Abstract
Background
Cepheid’s Xpert HIV‐1 Viral Load (Xpert VL), a simplified, automated, single-use quantitative assay used with the GeneXpert System, is not FDA-approved.
Objectives
Using stored plasma, we conducted a study to assess the ability of Xpert VL to quantify viral load relative to the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 (Cobas VL) and to examine the use of the Xpert VL as a qualitative diagnostic test.
Study Design
Following HIV-1 viral stock dilutions, we conducted a probit analysis to identify the concentration where 95% of specimens had quantified VLs. We also examined Xpert and Cobas log VL correlation in linearity panels; compared the proportion of 220 seroconverter specimens with virus detected using McNemar’s test; and tested specimens from persons with untreated, established HIV-1 infection (n=149) and uninfected persons (n=497). Furthermore, we examined Xpert VL as a qualitative test in seroconverter specimens with early (n=20) and later (n=68) acute infections.
Results
At 1.80 log10 copies/mL, 95% of specimens had quantifiable virus using Xpert VL. Xpert and Cobas VLs were highly correlated (R2=0.994). The proportion of seroconverter specimens with virus detected using Cobas and with Xpert VL was not statistically different (p=0.0578). Xpert VL detected 97.9% of established infections, and specificity was 99.80% (95% CI 98.87%−99.99%). Xpert VL detected 90% and 98.5% of early and later acute infections, respectively.
Conclusions
If approved, Xpert VL could allow U.S. laboratories that cannot bring on large, complex testing platforms to conduct HIV monitoring. An approval for diagnostic use may provide timely identification of HIV infections.
Keywords: HIV-1, nucleic acid test, viral load, diagnostic
Background
Viral load (VL) tests are used to monitor HIV infection to assess treatment adherence and viral suppression. (1–4) VL testing may occur at a local laboratory. However, due to testing complexity, specimens are often sent to referral laboratories, which may delay results. Though Food and Drug Administration (FDA)-approved VL tests exist,(5) therapeutic monitoring could be conducted more efficiently with fewer manual procedures using a simplified nucleic acid test (NAT). Use of a simplified NAT may expedite clinical decisions and counselling. In individuals failing to maintain viral suppression (e.g., ≥200 copies/mL (2.3 log10)), simplified NATs may alert clinicians to the need to change therapeutic regimens.(4)
A simplified quantitative assay, Xpert HIV‐1 Viral Load (Xpert VL; Cepheid, Sunnyvale, CA), targets the long terminal repeat region (LTR) of the HIV‐1 genome using reverse transcription real-time polymerase chain reaction in a self-contained cartridge that uses 1–1.2mLs of plasma. It provides results after 92 minutes with the GeneXpert System, which automates sample preparation, nucleic acid extraction, amplification, and detection of the target sequence. The Xpert VL includes reagents for the detection of HIV-1 RNA and internal controls for HIV-1 RNA quantitation and monitoring inhibitors in the RT and PCR reactions.(6) Xpert VL has not been approved by FDA, but 20 tests are available for use in the United States on this system.(7) The test was approved in the European Union.(8) The Xpert VL analytical range is from 1.6 to 7.0 log10 copies/mL.(9)
Simplified NATs are also needed for identification of acute infections, for use as supplemental tests after reactive screening tests, and as an aid in HIV diagnosis. Only one NAT, APTIMA HIV-1 RNA Qualitative Assay (APTIMA; Hologic Inc., San Diego, CA), has been FDA-approved for use as an aid in diagnosis, but it is a complex test that must be performed manually, and takes approximately five or more hours to perform. Other VL tests can be used for diagnosis if laboratory validation has occurred, or if ordered by a physician as part of clinical care.
Objectives
We conducted a study to assess whether the performance of the Xpert VL was comparable with the Roche COBAS AmpliPrep/COBAS TaqMan HIV-1 Version 2.0 (Cobas VL) for HIV-1 quantification, and to examine the use of Xpert VL as an aid in diagnosis.
Study Design
Detection of low concentrations of HIV using viral stock
We performed serial dilutions of a Virology Quality Assurance HIV-1 viral stock to obtain 4, 3, 2.7, 2.3, 2, 1.9, 1.8, 1.6, 1.3, and 0 log10 copies/ml, and determined the percent of 25 replicates detected at 1.3 and 1.6 log10 copies/mL (20 and 40 copies/mL, respectively). We also conducted a probit analysis to identify the concentration at which 95% of specimens had virus quantified with Xpert VL.
Reproducibility evaluation using linearity panel specimens
A technician tested specimens from the HIV-1 RNA AccuSpan linearity panel (Seracare, 2410–0221) with Xpert VL over three days using two kit lots. Another technician tested the panel over three days with a third kit lot. The linear panel product insert provided Cobas VL data. We calculated the average Xpert VL for each linearity panel member. Xpert VL values that were below or above the limit of quantification were considered to be at the limit of quantification (e.g., > 7.0 log10 copies/mL was set to 7.0 log10 copies/mL).
Linearity
Using the linearity panel specimens, we plotted the log VL for Xpert and Cobas VLs among those with a quantifiable VL, and calculated the correlation.
