Abstract
Helicobacter pylori colonizes the stomach in about half of the human population, leading to an increased risk of peptic ulcer disease and gastric cancer. H. pylori secretes an 88 kDa VacA toxin that contributes to pathogenesis. VacA assembles into oligomeric complexes in solution and forms anion-selective channels in cell membranes. Cryo-electron microscopy (cryo-EM) analyses of VacA oligomers in solution provided insights into VacA oligomerization but failed to reveal the structure of the hydrophobic N-terminal region predicted to be a pore-forming domain. In this study, we incubated VacA with liposomes and used single particle cryo-EM to analyze detergent-extracted VacA oligomers. A 3D structure of detergent-solubilized VacA hexamers revealed the presence of six α-helices extending from the center of the oligomers, a feature not observed in previous studies of water-soluble VacA oligomers. Cryo-electron tomography analysis and 2D averages of VacA associated with liposomes confirmed that central regions of the membrane-associated VacA oligomers can insert into the lipid bilayer. However, insertion is heterogenous, with some membrane-associated oligomers appearing only partially inserted and others sitting on top of the bilayer. These studies indicate that VacA undergoes a conformational change when contacting the membrane and reveal an α-helical region positioned to extend into the membrane. Although the reported VacA 3D structure does not represent a selective anion channel, our combined single particle 3D analysis, cryo-electron tomography, and modeling allow us to propose a model for the structural organization of the VacA N-terminus in the context of a hexamer as it inserts into the membrane.
Keywords: bacterial protein toxin, oligomerization, cryo-EM, cryo-ET, gastric cancer
Keywords: cryo-electron microscopy, cryo-electron tomography, bacteria, pathogenesis, pore-forming toxin
Introduction
Pore-forming toxins are secreted by many bacterial species. These toxins can function to kill other bacteria or act as virulence factors that aid in the colonization of eukaryotic hosts [1]. These proteins are typically secreted as soluble monomers, which bind to membranes of target cells. This leads to oligomerization and insertion at the membrane surface to form transmembrane oligomeric pores. Depending on the specific pore-forming toxin, the transition from a soluble protein to a membrane-inserted oligomer elicits a variety of downstream cellular effects, many of which are toxic to eukaryotic host cells. Pore-forming toxins are a group of proteins that are heterogeneous in size, domain number, target membranes, cell receptors, oligomerization states, and mechanisms of action. The two major classes of bacterial pore-forming toxins, termed α and β, are distinguished by the secondary structure of the transmembrane pore [2]. X-ray crystallography and electron microscopy (EM) experiments with soluble monomers, soluble oligomers, and oligomers in membrane-like environments (e.g., detergents, nanodiscs, and liposomes) have provided snapshots for how proteins from both classes oligomerize and undergo dynamic structural rearrangements to form active transmembrane pores [3].
The Gram-negative bacterium Helicobacter pylori, which colonizes the stomach in over half of the human population [4], secretes a protein called vacuolating cytotoxin A (VacA) [5–9]. H. pylori strains producing specific types of VacA (type s1) are associated with an increased risk of peptic ulcer disease and gastric cancer compared to strains producing alternate forms of the protein (type s2) [7–15]. Type s1 forms of VacA cause multiple alterations in gastric epithelial cells, including permeabilization of the plasma membrane, expansion of endosomal compartments (vacuolation), mitochondrial dysfunction, autophagy, and cell death [12, 14, 16–20]. VacA also can cause alterations in several types of immune cells [21–24] and has immunomodulatory effects in vivo [25, 26]. VacA forms anion-selective membrane channels in planar lipid bilayers, and patch clamping experiments indicate that VacA forms channels in the plasma membrane of intoxicated cells [27–30]. Therefore, VacA has been classified as a pore-forming toxin.
The mature, secreted VacA toxin (residues 1–821) consists of two regions: a 33 kDa N-terminal region (p33) and a 55 kDa C-terminal region (p55) [31–36]. The formation of VacA membrane channels and VacA cytotoxic effects are dependent on the presence of an N-terminal hydrophobic region (residues 6–27) [27–30]. The VacA N-terminal hydrophobic region contains multiple tandem GXXXG motifs (residues 14–26) required for channel activity and cytotoxic effects [37]. Thus, a VacA mutant protein lacking the N-terminal hydrophobic region (VacA Δ6–27) or one that contains alterations of the GXXXG motifs (G14A or G18A substitutions) does not form channels in planar lipid bilayers and lacks cell-vacuolating activity [10, 37–40]. Comparisons of oligomers formed in solution by wild-type VacA and VacA Δ6–27 using negative stain EM methods showed that the wild-type oligomers have a detectable central density that is not visible in the Δ6–27 oligomers [41, 42]. Further, computational modeling suggests that the glycine residues in the GXXXG motifs pack against small alanine or valine side chains to form a helical bundle that could form an anion-selective transmembrane pore [43]. Combined, these observations led to a model predicting that N-terminal residues of VacA are localized in or near the central region of the oligomers and constitute a central pore.
Recently determined sub-4 Å structures of soluble VacA hexamers showed that VacA protomers (p88) consist of a continuous right-handed β-helix that oligomerizes with neighboring protomers through salt bridges, side-chain hydrogen bonds, and extensive main-chain hydrogen bonds linking residues from both the p33 and p55 regions [44–46]. However, the lack of structured density in the center of the EM maps made it impossible to build a model for the N-terminus of p33, including the GXXXG motifs (residues 1–26). The first resolved residues of the p33 region in the EM maps (residues 27–37) correspond to an α-helical region buried in the oligomeric interface. It was proposed that in the context of membranes, this region might undergo a conformational change that would facilitate the GXXXG motifs to organize into a helical bundle to form a central pore [44, 45].
Previous studies using lipid flotation assays have shown that VacA can insert into membranes, and negative stain EM analysis revealed membrane-associated hexamers and heptamers [47]. Comparative analyses of membrane-associated VacA oligomers and water-soluble VacA oligomers revealed detectable density in the membrane-associated oligomers that was absent from the water-soluble oligomers [47]. These observations suggested that VacA might undergo a conformational change when in contact with the membrane. However, the two-dimensional (2D) classes of VacA oligomers in a lipid environment provided no secondary structural insight into the conformation of the GXXXG motifs and central region of VacA.
Collectively, previous studies suggest that VacA might be an α-helical pore-forming toxin, but thus far, there has been no experimental evidence demonstrating this. Moreover, the mechanisms by which VacA undergoes a dynamic rearrangement from a soluble monomer to binding membranes, oligomerizing, and forming a transmembrane pore have not been defined, limiting our understanding of this important virulence factor. Here, we used single particle cryo-electron microscopy (cryo-EM) and cryo-electron tomography (cryo-ET) to structurally analyze VacA in detergent-purified and liposome-bound states. These studies provide 2- and 3-dimensional (2D & 3D, respectively) snapshots of the structure of VacA associated with the membrane, showing that membrane association leads to structural changes in the central region of VacA oligomers and revealing a putative α-helical region that extends from the center of the oligomer.
Results
Isolation of VacA oligomers from membranes.
While the sub-4 Å resolution 3D structures and crosslinking studies of soluble VacA oligomers provided molecular details for how p88 protomers interact in the context of the oligomer [44–46], the structure of the N-terminal hydrophobic region (residues 1–26) was not resolved. The lack of an organized central density in the soluble VacA cryo-EM structures, combined with the observation that in negative stain 2D averages, the central region of membrane-bound VacA hexamers appears structurally distinct from the central region of soluble hexamers [47], suggests that the N-terminus of VacA changes conformation to become more structured in the presence of a lipid bilayer.
To experimentally test this hypothesis, we developed methods for binding VacA oligomers to in vitro-generated large unilamellar vesicles (LUVs), followed by extraction and isolation of the membrane-associated VacA particles, with the goal of determining a 3D structure of the extracted particles using single particle cryo-EM analysis. For these experiments, we used preparations of purified VacA that were acid-activated, because low pH causes VacA oligomers to dissociate into monomers in solution [46, 48–50]. Acid-activated VacA preparations were mixed with LUVs composed of eggDOPC/DOPS/cholesterol (ePC/PS/chol) (55/15/30 mol%). This membrane composition supports VacA anion-selective pore formation [29, 38], and VacA oligomers cannot be stripped from these LUVs using sodium carbonate extraction, a stringent biochemical assay that separates integral membrane proteins from the peripheral membrane and soluble proteins [47, 51]. These previous results indicate that VacA oligomers can insert and form active pores in membranes composed of this lipid composition.
After incubating VacA with LUVs, the next step was to isolate the membrane-associated oligomers. We tested the ability of n-dodecyl-β-D-maltoside (DDM, 0.02%), lauryl maltose neopentylglycol (LMNG, 0.002%), and 6-cyclohexyl-1-hexyl-β-D-maltoside (Cymal-6; 0.4%) to extract membrane-bound VacA to find a condition in which solubilized VacA oligomers remain stabilized as hexamers or heptamers, the oligomeric form of VacA predicted to interact with membrane [44, 45, 47]. As visualized by negative stain EM, soluble VacA forms ~10% single-layer oligomers (hexamers and heptamers) and ~90% double-layer oligomers (dodecamers and tetradecamers) [42]. In contrast, VacA oligomers associated with liposomes are hexamers or heptamers [47]. While negative stain analysis showed that solubilization of liposome-bound VacA with either LMNG or Cymal-6 led to the aggregation of the VacA oligomers (data not shown), images and subsequent 2D classification of DDM-solubilized VacA revealed that a little over half of the VacA oligomers were single-layer hexamers and heptamers (56%; Fig. S1).
3D structural analysis of detergent-solubilized VacA oligomers
To further investigate the structure of detergent-solubilized VacA oligomers, we analyzed the DDM-solubilized VacA particles using single particle cryo-EM. As expected from our initial negative stain analysis, 2D classification of vitrified particles revealed VacA hexamers and heptamers, as well as dodecamers and tetradecamers. After extensive iterative 2D and 3D classification steps to focus on hexamer and heptamers in the dataset, 3D structures of the DDM-solubilized VacA hexamers and heptamers appeared anisotropic and the particle orientation distribution maps revealed extreme preferred orientation of the particles in the vitrified ice layer (data not shown). To increase the range of particle orientations of detergent solubilized VacA in vitrified ice, we collected micrographs at a 30° tilt angle [52]. The 2D class averages of the DDM-solubilized VacA with en face orientation had clear density in the center of the oligomers (Fig. 1A). This was different from the previously observed cryo-EM analysis of VacA oligomers formed in solution, which did not have any defined central density in either the 2D class averages or in the resulting 3D structures [44, 45]. Thus, the 2D classes of DDM-solubilized VacA oligomers extracted from liposomes suggested that VacA oligomers have a structural conformation different from that of VacA oligomers that form in solution.
