Abstract
Saccharomyces cerevisiae requirement for reduced sulfur to synthesize methionine and cysteine during alcoholic fermentation, is mainly fulfilled through the sulfur assimilation pathway. Saccharomyces cerevisiae reduces sulfate into sulfur dioxide (SO2) and sulfide (H2S), whose overproduction is a major issue in winemaking, due to its negative impact on wine aroma. The amount of H2S produced is highly strain-specific and also depends on SO2 concentration, often added to grape must. Applying a bulk segregant analysis to a 96-strain-progeny derived from two strains with different abilities to produce H2S, and comparing allelic frequencies along the genome of pools of segregants producing contrasting H2S quantities, we identified two causative regions involved in H2S production in the presence of SO2. A functional genetic analysis allowed the identification of variants in four genes able to impact H2S formation, viz; ZWF1, ZRT2, SNR2, and YLR125W, and involved in functions and pathways not associated with sulfur metabolism until now. These data point out that, in wine fermentation conditions, redox status, and zinc homeostasis are linked to H2S formation while providing new insights into the regulation of H2S production, and a new vision of the interplay between the sulfur assimilation pathway and cell metabolism.
Keywords: QTL, Saccharomyces cerevisiae, wine, fermentation, yeast, aroma, H2S, ZWF1, ZRT2
The hydrogen sulfide production of Saccharomyces cerevisiae during alcoholic fermentation is regulated by zinc homeostasis and interconnected with cell metabolism.
Introduction
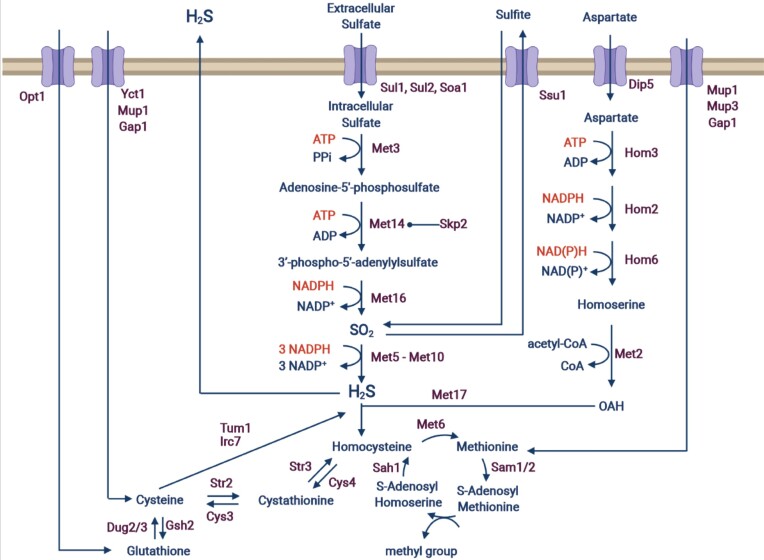
Sulfur is an essential element for cells, particularly as a component of sulfur-containing amino acids that are crucial for protein biosynthesis. While Saccharomyces cerevisiae, like other organisms, can utilize organic sulfur sources, its primary source is in the form of inorganic sulfate, which is naturally present in grape must. During the alcoholic fermentation process, yeast cells assimilate sulfur through the sulfur assimilation pathway (SAP) (Thomas and Surdin-Kerjan 1997) (Fig. 1). Briefly, extracellular sulfate is imported into the cell, where it is reduced with the consumption of two molecules of ATP and four molecules of NADPH per molecule of SO4. Hydrogen sulfide (H2S) will then be incorporated into carbon chains to produce, in the end, methionine and cysteine. Sulfur-containing amino acids are then consumed for cell growth, and in many essential biochemical reactions such as methyl group transfer, tRNA thiolation, S-adenosyl methionine synthesis, glutathione synthesis, and protection from oxidative stress (Walvekar and Laxman 2019).
Figure 1.
Global overview of sulfur metabolism. This image was created with BioRender.com.
H2S demand can be very high, depending on the environment S. cerevisiae has to cope with, but, in the wine industry, an excessive production of sulfide is not acceptable, given its unpleasant scent of rotten egg, cabbage, and onion.
For several reasons, an imbalance may at times occur between sulfide produced through SAP and its incorporation into carbon chains, which leads to an overproduction of H2S by yeast during fermentation. One of the most common and best-described circumstances is a nitrogen level sufficient for active growth and fermentation, but too low to provide carbon chains for S2− incorporation (Vos and Gray 1979, Jiranek et al. 1995, Mendes-Ferreira et al. 2009, Ugliano et al. 2009, 2011, Song et al. 2020, De Guidi et al. 2021). A lack or a suboptimal concentration of vitamins or cofactors needed for a proper SAP enzymatic activity can also result in the same outcome (Wang et al. 2003, Bohlscheid et al. 2007, 2011, Xing and Edwards 2019). Last, sulfur dioxide (SO2) addition, widely used in winemaking to prevent spoilage and oxidation, has been shown to impact H2S production (De Guidi et al. 2021).
In addition to targeted nutrient management, the use of specific strains with a low H2S production profile is nowadays one of the best ways to avoid reductive off-odors. One can exploit the natural diversity of S. cerevisiae for sulfide production (Spiropoulos et al. 2000) and desired phenotypes (Linderholm et al. 2010, Cordente et al. 2019) or, alternatively, engineer sulfite reductase by chemical mutagenesis (Cordente et al. 2009). Both strategies are valid, but with a low success rate, and both require intensive large-scale screening. In addition, a reduced sulfite reductase activity can cause a bottleneck effect and lead to a higher final sulfite concentration.
Another approach is to identify in a strain genes and their allelic variants, responsible for the desired characteristic, without any a priori. This approach makes it possible to introduce these desirable traits into as many genetic backgrounds as desired, and combine them with traits that are important in winemaking. Quantitative trait loci (QTL) mapping allows such strategy, and has been used to identify new genes involved in many technological properties such as wine aroma production, fermentative performances, sporulation efficiency, ethanol and temperature tolerance, flocculation, amino acid consumption, and nitrogen requirement, and even SO2 and H2S production (Brauer et al. 2006, Sinha et al. 2006, 2008, Hu et al. 2007, Marullo et al. 2007, Smith and Kruglyak 2008, Katou et al. 2009, Ambroset et al. 2011, Cubillos et al. 2011, 2013, 2017, Steyer et al. 2012, Brion et al. 2013, Huang et al. 2014, Jara et al. 2014, Wilkening et al. 2014, Noble et al. 2015, Eder et al. 2018, 2020, Haas et al. 2019, Ho et al. 2021, Villarroel et al. 2021). The role in sulfite production of the two genes MET2 and SKP2 has been evidenced and ultimately applied for the improvement of yeast starters (Blondin et al. 2017). MET2 encodes the homoserine O-trans-acetylase that catalyzes O-acetyl homoserine production from homoserine, providing the carbon chain into which S− is incorporated as a first step in methionine biosynthesis, therefore consuming H2S and reducing SO2 excretion. Skp2p is involved in the modulation of the SAP through the degradation of Met14p, a kinase located upstream the sulfide reductase (Met5p–Met10p). SKP2 allelic variants efficiently reduced H2S and SO2 excretion. Due to Skp2p position in the SAP (Fig. 1), the efficiency of the modulation of sulfite (and possibly sulfide) production, is most likely reduced in enological conditions, as SO2 from grape juice only enters midway the sulfur pathway.
More recently, the ecological significance of H2S production, associated with domestication features, has been elucidated, revealing that yeast strains that colonize different environments exhibited varying aptitudes to produce H2S (De Guidi et al. 2023).
In this study, we investigated the genetic basis of H2S production under winemaking conditions via QTL mapping. We selected two strains producing different amounts of H2S with different genetic background: one wine strain with moderate ability to produce H2S in the presence of SO2, that contained favorable MET2 and SKP2 alleles, and another low producer, previously identified as a hybrid between wine and wine–velum strains (Coi et al. 2017, Legras et al. 2018). After having measured the H2S production ability of 96 segregants, we used a bulk segregant analysis (BSA) strategy to detect QTLs explaining the variations in that trait. We detected two QTLs, and investigated the effects of allelic variations in seven genes using reciprocal CRISPR-Cas9 mediated allelic swaps. Four of these genes explained variations in H2S production through the activation of different mechanisms.
Materials and methods
Yeast strains used and growth conditions
Saccharomyces cerevisiae strains used in this work are listed in Table S1 (Supporting Information). 59A was obtained from our laboratory collection (MTF 1438), hLMD20 was a derivative of hLMD20, provided by Lallemand SAS, that harbors the JN17 allele of SKP2 (Noble et al. 2015). Yeast strains were grown at 28°C on yeast extract peptone dextrose medium (YPD; 2% glucose, 1% yeast extract, and 2% bactopeptone), supplemented with 2% agar where needed. Selective media were prepared by adding to agar medium 200 µg/ml geneticin (G418–Sigma A1720-5 G), 200 µg/ml nourseothricin (ClonNAT—Werner BioAgents) or 300 µg/ml hygromycin B (Invitrogen 10687010) according to the resistance to test.
