Abstract
With the rapid expansion of aging biology research, the identification and evaluation of longevity interventions in humans have become key goals of this field. Biomarkers of aging are critically important tools in achieving these objectives over realistic timeframes. However, the current lack of standards and consensus within the aging biomarker field hinders the further development and validation of these important tools for clinical applications. Here, we advance a framework for the terminology and characterization of biomarkers of aging, including classification and potential clinical use cases. We discuss validation steps and highlight ongoing challenges as potential areas in need of future research. This framework sets the stage for the future development of valid biomarkers of aging and their ultimate utilization in clinical trials and practice.
Introduction
Organisms change in various ways with the passage of time. Some of these changes reflect the execution of a genetic program of development, and others reflect the accumulated effects of experiences, exposures, and deleterious byproducts of life: collectively, these changes comprise aging (definition proposed by this work in Table 1). In the absence of a clear consensus on the biological definition of aging prior to this work1,2, its detrimental effects are broadly thought to be mediated by the negative consequences of biological, chemical, or physical processes, such as the accumulation of molecular damage3. Together, these events lead to the cumulative breakdown of physiological systems, loss of resilience, increased susceptibility to disease, and ultimately mortality4–6. Genetic, pharmacological, dietary, and lifestyle interventions, extend healthy lifespan and/or attenuate age-related functional decline in animal models7, suggesting that the biological processes underlying aging are amenable to modulation8,9. The geroscience hypothesis posits that targeting the aging process itself10, rather than the individual diseases of aging, may prevent, delay, reduce the severity of many age-related diseases in parallel9. In turn, this approach may modulate healthspan (Table 1)9. While the impact of interventions on longevity can be readily investigated in animal models with short lifespans, ethical, biological, and economic considerations challenge the translation of these findings to humans11,12. Hence, alternative means to quantify the accumulation of age-related molecular damage and clinical functional decline are required to test interventions targeting aging4,12. Moreover, lifespan (and its extension) alone may not be the most informative parameter in evaluating anti-aging interventions in humans: for instance, an intervention may significantly extend healthspan without a large impact on lifespan. For these reasons, the development of biomarkers that reflect the diverse biological processes underlying aging and its consequences and are ideally sensitive to interventions targeting aging are critically needed. Hereafter, we refer to these biomarkers as biomarkers of aging.
Table 1. Definition of terms utilized in this review, in order of appearance in the text.
Unless otherwise noted, terms are consensus working definitions proposed in the current work.
| Term | Definition |
|---|---|
| Aging | The process resulting from accumulation of consequences of life, such as molecular and cellular damage, that leads to increased risk of functional decline, chronic diseases, and ultimately mortality |
| Healthspan | The period of life prior to onset of chronic disease and disabilities of aging, i.e., in good health (extended from28) |
| Biomarker of aging | A quantitative parameter of an organism that either alone or in a composite predicts biological age and ideally its changes in response to interventions |
| Biological age | Conceptually, an individual’s age defined by the level of age-dependent biological changes, such as molecular and cellular damage accumulation. In practical use, this is often summarized as a number matching the chronological age where the average person in a reference population shares the individual’s level of age-dependent biological changes |
| Chronological age | An individual’s age defined by time elapsed since birth |
| Age acceleration (age deviation) | The difference between biological age and chronological age (originally defined by29); we propose adoption of the term age deviation (AgeDev) for this concept to distinguish it from an increased rate of aging and encompass changes in both directions |
| Geroprotector | An agent or intervention that increases healthspan or lifespan and ameliorates [tested] biomarkers of aging (extended from8,30) |
In the context of interventions, a biomarker is defined as a biological feature that can indicate processes of interest in a given individual. Such processes may be normal, pathologic, or in response to a given treatment or exposure13. The urgent need for biomarkers of aging to identify longevity interventions was recognized as early as the 1960s in response to the earlier discovery that aging is modifiable14 (Figure 1a). To address this need, a series of U.S. National Institute on Aging (NIA)-sponsored workshops and initiatives from 1981 to 2000 explored biomarkers of aging largely in animal models15,16. While it was previously deemed too early to constitute a definitive panel of biomarkers of aging for animal models or humans7,16, molecular and omic biomarkers of aging developed over the last decade represent promising candidates (Figure 1a)4,6,8,9,12. However, there is currently no consensus on evaluation and validation methods for these biomarkers8,17, nor is there any standardization of how such biomarkers are utilized, even in preclinical settings. To establish a foundation on which we may build biomarkers of aging up to their full potential, we propose a framework for classifying and assessing these biomarkers as tools to identify and evaluate longevity interventions. Our framework is grounded in prior advances in the field of aging and biomarker research, but extends them by engaging a multidisciplinary panel of experts to build consensus on key terminology (Table 1), the classification of biomarkers from a regulatory point of view (Figure 2), the explanation of certain use cases based on existing biomarkers (Table 2) and trials (Table 3), and the assessment of biomarkers, for instance using validated geroprotectors (Figure 3). Our goal is to take steps to address a critical unmet need in the field of aging research4: the evaluation biomarkers to assess changes in biological age (see Table 1 for definition; in contrast to chronological age). Defining common ground on these foundational issues will be key to systematically validating aging biomarkers and ultimately advancing them to the clinic.
Figure 1. Timeline of key events related to biomarkers of aging and types of common biomarkers of aging.
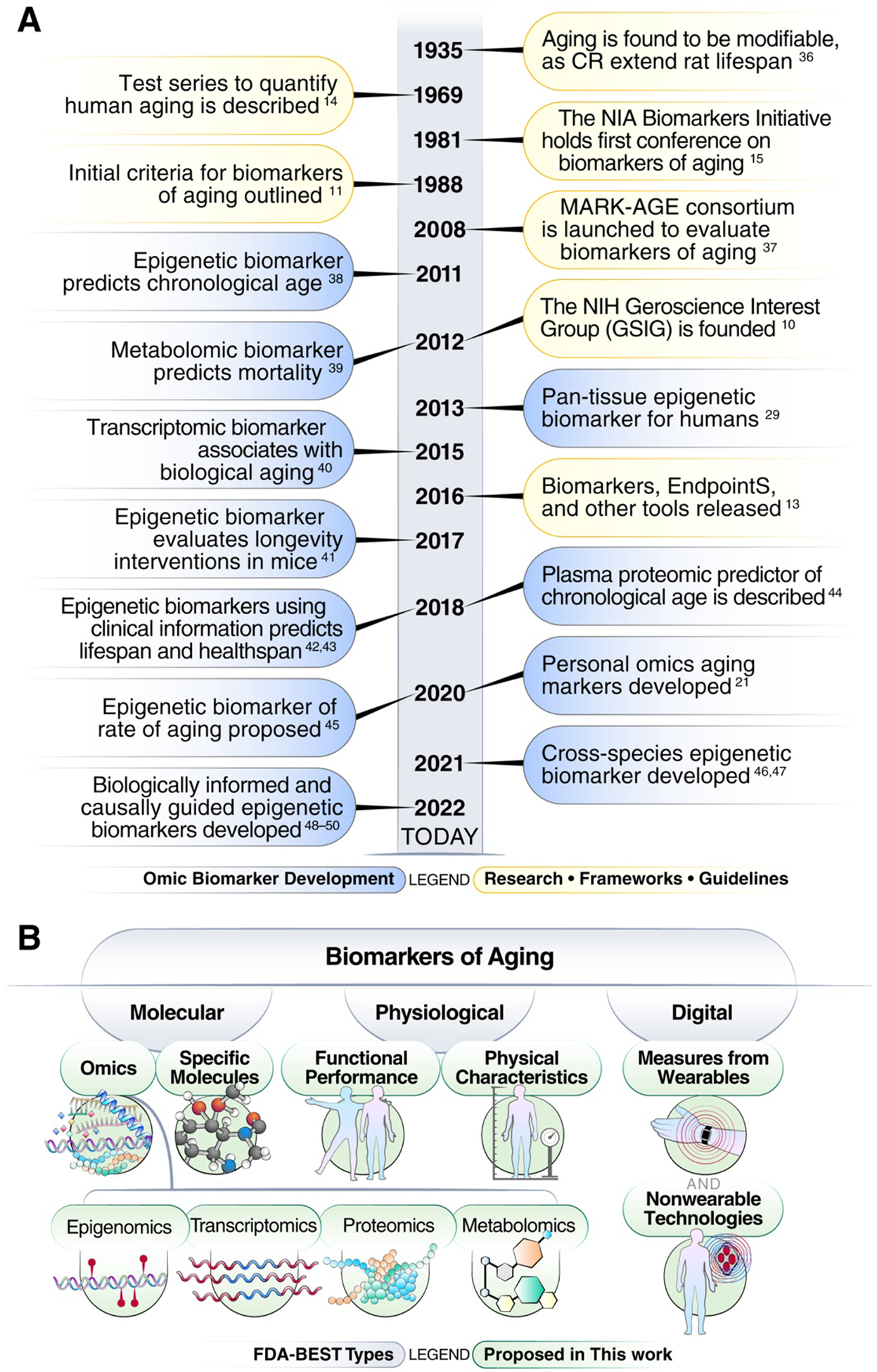
a. Timeline of landmark events related to biomarkers of aging. b. Common types and subtypes of biomarkers of aging, based on what they measure. Gray-shaded regions are based on the broad FDA-BEST types of biomarkers framework.
Figure 2. Categories of common and potential biomarkers of aging based on their application.
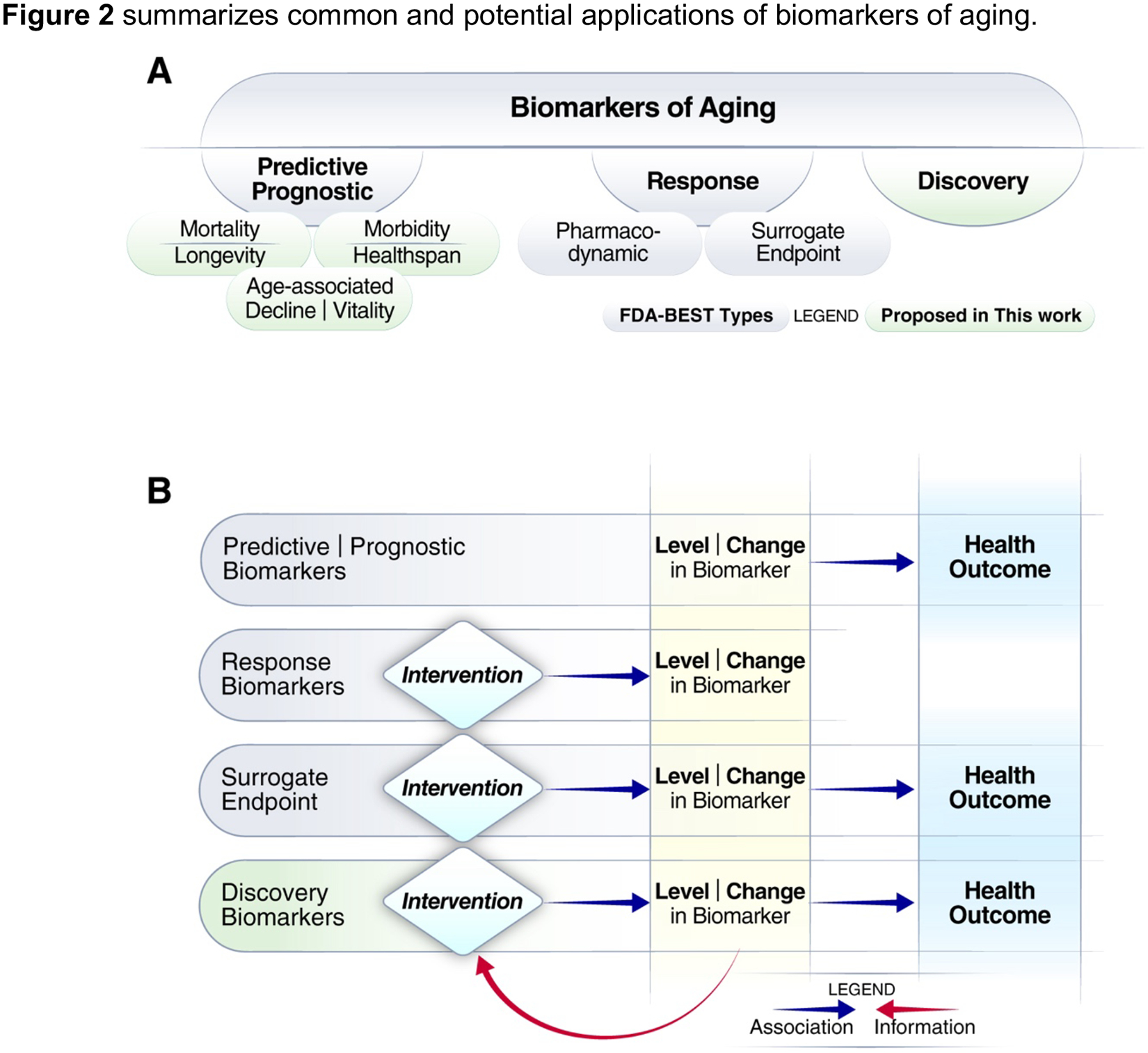
a. Classification of biomarkers of aging. Gray-shaded regions are based on the broad categories of biomarkers proposed by FDA-BEST. b. Relationships between biomarkers, interventions, and health outcomes, extended from Cummings and Kritchevsky (2022)52 to include discovery biomarkers and the information feedback loop.