Figure 1. Single particle cryo-EM analysis of DDM-solubilized hexameric VacA.
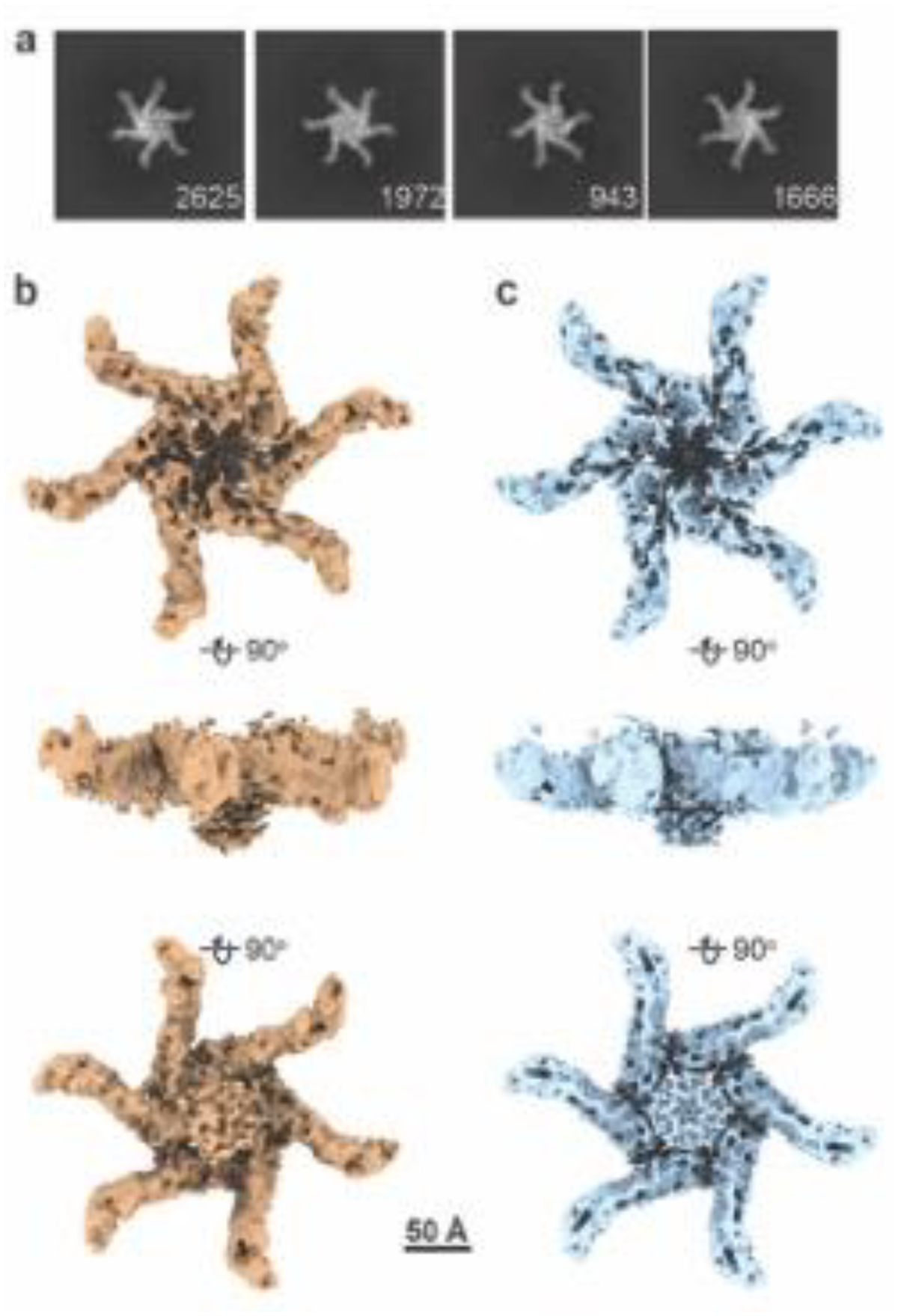
(A) Representative 2D class averages of VacA hexamers solubilized from LUVs (55/15/30 mol % eggPC/DOPS/Chol), based on micrographs taken at a 30° tilt. Numbers of particles are shown in bottom right corner of each class. Box size, 602 Å. (B, C) 3D structure of detergent-solubilized hexameric VacA with either C1 (B) or C6 symmetry (C) applied. Views of structures are rotated 90° around the x-axis. Scale bar, 50 Å.
To explore this observation in more detail, we used iterative and stringent 2D and 3D classification procedures (Fig. S2) and isolated a class of DDM-solubilized hexameric VacA that contained organized central density, either with no applied symmetry (Fig. 1B) or six-fold symmetry (Fig. 1C). While the resolutions of the structures are modest at 7.7 Å (C1) and 6.7 Å (C6) (Fig. S3 and Table 1), this structure has a central α-helical-like density protruding from each protomer (Fig. 1B, C). A comparison of the detergent-solubilized hexamer map with the structure of soluble VacA hexamers and dodecamers indicates that the central density extends from the face of the hexamer that was predicted to interact with membrane [44, 45]. Placement of the soluble VacA hexamer atomic model (PDB 6NYF) into the detergent-extracted VacA hexamer EM map shows that the majority of the soluble VacA model fits into the map and that the helical extensions seen in the DDM-solubilized hexamer map represent N-terminal regions of p88 not visible in the structures of soluble VacA hexamers (Fig. 2A). While the resolution of the central region of the DDM-solubilized oligomer is too low to build an atomic model directly into the EM density, we predicted the structure of the N-terminal residues of VacA (residues 5–35), a region that includes the three GXXXG motifs, using AlphaFold2 (AF2) [53, 54]. Placement of this model into each of the protruding densities in the VacA EM map showed that the helices form what looks like a central helical bundle (Fig. 2B). These results provide experimental evidence for the existence of a VacA α-helical pore-forming domain and allow us to build a composite model by combining the atomic model of the soluble hexamer (PDB 6NYF) with the computational model of residues 5–35 placed into the cryo-EM density map of a detergent-extracted VacA hexamer (Fig. 2C, D). In this model, the central α-helical bundle forms a pore ~30 Å in diameter and extends 34 Å perpendicular from what is predicted to be the membrane-facing surface of the oligomer [44, 45]. Notably, the pore formed by the α-helical bundle is wider than the ~13 Å diameter channel predicted in a previous computational model of the VacA N-terminus [43] and wider than what would be expected for the conformation of a selective anion channel.
Table 1.
Cryo-EM data collection and processing information
| Detergent-extracted hexameric VacA | ||
|---|---|---|
| Data collection and processing | No applied symmetry (C1) | Applied symmetry (C6) |
| Magnification | 105,000× | 105,000× |
| Voltage (kV) | 300 | 300 |
| Total electron dose (e−/Å2) | 51.61 | 51.61 |
| Tilt angle (°) | 30 | 30 |
| Defocus range (μm) | 0.5 to 4.0 | 0.5 to 4.0 |
| Pixel size (Å) | 0.86 | 0.86 |
| Symmetry imposed | C1 | C6 |
| EMDB Accension number | EMDB-43272 | EMDB-43271 |
| Initial particles (number) | 1,653,323 | 1,653,323 |
| Final particles (number) | 20,539 | 20,539 |
| Map resolution (Å) | 7.7 | 6.7 |
| FSC threshold | 0.143 | 0.143 |
Figure 2. Placement of soluble VacA hexamer structure and AlphaFold2 (AF2) model of the VacA amino-terminus into structure of DMM-solubilized hexameric VacA.
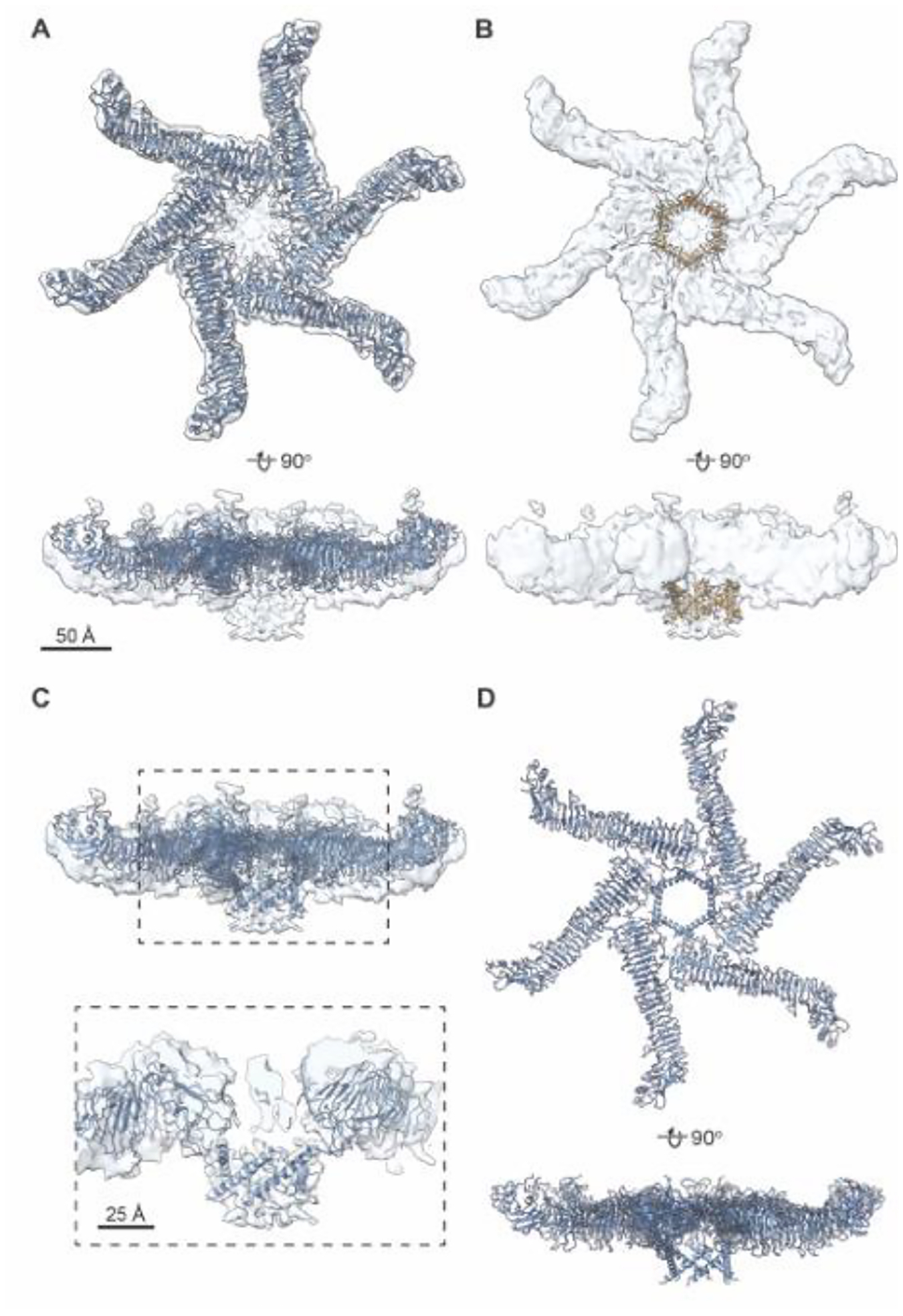
(A) Model of soluble hexamer VacA (PDB 6NYF, blue) fit into the 3D reconstruction of the DDM-solubilized VacA hexamer refined with C6 symmetry (opaque blue). Top, en face view. Bottom, view rotated around the x-axis 90°. Scale bar, 50 Å. (B) AF2 generated model of VacA amino acids 5–35 placed into the DDM-solubilized VacA hexamer 3D reconstruction (opaque blue). AF2 model is shown in goldenrod with one monomer highlighted in orange. Top, en face view. Bottom, view rotated around the x-axis 90°. Scale bar, 50 Å. (C) Composite model of a membrane associated VacA hexamer created by using the cryo-EM map of DDM-solubilized VacA hexamer (six-fold applied symmetry) to position the model of the soluble VacA hexamer (6NYF) with the computational model of VacA amino acids 5–35 (blue). Dashed box shows central region zoomed in below. (D). Composite 3D model of detergent-solubilized hexameric VacA described in (C) rotated 90° around the x-axis.
