Extended Data Fig. 3. The dependence of key crotonyl-CoA producing enzymes in GSCs.
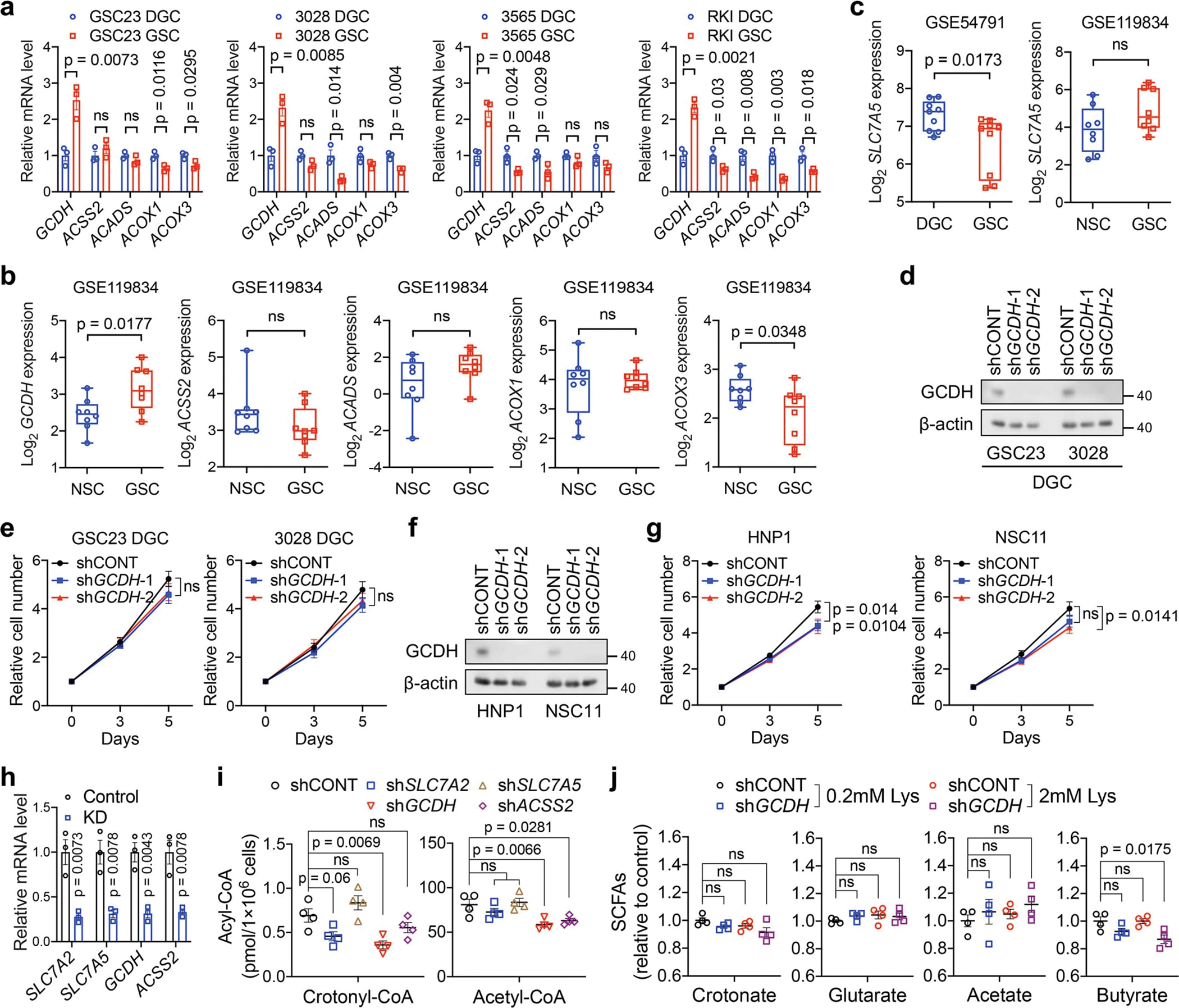
a, RT-qPCR analysis of key crotonyl-CoA-producing enzymes in paired GSCs and DGCs.
b, c, Expression levels of indicated genes from public datasets. n = 8 biologically independent cells in NSCs and GSCs from GSE119834. Three GSCs (MGG4, MGG6 and MGG8, each cell contains 3 replicates) and matched DGCs were queried in GSE54791.
d-g, IB analysis (d, f) and cell proliferation (e, g) in DGCs (d, e) or NSCs (f, g) with or without GCDH KD.
h, i, RT-qPCR analysis (h) and intracellular acyl-CoAs (i, n = 4 biologically independent samples) in GSC23 with or without indicated gene depletion.
j, Relative intracellular SCFAs (n = 4 biologically independent samples) in GSC23 with or without GCDH KD cultured in media with indicated concentrations of L-lysine.
Data are presented from three independent experiments in a, e, g and h. Representative of two independent experiments in d and f. Data are presented as mean ± SEM in a, e and g-j. Boxes represent data within the 25-to-75 percentiles in b and c. Whiskers depict the range of all data points. Horizontal lines within boxes represent mean values. Two-tailed unpaired t test for a-c and h, two-way ANOVA followed by multiple comparisons with adjusted p values for e and g, one-way ANOVA followed by multiple comparisons with adjusted p values for i and j. ns, not significant.
