Abstract
The present study sought to determine how usage of coreceptors by human immunodeficiency virus type 1 dictates cell tropism and depletion of CD4+ T cells in human lymphoid tissues cultured ex vivo. We found that coreceptor preferences control the marked, preferential depletion of coreceptor-expressing CD4+ lymphocytes. In addition, there was a strong, but not absolute, preference shown by CXCR4-using strains for lymphocytes and by CCR5-using strains for macrophages.
The hallmark of human immunodeficiency virus type 1 (HIV-1) disease is the progressive depletion of CD4+ lymphocytes. Different strains of HIV vary with respect to their target cell range and cytopathic potential. The molecular basis for differential cell tropism and virulence remained obscure until the discovery of select chemokine receptors that act as essential cofactors for cellular entry by HIV-1 (1). We previously reported that HIV-1 envelope glycoprotein (gp120) determinants controlling a preference for CXCR4 resulted in marked depletion of CD4+ T cells in human lymphoid histocultures, while those specifying a preference for CCR5 resulted in only mild depletion of such cells. These results suggested that either X4 viruses are intrinsically more cytopathic than R5 viruses or viruses with different coreceptor specificities target quantitatively distinct CD4+ T-cell pools. Our earlier study established that R5 HIV-1 variants exclusively deplete CCR5-expressing CD4+ lymphocytes, while X4 HIV-1 variants preferentially deplete CXCR4-expressing cells (5). However, the diverse HIV-1 isolates used in this work differed from each other by many parameters other than coreceptor usage that could influence cytopathicity.
The present study sought to establish a specific causative relationship among coreceptor usage, tropism, and CD4+ T-cell depletion in mature lymphoid tissue. Human tonsil histocultures were inoculated with pairs of recombinant strains of HIV-1 that differ exclusively in small regions of gp120 that control coreceptor preference. Three pairs of viruses based on an isogenic (NL4-3) viral backbone were studied: (i) NL4-3 (X4) and 49-5 (R5), virus chimeras that differ only in the gp120 V3 loop region that specifies strict reciprocal tropism for CXCR4 and CCR5, respectively (9, 12, 13); (ii) 134 (X4) and 126 (R5), site-directed mutants that differ in a single V3 amino acid residue that likewise dictates preference for CXCR4 or CCR5, respectively (3, 12); and 123 (X4) and USV3 (R5), chimeras that contain V3 loop segments derived from primary X4 and R5 viral isolates (references 3 and 12 and unpublished data).
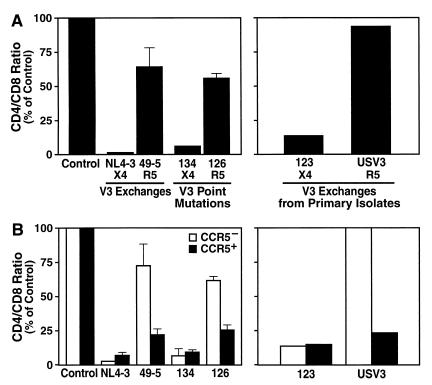
T-cell depletion and viral replication were measured 12 to 15 days following inoculation as described previously (4). Consistent with our earlier report (9), NL4-3 (X4) severely depleted these cultures of CD4+ T cells, while the paired virus 49-5 (R5) depleted these cells only mildly (Fig. 1A). Recombinant strain 134 (X4) also severely depleted these cells, while its paired strain, 126 (R5), which differs by a single amino acid within the V3 loop region, mildly depleted these cells (Fig. 1A). Viruses 123 and USV3, which encode V3 loop segments derived from primary viral isolates, likewise depleted CD4+ T cells according to coreceptor usage (Fig. 1A). These results confirm and extend previous observations by demonstrating that sequences within the V3 loop that control coreceptor preference dictate severe or mild CD4+ lymphocyte depletion. We have shown previously (9) and verified here for each virus pair (see Fig. 3) (data not shown) that the differential depletion effects occur despite comparable viral replication kinetics.
FIG. 1.
CD4+ T-cell depletion in human tonsil histocultures infected ex vivo by matched HIV-1 strains. (A) Left: CD4+ T-cell depletion as indicated by mean relative CD4/CD8 ratio on day 15 after infection by recombinant viruses NL4-3, 49-5, 134, and 126. For each data point, cells were pooled from 6 to 10 tissue blocks (mean plus standard error of the mean, n = 3) and analyzed by flow cytometry. Right: CD4 depletion on day 13 by viruses 123 and USV3, which were tested in a separate experiment. Presented are data from typical experiments with tissues from two to five donors. (B) CD4+ T-cell subset depletion data as indicated by the relative CD4/CD8 ratio for the CCR5− or CCR5+ T-cell subsets in the tissue samples presented in panel A.