Cryo-EM analysis of VacA oligomers associated small unilamellar vesicle lipid bilayers
Analysis of the dataset of solubilized VacA oligomers indicated that central α-helical-like protrusions were detectable in only one 3D class and attempts to further refine this structure did not improve the resolution of the map. This might reflect the instability of the central α-helical region during the process of detergent extraction and/or heterogeneity in the conformations of VacA oligomers interacting with the membrane. To address the possibilities of VacA instability and heterogeneity due to extraction from the membrane, we analyzed VacA oligomers in an intact membrane environment, using both 2D classification and cryo-ET analysis to analyze VacA bound to liposomes.
First, we visualized VacA associated with eggPC/DOPS/Cholesterol (55/15/30 mol %) liposomes. For this analysis, we used small unilamellar vesicles (SUVs, ~20–40 nm in diameter) instead of the larger LUVs to make it easier to visualize individual oligomers in the bilayer. As described above, VacA was first acid-activated to disrupt the soluble oligomers before mixing with lipids. Cryo-EM images of VacA bound to SUVs showed views of VacA bound to the rim of the vesicle (Fig. 3A). 2D classification of 1,148 VacA bound to SUVs revealed two main membrane-associated VacA types. In some 2D classes, the inner and outer leaflets of the bilayer appear intact (Fig. 3B, middle panel), while in other averages, the bilayer leaflets appear to lack continuous density (Fig 3B). These results indicate that the central region of VacA oligomers can insert into the bilayer, but we did not detect convincing evidence of a VacA channel completely spanning the bilayer. Interestingly, we also noticed that the p55 “arms” are not crisply defined in the 2D averages of VacA oligomers bound to the SUVs, and there is space between the “arms” and the membrane, suggesting flexibility in the orientation of p55 “arms” when oligomers are bound to membrane (Fig. 3B). To examine this apparent flexibility in more detail, we performed symmetry expansion on the particles that contributed to the 3D structure of the VacA hexamer with an ordered central density and used these particles for 3D variability analysis (3DVA). For this analysis, three structural intermediates were calculated. The particles were submitted to 3DVA with a filter resolution of 10 Å. Three eigenvector modes (Mode 0, 1, and 2) were indicated, each of which consisted of a series of 20 volumes that were used for making a movie showing the variability in the position of the p55 “arms” (Movie 1). From the volume series, the first and last frames (frames 0 and 19) represent the negative and positive values along the reaction coordinate for each variability component (Movie 1 and Fig. S4). These results are consistent with structural heterogeneity both in how the central region of VacA is associated with membrane and the orientation of p55 regions of the VacA oligomer in relation to the membrane. Both observations provide insight into why we found only one 3D class of detergent-solubilized VacA with clear central densities. We speculate that the p55 regions may exhibit less flexibility upon VacA contact with membranes that have less curvature than the SUVs used in these experiments.
Figure 3. Cryo-EM analysis of VacA oligomers bound to small unilamellar vesicles (SUVs).
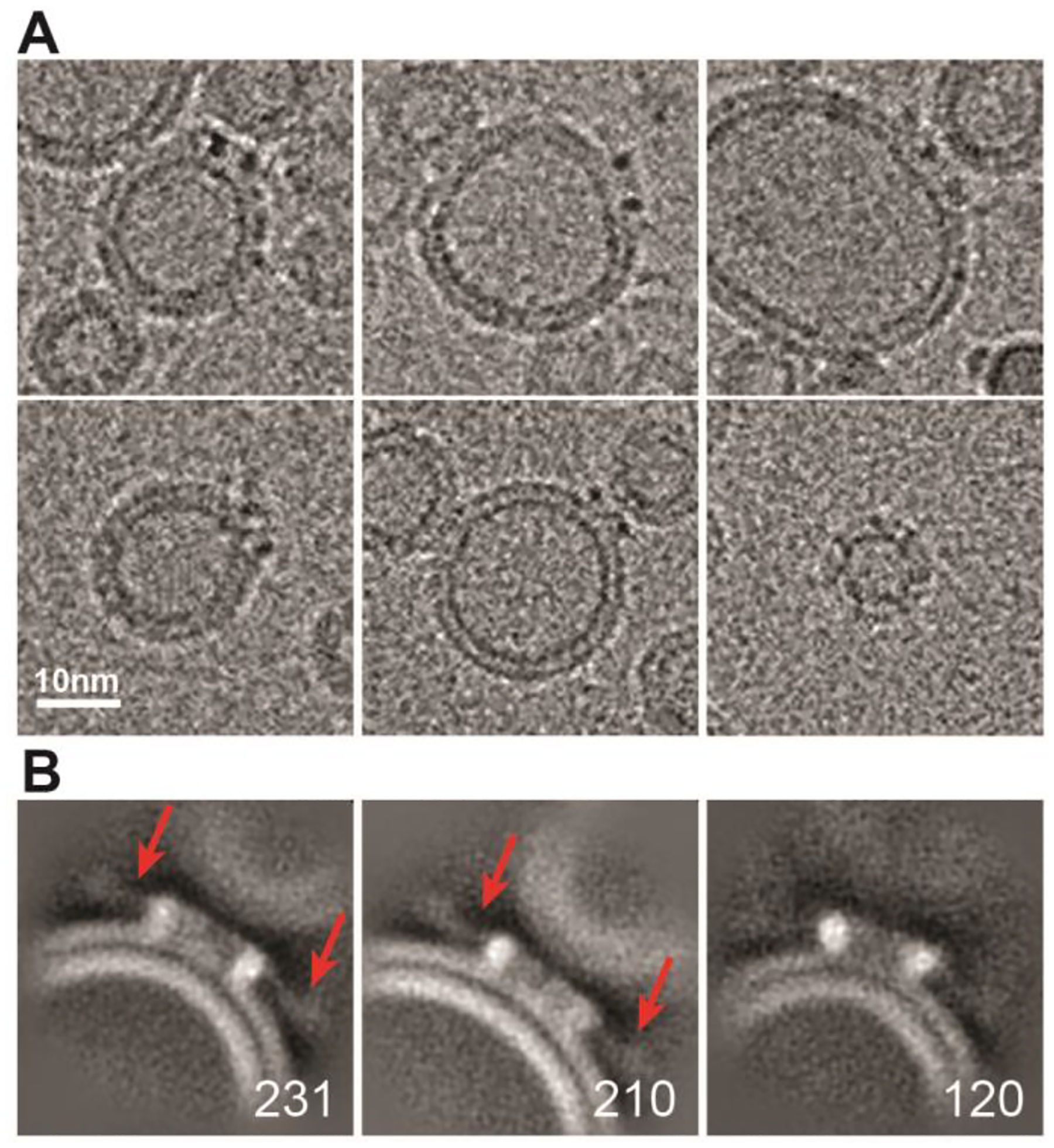
(A) Representative images of VacA oligomers bound to SUVs. Scale bar, 10 nm. (B) 2D classes averages of VacA oligomers bound to SUVs. Red arrows in first two panels point to location of the fuzzy p55 “arms”. Numbers of particles in each class are shown in bottom right corner. Box size, 364 Å.
As an alternative approach for analyzing the central region of the membrane-associated VacA oligomers, we used cryo-electron tomography (cryo-ET). Acid-activated VacA was bound to ~200 nm eggPC/DOPS/Cholesterol (55/15/30 mol %) liposomes and vitrified for cryo-ET analysis (Fig. 4A). Individual VacA oligomers were visible in the tomograms, and although the flexibility of the oligomeric arms made subtomogram averaging difficult (Fig. S5), slices through the tomograms clearly showed VacA oligomers bound to the LUVs (Fig. 4A, B). Closer examination of a slice of the resulting tomograms showed VacA oligomers with varying amounts of density inserted into the membrane (Fig. 4C). These included oligomers that appear to be simply sitting on top of the bilayer (Fig. 4C, panel marked with *), oligomers that have a central density that crosses one layer of the bilayer (Fig. 4C, panels marked with ^), and oligomers that have a central region that spans both bilayers (Fig. 4C, panels marked with #). Measuring the width between the densities showed a range spanning from 2.0 – 7.5 nm (Fig. 4D). The p55 arms are not visible in these panels because specific slices of the tomogram were chosen where it was easiest to see VacA insertion into the membrane. These results provide further evidence for heterogeneity in the conformations of the N-terminal α-helix in membrane-associated VacA, including verification that some of the membrane-associated VacA oligomers insert into the bilayer.
