Summary
Natural Killer (NK) cells are commonly absent from many human solid tumors, which presents a unique problem for generating a potent anti-tumor response. Understanding the mechanisms that lead to this paucity of cytolytic NK cells remains an unmet challenge. In this study, we investigated an inverse correlation between the abundance of monocyte-derived macrophages (mo-macs) and NK cells in human non-small cell lung cancer (NSCLC) lesions. We show that the Triggering Receptor Expressed on Myeloid Cells 2 (TREM2) governs a pro-tumorigenic gene program, encoding a lipid and cholesterol metabolism machinery, that is used by NSCLC-infiltrating mo-macs that have engulfed cell debris. Notably, genetic deletion of Trem2 rescued NK cell accumulation and enabled NK cell-mediated regression of lung tumors. By generating single-cell mRNA profiles of mo-macs, we identified the IL-18/IL-18BP decoy interaction as a potential mechanism through which TREM2+ mo-macs impair NK cell activation; indeed, we found that the NK cell-dependent elimination of lung tumors in the absence of the TREM2 program was dependent on IL-18 signaling. In addition, the successful activation of NK cells was also critically dependent on IL-15. Importantly, antibody-based neutralization of TREM2 synergized with an NK cell activating agent to further inhibit the growth of lung tumors. In sum, our findings identify a novel immune axis in NSCLC, in which TREM2+ mo-macs suppress the accumulation and cytolytic activity of NK cells. Accordingly, the dual combination of TREM2 blockade and secondary enhancement of NK cell function represents a novel therapeutic design that significantly boosts anti-tumor immunity.
Introduction
The native anti-tumor immune response is constrained by several variables, including the dysfunction of chronically stimulated, exhausted T cells and the unique ability for tumor cells to escape recognition and killing by T cells by modulating the cell surface expression of immunogenic tumor antigens in a process known as ‘immuno-editing’ (Dunn et al., 2004). While various modalities of immunotherapy seek to leverage our knowledge of the pathways involved in the generation of dysfunctional T cells – either attempting to reverse or reinvigorate them – designing tools that empower other effector immune cells, such as Natural Killer (NK) cells, that are not dependent on tumor antigen-specific recognition represents an attractive alternative. But, while NK cells have been linked to better control of tumor growth and metastases, they are scarce in both human and murine tumor lesions (Gasser et al., 2005; Raulet et al., 2013). The reasons why they are so often excluded from the tumor bed and why the NK cells that do infiltrate exhibit a highly dysfunctional phenotype with low cytolytic potential remain unclear. At least with regards to the latter, it has been reported that some tumors actively evade NK cell-mediated killing by either down-regulating or shedding activating surface ligands (e.g., NKG2D ligands) or up-regulating inhibitory ones (Smyth et al., 2002; Hayakawa et al., 2006). Interestingly, known inhibitory receptors that mediate and maintain NK cell de-activation also recognize elements of the MHC class I machinery, including HLA-A, -B, -C, and -E (Parham and Guethlein, 2018), which are already often suppressed by tumors so as to escape pressure from tumor-infiltrating CD8 T cells (Garrido et al., 2016; McGranahan et al., 2017). It follows that, while NK cells accumulate and exert greater control over lesions with low MHC-I, many of these tumors persist, suggesting that functionally redundant modes of suppression dampen NK cell cytotoxicity (Shimasaki et al., 2020; Cózar et al., 2021). Which cellular players of the tumor microenvironment (TME) contribute to these immunosuppressive networks – and specifically, to the paucity and dysfunction of NK cells – has yet to be fully elucidated.
Our groups and others have found that many human tumor lesions are deprived of NK cells and are heavily infiltrated by monocyte-derived macrophages (mo-macs) (Lavin et al., 2017; Leader et al., 2021; Mulder et al., 2021). Mo-macs are purportedly immunosuppressive (DeNardo et al., 2009; Masetti et al., 2021), and targeting mo-macs to modulate their phenotype or their recruitment to inflamed tissues and elicit a stronger anti-tumor immune response has proven difficult, in part due to a limited understanding of the heterogeneity of macrophage states that are present in the TME (Lavin et al., 2015). Therefore, the need to dissect and characterize the role of these programs in the generation and maintenance of an anti-tumor response – or conversely, in tumor progression – is ever more vital (Park et al., 2022). For instance, we demonstrated that, during tumor inception, tissue-resident alveolar macrophages (AMs) physically interact with early non-small cell lung cancer (NSCLC) tumor cells, promote epithelial-mesenchymal transition, and recruit regulatory T cells (Tregs) to the early tumor niche (Casanova-Acebes et al., 2021). During later stages of tumor growth, AM are reorganized to the periphery of the tumor (Casanova-Acebes et al., 2021; Loyher et al., 2018). Defining the molecular signature of mo-macs that supplant AM during later stages of tumor growth and characterizing their functional roles and how to modulate their immunosuppressive functions may augment the effect of cancer immunotherapy.
We demonstrated that, in both human and murine lung tumors, mo-macs express the Triggering Receptor Expressed on Myeloid Cells 2 (TREM2) (Lavin et al., 2017; Casanova-Acebes et al., 2021; Leader et al., 2021). Though originally described in microglia as a phagocytic receptor for myelin debris (Guerreiro et al., 2013; Sims et al., 2017; Keren-Shaul et al., 2017; Zhou et al., 2020; Wang et al., 2020), analogous populations of TREM2+ mo-macs have been identified in multiple human and murine tumors. In models of sarcoma and ovarian cancer, genetic deletion of Trem2 or antibody-mediated blockade of the receptor generated an enhanced CD8 T cell response that delayed tumor growth and synergized with immune checkpoint blockade (Molgora et al., 2020; Katzenelenbogen et al., 2020; Binnewies et al., 2021). These findings suggested an immunosuppressive role for TREM2 in anti-tumor immunity. However, the most recent work on hepatocellular carcinoma showed that TREM2+ macrophages may, in fact, limit tumor progression in the hepatic TME, suggesting that the functional contribution of these phagocytes to anti-tumor immunity may be tissue-specific (Perugorria et al., 2019; Esparza-Baquer et al., 2021).
Therefore, in this study, we probed the role that TREM2+ mo-macs play in the development of non-small cell lung cancer (NSCLC), using a KrasG12D/+ p53−/− (KP) model of primary lung adenocarcinoma (DuPage et al., 2009), with the initial hypothesis that they may underscore the lack of NK cells in primary lung tumors. We first show that phagocytic sensing of tumor cell-derived antigens triggers the TREM2 program in murine mo-macs. Then, by profiling TREM2-proficient and - deficient myeloid cells of tumor-bearing, bone marrow chimeric mice by single-cell RNA sequencing (scRNAseq), we show that TREM2 controls the transition of mo-macs towards a pro-tumorigenic state. Consistent with these findings, we found that TREM2+ mo-macs suppress NK cell recruitment and activation by disabling IL-18 signaling. Strikingly, we found that the enhanced NK cell activity following TREM2 blockade is also critically dependent on IL-15. Indeed, genetically ablating Trem2 or inhibiting it with a blocking antibody for TREM2 unleashed NK cell immunity and significantly eliminated tumors. Finally, we show that combining TREM2 blockade with an NK cell activating agent yields an even greater anti-tumor immune response, thus framing a novel therapeutic design with high translational potential.
Results
NK cells poorly infiltrate NSCLC lesions, unlike TREM2+ mo-macs
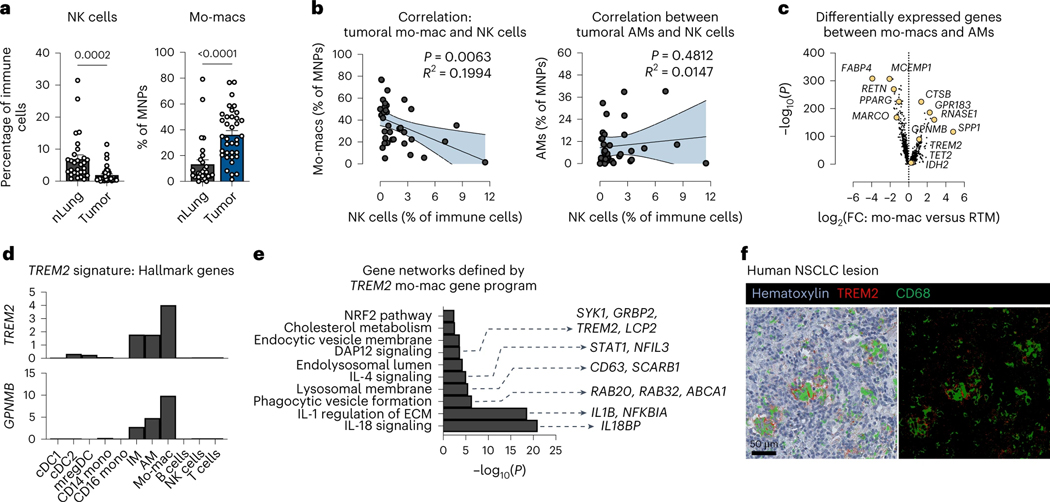
We previously generated a large single-cell RNA sequencing (scRNAseq) dataset – and accompanied proteomic data by CITE-seq – of immune cells from resected tumor lesions and patient-matched, non-involved lung tissues of a cohort of 35 treatment-naïve NSCLC patients (Leader et al., 2021). From this atlas, we found that NSCLC lesions are significantly deprived of NK cells, compared to non-involved, adjacent lung tissue (nLung) (Fig. 1a, left); instead, they are heavily enriched with mo-macs (Fig. 1a, right) that are transcriptomically distinct from the pool of tissue-resident AM, which were preferentially enriched in nLung (Leader et al., 2021). Accordingly, the frequency of intratumoral NK cells was inversely correlated with the frequency of mo-macs (R2=0.0397, p-value=0.0063), but not with AM, indicating a potential role for mo-macs – among tumor-enriched mononuclear phagocytes (MNPs) (Leader et al., 2021) – in contributing to the relative absence of NK cells in the TME (Fig. 1b).
Figure 1. TREM2+ monocyte-derived macrophages are major constituents of an NK cell-deprived microenvironment.
a) Cell frequency of NK cells identified by scRNAseq profiling as a percentage of immune cells (left) and the cell frequency of monocyte-derived macrophages (mo-macs) as a percentage of mononuclear phagocytes (MNPs) (right) in resected tumors (tumor) and patient-matched, non-involved adjacent lung tissues (nLung) from human non-small cell lung cancer (NSCLC) samples. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
b) Correlation between the frequency of tumor-infiltrating mo-macs (left) or alveolar macrophages (AMs) (right) and of intratumoral NK cells per patient. (Pearson correlation test was performed at a 95% confidence interval)
c) Identification of the TREM2 gene program as a universal mo-mac cell state among differentially expressed genes that distinguish mo-macs from AMs.
d) Expression of hallmark TREM2 program genes, showing specificity to mo-macs in the tumor microenvironment, relative to other immune cells identified by scRNAseq.
e) Gene set enrichment analysis of gene networks that form amongst expressed genes in the TREM2 program from (c).
f) Multiplex immunohistochemistry (mIHC) of human NSCLC lesions for TREM2 (red) and CD68 (green), shown with hematoxylin counterstain (left) and on a black negative background (right). Scale bar, 50 μm.
To better characterize mo-macs in the TME, we computed differentially expressed genes that distinguished them from the AMs; these included TREM2, GPNMB, SPP1, CTSB, RNASE1, and GPR183 (Fig. 1c, Supplementary Table 1). While no lymphoid cells express TREM2 (Heng et al., 2008), the latent expression of the TREM2 signature genes by AMs in human NSCLC may indicate that intratumoral cues that induce this signature in recruited mo-macs can also impact the phenotype of the local population of AMs that otherwise express negligible levels of TREM2 at the steady-state (Heng et al., 2008) (Fig. 1d). The converse – the potential induction of AM gene programs in recruited mo-macs – may also be probable; however, the de-enrichment of AMs from the intratumoral terrain of the TME and the lack of AM-specific genes (e.g., MARCO, PPARG, AXL, IL18) expressed by mo-macs, indicate that microenvironmental cues may favor the former scenario, in which tumor-derived signals trigger mo-mac-descriptive gene programs, like the TREM2 program (Supplementary Table 1). Gene set enrichment analysis of the TREM2 program identified active signaling pathways, such as IL-18 signaling, IL-1 signaling, and phagocytic vesicle formation, that underscored a unique efferocytic state in mo-macs (Fig. 1e). We confirmed the presence of these TREM2+ macrophages in human NSCLC lesions by immunohistochemical (IHC) co-staining of TREM2 and CD68 proteins (Fig. 1f), motivating us to dissect this signature further to understand how it may modulate the composition and immunophenotype of the TME.