Stable haploid parental strain hLMD20
To obtain a stable haploid strain to be used as a parental strain, LMD20 was deleted for HO by geneticin resistance cassette insertion (KanMX4). KanMX4 cassette was amplified by high fidelity PCR (with Phusion High Fidelity Taq polymerase; Thermo Fischer, Illkirch, France) from the plasmid pUG6 using the couple of primers HOKOMXfw—HOKOMXTTGG-rev (Table S2, Supporting Information), having respectively 20 and 22 bases complementary to the plasmid (upstream the promoter and downstream the terminator of the antibiotic cassette) and the 40-bp complementary to the HO gene to be deleted. Deletion mutants (LMD20ΔHO/HO) were selected on YPD agar + G418 and confirmed by PCR with the primers dHOVerifFOR—HOdelVerifREV located upstream and downstream of the insertion region. A two-steps CRISPR-Cas9 system was then used to remove the kanMX4 cassette from HO in hLMD20 ΔHO::kanMX4. Yeast strains were transformed with a modified pCfB2312 plasmid carrying the selection marker for hygromycin and CAS9. Then, transformation with a pMEL15 plasmid containing the gRNA for kanMX4 and an inactive copy of HO (ho) as repair fragment allowed to obtain a stable haploid (hLMD20) without any integrated resistance cassette. All the plasmids used in this work are listed in Table S3 (Supporting Information).
Segregants
Haploid parents (59A and hLMD20) were mated on solid medium. The resulting diploid hybrids were checked for the presence of an (a/α) MAT genotype by multiplex PCR [Table 2 (Supporting Information) for the primers used]. One diploid cross (59A × hLMD20) was grown on PRE5 and cells were transferred at mid-exponential growth phase to SPO2 medium for sporulation (Codon et al. 1995). The presence of asci was verified with an optic microscope. After asci digestion with β-glucuronidase, spores were teased apart and isolated with the Singer Instrument MSM300 micromanipulator tetrad dissection system (Watchet, UK). 96 segregants were then isolated and phenotyped for their H2S production during alcoholic fermentation.
Phenotyping
Fermentation conditions
Fermentations were conducted in 300 ml glass bioreactors in synthetic musts (SM) (Bely et al. 1990) prepared with 200 g/l of a 1:1 mix of glucose and fructose, 200 mg/l assimilable nitrogen (amino acids and ammonium), one-fourth of original recipe anaerobic factor amount, and 60 mg/l SO2 added as potassium metabisulfite (K2S2O5). When H2S production in response to different zinc concentrations during fermentation was evaluated, SMs (that generally contain 0.91 mg/l zinc) were prepared with either a zinc deficiency (0.091 mg/l, corresponding to 1/10 of the control amount) or an excess (2.73 mg/l, corresponding to three times the control amount).
For each strain, an overnight YPD preculture was diluted 100-fold in SM and grown at 28°C for 24 h. Cell population was then determined with an electronic particle counter (Beckman Coulter) and 250 ml of SM were inoculated at a concentration of 1 × 106 cells/ml. Fermentations were carried out at 28°C, under permanent stirring (280 rpm) and daily followed by weight loss, until the theoretical percentage of sugar consumed reached 95% (87.4 g CO2/l produced for a must containing 200 g/l of sugar). The 96 segregants were separated into two subsets of 48 strains, and each subset was tested in three independent runs (blocks) that included the two parental strains in duplicate or triplicate.
H2S and SO2 quantification
The total amount of H2S produced during each fermentation was channeled into a zinc-based trap thanks to special glassware, accumulated as zinc sulfide and quantified with a sulfide-specific fluorescent probe, as previously described (De Guidi et al. 2021). Total SO2 in fermented SM was quantified by titration with TitraEVO (Ref.120700, Laboratoires Dujardin-Salleron).
Central carbon metabolism products quantification
Central carbon metabolism products (ethanol, glycerol, succinate, α-ketoglutarate, and acetate) concentrations were determined by HPLC (HPLC 1260 Infinity, Agilent™ Technologies, Santa Clara, California) on a Phenomenex Rezex ROA column (Phenomenex™, Le Pecq, France) at 60°C, from samples collected at the end of fermentation. The column was eluted with 2.5 mM H2SO4 at a 0.6 ml/min flow rate. Organic acids were analysed with a UV detector (Agilent™ Technologies) at 210 nm; the other compounds were quantified with a refractive index detector (Agilent™ Technologies). Analysis was carried out with the Agilent™ ChemStation software package.
QTL mapping
Given the high number of fermentations required to analyze H2S production in the segregant population, the experiments were split into blocks. To neutralize variations between blocks, data were normalized by calculating the z score of each sample in relation to the mean and standard deviation of the block as follows:
 |
where zij is the z score of the sample i in the block j, xij is the quantity (in µg) of H2S produced from the sample i in the fermentation carried in the block j, μj is the mean of H2S produced by all the samples in block j, and σj the standard deviation within block j. The mean z score was used to rank each segregant. After normalization, no differences were observed between blocks for the low producing strain 59A, while small differences could still be observed for the high H2S producer. We chose two bulks of 16 segregants with the most extreme production in order to maximize the QTL detection power (Magwene et al. 2011). The transgressive index of the phenotype was calculated as previously described (Marullo et al. 2006).
Genomic DNA extraction for sequencing
For each segregant in the two bulks, an independent preculture was prepared, and genomic DNA was isolated from liquid cultures in the stationary phase (after 48 h of growth on YPD at 28°C). The number of cells per ml was evaluated with an electronic particle counter (Multisizer 3 counter; Beckman Coulter) and, for each bulk, an equal number of cells for each of the strains was mixed, to obtain two separate populations of high and low producers. DNA was extracted using a traditional method involving phenol–chloroform extraction, as described previously (Saubin et al. 2020). After nucleic acid extraction, samples were combined with a DNA absorption solution (50 µl of 5 M NaCl, 15 µl of Perkin-Elmer Chemagen CMG 252-A beads, 250 µl 7.8 M guanidinium chloride, 800 µl isopropanol). Subsequently, the metal beads with DNA attached to their silica surfaces were collected using the DynaMag™-2 Magnet tube holder (12321D-DynaMag-2—Invitrogen), washed twice with AMMLAV/E buffer (10 mM Tris pH 8, 0.1 mM EDTA, 60 mM potassium acetate, 65% ethanol), and twice with 75% ethanol. The DNA was desorbed in an aqueous solution.
To assess DNA purity, we measured OD ratios at 260 nm/280 nm and 260 nm/230 nm using a NanoDrop 1000 spectrophotometer (Thermo Scientific, Illkirch, France). For quantification, we employed the QuantiFluor kit, dsDNA system (Promega), and subsequently stored DNA at −20°C.
Genome sequence and analysis
DNA samples of the two bulks, and of hLMD20, were processed separately to generate libraries of 500 bp inserts. After passing quality control, the libraries were sequenced with DNBseq technology using BGISEQ-500 platform, generating paired-end reads of 2 × 150 bp.
For each library, low-quality reads were processed and filtered using the FASTX Toolkit v0.0.13.2 and TRIMMOMATIC v0.36 (Bolger et al. 2014) with the following parameters (LEADING:10 HEADCROP:5 SLIDINGWINDOW:4:15 MINLEN:50).
Reads were then mapped to the 59A genome, used as reference, with BWA v0.6.2 with default parameters (Li and Durbin 2009) and genotyping performed with samtools v1.11. We obtained a variant file with the sequencing depth of each allelic variant that was considered as a proxy to allelic frequency.
Afterwards, sequence positions were filtered for quality criteria: sufficient coverage position as well as genotyping and mapping quality (MQ > −20) were kept. The impact of the different SNPs was evaluated with SNPeffect 4.3 (Cingolani et al. 2012). Only damaging SNPs were retained for the selection of candidate genes in each QTL.
Block regression mapping
The statistical method Block Regression Mapping (BRM) (Huang et al. 2020) was chosen for QTL mapping based on bulked segregant analysis by deep sequencing, run in R environment (R version 4.0.2; R Core Team 2020). BRM estimates the probability of a significant deviation of the allelic frequencies (F) from the neutral situation (F = 0.5) and estimates a significance threshold given the recombination rate and the length of the chromosome, taking in account multiple testing. To find candidate QTL peaks, the genome is first divided into small blocks of equal size of 1 kb. Based on the allelic frequency data, the algorithm calculates the average allele frequency of each block in each pool, the average allele frequency in the population of each block, along with the allele frequency difference (AFD) between two pools, in each block. In a second step, BRM infers the AFD threshold of the 5% overall significance level at every genomic position (5% risk of concluding that a difference exists when there is no actual difference). In the last step, BRM identifies possible QTL positions (with significant AFD) and calculates chromosomal coordinates corresponding to a 95% confidence interval of each QTL. The potential impact of each SNP was assessed using snpEffect v4.3 (Cingolani et al. 2012).
