Abstract
The search for biomarkers that quantify biological aging (particularly ‘omics’-based biomarkers) has intensified in recent years. Such biomarkers could predict aging-related outcomes and could serve as surrogate endpoints for the evaluation of interventions promoting healthy aging and longevity. However, no consensus exists on how biomarkers of aging should be validated prior to their translation to the clinic. Here, we review current efforts to evaluate the predictive validity of omics biomarkers of aging in population studies, discuss challenges in comparability and generalizability, and provide recommendations to facilitate future validation of biomarkers of aging. Finally, we discuss how systematic validation can accelerate clinical translation of biomarkers of aging and their use in gerotherapeutic clinical trials.
Introduction
Aging is the strongest risk factor for most chronic diseases, physical and cognitive impairment, and death. Despite this, our approach to understanding and treating aging-associated diseases has largely overlooked the biology underlying the aging process. The geroscience hypothesis posits that targeting aging itself has the potential to forestall multiple aging-associated disease processes simultaneously. As the aging population continues to grow across the globe, the promise of therapeutic targeting of aging to extend healthy lifespan has come into ever-sharper focus. To achieve this goal, there is growing interest in biomarkers that can quantitatively assess biological age and may ultimately serve as surrogate endpoints for aging-associated outcomes in clinical studies.
Many existing biomarkers of aging were initially developed to predict chronological age, although it was found that the deviation between their predicted and the true chronological age (‘AgeDev’) was associated with age-related outcomes and disease. More recent biomarkers of aging focus instead on prediction of biological age – that is, the level of age-dependent molecular and cellular damage accumulation and their consequences at a certain point in time – and/or health outcomes, rather than chronological age. Of note, in practical use, biological age is often summarized as a number (in units of time), just like chronological age. Regardless of the development strategy, most current biomarkers of aging predict aging-related outcomes and identify factors associated with the pace of aging, in retrospective epidemiological studies 1-7. In addition, they have started to provide clues on the biological mechanisms of aging. Despite these advances, the validity and usefulness of biomarkers of aging is still not widely acknowledged by biomedical scientists 1. In contrast to biomarkers of various specific diseases, there are currently no recommended guidelines for standardizing development, measurement, or validation of biomarkers of aging by regulatory bodies such as the Food and Drug Administration (FDA) or European Medicines Agency (EMA).
Validation is the multistep process by which the characteristics of biomarkers are defined, including the conditions under which they prove reliable and accurate, and their ability to predict relevant outcomes 8-10. In the context of aging biomarkers, this process requires a wide range of expertise in areas such as the biological mechanisms of aging, including conserved pathways and mechanisms in model systems and in humans; the design and construction of composite biomarkers, the design, execution and analysis of epidemiological studies that collect and store biological specimens and assess age-related predictors and outcomes in representative populations (including biobanks and cohorts), and the validation of biomarkers across multiple, diverse population samples. Thus, collaboration between basic scientists and clinical investigators is essential for successfully navigating this process.
We previously proposed a consensus framework for classification and evaluation of aging biomarkers 1. Now, we address biomarker validation as the next step in the clinical translation process. First, we review current efforts to validate predictive biomarkers of aging using population-based cohort studies and discuss challenges encountered during this process. We primarily focus on biomarkers that are: (1) blood-based, as blood is non-invasively obtained, widely accessible, and in constant contact with other tissues, potentially providing information about the biological age of the entire organism (although this is still under active exploration 11-13 ); (2) composite, as panels of molecular biomarkers are more likely to capture systemic effects of the complex aging process than single (molecule) biomarkers12-16; and (3) based on omic assays, as the rapid expansion of high-throughput omic technologies and artificial intelligence (Al) methods are expected to substantially advance the performance and translational value of the next generation of aging biomarkers 15.To facilitate and enhance rigor in the validation process 17, we provide guidelines for standardization and harmonization of biomarkers across populations with unique characteristics, and we make recommendations on the metrics that should be used to report their predictive performance.
