Abstract
The aggregation of proteins into amyloid-like fibrils is seen in many neurodegenerative diseases. Recent years have seen much progress in our understanding of these misfolded protein inclusions, thanks to advances in techniques such as solid-state nuclear magnetic resonance (ssNMR) spectroscopy and cryogenic electron microscopy (cryo-EM). However, multiple repeat-expansion-related disorders have presented special challenges to structural elucidation. This review discusses the special role of ssNMR analysis in the study of protein aggregates associated with CAG repeat expansion disorders. In these diseases, the misfolding and aggregation affect mutant proteins with expanded polyglutamine segments. The most common disorder, Huntington's disease (HD), is connected to the mutation of the huntingtin protein. Since the discovery of the genetic causes for HD in the 1990s, steady progress in our understanding of the role of protein aggregation has depended on the integrative and interdisciplinary use of multiple types of structural techniques. The heterogeneous and dynamic features of polyQ protein fibrils, and in particular those formed by huntingtin N-terminal fragments, have made these aggregates into challenging targets for structural analysis. ssNMR has offered unique insights into many aspects of these amyloid-like aggregates. These include the atomic-level structure of the polyglutamine core, but also measurements of dynamics and solvent accessibility of the non-core flanking domains of these fibrils' fuzzy coats. The obtained structural insights shed new light on pathogenic mechanisms behind this and other protein misfolding diseases.
Introduction
Protein aggregation diseases are human disorders associated with, and hallmarked by, the formation of protein aggregates in afflicted patients [1]. These aggregates often share a common structural classification: an amyloid or amyloid-like fibrillar structure, representing long filaments with a characteristic ‘cross-β’ type architecture. For many protein aggregation diseases we now have a detailed understanding of the structures formed by aggregated proteins. The current article will review recent developments in the structural biology of polyglutamine (polyQ) protein aggregation, with a focus on the role played by magic angle spinning (MAS) solid-state nuclear magnetic resonance (ssNMR) spectroscopy.
polyQ diseases
The polyQ protein diseases are CAG repeat expansion disorders: inherited neurodegenerative diseases connected to the genetic expansion of CAG codon repeats in various human genes [2]. This family includes several different ataxias and Huntington's disease (HD). In each disorder, a particular gene that harbors a CAG repeat element is mutated. Individuals in which the CAG repeat is expanded beyond a certain threshold (often in the range of 35–50 CAG repeats) are at risk for disease. Notably, the age of onset is inversely correlated to the repeat length, with juvenile cases stemming from long expansion lengths. Naturally, the mutated genes yield mutant proteins featuring an expanded polyQ segment somewhere in the mutant protein (Figure 1A–C). These mutant proteins, or fragments thereof, form inclusions or aggregates [3]. The aggregation propensity is correlated to the polyQ length [4]. Most knowledge of the molecular mechanisms, the mutant proteins, and the aggregates is available for the most common polyQ disorder: HD [5]. This review will focus on HD, with readers referred to other literature for more information on other polyQ disorders [2,6,7].
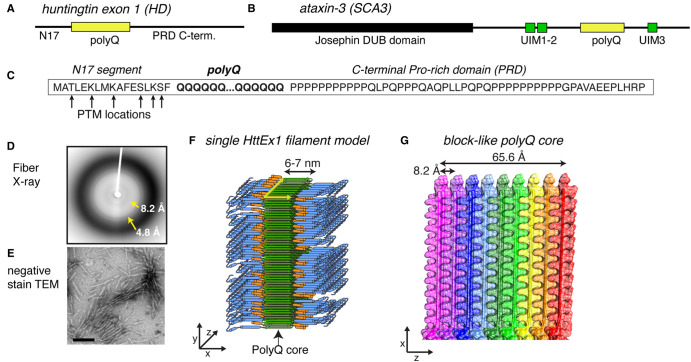
Figure 1. Summary of HttEx1 polyQ protein structures.
(A,B) Domain architecture of protein (fragments) from the most common polyQ diseases: HttEx1 from Huntington's disease (HD) and ataxin-3 from spinocerebellar ataxia 3 (SCA3). The polyQ segment is shown in yellow, alongside neighboring flanking domains with and without folded domains. HttEx1 is disordered in its monomeric state. (C) Sequence of HttEx1, with the location of prominent post-translational modifications indicated. The three key parts of HttEx1 are marked: N17, polyQ, and PRD. (D,E) X-ray fiber diffraction and negative stain transmission EM of aggregated polyQ/HttEx1. Adapted with permission from Hoop et al. [28]. (F) Structural model of aggregated HttEx1 from ssNMR and other techniques. (G) The polyQ core showing the block-like architecture that buries most of the polyQ segments. (F,G) Adapted with permission from Boatz et al. [33], under its CC-BY license.
In HD, the mutated protein is huntingtin (Htt), with the CAG repeat in its first exon (HttEx1; Figure 1A,C). Full-length Htt exceeds 3000 amino acids, and its structure in the non-aggregated state has been solved by cryo-EM [8,9]. However, in patients and model animals, the aggregating species is not the full-length protein, but rather N-terminal fragments containing the expanded polyQ [10]. Much research focuses on fragments that coincide with HttEx1, as it appears to play an important pathogenic role [11,12], even though other types of N-terminal fragments may be relevant as well. Until now the precise mechanism by which the HD mutation (and mutant protein) can lead to disease, remains uncertain. That said, many studies of HD pathogenesis focus on HttEx1 as a representative model for the suspected proteinaceous disease-causing agent, invoking a gain-of-toxic-function mechanism associated with HttEx1 misfolding and aggregation.
Earlier structural studies of polyQ protein aggregates
Since the 1990s, there has been much interest in the structure of aggregated polyQ proteins. Electron microscopy (EM; Figure 1E) and atomic force microscopy (AFM) showed the aggregates to be elongated filaments [3,13]. A noted feature of HttEx1 fibrils is their heterogeneous (or fuzzy) appearance, and propensity for higher-order clustering. X-ray fibril diffraction showed that aggregated polyQ proteins display the characteristic cross-β signature common for amyloid-forming polypeptides (Figure 1D) [14–16]. The cross-β pattern of polyQ aggregates suggests a 4.8 Å strand-to-strand distance in the hydrogen bonding direction, and a 8.2 Å sheet-to-sheet distance between the sheets [14]. Thus, polyQ protein fibrils display the hallmarks of amyloid or amyloid-like fibrils.
Cryo-EM
Recent years have seen dramatic improvements in cryo-EM, including in its application to amyloid fibrils [17]. In the HD field, the structure of the un-aggregated full-length Htt was determined by single-particle averaging [8,9]. In it, HttEx1 and its polyQ segment are not visible due to inherent disorder and flexibility. Aggregates formed by the HttEx1 fragment have been subjected to both cryo-EM and electron tomography (ET), in cells and in vitro [18–20]. These studies provided a view of the supramolecular architecture of HttEx1 fibrils and their polymorphism, and permitted a comparison between cellular aggregates and those prepared from pure proteins. The cryo-ET studies provide a compelling perspective on the way the HttEx1 fibrils interact with each other and with subcellular compartments and membranes. They also report a high degree of similarity between HttEx1 fibrils aggregated in a cellular context and in vitro. Individual fibrils have a width of multiple nm, but these fibrils cluster into larger assemblies, which become visible as puncta in fluorescence microscopy studies. Similar conclusions were drawn by recent super-resolution studies, which visualized nm-sized filaments alongside the larger clusters typically detected by (confocal) microscopy [21]. Unlike dramatic atomic-resolution structures obtained for other amyloid fibrils [17], no such high-resolution EM structures are available for any polyQ protein fibrils. A recent paper [20] provided the first results from cryo-EM analysis of HttEx1 fibrils, yielding improved insights into their architecture and their polymorphism (more below; Figure 3D). However, the obtained data did not permit an atomic-level structure to be determined, due to fibril heterogeneity and disorder. Another challenge for cryo-EM analysis is that the fibrils lack a clear and systematic twist, which is helpful for cryo-EM image analysis and atomic structure reconstruction.
