Abstract
The development of the olfactory system is influenced by sensory inputs, and it maintains neuronal generation and plasticity throughout the lifespan. The olfactory bulb contains a higher proportion of interneurons than other brain regions, particularly during the early postnatal period of neurogenesis. Although the relationship between sensory stimulation and olfactory bulb development during the postnatal period has been well studied, the molecular mechanisms have yet to be identified. In this study, we used western blotting and immunohistochemistry to analyze the expression of the transcription factor Npas4, a neuron-specific immediate-early gene that acts as a developmental regulator in many brain regions. We found that Npas4 is highly expressed in olfactory bulb interneurons during the early postnatal stages and gradually decreases toward the late postnatal stages. Npas4 expression was observed in all olfactory bulb layers, including the rostral migratory stream, where newborn neurons are generated and migrate to the olfactory bulb. Under sensory deprivation, the olfactory bulb size and the number of olfactory bulb interneurons were reduced. Furthermore, Npas4 expression and the expression of putative Npas4 downstream molecules were decreased. Collectively, these findings indicate that Npas4 expression induced by sensory input plays a role in the formation of neural circuits with excitatory mitral/tufted cells by regulating the survival of olfactory bulb interneurons during the early stages of postnatal development.
Keywords: Npas4, Immediate-early gene, Olfactory bulb interneurons, Olfactory bulb development, Postnatal development, Unilateral naris occlusion
INTRODUCTION
Sensory input during early postnatal development is essential for the normal development of the sensory systems in the brain [1-5]. After birth, sensory experiences modulate both the functional and structural properties of neural circuits in different sensory systems, including the visual, somatosensory, olfactory, and auditory systems [6-9]. Deprivation of sensory stimulation in early postnatal development induces disturbances in both synaptic connections and an imbalance in the excitation and inhibition of sensory systems [10-13]. The development of γ-aminobutyric acid (GABA)-releasing inhibitory interneurons (GABAergic interneurons) occurs during the early postnatal period and is completed by the end of adolescence [14-17]. Thus, the abnormal formation of interneurons during early postnatal development leads to neurodevelopmental disorders, such as autism spectrum disorder or Tourette’s syndrome [18, 19].
Interestingly, neuronal immediate-early genes (IEGs), which play essential roles in neuronal development, plasticity, and cognitive function, are highly expressed in the early postnatal period [20-22]. The neural activity-induced effects on sensory system development may be due to changes in the expression of various genes, half of which are transcription factors [23]. IEGs play essential roles in brain development and neuronal plasticity, and many IEGs, such as c-Fos, Ark, Nr4a1, and Npas4, are expressed in various brain regions [20-22]. Npas4 (Neuronal Per/Arnt/Sim (PAS) domain protein 4) is a member of the helix-loop-helix class of transcriptional regulators involved in numerous developmental events via neuronal activity [24, 25]. In particular, research shows that Npas4 is expressed in diverse interneurons, including somatostatin, parvalbumin, and olfactory bulb interneurons and regulated by neuronal activity [26-28]. It has been reported that Npas4 plays a role in the development of inhibitory synapses in hippocampal neurons and the visual system [29]. In addition, Npas4 controls the balance between excitatory and inhibitory neurons and visual cortical plasticity [30]. It interacts with several promoters that are regulated in a neuronal activity-dependent manner [31]. Furthermore, Npas4 has neuroprotective effects in the hippocampus in vivo and in vitro, and Npas4 knock-out mice are more susceptible to neuronal death than control mice due to neurodegeneration [32-34]. However, few studies have investigated whether Npas4 is involved in sensory stimulation-dependent development and its role in early postnatal development when the sensory system is established.
The olfactory bulb (OB) processes odor information detected by olfactory receptors on the olfactory sensory neurons and transmits it to the brain. The mammalian OB expresses more abundant and diverse interneurons innervating excitatory neurons than other brain regions [35]. Most interneurons in the OB are GABAergic and are generated throughout life until the adult stage [36-40]. More than half of the OB interneurons are generated during the initial 2 weeks of the early postnatal period, in the subventricular zone (SVZ), and migrate into the OB along the rostral migratory stream (RMS) [38, 41, 42]. Newborn interneurons differentiate into GABAergic interneurons and form neural circuits for excitatory mitral/tufted cells in the OB [36-40]. For this reason, OB interneurons are a good model for studying the modification of neural circuits by sensory stimulation from the external environment during the early postnatal period [43-46]. Research on the development of OB interneurons is essential for understanding complicated brain physiology.
In this study, we aimed to confirm that Npas4 is temporally expressed in the overall interneurons in the OB layers, and sensory stimulation-dependent Npas4 expression using unilateral naris occlusion occurs during the early postnatal development period. We further investigated whether the putative Npas4 downstream molecules are affected by olfactory sensory input-dependent Npas4 expression during early postnatal development. Overall, we propose that Npas4 expression by olfactory stimulation contributes to the development of OB interneurons and the proper formation of neural circuits in excitatory mitral/tufted cells during early postnatal development.
MATERIALS AND METHODS
Animals
All experimental protocols were approved by the Institutional Animal Care and Use Committee of the Daegu Gyeongbuk Institute of Science and Technology (DGIST) (DGIST-IACUC-21041902-0001). All applicable guidelines for the care and use of laboratory animals from the National Institutes of Health. Male C57BL/6 mice (postnatal day 28, 56 days old) were purchased from KOATECH (Daegu, South Korea). On postnatal day 0, 14-day-old male mice were obtained from pups born to pregnant C57BL/6 female mice timed pregnant 19, (TP19). Postnatal day 14 mice were housed and handled according to the Laboratory Animal Resource Center of DGIST, maintained at a temperature of 20°C±2°C and a 14-h light, 10-h dark cycle, with access to food and water. Only male animals were used in this study.
Tissue preparation
Tissue for western blot
Mice were anesthetized by intraperitoneal injection of 1.25% avertin (0.02 ml per gram of body weight) and transcardially perfused with 50 ml of 0.9% saline (9 g of saline in distilled water, 4°C). Following perfusion, the heads were divided in the sagittal direction, and the olfactory bulbs were dissected on ice.
Tissue for immunohistochemistry
Mice were anesthetized by intraperitoneal injection of 1.25% avertin (0.02 ml per gram of body weight) and transcardially perfused with 50 ml of 0.9% saline (4°C) and 40 ml of fresh 4% paraformaldehyde (PFA) in 0.1 M phosphate buffer (PB) (4°C). Following perfusion, the brains were dissected and post-fixed with 4% PFA for 16~20 hours at 4°C. The fixed brains were washed with water and dehydrated with 70%, 80%, 85%, 90%, 95%, and 100% ethanol. Brains were embedded twice in xylene, paraffin solvent, and paraffin wax. Paraffin blocks of the brain were cut into serial sections (sagittal or coronal, 5~7 μm) with a microtome (Leica, Germany), and each section was preserved on a MUTO saline-coated microslide (MUTO, Japan).
Western blot
The isolated olfactory bulbs were lysed in RIPA radioimmunoprecipitation assay buffer (Thermo Scientific, USA) with a protease and phosphatase inhibitor cocktail (Thermo Scientific, USA) on ice. The tissues were mechanically homogenized and samples were incubated on ice for 10 min and centrifuged for 20 min at 4°C and 13,300 rpm. Proteins in the supernatant were quantified using a BCA assay. Then 60 μg of proteins were separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Next, 8% separating gels were used to resolve Npas4 and β-actin. Separated proteins were transferred to a 0.45 μm polyvinylidene difluoride membrane (Millipore, USA). The membranes were blocked with 5% bovine serum albumin (BSA) (Sigma-Aldrich, USA) in Tris-buffered saline with 0.1% Tween 20 (TBS-T) and then incubated with primary antibodies at an appropriate concentration (Table 1). diluted in 3% BSA/TBS-T for 16~20 hours at 4°C in a shaking incubator. After washing for 30 min with TBS-T, the membranes were incubated with horseradish peroxidase (HRP) conjugated anti-mouse and anti-rabbit antibodies in 3% BSA/TBS-T for 1 h at RT. After washing for 30 min with TBS-T, the immunoblots were visualized using an enhanced chemiluminescence substrate kit (Thermo Scientific, USA).
Table 1.
Information on primary antibodies used in western blot
| Antibodies | Source | Catalog no. | Working concentration |
|---|---|---|---|
| Anti- Npas4 | Novusbio | NBP2-47252 | WB 1:1000 |
| Anti- β-actin | Santa Cruz | sc-47778 | WB 1:20000 |
| Anti- GAD67 | Sigma-Aldrich | G5419 | WB 1:10000 |
| Anti- VGLUT1 | Atlas antibodies | Amab91041 | WB 1:1000 |
| Anti- TH | Merck | MAB318 | WB 1:1000 |
| Anti- Calretinin | Merck | MAB1568 | WB 1:3000 |
Immunoprecipitation
To measure levels of phosphorylated Npas4 from postnatal mouse olfactory bulb, the isolated olfactory bulbs were lysed in RIPA radioimmunoprecipitation assay buffer (Thermo Scientific, USA) with a protease and phosphatase inhibitor cocktail (Thermo Scientific, USA) on ice. The tissues were mechanically homogenized and samples were incubated on ice for 10 min and centrifuged for 20 min at 4°C and 13,300 rpm. Proteins in the supernatant were quantified using a BCA assay. Equal amounts of protein (2 mg) from lysates of OBs were incubated with 15 μl of antibody against phosphoserine/threonine/tyrosine antibody (#61-8300, Thermo Scientific, USA) for 2 h at room temperature, and then incubated with Protein-A/G agarose beads (#20421, Thermo Scientific, USA) for 16 h at 4°C. And then its supernatant was washed three times with PBS and loaded with SDS-PAGE to detect phosphorylated Npas4.