Phylogeny
Maximum-likelihood phylogeny of ZRT2 alleles from hLMD20, 59A, genomes from Evolya and Genowine projects (Coi et al. 2017, Legras et al. 2018) and other available genomes (SGD 2021) was estimated using RAXML (Stamatakis 2014).
Gene expression analysis
Transcriptomic analyses were performed to understand the impact on cellular physiology of allelic variants in different genetic backgrounds. We analyzed gene expression in each parent in comparison to the strain containing the other parental allele during alcoholic fermentation. Samples were collected when 35 g/l CO2 had been produced as a result of alcoholic fermentation, which corresponded to a maximum in sulfide production for strain LMD20, from which hLMD20 is derived (Figure S1, Supporting Information).
Fermentations were performed in triplicate, in 1.2 l bioreactor systems, in the same fermentation conditions as previous experiments in this work (i.e. SM 200 with 60 mg/l SO2, 28°C). RNAs were extracted as described before (Rollero et al. 2016). Agilent 8 × 15 k gene expression microarrays (Design ID 038619, including 40 EC1118-specific genes; Agilent Technologies) were used according to the manufacturer’s instructions. Hybridization and scanning were performed as described previously (Rollero et al. 2016) with 4000B laser Scanner (Axon Instruments).
For each gene, the transcriptome of the strains carrying the alternative parental allele was compared to the one with the original allele. The limma package was used to import and normalize the global microarray data (quantile method for normalization between arrays). The Benjamini–Hochberg correction was used for multiple comparisons, and the false discovery rate (FDR) threshold was set at 5% (Ritchie et al. 2015).
CRISPR-Cas9 targeted genome modifications (allelic replacement)
Both two parental strains (59A and hLMD20) underwent a two-step allelic replacement, for each of the candidate genes.
Allele deletions
For each modification, the entire gene (from ATG to stop codon) was replaced in a different transformation experiment by the nourseothricin resistance cassette (natMX), amplified by PCR from pAG25 plasmid, with the couple of primers gene_name_del_UP and gene_name_del_DW (Table S2, Supporting Information) and Phusion High Fidelity Taq polymerase (Thermo Fischer). The resulting antibiotic cassette contained 80 base pairs of upstream and downstream homology arms for the gene to be deleted, and was used to transform yeast cells that replaced the target gene through homologous recombination.
Allelic replacement
Mutants were then transformed with a plasmid derived from pCfB2312 containing CAS9, an hygromycin resistance cassette and the guide RNA targeting natMX cassette. For each deleted gene, one parent received, as a repair fragment, the alternative variant of the gene under investigation, synthesized by PCR performed with Phusion High Fidelity Taq polymerase (Thermo Fischer) with primers gene_name_A and gene_name_D (Table S2, Supporting Information) from the other parent. These primers were designed to amplify the gene of interest with an average of 325 bp upstream and downstream homology, to include the promoting region. For each candidate gene, three independent allelic swap transformations were performed, and H2S produced during alcoholic fermentation was assessed for each resulting strain.
All transformations were performed following the lithium acetate method (Gietz and Schiestl 2007), adapting volumes to a 96-well round (U) Bottom Plate well.
Statistical analysis
Statistical analyses were conducted in the R environment (R version 4.0.2 (2020–06-22) (R Core Team 2020).
ANOVA was performed to test the effect of the genetic modification in each strain (allelic replacements or deletions). For all the experiments, when the impact of one factor was significant, differences between modalities were evaluated by post hoc testing (Tukey’s HSD multiple-comparison test, P-value < .05).
Results
Production of a population of segregants and phenotyping for H2S production
To unravel the genetic bases explaining wine strains differences in H2S production in the presence of SO2, we selected two haploid strains derived from yeasts used in winemaking: 59A is a stable haploid descendant of EC1118 whose H2S production remains low despite the presence of SO2, while, on the contrary, hLMD20 produces more H2S in presence of SO2 (Fig. 2).
Figure 2.
Total H2S production by the two parental strains (59A and LMD20) in SM containing 0 or 60 mg/l of SO2.
The variability of total H2S production under fermentation in the presence of SO2 (Fig. 3) was assessed for 96 segregants generated from the crossing of hLMD20 and 59A, and for parental strains, using the trapping/colorimetric method previously described (De Guidi et al. 2021).
Figure 3.
Standard score representation of the cumulated H2S production during alcoholic fermentation for each segregant in comparison with the parental strains. Dark red bars: low H2S producers’ bulk; blue bars: high H2S producers’ bulk; black: parents.
The trait exhibited a continuous distribution, confirming its multigenic nature. Furthermore, some segregants produced up to three times more H2S than the higher-producing parent and up to 2.5 times less than the lower-producing one, with a total of 50 segregants in which H2S production was more extreme than that of the parental strains. A transgressive index of 3.12% and 2.08% for the low H2S and high H2S phenotypes, respectively, suggests the presence of alleles with opposite effects at multiple loci in the genome of each parental strain.
QTL identification
To link these phenotypic differences to the genetic variation between parental strains, we performed a BSA on the progeny. We selected two groups of 16 segregants with the most extreme phenotypes, i.e. low and high sulfide production.
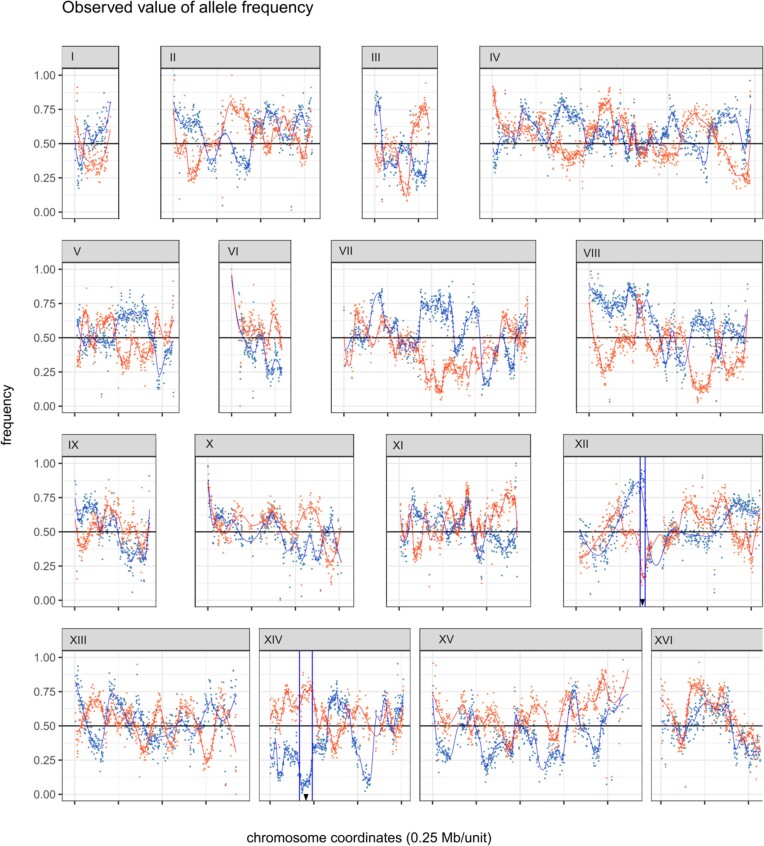
After processing the sequencing data and performing genotyping, we detected 22 685 variant positions between the two parental strains. Figure 4 shows the frequencies of each parental variant across the genome of the two bulks. To detect possible departures from neutrality at specific loci, we used the LOESS method of local regression (Jacoby 2000) applied to blocks of equal size as implemented in the BRM software (Huang et al. 2020). Allelic departures from normality of AFD could be detected for two genomic regions that reached the theoretical threshold inferred by BRM (± 0.6947), indicating two potential QTLs (Table 1 and Fig. 4; Figure S2, Supporting Information). The first one (QTL_chrXII), was located on chromosome XII, between YLR119W and YLR135W, in a 28-kb region. The positive direction of the peak means that the low sulfide producer bulk inherited the SNPs from the low H2S producer parent (59A). The second genomic region (QTL_chrXIV), located on chromosome XIV was wider (73 kb), spanning from YNL250W to the origin of replication ARS1413, with a maximum at YNL234W. In this case, the allelic variants from the high sulfide parent (hLMD20), conferred a lower production to the progeny. A third region located on chromosome VII almost reached the threshold but was not explored here.
Figure 4.
Frequencies of low-H2S-producer-parent allelic variants along the genome of the low H2S producer bulk, in red, and high H2S producer bulk, in blue. The limits of each QTL region identified on chromosome XII and XIV are indicated within two blue vertical lines and the peak of the QTL with a black arrow. Chromosome coordinates refer to the 59A genome.
Table 1.