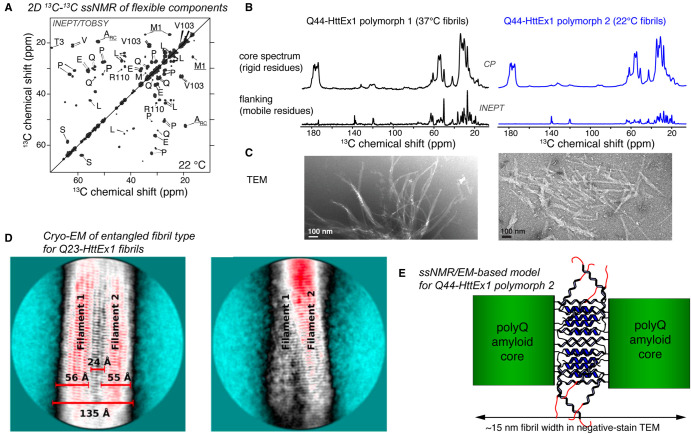
Figure 3. Studies of the HttEx1 flanking domains and their interactions.
(A) 2D ssNMR spectrum obtained by scalar-recoupling-based ssNMR spectroscopy, showing the flexible flanking domains of HttEx1 fibrils. (B) Comparison of 1D 13C traditional (top; CP) and scalar (bottom; INEPT) ssNMR spectra for different HttEx1 polymorphs, revealing primarily differences in flanking domain motion in the bottom spectra. (C) Negative-stain TEM of the same Q44-HttEx1 fibril types, which differed in their fibril widths. (D) Cryo-EM data on a sub-class of Q23-HttEx1 fibrils, showing an architecture mediated by flanking domain interactions. The two images represent different views on the same fibril type. (E) Model of flanking domain entanglement in protofilament interactions in wider HttEx1 fibrils (e.g. type 2 shown in B,C), deduced from combined analysis of negative stain TEM and ssNMR data. (A–C,E) Adapted with permission from Lin et al. [32], under the CC license. Panel (D) Reprinted with permission from Nazarov et al. [20] Copyright 2022 American Chemical Society.
ssNMR of polyQ amyloid cores
Since more than 15 years, ssNMR has been used to determine the structures of various pathogenic and functional amyloids [22–24]. Those structure determinations incorporated also constraints from EM (and other) techniques to generate full atomic-resolution structures. At the current time, no (peer-reviewed) atomic structure for polyQ or HttEx1 fibrils has yet been published (although a preprinted report on their integrative structure determination was posted in 2023 [25], but I here focus on the peer-reviewed literature). Historically, the literature on ssNMR of polyQ aggregates began in 2011, with two reports from different groups [26,27]. Combined, these early studies identified several key features of polyQ amyloid structure. Independent of the polyQ length, or the presence or absence of Htt-derived flanking sequences, the glutamine residues in the polyQ core gave a characteristic doubled set of ssNMR signals (illustrated in Figure 2A,B, from [28]). As discussed in a later study [29], these signals are atypical for normal glutamine conformations, and seem to represent a ssNMR signature for polyQ amyloid. Strikingly, two sets of signals make up this signature, being present in equal intensities. When targeted residue-specific isotope labeling is applied, this peak doubling is not removed, independent of the labeled site (Figure 2A, top) [26,28,30]. Thus, it is apparent that each of two sets of peaks represents the summed signals of many individual glutamine residues at different sequence positions in the protein. This contrasts with non-repetitive (amyloid) proteins, in which one aims to observe one peak per sequence position (i.e. per residue). Yet, the ssNMR (13C) peak linewidth of fully labeled polyQ proteins does not differ dramatically from aggregates in which isolated residues were labeled (Figure 2A top vs bottom; and refs. [26,28–31]). This means that there is a high degree of structural order and homogeneity in the fibril core, despite the morphological heterogeneity and disorder seen by cryo-EM (above). The recent literature contains a series of papers from different ssNMR groups that continue to show the remarkable consistency of this polyQ ssNMR signature in many polyQ and HttEx1 fibril polymorphs (Figures 2A and 3B) [28,30,32–37]. This is notably different from ssNMR studies of e.g. Aβ or α-synuclein polymorphs, where ssNMR signals of the amyloid core residues vary significantly between preparations and between research groups [38–40]. Thus, ssNMR suggests that polyQ-driven aggregation typically yields a reproducible, highly ordered, and characteristic amyloid core architecture [28,41].
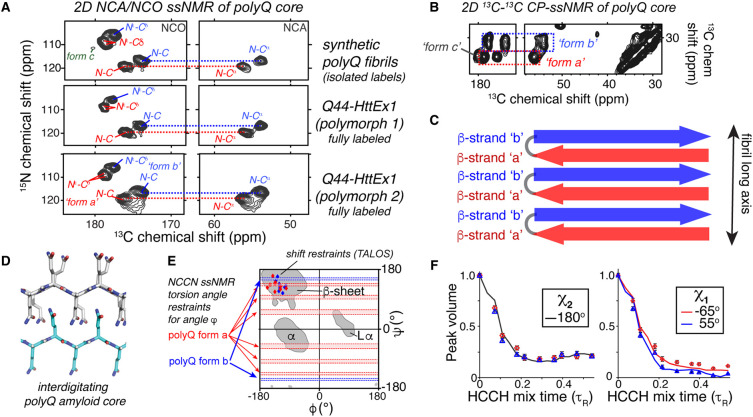
Figure 2. Example ssNMR data on the structure of polyQ amyloid cores.
(A) Comparison of 2D 13C-15N ssNMR spectra (NCA and NCO spectra) for three different polyQ aggregates, showing identical chemical shifts for the polyQ core. The ssNMR signature of the polyQ core features two sets of signals, coined forms a and b (marked in red and blue, respectively), in equal intensities. Note that the bottom samples are fully labeled HttEx1 protein fibrils, whilst the top shows a sample in which only two glutamines are selectively 13C,15N-labeled. (B) Part of a 2D 13C-13C CP-DARR ssNMR spectrum showing the same two glutamine conformers, in equal intensities. A smaller third peak (form c) is attributed to residues outside the buried core. (C) Schematic showing the molecular origin of the red/blue conformers (forms a/b) in the antiparallel β-sheets of polyQ amyloid. (D) Published schematic showing the glutamine side chain interdigitation in the core, based on ssNMR structural analysis. (E) PolyQ torsion angle ssNMR restraints for the backbone dihedral angles, shown in a Ramachandran plot. Horizontal red/blue areas mark permitted regions based on dipolar recoupling measurements of the Ψ angle (NCCN experiments), while the diamonds mark chemical-shift-based estimates from the TALOS program. (F) ssNMR data curves that constrain the side chain torsion angles χ1 and χ2 of polyQ amyloid, based on HCCH-type dihedral angle measurements. The red and blue datapoints represent the two Gln conformers, along with lines showing the best-fit dihedral angles. (A,B–F) Adapted with permission from Hoop et al. [28].
The determination of protein structures by ssNMR typically depends on measurements of distances between specific residues, using dipolar recoupling techniques [42]. However, the glutamine residues in the polyQ core all behave the same — a particular glutamine has a 50:50 chance to adopt one of the two hallmark signals. Even residue-specific labeling (achievable by solid-phase peptide synthesis [26,30] or using Amber-codon approaches [43]) yields the same pattern (Figure 2A). Therefore, distance measurements as a means toward an atomic-resolution structure are at best highly challenging. Does this mean that ssNMR structure determination is not feasible in polyQ amyloid? Previous structural studies of amyloidogenic polypeptides [44–46] deployed measurements of another structural parameter: torsion angles [47]. Specialized ssNMR experiments allow the determination of the relative orientations of neighboring chemical bonds (e.g. N–H, C–H or C–N bonds), based on the recoupling of dipolar interactions or chemical shift anisotropy tensors. This allows one to constrain the backbone (Ramachandran angles) and side-chain torsion angles (rotamer states). For a more in-depth discussion of these experiments, readers are referred to a recent review article [47]. In polyQ amyloids, these experiments were applied to backbone and side-chain dihedral angles of glutamine residues within the amyloid core (Figure 2E,F) [25,28]. These torsion angle measurements showed unambiguously that the two signals of the polyQ signature stem from two different glutamine conformations. Both conformations reflect β-sheet structure, but they differ in their Ramachandran angles (Figure 2E) and rotamer states (Figure 2F). These findings were based on the application of so-called NCCN and HCCH dihedral angle measurements, where dipolar interactions were recoupled between carbons, nitrogens and protons [28,47]. Antiparallel β-sheets are inherently expected (and observed) to assemble by alternating β-strands with different backbone conformations (Figure 2C; red/blue arrows) [48]. This alternating pattern offers an elegant explanation of the observation that two signals are typically observed in equal intensities in polyQ amyloids, independent of the sample type (Figure 2A). Moreover, 2D ssNMR and IR studies using mixtures of isotope-labeled proteins provided evidence that polyQ-based aggregates (with long polyQ segments) contain β-hairpin structures (Figure 2C) [28,49]. Such experiments exemplify the way that targeted isotope labeling can be deployed to distinguish intra- from intermolecular interactions. The β-hairpin motifs could be recognized by detecting the presence of backbone-backbone interactions in samples where the 13C-labeled protein was diluted with non-13C proteins. The detected interactions could only be explained by intramolecular hydrogen bonding as found in β-hairpin structures.