Immunohistochemistry
The brain sections were deparaffinized with xylene and rehydrated using descending grades of ethanol (100%, 95%, 85%, and 70% DW). The sections were permeabilized with PBS-T (0.01% Triton-X in PBS) for 15 min and then incubated in 3% H2O2/PBS for 13 min to remove endogenous peroxidase activity in the tissue. Antigen retrieval was carried out by boiling 0.01 M citrate buffer/DW with 0.05% Tween 20 for 30 sec using a microwave and then cooling for 30 min at room temperature (RT). The sections were washed in PBS and blocked with 4% normal donkey serum (Jackson ImmunoResearch, USA) in PBS for 1 h at RT to minimize nonspecific labeling. After blocking, the sections were incubated in a humid chamber overnight at 4°C with primary antibodies in an appropriate concentration (Table 2), diluted in 4% normal donkey serum, and then incubated for 1 h at RT. Sections were washed with PBS for 1 h and incubated with secondary antibodies in 4% NDS (1:300) for 1 h at RT. After washing with PBS, sections were mounted using a mounting solution containing DAPI (Vector Laboratories, UK) and cover glasses (Marienfeld Superior, Germany). Images were captured using a high-resolution confocal microscope LSM780 (Carl Zeiss, Germany) and processed using the ImageJ software (NIH, USA).
Table 2.
Information on primary antibodies used in immunohistochemistry
| Antibodies | Source | Catalog no. | Working concentration |
|---|---|---|---|
| Anti- Npas4 | Invitrogen | PA5-39300 | IHC 1:200 |
| Anti- GAD67 | Sigma-Aldrich | MAB5406 | IHC 1:100 |
| Anti- Calretinin | Sigma-Aldrich | MAB1568 | IHC 1:100 |
| Anti- TH | Novusbio | NBP2-46647 | IHC 1:200 |
RNA extraction and quantification of mRNA level (RT-qPCR)
Total RNA was extracted from the OBs using a TRIzol reagent (Ambion). A mixture of 1 μg of total RNA, distilled water, and oligo dT 2 μl was used to produce 15 μl and incubated for 5 min at 70°C. M-MLV 5X buffer (Promega) 5 μl, dNTP 5 μl, 0.625 μl RNA inhibitor, and M-MLV reverse transcriptase (Promega) 1 μl were added before incubation at 42°C for 1 h. RT-qPCR was performed using cDNA and each primer from Table 3 using the GoTaq® DNA Polymerase kit (Promega). The reaction was run in a PCR machine (T100TM Thermal Cycler, Bio-Rad) with the following parameters: 95°C (30 sec), 55 (30 sec), 72°C (30 sec), and 35 cycles. Electrophoresis bands were detected using ChemiXRS+ (Bio-Rad) and scanned using ImageJ software (NIH, USA).
Table 3.
List of RT-PCR primer sequences
| Gene | Accession # | Forward sequence | Reverse sequence |
|---|---|---|---|
| Npas4 | NM_153553.5 | TCATGAGTCTTGCCTGCATC | GAGGGACTTGGAGGTGTTGA |
| BDNF | NM_001048139.1 | TTCATACTTTGGTTGCATGA | TTCAGTTGGCCTTTGGATAC |
| HDAC9 | NM_001271386.1 | TCAGAGGTTCCTATGGGCCTG | TGGAGACGTTCCACTGAGGG |
| Nur77 | NM_010444.2 | CCACCTCTCCGAACCGTGACA | GAGAAGATTGGTAGGGAGGC |
| Mdm2 | NM_001288586.2 | GAATCCTCCCCTTCCATCACAC | AAGCCTTCTTCTGCCTGAGC |
| Dcx | NM_001110222.1 | TACGTTTCTACCCAATGGGG | CTGCTTTCCATCAAGGGTGTA |
| Syt10 | NM_018803.2 | TTCGCGGGTCAGGTGGAGTG | TTGGGTGCTGGCTTTTCATTTTC |
| β-actin | NM_007393.5 | TGTGATGGTGGGAATGGGTCAG | TTTGATGTCACGCACGATTTCC |
Statistical analysis
Statistical analyses and graph plotting were performed using the Prism software (GraphPad Software, USA). A one-way ANOVA analysis was used to determine the statistical significance of Npas4 protein levels during the postnatal period. One-tailed paired t-tests were used to compare the open and closed sides of the naris muscles. The Bonferroni test was used for multiple comparisons. All statistical data were presented as the mean±SEM (standard error of the mean). Statistical significance was set at p<0.05.
RESULTS
Time-specific Npas4 expression during early postnatal development in overall OB layers
To investigate Npas4 expression in the OB during early postnatal period, we conducted immunoblotting using lysates from mouse OBs at P0, P14, P28, and P56. Postnatal OB neurogenesis is known to peak for 2 weeks after birth and then decline gradually, with minimal neurogenesis occurring in adulthood [35]. Based on this study [35], we categorized four developmental stages: P0, immediately after birth; P14, the period of rapid postnatal neurogenesis and integration of newborn OB interneurons; P28, the adolescent stage with decreasing postnatal neurogenesis; and P56, the young adult stage. Interestingly, Npas4 expression differed according to the postnatal period (Fig. 1A). The expression level of Npas4 was highest at P0 and decreased toward P56 (Fig. 1B). We observed that the Npas4 band appeared as two distinct bands, suggesting that Npas4 may exist in both mature and immature forms via post-translational modifications. For a more detailed comparison, we separately analyzed the upper and lower Npas4 bands in the immunoblot. The expression of the upper band of Npas4 tended to decrease during postnatal development (Fig. 1C). In contrast, expression of the lower band of Npas4 did not change during the postnatal period (Fig. 1D). Recently, it was reported that mitogen-activated protein kinase (MAPK) phosphorylates multiple sites on Npas4 and increases its activity and interactions with other proteins in striatal neurons [47]. Thus, Npas4 in the OB may be modified by various post-translational modifications, consistent with previous reports. To investigate whether the upper band of Npas4 observed in the immunoblot is phosphorylated Npas4, we conducted immunoprecipitation. First, phosphorylated proteins were pulled down from P0, P14, P28, and P56 mouse OBs lysates using antibody against phosphoserine/threonine/tyrosine. Npas4 antibody was then used to detect phosphorylated Npas4. We observed a similar trend in the expression of phosphorylated Npas4, showing a decrease in intensity similar to the reduction observed in the upper band of Npas4 as P56 progressed (Fig. 1E, F).
Fig. 1.
Expression of Npas4 in the mouse postnatally developing olfactory bulb. Expression levels of Npas4 during postnatal mouse olfactory bulb development (n=3 for each age group) using Npas4 and β-actin antibodies. (A) Immunoblot of Npas4 expression (band at approximately 100 kDa) at different developmental stages. Actin (43 kDa band) is used as a loading control. (B) Quantitative analysis of overall Npas4 protein expression in immunoblot images. (C) and (D) Quantitative analysis of Npas4 protein expression in the upper and lower parts. Npas4 protein levels are normalized by the level of β-actin expression. (E) Expression levels of phosphorylated Npas4 during postnatal mouse olfactory bulb development (n=4 for each age group) using Npas4 antibodies. Lysates were immunoprecipitated with phosphoserine/threonine/tyrosine antibody and then probed with Npas4 antibody. (F) Quantitative analysis of overall phosphorylated Npas4 protein expression in immunoblot images. Error bars represent the mean±SE, and significance is determined using a one-way ANOVA with Bonferroni’s multiple comparisons tests. Individual animals are labeled with black circles. *p<0.05, **p<0.01, ***p<0.001, ****p<0.0001.
Overall, Npas4 was expressed differently during the postnatal period and modified by phosphorylation, being higher in P0 and P14 than in P28 and P56 mouse OBs. This pattern is similar to the OB postnatal neurogenesis for 2 weeks. This data suggests that Npas4 may play a more critical role in the early postnatal stages than in the late postnatal stages.
Next, we performed immunohistochemistry to show the Npas4 expression patterns during postnatal development in each layer of the mouse OB. The OB is divided into several layers, with different types of OB neurons and cell bodies in each layer. There are excitatory projection neurons, such as mitral cells and tufted cells (M/TCs), whose somas are located in the mitral cell layer (MCL) and the external plexiform layer (EPL). Granule cells (GCs) and periglomerular cells (PGCs) are OB interneurons, and their cell bodies are located in the granule cell layer (GCL) and glomerular layer (GL), respectively. Therefore, it is possible to determine the types of neurons that express Npas4. To confirm the expression pattern of Npas4 in OB layers, we co-stained Npas4 with the mitral/tufted cell marker Tbr2. For a more precise comparison of Npas4 expression patterns, we quantified Npas4 expression in individual cells in each layer of the OB (Table 4) and Npas4 intensity was measured. A rectangular region of interest (ROI) of the same size was randomly placed in the image data of each layer, and the ratio of Npas4 expressing cells to the total cells inside the ROI was counted. In P0 mouse OB, Npas4 was ubiquitously expressed in the GL, EPL, MCL, and GCL (Fig. 2A). Notably, in the GCL, most cells expressed Npas4, and the intensity of the signals was higher than that in the other layers (Fig. 2E). In P14 mouse OB, as in P0, Npas4 expression was observed in all OB layers (Fig. 2B, E). Similarly, P28 and P56 mouse OBs showed Npas4 expression in all layers of the OB (Fig. 2C, D). However, Npas4 expression decreased relatively at P28 and P56 compared to P0 and P14 (Fig. 2E) and some cells in the GCL showed strong and distinct signals compared to other cells (Fig. 2C, D). Also, we confirmed not only that some Tbr2+ cells expressed Npas4 at P0 and P14, but it rapidly decreased during P28 and P56 but also the proportion of Tbr2+ cells expressing Npas4 was not notably high (Table 5). As a result, the overall Npas4 signal in the OB gradually decreased during the postnatal period consistent with our immunoblotting results (Fig. 1A) and we could observe the possibility that Npas4 is more likely to be expressed at a higher proportion in inhibitory interneurons than in excitatory mitral/tufted cells.