Table 2. A select list of human predictive biomarkers of aging associated with various age-related conditions and their commercial applications.
Refer to the supplementary materials for the commercial application details.
| Biomarker of aging | Biomarker type | Age-related conditions | Commercial application |
|---|---|---|---|
| DNAmAge (Horvath29, Hannum53) | Epigenetic clocks, based on a set of DNA methylation measures associated with chronological age | Associated with multiple aging diseases and time-to-death, based on meta-analyses54,55 | Licensed for estimating chronological age |
| GlycanAge56 | A panel of molecular measures based on glycans attached to Immunoglobulin G (IgG) antibodies associated with chronological age | Associated with multiple diseases57 | Commercially used to track responses to lifestyle changes |
| PhenoAge42 and GrimAge43 | Epigenetic clocks, based on a set of DNA methylation measures associated with “clinical phenotypic age measures” (a panel of age-associated molecular and physiological biomarkers, measured in blood) | Higher association with multiple aging-related diseases and time-to-death, compared to previous DNAm biomarkers, and associated with healthspan42,43. Associated with multiple age-related clinical phenotypes (walking speed, frailty, and cognitive functions)58 | Licensed for optimizing life insurance |
| DunedinPoAm and DunedinPACE59 | Epigenetic clocks, based on a set of DNA methylation measures associated with “pace of aging measures” (a panel of age-associated molecular and physiological biomarker measurements of different organ systems) | Associated with the incidence of multiple chronic diseases, including dementia, disability, and mortality59,60 | Licensed for tracking the rate of aging |
| Multi-omic biological age estimation based on KDM61 | KDM applied to over 900 principal component transformed biomarkers (metabolites, proteins, genomics, and clinical measures) | Positively and negatively modulated by “healthy” and “unhealthy” behaviors/health conditions (e.g., type 2 diabetes), respectively61 | Licensed for tracking biological age |
| Aging.AI, Deep Transcriptomic and Proteomic Clocks | AI-based blood clocks, based on hematological parameters, transcriptomic and proteomic data | Associated with all-cause mortality62 and wasting63 | Commercially available for use in clinical trials |
Table 3.
A list of recently completed ongoing registered clinical trials or post hoc analyses using epigenetic biomarkers of aging with a focus on longevity.
| Type | Study | Intervention | Title | Design, N, age range, (m/f) | Primary Outcome Measure | Biomarker | Biomarker Outcome Measure | Result |
|---|---|---|---|---|---|---|---|---|
| Lifestyle | CALERIE | CR for 2 years | Comprehensive Assessment of Long-Term Effects of Reducing Intake of Energy | RCT, 218, 21–50 | Change in core body temperature and metabolic rate at 24 months compared to baseline | DunedinPACE, GrimAge, PhenoAge (blood chemistry), Horvath and Hannum clocks | Post hoc analysis | Significant reduction of DunedinPACE and PhenoAge (blood chemistry), no significant effects for other biomarkers of aging72,73 |
| DAMA | Plant-food rich diet, exercise | Diet Exercise and Mammography Trial | RCT, 219, 50–69 (f) | Change in mammographic breast density | GrimAge | Post hoc analysis | Dietary intervention: 0.66 years ↓ (GrimAge)74 | |
| MDL | Diet, exercise, stress management, phytonutrient and probiotic supplements | Methylation Diet and Lifestyle Study | RCT, 44, 50–72 (m) | Health-related quality of life | Horvath clock | Exploratory | 3.2 years ↓75 | |
| Tirol-GESUND | Intermittent fasting or smoking cessation | TirolGESUND: General Exercise, Smoking Undone, and Nutrition Diet | BCS, 156, 30–60 (f) | Epigenetic biomarkers of aging and disease risk | WID-REA, -RIA, pcgtAge, and WID-SOLA | Primary | Not yet reported | |
| Pharmacological | Dasatinib/Quercetin | Dasatinib and quercetin | Safety and Effectiveness of Quercetin & Dasatinib on Epigenetic Aging | BCS, 25*, > 40 | Epigenetic clock | DNAm (exact biomarker not defined) | Primary | Not yet reported |
| RAPA | Rapamycin | Topical-RAPA Use in Inflammation Reversal and Re-setting the Epigenetic Clock | RCT, 50*, 65–95 | Epigenetic clock | Horvath clock | Primary | Not yet reported | |
| SGLT2i | Dapagliflozin | SGLT2 Inhibition in Older Obese Adults With Prediabetes | RCT, 20*, > 60 | Advanced glycation end products in urine | DNAm (exact biomarker not defined) | Secondary | Not yet reported | |
| TRIIMX | Growth hormone for 1 year | Thymus Regeneration, Immunorestoration, and Insulin Mitigation Extension | RCT, 85*, 40–80 | Epigenetic clock, thymus regeneration | GrimAge | Primary | Not yet reported | |
| Plasmapheresis | PLASMA | Young plasma | The Plasma for Alzheimer SymptoM Amelioration (PLASMA) Study | BCS, 18, 60–95 | Adverse effects as a measure of safety and tolerability | GrimAge, Horvath, Hannum and Skin and Blood76 clocks, PhenoAge, DNAmTL77 | Post hoc analysis | 0.86 years ↓ (GrimAge), no change in other clocks78 |
| Plasmapheresis | Young plasma | Effects of Plasmapheresis on Aging Biomarkers | O, 41*, 40–60 | Epigenetic clock | DNAm (exact biomarker not defined) | Primary | Not yet reported | |
| RESET-YOUTH | Young plasma | Reversing Epigenetic and Other Markers of Senescence by Transfusing Young Plasma To Older Human Subjects | BCS, 2120*, > 40 (m) | Epigenetic clock | DNAm (exact biomarker not defined) | Primary | Planned | |
| Supplement | AC11 | AC-11 Supplement for 2 months | AC-11 Supplement and Biological Aging | BCS, 32*, > 55 | Epigenetic clock, telomere length | DNAm (exact biomarker not defined) | Primary | Not yet reported |
| D-SUNNY | Vitamin D for 4 months | Vitamin D Supplementation in Overweight/Obese African American Adults and Youth | RCT, 74, 13–4 | Cardiovascular phenotypes, Dose-response | Horvath and Hannum age deviation | Post hoc analysis | 1.85 years ↓ (Horvath age deviation) compared to placebo79 | |
| NMN | Nicotinamide mononucleotide | To Evaluate the Efficacy and Safety of NMN as an Anti-ageing Supplement in Middle Aged and Older Adults | RCT, 90, 40–65 | Cellular NAD+ levels, walking test, health questionnaire | Aging.Ai 3.0 calculator (https://www.aging.ai) | Exploratory | Maintenance of blood biological age compared to placebo80 | |
| Rejuvant | Alpha-ketoglutarate | Rejuvant™ Safety and Biomarker Study | RCT, 100, 45–75 | c-reactive protein levels | DNAm (exact biomarker not defined) | Exploratory | Not yet reported |
Table is ordered by intervention type (lifestyle, pharmacological, plasmapheresis, and supplement) and alphabetically. Most clinical studies to-date have used epigenetic clocks such as the Horvath Clock. Abbreviations: N, number of participants; m, male participants only; f, female participants only; RCT, randomized controlled trial; BCS, baseline-controlled study; O, observational. Symbols: *, estimated.
Figure 3. Relation between biomarkers of aging and geroprotectors.
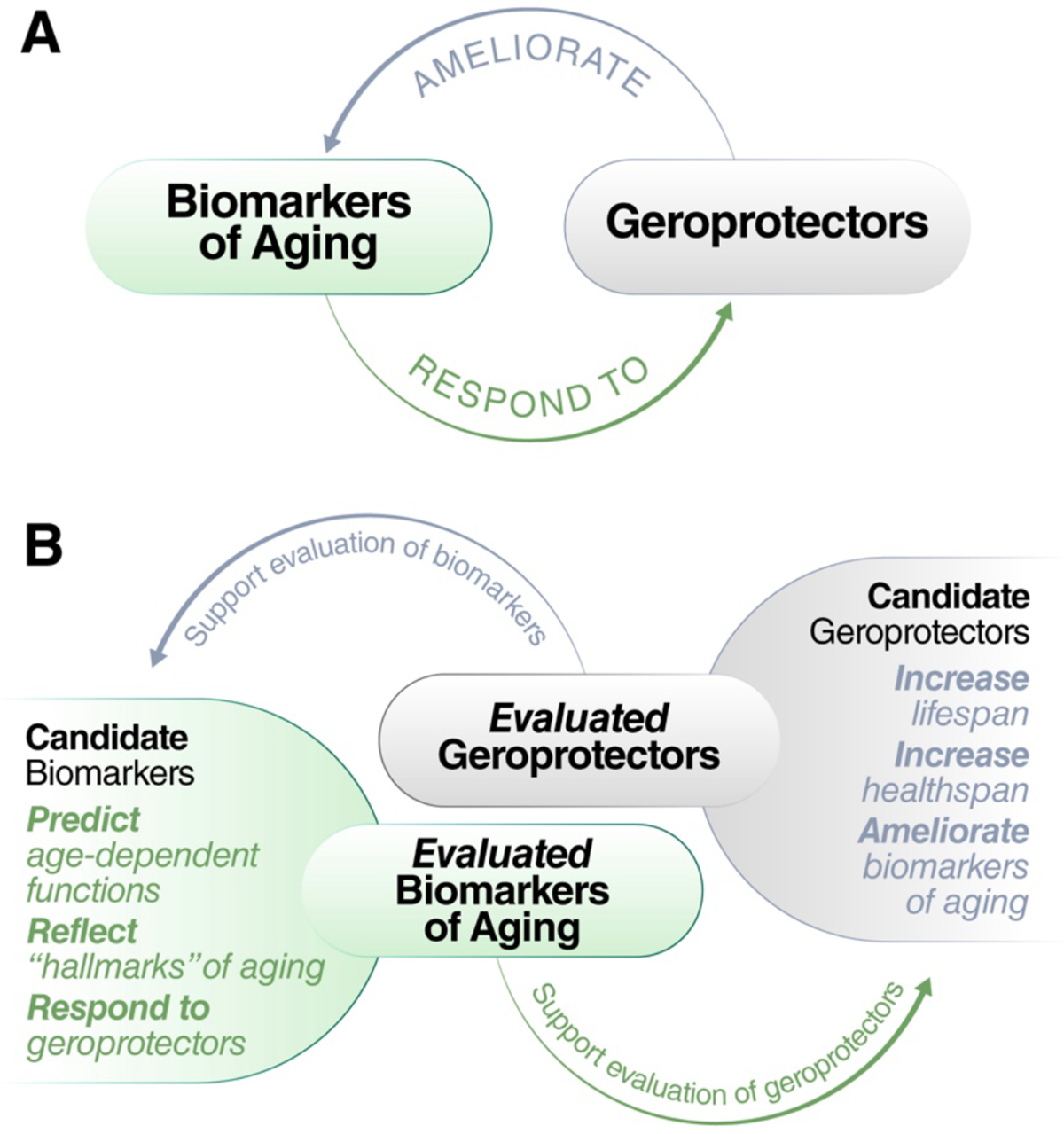
Biomarkers of aging and geroprotectors may appear to have a circular relationship (a). However, development of each is useful to the other: evaluated geroprotectors can be used to develop and benchmark biomarkers of aging, while evaluated biomarkers of aging may be used to predict or test the response to candidate geroprotectors (b).
Terminology and conceptual considerations for biomarkers of aging
Aging biology as a field suffers from a lack of consensus on the biological nature of aging, and various scientists use the term “aging” to refer to different processes1,2,12. The current evidence suggests that aging involves deleterious changes associated with life and results in cumulative breakdown of multiple physiological systems12,18. However, the mechanisms underlying these changes are not uniform across time, cell types19, organ systems20, individuals12,21, and populations22. This renders it challenging to define a single and highly generalizable molecule, method, or assay that measures “aging” because this process involves multiple potentially discordant systems, as well as the loss of their communication and interactions. Moreover, aging may be influenced by individual variabilities, such as the interplay of genetics, lifetime exposures and other factors such as disease12,23. Given these complexities, many definitions of aging have been proposed. Crucially, multiple notions of biological aging may be valid for different aspects of this process. For instance, a definition that focuses on the age-related loss of health may be used by demographers and geriatricians, but is less useful for biologists studying the basic mechanisms underlying aging. Similarly, different biomarkers may capture diverse aspects of aging, as has recently been proposed in cross-comparison studies investigating multiple molecular biomarkers of aging24–27. These issues highlight the important need for further research and systematic evaluation to provide insights into the underlying mechanisms of aging and their relationships with various biomarkers of aging.