ssNMR side-chain dihedral angle measurements identified differences in the χ1 torsion angles of the two dominant glutamine conformers (Figure 2F) [28]. These were experiments where the relative orientations of the C–H bonds in the side chain were measured, using dipolar recoupling experiments. These measurements also showed that the χ2 angle was close to 180° for both conformers. Similar angular constraints were also reported by Raman spectroscopy [50]. Combining these structural insights, one can build a picture of the internal structure of the polyQ amyloid core, with glutamine side chains extended into an interdigitated ‘steric zipper’ interface (Figure 2D) [25,28]. The steric zipper concept describes a hallmark feature of amyloids, which is that they often feature a dehydrated, highly stable core structure in which amino acid sidechains interdigitate into a tight zipper-like configuration [51]. A related concept is the idea of polyQ polar zippers, as originally proposed by Perutz et al. [16]: ‘linking β-strands together into sheets or barrels by networks of hydrogen bonds between their main-chain amides and between their polar side chains’. However, back in 1994, these authors lacked the atomic-resolution data to build an experiment-based atomic-level structure for polyQ. Notably, other glutamine-rich amyloids feature such steric or polar zippers [45,51–55], but it is important to stress that those fibrils typically feature parallel β-sheets, whilst the antiparallel structure (Figure 2C) of the polyQ fibril core makes it qualitatively different. The use of ssNMR now enables structural measurements that can elevate general amyloid concepts to an atomic-level understanding of core of polyQ amyloids, shared by polyQ model polypeptides and disease-relevant HttEx1 fibrils.
A strength and limitation of ssNMR is its sensitivity to sub-nm structural features. On the one hand, this means that ssNMR can provide atomic-level details even in absence of high-order structural homogeneity (unlike X-ray diffraction). However, it also means that (fibril) structures determined by ssNMR rely on the integration of sub-nm information from ssNMR with longer-range information from techniques such as EM, ET, or AFM. Thus, ssNMR fibril structures are nice examples of ‘integrative structural biology’ [56]. EM and AFM studies showed the filaments of polyQ-expanded HttEx1 to be several nm in width, both in vitro and in cells. Combining this with inter-sheet and inter-strand distances known from X-ray fiber diffraction (above) and structural ssNMR details, one can start to build a representative architecture (Figures 1F,G and 3) [25,32,33]. Zooming out beyond the ‘zippered’ side chains, and antiparallel β-sheets, the ssNMR data are only explainable by a type of block-like architecture, with multiple β-sheets stacked side by side (Figure 1G) [28,32,33]. Notably, this type of architecture was previously suggested for polyQ aggregates, based on lower-resolution fiber diffraction and mutational studies [14,57]. More recently, cryo-EM analysis provided more corroboration of this key aspect of the HttEx1 fibril architecture (Figure 3D) [20]. As noted above, this EM analysis did not achieve atomic resolution due to fibril heterogeneity, but it did support key features deduced previously from ssNMR-based analyses. Thus, combining the insights from different techniques (with a key role for ssNMR), we have obtained a good understanding of the internal architecture of polyQ amyloid.
Beyond structure alone: dynamics, surfaces and more
MAS ssNMR offers various other valuable types of information, beyond structural restraints alone. Like liquid-state NMR, ssNMR is sensitive to molecular motions, allowing a variety of dynamics measurements and spectral editing techniques [58,59]. Direct detection of molecular motion can be done by measuring ssNMR relaxation and (dipolar) order parameters, which probe different regimes of motion. Such dynamic measurements reveal that the dominant signals seen for the polyQ amyloid core stem from residues that are highly rigid, akin to a crystal-like order [26–28,30]. For instance, this was detected in so-called DIPSHIFT measurements of dipolar order parameters, comparing the dipolar couplings of C–H or N–H bonds to those expected for completely rigid molecules [30,32]. Parts of the protein outside the (polyQ) core display lower order parameters and different relaxation properties, hinting at solvent exposure. This is most apparent for the flanking domains, which will be discussed below (Figure 3). Comparisons of domain motions were facilitated in part by the straightforward recognition of characteristic amino acid signals (e.g. Pro) that are unique to specific protein domains. A bigger challenge is generated by the polyQ core itself, as it is made up out of dozens of identical residues with highly similar chemical shifts. As such, their signals are dominated by the rigid core, making the detection of e.g. surface residues challenging. A recent study [34] deployed specialized ssNMR spectral editing techniques to selectively detect glutamines on the fibril surface, even against a larger background of residues in the fibril core. This was based on an approach designed to detect residues experiencing intermediate motion, by suppressing signals from the most rigid (core) residues that normally dominate (cross-polarization) ssNMR spectra. Another powerful approach, based on somewhat similar principles, achieves a type of water-edited spectroscopy in which one can determine the solvent-proximity of residues within protein fibrils. Such solvent-proximity measurements have been applied to polyQ peptide and HttEx1 fibrils in many studies [26,27]. Indeed, the insights into dynamics and surface exposure (enabled by ssNMR) were crucial to the development of structural models in which the large majority of polyQ segment residues form a rigid dehydrated core (Figure 1F,G), with few solvent-exposed glutamines on the surface. These models explain the formation of multiple-nm-wide fibrils containing tightly-packed interdigitating β-sheets, which are impenetrable to the stains used in negative stain TEM (Figures 1G and 2D).
Going beyond the (polyQ) core: HttEx1 flanking domains
The polypeptide chain beyond the polyQ segment cannot be ignored. In HD, the HttEx1 flanking segments have dramatic effects on both aggregation propensity and kinetics [26,60,61], and they determine also the fate and effects of HttEx1 aggregates in cells [62–64]. The N-terminal segment (known as N17 or HttNT) boosts aggregation, whilst the C-terminal proline-rich domain (PRD) delays it (Figure 1A,C). The flanking domains (in particular N17) contain residues that are impacted by post-translational modification (PTM): phosphorylation, acetylation, and ubiquitination (Figure 1C). The interest in these PTMs is substantial, as they prevent aggregation, reduce toxicity (in animal models) and facilitate degradation processes [65–67]. Also chaperone proteins that are able to disaggregate pre-formed amyloid, seem to target non-core flanking segments as essential binding sites [63,68]. MAS ssNMR has been used to elucidate the structural fate of these flanking segments in a variety of HttEx1 fibril types [26,32–35,37,69,70]. These studies showed the flanking domains to lack the β-sheet structure and rigidity of the amyloid core. Other ssNMR experiments probed the solvent-exposure of the flanking segments, showing them to be much more solvent-exposed than the polyQ amyloid core.