Table 4.
The ratio of Npas4-expressing cells to the total cells in each layer of OB
| Layers Stages |
GL1 (Npas4+/total) | GL2 (Npas4+/total) | GL3 (Npas4+/total) | GL ratio average |
|---|---|---|---|---|
| P0 | 35/43 | 21/29 | 35/45 | 0.771956333 |
| P14 | 9/21 | 8/11 | 10/16 | 0.593614667 |
| P28 | 12/29 | 14/29 | 22/38 | 0.491833 |
| P56 | 20/34 | 19/52 | 13/21 | 0.524222667 |
| Layers Stages |
EPL1 (Npas4+/total) | EPL2 (Npas4+/total) | EPL3 (Npas4+/total) | EPL ratio average |
| P0 | 7/9 | 5/10 | 6/10 | 0.625926 |
| P14 | 10/16 | 10/15 | 13/26 | 0.597222333 |
| P28 | 1/9 | 7/8 | 6/14 | 0.471560667 |
| P56 | 6/11 | 6/12 | 4/10 | 0.481818333 |
| Layers Stages |
MCL1 (Npas4+/total) | MCL2 (Npas4+/total) | MCL3 (Npas4+/total) | MCL ratio average |
| P0 | 13/16 | 13/15 | 9/14 | 0.774008 |
| P14 | 30/34 | 16/18 | 26/34 | 0.845316 |
| P28 | 12/33 | 10/18 | 13/23 | 0.494803 |
| P56 | 21/38 | 10/22 | 10/33 | 0.436735667 |
| Layers Stages |
GCL1 (Npas4+/total) | GCL2 (Npas4+/total) | GCL3 (Npas4+/total) | GCL ratio average |
| P0 | 28/35 | 23/25 | 25/30 | 0.851111 |
| P14 | 24/35 | 22/25 | 28/35 | 0.788571333 |
| P28 | 24/52 | 15/33 | 13/32 | 0.440777667 |
| P56 | 13/25 | 8/30 | 12/35 | 0.376508 |
Fig. 2.
Expression pattern of Npas4 in mouse olfactory bulbs for the postnatal developing period. (A~D) Representative images of the P0, P14, P28, and P56 olfactory bulbs. Cells are co-stained with the Npas4, Tbr2, and DAPI and imaged using confocal microscopy. Blue represents nuclear DNA stained with DAPI, red represents Tbr2, and green represents Npas4 (primarily expressed in the nucleus). Each layer of the olfactory bulb glomerular layer (GL), external plexiform layer (EPL), mitral cell layer (MCL), and granule cell layer (GCL) is indicated in each image. (E) A rectangular region of interest (ROI) of the same size was randomly placed in the image data of each layer, and the ratio of Npas4 expressing cells to the total cells and the ratio of Npas4 expressing cells to the Tbr2+ cells inside the ROI was counted and the intensity of Npas4 expressing cells was measured. The counts for Npas4+ cells and total cells are all listed in Table 4. The counts for Npas4+ cells and Tbr2+ cells are all listed in Table 5. Scale bar: 100 μm.
Table 5.
The ratio of Npas4-expressing cells to the Tbr2+ cells count in each layer of OB
| Layers Stages |
GL1 (Npas4+/Tbr2) | GL2 (Npas4+/Tbr2) | GL3 (Npas4+/Tbr2) | GL ratio average |
|---|---|---|---|---|
| P0 | 1/4 | 1/5 | 1/6 | 0.205555 |
| P14 | 3/11 | 4/10 | 3/9 | 0.335353 |
| P28 | 1/11 | 1/6 | 1/6 | 0.141414 |
| P56 | 1/9 | 3/8 | 1/7 | 0.209656 |
| Layers Stages |
EPL1 (Npas4+/Tbr2) | EPL2 (Npas4+/Tbr2) | EPL3 (Npas4+/Tbr2) | EPL ratio average |
| P0 | 1/4 | 2/4 | 1/3 | 0.361111 |
| P14 | 2/17 | 4/17 | 4/16 | 0.200980 |
| P28 | 1/4 | 1/5 | 0/3 | 0.15 |
| P56 | 0/4 | 0/2 | 1/3 | 0.111111 |
| Layers Stages |
MCL1 (Npas4+/Tbr2) | MCL2 (Npas4+/Tbr2) | MCL3 (Npas4+/Tbr2) | MCL ratio average |
| P0 | 4/19 | 3/18 | 6/15 | 0.259064 |
| P14 | 2/17 | 2/9 | 2/8 | 0.196623 |
| P28 | 1/7 | 1/8 | 1/6 | 0.144841 |
| P56 | 1/5 | 1/6 | 1/12 | 0.15 |
| Layers Stages |
GCL1 (Npas4+/Tbr2) | GCL2 (Npas4+/Tbr2) | GCL3 (Npas4+/Tbr2) | GCL ratio average |
| P0 | 5/9 | 8/16 | 5/12 | 0.490740 |
| P14 | 1/4 | 2/5 | 1/5 | 0.283333 |
| P28 | 0/3 | 1/4 | 0/2 | 0.083333 |
| P56 | 0/1 | 0/2 | 0/1 | 0 |
OB interneurons express the Npas4 protein during early postnatal development
Based on the above results, it has been confirmed that Npas4 is more likely to be expressed at a higher proportion in interneurons during the early postnatal period, from P0 to P14, known as explosive development of OB interneurons. Since these results suggested that Npas4 is involved in the development of OB interneurons, we investigated whether Npas4 is expressed in OB interneurons and which cell types express Npas4 among the various subtypes of OB interneurons in P14 mice. We stained OB layers using the GABAergic interneuron marker Glutamic Acid Decarboxylase 67 (GAD67) and Npas4 antibody to verify Npas4 expression of OB interneurons in P14. Npas4 was expressed in the majority of GAD67-positive interneurons in the GL, MCL, and GCL of P14 mice (Fig. 3A). OB interneurons can be classified into several subtypes based on gene expression [48]. To clarify Npas4 expression in the subtypes of OB interneurons, we marked OB interneurons with a PGC marker, tyrosine hydroxylase (TH), and a GCs marker, calretinin (CALR). We found that Npas4 was expressed at a high proportion in not only TH-positive PGCs of the GL but also CALR-positive of the GCs (Fig. 3B, Table 6, 7). Together, these results indicated that during the explosive development of OB interneurons in the early postnatal period, Npas4 is expressed in the majority of the interneuron population.
Fig. 3.
OB interneurons express Npas4 during the postnatal developing period. (A) Co-stained with GABAergic interneuron markers GAD67 and NPAS4 in P14 mouse OB. GABAergic interneurons expressing Npas4 in the glomerular layer (GL), mitral cell layer (MCL), and granule cell layer (GCL) were labeled with antibodies for Npas4 and GAD67; red represents GAD67, and green represents Npas4. Squared areas are magnified, and signals are shown in red and green, respectively. The closed arrow represents GABAergic interneurons expressing Npas4 protein. The ratio of Npas4 expressing cells to the GAD67+ cells in the image data of each layer. (B) Co-stained with tyrosine hydroxylase and Npas4, and calretinin and Npas4 in P14 mouse OB. TH-positive interneurons expressing the Npas4 protein in the GL were labeled with antibodies for Npas4 and TH. CALR-positive interneurons expressing Npas4 protein in the GCL are labeled with antibodies for Npas4 and calretinin; red represents TH-positive (primarily expressed in the cytoplasm) and CALR-positive interneurons; green represents Npas4. Squared areas are magnified, and signals are shown in red and green, respectively. The closed arrow represents TH-positive interneurons and CALR-positive interneurons expressing Npas4. The ratio of Npas4 expressing cells to the TH+ and CALR+ cells in the image data of each layer. The closed arrow represents TH-positive interneurons and CALR-positive interneurons expressing Npas4. The counts for Npas4+ cells, GAD67+ cells, TH+ cells, and CALR+ cells are all listed in Table 6 and Table 7. Scale bar: 50 μm and 10 μm.
Table 6.
The ratio of Npas4-expressing cells to the GAD67+ cells count in each layer of OB
| Layers Stages |
GL1 (Npas4+/GAD67) | GL2 (Npas4+/GAD67) | GL3 (Npas4+/GAD67) | GL ratio average |
|---|---|---|---|---|
| P14 | 18/22 | 28/31 | 29/30 | 0.896024 |
| Layers Stages |
MCL1 (Npas4+/GAD67) | MCL2 (Npas4+/GAD67) | MCL3 (Npas4+/GAD67) | MCL ratio average |
| P14 | 19/21 | 24/28 | 22/25 | 0.880634 |
| Layers Stages |
GCL1(Npas4+/GAD67) | GCL2 (Npas4+/GAD67) | GCL3 (Npas4+/GAD67) | GCL ratio average |
| P14 | 26/27 | 28/30 | 39/40 | 0.957098 |
Table 7.