Efferocytosis of tumor cell-derived antigens induces the TREM2 gene program in mo-macs
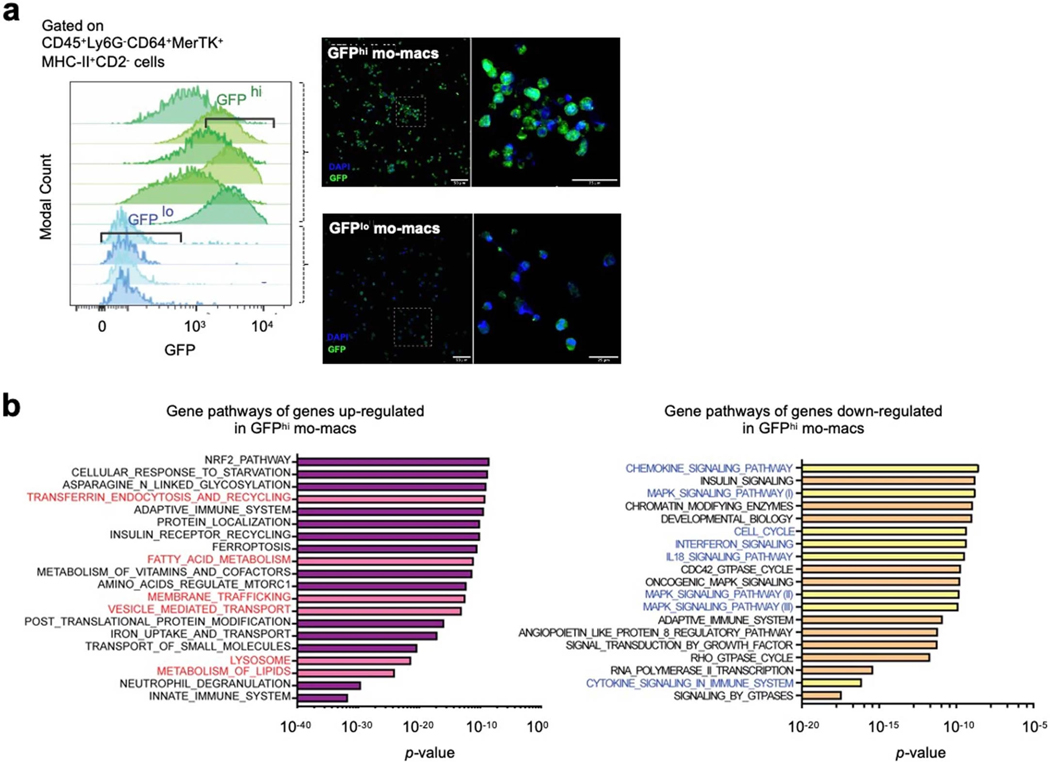
Our group previously published single-cell transcriptomic profiles of mononuclear phagocytes in a murine model of lung cancer (Casanova-Acebes et al., 2021). Homology analysis of gene expression across murine and human macrophage clusters showed that analogous TREM2+ mo-macs are significantly enriched in tumor-bearing lungs, relative to naïve lungs (Casanova-Acebes et al., 2021). In order to study the dynamic of this population and its functional contribution to anti-tumor immunity, we leveraged the same orthotopic model of primary lung adenocarcinoma (DuPage et al., 2009), by injecting GFP-expressing KP cells (KP-GFP) to use GFP as a surrogate tumor antigen and track the localization of TREM2+ cells, with respect to GFP+ cells. By confocal microscopy, we found that TREM2+ cells progressively accumulate within growing KP-GFP lung tumors over a 28-day period (Fig. 2a, top), resembling their intratumoral enrichment that we observed in human NSCLC.
Figure 2. Uptake of cellular debris induces the TREM2 gene program in mo-macs.
a) (top) Confocal immunofluorescence IF) imaging of murine KP-GFP tumor-bearing lungs depicting GFP+ tumor cells (green) and TREM2+ cells (red) at different timepoints during tumor progression (day 7, 14, 21, and 28 post-inoculation). (bottom, left) Confocal IF imaging of KP-GFP tumor-bearing lungs after 28 days of engraftment and growth showing intracellular staining of nuclei (DAPI) and GFP (green) in TREM2+ cells (red). Middle and right images represent magnification of the region of interest (ROI, white outline); middle panel depicts cells staining for both DAPI and GFP (white arrow), and the right panel depicts cells staining for both GFP and TREM2 (white arrow). Scale bar, 50 μm. (bottom, right) Pictorial diagram showing relationship between efferocytosis and TREM2.
b) Volcano plot of differentially expressed genes between GFPlo and GFPhi mo-macs. Statistical significance defined based on an adjusted P < 0.01. Significant genes are highlighted in either red (up-regulated upon tumor antigen capture) or blue (down-regulated upon tumor antigen capture). Data representative of two independent experiments.
c) Expression of Trem2 signature genes and other select genes encoding machinery involved in phagocytosed cargo transport, antigen processing, and lipid metabolism by GFPlo and GFPhi mo-macs. Values shown as the logarithm of fold change of absolute number of transcripts relative to the mean value across replicates.
d) Absolute expression (transcripts per million) of genes encoding LDL receptor, NPC1, and PLIN2 in GFPlo and GFPhi mo-macs. Paired values shown for paired GFPlo and GFPhi mo-macs that were purified from the same biological replicate. Data from two independent experiments. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
e) Absolute expression (transcripts per million) of genes encoding GPR183 and MARCKSL1 in GFPlo and GFPhi mo-macs. Paired values shown for paired GFPlo and GFPhi mo-macs that were purified from the same biological replicate. Data from two independent experiments. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
f) Relative qPCR quantification of Trem2, Gpnmb, and Apoe transcripts from in vitro differentiated bone marrow-derived macrophages (BMDMs) with or without (control) exposure to apoptotic KP-GFP cells. Briefly, BMDMs were differentiated and polarized with IL-4 and GM-CSF. KP-GFP cells were exposed to ultraviolet (UV) light for 30 min, left to undergo apoptosis overnight, and then added to BMDMs. After a four hour co-culture period, CD45+ CD64+ MerTK+ GFP+ cells were sorted by flow cytometry for PCR analysis. BMDMs cultured in the absence of apoptotic tumor cells were used as a negative control. Transcripts are shown as the two-fold difference in gene expression (ΔCt) over Ptprc expression. Data representative of two independent experiments. Values are shown as mean transcripts per million (TPM) ± standard error of mean (S.E.M.). (Unpaired two-tailed t-test at a 95% confidence interval)
TREM2 has been shown to recognize a variety of ligands – from lipopolysaccharides to sulfated glycosaminoglycans and different lipidic species – and signals via the DAP12 adaptor protein (Daws et al., 2003; Cannon et al., 2012). Given that it has also been shown to sense phosphatidylserine on the inverted membranes of dying cells (Takahashi et al., 2005; Hsieh et al., 2009; Lue et al., 2015; Filipello et al., 2018; Shirotani et al., 2019; Zhang et al., 2019), we hypothesized that either the uptake or phagocytic sensing of tumor cell-derived antigens from apoptotic tumor cells may induce the TREM2 program. Confocal imaging of GFP and TREM2+ cells showed that many of these phagocytes indeed display intracellular GFP signal (Fig. 2a, bottom left), supporting our hypothesis that efferocytosis – including the sensing of the debris that is engulfed – is associated with the expression of TREM2 (Fig. 2a, bottom right).
To test this hypothesis, we sought to characterize the GFP− and GFP+ subsets of mo-macs. Towards this end, we purified mo-macs from tumor-bearing lungs, based on previously identified markers, and profiled them using bulk RNA sequencing. From digested tumor-bearing lungs, the intracellular intensity of GFP in mo-macs was not discretely positive or negative, since these cells are likely actively engaged in antigen uptake and digestion upon entering the TME; so instead, we sorted GFPlo and GFPhi mo-macs (Extended Data Fig. 1a). Sequencing these mo-mac subsets showed that efferocytosis of antigens induces significant changes to the transcriptome (Fig. 2b, Supplementary Table 3). Significant differentially expressed genes included hallmark genes belonging to the TREM2 gene program (e.g., Trem2, Lpl, Syngr1, Apoe). Notably, genes more highly expressed by GFPlo mo-macs included co-stimulatory ligands, MHC class II molecules, and cytokines and chemokines critical for the recruitment and activation of CD8 T cells (i.e., Cd40, Il15, Il12b, Cx3cl1, Ccl5) (Fig. 2b). In contrast, GFPhi mo-macs expressed higher levels of inhibitory genes, including Cd274, which encodes PD-L1 (Fig. 2b).
In line with our hypothesis that the induction of the TREM2 gene program is driven by efferocytosis, we evaluated the expression of genes encoding machinery involved in this process. Specifically, we observed that lipid metabolism genes (Apoe, Lipa, Plin2, Lpl, Hilpda), genes involved in antigen uptake and trafficking (Lrp12, Rab7, Vat1, Mfge8) and genes encoding machinery proteins for degrading phagocytosed cargo (Ctsb, Ctsd, Ctss) were enriched in GFPhi mo-macs (Fig. 2c). Pathways governing these genes included oxidative phosphorylation, fatty acid metabolism, and lysosomal degradation (Extended Data Fig. 1c, left), whereas pathways active in GFPlo mo-macs are immunogenic (Extended Data Fig. 1c, right), concordant with the list of differentially expressed genes (Fig. 2b). Notably, significant changes were also observed for cholesterol metabolism genes encoding transporters (Ldlr), traffickers (Npc1), and packagers (Plin2), indicating that potential subcellular changes seen during the acquisition of the TREM2 gene program include cholesterol trafficking (Fig. 2d). Interestingly, GFPhi mo-macs also down-regulated their expression of Gpr183 and Marcksl1, suggesting that upon acquiring the TREM2 program, these phagocytes suppress their response to migratory cues that initially recruited them to the TME (Fig. 2e). Consistent with our ex vivo results, in vitro GFP+ mo-macs (mo-macs cultured with apoptotic tumor cells) up-regulated the Trem2 signature genes, compared to GFP− mo-macs (control cells) by qPCR (Fig. 2f). Altogether, our results identify efferocytosis as a trigger of the TREM2 program that critically limits the cell-intrinsic immunogenicity of mo-macs.
TREM2 promotes the transition of mo-macs towards a pro-tumorigenic state
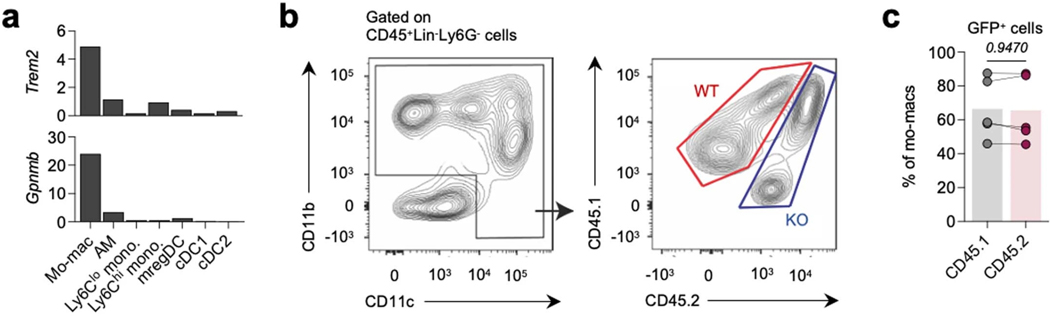
In order to determine the functional role of TREM2 signaling in tumor-infiltrating mo-macs, we challenged wild-type (WT) and complete Trem2−/− (KO) mice with KP-GFP cells and assessed survival and tumor burden in the lungs at 21 and 28 days post-inoculation. KO mice exhibited significantly improved survival, and histological quantification of lung tumors showed a striking reduction of tumor burden in KO mice, compared to WT littermates, as early as 21 days post-inoculation with tumor cells (Figure 3a,b). We performed scRNAseq of the MNP compartment from the tumor-bearing lungs of WT and KO mice to identify potential shifts in antigen-presenting and tissue sentinel cells that might underlie the tumor reduction phenotype. Unsupervised clustering identified tissue-resident AMs, mo-macs, inflammatory (Ly6Chi, Ly6c2-expressing) and patrolling (Ly6Clo, Ly6c2-non-expressing) monocytes, and conventional dendritic cells (cDC), including type I cDCs (cDC1), type II cDCs (cDC2), and mature cDCs (mregDC). Tumor-bearing lungs of KO mice were more enriched with AMs and inflammatory monocytes, while exhibiting an attrition of Trem2-expressing mo-macs (Fig. 3c). Flow cytometric quantification of mo-macs in the tumor-bearing lungs of WT and KO mice confirmed this reduction (Fig. 3d). Interestingly, while the frequency of AMs did increase, the absolute number of AMs was not significantly different between tumor-bearing WT and KO mice, suggesting that the reduced infiltration of tumor-bearing lungs by monocyte-derived cells was a major, but likely not the only, driver of changes to the MNP landscape, in the absence of functional TREM2 (Fig. 3e). By both scRNAseq and flow cytometry, we also found that cDC1 were enriched in frequency and absolute numbers in KO mice (Fig. 3c,f), reflecting a preferential enrichment for more cDC with potent cross-presentation potential in KO mice. Given that Trem2 expression is not detected in other MNPs, including the cDCs (Extended Data Fig. 2a), we surmised that shifts in the cDC pool from the tumor-bearing lungs of KO mice likely reflect the activity of TREM2-deficient mo-macs.