A key early finding was that the aggregation-enhancing N17 segment was partly α-helical in HttEx1-like peptide fibrils [26,31]. Later studies of various HttEx1 fibril types showed that different fibril polymorphs differ in their N17 structure and dynamics [32,33,35,37]. The flexibility or mobility was even more striking for the C-terminal PRD. In ssNMR and electron spin resonance studies parts of the PRD were remarkably flexible, behaving similar to an intrinsically disordered protein (Figure 3A,B) [69,71]. Notably, proximal to the polyQ core, the PRD becomes more and more ordered, leading to a gradient of flexibility along the C-terminal end of HttEx1, as can be detected via NMR relaxation measurements [69]. Such ssNMR studies were previously also applied to other fibrils featuring a ‘fuzzy coat’ of flexible polypeptide segments outside the fibril core [38,72]. This includes MAS ssNMR experiments that are based on similar principles as common in liquid-state NMR: the use of through-bond or scalar-interactions to transfer magnetization (signal) between nuclei (INEPT experiments). In their normal implementation, rigid parts of the sample (e.g. the fibril core) are invisible in such experiments, due to the short 1H T2 relaxation times (in absence of 1H decoupling). This suppresses all signals, except those from highly flexible protein segments (Figure 3A,B) [58,73]. Such MAS ssNMR experiments probed the secondary structure, dynamics and interactions of the flexible flanking domains of HttEx1 fibrils [32,35,69]. This revealed the rigid polyQ core to be decorated with flexible, exposed, flanking domains (Figure 1F,G). This molecular architecture manifests in EM as a fuzzy coat, or a bottle-brush type architecture [71], a feature also seen in other pathogenic amyloid fibrils [74,75].
ssNMR views of supramolecular HttEx1 polymorphism
One important aspect of the fuzzy coat of HttEx1 fibrils is that it plays a striking role in HttEx1 fibril polymorphism [32,37]. Amyloid polymorphism describes the observation that most amyloid-forming polypeptides form an array of different kinds of fibrils depending on conditions. The interest in these ‘polymorphs’ stems from the fact that their different structures translate into differences in biological properties such as fibril stability and toxicity [40]. Moreover, once formed, certain fibril types are able to propagate themselves by recruiting soluble protein monomers and catalyzing their transformation into the amyloid state. The existence of fibril polymorphs can offer a rationale for conflicting findings regarding the cellular toxicity of protein aggregates. Even if certain types of fibril assemblies are measured to have low cytotoxicity [76], other polymorphs may be much more pathogenic, complicating direct comparisons unless one validates the polymorph structure between measurements.
The sensitivity of ssNMR to atomic-level details has made it a valuable tool for detecting and dissecting amyloid polymorphs. Commonly, amyloid polymorphs have distinct ssNMR spectra with peaks arising at different chemical shift values [38–40]. A striking feature of HttEx1 fibrils is that different preparations do not differ in peak positions, even when they can look dramatically different by EM, have different polyQ lengths, or are prepared by different research groups [26–29,32,33,35,37]. What does this mean? When HttEx1 is aggregated under different conditions, EM measurements and antibody binding assays can show differences in the fibrils [32,37,77]. Yet, ssNMR spectra on these samples yield very similar spectra in which peak positions do not change, arguing that the local atomic-level structure is not different. Then how can these be distinct polymorphs? Clues are found in dynamics seen by ssNMR: different polymorphs differ in the dynamics and solvent-accessibility of flanking domains. This pointed to a model in which the HttEx1 aggregates tend to contain very similarly structured fibril cores (on the atomic level; Figure 2A) but supramolecular interactions within and between protofilaments differ (Figure 3D,E). Here, the term protofilament refers to smaller filaments that wrap around each other to form wider fibrils. The ssNMR data make sense if HttEx1 fibrils are assembled from thinner protofilaments (∼4–6 nm wide), with varying interactions between the protofilaments mediated by flanking domains (and in particular the PRD). This idea is reminiscent of the subsequently introduced concept of ‘supramolecular’ or ‘ultrastructural’ polymorphism, in which tau fibrils were found to contain similarly structured protofilaments engaged in a variety of supramolecular interactions [78]. Crucially, this model has found direct support from recent cryo-EM analysis of HttEx1 fibril structure (Figure 3D) [20]. Aside from differences in the flanking segments (and their interactions), the polyQ literature (and ssNMR studies therein) also suggest changes or variations in the supramolecular architecture of the polyQ segment itself. As discussed above, ssNMR on expanded polyQ protein aggregates revealed β-hairpins within the fibril core [28]. Shorter polyQ peptides or proteins appear to lack the β-hairpin, even if they still form the same type of antiparallel β-sheet architecture [41]. Thus, it has been suggested that there is a polyQ-dependent switch in aggregation mechanisms, which may explain the polyQ threshold phenomenon in the respective diseases [29,41,79]. Notably, as yet, ssNMR has been one of the few techniques that was able to structurally analyze these types of hairpin structures in polyQ fibrils [28,29,49,80,81].
Placing polyQ protein structures in context
The above has summarized how the unique abilities of ssNMR reveal structural features of polyQ amyloid proteins. But how do these structural insights enhance our understanding of disease mechanisms? There is much debate about the role of HttEx1 aggregates in HD pathogenesis. One the one hand, studies show the toxic effect of pre-formed aggregates on neuronal cells, arguing for their toxic properties [82,83]. Other studies suggest that cellular inclusions (visible by confocal microscopy) are benign and may act as a rescue mechanism [76]. One way to unify these findings is to argue that different types of aggregates (polymorphs) are formed in different studies, with polymorph-dependent differences in toxicity. What would this mean considering the discussed structural data that suggest a common core structure, but differences in flanking domain exposure? Postulated mechanisms for aggregate toxicity have a common theme: the propensity for misfolded proteins to engage with cellular components. These components could be essential proteins, cellular membranes, or components of the proteostasis network. Notably, it is increasingly clear that many such interactions are mediated by HttEx1 flanking segments, rather than the polyQ domain [63,64]. Thus, the disposition, flexibility and exposure of flanking domains (which vary among HttEx1 polymorphs) are parameters likely to dictate the propensity for cellular interactions, the aggregates’ interactome, and thus toxic mechanisms. The identification of the polymorph-dependence of flanking domain exposure, by ssNMR, therefore is essential to understand this disease mechanism.
There is also much interest in PTMs that affect HttEx1, before and after aggregation. PTMs are implicated in reducing aggregation, toxicity and degradation of misfolded proteins [31,65,67,84]. ssNMR-based structural models of HttEx1 aggregates show that the PTM sites (in N17; Figure 1C) may be outside the core, but nonetheless are sequestered due to being surrounded by longer PRD segments. This limits their accessibility to kinases [66,67], ubiquitinases [65] and other N17-targeting proteins (including chaperones [85]). Naturally, ssNMR can probe the impact of PTMs and PTM-mimicking mutations directly. One exploratory study did so, showing no large change in conformation, but again suggesting a stronger role for changes in dynamics and aggregate stability [31].
The focus above has been on our understanding the end-products of the misfolding and aggregation process. This process is complex and multifaceted, with roles for the polyQ as well as flanking segments [86]. Aside from mature fibrils, studies have reported soluble oligomers and other non-fibrillar species [87–89]. Understanding features of the end-products helps us understand the possible role of different HttEx1 segments in the aggregation pathway, for instance by offering support for α-helical N17 stabilization [26]. Moreover, ssNMR can be a combined with mutations or modifications that test hypotheses about the aggregation mechanism. This is exemplified by ssNMR studies of polyQ peptides outfitted with β-hairpin stabilizing modifications, which tested the possible role of this folding motif in the aggregation process [29,80,81]. Such β-hairpin-stabilizing modifications boost aggregation, whilst ssNMR measurements showed that this occurred without a fundamental change in the internal aggregate structure.
