Abstract
Retrovirus plus-strand synthesis is primed by a cleavage remnant of the polypurine tract (PPT) region of viral RNA. In this study, we tested replication properties for Moloney murine leukemia viruses with targeted mutations in the PPT and in conserved sequences upstream, as well as for pools of mutants with randomized sequences in these regions. The importance of maintaining some purine residues within the PPT was indicated both by examining the evolution of random PPT pools and from the replication properties of targeted mutants. Although many different PPT sequences could support efficient replication and one mutant that contained two differences in the core PPT was found to replicate as well as the wild type, some sequences in the core PPT clearly conferred advantages over others. Contributions of sequences upstream of the core PPT were examined with deletion mutants. A conserved T-stretch within the upstream sequence was examined in detail and found to be unimportant to helper functions. Evolution of virus pools containing randomized T-stretch sequences demonstrated marked preference for the wild-type sequence in six of its eight positions. These findings demonstrate that maintenance of the T-rich element is more important to viral replication than is maintenance of the core PPT.
Upon entering cells, retroviruses convert their single-stranded RNA genomes to double-stranded DNA. As minus-strand DNA is synthesized, the RNase H activity of reverse transcriptase (RT) degrades the RNA in the RNA/DNA duplex. However, one portion of the RNA, the polypurine tract (PPT), is resistant to this RNase H degradation. The PPT RNA is subsequently used as the primer for plus-strand synthesis (2).
The region of the retroviral genome required for plus-strand priming was initially characterized by Sorge and Hughes, who noted that more than 9 but not more than 29 bases upstream of the primer cleavage site are required for avian sarcoma virus replication (30). In a previous study, we established that sequences as far upstream as −28 (where −1 refers to the base immediately upstream of the cleavage site) are required for Moloney murine leukemia virus (Mo-MLV) plus-strand priming and that a T-rich stretch in this region is critical (24). Noad et al. have similarly established that T-rich sequences upstream of the PPT are required for plus-strand priming for the pararetrovirus cauliflower mosaic virus (16). Additionally, Ilyinskii et al. have demonstrated that the T stretch upstream of the simian immunodeficiency virus (SIV) PPT is required for SIV replication (11), and a T stretch upstream of the Ty1 PPT is important for plus-strand priming and transposition of that yeast retroelement (33).
Several reports have examined roles of sequences within the PPT in plus-strand priming: most using model templates in purified reactions. Rattray and Champoux demonstrated that when PPTs with mutations at position −1, −2, −4, or −7 were tested, additional cleavage sites appeared, suggesting that the integrity of these positions is necessary for Mo-MLV cleavage specificity (22). Powell and Levin showed that the cleavage site-proximal half of the human immunodeficiency virus type 1 (HIV-1) PPT is required for plus-strand priming, while the cleavage site-distal half of the PPT is expendable in vitro (20). Similarly, the PPT's cleavage site-proximal G stretch is important for plus-strand priming during the replication of both cauliflower mosaic virus and Mo-MLV, although individual targeted mutations within this region are tolerated by Mo-MLV (16, 24). In this study, we examined how much genetic variation within the PPT and its upstream T stretch was compatible with replication by determining which sequences persisted in replicating Mo-MLV populations when these regions initially contained randomized sequences.
MATERIALS AND METHODS
Plasmid construction.
3′ untranslated region mutations were introduced into a Mo-MLV provirus plasmid, pMLV-neo (24), or the packaging mutant pMLV Ψ− (17). Mutations were introduced using PCR-mediated site-directed mutagenesis and other standard techniques. Sequences of all 3′ untranslated regions were confirmed by dideoxy sequencing (Sequenase II kit; U.S. Biochemical). Oligonucleotides that were synthesized in the University of Michigan Biomedical Research Core Facility and that contained the indicated mixtures of nucleotides at specified positions were used to synthesize the mutant pools. Ligation mixtures for generating plasmid pools were introduced into Escherichia coli DH5α cells by electroporation. Calculated pool sizes represent the total number of colonies obtained multiplied by the percentage of colonies expected to contain correct plasmids. This percentage was based on screening 10 to 30 individual colonies from the transformation plates for each pool to determine the percentage that contained a pool plasmid instead of the highly replication-defective parental plasmid. Bacterial colonies were then pooled and propagated to produce the pooled plasmid preparations subsequently used to transform mammalian cells. Note that several of the pools used here contained significantly fewer members than would be required to represent the theoretical genetic complexity possible from the degenerate oligonucleotides used in mutagenesis. Instances where this is the case are indicated in the text. In some instances, degenerate positions within the oligonucleotides were inadvertently biased toward particular residues rather than containing all intended substitutions at equal levels. However, high levels of sequence heterogeneity were nonetheless present within all pools, as confirmed by sequencing several individual members of each pool (Table 1, parts A, D, F, H, and J, and data not shown). These sequencing analyses suggested that for all pools, for every 20 pool members, there were at least 17 different individual sequences, and in most instances even less sequence repetition was observed. Note that two different degenerate oligonucleotide preparations, each with its own biases in substitutions, were used to generate the GATC PPT pools; one preparation was used to synthesize the 530-member pool, and the other was used to generate the larger pools.
TABLE 1.
Sequences of degenerate pools before and after passage