Xpert VL sensitivity using HIV-1 seroconverter specimens
We tested seroconversion panels from 25 antiretroviral-naïve persons (n=220 specimens) obtained from Zeptometrix, Inc. (Buffalo, NY, USA) and BBI-SeraCare Diagnostics (Milford, MA, USA) to examine assay sensitivity during seroconversion.(10, 11) These specimens were presumed to be infected with subtype B HIV-1. They had at least one specimen with indeterminate or positive results on the HIV-1 Western blot (WB) and at least one specimen with a positive result on an HIV-1 RNA test.
For seroconverter specimens, testing with the Cobas VL was performed on a different aliquot, approximately 10 years before Xpert VL testing. We compared the proportion of specimens with virus detected with Cobas and Xpert VLs using McNemar’s test, and calculated the Kappa coefficient. We repeated this analysis after raising the threshold for detection to 1.6 log10 copies/mL. We described the proportion of specimens with an Xpert VL ≥2.3 and 3.0 log10 copies/ml when the Cobas VL was at least 2.3 and 3.0 log10 copies/ml, respectively. The threshold recommended for resistance testing is 3.0 log10 copies/mL (1,000 copies/mL).(4)
Detection of established HIV-1 infection in specimens from antiretroviral-treated and -untreated persons
We tested 149 plasma specimens from persons with established HIV-1 infection (i.e., antibody positive) who were not being treated with antiretrovirals (12, 13). Parallel serum specimens from the same individuals tested positive with an antigen/antibody test, Western blot, and a NAT. We also tested plasma from 73 antiretroviral-treated persons with HIV-1-positive results using antigen/antibody and HIV-1/HIV-2 antibody differentiation tests, (14) presumed to be subtype B. Specimens were tested with Xpert and Cobas VLs in parallel.
HIV-1 non- B subtypes and Group O
We tested 97 specimens with positive HIV-1 antibody and NAT results from Cameroon with Xpert and Cobas VLs in parallel. The samples were previously sequenced in the envelope gp41 and/or gag p17 regions and subtyped as A (n=23), HIV circulating recombinant form (CRF) 01_AE (n=2), CRF02_AG (n=51), CRF09_cpx (n=1), CRF11_cpx (n=7), D (n=1), F1 (n=1), F2 (n=4), G (n=5) and Group O (n=2).(15) We examined the correlation between the Xpert and Cobas VLs among specimens with a quantifiable VL.
Bland Altman analysis
We conducted a Bland Altman analysis on 374 specimens with quantifiable VLs from seroconverters, established HIV-1 infections, and HIV-1 non-B subtype and group O infections.
Assay specificity
We tested specimens (n=497) that were nonreactive by an antigen/antibody test, Western blot and NAT to evaluate specificity. (16, 17)
Qualitative diagnostic use of Xpert VL in specimens from seroconverters and persons with established infections
We evaluated qualitative Xpert VL results among a subset of specimens described above: (1) 20 seroconverter specimens reactive by NAT and non-reactive by an antigen/antibody test; (2) 68 seroconverter specimens reactive by an antigen/antibody test and NAT but negative or indeterminate with an HIV-1/HIV-2 differentiation antibody test; and (3) 143 specimens from persons with established infection not on treatment. These specimens were reactive using APTIMA, the only HIV-1 NAT approved for diagnostic use. Xpert VL results that were quantifiable or detectable but not quantifiable (<40 copies/mL) were considered reactive.
Technical errors
We examined the proportion of Xpert VL technical errors within the specimen sets utilized for this study.
Analyses were conducted using SAS version 9.4 (Cary, NC).
Results
Detection of low concentrations of HIV using viral stock
At 1.3 log10 copies/mL, 23/25 (92%) diluted viral stock replicates had Xpert VL results with virus detected but not in the quantifiable range, and two (8%) did not have virus detected. At 1.6 log10 copies/mL, Xpert VL detected virus in all replicates; 16 were detectable but not quantifiable and the average VL of the others was 1.7 log10 copies/mL. At 1.80 log10 copies/mL (63.10 copies/mL), 95% CI (1.74–1.90 log10 copies/mL) 95% of Xpert VL results were quantifiable among replicates.
Reproducibility and linearity evaluations using linearity panel specimens
Table 1 reports the average Xpert VL at each concentration of Cobas VL. Cobas and Xpert VLs were highly correlated (R2=0.994) (Figure 1).
Table 1.
Average Xpert VL and standard deviation using linear panel specimens previously tested with Cobas VL
| Linearity Panel Member and Average Cobas VL (log10 copies/mL) | Average Xpert VLa (log10 copies/mL) and Standard Deviation |
|---|---|
| 1) 7.91 | 7.00 (0) |
| 2) 6.72 | 6.91 (0.03) |
| 3) 5.85 | 5.87 (0.03) |
| 4) 4.96 | 4.92 (0.05) |
| 5) 4.22 | 3.93 (0.05) |
| 6) 3.07 | 2.88 (0.08) |
| 7) 1.91 | 1.81 (0.16) |
| 8) < 1.30 | 0.53 (0.80) |
| 9) Target not detected | 0 (0) |
| 10) Target not detected | 0.18 (0.53) |
9 iterations
Figure 1.