Figure 4. Cryo-ET analysis of VacA associated with liposomes.
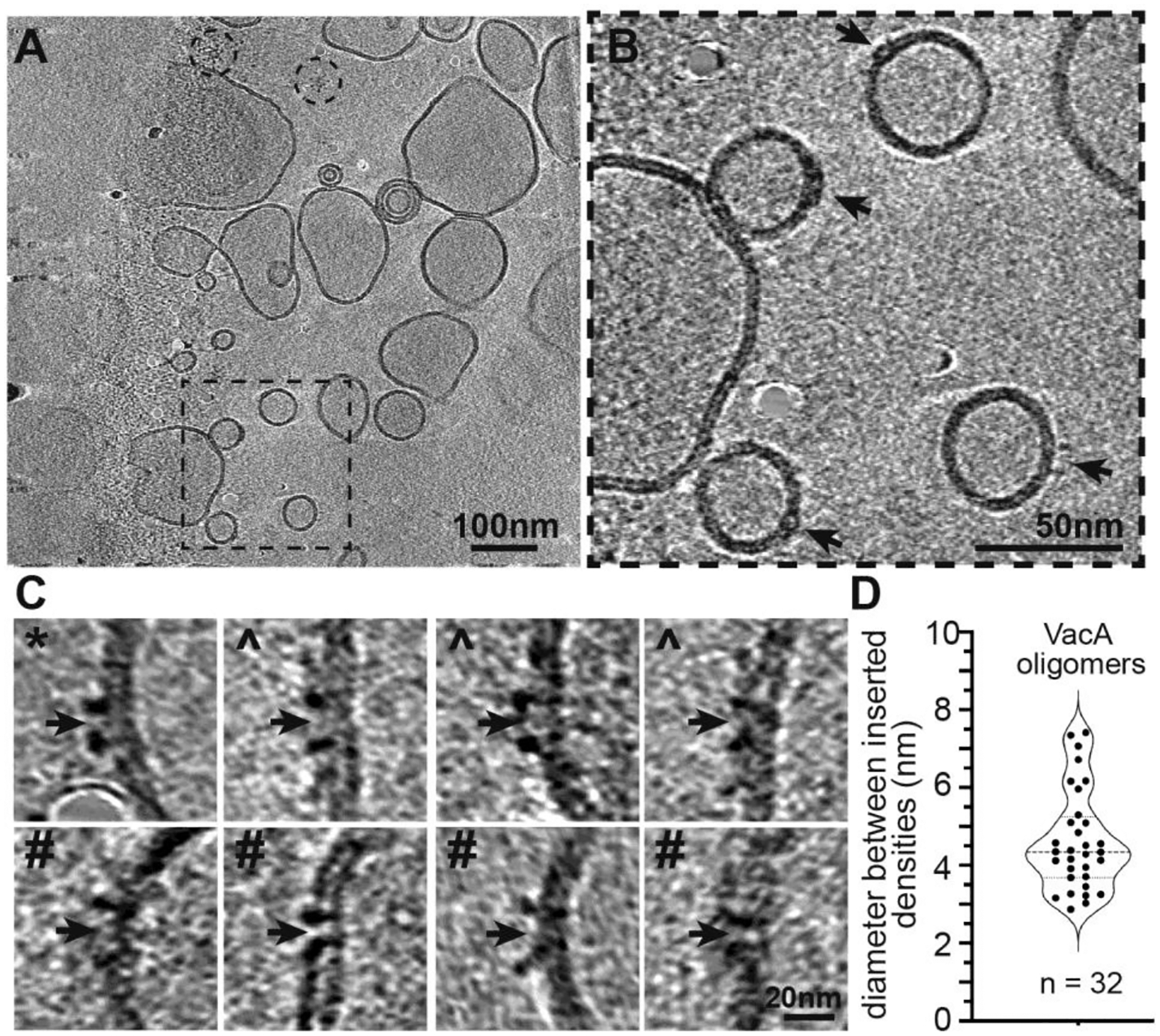
(A) Representative slice through a tomogram of VacA bound to liposomes. Black dashed box shows the area enlarged in panel B. Dashed circles indicate location of two VacA dodecamers not bound to liposomes. Scale bar = 100 nm. (B) Enlarged area of a slice through the tomogram shown in panel (A). Black arrows indicate VacA binding events on the liposomes. Scale bar, 50 nm. (C) Representative images of VacA oligomers bound to liposomes from a slice of cryo-ET tomograms. Centers of pore insertion are indicated with black arrows. * marks VacA oligomers associated with bilayer; ^ marks VacA oligomer crossing one layer of bilayer; # marks VacA oligomer fully spanning bilayer. Scale bar, 20 nm. (D) Violin plot showing the diameter (nm) between densities inserted in the membrane of VacA oligomers. N = 32.
Discussion
Recent cryo-EM structures of soluble VacA hexamers, heptamers, dodecamers, and tetradecamers revealed the structural basis of VacA oligomerization [44, 45]; however, none of these structures had central densities that could be used to assign residues for the N-terminal GXXXG motif residues (amino acids 14–26), which are critical for VacA channel activity. Our studies of VacA extracted from LUVs with detergent and VacA bound to vesicles reveal that membrane-associated VacA oligomers can adopt a conformational state different from VacA oligomers in solution. While our previous study showed negative stain EM 2D class averages of VacA hexamers on LUVs [47], there has not been a definitive observation of the structural organization of the central region without the presence of stain. Here, we show that 2D classes of detergent-solubilized VacA hexamers in vitrified ice have an extra central density that was not observed in 2D classes and 3D structures of soluble VacA oligomers in vitrified ice. This suggests that VacA oligomers can adopt a different structural conformation in the presence of detergent and/or membrane than what is seen from structures of water-soluble VacA oligomers [44, 45]. 3D reconstructions of the VacA complexes extracted from membranes using DDM show that for one 3D class, there are central α-helical densities that are ~30 Å in length. These densities extend from a VacA oligomeric surface that has been predicted to be the membrane-associated face of VacA oligomers [44, 45, 47]. At the resolution of the EM map (6.7 Å), α-helices appear as tubular densities, and modeling of the N-terminus of VacA (residues 5–35) predicts that this region forms an α-helix. The modeled structure of VacA residues 5–35 can be placed into the tube-like structures in the EM map, and when combined with the atomic model of the soluble VacA hexamer, we generated a composite model. This model suggests that VacA oligomers can form a central α-helical bundle when associated with membranes. Interestingly, 2D class averages of VacA oligomers bound to SUVs and cryo-ET analysis of VacA oligomers bound to liposomes show that central regions of the VacA oligomer appear to pierce into the bilayer – sometimes just into the outer leaflet of the SUV lipid bilayer, but sometimes traversing both lipid leaflets. This may indicate that VacA pore formation occurs in multiple steps, requiring multiple conformational changes, or alternatively, the artificial lipids and in vitro binding of VacA to the liposomes may not accurately recapitulate the environmental cues that stimulate VacA pore formation in intact cells. These observations lead us to propose that membrane-associated VacA oligomers undergo a conformational change to form a structured α-helical central region that forms a fully formed pore or channel only under the proper environmental conditions.
A previous study used computational modeling approaches to propose a model for the N-terminal region of hexameric VacA (residues 7–29) [43]. Like our AF2 model of residues 5–35 placed into the protruding central densities of the cryo-EM map (Fig. 5A, B), the previous computational model consists of six α-helices organized into the overall channel structure (Fig. 5C). Our current AF2-based model of the VacA amino-terminus, placed in the EM map, has a significantly larger diameter (~30 Å) compared to the diameter of the previous model (13 Å). Despite the difference in pore diameters and the number of residues included in each model, the length of the α-helices in both our AF2-based model and the previous model is approximately the same (34 Å versus 35 Å respectively, Fig. 5B, C). However, unlike the previous model [43], the α-helical bundle seen in our EM map could not represent a selective chloride channel because the side chains are too far apart to confer anion specificity. It is possible that in the context of the ideal cellular environment, following an initial insertion into the membrane, the VacA α-helices might organize into a more compact and canonical channel structure with a diameter similar to that of the previous predicted model. A rearrangement of the helices in our model to the angle and proximity of the previous model would increase the channel length to ~45 Å. Assuming a membrane bilayer width of ~50 Å, if the extended α-helical regions seen our map organize into a more channel-like conformation, the α-helical bundle would almost be long enough span the entire membrane. Interestingly, the first N-terminal residues visible in the structure of soluble VacA oligomers were sandwiched between the protomers and spanned Leu27 to Lys44, with Gly29-Asn39 forming a short α-helix and the other residues adopting no defined secondary structure [44, 45] (Fig. 6). Our composite model of a membrane-associated VacA hexamer suggests that upon interaction with membrane, the small α-helix (Gly29-Asn39) seen in the soluble VacA hexamer map likely undergoes a conformational change, “swinging” in towards the center of the oligomer (Fig. 6C). This would not only allow for organization of an N-terminal α-helical bundle that could under the proper cellular conditions form an anion channel, but also would provide the extra length required to completely span a membrane (Fig. 5 and 6). When the AF2 model is extended to span a longer stretch of the VacA N-terminus (residues 1–45, encompassing residues 36–45 predicted to swing into the center of the oligomer when associated with membrane) reveals that residues 2–38 could form a continuous α-helix, with residues 39–45 being unstructured (data not shown). Therefore, it is possible that VacA could span the lipid bilayer with the length of an α-helix composed of residues 2–38, along with an unstructured region that includes residues 39–45. However, while it is interesting to speculate how VacA might span the membrane, the structural conformation of the VacA hexamer as a selective anion channel remains to be experimentally determined.
Figure 5. Comparison of models of the N-terminus of VacA.