| Source of sequencesa | Sequenceb | Data |
|---|---|---|
| Wild typebc | TTTTATTTAGTCTCCAGAAAAAGGGGGG AAT | 5 |
| A (founding members of 179-member purine PPT poolc) | ||
| TTTTATTTAGTCTCCAGAGAGGGGGAGA AAT | 6 | |
| TTTTATTTAGTCTCCAGAAAAGGGAGGG AAT | 6 | |
| TTTTATTTAGTCTCCAGAAGGGGAGGGG AAT | 6 | |
| TTTTATTTAGTCTCCAGAAGGGGGAGGG AAT | 6 | |
| TTTTATTTAGTCTCCAGAAGAGGGAAGG AAT | 9 | |
| TTTTATTTAGTCTCCAGAAGGAGGGGAA AAT | 9 | |
| TTTTATTTAGTCTCCAGAGAGGGAGGGA AAT | 9 | |
| TTTTATTTAGTCTCCAGGGGAGGGGGGA AAT | 9 | |
| TTTTATTTAGTCTCCAG–AGAGGGGGAG AAT | 9 | |
| TTTTATTTAGTCTCCAGAGAGAGGAAGG AAT | 9 | |
| TTTTATTTAGTCTCCAGAAGGGGAGGAA AAT | 9 | |
| TTTTATTTAGTCTCCAGAAGGAAGGGAG AAT | 9 | |
| TTTTATTTAGTCTCCAGAGGGGAGGGGG AAT | 9 | |
| TTTTATTTAGTCTCCAGGGGAGGAAGGG AAT | 12 | |
| TTTTATTTAGTCTCCAGAGGGGGGGAGG AAT | 12 | |
| TTTTATTTAGTCTCCAGGGGAGGGGGAG AAT | 12 | |
| TTTTATTTAGTCTCCAGGGAGAGAGGAG AAT | 12 | |
| TTTTATTTAGTCTCCAGAGGGAGGGGAA AAT | 15 | |
| TTTTATTTAGTCTCCAGAGGGGGAGAAA AAT | 21 | |
| TTTTATTTAGTCTCCAGAGGGGAAGAGG AAT | 21 | |
| TTTTATTTAGTCTCCAGAGGGGAGGAAA AAT | 24 | |
| TTTTATTTAGTCTCCAGGGGAAGGGAAA AAT | 29 | |
| TTTTATTTAGTCTCCAGGGGGAAGGAGA AAT | >60 | |
| TTTTATTTAGTCTCCAGGGGGGGAAGAG AAT | >60 | |
| TTTTATTTAGTCTCCAGAGGGGAGGAGG AAT | >60 | |
| TTTTATCTAGTCTCCAGGGAGGGGAGGA AAT | >60 | |
| TTTTATTTAGTCTCCAGAGGGGGGAGAG AAT | >60 | |
| B (179-member purine PPT pool clones after first passaged) | ||
| TTTTATTTAGTCTCCAGAAGAAGGAGGG AAT | 10 | |
| TTTTATTTAGTCTCCAGAGAGGGGGAGA AAT | 2 | |
| TTTTATTTAGTCTCCAGAAAAGAGGAGG AAT | 4 | |
| TTTTATTTAGTCTCCAGAGGGGGGGGGG AAT | 1 | |
| TTTTATTTAGTCTCCAGGGAGGGGGGGA AAT | 1 | |
| TTTTATTTAGTCTCCAGAAGGGGGGGGA AAT | 1 | |
| TTTTATTTAGTCTCCAGGAAGGGGGGGG AAT | 1 | |
| TTTTATTTAGTCTCCAGAAGAAGGAAGG AAT | 1 | |
| C (predominant purine PPT pool sequences after passagec) | ||
| 179-member pool | TTTTATTTAGTCTCCAGAAGAAGGAGGG AAT | 5 |
| 500-member pool | TTTTATTTAGTCTCCAGAAAGAGGGGGG AAT | ∼5 |
| 6,300-member pool | TTTTATTTAGTCTCCAGRRRRRGGGGGG AAT | ∼5 |
| D (founding members of 530-member GATC PPT poolc) | ||
| TTTTATTTAGTCTCCAGAGATAGGGAGG AAT | 6 | |
| TTTTATTTAGTCTCCAGGGAGATGGAAA AAT | 12 | |
| TTTTATTTAGTCTCCAGCGGGAAGTGAG AAT | 12 | |
| TTTTATTTAGTCTCCAGAAGGAAAGGGG AAT | 12 | |
| TTTTATTTAGTCTCCAGAATAAAAGAGG AAT | 33 | |
| TTTTATTTAGTCTCCAGGACCGAATAAC AAT | >60 | |
| TTTTATTTAGTCTCCAGCGATTTAAAGG AAT | >60 | |
| TTTTATTTAGTCTCCAG–GAATAGCTTG AAT | >60 | |
| TTTTATTTAGTCTCCAGATTAGATAAGC AAT | >60 | |
| TTTTATTTAGTCTCCAGATTTAGAAGAA AAT | >60 | |
| TTTTATTTAGTCTCCAGCGGGAATAAAA AAT | >60 | |
| TTTTATTTAGTCTCCAGTTAAGAAGACA AAT | >60 | |
| TTTTATTTAGTCTCCAG––AGAAAAACT AAT | >60 | |
| E (predominant GATC PPT pool sequences after passagec) | ||
| Subset of 530-member pool | TTTTATTTAGTCTCCAGGTAAAGGGATA AAT | 7.5 |
| 530-member pool | TTTTATTTAGTCTCCAGARRWAGGGRRR AAT | NDe |
| 4,000-member pool | TTTTATTTAGTCTCCAGGGAGAGGGGGG AAT | ND |
| 5,000-member pool | TTTTATTTAGTCTCCAGAAAAAGGGGGG AAT | ND |
| F (founding members of GATC T-stretch pool) | ||
| ATTCTTCGAGTCTCCAGAAAAAGGGGGG AAT | ||
| ATTGTTCTAGTCTCCAGAAAAAGGGGGG AAT | ||
| CTTCTTGAAGTCTCCAGAAAAAGGGGGG AAT | ||
| GCCCACGTAGTCTCCAGAAAAAGGGGGG AAT | ||
| GTCTGCTTAGTCTCCAGAAAAAGGGGGG AAT | ||
| G (predominant GATC T-stretch pool sequences after passage) | ||
| Expt 1 | TTTTATWTAGTCTCCAGAAAAAGGGGGG AAT | |
| Expt 2 | TTTTATTYAGTCTCCAGAAAAAGGGGGG AAT | |
| H (founding members of TC T-stretch pool) | ||
| CTCCACCTAGTCTCCAGAAAAAGGGGGA AAT | ||
| TCCCGCCCAGTCTCCAGAAAAAGGGGGA AAT | ||
| CTCCATTCAGTCTCCAGAAAAAGGGGGA AAT | ||
| CTTCACTCAGTCTCCAGAAAAAGGGGGA AAT | ||
| TCCCGCCTAGTCTCCAGAAAAAGGGGGA AAT | ||
| I (TC T-stretch pool clones after first passaged) | ||
| TTTTATTCAGTCTCCAGAAAAAGGGGGA AAT | 4 | |
| TTTTATTCAGTCTCCAGAAAAAGGGGGG AAT | 1 | |
| J (founding members of TA T-stretch pool) | ||
| TTATTTAAAGTCTCCAGAAAAAGGGGGA AAT | ||
| ATTAATAAAGTCTCCAGAAAAAGGGGGA AAT | ||
| TAATTTTAAGTCTCCAGAAAAAGGGGGA AAT | ||
| ATTATATAAGTCTCCAGAAAAAGGGGGA AAT | ||
| ATAAAAATAGTCTCCAGAAAAAGGGGGA AAT | ||
| K (TA T-stretch pool clones after first passaged) | ||
| TTTTATTTAGTCTCCAGAAAAAGGGGGA AAT | 6 | |
| ATTTATTTAGTCTCCAGAAAAAGGGGGA GAT | 1 | |
| TTTTATTAAGTCTCCAGAAAAAGGGGGA AAT | 1 |
Founding members are sequences of clones that were part of the original transfected pool. Clones after first passage are sequences of individual clones after one passage of the pool. Predominant sequences are pool sequences when no further evolution of the pool was detectable.
Sequences varied in pools are underlined; positions that differ from wild type are in boldface. The space in each sequence denotes the PPT/U3 junction. R is any purine, Y is any pyrimidine, and W is T or A.
Data column shows replication efficiency measured as a function of days after infection that viral spread was detectable.
Data column shows the number of separate clones analyzed that possessed this sequence.
ND, not determined.
Tandem PPT constructs have been described previously (24). The PPT2 insertion is a duplication of sequences upstream of and including the PPT (positions −1 to −41) followed by a mutant att sequence. Deletions were introduced upstream of PPT1 or PPT2 by PCR mutagenesis.
Cells.
NIH 3T3 cells, Rat2 cells, and XC cells and derivatives were grown in Dulbecco's modified Eagle's medium supplemented with 10% calf serum (Gibco). 293T cells were grown in Dulbecco's modified Eagle's medium with 10% fetal bovine serum (HyClone). Puromycin-resistant 3T3 cells were selected in puromycin (6 μg/ml; Sigma).
Replication assays of targeted mutants.
Forty percent confluent 6-cm-diameter plates of 293T cells were cotransfected with pMLV-neo derivatives and the LacZ reporter plasmid pCH110 (8), using CaPO4 (15) or Lipofectamine as instructed by the manufacturer (Gibco). Two days posttransfection, supernatants were harvested and cells were stained with 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal) to determine transfection efficiency as previously described (17). Five to 20% of the cells typically stained blue in this assay.
Virus from transient transfections was polyethylene glycol precipitated (18) before quantification by RT DNA polymerase levels (32). Serial dilutions of wild-type virus were assayed to establish a standard curve for each experiment. All values were within the linear range of the assay as previously described (24).