Figure 3. TREM2 deficiency restricts tumor growth by remodeling the composition of MNPs.
a) Kaplan Meier survival curve of KP-GFP tumor-bearing WT (n = 8) and KO (n = 9) mice.
b) Representative H&E images of tumor-bearing lungs of WT and KO mice at day 21 (WT, n = 4; KO, n = 4) and at day 28 (WT, n = 11; KO, n = 6) post-inoculation of tumor cells (left). Quantification of the tumor area as a percent of the total area of the lung cross-section (right) is shown. Data are representative of three independent experiments. (Multiple unpaired two-sampled t-tests)
c) Clustering of mononuclear phagocytes (MNPs) based on scRNAseq profiling (CD45+ CD3− CD19− NK1.1− Ly6G− CD11b+ CD11c+) from the tumor-bearing lungs of WT (n = 2) and KO (n = 2) mice, revealing cell frequency changes among Ly6Chi (inflammatory) and Ly6Clo (patrolling) monocytes, alveolar macrophages (AM), and Trem2+ mo-macs. Bar plot shows the composition of the monocyte-macrophage compartment in WT and KO mice. Frequencies for each group are shown as the mean of each pair of duplicates.
d) Flow cytometric quantification of mo-macs, represented as relative frequency among CD45+ immune cells (left) and as absolute number of cells per lung (right) from tumor-bearing lungs of WT and KO mice at day 28 post-inoculation. The results shown are from one experiment, representative of five independent experiments. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
e) Flow cytometric quantification of AMs, represented as relative frequency among CD45+ immune cells (left) and as the absolute number of cells per lung (right), from tumor-bearing lungs of WT and KO mice at day 28 post-inoculation. The results shown are from one experiment, representative of five independent experiments. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
f) Flow cytometric quantification of cDC1, represented as the absolute number of cells per lung from tumor-bearing lungs of WT and KO mice at day 28 post-inoculation of tumor cells. The results shown are from one experiment, representative of five independent experiments. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
g) Sub-lethally irradiated CD45.1 WT recipient mice were reconstituted with CD45.1 WT (WT, n = 6), CD45.2 KO (KO, n = 6), or a 1:1 mixture of both CD45.1 WT and CD45.2 KO bone marrow cells (chimera, n = 6). Full donor marrow engraftment was confirmed after seven weeks.. Shown are representative H&E images of tumor-bearing lungs of WT, KO, and chimeric mice (left) at day 28 post-tumor cell inoculation. Quantification of the tumor area as a percent of the total area of the lung cross-section (right) is shown. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
h) Genes of the TREM2 gene program whose expression defines the TREM2 cell state. Frequencies of mo-macs in the TREM2hi and TREM2lo cell states, represented as a relative frequency among MNPs from the purified CD45.1 WT and CD45.2 KO fractions of MNPs from KP-GFP tumor-bearing chimeric mice. Paired values shown for paired CD45.1 WT and CD45.2 KO cells that were purified from the same biological replicate. (mean ± standard error of mean (S.E.M.); paired two-tailed t-test at a 95% confidence interval)
Alternatively, we considered the possibility that the snapshots of the MNP compartment shown by scRNAseq were a result of reduced tumor growth. To account for this, we generated bone marrow chimeric mice by reconstituting sub-lethally irradiated CD45.1 WT recipient mice with either (i) CD45.1 WT marrow (“WT”), (ii) CD45.2 Trem2−/− marrow (“KO”), or (iii) a 1:1 mixture of both genotypes (“Chimera”), so that in tumor-challenged chimeric mice, both TREM2-proficient and -deficient cells are exposed to the same TME, allowing us to control for the differences in tumor burden that we previously observed. As expected, KO mice exhibited significantly reduced tumor burden, relative to WT mice. Strikingly, chimeric mice exhibited an ‘intermediate’ phenotype, suggesting that the relative abundance of mo-macs highly enriched for the Trem2 signature drives tumor progression (Fig. 3g). In the chimeric mice, we observed no differences in the frequency of GFP+ cells among mo-macs (Extended Data Fig. 2b,c), indicating that additional means of uptake may be present.
To assess cell-intrinsic differences between CD45.1 WT and CD45.2 KO phagocytes, we performed scRNAseq of purified CD45.1 WT and CD45.2 KO MNPs from the tumor-bearing lungs of the chimeric mice. Unsupervised clustering identified a mix of CD45.1 and CD45.2 mo-macs in the TME of chimeric mice; however, examination of the expression of genes belonging to the TREM2 gene program (Gpnmb, Spp1, Lipa, Lpl, Plin2, Fabp5, Syngr1, Gpr183, Cd9, Sgk1, Apoe, Mfge8, Il18bp, Lilrb4a) – with the exception of Trem2 itself – enabled us to describe the role of TREM2 in the transcriptomic architecture of mo-macs in a tumor burden-controlled setting (Fig. 3h, left). We could partition mo-macs into those highly enriched for the expression of the aforementioned genes, in which case we denoted these cells as those in the TREM2hi state, whereas those lowly enriched for the expression of these genes were annotated as cells in a TREM2lo state. Strikingly, mo-macs in the TREM2hi state were dominantly represented by CD45.1 WT cells, whereas mo-macs in the TREM2lo state were more significantly represented by CD45.2 KO cells (Fig. 3h, middle and right), suggesting that the expression of TREM2 acts as a master regulator of the broader TREM2 gene program and that its genetic deletion interrupts the molecular transition of mo-macs towards a pro-tumorigenic state. Importantly, given that monocytes do not express Trem2 and that TREM2 is expressed once monocytes differentiate into mo-macs in tumor lesions (Extended Data Fig. 2a), there may be a temporal element to the acquisition of and enrichment for the TREM2 program that is initially coupled to the degree of efferocytic sensing. Collectively, our results highlight a crucial, cell-intrinsic role for TREM2 in governing the mo-mac transition towards the TREM2hi state, which we find has major consequences for anti-tumor immunity.
TREM2+ mo-macs restrict NK cell-mediated anti-tumor immunity
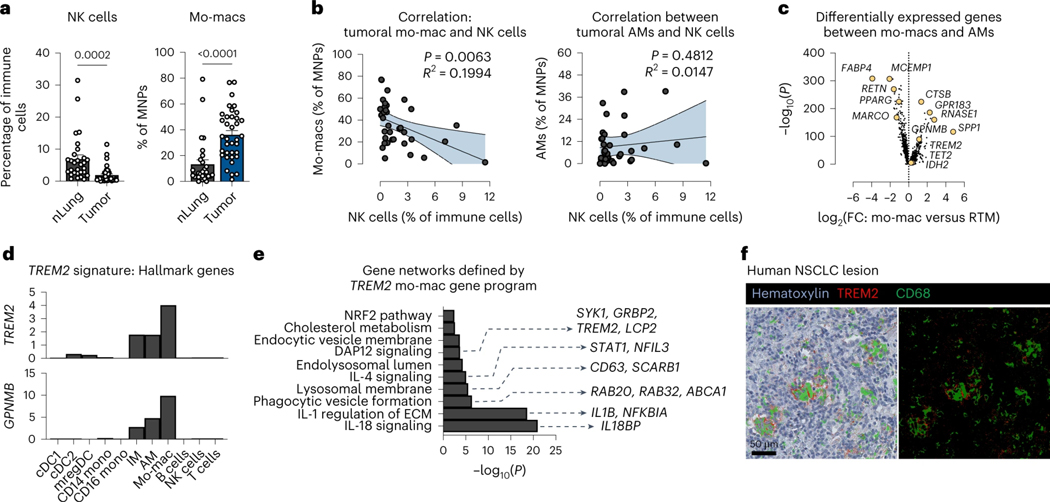
We then sought to understand how TREM2 deficiency modulates the effector immune cells that might underlie the tumor reduction phenotype in KO mice. Flow cytometry of T cells revealed a marked expansion of antigen-specific, polyfunctional CD8 T cells in the tumor-bearing lungs and tumor-draining lymph nodes (tdLN) of KO mice challenged with OVA-expressing KP-GFP cells (KP-GFP-OVA), compared to KP-GFP-OVA tumor-bearing WT mice (Fig. 4a and Extended Data Fig. 3a). We also observed an expansion of NK cells that were significantly more activated and cytolytic in the tumor-bearing lungs of KO mice, compared to WT mice (Fig. 4b and Extended Data Fig. 3b); this was confirmed by staining for the bona fide NK cell marker NKp46 in murine KP-GFP tumors from WT and KO mice (Fig. 4c). Moreover, the immunohistochemistry also indicated that, in the absence of functional TREM2, NK cells accumulate within tumors, indicating a spatially relevant increase in the NK cell content of tumor-bearing lungs in KO mice (Fig. 4c).
Figure 4. Therapeutic effect of TREM2 deficiency is critically dependent on intact IL-18 and IL-15 signaling.
a) Flow cytometric quantification of OVA-tetramer+ (antigen-specific) CD8 T cells, represented as the relative frequency among total CD8 T cells, in the lungs (left) and tumor-draining lymph nodes (tdLN) (right) of KP-GFP-OVA tumor-bearing WT and KO mice. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
b) Flow cytometric quantification of NK cells, represented as the relative frequency of CD45+ immune cells (left) and as absolute number of cells per lung (right), in the KP-GFP tumor-bearing lungs of WT and KO mice. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
c) Representative immunohistochemistry (IHC) staining of NKp46 for NK cells in a KP-GFP tumor from a KO mouse (left). Arrows mark positively-stained cells. Quantification of NKp46+ cells in a representative tissue section per lung sample from WT (n = 6) and KO (n = 6) mice (right).
d) Representative H&E images of tumor-bearing lungs of WT, isotype antibody (n = 4), KO, isotype antibody (n = 4), KO mice depleted of CD8 T cells (KO+αCD8, n = 5), KO mice depleted of NK cells (KO+αNK, n = 8) at day 28 post-inoculation of tumor cells (left). Quantification of the tumor area as a percent of the total area of the lung cross-section (right) is shown. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
e) Flow cytometric quantification of RAE-1+H60+ tumor cells, as a relative frequency of total KP-GFP tumor cells, in the tumor-bearing lungs of WT and KO mice. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
f) Expression of the Il18bp (left) and Il18 (right) transcripts, as defined as the mean number of unique molecular identifiers (UMI), by AMs, Trem2hi mo-macs, and Trem2lo mo-macs, as identified by unsupervised clustering of scRNAseq profiling of MNPs in KP-GFP tumor-bearing WT and KO mice.
g) Experimental schematic of in vitro co-culture study of mo-macs generated from the bone marrow of WT and KO mice and of splenic NK cells isolated from wild-type and Il18r1 knockout mice (Il18r1−/−). In brief, after mo-macs were fed with apoptotic KP-GFP cells for 4 hours, splenic NK cells were isolated and cultured with mo-macs at a 1:10 ratio. These cells were then stimulated with recombinant IL-18 and IL-12 with Brefeldin A for 4 hours, after which NK cells were collected for flow cytometric analysis.
h) Flow cytometric quantification of CD69+ activated NK cells (left) and of Ki-67+ proliferative NK cells (right), shown as a frequency of total NK cells. The results shown are from one experiment, representative of three independent experiments. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
i) Quantification of the tumor area, as a percent of the total area of the lung cross-section, of tumor-bearing lungs of WT, isotype antibody (n = 6), KO, isotype antibody (n = 6), and KO mice that received the IL-18 neutralizing antibody (n = 8). The results shown are from two independent experiment. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
j) Flow cytometric quantification of NK cells, as a relative frequency of CD45+ immune cells (left), and of IFN-ɣ-producing NK cells, as a relative frequency among total NK cells (right). (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
To determine which of these shifts in effector immune cells reflects a necessary change to facilitate the therapeutic benefit seen in KO mice, we depleted either CD8 T cells or NK cells from tumor-challenged KO mice. Interestingly, while depleting CD8 T cells only partially reversed the decrease in tumor burden in tumor-bearing KO mice, NK cell depletion completely abrogated the tumor reduction phenotype (Fig. 4d). Moreover, only NK cell depletion reversed the expansion of cDC1 in KO mice (Extended Data Fig. 3c). Indeed, in tumor-bearing WT and KO mice deprived of NK cells, the NK cell-dependent reduction of tumor burden could only be observed in the KO mice, suggesting that the significance of the NK cell response to KP-GFP lung tumors is only realized in the absence of functional TREM2+ mo-macs (Extended Data Fig. 3d,e). In line with these results, we found a reduction in the proportion of KP-GFP tumor cells that express NKG2D ligands (RAE-1 and H60) in KO mice, compared to WT mice, reflecting the NK cell-driven elimination of tumor cells that is enabled by TREM2 deficiency (Fig. 4e). For these reasons, we postulated that TREM2+ mo-macs do indeed play an active role in the paucity and poor cytolytic activity of NK cells in the TME of lung tumors.