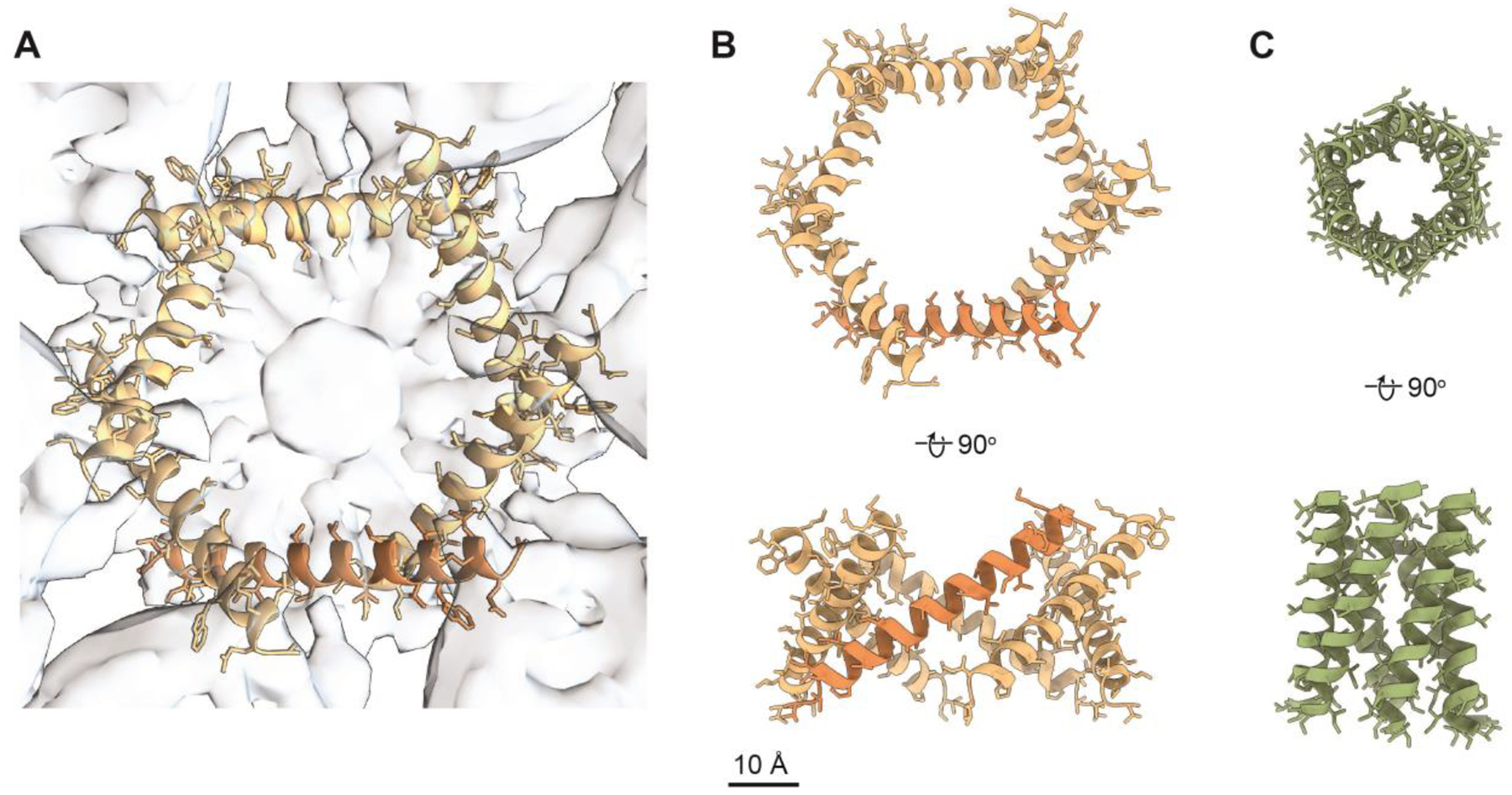
(A) Computational model of VacA amino acids 5–35 placed into the 3D reconstruction of DDM-solubilized VacA hexamers refined with C6 symmetry (opaque blue). AF2-based model is shown in goldenrod with one monomer highlighted in orange. 3D reconstruction is shown in opaque blue. (B) AF2-based model of VacA amino acids 5–35 and (C) a computationally predicted model of VacA amino acids 7–29 (PDB 1SEW). Both models are shown rotated 180° around the x-axis. PDB 1SEW is shown in light green. Scale bar, 10 Å.
Figure 6. Model for how the N-terminus of VacA rearranges during transition from soluble to membrane-associated protein.
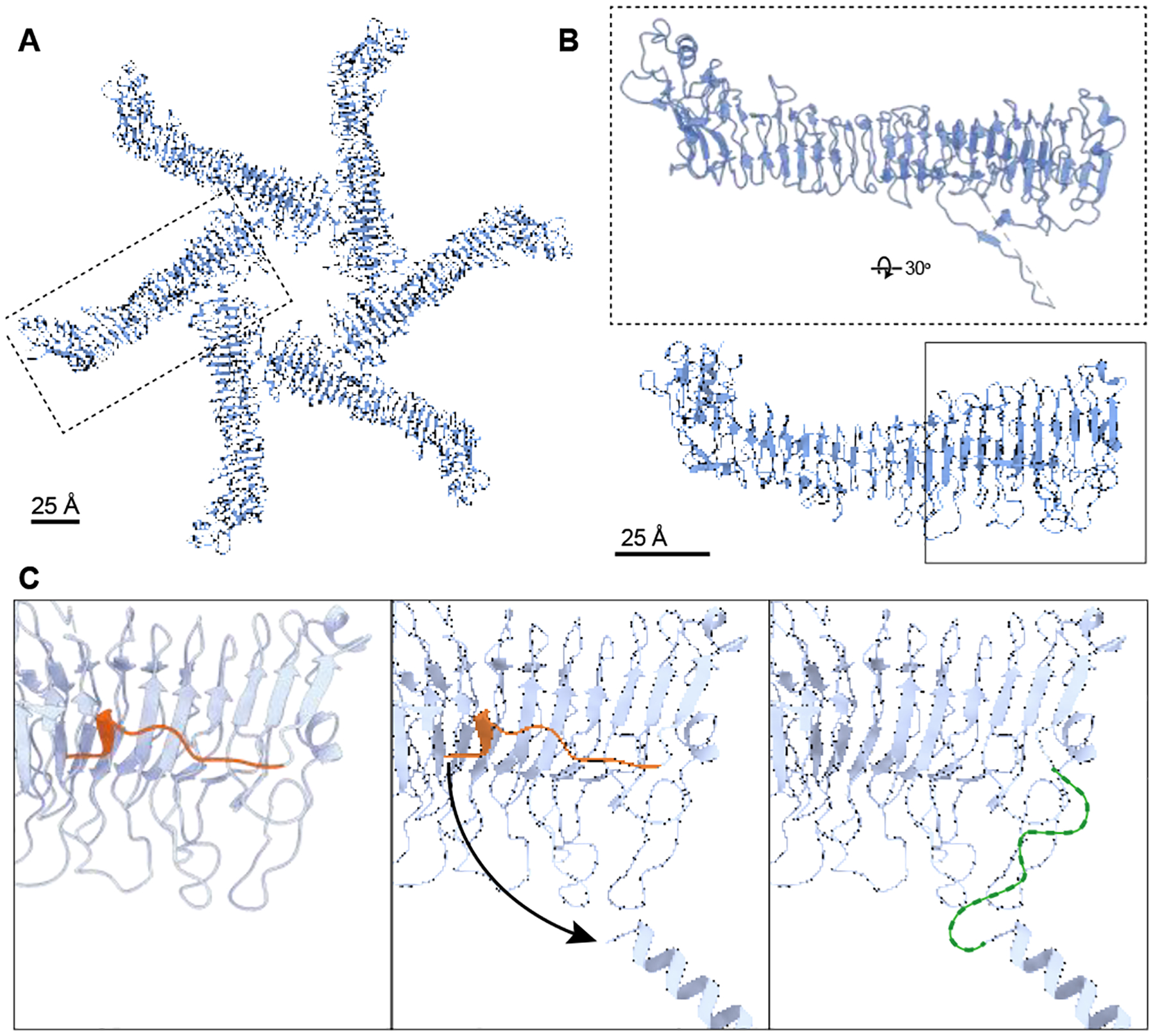
(A) Model of soluble hexamer VacA (PDB 6NYF, blue) with one protomer marked with a dashed box. Scale bar, 25 Å. (B) VacA protomer dashed boxed in A, rotated 30° around x-axis to show location of amino acids 36–45, which form a short α-helix and an unstructured region. Solid box shows region of model that is shown in (C). Scale bar, 25 Å. (C) Zoomed-in region of VacA p33 that is predicted to undergo a conformational change when associated with membranes. Left panel: When in solution, VacA residues 36–45 form a short α-helix and an unstructured region (orange) abutted against the p33 domain. Middle panel: When VacA associates with membrane, we predict that residues 5–35 form a continuous α-helix and, at a minimum, residues 36–45 (orange) change conformation allowing the N-terminal helix to orient into the center of the oligomer (arrow depicts direction). Right panel: This change in conformation makes it possible for six N-terminal α-helixes to form an α-helical bundle in the center of the hexamer. The resolution of the EM map of detergent-solubilized VacA is not at a high enough resolution to resolve amino acids 36–45 in the map. Thus, these residues are depicted as an undefined loop (green).
Higher resolution structures of membrane-associated VacA oligomers are still needed to fully characterize the structural organization of the pore and understand which regions of p33 are involved in facilitating the conformational changes that occur when VacA inserts and oligomerizes in association with the membrane. For example, while the three GXXXG motifs are required for active pore formation [37], lipid flotation assays show that the VacA Δ6–27, a mutant lacking all three N-terminal GXXXG repeats, can still insert into the lipid bilayer [47]. This result suggests that there are additional regions of p33, other than just the N-terminal residues, that insert into the membrane and may structurally contribute to pore formation. Notably, the flotation assays also showed that p55 alone does not co-sediment with the membrane [47], which suggests that p33 is the major driver of membrane insertion. Our observation that the p55 “arms” of VacA oligomers adopt flexible angles in relation to the membrane and have continuous conformational flexibility when examined by 3DVA analysis (Figs. 3B and S4, Movie 1) provides additional evidence that p55 may not interact tightly with the membrane.
While our current analysis shows that structured central density is visible when VacA is membrane-associated, the resolution of the 3D maps is too low to build a molecular model for how specific residues are organized within this density. There are many cellular factors that influence VacA’s activity in the context of the cell that could affect the structure of VacA oligomers. These include interaction with cellular receptors, association with lipid bilayers containing cholesterol and/or sphingomyelin, as well as changes in pH that occur in the endosome during endocytosis [55–59]. Although we tried adding sphingomyelin into liposomes used to extract VacA oligomers with the goal of improving structural homogeneity, we did not see any obvious change in results (data not shown). Thus, to gain more detailed molecular snapshots of how VacA forms an active pore, it will be necessary to visualize and determine structures of VacA oligomers bound to cellular binding partners and/or in a lipid environment closer to its cellular context. The methods and results presented in the current study provide an important foundation to guide future in situ structural studies.