Despite these complexities, it is necessary to establish common working terminology for biomarker research. Relevant terms, definitions, and conceptual considerations are briefly outlined in Table 1.
Classifications and applications of biomarkers of aging
Classifications of biomarkers of aging
Multiple principal categories based on the types of associated measurements have been proposed for biomarkers of aging. These include molecular, biological, functional, clinical, and phenotypic biomarkers of aging31–33. To increase consistency with the broader biomarker research field, we propose to adapt and extend definitions from the U.S. Food and Drug Administration (FDA) that pertain to interventions, specifically the FDA-NIH Biomarkers, EndpointS, and other Tools (BEST) classification for biomarkers of aging. The FDA-BEST glossary was developed with the aim of harmonizing terms used in translational medicine to improve consistency and align expectations. This glossary broadly classifies biomarkers as molecular, physiological, histologic, or radiographic13. Molecular biomarkers of aging, perhaps the largest class of such biomarkers, can be based on omics (e.g., epigenomics, proteomics or metabolomics), or specific individual molecules (e.g., circulating levels of interleukin-6 or insulin-like growth factor 1, or composites of blood markers). Physiological biomarkers of aging can be measures of functional performance (e.g., cardiorespiratory fitness, VO2 max, gait speed, timed walking distance, grip strength, or cognitive function) or physical characteristics (e.g., body-mass index or weight-height ratio)31. The FDA has accepted some of these surrogates, such as 6-minute walk distance34, for approving therapies for disease indications. However, they are not currently included in the FDA’s Table of Surrogate Endpoints, which are the basis of drug approval or licensure (https://www.fda.gov/drugs/development-resources/table-surrogate-endpoints-were-basis-drug-approval-or-licensure). Presumably, surrogates accepted by the FDA and other authorities for supporting disease indications could be used as primary endpoints for disease-prevention or healthspan trials.
Other types of biomarkers are also being increasingly put forward for aging applications. For instance, digital biomarkers have recently been proposed. This type of biomarker is garnering increased attention due to advances in digital health technologies (DHTs), including both wearable and nonwearable technologies. Although current digital biomarkers of aging often overlap with functional and physical biomarkers23,35, considering them as orthogonal entities may broaden the applications of DHTs to include other biomarkers such as longitudinal monitoring biomarkers in humans23 and animal models35. Histologic and radiographic biomarkers comprise another class of tools, that have not been utilized as widely in aging research for several reasons: (1) as they often require specialized equipment, expertise, and time, they can be more difficult to measure; (2) they often measure the characteristics of only specific tissues; and (3) computational methods for handling these data are less developed. These biomarkers potentially stand to become more widely used given recent advances addressing these issues.
Adapting the BEST framework, we propose a classification of common biomarkers of aging (Figure 1b). Note that a biomarker could belong to multiple classes (e.g., molecular and physiological or functional and digital). As they are the most developed class of aging biomarkers to date, we focus mainly on molecular biomarkers of aging.
Clinical applications of biomarkers of aging
In addition to classification by type, biomarkers may also be categorized based on their clinical application. Such categories include response, predictive, investigational, mechanistic/underlying biology, surrogate, and disease outcomes33,51,52. Again, we propose an adaptation and extension of the FDA-BEST classification, which defines the categories of clinical susceptibility/risk, diagnostic, monitoring, prognostic, predictive, response, and safety13, for application to biomarkers of aging. Among the listed categories of biomarkers, predictive and response biomarkers are currently the most relevant in the context of aging research, although it should be noted that no aging biomarkers of any category have been approved by U.S. regulators for clinical applications.
Predictive biomarkers
In intervention studies or clinical care, a predictive biomarker can enrich or identify individuals who may be most likely to experience beneficial or detrimental effects from a certain treatment or exposure13. The term is also occasionally used in epidemiological research to describe biomarkers that help to identify individuals more likely to experience a certain event (e.g., death) than others in the absence of intervention (Figure 2a). In 2019, the NIA Predictive Biomarkers Initiative (https://www.predictivebiomarkers.org) was launched to aid the development and validation of both existing and novel predictive biomarkers for age-related disease, with core goals of analytical validation of high-throughput assays and evaluation of associations of outcomes in longitudinal studies. Additionally, many recent studies have applied various biomarkers of aging to predict healthspan, lifespan, or other age-related conditions and diseases (see Table 2 for a selected list with commercial applications). For an additional list of proposed biomarkers of aging, we refer readers to the MARK-AGE project37, the Digital Aging Atlas (https://www.ageing-map.org/), and the TAME Biomarkers Workgroup51.
Prognostic biomarkers
Recent studies have also explored the potential application of biomarkers of aging as prognostic biomarkers of age-related diseases64,65. Prognostic biomarkers are similar to predictive biomarkers but are applied in already diseased individuals to predict disease course and/or future outcomes. For instance, some biomarkers developed broadly in the context of aging have been shown to be associated with progression of some age-related diseases, such as Alzheimer’s disease60 and cancer64,65. On the other hand, specific prognostic biomarkers of individual age-related diseases have been increasingly identified and tested (e.g., plasma phosphorylated-tau181 as a prognostic biomarker of Alzheimer’s disease66). However, few studies have proposed or developed prognostic biomarkers of aging to predict progression or outcomes of the aging process as a whole.
Response biomarkers
A response biomarker indicates the biological reaction of an individual to an exposure or intervention. Response biomarkers may indicate uptake or metabolism of a drug (pharmacodynamic biomarker) or a change in a biological pathway caused by the intervention13. Such biomarkers may be used to establish proof-of-concept, assist in dose selection or measure the response to medical products or environmental agents13. For example, interleukin-613 and oxidative stress biomarkers67 may be used as pharmacodynamic biomarkers when evaluating inflammation associated with the use of tobacco. Additionally, response biomarkers in pathways associated with an outcome may become candidate surrogate endpoint biomarkers, predictive of a clinical outcome.
Surrogate endpoint biomarkers
After appropriate validation, surrogate endpoint biomarkers may be used in clinical trials as a substitute for a direct measure of how a patient or participant feels, functions, or survives13 (Figure 2). In other words, they may serve as the primary efficacy endpoints in large, well-controlled trials intended to support regulatory approval of an intervention. For instance, blood pressure reduction is an FDA-validated surrogate endpoint for reduction in rates of stroke13,68. Surrogate endpoints are particularly useful when the actual desired clinical endpoint is challenging to measure or expected to manifest long after an intervention is initiated. For this reason, surrogate endpoints are highly relevant to aging, where age-related disease(s) of interest or mortality would be primary endpoints12,13,52. However, FDA acceptance of a surrogate endpoint is considerably challenging. Based on how well a potential surrogate endpoint is validated, U.S. regulation recognizes “validated”, “likely”, or “candidate” surrogate endpoints. Clinical trials are needed to show that surrogate endpoints are predictive of, or correlate with, clinical outcomes. Establishing that a biomarker is responsive to an intervention (response biomarker) is the first step toward identifying a surrogate endpoint biomarker. This is not the only requirement, and examples of failed surrogate endpoints exist52. There are not yet any formally validated or likely surrogate endpoint biomarkers of aging — in part because they have only recently been described. The challenge of validating surrogate endpoints is compounded by the lack of consensus on interventions that improve clinical outcomes relevant to healthspan or lifespan. The correlation of candidate surrogate endpoints with clinical responses to effective interventions is generally required for full validation. Nonetheless, the investigation of biomarkers of aging, as response and potential candidate surrogate endpoint biomarkers has rapidly increased over recent years51,69,70. In particular, epigenetic biomarkers commonly termed ‘clocks’ have been increasingly used as candidate biomarkers of aging in clinical trials with a focus on longevity or rejuvenation. Table 3 lists recently completed or ongoing registered clinical trials that evaluate epigenetic aging signatures as response biomarkers, either as predefined outcome measures or during post hoc analyses. Further studies will be required to confirm whether these biomarkers are associated with or predictive of later clinical outcomes and thereby represent candidate surrogate endpoint biomarkers.
Discovery biomarkers
Biomarkers of aging that can be linked to biological pathways may provide practical utility for the identification of novel therapeutic targets and longevity interventions (Figure 2b). Additionally, it has been demonstrated that the use of large-scale omics data in combination with artificial intelligence models can aid in the identification of novel targets71. Once validated, such discovery biomarkers, possibly in combination with computational models, may reduce the prohibitive cost and time of the drug discovery process for diseases of aging.
Criteria for the assessment of biomarkers of aging
Over the past several decades of aging research, several criteria for an ideal biomarker of aging have been proposed11,15,69,81: (1) measurement of the biomarker should be minimally invasive and reliable, i.e., it should be possible to conduct longitudinal measurements with little technical variability; (2) the biomarker should be relevant to aging; (3) the biomarker should predict functional aspects of aging, e.g., mortality, better than chronological age; and (4) the biomarker should ideally be responsive to longevity interventions. We expand on these concepts below. Importantly, the criteria explored here are, as a whole, neither necessary nor sufficient for the validation of biomarkers of aging. Rather, they represent a framework for the characterization and assessment of aging biomarkers to assess the extent to which a candidate biomarker may be feasible, valid, and useful for a specific context of use. As mentioned above, it may be unrealistic to identify a single biomarker that captures all aspects of biological aging and satisfies all criteria. Each biomarker of aging has advantages and limitations, which may be evaluated using this framework.
Feasibility and validity
To allow for repeated measurements in animals during studies of longevity interventions and a subsequent translation to human trials at later stages, the feasibility criteria state that the measurement should be (1) nonlethal to model animals and minimally invasive to humans, (2) repeatable (to allow for monitoring in longitudinal studies) and (3) measurable during a short time relative to the organism’s lifespan11,15,82. The criterion of non-age-accelerating – i.e., the act of measuring the biomarker itself should not accelerate biological aging – has additionally been proposed for biomarkers of aging16. Feasibility criteria are relatively straightforward to establish as they relate to the practical aspects of making biological/clinical measurements.
Criteria of validity comprise a more complicated set of considerations, which we explore in detail below. Briefly, the first validity criterion posits that a biomarker of aging should be age-sensitive15. While this may appear trivial, some considerations are warranted when applied to biomarkers of aging: for instance, a biomarker perfectly correlated with chronological age would be ideal for purposes such as forensics, but it may be less informative for assessment of longevity interventions (i.e., “paradox of biomarkers”)83. Furthermore, a recent study suggested that when the performance of a biomarker of chronological age approaches near-perfect predictive accuracy, its association with mortality attenuates83. Therefore, a biomarker of aging suitable for the evaluation of longevity interventions is expected to show a strong – but not perfect – correlation with chronological age. The implicit assumption underlying this criterion is that a biomarker of aging measures biological rather than chronological age. This assumption was the foundation of the first formally proposed definition of a biomarker of aging as a single or composite parameter capable of predicting aging-associated outcomes better than chronological age alone11.
Since the time of this original definition, advances in big data and machine learning have facilitated the identification of features, or a composite thereof, that predict biological age. However, these data-driven approaches create new challenges in evaluating validity: in the absence of underlying biological or interpretable models, algorithms are likely to detect a mix of features that can cause aging or be caused by the aging process. Among those features that are not directly causal, some may still be relevant to biological age, but some may simply be uninformative correlates of chronological age. Depending on factors underlying study design and model development, models may also include features that are specific to a given study population but not generalizable, and/or capture technical noise or batch effects driven by measurement errors. The interpretation of any algorithmic biomarker of aging depends heavily on understanding the contribution of individual features, and biomarkers of aging that help to mechanistically understand the aging process should ideally retain only features that cause or are directly caused by aging84. Despite such challenges, many existing biomarkers of aging perform very well in predicting chronological age, future mortality, and possibly response to interventions, but we must remain cautious about extending their interpretation as direct measures of biological aging.
Age-sensitivity criteria
Validity criteria may be further expanded into two related criteria: a biomarker of aging should (1) correlate with age-sensitive features across multiple domains after adjusting for chronological age, and (2) be a good predictor of all-cause mortality7,16. Adaptation of these criteria for functional states during aging, i.e., functional aging, has been formalized as predicting physiological, cognitive, and physical function in an age-coherent way, doing so better than chronological age, and predicting the years of remaining functionality better than chronological age16. While these criteria are not quantitative, they provide additional features against which a candidate biomarker may be assessed. At the heart of these extended criteria is the observation that aging is a multisystem process, and therefore, a biomarker of aging should be an indicator of biological processes or responses relating to multiple physiological systems or should encompass a combination of various biomarkers of different physiological systems17,85,85,86. Unsurprisingly, composite multisystem biomarkers are more robust and show a stronger correlation with other features of aging than single biomarkers of aging25,86,87. This is likely for two reasons: first, integrative biomarkers condense signal while eliminating, or at least minimizing, measurement error and noise. Second, integrative biomarkers incorporate heterogeneous aspects of the aging process and thus tend to arrive closer to some ‘consensus’ signal88. However, as mentioned above, the substance of the algorithms, including weights assigned to different biomarkers/processes/systems, is not necessarily biologically informed, and the results should accordingly be interpreted with caution.