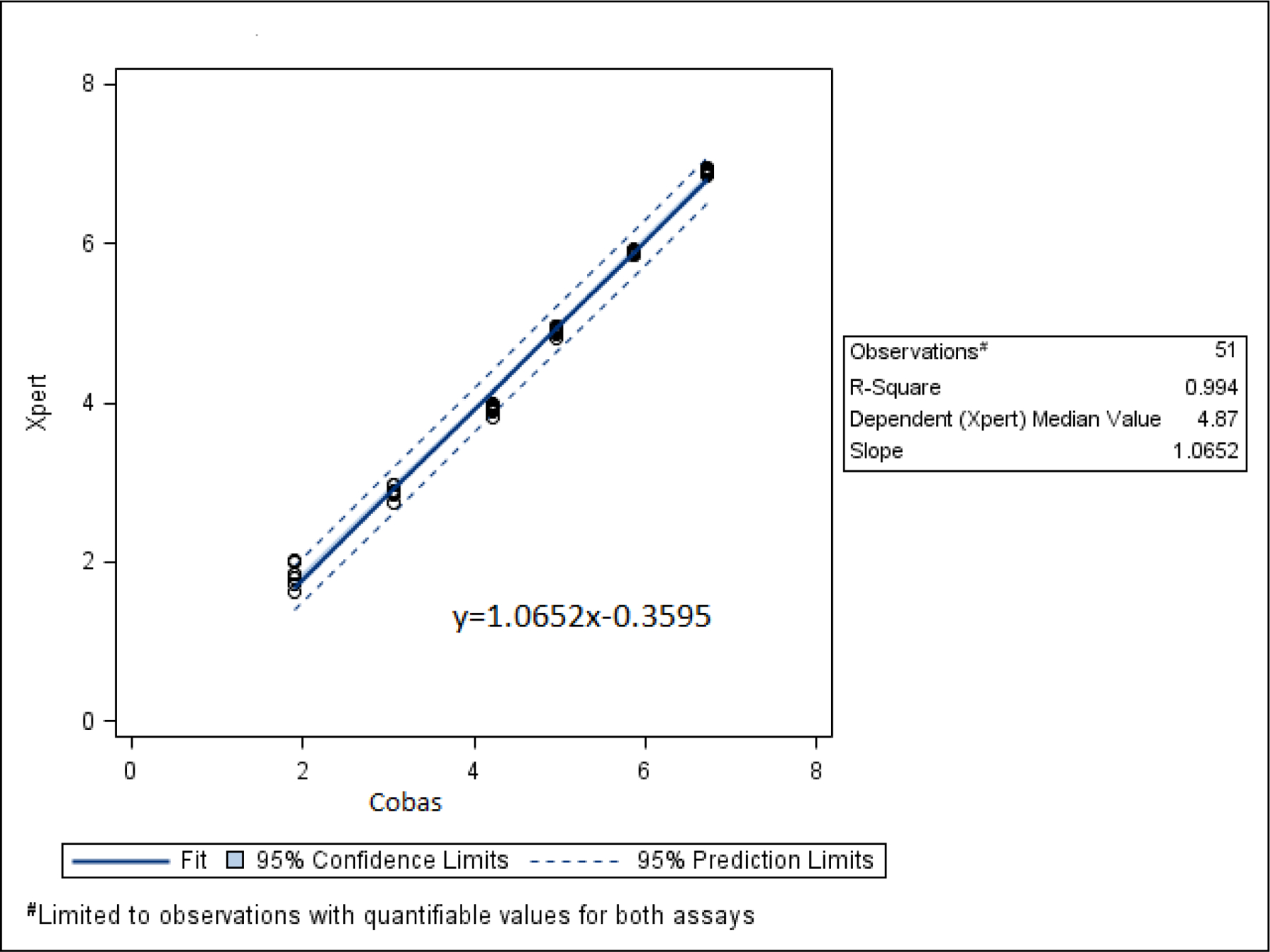
Linear regression of Xpert and Cobas log10 VLs among linearity panel specimens with VLs that were quantifiable by both tests. The Xpert and Cobas VLs were highly correlated with an R2 value of 0.994.
Xpert VL sensitivity using seroconverter specimens
Of 152 seroconverter specimens with virus detected by Cobas VL, 144 (94.7%) had virus detected by Xpert VL (Table 2).
Table 2.
Virus detection using Cobas VL and Xpert VL in seroconverter specimens
| Virus detected using Xpert VL | Virus not detected using Xpert VL | Total | |
|---|---|---|---|
| Virus detected using Cobas VL | 144 (94.7%) | 8 (5.3%)a | 152 |
| Virus not detected using Cobas VL | 2 (3.3%)b | 59 (96.7%) | 61 |
Two had virus detected that was not in the quantifiable range (Cobas VL <1.3 log10 copies/mL or 20 copies/mL) and six had Cobas VL between 1.37 and 1.62 log10 copies/mL.
Both had detectable but not quantifiable Xpert VL results (i.e., <1.6 log10 copies/mL or 40 copies/mL).
Cobas and Xpert VL test results were not statistically different (p=0.0578). The Kappa coefficient was 0.89 95% C.I. (0.82, 0.96). When the threshold was raised to 1.6 log10 copies/mL, the Kappa coefficient was 0.94 95% C.I. (0.89, 0.99).
There were Xpert VL errors among seroconverter specimens, including three assay-specific termination errors, a digital temperature reading error, and two operator errors. There was also an invalid result. These specimens lacked volume for repeat testing.