FIG. 3.
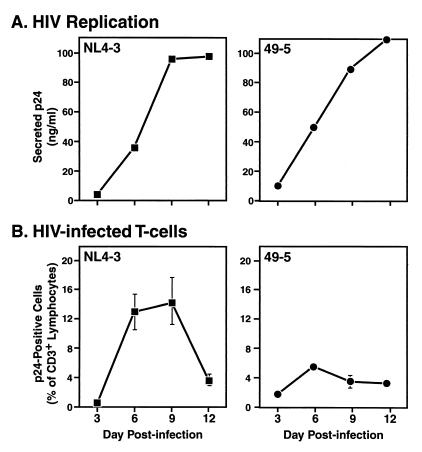
Kinetic analysis of HIV-1 replication and lymphocyte infection in human tonsil histocultures. (A) HIV-1 p24 concentration in the culture medium of NL4-3- and 49-5-infected histocultures (Fig. 2) and sampled at various time points. (B) Frequencies of productively infected lymphocytes in these cultures as demonstrated by flow cytometry of cells isolated from infected tissues and coimmunostained with MAbs recognizing CD3 and p24.
To test the hypothesis that target cell availability influences the magnitude of cellular depletion by each viral strain, we used flow cytometry to determine the relative prevalence of potential target cells as defined by CXCR4 and CCR5 expression. CXCR4 was expressed on the overwhelming majority of CD4+ T cells in resting tissues (mean, 88.5% ± 1.6%, n = 25), whereas CCR5 expression was restricted to a much smaller subset of these cells (mean, 10.4% ± 0.8%, n = 25) (reference 5 and data not shown). We determined whether these expression profiles are linked to preferential depletion by inoculating histocultures with viruses that vary only in coreceptor phenotype. Since CXCR4 is widely expressed on both CCR5+ and CCR5− cells, our hypothesis predicts that X4 viruses would deplete cells in both subsets, whereas R5 viruses would deplete preferentially within the CCR5+ subset. We therefore focused our analysis on quantitation of the CCR5+ and CCR5− subsets of CD4+ lymphocytes following infection. Indeed, all three of the X4 viruses (NL4-3, 134, and 123) massively and comparably depleted both CCR5− and CCR5+ cells, while the R5 viruses (49-5, 126, and USV3) caused depletion preferentially within the CCR5+ subset and comparatively modest depletion within the CCR5− subset (Fig. 1B). We speculate, but cannot prove, that the partial depletion of CCR5− cells by R5 viruses is explained by subthreshold levels of CCR5 expression on some cells, since previous work established that certain CD4+ lymphocytes that do not express CCR5 at levels detectable by flow cytometric methods were infectable by R5 viruses (11). Nonetheless, in all of our experiments, depletion within the CCR5+ subset by R5 viruses exceeded that in the CCR5− subset by twofold or more. These results together demonstrate that envelope-determined coreceptor preferences direct the selective depletion of cognate coreceptor-expressing CD4+ lymphocytes in human lymphoid histocultures.
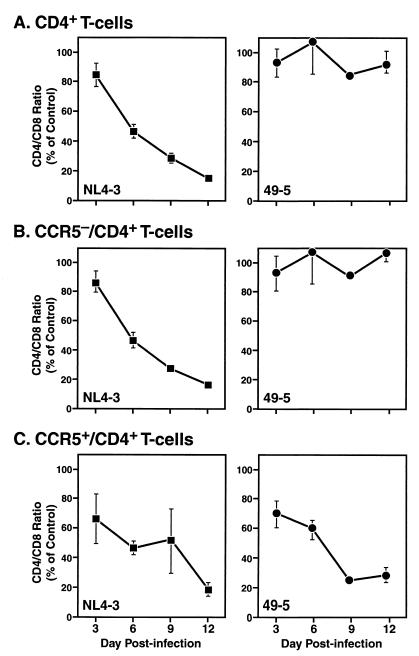
Despite a marked difference in the frequencies of potential cell targets for X4 and R5 viruses and the selective depletion of targets by each virus type, the replication kinetics of these viruses based on virus production were quite similar. Although the absolute levels of virus replication varied among individual tissue donors, there were no consistent differences in the peak levels of viral replication or the kinetics of virus accumulation in the culture medium upon inoculation of any particular tissue specimen by pairs of X4 and R5 strains. To elucidate further the relationship between virus-induced CD4+ T-cell depletion and virus replication, we performed concurrent kinetic measurements of these parameters in histocultures infected by NL4-3 or 49-5. In a typical experiment, NL4-3 progressively depressed the overall CD4/CD8 ratio with early effects evident by day 3 and progressive effects on days 6 and 12 (Fig. 2A, left). In contrast, the effect of 49-5 on CD4/CD8 ratio was very modest at all time points (Fig. 2A, right).