Integrative biomarkers of systemic biological aging fall into two broad categories: measures of the progress of aging, i.e., the extent of biological damage accumulation/deterioration and corresponding loss of integrity/resilience-capacity of tissues and organ systems, and measures of the pace of aging, i.e., the rate at which this progress accumulates. Biomarkers of the progress of aging typically estimate biological age. Variation in the progress of aging between individuals of the same chronological age is quantified as the difference between an individual’s chronological age and their biomarker-estimated biological age. This is sometimes referred to as age acceleration, but we propose the more informative and straightforward term ‘age deviation’ (AgeDev, Table 1)29. The association of AgeDev and aging-associated outcomes such as morbidity, disability, and mortality is the established criterion for evaluating proposed metrics of biological aging89–92. Alternatively, biomarkers of aging could be designed to directly correspond to AgeDev by controlling for chronological age in the design process (e.g., GrimAge43 and DunedinPACE59). Notably, the criterion of age sensitivity primarily applies to biomarkers of biological age, where the biomarker should, on average, exhibit higher scores with increasing age. By contrast, biomarkers of the rate of aging do not need to correlate with chronological age, although they may be considered ‘age-sensitive by design’.
Mechanistic criteria
Mechanistic criteria relate to the underlying biology of aging. An improved understanding of the cellular and molecular “hallmarks” or “pillars” of aging, as well as an improved understanding of the nature of aging, suggests that underlying factors contribute to age-associated physiological decline and together determine aging phenotypes33,93. Accordingly, a valid biomarker of aging should reflect these underlying cellular and molecular processes94. A new generation of biologically informed (e.g., preliminary evidence from PRC2 clock48 and deconstructed clocks50) or causally guided (e.g., preliminary evidence from DamAge clock49) epigenetic clocks, as well as biomarkers of aging based on plasma proteomics (e.g., PROage95 and the proteomic clock96) are examples of efforts toward more mechanistic biomarkers of aging. In addition, some longevity interventions have been found to modify omic biomarkers of aging, suggesting that these biomarkers could reflect the underlying mechanisms48,97.
Generalizability criteria
Generalizability broadly refers to the ability of biomarkers to function across different applications. For instance, a biomarker of aging may be specific to an individual cell type (e.g., naive CD4+ T cells or oocytes), organ (e.g., liver or brain; for a recent review, see98), organ system (immune or nervous system), species (e.g., mouse or human), or human population, or may be more broadly applicable. In line with the cellular view of aging promoted by the geroscience hypothesis and the hallmarks of aging, biomarkers that measure underlying common molecular processes of cellular aging might capture aging-associated molecular and cellular damage common to multiple cell/tissue types or species. Generalizability of biomarkers between different tissues and between in vivo and in vitro settings may therefore be helpful to support mechanistic studies.
As features of biological aging are conserved across multiple species93, aging biomarkers have also been developed for model organisms. In fact, model organisms provide an opportunity to better understand various aspects of biomarkers, including their development, validation, and application. Several epigenetic biomarkers have been developed for mice, including blood and multitissue epigenetic clocks41,99,100. These biomarkers were validated by analyzing dietary (e.g., calorie restriction), genetic (e.g., growth hormone receptor knockout) and pharmacological (e.g., rapamycin) interventions41,99,100. Recognizing the value of cross-species biomarkers, the American Federation of Aging Research has proposed an additional criterion applicability in both humans and model organisms for a ‘true’ biomarker of aging31,101. Examples of dual-species102 and preliminary panmammalian47 epigenetic biomarkers of age have been reported. The cross-species translatability criterion favors molecular, cellular or subcellular level biomarkers12 and disfavors organism-specific biomarkers such as the highly accurate PhotoAgeClock, which is based on changes in human eye corners12, or the FRIGHT clock, which uses measures of frailty in mice103. However, the requirement for cross-species generalizability may not always be applicable, as there are certain aspects of biological aging that may differ between models/species. For example, telomere shortening is detected in human somatic tissues with aging, but not in most strains of laboratory mice due to their much greater overall telomere length37,104; similarly, higher fasting blood glucose is associated with increased mortality in primates but not in mice105. Discarding potential biomarker candidates due to a lack of translatability may therefore result in exclusion of interesting human biomarkers37. However, cross-species translatability offers the advantage of applying the same biomarkers in preclinical and clinical studies.
Moving to clinical considerations, a key aspect of human aging biomarker generalizability is the ability of biomarkers to function across different populations of humans. In other words, if a biomarker is derived and validated in one population, its utility should ideally extend to other populations, although this may be a challenge for some aging biomarkers. At the very least, it should be understood what limits the wider utility of a given aging biomarker. It is thus critical to validate proposed biomarkers across countries, sexes, ethnicities, and other demographic factors. Studies in nonindustrialized populations106,107 are also crucial to establishing biomarkers as being universally valid in humans.
Response criteria
Generally, response criteria posit that a biomarker of aging should be responsive to both accelerated and decelerated aging51. Biomarkers of aging are expected to indicate higher age in models of accelerated aging108. Likewise, known factors with systemic negative effects on longevity, such as chronic stress109 or poverty110, are expected to increase scores of biomarkers of aging. As these and other models of accelerated aging are further developed and understood111, biomarkers of aging can be evaluated using these conditions. The association of conditions negatively impacting longevity with biomarkers of aging could help to further characterize these biomarkers112, for instance, by linking them to known underlying pathomechanisms.
At the opposite end of the spectrum, geroprotectors are agents or interventions that potentially slow, inhibit, or reverse biological aging8,30. For a subset of candidate geroprotectors, sufficient scientific evidence supporting their efficacy has accumulated to warrant the initiation of human clinical longevity intervention trials, such as caloric restriction (CALERIE72), exercise (DAMA74, DO-HEALTH113, and Generation-100114 trials), mTOR inhibitors115–117, and plasmapheresis78. As geroprotective properties and mechanisms of certain agents and interventions are becoming more evident, their effects on biomarkers of aging are being increasingly evaluated. Geroprotectors with large effects on multiple physiological systems are expected to reduce scores of biomarkers of aging which correspond to biological age.
As one of the proposed criteria for a candidate geroprotector is reduction in biological age assessed with biomarkers8,30,81, the relationship between biomarkers of aging and geroprotectors may seem circular (Figure 3a). Importantly, there exist many candidate biomarkers of aging and many proposed geroprotectors, and the accumulated evidence supporting either could be leveraged to validate the other (Figure 3b). Candidate geroprotectors may be evaluated by examining their effect on healthspan, lifespan, or evaluated biomarkers. In turn, candidate biomarkers of aging could be assessed based on their response to evaluated geroprotectors (Figure 3b). Indeed, this is currently the common practice for analogous biomarkers developed for use in model organisms, where numerous lifespan-extending interventions have been unequivocally identified41.
Cost considerations
To implement biomarkers of aging in large-scale settings, including investigations of large biobanks, large clinical trials, population studies, or genome-wide screens, the cost of measurement must be relatively low. Most established biomarkers of aging currently remain cost-prohibitive for such purposes. Recent advances in cost reduction for measuring biomarkers of aging involve the development of probabilistic statistical frameworks and low-pass sequencing approaches applied to single cells and bulk samples (e.g., scAge118) and targeted high-throughput analyses of subsets of established biomarker loci (e.g., DNAm profiling of epigenetic clock CpG sites only, rather than unbiased or genome-wide profiling, as in preliminary evidence from TIME-seq119). The development of ultrahigh-throughput sequencing technologies is also expected to lead to reduced sequencing costs in the near future, thus enabling the application of these and other methods in a cost-effective manner.
Validation of biomarkers of aging
Biomarkers must undergo both analytical and clinical validation to ensure they are adequate for their intended use (Figure 4). Validation is an intensive process with many factors that must be carefully considered. Although an exhaustive review of biomarker validation is beyond the scope of the present work, we outline some of the key considerations for such validation efforts.
Figure 4. Analytical and clinical validation.
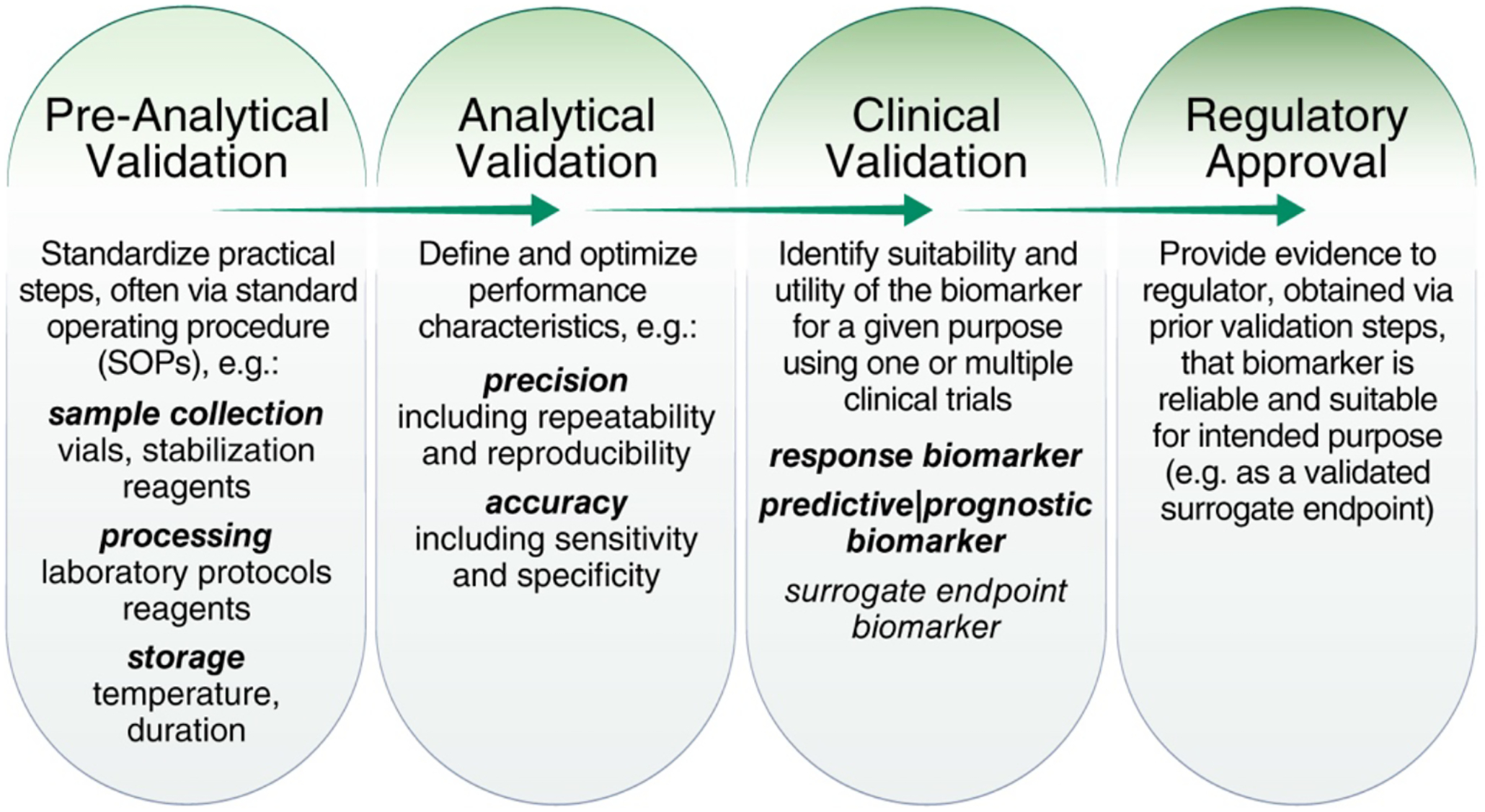
Adapted from Dobbin et al. (2016)125 by including details on individual validation steps.
Analytical validation
It is important that a biomarker meets technical specifications and standards. Analytical validation seeks to determine that the biomarker and potential resulting test exhibit adequate precision13. Quantification of precision can be performed under repeatability (same laboratory, operator, and equipment) and reproducibility conditions (different laboratory, operator, and equipment), and assesses the agreement of independent test results120. These analytical aspects are important to ensure that the error is minimal and within an acceptable range. Ultimately, this should enable test performance with high signal (biological variation) but low noise (technical variation), which is particularly important for longitudinal studies evaluating changes in biomarkers of aging in response to longevity interventions. Here, technical variability affects both the treatment and control groups, as well as both baseline and follow-up measurements. Thus, if technical variability is high in such studies, detection of biological variation may be limited59,121.