After virus was quantified, amounts equivalent to 100 μl of wild-type virus were used to infect 10% confluent 6-cm-diameter plates of 3T3 cells. Infections were performed in a total volume of 800 μl in the presence of 8 μg of hexadimethrine bromide (Polybrene; Sigma) per ml for 2 h at 37°C. Culture medium was sampled, and cells were passaged 1:10 every 3 days thereafter. Virus spread was monitored by assaying RT activity (7). The infectivity of each mutant was tested at least twice.
Replication assays of degenerate pools.
Degenerate pool plasmids were transfected into 293T cells, and virus was quantified by RT activity as described above. The number of infectious virions per volume of supernatant was also determined by XC assay as previously described (25). Briefly, 10% confluent 10-cm-diameter plates of 3T3 cells were infected with serial dilutions of virus. After 2 days, supernatants were removed and infected cells were UV irradiated for 10 s by exposure to the sterilizing lamp in a biosafety cabinet. Media containing ∼106 XC cells was added; 2 days later, cells were fixed by adding 0.5 ml of formaldehyde and stained with hematoxylin. Plaques were counted using a 10× magnification dissecting microscope.
After quantification of virus, 3T3 cells were infected as above with the equivalent of enough virus to saturate the pools (approximately three times as many PFU, as determined by XC assay, as the size of the pool used). Once cells infected with pooled viruses showed detectable virus spread by RT assay, subsequent infections with the surviving pool viruses were performed by infecting fresh 3T3 cells with the equivalent of 1 to 20 μl of wild-type virus and subsequent passaging of the cells every 3 days as described above. For larger pool sizes, it was necessary to infect multiple plates of 3T3 cells for the first passage in order to ensure complete representation of the pool. In this case, after the first passage, viruses from all plates were pooled before being quantified and analyzed.
Preparation and analysis of viral DNA.
Low-molecular-weight DNA was extracted from Rat2 cells 24 h postinfection (9). PPT region DNA was analyzed by PCR using primers specific for env and U3. For sequencing, ClaI-to-NheI restriction fragments of these PCR products were introduced into pUC19 and individual subclones were sequenced or, where indicated, PCR products were sequenced directly without subcloning.
PPT use for the tandem PPT constructs was analyzed by Southern blotting. Low-molecular-weight DNA was digested with EcoRV, phenol chloroform extracted, ethanol precipitated, and resuspended in 70% formamide–10 mM EDTA. After heating to 95°C, samples were separated on 5% polyacrylamide–8 M urea gels, electrotransferred to nylon (HyBond) at 80 V for 2 h, and hybridized with a 32P-labeled probe generated with a Rediprime II random primer kit (Amersham) under standard hybridization conditions (26). The probe was an NheI/XbaI restriction fragment from U3 of pMLV-neo.
RESULTS
Proviral clones with targeted alterations to the Mo-MLV PPT region.
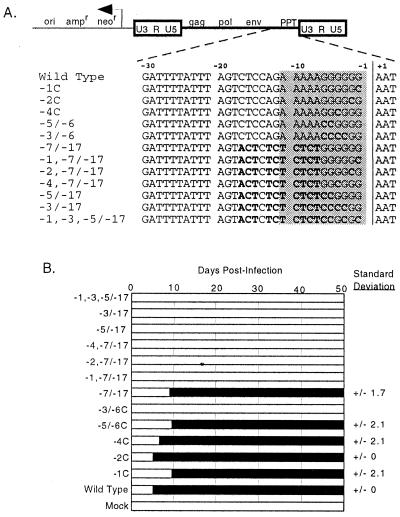
Most previously described Mo-MLV PPT point mutants retained at least partial replication function, and even substitutions in the entire cleavage site-distal half of the PPT (the −7/−17 substitution mutant) remained capable of limited replication (24). Thus, with the aim of further defining which PPT residues were mandatory for replication, additional targeted mutations were introduced into the PPT of an infectious proviral clone. Each of the mutations described in Fig. 1A was studied both as a single mutation and in the context of the −7/−17 substitution. Mutations tested included previously examined changes in the highly conserved −2 and −4 positions, a point mutation at −1, and blocks of mutations at −5 and −6 and at −3 to −6. In each of these positions, the wild-type G was substituted with C. In the context of the −7/−17 substitutions, these reduced the purine content of the remaining PPT to five, four, or two bases, respectively. An additional mutation (the −1,−3,−5/−17 mutation), with substitutions at −1, −3, −5, and −6, was introduced into the −7/−17 background. This mutation left only the highly conserved −2 and −4 positions of the PPT intact.
FIG. 1.
Targeted 3′ untranslated region mutations. (A) The mutations. Shown at the top is the structure of the pMLV-neo proviral plasmid. Dashed lines indicate the position of sequences shown below. For mutants, differences from wild type are boldfaced. The core PPT is shaded. The vertical line indicates the PPT/U3 boundary and the normal site of plus-strand primer cleavage. (B) Replication efficiency of mutants. The leftmost edge of the black bars indicates the time point at which virus spread was first detected. Standard deviations, in days, of time points when replication was detected are shown at the right for those mutants that replicated.
Some of the core PPT is required for viral replication.
To examine replication efficiencies of these mutants, proviral plasmids were transiently transfected into 293T cells. Virus was harvested and used to infect NIH 3T3 cells, and viral spread was monitored every 3 days. Results are summarized in Fig. 1B. Consistent with earlier findings (24), the −2C and −4C mutants replicated with little or no delay relative to wild type, and the −7/−17 mutant replicated with an average delay of 4 days. When either −2C or −4C was combined with the −7/−17 mutation, virus spread remained undetectable for more than 47 days. Similar results were observed with the −1C and −5/−6C mutants. These two mutants replicated with modest delays when present alone, but replication was not detectable throughout 47 days of infected cell passage for virus harboring these changes in the context of the −7/−17 mutation. Replication was not detectable for virus with the −3/−6C mutation either alone or in the context of the −7/−17 mutation. The −1,−3,−5/−17 mutant also failed to replicate. For mutants that did replicate, sequence analysis of viral DNAs after replication revealed that no changes to the original sequences had occurred within the sequenced 3′ untranslated region.
Generation and evolution of random sequence PPT pools.
Degenerate mutant pools were used to further examine sequence requirements within and upstream of the PPT. Table 1 presents a compilation of the genome regions subjected to randomization in this study. The table summarizes the sequence properties of these mutant pools both before and after virus passage, as determined in the experiments described below.
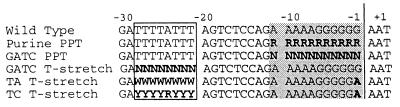
The first pools were created to address whether a specific sequence of purines is required in the PPT or if a structure conferred by any purine-rich sequence is sufficient (3, 20). Experimental evidence suggests that for both HIV-1 and Mo-MLV, RNase H cleavage specificity is determined by the structure of the nucleic acid substrate when it is bound by RT and that subtle sequence differences might affect the structure enough to alter substrate recognition by RNase H (5).
In these randomized purine PPT pools, positions −1 to −11 of the PPT core were randomized such that each position contained either a G or an A (Fig. 2, purine PPT). Mutagenized plasmid pools of various sizes—some containing significantly fewer members than would be required to account for the total possible genetic complexity (2,048 different sequences)—were transiently transfected into 293T cells. Virus was collected and used to infect NIH 3T3 cells, and viral spread was monitored as described above. Once the culture showed detectable viral spread, virus was harvested and quantified by RT assay, and equal amounts of wild-type and pool virus were used to infect fresh cells. The time from infection of NIH 3T3 cells until virus spread was detectable was considered a single passage of the pool. The time required to complete a passage under these conditions (roughly 5 to 8 days) suggests that each passage consisted of several successive rounds of viral replication. After each passage, a PCR product of pooled surviving viral DNA was sequenced. Pools were passaged in this way until no evidence of further evolution of the initially randomized region was detectable.