QTL detected by BSA using BRM software.
| CHR | QTL Peak (Position) | AFD Peak | Peak direction | Start | End | Kb |
|---|---|---|---|---|---|---|
| XII | 382 337 (YLR129W) | 0.7508 | + | 365 894 (YLR119W) | 394 323 (YLR135W) | 28 |
| XIV | 200 384 (YNL234W) | −0.6969 | – | 168 137 (YNL250W) | 240 787 (ARS1413) | 73 |
Within these QTLs, we searched for potentially impacting variants using the SnpEff software, not taking in account those located in intergenic regions. In QTL_chrXII, we found 14 SNPs that were categorized as missense variants with moderate effects and one that caused a frameshift and was classified as highly impacting. In QTL_chrXIV, our analysis revealed the presence of 21 missense mutations and one premature stop codon (as detailed in Table S4, Supporting Information). Given QTL_chrXIV size, we focused on potentially impacted genes located close to the peak of the maximum allelic frequency shift.
We evaluated the effect of seven of the genes with impacting mutations, five on chromosome XII (SRN2, YLR125W, YLR126C, ZRT2, and ACE2) and two on chromosome XIV (ATG2 and ZWF1). The amino acid changes caused by allelic variation of the four genes that led to the modulation of H2S production are given in Table 2. No significant effect was observed for the three remaining genes (ACE2, YLR126C, and ATG2).
Table 2.
Amino acid changes between parental alleles detected in the candidate genes proven to have an impact on H2S production.
| Gene | AA position | S288C | 59A | hLMD20 | |
|---|---|---|---|---|---|
| CHR XIV | ZWF1 | 58 | Glu | – | Glu |
| 358 | Ala | Ala | Val | ||
| CHR XII | SRN2 | 133 | Glu | Glu | Gly |
| 180 | Asn | Asp | Asn | ||
| YLR125W | 34 | Asn | fs | Asn | |
| ZRT2 | 19 | Gly | Asp | Gly | |
| 322 | Lys | Glu | Lys | ||
| 325 | Met | Thr | Met |
ZWF1 and the activity of PPP modulate H2S production
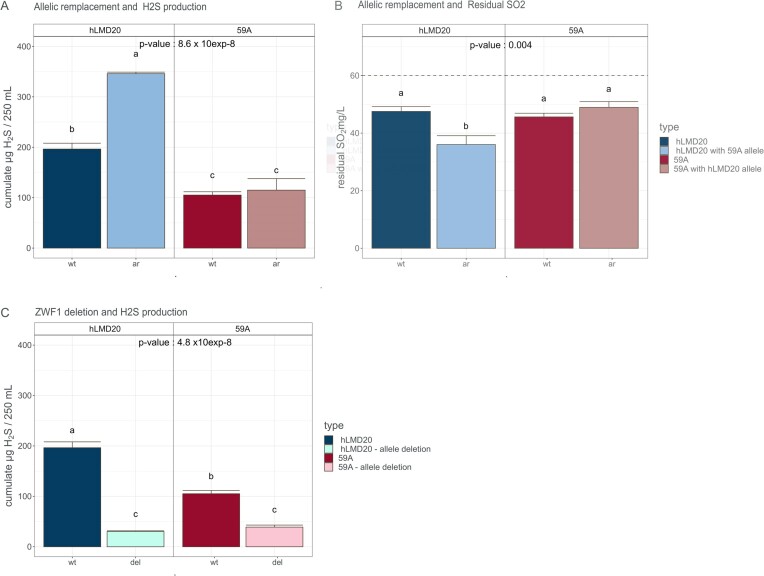
The allelic replacement of ZWF1 hLMD20 by ZWF1 59A led to a 76% increase in H2S production in hLMD20 (Fig. 5A). The differences in residual sulfite contents at the end of alcoholic fermentation were symmetrical to the production of H2S (Fig. 5B) and suggest a lower sulfite reductase activity. ZWF1 deletion also led to a decrease in sulfide biosynthesis in both parental strains (−85% for hLMD20 and −63% for 59A—Fig. 5C), which suggests that the lower production associated to ZWF1 hLMD20 is due to a less active version of the protein. As ZWF1 codes the glucose-6-phosphate dehydrogenase, we could expect that differences in enzyme activity in parental strains should lead to a difference in the production of NADPH required for the reduction of sulfate into sulfite through Met16 activity and then sulfite to sulfide by the heterotetramer Met5–Met10.
Figure 5.
Effect of different allelic versions of ZWF1 on total H2S production (A) and residual SO2 content at the end of fermentation (B) Effect of deletion of ZWF1 on total H2S production (C). For each panel, different lowercase letters (a, b, and c if present) on the top of the bars, indicate statistically significant differences between modalities after Tukey’s multiple comparison of means at 95% family-wise confidence level. wt: wild-type strain; ar: parental strain after allelic replacement (i.e. carrying the alternative allele of the other parent); del: deletion, strain without the gene under investigation. Results are given as the mean of three replicates ± standard error. The dashed line indicates the initial content of the grape must (60 mg/l).
If the decrease in H2S production resulted from a reduced activity in the PPP attributed to decreased Zwf1 activity, with a consequent decrease in the NADPH pool, we should observe an impact on other metabolites, whose synthesis is known to be sensitive to cellular redox balance, such as acetate, α-ketoglutarate, pyruvate, and succinate. Indeed, in 59A, the introduction of ZWF1 hLMD20 led to an increase in acetate (from 0.61 to 0.82 mg/l; F3,11 = 46.87, P-value = 1.48 × 10−6), while in hLMD20, ZWF1 59A triggered a decrease in α-ketoglutarate (from 0.13 to 0.07 mg/l; F3,11 = 26.58, P-value = 2.44 × 10−5) and pyruvate (from 0.31 to 0.22 mg/l; F3,11 = 16.93, P-value = 1.96 × 10−4) (Figure S3, Supporting Information). These allelic replacements did not cause any significant variation in the production of other main compounds of the central carbon metabolism (ethanol and glycerol), even though it has to be noted that fermentations were stopped when only 95% of sugars were consumed.
To better explore how this allelic exchange impacts cell physiology, we compared the transcriptome of each parental strain with its corresponding strain carrying the alternative ZWF1 allele. This allelic change had a high impact on gene expression, as 770 and 1895 genes were differentially expressed between hLMD20 and hLMD20-ZWF1 59A, and between 59A and 59A-ZWF1 hLMD20, respectively (Table 3), with 21% of differentially expressed genes shared among strains. The replacement of the 59A allele of ZWF1 by the hLMD20 allele induced a strong activation of 22 of the genes of the SAP pathway (GO Term: sulfur compound metabolic process; P-value 9.74 × 10−08; Table S6A, Supporting Information). In addition, the induction of the NADP dependent aldehyde dehydrogenase ALD6 is noteworthy, in line with reduced NADPH availability. By contrast, the introduction of the 59A allele in hLMD20 led to an induction of several genes in the PPP, and to a repression of genes involved in sulfur compound metabolic process GO category (P-value 3.69 × 10−09; Table S6D, Supporting Information). Moreover, no changes in ZWF1 expression were detected between hLMD-ZWF1 59A and 59A-ZWF1 hLMD20, supporting the hypothesis that the phenotype observed was due to a different activity of the protein rather than to a difference in expression.
Table 3.
Number of differentially expressed genes with fold change > 0.5 and FDR threshold of 5%.
| ZWF1 | ZRT2 | YLR125W | ||
|---|---|---|---|---|
| hLMD20 wt vs. hLMD20 + allele 59A | Upregulated | 425 | 510 | 302 |
| Downregulated | 345 | 488 | 295 | |
| 59A wt vs. 59A + allele hLMD20 | Upregulated | 914 | 369 | 149 |
| Downregulated | 981 | 406 | 101 |
All these metabolic and transcriptomic data are consistent with a difference between strains in the availability of NADPH caused by ZWF1 allelic variations that leads to differences in H2S production.
Zinc uptake modulates H2S production
In QTL_chrXII, ZRT2 is close to the maximum AFD of the region. The exchange of alleles in parental strains induced a symmetrical and opposite variation: for hLMD20-ZRT2 59A H2S production decreased by 37%, whereas ZRT2 hLMD20 increased H2S production by 160% in 59A (Fig. 6).
Figure 6.
Effect of different allelic versions of ZRT2 in each parental strain on total H2S production. Different lowercase letters (a, b, and c) on the top of the bars, indicate statistically significant differences after Tukey’s multiple comparison of means at 95% family-wise confidence level. wt: wild-type strain; ar: parental strain after allelic replacement (i.e. carrying the alternative allele of the other parent). Results are given as the mean of three replicates ± standard error.
As ZRT2 codes a low-affinity zinc transporter, active at the zinc concentration present in this synthetic media, we evaluated whether different zinc concentrations in the fermentation media might lead to differences in H2S total production. The response of the different strains carrying their own allele or the other parental allele is presented in Fig. 7. The H2S production of strains carrying the ZRT2 hLMD20 was correlated to zinc concentration in the media. However, strains carrying ZRT2 59A were less sensitive to variations in zinc concentration.
Figure 7.
Effect of different allelic versions of ZRT2 in each parental strain on total H2S production in response to different zinc concentrations. Results are given as the mean of three replicates with a standard error. The significance of the effect of Zn on H2S production has been evaluated globally for all strains (P-value = .006). Results are given as the mean of three replicates ± standard error.