Additionally, analytical validation seeks to determine the accuracy of the measurement, i.e., how close the measured value is to the true value13,120,122. For instance, sensitivity and specificity are two common measures of accuracy13, but applying them to biomarkers of aging poses several challenges: there is as of yet no consensus as to how thresholds for positive or negative results could be set, and how one would determine whether the true biological age was “correctly” determined. The existing analytical validation framework based on sensitivity and specificity most frequently supposes that the target information is dichotomous (e.g., old vs. young), which is not appropriate in the context of a continuous process such as aging. Further work to form a clear consensus on how to establish analytical validation of accuracy for continuous biomarkers of aging is needed.
Ultimately, for clinical implementation of a molecular biomarker of aging as a test, three essential practical components need to be considered during analytical validation123: (1) sampling and source materials, including biological sample collection, storage, and processing conditions (pre-analytical); (2) assays for obtaining the measurement, for instance, methylation microarrays or next generation sequencing; and (3) methods and criteria for interpreting the measurements, such as algorithms including simple thresholding, linear models, or more complex models based on deep learning. These factors must be considered as a whole and should be standardized. In the context of omic biomarkers of aging, it has been shown individual components (e.g., DNA methylation levels for epigenetic clocks) may carry relatively high technical noise124, but this could potentially be overcome via computational approaches to bolster reliability121 or judicious choice of reliable probes59.
Clinical validation
Clinical validity and utility are established through application in human clinical trials. Clinical validation seeks to establish whether a biomarker can indeed identify the outcome of interest and determines how useful it is for clinical decision making13. Validation of a biomarker as a surrogate endpoint requires clinical evidence that a change in the surrogate endpoint predicts a specific clinical benefit13. For instance, a reduction in epigenetic age is only meaningful if it is indeed associated with a positive clinical change, such as a reduction in the risk of age-related diseases, frailty, mortality, or similar changes.
The process of clinical validation depends on the intended purpose of the proposed biomarker13. For example, predictive or prognostic biomarkers could help to estimate the magnitude of the potential benefit of a geroprotective treatment or an individual’s risk of developing a specific age-related clinical outcome or their risk of death. A response biomarker of aging provides proof-of-concept that a longevity intervention has an expected biological effect. Select candidate biomarkers, such as methylation-based biomarkers, have performed well as predictive and prognostic biomarkers (Table 2) and demonstrated responsiveness to geroprotective interventions (Table 3). However, further validation is needed to establish robust associations of these biomarkers with clinical endpoints to enable their consideration as surrogate endpoint biomarkers for longevity interventions. A key step along this road is validation of biomarkers across multiple cohorts of humans. Several efforts have begun to address this need, but challenges accessing human data while maintaining participant privacy remain and should be addressed. Nevertheless, it is increasingly clear that validated biomarkers of aging have strong potential to become key tools for clinical trials and practice.
Ongoing challenges and directions for future research
Biomarkers of aging have revolutionized preclinical aging biology research and stand to have an equal or greater impact on clinical trials. To realize this potential, several key challenges require ongoing collective attention from the field. These range from fundamental conceptual challenges rooted in the basic biology of aging to technical and clinical considerations. In the current work we overcome some of these challenges, such as harmonizing terminology, classification, potential use cases, and validation steps. However, there remain ongoing challenges that may be areas of particular interest for researchers to advance the field of aging research.
At the most fundamental level, the biological nature of aging remains incompletely understood, and we lack consensus on which molecular, cellular, physiological, or clinical changes are causal to aging rather than simply associated with it. Disease, cumulative impacts of industrialized lifestyles, or other features – perhaps unrelated to aging entirely – underlying the sampled population may confound biomarker measurements. Biological differences between men and women and their potential underlying factors also must be addressed: women tend to live longer but may experience worse health at the end of life126. Similarly, ethnicity and a host of environmental conditions are likely important determinants of aging. As we continue to develop our understanding of what aging is and what it is not, measurement of parameters such as vitality or resilience, which are being increasingly considered in aging studies127, may represent a pragmatic, complementary health-focused rather than disease-focused strategy for biomarker development. Regardless, these considerations speak to the significant need for further research aimed at understanding the biological nature of aging at a very fundamental level.
Moving to the development of aging biomarkers, a key consideration for any biomarker needs to be addressed: what assumptions are made to identify its feature(s)? Many biomarkers of aging are developed to estimate biological age by training a statistical model to predict the chronological age of the sample donor; the difference between biomarker-predicted biological age and chronological age is then referred to as the age deviation (AgeDev). However, reliance on chronological age in biomarker development may be a less than ideal strategy, as discussed in the section ‘Feasibility and Validity’. Inclusion of relevant age-related functional parameters and outcomes to train prediction models, as implemented in recent epigenetic biomarkers (e.g., PhenoAge, GrimAge, or DunedinPACE) could improve biomarkers of aging12.
An important conceptual distinction needs to be made between biomarkers of biological age, and biomarkers of the rate of aging. The former aim to capture the extent of aging that has occurred in an individual to date, while the latter estimate how fast aging processes are occurring at a given point in time128. Biomarkers of the rate of aging relate to markers of biological age as speedometers do to odometers129,130; thus they may be complementary. For instance, a high AgeDev could indicate either that the individual is currently aging faster and therefore has an increased biological age, or it could reflect downstream ramifications of faster aging earlier in life, even though the sampled individual is currently aging at a normal or even slower pace. Speedometer biomarkers could directly inform on the aging rate at a given point in time, and may also be more sensitive to intervention-induced changes in aging processes particularly over short follow-up intervals131. However, speedometer biomarkers on their own do not provide the whole picture, as they do not give an indication of the baseline biological age of the subject. These biomarkers may also be oversensitive to short-term perturbations. Hence, simultaneously measuring both AgeDev (‘odometer’) and the pace of aging (‘speedometer’) could improve prediction of future outcomes, and future research may consider their dual evaluation.
Moving to more practical considerations, the biological sample used for measuring aging is critical. For instance, biomarkers measured in blood that depend on material from cellular sources (e.g., blood cell DNA methylation or gene expression profiles) could primarily reflect changes in the hematopoietic system but not in other organ systems with lower turnover rates and different cell types. Studies have suggested that cell type-specific aging may occur19,20,132,133, and hence choosing the appropriate sample type and biomarker is needed. In this regard, cell-type and tissue-specific biomarkers may prove valuable. Moreover, the temporal dynamics, including ‘normal’134 and stress-induced fluctuations135, of many biomarkers of aging remain to be studied, as we are only beginning to understand how to separate signal from noise in these biomarkers. For potential clinical biomarkers of aging, these challenges may be addressed by standardizing measurements of the biomarkers and clinical outcomes, banking samples for future measurement, and reducing barriers to data access while preserving patient privacy. Furthermore, investigators should aim to apply multiple biomarkers within the same cohort. In addition to improving our understanding of fundamental aspects of aging, this would also enable further validation of biomarkers of aging and ultimately advance their clinical use.
Validation of biomarkers of aging for use as surrogate outcomes in clinical trials is highly desirable to reduce sample numbers and trial duration. An essential aspect requiring further consideration within the community will be establishing consensus on acceptable clinical outcomes for aging trials. Although a detailed discussion is beyond the scope of this perspective (for a recent review, see52), endpoints must be linked, in part, to the fundamental biology of aging, must be objectively quantifiable, and should evaluate the effect of an agent or intervention on how the patient or participant “functions, feels, or survives” in a manner inherently meaningful to participants (or patients), clinicians, and regulatory officials52. In the context of aging research, both broad (e.g., disability-free survival or incident frailty) and specific (e.g., age-related diseases or conditions) potential endpoints have been proposed to meet this definition. Total mortality is generally not considered a candidate clinical endpoint for modern trials given issues of feasibility (sample size, duration) and conceptual challenge for all-cause vs. cause-specific death as a clinical indicator of aging as a process52. The latter point is important, as many biomarkers of aging are developed to predict total mortality, even though it is unlikely to serve as a primary outcome for geroprotector trials. Notably, most of the above clinical outcomes primarily occur in later life and are not ideally suited to prevention trials in young populations with low incidence of any of these outcomes. However, validated biomarkers of aging may one day enable the prediction of responsiveness to geroprotective interventions applied years before the clinical manifestations of aging.
Conclusion
Recent decades have seen considerable progress in our understanding of the aging process and how we research and quantify it. While insights into biological hallmarks of aging continue to expand93, it is vital to characterize what specific biomarkers of aging measure and in what settings they might find suitable applications. Biomarkers of aging offer great potential for enabling human longevity intervention trials and personalized clinical decision making. Longevity interventions may prove to be useful when applied early in life131,136, but given the relatively long human lifespan, trials with lifespan-focused outcomes would be impractical. Biomarkers of aging could therefore serve several important roles in the field of aging research: (1) to give an early indication of whether an intervention increases healthspan and/or lifespan; (2) to identify individuals who might benefit from a treatment; (3) to prioritize candidate interventions for longer-term assessment; and (4) as validated surrogate endpoints for regulatory and clinical purposes if short-term changes in aging biomarkers are shown to be predictive of longer-term outcomes. To facilitate validation and ultimate clinical application of biomarkers of aging, a robust framework for their evaluation is needed. Our hope is that the consensus terminology, classification, evaluation criteria, and identified challenges and future directions for research presented in this work represent a solid first step toward achieving this goal.
Supplementary Material
Contributor Information
Mahdi Moqri, Institute for Stem Cell Biology and Regenerative Medicine, Stanford School of Medicine, Stanford, CA, USA.
Chiara Herzog, European Translational Oncology Prevention and Screening Institute, Universität Innsbruck, Austria.
Jesse R. Poganik, Division of Genetics, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA
Jamie Justice, Sticht Center for Healthy Aging and Alzheimer’s Prevention, Wake Forest School of Medicine, Winston-Salem, NC, USA.
Daniel Belsky, Department of Epidemiology & Butler Columbia Aging Center, Columbia University Mailman School of Public Health, New York, NY, USA.
Albert Higgins-Chen, Department of Psychiatry, Yale University, New Haven, CT, USA.
Alexey Moskalev, Institute of Biology of Aging, Nizhny Novgorod State University, Russia.
Georg Fuellen, Institute for Biostatistics and Informatics in Medicine and Ageing Research, Rostock University Medical Center, Rostock, Germany.
Alan A. Cohen, Butler Columbia Aging Center, Department of Environmental Health Sciences, Columbia University Mailman School of Public Health, New York, NY, USA
Ivan Bautmans, Gerontology Department and Frailty in Ageing Research Department, Vrije Universiteit Brussel, Brussels, Belgium.
Martin Widschwendter, European Translational Oncology Prevention and Screening Institute, Universität Innsbruck, Austria.
Jingzhong Ding, Gerontology and Geriatric Medicine, Wake Forest School of Medicine, Winston-Salem, NC, USA.
Alexander Fleming, Kitalys Institute, Charlottesville, VA, USA.
Joan Mannick, Tornado Therapeutics, New York, NY, USA.
Jing-Dong Jackie Han, Peking-Tsinghua Center for Life Sciences, Academy for Advanced Interdisciplinary Studies, Center for Quantitative Biology, Peking University, Beijing, China.
Alex Zhavoronkov, Insilico Medicine Hong Kong, Pak Shek Kok, New Territories, Hong Kong SAR, China.
Nir Barzilai, Albert Einstein College of Medicine, Bronx, NY, USA.
Matt Kaeberlein, Department of Laboratory Medicine and Pathology, University of Washington, Seattle, WA, USA.
Steven Cummings, San Francisco Coordinating Center, California Pacific Medical Center Research Institute and the Department of Epidemiology and Biostatistics, University of California, San Francisco, CA, USA.
Brian Kennedy, Healthy Longevity Translational Research Programme, Yong Loo Lin School of Medicine, National University of Singapore, Singapore.
Luigi Ferrucci, National Institute on Aging, Baltimore, MD, USA.
Steve Horvath, Altos Labs, San Diego, CA, USA.
Eric Verdin, Buck Institute for Research on Aging, Novato, CA, USA.
Andrea B. Maier, Department of Human Movement Sciences, @AgeAmsterdam, Amsterdam Movement Sciences, Faculty of Behavioural and Movement Sciences, Vrije Universiteit Amsterdam, Amsterdam, Netherlands; Healthy Longevity Translational Research Program, Yong Loo Lin School of Medicine, National University of Singapore, Centre for Healthy Longevity, @AgeSingapore, National University Health System, Singapore
Michael P. Snyder, Department of Genetics, School of Medicine, Stanford University, Stanford, CA, USA
Vittorio Sebastiano, Department of Obstetrics and Gynecology, Stanford School of Medicine, Stanford University, Stanford, CA, USA.