FIG. 2.
Composition of degenerate pools. Boldface lettering indicates positions that differed from wild type. R indicates any purine, W indicates T or A, Y indicates any pyrimidine, and N indicates any nucleotide. The core PPT is shaded, and the T stretch is boxed. The vertical line indicates the site of wild-type plus-strand primer cleavage.
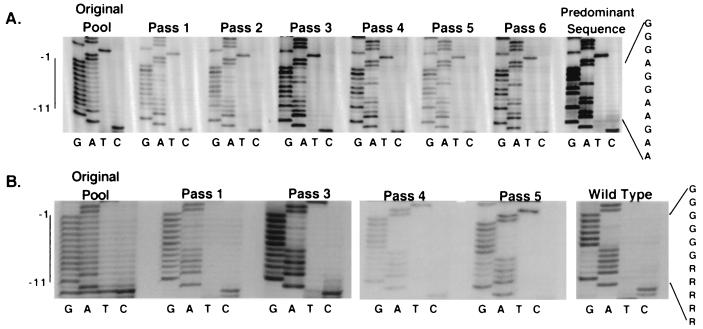
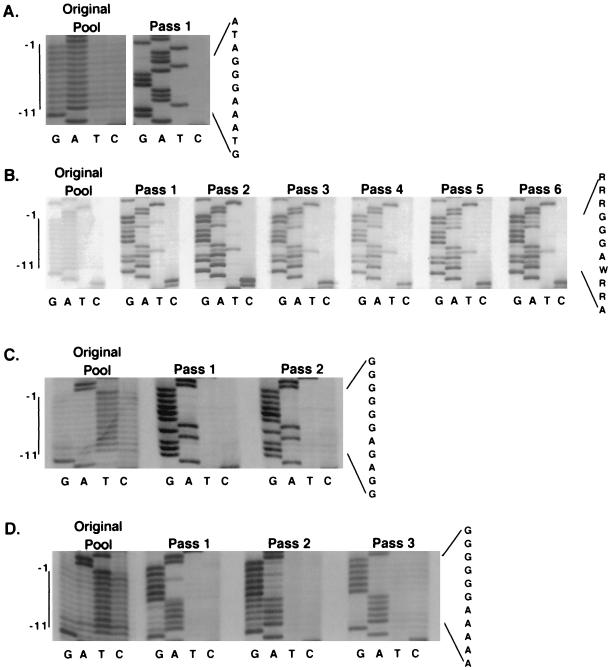
Three purine PPT pools were created of 179, ∼500, and ∼6,300 members. Five evolution experiments involving serial passages as described above were performed: two with the 179-member pool (Table 1, part A), two with the ∼500-member pool, and one with the pool containing ∼6,300 members. Note that the first two pools contained significantly fewer members than would be required to saturate the possible genetic complexity and thus might allow the survival of variants less fit than the optimal sequences that would dominate the population after replication of a fully representative pool. Replication of each pool was detected at a time point similar to that observed for wild type, suggesting that many sequences in the pools were capable of supporting replication. To examine the evolution of pool populations over sequential passages, the PPT region of pooled viral DNAs was amplified by PCR, and the PCR products were sequenced directly. Sequencing of pooled viral DNA from six passages for the 179-member pool, which contained less than 1/10 of the total possible sequences in the randomized region, is shown in Fig. 3A. The sequence that predominated in the pool after these passages differed from wild type only at the −4 and −9 positions (Table 1, part C), and the same sequence predominated in a second independent evolution of this pool.
FIG. 3.
Evolution of randomized purine PPT pools. (A) Evolution of 179-member pool. Direct sequencing of uncloned PCR products containing the PPT region amplified from pools of unintegrated viral DNAs harvested from Rat2 cells infected with pool virus or of plasmid PCR products. At the left is the sequence of a PCR product of the original purine PPT plasmid pool, with the position of the core PPT indicated. Sequences of six successive passages of the pool are then shown. The sequencing reaction on the right is of a PCR product from a plasmid clone containing the predominant sequence that emerged after serial passage. At the far right is the predominant sequence. (B) Evolution of ∼6,300-member purine PPT pool. The original plasmid pool is on the left. Sequences of PCR products from viral DNA after passages 1, 3, 4, and 5 are shown, followed by the sequence of a wild-type PCR product. The predominant sequence is shown at the right.
The two evolution experiments with the ∼500-member pool both yielded a sequence that differed from the one which dominated the 179-member pool. This sequence differed from wild type only at the −8 position (Table 1, part C).
As shown in Fig. 3B, the ∼6,300-member purine PPT pool, which should contain all possible sequences, evolved to wild type from positions −1 to −6. However, little sequence preference was detected from −7 to −11, with A and G both still present within the population in each of these positions after five passages (Fig. 3B; Table 1, part C). Sequencing gels that tracked the evolution of the pools were quantified by phosphorimager. These data quantitatively confirmed visual inspection-based interpretations of evolutionary trends in different portions of the PPT during pool passage (data not shown).
Similar conclusions about the relative importance of wild-type sequences in specific PPT positions could be made from two experiments that examined individual purine PPT pool members: some that were isolated from the pool that survived one replicative passage, and others that were founding members of the plasmid pool. When individual virus sequences were cloned after a single passage of the 179-member pool and 21 of these clones were sequenced, over 80% were found to contain the wild-type sequence at positions −1, −2, −5, −6, −10, and −11 (Table 1, part B). In a separate experiment, the replication efficiencies of 27 founding members of the purine PPT pool were examined individually. As evident from variations in detectable virus spread (indicated in the data column for part A in Table 1), sequences that were compatible with efficient replication bore a stronger resemblance to wild type than those in viruses which replicated poorly or not at all.
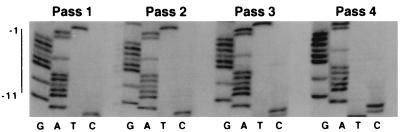
Since the 179-member pool was significantly smaller than the total potential pool size of 2,048, it may not have included the wild-type PPT. Therefore, to determine how well the predominant sequence from this pool replicated relative to wild type, NIH 3T3 cells were coinfected with equivalent amounts of virus containing the wild-type PPT and virus containing this predominant sequence PPT. The sequence of the surviving virus was examined through four passages (Fig. 4). The predominant sequence was present in amounts equivalent to wild-type levels throughout four passages of the virus. These findings suggest that virus with this sequence replicated with the same efficiency as wild type in this culture system. Note, however, that because these experiments were not carried out at low multiplicity of infection, the possibility of trans-acting differences could not be ruled out conclusively.
FIG. 4.
Competition between the wild-type sequence and the 179-member purine PPT pool predominant sequence. The predominant sequence differed from the wild-type sequence at positions −4 and −9. Sequences of PCR-amplified PPT regions of unintegrated viral DNA harvested from Rat2 cells infected with competition virus from passages 1 through 4 were determined. The position of the core PPT is indicated on the left.
Requirement for purines in the PPT.
Some retroviruses, including human adult T-cell leukemia virus, Mason-Pfizer monkey virus, and simian retrovirus type 1, contain pyrimidines within their PPTs (21, 28, 29). Thus, a second random sequence pool in which PPT positions −1 to −11 were any of the four bases (Fig. 2, GATC PPT) was created to address whether or not pyrimidines would be permitted at any positions in the Mo-MLV PPT. Three mutant plasmid pools of approximately 530, 4,000 and 5,000 members were generated. The theoretical genetic complexity of this pool was ∼4.2 × 106 sequences. Thus, each of the studied pools contained only a small subset of possible sequences, and the pools may or may not have included the wild type. PPT region sequences of randomly selected prepassage GATC PPT pool members are shown in Table 1 (part D).