Challenges and future directions
Most ssNMR studies discussed above were performed using proteins or peptides with isotope enrichment (13C and/or 15N). This enables multidimensional spectroscopy and speeds up NMR analysis. Until now, this has limited the ssNMR to aggregates prepared from purified proteins, as the production of isotope-enriched materials in neuronal cells or model animals remains challenging. An important future goal is to test directly whether aggregates formed in cells, animals, or patients are different in structure. One way is to pursue ssNMR analysis of unlabeled proteins. The literature includes many studies in which the signature peaks of polyQ amyloids were detected in absence of isotope labeling [26,81,90,91]. One particularly promising novel approach is the combination of ssNMR with dynamic nuclear polarization (DNP), which boosts the ssNMR sensitivity by orders of magnitude, making 2D DNP-ssNMR experiments on unlabeled proteins practical [90,91]. An alternative approach has been deployed in other amyloid studies [92]: using unlabeled ex vivo aggregates as seeds for the aggregation of isotope-labeled proteins. Such experiments could test whether the seeded aggregates feature polyQ core signatures, or if this may open up different aggregation pathways [93]. Fortunately, the study of polyQ proteins will also benefit from ongoing advances in ssNMR technology. These developments include the increasing availability of higher-field ssNMR magnets, ever-faster MAS spinning rates, 1H-detected MAS NMR, and cryogenically cooled MAS NMR probes. The application of these novel technologies will enable the study of (much) smaller sample sizes and the detection of more subtle structural/dynamic effects. Especially the former can be valuable to pursue ex vivo samples, in-cell studies and difficult-to-prepare samples (e.g. based on segmental- or residue-specific labeling techniques[43,94]).
An obvious (and necessary) future direction is the ssNMR analysis of other polyQ disease proteins. Until now, ssNMR data are limited to polyQ model peptides and derivatives of the HttEx1 protein. MAS ssNMR analysis of polyQ-expanded ataxin-1 or ataxin-3 aggregates would be very interesting, even just to see if such aggregates contain a common polyQ core structure. Naturally, the flanking segments would differ completely from those in HttEx1. It is likely that those proteins also yield heterogeneous aggregates, with disordered regions, leaving again an important role for ssNMR analysis. Even in the context of HD, there are many open questions related to the impact of PTMs, different polyQ lengths, Htt fragments, interactions with membranes, chaperones and other proteins.
In conclusion, this mini-review set out to provide a basic overview of key contributions of ssNMR to our understanding of the polyQ protein aggregates. This research area illustrates how ssNMR can play an essential role in the study of amyloid fibrils, illuminating aspects not easily determined by other structural techniques. The ability to probe the fibrils’ dynamic fuzzy coat (flanking domains) was discussed in some detail, and how this is connected to important biological questions relevant to disease. Many open questions remain to be answered, with a continued role for ssNMR as a key technique in the integrative structural biology of polyQ disease proteins, as well as other protein aggregates and biomolecular complexes.
Perspectives
Protein aggregation into amyloid-like fibrils is a hallmark feature of the CAG repeat expansion, or polyglutamine expansion, disorders — a whole family of incurable inherited neurodegenerative diseases. The structural fate of the mutant proteins in their misfolded state is an essential part of the pathogenic mechanism.
ssNMR spectroscopy has been an essential technique for understanding the structure of aggregated polyQ proteins, and in particular the mutant huntingtin exon 1 from HD. Surprising structural features of the polymorphic huntingtin aggregates shed light on their biological properties.
Future applications of ssNMR promise to widen our understanding of other polyglutamine disease proteins, further deepen our knowledge of the molecular disease mechanisms and pave the way for better diagnostics and treatments.
Abbreviations
- Aβ
amyloid-β
- AFM
atomic force microscopy
- CP
cross polarization
- cryo-EM
cryogenic electron microscopy
- DNP
dynamic nuclear polarization
- EM
electron microscopy
- ET
electron tomography
- HD
Huntington's disease
- Htt
huntingtin
- HttEx1
huntingtin exon 1
- MAS
magic angle spinning
- N17
N-terminal segment of HttEx1
- NMR
nuclear magnetic resonance
- polyQ
polyglutamine
- PRD
proline-rich domain
- PTM
post-translational modification
- ssNMR
solid-state nuclear magnetic resonance
Competing Interests
The author declares that there are no competing interests associated with this manuscript.
Funding
This research on polyQ amyloid studies in the Van der Wel group has been supported by the NIH/NIGMS [R01GM112678], CampagneTeam Huntington, and the CHDI Foundation [A-15744].
Open Access
Open access for this article was enabled by the participation of University of Groningen in an all-inclusive Read & Publish agreement with Portland Press and the Biochemical Society under a transformative agreement with Individual.
Author Contribution
P.C.A.v.d.W. wrote the manuscript.
References
- 1.Chiti, F. and Dobson, C.M. (2017) Protein misfolding, amyloid formation, and human disease: a summary of progress over the last decade. Annu. Rev. Biochem. 86, 27–68 10.1146/annurev-biochem-061516-045115 [DOI] [PubMed] [Google Scholar]
- 2.Bunting, E.L., Hamilton, J. and Tabrizi, S.J. (2022) Polyglutamine diseases. Curr. Opin. Neurobiol. 72, 39–47 10.1016/j.conb.2021.07.001 [DOI] [PubMed] [Google Scholar]
- 3.Scherzinger, E., Lurz, R., Turmaine, M., Mangiarini, L., Hollenbach, B., Hasenbank, R.et al. (1997) Huntingtin-encoded polyglutamine expansions form amyloid-like protein aggregates in vitro and in vivo. Cell 90, 549–558 10.1016/s0092-8674(00)80514-0 [DOI] [PubMed] [Google Scholar]
- 4.Kar, K., Jayaraman, M., Sahoo, B., Kodali, R. and Wetzel, R. (2011) Critical nucleus size for disease-related polyglutamine aggregation is repeat-length dependent. Nat. Struct. Mol. Biol. 18, 328–336 10.1038/nsmb.1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bates, G.P., Dorsey, R., Gusella, J.F., Hayden, M.R., Kay, C., Leavitt, B.R.et al. (2015) Huntington disease. Nat. Rev. Dis. Primers 1, 15005 10.1038/nrdp.2015.5 [DOI] [PubMed] [Google Scholar]
- 6.McLoughlin, H.S., Moore, L.R. and Paulson, H.L. (2020) Pathogenesis of SCA3 and implications for other polyglutamine diseases. Neurobiol. Dis. 134, 104635 10.1016/j.nbd.2019.104635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barbosa Pereira, P.J., Manso, J.A. and Macedo-Ribeiro, S. (2023) The structural plasticity of polyglutamine repeats. Curr. Opin. Struct. Biol. 80, 102607 10.1016/j.sbi.2023.102607 [DOI] [PubMed] [Google Scholar]
- 8.Guo, Q., Huang, B., Cheng, J., Seefelder, M., Engler, T., Pfeifer, G.et al. (2018) The cryo-electron microscopy structure of huntingtin. Nature 555, 117–120 10.1038/nature25502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Harding, R.J., Deme, J.C., Hevler, J.F., Tamara, S., Lemak, A., Cantle, J.P.et al. (2021) Huntingtin structure is orchestrated by HAP40 and shows a polyglutamine expansion-specific interaction with exon 1. Commun. Biol. 4, 1374 10.1038/s42003-021-02895-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.DiFiglia, M., Sapp, E., Chase, K.O., Davies, S.W., Bates, G.P., Vonsattel, J.P.et al. (1997) Aggregation of huntingtin in neuronal intranuclear inclusions and dystrophic neurites in brain. Science 277, 1990–1993 10.1126/science.277.5334.1990 [DOI] [PubMed] [Google Scholar]
