pairs. Cutting threshold is the same as in Table S3A. (C) Analysis approach.
Highlights.
Deconstruction of early skin development by scRNA-seq and in situ RNA staining
Fibroblasts’ diversification towards functional lineages starts already at E12.5
The PCM forms around E13.5 in close association with muscle-supportive fibroblasts
Unexpected epithelial heterogeneity at E12.5 including a signaling-rich periderm
Acknowledgements
We would like to thank Adelheid Elbe-Buerger for valuable scientific discussions; Hong Qian and Lakshmi Sandhow for generously sharing Ebf2-tracing mice; Thibault Bouderlique for sharing his expertise on embryo dissections; Stefania Giacomello (from National Bioinformatics Infrastructure Sweden at SciLifeLab), Simon Joost and Christoph Ziegenhain for advice on computational analysis; Rickard Sandberg and Hao Yuan for help with implementing the online tool; Alexandra Are and Xiaoyan Sun for help with mice and stainings; and Makbule Sagici for embedding and cutting paraffin samples. This work was supported by grants from the Swedish Research Council, Swedish Foundation for Strategic Research, Center for Innovative Medicine (CIMED), Swedish Cancer Society, Karolinska Institutet (StratRegen SFO, Consolidator Grant), and Ragnar Söderberg Foundation to MK, Karolinska Institutet KID funding to TJ and KA, and a Wenner-Gren postdoctoral fellowship to TD. The G.D. laboratory was supported by AIRC (MFAG 2018 ID: 21640) and a Chan Zuckerberg Initiative Grant (DAF2020-217532). M.R. was supported by grants from NIH/NIAMS (R01AR071047, R01AR073259, R01AR077593, R01AR079475) and New York State Department of Health (NYSTEM-C32561GG). P.C. and Å.B. were supported by the Knut and Alice Wallenberg Foundation as part of the National Bioinformatics Infrastructure Sweden at SciLifeLab. All single-cell experiments were performed at the Eukaryotic Single Cell Genomics core facility, SciLifeLab, Stockholm. The authors also acknowledge support from the National Genomics Infrastructure in Stockholm funded by Science for Life Laboratory, the Knut and Alice Wallenberg Foundation and the Swedish Research Council, and SNIC/Uppsala Multidisciplinary Center for Advanced Computational Science for assistance with massively parallel sequencing and access to the UPPMAX computational infrastructure. Imaging was performed at the LCI facility/Nikon Center of Excellence, Karolinska Institutet, supported by grants from the Knut and Alice Wallenberg Foundation, Swedish Research Council, KI infrastructure, Centre for Innovative Medicine and Jonasson center at the Royal Institute of Technology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of Interest
The authors declare no competing interests. Tina Jacob currently works at Schain Research AB, 11151 Stockholm, Sweden. Tim Dalessandri currently works at XNK Therapeutics AB, 14157 Huddinge, Sweden. Paulo Czarnewski currently works at DeepLife, 75014 Paris, France.
References
- 1.De Falco M, Pisano MM, and De Luca A (2014). Embryology and Anatomy of the Skin. In Current Clinical Pathology. Skin cancer, Baldi A, ed. (Springer Science+Business Media; ), pp. 1–15. 10.1007/978-1-4614-7357-2. [DOI] [Google Scholar]
- 2.Biggs LC, and Mikkola ML (2014). Early inductive events in ectodermal appendage morphogenesis. Semin. Cell Dev. Biol 25–26, 11–21. 10.1016/j.semcdb.2014.01.007. [DOI] [PubMed] [Google Scholar]
- 3.Sennett R, and Rendl M (2012). Mesenchymal-epithelial interactions during hair follicle morphogenesis and cycling. Semin. Cell Dev. Biol 23, 917–927. 10.1016/j.semcdb.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Saxena N, Mok KW, and Rendl M (2019). An updated classification of hair follicle morphogenesis. Exp. Dermatol 28, 332–344. 10.1111/exd.13913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schmidt-Ullrich R, and Paus R (2005). Molecular principles of hair follicle induction and morphogenesis. BioEssays 27, 247–261. 10.1002/bies.20184. [DOI] [PubMed] [Google Scholar]
- 6.Fuchs E (2007). Scratching the surface of skin development. Nature 445, 834–842. 10.1038/nature05659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu MS, Borrelli MR, Hong WX, Malhotra S, Cheung ATM, Ransom RC, Rennert RC, Morrison SD, Lorenz HP, and Longaker MT (2018). Embryonic skin development and repair. Organogenesis 14, 46–63. 10.1080/15476278.2017.1421882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sotiropoulou PA, and Blanpain C (2012). Development and homeostasis of the skin epidermis. Cold Spring Harb. Perspect. Biol 4, 1–9. 10.1101/cshperspect.a008383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mok KW, Saxena N, Heitman N, Grisanti L, Srivastava D, Muraro MJ, Jacob T, Sennett R, Wang Z, Su Y, et al. (2019). Dermal Condensate Niche Fate Specification Occurs Prior to Formation and Is Placode Progenitor Dependent. Dev. Cell 48, 32–48.e5. 10.1016/j.devcel.2018.11.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta K, Levinsohn J, Linderman G, Chen D, Sun TY, Dong D, Taketo MM, Bosenberg M, Kluger Y, Choate K, et al. (2019). Single-Cell Analysis Reveals a Hair Follicle Dermal Niche Molecular Differentiation Trajectory that Begins Prior to Morphogenesis. Dev. Cell 48, 17–31.e6. 10.1016/j.devcel.2018.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ge W, Tan SJ, Wang SH, Li L, Sun XF, Shen W, and Wang X (2020). Single-cell transcriptome profiling reveals dermal and epithelial cell fate decisions during embryonic hair follicle development. Theranostics 10, 7581–7598. 10.7150/thno.44306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Biggs LC, Mäkelä OJM, Myllymäki SM, Das Roy R, Närhi K, Pispa J, Mustonen T, and Mikkola ML (2018). Hair follicle dermal condensation forms via FGF20 primed cell cycle exit, cell motility, and aggregation. Elife 7, 1–33. 10.7554/eLife.36468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Driskell RR, Lichtenberger BM, Hoste E, Kretzschmar K, Simons BD, Charalambous M, Ferron SR, Herault Y, Pavlovic G, Ferguson-Smith AC, et al. (2013). Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature 504, 277–281. 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rinkevich Y, Walmsley GG, Hu MS, Maan ZN, Newman AM, Drukker M, Januszyk M, Krampitz GW, Gurtner GC, Lorenz HP, et al. (2015). Identification and isolation of a dermal lineage with intrinsic fibrogenic potential. Science 348. 10.1126/science.aaa2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.La Manno G, Soldatov R, Zeisel A, Braun E, Hochgerner H, Petukhov V, Lidschreiber K, Kastriti ME, Lönnerberg P, Furlan A, et al. (2018). RNA velocity of single cells. Nature 560, 494–498. 10.1038/s41586-018-0414-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Barresi MJF, and Gilbert SF (2020). Developmental Biology 12th Editi. (Oxford University Press; ). [Google Scholar]