The comparison of the transcriptome of strains carrying the two alternative alleles revealed that the reduction in H2S production in hLMD20-ZRT2 59A corresponded to a downregulation of SAP genes (Figure S4, Supporting Information) and a strong upregulation of ZRT2 allele (GO Term: sulfur compound biosynthetic process, P-value = 5.81 × 10−10; Table S8D, Supporting Information). By contrast, 59A-ZRT2 hLMD20 presented a less pronounced response with the upregulation of the SAM biosynthetic process (P-value: .017; Table S8A, Supporting Information). This response is similar to the response of yeast to a moderate zinc deficiency, which results in ZRT2 upregulation and SAP genes downregulation (Eide 2009). This suggests that ZRT2 59A transports less efficiently the divalent ion inside the cell, although it is supposed to act as the main transporter in these conditions.
By comparing the ZRT2 gene sequence of the parental strains, 59A and hLMD20, with that of 88 other strains (Coi et al. 2017, Legras et al. 2018, SGD 2021), we observed that ZRT2 59A allele had the same sequence as ZRT2 of velum strains, while ZRT2 hLMD20 had a typical wine-strain sequence (Fig. 8). ZRT2 59A allele presents three major mutations leading to changes in electrical charge and hydrophobicity: D19G, E322K, and T325M. According to a secondary structure prediction performed with PROTTER (Omasits et al. 2014) (Figure S6, Supporting Information), all three mutations impact amino acids located in the N-terminal or in a loop, both oriented to the extracellular compartment, which suggests that they could be involved in zinc ion sensing or binding.
Figure 8.

Maximum-likelihood phylogeny of ZRT2 hLMD20 and ZRT2 59A (in red) among 90 strains of different origins.
An uncharacterized novel mechanism impacting H2S production
Among the highly impacted genes encountered in the QTL located on chromosome XII, YLR125W, whose function is unknown, was present in a truncated form in the genome of 59A. YLR125W hLMD20 replacement with the truncated YLR125W 59A led to a 21% decrease in H2S production, while the replacement of YLR125W 59A by the full-length YLR125W hLMD20 led to an increase of 81% (Fig. 9A). As this phenotype could result from the inactivation of the short-version allele, we compared the result of the allelic exchange with that of parental strains deleted for YLR125W. Surprisingly, the deletion effect was only visible in the 59A genetic background, for which the magnitude of the deletion (nearly 2.5-fold higher H2S production) was even higher than the presence of the full-length allele (Fig. 9B).
Figure 9.
Effect on total H2S production of different allelic versions of YLR125W (A) and of its deletion (B) in each parental strain. For each panel, different lowercase letters (a, b, and c if present) on the top of the bars, indicate statistically significant differences after Tukey’s multiple comparison of means at 95% family-wise confidence level. wt: wild-type strain; ar: parental strain after allelic replacement (i.e. carrying the alternative allele of the other parent); del: deletion, strain without the gene under investigation. Results are given as the mean of three replicates ± standard error.
To understand the possible mechanisms involved in this trait, we also compared gene expression in each parental strain following allelic swapping. Interestingly, the allelic exchange in 59A led to less variation in expression without any GO term associated with sulfur metabolism (Table 3; Table S7A, Supporting Information). Conversely, in hLMD20-YLR125W59A, genes of the sulfur biosynthesis GO category were repressed (P-value 4.01 × 10−08), as well as genes of vitamin metabolism, including that of thiamine (P-value 1.29 × 10−02) (Table S7C, Supporting Information).
ESCRT machinery participates in the modulation of H2S production
A fourth gene was found to be involved in the regulation of total H2S production between parental strains. The evaluation of SRN2 allelic swaps performed in each parent led to opposite and symmetrical effects (Fig. 10). In hLMD20, the replacement of SRN2 hLMD20 by SRN2 59A decreased H2S production by 33%, while in 59A the replacement of the wild type allele by SRN2 hLMD20 increased this production by 83%. Interestingly, Srn2, a component of the ESCRT machinery involved in cytoplasmic membrane remodeling, participates in the degradation of the general amino acid permease Gap1 and the high affinity methionine permease Mup1. A E133G substitution is found in SRN2 hLMD20 (Table 2), whereas this E residue is conserved in yeast, fly, and human versions of the gene (Kostelansky et al. 2006).
Figure 10.
Effect of different allelic versions of SRN2 in each parental strain on total H2S production. Different lowercase letters (a, b, and c) on the top of the bars, indicate statistically significant differences after Tukey’s multiple comparison of means at 95% family-wise confidence level. wt: wild-type strain; ar: parental strain after allelic replacement (i.e. carrying the alternative allele of the other parent). Results are given as the mean of three replicates ± the standard error.
Cumulate effect of QTL_XII genes’ cluster
QTL mapping serves to elucidate the genetic bases of different mechanisms, but it can also be the first step for a targeted genetic improvement. Before starting backcrossing programs, it is wise to evaluate the effect of introgression of such a wide region as the one in chromosome XII, in the genome of the target strain to improve. Therefore, we evaluated the effect of the whole QTL identified on chromosome XII in both parental strains, applying the same double step strategy as for single-gene allelic replacements. Due to the presence of essential genes in the region, we modified hLMD20 ZRT2 59A, transformant 1, to first delete the region from upstream SRN2 to downstream YLR125W and subsequently replace it with the one amplified from 59A. As for all the candidate genes evaluated individually, the same modification was carried out on the other parent strain (59A ZRT2 hLMD20 deleted for its SRN2–YLR125W region, replaced with the one amplified from hLMD20). As shown in Fig. 11, we obtained a decrease in H2S production in hLMD20 when the region from 59A was present (−34%). Similar to the mirror effect observed in single-gene allelic replacements, 59A modification produced an increase in H2S production of 57%. However, the effect of the three genes was not cumulative, since the magnitude of the decrease due to the whole region swapping was not higher than the single-gene ones.
Figure 11.
Effect of different allelic versions of the region between SRN2 and ZRT2 in each parental strain on total H2S production. wt: wild-type strain; ar: parental strain after allelic replacement (i.e. carrying the alternative allele of the other parent). Results are given as the mean of two replicates ± standard error.
Discussion
H2S is a key metabolite essential for the synthesis of sulfur-containing amino-acids. In S. cerevisiae, its production results from SAP activity and, in the most simplistic way, from the successive activity of sulfate and sulfide reductases. Its yield depends on complex metabolic interactions, and is affected by many factors. Some of them are of considerable relevance in oenological conditions, as SO2, nitrogen, and vitamin contents of grape must. Yeast strains display variable ability to produce H2S (Spiropoulos et al. 2000) and, given H2S negative impact on wine aroma, those characterized by an excessively high production are avoided. While the SAP has been known for more than 25 years (Thomas and Surdin-Kerjan 1997), efforts have recently been made into the exploration of the genetic basis of these differences in wine or vineyard yeast isolates. Among them, the allelic variation in four SAP genes (viz; MET10, MET2, MET5, and SKP2) and one gene coding for the β-lyase TUM1 have been found to explain differences in H2S production (Cordente et al. 2009, Linderholm et al. 2010, Huang et al. 2014, 2016, Noble et al. 2015). However, these genes are directly involved with H2S production and do not tell us more about the interplay between SAPs and cell metabolism.
Using a quantitative genetic approach, that limits preset choice about the targets potentially involved, we describe here for the first time four different mechanisms that participate in the variation in H2S production, and tightly connect this production to cell metabolism.
The first mechanism described here is caused by differences in the activity of the pentose phosphate pathway (PPP), generated by a lower activity of the glucose-6-phosphate dehydrogenase, coded by ZWF1. Indeed, the reduction of one mole of sulfate requires six moles of NADPH for the synthesis of methionine. It has been shown recently from metabolic flux modeling that the decrease in NADPH availability results in a lower production of sulfur-containing compounds and to a global derepression of the sulfate assimilation pathway (Celton et al. 2012), quite in line with our results. However, this PPP modulation has one cost: a moderate increase in acetate production.
Besides the somehow expected interaction between NADPH availability and H2S production, we show for the first time that zinc availability mediated by low affinity transporter interacts with H2S production under wine making conditions.
A relation between cell Zn2+ content and sulfide synthesis has been suggested from the analysis of the adaptive response to zinc depletion (Eide 2009). In addition, it has been shown recently that the methionine synthase Met6p achieves, along with 19 other proteins, 90% of cell zinc requirements (Wang et al. 2018). Yeast cells contain three divalent Zn2+ ion transporters: the high affinity transporter Zrt1, the vacuolar transporter Zrt3 and the low affinity transporter Zrt2 (Zhao and Eide 1996a,b) that is especially active at moderate zinc concentrations (such as that of the grape must concentration mimicked by our SM, i.e; 14 µM zinc). These transporters are regulated by the transcription factor of Zap1. Under moderate zinc deficiency, ZAP1 is overexpressed to compensate for the shortage of zinc, which leads to the upregulation of ZRT2 and MET30 (Eide 2009, Wu et al. 2009). Met30 is responsible for the degradation of the transcriptional activator Met4 that regulates genes of the sulfur amino acid pathway with Cbf1, Met28, Met31, and Met32. Therefore, an upregulation of Zap1 induces a downregulation of SAP genes (Eide 2009), and thus a decrease in H2S production.