In summary, we have shown that VacA can assemble into hexameric and heptameric complexes in the presence of membrane, and there are differences structural organization of the N-terminus of the VacA protomers when comparing the structure of membrane-associated and soluble VacA oligomers. Our 3D analysis of detergent-solubilized VacA hexamers allowed us to visualize α-helices protruding from the center of the oligomers, providing the first structural evidence supporting VacA classification as an α-helical pore-forming toxin.
Materials and Methods
Expression and purification of H. pylori VacA
The VacA s1/i1/m1 protein used in all experiments contains a strep tag at residue 808. [36]. An H. pylori strain secreting this strep-tagged VacA protein was grown in Brucella-broth supplemented with cholesterol lipid concentrate (Gibco) (BB-cholesterol) for 48 hours. VacA was then purified from H. pylori culture supernatant using the strep tag as previously described [36, 46].
LUV formation and binding to VacA
Before addition of purified VacA to detergent or membrane environments, the VacA was acid-activated to dissociate oligomers into monomers by dropwise addition of 0.5M HCl to pH 4.0.
Egg PC (L-α-phosophatidylcholine), DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine sodium salt), and ovine wool cholesterol (Avanti Polar Lipids) were mixed in chloroform at a 55/15/30 eggPC/DOPS/cholesterol mol % ratio. The chloroform was evaporated under N2 stream and dried overnight in a vacuum desiccator. Dried lipids were resuspended to 1 mg/mL with 10 mM HEPES pH 7.3, 100 mM KCl and incubated at room temperature for 30 minutes. To form LUVs, the resuspended lipids underwent 3 cycles of freezing in a dry ice ethanol bath and thawing in a 37°C water bath. They were then extruded 11 times through an 0.8 μm polycarbonate membrane (Millipore Sigma) using an Avanti Mini Extruder. For samples that would be detergent extracted, LUVs were mixed with acid-activated VacA at a lipid to protein ratio (LPR) of 25:1 (w:w) for 30 minutes at room temperature. For samples that were plunge frozen for cryo-ET analysis, LUVs were mixed with acid-activated VacA at a 20:1 LPR and incubated for 1 hour at room temperature.
Extraction of VacA from LUVs with detergents
The sample of VacA bound to LUVs was pelleted at 50,000xg for 45 minutes at 7°C. The pellet was washed 3 times with 10 mM HEPES pH 7.3, 100 mM KCl and resuspended in 10 mM HEPES, pH 7.3, 100 mM KCl, 2% DDM for 1 hour at room temperature. The resuspended sample was centrifuged at 50,000xg for 45 minutes at 7°C. The supernatant containing solubilized VacA in detergent micelles was collected and bound to Strep-Tactin resin (IBA Lifesciences) for 30 minutes at 4°C, washed 4 times with 10 mM HEPES pH 7.3, 100 mM KCl, 0.02% DDM, and eluted with 10 mM HEPES pH 7.3, 100 mM KCl, 0.02% DDM, 5 mM D-Desthiobiotin.
Negative stain EM of VacA extracted from LUVs
Negative stain EM was carried out using established methods [60]. 400-mesh copper grids covered with carbon-coated collodion film (Electron Microscopy Sciences) were glow discharged for 30 s at 5 mA in a PELCO easiGlow glow discharge unit (Ted Pella). The samples of VacA detergent-solubilized from LUVs (3 μL of 10–100μg/mL) were adsorbed to the grids and incubated for 1 minute at room temperature. The grids were then washed twice with water, negatively stained with 0.7% (w/v) uranyl formate solution and blotted until dry. Transmission electron microscopy (TEM) images were obtained on a FEI Morgagni electron microscope (Thermo Fisher Scientific) run at 100 keV at a nominal magnification of 22,000x (2.1Å/pixel).
A negative stain dataset of the DDM-extracted VacA particles was collected using a Tecnai Spirit T12 transmission electron microscope (Thermo Fisher Scientific) operated at 120 kV and at a nominal magnification of 26,000x (2.3 Å/pixel). Images were acquired with Leginon [61] on a 4K x 4K Rio complementary metal-oxide semiconductor camera (Gatan) at −1.5μm defocus value. Contrast transfer function (CTF) estimation was performed using gCTF [62]. Particles were picked using crYOLO’s general negative stain model and analyzed with the crYOLO box manager using SBGrid [63, 64]. Using RELION 3.0.8, particles were extracted with a 618 Å box size and 2D classified into 20 classes with a 400 Å mask size [65, 66]. 2D class averages were generated using 4,662 VacA particles.
Cryo-EM grid preparation and tilted data collection of VacA extracted from LUVs with DDM
DDM-extracted VacA oligomers (3.5 μL, final DDM concentration of 0.02%) were applied two times to Quantifoil R 1.2/1.3 200 mesh copper grids with an ultrathin continuous carbon layer (Electron Microscopy Science) that were glow discharged for 30 seconds at 5 mA. There was a wait time of 30 seconds for each application. The sample was vitrified by plunge freezing into liquid ethane using a Vitrobot Mark IV plunger (Thermo Fisher Scientific). The chamber was set to 100% humidity/4°C, and grids were blotted for 4.5 seconds with a blot force of 1.
Images were collected on a Titan Krios G4i transmission electron microscope (Thermo Fisher Scientific) operated at 300 kV and equipped with a K3 direct detection camera with a BioQuantum energy filter with a slit width of 20 eV (Gatan). Images were collected at a nominal magnification of 105,000x (0.86Å/pixel), and the stage was tilted 30°for data collection. 12,828 movies were recorded for the DDM extracted VacA oligomers using EPU (Thermo Fisher Scientific) with a total dose of 51.61 e-/Å2 and a −2.5 μm defocus.
Cryo-EM single particle analysis image processing of VacA oligomers extracted from LUVs with DDM
All image processing and 3D refinements were done in cryoSPARC v3.3X [67]. 12,828 movies were corrected for local beam-induced drift using patch motion correction, and the local CTF parameters were estimated using patch CTF estimation. Exposures were curated to include only those with a CTF estimation of ≤5Å, resulting in 10,545 selected micrographs. About 1,000 particles were manually selected from micrographs and used as templates for template-based picking of particles. After template-based picking, 1,653,323 particles were extracted in 700-pixel2 boxes (602 Å x 602 Å) and binned to 300-pixel2 boxes (SFig. 2). Two rounds of 2D classification were performed to remove heptameric oligomers and obvious double layer VacA particles. Following these rounds of 2D classification, 239,955 particles from obvious VacA containing 2D classes were used for a two-class ab initio 3D reconstruction. To further sort the particles, the 133,711 particles from the ab initio 3D reconstruction containing VacA particles were heterogeneously refined using both ab initio 3D reconstructions (the 3D hexameric VacA volume composed of the 133,711 particles and the 3D “junk” volume) as input volumes. The resulting 3D reconstruction was hexameric VacA, but the density also appeared to contain double-layer VacA particles. To sort hexamers from dodecamers, the 102,343 particles that contributed to this VacA refinement underwent 3D classification into 10 classes. After the exclusion of the five classes whose reconstructions represented VacA dodecamers, the remaining 59,955 particles were used as input into a two-class ab initio reconstruction. Both 3D reconstructions exhibited the overall β-strand secondary structure expected in the VacA “arms”; however, one 3D class also had clear central protruding central density. The 20,539 particles from the ab initio exhibiting the structured central density were used for C1 and C6 non-uniform refinements. The masked Fourier Shell Correlation (FSC) resolutions for the C1 and C6 non-uniform refinements were 7.7 Å and 6.7 Å, respectively (Table 1).
For 3DVA analysis, the 20,539 particles in the VacA hexamer structure were symmetry expanded with C6 symmetry. The symmetry expanded particles (123,234) were then used in the 3DVA clustering routine in cryoSPARC [67]. Three structural intermediates were calculated with a filter resolution of 10 Å. Three eigenvector modes were indicated, each of which consisted of a series of 20 volumes that were used for making movies (Movie 1).
AlphaFold2 prediction of VacA N-terminal residues and placement in EM density map
The VacA N-terminal region of amino acids 5–35 were input into AlphaFold2 using ColabFold [53, 54]. Templates were detected in pdb70, mmseqs2_uniref_env was the MSA mode used, and an unpaired-paired pair mode was used. The AF2 model (residues 1–45) referenced in the discussion was generated in the same way. Six identical copies of the AF2 monomeric VacA 5–35 model were then placed into the central density of the EM map of DDM-solubilized VacA hexamer (C6 symmetry). Each AF2 predicted helix was placed into the density using the apparent pitch of the helices in the EM density. The resolution of the EM map was not high enough to confidently model the location of specific amino-acids or the angles of the amino-acid side chains.
VacA binding to SUVs
Egg PC (L-a-phosophatidylcholine 840051C), DOPS (1,2-dioleoyl-sn-glycero-3-phospho-L-serine sodium salt 840035C), and ovine wool cholesterol (Avanti Polar Lipids) were mixed in chloroform at a 55/15/30 eggPC/DOPS/cholesterol mol % ratio. The solvent was evaporated under N2 stream and dried overnight in a vacuum desiccator. Dried lipids were resuspended to 2 mg/mL with 10 mM HEPES pH 7.3, 100 mM KCl. Lipids were vortexed for 2 minutes and sonicated in a water bath sonicator for 50 minutes until the solution was clear. Prior to binding of SUVs with VacA, SUVs were prewarmed in a 37°C incubator for 15 minutes. VacA was diluted to 0.5 mg/mL in 10 mM HEPES pH 7.3, 100 mM KCl, acid-activated with HCl, and mixed slowly with prewarmed SUVs at a LPR of 20:1. The mixture was then incubated at 37°C for 15 minutes.
Cryo-EM single particle grid preparation and data collection of VacA bound to SUVs
3.5 μL of VacA bound to SUV samples was applied to Quantifoil R 1.2/1.3 200 mesh copper grids with an ultrathin continuous carbon layer that were glow discharged for 30 seconds at 5 mA (Electron Microscopy Science). Following a 4.5 second blotting at blot force 1 and a 30 second wait time, the sample was plunge frozen in liquid ethane using a Vitrobot Mark IV plunger (Thermo Fisher Scientific). The chamber was set to 100% humidity/4°C. 3,800 VacA wild-type SUV movies were recorded using Leginon on a Talos Arctica transmission electron microscope (Thermo Fisher Scientific) operated at 200 keV and equipped with a K2 direct detection camera (Gatan) (Suloway, 2005). The pixel size of the collection was 0.91 Å/pixel, and the dataset was collected with a total dose of ~60e-/Å2 and a −2 to −3.5 μm defocus range.