Vadim N. Gladyshev, Department of Medicine, Brigham and Women’s Hospital, Harvard Medical School, Boston, MA, USA
References
- 1.Cohen AA, Kennedy BK, Anglas U, Bronikowski AM, Deelen J, Dufour F, Ferbeyre G, Ferrucci L, Franceschi C, Frasca D, et al. (2020). Lack of consensus on an aging biology paradigm? A global survey reveals an agreement to disagree, and the need for an interdisciplinary framework. Mech. Ageing Dev 191, 111316. 10.1016/j.mad.2020.111316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gladyshev VN, Anderson B, Barlit H, Barré B, Beck S, Behrouz B, Belsky DW, Boulahouache L, Chaix A, Chamoli M, et al. (2023). Disagreement on foundational principles of biological aging [Manuscript under review]. [DOI] [PMC free article] [PubMed]
- 3.Gladyshev VN, Kritchevsky SB, Clarke SG, Cuervo AM, Fiehn O, de Magalhães JP, Mau T, Maes M, Moritz RL, Niedernhofer LJ, et al. (2021). Molecular damage in aging. Nat. Aging 1, 1096–1106. 10.1038/s43587-021-00150-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lee MB, and Kaeberlein M (2018). Translational geroscience: From invertebrate models to companion animal and human interventions. Transl. Med. Aging 2, 15–29. 10.1016/j.tma.2018.08.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wagner K-H, Cameron-Smith D, Wessner B, and Franzke B (2016). Biomarkers of Aging: From Function to Molecular Biology. Nutrients 8, 338. 10.3390/nu8060338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rutledge J, Oh H, and Wyss-Coray T (2022). Measuring biological age using omics data. Nat. Rev. Genet 23, 715–727. 10.1038/s41576-022-00511-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Miller R (2001). Biomarkers and the Genetics of Aging in Mice. In Cells and Surveys: Should Biological Measures Be Included in Social Science Research?, V. J Finch CE Kinsella K, ed. (National Academies Press (US)). [PubMed] [Google Scholar]
- 8.Partridge L, Fuentealba M, and Kennedy BK (2020). The quest to slow ageing through drug discovery. Nat. Rev. Drug Discov 19, 513–532. 10.1038/s41573-020-0067-7. [DOI] [PubMed] [Google Scholar]
- 9.Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, et al. (2014). Geroscience: Linking Aging to Chronic Disease. Cell 159, 709–713. 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sierra F, and Kohanski R (2017). Geroscience and the trans-NIH Geroscience Interest Group, GSIG. GeroScience 39, 1–5. 10.1007/s11357-016-9954-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Baker GT, and Sprott RL (1988). Biomarkers of aging. Exp. Gerontol 23, 223–239. 10.1016/0531-5565(88)90025-3. [DOI] [PubMed] [Google Scholar]
- 12.Galkin F, Mamoshina P, Aliper A, de Magalhães JP, Gladyshev VN, and Zhavoronkov A (2020). Biohorology and biomarkers of aging: current state-of-the-art, challenges and opportunities. Ageing Res. Rev 60, 101050. 10.1016/j.arr.2020.101050. [DOI] [PubMed] [Google Scholar]
- 13.FDA-NIH Biomarker Working Group (2016). Glossary. In BEST (Biomarkers, EndpointS, and other Tools) Resource (Food and Drug Administration (US)). [PubMed] [Google Scholar]
- 14.Comfort A (1969). TEST-BATTERY TO MEASURE AGEING-RATE IN MAN. The Lancet 294, 1411–1415. 10.1016/s0140-6736(69)90950-7. [DOI] [PubMed] [Google Scholar]
- 15.Reff ME, and Schneider EL (1981). Biological Markers of Aging: Proceedings of Conference on Nonlethal Biological Markers of Physiological Aging on June 19 and 20, 1981. (U.S. Department of Health and Human Services, National Institutes of Health, Public Health Service; ). [Google Scholar]
- 16.Butler RN, Sprott R, Warner H, Bland J, Feuers R, Forster M, Fillit H, Harman SM, Hewitt M, Hyman M, et al. (2004). Aging: The Reality: Biomarkers of Aging: From Primitive Organisms to Humans. J. Gerontol. Ser. A 59, B560–B567. 10.1093/gerona/59.6.b560. [DOI] [PubMed] [Google Scholar]
- 17.Lara J, Cooper R, Nissan J, Ginty AT, Khaw K-T, Deary IJ, Lord JM, Kuh D, and Mathers JC (2015). A proposed panel of biomarkers of healthy ageing. BMC Med. 13, 222. 10.1186/s12916-015-0470-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gladyshev VN (2016). Aging: progressive decline in fitness due to the rising deleteriome adjusted by genetic, environmental, and stochastic processes. Aging Cell 15, 594–602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Buckley MT, Sun ED, George BM, Liu L, Schaum N, Xu L, Reyes JM, Goodell MA, Weissman IL, Wyss-Coray T, et al. (2023). Cell-type-specific aging clocks to quantify aging and rejuvenation in neurogenic regions of the brain. Nat. Aging 3, 121–137. 10.1038/s43587-022-00335-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nie C, Li Y, Li R, Yan Y, Zhang D, Li T, Li Z, Sun Y, Zhen H, Ding J, et al. (2022). Distinct biological ages of organs and systems identified from a multi-omics study. Cell Rep. 38, 110459. 10.1016/j.celrep.2022.110459. [DOI] [PubMed] [Google Scholar]
- 21.Ahadi S, Zhou W, Rose SMS-F, Sailani MR, Contrepois K, Avina M, Ashland M, Brunet A, and Snyder M (2020). Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med 26, 83–90. 10.1038/s41591-019-0719-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cohen AA, Legault V, Fuellen G, Fülöp T, Fried LP, and Ferrucci L (2018). The risks of biomarker-based epidemiology: Associations of circulating calcium levels with age, mortality, and frailty vary substantially across populations. Exp. Gerontol 107, 11–17. 10.1016/j.exger.2017.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pyrkov TV, Sokolov IS, and Fedichev PO (2021). Deep longitudinal phenotyping of wearable sensor data reveals independent markers of longevity, stress, and resilience. Aging 13, 7900–7913. 10.18632/aging.202816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jansen R, Han LK, Verhoeven JE, Aberg KA, van den Oord EC, Milaneschi Y, and Penninx BW (2021). An integrative study of five biological clocks in somatic and mental health. eLife 10, e59479. 10.7554/elife.59479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Belsky DW, Moffitt TE, Cohen AA, Corcoran DL, Levine ME, Prinz JA, Schaefer J, Sugden K, Williams B, Poulton R, et al. (2017). Eleven Telomere, Epigenetic Clock, and Biomarker-Composite Quantifications of Biological Aging: Do They Measure the Same Thing? Am. J. Epidemiol 187, 1220–1230. 10.1093/aje/kwx346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reimann B, Martens DS, Wang C, Ghantous A, Herceg Z, Plusquin M, and Nawrot TS (2022). Interrelationships and determinants of aging biomarkers in cord blood. J. Transl. Med 20, 353. 10.1186/s12967-022-03541-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li X, Ploner A, Wang Y, Magnusson PK, Reynolds C, Finkel D, Pedersen NL, Jylhävä J, and Hägg S (2020). Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. eLife 9, e51507. 10.7554/elife.51507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaeberlein M (2018). How healthy is the healthspan concept? GeroScience 40, 361–364. 10.1007/s11357-018-0036-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Horvath S (2013). DNA methylation age of human tissues and cell types. Genome Biol 14, R115. 10.1186/gb-2013-14-10-r115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moskalev A, Chernyagina E, Kudryavtseva A, and Shaposhnikov M (2017). Geroprotectors: A Unified Concept and Screening Approaches. Aging Dis. 8, 354–363. 10.14336/ad.2016.1022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xia X, Chen W, McDermott J, and Han J-DJ (2017). Molecular and phenotypic biomarkers of aging. F1000Research 6, 860. 10.12688/f1000research.10692.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hartmann A, Hartmann C, Secci R, Hermann A, Fuellen G, and Walter M (2021). Ranking Biomarkers of Aging by Citation Profiling and Effort Scoring. Front. Genet 12, 686320. 10.3389/fgene.2021.686320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Moskalev Alexey ed. (2019). Biomarkers of Human Ageing (Springer; ). [Google Scholar]
- 34.Agarwala P, and Salzman SH (2020). Six-Minute Walk Test Clinical Role, Technique, Coding, and Reimbursement. Chest 157, 603–611. 10.1016/j.chest.2019.10.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Baran SW, Lim MA, Do JP, Stolyar P, Rabe MD, Schaevitz LR, and Cadena SM (2021). Digital Biomarkers Enable Automated, Longitudinal Monitoring in a Mouse Model of Aging. J. Gerontol. Ser. A 76, glab024. 10.1093/gerona/glab024. [DOI] [PubMed] [Google Scholar]
- 36.McCay CM, Crowell MF, and Maynard LA (1935). The Effect of Retarded Growth Upon the Length of Life Span and Upon the Ultimate Body Size. J. Nutr 10, 63–79. 10.1093/jn/10.1.63. [DOI] [PubMed] [Google Scholar]
- 37.Bürkle A, Moreno-Villanueva M, Bernhard J, Blasco M, Zondag G, Hoeijmakers JHJ, Toussaint O, Grubeck-Loebenstein B, Mocchegiani E, Collino S, et al. (2015). MARK-AGE biomarkers of ageing. Mech. Ageing Dev 151, 2–12. 10.1016/j.mad.2015.03.006. [DOI] [PubMed] [Google Scholar]
- 38.Bocklandt S, Lin W, Sehl ME, Sanchez FJ, Sinsheimer JS, Horvath S, and Vilain E (2011). Epigenetic predictor of age. PLoS One 6, e14821. 10.1371/journal.pone.0014821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Menni C, Kastenmüller G, Petersen AK, Bell JT, Psatha M, Tsai P-C, Gieger C, Schulz H, Erte I, John S, et al. (2013). Metabolomic markers reveal novel pathways of ageing and early development in human populations. Int. J. Epidemiol 42, 1111–1119. 10.1093/ije/dyt094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Peters MJ, Joehanes R, Pilling LC, Schurmann C, Conneely KN, Powell J, Reinmaa E, Sutphin GL, Zhernakova A, Schramm K, et al. (2015). The transcriptional landscape of age in human peripheral blood. Nat. Commun 6, 8570. 10.1038/ncomms9570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Petkovich DA, Podolskiy DI, Lobanov AV, Lee S-G, Miller RA, and Gladyshev VN (2017). Using DNA Methylation Profiling to Evaluate Biological Age and Longevity Interventions. Cell Metab. 25, 954–960.e6. 10.1016/j.cmet.2017.03.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Levine ME, Lu AT, Quach A, Chen BH, Assimes TL, Bandinelli S, Hou L, Baccarelli AA, Stewart JD, Li Y, et al. (2018). An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591. 10.18632/aging.101414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lu AT, Quach A, Wilson JG, Reiner AP, Aviv A, Raj K, Hou L, Baccarelli AA, Li Y, Stewart JD, et al. (2019). DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327. 10.18632/aging.101684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tanaka T, Biancotto A, Moaddel R, Moore AZ, Gonzalez-Freire M, Aon MA, Candia J, Zhang P, Cheung F, Fantoni G, et al. (2018). Plasma proteomic signature of age in healthy humans. Aging Cell 17, e12799. 10.1111/acel.12799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belsky DW, Caspi A, Arseneault L, Baccarelli A, Corcoran DL, Gao X, Hannon E, Harrington HL, Rasmussen LJ, Houts R, et al. (2020). Quantification of the pace of biological aging in humans through a blood test, the DunedinPoAm DNA methylation algorithm. eLife 9, e54870. 10.7554/elife.54870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Horvath S, Haghani A, Macoretta N, Ablaeva J, Zoller JA, Li CZ, Zhang J, Takasugi M, Zhao Y, Rydkina E, et al. (2022). DNA methylation clocks tick in naked mole rats but queens age more slowly than nonbreeders. Nat. Aging 2, 46–59. 10.1038/s43587-021-00152-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu AT, Fei Z, Haghani A, Robeck TR, Zoller JA, Li CZ, Lowe R, Yan Q, Zhang J, Vu H, et al. (2022). Universal DNA methylation age across mammalian tissues. bioRxiv, 2021.01.18.426733. 10.1101/2021.01.18.426733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Moqri M, Cipriano A, Nachun D, Murty T, Brandine G de S, Rasouli S, Tarkhov A, Aberg KA, van den Oord E, Zhou W, et al. (2022). PRC2 clock: a universal epigenetic biomarker of aging and rejuvenation. bioRxiv, 2022.06.03.494609. 10.1101/2022.06.03.494609. [DOI] [Google Scholar]