Most seroconverter specimens had concordant results at the 2.3 and 3.0 log10 thresholds with few exceptions. There were 9/213 (4.2%) specimens with Cobas VL ≥2.3 log10 copies/mL that were <2.3 with Xpert VL. Xpert VL ranged from < 1.6 to 2.1 log10 copies/mL. Likewise, 15/213 (7%) specimens had a Cobas VL that was ≥3.0 log10 copies/mL but <3.0 with Xpert VL. Xpert VL were between 1.6 and 2.9 log10 copies/mL.
Established HIV-1 infection
Xpert VL detected most untreated, established infections (140/143, 97.9%). The Cobas VLs of the specimens with virus not detected by Xpert VL were 1.3, 4.7 and 5.5 log10 copies/mL. There were five invalid results and an error; none had repeat testing.
Of 71 specimens from persons with treated, established infections 34 (47.9%) had detectable Cobas VL results, 24 (70.6%) of which had detectable Xpert VL results. Of those with undetectable Xpert VL results, 9 had detectable but not quantifiable Cobas VLs and 1 had a VL of 1.5 log10 copies/mL. Of 37 treated, established infections not detected by Cobas VL, 8 (21.6%) were detected but not quantified using Xpert VL. Two specimens with invalid Xpert VL results did not have volume for repeat testing.
HIV-1 non- B subtypes and Group O
Xpert and Cobas VLs among specimens with non-B subtypes and Group O infections were correlated at R2 of 0.7832. Xpert VL values overestimated Cobas results in these specimens (data not shown).
Bland Altman Analysis
Based on Bland Altman analysis, the difference between Xpert and Cobas VLs was evenly distributed around zero at all mean concentrations of virus, and typically was within one log, though most were within half a log. (Figure 2)
Figure 2.
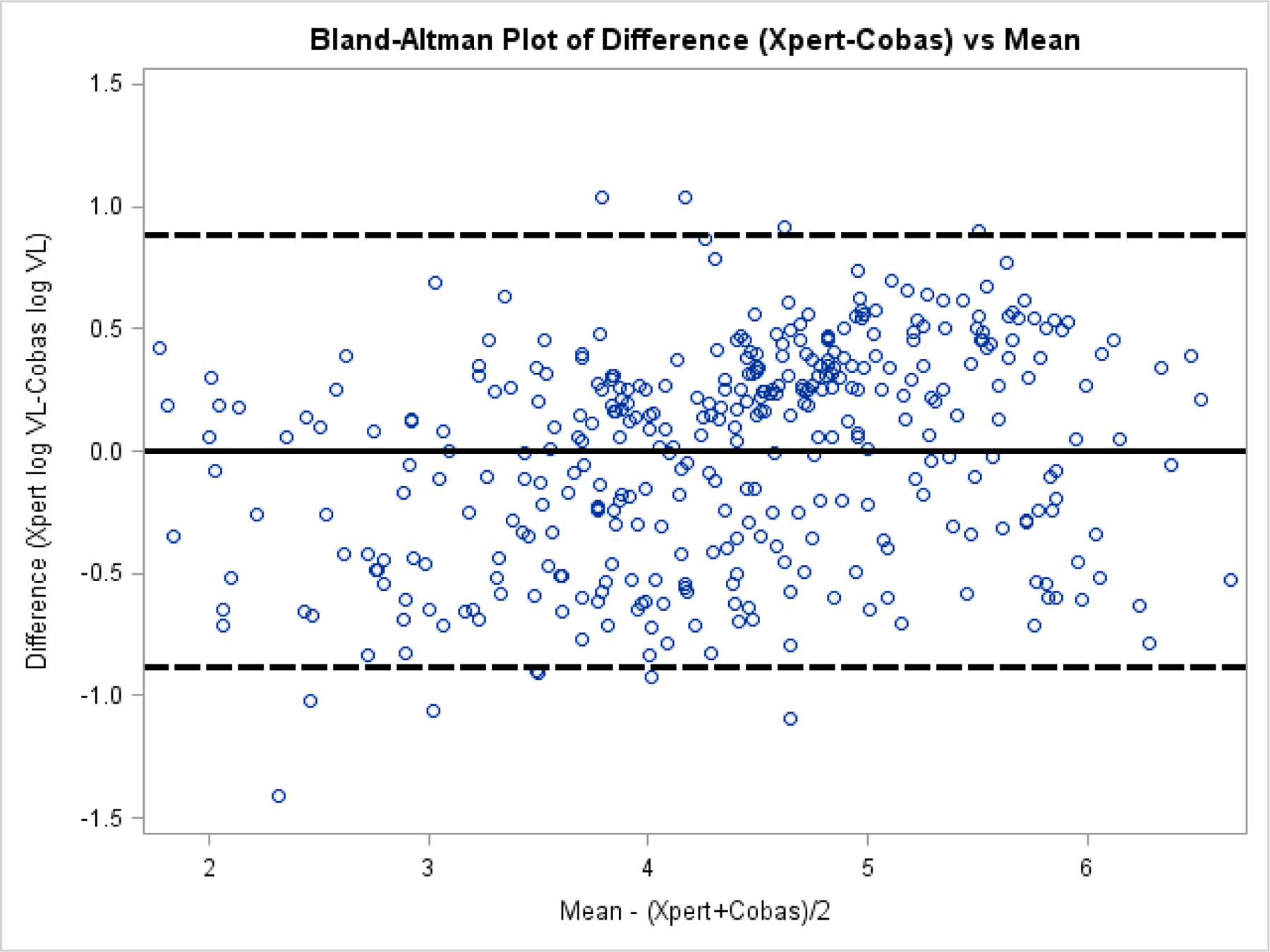
Bland Altman plot of the difference of Cepheid Xpert log10 VL and the Cobas log10 VL against the mean log10 VL of specimens with quantifiable VLs from seroconverters, persons with established HIV-1 infections, and persons with HIV-1 non-B subtype and group O infections. The difference between Xpert and Cobas VLs was evenly distributed around zero at all mean concentrations of virus, and typically was within one log, though most were within half a log. The solid line is the mean of the difference. The hatched lines are at + and – 2 standard deviations of the mean of the difference.