- 17.Yamashita S, Miyaki S, Kato Y, Yokoyama S, Sato T, Barrionuevo F, Akiyama H, Scherer G, Takada S, and Asahara H (2012). L-Sox5 and Sox6 proteins enhance chondrogenic miR-140 MicroRNA expression by strengthening dimeric Sox9 activity. J. Biol. Chem 287, 22206–22215. 10.1074/jbc.M112.343194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zuo C, Wang L, Kamalesh RM, Bowen ME, Moore DC, Dooner MS, Reginato AM, Wu Q, Schorl C, Song Y, et al. (2018). SHP2 regulates skeletal cell fate by modifying SOX9 expression and transcriptional activity. Bone Res 6. 10.1038/s41413-018-0013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bartoletti G, Dong C, Umar M, and He F (2020). Pdgfra regulates multipotent cell differentiation towards chondrocytes via inhibiting Wnt9a/beta-catenin pathway during chondrocranial cartilage development. Dev. Biol 466, 36–46. 10.1016/j.ydbio.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bosserhoff AK, and Buettner R (2003). Establishing the protein MIA (melanoma inhibitory activity) as a marker for chondrocyte differentiation. Biomaterials 24, 3229–3234. 10.1016/S0142-9612(03)00184-4. [DOI] [PubMed] [Google Scholar]
- 21.Chen D, Jarrell A, Guo C, Lang R, and Atit R (2012). Dermal beta-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development 139, 1522–1533. 10.1242/dev.076463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Andl T, Reddy ST, Gaddapara T, and Millar SE (2002). WNT signals are required for the initiation of hair follicle development. Dev. Cell 2, 643–653. 10.1016/S1534-5807(02)00167-3. [DOI] [PubMed] [Google Scholar]
- 23.Enshell-Seijffers D, Lindon C, Kashiwagi M, and Morgan B. a. (2010). β-catenin Activity in the Dermal Papilla Regulates Morphogenesis and Regeneration of Hair. Dev. Cell 18, 633–642. 10.1016/j.devcel.2010.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tsai SY, Sennett R, Rezza A, Clavel C, Grisanti L, Zemla R, Najam S, and Rendl M (2014). Wnt/β-catenin signaling in dermal condensates is required for hair follicle formation. Dev. Biol 385, 179–188. 10.1016/j.ydbio.2013.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu J, and Hsu W (2013). Epidermal Wnt Controls Hair Follicle Induction by Orchestrating Dynamic Signaling Crosstalk between the Epidermis and Dermis. J. Invest. Dermatol 133, 890–898. 10.1038/jid.2012.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.DasGupta R, and Fuchs E (1999). Multiple roles for activated LEF/TCF transcription complexes during hair follicle development and differentiation. Development 126, 4557–4568. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Y, Tomann P, Andl T, Gallant NM, Huelsken J, Jerchow B, Birchmeier W, Paus R, Piccolo S, Mikkola ML, et al. (2009). Reciprocal Requirements for EDA/EDAR/NF-κB and Wnt/β-Catenin Signaling Pathways in Hair Follicle Induction. Dev. Cell 17, 49–61. 10.1016/j.devcel.2009.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Song Y, Boncompagni AC, Kim SS, Gochnauer HR, Zhang Y, Loots GG, Wu D, Li Y, Xu M, and Millar SE (2018). Regional Control of Hairless versus Hair-Bearing Skin by Dkk2. Cell Rep. 25, 2981–2991.e3. 10.1016/j.celrep.2018.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Galbiati F, Volonte D, Brown AMC, Weinstein DE, Ben-Ze’ev A, Pestell RG, and Lisantia MP (2000). Caveolin-1 expression inhibits Wnt/β-catenin/Lef-1 signaling by recruting β-catenin to caveolae membrane domains. J. Biol. Chem 275, 23368–23377. 10.1074/jbc.M002020200. [DOI] [PubMed] [Google Scholar]
- 30.Pentinmikko N, Iqbal S, Mana M, Andersson S, Cognetta AB, Suciu RM, Roper J, Luopajärvi K, Markelin E, Gopalakrishnan S, et al. (2019). Notum produced by Paneth cells attenuates regeneration of aged intestinal epithelium. Nature 571, 398–402. 10.1038/s41586-019-1383-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Benias PC, Wells RG, Sackey-Aboagye B, Klavan H, Reidy J, Buonocore D, Miranda M, Kornacki S, Wayne M, Carr-Locke DL, et al. (2018). Structure and distribution of an unrecognized interstitium in human tissues. Sci. Rep 8, 1–8. 10.1038/s41598-018-23062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Merrick D, Sakers A, Irgebay Z, Okada C, Calvert C, Morley MP, Percec I, and Seale P (2019). Identification of a mesenchymal progenitor cell hierarchy in adipose tissue. Science 364. 10.1126/science.aav2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eliazer S, Muncie JM, Christensen J, Sun X, D’Urso RS, Weaver VM, and Brack AS (2019). Wnt4 from the Niche Controls the Mechano-Properties and Quiescent State of Muscle Stem Cells. Cell Stem Cell 25, 654–665.e4. 10.1016/j.stem.2019.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Foulstone EJ, Savage PB, Crown AL, Holly JMP, and Stewart CEH (2003). Role of insulin-like growth factor binding protein-3 (IGFBP-3) in the differentiation of primary human adult skeletal myoblasts. J. Cell. Physiol 195, 70–79. 10.1002/jcp.10227. [DOI] [PubMed] [Google Scholar]
- 35.Murphy MM, Lawson JA, Mathew SJ, Hutcheson DA, and Kardon G (2011). Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637. 10.1242/dev.064162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Light N, and Champion AE (1984). Characterization of muscle epimysium, perimysium and endomysium collagens. Biochem. J 219, 1017–1026. 10.1042/bj2191017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kjær M (2004). Role of Extracellular Matrix in Adaptation of Tendon and Skeletal Muscle to Mechanical Loading. Physiol. Rev 84, 649–698. 10.1152/physrev.00031.2003. [DOI] [PubMed] [Google Scholar]
- 38.Sasse J, von der Mark H, Kühl U, Dessau W, and von der Mark K (1981). Origin of collagen types I, III, and V in cultures of avian skeletal muscle. Dev. Biol 83, 79–89. 10.1016/S0012-1606(81)80010-3. [DOI] [PubMed] [Google Scholar]
- 39.Pace RA, Peat RA, Baker NL, Zamurs L, Mörgelin M, Irving M, Adams NE, Bateman JF, Mowat D, Smith NJC, et al. (2008). Collagen VI glycine mutations: Perturbed assembly and a spectrum of clinical severity. Ann. Neurol 64, 294–303. 10.1002/ana.21439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cescon M, Gattazzo F, Chen P, and Bonaldo P (2015). Collagen VI at a glance. J. Cell Sci 128, 3525–3531. 10.1242/jcs.169748. [DOI] [PubMed] [Google Scholar]
- 41.Purslow PP (2002). The structure and functional significance of variations in the connective tissue within muscle. Comp. Biochem. Physiol. - A Mol. Integr. Physiol 133, 947–966. 10.1016/S1095-6433(02)00141-1. [DOI] [PubMed] [Google Scholar]
- 42.Tidball JG (1991). Force transmission across muscle cell membranes. J. Biomech 24, 43–52. 10.1016/0021-9290(91)90376-X. [DOI] [PubMed] [Google Scholar]