To identify what modes of communication facilitate this process, we leveraged the molecular profiles of Trem2hi and Trem2lo mo-macs from the scRNAseq of CD45.1 WT and CD45.2 KO MNPs in the lungs of tumor-bearing chimeric mice to identify potential molecules that could impact NK cell recruitment and activation. Among differentially expressed genes, we observed that Trem2hi mo-macs express uniquely high levels of Il18bp, compared to Trem2lo mo-macs and AM (Fig. 4f, left). The Il18bp gene encodes the decoy protein IL-18BP that intercepts IL-18 and prevents signaling in cells expressing the IL-18 receptor. AM are the main expressors of Il18 (Fig. 4f, right), highlighting the potential role of IL-18 signaling – or rather, the lack thereof possibly due to the reorganization of AM to the peripheral tumor bed (Casanova-Acebes et al., 2021) – in the regulation of NK cells by TREM2+ mo-macs. These results aligned with the network analysis of human TREM2+ mo-macs (Fig. 1e).
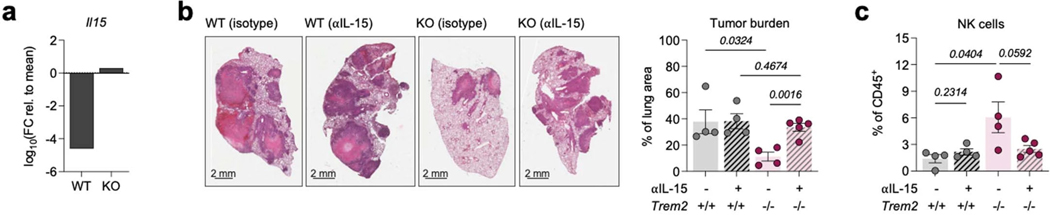
To functionally test this, we leveraged an in vitro co-culture assay, where we generated mo-macs from WT and KO mice and cultured them with splenic NK cells from WT and Il18r1 knockout mice that were stimulated with IL-18 and IL-12 (Fig. 4g). We assessed NK cell activation based on the expression of the activation marker CD69 and proliferative potential (expression of Ki-67); the absence of TREM2 on mo-macs significantly enhanced the activation of NK cells, but this positive response was abrogated in the absence of the IL-18 receptor on NK cells, despite the deletion of Trem2 on mo-macs (Fig. 4h). We surmised that, in accordance with our scRNAseq analyses, this reflects the possible role of secreted IL-18BP in interfering with the available IL-18 that we introduced to the system. Moreover, as IL-12 was included in the stimulus cocktail for all conditions, the significant decline in response – in the absence of the IL-18 receptor on NK cells – suggested that IL-12 alone, though it may still contribute to the shaping of the CD8 T cell response, likely does not play a major role in the regulation of NK cells by TREM2+ mo-macs. Given these results, we next sought to determine whether this mechanism is relevant in vivo. We modeled conditions reflecting IL-18 interference by treating KP-GFP-challenged KO mice with a neutralizing antibody. IL-18 blockade in KO mice partially reversed the tumor reduction phenotype, compared to untreated KO mice (Fig. 4i); it also tampered with the recruitment and activation of NK cells and the activation of CD8 T cells (Fig. 4j and Extended Data Fig. 3f), suggesting that IL-18 signaling contributes to the activation of NK cells that is suppressed by TREM2 on mo-macs.
IL-18 is well known to require cooperative signaling with cytokines expressed by cDC, such as the trans-presented cytokine IL-15, to enhance and sustain the cytolytic NK cell response (Chaix et al., 2008; Nielsen et al., 2016). So, we sought to determine whether the NK cell enhancement in the absence of TREM2 relied upon this example of cooperative cytokine signaling. From our single-cell profiling of MNPs in the tumor-bearing lungs of WT and KO mice, we documented an upregulation of the Il15 transcript by the mature mregDCs in KO mice (Extended Data Fig. 4a). Therefore, we chose to examine the role of IL-15 in the tumor reduction phenotype in KO mice. We blocked IL-15 in tumor-bearing KO mice, using an IL-15 neutralizing antibody, to determine whether IL-15 signaling itself is an essential component of the TREM2+ mo-mac/NK cell mechanism. IL-15 blockade completely abrogated both the tumor reduction phenotype and expansion of NK cells in KO lungs (Extended Data Fig. 4b,c), suggesting that upon expansion of cDC1 into the TME of KO mice, IL-15 is needed to sustain the NK cell response.
TREM2 blockade and activation of NK cells represents a novel therapeutic strategy
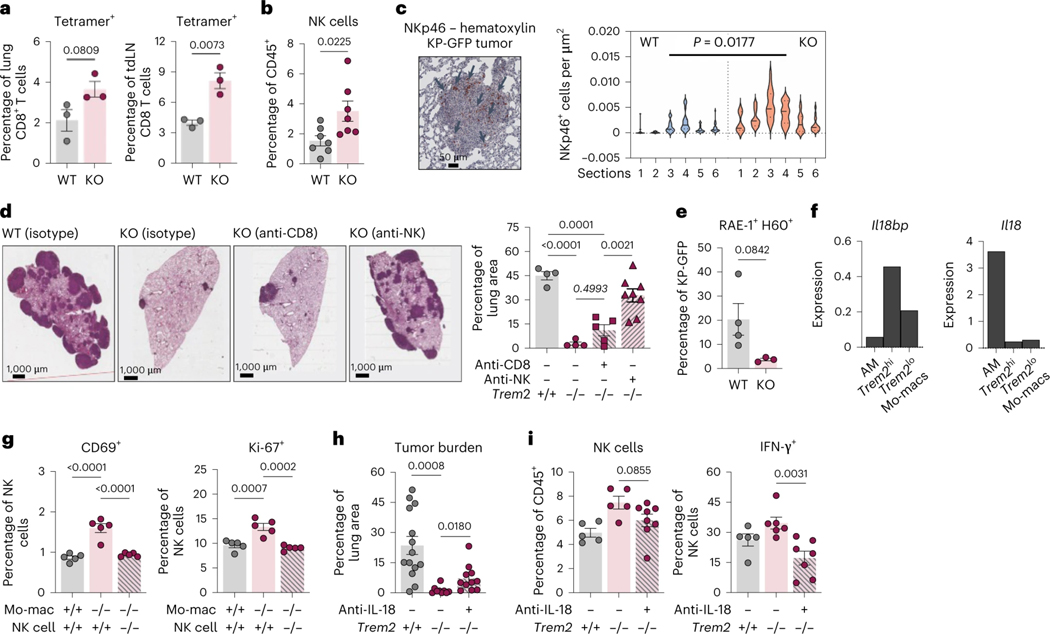
To evaluate the therapeutic and translational potential of targeting TREM2+ mo-macs in the lung TME, we tested a non-depleting TREM2 blocking antibody (αTREM2) in KP-GFP-challenged WT mice, as previously described (Molgora et al., 2020). Treatment resulted in a strong reduction of tumor growth in WT mice, comparable to the effect observed in KO mice (Fig. 5a). The recruitment and activation of NK cells in αTREM2-treated mice was also comparable to those in the tumor-bearing lungs of KO mice (Fig. 5b). We reasoned that leveraging strategies to boost any feature of NK cell immunity could enhance the anti-tumor effect of TREM2 blockade. As a proof-of-principle, we assessed the utility of blocking TREM2 signaling in combination with agents that release the breaks on NK cell activation, by preventing the degradation of NK cell-activating ligands. The human MHC class I polypeptide-related sequence A (MIC-A), for example, is an activating ligand that interacts with NKG2D on NK cells and relays a positive signal that boosts NK cell-derived IFN-ɣ production and the release of granzymes. However, while MIC-A expression is conserved across many human tumors, these lesions are able to escape NK cell recognition by cleaving MIC-A with ADAMs/MMPs and shedding it from the surface (De Andrade et al., 2018). An antibody that stabilizes MIC-A by shielding the extracellular domains from proteolytic cleavage was recently shown to be a promising NK cell-based immunotherapeutic (De Andrade et al., 2018). Therefore, we explored its synergistic potential in combination with TREM2 deficiency by challenging WT and KO mice with the metastatic melanoma B16-F10 cell line that is engineered to expresses MIC-A (B16-F10-MICA). Mice then either received isotype control or the MIC-A stabilizing antibody (αMICA). A final group of mice received both the αMICA and an NK cell-depleting antibody. Treatment with αMICA significantly reduced metastatic lesions in the lungs of WT mice and such therapeutic effect was substantially enhanced in KO mice, confirming the dependence of the anti-tumor response to lung tumors on NK cells and indicating a synergy between both TREM2 deficiency and the stabilization of NK cell activation in vivo; in addition, the therapeutic effect of the combination treatment was completely abrogated by NK cell depletion, indicating the efficacy of the treatment and reinforcing the idea that TREM2+ mo-macs hamper NK cell quality (Fig. 5c), highlighting a robust axis of immunity between TREM2+ mo-macs and NK cells that oversees the lung environment, whether that be in response to primary lung tumors or metastatic melanoma lesions in the lungs (Fig. 5d).
Figure 5. Therapeutic inhibition of TREM2 synergizes with an NK cell stabilizing agent to promote superior NK cell immunity and tumor elimination.
a) Representative H&E images of tumor-bearing lungs of WT, isotype antibody (n = 5), KO, isotype antibody (n = 5), KO that received the TREM2 blocking antibody (αTREM2) (n = 5) (left). Quantification of the tumor area as a percent of the total area of the lung cross-section (right) is shown. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
b) Flow cytometric quantification of NK cells, as a relative frequency of CD45+ immune cells (left), and KLRG1-expressing NK cells, as a relative frequency of total NK cells (right), in the tumor-bearing lungs of WT mice and KO mice that were treated with either an isotype control or the αTREM2 antibody. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
c) Representative H&E images of tumor-bearing lungs of WT, isotype antibody (n = 5), KO, isotype antibody (n = 5), WT that received the MIC-A stabilizing antibody (αMICA) (n = 5), KO that received the αMICA (n = 6), and KO that received both αMICA and an NK cell-depleting antibody (n = 5) (left). Quantification of the tumor area as a percent of the total area of the lung cross-section from tumor-bearing lungs of these mice is shown (right). (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
d) Graphical summary of the regulation of NK cells by TREM2+ mo-macs in the tumor microenvironment.
Discussion
A scarcity of intratumoral cytolytic NK cells is a feature of the TME, not just in NSCLC but in many other cancer types (Izawa et al., 2011; Jin et al., 2014; Sconocchia et al., 2014; Stankovic et al., 2014), suggesting that the depletion of NK cells enables tumor growth. Yet, identifying the cellular networks behind this ‘NK cell desert’ phenotype in the tumor topography has continued to be a major obstacle. Unleashing NK cell-mediated elimination of target tumor cells provides the unique advantage of occurring independently of the repertoire of tumor antigens; therefore, this could still eliminate tumors that escaped T cell surveillance. In the present study, we report that among the various mononuclear phagocytes that instruct effector cell activity, the pool of mo-macs expressing TREM2 is uniquely abundant in tumor lesions and inversely correlated in frequency with that of NK cells. This motivated our hypothesis that these macrophages may disable NK cell recruitment and activation in NSCLC lesions. We leveraged public human scRNAseq dataset to elucidate and refine the full transcriptomic signature of TREM2+ mo-macs and used a murine model of lung adenocarcinoma to show that murine TREM2+ mo-macs, like their homologous human counterparts, accumulate with tumor growth and efficiently infiltrate lesions, unlike the tissue-resident AMs (Casanova-Acebes et al., 2021). Of note, in contrast to the expression of TREM2 signature genes by AMs – albeit to a lower extent than mo-macs – in human NSCLC, the lack of AM genes (i.e., MARCO, PPARG, FABP4) expressed by mo-macs may suggest that the homeostatic signals that otherwise promote ‘proper’ differentiation of tissue-infiltrating monocyte-derived cells into AMs do not seem to promote the AM program in recruited macrophages during tumorigenesis (Park et al., 2022). This particular difference emphasizes both the lack of homeostatic cues due to the loss of niche integrity and supports the ontogenic distinction between AMs and mo-macs in lung tumors, as we previously demonstrated by fate-mapping MNPs in murine lung tumors (Casanova-Acebes et al., 2021, Loyher et al., 2018).