In an initial experiment, cells were infected with virus containing a subset (roughly 100 members) of the 530-member pool, as determined by XC assay. Unlike purine PPT pool virus, which replicated as efficiently as wild type, viral replication by this GATC PPT pool subset was not detectable until 17 days after infection, a 9-day delay relative to wild type. After this first passage of the pool, a single PPT sequence was detectable (Fig. 5A; Table 1, part E). This sequence bears little overall resemblance to wild type, containing pyrimidines at positions −2 and −10. However, the central part of the PPT, positions −4 to −9, were identical to wild type. Coinfection with equal amounts of this virus and of wild-type virus yielded only wild-type sequences after a single passage (data not shown), indicating that this sequence was a poor competitor with the wild-type sequence.
FIG. 5.
Evolution of the GATC PPT pools. Sequences of PCR-amplified PPT regions of unintegrated viral DNA harvested from Rat2 cells infected with GATC PPT pool virus and of PCR products of the original plasmid pools were determined. (A) Evolution of a subset of the 530-member GATC PPT pool. The original GATC PPT plasmid pool is shown on the left. Sequencing of the pool after a single passage is shown on the right. On the far right is the predominant sequence after passage. At the far left, the position of the core PPT is indicated. (B) Evolution of the complete 530-member GATC pool. On the left is this GATC PPT plasmid pool. Six passages of the pool are then shown. The predominant sequence postpassage is shown at the right. (C) Evolution of the 4,000-member GATC pool. On the left is this GATC PPT plasmid pool before transfection. Two passages of the pool are then shown. Shown on the right is the predominant sequence after passage. (D) Evolution of the 5,000-member GATC pool. On the left is this pool before transfection. Three passages of the pool are shown. Shown on the right is the predominant postpassage sequence.
In a second experiment that involved infection with the complete 530-member pool, spread of the pooled virus was detectable with a 3-day delay relative to wild type. Unlike the severely delayed subpopulation above, the virus which persisted was not homogeneous after the first couple of passages of this larger pool (Fig. 5B). In this pool as well as in the 4,000- and 5,000-member pools (Fig. 5C and D), pyrimidines were not detectable in most positions after a single passage of the virus. Purines were strongly favored at all positions of the PPT, but discrimination between the purines occurred more slowly than did discrimination against pyrimidines. Predominant sequences of these pools are shown in Table 1 (part E). The finding that each subset pool generated the same trend in sequence selection suggests that the conclusions drawn may hold true for the entire GATC PPT pool.
Eighteen members of the 530-member prepassage pool were assayed individually for replication efficiency. Consistent with the results of the pool after passaging, only those sequences with a significant number of purines were capable of replicating (Table 1, part D).
Importance of the T-stretch upstream of the PPT.
We have previously shown that a mutation which obliterates the T stretch upstream of the Mo-MLV PPT eliminates viral replication and decreases the efficiency of plus-strand priming (24). To examine how much variation in this region was compatible with Mo-MLV replication, a T-stretch degenerate pool was tested. A pool of approximately 7,000 sequences was created in an infectious proviral clone, in which each position from −21 through −28 was replaced with any of the four bases (Fig. 2, GATC T-stretch). The theoretical size of this pool was about 6.5 × 104. Note, however, that T was overrepresented in all randomized positions of the pool (Fig. 5, original pool sequences), presumably as an unplanned consequence of the oligonucleotide synthesis process. Thus, T-rich sequences were more prevalent in the population than would be predicted if the pool were equally randomized. Nonetheless, sequencing individual members of the prepassage pool revealed that the intended genetic variation was well represented (Table 1, part F). These degenerate T-stretch pools were passaged as described above for the PPT pools.
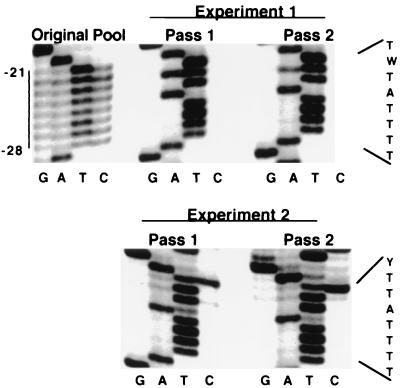
Shown in Fig. 6 is the evolution of a subset of sequences (roughly 1,000) from the 7,000-member pool. In contrast to the slow evolution seen for the PPT pools, the only sequences detected in the −23/−28 region of the first-passage survivors were wild type in two independent experiments, suggesting that very little sequence variation in this region was compatible with replication. Somewhat more variation was observed in the −21 and −22 positions. The sequence of these positions remained heterogeneous, with a T/C mix in the −22 position after passaging in one evolution experiment and a T/A mix in the −21 position observed in an independent evolution experiment (Fig. 6; Table 1, part G).
FIG. 6.
Evolution of T-stretch pools. Results from two independent evolution experiments performed with subsets of the 7,000-member T-stretch pool are presented. At the left in the first row is the sequence of the T-stretch plasmid pool before transfection, with the position of the T stretch indicated. Two successive passages for each of two evolution experiments are shown. Shown at the right is the predominant sequence that emerged in each experiment.
To confirm this rapid evolution toward the wild-type sequence in the −23/−28 region, two additional T-stretch pools were constructed. The TA pool contained either A or T at every position from −21 to −28 (Fig. 2, TA T-stretch), while the TC pool contained an A or G at −24 and a T or C at every other position in this region (Fig. 2, TC T-stretch). Both of these pools were genetically marked with a −1A substitution to rule out the possibility that what appeared to be rapid evolution could have been due to wild-type virus contamination. The plasmid pools tested were severalfold larger than the theoretical complexity (256 members). Sequencing the pooled plasmids before virus passage as well as sequencing individual members of the prepassage pools revealed that the intended genetic variation was well represented (Table 1, parts H and J, and data not shown). These pools were passaged as described above.
As with the completely degenerate pool, these two pools evolved to essentially a single sequence within a single passage. In six of eight single clones randomly selected from the first passage of the TA pool, the T-stretch sequence was completely wild type (Table 1, part K). In the other two clones, the only differences from wild type were that the −21 or −28 position was A instead of T. In five of five clones from the TC pool, the T stretch was wild type in all positions except −21, which was a C (Table 1, part I). These data are consistent with those of the completely degenerate T-stretch pool and indicate that whereas the −21 and −22 positions of the T stretch can vary, there is strong selective pressure for maintaining wild-type sequences in the −23/−28 region.
Effects of T-stretch mutations on trans-acting replication functions.
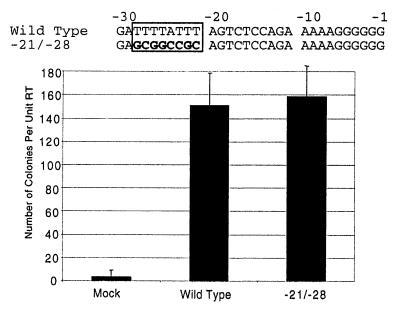
We previously reported that replication defects of T-stretch mutants are greater than can be accounted for by the magnitude of effects on plus-strand priming (24). Thus, to test whether or not mutations in the T-stretch affected virus protein production, we introduced the −21/−28 mutation (24) (Fig. 7), which contains a GC-rich sequence in place of the T stretch, into a Ψ− packaging-defective proviral plasmid. This construct was used to examine possible effects of T-stretch alterations on helper functions. This T-stretch-defective Ψ− construct and a Ψ− wild-type provirus plasmid were individually cotransfected with a puromycin resistance-conferring retroviral vector into 293T cells to generate vector-containing virions. No differences in virion content in the media of cells transfected with wild type versus those with the −21/−28 mutant helper were detectable (data not shown), suggesting that production of the Gag-Pol polyprotein was not affected by the T-stretch mutation. To examine possible effects on envelope production or other functional properties, the infectivity of the virions was tested. Equivalent amounts of virus were used to infect NIH 3T3 cells, and puromycin-resistant titers were determined. Equivalent numbers of puromycin-resistant colonies were obtained from vectors expressed with wild type or with −21/−28 mutant helpers (Fig. 7). Thus, trans-acting helper functions were not affected to a detectable extent by this T-stretch mutation.