- 43.Gatchalian CL, Schachner M, and Sanes JR (1989). Fibroblasts that proliferate near denervated synaptic sites in skeletal muscle synthesize the adhesive molecules tenascin(J1), N-CAM, fibronectin, and a heparan sulfate proteoglycan. J. Cell Biol 108, 1873–1890. 10.1083/jcb.108.5.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gillies AR, and Lieber RL (2011). Structure and function of the skeletal muscle extracellular matrix. Muscle and Nerve 44, 318–331. 10.1002/mus.22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kühl U, Öcalan M, Timpl R, Mayne R, Hay E, and von der Mark K (1984). Role of muscle fibroblasts in the deposition of type-IV collagen in the basal lamina of myotubes. Differentiation 28, 164–172. 10.1111/j.1432-0436.1984.tb00279.x. [DOI] [PubMed] [Google Scholar]
- 46.Sanderson RD, Fitch JM, Linsenmayer TR, and Mayne R (1986). Fibroblasts promote the formation of a continuous basal lamina during myogenesis in vitro. J. Cell Biol 102, 740–747. 10.1083/jcb.102.3.740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Kolehmainen M, Salopuro T, Schwab US, Kekäläinen J, Kallio P, Laaksonen DE, Pulkkinen L, Lindi VI, Sivenius K, Mager U, et al. (2008). Weight reduction modulates expression of genes involved in extracellular matrix and cell death: The GENOBIN study. Int. J. Obes 32, 292–303. 10.1038/sj.ijo.0803718. [DOI] [PubMed] [Google Scholar]
- 48.Joost S, Annusver K, Jacob T, Sun X, Dalessandri T, Sivan U, Sequeira I, Sandberg R, and Kasper M (2020). The Molecular Anatomy of Mouse Skin during Hair Growth and Rest. Cell Stem Cell 26, 441–457.e7. 10.1016/j.stem.2020.01.012. [DOI] [PubMed] [Google Scholar]
- 49.Freytag SO, Paielli DL, and Gilbert JD (1994). Ectopic expression of the CCAAT/enhancer-binding protein α promotes the adipogenic program in a variety of mouse fibroblastic cells. Genes Dev. 8, 1654–1663. 10.1101/gad.8.14.1654. [DOI] [PubMed] [Google Scholar]
- 50.Tontonoz P, Hu E, Graves RA, Budavari AI, and Spiegelman BM (1994). mPPARg2: tissue-specific regulator of an adipocyte enhancer. Genes Dev. 8, 1223–1234. 10.4324/9781843143215-76. [DOI] [PubMed] [Google Scholar]
- 51.Benjamin M (2009). The fascia of the limbs and back - A review. J. Anat 214, 1–18. 10.1111/j.1469-7580.2008.01011.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Distel RJ, Ro HS, Rosen BS, Groves DL, and Spiegelman BM (1987). Nucleoprotein complexes that regulate gene expression in adipocyte differentiation: direct participation of c-fos. Cell 49, 835–844. 10.1016/0092-8674(87)90621-0. [DOI] [PubMed] [Google Scholar]
- 53.Ferrero R, Rainer P, and Deplancke B (2020). Toward a Consensus View of Mammalian Adipocyte Stem and Progenitor Cell Heterogeneity. Trends Cell Biol. 30, 937–950. 10.1016/j.tcb.2020.09.007. [DOI] [PubMed] [Google Scholar]
- 54.Wojciechowicz K, Gledhill K, Ambler CA, Manning CB, and Jahoda CAB (2013). Development of the Mouse Dermal Adipose Layer Occurs Independently of Subcutaneous Adipose Tissue and Is Marked by Restricted Early Expression of FABP4. PLoS One 8, 1–15. 10.1371/journal.pone.0059811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Seale P, Sabourin LA, Girgis-Gabardo A, Mansouri A, Gruss P, and Rudnicki MA (2000). Pax7 is required for the specification of myogenic satellite cells. Cell 102, 777–786. 10.1016/S0092-8674(00)00066-0. [DOI] [PubMed] [Google Scholar]
- 56.Relaix F, Montarras D, Zaffran S, Gayraud-Morel B, Rocancourt D, Tajbakhsh S, Mansouri A, Cumano A, and Buckingham M (2006). Pax3 and Pax7 have distinct and overlapping functions in adult muscle progenitor cells. J. Cell Biol 172, 91–102. 10.1083/jcb.200508044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zammit PS (2008). All muscle satellite cells are equal, but are some more equal than others? J. Cell Sci 121, 2975–2982. 10.1242/jcs.019661. [DOI] [PubMed] [Google Scholar]
- 58.Zammit PS, Partridge TA, and Yablonka-Reuveni Z (2006). The skeletal muscle satellite cell: The stem cell that came in from the cold. J. Histochem. Cytochem 54, 1177–1191. 10.1369/jhc.6R6995.2006. [DOI] [PubMed] [Google Scholar]
- 59.Grzelkowska-Kowalczyk K (2016). The Importance of Extracellular Matrix in Skeletal Muscle Development and Function. In Composition and Function of the Extracellular Matrix in the Human Body, Travascio F, ed. (IntechOpen; ). [Google Scholar]
- 60.Schiaffino S, and Reggiani C (2011). Fiber types in Mammalian skeletal muscles. Physiol. Rev 91, 1447–1531. 10.1152/physrev.00031.2010. [DOI] [PubMed] [Google Scholar]
- 61.Rubenstein AB, Smith GR, Raue U, Begue G, Minchev K, Ruf-Zamojski F, Nair VD, Wang X, Zhou L, Zaslavsky E, et al. (2020). Single-cell transcriptional profiles in human skeletal muscle. Sci. Rep 10, 1–15. 10.1038/s41598-019-57110-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Pisconti A, Bernet JD, and Olwin BB (2012). Syndecans in skeletal muscle development, regeneration and homeostasis. Muscles. Ligaments Tendons J 2, 1–9. [PMC free article] [PubMed] [Google Scholar]
- 63.Hayward LJ, and Schwartz RJ (1986). Sequential expression of chicken actin genes during myogenesis. J. Cell Biol 102, 1485–1493. 10.1083/jcb.102.4.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Holstege G, and Blok BF (1989). Descending pathways to the cutaneus trunci muscle motoneuronal cell group in the cat. J. Neurophysiol 62, 1260–1269. 10.1152/jn.1989.62.6.1260. [DOI] [PubMed] [Google Scholar]
- 65.Theriault E, and Diamond J (1988). Nociceptive cutaneous stimuli evoke localized contractions in a skeletal muscle. J. Neurophysiol 60, 446–462. 10.1152/jn.1988.60.2.446. [DOI] [PubMed] [Google Scholar]
- 66.Petruska JC, Barker DF, Garraway SM, Trainer R, Fransen JW, Seidman PA, Soto RG, Mendell LM, and Johnson RD (2014). Organization of sensory input to the nociceptive-specific cutaneous trunk muscle reflex in rat, an effective experimental system for examining nociception and plasticity. J Comp Neurol 522, 1048–1071. 10.1002/cne.23461.Organization. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan-Sindhunata MB, Mathijssen IB, Smit M, Baas F, De Vries JI, Van Der Voorn JP, Kluijt I, Hagen MA, Blom EW, Sistermans E, et al. (2015). Identification of a Dutch founder mutation in MUSK causing fetal akinesia deformation sequence. Eur. J. Hum. Genet 23, 1151–1157. 10.1038/ejhg.2014.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Reist NE, Werle MJ, and McMahan UJ (1992). Agrin released by motor neurons induces the aggregation of acetylcholine receptors at neuromuscular junctions. Neuron 8, 865–868. 10.1016/08966273(92)90200-W. [DOI] [PubMed] [Google Scholar]