- 11.Landles, C., Sathasivam, K., Weiss, A., Woodman, B., Moffitt, H., Finkbeiner, S.et al. (2010) Proteolysis of mutant huntingtin produces an exon 1 fragment that accumulates as an aggregated protein in neuronal nuclei in Huntington disease. J. Biol. Chem. 285, 8808–8823 10.1074/jbc.M109.075028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sathasivam, K., Neueder, A., Gipson, T.A., Landles, C., Benjamin, A.C., Bondulich, M.K.et al. (2013) Aberrant splicing of HTT generates the pathogenic exon 1 protein in Huntington disease. Proc. Natl Acad. Sci. U.S.A. 110, 2366–2370 10.1073/pnas.1221891110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dahlgren, P.R., Karymov, M.A., Bankston, J., Holden, T., Thumfort, P., Ingram, V.M.et al. (2005) Atomic force microscopy analysis of the Huntington protein nanofibril formation. Nanomedicine 1, 52–57 10.1016/j.nano.2004.11.004 [DOI] [PubMed] [Google Scholar]
- 14.Sharma, D., Shinchuk, L.M., Inouye, H., Wetzel, R. and Kirschner, D.A. (2005) Polyglutamine homopolymers having 8-45 residues form slablike beta-crystallite assemblies. Proteins 61, 398–411 10.1002/prot.20602 [DOI] [PubMed] [Google Scholar]
- 15.Sikorski, P. and Atkins, E. (2005) New model for crystalline polyglutamine assemblies and their connection with amyloid fibrils. Biomacromolecules 6, 425–432 10.1021/bm0494388 [DOI] [PubMed] [Google Scholar]
- 16.Perutz, M.F., Johnson, T., Suzuki, M. and Finch, J.T. (1994) Glutamine repeats as polar zippers: their possible role in inherited neurodegenerative diseases. Proc. Natl Acad. Sci. U.S.A. 91, 5355–5358 10.1073/pnas.91.12.5355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fitzpatrick, A.W.P. and Saibil, H.R. (2019) Cryo-EM of amyloid fibrils and cellular aggregates. Curr. Opin. Struct. Biol. 58, 34–42 10.1016/j.sbi.2019.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galaz-Montoya, J.G., Shahmoradian, S.H., Shen, K., Frydman, J. and Chiu, W. (2021) Cryo-electron tomography provides topological insights into mutant huntingtin exon 1 and polyQ aggregates. Commun. Biol. 4, 849 10.1038/s42003-021-02360-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bäuerlein, F.J.B., Saha, I., Mishra, A., Kalemanov, M., Martínez-Sánchez, A., Klein, R.et al. (2017) In situ architecture and cellular interactions of PolyQ inclusions. Cell 171, 179–187.e10 10.1016/j.cell.2017.08.009 [DOI] [PubMed] [Google Scholar]
- 20.Nazarov, S., Chiki, A., Boudeffa, D. and Lashuel, H.A. (2022) Structural basis of huntingtin fibril polymorphism revealed by cryogenic electron microscopy of exon 1 HTT fibrils. J. Am. Chem. Soc. 144, 10723–10735 10.1021/jacs.2c00509 [DOI] [PubMed] [Google Scholar]
- 21.Duim, W.C., Jiang, Y., Shen, K., Frydman, J. and Moerner, W.E. (2014) Super-resolution fluorescence of huntingtin reveals growth of globular species into short fibers and coexistence of distinct aggregates. ACS Chem. Biol. 9, 2767–2778 10.1021/cb500335w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tycko, R. (2011) Solid-state NMR studies of amyloid fibril structure 62, 279–299 10.1146/annurev-physchem-032210-103539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.van der Wel, P.C.A. (2017) Insights into protein misfolding and aggregation enabled by solid-state NMR spectroscopy. Solid State Nucl. Magn. Reson. 88, 1–14 10.1016/j.ssnmr.2017.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Loquet, A., El Mammeri, N., Stanek, J., Berbon, M., Bardiaux, B., Pintacuda, G.et al. (2018) 3D structure determination of amyloid fibrils using solid-state NMR spectroscopy. Methods 138–139: 26–38 10.1016/j.ymeth.2018.03.014 [DOI] [PubMed] [Google Scholar]
- 25.Bagherpoor Helabad, M., Matlahov, I., Daldrop, J.O., Jain, G., Van Der Wel, P.C.A. and Miettinen, M.S. (2023) Integrative determination of the atomic structure of mutant huntingtin exon 1 fibrils from Huntington's disease. bioRxiv 10.1101/2023.07.21.549993 [DOI]
- 26.Sivanandam, V.N., Jayaraman, M., Hoop, C.L., Kodali, R., Wetzel, R. and van der Wel, P.C.A. (2011) The aggregation-enhancing huntingtin N-terminus is helical in amyloid fibrils. J. Am. Chem. Soc. 133, 4558–4566 10.1021/ja110715f [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider, R., Schumacher, M.C., Mueller, H., Nand, D., Klaukien, V., Heise, H.et al. (2011) Structural characterization of polyglutamine fibrils by solid-state NMR spectroscopy. J. Mol. Biol. 412, 121–136 10.1016/j.jmb.2011.06.045 [DOI] [PubMed] [Google Scholar]
- 28.Hoop, C.L., Lin, H.-K., Kar, K., Magyarfalvi, G., Lamley, J.M., Boatz, J.C.et al. (2016) Huntingtin exon 1 fibrils feature an interdigitated β-hairpin-based polyglutamine core. Proc. Natl Acad. Sci. U.S.A. 113, 1546–1551 10.1073/pnas.1521933113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kar, K., Hoop, C.L., Drombosky, K.W., Baker, M.A., Kodali, R., Arduini, I.et al. (2013) β-hairpin-mediated nucleation of polyglutamine amyloid formation. J. Mol. Biol. 425, 1183–1197 10.1016/j.jmb.2013.01.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hoop, C.L., Lin, H.-K., Kar, K., Hou, Z., Poirier, M.A., Wetzel, R.et al. (2014) Polyglutamine amyloid core boundaries and flanking domain dynamics in huntingtin fragment fibrils determined by solid-state nuclear magnetic resonance. Biochemistry 53, 6653–6666 10.1021/bi501010q [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mishra, R., Hoop, C.L., Kodali, R., Sahoo, B., van der Wel, P.C.A. and Wetzel, R. (2012) Serine phosphorylation suppresses huntingtin amyloid accumulation by altering protein aggregation properties. J. Mol. Biol. 424, 1–14 10.1016/j.jmb.2012.09.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lin, H.-K., Boatz, J.C., Krabbendam, I.E.. Kodali, R., Hou, Z., Wetzel, R.et al. (2017) Fibril polymorphism affects immobilized non-amyloid flanking domains of huntingtin exon1 rather than its polyglutamine core. Nat. Commun. 8, 15462 10.1038/ncomms15462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Boatz, J.C., Piretra, T., Lasorsa, A., Matlahov, I., Conway, J.F. and van der Wel, P.C.A. (2020) Protofilament structure and supramolecular polymorphism of aggregated mutant huntingtin exon 1. J. Mol. Biol. 432, 4722–4744 10.1016/j.jmb.2020.06.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Matlahov, I., Boatz, J.C. and Van Der Wel, P.C.A. (2022) Selective observation of semi-rigid non-core residues in dynamically complex mutant huntingtin protein fibrils. J. Struct. Biol.: X 6, 100077 10.1016/j.yjsbx.2022.100077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Isas, J.M., Langen, R. and Siemer, A.B. (2015) Solid-state nuclear magnetic resonance on the static and dynamic domains of huntingtin exon-1 fibrils. Biochemistry 54, 3942–3949 10.1021/acs.biochem.5b00281 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Isas, J.M., Langen, A., Isas, M.C., Pandey, N.K. and Siemer, A.B. (2017) Formation and structure of wild type huntingtin exon-1 fibrils. Biochemistry 56, 3579–3586 10.1021/acs.biochem.7b00138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Isas, J.M., Pandey, N.K., Xu, H., Teranishi, K., Okada, A.K., Fultz, E.K.et al. (2021) Huntingtin fibrils with different toxicity, structure, and seeding potential can be interconverted. Nat. Commun. 12, 4272 10.1038/s41467-021-24411-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Heise, H., Hoyer, W., Becker, S., Andronesi, O.C., Riedel, D. and Baldus, M. (2005) Molecular-level secondary structure, polymorphism, and dynamics of full-length alpha-synuclein fibrils studied by solid-state NMR. Proc. Natl Acad. Sci. U.S.A. 102, 15871–15876 10.1073/pnas.0506109102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gath, J., Bousset, L., Habenstein, B., Melki, R., Böckmann, A. and Meier, B.H. (2014) Unlike twins: an NMR comparison of two α-synuclein polymorphs featuring different toxicity. PLoS ONE 9, e90659 10.1371/journal.pone.0090659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tycko, R. (2015) Amyloid polymorphism: structural basis and neurobiological relevance. Neuron 86, 632–645 10.1016/j.neuron.2015.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matlahov, I. and van der Wel, P.C. (2019) Conformational studies of pathogenic expanded polyglutamine protein deposits from Huntington's disease. Exp. Biol. Med. 244, 1584–1595 10.1177/1535370219856620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Comellas, G. and Rienstra, C.M. (2013) Protein structure determination by magic-angle spinning solid-state NMR, and insights into the formation, structure, and stability of amyloid fibrils. Annu. Rev. Biophys. 42, 515–536 10.1146/annurev-biophys-083012-130356 [DOI] [PubMed] [Google Scholar]
