Abstract
We characterized hepatitis B virus (HBV) isolates from sera of 21 hepatitis B virus surface antigen-positive apes, members of the families Pongidae and Hylobatidae (19 gibbon spp., 1 chimpanzee, and 1 gorilla). Sera originate from German, French, Thai, and Vietnamese primate-keeping institutions. To estimate the phylogenetic relationships, we sequenced two genomic regions, one located within the pre-S1/pre-S2 region and one including parts of the polymerase and the X protein open reading frames. By comparison with published human and ape HBV isolates, the sequences could be classified into six genomic groups. Four of these represented new genomic groups of gibbon HBV variants. The gorilla HBV isolate was distantly related to the chimpanzee isolate described previously. To confirm these findings, the complete HBV genome from representatives of each genomic group was sequenced. The HBV isolates from gibbons living in different regions of Thailand and Vietnam could be classified into four different phylogenetically distinct genomic groups. The same genomic groups were found in animals from European zoos. Therefore, the HBV infections of these apes might have been introduced into European primate-keeping facilities by direct import of already infected animals from different regions in Thailand. Taken together, our data suggest that HBV infections are indigenous in the different apes. One event involving transmission between human and nonhuman primates in the Old World of a common ancestor of human HBV genotypes A to E and the ape HBV variants might have occurred.
Human hepatitis B virus (HBV), the prototype member of the genus Hepadnaviridae, can infect different primates. In nature, HBV infections occur mainly in humans, although they have been also documented in chimpanzees (16), gibbons (8, 10), and orangutans (17, 18). Other members of the family Orthohepadnaviridae infect rodents, e.g., woodchucks (3) and ground squirrels (14, 15). A new hepadnavirus was recently isolated from a woolly monkey, a New World primate (woolly monkey HBV [WMHBV]) (7). This virus is distantly related to the human HBV group and is therefore considered to represent a progenitor of the human viruses.
HBV shows a remarkable genetic variability, which is reflected in the occurrence of at least six human HBV genotypes and two nonhuman primate HBV genotypes, consisting of a chimpanzee HBV isolate and a gibbon HBV isolate (9, 10). All these primate HBV genotypes are defined by an intergenotype variation of 8% based on complete viral genomes (12). The different human HBV genotypes show characteristic geographical distributions (11).
There are only a few reports about the occurrence of HBV infections in ape populations (6, 17). It is still a matter of debate whether these infections are of human origin or indigenous to the different apes (7, 17). At present, the complete HBV sequence from ape isolates has been analyzed only for one chimpanzee and one gibbon (10, 16). These viruses might as well represent human viruses introduced “artificially” into these animals (7). In order to answer this question, we analyzed in this study HBV isolates from HBV surface antigen (HBsAg)-positive apes kept in different primate-keeping facilities in Germany, France, Thailand, and Vietnam. A correlation between the geographic origins of the apes and their HBV sequences was investigated by analyzing parts of the viral genome. In addition, we tried to find chains of HBV transmission within the different primate-keeping institutions. The sequences of two short HBV genomic regions were analyzed to characterize phylogenetically closely related viral isolates. One region (A) encompasses the pre-S1 genomic region, and the second region (B) includes an area that codes for the viral polymerase and a part of the X protein. However, conclusions concerning possible new genotypes can only be drawn on the basis of complete genomes as defined by Okamoto et al. (12). Thus, the complete genomes of representatives of each genomic group were then fully sequenced in order to analyze the intergenotype variation.
Sera from 19 gibbon spp., 1 chimpanzee, and 1 gorilla were part of the primate serum bank stored at the Zoological and Botanical Garden Wilhelma, Stuttgart, Germany. Sera were collected during routine veterinary procedures in different zoos and primate-keeping facilities in Europe, Thailand, and Vietnam (Table 2). All sera were positive for at least one of the HBV serologic parameters HBsAg, HBeAg, anti-HBs, and anti-HBc, as determined by microparticle enzyme immunoassays using commercial test kits (IMx-HBsAg, IMs-HBe2, IMx-AUSAB, and IMx-CORE; Abbott Laboratories). HBV DNA, detected by PCR, was present in all samples.
TABLE 2.