FIG. 2.
Kinetic analysis of overall and subset-specific T-cell depletion in human tonsil histocultures in NL4-3- and 49-5-infected tonsillar tissue (left and right panels, respectively). Mean values (± standard errors of the means) from a representative experiment (n = 3) are shown.
Stratification by CCR5 expression revealed a more complex pattern. In NL4-3-infected cultures both CCR5− (Fig. 2B, left) and CCR5+ (Fig. 2C, right) lymphocytes were lost markedly and progressively over the 12-day period with kinetics that paralleled the overall depletion of CD4+ T cells (Fig. 1A). In contrast, 49-5 had minimal effects on the CCR5− population (Fig. 2B, right) but severely and progressively depleted CCR5+ cells (Fig. 2C, right). In each of these cases, the kinetics of depletion of CD4+ T cells overall corresponded to the kinetics of depletion within the CCR5− population, which reflects the small contribution of the minor CCR5+ pool. These kinetic analyses provide further evidence that R5 viruses deplete T cells in a coreceptor-dependent process leading to preferential loss of CCR5+ cells. It should be noted that, in some experiments with 49-5, partial loss of CCR5+ cells was detected by day 3 without further loss on subsequent days (data not shown). This donor-specific effect suggests that under some conditions CCR5 expression is necessary but not sufficient for CD4+ T-cell depletion by R5 viruses and that cellular properties other than coreceptor expression may also influence susceptibility to depletion.
In view of the coreceptor-dependent depletion of major and minor T-cell subsets by X4 and R5 strains, respectively, the relationship among cellular coreceptor expression, productive viral infection, and cell killing is not immediately evident. In particular, R5 and X4 viruses differentially depleted CD4+ T cells yet exhibited similar replication profiles. Also, we previously found that the frequencies of productively infected lymphocytes were not consistently different in X4 and R5 virus-infected cultures as assessed at day 12 postinfection (9). To clarify this apparent discrepancy, we concurrently examined viral output and infected T-cell frequencies in infected cultures. As described earlier, in a typical experiment NL4-3- and 49-5-infected tonsil histocultures produced nearly identical amounts of virus with very similar kinetics as assessed by the p24 content of culture supernatants (Fig. 3A). To measure the frequency of infected cells in these cultures, cells were harvested at various time points, coimmunostained with monoclonal antibodies (MAbs) to CD3 and p24, and analyzed by flow cytometry. By this measurement, at days 6 to 9 postinfection, NL4-3 productively infected a relatively large proportion of the T cells, reaching approximately 15% of total CD3+ lymphocytes and declining by the end of the experiment (Fig. 3B). In contrast, 49-5 infected fewer T cells, reaching approximately 5% of CD3+ cells (Fig. 3B).