Cryo-EM single particle data processing of VacA bound to SUVs
All frames were aligned using MotionCor2 [68]. CTFFIND4 was used to estimate CTF values [69]. 1,158 VacA bound SUVs were manually selected in RELION 3.1.4, followed by extraction into 364 Å x 364 Å box sizes [65, 66]. 2D classification of VacA bound SUV particles was performed using cisTEM [70].
Cryo-ET grid preparation and data collection of VacA bound to LUVs
VacA bound to LUVs was mixed at a 5:3 sample to fiducial ratio with 10 nm BSA gold tracer (Electron Microscopy Sciences) concentrated 100x in 10mM HEPES pH 7.3, 100 mM KCl. 3.5 μL of the VacA bound to LUVs and the fiducial mixture was applied two times to Quantifoil R 2/2 200 mesh copper grids with an ultrathin continuous carbon layer. The grids were glow discharged for 30 seconds at 5 mA. For each application, there was a 30-second wait time. A Vitrobot Mark IV plunger (Thermo Fisher Scientific) was used to vitrify the sample in liquid ethane. The chamber was set to 100% humidity at 4°C, and grids were blotted for 5.5 seconds with a blot force of 1.
41 tilt series were collected on a Krios G4i transmission electron microscope (Thermo Fisher Scientific) operated at 300 keV and equipped with a K3 direct detection camera and BioQuantum energy filter with a slit width of 20 eV (Gatan). Images were collected at a nominal magnification of 42,000x (2.1 Å/pixel). Tilt-series were collected using SerialEM 3.8 software with a dose-symmetric tilt scheme ranging from −60° to +60° [71, 72]. Tilt series were collected with a total dose of ~100e/Å2 and with a defocus range of −4 to −5 μm.
Cryo-ET data processing, measurement, and segmentation of VacA bound to LUVs
VacA wild-type LUV tilt series were aligned using fiducial-based alignment in IMOD 4.11.11 using eTomo [71]. VacA insertion diameters were measured manually in IMOD 4.11.11 [71]. Segmentation was performed in EMAN2 [73]. Following manual segmentation of LUV membrane and VacA particles, neural networks were iteratively trained to segment membrane and VacA particles.
Supplementary Material
Highlights.
VacA oligomers undergo a conformational change when associated with membrane
The VacA N-terminus forms an α-helical bundle
Cryo-electron tomography shows that the central region of VacA oligomers inserts into membrane
Acknowledgements
We thank the Cover and Ohi labs for helpful discussions, as well as cryo-EM advice provided by Ashleigh Raczkowski, Vinson Lam, and Alexandra Rizo. The UM Cryo-EM Facility has received generous support from the U-M Life Sciences Institute, the U-M Biosciences Initiative, and the Arnold and Mabel Beckman Foundation.
Funding
This work was supported by NIH AI039657 (to T.L.C.), NIH AI118932 (to T.L.C and M.D.O.), NIH CA116087 (to T.L.C), NIH S10OD030275 (to M.D.O.), NIH T32GM08320 (to A.L.E.), NIH T32GM007315 (to S.C.), NIH F31GM139291 (to R.M.T), American Heart Association grant 905705 (to S.C.), and the Department of Veterans Affairs 5I01BX004447 (to T.L.C.).
Data Availability
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-43272 (C1 hexamer) and EMD-43271 (C6 hexamer).
References
- [1].Dal Peraro M, van der Goot FG. Pore-forming toxins: ancient, but never really out of fashion. Nature reviews. 2016;14:77–92. [DOI] [PubMed] [Google Scholar]
- [2].Li Y, Li Y, Mengist HM, Shi C, Zhang C, Wang B, et al. Structural Basis of the Pore-Forming Toxin/Membrane Interaction. Toxins. 2021;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Mondal AK, Lata K, Singh M, Chatterjee S, Chauhan A, Puravankara S, et al. Cryo-EM elucidates mechanism of action of bacterial pore-forming toxins. Biochim Biophys Acta Biomembr. 2022;1864:184013. [DOI] [PubMed] [Google Scholar]
- [4].Hooi JKY, Lai WY, Ng WK, Suen MMY, Underwood FE, Tanyingoh D, et al. Global Prevalence of Helicobacter pylori Infection: Systematic Review and Meta-Analysis. Gastroenterology. 2017;153:420–9. [DOI] [PubMed] [Google Scholar]
- [5].Cover TL, Blaser MJ. Purification and characterization of the vacuolating toxin from Helicobacter pylori. The Journal of Biological Chemistry. 1992;267:10570–5. [PubMed] [Google Scholar]
- [6].Atherton JC, Cao P, Peek RM Jr., Tummuru MK, Blaser MJ, Cover TL. Mosaicism in vacuolating cytotoxin alleles of Helicobacter pylori. Association of specific vacA types with cytotoxin production and peptic ulceration. The Journal of Biological Chemistry. 1995;270:17771–7. [DOI] [PubMed] [Google Scholar]
- [7].Cover TL, Blanke SR. Helicobacter pylori VacA, a paradigm for toxin multifunctionality. Nature Reviews. 2005;3:320–32. [DOI] [PubMed] [Google Scholar]
- [8].Foegeding NJ, Caston RR, McClain MS, Ohi MD, Cover TL. An Overview of Helicobacter pylori VacA Toxin Biology. Toxins. 2016;8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McClain MS, Beckett AC, Cover TL. Helicobacter pylori Vacuolating Toxin and Gastric Cancer. Toxins. 2017;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].McClain MS, Cao P, Iwamoto H, Vinion-Dubiel AD, Szabo G, Shao Z, et al. A 12-amino-acid segment, present in type s2 but not type s1 Helicobacter pylori VacA proteins, abolishes cytotoxin activity and alters membrane channel formation. Journal of Bacteriology. 2001;183:6499–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Letley DP, Rhead JL, Twells RJ, Dove B, Atherton JC. Determinants of non-toxicity in the gastric pathogen Helicobacter pylori. The Journal of biological chemistry. 2003;278:26734–41. [DOI] [PubMed] [Google Scholar]
- [12].Genisset C, Puhar A, Calore F, de Bernard M, Dell’Antone P, Montecucco C. The concerted action of the Helicobacter pylori cytotoxin VacA and of the v-ATPase proton pump induces swelling of isolated endosomes. Cellular Microbiology. 2007;9:1481–90. [DOI] [PubMed] [Google Scholar]
- [13].Gangwer KA, Shaffer CL, Suerbaum S, Lacy DB, Cover TL, Bordenstein SR. Molecular evolution of the Helicobacter pylori vacuolating toxin gene vacA. Journal of Bacteriology. 2010;192:6126–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Calore F, Genisset C, Casellato A, Rossato M, Codolo G, Esposti MD, et al. Endosome-mitochondria juxtaposition during apoptosis induced by H. pylori VacA. Cell Death and Differentiation. 2010;17:1707–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Cover TL. Helicobacter pylori Diversity and Gastric Cancer Risk. mBio. 2016;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Jain P, Luo ZQ, Blanke SR. Helicobacter pylori vacuolating cytotoxin A (VacA) engages the mitochondrial fission machinery to induce host cell death. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:16032–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Radin JN, Gonzalez-Rivera C, Ivie SE, McClain MS, Cover TL. Helicobacter pylori VacA induces programmed necrosis in gastric epithelial cells. Infection and Immunity. 2011;79:2535–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Kern B, Jain U, Utsch C, Otto A, Busch B, Jimenez-Soto L, et al. Characterization of Helicobacter pylori VacA-containing vacuoles (VCVs), VacA intracellular trafficking and interference with calcium signalling in T lymphocytes. Cellular Microbiology. 2015;17:1811–32. [DOI] [PubMed] [Google Scholar]
- [19].Kim IJ, Lee J, Oh SJ, Yoon MS, Jang SS, Holland RL, et al. Helicobacter pylori Infection Modulates Host Cell Metabolism through VacA-Dependent Inhibition of mTORC1. Cell Host & Microbe. 2018;23:583–93 e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Capurro MI, Greenfield LK, Prashar A, Xia S, Abdullah M, Wong H, et al. VacA generates a protective intracellular reservoir for Helicobacter pylori that is eliminated by activation of the lysosomal calcium channel TRPML1. Nat Microbiol. 2019;4:1411–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Gebert B, Fischer W, Weiss E, Hoffmann R, Haas R. Helicobacter pylori vacuolating cytotoxin inhibits T lymphocyte activation. Science (New York, NY. 2003;301:1099–102. [DOI] [PubMed] [Google Scholar]
- [22].Sundrud MS, Torres VJ, Unutmaz D, Cover TL. Inhibition of primary human T cell proliferation by Helicobacter pylori vacuolating toxin (VacA) is independent of VacA effects on IL-2 secretion. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7727–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Oertli M, Noben M, Engler DB, Semper RP, Reuter S, Maxeiner J, et al. Helicobacter pylori gamma-glutamyl transpeptidase and vacuolating cytotoxin promote gastric persistence and immune tolerance. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:3047–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Engler DB, Reuter S, van Wijck Y, Urban S, Kyburz A, Maxeiner J, et al. Effective treatment of allergic airway inflammation with Helicobacter pylori immunomodulators requires BATF3-dependent dendritic cells and IL-10. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:11810–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Altobelli A, Bauer M, Velez K, Cover TL, Muller A. Helicobacter pylori VacA Targets Myeloid Cells in the Gastric Lamina Propria To Promote Peripherally Induced Regulatory T-Cell Differentiation and Persistent Infection. mBio. 2019;10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Kyburz A, Fallegger A, Zhang X, Altobelli A, Artola-Boran M, Borbet T, et al. Transmaternal Helicobacter pylori exposure reduces allergic airway inflammation in offspring through regulatory T cells. J Allergy Clin Immunol. 2019;143:1496–512 e11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Szabo I, Brutsche S, Tombola F, Moschioni M, Satin B, Telford JL, et al. Formation of anion-selective channels in the cell plasma membrane by the toxin VacA of Helicobacter pylori is required for its biological activity. The EMBO Journal. 1999;18:5517–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Tombola F, Carlesso C, Szabo I, de Bernard M, Reyrat JM, Telford JL, et al. Helicobacter pylori vacuolating toxin forms anion-selective channels in planar lipid bilayers: possible implications for the mechanism of cellular vacuolation. Biophysical Journal. 1999;76:1401–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Czajkowsky DM, Iwamoto H, Cover TL, Shao Z. The vacuolating toxin from Helicobacter pylori forms hexameric pores in lipid bilayers at low pH. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:2001–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Iwamoto H, Czajkowsky DM, Cover TL, Szabo G, Shao Z. VacA from Helicobacter pylori: a hexameric chloride channel. FEBS letters. 1999;450:101–4. [DOI] [PubMed] [Google Scholar]