Survey of the geographic origins of the HBV-infected apes and the phylogenetic classification of the corresponding virus isolates, based on sequence analysis of fragment Aa
| Species | Isolateb | Genomic group | Origin |
|---|---|---|---|
| Western lowland gorilla (Gorilla gorilla gorilla) | gor97 | chimp (6% divergence) | Leipzig, Germany (wild-caught animal from Cameroon) |
| Chimpanzee (Pan troglodytes) | chimp82 | gibI | Welzheim, Germany |
| White-handed gibbon (Hylobates lar) | gib644 | gibIIc | Wuppertal, Germany |
| gib645 | gibIIc | ||
| gib700 | gibIIc | ||
| Silvery Javan gibbon (Hylobates moloch) | gib156 | gibIV | Berlin, Germany |
| Capped gibbon (Hylobates pileatus) | gib160 | gibIII | |
| Capped gibbon (Hylobates pileatus) | gib731 | gibIII | Mulhouse, France |
| Gibbon sp. | gib759 | gibId | Phuket, Thailand |
| gib760 | gibId | ||
| gib761 | gibId | ||
| gib762 | gibI | ||
| gib763 | gibI | ||
| gib764 | gibI | ||
| White-handed gibbon (Hylobates lar) | gib174 | gibIII (region B not analyzed) | Pata, Thailand |
| Capped gibbon (Hylobates pileatus) | gib157 | gibIII | Chiang Mai, Thailand |
| Black gibbon (Hylobates concolor) | gib151 | gibV | Dusit, Thailand |
| gib153 | gibIV | ||
| gib154 | gibIV | ||
| White-checked gibbon (Hylobates leucogenys) | gib751 | gibIV | |
| White-checked gibbon (Hylobates leucogenys) | gib824 | gibV | Cuc Phuong, Vietnam |
HBV DNA was extracted from serum as described previously (5). Primers used for amplification of overlapping fragments spanning the HBV genome are given in Table 1. Nested PCR was carried out (5) for 35 cycles, with denaturation at 95°C for 15 s, annealing at 57°C for 15 s, and extension at 72°C for 70 s, as described previously. Fragments 2 and 4 were amplified from all samples to determine genomic groups of regions A and B. We were able to amplify these target sequences from almost all ape sera with the exception of that from gibbon 174. Here, we could amplify fragment 2 only. Fragments 1 and 3 were amplified additionally from one representative of each genomic group to sequence the complete genome.
TABLE 1.
Primers used for amplification and sequencinga
| Fragmentsb | Oligonucleotides used for:
|
Sequencing primersc | |
|---|---|---|---|
| First-round amplification | Second-round amplification | ||
| 1 | 3207-21f/1276-22r | 57-22f/1196-22r | 57-22f, 433-22r, 412-22f, 720-21r, 703-20f, 1196-22r |
| 2 | 2360-21f/477-23r | 2382-21f/433-22r | 2382-21f, 2850-22r, 57-22f, 433-22r |
| 2724-25f, 3221-21r (region A) | |||
| 3 | 1394-23f/2441-22r | 1425-23f/2441-22r | 1425-23f, 1789-20r, 1785-24f, 1959-23r, 2045-23f, 2441-22r |
| 3.I | 1099-23f/1789-20r | 1253-21f/1789-20r | 1253-21f, 1425-23f, 1789-20r |
| 3.II | 1785-24f/2850-22r | 1785-24f/2441-22r | 1785-24f, 1959-23r, 2045-23f, 2441-22r |
| 4 | 685-20f/1701-20r | 703-20f/1673-24r | 703-20f, 1196-22r |
| 1099-23f, 1673-24r (region B) | |||
Sequences of oligonucleotides: 57-22f, CTGCTGGTGGCTCCAGTTCAGG; 703-20f, CGTAGGGCTTTCCCCCACTG; 1099-23f, TCGCCAACTTACAAGGCCTTTCT; 1196-22r, GTTGCGTCAGCAAACACTTGGC; 1253-21f, CTCCTCTGCCGATCCATACTG; 1785-24f, GGCATAAATTGGTCTGCGCACCAG; 1789-20r, TGCCTACAGCCTCCTAGTAC; 1959-23r, GGCAAAAAAGAGAGTAACTCCAC; 2441-22r, GAGATTGAGATCTTCTGCGACG. Sequences of the other oligonucleotides are given in reference 5. Regions 2 and 4 were amplified and regions A and B were analyzed for all HBV isolates. Amplification and sequence analysis of the whole genome were done with representatives of each genomic group (see Table 2).