The ratio of Npas4-expressing cells to the TH+ cells in GL and CALR+ cells in GCL
| Layers Stages |
GL1 (Npas4+/TH+) | GL2 (Npas4+/TH+) | GL3 (Npas4+/TH+) | GL ratio average |
|---|---|---|---|---|
| P14 | 6/8 | 5/7 | 7/9 | 0.747354 |
| Layers Stages |
GCL1 (Npas4+/CALR+) | GCL2 (Npas4+/CALR+) | GCL3 (Npas4+/CALR+) | GCL ratio average |
| P14 | 22/29 | 17/24 | 15/21 | 0.727079 |
It was reported that in the postnatal first and second week, newborn OB interneuron precursors (neuroblasts) are generated from the subventricular zone (SVZ). SVZ-derived precursors migrate to OB through the rostral migratory stream (RMS) and then differentiate into OB interneurons [38]. To check the relationship between Npas4 expression and SVZ-derived OB interneuron precursor cells during early postnatal period, we co-stained Npas4 with the RMS marker DCX antibody to examine the expression pattern. DCX was expressed throughout the entire OB layer, showing a pattern similar to Npas4 expression, with higher levels at P0 and P14 gradually decreasing by P28 and P56 (Fig. 4A, B). Since neurogenesis for OB interneurons continuously occurs in the adult stages, despite the overall decrease in DCX expression at P28 and P56, it was observed to remain relatively strong expression in the RMS (Fig. 4A, B). Furthermore, DCX-positive OB interneuron precursor cells exhibited high levels of Npas4 expression at P0 and P14 and reduced toward P28 and P56 and the expression of Npas4 in DCX-positive neuroblasts remains strong in the RMS, where adult neurogenesis occurs (Fig. 4C, D, Table 8).
Fig. 4.
The expression pattern of cells expressing Npas4 and DCX in OB layer during the postnatal developing period. (A) Representative images of the P0, P14, P28, and P56 olfactory bulbs. Cells are co-stained with the DCX, Tbr2, and DAPI and imaged using confocal microscopy. Blue represents nuclear DNA stained with DAPI, red represents DCX, and green represents Tbr2. Each layer of the olfactory bulb glomerular layer (GL), external plexiform layer (EPL), mitral cell layer (MCL), and granule cell layer (GCL) is indicated in each image. (B) A rectangular region of interest (ROI) of the same size was randomly placed in the image data of each layer and the intensity of DCX expressing cells was measured. (C) Representative images of the P0, P14, P28, and P56 olfactory bulbs. Cells are co-stained with the DCX, Npas4, and DAPI and imaged using confocal microscopy. Blue represents nuclear DNA stained with DAPI, red represents DCX, and green represents Npas4. (D) A rectangular region of interest (ROI) of the same size was randomly placed in the image data of each layer, and the ratio of Npas4 expressing cells to the DCX+ cells inside the ROI was counted. The counts for Npas4+ cells and DCX+ cells are all listed in Table 8. Scale bar: 100 μm.
Table 8.
The ratio of Npas4-expressing cells to the DCX+ cell count in each layer of OB
| Layers Stages |
GL1 (Npas4+/DCX+) | GL2 (Npas4+/DCX+) | GL3 (Npas4+/DCX+) | GL (%) average |
|---|---|---|---|---|
| P0 | 4/7 | 3/10 | 5/9 | 47.6 |
| P14 | 6/16 | 7/19 | 5/17 | 34.6 |
| P28 | 2/8 | 3/8 | 2/7 | 30.4 |
| P56 | 1/7 | 3/11 | 3/13 | 21.5 |
| Layers Stages |
MCL1 (Npas4+/DCX+) | MCL2 (Npas4+/DCX+) | MCL3 (Npas4+/DCX+) | MCL (%) average |
| P0 | 5/7 | 7/11 | 6/8 | 70.0 |
| P14 | 8/11 | 7/12 | 8/15 | 61.5 |
| P28 | 3/8 | 4/10 | 2/12 | 13.8 |
| P56 | 5/13 | 5/15 | 6/16 | 36.4 |
| Layers Stages |
GCL1 (Npas4+/DCX+) | GCL2 (Npas4+/DCX+) | GCL3 (Npas4+/DCX+) | GCL (%) average |
| P0 | 19/23 | 21/25 | 14/20 | 78.9 |
| P14 | 22/31 | 30/42 | 25/37 | 68.2 |
| P28 | 7/16 | 8/19 | 6/17 | 40.4 |
| P56 | 7/19 | 10/21 | 7/18 | 41.1 |
| Layers Stages |
RMS1 (Npas4+/DCX+) | RMS2 (Npas4+/DCX+) | RMS3 (Npas4+/DCX+) | RMS (%) average |
| P0 | 10/19 | 6/16 | 9/16 | 48.8 |
| P14 | 45/56 | 43/59 | 39/49 | 78.2 |
| P28 | 18/27 | 25/38 | 23/32 | 68.1 |
| P56 | 31/43 | 41/57 | 35/47 | 72.8 |
Taken together, these results indicate that during the early postnatal period, known as a critical time for OB interneuron development, Npas4 is expressed in the majority of inhibitory neurons and continues to be expressed in the RMS where adult neurogenesis persists. Also, the expression pattern of Npas4 during early postnatal OB development is consistent with the period in which newborn SVZ-produced neuroblast DCX-positive cells migrate along the RMS toward the OB.
Sensory stimulation-dependent OB development and Npas4 expression during early postnatal development
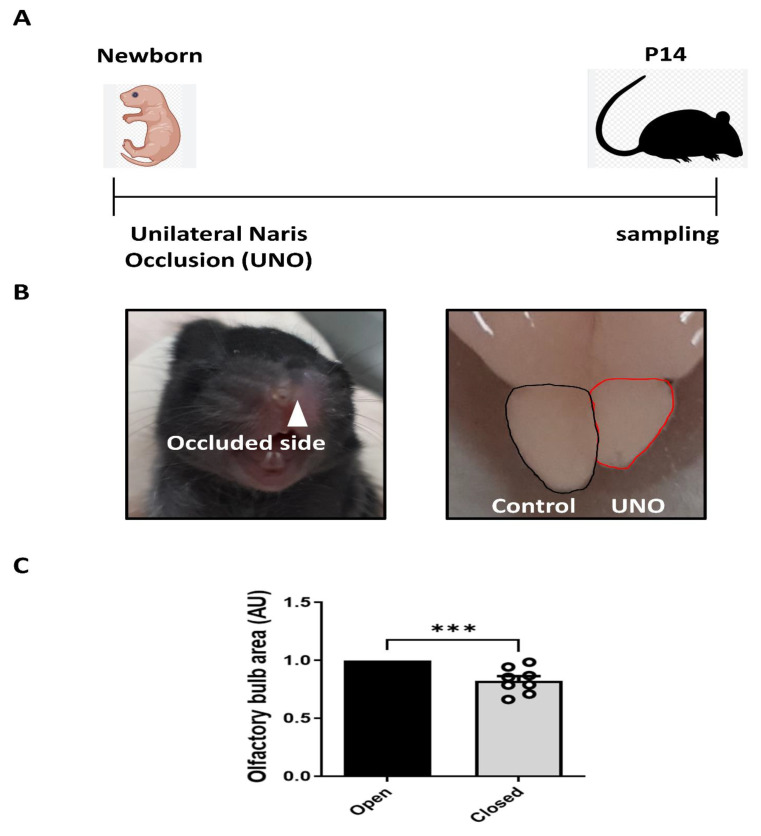
It has been reported that odor stimulation is essential for the development of the OB [48-50]. To determine whether stimulus-induced neuronal activity regulates OB development and Npas4 expression during the postnatal period, we performed a sensory deprivation test by unilateral naris occlusion in P0 mice and examined olfactory tissues at P14 (Fig. 5A, B), because most OB interneuron development occurs approximately 2 weeks after birth [35], and the level of Npas4 at P14 was significantly higher than that at the P28 and P56 stages seen above. To confirm the effect of sensory input on OB development during the early postnatal period, we measured and compared the dorsal area of the OB between the control and occluded sides. When measuring the OB dorsal area at P14, the ipsilateral occluded-side OB dorsal area was noted to have decreased by approximately 17.4% compared to the control side (Fig. 5C).
Fig. 5.
Effect of unilateral naris occlusion on the olfactory bulb’s development. (A) Schematic diagram of unilateral naris occlusion during postnatal olfactory bulb development. (B) Representative images of the mouse and the olfactory bulb occluded side. (C) The relative level of olfactory bulb size in open naris vs. closed naris. Error bars represent the mean±SE, and significance is determined using a t-test. (n=8, ***p<0.001).
Many IEGs and transcription factors are expressed in the OB, more so in the early stages than in the adult stages [51-54]. Since sensory stimulation significantly influences OB development, we examined the effect of odor deprivation on Npas4 expression during early postnatal OB development. To demonstrate the change in the Npas4 expression pattern caused by sensory deprivation, we performed immunoblotting and RT-qPCR analyses. We observed that Npas4 protein (Fig. 6A, B) but also Npas4 mRNA levels significantly decreased on the occluded side (Fig. 6C, D). To date, we have confirmed that Npas4 is expressed in the OB layers during the early postnatal period and that olfactory stimulation affects both OB development and Npas4 expression.
Fig. 6.
Effect of unilateral naris occlusion on Npas4 expression during postnatal olfactory bulb development. (A) Immunoblot of Npas4 in P14 olfactory bulbs isolated from the open and closed sides of the naris. (B) The relative level of Npas4 normalized by β-actin level (n=6, ****p<0.0001). (C) RT-qPCR products of Npas4 mRNAs from the open and closed sides of the naris in P14. (D) Quantification of the intensity of Npas4 normalized by β-actin level (n=6, **p<0.01).