FIG. 7.
Effect of the −21/−28 T-stretch on helper functions. Sequences of the wild type and mutant are compared at the top. The T stretch is boxed in black, and differences from wild type are shown in boldface Ψ− proviral clones containing a wild-type T stretch or the −21/−28 mutation were cotransfected with a puromycin resistance-conferring retroviral vector. Virus was collected, quantified by RT activity, and used to infect NIH 3T3 cells. Shown is the average number of puromycin-resistant colonies per unit of RT for three independent experiments.
Effects of deletions upstream of the PPT on plus-strand priming.
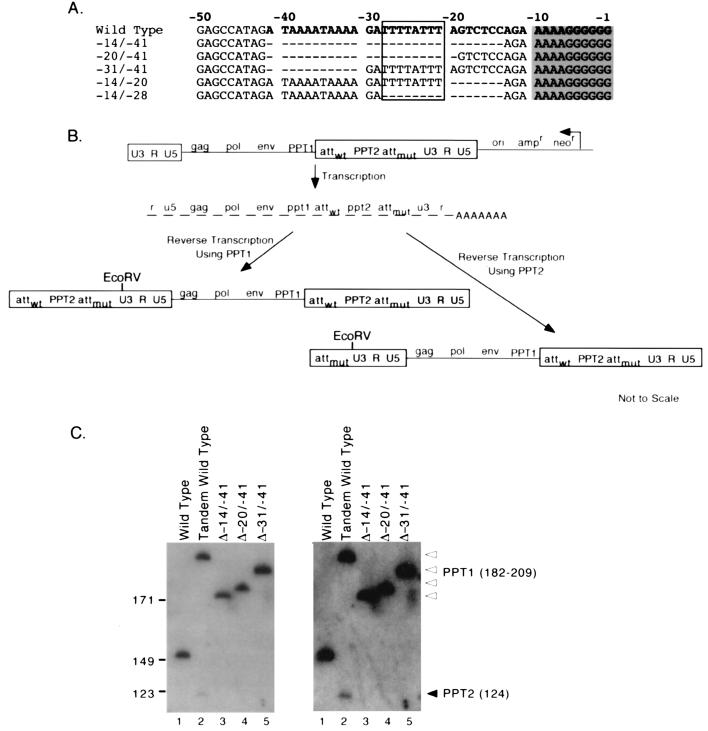
To test the importance of sequences upstream of the core PPT for plus-strand priming and replication, five deletion mutations were introduced into the 3′ untranslated region between the end of env and the PPT (Fig. 8A). In addition to the T stretch described above, this region of Mo-MLV contains an A stretch from −31 to −39 (Fig. 8A). The deletions that were constructed are shown in Fig. 8A. The replication efficiencies of these deletion mutants were determined as described above. Consistent with results published while this work was in progress (1), the Δ−14/−41 mutant, which contained a deletion of the entire upstream sequence, replicated with a significant delay (12 days later than wild type) without reversion in this region (data not shown). Similar results were found with the Δ−14/−20 and Δ−14/−28 deletions. Replication time courses for Δ−20/−41 and Δ−31/−41 were similar to those of the other deletion mutants; however, PCR of viral DNA revealed that these deletions were no longer present (data not shown). This most likely indicates reversion by patch repair as previously observed for different mutants in this region (24). These putative revertants were not characterized further.
FIG. 8.
Effects of deletions upstream of the core PPT. (A) Deletions introduced upstream of the core PPT. Nucleotides shown in boldface are the same in both PPT1 and PPT2 of the wild-type tandem PPT construct. Dashes indicate position of deletions. When deletions were placed in tandem PPT constructs, they were introduced into PPT2. The core PPT is shaded, and the T stretch is boxed. (B) Schematic representation of the structure and predicted products of the tandem PPT construct. The second PPT inserted in U3 (PPT2) and mutated att (attmut) are shown. The first line indicates the structure of the transfected proviral construct. Boxed regions indicate LTRs. The structural organization of the encapsidated RNAs is indicated on the second line. The final two lines indicate the predicted structures of the reverse transcription products; the single RNA species on the second line generates two different DNA products that differ in their upstream LTRs. The EcoRV site used to generate end products for Southern blots is shown. Note that deletions introduced into PPT2 will alter the sizes of the reverse transcription product that result from PPT1 use but that products which result when PPT2 primes plus-strand synthesis will be the same size for all deletion mutants and for wild type. Drawings are not to scale. (C) Southern blot of EcoRV-digested nonintegrated viral DNA products. Marker (left) and product (right) lengths are indicated in base pairs. PPT1 bands are indicated with open arrowheads, and the PPT2 band is indicated with a filled arrowhead. PPT1 product sizes are as follows: 209 bp for wild-type tandem PPT1 products, 199 bp for Δ−31/−41, 188 bp for Δ−20/−41, and 182 bp for Δ−14/−41. PPT2 products are 124 bp for all constructs. The wild-type (single PPT) product is 144 bp. The panel on the right is a darker exposure of the one at the left.
Three deletion mutations (Δ−14/−41, Δ−20/−41, and Δ−31/−41) were introduced into PPT2 of the tandem PPT system that we have previously described, to examine the mutations' effects on plus-strand priming (24). In these tandem PPT vectors, a second PPT is inserted into U3 downstream of the endogenous PPT (Fig. 8B). In this system, the viral RNA contains two PPTs, either of which can prime plus-strand synthesis. Because the choice of PPT determines the left edge of the final double-stranded DNA, the relative use of the two PPTs can be determined by examining the upstream long terminal repeat (LTR) of reverse transcription products. The ectopic PPT, PPT2, includes 30 bases of sequence upstream of the core PPT. We have shown previously that if both PPTs contain wild-type sequences, PPT1 is used about 70% of the time and PPT2 is used 30% of the time (24). Since the wild-type PPT1 was present in all constructs, all three PPT2 deletion mutants replicated with wild-type efficiency as expected, and none contained changes to the original sequence in this region after replication.
To visualize the PPT1 and PPT2 products for all deletion mutants, Southern blotting was performed on nonintegrated reverse transcription products digested with EcoRV to generate LTR end fragments. As shown in Fig. 8C, PPT2 use was detectable at a low level for the parental construct but not for any of the deletion mutants, which suggested that for each deletion mutant, sequences or structures upstream of the PPT which were disrupted are important for plus-strand priming. Note that because the level of priming from the wild-type PPT2 was low, the extent to which the deletions affected plus-strand priming could not be quantified precisely. However, these observations were consistent with confirmatory findings from experiments with reiterative primer extension performed as previously described (24) (data not shown).
DISCUSSION
In this study we examined the replicative advantages of maintaining wild-type sequences within specific regions of the Mo-MLV 3′ untranslated region. We observed that maintenance of the T-stretch core, which consists of nucleotides from −23 to −28 upstream of the site of plus-strand cleavage, was highly selected during viral replication. Results indicated that maintenance of the −1/−6 cleavage site-proximal half of the PPT conferred a selective advantage during viral replication as well, albeit at a lower level than conservation of the T stretch. High purine content in the cleavage site-distal half of the PPT was also advantageous. However, this latter region was far more tolerant of variation from the wild-type sequence than was the core PPT's cleavage site-proximal half.