Two hypotheses could be made to explain the relation between Zn2+ concentration and H2S production. One could stem from a detoxification mechanism associated with an excess of metal ions, while the other might be the activation of Met6p favoring an intense methionine synthesis, and thus stimulating growth. These differences in zinc transport in connection with SAP may also have an ecological ground. Indeed, it is noteworthy that the ZRT2 allele of 59A is a wine–velum-type allele, whereas the allele of hLMD20 is a classical wine allele (the glycine in position 19 and the methionine in 325 are specific to wine strains). According to (Yampolsky and Stoltzfus 2005), the mutation permitting the substitution of threonine to methionine, such the one in position 325, is not very frequent. Moreover, it has recently been shown that wine and wine–velum yeast isolates have extremely different alleles for the high affinity transporter ZRT1, very likely resulting from repeated positive selection (Coi et al. 2017), as well as quite different phylogeny for ZRT3 (unpublished results).
Zinc is an essential element for vine development (Volschenk et al. 1996), and dithiocarbamate-based fungicides are also a significant source of zinc in vineyards. However, few works report the variability in zinc content of grapes and grape juice (Olalla et al. 2004, Dani et al. 2012, Ribéreau-Gayon et al. 2012, Daccak et al. 2020).
Zinc also plays a crucial role in supporting yeast balanced development, metabolic processes, and overall physiological functions. The activator role of zinc for wort fermentation has long been known in brewery (reviewed by Gibson 2011), but has been less described for wine fermentations (De Nicola et al. 2009). Interestingly, zinc is taken up in the first hours of the grape must fermentation, which it activates. In addition, Coi et al. (2017) evidenced the presence of highly different alleles of the zinc transporters Zrt1, Zrt2, and Zrt3 between velum and wine yeasts, but did not observe a clear phenotype caused by allelic swaps of wine and flor alleles of ZRT1 on velum growth. This suggests a possibly different management of zinc in velum and wine strains, in association with the ecology of these strains.
We show here for the first time the role of YLR125W on the modulation of H2S production. Surprisingly, the role of this gene could not be determined from the comparison of the deleted and wild type form, but from the presence of a truncated allele. Furthermore, the contribution of this gene to the sulfate assimilation pathway is in line with its regulation by Met31, Met32, and Cbf1 (Moxley et al. 2009) as well as its genetics interaction with Atg1 and Pho85 (Ptacek et al. 2005), and with its repression in case of leucine or sulfur limitation (Saldanha et al. 2004, Tai et al. 2005). Interestingly, when mining for the presence of this truncated allele of YLR125W, we only found it in EC1118, and in none of the 1006 sequenced strains (Peter et al. 2018), however, we could detect the high frequency of a truncated, but different, version of this gene in the groups of sake strain and Asian beverages.
The fourth mechanism participating in the modulation of H2S production is the ESCRT system. The comparison of the two allelic sequences of SRN2 to the known structure of the Srn2 complexes in yeast, fly and human enabled us to identify a mutation in the high H2S-producing parent, located in a conserved residue of the protein. This mutation might impact the assembly, or at least the stability of the complex itself, and suggests a loss of function of the hLMD20 allele. It is worth mentioning that the reference strain S288C is 30 amino acids longer, with at the N-terminal, which causes a 30aa shift in the amino acid numbering in the strains studied in comparison to S288C.
An improper functioning of the ESCRT-I complex means that the machinery of degradation of certain membrane proteins such as Mup1 and Gap1 might be impaired, and these proteins might not be sorted to the vacuole. This could lead to an improved amino acids uptake, including sulfur-containing ones, methionine and cysteine, and a higher cellular cysteine concentration can lead to an increase in H2S production (Jiranek et al. 1995, Winter et al. 2014, Huang et al. 2016).
Besides the complementarity of these four different mechanisms permitting H2S production modulation, one remarkable result is the localization of three of four of these genes in the QTL_chrXII region, within 18.5 kb regions on chromosome XII (Fig. 12). The presence of several genes with opposite effects have been mentioned in a single QTL (Ben-Ari et al. 2006, Brice et al. 2014); however, it is to our knowledge the first time that three genes acting similarly are found in a single small genomic region. This suggests the presence of a putative cluster involved in sulfur or zinc metabolism.
Figure 12.
Overview of all genes present in the QTL_chrXII region on chromosome XII. Genes in yellow impact total H2S production. Genes marked with an asterisk contain one or more nonsynonymous mutations in the ORF.
From a fermentation industry perspective, this work sheds new light on the significance of zinc content in the must for alcoholic fermentation. Zinc was already known to be an important factor for fermentation speed of high gravity musts for brewery (Gibson 2011); however, zinc content is rarely considered in wine fermentation, very likely because of the naturally high zinc concentrations of the grape must (Ribéreau-Gayon et al. 2012); In addition, the relation between zinc content and H2S production in a wine making environment has never been described yet. Given that the Zrt2 transporter is also inhibited by copper (Zhao and Eide 1996b), the impact of Cu2+, extensively used for vine sanitary management, on H2S production should be investigated. In addition, this work also shows PPP importance for H2S production. We discovered here an allele of ZWF1 allowing a low NADPH production of with a consequently low H2S production. As the NADPH content is strictly regulated, the impact of this allele of ZWF1 in different genetic backgrounds should be evaluated, to allow further genetic improvement of yeast starters.
Altogether, our results indicate that sulfide production in the cell is closely connected to cell physiology (Fig. 13), through the NADPH and Zn2+ availability, through the status of the protein degradation machinery, and through an unknown mechanism associated with YLR125W, that remains to be discovered.
Figure 13.
Updated proposal of the global regulation of total H2S production and sulfur metabolism during fermentation. This image was created with BioRender.com.
Supplementary Material
Acknowledgments
The authors thank Martine Pradal for her guidance with the RNA microarray, and Faiza Macna for the HPLC analysis.
Contributor Information
Irene De Guidi, SPO, Université de Montpellier, INRAE, Institut Agro, Montpellier 34060, France.
Céline Serre, SPO, Université de Montpellier, INRAE, Institut Agro, Montpellier 34060, France.
Jessica Noble, Lallemand SAS, Blagnac 31702, France.
Anne Ortiz-Julien, Lallemand SAS, Blagnac 31702, France.
Bruno Blondin, SPO, Université de Montpellier, INRAE, Institut Agro, Montpellier 34060, France.
Jean-Luc Legras, SPO, Université de Montpellier, INRAE, Institut Agro, Montpellier 34060, France.
Conflict of interest
None declared.
Funding
This work has received funding from the European Union’s Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie Actions (grant agreement number 764364) and from the French National Research Agency (funding agreement number ANR-21-PRRD-0008-01).
Data Availability
Raw genomic data are available from ENA under bioprojects PRJEB67897 and PRJEB6529 (SAMEA3730514). Transcriptome data are available at GEO database under number GSE24631. Experimental data are available at DOI: https://doi.org/10.57745/DV6ZZ1.