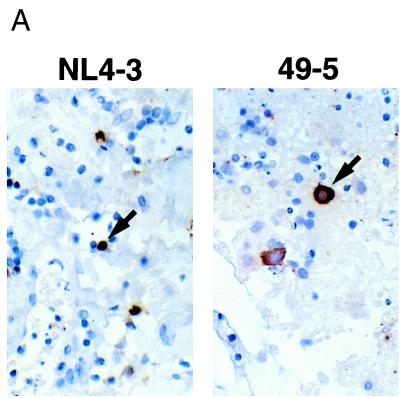
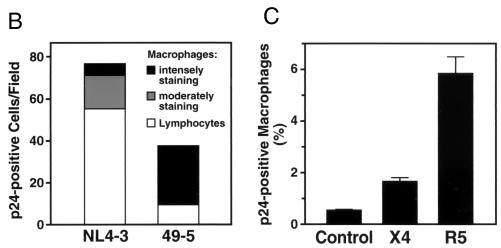
The difference in T-cell infection efficiency exhibited by these two viruses corresponds to the more aggressive depletion effect of X4 viruses compared to that of R5 viruses, but it also represents a paradox in the context of their very similar viral output profiles (Fig. 3A). One hypothesis to account for this paradox is that macrophages constitute an additional source of virus output that is not reflected in the analysis of infected T cells. We therefore used immunohistochemistry to visualize the full spectrum of infected cells in these histocultures. Immunostaining of formalin-fixed tonsil sections for the macrophage-specific antigen CD68 (6) revealed large numbers of macrophages distributed throughout the tissue (reference 4 and data not shown). Immunostaining for p24 revealed striking differences between the X4 and R5 virus-infected cultures. NL4-3-infected tissue demonstrated a predominance of p24-positive cells that appeared to be lymphocytes based on their small size and high nucleus/cytoplasm ratio (Fig. 4A, left, and 4B). In contrast, in 49-5-infected tissue many of the p24-positive cells appeared to be macrophages, based on their large size and abundant cytoplasm (Fig. 4A, right). Interestingly, some of these macrophage-like cells were also p24-positive in the NL4-3-infected cultures, and these were distinguishable as moderately and intensely stained cells. Likewise, a small number of intensely staining p24-positive lymphocytes was observed in the 49-5-infected samples. To compare target cell frequencies quantitatively, p24-positive lymphocytes and macrophage-like cells in the two cultures were counted by visual inspection (Fig. 4B), which confirmed the strong but not absolute preference of an X4 strain for lymphocytes and of an R5 strain for macrophage-like cells. Thus, as analyzed by immunohistochemistry, productive infection of both lymphocytes and macrophage-like cells appears to contribute to the total output of virus in HIV-1-infected histocultures.
FIG. 4.
Immunohistochemical and flow cytometric detection of intracellular HIV-1 p24 in infected tonsil histocultures. (A) Representative examples of p24-positive lymphocytes and macrophages in tonsil tissue infected ex vivo with NL4-3 and 49-5. Arrows indicate a typical lymphocyte (left) or macrophage (right). (B) Quantitation by counting p24-positive cells in these NL4-3- and 49-5-infected tissue sections that have comparable cross-sectional areas. (C) Frequencies of productively infected macrophages following a matched inoculation by a pair of isogenic viruses, NL4-3 (X4) and NL107 (R5). On day 9, cultures were coimmunostained with MAbs recognizing CD14 (phycoerythrin conjugated), CD68 (phycoerythrin conjugated), and p24 (fluorescein-isothiocyanate conjugated). NL107 is isogenic with NL4-3 but harbors the V1-V3 loop gp120 segments from BaL in the backbone of NL4-3 (13). Results are means of triplicate samples (plus standard errors of the means).
We also developed a flow cytometric approach to identify and quantify macrophage-like cell types in HIV-infected lymphoid tissues. Macrophages were identified by immunostaining cells dispersed from histocultures for markers that distinguish T cells (CD3) from macrophages (CD14 and CD68). Cytospin and transmission electron microscopic analysis of cells sorted by positive staining for these markers validated this separation technique (data not shown). To analyze infection in this population, cells from infected tonsil histocultures were also immunostained for viral p24. These experiments demonstrated that the R5 virus infected a significantly higher proportion of macrophages in these tissues than did the matched X4 virus (Fig. 4C); similar results were obtained using two other distinct pairs of isogenic X4 and R5 viruses (data not shown). These results confirm that macrophages serve as significant cellular hosts for productive infection by R5 viruses, presumably contributing to the total viral output. The relative contribution macrophages make to replication of X4 viruses in these lymphoid tissues appears to be smaller (Fig. 4). Additionally, other cells not identified in this analysis (e.g., dendritic cells) may also contribute to total viral output in tonsil histocultures.
In the present studies of matched recombinant viruses, we confirmed that X4 HIV-1 strains are more pathogenic toward the overall CD4+ T-cell population than are isogenic R5 strains. We also verified that CXCR4 is expressed very widely among CD4+ T cells in these lymphoid cultures, while CCR5 is expressed on fewer cells. Correspondingly, all X4 strains depleted CD4+ T cells broadly in these experiments, while their matched R5 counterparts potently and preferentially depleted CD4+ T cells within the smaller CCR5+ pool. Additionally, the overall frequency of productively infected lymphocytes in X4 HIV-1-infected tissues is higher than in R5 HIV-1-infected tissues, suggesting that the measured frequencies of productively infected lymphocytes are proportional and related to the cumulative attrition of infected cells. These experiments strongly support the hypothesis that X4 viruses infect lymphocytes at high frequencies and exhibit high overall virulence because CXCR4-expressing targets are abundant, while R5 viruses are also pathogenic for cell targets but exhibit lower overall virulence because CCR5-expressing cells are much less abundant. Furthermore, they prove that sequences in gp120 controlling coreceptor specificity alone are sufficient to determine which subset of CD4+ T cells is depleted by each virus.