Fragments 3.I and 3.II were analyzed when amplification of fragment 3 (spanning the “nick” of the HBV genome) failed because of low serum HBV DNA concentration.
Primers in boldface were used for amplification of regions A and B, respectively.
To determine the phylogenetic relationships, we sequenced two genomic regions, one located within the pre-S1/pre-S2 region (region A) and one including parts of the polymerase and the X protein open reading frame (region B). Nucleotide sequences were determined for both strands with the PRISM Ready Reaction DyeDeoxy Terminator Cycle sequencing kit on an ABI 373A DNA sequencer (Applied Biosystems, Weiterstadt, Germany) using PCR primers and internal oligonucleotides (Table 1).
Sequence analysis was performed using the program package GCG (Wisconsin sequence analysis package, version 8.1, University of Wisconsin, Madison). Some phylogenetic analyses were done with programs from the PHYLIP package, version 3.57c (J. Felsenstein, Department of Genetics, University of Washington, Seattle, 1993). The sequences obtained in our study (Table 2) were compared to published human, chimpanzee, and gibbon sequences.
Sequence analysis of the pre-S1/pre-S2 region (region A).
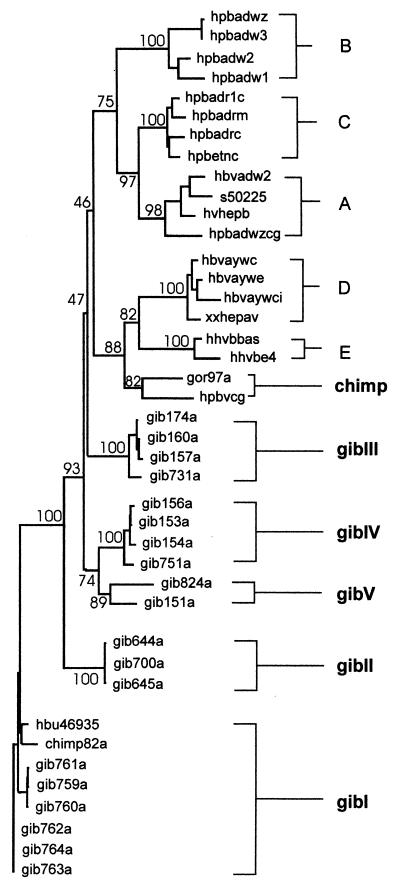
As described previously for the classification of human HBV isolates (4), the sequence analysis of region A allowed a rough estimate of the phylogenetic relationships among HBV isolates. A dendrogram of the sequences of region A including human and gibbon HBV isolates is given in Fig. 1. Here, human, chimpanzee, and gibbon HBV isolates clustered in different groups. All isolates obtained from gibbons were related to the gibbon HBV sequence (hbu46935; EMBL accession no. U46935) published by Norder et al. (10). Furthermore, the isolate from the gorilla (gor97a) was found to be distantly related to the chimpanzee HBV isolate (hpbvcg; EMBL accession no. D00220) described by Vaudin et al. (16). Surprisingly, the HBV isolate from the chimpanzee analyzed here (chimp82) was found within a gibbon HBV genomic group (gibI). The robustness of this tree was confirmed by bootstrap resampling with 100 data sets. Here, the gibbon HBV sequences were found within five distinct genomic groups (gibI to gibV) with intergroup distances of 7 to 11 changes per 100 nucleotides, compared to intergenotype distances of 8 to 22 changes per 100 nucleotides for human HBV genotypes.
FIG. 1.
Phylogram based on nucleotide sequences of fragment A, encompassing a part of the pre-S1 gene, using the Kimura two-parameter matrix and the neighbor-joining method. The numbers represent the percentages of bootstrap replicates (of 100 total) for each node. Analysis of this region allows a rough estimate of phylogenetic relationships, as all human sequences cluster within their respective genomic groups. A to E, human HBV genotypes; chimp and gibI to gibV, ape HBV genomic groups.
Sequence analysis of the complete HBV genome from representatives of gibbon HBV genomic groups I to V.