Specificity
Xpert VL specificity was high (99.80%; 95% confidence interval (CI) 98.87%−99.99%). The only Xpert VL false positive result was detectable but not quantifiable. Eight specimens with insufficient volume for repeat testing gave errors. Three specimens with initial errors did not have virus detected on repeat.
Qualitative diagnostic use of Xpert VL
Of 20 seroconverter specimens with virus detected by NAT that had negative results on the antigen/antibody test, 18 (90%) had virus detected using Xpert VL. In seroconverter specimens with reactive results on an antigen/antibody test and NAT, and negative or indeterminate results on an HIV-1/HIV-2 antibody differentiation test, 67 of 68 (98.5%) had virus detected by Xpert VL. Xpert VL detected most (97.9%) untreated, established HIV-1 infections.
Technical errors
No technical errors or invalid results were reported within the viral stock, linearity panel or non-B subtype specimen sets. The other specimen sets had errors or invalid results in 2.2% to 4% of specimens: (2.2%, 11/497) uninfected specimens, (2.7%, 2/73) established HIV-1 infections on treatment, 3.2% (7/220) seroconverter specimens, 4% (6/149) established HIV-1 infections not on treatment.
Discussion
As demonstrated in similar studies examining the performance of Xpert VL, including among diverse subtypes, Xpert VL results were highly correlated and concordant with Cobas VL. (18–22) In our small panel of non-B subtypes and Group Os, Xpert overestimated Cobas VL. Lower concordance has been reported for the CRF02_AG subtype variant, which accounted for almost half of the non-B subtypes evaluated in our study.(22) We were unable to assess the performance of Xpert VL with subtype C specimens, though studies conducted in areas where it is predominant suggest that it performs well.(23)
Xpert VL detected almost 95% of early infections. Seroconverter specimens with virus not detected using Xpert VL had VL levels under 2.0 log10 copies/mL (100 copies/mL). Most replicates (92%) had detectable VL at 1.3 log10 copies/mL, close to the limit of detection reported by the manufacturer and the World Health Organization (6, 9) The concentration of virus at which 95% of replicates were quantified was slightly above the limit of quantification from the package insert, 1.6 log10 copies/mL. This difference in analytical sensitivity estimates may be due to a dilution error. Xpert VL results were reproducible.
There was a small degree of misclassification at the threshold for treatment failure, 2.3 log10 copies/mL. Cobas VL targets both gag and LTR regions of the genome, instead of LTR only, which may account for some discrepant results. Approximately 4% of seroconverters above this threshold by Cobas VL were below it with Xpert VL. For the misclassified specimens, follow-up VL testing may have identified treatment failure if it existed. Viral suppression below the level of detection of the HIV RNA assay used is optimal, so persons with detectable virus would likely be monitored closely by their provider.(4) At 3.0 log10 copies/mL, (4) one study reported 3% misclassification by Xpert VL compared to Cobas VL and Abbott HIV-1 RealTime (Abbott HIV-1 RT) (Abbott Molecular, Des Plaines, Illinois) (20) while another reported 8% misclassification relative to Abbott.(19) In our study, 7% of seroconverters with a Cobas VL at or above 3.0 log10 copies/mL had VLs below that level with Xpert VL. Of 15 misclassified specimens, five had Xpert VL results between 2.7 to 3.0 log10 copies/mL, which may be sufficient for amplification for resistance testing.(4)
Most untreated, established infections (98%) were detected by Xpert VL, though two specimens with Cobas VL results near 5.0 log10 copies/mL did not have virus detected using Xpert VL. Specimen mix-up could not be ruled out, but repeat testing was not possible. Alternatively, sequence variation in the target binding sites or other molecular factors could account for the discordance. Specimens with virus not detected by Xpert VL from persons with established infections treated with antiretrovirals had low Cobas VL results (1.5 log10 copies/mL or less) as did specimens with detectable virus when using Xpert VL but not Cobas VL. Optimal treatment results in a sustained, undetectable viral load, but isolated, low level detectable virus is not predictive of failure, thus variation in test results within and across tests in specimens with low viral load may not be clinically meaningful.(4)
The Xpert VL performed well as a qualitative test, suggesting it could be used as an aid in the diagnosis of HIV-1 infection at multiple points in a testing algorithm. It may be used as a screening test for high-risk persons or persons with a recent exposure, as it detected most infections in seroconverters. It has high specificity, though further testing would be needed to resolve false-positive results. Xpert VL could be used as a supplemental test based on the high proportion of specimens from seroconverter and established infection specimens reactive on an antigen/antibody test and with virus detected. Using a NAT as the second test in the laboratory algorithm may expedite patient receipt of results and decrease time to treatment and VL suppression. In the rare case that the Xpert VL is negative after a reactive antigen/antibody test result, an additional antibody test may be needed.(24, 25) Outside the U.S., Cepheid produces the Xpert HIV-1 Qual assay, which does not quantify detectable virus. When used on serum from uninfected persons and persons with acute infection, it correctly classified the infection status of all specimens tested.(26)
Simplified NATs can reduce the time for patients’ receipt of results; a sexual health clinic reduced receipt of results by eight days when using the Xpert HIV-1 Qual assay for CT/NG NAT.(27) Diagnostic use of the Xpert VL is desirable, though its run time may not be conducive to real-time provision of results. When a “sample-first” clinic flow approach was adopted only around one-fifth of patients received results from the CT/NG NAT before they left the office.(28) Nevertheless, Xpert VL results are likely to be available before manual NAT alternatives.