We show that these TREM2+ mo-macs have significant intracellular GFP signal, captured from KP-GFP tumor cells, but at comparable levels to TREM2-deficient mo-macs, indicating two notable features about the biology of the TREM2 program in these cells: first, the TREM2-deficient phagocytes remain able to capture tumor cell-derived antigens, and second, given that the rate of GFP uptake is similar between TREM2-proficient and -deficient mo-macs, it is likely that the sensing of lipidic cues amongst the cell debris – not necessarily the physical uptake of these materials – induces the TREM2 program. Importantly, the acquisition of the TREM2 program by mo-macs mutes pro-inflammatory pathways by downregulating co-stimulation genes and upregulating those encoding immune checkpoint molecules. The identification of lipid metabolism pathways based on the program-specific genes and specific changes in the expression of genes encoding cholesterol trafficking machinery prompted us to describe these mo-macs enriched in the TREM2 program as having entered a dysfunctional cholesterolemic state, but the extent to which we further investigate their sub-cellular properties will be the focus of future studies. We suspect that TREM2+ mo-macs divert cholesterol to excess lipid droplets, which have been shown to have immunoregulatory properties associated with the resource partitioning of free cholesterol via apolipoprotein E (encoded by Apoe), suppression of antigen presentation, and response to interferon in cDC1 (Bonacina et al., 2018; Gouna et al., 2021). But additional work will still be necessary to fully understand how this lipidic state regulates the immunoregulatory potential of intratumoral TREM2+ mo-macs (van Eijk et al., 2021). Recent molecular work suggests that tumor cell-derived glucosyl-ceramide stimulates a reshuffling of lipid composition in macrophages by inducing cholesterol dyshomeostasis and promoting ER stress, both of which result in the expression of XBP1. Deleting the Xbp1 gene in intratumoral macrophages reversed this phenotype and restrained growth of subcutaneous melanoma lesions (Di Conza et al., 2021). TREM2 itself has also been shown to regulate cholesterol metabolism in microglia (Nugent et al., 2020), so an understanding of how intratumoral TREM2+ mo-macs utilize their intracellular cholesterol depots, in contrast to the ways their tissue-resident counterparts may do so, would truly be essential for exploring other methods of skewing the phenotype of these cells.
Genetic ablation of Trem2 significantly reduced lung tumor burden in both our KP model of lung adenocarcinoma and in the B16-F10 model of metastatic melanoma. At the cellular level, deleting Trem2 disrupted the cell state transition of mo-macs towards a pro-tumorigenic lipidic state that involved the upregulation of Il18bp, encoding the decoy protein that intercepts IL-18 and has been shown to suppress anti-tumor NK cell responses (Zhou et al., 202a). Interestingly, as AM are a source of IL-18 (Jordan et al., 2001; Pechkovsky et al., 2006), given their topological organization at the tumor periphery, it stands to reason that the exclusion of NK cells at the tumor bed may reflect NK cells simply ‘following the crumbs’, in lieu of actually infiltrating the lesions. Based on our findings, we surmise that TREM2+ mo-macs exacerbate the NK cell problem by secreting IL-18BP and ‘sweeping up the crumbs’ along the peritumoral bed. The absence of TREM2 corresponded to both the loss of Il18bp expression and an expansion of NK cells and enhanced NK cell function that was dependent on IL-15, which is highly expressed by mregDCs. While the recruitment of NK cells by mregDCs and their role in anti-tumor immunity is well-appreciated (Barry et al., 2018; Böttcher et al., 2018; Mattiuz et al., 2021), it was striking to find that this axis was, in fact, tightly regulated by TREM2+ mo-macs in NSCLC. We showed that IL-15 was indeed crucial for the successful maintenance of the NK cell response in KO mice. Altogether, these results position TREM2+ mo-macs as ‘bouncers’ of the TME, keeping NK cells from effectively killing tumor cells. The enrichment of NK cells and cDC1 has been associated with response to PD-1 blockade in melanoma patients, so answering whether TREM2+ mo-macs also define response to immune checkpoint blockade in NSCLC patients could be additional, powerful evidence that these macrophages should be studied in greater detail in different tissues, as it has become increasingly clear that TREM2 on either tissue-resident macrophages or mo-macs has pleiotropic functions that are documented in contexts, such as non-alcoholic steatohepatitis or Alzheimer’s disease.
In our report, we demonstrate that combining TREM2 blockade with exogenous NK cell-enhancing agents does indeed further boost the anti-tumor response, and this finding highlights a new therapeutic strategy for targeting effector immune cells in the TME without concerns for revitalizing an antigen-specific adaptive immune response that is so heavily dependent on presentation of immunogenic tumor antigens. In summary, we propose that TREM2 on mo-macs is a major immune checkpoint that mutes NK cell-mediated anti-tumor immunity in NSCLC. The ability for TREM2+ mo-macs to regulate NK cells provides a strong rationale for the clinical development of combinatorial therapies that concurrently inhibit TREM2 and enhance NK cells, including the αMIC-A stabilizing agent that we used in our study and is currently in clinical trials to manage solid tumors (NCT05117476).
Methods
Materials Availability
The study did not generate new unique reagents.
Data and Code Availability
All murine sequencing data will be made publicly available (GSE184304, GSE184309, GSE184317). The human dataset is available at the Sequence Read Archive (SRA) with BioProject accession PRJNA609924.
Mouse strains and tumor cell lines
C57BL/6J-Trem2em2Adiuj/J (Trem2−/−) and B6.129P2-Il18r1tm1Aki/J (Il18r1−/−) mice were purchased from Jackson Laboratory. Both these and wild-type C57BL/6 mice used as controls for experimentation were either bred at the Icahn School of Medicine at Mount Sinai or purchased from Jackson and housed for a minimum of seven days before experimental use. For bone marrow transplantation, B6.SJL-PtprcaPepcb/BoyJ (CD45.1) mice were also purchased from Jackson. All mice were maintained at specific pathogen-free (SPF) conditions. Mice within experiments were age and sex-matched. All studies performed on mice were done in accordance with the Institutional Animal Care and Use Committee (IACUC) at the Icahn School of Medicine at Mount Sinai.
Murine tumor models
To model primary lung adenocarcinoma, mice were intravenously injected via the tail vein with KrasG12D/+p53−/− (KP) lung epithelial cells, KP cells engineered to express GFP (KP-GFP), or KP cells expressing both GFP and OVA (KP-GFP-OVA) (5×105 cells per mouse in 250μL PBS). KP cells were grown in complete cell culture medium (RPMI+10%FBS+1%P/S) and were detached for use at 70% confluence using 0.25% trypsin. The tumor cells were originally derived from KP mice (DuPage et al., 2009) generated by crossing LSL-KrasG12D/+ mice (Jackson Laboratories) with p53fl/fl mice (Jackson Laboratories). Tumor-bearing lungs were analyzed at 22 or 28 days post-injection of KP or KP-GFP tumors and at seven to eight weeks post-injection of KP-GFP-OVA tumors, except when otherwise indicated. For αMICA experiments, the C57BL/6-derived metastatic melanoma cell line B16-F10 expressing human MIC-A was provided by L. Ferrari De Andrade (Icahn School of Medicine at Mount Sinai) and intravenously injected at 2.5×105 cells per mouse. Mice were analyzed at 21 days post-injection, and tumor quantification was performed on hematoxylin & eosin (H&E)-stained slides of formalin-fixed paraffin-embedded (FFPE) 4 μm lung tissue sections. Slides were scanned using an Olympus digital scanner and analyzed using the Panoramic viewer and QuPath software.
Antibody-mediated blockade and depletion studies
To deplete CD8 T cells and NK cells, mice were intraperitoneally injected with anti-CD8α (BioXCell, clone 2.43) and anti-NK1.1 (BioXCell, clone PK136), respectively. Blocking antibodies used for experimentation include αIL-18 (BioXCell, clone YIGIF74–1G7), αIL-15 (BioXCell, clone AIO.3), and the corresponding isotype antibodies for both. The αTREM2 antibody and its isotype control, both of which were provided by M. Colonna (Washington University School of Medicine at St. Louis), were used to assess therapeutic blockade of TREM2. For each treatment regimen, mice received an initial dose at 200μg/mouse at 14 days post-injection of tumor cells, followed by subsequent 100μg every other day. The MIC-A stabilizing antibody, provided by F. De Andrade (Icahn School of Medicine at Mount Sinai), was given as single doses of 200μg/mouse on days 6, 9, 13, 15, and 19 post-tumor challenge.
Bone marrow reconstitution
Donor bone marrow cells were isolated from femur exudates of WT and Trem2−/− mice bred at the Icahn School of Medicine at Mount Sinai. Transplantation was performed by retro-orbitally injecting 5×105 donor cells into sub-lethally irradiated (twice, 6.5 Gy, six hours apart). Recipients were supplemented with sulfamethoxazole/trimethoprim (STI) for three weeks. Successful reconstitution was assessed by flow cytometric analysis of peripheral blood.
Flow cytometry and fluorescence-activated cell sorting
Single-cell suspensions from perfused murine lungs were obtained upon lung tissue digestion with collagenase IV (0.25 mg ml−1; Sigma) at 37 °C for 30 min in agitation (90 rpm) followed by passing through a 70μm cell strainer and red blood cell lysis (RBC lysis buffer, BioLegend) for 5 min at room temperature (RT). For flow cytometry or FACS, cells were stained in FACS buffer (phosphate-buffered saline (PBS) supplemented with 2% bovine serum albumin (BSA) and 5 mM EDTA) with different combination of the following monoclonal antibodies (1:200 dilution) specific to: CD45 (clone 30-F11, BioLegend), CD45.1 (clone A20, BioLegend), CD45.2 (clone 104, BioLegend), B220 (clone RA3–6B, BioLegend,), CD19 (clone eBio1D3, eBiosciences), Ly6G (clone 1A8, BioLegend), CD64 (clone X54–5/7.1, BioLegend), MERTK (clone 2B10C42, BioLegend) CD2 (clone RM2–5, BioLegend), Siglec F (clone E50–2440, BD Pharmingen), MHC I-A/I-E (clone M5/114.15.2, eBiosciences), CD11b (clone M1/70, eBiosciences), CD11c (clone N418, Invitrogen), CD24 (clone M1/69, Invitrogen), CD103 (clone 2E7, BioLegend), XCR1 (clone ZET, BioLegend), CD107a (clone 1D4B, BioLegend), CD172A (clone P84, BioLegend), CD3 (clone 145–2C11, eBiosciences), CD8a (clone 53–6.7), CD4 (clone GK1.5 eBiosciences), CD44 (clone IM7, BioLegend) PD1 (clone RMP1–30, BioLegend) TCF1 (clone 7F11A10, BioLegend), Granzyme B (clone GB11, Thermo Fisher), NK1.1 (clone PK136, BioLegend), NKp46 (clone 29A1.4, BioLegend), CD49b (clone DX5, Thermo Fisher), CD49A (clone HMa1, BioLegend) KLRG1 (clone MAFA, BioLegend), NKG2D (clone C7, BioLegend), TNF (clone MP6-XT22, eBiosciences), GZMB (clone GB11, eBiosciences) and/ or IFN-γ (clone XMG1.2, eBiosciences). For intracellular staining, cells were fixed with either BD Cytofix/Cytoperm kit for cytokine stains or with the eBiosciences Foxp3 transcription factor kit for intranuclear stains, both according to the manufacturer’s instructions. For T-cell cytokine stains, cells were incubated with 10 μg/ml brefeldin A, 0.2 μg/ml ionomycin and 0.5 μg/ml phorbol myristate acetate (PMA) (Sigma) for 3 hours at 37 °C followed by staining of surface markers and fixation. For flow cytometry, cells were analyzed in a BD LSR Fortessa analyzer (BD Biosciences). For FACS, cells were prepared, stained, and purified using a BD FACSAria sorter (BD Biosciences). Flow cytometry data was acquired using FACS Diva software v.7 (BD), and the data obtained were analyzed using FlowJo (LLC).