- 43.Urbanek, A., Morato, A., Allemand, F., Delaforge, E., Fournet, A., Popovic, M.et al. (2018) A general strategy to access structural information at atomic resolution in polyglutamine homorepeats. Angew. Chem. Int. Ed. 57, 3598–3601 10.1002/anie.201711530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaroniec, C.P., MacPhee, C.E., Bajaj, V.S., McMahon, M.T., Dobson, C.M. and Griffin, R.G. (2004) High-resolution molecular structure of a peptide in an amyloid fibril determined by magic angle spinning NMR spectroscopy. Proc. Natl Acad. Sci. U.S.A. 101, 711–716 10.1073/pnas.0304849101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.van der Wel, P.C.A., Lewandowski, J.R. and Griffin, R.G. (2010) Structural characterization of GNNQQNY amyloid fibrils by magic angle spinning NMR. Biochemistry 49, 9457–9469 10.1021/bi100077x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fitzpatrick, A.W.P., Debelouchina, G.T., Bayro, M.J., Clare, D.K., Caporini, M.A., Bajaj, V.S.et al. (2013) Atomic structure and hierarchical assembly of a cross-β amyloid fibril. Proc. Natl Acad. Sci. U.S.A. 110, 5468–5473 10.1073/pnas.1219476110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.van der Wel, P.C.A. (2021) Dihedral angle measurements for structure determination by biomolecular solid-state NMR spectroscopy. Front. Mol. Biosci. 8, 791090 10.3389/fmolb.2021.791090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nielsen, J.T., Bjerring, M., Jeppesen, M.D., Pedersen, R.O., Pedersen, J.M., Hein, K.L.et al. (2009) Unique identification of supramolecular structures in amyloid fibrils by solid-state NMR spectroscopy. Angew. Chem. Int. Ed. 48, 2118–2121 10.1002/anie.200804198 [DOI] [PubMed] [Google Scholar]
- 49.Buchanan, L.E., Carr, J.K., Fluitt, A.M., Hoganson, A.J., Moran, S.D., de Pablo, J.J.et al. (2014) Structural motif of polyglutamine amyloid fibrils discerned with mixed-isotope infrared spectroscopy. Proc. Natl Acad. Sci. U.S.A. 111, 5796–5801 10.1073/pnas.1401587111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Punihaole, D., Workman, R.J., Hong, Z., Madura, J.D. and Asher, S.A. (2016) Polyglutamine fibrils: new insights into antiparallel β-sheet conformational preference and side chain structure. J. Phys. Chem. B 120, 3012–3026 10.1021/acs.jpcb.5b11380 [DOI] [PubMed] [Google Scholar]
- 51.Nelson, R. and Eisenberg, D. (2006) Recent atomic models of amyloid fibril structure. Curr. Opin. Struct. Biol. 16, 260–265 10.1016/j.sbi.2006.03.007 [DOI] [PubMed] [Google Scholar]
- 52.Hervas, R., Rau, M.J., Park, Y., Zhang, W., Murzin, A.G., Fitzpatrick, J.A.J.et al. (2020) Cryo-EM structure of a neuronal functional amyloid implicated in memory persistence in Drosophila. Science 367, 1230–1234 10.1126/science.aba3526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chan, J.C.C., Oyler, N.A., Yau, W.-M. and Tycko, R. (2005) Parallel beta-sheets and polar zippers in amyloid fibrils formed by residues 10-39 of the yeast prion protein Ure2p. Biochemistry 44, 10669–10680 10.1021/bi050724t [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lee, M., Ghosh, U., Thurber, K.R., Kato, M. and Tycko, R. (2020) Molecular structure and interactions within amyloid-like fibrils formed by a low-complexity protein sequence from FUS. Nat. Commun. 11, 5735 10.1038/s41467-020-19512-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wiegand, T. and Meier, B.H. (2020) Asparagine and glutamine side-chains and ladders in HET-s(218–289) amyloid fibrils studied by fast magic-angle spinning NMR. Front. Mol. Biosci. 7, 582033 10.3389/fmolb.2020.582033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Rout, M.P. and Sali, A. (2019) Principles for integrative structural biology studies. Cell 177, 1384–1403 10.1016/j.cell.2019.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Thakur, A.K. and Wetzel, R. (2002) Mutational analysis of the structural organization of polyglutamine aggregates. Proc. Natl Acad. Sci. U.S.A. 99, 17014–17019 10.1073/pnas.252523899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Matlahov, I. and Van der Wel, P.C.A. (2018) Hidden motions and motion-induced invisibility: dynamics-based spectral editing in solid-state NMR. Methods 148, 123–135 10.1016/j.ymeth.2018.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Schanda, P. and Ernst, M. (2016) Studying dynamics by magic-angle spinning solid-state NMR spectroscopy: principles and applications to biomolecules. Prog. Nucl. Magn. Reson. Spectrosc. 96, 1–46 10.1016/j.pnmrs.2016.02.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Darnell, G., Orgel, J.P.R.O., Pahl, R. and Meredith, S.C. (2007) Flanking polyproline sequences inhibit beta-sheet structure in polyglutamine segments by inducing PPII-like helix structure. J. Mol. Biol. 374, 688–704 10.1016/j.jmb.2007.09.023 [DOI] [PubMed] [Google Scholar]
- 61.Thakur, A.K., Jayaraman, M., Mishra, R., Thakur, M., Chellgren, V.M., Byeon, I.-J.L.et al. (2009) Polyglutamine disruption of the huntingtin exon 1 N terminus triggers a complex aggregation mechanism. Nat. Struct. Mol. Biol. 16, 380–389 10.1038/nsmb.1570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shen, K., Calamini, B., Fauerbach, J.A., Ma, B., Shahmoradian, S.H., Serrano Lachapel, I.L.et al. (2016) Control of the structural landscape and neuronal proteotoxicity of mutant Huntingtin by domains flanking the polyQ tract. eLife 5, e18065 10.7554/eLife.18065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Monsellier, E., Redeker, V., Ruiz-Arlandis, G., Bousset, L. and Melki, R. (2015) Molecular interaction between the chaperone Hsc70 and the N-terminal flank of huntingtin exon 1 modulates aggregation. J. Biol. Chem. 290, 2560–2576 10.1074/jbc.M114.603332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Burke, K.A., Kauffman, K.J., Umbaugh, C.S., Frey, S.L. and Legleiter, J. (2013) The interaction of polyglutamine peptides with lipid membranes is regulated by flanking sequences associated with huntingtin. J. Biol. Chem. 288, 14993–15005 10.1074/jbc.M112.446237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Juenemann, K., Wiemhoefer, A. and Reits, E.A. (2015) Detection of ubiquitinated huntingtin species in intracellular aggregates. Front Mol Neurosci 8, 1 10.3389/fnmol.2015.00001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Gu, X., Greiner, E.R., Mishra, R., Kodali, R.B., Osmand, A.P., Finkbeiner, S.et al. (2009) Serines 13 and 16 are critical determinants of full-length human mutant huntingtin induced disease pathogenesis in HD mice. Neuron 64, 828–840 10.1016/j.neuron.2009.11.