This study had some limitations. We used stored specimens, so specimen degradation may have occurred. It is unclear whether assay errors took place due to specimen storage duration or conditions, though specimens were handled similarly for the VL tests being compared. Error rates as high as 4%, similar to those observed in some specimen sets in our study, have been reported. (29) For the quantitative assessment, this study was limited to one comparator assay, Cobas VL, which may be less accurate at lower viral loads than other comparators, such as the Abbott RealTime HIV-1 assay.(30)
Availability of the Xpert VL in the United States is contingent on FDA review, a process that may become simpler.(31) FDA approval could allow laboratories that cannot bring on a large testing platform to conduct HIV VL monitoring. Further, approval of the test for diagnostic use may provide timely identification of infections, including during the acute stage.
Highlights.
Xpert HIV‐1 Viral Load is a simplified, automated, single-use quantitative assay.
Cobas and Xpert VLs were highly correlated (R2 = 0.994).
Xpert VL detected 97.9 % of established infections, and specificity was 99.80 %.
Xpert VL detected 90 % and 98.5 % of early and later acute infections, respectively.
Xpert VL performs proficiently for quantitative and qualitative uses.
Acknowledgements
We thank Cheryl Jennings at Rush University for providing the HIV-1 VQA stock.
The findings and conclusions in this report are those of the authors and do not necessarily represent the views of the Centers for Disease Control and Prevention.
Funding:
Testing supplies were provided by Cepheid to CDC as part of a federal notice of collaboration for manufacturers of nucleic acid tests. Cepheid did not have a role in the collection, analysis and interpretation of data, in writing the report, or in the decision to submit the article for publication. Ms. Smith was supported in part by an appointment to the Research Participation Program at the CDC administered by the Oak Ridge Institute for Science and Education through an interagency agreement between the U.S. Department of Energy and CDC.
Footnotes
Author credit
Laura Wesolowski conceptualization; methodology; software; formal analysis; data curation; Writing – Original Draft; Writing – Review & Editing; Visualization; Supervision; Project Administration
Silvina Masciotra conceptualization; methodology; software; formal analysis; resources; data curation; Writing – Original Draft; Writing – Review & Editing; Supervision; Project Administration
Kevin Delaney conceptualization; methodology; software; formal analysis; data curation; Writing – Original Draft; Writing – Review & Editing; Visualization; Supervision; Project Administration
William Switzer conceptualization; methodology; resources; Writing – Original Draft; Writing –Review & Editing; Supervision
S. Michele Owen conceptualization; methodology; Writing – Original Draft; Writing – Review & Editing; Supervision
William Fowler validation; investigation; resources; data curation; Writing – Review & Editing
Wei Luo validation; investigation; resources; data curation; Writing – Review & Editing
Vickie Sullivan investigation; resources; data curation; Writing – Review & Editing
Tara Smith investigation; resources; data curation; Writing – Review & Editing
Rebecca Rossetti investigation; resources data curation; Writing – Review & Editing
Steven Ethridge investigation, resources, data curation; Writing – Review & Editing
Pollyanna Chavez Conceptualization; Writing – Original Draft; Writing – Review & Editing
Emeka Oraka software; formal analysis; data curation; Writing – Review & Editing; Visualization
Disclosure of relationship: The Centers for Disease Control and Prevention (CDC) and our content experts wish to disclose that they have no financial interests or other relationships with the manufacturers of commercial products, suppliers of commercial services, or commercial supporters. Trade names are used for identification purposes only. Their use does not imply endorsement by CDC.
Competing interests: None declared
Ethical approval: The study was considered to be research not involving identifiable human subjects by the National Center for HIV/AIDS, Viral Hepatitis, STD and TB Prevention, CDC.