In vitro culture of bone marrow-derived macrophages
Bone marrow cells were isolated by flushing murine femurs, tibias, and humeri with PBS (0.5% BSA, 2 nM EDTA, 1% P/S). Cells were strained through a 70-μm filter and centrifuged before resuspension in 1X Red Blood Cell (RBC) lysis buffer (BioLegend) for 5 min at RT.
Total bone marrow cells were plated in non-tissue culture treated dishes with RPMI medium, supplemented with 10% fetal calf serum (FCS), 1% P/S, and 200 ng/mL of murine recombinant colony stimulating factor 1 (CSF1) (Peprotech) (differentiation media). After 4 days of differentiation, non-adherent cells were aspirated and washed with PBS to enrich for highly adherent cells. Fresh media was added and supplemented with 200 ng/mL of recombinant murine GM-CSF (Peprotech). Monocyte-derived cells were then cultured with GM-CSF for 24 hrs, then media was refreshed prior to supplementation with 200 ng/ml of recombinant murine IL-4 (Shenandoah Biotechnology) for 24 hrs. This was then substituted for differentiation media. On day 7, apoptotic KP-GFP cells (generated by exposure to UV irradiation for 12 hrs) were added to mo-mac cultures for 4 hrs. Some macrophages were left unexposed as controls. Cells were then detached using cold PBS supplemented with EDTA and gentle pipetting. Mo-macs were FACS-sorted into 500 μL of Trizol for downstream analysis.
To assess the effect of TREM2 deficiency on the ability for mo-macs to directly influence the activation and proliferation of NK cells, splenocytes from either WT or Il18r1−/− mice were enriched for NK cells via negative selection (Miltenyi Biotec) and splenic NK cells were added to cultures of fed mo-macs generated from either WT or Trem2−/− mice in a 1:10 ratio. The cell mixture was stimulated with recombinant IL-18 (1 nM) (R&D Systems, Cat. # 9139-IL) and IL-12 (10 ng/mL) (R&D Systems, Cat. # 419-ML) with Brefeldin A (5 mg/mL, 1:250). After 4 hours, the NK cells were collected for flow cytometry.
Ultra-low Input Bulk RNA-sequencing
For ultra-low input bulk RNA-sequencing, 104–105 GFPhi and GFPlo mo-macs were sorted from digested KP-GFP tumor-bearing lungs by FACS into 500μL of Tryzol. RNA was then extracted using the RNeasy Micro Kits (Qiagen), according to manufacturer’s instructions. RNA was quantified and quality controlled using a Qubit and a Bioanalyzer. Between 0.5–1 ng of RNA was retrotranscribed and processed to cDNA libraries using the Smart-Seq v4 Ultra Low Input RNA Kit for Sequencing (Takara Bio). Libraries were sequenced on an Illumina NextSeq 550 system.
Differential gene expression testing
Raw bulk RNA sequencing data was processed and aligned to genome reference mm10 using the R package Rsubread with default parameters. Gene expression normalization and differential expression analyses were conducted using DESeq2, with genotype, antigen condition and replicate as design matrix input. To account for batch effects across replicates, limma batch effect correction was applied on the normalized counts prior to gene expression heatmap visualizations. Differentially expressed genes were called using log2(fold change)>1 and adjusted p-value<0.1. Volcano plots were generated using R packages ggplot and seriation.
Sample preparation for scRNAseq
Single-cell suspensions from lung tissues were obtained, as described above. For scRNAseq, these cells were suspended and stained in 100μL of multiplex hashing antibodies at 4°C for 20 minutes. Stained cells were washed three times in PBS+0.5% BSA to remove unbound antibodies. Washed cells were resuspended in 150μL of wash buffer and counted using a Nexcelom Cellometer Auto2000. Hashed samples were pooled in equal amounts of live cells. Volume was adjusted to achieve a target of 2×106 cells/mL. Hashed samples were loaded onto 10X Genomics NextGen 5’ v1.1 assay, as per the manufacturer’s instructions, for a target cell recovery of 20,000 cells/lane. Libraries were constructed, as per the manufacturer’s instructions. During cDNA amplification, hashtag oligonucleotides (HTO) were enriched during cDNA amplification with the addition of 3 pmol of HTO Additive primer (5’GTGACTGGAGTTCAGACGTGTGCTC). This PCR product was isolated from the mRNA-derived cDNA via SPRISelect size selection, and libraries were made as per the New York Genome Center Hashing protocol. All libraries were quantified via Agilent 2100 hsDNA Bioanalyzer and KAPA library quantification kit (Roche, Cat. No. 0796014001.) Gene expression libraries were sequenced at a targeted depth of 25,000 reads per cells, and HTO libraries were sequenced at a targeted read depth of 1,000 reads per cell. All libraries were sequenced on the Illumina NovaSeq S2 100 cycle kit with run parameters set to 28×8×0×60 (R1xi7xi5xR2).
scRNAseq analysis
After library demultiplexing, gene-expression libraries were aligned to the mm10 reference transcriptome and count matrices were generated using the default Cell Ranger 2.1 workflow, using the ‘raw’ matrix output. Where applicable, doublets were removed based on co-staining of distinct sample-barcoding (‘Hashing’) antibodies (maximum staining antibody counts/second-most staining antibody counts = less than 5). Following alignment, cell barcodes corresponding to cells that contained more than 500 UMIs were extracted. From among these, cells whose transcripts constituted more than 25% mitochondrial genes were filtered from downstream analyses. The R package Seurat was used to scale the data, transform via a log normalization method, adjust for batch correction, cluster cells based on shared nearest neighbors, and perform dimensionality reduction based on the first 15 principal components. Gene module analyses was performed, based on the identification of groups of highly correlated genes, according to the Pearson correlation matrix of the most variable genes. This was done using the R package scDissector. Highly correlated gene were assigned into groups by hierarchical clustering. Differentially expressed genes were identified using the FindMarkers function in Seurat. Average UMI expression values were imputed to determine log fold change differences between cell types to further the analysis of markers of interest.
scRNAseq of human NSCLC lesions and non-involved lung tissues
A scRNAseq dataset of immune cells isolated from resected NSCLC lesions and matched, non-involved adjacent lung tissues from 35 patients was used to probe the relative distribution of NK cells and mo-macs and to identify a conserved molecular program across mo-mac subsets (Leader et al., 2021). Human samples for this particular study were collected by the Cancer BioRepository at the Icahn School of Medicine at Mount Sinai.
Multiplex imaging of human and murine tissue sections
Formalin-fixed paraffin-embedded (FFPE) tissue sections (4μm) were stained using the Multiplexed Immunohistochemical Consecutive Staining on a Single Slide (MICSSS) protocol, as previously described (Remark et al., 2016). Briefly, slides were baked at 50°C overnight, deparaffinized in xylene and rehydrated in decreasing concentration of ethanol (100%, 90%, 70%, 50% and dH2O). Sample slides were incubated in pH 6 or pH 9 buffers at 95°C for 30min for antigen retrieval, then in 3% hydrogen peroxide for 15min and in serum-free protein block solution (Dako) for 30min. Primary antibody staining was performed using the optimized dilution during 1 hour at RT or at 4°C overnight followed by signal amplification using associated secondary antibody conjugated to horseradish peroxidase (HRP) during 30min. Chromogenic revelation was performed using AEC (Vector). Tissue sections were counterstained with hematoxylin, mounted with a glycerol-based mounting medium and finally scanned to obtain digital images (Aperio AT2, Leica). Then, the same slides were bleach and stained again as previously described (Remark et al., 2016). Primary antibodies were anti-human CD68 (clone KP1, Dako) and anti-human TREM2 (D8I4C, Cell Signaling Technology) and anti-mouse NKp46 (AF2225, BioLegend). Quantification of positively stained cells were performed using the positive cell detection algorithm in the QuPath software with default settings.
Immunofluorescence imaging of murine tissue sections
Perfused lungs isolated from tumor-bearing mice were fixed overnight at 4 °C in paraformaldehyde (PFA), cryopreserved using the sucrose gradient method (2 hrs of successive incubations in 10, 20 and 30% sucrose in PBS), then embedded and frozen in Optimal Cutting Temperature compound Tissue-Plus™ (OCT, Thermo Fisher). Frozen tissue sections were thawed and permeabilized with 0.1% Triton X-100 for 10 min. Blocking was performed using 10% BSA + 2% normal goat serum in PBS for 1 hr. Sections were incubated with primary antibodies for 3 days at 4 °C, then with secondary antibodies for 1 hr at RT. Mounting was done using ProLong Gold Antifade Reagent with DAPI (Invitrogen, P36931), anti-TREM2 (clone AF1729, R&D Systems, 1:100) and anti-GFP (clone ab13970, Abcam, 1:200) were used as primary or directly conjugated antibodies respectively. AlexaFluor Donkey anti-sheep 555 (Abcam ab150178) was used as secondary antibody for TREM2 staining. Imaging was performed using a Zeiss 780 Confocal Microscope. Quantification of fluorescence intensity and post-acquisition processing of images was performed using ImageJ software and ZEN Black Imaging Software (Zeiss). For imaging of FACS-sorted mo-macs, between 103–104 GFPhi and GFPlo mo-macs were FACS-sorted and centrifuged into Alcian-blue-treated coverslips and fixed in 1% PFA. Cells were permeabilized in 0.1 % Triton-X in PBS for 10 min at RT. Coverslips were washed with PBS and stained with an anti-GFP antibody for 2 hrs at room temperature. Next Coverslips were washed with PBS again (2x) and stained with 4′,6-diamidino-2-phenylindole (DAPI; 1 ng ml−1) for 5 min. Finally, coverslips were mounted using Prolong Gold Anti-Fade, and sections were imaged on a Zeiss 780 Confocal Microscope.
Quantitative PCR
RNA was retrotranscribed to cDNA using the RNA to cDNA EcoDry™ Premix (Double Primed) kit (Takara). Transcripts were quantified using the system AB7900-FAST-384 with a two-step reverse-transcription qPCR process. KiCqStart® SYBR® Green Primers pre-designed and customized were purchased to Sigma Aldrich, USA. Quantitative PCRs using primers for CD45 mRNA was conducted in each plate to provide a normalization reference. Quantification of relative gene expression was calculated by the comparative Ct method (2−ΔΔCt), relative to CD45 Ct. Primer sequences are provided down below (5’ to 3’):
Apoe → F:CTGACAGGATGCCTAGCCG, R:CGCAGGTAATCCCAGAAGC;
Gpnmb→F:CATTCCCATCTCGAAGGTGAAA, R:AAATGGCAGAGTCGTTGAGGA;
Trem2 → F:CTGGAACCGTCACCATCACTC, R:CGAAACTCGATGACTCCTCGG;
Extended Data
Extended Data Figure 1. Purification and active signaling pathways in GFPlo and GFPhi mo-macs.
a) Flow cytometry sort strategy (left) for purifying GFPhi and GFPlo mo-macs from single-cell suspensions of digested KP-GFP tumor-bearing lungs at four weeks post-inoculation of KP-GFP cells. Histograms show the fluorescence spectrum of GFP in mo-macs from tumor-bearing lungs, comprised of GFPhi (green) and GFPlo (blue) mo-macs. Each row represents an individual replicate. Representative confocal immunofluorescence (IF) imaging of FACS-sorted GFPlo and GFPhi mo-macs (right). For each pair of images, a broad-field view with a region of interest (ROI, white outline) (left) and a magnification of the ROI (right) are shown.
b) Gene set enrichment analysis (GSEA) (Broad Institute) for pathways annotated in the KEGG and Reactome databases performed on the significantly differentially regulated genes in GFPhi mo-macs, relative to GFPlo mo-macs. The top 15 most significantly enriched terms are shown, with notable pathways highlighted in red for up-regulated genes in GFPhi mo-macs (left) and up-regulated genes in GFPlo mo-macs (right).