References
- Ambroset C, Petit M, Brion C et al. Deciphering the molecular basis of wine yeast fermentation traits using a combined genetic and genomic approach. G3 genes genomes Genet. 2011;1:263–81. 10.1534/g3.111.000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bely M, Sablayrolles J-M, Barre P. Automatic detection of assimilable nitrogen deficiencies during alcoholic fermentation in oenological conditions. J Ferment Bioeng. 1990;70:246–52. 10.1016/0922-338X(90)90057-4. [DOI] [Google Scholar]
- Ben-Ari G, Zenvirth D, Sherman A et al. Four linked genes participate in controlling sporulation efficiency in budding yeast. PLoS Genet. 2006;2:1815–23. 10.1371/journal.pgen.0020195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blondin B, Noble J, Sanchez I. Method for controlling the production of sulphites, of hydrogen sulphide and of acetaldehyde by yeasts. 2017, US patent 9,551,000 B2, 1–7. [Google Scholar]
- Bohlscheid JC, Fellman JK, Wang XD et al. The influence of nitrogen and biotin interactions on the performance of Saccharomyces in alcoholic fermentations. J Appl Microbiol. 2007;102:390–400. 10.1111/j.1365-2672.2006.03180.x. [DOI] [PubMed] [Google Scholar]
- Bohlscheid JC, Osborne JP, Ross CF et al. Interactive effects of selected nutrients and fermentation temperature on H2S production by wine strains of Saccharomyces. J Food Qual. 2011;34:51–5. 10.1111/j.1745-4557.2010.00365.x. [DOI] [Google Scholar]
- Bolger AM, Lohse M, Usadel B. Trimmomatic. A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–20. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brauer MJ, Christianson CM, Pai DA et al. Mapping novel traits by array-assisted bulk segregant analysis in Saccharomyces cerevisiae. Genetics. 2006;173:1813–6. 10.1534/genetics.106.057927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brice C, Sanchez I, Bigey F et al. A genetic approach of wine yeast fermentation capacity in nitrogen-starvation reveals the key role of nitrogen signaling. BMC Genomics. 2014;15:495. 10.1186/1471-2164-15-495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brion C, Ambroset C, Sanchez I et al. Differential adaptation to multi-stressed conditions of wine fermentation revealed by variations in yeast regulatory networks. BMC Genomics. 2013;14:681. 10.1186/1471-2164-14-681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Celton M, Sanchez I, Goelzer A et al. A comparative transcriptomic, fluxomic and metabolomic analysis of the response of Saccharomyces cerevisiae to increases in NADPH oxidation. BMC Genomics. 2012;13:317. 10.1186/1471-2164-13-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani P, Platts A, Wang LL et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff. Fly. 2012;6:80–92. 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Codon AC, Gasent-Ramirez JM, Benitez T. Factors which affect the frequency of sporulation and tetrad formation in Saccharomyces cerevisiae baker's yeasts. Appl Environ Microb. 1995;61:630–8. 10.1128/aem.61.2.630-638.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coi AL, Bigey F, Mallet S et al. Genomic signatures of adaptation to wine biological ageing conditions in biofilm-forming flor yeasts. Mol Ecol. 2017;26:2150–66. 10.1111/mec.14053. [DOI] [PubMed] [Google Scholar]
- Cordente AG, Borneman AR, Bartel C et al. Inactivating mutations in Irc7p are common in wine yeasts, attenuating carbonsulfur β-lyase activity and volatile sulfur compound production. Appl Environ Microb. 2019;85:1–14. 10.1128/AEM.02684-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordente AG, Heinrich A, Pretorius IS et al. Isolation of sulfite reductase variants of a commercial wine yeast with significantly reduced hydrogen sulfide production. FEMS Yeast Res. 2009;9:446–59. 10.1111/j.1567-1364.2009.00489.x. [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Billi E, Zörgö E et al. Assessing the complex architecture of polygenic traits in diverged yeast populations. Mol Ecol. 2011;20:1401–13. 10.1111/j.1365-294X.2011.05005.x. [DOI] [PubMed] [Google Scholar]
- Cubillos FA, Brice C, Molinet J et al. Identification of nitrogen consumption genetic variants in yeast through QTL mapping and bulk segregant RNA-seq analyses. G3 Genes Genomes Genet. 2017;7:1693–705. 10.1534/g3.117.042127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cubillos FA, Parts L, Salinas F et al. High-resolution mapping of complex traits with a four-parent advanced intercross yeast population. Genetics. 2013;195:1141–55. 10.1534/genetics.113.155515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daccak D, Coelho ARF, Marques AC et al. Grape enrichment with zinc for vinification: mineral analysis with atomic absorption spectrophotometry, XRF and tissue analysis. The 1st International Electronic Conference on Plant Science. MDPI. Basel Switzerland, 2020, 84. https://www.mdpi.com/2673-9976/4/1/84. [Google Scholar]
- Dani C, Oliboni LS, Pra D et al. Mineral content is related to antioxidant and antimutagenic properties of grape juice. Genet Mol Res. 2012;11:3154–63. 10.4238/2012.September.3.4. [DOI] [PubMed] [Google Scholar]
- De Guidi I, Farines V, Legras JL et al. Development of a new assay for measuring H2S production during alcoholic fermentation: application to the evaluation of the main factors impacting H2S production by three Saccharomyces cerevisiae wine strains. Fermentation. 2021;7:213. 10.3390/fermentation7040213. [DOI] [Google Scholar]
- De Guidi I, Galeote V, Blondin B et al. How copper based grape pest management has impacted wine aroma. BioRxiv. 2023. https://doi.org/10.1101/2023.11.12.566766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Nicola R, Hall N, Bollag T et al. Zinc accumulation and utilization by wine yeasts. Int J Wine Res. 2009;1:85–94. 10.2147/IJWR.S4570. [DOI] [Google Scholar]
- Eder M, Nidelet T, Sanchez I et al. QTL mapping of modelled metabolic fluxes reveals gene variants impacting yeast central carbon metabolism. Sci Rep. 2020;10:2162. 10.1038/s41598-020-57857-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eder M, Sanchez I, Brice C et al. QTL mapping of volatile compound production in Saccharomyces cerevisiae during alcoholic fermentation. BMC Genomics. 2018;19:166. 10.1186/s12864-018-4562-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eide DJ. Homeostatic and adaptive responses to zinc deficiency in Saccharomyces cerevisiae. J Biol Chem. 2009;284:18565–9. 10.1074/jbc.R900014200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson BR. 125th anniversary review: improvement of higher gravity brewery fermentation via wort enrichment and supplementation. J Inst Brew. 2011;117:268–84. 10.1002/j.2050-0416.2011.tb00472.x. [DOI] [Google Scholar]
- Gietz RD, Schiestl RH. High-efficiency yeast transformation using the LiAc/SS carrier DNA/PEG method. Nat Protoc. 2007;2:31–4. 10.1038/nprot.2007.13. [DOI] [PubMed] [Google Scholar]
- Haas R, Horev G, Lipkin E et al. Mapping ethanol tolerance in budding yeast reveals high genetic variation in a wild isolate. Front Genet. 2019;10:998. 10.3389/fgene.2019.00998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho PW, Piampongsant S, Gallone B et al. Massive QTL analysis identifies pleiotropic genetic determinants for stress resistance, aroma formation, and ethanol, glycerol and isobutanol production in Saccharomyces cerevisiae. Biotechnol Biofuels. 2021;14:1–18. 10.1186/s13068-021-02059-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu XH, Wang MH, Tan T et al. Genetic dissection of ethanol tolerance in the budding yeast Saccharomyces cerevisiae. Genetics. 2007;175:1479–87. 10.1534/genetics.106.065292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C, Roncoroni M, Gardner RC. MET2 affects production of hydrogen sulfide during wine fermentation. Appl Microbiol Biotechnol. 2014;98:7125–35. 10.1007/s00253-014-5789-1. [DOI] [PubMed] [Google Scholar]
- Huang CW, Walker ME, Fedrizzi B et al. The yeast TUM1 affects production of hydrogen sulfide from cysteine treatment during fermentation. FEMS Yeast Res. 2016;16:fow100. 10.1093/femsyr/fow100. [DOI] [PubMed] [Google Scholar]
- Huang L, Tang W, Bu S et al. BRM: a statistical method for QTL mapping based on bulked segregant analysis by deep sequencing. Bioinformatics. 2020;36:2150–6. 10.1093/bioinformatics/btz861. [DOI] [PubMed] [Google Scholar]
- Jacoby WG. Loess: a nonparametric, graphical tool for depicting relationships between variables. Elect Stud. 2000;19:577–613. 10.1016/S0261-3794(99)00028-1. [DOI] [Google Scholar]
- Jara M, Cubillos FA, García V et al. Mapping genetic variants underlying differences in the central nitrogen metabolism in fermenter yeasts. PLoS ONE. 2014;9:e86533. 10.1371/journal.pone.0086533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiranek V, Langridge P, Henschke PA. Regulation of hydrogen sulfide liberation in wine-producing Saccharomyces cerevisiae strains by assimilable nitrogen. Appl Environ Microb. 1995;61:461–7. 10.1128/aem.61.2.461-467.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katou T, Namise M, Kitagaki H et al. QTL mapping of sake brewing characteristics of yeast. J Biosci Bioeng. 2009;107:383–93. 10.1016/j.jbiosc.2008.12.014. [DOI] [PubMed] [Google Scholar]
- Kostelansky MS, Sun J, Lee S et al. Structural and functional organization of the ESCRT-I trafficking complex. Cell. 2006;125:113–26. 10.1016/j.cell.2006.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legras J-L, Galeote V, Bigey F et al. Adaptation of S. cerevisiae to fermented food environments reveals remarkable genome plasticity and the footprints of domestication. Mol Biol Evol. 2018;35:1712–27. 10.1093/molbev/msy066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–60. 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linderholm A, Dietzel K, Hirst M et al. Identification of MET10-932 and characterization as an allele reducing hydrogen sulfide formation in wine strains of Saccharomyces cerevisiae. Appl Environ Microb. 2010;76:7699–707. 10.1128/AEM.01666-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magwene PM, Willis JH, Kelly JK. The statistics of bulk segregant analysis using next generation sequencing. PLoS Comput Biol. 2011;7:1–9. 10.1371/journal.pcbi.1002255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marullo P, Aigle M, Bely M et al. Single QTL mapping and nucleotide-level resolution of a physiologic trait in wine Saccharomyces cerevisiae strains. FEMS Yeast Res. 2007;7:941–52. 10.1111/j.1567-1364.2007.00252.x. [DOI] [PubMed] [Google Scholar]
- Marullo P, Bely M, Masneuf-Pomarède I et al. Breeding strategies for combining fermentative qualities and reducing off-flavor production in a wine yeast model. FEMS Yeast Res. 2006;6:268–79. 10.1111/j.1567-1364.2006.00034.x [DOI] [PubMed] [Google Scholar]
- Mendes-Ferreira A, Barbosa C, Falco V et al. The production of hydrogen sulphide and other aroma compounds by wine strains of Saccharomyces cerevisiae in synthetic media with different nitrogen concentrations. J Ind Microbiol Biotechnol. 2009;36:571–83. 10.1007/s10295-009-0527-x. [DOI] [PubMed] [Google Scholar]
- Moxley JF, Jewett MC, Antoniewicz MR et al. Linking high-resolution metabolic flux phenotypes and transcriptional regulation in yeast modulated by the global regulator Gcn4p. Proc Natl Acad Sci USA. 2009;106:6477–82. 10.1073/pnas.0811091106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noble J, Sanchez I, Blondin B. Identification of new Saccharomyces cerevisiae variants of the MET2 and SKP2 genes controlling the sulfur assimilation pathway and the production of undesirable sulfur compounds during alcoholic fermentation. Microb Cell Fact. 2015;14:68. 10.1186/s12934-015-0245-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olalla M, Fernández J, Cabrera C et al. Nutritional study of copper and zinc in grapes and commercial grape juices from Spain. J Agric Food Chem. 2004;52:2715–20. 10.1021/jf030796w. [DOI] [PubMed] [Google Scholar]
- Omasits U, Ahrens CH, Müller S et al. Protter: interactive protein feature visualization and integration with experimental proteomic data. Bioinformatics. 2014;30:884–6. 10.1093/bioinformatics/btt607. [DOI] [PubMed] [Google Scholar]
- Peter J, De Chiara M, Friedrich A et al. Genome evolution across 1,011 Saccharomyces cerevisiae isolates. Nature. 2018;556:339–44. 10.1038/s41586-018-0030-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J, Devgan G, Michaud G et al. Global analysis of protein phosphorylation in yeast. Nature. 2005;438:679–84. 10.1038/nature04187. [DOI] [PubMed] [Google Scholar]
- R Core Team . R: a language and environment for statistical computing. 2020. https://www.r-project.org/ (25 September 2023, date last accessed).