These principles should be relevant for other lymphoid organs as well (4, 9). Moreover, no consistent differences were observed between pathologic tonsil specimens and nonpathologic spleen specimens in the expression of various common markers of activation (data not shown), suggesting that a particular inflammatory state is not required for these viral properties to be manifest. In addition, CXCR4 and CCR5 expression patterns were comparable in these tissues (data not shown). However, there may be differences in relative activation status and/or coreceptor expression in other lymphoid tissues not examined here, such as gut-associated lymphoid tissue or thymus tissue. It is reasonable to expect that in these tissue contexts as well, the level of pathogenicity demonstrated by X4 and R5 viruses would be governed by the relative expression levels of CXCR4 and CCR5. For example, the level of CCR5 is significantly higher in gut-associated lymphoid tissue than in peripheral blood (7), and this tissue in rhesus monkeys was found to be highly susceptible to the pathogenic effects of an R5 simian immunodeficiency virus strain (14). In contrast, CCR5 expression in the human thymus is very low, and R5 viruses replicate poorly in this tissue and cause minimal cytopathic effects (2).
Despite differences in the pattern of selective CD4 depletion, X4 and R5 strains produced comparable amounts of virus with similar kinetics, in contrast to other reports based on xenotransplant model systems (2, 8, 10). The presence of significant numbers of tissue macrophages and dendritic cells is one feature that distinguishes the histoculture model (4). Indeed, we detected large, p24-positive macrophage-like cells in both R5- and X4-infected tissues by immunohistochemistry and flow cytometry. It is possible that this p24 staining represented cells that had endocytosed infected lymphocytes rather than those directly infected by HIV-1. By this interpretation, one would expect a higher proportion of p24-positive macrophage-like cells in X4-infected cultures, given the higher level of infection of T cells by these strains. However, a larger proportion of macrophage-like cells were found to be p24-positive in R5-infected cultures than in X4-infected cultures, making it likely that the majority of large, p24-positive cells represent direct and productive infection. Thus, macrophages may contribute to viral output for both virus types but make a greater contribution for R5 strains. Therefore, the combined viral output from infected T cells, macrophages, and possibly other related cell types not identified in these analyses could account for the overall similarities in replication kinetics. Unfortunately, current technology has not yet permitted a robust and direct determination of the actual sources of virus production in these histocultures. Nonetheless, one interesting speculation is that the relative contribution of cell types other than CD4+ T cells (e.g., macrophages) to viral load may increase over time in conjunction with progressive loss of specific T cells that are susceptible to the cytopathic effects of HIV-1.
These data provide compelling reasons to continue anti-HIV-1 therapeutic efforts aimed at developing antagonists to both CCR5 and CXCR4. Because macrophages can be infected detectably by both X4 and R5 strains, such antagonists may be effective not only in preventing CD4 depletion but also in limiting viral replication in alternative non-T-cell reservoirs.
Acknowledgments
J.-C.G. and M.L.P. contributed equally to this work.
We thank Bruce Chesebro and Malcolm Martin for kindly providing plasmids and Dee J. Holthe, Cecilia Stewart, Sharon Hall, Claudette Delphis, Ursula Perotti, Mark Weinstein, and the surgical staffs at Kaiser Hospitals (San Rafael and San Francisco) and San Francisco General Hospital for generous assistance in obtaining posttonsillectomy samples. We acknowledge the technical assistance of Eric Wieder and Lisa Gibson in these experiments and the assistance of Heather Gravois, John Carroll, and Neile Shea in the preparation of the manuscript.
M.L.P. was supported by the California Universitywide AIDS Research Program, the Biomedical Sciences Graduate Program, and the National Institutes of Health Medical Scientist Training Program at UCSF. D.A.E. was supported by the National Institutes of Health Medical Scientist Training Program at UCSF. B.S. was supported by the Boehringer Ingelheim Fund, and R.F.S. was supported in part by the UCSF-GIVI CFAR. This work was supported in part by NIH grant R01-AI43695 (M.A.G.), the J. David Gladstone Institutes (M.A.G.), and the NASA/NIH Center for Three-Dimensional Tissue Culture.