Extended Data Figure 2. scRNAseq profiling of mo-macs in chimeric mice.
a) Expression of Trem2 and Gpnmb, as hallmark genes of the TREM2 gene program, across cell subtypes (mo-macs, AMs, Ly6Clo and Ly6Chi monocytes, mregDCs, cDC1, and cDC2) identified by unsupervised clustering.
b) Flow cytometry sort strategy to purify CD45.1 WT and CD45.2 KO fractions of MNPs from KP-GFP tumor-bearing chimeric mice.
c) Flow cytometric quantification of GFP+ mo-macs, represented as relative frequency among total mo-macs from the purified CD45.1 WT and CD45.2 KO fractions of MNPs from KP-GFP tumor-bearing mice. Paired values shown for paired CD45.1 WT and CD45.2 KO cells that were purified from the same biological replicate. (mean ± standard error of mean (S.E.M.); paired two-tailed t-test at a 95% confidence interval)
Extended Data Figure 3. Contributions of CD8 T cells and NK cells to tumor regression in KO mice.
a) Flow cytometric quantification of IFN-ɣ/TNF-α-producing CD8 T cells, as a relative frequency of total antigen-specific CD8 T cells, in the tumor-bearing lungs (left) and in the tumor-draining lymph nodes (tdLN) (right) of KP-GFP tumor-bearing WT and KO mice. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
b) Flow cytometric quantification of NK cells expressing activation/degranulation markers, as a relative frequency of total NK cells. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
c) Flow cytometric quantification of type 1 conventional dendritic cells (cDC1), as a relative frequency of CD45+ immune cells, in the tumor-bearing lungs of WT mice and KO mice treated with either an isotype control or CD8 (right) or NK cell (left)-depleting antibody. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
d) Quantification of the tumor area as a percent of the total area of the lung cross-section from tumor-bearing lungs of WT and KO mice treated with either an isotype control of NK cell-depleting antibody.
e) Flow cytometric quantification of NK cells, as a relative frequency of CD45+ immune cells. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
f) Flow cytometric quantification of IFN-ɣ-producing (left) and TNF-α-producing (right) CD8 T cells in the tumor-bearing lungs of WT mice with isotype antibody, KO mice with isotype antibody, and KO mice given IL-18 neutralizing antibody. Data shown as a relative frequency among total CD8 T cells. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
Extended Data Figure 4. Cooperative IL-15 signaling is needed to confer the therapeutic benefit of a TREM2-deficient setting.
a) Relative expression of Il15 by mature mregDCs, as determined by scRNAseq profiling of MNPs in the tumor-bearing lungs of WT and KO mice (Figure 3C).
b) Representative H&E images of tumor-bearing lungs of WT, isotype antibody (n = 4), WT mice that received the IL-15 neutralizing antibody (n = 4), KO, isotype antibody (n = 4), KO mice that received the IL-15 neutralizing antibody (n = 5) (left). Quantification of the tumor area as a percent of the total area of the lung cross-section (right) is shown. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
c) Flow cytometric quantification of NK cells, as a relative frequency of CD45+ immune cells, in the KP-GFP tumor-bearing lungs of isotype control WT and KO mice and WT and KO mice treated with the αIL-15 antibody. (mean ± standard error of mean (S.E.M.); unpaired two-tailed t-test at a 95% confidence interval)
Supplementary Material
Table S1. Differentially expressed genes between tissue-resident alveolar macrophages and monocyte-derived macrophages in NSCLC.
Table S2. Gene expression across dendritic cell, monocyte, macrophage, and lymphoid cell subsets in NSCLC.
Table S3. Differentially expressed genes between CD45.1 vs CD45.2 monocyte-derived macrophages identified by scRNAseq of chimeric mice (day 28 post-inoculation of tumor cells).
Acknowledgements
We thank members of the Merad and Brown laboratories at the Marc and Jennifer Lipschultz Precision Immunology Institute at Mount Sinai and the Tisch Cancer Institute for insightful discussions and feedback; we would specifically like to thank Matthew J. Lin and Joshua Brody for lending us flow cytometry antibodies against the NKG2D ligands; the Mount Sinai Flow Cytometry Core, the Human Immune Monitoring Center, and the Mount Sinai Biorepository for support. Data in this paper were used in a dissertation as partial fulfillment of the requirements for a PhD degree at the Graduate School of Biomedical Sciences at Mount Sinai.
Footnotes
Competing Interests
LFA is a named inventor of the MICA therapeutic antibody in a filed patient (US20200165343A1) and has received royalty payments in relation to this therapeutic. MC receives research support from NGM Biotechnology and Vigil Neuro, is a scientific advisory board member of NGM Biotechnology and Vigil Neuro and has a patent for TREM2 pending.
References
- 1.de Andrade LF, Tay RE, Pan D, Luoma AM, Ito Y, Badrinath S, Tsoucas D, Franz B, May KF, Harvey CJ, et al. (2018). Antibody-mediated inhibition of MICA and MICB shedding promotes NK cell–driven tumor immunity. Science 359, 1537–1542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barry KC, Hsu J, Broz ML, Cueto FJ, Binnewies M, Combes AJ, et al. (2018). A natural killer-dendritic cell axis defines checkpoint therapy-responsive tumor microenvironments. Nat. Med. 24, 1178–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Binnewies M, Pollack JL, Rudolph J, Dash S, Abushawish M, Lee T, Jahchan NS, Canaday P, Lu E, Norng M, et al. (2021). Targeting TREM2 on tumor-associated macrophages enhances immunotherapy. Cell Rep. 37, 109844. [DOI] [PubMed] [Google Scholar]
- 4.Bonacina F, Coe D, Wang G, Longhi MP, Baragetti A, Moregola A, Garlaschelli K, Uboldi P, Pellegatta F, Grigore L, et al. (2018). Myeloid apolipoprotein E controls dendritic cell antigen presentation and T cell activation. Nat. Comm. 9, 3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bonnardel J, T’Jonck W, Gaublomme D, Browaeys R, Scott CL, Martens L, Vanneste B, De Prijck S, Nedospasov SA, Kremer A, et al. (2019). Stellate Cells, Hepatocytes, and Endothelial Cells Imprint the Kupffer Cell Identity on Monocytes Colonizing the Liver Macrophage Niche. Immunity 51, 638–654.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Böttcher JP, Bonavita E, Chakravarty P, Blees H, Cabeza-Cabrerizo M, Sammicheli S, Rogers NC, Sahai E, Zelenay S, and Reis e Sousa C. (2018). NK Cells Stimulate Recruitment of cDC1 into the Tumor Microenvironment Promoting Cancer Immune Control. Cell 172, 1022–1037.e14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Browaeys R, Saelens W, and Saeys Y. (2020). NicheNet: modeling intercellular communication by linking ligands to target genes. Nat. Methods 17, 159–162. [DOI] [PubMed] [Google Scholar]
- 8.Cannon JP, O’Driscoll M, and Litman GW (2012). Specific lipid recognition is a general feature of CD300 and TREM molecules. Immunogenetics 64, 39–47. [DOI] [PubMed] [Google Scholar]
- 9.Casanova-Acebes M, Dalla E, Leader AM, LeBerichel J, Nikolic J, Morales BM, Brown M, Chang C, Troncoso L, Chen ST, et al. (2021). Tissue-resident macrophages provide a pro-tumorigenic niche to early NSCLC cells. Nature 595, 578–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chaix J, Tessmer MS, Hoebe K, Fuséri N, Ryffel B, Dalod M, Alexopoulou L, Beutler B, Brossay L, Vivier E, et al. (2008). Cutting Edge: Priming of NK Cells by IL-18. The Journal of Immunology 181, 1627–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cohen M, Giladi A, Barboy O, Hamon P, Li B, Zada M, Gurevich-Shapiro A, Beccaria CG, David E, Maier B, et al. (2022). The interaction of CD4+ helper T cells with dendritic cells shapes the tumor microenvironment and immune checkpoint blockade response. Nature Caancer 3, 303–317. [DOI] [PubMed] [Google Scholar]
- 12.Conza GD, Di Conza G, Tsai C-H, Gallart-Ayala H, Yu Y-R, Franco F, Zaffalon L, Xie X, Li X, Xiao Z, et al. (2021). Tumor-induced reshuffling of lipid composition on the endoplasmic reticulum membrane sustains macrophage survival and pro-tumorigenic activity. Nature Immunology 22, 1403–1415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cózar B, Greppi M, Carpentier S, Narni-Mancinelli E, Chiossone L, and Vivier E. (2021). Tumor-Infiltrating Natural Killer Cells. Cancer Discov. 11, 34–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cursons J, Souza-Fonseca-Guimaraes F, Foroutan M, Anderson A, Hollande F, Hediyeh-Zadeh S, et al. (2019). A gene signature predicting natural killer cell infiltration and improved survival in melanoma patients. Cancer Immunol. Res. 7, 1162–74. [DOI] [PubMed] [Google Scholar]
- 15.Daws MR, Sullam PM, Niemi EC, Chen TT, Tchao NK, and Seaman WE (2003). Pattern Recognition by TREM-2: Binding of Anionic Ligands. The Journal of Immunology 171, 594–599. [DOI] [PubMed] [Google Scholar]
- 16.DeNardo DG, Barreto JB, Andreu P, Vasquez L, Tawfik D, Kolhatkar N, and Coussens LM (2009). CD4(+) T cells regulate pulmonary metastasis of mammary carcinomas by enhancing protumor properties of macrophages. Cancer Cell 16, 91–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Pilato M, Kfuri-Rubens R, Pruessmann JN, Ozga AJ, Messemaker M, Cadilha BL, Sivakumar R, Cianciaruso C, Warner RD, Marangoni F, et al. (2021). CXCR6 positions cytotoxic T cells to receive critical survival signals in the tumor microenvironment. Cell 184, 4512–4530.e22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dunn GP, Bruce AT, Ikeda H, Old LJ, and Schreiber RD (2002). Cancer immunoediting: from immunosurveillance to tumor escape. Nat. Immunol. 3, 991–998. [DOI] [PubMed] [Google Scholar]
- 19.DuPage M, Dooley AL, and Jacks T. (2009). Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat. Protoc. 4, 1064–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Eijk M, and Aerts JMFG (2021). The Unique Phenotype of Lipid-Laden Macrophages. Int. J. Mol. Sci. 22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Esparza-Baquer A, Labiano I, Sharif O, Agirre-Lizaso A, Oakley F, Rodrigues PM, Zhuravleva E, O’Rourke CJ, Hijona E, Jimenez-Agüero R, et al. (2021). TREM-2 defends the liver against hepatocellular carcinoma through multifactorial protective mechanisms. Gut 70, 1345–1361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Filipello F, Morini R, Corradini I, Zerbi V, Canzi A, Michalski B, Erreni M, Markicevic M, Starvaggi-Cucuzza C, Otero K, et al. (2018). The Microglial Innate Immune Receptor TREM2 Is Required for Synapse Elimination and Normal Brain Connectivity. Immunity 48, 979–991.e8. [DOI] [PubMed] [Google Scholar]
- 23.Gasser S, Orsulic S, Brown EJ, and Raulet DH (2005). The DNA damage pathway regulates innate immune system ligands of the NKG2D receptor. Nature 436, 1186–1190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giladi A, and Amit I. (2018). Single-Cell Genomics: A Stepping Stone for Future Immunology Discoveries. Cell 172, 14–21. [DOI] [PubMed] [Google Scholar]
- 25.Gouna G, Klose C, Bosch-Queralt M, Liu L, Gokce O, Schifferer M, Cantuti-Castelvetri L, and Simons M. (2021). TREM2-dependent lipid droplet biogenesis in phagocytes is required for remyelination. J. Exp. Med. 218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guerreiro R, Wojtas A, Bras J, Carrasquillo M, Rogaeva E, Majounie E, Cruchaga C, Sassi C, Kauwe JSK, Younkin S, et al. (2013). TREM2 Variants in Alzheimer’s Disease. New England Journal of Medicine 368, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hannedouche S, Zhang J, Yi T, Shen W, Nguyen D, Pereira JP, Guerini D, Baumgarten BU, Roggo S, Wen B, et al. (2011). Oxysterols direct immune cell migration via EBI2. Nature 475, 524–527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harms RZ, Creer AJ, Lorenzo-Arteaga KM, Ostlund KR, and Sarvetnick NE (2017). Interleukin (IL)-18 Binding Protein Deficiency Disrupts Natural Killer Cell Maturation and Diminishes Circulating IL-18. Front. Immunol. 8, 1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hayakawa Y, and Smyth MJ (2006). NKG2D and cytotoxic effector function in tumor immune surveillance. Seminars in Immunology 18, 176–185. [DOI] [PubMed] [Google Scholar]
- 30.Heng TSP, Painter MW, The Immunological Genome Project Consortium, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, et al. (2008). The Immunological Genome Project: networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094. [DOI] [PubMed] [Google Scholar]