- 49.Ying K, Liu H, Tarkhov AE, Lu AT, Horvath S, Kutalik Z, Shen X, and Gladyshev VN (2022). Causal Epigenetic Age Uncouples Damage and Adaptation. bioRxiv, 2022.10.07.511382. 10.1101/2022.10.07.511382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levine ME, Higgins-Chen A, Thrush K, Minteer C, and Niimi P (2022). Clock Work: Deconstructing the Epigenetic Clock Signals in Aging, Disease, and Reprogramming. bioRxiv, 2022.02.13.480245. 10.1101/2022.02.13.480245. [DOI] [Google Scholar]
- 51.Justice JN, Ferrucci L, Newman AB, Aroda VR, Bahnson JL, Divers J, Espeland MA, Marcovina S, Pollak MN, Kritchevsky SB, et al. (2018). A framework for selection of blood-based biomarkers for geroscience-guided clinical trials: report from the TAME Biomarkers Workgroup. GeroScience 40, 419–436. 10.1007/s11357-018-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cummings SR, and Kritchevsky SB (2022). Endpoints for geroscience clinical trials: health outcomes, biomarkers, and biologic age. GeroScience 44, 2925–2931. 10.1007/s11357-022-00671-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hannum G, Guinney J, Zhao L, Zhang L, Hughes G, Sadda S, Klotzle B, Bibikova M, Fan JB, Gao Y, et al. Genome-wide methylation profiles reveal quantitative views of human aging rates. Mol Cell 49, 359–367. 10.1016/j.molcel.2012.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Fransquet PD, Wrigglesworth J, Woods RL, Ernst ME, and Ryan J (2019). The epigenetic clock as a predictor of disease and mortality risk: a systematic review and meta-analysis. Clin. Epigenetics 11, 62. 10.1186/s13148-019-0656-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chen BH, Marioni RE, Colicino E, Peters MJ, Ward-Caviness CK, Tsai P-C, Roetker NS, Just AC, Demerath EW, Guan W, et al. (2016). DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8, 1844–1859. 10.18632/aging.101020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Jurić J, Kohrt WM, Kifer D, Gavin KM, Pezer M, Nigrovic PA, and Lauc G (2020). Effects of estradiol on biological age measured using the glycan age index. Aging 12, 19756–19765. 10.18632/aging.104060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Macdonald-Dunlop E, Taba N, Klarić L, Frkatović A, Walker R, Hayward C, Esko T, Haley C, Fischer K, Wilson JF, et al. (2022). A catalogue of omics biological ageing clocks reveals substantial commonality and associations with disease risk. Aging 14, 623–659. 10.18632/aging.203847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCrory C, Fiorito G, Hernandez B, Polidoro S, O’Halloran AM, Hever A, Cheallaigh CN, Lu AT, Horvath S, Vineis P, et al. (2020). GrimAge Outperforms Other Epigenetic Clocks in the Prediction of Age-Related Clinical Phenotypes and All-Cause Mortality. J. Gerontol. Ser. A 76, 741–749. 10.1093/gerona/glaa286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Belsky DW, Caspi A, Corcoran DL, Sugden K, Poulton R, Arseneault L, Baccarelli A, Chamarti K, Gao X, Hannon E, et al. (2022). DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 11, e73420. 10.7554/elife.73420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sugden K, Caspi A, Elliott ML, Bourassa KJ, Chamarti K, Corcoran DL, Hariri AR, Houts RM, Kothari M, Kritchevsky S, et al. (2022). Association of Pace of Aging Measured by Blood-Based DNA Methylation With Age-Related Cognitive Impairment and Dementia. Neurology 99, e1402–e1413. 10.1212/wnl.0000000000200898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Earls JC, Rappaport N, Heath L, Wilmanski T, Magis AT, Schork NJ, Omenn GS, Lovejoy J, Hood L, and Price ND (2019). Multi-Omic Biological Age Estimation and Its Correlation With Wellness and Disease Phenotypes: A Longitudinal Study of 3,558 Individuals. J. Gerontol. Ser. A 74, S52–S60. 10.1093/gerona/glz220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mamoshina P, Kochetov K, Putin E, Cortese F, Aliper A, Lee W-S, Ahn S-M, Uhn L, Skjodt N, Kovalchuk O, et al. (2018). Population Specific Biomarkers of Human Aging: A Big Data Study Using South Korean, Canadian, and Eastern European Patient Populations. J. Gerontol. Ser. A 73, 1482–1490. 10.1093/gerona/gly005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mamoshina P, Volosnikova M, Ozerov IV, Putin E, Skibina E, Cortese F, and Zhavoronkov A (2018). Machine Learning on Human Muscle Transcriptomic Data for Biomarker Discovery and Tissue-Specific Drug Target Identification. Front. Genet 9, 242. 10.3389/fgene.2018.00242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zhu T, Gao Y, Wang J, Li X, Shang S, Wang Y, Guo S, Zhou H, Liu H, Sun D, et al. (2019). CancerClock: A DNA Methylation Age Predictor to Identify and Characterize Aging Clock in Pan-Cancer. Front. Bioeng. Biotechnol 7, 388. 10.3389/fbioe.2019.00388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chen L, Ganz PA, and Sehl ME (2022). DNA Methylation, Aging, and Cancer Risk: A Mini-Review. Front. Bioinforma 2, 847629. 10.3389/fbinf.2022.847629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Simrén J, Leuzy A, Karikari TK, Hye A, Benedet AL, Lantero-Rodriguez J, Mattsson-Carlgren N, Schöll M, Mecocci P, Vellas B, et al. (2021). The diagnostic and prognostic capabilities of plasma biomarkers in Alzheimer’s disease. Alzheimers Dement. 17, 1145–1156. 10.1002/alz.12283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Christensen CH, Chang JT, Rostron BL, Hammad HT, van Bemmel DM, Valle-Pinero AYD, Wang B, Mishina EV, Faulcon LM, DePina A, et al. (2021). Biomarkers of Inflammation and Oxidative Stress among Adult Former Smoker, Current E-Cigarette Users—Results from Wave 1 PATH Study. Cancer Epidemiol. Biomarkers Prev 30, 1947–1955. 10.1158/1055-9965.epi-21-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Fleming TR, and Powers JH (2012). Biomarkers and surrogate endpoints in clinical trials. Stat. Med 31, 2973–2984. 10.1002/sim.5403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Justice JN, and Kritchevsky SB (2020). Aging and Geroscience: Putting epigenetic biomarkers to the test for clinical trials. eLife 9:e58592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Longo VD, Antebi A, Bartke A, Barzilai N, Brown-Borg HM, Caruso C, Curiel TJ, Cabo R, Franceschi C, Gems D, et al. (2015). Interventions to Slow Aging in Humans: Are We Ready? Aging Cell 14, 497–510. 10.1111/acel.12338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Pun FW, Leung GHD, Leung HW, Liu BHM, Long X, Ozerov IV, Wang J, Ren F, Aliper A, Izumchenko E, et al. (2022). Hallmarks of aging-based dual-purpose disease and age-associated targets predicted using PandaOmics AI-powered discovery engine. Aging 14, 2475–2506. 10.18632/aging.203960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Waziry R, Corcoran D, Huffman K, Kobor M, Kothari M, Kraus V, Kraus W, Lin D, Pieper C, Ramaker M, et al. (2021). Effect of Long-Term Caloric Restriction on DNA Methylation Measures of Biological Aging in Healthy Adults: CALERIE™ Trial Analysis. medRxiv, 2021.09.21.21263912. 10.1101/2021.09.21.21263912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kwon D, and Belsky DW (2021). A toolkit for quantification of biological age from blood chemistry and organ function test data: BioAge. GeroScience 43, 2795–2808. 10.1007/s11357-021-00480-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fiorito G, Caini S, Palli D, Bendinelli B, Saieva C, Ermini I, Valentini V, Assedi M, Rizzolo P, Ambrogetti D, et al. (2021). DNA methylation-based biomarkers of aging were slowed down in a two-year diet and physical activity intervention trial: the DAMA study. Aging Cell 20, e13439. 10.1111/acel.13439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Fitzgerald KN, Hodges R, Hanes D, Stack E, Cheishvili D, Szyf M, Henkel J, Twedt MW, Giannopoulou D, Herdell J, et al. (2021). Potential reversal of epigenetic age using a diet and lifestyle intervention: a pilot randomized clinical trial. Aging 13, 9419–9432. 10.18632/aging.202913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Horvath S, Oshima J, Martin GM, Lu AT, Quach A, Cohen H, Felton S, Matsuyama M, Lowe D, Kabacik S, et al. (2018). Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 10, 1758–1775. 10.18632/aging.101508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lu AT, Seeboth A, Tsai P-C, Sun D, Quach A, Reiner AP, Kooperberg C, Ferrucci L, Hou L, Baccarelli AA, et al. (2019). DNA methylation-based estimator of telomere length. Aging 11, 5895–5923. 10.18632/aging.102173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Clement J, Yan Q, Agrawal M, Coronado RE, Sturges JA, Horvath M, Lu AT, Brooke RT, and Horvath S (2022). Umbilical cord plasma concentrate has beneficial effects on DNA methylation GrimAge and human clinical biomarkers. Aging Cell 21, e13696. 10.1111/acel.13696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chen L, Dong Y, Bhagatwala J, Raed A, Huang Y, and Zhu H (2018). Effects of Vitamin D3 Supplementation on Epigenetic Aging in Overweight and Obese African Americans With Suboptimal Vitamin D Status: A Randomized Clinical Trial. J. Gerontol. Ser. A 74, 91–98. 10.1093/gerona/gly223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Yi L, Maier AB, Tao R, Lin Z, Vaidya A, Pendse S, Thasma S, Andhalkar N, Avhad G, and Kumbhar V (2022). The efficacy and safety of β-nicotinamide mononucleotide (NMN) supplementation in healthy middle-aged adults: a randomized, multicenter, double-blind, placebo-controlled, parallel-group, dose-dependent clinical trial. GeroScience, 1–15. 10.1007/s11357-022-00705-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Moskalev A, Chernyagina E, Tsvetkov V, Fedintsev A, Shaposhnikov M, Krut’ko V, Zhavoronkov A, and Kennedy BK (2016). Developing criteria for evaluation of geroprotectors as a key stage toward translation to the clinic. Aging Cell 15, 407–415. 10.1111/acel.12463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sprott RL (1999). Biomarkers of Aging. J. Gerontol. Ser. A 54, B464–B465. 10.1093/gerona/54.11.b464. [DOI] [PubMed] [Google Scholar]
- 83.Zhang Q, Vallerga CL, Walker RM, Lin T, Henders AK, Montgomery GW, He J, Fan D, Fowdar J, Kennedy M, et al. (2019). Improved precision of epigenetic clock estimates across tissues and its implication for biological ageing. Genome Med. 11, 54. 10.1186/s13073-019-0667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hertel J, Frenzel S, König J, Wittfeld K, Fuellen G, Holtfreter B, Pietzner M, Friedrich N, Nauck M, Völzke H, et al. (2019). The informative error: A framework for the construction of individualized phenotypes. Stat. Methods Med. Res 28, 1427–1438. 10.1177/0962280218759138. [DOI] [PubMed] [Google Scholar]
- 85.Engelfriet PM, Jansen EHJM, Picavet HSJ, and Dollé MET (2013). Biochemical Markers of Aging for Longitudinal Studies in Humans. Epidemiol. Rev 35, 132–151. 10.1093/epirev/mxs011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wey TW, Roberge É, Legault V, Kemnitz JW, Ferrucci L, and Cohen AA (2019). An Emergent Integrated Aging Process Conserved Across Primates. J. Gerontol. Ser. A 74, 1689–1698. 10.1093/gerona/glz110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Li Q, Wang S, Milot E, Bergeron P, Ferrucci L, Fried LP, and Cohen AA (2015). Homeostatic dysregulation proceeds in parallel in multiple physiological systems. Aging Cell 14, 1103–1112. 10.1111/acel.12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Cohen AA, Leblanc S, and Roucou X (2021). Robust Physiological Metrics From Sparsely Sampled Networks. Front. Physiol 12, 624097. 10.3389/fphys.2021.624097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Bell CG, Lowe R, Adams PD, Baccarelli AA, Beck S, Bell JT, Christensen BC, Gladyshev VN, Heijmans BT, Horvath S, et al. (2019). DNA methylation aging clocks: challenges and recommendations. Genome Biol. 20, 249. 10.1186/s13059-019-1824-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Levine ME (2020). Assessment of Epigenetic Clocks as Biomarkers of Aging in Basic and Population Research. J. Gerontol. Ser. A 75, 463–465. 10.1093/gerona/glaa021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Maddock J, Castillo-Fernandez J, Wong A, Cooper R, Richards M, Ong KK, Ploubidis GB, Goodman A, Kuh D, Bell JT, et al. (2019). DNA methylation age and physical and cognitive ageing. J. Gerontol. Ser. A 75, 504–511. 10.1093/gerona/glz246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McCrory C, Fiorito G, McLoughlin S, Polidoro S, Cheallaigh CN, Bourke N, Karisola P, Alenius H, Vineis P, Layte R, et al. (2019). Epigenetic Clocks and Allostatic Load Reveal Potential Sex-Specific Drivers of Biological Aging. J. Gerontol. Ser. A 75, 495–503. 10.1093/gerona/glz241. [DOI] [PubMed] [Google Scholar]