REFERENCES
- 1.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Berkowitz R D, Alexander S, Bare C, Linquist-Stepps V, Bogan M, Moreno M E, Gibson L, Wieder E D, Kosek J, Stoddart C A, McCune J M. CCR5- and CXCR4-utilizing strains of human immunodeficiency virus type 1 exhibit differential tropism and pathogenesis in vivo. J Virol. 1998;72:10108–10117. doi: 10.1128/jvi.72.12.10108-10117.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chesebro B, Wehrly K, Nishio J, Perryman S. Mapping of independent V3 envelope determinants of human immunodeficiency virus type 1 macrophage tropism and syncytium formation in lymphocytes. J Virol. 1996;70:9055–9059. doi: 10.1128/jvi.70.12.9055-9059.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Glushakova S, Baibakov B, Zimmerberg J, Margolis L B. Experimental HIV infection of human lymphoid tissue: correlation of CD4+ T cell depletion and virus syncytium-inducing/non-syncytium-inducing phenotype in histocultures inoculated with laboratory strains and patient isolates of HIV type 1. AIDS Res Hum Retrovir. 1997;13:461–471. doi: 10.1089/aid.1997.13.461. [DOI] [PubMed] [Google Scholar]
- 5.Grivel J-C, Margolis L B. CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue. Nat Med. 1999;5:344–346. doi: 10.1038/6565. [DOI] [PubMed] [Google Scholar]
- 6.Herndier B G, Werner A, Arnstein P, Abbey N W, Demartis F, Cohen R L, Shuman M A, Levy J A. Characterization of a human Kaposi's sarcoma cell line that induces angiogenic tumors in animals. AIDS. 1994;8:575–581. doi: 10.1097/00002030-199405000-00002. [DOI] [PubMed] [Google Scholar]
- 7.Lapenta C, Boirivant M, Marini M, Santini S M, Logozzi M, Viora M, Belardelli F, Fais S. Human intestinal lamina propria lymphocytes are naturally permissive to HIV-1 infection. Eur J Immunol. 1999;29:1202–1208. doi: 10.1002/(SICI)1521-4141(199904)29:04<1202::AID-IMMU1202>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.Mosier D E. Human immunodeficiency virus infection of human cells transplanted to severe combined immunodeficient mice. Adv Immunol. 1996;63:79–125. doi: 10.1016/s0065-2776(08)60855-x. [DOI] [PubMed] [Google Scholar]
- 9.Penn M L, Grivel J C, Schramm B, Goldsmith M A, Margolis L. CXCR4 utilization is sufficient to trigger CD4+ T cell depletion in HIV-1-infected human lymphoid tissue. Proc Natl Acad Sci USA. 1999;96:663–668. doi: 10.1073/pnas.96.2.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Picchio G R, Gulizia R J, Wehrly K, Chesebro B, Mosier D E. The cell tropism of human immunodeficiency virus type 1 determines the kinetics of plasma viremia in SCID mice reconstituted with human peripheral blood leukocytes. J Virol. 1998;72:2002–2009. doi: 10.1128/jvi.72.3.2002-2009.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Riley J L, Levine B L, Craighead N, Francomano T, Kim D, Carroll R G, June C H. Naïve and memory CD4 T cells differ in their susceptibilities to human immunodeficiency virus type 1 infection following CD28 costimulation: implications for transmission and pathogenesis. J Virol. 1998;72:8273–8280. doi: 10.1128/jvi.72.10.8273-8280.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Speck R F, Wehrly K, Platt E J, Atchison R E, Charo I F, Kabat D, Chesebro B, Goldsmith M A. Selective employment of chemokine receptors as human immunodeficiency virus type 1 coreceptors determined by individual amino acids within the envelope V3 loop. J Virol. 1997;71:7136–7139. doi: 10.1128/jvi.71.9.7136-7139.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toohey K, Wehrly K, Nishio J, Perryman S, Chesebro B. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology. 1995;213:70–79. doi: 10.1006/viro.1995.1547. [DOI] [PubMed] [Google Scholar]
- 14.Veazey R S, DeMaria M, Chalifoux L V, Shvetz D E, Pauley D R, Knight H L, Rosenzweig M, Johnson R P, Desrosiers R C, Lackner A A. Gastrointestinal tract as a major site of CD4+ T cell depletion and viral replication in SIV infection. Science. 1998;280:427–431. doi: 10.1126/science.280.5362.427. [DOI] [PubMed] [Google Scholar]