- 31.Hsieh CL, Koike M, Spusta SC, Niemi EC, Yenari M, Nakamura MC, and Seaman WE (2009). A role for TREM2 ligands in the phagocytosis of apoptotic neuronal cells by microglia. Journal of Neurochemistry 109, 1144–1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iizasa E. ‘ichi, Chuma Y, Uematsu T, Kubota M, Kawaguchi H, Umemura M, Toyonaga K, Kiyohara H, Yano I, Colonna M, et al. (2021). TREM2 is a receptor for non-glycosylated mycolic acids of mycobacteria that limits anti-mycobacterial macrophage activation. Nat. Commun. 12, 2299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Imaoka H, Hoshino T, Takei S, Kinoshita T, Okamoto M, Kawayama T, Kato S, Iwasaki H, Watanabe K, and Aizawa H. (2008). Interleukin-18 production and pulmonary function in COPD. Eur. Respir. J. 31, 287–297. [DOI] [PubMed] [Google Scholar]
- 34.Imaoka H, Gauvreau GM, Watson RM, Smith SG, Dua B, Baatjes AJ, Howie K, Hoshino T, Killian KJ, Aizawa H, et al. (2011). Interleukin-18 and interleukin-18 receptor- expression in allergic asthma. European Respiratory Journal 38, 981–983. [DOI] [PubMed] [Google Scholar]
- 35.Izawa S, Kono K, Mimura K, Kawaguchi Y, Watanabe M, Maruyama T, et al. (2011). H2O2 production within tumor microenvironment inversely correlated with infiltration of CD56dim NK cells in gastric and esophageal cancer: possible mechanism of NK cell dysfunction. Cancer Immunol. Immunother. 60, 1801–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin S, Deng Y, Hao JW, Li Y, Liu B, Yu Y, et al. (2014). NK cell phenotypic modulation in lung cancer environment. PLoS One 9, e109976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jordan JA, Guo RF, Yun EC, Sarma V, Warner RL, Crouch LD, Senaldi G, Ulich TR, and Ward PA (2001). Role of IL-18 in acute lung inflammation. J. Immunol. 167, 7060–7068. [DOI] [PubMed] [Google Scholar]
- 38.Katzenelenbogen Y, Sheban F, Yalin A, Yofe I, Svetlichnyy D, Jaitin DA, Bornstein C, Moshe A, Keren-Shaul H, Cohen M, et al. (2020). Coupled scRNA-Seq and Intracellular Protein Activity Reveal an Immunosuppressive Role of TREM2 in Cancer. Cell 182, 872–885.e19. [DOI] [PubMed] [Google Scholar]
- 39.Lavin Y, Mortha A, Rahman A, and Merad M. (2015). Regulation of macrophage development and function in peripheral tissues. Nat. Rev. Immunol. 15(12), 731–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lavin Y, Kobayashi S, Leader A, Amir E-AD, Elefant N, Bigenwald C, Remark R, Sweeney R, Becker CD, Levine JH, et al. (2017). Innate Immune Landscape in Early Lung Adenocarcinoma by Paired Single-Cell Analyses. Cell 169, 750–765.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Leader AM, Grout JA, Maier BB, Nabet BY, Park MD, Tabachnikova A, Chang C, Walker L, Lansky A, Le Berichel J, et al. (2021). Single-cell analysis of human non-small cell lung cancer lesions refines tumor classification and patient stratification. Cancer Cell 39, 1594–1609.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Loyher P-L et al. Macrophages of distinct origins contribute to tumor development in the lung. J. Exp. Med 215, 2536–2553 (2018) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lue L-F, Schmitz C, and Walker DG (2015). What happens to microglial TREM2 in Alzheimer’s disease: Immunoregulatory turned into immunopathogenic? Neuroscience 302, 138–150. [DOI] [PubMed] [Google Scholar]
- 44.Maier B, Leader AM, Chen ST, Tung N, Chang C, LeBerichel J, Chudnovskiy A, Maskey S, Walker L, Finnigan JP, et al. (2020). A conserved dendritic-cell regulatory program limits antitumour immunity. Nature 580, 257–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Masetti M, Carriero R, Portale F, Marelli G, Morina N, Pandini M, Iovino M, Partini B, Erreni M, Ponzetta A, et al. (2022). Lipid-loaded tumor-associated macrophages sustain tumor growth and invasiveness in prostate cancer. J. Exp. Med. 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mattiuz R, Brousse C, Ambrosini M, Cancel JC, Bessou G, Mussard J, Sanlaville A, Cauz C, Bendriss-Vermare N, Valladeau-Guilemond J, et al. (2021). Type 1 conventional dendritic cells and interferons are required for spontaneous CD4+ and CD8+ T-cell protective responses to breast cancer. CTI 10, e1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Molgora M, Esaulova E, Vermi W, Hou J, Chen Y, Luo J, Brioschi S, Bugatti M, Omodei AS, Ricci B, et al. (2020). TREM2 Modulation Remodels the Tumor Myeloid Landscape Enhancing Anti-PD-1 Immunotherapy. Cell 182, 886–900.e17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Molgora M, Cortez VS, and Colonna M. (2021). Killing the Invaders: NK Cell Impact in Tumors and Anti-Tumor Therapy. Cancers 13, 595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Mulder K, Patel AA, Kong WT, Piot C, Halitzki E, Dunsmore G, Khalilnezhad S, Irac SE, Dubuisson A, Chevrier M, et al. (2021). Cross-tissue single-cell landscape of human monocytes and macrophages in health and disease. Immunity 54, 1883–1900.e5. [DOI] [PubMed] [Google Scholar]
- 50.Nielsen CM, Wolf A-S, Goodier MR, and Riley EM (2016). Synergy between Common γ Chain Family Cytokines and IL-18 Potentiates Innate and Adaptive Pathways of NK Cell Activation. Frontiers in Immunology 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Nugent AA, Lin K, van Lengerich B, Lianoglou S, Przybyla L, Davis SS, Llapashtica C, Wang J, Kim DJ, Xia D, et al. (2020). TREM2 Regulates Microglial Cholesterol Metabolism upon Chronic Phagocytic Challenge. Neuron 105, 837–854.e9.. [DOI] [PubMed] [Google Scholar]
- 52.Park MD, Silvin A, Ginhoux F. & Merad M. Macrophages in health and disease. Cell 185, 4259–4279 (2022) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pechkovsky DV, Goldmann T, Vollmer E, Müller-Quernheim J. & Zissel G. Interleukin-18 expression by alveolar epithelial cells type II in tuberculosis and sarcoidosis. FEMS Immunol. Med. Microbiol 46, 30–38 (2006) [DOI] [PubMed] [Google Scholar]
- 54.Perugorria MJ, Esparza-Baquer A, Oakley F, Labiano I, Korosec A, Jais A, Mann J, Tiniakos D, Santos-Laso A, Arbelaiz A, et al. (2019). Non-parenchymal TREM-2 protects the liver from immune-mediated hepatocellular damage. Gut 68, 533–546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Preuss I, Ludwig M-G, Baumgarten B, Bassilana F, Gessier F, Seuwen K, and Sailer AW (2014). Transcriptional regulation and functional characterization of the oxysterol/EBI2 system in primary human macrophages. Biochem. Biophys. Res. Commun. 446, 663–668. [DOI] [PubMed] [Google Scholar]
- 56.Raulet DH, Gasser S, Gowen BG, Deng W, and Jung H. (2013). Regulation of ligands for the NKG2D activating receptor. Annu. Rev. Immunol. 31, 413–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rautela J, and Huntington ND (2017). IL-15 signaling in NK cell cancer immunotherapy. Curr. Opin. Immunol. 44, 1–6. [DOI] [PubMed] [Google Scholar]
- 58.Remark R, Merghoub T, Grabe N, Litjens G, Damotte D, Wolchok JD, Merad M, and Gnjatic S. (2016). In-depth tissue profiling using multiplexed immunohistochemical consecutive staining on single slide. Sci Immunol 1, aaf6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rutkowska A, O’Sullivan SA, Christen I, Zhang J, Sailer AW, and Dev KK (2016). The EBI2 signaling pathway plays a role in cellular crosstalk between astrocytes and macrophages. Scientific Reports 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Sautès-Fridman C, Petitprez F, Calderaro J, and Fridman WH (2019). Tertiary lymphoid structures in the era of cancer immunotherapy. Nat. Rev. Cancer 19, 307–325. [DOI] [PubMed] [Google Scholar]
- 61.Sconocchia G, Eppenberger S, Spagnoli GC, Tornillo L, Droeser R, Caratelli S, et al. (2014). NK cells and T cells cooperate during the clinical course of colorectal cancer. Oncoimmunology 3, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shimasaki N, Jain A, and Campana D. (2020). NK cells for cancer immunotherapy. Nat. Rev. Drug Discov. 19, 200–218. [DOI] [PubMed] [Google Scholar]
- 63.Shirotani K, Hori Y, Yoshizaki R, Higuchi E, Colonna M, Saito T, Hashimoto S, Saito T, Saido TC, and Iwata N. (2019). Aminophospholipids are signal-transducing TREM2 ligands on apoptotic cells. Sci. Rep. 9, 7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sims R, van der Lee SJ, Naj AC, Bellenguez C, Badarinarayan N, Jakobsdottir J, Kunkle BW, Boland A, Raybould R, Bis JC, et al. (2017). Rare coding variants in PLCG2, ABI3, and TREM2 implicate microglial-mediated innate immunity in Alzheimer’s disease. Nat. Genet. 49, 1373–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Smyth MJ, Hayakawa Y, Takeda K, and Yagita H. (2002). New aspects of natural-killer-cell surveillance and therapy of cancer. Nat. Rev. Cancer 2, 850–861. [DOI] [PubMed] [Google Scholar]
- 66.Stankovic B, Bjørhovde HAK, Skarshaug R, Aamodt H, Frafjord A, Müller E, et al. (2018). Immune cell composition in human non-small cell lung cancer. Front Immunol. 9, 3101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Takahashi K, Rochford CDP, and Neumann H. (2005). Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J. Exp. Med. 201, 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Tang W, Lv B, Yang B, Chen Y, Yuan F, Ma L, Chen S, Zhang S, and Xia J. (2019). TREM2 acts as a tumor suppressor in hepatocellular carcinoma by targeting the PI3K/Akt/β-catenin pathway. Oncogenesis 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thommen DS, Koelzer VH, Herzig P, Roller A, Trefny M, Dimeloe S, Kiialainen A, Hanhart J, Schill C, Hess C, et al. (2018). A transcriptionally and functionally distinct PD-1+ CD8+ T cell pool with predictive potential in non-small-cell lung cancer treated with PD-1 blockade. Nat. Med. 24, 994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Vanhersecke L, Brunet M, Guégan J-P, Rey C, Bougouin A, Cousin S, Moulec SL, Besse B, Loriot Y, Larroquette M, et al. (2021). Mature tertiary lymphoid structures predict immune checkpoint inhibitor efficacy in solid tumors independently of PD-L1 expression. Nat Cancer 2, 794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Wang S, Mustafa M, Yuede CM, Salazar SV, Kong P, Long H, Ward M, Siddiqui O, Paul R, Gilfillan S, et al. (2020). Anti-human TREM2 induces microglia proliferation and reduces pathology in an Alzheimer’s disease model. J. Exp. Med. 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yofe I, Dahan R, and Amit I. (2020). Single-cell genomic approaches for developing the next generation of immunotherapies. Nat. Med. 26, 171–177. [DOI] [PubMed] [Google Scholar]
- 73.York AG, and Bensinger SJ (2013). Subverting sterols: rerouting an oxysterol-signaling pathway to promote tumor growth. J. Exp. Med. 210, 1653–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zhang S, Weinberg S, DeBerge M, Gainullina A, Schipma M, Kinchen JM, Ben-Sahra I, Gius DR, Yvan-Charvet L, Chandel NS, et al. (2019). Efferocytosis Fuels Requirements of Fatty Acid Oxidation and the Electron Transport Chain to Polarize Macrophages for Tissue Repair. Cell Metab. 29, 443–456.e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhou T, Damsky W, Weizman O-E, McGeary MK, Hartmann KP, Rosen CE, Fischer S, Jackson R, Flavell RA, Wang J, et al. (2020a). IL-18BP is a secreted immune checkpoint and barrier to IL-18 immunotherapy. Nature 583, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Zhou Y, Song WM, Andhey PS, Swain A, Levy T, Miller KR, Poliani PL, Cominelli M, Grover S, Gilfillan S, et al. (2020b). Human and mouse single-nucleus transcriptomics reveal TREM2-dependent and TREM2-independent cellular responses in Alzheimer’s disease. Nat. Med. 26, 981. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Differentially expressed genes between tissue-resident alveolar macrophages and monocyte-derived macrophages in NSCLC.
Table S2. Gene expression across dendritic cell, monocyte, macrophage, and lymphoid cell subsets in NSCLC.
Table S3. Differentially expressed genes between CD45.1 vs CD45.2 monocyte-derived macrophages identified by scRNAseq of chimeric mice (day 28 post-inoculation of tumor cells).
Data Availability Statement
All murine sequencing data will be made publicly available (GSE184304, GSE184309, GSE184317). The human dataset is available at the Sequence Read Archive (SRA) with BioProject accession PRJNA609924.