- 69.Jin S, Guerrero-Juarez CF, Zhang L, Chang I, Ramos R, Kuan CH, Myung P, Plikus MV, and Nie Q (2021). Inference and analysis of cell-cell communication using CellChat. Nat. Commun 12, 1–20. 10.1038/s41467-021-21246-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Joost S, Jacob T, Sun X, Annusver K, La Manno G, Sur I, and Kasper M (2018). Single-Cell Transcriptomics of Traced Epidermal and Hair Follicle Stem Cells Reveals Rapid Adaptations during Wound Healing. Cell Rep. 25, 585–597.e7. 10.1016/j.celrep.2018.09.059. [DOI] [PubMed] [Google Scholar]
- 71.Kriehuber E, Breiteneder-Geleff S, Groeger M, Soleiman A, Schoppmann SF, Stingl G, Kerjaschki D, and Maurer D (2001). Isolation and characterization of dermal lymphatic and blood endothelial cells reveal stable and functionally specialized cell lineages. J. Exp. Med 194, 797–808. 10.1084/jem.194.6.797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kidoya H, Naito H, Muramatsu F, Yamakawa D, Jia W, Ikawa M, Sonobe T, Tsuchimochi H, Shirai M, Adams RH, et al. (2015). APJ Regulates Parallel Alignment of Arteries and Veins in the Skin. Dev. Cell 33, 247–259. 10.1016/j.devcel.2015.02.024. [DOI] [PubMed] [Google Scholar]
- 73.Poliakov A, Cotrina M, and Wilkinson DG (2004). Diverse roles of eph receptors and ephrins in the regulation of cell migration and tissue assembly. Dev. Cell 7, 465–480. 10.1016/j.devcel.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 74.Wigle JT, and Oliver G (1999). Prox1 function is required for the development of the murine lymphatic system. Cell 98, 769–778. 10.1016/S0092-8674(00)81511-1. [DOI] [PubMed] [Google Scholar]
- 75.Ordóñez NG (2012). Immunohistochemical endothelial markers: A review. Adv. Anat. Pathol 19, 281–295. 10.1097/PAP.0b013e3182691c2a. [DOI] [PubMed] [Google Scholar]
- 76.Alitalo K, Tammela T, and Petrova TV (2005). Lymphangiogenesis in development and human disease. Nature 438, 946–953. 10.1038/nature04480. [DOI] [PubMed] [Google Scholar]
- 77.Betterman KL, and Harvey NL (2018). Histological and Morphological Characterization of Developing Dermal Lymphatic Vessels. In Lymphangiogenesis - Methods and Protocols, Oliver G and Kahn ML, eds. (Springer Science+Business Media; ), pp. 19–36. [DOI] [PubMed] [Google Scholar]
- 78.Ribatti D, Vacca A, and Presta M (2000). The discovery of angiogenic factors: A historical review. Gen. Pharmacol. Vasc. Syst 35, 227–231. 10.1016/S0306-3623(01)00112-4. [DOI] [PubMed] [Google Scholar]
- 79.Betsholtz C (2018). Cell–cell signaling in blood vessel development and function. EMBO Mol. Med 10, 2–5. 10.15252/emmm.201708610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Ruhrberg C, Gerhardt H, Golding M, Watson R, Ioannidou S, Fujisawa H, Betsholtz C, and Shima DT (2002). Spatially restricted patterning cues provided by heparin-binding VEGF-A control blood vessel branching morphogenesis. Genes Dev. 16, 2684–2698. 10.1101/gad.242002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Mignatti P, and Rifkin DBB (1996). Plasminogen activators and matrix metalloproteinases in angiogenesis. Enzym. protein2 49, 117–137. 10.1159/000468621. [DOI] [PubMed] [Google Scholar]
- 82.Detmar M (2000). The role of VEGF and thrombospondins in skin angiogenesis. J. Dermatol. Sci 24, 78–84. 10.1016/S0923-1811(00)00145-6. [DOI] [PubMed] [Google Scholar]
- 83.Luttun A, Tjwa M, and Carmeliet P (2002). Placental Growth Factor (PIGF) and Its Receptor Flt-1 (VEGFR-1). Ann. N.Y. Acad. Sci 979, 80–93. [DOI] [PubMed] [Google Scholar]
- 84.Cao R, Bråkenhielm E, Li X, Pietras K, Widenfalk J, Östman A, Eriksson U, and Cao Y (2002). Angiogenesis stimulated by PDGF-CC, a novel member in the PDGF family, involves activation of PDGFR-aa and -ap receptors. FASEB J. 16, 1575–1583. 10.1096/fj.02-0319com. [DOI] [PubMed] [Google Scholar]
- 85.David L, Feige JJ, and Bailly S (2009). Emerging role of bone morphogenetic proteins in angiogenesis. Cytokine Growth Factor Rev. 20, 203–212. 10.1016/j.cytogfr.2009.05.001. [DOI] [PubMed] [Google Scholar]
- 86.Goumans M-J, and Mummery C (2000). Function analysis of the TGFbeta receptor/Smad pathway through gene ablation in mice. Int. J. Dev. Biol 44, 253–265. [PubMed] [Google Scholar]
- 87.Newman AC, Nakatsu MN, Chou W, Gershon PD, and Hughes CCW (2011). The requirement for fibroblasts in angiogenesis: Fibroblast-derived matrix proteins are essential for endothelial cell lumen formation. Mol. Biol. Cell 22, 3791–3800. 10.1091/mbc.E11-05-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Shima DT, and Mailhos C (2000). Vascular developmental biology: Getting nervous. Curr. Opin. Genet. Dev 10, 536–542. 10.1016/S0959-437X(00)00124-6. [DOI] [PubMed] [Google Scholar]
- 89.Malhotra R, Stenn KS, Fernandez LA, and Braverman IM (1989). Angiogenic properties of normal and psoriatric skin associate with epidermis, not dermis. Lab Invest 61, 162–165. [PubMed] [Google Scholar]
- 90.Soriano P (1994). Abnormal kidney development and hematological disorders in PDGF β-receptor mutant mice. Genes Dev. 8, 1888–1896. 10.1101/gad.8.16.1888. [DOI] [PubMed] [Google Scholar]
- 91.Levéen P, Pekny M, Gebre-Medhin S, Swolin B, Larsson E, and Betsholtz C (1994). Mice deficient for PDGF B show renal, cardiovascular, and hematological abnormalities. Genes Dev. 8, 1875–1887. 10.1101/gad.8.16.1875. [DOI] [PubMed] [Google Scholar]
- 92.Gerhardt H, and Betsholtz C (2003). Endothelial-pericyte interactions in angiogenesis. Cell Tissue Res. 314, 15–23. 10.1007/s00441-003-0745-x. [DOI] [PubMed] [Google Scholar]
- 93.Muhl L, Genové G, Leptidis S, Liu J, He L, Mocci G, Sun Y, Gustafsson S, Buyandelger B, Chivukula IV, et al. (2020). Single-cell analysis uncovers fibroblast heterogeneity and criteria for fibroblast and mural cell identification and discrimination. Nat. Commun 11, 1–18. 10.1038/s41467-020-17740-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Stratman AN, Schwindt AE, Malotte KM, and Davis GE (2010). Endothelial-derived PDGF-BB and HB-EGF coordinately regulate pericyte recruitment during vasculogenic tube assembly and stabilization. Blood 116, 4720–4730. 10.1182/blood-2010-05-286872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Benjamin LE, Hemo I, and Keshet E (1998). A plasticity for blood vessel remodelling is defined by pericyte coverage of the performed endothelial network and is regulated by PDGF-B and VEGF. Development 125, 1591–1598. [DOI] [PubMed] [Google Scholar]
- 96.Reynolds LP, Grazul-Bilska AT, and Redmer DA (2000). Angiogenesis in the corpus luteum. Endocrine 12, 1–9. 10.1385/endo:12:1:1. [DOI] [PubMed] [Google Scholar]
- 97.Amselgruber WM, Schäfer M, and Sinowatz F (1999). Angiogenesis in the bovine corpus luteum: An immunocytochemical and ultrastructural study. Anat. Histol. Embryol 28, 157–166. 10.1046/j.1439-0264.1999.00195.x. [DOI] [PubMed] [Google Scholar]