Next, to clarify which types of OB neurons are affected by sensory deprivation during development, we examined the levels of both glutamic acid decarboxylase (GAD67), a marker of GABAergic interneurons, and vesicular glutamate transporter 1 (VGLUT1), a marker of glutamatergic neurons. Because it was observed that during the early postnatal OB interneuron development period, Npas4 expression was more prominent in inhibitory neurons than excitatory neurons, and confirmed through staining with Tbr2, GAD67 (Fig. 2, 3A). Furthermore, it was found to be highly expressed in various subtypes of OB interneurons by staining TH and CALR (Fig. 3B). Immunoblot analysis revealed that GAD67 expression was significantly reduced, but the expression of VGLUT1 was unchanged (Fig. 7A, B), indicating that olfactory stimulation in early postnatal OB interneuron development had a greater effect on inhibitory neurons than on excitatory neurons. Furthermore, to investigate which subtypes of OB interneurons are affected by sensory deprivation, we tested the expression levels of TH, a PGC marker, and CALR, a GC marker. TH protein levels were decreased by sensory deprivation, whereas CALR protein levels were slightly increased on the occluded side compared to the open side (Fig. 7C, D). These results implicate that olfactory sensory input at postnatal period might significantly influence the proper development of OB interneurons and their subtype composition.
Fig. 7.
Effect of unilateral naris occlusion on OB interneurons during postnatal olfactory bulb development. (A) Immunoblot of GAD67 and VGLUT1 in P14 olfactory bulbs isolated from the open and closed sides of the naris. (B) The relative levels of GAD67 and VGLUT1 normalized by β-actin level (n=11, ***p<0.001). (C) Immunoblots of tyrosine hydroxylase (TH) and calretinin (CALR) normalized by β-actin level in P14 olfactory bulbs isolated from the open and closed sides of the naris. (D) The relative levels of TH and CALR were normalized by β-actin level (n=11, **p<0.01, ****p<0.0001).
We observed in the previous findings that Npas4 is highly expressed in DCX-positive cells, SVZ-derived OB interneuron precursor cells, and also co-localizes with DCX in the RMS, where neurogenesis continues to occur in the adult stage (Fig. 4C, D). If the DCX-positive neuroblast cells in the RMS fail to migrate into the OB, they may fail to differentiate, leading to underdevelopment of the OB. To investigate whether defects in neuronal migration contribute to the developmental abnormalities of OB after sensory deprivation, we co-stained DCX, Tbr2, and Npas4 in the OB layers under sensory deprivation conditions to examine their expression patterns (Fig. 8A, B). Under sensory deprivation conditions, there was no significant change in the intensity of DCX observed in the OB layers (Fig. 8C), indicating minimal impact on neuronal migration, as confirmed by the positioning of cells expressing Tbr2 (Fig. 8A). Moreover, there was no variation in the number of DCX-positive OB interneuron precursor cells expressing Npas4 across the OB layers (Fig. 8D, Table 9). Conversely, a decrease in Npas4 intensity was noted all OB layers was observed, along with a trend of reduction in the number of cells expressing Npas4 (Fig. 8C, D, Table 10). Thus, the decrease in Npas4 due to sensory deprivation does not appear to affect neuronal migration or OB layer formation but may potentially influence the decrease in cell numbers and abnormal OB interneuron composition.
Fig. 8.
The expression pattern of cells expressing Npas4 and DCX in OB layer during the postnatal developing period under the unilateral naris occlusion. (A) Representative images in P14 olfactory bulbs isolated from the open and closed sides of the naris. Cells are co-stained with the DCX, Tbr2, and DAPI and imaged using confocal microscopy. Blue represents nuclear DNA stained with DAPI, red represents DCX, and green represents Tbr2. Each layer of the olfactory bulb glomerular layer (GL), external plexiform layer (EPL), mitral cell layer (MCL), and granule cell layer (GCL) is indicated in each image. (B) Representative images in P14 olfactory bulbs isolated from the open and closed sides of the naris. Cells are co-stained with the DCX, Npas4, and DAPI and imaged using confocal microscopy. Blue represents nuclear DNA stained with DAPI, red represents DCX, and green represents Npas4. (C) A rectangular region of interest (ROI) of the same size was randomly placed in the image data of each layer and the intensity of DCX expressing cells and Npas4 expressing cells was measured. in the open and closed side of the naris. (D) A rectangular region of interest (ROI) of the same size was randomly placed in the image data of each layer, and the ratio of Npas4 expressing cells to the DCX+ cells and total cells inside the ROI was counted. The counts for Npas4+ cells and DCX+ cells are all listed in Table 9 and Table 10. Scale bar: 100 μm.
Table 9.
The ratio of Npas4-expressing cells to the DCX+ cells in open and closed sides of naris
| Layers Stages |
GL1 (Npas4+/DCX+) | GL2 (Npas4+/DCX+) | GL3 (Npas4+/DCX+) | GL (%) average |
|---|---|---|---|---|
| Open | 6/17 | 9/20 | 6/15 | 40.1 |
| Closed | 9/25 | 11/25 | 9/21 | 41.0 |
| Layers Stages |
MCL1 (Npas4+/DCX+) | MCL2 (Npas4+/DCX+) | MCL3 (Npas4+/DCX+) | MCL (%) average |
| Open | 13/20 | 13/23 | 12/21 | 59.6 |
| Closed | 14/26 | 13/21 | 14/22 | 59.8 |
| Layers Stages |
GCL1 (Npas4+/DCX+) | GCL2 (Npas4+/DCX+) | GCL3 (Npas4+/DCX+) | GCL (%) average |
| Open | 15/25 | 19/30 | 13/20 | 62.8 |
| Closed | 13/20 | 15/23 | 15/27 | 61.9 |
| Layers Stages |
RMS1 (Npas4+/DCX+) | RMS2 (Npas4+/DCX+) | RMS3 (Npas4+/DCX+) | RMS (%) average |
| Open | 41/53 | 41/59 | 36/47 | 74.5 |
| Closed | 27/36 | 33/42 | 36/45 | 77.9 |
Table 10.
The ratio of Npas4-expressing cells to the total cells in open and closed sides of naris
| Layers Stages |
GL1 (Npas4+/total) | GL2 (Npas4+/total) | GL3 (Npas4+/total) | GL (%) average |
|---|---|---|---|---|
| Open | 17/28 | 20/35 | 15/26 | 58.5 |
| Closed | 25/56 | 26/55 | 21/46 | 45.9 |
| Layers Stages |
MCL1 (Npas4+/total) | MCL2 (Npas4+/total) | MCL3 (Npas4+/total) | MCL (%) average |
| Open | 20/35 | 23/38 | 21/36 | 58.7 |
| Closed | 26/52 | 21/45 | 22/46 | 48.2 |
| Layers Stages |
GCL1 (Npas4+/total) | GCL2 (Npas4+/total) | GCL3 (Npas4+/total) | GCL (%) average |
| Open | 25/35 | 30/39 | 20/28 | 73.3 |
| Closed | 20/32 | 23/38 | 27/48 | 59.8 |
| Layers Stages |
RMS1 (Npas4+/total) | RMS2 (Npas4+/total) | RMS3 (Npas4+/total) | RMS (%) average |
| Open | 53/67 | 59/75 | 47/60 | 78.7 |
| Closed | 36/55 | 42/65 | 45/69 | 65.1 |
Taken together, these results indicated that sensory stimulation plays a crucial role in OB development and Npas4 expression during early postnatal OB development. In addition, OB development by sensory input is mainly due to the development of OB interneurons. Different responses of OB interneuron composition in sensory deprivation conditions suggest that Npas4 might affect the regulation of the balance of OB interneuron circuit formation and neuroprotection, or have different molecular machinery for each subtype of OB interneurons.
Effect of sensory stimulation on the expression of putative Npas4 downstream molecules during early postnatal development
It has been reported that Npas4 has various functions in many brain regions (Table 11) [55-57], mainly related to neurogenesis, plasticity, and neuroprotection. In addition, Npas4 CHIP-seq and RNA-seq data suggest that Bdnf, Hdac9, and Nur77 are putative downstream molecules of Npas4 [32]. Thus, we hypothesized that aberrant OB development was due to the failure to adequately regulate the presumed downstream molecules of Npas4, resulting from decreased Npas4 expression by sensory deprivation during postnatal OB interneuron development. To test this possibility, we measured the mRNA levels of putative Npas4 target molecules (Fig. 9A). We observed that the mRNA expression levels of Bdnf, Hdac9, and Nur77, which are presumed to be downstream molecules of Npas4 and involved in cell survival and neural circuit formation [32, 34, 52, 58, 59], decreased on the unilateral naris occlusion side (Fig. 9B). Also, another putative downstream molecule of Npas4, the mRNA levels of Mdm2 and Dcx, which regulate sensory input-dependent dendritic spines of OB interneurons [60], did not change on the occluded side (Fig. 9C). In addition, for Syt10, which is known to promote the release of IGF-1 in OB [61], mRNA levels did not change (Fig. 9C).
Table 11.
Functions of Npas4 in brain regions
| Region of Npas4 expression | Functions of Npas4 | Reference |
|---|---|---|
| Whole brain | Npas4 KO mouse die earlier due to progressive brain damages. | Ooe et al., 2009 [32] |
| Cerebral cortex | Npas4 has a neuroprotection role in ischemic stroke. | Choy et al., 2015 [55]; Choy et al., 2016 [56] |
| Hippocampus | Npas4 regulates the expression of BDNF and c-fos in CA3 region. Npas4 and its downstream molecule Syt10 have a protective effect in the hippocampus. |
Woitecki et al., 2016 [33]; Ramamoorthi et al., 2011 [57] |
| Granule cells in OB | Npas4 upregulates the activity-dependent OB GC distal dendrite density. | Yoshihara et al., 2014 [28] |
| Medial ganglionic eminence (MGE) primary culture neurons, Cortical primary culture neurons | Npas4 has cell type-specific transcriptional functions. | Spiegel et al., 2014 [25] |
| Neuro2a cells | Npas4 has an important role in plasticity of neurons. | Yun et al., 2013 [59] |
Fig. 9.