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cariulo, C., Azzollini, L., Verani, M., Martufi, P., Boggio, R., Chiki, A.et al. (2017) Phosphorylation of huntingtin at residue T3 is decreased in Huntington's disease and modulates mutant huntingtin protein conformation. Proc. Natl Acad. Sci. U.S.A. 10, 201705372 10.1073/pnas.1705372114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Scior, A., Buntru, A., Arnsburg, K., Ast, A., Iburg, M., Juenemann, K.et al. (2018) Complete suppression of Htt fibrilization and disaggregation of Htt fibrils by a trimeric chaperone complex. EMBO J. 37, 282–299 10.15252/embj.201797212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Caulkins, B.G., Cervantes, S.A., Isas, J.M. and Siemer, A.B. (2018) Dynamics of the proline-rich C-terminus of huntingtin exon-1 fibrils. J. Phys. Chem. B 122, 9507–9515 10.1021/acs.jpcb.8b09213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Falk, A.S., Bravo-Arredondo, J.M., Varkey, J., Pacheco, S., Ralf, L. and Siemer, A.B. (2020) Structural model of the proline-rich domain of huntingtin exon-1 fibrils. Biophys. J. 119, 2019–2028 10.1016/j.bpj.2020.10.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bugg, C.W., Isas, J.M., Fischer, T., Patterson, P.H. and Langen, R. (2012) Structural features and domain organization of huntingtin fibrils. J. Biol. Chem. 287, 31739–31746 10.1074/jbc.M112.353839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Siemer, A.B., Arnold, A.A., Ritter, C., Westfeld, T., Ernst, M., Riek, R.et al. (2006) Observation of highly flexible residues in amyloid fibrils of the HET-s prion. J. Am. Chem. Soc. 128, 13224–13228 10.1021/ja063639x [DOI] [PubMed] [Google Scholar]
- 73.Andronesi, O.C., Becker, S., Seidel, K., Heise, H., Young, H.S. and Baldus, M. (2005) Determination of membrane protein structure and dynamics by magic-angle-spinning solid-state NMR spectroscopy. J. Am. Chem. Soc. 127, 12965–12974 10.1021/ja0530164 [DOI] [PubMed] [Google Scholar]
- 74.Bhopatkar, A.A. and Kayed, R. (2023) Flanking regions, amyloid cores, and polymorphism: the potential interplay underlying structural diversity. J. Biol. Chem. 299, 105122 10.1016/j.jbc.2023.105122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ulamec, S.M., Brockwell, D.J. and Radford, S.E. (2020) Looking beyond the core: the role of flanking regions in the aggregation of amyloidogenic peptides and proteins. Front. Neurosci. 14, 611285 10.3389/fnins.2020.611285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Arrasate, M., Mitra, S., Schweitzer, E.S., Segal, M.R. and Finkbeiner, S. (2004) Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature 431, 805–810 10.1038/nature02998 [DOI] [PubMed] [Google Scholar]
- 77.Legleiter, J., Lotz, G.P., Miller, J., Ko, J., Ng, C., Williams, G.L.et al. (2009) Monoclonal antibodies recognize distinct conformational epitopes formed by polyglutamine in a mutant huntingtin fragment. J. Biol. Chem. 284, 21647–21658 10.1074/jbc.M109.016923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Fitzpatrick, A.W.P., Falcon, B., He, S., Murzin, A.G., Murshudov, G., Garringer, H.J.et al. (2017) Cryo-EM structures of tau filaments from Alzheimer's disease. Nature 547, 185–190 10.1038/nature23002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Kandola, T., Venkatesan, S., Zhang, J., Lerbakken, B.T., Von Schulze, A., Blanck, J.F.et al. (2023) Pathologic polyglutamine aggregation begins with a self-poisoning polymer crystal. eLife 12, RP86939 10.7554/eLife.86939.3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kar, K., Baker, M.A., Lengyel, G.A., Hoop, C.L., Kodali, R., Byeon, I.-J.et al. (2017) Backbone engineering within a latent β-hairpin structure to design inhibitors of polyglutamine amyloid formation. J. Mol. Biol. 429, 308–323 10.1016/j.jmb.2016.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Parlato, R., Volarić, J., Lasorsa, A., Bagherpoor Helabad, M., Kobauri, P., Jain, G.et al. (2024) Photocontrol of the β-hairpin polypeptide structure through an optimized azobenzene-based amino acid analogue. J. Am. Chem. Soc. 146, 2062–2071 10.1021/jacs.3c11155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Yang, W., Dunlap, J.R., Andrews, R.B. and Wetzel, R. (2002) Aggregated polyglutamine peptides delivered to nuclei are toxic to mammalian cells. Hum. Mol. Genet. 11, 2905–2917 10.1093/hmg/11.23.2905 [DOI] [PubMed] [Google Scholar]
- 83.Kar, K., Arduini, I., Drombosky, K.W., van der Wel, P.C.A. and Wetzel, R. (2014) D-polyglutamine amyloid recruits L-polyglutamine monomers and kills cells. J. Mol. Biol. 426, 816–829 10.1016/j.jmb.2013.11.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Chaibva, M., Jawahery, S., Pilkington, A.W., Arndt, J.R., Sarver, O., Valentine, S.et al. (2016) Acetylation within the first 17 residues of huntingtin exon 1 alters aggregation and lipid binding. Biophys. J. 111, 349–362 10.1016/j.bpj.2016.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Shen, K. and Frydman, J. (2013) The interplay between the chaperonin TRiC and N-terminal region of Huntingtin mediates Huntington's disease aggregation and pathogenesis. In Protein Quality Control in Neurodegenerative Diseases (Marimoto, R.I. and Christen, Y., eds), Springer, Berlin, Germany. [Google Scholar]
- 86.Jayaraman, M., Mishra, R., Kodali, R., Thakur, A.K., Koharudin, L.M.I., Gronenborn, A.M.et al. (2012) Kinetically competing huntingtin aggregation pathways control amyloid polymorphism and properties. Biochemistry 51, 2706–2716 10.1021/bi3000929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peskett, T.R., Rau, F., O'Driscoll, J., Patani, R., Lowe, A.R. and Saibil, H.R. (2018) A liquid to solid phase transition underlying pathological huntingtin exon1 aggregation. Mol. Cell 70, 588–601.e6 10.1016/j.molcel.2018.04.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Ceccon, A., Tugarinov, V., Ghirlando, R. and Clore, G.M. (2020) Abrogation of prenucleation, transient oligomerization of the Huntingtin exon 1 protein by human profilin I. Proc. Natl Acad. Sci. U. S.A. 117, 5844–5852 10.1073/pnas.1922264117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Legleiter, J., Mitchell, E., Lotz, G.P., Sapp, E., Ng, C., DiFiglia, M.et al. (2010) Mutant huntingtin fragments form oligomers in a polyglutamine length-dependent manner in vitro and in vivo. J. Biol. Chem. 285, 14777–14790 10.1074/jbc.M109.093708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Smith, A.N., Harrabi, R., Halbritter, T., Lee, D., Aussenac, F., van der Wel, P.C.A.et al. (2023) Fast magic angle spinning for the characterization of milligram quantities of organic and biological solids at natural isotopic abundance by 13C–13C correlation DNP-enhanced NMR. Solid State Nucl. Magn. Reson. 123, 101850 10.1016/j.ssnmr.2022.101850 [DOI] [PubMed] [Google Scholar]
- 91.Smith, A.N., Märker, K., Piretra, T., Boatz, J.C., Matlahov, I., Kodali, R.et al. (2018) Structural fingerprinting of protein aggregates by dynamic nuclear polarization-enhanced solid-state NMR at natural isotopic abundance. J. Am. Chem. Soc. 140, 14576–14580 10.1021/jacs.8b09002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Paravastu, A.K., Qahwash, I., Leapman, R.D., Meredith, S.C. and Tycko, R. (2009) Seeded growth of beta-amyloid fibrils from Alzheimer's brain-derived fibrils produces a distinct fibril structure. Proc. Natl Acad. Sci. U.S.A. 106, 7443–7448 10.1073/pnas.0812033106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nekooki-Machida, Y., Kurosawa, M., Nukina, N., Ito, K., Oda, T. and Tanaka, M. (2009) Distinct conformations of in vitro and in vivo amyloids of huntingtin-exon1 show different cytotoxicity. Proc. Natl Acad. Sci. U.S.A. 106, 9679–9684 10.1073/pnas.0812083106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Schubeis, T., Nagaraj, M. and Ritter, C. (2017) Segmental isotope labeling of insoluble proteins for solid-state NMR by protein trans-splicing. Methods Mol. Biol. 1495, 147–160 10.1007/978-1-4939-6451-2_10 [DOI] [PubMed] [Google Scholar]