- 98.Henri S, Poulin LF, Tamoutounour S, Ardouin L, Guilliams M, De Bovis B, Devilard E, Viret C, Azukizawa H, Kissenpfennig A, et al. (2010). CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med 207, 189–206. 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Haniffa M, Shin A, Bigley V, McGovern N, Teo P, See P, Wasan PS, Wang XN, Malinarich F, Malleret B, et al. (2012). Human Tissues Contain CD141 hi Cross-Presenting Dendritic Cells with Functional Homology to Mouse CD103 + Nonlymphoid Dendritic Cells. Immunity 37, 60–73. 10.1016/j.immuni.2012.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Merad M, and Manz MG (2009). Dendritic cell homeostasis. Blood 113, 3418–3427. 10.1182/blood-2008-12-180646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Nussenzweig BMC, Steinman RM, Gutchinov B, and Cohn ZA (1980). Development Cells Are Accessory Cells for the of Anti-Trinitrophenyl. October 152, 1070–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Pierre P, Turley SJ, Gatti E, Hull M, Meltzer J, Mirza A, Inaba K, Steinman RM, and Mellman I (1997). Developmental regulation of invariant chain proteolysis controls MHC class II trafficking in mouse dendritic cells. Nature 388, 787–792. 10.1016/S0092-8674(00)81458-0. [DOI] [PubMed] [Google Scholar]
- 103.Steinman RM, Gutchinov B, Witmer MD, and Nussenzweig MC (1978). Dendritic cells are the principal stimulators of the primary mixed leukocyte reaction in mice. Proc. Natl. Acad. Sci 75, 5132–5136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Förster R, Schubel A, Breitfeld D, Kremmer E, Renner-Müller I, Wolf E, and Lipp M (1999). CCR7 coordinates the primary immune response by establishing functional microenvironments in secondary lymphoid organs. Cell 99, 23–33. 10.1016/S0092-8674(00)80059-8. [DOI] [PubMed] [Google Scholar]
- 105.Mildner A, and Jung S (2014). Development and function of dendritic cell subsets. Immunity 40, 642–656. 10.1016/j.immuni.2014.04.016. [DOI] [PubMed] [Google Scholar]
- 106.Tamoutounour S, Guilliams M, MontananaSanchis F, Liu H, Terhorst D, Malosse C, Pollet E, Ardouin L, Luche H, Sanchez C, et al. (2013). Origins and functional specialization of macrophages and of conventional and monocyte-derived dendritic cells in mouse skin. Immunity 39, 925–938. 10.1016/j.immuni.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 107.Hashimoto D, Miller J, and Merad M (2011). Dendritic Cell and Macrophage Heterogeneity In Vivo. Immunity 35, 323–335. 10.1016/j.immuni.2011.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Gautier EL, Shay T, Miller J, Greter M, Jakubzick C, Ivanov S, Helft J, Chow A, Elpek KG, Gordonov S, et al. (2012). Gene-expression profiles and transcriptional regulatory pathways that underlie the identity and diversity of mouse tissue macrophages. Nat. Immunol 13, 1118–1128. 10.1038/ni.2419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hoeffel G, Wang Y, Greter M, See P, Teo P, Malleret B, Leboeuf M, Low D, Oller G, Almeida F, et al. (2012). Adult Langerhans cells derive predominantly from embryonic fetal liver monocytes with a minor contribution of yolk sac-derived macrophages. J. Exp. Med 209, 1167–1181. 10.1084/jem.20120340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Hoeffel G, Chen J, Lavin Y, Low D, Almeida FF, See P, Beaudin AE, Lum J, Low I, Forsberg EC, et al. (2015). C-Myb+ Erythro-Myeloid Progenitor-Derived Fetal Monocytes Give Rise to Adult Tissue-Resident Macrophages. Immunity 42, 665–678. 10.1016/j.immuni.2015.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ginhoux F, and Merad M (2010). Ontogeny and homeostasis of Langerhans cells. Immunol. Cell Biol 88, 387–392. 10.1038/icb.2010.38. [DOI] [PubMed] [Google Scholar]
- 112.Chorro L, Sarde A, Li M, Woollard KJ, Chambon P, Malissen B, Kissenpfennig A, Barbaroux JB, Groves R, and Geissmann F (2009). Langerhans cell (LC) proliferation mediates neonatal development, homeostasis, and inflammation-associated expansion of the epidermal LC network. J. Exp. Med 206, 3089–3100. 10.1084/jem.20091586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dwyer DF, Barrett NA, Austen KF, Kim EY, Brenner MB, Shaw L, Yu B, Goldrath A, Mostafavi S, Regev A, et al. (2016). Expression profiling of constitutive mast cells reveals a unique identity within the immune system. Nat. Immunol 17, 878–887. 10.1038/ni.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Gentek R, Ghigo C, Hoeffel G, Bulle MJ, Msallam R, Gautier G, Launay P, Chen J, Ginhoux F, and Bajénoff M (2018). Hemogenic Endothelial Fate Mapping Reveals Dual Developmental Origin of Mast Cells. Immunity 48, 1160–1171.e5. 10.1016/j.immuni.2018.04.025. [DOI] [PubMed] [Google Scholar]
- 115.Hayashi C, Sonoda T, Nakano T, Nakayama H, and Kitamura Y (1985). Mast-cell precursors in the skin of mouse embryos and their deficiency in embryos of Sl/Sld genotype. Dev. Biol 109, 234–241. 10.1016/0012-1606(85)90363-X. [DOI] [PubMed] [Google Scholar]
- 116.Kuhlbrodt K, Herbarth B, Sock E, Hermans-Borgmeyer I, and Wegner M (1998). Sox10, a novel transcriptional modulator in glial cells. J. Neurosci 18, 237–250. 10.1523/jneurosci.18-01-00237.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Martin P (1990). Tissue patterning in the developing mouse limb. Int. J. Dev. Biol 34, 323–336. 10.1387/ijdb.1702679. [DOI] [PubMed] [Google Scholar]
- 118.Jenkins BA, and Lumpkin EA (2017). Developing a sense of touch. Dev. 144, 4048–4090. 10.1242/dev.120402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Peng K, Sant D, Andersen N, Silvera R, Camarena V, Piñero G, Graham R, Khan A, Xu XM, Wang G, et al. (2020). Magnetic separation of peripheral nerve-resident cells underscores key molecular features of human Schwann cells and fibroblasts: an immunochemical and transcriptomics approach. Sci. Rep 10, 1–20. 10.1038/s41598-020-74128-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Frank E, and Sanes JR (1991). Lineage of neurons and glia in chick dorsal root ganglia: Analysis in vivo with a recombinant retrovirus. Development 111, 895–908. [DOI] [PubMed] [Google Scholar]
- 121.Yamagishi SI, Ogasawara S, Mizukami H, Yajima N, Wada RI, Sugawara A, and Yagihashi S (2008). Correction of protein kinase C activity and macrophage migration in peripheral nerve by pioglitazone, peroxisome proliferator activated-γ- ligand, in insulin-deficient diabetic rats. J. Neurochem 104, 491–499. 10.1111/j.1471-4159.2007.05050.x. [DOI] [PubMed] [Google Scholar]
- 122.Allen SJ, and Dawbarn D (2006). Clinical relevance of the neurotrophins and their receptors. Clin. Sci 110, 175–191. 10.1042/CS20050161. [DOI] [PubMed] [Google Scholar]
- 123.Roosterman D, Goerge T, Schneider SW, Bunnett NW, and Steinhoff M (2006). Neuronal control of skin function: The skin as a neuroimmunoendocrine organ. Physiol. Rev 86, 1309–1379. 10.1152/physrev.00026.2005. [DOI] [PubMed] [Google Scholar]