We have previously demonstrated that viruses which possess only the cleavage site-proximal half of the PPT can replicate with relatively modest delays (24). To assess the replication efficiency of viruses with even fewer wild-type PPT residues, additional targeted changes were introduced in the context of the −7/−17 mutation. The results suggested that whereas many individual changes within the cleavage site-proximal half of the PPT are tolerated, the entire cleavage site-proximal five or six bases are necessary for replication in the absence of the cleavage site-distal half of the PPT.
Evolution of randomized purine PPT pools further illustrated sequence biases within the PPT. Wild-type sequences at positions −2, −5, −6, −10, and −11 were strongly preferred. Selection for wild type at all other PPT positions appeared to be significantly weaker, as these positions were slower to evolve toward wild type. A random purine PPT mutant that differed from wild type at positions −4 and −9 was shown to replicate as well as the wild-type virus during extended passage in mixing experiments. This demonstrates that PPT sequences other than wild type can be fully infectious, at least under the relatively high-multiplicity-of-infection conditions examined here.
Comparison of the PPT sequences of many different retroviruses and retroelements reveals that the core PPTs of some of these include pyrimidine residues (mostly T's) (22, 24). Thus, replication was tested for Mo-MLV PPT pools in which all four bases were included at each PPT position. Consistent with results from the degenerate purine pools, these fully degenerate GATC PPT pools showed rapid evolution toward sequences with wild-type residues at most positions. After two passages of a small GATC PPT pool, T's remained detectable at positions −2 and −10, while surviving members of larger GATC pools contained no detectable PPT pyrimidines after a single passage. These observations indicate a stronger preference for purine content in the Mo-MLV PPT than is the case for many other retroviruses and retroelements. Not only do some of these other elements naturally possess pyrimidines within their core PPT regions, but tolerance during replication for variation that includes pyrimidines has also been reported (13).
It has been suggested that a unique structure within the PPT is responsible for its recognition during plus-strand primer generation (for a review, see reference 10). Although binding of RT to nucleic acids alters the nucleic acids' structures, at least in the case of DNA/DNA duplexes (12), inherent structural differences between PPT duplexes and other RNA/DNA duplexes that form during replication might contribute to the resistance of the PPT to RNase H cleavage. Findings here demonstrate that the specific sequence of purines within the PPT, and not mere purine richness, is important to Mo-MLV PPT function. Therefore, if the structure of the nucleic acids is important to the PPT's function during replication, it must be a structure more distinct than that which can form from any repeated purine sequence.
When RT binds nucleic acids, most contacts between the enzyme and nucleic acids occur in the DNA polymerase domain, with the limited contacts that form in the RNaseH domain itself less important to substrate positioning (12). Thus, it has been suggested that proper placement of the RNA/DNA substrate into the RNase H active site may be highly sensitive to subtle changes in substrate regions that are relatively free of protein contacts (5). In the context of RT bound for PPT primer generation, this region would roughly coincide with the cleavage site-proximal half of the PPT, which studies reported here found to be especially conserved.
Requirements for sequences upstream of the PPT in viral replication were also examined in this study. Particular attention was paid to investigating roles of the T stretch which we have previously shown to be required for optimal replicative plus-strand priming (24). In the present study, the striking extent to which selectivity for wild-type sequences was observed in the T stretch, relative to advantages for wild-type sequences in the core PPT, was especially evident from comparisons of sequences that persisted through passage of pooled viruses randomized in these regions. Even after several passages, each consisting of several successive rounds of viral replication, significant sequence variation was observed in randomized PPT pools (for example, Fig. 5B). Among 21 individual survivors of a randomized core PPT pool's first passage (Table 1, part B), only two single positions (−2 and −5) were conserved, with no contiguous patches of even two wild-type residues found in common among all survivors. In contrast, all 13 sequenced survivors of single passages of T-stretch pools were identical to wild type in all six positions from −22 to −27 (Table 1, parts I and K).
Deletions in the 30 bases upstream of the PPT, as shown in tests involving the tandem PPT system, interfered with efficient plus-strand priming. Nonetheless, the Δ−14/−41 mutant with a deletion in this region was capable of supporting viral replication when present as a single PPT. This is consistent with results that Bacharach et al. published while this work was in progress (1). The results of Bacharach et al. suggest that deletions in this genome region affect the site of plus-strand priming and/or removal of the plus-strand primer. We observed that alterations to the T-rich region did not affect helper functions, suggesting that this sequence contributed to viral replication solely in cis. These findings are also consistent with the recent report by Bacharach et al. (1).
All replication roles of the 3′ sequences upstream of the Mo-MLV PPT are not yet understood, even for the core PPT and upstream T stretch. A model has been proposed which suggests that the T stretch located in PPT upstream sequences might function in a DNA polymerase-dependent step of the plus-strand priming process (16). Mo-MLV RT has been shown by footprinting to contact from −27 to +6 of the template strand (numbered relative to the site of polymerization) and from −26 to −1 of the primer strand when bound in the configuration to polymerize DNA (34). Thus, the T stretch studied here is likely to contact RT when it is bound to the PPT region. Mutations in the DNA polymerase domain of RT have been shown to affect RNase H cleavage (4, 6, 14, 19, 23, 27, 31). Thus, it is reasonable to propose that contacts made by the DNA polymerase domain upstream of the PPT during DNA polymerization might contribute to the specificity or efficiency of RNase H cleavage at the PPT.
ACKNOWLEDGMENTS
We acknowledge Monica Roth for providing XC cells and advice on their use; David Peterson, Alison Dormer, Deanna Kulpa-Stom, and Rachel Westfall for assistance at various stages of this project; and Julie Pfeiffer, Rosa Yu, and Terry Dixon for helpful reading of the manuscript.
This research was supported by NIH grant R29 CA69300.