Effect of unilateral naris occlusion on Npas4 putative downstream molecules’ mRNA levels during postnatal olfactory bulb development. (A) Putative downstream molecules of the Npas4-Network interaction image from STRING: functional protein association networks. (B) RT-qPCR products of Bdnf, Hdac9, Nur77, Mdm2, Dcx, and Syt10 mRNAs from open and closed sides of the naris in P14. (B) and (C) Quantification of the intensity of Bdnf, Hdac9, Nur77, Mdm2, Dcx, and Syt10 normalized by β-actin level (n=6, *p<0.05, **p<0.01).
Taken together, these results suggest that the olfactory stimulation-dependent Npas4 expression is involved in the developmental processes of OB interneurons by the establishment of neural circuits with excitatory projection cells or the survival of OB interneurons during the early stages of postnatal development.
DISCUSSION
In this study, we suggested that Npas4 is strongly expressed in OB interneurons during the postnatal period of early explosive OB development triggered by sensory-input, which implicates its potential involvement in the composition and survival of OB interneurons.
In previous reports and our result in Figure 2, Npas4 was predominantly expressed in inhibitory neurons, but it was also observed to be expressed at a lower ratio in the mitral/tufted cell layer. [20]. Therefore, changes in Npas4 expression may be measured in excitatory projection neurons under our experiment condition. However, the OB has a substantial proportion of interneurons to excitatory neurons (ratio 100:1) compared to other brain regions (ratio 1:5) [35] and most excitatory projection neurons in the OB are generated during embryonic stages [62]. Therefore, we assume that Npas4 expressed in excitatory neurons during postnatal period can be expected to be involved in neuronal functions and not in the development of excitatory neurons.
Sensory experience regulates the development of various nervous systems in the brain [63]. Similarly, the olfactory sensory experience facilitates the development, dendritic morphogenesis, spine formation, and survival of OB interneurons [64, 65]. In this study, the Npas4 expression in P14 olfactory bulb was reduced in the absence of olfactory stimulation, but the neuronal development tendency of the two interneurons was different. The number of OB interneurons expressed GAD67 was reduced, and the level of TH-positive cells was decreased. In contrast, the number of CALR-positive cells increased, resulting in a change in the OB interneuron ratio on the occluded side. Despite significantly reduced Npas4 expression, the underlying cause of the differing developmental patterns among interneurons during the early developmental stages remains unknown. One possibility is the differences in the synaptic organization among interneurons. Periglomerular cells receive direct input from olfactory sensory neurons, whereas inputs to granule cells are received from synapses from both mitral and tufted cells. The differences in synaptic organization may affect the developmental responsiveness of interneurons involved in Npas4 expression. Another possibility is the difference in the position of the two interneurons. PGCs, TH-positive interneurons, are usually located in the GL, the most superficial layer of the OB. In contrast, GCs, CALR-positive interneurons, are located in the GCL. The GCL is a deep layer of the OB. We assume that the effect of sensory stimulation on neuronal activation may affect OB interneuron subtypes differently, depending on spatial and positional differences. Therefore, it is expected that there will be differences in the activity and function of Npas4.
Npas4 is a transcription factor translated from neuronal activity-dependent IGEs. It has been reported that Npas4 regulates not only the transcription of various genes that control inhibitory synapse development and synaptic plasticity [47], but also the expression of several downstream molecules, including BDNF, HDAC9, and Nur77 [28, 32]. Recently, it has been reported that BDNF regulates the formation of neural circuits and dendrite morphology in developing GABAergic interneurons, including TH-positive and CALR-positive cells, by enhancing synaptic activity [66], and can be synthesized in various GABAergic cells [67]. In addition, activity-dependent secretion of BDNF is critical for regulating adult OB interneuron neurogenesis [68]. These results indicate that olfactory sensory input during postnatal period might significantly influence proper development [69]. BDNF is expressed in the RMS and suggested as a chemoattractant for SVZ neuroblasts migration [69]. Migrating neuroblasts in RMS directly contact to blood vessels, and BDNF released from vascular endothelial cells modulates migration in RMS. In the nasal occlusion experiments, it was confirmed that BDNF is a putative downstream molecule of Npas4. Therefore, BDNF is expected to be involved in synapse formation and development of OB interneurons by regulating the migration of neuroblasts in the RMS through interaction with Npas4. HDACs are enzymes that remove acetyl groups and regulate DNA expression. HDACs are reported to be involved in the expression of TH in the OB and RMS [70] and neuronal rewiring with BDNF [71]. Furthermore, HDAC2 may modulate inhibitory circuit activity by enhancing synaptic plasticity in parvalbumin interneurons in the mouse visual cortex [72]. Nuclear receptor 4A1 (NR4a1, also known as Nur77) is involved in the cell cycle, inflammation, and apoptosis [73]. Recently, Nur77 was shown to regulate the transcription of synaptic formation molecules in inhibitory GABAergic interneurons of the forebrain [74]. It is necessary to promote and complete striatal developmental maturation by altering cholinergic interneuronal activity [75]. Our study did not directly demonstrate the downstream localization of BDNF, HDAC9, and Nur77 molecules to Npas4. However, based on previously published results, there is sufficient evidence to suggest that Npas4 interacts with the aforementioned molecules and could provide various information and research topics to explain the results of our experiment. These reports suggest that Npas4 expression induced by olfactory stimulation during early postnatal OB development may play an essential role in correctly forming synapses and circuits of OB interneurons by regulating putative downstream molecules of Npas4.
It was reported that Npas4 regulates Mdm2 expression to ubiquitinate and degrade DCX during dendritic spine development in newborn OB GCs after sensory stimulation in at P21, a period similar to our experimental condition at P28, during adolescence [28]. Also, it was known that phosphorylation of Npas4 enhances the transcriptional activity of Npas4 and BDNF promoter activity resulting in regulation of synaptic plasticity and enhancement of reward-related learning and memory [47]. Under our test conditions, phosphorylated Npas4 showed strong expression from P0 to P14, but there were no changes in the expression of Mdm2 and Dcx mRNA, as well as in the intensity of DCX. Although our experimental results differ from those obtained in other experiments conducted during P21 adolescence and adult stages, we propose the possibility that Npas4 may have another function in neuronal composition and neuroprotection during the OB interneuron developmental process.
In summary, our results provide evidence that Npas4 expression in newborn OB interneurons contributes to the formation of neural circuits with mitral/tufted cells during early postnatal OB development by regulating their survival under sensory input-dependent conditions (Fig. 10).
Fig. 10.
Schematic summary of sensory-input dependent Npas4 expression in the olfactory bulb during postnatal development.
ACKNOWLEDGEMENTS
Kwon, O.-H., Choe, J., Kim, D., Kim, S., and Moon, C. designed the study; Kim, D., and Kim, S. prepared the samples and performed the experiments; Kwon, O.-H., Choe, J., and Kim, S. analyzed the data; Kwon, O.-H., and Moon, C. managed the project and wrote the initial manuscript. All authors discussed the results and commented on the final manuscript. This work was supported by the Ministry of Education (2020R1A6A1A03040516; RS-2023-00239274) and the Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Science and ICT (2020R1A2C2005174).