- 124.Ansel JC, Kaynard AH, Armstrong CA, Olerud J, Bunnett N, and Payan D (1996). Skin-Nervous system interactions. J. Invest. Dermatol 106, 198–204. 10.1111/1523-1747.ep12330326. [DOI] [PubMed] [Google Scholar]
- 125.Botchkarev VA, Yaar M, Peters EMJ, Raychaudhuri SKSP, Botchkareva NV, Marconi A, Raychaudhuri SKSP, Paus R, and Pincelli C (2006). Neurotrophins in skin biology and pathology. J. Invest. Dermatol 126, 1719–1727. 10.1038/sj.jid.5700270. [DOI] [PubMed] [Google Scholar]
- 126.Di Marco E, Marchisio PC, Bondanza S, Franzi AT, Cancedda R, and De Luca M (1991). Growth-regulated synthesis and secretion of biologically active nerve growth factor by human keratinocytes. J. Biol. Chem 266, 21718–21722. 10.1016/s0021-9258(18)54695-0. [DOI] [PubMed] [Google Scholar]
- 127.Roggenkamp D, Falkner S, Stäb F, Petersen M, Schmelz M, and Neufang G (2012). Atopic keratinocytes induce increased neurite outgrowth in a coculture model of porcine dorsal root ganglia neurons and human skin cells. J. Invest. Dermatol 132, 1892–1900. 10.1038/jid.2012.44. [DOI] [PubMed] [Google Scholar]
- 128.Movafagh S, Hobson JP, Spiegel S, Kleinman HK, Zukowska Z, Movafagh S, Hobson JP, Spiegel S, Kleinman HK, and Zukowska Z (2006). Neuropeptide Y induces migration, proliferation, and tube formation of endothelial cells bimodally via Y1, Y2, and Y5 receptors. FASEB J. 20, 1924–1926. 10.1096/fj.05-4770fje. [DOI] [PubMed] [Google Scholar]
- 129.Paus R, Theoharides TC, and Arck PC (2006). Neuroimmunoendocrine circuitry of the “brain-skin connection.” Trends Immunol. 27, 32–39. 10.1016/j.it.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 130.Chang AY, Wanat KA, and Seykora JT (2014). Dermatopathology. Melanocytes and Vitiligo (and Hair Graying). In Pathobiology of Human Disease : A Dynamic Encyclopedia of Disease Mechanisms, McManus LM, ed. (Elsevier Science & Technology; ), pp. 1148–1157. [Google Scholar]
- 131.Ostrowski SM, and Fisher DE (2018). Chapter 20. Pigmentation and Melanocyte Biology. In Fitzpatrick’s Dermatology, Kang S, Amagai M, Bruckner AL, Enk AH, Margolis DJ, McMichael AJ, and Orringer JS, eds. (McGraw-Hill Education / Medical; ). [Google Scholar]
- 132.Cichorek M, Wachulska M, Stasiewicz A, and Tyminska A (2013). Skin melanocytes: biology and development. Postep. Dermatologii i Alergol 30, 30–41. 10.5114/pdia.2013.33376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yoshida H, Kunisada T, Kusakabe M, Nishikawa SSI, and Nishikawa SSI (1996). Distinct stages of melanocyte differentiation revealed by analysis of nonuniform pigmentation patterns. Development 122, 1207–1214. [DOI] [PubMed] [Google Scholar]
- 134.Mayer TC (1973). The migratory pathway of neural crest cells into the skin of mouse embryos. Dev. Biol 34, 39–46. 10.1016/0012-1606(73)90337-0. [DOI] [PubMed] [Google Scholar]
- 135.Hirobe T (1984). Histochemical survey of the distribution of the epidermal melanoblasts and melanocytes in the mouse during fetal and postnatal periods. Anat. Rec 208, 589–594. 10.1002/ar.1092080414. [DOI] [PubMed] [Google Scholar]
- 136.Suzuki I, Tada A, Ollmann MM, Barsh GS, Im S, Lamoreux ML, Hearing VJ, Nordlund JJ, and Abdel-Malek ZA (1997). Agouti signaling protein inhibits melanogenesis and the response of human melanocytes to α-melanotropin. J. Invest. Dermatol 108, 838–842. 10.1111/1523-1747.ep12292572. [DOI] [PubMed] [Google Scholar]
- 137.Reddy S, Andl T, Bagasra a, Lu MM, Epstein DJ, Morrisey EE, and Millar SE (2001). Characterization of Wnt gene expression in developing and postnatal hair follicles and identification of Wnt5a as a target of Sonic hedgehog in hair follicle morphogenesis. Mech. Dev 107, 69–82. 10.1016/S0925-4773(01)00452-X. [DOI] [PubMed] [Google Scholar]
- 138.Richardson RJ, Hammond NL, Coulombe PA, Saloranta C, Nousiainen HO, Salonen R, Berry A, Hanley N, Headon D, Karikoski R, et al. (2014). Periderm prevents pathological epithelial adhesions during embryogenesis. J. Clin. Invest 124, 3891–3900. 10.1172/JCI71946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.M’Boneko V, and Merker HJ (1988). Development and morphology of the periderm of mouse embryos (days 9–12 of gestation). Acta Anat. (Basel) 133, 325–336. [DOI] [PubMed] [Google Scholar]
- 140.Hammond NL, Dixon J, and Dixon MJ (2019). Periderm: Life-cycle and function during orofacial and epidermal development. Semin. Cell Dev. Biol 91, 75–83. 10.1016/j.semcdb.2017.08.021. [DOI] [PubMed] [Google Scholar]
- 141.McGowan KM, and Coulombe PA (1998). Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol 143, 469–486. 10.1083/jcb.143.2.469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Morita K, Furuse M, Yoshida Y, Itoh M, Sasaki H, Tsukita S, and Miyachi Y (2002). Molecular architecture of tight junctions of periderm differs from that of the maculae occludentes of epidermis. J. Invest. Dermatol 118, 1073–1079. 10.1046/j.1523-1747.2002.01774.x. [DOI] [PubMed] [Google Scholar]
- 143.De La Garza G, Schleiffarth JR, Dunnwald M, Mankad A, Weirather JL, Bonde G, Butcher S, Mansour TA, Kousa YA, Fukazawa CF, et al. (2013). Interferon regulatory factor 6 promotes differentiation of the periderm by activating expression of grainyhead-like 3. J. Invest. Dermatol 133, 68–77. 10.1038/jid.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Dale BA, Holbrook KA, Kimball JR, Hoff M, and Sun TT (1985). Expression of epidermal keratins and filaggrin during human fetal skin development. J. Cell Biol 101, 1257–1269. 10.1083/jcb.101.4.1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Morita R, Sanzen N, Sasaki H, Hayashi T, Umeda M, Yoshimura M, Yamamoto T, Shibata T, Abe T, Kiyonari H, et al. (2021). Tracing the origin of hair follicle stem cells. Nature 594, 547–552. 10.1038/s41586-021-03638-5. [DOI] [PubMed] [Google Scholar]
- 146.Lechler T, and Fuchs E (2005). Asymmetric cell divisions promote stratification and differentiation of mammalian skin. Nature 437, 275–280. 10.1038/nature03922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Damen M, Wirtz L, Soroka E, Khatif H, Kukat C, Simons BD, and Bazzi H (2021). High proliferation and delamination during skin epidermal stratification. Nat. Commun 12, 1–16. 10.1038/s41467-021-23386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Cockburn K, Annusver K, Gonzalez DG, Ganesan S, May DP, Mesa KR, Kawaguchi K, Kasper M, and Greco V (2022). Gradual differentiation uncoupled from cell cycle exit generates heterogeneity in the epidermal stem cell layer. Nat. Cell Biol 24, 1692–1700. 10.1038/s41556-022-01021-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Blanpain C, Lowry WE, Pasolli HA, and Fuchs E (2006). Canonical notch signaling functions as a commitment switch in the epidermal lineage. Genes Dev. 20, 3022–3035. 10.1101/gad.1477606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Rangarajan A, Talora C, Okuyama R, Nicolas M, Mammucari C, Oh H, Aster JC, Krishna S, Metzger D, Chambon P, et al. (2001). Notch signaling is a direct determinant of keratinocyte growth arrest and entry into differentiation. EMBO J. 20, 3427–3436. 10.1093/emboj/20.13.3427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Miroshnikova YA, Le HQ, Schneider D, Thalheim T, Rübsam M, Bremicker N, Polleux J, Kamprad N, Tarantola M, Wang I, et al. (2018). Adhesion forces and cortical tension couple cell proliferation and differentiation to drive epidermal stratification. Nat. Cell Biol 20, 69–80. 10.1038/s41556-017-0005-z. [DOI] [PubMed] [Google Scholar]