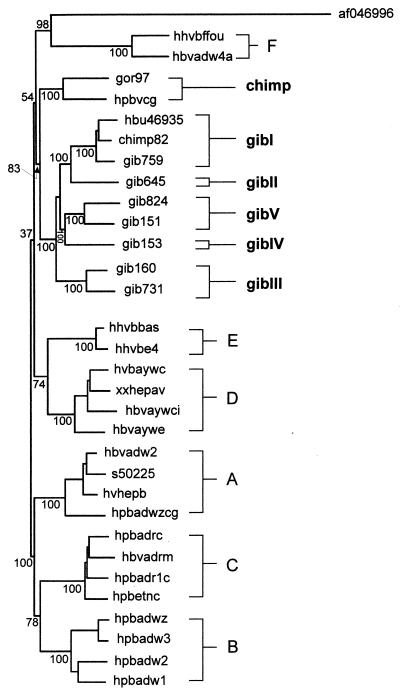
The complete genome of at least one representative of each group was sequenced. The results of phylogenetic analysis of complete viral genomes were similar to the ones obtained by analyzing region A (Fig. 2). There is one branch leading to the nonhuman primate HBV isolates, including the isolates from a naturally infected chimpanzee (hpbvcg) (16) and from a gorilla (whose HBV isolate was analyzed in this work). The other branch leads to the human HBV genotypes A to E. A third one leads to human HBV genotype F and to WMHBV (af046996). All gibbon HBV sequences are found within one monophyletic group, and within this gibbon HBV group, five genomic groups can be distinguished. Distances among these groups are between 6.3 and 7.9 changes per 100 nucleotides, thus slightly lower than distances among genotypes, which are defined by a distance of at least 8% (12). Intragroup distances were between 2.5 and 4.6%, comparable to intragroup distances within human genotypes. A summary of the intra- and intergroup distances of human HBV, chimpanzee HBV, WMHBV, and gibbon HBV variants is given in Table 3.
FIG. 2.
Phylogram based on complete nucleotide sequences, using the Kimura two-parameter matrix and the neighbor-joining method. The numbers represent the percentages of bootstrap replicates (of 100 total) for the nodes. A to F, human HBV genotypes; chimp and gibI to gibV, ape HBV genomic groups.
TABLE 3.
Unweighted distances between representatives of all known human HBV genotypes, the chimpanzee and gibbon HBV genomic groups, and WMHBV
| Genotype | Unweighted distancea (exchanges/100 nucleotides) from genotype:
|
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A (n = 4) | B (n = 4) | C (n = 4) | D (n = 4) | E (n = 2) | F (n = 2) | Chimpb (n = 2) | gibIc (n = 3) | gibII (n = 1) | gibIII (n = 2) | gibIV (n = 1) | gibV (n = 2) | WMHBV (n = 1) | |
| A | 3.7 | 9.4 | 9.5 | 9.3 | 9.9 | 14.1 | 9.8 | 10.5 | 10.8 | 10.4 | 10.2 | 10.6 | 22.6 |
| B | 4.2 | 9.1 | 10.1 | 10.8 | 13.7 | 10.2 | 10.5 | 11 | 10.2 | 10.2 | 10 | 21.8 | |
| C | 3.3 | 10.4 | 10.6 | 13.9 | 10.6 | 10.4 | 11 | 10.4 | 10.3 | 10.4 | 22 | ||
| D | 3.9 | 7.7 | 13.5 | 10.1 | 10.9 | 11.2 | 11 | 11 | 10.9 | 22.1 | |||
| E | 1.4 | 13.4 | 9 | 10.2 | 10.2 | 10.2 | 10.2 | 10.4 | 22 | ||||
| F | 5 | 12.9 | 12.8 | 13.3 | 13.4 | 13.1 | 13.7 | 23.4 | |||||
| chimp | 6 | 9 | 9.2 | 9.4 | 9.4 | 9.5 | 22.3 | ||||||
| gibI | 2.5 | 6.3 | 7.5 | 7.4 | 7.5 | 22.3 | |||||||
| gibII | NAd | 7.8 | 7.5 | 7.9 | 22.5 | ||||||||
| gibIII | 3.8 | 7.3 | 7.5 | 22.8 | |||||||||
| gibIV | NA | 6.8 | 22.2 | ||||||||||
| gibV | 4.6 | 22.4 | |||||||||||
Intragroup differences are in boldface.
Includes the isolate from a chimpanzee (16) and the HBV isolate from a naturally infected gorilla.
Includes the gibbon HBV isolate described by Norder et al. (10) and the HBV sequence from a chimpanzee analyzed in this work.
NA, not applicable. The whole genome of only one isolate was analyzed so no intragroup distance can be given.
Isolate gib731 (group gibIII) had a 236-nucleotide deletion, from nucleotide 1965 to 2200, within the core gene, which should lead to the formation of an altered, truncated core protein.