- 93.López-Otín C, Blasco MA, Partridge L, Serrano M, and Kroemer G (2023). Hallmarks of aging: An expanding universe. Cell. 10.1016/j.cell.2022.11.001. [DOI] [PubMed] [Google Scholar]
- 94.Guerville F, Barreto PDS, Ader I, Andrieu S, Casteilla L, Dray C, Fazilleau N, Guyonnet S, Langin D, Liblau R, et al. (2020). Revisiting the Hallmarks of Aging to Identify Markers of Biological Age. J. Prev. Alzheimers Dis 7, 56–64. 10.14283/jpad.2019.50. [DOI] [PubMed] [Google Scholar]
- 95.Tanaka T, Basisty N, Fantoni G, Candia J, Moore AZ, Biancotto A, Schilling B, Bandinelli S, and Ferrucci L (2020). Plasma proteomic biomarker signature of age predicts health and life span. eLife 9, e61073. 10.7554/elife.61073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lehallier B, Gate D, Schaum N, Nanasi T, Lee SE, Yousef H, Losada PM, Berdnik D, Keller A, Verghese J, et al. (2019). Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med 25, 1843–1850. 10.1038/s41591-019-0673-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Aminzadeh-Gohari S, Kofler B, and Herzog C (2022). Dietary restriction in senolysis and prevention and treatment of disease. Crit. Rev. Food Sci. Nutr, 1–27. 10.1080/10408398.2022.2153355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Aging Biomarker Consortium, Bao H, Cao J, Chen M, Chen M, Chen W, Chen X, Chen Y, Chen Y, Chen Y, et al. (2023). Biomarkers of aging. Sci. China Life Sci 66, 893–1066. 10.1007/s11427-023-2305-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Meer MV, Podolskiy DI, Tyshkovskiy A, and Gladyshev VN (2018). A whole lifespan mouse multi-tissue DNA methylation clock. eLife 7, e40675. 10.7554/eLife.40675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Thompson MJ, Chwiałkowska K, Rubbi L, Lusis AJ, Davis RC, Srivastava A, Korstanje R, Churchill GA, Horvath S, and Pellegrini M (2018). A multi-tissue full lifespan epigenetic clock for mice. Aging 10, 2832–2854. 10.18632/aging.101590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Johnson TE (2006). Recent results: Biomarkers of aging. Exp. Gerontol 41, 1243–1246. 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 102.Horvath S, Lu AT, Haghani A, Zoller JA, Li CZ, Lim AR, Brooke RT, Raj K, Serres-Armero A, Dreger DL, et al. (2022). DNA methylation clocks for dogs and humans. Proc. Natl. Acad. Sci 119, e2120887119. 10.1073/pnas.2120887119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schultz MB, Kane AE, Mitchell SJ, MacArthur MR, Warner E, Vogel DS, Mitchell JR, Howlett SE, Bonkowski MS, and Sinclair DA (2020). Age and life expectancy clocks based on machine learning analysis of mouse frailty. Nat. Commun 11, 4618. 10.1038/s41467-020-18446-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Calado RT, and Dumitriu B (2013). Telomere Dynamics in Mice and Humans. Semin. Hematol 50, 165–174. 10.1053/j.seminhematol.2013.03.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Palliyaguru DL, Shiroma EJ, Nam JK, Duregon E, Teixeira CVL, Price NL, Bernier M, Camandola S, Vaughan KL, Colman RJ, et al. (2021). Fasting blood glucose as a predictor of mortality: Lost in translation. Cell Metab. 33, 2189–2200.e3. 10.1016/j.cmet.2021.08.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Horvath S, Gurven M, Levine ME, Trumble BC, Kaplan H, Allayee H, Ritz BR, Chen B, Lu AT, Rickabaugh TM, et al. (2016). An epigenetic clock analysis of race/ethnicity, sex, and coronary heart disease. Genome Biol. 17, 171. 10.1186/s13059-016-1030-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Kraft TS, Stieglitz J, Trumble BC, Garcia AR, Kaplan H, and Gurven M (2020). Multi-system physiological dysregulation and ageing in a subsistence population. Philos. Trans. R. Soc. B Biol. Sci 375, 20190610. 10.1098/rstb.2019.0610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Kõks S, Dogan S, Tuna BG, González-Navarro H, Potter P, and Vandenbroucke RE (2016). Mouse models of ageing and their relevance to disease. Mech. Ageing Dev 160, 41–53. 10.1016/j.mad.2016.10.001. [DOI] [PubMed] [Google Scholar]
- 109.Yegorov YE, Poznyak AV, Nikiforov NG, Sobenin IA, and Orekhov AN (2020). The Link between Chronic Stress and Accelerated Aging. Biomedicines 8, 198. 10.3390/biomedicines8070198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Crimmins EM, Kim JK, and Seeman TE (2009). Poverty and Biological Risk: The Earlier “Aging” of the Poor. J. Gerontol. Ser. A 64A, 286–292. 10.1093/gerona/gln010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Gonzalo S, Kreienkamp R, and Askjaer P (2017). Hutchinson-Gilford Progeria Syndrome: A premature aging disease caused by LMNA gene mutations. Ageing Res. Rev 33, 18–29. 10.1016/j.arr.2016.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Ashapkin VV, Kutueva LI, Kurchashova SY, and Kireev II (2019). Are There Common Mechanisms Between the Hutchinson–Gilford Progeria Syndrome and Natural Aging? Front. Genet 10, 455. 10.3389/fgene.2019.00455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bischoff-Ferrari HA, de G.R.C. Molino C, Rival S, Vellas B, Rizzoli R, Kressig RW, Kanis JA, Manson JE, Dawson-Hughes B, Orav EJ, et al. (2021). DO-HEALTH: Vitamin D3 - Omega-3 - Home exercise - Healthy aging and longevity trial - Design of a multinational clinical trial on healthy aging among European seniors. Contemp. Clin. Trials 100, 106124. 10.1016/j.cct.2020.106124. [DOI] [PubMed] [Google Scholar]
- 114.Stensvold D, Viken H, Steinshamn SL, Dalen H, Støylen A, Loennechen JP, Reitlo LS, Zisko N, Bækkerud FH, Tari AR, et al. (2020). Effect of exercise training for five years on all cause mortality in older adults—the Generation 100 study: randomised controlled trial. BMJ 371, m3485. 10.1136/bmj.m3485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Mannick JB, Morris M, Hockey H-UP, Roma G, Beibel M, Kulmatycki K, Watkins M, Shavlakadze T, Zhou W, Quinn D, et al. (2018). TORC1 inhibition enhances immune function and reduces infections in the elderly. Sci. Transl. Med 10. 10.1126/scitranslmed.aaq1564. [DOI] [PubMed] [Google Scholar]
- 116.Mannick JB, Giudice GD, Lattanzi M, Valiante NM, Praestgaard J, Huang B, Lonetto MA, Maecker HT, Kovarik J, Carson S, et al. (2014). mTOR inhibition improves immune function in the elderly. Sci. Transl. Med 6, 268ra179. 10.1126/scitranslmed.3009892. [DOI] [PubMed] [Google Scholar]
- 117.Mannick JB, Teo G, Bernardo P, Quinn D, Russell K, Klickstein L, Marshall W, and Shergill S (2021). Targeting the biology of ageing with mTOR inhibitors to improve immune function in older adults: phase 2b and phase 3 randomised trials. Lancet Healthy Longev. 2, e250–e262. 10.1016/s2666-7568(21)00062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Trapp A, Kerepesi C, and Gladyshev VN (2021). Profiling epigenetic age in single cells. Nat. Aging 1, 1189–1201. 10.1038/s43587-021-00134-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Griffin PT, Kane AE, Trapp A, Li J, Arnold M, Poganik JR, McNamara MS, Meer MV, Hoffman N, Amorim J, et al. (2022). TIME-Seq Enables Scalable and Inexpensive Epigenetic Age Predictions. bioRxiv, 2021.10.25.465725. 10.1101/2021.10.25.465725. [DOI] [Google Scholar]
- 120.Taylor BN, and Kuyatt CE (1994). Guidelines for Evaluating and Expressing the Uncertainty of NIST Measurement Results.
- 121.Higgins-Chen AT, Thrush KL, Wang Y, Minteer CJ, Kuo P-L, Wang M, Niimi P, Sturm G, Lin J, Moore AZ, et al. (2022). A computational solution for bolstering reliability of epigenetic clocks: implications for clinical trials and longitudinal tracking. Nat. Aging 2, 644–661. 10.1038/s43587-022-00248-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.International Organization for Standardization (1994). Accuracy (trueness and precision) of measurement methods and results — Part 1: General principles and definitions.
- 123.Center for Drug Evaluation and Research, Food and Drug Administration (2018). Biomarker Qualification: Evidentiary Framework Guidance for Industry and FDA Staff.
- 124.Sugden K, Hannon EJ, Arseneault L, Belsky DW, Corcoran DL, Fisher HL, Houts RM, Kandaswamy R, Moffitt TE, Poulton R, et al. (2020). Patterns of Reliability: Assessing the Reproducibility and Integrity of DNA Methylation Measurement. Patterns 1, 100014. 10.1016/j.patter.2020.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Dobbin KK, Cesano A, Alvarez J, Hawtin R, Janetzki S, Kirsch I, Masucci GV, Robbins PB, Selvan SR, Streicher HZ, et al. (2016). Validation of biomarkers to predict response to immunotherapy in cancer: Volume II — clinical validation and regulatory considerations. J. Immunother. Cancer 4, 77. 10.1186/s40425-016-0179-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hägg S, and Jylhävä J (2021). Sex differences in biological aging with a focus on human studies. eLife 10, e63425. 10.7554/elife.63425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bautmans I, Knoop V, Thiyagarajan JA, Maier AB, Beard JR, Freiberger E, Belsky D, Aubertin-Leheudre M, Mikton C, Cesari M, et al. (2022). WHO working definition of vitality capacity for healthy longevity monitoring. Lancet Healthy Longev. 3, e789–e796. 10.1016/s2666-7568(22)00200-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Moffitt TE, Belsky DW, Danese A, Poulton R, and Caspi A (2016). The Longitudinal Study of Aging in Human Young Adults: Knowledge Gaps and Research Agenda. J. Gerontol. A. Biol. Sci. Med. Sci 72, 210–215. 10.1093/gerona/glw191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Belsky DW, Caspi A, Houts R, Cohen HJ, Corcoran DL, Danese A, Harrington H, Israel S, Levine ME, Schaefer JD, et al. (2015). Quantification of biological aging in young adults. Proc. Natl. Acad. Sci 112, E4104–E4110. 10.1073/pnas.1506264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Miller RA, Li X, and Garcia G (2022). Aging Rate Indicators: Speedometers for Aging Research in Mice. Aging Biol. 1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Fontana L, Kennedy BK, Longo VD, Seals D, and Melov S (2014). Medical research: Treat ageing. Nature 511, 405–407. 10.1038/511405a. [DOI] [PubMed] [Google Scholar]
- 132.Zhang MJ, Pisco AO, Darmanis S, and Zou J (2021). Mouse aging cell atlas analysis reveals global and cell type-specific aging signatures. eLife 10, e62293. 10.7554/elife.62293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Barrett JE, Herzog C, Kim YN, Bartlett TE, Jones A, Evans I, Cibula D, Zikan M, Bjorge L, Harbeck N, et al. Susceptibility to hormone-mediated cancer is reflected by different tick rates of the epithelial and general epigenetic clock. Genome Biol 23, 52. 10.1186/s13059-022-02603-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Komaki S, Ohmomo H, Hachiya T, Sutoh Y, Ono K, Furukawa R, Umekage S, Otsuka-Yamasaki Y, Minabe S, Takashima A, et al. (2022). Evaluation of short-term epigenetic age fluctuation. Clin. Epigenetics 14, 76. 10.1186/s13148-022-01293-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Poganik JR, Zhang B, Baht GS, Tyshkovskiy A, Deik A, Kerepesi C, Yim SH, Lu AT, Haghani A, Gong T, et al. (2023). Biological age is increased by stress and restored upon recovery. Cell Metab. 35, 807–820.e5. 10.1016/j.cmet.2023.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Shindyapina AV, Cho Y, Kaya A, Tyshkovskiy A, Castro JP, Deik A, Gordevicius J, Poganik JR, Clish CB, Horvath S, et al. (2022). Rapamycin treatment during development extends life span and health span of male mice and Daphnia magna. Sci. Adv 8, eabo5482. 10.1126/sciadv.abo5482. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