- 152.Lin ZZ, Jin S, Chen J, Li Z, Lin ZZ, Tang L, Nie Q, and Andersen B (2020). Murine interfollicular epidermal differentiation is gradualistic with GRHL3 controlling progression from stem to transition cell states. Nat. Commun 11, 1–15. 10.1038/s41467-020-19234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Rhee H, Polak L, and Fuchs E (2006). Lhx2 maintains stem cell character in hair follicles. Science 312, 1946–1949. 10.1126/science.1128004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Levy V, Lindon C, Harfe BD, and Morgan B. a. (2005). Distinct stem cell populations regenerate the follicle and interfollicular epidermis. Dev. Cell 9, 855–861. 10.1016/j.devcel.2005.11.003. [DOI] [PubMed] [Google Scholar]
- 155.Ahtiainen L, Lefebvre S, Lindfors PH, Renvoisé E, Shirokova V, Vartiainen MK, Thesleff I, and Mikkola ML (2014). Directional Cell Migration, but Not Proliferation, Drives Hair Placode Morphogenesis. Dev. Cell 28, 588–602. 10.1016/j.devcel.2014.02.003. [DOI] [PubMed] [Google Scholar]
- 156.Närhi K, Järvinen E, Birchmeier W, Taketo MM, Mikkola ML, Thesleff I, Narhi K, Jarvinen E, Birchmeier W, Taketo MM, et al. (2008). Sustained epithelial b-catenin activity induces precocious hair development but disrupts hair follicle down-growth and hair shaft formation. Development 135, 1019–1028. 10.1242/dev.016550. [DOI] [PubMed] [Google Scholar]
- 157.Fliniaux I, Mikkola ML, Lefebvre S, and Thesleff I (2008). Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Dev. Biol 320, 60–71. 10.1016/j.ydbio.2008.04.023. [DOI] [PubMed] [Google Scholar]
- 158.Xiao Y, Woo W-M, Nagao K, Li W, Terunuma A, Mukouyama Y, Oro AE, Vogel JC, and Brownell I (2013). Perivascular hair follicle stem cells associate with a venule annulus. J. Invest. Dermatol 133, 2324–2331. 10.1038/jid.2013.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Brownell I, Guevara E, Bai CB, Loomis CA, and Joyner AL (2011). Nerve-derived sonic hedgehog defines a niche for hair follicle stem cells capable of becoming epidermal stem cells. Cell Stem Cell 8, 552–565. 10.1016/j.stem.2011.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Adachi T, Kobayashi T, Sugihara E, Yamada T, Ikuta K, Pittaluga S, Saya H, Amagai M, and Nagao K (2015). Hair follicle–derived IL-7 and IL-15 mediate skin-resident memory T cell homeostasis and lymphoma. Nat. Med 21, 1272–1279. 10.1038/nm.3962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Wang ECE, Dai Z, Ferrante AW, Drake CG, and Christiano AM (2019). A Subset of TREM2 + Dermal Macrophages Secretes Oncostatin M to Maintain Hair Follicle Stem Cell Quiescence and Inhibit Hair Growth. Cell Stem Cell 24, 654–669.e6. 10.1016/j.stem.2019.01.011. [DOI] [PubMed] [Google Scholar]
- 162.Sennett R, Wang Z, Rezza A, Grisanti L, Roitershtein N, Sicchio C, Mok KW, Heitman NJ, Clavel C, Ma’ayan A, et al. (2015). An Integrated Transcriptome Atlas of Embryonic Hair Follicle Progenitors, Their Niche, and the Developing Skin. Dev. Cell 34, 577–591. 10.1016/j.devcel.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Fan X, Wang D, Burgmaier JE, Teng Y, Romano RA, Sinha S, and Yi R (2018). Single Cell and Open Chromatin Analysis Reveals Molecular Origin of Epidermal Cells of the Skin. Dev. Cell 47, 21–37.e5. 10.1016/j.devcel.2018.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Sulic A, Roy R. Das, Papagno V, Lan Q, and Saikkonen R (2022). Transcriptomic landscape of early hair follicle and epidermal development. Prepr. bioRxiv 10.1101/2022.11.03.515012. [DOI] [PubMed] [Google Scholar]
- 165.Atit R, Sgaier SK, Mohamed OA, Taketo MM, Dufort D, Joyner AL, Niswander L, and Conlon RA (2006). Β-Catenin Activation Is Necessary and Sufficient To Specify the Dorsal Dermal Fate in the Mouse. Dev. Biol 296, 164–176. 10.1016/j.ydbio.2006.04.449. [DOI] [PubMed] [Google Scholar]
- 166.Amini-Nik S, Glancy D, Boimer C, Whetstone H, Keller C, and Alman BA (2011). Pax7 expressing cells contribute to dermal Wound repair, regulating scar size through a β-catenin mediated process. Stem Cells 29, 1371–1379. 10.1002/stem.688. [DOI] [PubMed] [Google Scholar]
- 167.Lepper C, Partridge TA, and Fan CM (2011). An absolute requirement for pax7-positive satellite cells in acute injury-induced skeletal muscle regeneration. Development 138, 3639–3646. 10.1242/dev.067595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Foster DS, Januszyk M, Yost KE, Chinta MS, Gulati GS, Nguyen AT, Burcham AR, Salhotra A, Ransom RC, Henn D, et al. (2021). Integrated spatial multiomics reveals fibroblast fate during tissue repair. Proc. Natl. Acad. Sci. USA 118, 1–10. 10.1073/pnas.2110025118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Phan QM, Sinha S, Biernaskie J, and Driskell RR (2021). Single-cell transcriptomic analysis of small and large wounds reveals the distinct spatial organization of regenerative fibroblasts. Exp. Dermatol 30, 92–101. 10.1111/exd.14244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Guerrero-Juarez CF, Dedhia PH, Jin S, Ruiz-Vega R, Ma D, Liu Y, Yamaga K, Shestova O, Gay DL, Yang Z, et al. (2019). Single-cell analysis reveals fibroblast heterogeneity and myeloid-derived adipocyte progenitors in murine skin wounds. Nat. Commun 10, 1–17. 10.1038/s41467-018-08247-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Sun X, Annusver K, Dalessandri T, and Kasper M (2022). Rare Gli1+ perivascular fibroblasts promote skin wound repair. Prepr. bioRxiv 10.1101/2022.05.16.491785. [DOI] [Google Scholar]