The HBV sequences obtained from gibbons 759, 760, and 761 were almost identical in both regions A and B. Short distances were due to ambiguities only. Assuming a hepadnaviral mutation rate for nucleotide substitutions of between 1.75 × 10−5 and 7.62 × 10−5 substitutions per site per year (13), this sequence identity indicates a recent transmission event among these apes. A second viral transmission was found in a gibbon family group living in a German zoo. The HBV isolates of all three individuals (gibbons 644, 645, and 700) were nearly identical in regions A and B and cluster within genomic group gibII. Low distances were again caused by ambiguous nucleotide positions. The transmission events are indicated in Table 2.
There are some reports about the occurrence of HBV infections in ape populations (6, 17). The question of whether these infections were of human origin or indigenous to the different ape populations is still a matter of debate (7, 17, 18). We found strong evidence for the presumption that these HBV infections are indigenous to the different nonhuman primate populations by analyzing a large number of ape HBV sequences. All isolates analyzed in this work cluster within one monophyletic group, including the previously described chimpanzee and gibbon sequences. Phylogenetic analysis shows that there is one branch leading to human HBV genotypes A to E, a second leading to the ape HBV genomic groups, and a third one leading to human HBV genotype F and to the New World WMHBV. All ape isolates were from animals of different geographic origins and include different species. These facts are consistent with the presumption that there was a single transmission event between humans and apes in the Old World involving a common ancestor of human HBV genotypes A to E and the ape viruses. Prior to this, the branch leading to the New World HBV isolates, WMHBV, and human HBV genotype F might have split off. Norder et al. (11) found indications that group F represented the genomic group of HBV among populations with origins in the New World.
With few exceptions, gibbon isolates from the different Thai and the Vietnamese locations could be classified into four genomic groups (gibI, -III, -IV, and -V, with genetic distances between 6.3 and 7.9%), corresponding to the geographical origins of the apes. This finding is analogous to the finding by Norder et al. (12) of different geographical distributions of the different human HBV genotypes. Thus, genotype definition (12) should be discussed for the gibbon HBV variants.
Genomic gibbon HBV group gibII represents nearly identical isolates from a family group living in a German zoo. The HBV isolates from the primates living in other European zoos could be integrated into the Thai genomic groups gibI, gibIII, and gibIV. Therefore, the HBV infection of these apes might be introduced by captured wild apes which were already infected. Surprisingly, the sequence of the HBV isolate of the chimpanzee analyzed here showed a very close phylogenetic relationship to the gibbon isolates within group gibI. Most likely, this reflects an infection of this chimpanzee by a gibbon HBV variant, as this animal was kept with some gibbons in another zoo. Unfortunately, we did not get serum samples from apes of this zoo. However, this is the first probable “quasinatural” interspecies transmission of HBV. The infectivity of a gibbon HBV variant to a chimpanzee was documented by Mimms et al. (8). In addition, different experimental studies revealed that human HBV isolates can infect gibbons and chimpanzees (1, 2). In general, these results imply that nonhuman primate HBV variants may be transmittable to humans. At least they infect representatives of the families Pongidae and Hylobatidae. However, at present it remains to be elucidated if nonhuman primate HBV variants are transmittable to humans and whether they cause disease in humans. We think that this is likely, because the transmission of human HBV variants to nonhuman primates is well documented. Thus, we strongly recommend routine screening for HBV markers and HBV vaccination of zoo-kept apes, primate-handling zoo keepers, and veterinarians.
Acknowledgments
S.G. and J.-O.H. contributed equally to this work.
We thank Rainer Thomssen for helpful discussion and personal support.
J.-O.H. was supported by the Grimminger Foundation for Zoonosis Research, Stuttgart, Germany.