References
- 1.Carvell GE, Simons DJ. Abnormal tactile experience early in life disrupts active touch. J Neurosci. 1996;16:2750–2757. doi: 10.1523/JNEUROSCI.16-08-02750.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frasnelli J, Collignon O, Voss P, Lepore F. Crossmodal plasticity in sensory loss. Prog Brain Res. 2011;191:233–249. doi: 10.1016/B978-0-444-53752-2.00002-3. [DOI] [PubMed] [Google Scholar]
- 3.Grubb MS, Thompson ID. The influence of early experience on the development of sensory systems. Curr Opin Neurobiol. 2004;14:503–512. doi: 10.1016/j.conb.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell DE. The extent of visual recovery from early monocular or binocular visual deprivation in kittens. J Physiol. 1988;395:639–660. doi: 10.1113/jphysiol.1988.sp016939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Xu H, Kotak VC, Sanes DH. Conductive hearing loss disrupts synaptic and spike adaptation in developing auditory cortex. J Neurosci. 2007;27:9417–9426. doi: 10.1523/JNEUROSCI.1992-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Argandoña EG, Lafuente JV. Effects of dark-rearing on the vascularization of the developmental rat visual cortex. Brain Res. 1996;732:43–51. doi: 10.1016/0006-8993(96)00485-4. [DOI] [PubMed] [Google Scholar]
- 7.Briner A, De Roo M, Dayer A, Muller D, Kiss JZ, Vutskits L. Bilateral whisker trimming during early postnatal life impairs dendritic spine development in the mouse somatosensory barrel cortex. J Comp Neurol. 2010;518:1711–1723. doi: 10.1002/cne.22297. [DOI] [PubMed] [Google Scholar]
- 8.Kral A, Eggermont JJ. What's to lose and what's to learn: development under auditory deprivation, cochlear implants and limits of cortical plasticity. Brain Res Rev. 2007;56:259–269. doi: 10.1016/j.brainresrev.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 9.Rochefort C, Gheusi G, Vincent JD, Lledo PM. Enriched odor exposure increases the number of newborn neurons in the adult olfactory bulb and improves odor memory. J Neurosci. 2002;22:2679–2689. doi: 10.1523/JNEUROSCI.22-07-02679.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Butz M, Wörgötter F, van Ooyen A. Activity-dependent structural plasticity. Brain Res Rev. 2009;60:287–305. doi: 10.1016/j.brainresrev.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 11.Fox K, Wong RO. A comparison of experience-dependent plasticity in the visual and somatosensory systems. Neuron. 2005;48:465–477. doi: 10.1016/j.neuron.2005.10.013. [DOI] [PubMed] [Google Scholar]
- 12.House DR, Elstrott J, Koh E, Chung J, Feldman DE. Parallel regulation of feedforward inhibition and excitation during whisker map plasticity. Neuron. 2011;72:819–831. doi: 10.1016/j.neuron.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pouille F, Marin-Burgin A, Adesnik H, Atallah BV, Scanziani M. Input normalization by global feedforward inhibition expands cortical dynamic range. Nat Neurosci. 2009;12:1577–1585. doi: 10.1038/nn.2441. [DOI] [PubMed] [Google Scholar]
- 14.Benes FM, Vincent SL, Molloy R, Khan Y. Increased interaction of dopamine-immunoreactive varicosities with GABA neurons of rat medial prefrontal cortex occurs during the postweanling period. Synapse. 1996;23:237–245. doi: 10.1002/(SICI)1098-2396(199608)23:4<237::AID-SYN1>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 15.Caballero A, Flores-Barrera E, Cass DK, Tseng KY. Differential regulation of parvalbumin and calretinin interneurons in the prefrontal cortex during adolescence. Brain Struct Funct. 2014;219:395–406. doi: 10.1007/s00429-013-0508-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kilb W. Development of the GABAergic system from birth to adolescence. Neuroscientist. 2012;18:613–630. doi: 10.1177/1073858411422114. [DOI] [PubMed] [Google Scholar]
- 17.Vincent SL, Pabreza L, Benes FM. Postnatal maturation of GABA-immunoreactive neurons of rat medial prefrontal cortex. J Comp Neurol. 1995;355:81–92. doi: 10.1002/cne.903550110. [DOI] [PubMed] [Google Scholar]
- 18.Ashwin C, Chapman E, Howells J, Rhydderch D, Walker I, Baron-Cohen S. Enhanced olfactory sensitivity in autism spectrum conditions. Mol Autism. 2014;5:53. doi: 10.1186/2040-2392-5-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marco EJ, Hinkley LB, Hill SS, Nagarajan SS. Sensory processing in autism: a review of neurophysiologic findings. Pediatr Res. 2011;69(5 Pt 2):48R–54R. doi: 10.1203/PDR.0b013e3182130c54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bepari AK, Watanabe K, Yamaguchi M, Tamamaki N, Takebayashi H. Visualization of odor-induced neuronal activity by immediate early gene expression. BMC Neurosci. 2012;13:140. doi: 10.1186/1471-2202-13-140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kosofsky BE, Genova LM, Hyman SE. Postnatal age defines specificity of immediate early gene induction by cocaine in developing rat brain. J Comp Neurol. 1995;351:27–40. doi: 10.1002/cne.903510104. [DOI] [PubMed] [Google Scholar]
- 22.McCormack MA, Rosen KM, Villa-Komaroff L, Mower GD. Changes in immediate early gene expression during postnatal development of cat cortex and cerebellum. Brain Res Mol Brain Res. 1992;12:215–223. doi: 10.1016/0169-328X(92)90087-R. [DOI] [PubMed] [Google Scholar]
- 23.Pérez-Cadahía B, Drobic B, Davie JR. Activation and function of immediate-early genes in the nervous system. Biochem Cell Biol. 2011;89:61–73. doi: 10.1139/O10-138. [DOI] [PubMed] [Google Scholar]
- 24.Ooe N, Saito K, Mikami N, Nakatuka I, Kaneko H. Identification of a novel basic helix-loop-helix-PAS factor, NXF, reveals a Sim2 competitive, positive regulatory role in dendritic-cytoskeleton modulator drebrin gene expression. Mol Cell Biol. 2004;24:608–616. doi: 10.1128/MCB.24.2.608-616.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spiegel I, Mardinly AR, Gabel HW, Bazinet JE, Couch CH, Tzeng CP, Harmin DA, Greenberg ME. Npas4 regulates excitatory-inhibitory balance within neural circuits through cell-type-specific gene programs. Cell. 2014;157:1216–1229. doi: 10.1016/j.cell.2014.03.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shepard R, Heslin K, Hagerdorn P, Coutellier L. Downregulation of Npas4 in parvalbumin interneurons and cognitive deficits after neonatal NMDA receptor blockade: relevance for schizophrenia. Transl Psychiatry. 2019;9:99. doi: 10.1038/s41398-019-0436-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yang J, Serrano P, Yin X, Sun X, Lin Y, Chen SX. Functionally distinct NPAS4-expressing somatostatin interneuron ensembles critical for motor skill learning. Neuron. 2022;110:3339–3355.e8. doi: 10.1016/j.neuron.2022.08.018. [DOI] [PubMed] [Google Scholar]
- 28.Yoshihara S, Takahashi H, Nishimura N, Kinoshita M, Asahina R, Kitsuki M, Tatsumi K, Furukawa-Hibi Y, Hirai H, Nagai T, Yamada K, Tsuboi A. Npas4 regulates Mdm2 and thus Dcx in experience-dependent dendritic spine development of newborn olfactory bulb interneurons. Cell Rep. 2014;8:843–857. doi: 10.1016/j.celrep.2014.06.056. [DOI] [PubMed] [Google Scholar]
- 29.Lin Y, Bloodgood BL, Hauser JL, Lapan AD, Koon AC, Kim TK, Hu LS, Malik AN, Greenberg ME. Activity-dependent regulation of inhibitory synapse development by Npas4. Nature. 2008;455:1198–1204. doi: 10.1038/nature07319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maya-Vetencourt JF, Tiraboschi E, Greco D, Restani L, Cerri C, Auvinen P, Maffei L, Castrén E. Experience-dependent expression of NPAS4 regulates plasticity in adult visual cortex. J Physiol. 2012;590:4777–4787. doi: 10.1113/jphysiol.2012.234237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, Markenscoff-Papadimitriou E, Kuhl D, Bito H, Worley PF, Kreiman G, Greenberg ME. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ooe N, Motonaga K, Kobayashi K, Saito K, Kaneko H. Functional characterization of basic helix-loop-helix-PAS type transcription factor NXF in vivo: putative involvement in an "on demand" neuroprotection system. J Biol Chem. 2009;284:1057–1063. doi: 10.1074/jbc.M805196200. [DOI] [PubMed] [Google Scholar]
- 33.Woitecki AM, Müller JA, van Loo KM, Sowade RF, Becker AJ, Schoch S. Identification of synaptotagmin 10 as effector of NPAS4-mediated protection from excitotoxic neurodegeneration. J Neurosci. 2016;36:2561–2570. doi: 10.1523/JNEUROSCI.2027-15.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang SJ, Zou M, Lu L, Lau D, Ditzel DA, Delucinge-Vivier C, Aso Y, Descombes P, Bading H. Nuclear calcium signaling controls expression of a large gene pool: identification of a gene program for acquired neuroprotection induced by synaptic activity. PLoS Genet. 2009;5:e1000604. doi: 10.1371/journal.pgen.1000604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bayer SA. 3H-thymidine-radiographic studies of neurogenesis in the rat olfactory bulb. Exp Brain Res. 1983;50:329–340. doi: 10.1007/BF00239197. [DOI] [PubMed] [Google Scholar]