- Ribéreau-Gayon P, Glories Y, Maujean A et al. L'extrait Sec et Les matières Minérales. In: Traité d'oenologie—Tome 2 Chimie Du Vin Stabilisation et Traitement, Paris: Dunod, 2012, 117–40. ISBN 978-2-10-076561-4. [Google Scholar]
- Ritchie ME, Phipson B, Wu D et al. Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rollero S, Mouret JR, Sanchez I et al. Key role of lipid management in nitrogen and aroma metabolism in an evolved wine yeast strain. Microb Cell Fact. 2016;15:1–15. 10.1186/s12934-016-0434-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saldanha AJ, Brauer MJ, Botstein D. Nutritional homeostasis in batch and steady-state culture of yeast. MBoC. 2004;15:4089–104. 10.1091/mbc.e04-04-0306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saubin M, Devillers H, Proust L et al. Investigation of genetic relationships between Hanseniaspora species found in grape musts revealed interspecific hybrids with dynamic genome structures. Front Microbiol. 2020;10:2960. 10.3389/fmicb.2019.02960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SGD .“Saccharomyces Genome Database | SGD”. 2021. https://www.yeastgenome.org/ (25 July 2023, date last accessed). [DOI] [PMC free article] [PubMed]
- Sinha H, David L, Pascon RC et al. Sequential elimination of major-effect contributors identifies additional quantitative trait loci conditioning high-temperature growth in yeast. Genetics. 2008;180:1661–70. 10.1534/genetics.108.092932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha H, Nicholson BP, Steinmetz LM et al. Complex genetic interactions in a quantitative trait locus. PLoS Genet. 2006;2:140–7. 10.1371/journal.pgen.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith EN, Kruglyak L. Gene–environment interaction in yeast gene expression. PLoS Biol. 2008;6:e83. 10.1371/journal.pbio.0060083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Gibney PA, Cheng L et al. Yeast assimilable nitrogen concentrations influence yeast gene expression and hydrogen sulfide production during cider fermentation. Front Microbiol. 2020;11:1264. 10.3389/fmicb.2020.01264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spiropoulos A, Tanaka J, Flerianos I et al. Characterization of hydrogen sulfide formation in commercial and natural wine isolates of Saccharomyces. Am J Enol Vitic. 2000;51:233–48. 10.5344/ajev.2000.51.3.233. [DOI] [Google Scholar]
- Stamatakis A. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30:1312–3. 10.1093/bioinformatics/btu033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steyer D, Ambroset C, Brion C et al. QTL mapping of the production of wine aroma compounds by yeast. BMC Genomics. 2012;13:573. 10.1186/1471-2164-13-573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tai SL, Boer VM, Daran-Lapujade P et al. Two-dimensional transcriptome analysis in chemostat cultures. J Biol Chem. 2005;280:437–47. 10.1074/jbc.M410573200. [DOI] [PubMed] [Google Scholar]
- Thomas D, Surdin-Kerjan Y. Metabolism of sulfur amino acids in Saccharomyces cerevisiae. Microbiol Mol Biol Rev. 1997;61:503–32. 10.1128/mmbr.61.4.503-532.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ugliano M, Fedrizzi B, Siebert T et al. Effect of nitrogen supplementation and Saccharomyces species on hydrogen sulfide and other volatile sulfur compounds in Shiraz fermentation and wine. J Agric Food Chem. 2009;57:4948–55. 10.1021/jf8037693. [DOI] [PubMed] [Google Scholar]
- Ugliano M, Kolouchova R, Henschke PA. Occurrence of hydrogen sulfide in wine and in fermentation: influence of yeast strain and supplementation of yeast available nitrogen. J Ind Microbiol Biotechnol. 2011;38:423–9. 10.1007/s10295-010-0786-6. [DOI] [PubMed] [Google Scholar]
- Villarroel CA, Bastías M, Canessa P et al. Uncovering divergence in gene expression regulation in the adaptation of yeast to nitrogen scarcity. mSystems. 2021;6:1e00466–21. 10.1128/mSystems.00466-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volschenk CG, Hunter JJ, Watts JE. The effect of different zinc levels on the growth of grapevines. J Plant Nutr. 1996;19:827–37. 10.1080/01904169609365165. [DOI] [Google Scholar]
- Vos PJA, Gray RS. The origin and control of hydrogen sulfide during fermentation of grape must. Am J Enol Vitic. 1979;30:187–97. 10.5344/ajev.1979.30.3.187. [DOI] [Google Scholar]
- Walvekar AS, Laxman S. Methionine at the heart of anabolism and signaling: perspectives from budding yeast. Front Microbiol. 2019;10:2624. 10.3389/fmicb.2019.02624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XD, Bohlscheid JC, Edwards CG. Fermentative activity and production of volatile compounds by Saccharomyces grown in synthetic grape juice media deficient in assimilable nitrogen and/or pantothenic acid. J Appl Microbiol. 2003;94:349–59. 10.1046/j.1365-2672.2003.01827.x. [DOI] [PubMed] [Google Scholar]
- Wang Y, Weisenhorn E, MacDiarmid CW et al. The cellular economy of the Saccharomyces cerevisiae zinc proteome. Metallomics. 2018;10:1755–76. 10.1039/C8MT00269J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkening S, Lin G, Fritsch ES et al. An evaluation of high-throughput approaches to QTL mapping in Saccharomyces cerevisiae. Genetics. 2014;196:853–65. 10.1534/genetics.113.160291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter G, Cordente AG, Curtin C. Formation of hydrogen sulfide from cysteine in Saccharomyces cerevisiae BY4742: genome wide screen reveals a central role of the vacuole. PLoS ONE. 2014;9:1–20. 10.1371/journal.pone.0113869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu C-Y, Roje S, Sandoval FJ et al. Repression of sulfate assimilation is an adaptive response of yeast to the oxidative stress of zinc deficiency. J Biol Chem. 2009;284:27544–56. 10.1074/jbc.M109.042036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xing H, Edwards CG. Hydrogen sulphide production by Saccharomyces cerevisiae UCD 522 in a synthetic grape juice medium deficient of thiamin (vitamin B1) and/or pyridoxine (vitamin B6). Lett Appl Microbiol. 2019;69:379–84. 10.1111/lam.13217. [DOI] [PubMed] [Google Scholar]
- Yampolsky LY, Stoltzfus A. The exchangeability of amino acids in proteins. Genetics. 2005;170:1459–72. 10.1534/genetics.104.039107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao H, Eide D. The ZRT2 gene encodes the low affinity zinc transporter in Saccharomyces cerevisiae. J Biol Chem. 1996a;271:23203–10. 10.1074/jbc.271.38.23203. [DOI] [PubMed] [Google Scholar]
- Zhao H, Eide D. The yeast ZRT1 gene encodes the zinc transporter protein of a high-affinity uptake system induced by zinc limitation. Proc Natl Acad Sci USA. 1996b;93:2454–8. 10.1073/pnas.93.6.2454. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw genomic data are available from ENA under bioprojects PRJEB67897 and PRJEB6529 (SAMEA3730514). Transcriptome data are available at GEO database under number GSE24631. Experimental data are available at DOI: https://doi.org/10.57745/DV6ZZ1.