REFERENCES
- 1.Alter H J, Purcell R H, Gerin J L, London W T, Kaplan P M, McAuliffe V J, Wagner J, Holland P V. Transmission of hepatitis B to chimpanzees by hepatitis B surface antigen-positive saliva and semen. Infect Immun. 1977;16:928–933. doi: 10.1128/iai.16.3.928-933.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bancroft W H, Snitbhan R, Scott R M, Tingpalapong M, Watson W T, Tanticharoenyos P, Karwacki J J, Srimarut S. Transmission of hepatitis B virus to gibbons by exposure to human saliva containing hepatitis B surface antigen. J Infect Dis. 1977;135:79–85. doi: 10.1093/infdis/135.1.79. [DOI] [PubMed] [Google Scholar]
- 3.Galibert F, Chen T N, Mandart E. Nucleotide sequence of a cloned woodchuck hepatitis virus genome: comparison with the hepatitis B virus sequence. J Virol. 1982;41:51–65. doi: 10.1128/jvi.41.1.51-65.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grethe S. Neue genetische Varianten des Hepatitis B Virus—Bedeutung für Pathogenese, Diagnostik, Infektkettenanalyse und Immunprophylaxe. Vol. 1. Germany: Cuvillier, Göttingen; 1997. [Google Scholar]
- 5.Grethe S, Monazahian M, Bohme I, Thomssen R. Characterization of unusual escape variants of hepatitis B virus isolated from a hepatitis B surface antigen-negative subject. J Virol. 1998;72:7692–7696. doi: 10.1128/jvi.72.9.7692-7696.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kalter S S, Heberling R L, Cooke A W, Barry J D, Tian P Y, Northam W J. Viral infections of nonhuman primates. Lab Anim Sci. 1997;47:461–467. [PubMed] [Google Scholar]
- 7.Lanford R E, Chavez D, Brasky K M, Burns III R B, Rico-Hesse R. Isolation of a hepadnavirus from the woolly monkey, a New World primate. Proc Natl Acad Sci USA. 1998;95:5757–5761. doi: 10.1073/pnas.95.10.5757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mimms L T, Solomon L R, Ebert J W, Fields H. Unique preS sequence in a gibbon-derived hepatitis B virus variant. Biochem Biophys Res Commun. 1993;195:186–191. doi: 10.1006/bbrc.1993.2028. [DOI] [PubMed] [Google Scholar]
- 9.Norder H, Courouce A M, Magnius L O. Complete genomes, phylogenetic relatedness, and structural proteins of six strains of the hepatitis B virus, four of which represent two new genotypes. Virology. 1994;198:489–503. doi: 10.1006/viro.1994.1060. [DOI] [PubMed] [Google Scholar]
- 10.Norder H, Ebert J W, Fields H A, Mushahwar I K, Magnius L O. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–223. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 11.Norder H, Hammas B, Lee S D, Bile K, Courouce A M, Mushahwar I K, Magnius L O. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341–1348. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- 12.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo R I, Imai M, Miyakawa Y, Mayumi M. Typing hepatitis B virus by homology in nucleotide sequence: comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–2583. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 13.Orito E, Mizokami M, Ina Y, Moriyama E N, Kameshima N, Yamamoto M, Gojobori T. Host independent evolution and a genetic classification of the hepadnavirus family based on nucleotide sequences. Proc Natl Acad Sci USA. 1989;86:7059–7062. doi: 10.1073/pnas.86.18.7059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seeger C, Ganem D, Varmus H E. Nucleotide sequence of an infectious molecularly cloned genome of ground squirrel hepatitis virus. J Virol. 1984;51:367–375. doi: 10.1128/jvi.51.2.367-375.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Testut P, Renard C A, Terradillos O, Vitvitski-Trepo L, Tekaia F, Degott C, Blake J, Boyer B, Buendia M A. A new hepadnavirus endemic in arctic ground squirrels in Alaska. J Virol. 1996;70:4210–4219. doi: 10.1128/jvi.70.7.4210-4219.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaudin M, Wolstenholme A J, Tsiquaye K N, Zuckerman A J, Harrison T J. The complete nucleotide sequence of the genome of a hepatitis B virus isolated from a naturally infected chimpanzee. J Gen Virol. 1988;69:1383–1389. doi: 10.1099/0022-1317-69-6-1383. [DOI] [PubMed] [Google Scholar]
- 17.Warren K S, Niphuis H, Heriyanto, Verschoor E J, Swan R A, Heeney J L. Seroprevalence of specific viral infections in confiscated orangutans (Pongo pygmaeus) J Med Primatol. 1998;27:33–37. doi: 10.1111/j.1600-0684.1998.tb00066.x. [DOI] [PubMed] [Google Scholar]
- 18.Warren K S, Heeney J L, Swan R A, Heriyanto, Verschoor E J. A new group of hepadnaviruses naturally infecting orangutans (Pongo pygmaeus) J Virol. 1999;73:7860–7865. doi: 10.1128/jvi.73.9.7860-7865.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]