- [31].Telford JL, Ghiara P, Dell’Orco M, Comanducci M, Burroni D, Bugnoli M, et al. Gene structure of the Helicobacter pylori cytotoxin and evidence of its key role in gastric disease. The Journal of Experimental Medicine. 1994;179:1653–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Torres VJ, McClain MS, Cover TL. Interactions between p-33 and p-55 domains of the Helicobacter pylori vacuolating cytotoxin (VacA). The Journal of Biological Chemistry. 2004;279:2324–31. [DOI] [PubMed] [Google Scholar]
- [33].Torres VJ, Ivie SE, McClain MS, Cover TL. Functional properties of the p33 and p55 domains of the Helicobacter pylori vacuolating cytotoxin. The Journal of Biological Chemistry. 2005;280:21107–14. [DOI] [PubMed] [Google Scholar]
- [34].Gangwer KA, Mushrush DJ, Stauff DL, Spiller B, McClain MS, Cover TL, et al. Crystal structure of the Helicobacter pylori vacuolating toxin p55 domain. Proceedings of the National Academy of Sciences of the United States of America. 2007;104:16293–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Gonzalez-Rivera C, Gangwer KA, McClain MS, Eli IM, Chambers MG, Ohi MD, et al. Reconstitution of Helicobacter pylori VacA toxin from purified components. Biochemistry. 2010;49:5743–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Gonzalez-Rivera C, Campbell AM, Rutherford SA, Pyburn TM, Foegeding NJ, Barke TL, et al. A Nonoligomerizing Mutant Form of Helicobacter pylori VacA Allows Structural Analysis of the p33 Domain. Infection and Immunity. 2016;84:2662–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].McClain MS, Iwamoto H, Cao P, Vinion-Dubiel AD, Li Y, Szabo G, et al. Essential role of a GXXXG motif for membrane channel formation by Helicobacter pylori vacuolating toxin. The Journal of Biological Chemistry. 2003;278:12101–8. [DOI] [PubMed] [Google Scholar]
- [38].Vinion-Dubiel AD, McClain MS, Czajkowsky DM, Iwamoto H, Ye D, Cao P, et al. A dominant negative mutant of Helicobacter pylori vacuolating toxin (VacA) inhibits VacA-induced cell vacuolation. The Journal of Biological Chemistry. 1999;274:37736–42. [DOI] [PubMed] [Google Scholar]
- [39].Ye D, Blanke SR. Mutational analysis of the Helicobacter pylori vacuolating toxin amino terminus: identification of amino acids essential for cellular vacuolation. Infection and Immunity. 2000;68:4354–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Ye D, Blanke SR. Functional complementation reveals the importance of intermolecular monomer interactions for Helicobacter pylori VacA vacuolating activity. Molecular Microbiology. 2002;43:1243–53. [DOI] [PubMed] [Google Scholar]
- [41].El-Bez C, Adrian M, Dubochet J, Cover TL. High resolution structural analysis of Helicobacter pylori VacA toxin oligomers by cryo-negative staining electron microscopy. Journal of Structural Biology. 2005;151:215–28. [DOI] [PubMed] [Google Scholar]
- [42].Chambers MG, Pyburn TM, Gonzalez-Rivera C, Collier SE, Eli I, Yip CK, et al. Structural analysis of the oligomeric states of Helicobacter pylori VacA toxin. Journal of Molecular Biology. 2013;425:524–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Kim S, Chamberlain AK, Bowie JU. Membrane channel structure of Helicobacter pylori vacuolating toxin: role of multiple GXXXG motifs in cylindrical channels. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:5988–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Su M, Erwin AL, Campbell AM, Pyburn TM, Salay LE, Hanks JL, et al. Cryo-EM Analysis Reveals Structural Basis of Helicobacter pylori VacA Toxin Oligomerization. Journal of Molecular Biology. 2019;431:1956–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhang K, Zhang H, Li S, Pintilie GD, Mou TC, Gao Y, et al. Cryo-EM structures of Helicobacter pylori vacuolating cytotoxin A oligomeric assemblies at near-atomic resolution. Proceedings of the National Academy of Sciences of the United States of America. 2019;116:6800–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Caso GC, McClain MS, Erwin AL, Truelock MD, Campbell AM, Leasure CS, et al. Functional Properties of Oligomeric and Monomeric Forms of Helicobacter pylori VacA Toxin. Infection and Immunity. 2021;89:e0034821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Pyburn TM, Foegeding NJ, Gonzalez-Rivera C, McDonald NA, Gould KL, Cover TL, et al. Structural organization of membrane-inserted hexamers formed by Helicobacter pylori VacA toxin. Molecular Microbiology. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].de Bernard M, Papini E, de Filippis V, Gottardi E, Telford J, Manetti R, et al. Low pH activates the vacuolating toxin of Helicobacter pylori, which becomes acid and pepsin resistant. The Journal of Biological Chemistry. 1995;270:23937–40. [DOI] [PubMed] [Google Scholar]
- [49].Cover TL, Hanson PI, Heuser JE. Acid-induced dissociation of VacA, the Helicobacter pylori vacuolating cytotoxin, reveals its pattern of assembly. The Journal of Cell Biology. 1997;138:759–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Molinari M, Galli C, de Bernard M, Norais N, Ruysschaert JM, Rappuoli R, et al. The acid activation of Helicobacter pylori toxin VacA: structural and membrane binding studies. Biochemical and Biophysical Research Communications. 1998;248:334–40. [DOI] [PubMed] [Google Scholar]
- [51].Fossati M, Goud B, Borgese N, Manneville JB. An investigation of the effect of membrane curvature on transmembrane-domain dependent protein sorting in lipid bilayers. Cellular Logistics. 2014;4:e29087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Tan YZ, Baldwin PR, Davis JH, Williamson JR, Potter CS, Carragher B, et al. Addressing preferred specimen orientation in single-particle cryo-EM through tilting. Nature Methods. 2017;14:793–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Varadi M, Anyango S, Deshpande M, Nair S, Natassia C, Yordanova G, et al. AlphaFold Protein Structure Database: massively expanding the structural coverage of protein-sequence space with high-accuracy models. Nucleic Acids Research. 2022;50:D439–D44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Mirdita M, Schutze K, Moriwaki Y, Heo L, Ovchinnikov S, Steinegger M. ColabFold: making protein folding accessible to all. Nature Methods. 2022;19:679–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Sewald X, Gebert-Vogl B, Prassl S, Barwig I, Weiss E, Fabbri M, et al. Integrin subunit CD18 Is the T-lymphocyte receptor for the Helicobacter pylori vacuolating cytotoxin. Cell Host & Microbe. 2008;3:20–9. [DOI] [PubMed] [Google Scholar]
- [56].Gupta VR, Patel HK, Kostolansky SS, Ballivian RA, Eichberg J, Blanke SR. Sphingomyelin functions as a novel receptor for Helicobacter pylori VacA. PLoS Pathogens. 2008;4:e1000073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Gupta VR, Wilson BA, Blanke SR. Sphingomyelin is important for the cellular entry and intracellular localization of Helicobacter pylori VacA. Cellular Microbiology. 2010;12:1517–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Raghunathan K, Foegeding NJ, Campbell AM, Cover TL, Ohi MD, Kenworthy AK. Determinants of Raft Partitioning of the Helicobacter pylori Pore-Forming Toxin VacA. Infection and Immunity. 2018;86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Foegeding NJ, Raghunathan K, Campbell AM, Kim SW, Lau KS, Kenworthy AK, et al. Intracellular Degradation of Helicobacter pylori VacA Toxin as a Determinant of Gastric Epithelial Cell Viability. Infection and Immunity. 2019;87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Ohi M, Li Y, Cheng Y, Walz T. Negative Staining and Image Classification - Powerful Tools in Modern Electron Microscopy. Biol Proced Online. 2004;6:23–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Suloway C, Pulokas J, Fellmann D, Cheng A, Guerra F, Quispe J, et al. Automated molecular microscopy: the new Leginon system. Journal of Structural Biology. 2005;151:41–60. [DOI] [PubMed] [Google Scholar]
- [62].Zhang K Gctf: Real-time CTF determination and correction. Journal of Structural Biology. 2016;193:1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Wagner T, Merino F, Stabrin M, Moriya T, Antoni C, Apelbaum A, et al. SPHIRE-crYOLO is a fast and accurate fully automated particle picker for cryo-EM. Commun Biol. 2019;2:218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Morin A, Eisenbraun B, Key J, Sanschagrin PC, Timony MA, Ottaviano M, et al. Collaboration gets the most out of software. eLife. 2013;2:e01456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Scheres SH. RELION: implementation of a Bayesian approach to cryo-EM structure determination. Journal of Structural Biology. 2012;180:519–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Zivanov J, Nakane T, Forsberg BO, Kimanius D, Hagen WJ, Lindahl E, et al. New tools for automated high-resolution cryo-EM structure determination in RELION-3. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nature methods. 2017;14:290–6. [DOI] [PubMed] [Google Scholar]
- [68].Zheng SQ, Palovcak E, Armache JP, Verba KA, Cheng Y, Agard DA. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nature Methods. 2017;14:331–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Rohou A, Grigorieff N. CTFFIND4: Fast and accurate defocus estimation from electron micrographs. Journal of Structural Biology. 2015;192:216–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Grant T, Rohou A, Grigorieff N. cisTEM, user-friendly software for single-particle image processing. eLife. 2018;7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Mastronarde DN. Automated electron microscope tomography using robust prediction of specimen movements. Journal of Structural Biology. 2005;152:36–51. [DOI] [PubMed] [Google Scholar]
- [72].Turonova B, Hagen WJH, Obr M, Mosalaganti S, Beugelink JW, Zimmerli CE, et al. Benchmarking tomographic acquisition schemes for high-resolution structural biology. Nature Communications. 2020;11:876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Chen M, Dai W, Sun SY, Jonasch D, He CY, Schmid MF, et al. Convolutional neural networks for automated annotation of cellular cryo-electron tomograms. Nature Methods. 2017;14:983–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The cryo-EM maps have been deposited in the Electron Microscopy Data Bank under accession codes EMD-43272 (C1 hexamer) and EMD-43271 (C6 hexamer).