References
- 1.O’Brien WA, Hartigan PM, Martin D, Esinhart J, Hill A, Benoit S, Rubin M, Simberkoff MS, Hamilton JD. 1996. Changes in plasma HIV-1 RNA and CD4+ lymphocyte counts and the risk of progression to AIDS. Veterans Affairs Cooperative Study Group on AIDS. N Engl J Med 334:426–31. [DOI] [PubMed] [Google Scholar]
- 2.Ruiz L, Romeu J, Clotet B, Balague M, Cabrera C, Sirera G, Ibanez A, Martinez-Picado J, Raventos A, Tural C, Segura A, Foz M. 1996. Quantitative HIV-1 RNA as a marker of clinical stability and survival in a cohort of 302 patients with a mean CD4 cell count of 300 × 10(6)/l. AIDS 10:F39–44. [DOI] [PubMed] [Google Scholar]
- 3.Saag MS, Holodniy M, Kuritzkes DR, O’Brien WA, Coombs R, Poscher ME, Jacobsen DM, Shaw GM, Richman DD, Volberding PA. 1996. HIV viral load markers in clinical practice. Nat Med 2:625–9. [DOI] [PubMed] [Google Scholar]
- 4.Panel on Antiretroviral Guidelines for Adults and Adolescents. 2019. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents., on Department of Health and Human Services. http://aidsinfo.nih.gov/contentfiles/lvguidelines/adultandadolescentgl.pdf. Accessed 08/23/2019.
- 5.U.S. Food and Drug Administration. Infectious Disease Tests: HIV-1 Nucleic Acid Testing. https://www.fda.gov/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/ucm126582.htm. Accessed 07/03/2018.
- 6.World Health Organization. 2017. WHO Prequalification of In Vitro Diagnostics: Product Xpert HIV-1 Viral Load with GeneXpert Dx. https://www.who.int/diagnostics_laboratory/evaluations/pq-list/hiv-vrl/170720_final_pq_report_pqdx_0192_0193_0194_0195_070-00.pdf?ua=1. Accessed 08/23/2019.
- 7.Cepheid. 2017. Cepheid Announces FDA Clearance Of Xpert® Xpress Flu And Xpert Xpress Flu/RSV http://www.cepheid.com/us/about-us/news-events/press-releases/217-cepheid-announces-fda-clearance-of-xpert-xpress-flu-and-xpert-xpress-flu-rsvCepheid Announces FDA Clearance Of Xpert® Xpress Flu And Xpert Xpress Flu/RSV Accessed 10/02/2017.
- 8.PRNewswire. 2014. Cepheid and FIND Announce European Approval of Xpert HIV-1 Viral Load. http://www.prnewswire.com/news-releases/cepheid-and-find-announce-european-approval-of-xpert-hiv-1-viral-load-300014112.html. Accessed 10/02/2017.
- 9.Cepheid. 2019. Xpert HIV-1 Viral Load Data Sheet. http://www.cepheid.com/administrator/components/com_productcatalog/library-files/f3d173a26b0ba1763775f9d1c9606297-e9f319c1b7c611fc41fe17cfa76f19ff-Xpert-HIV-1-Viral-Load-Datasheet-3038-02.pdf. Accessed 01/17/2019.
- 10.Masciotra S, McDougal JS, Feldman J, Sprinkle P, Wesolowski L, Owen SM. 2011. Evaluation of an alternative HIV diagnostic algorithm using specimens from seroconversion panels and persons with established HIV infections. J Clin Virol 52 Suppl 1:S17–22. [DOI] [PubMed] [Google Scholar]
- 11.Nasrullah M, Wesolowski LG, Meyer WA 3rd, Owen SM, Masciotra S, Vorwald C, Becker WJ, Branson BM. 2013. Performance of a fourth-generation HIV screening assay and an alternative HIV diagnostic testing algorithm. AIDS 27:731–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wesolowski LG, Delaney KP, Hart C, Dawson C, Owen SM, Candal D, Meyer WA 3rd, Ethridge SF, Branson BM. 2011. Performance of an alternative laboratory-based algorithm for diagnosis of HIV infection utilizing a third generation immunoassay, a rapid HIV-1/HIV-2 differentiation test and a DNA or RNA-based nucleic acid amplification test in persons with established HIV-1 infection and blood donors. J Clin Virol 52 Suppl 1:S45–9. [DOI] [PubMed] [Google Scholar]
- 13.Chavez P, Wesolowski L, Patel P, Delaney K, Owen SM. 2011. Evaluation of the performance of the Abbott ARCHITECT HIV Ag/Ab Combo Assay. J Clin Virol 52 Suppl 1:S51–5. [DOI] [PubMed] [Google Scholar]
- 14.Delaney KP, Katz D, Ure G, Wesolowski LG, McDougal S, Chavez PR, Bowles K, Mcmahan V, Ethridge SF, Swenson P, Pitasi M, DiNenno EA, Owen SM, Stekler J. 2016. Evaluation of New HIV Testing Technologies in a Clinical Setting with High Incidence: Rationale, Study Design and Preliminary Results from Project DETECT. HIV Diagnostics Conference. [Google Scholar]
- 15.Fonjungo PN, Mpoudi EN, Torimiro JN, Alemnji GA, Eno LT, Lyonga EJ, Nkengasong JN, Lal RB, Rayfield M, Kalish ML, Folks TM, Pieniazek D. 2002. Human immunodeficiency virus type 1 group m protease in Cameroon: genetic diversity and protease inhibitor mutational features. J Clin Microbiol 40:837–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Delaney KP, Branson BM, Uniyal A, Phillips S, Candal D, Owen SM, Kerndt PR. 2011. Evaluation of the performance characteristics of 6 rapid HIV antibody tests. Clin Infect Dis 52:257–63. [DOI] [PubMed] [Google Scholar]