REFERENCES
- 1.Bacharach E, Gonsky J, Lim D, Goff S P. Deletion of a short, untranslated region adjacent to the polypurine tract in Moloney murine leukemia virus leads to formation of aberrant 5′ plus-strand DNA ends in vivo. J Virol. 2000;74:4755–4764. doi: 10.1128/jvi.74.10.4755-4764.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Champoux J J. Roles of ribonuclease H in reverse transcription. In: Skalka A M, Goff S P, editors. Reverse transcriptase. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1993. pp. 103–117. [Google Scholar]
- 3.Fedoroff O Y, Ge Y, Reid B R. Solution structure of r(gaggacug):d(CAGTCCTC) hybrid: implications for the initiation of HIV-1 (+)-strand synthesis. J Mol Biol. 1997;269:225–239. doi: 10.1006/jmbi.1997.1024. [DOI] [PubMed] [Google Scholar]
- 4.Gao H-Q, Boyer P L, Arnold E, Hughes S H. Effects of mutations in the polymerase domain on the polymerase, RNase H and strand transfer activities of human immunodeficiency virus type 1 reverse transcriptase. J Mol Biol. 1998;277:559–572. doi: 10.1006/jmbi.1998.1624. [DOI] [PubMed] [Google Scholar]
- 5.Gao H-Q, Sarafianos S G, Arnold E, Hughes S H. Similarities and differences in the RNaseH activities of human immunodeficiecy virus type 1 reverse transcriptase and Moloney murine leukemia virus reverse transcriptase. J Mol Biol. 1999;294:1097–1113. doi: 10.1006/jmbi.1999.3325. [DOI] [PubMed] [Google Scholar]
- 6.Ghosh M, Williams J, Powell M D, Levin J G, LeGrice S F J. Mutating a conserved motif of the HIV-1 reverse transcriptase palm subdomain alters primer utilization. Biochemistry. 1997;36:5758–5768. doi: 10.1021/bi963045e. [DOI] [PubMed] [Google Scholar]
- 7.Goff S P, Traktman P, Baltimore D. Isolation and properties of Moloney murine leukemia virus mutants: use of a rapid assay for release of virion reverse transcriptase. J Virol. 1981;38:239–248. doi: 10.1128/jvi.38.1.239-248.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hall C V, Jacob P E, Ringold G M, Lee F. Expression and regulation of Escherichia coli lacZ gene fusions in mammalian cells. J Mol Appl Genet. 1983;2:101–109. [PubMed] [Google Scholar]
- 9.Hirt B. Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol. 1967;26:365–369. doi: 10.1016/0022-2836(67)90307-5. [DOI] [PubMed] [Google Scholar]
- 10.Hughes S H, Arnold E, Hostomsky Z. RNase H of retroviral reverse transcriptase. In: Crouch R J, Tuolme J J, editors. Ribonucleases H. Paris, France: INSERM Editions; 1998. pp. 195–224. [Google Scholar]
- 11.Ilyinskii P O, Desrosiers R C. Identification of a sequence element immediately upstream of the polypurine tract that is essential for replication of simian immunodeficiency virus. EMBO J. 1998;17:3766–3774. doi: 10.1093/emboj/17.13.3766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobo-Molina A, Ding J, Nanni R G, Clark A D J, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Crystal structure of human immunodeficiency virus type 1 reverse transcriptase coplexed with double-stranded DNA at 3.0A resolution shows bent DNA. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lauermann V, Hughes S H, Pedent K W C. Maintenance of an unusual polypurine tract in HIV-2: stability to passage in culture. Virology. 1997;236:208–212. doi: 10.1006/viro.1997.8721. [DOI] [PubMed] [Google Scholar]
- 14.Loya S, Gao H-Q, Avidan O, Boyer P L, Hughes S H, Hizi A. Subunit-specific mutagenesis of the cysteine 280 residue of the reverse transcriptase of human immunodeficiency virus type 1: effects on sensitivity to a specific inhibitor of the RNase H activity. J Virol. 1997;71:5668–5672. doi: 10.1128/jvi.71.7.5668-5672.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Miller A D, Miller D G, Garcia J V, Lynch C M. Use of retroviral vectors for gene transfer and expression. Methods Enzymol. 1993;217:581–599. doi: 10.1016/0076-6879(93)17090-r. [DOI] [PubMed] [Google Scholar]
- 16.Noad R J, Al-Kaff N S, Turner D S, Covey S N. Analysis of polypurine tract-associated DNA plus-strand priming in vivo utilizing a plant pararetroviral vector carrying redundant ectopic priming elements. J Biol Chem. 1998;273:32568–32575. doi: 10.1074/jbc.273.49.32568. [DOI] [PubMed] [Google Scholar]
- 17.Pfeiffer J K, Topping R S, Shin N-H, Telesnitsky A. Altering the intracellular environment increases the frequency of tandem repeat deletion during Moloney murine leukemia virus reverse transcription. J Virol. 1999;73:8441–8447. doi: 10.1128/jvi.73.10.8441-8447.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Powell M D, Ghosh M, Jacques P S, Howard K J, LeGrice S F J, Levin J G. Alanine-scanning mutations in the “primer grip” of p66 HIV-1 reverse transcriptase result in selective loss of RNA priming activity. J Biol Chem. 1997;272:13262–13269. doi: 10.1074/jbc.272.20.13262. [DOI] [PubMed] [Google Scholar]
- 20.Powell M D, Levin J G. Sequence and structural determinants required for priming of plus-strand DNA synthesis by the human immunodeficiency virus type 1 polypurine tract. J Virol. 1996;70:5288–5296. doi: 10.1128/jvi.70.8.5288-5296.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Power M D, Marx P A, Bryant M L, Gardner M B, Barr P J, Luciw P A. Nucleotide sequence of SRV-1, a type D simian acquired immune deficiency syndrome retrovirus. Science. 1986;231:1567–1572. doi: 10.1126/science.3006247. [DOI] [PubMed] [Google Scholar]
- 22.Rattray A J, Champoux J J. Plus-strand priming by Moloney murine leukemia virus: the sequence features important for cleavage by RNase H. J Mol Biol. 1989;208:445–456. doi: 10.1016/0022-2836(89)90508-1. [DOI] [PubMed] [Google Scholar]
- 23.Rausch J W, Le Grice S F J. Substituting a conserved residue of the ribonuclease H domain alters substrate hydrolysis by retroviral reverse transcriptase. J Biol Chem. 1997;272:8602–8610. doi: 10.1074/jbc.272.13.8602. [DOI] [PubMed] [Google Scholar]
- 24.Robson N D, Telesnitsky A. Effects of 3′ untranslated region mutations on plus-strand priming during Moloney murine leukemia virus replication. J Virol. 1999;73:948–957. doi: 10.1128/jvi.73.2.948-957.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rowe W P, Pugh W E, Hartley J W. Plaque assay techniques for murine leukemia viruses. Virology. 1970;42:1136–1139. doi: 10.1016/0042-6822(70)90362-4. [DOI] [PubMed] [Google Scholar]
- 26.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 27.Schultz S J, Champoux J J. RNase H domain of Moloney murine leukemia virus reverse transcriptse retains activity but requires the polymerase domain for specificity. J Virol. 1996;70:8630–8638. doi: 10.1128/jvi.70.12.8630-8638.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Seiki M, Hattori S, Hirayama Y, Yoshida M. Human adult T-cell leukemia virus: complete nucleotide sequence of the provirus genome integrated in leukemia cell DNA. Proc Natl Acad Sci USA. 1983;80:3618–3622. doi: 10.1073/pnas.80.12.3618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sonigo P, Barker C, Hunter E, Wain-Hobson S. Nucleotide sequence of Mason-Pfizer monkey virus: an immunosuppressive D-type retrovirus. Cell. 1986;45:375–385. doi: 10.1016/0092-8674(86)90323-5. [DOI] [PubMed] [Google Scholar]
- 30.Sorge J, Hughes S H. Polypurine tract adjacent to the U3 region of the Rous sarcoma virus genome provides a cis-acting function. J Virol. 1982;43:482–488. doi: 10.1128/jvi.43.2.482-488.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tanese N, Goff S P. Domain structure of the Moloney murine leukemia virus reverse transcriptase: mutational analysis and separate expression of the DNA polymerase and RNase H activities. Proc Natl Acad Sci USA. 1988;85:1777–1781. doi: 10.1073/pnas.85.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Telesnitsky A, Blain S, Goff S P. Assays for retroviral reverse transcriptase. Methods Enzymol. 1995;262:347–362. doi: 10.1016/0076-6879(95)62029-x. [DOI] [PubMed] [Google Scholar]
- 33.Wilhelm M, Heyman T, Boutabout M, Wilhelm F-X. A sequence immediately upstream of the plus-strand primer is essential for plus-strand DNA synthesis of the Saccharomyces cerevisiae Ty1 retrotransposon. Nucleic Acids Res. 1999;27:4547–4552. doi: 10.1093/nar/27.23.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wohrl B M, Georgiadis M M, Telesnitsky A, Hendrickson W A, LeGrice S F J. Footprint analysis of replicating murine leukemia virus reverse transcriptase. Science. 1995;267:96–99. doi: 10.1126/science.7528942. [DOI] [PubMed] [Google Scholar]