- 172.Phan QM, Fine GM, Salz L, Herrera GG, Wildman B, Driskell IM, and Driskell RR (2020). Lef1 expression in fibroblasts maintains developmental potential in adult skin to regenerate wounds. Elife 9, 1–19. 10.7554/ELIFE.60066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Driskell RR, Jahoda CABB, Chuong C-MM, Watt FM, and Horsley V (2014). Defining dermal adipose tissue. Exp. Dermatol 23, 629–631. 10.1111/exd.12450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Collins C. a., Kretzschmar K, and Watt FM (2011). Reprogramming adult dermis to a neonatal state through epidermal activation of -catenin. Development 138, 5189–5199. 10.1242/dev.064592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Donati G, Proserpio V, Lichtenberger BM, Natsuga K, Sinclair R, Fujiwara H, and Watt FM (2014). Epidermal Wnt/β-catenin signaling regulates adipocyte differentiation via secretion of adipogenic factors. Proc. Natl. Acad. Sci. USA 111, E1501–9. 10.1073/pnas.1312880111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Lichtenberger BM, Mastrogiannaki M, and Watt FM (2016). Epidermal β-catenin activation remodels the dermis via paracrine signalling to distinct fibroblast lineages. Nat. Commun 7, 10537. 10.1038/ncomms10537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Donati G, Rognoni E, Hiratsuka T, Liakath-Ali K, Hoste E, Kar G, Kayikci M, Russell R, Kretzschmar K, Mulder KW, et al. (2017). Wounding induces dedifferentiation of epidermal Gata6 + cells and acquisition of stem cell properties. Nat. Cell Biol 19, 603–613. 10.1038/ncb3532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Qian H, Badaloni A, Chiara F, Stjernberg J, Polisetti N, Nihlberg K, Consalez GG, and Sigvardsson M (2013). Molecular Characterization of Prospectively Isolated Multipotent Mesenchymal Progenitors Provides New Insight into the Cellular Identity of Mesenchymal Stem Cells in Mouse Bone Marrow. Mol. Cell. Biol 33, 661–677. 10.1128/mcb.01287-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Stuart T, Butler A, Hoffman P, Hafemeister C, Papalexi E, Mauck WM, Hao Y, Stoeckius M, Smibert P, and Satija R (2019). Comprehensive Integration of Single-Cell Data. Cell 177, 18881902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Bergen V, Lange M, Peidli S, Wolf FA, and Theis FJ (2020). Generalizing RNA velocity to transient cell states through dynamical modeling. Nat. Biotechnol 38, 1408–1414. 10.1038/s41587-020-0591-3. [DOI] [PubMed] [Google Scholar]
- 181.Wolf FA, Angerer P, and Theis FJ (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19. 10.1111/1462-2920.13787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Polański K, Young MD, Miao Z, Meyer KB, Teichmann SA, and Park JE (2020). BBKNN: Fast batch alignment of single cell transcriptomes. Bioinformatics 36, 964–965. 10.1093/bioinformatics/btz625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Lange M, Bergen V, Klein M, Setty M, Reuter B, Bakhti M, Lickert H, Ansari M, Schniering J, Schiller HB, et al. (2022). CellRank for directed single-cell fate mapping. Nat. Methods 19, 159–170. 10.1038/s41592-021-01346-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. (2012). Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682. 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, et al. (2010). A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat. Neurosci 13, 133–140. 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Andrews TS, and Hemberg M (2019). M3Drop: Dropout-based feature selection for scRNASeq. Bioinformatics 35, 2865–2867. 10.1093/bioinformatics/bty1044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Traag VA, Waltman L, and van Eck NJ (2019). From Louvain to Leiden: guaranteeing well-connected communities. Sci. Rep 9, 1–12. 10.1038/s41598-019-41695-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Ramilowski JA, Goldberg T, Harshbarger J, Kloppman E, Lizio M, Satagopam VP, Itoh M, Kawaji H, Carninci P, Rost B, et al. (2015). A draft network of ligand-receptor-mediated multicellular signalling in human. Nat. Commun 6. 10.1038/ncomms8866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Cabello-Aguilar S, Alame M, Kon-Sun-Tack F, Fau C, Lacroix M, and Colinge J (2020). SingleCellSignalR: inference of intercellular networks from single-cell transcriptomics. Nucleic Acids Res. 48, e55. 10.1093/nar/gkaa183. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S4. Oligonucleotides, Related to STAR Methods and Key Resources Table.
Table S1. Marker genes for populations and subpopulations, Related to Figures 2, 3, 5, 6 and 7. (A) Marker genes for the major populations. Marker genes had to fulfil the following criteria: (i) Upregulated in the cluster of interest (log fold change ≥ 0.25), (ii) Expressed in at least 20% of cells in the cluster of interest, and (iii) False discovery rate (FDR)-adjusted p-value ≤ 0.05. (B) Marker gene details for the major populations incl. adjusted p-value and log-fold change compared to all other cells. Cutting thresholds are the same as in Table S1A. (C) Marker genes for the fibroblast subpopulations. Cutting thresholds are the same as in Table S1A. (D) Marker gene details for the fibroblast subpopulations incl. adjusted p-value and log-fold change compared to all other fibroblasts. Cutting thresholds are the same as in Table S1A. (E) Marker genes for the epidermis subpopulations. Cutting thresholds are the same as in Table S1A. (F) Marker gene details for the epidermis subpopulations incl. adjusted p-value and log-fold change compared to all other epidermal cells. Cutting thresholds are the same as in Table S1A. (G) All considered transcription factors.
Table S2. Contribution of metadata categories to populations and subpopulations, Related to Figures 2, 3, 5, 6 and 7. (A) Contribution of metadata categories to the major populations. (B) Contribution of metadata categories to the fibroblast subpopulations. (C) Contribution of metadata categories to the epidermal subpopulations. (D) Contribution of metadata categories to the cells excluded from the final dataset.
Table S3. Receptor-Ligand-Pairs between all major, fibroblast and epidermis subclusters, Related to Figures 5, 6, S5, S7 and STAR Methods. (A) Identified receptor-ligand pairs incl. functional annotation. Receptor-ligand pairs with FDR-corrected p-value < 0.05 were included. (B) Scoring of identified receptor-ligand pairs. Cutting threshold is the same as in Table S3A. (C) Analysis approach.
Data Availability Statement
Single-cell RNA-seq data have been deposited on ArrayExpress under the accession number E-MTAB-11920. The original code has been deposited on GitHub (https://github.com/kasperlab; DOI for code release at publication: https://doi.org/10.5281/zenodo.8152645), and input files for the analysis pipelines and the annotated and analyzed sequencing data have been deposited at Zenodo (https://doi.org/10.5281/zenodo.7805311). The raw microscopy data that support the findings of this study are available from the lead contact upon reasonable request. Any additional information required to reanalyze the data reported in this paper is available from the Lead Contact upon request.