- 17.Delaney KP, Ethridge SF, Adams S, Luo W, Masciotra S, Wesolowski LG, Owen SM. 2016. Evaluation of Newly Approved HIV Antigen-Antibody Tests Individually and When Used in the CDC/APHL HIV Diagnostic Algorithm HIV Diagnostics Conference. [Google Scholar]
- 18.Gueudin M, Baron A, Alessandri-Gradt E, Lemee V, Mourez T, Etienne M, Plantier JC. 2016. Performance Evaluation of the New HIV-1 Quantification Assay, Xpert HIV-1 Viral Load, on a Wide Panel of HIV-1 Variants. J Acquir Immune Defic Syndr 72:521–6. [DOI] [PubMed] [Google Scholar]
- 19.Ceffa S, Luhanga R, Andreotti M, Brambilla D, Erba F, Jere H, Mancinelli S, Giuliano M, Palombi L, Marazzi MC. 2016. Comparison of the Cepheid GeneXpert and Abbott M2000 HIV-1 real time molecular assays for monitoring HIV-1 viral load and detecting HIV-1 infection. J Virol Methods 229:35–9. [DOI] [PubMed] [Google Scholar]
- 20.Gous N, Scott L, Berrie L, Stevens W. 2016. Options to Expand HIV Viral Load Testing in South Africa: Evaluation of the GeneXpert(R) HIV-1 Viral Load Assay. PLoS One 11:e0168244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nash M, Huddart S, Badar S, Baliga S, Saravu K, Pai M. 2018. Performance of the Xpert HIV-1 Viral Load Assay: a Systematic Review and Meta-analysis. J Clin Microbiol 56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Avidor B, Matus N, Girshengorn S, Achsanov S, Gielman S, Zeldis I, Schweitzer I, Adler A, Turner D. 2017. Comparison between Roche and Xpert in HIV-1 RNA quantitation: A high concordance between the two techniques except for a CRF02_AG subtype variant with high viral load titters detected by Roche but undetected by Xpert. J Clin Virol 93:15–19. [DOI] [PubMed] [Google Scholar]
- 23.Swathirajan CR, Vignesh R, Boobalan J, Solomon SS, Saravanan S, Balakrishnan P. 2017. Performance of point-of-care Xpert HIV-1 plasma viral load assay at a tertiary HIV care centre in Southern India. 66:1379–1382. [DOI] [PubMed] [Google Scholar]
- 24.Owen SM, Yang C, Spira T, Ou CY, Pau CP, Parekh BS, Candal D, Kuehl D, Kennedy MS, Rudolph D, Luo W, Delatorre N, Masciotra S, Kalish ML, Cowart F, Barnett T, Lal R, McDougal JS. 2008. Alternative algorithms for human immunodeficiency virus infection diagnosis using tests that are licensed in the United States. J Clin Microbiol 46:1588–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Patel P, Mackellar D, Simmons P, Uniyal A, Gallagher K, Bennett B, Sullivan TJ, Kowalski A, Parker MM, LaLota M, Kerndt P, Sullivan PS, Centers for Disease C, Prevention Acute HIVISG. 2010. Detecting acute human immunodeficiency virus infection using 3 different screening immunoassays and nucleic acid amplification testing for human immunodeficiency virus RNA, 2006–2008. Arch Intern Med 170:66–74. [DOI] [PubMed] [Google Scholar]
- 26.Michaeli M, Wax M, Gozlan Y, Rakovsky A, Mendelson E, Mor O. 2016. Evaluation of Xpert HIV-1 Qual assay for resolution of HIV-1 infection in samples with negative or indeterminate Geenius HIV-1/2 results. J Clin Virol 76:1–3. [DOI] [PubMed] [Google Scholar]
- 27.Whitlock GG, Gibbons DC, Longford N, Harvey MJ, McOwan A, Adams EJ. 2018. Rapid testing and treatment for sexually transmitted infections improve patient care and yield public health benefits. Int J STD AIDS 29:474–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Harding-Esch EM, Nori AV, Hegazi A, Pond MJ, Okolo O, Nardone A, Lowndes CM, Hay P, Sadiq ST. 2017. Impact of deploying multiple point-of-care tests with a ‘sample first’ approach on a sexual health clinical care pathway. A service evaluation. Sex Transm Infect 93:424–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ndlovu Z, Fajardo E, Mbofana E, Maparo T, Garone D, Metcalf C, Bygrave H, Kao K, Zinyowera S. 2018. Multidisease testing for HIV and TB using the GeneXpert platform: A feasibility study in rural Zimbabwe. PLoS One 13:e0193577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Amendola A, Marsella P, Bloisi M, Forbici F, Angeletti C, Capobianchi MR. 2014. Ability of two commercially available assays (Abbott RealTime HIV-1 and Roche Cobas AmpliPrep/Cobas TaqMan HIV-1 Version 2.0) to quantify low HIV-1 RNA Levels (<1,000 copies/milliliter): comparison with clinical samples and NIBSC working reagent for nucleic acid testing assays. J Clin Microbiol 52:2019–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Orr E 2018. FDA Panel Backs Eased HIV Test Regs. https://medtech.pharmaintelligence.informa.com/MT123001/FDA-Panel-Backs-Eased-HIV-Test-Regs. Accessed [Google Scholar]