- 36.Adam Y, Mizrahi A. Circuit formation and maintenance--perspectives from the mammalian olfactory bulb. Curr Opin Neurobiol. 2010;20:134–140. doi: 10.1016/j.conb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 37.Kaneko N, Marín O, Koike M, Hirota Y, Uchiyama Y, Wu JY, Lu Q, Tessier-Lavigne M, Alvarez-Buylla A, Okano H, Rubenstein JL, Sawamoto K. New neurons clear the path of astrocytic processes for their rapid migration in the adult brain. Neuron. 2010;67:213–223. doi: 10.1016/j.neuron.2010.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lledo PM, Merkle FT, Alvarez-Buylla A. Origin and function of olfactory bulb interneuron diversity. Trends Neurosci. 2008;31:392–400. doi: 10.1016/j.tins.2008.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sakamoto M, Imayoshi I, Ohtsuka T, Yamaguchi M, Mori K, Kageyama R. Continuous neurogenesis in the adult forebrain is required for innate olfactory responses. Proc Natl Acad Sci U S A. 2011;108:8479–8484. doi: 10.1073/pnas.1018782108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Whitman MC, Greer CA. Adult neurogenesis and the olfactory system. Prog Neurobiol. 2009;89:162–175. doi: 10.1016/j.pneurobio.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mirich JM, Williams NC, Berlau DJ, Brunjes PC. Comparative study of aging in the mouse olfactory bulb. J Comp Neurol. 2002;454:361–372. doi: 10.1002/cne.10426. [DOI] [PubMed] [Google Scholar]
- 42.Rall W, Shepherd GM, Reese TS, Brightman MW. Dendrodendritic synaptic pathway for inhibition in the olfactory bulb. Exp Neurol. 1966;14:44–56. doi: 10.1016/0014-4886(66)90023-9. [DOI] [PubMed] [Google Scholar]
- 43.Katz LC, Shatz CJ. Synaptic activity and the construction of cortical circuits. Science. 1996;274:1133–1138. doi: 10.1126/science.274.5290.1133. [DOI] [PubMed] [Google Scholar]
- 44.Lepousez G, Valley MT, Lledo PM. The impact of adult neurogenesis on olfactory bulb circuits and computations. Annu Rev Physiol. 2013;75:339–363. doi: 10.1146/annurev-physiol-030212-183731. [DOI] [PubMed] [Google Scholar]
- 45.Nithianantharajah J, Hannan AJ. Enriched environments, experience-dependent plasticity and disorders of the nervous system. Nat Rev Neurosci. 2006;7:697–709. doi: 10.1038/nrn1970. [DOI] [PubMed] [Google Scholar]
- 46.Sanes JR, Lichtman JW. Induction, assembly, maturation and maintenance of a postsynaptic apparatus. Nat Rev Neurosci. 2001;2:791–805. doi: 10.1038/35097557. [DOI] [PubMed] [Google Scholar]
- 47.Funahashi Y, Ariza A, Emi R, Xu Y, Shan W, Suzuki K, Kozawa S, Ahammad RU, Wu M, Takano T, Yura Y, Kuroda K, Nagai T, Amano M, Yamada K, Kaibuchi K. Phosphorylation of Npas4 by MAPK regulates reward-related gene expression and behaviors. Cell Rep. 2019;29:3235–3252.e9. doi: 10.1016/j.celrep.2019.10.116. [DOI] [PubMed] [Google Scholar]
- 48.Takahashi H, Yoshihara S, Tsuboi A. The functional role of olfactory bulb granule cell subtypes derived from embryonic and postnatal neurogenesis. Front Mol Neurosci. 2018;11:229. doi: 10.3389/fnmol.2018.00229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Brunjes PC. Unilateral odor deprivation: time course of changes in laminar volume. Brain Res Bull. 1985;14:233–237. doi: 10.1016/0361-9230(85)90088-7. [DOI] [PubMed] [Google Scholar]
- 50.Frazier LL, Brunjes PC. Unilateral odor deprivation: early postnatal changes in olfactory bulb cell density and number. J Comp Neurol. 1988;269:355–370. doi: 10.1002/cne.902690304. [DOI] [PubMed] [Google Scholar]
- 51.Bloodgood BL, Sharma N, Browne HA, Trepman AZ, Greenberg ME. The activity-dependent transcription factor NPAS4 regulates domain-specific inhibition. Nature. 2013;503:121–125. doi: 10.1038/nature12743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Johnson RS, Spiegelman BM, Papaioannou V. Pleiotropic effects of a null mutation in the c-fos proto-oncogene. Cell. 1992;71:577–586. doi: 10.1016/0092-8674(92)90592-Z. [DOI] [PubMed] [Google Scholar]
- 53.Korte M, Carroll P, Wolf E, Brem G, Thoenen H, Bonhoeffer T. Hippocampal long-term potentiation is impaired in mice lacking brain-derived neurotrophic factor. Proc Natl Acad Sci U S A. 1995;92:8856–8860. doi: 10.1073/pnas.92.19.8856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mori K, Nagao H, Yoshihara Y. The olfactory bulb: coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- 55.Choy FC, Klaric TS, Koblar SA, Lewis MD. The role of the neuroprotective factor Npas4 in cerebral ischemia. Int J Mol Sci. 2015;16:29011–29028. doi: 10.3390/ijms161226144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choy FC, Klaric TS, Leong WK, Koblar SA, Lewis MD. Reduction of the neuroprotective transcription factor Npas4 results in increased neuronal necrosis, inflammation and brain lesion size following ischaemia. J Cereb Blood Flow Metab. 2016;36:1449–1463. doi: 10.1177/0271678X15606146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ramamoorthi K, Fropf R, Belfort GM, Fitzmaurice HL, Mckinney RM, Neve RL, Otto T, Lin Y. Npas4 regulates a transcriptional program in CA3 required for contextual memory formation. Science. 2011;334:1669–1675. doi: 10.1126/science.1208049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Xiao G, Sun T, Songming C, Cao Y. NR4A1 enhances neural survival following oxygen and glucose deprivation: an in vitro study. J Neurol Sci. 2013;330:78–84. doi: 10.1016/j.jns.2013.04.010. [DOI] [PubMed] [Google Scholar]
- 59.Yun J, Nagai T, Furukawa-Hibi Y, Kuroda K, Kaibuchi K, Greenberg ME, Yamada K. Neuronal Per Arnt Sim (PAS) domain protein 4 (NPAS4) regulates neurite outgrowth and phosphorylation of synapsin I. J Biol Chem. 2013;288:2655–2664. doi: 10.1074/jbc.M112.413310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Yoshihara S, Takahashi H, Nishimura N, Naritsuka H, Shirao T, Hirai H, Yoshihara Y, Mori K, Stern PL, Tsuboi A. 5T4 glycoprotein regulates the sensory input-dependent development of a specific subtype of newborn interneurons in the mouse olfactory bulb. J Neurosci. 2012;32:2217–2226. doi: 10.1523/JNEUROSCI.5907-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cao P, Maximov A, Südhof TC. Activity-dependent IGF-1 exocytosis is controlled by the Ca(2+)-sensor synaptotagmin-10. Cell. 2011;145:300–311. doi: 10.1016/j.cell.2011.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tufo C, Poopalasundaram S, Dorrego-Rivas A, Ford MC, Graham A, Grubb MS. Development of the mammalian main olfactory bulb. Development. 2022;149:dev200210. doi: 10.1242/dev.200210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Voss MW, Vivar C, Kramer AF, van Praag H. Bridging animal and human models of exercise-induced brain plasticity. Trends Cogn Sci. 2013;17:525–544. doi: 10.1016/j.tics.2013.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Livneh Y, Feinstein N, Klein M, Mizrahi A. Sensory input enhances synaptogenesis of adult-born neurons. J Neurosci. 2009;29:86–97. doi: 10.1523/JNEUROSCI.4105-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Saghatelyan A, Roux P, Migliore M, Rochefort C, Desmaisons D, Charneau P, Shepherd GM, Lledo PM. Activity-dependent adjustments of the inhibitory network in the olfactory bulb following early postnatal deprivation. Neuron. 2005;46:103–116. doi: 10.1016/j.neuron.2005.02.016. [DOI] [PubMed] [Google Scholar]
- 66.Nieto-Estévez V, Defterali Ç, Vicario C. Distinct effects of BDNF and NT-3 on the dendrites and presynaptic boutons of developing olfactory bulb GABAergic interneurons in vitro. Cell Mol Neurobiol. 2022;42:1399–1417. doi: 10.1007/s10571-020-01030-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Barreda Tomás FJ, Turko P, Heilmann H, Trimbuch T, Yanagawa Y, Vida I, Münster-Wandowski A. BDNF expression in cortical GABAergic interneurons. Int J Mol Sci. 2020;21:1567. doi: 10.3390/ijms21051567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bath KG, Mandairon N, Jing D, Rajagopal R, Kapoor R, Chen ZY, Khan T, Proenca CC, Kraemer R, Cleland TA, Hempstead BL, Chao MV, Lee FS. Variant brain-derived neurotrophic factor (Val66Met) alters adult olfactory bulb neurogenesis and spontaneous olfactory discrimination. J Neurosci. 2008;28:2383–2393. doi: 10.1523/JNEUROSCI.4387-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chiaramello S, Dalmasso G, Bezin L, Marcel D, Jourdan F, Peretto P, Fasolo A, De Marchis S. BDNF/ TrkB interaction regulates migration of SVZ precursor cells via PI3-K and MAP-K signalling pathways. Eur J Neurosci. 2007;26:1780–1790. doi: 10.1111/j.1460-9568.2007.05818.x. [DOI] [PubMed] [Google Scholar]
- 70.Akiba Y, Cave JW, Akiba N, Langley B, Ratan RR, Baker H. Histone deacetylase inhibitors de-repress tyrosine hydroxylase expression in the olfactory bulb and rostral migratory stream. Biochem Biophys Res Commun. 2010;393:673–677. doi: 10.1016/j.bbrc.2010.02.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sada N, Fujita Y, Mizuta N, Ueno M, Furukawa T, Yamashita T. Inhibition of HDAC increases BDNF expression and promotes neuronal rewiring and functional recovery after brain injury. Cell Death Dis. 2020;11:655. doi: 10.1038/s41419-020-02897-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nott A, Cho S, Seo J, Tsai LH. HDAC2 expression in parvalbumin interneurons regulates synaptic plasticity in the mouse visual cortex. Neuroepigenetics. 2015;1:34–40. doi: 10.1016/j.nepig.2014.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pei L, Castrillo A, Tontonoz P. Regulation of macrophage inflammatory gene expression by the orphan nuclear receptor Nur77. Mol Endocrinol. 2006;20:786–794. doi: 10.1210/me.2005-0331. [DOI] [PubMed] [Google Scholar]
- 74.Huang M, Pieraut S, Cao J, de Souza Polli F, Roncace V, Shen G, Maximov A. Nr4a1 regulates inhibitory circuit structure and function in the mouse brain. bioRxiv. 2022 doi: 10.1101/2022.06.14.496205. doi: 10.1101/2022.06.14.496205. [DOI] [Google Scholar]
- 75.Cirnaru MD, Melis C, Fanutza T, Naphade S, Tshilenge KT, Muntean BS, Martemyanov KA, Plotkin JL, Ellerby LM, Ehrlich ME. Nuclear receptor Nr4a1 regulates striatal striosome development and dopamine D1 receptor signaling. eNeuro. 2019;6:ENEURO.0305-19.2019. doi: 10.1523/ENEURO.0305-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]