Current status of validation efforts
Ideally, a biomarker measure should be robust against random and systematic sources of variability arising from technical and pre-analytical sources or application to different populations. Also, extensive information should be available on covariates to be considered to optimize their performance. We briefly outline some important types of conceptual and technical considerations and terminology important for biomarker validation in Box 1. Overall, a comprehensive process that encompasses multiple types of validation is desirable to establish reliability, accuracy, and clinical utility of a biomarker of aging.
Box 1. Types of biomarker validation relevant to biomarkers of aging.
Biological validation evaluates the extent to which the measurement reflects the fundamental knowledge about the biology of aging. Biomarkers can be particularly insightful if they lie within a pathway that is causal to, rather than merely associated with, aging.
Cross-species validation involves assessing the functionality of a biomarker in multiple species. If a pathway associated with a biomarker is phylogenetically conserved, it is more likely to be connected with aging as a universal phenomenon1,87
Predictive validation involves unbiased testing of the performance of the predictive model underlying the biomarker to predict a future aging-associated outcome. For instance, hazard ratios or time-to-event may be evaluated. Ideally, a true external predictive validation is carried out using independent data that was not used to train the model (often using machine learning or statistical methods). In the context of aging biomarkers, most predictive validation has been performed using retrospective analysis, but future studies should consider performing predictive validation by tracking aging-associated outcomes prospectively.
Analytical validation assesses the accuracy and reliability of the methods used to measure the biomarker, including sample collection and storage methods, analytical assays, and covariates considered. This process aims to establish standard measurement practices and determine the precision, sensitivity, specificity, and reproducibility of the assay.
Clinical validation aims to determine the clinical utility of a biomarker, i.e., whether using that biomarker in a given setting allows for a better understanding of the ongoing disease or process that may contribute to better health outcomes. For instance, clinical validation of an aging biomarker may involve establishing that the biomarker has better predictive power for aging-associated outcomes than does chronological age.
To date, predictive validation of aging biomarkers (for their association with age-related outcomes) has mostly relied on data previously collected in observational cohort studies. This process is currently the most active area of research in the aging biomarker validation space as an important prerequisite to further validation and ultimate clinical use. Cohort studies typically collect samples and clinical data on health and functional status at multiple points in time and allow assessment of association and predictivity of biomarkers for multiple health outcomes across different populations, as well as the identification of relevant covariates. We focus on cross-population validation (that is, validation in more than one cohort) because it is the most robust approach for validation of blood-based biomarkers of aging in observational studies. To contextualize recommendations outlined in later sections, we first outline the current state biomarker validation efforts (including different data sources) and discuss challenges to progress in this field.
Application of different data sources and study designs
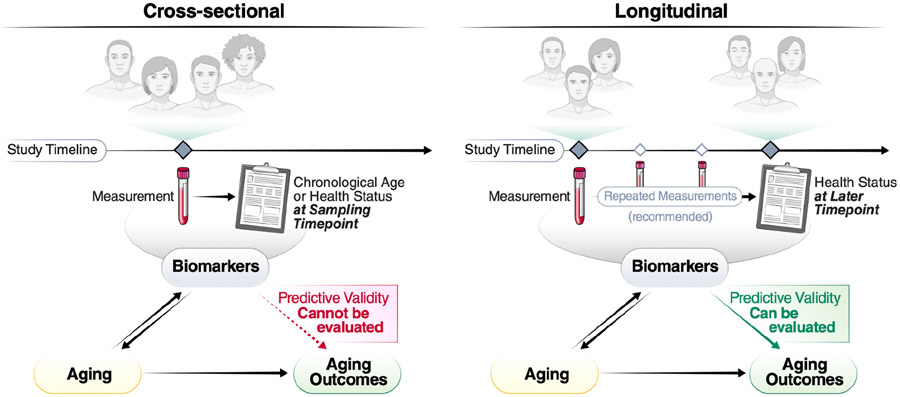
The development of early biomarkers of aging was facilitated by open-access availability of large datasets (such as those stored within the Gene Expression Omnibus [GEO] 18), many of which are derived from cross-sectional studies. Cross-sectional studies provide a snapshot in time of variable measurements and corresponding phenotypic data (Figure 1a). Such studies identified many biomarkers that correlate with chronological age. These include several soluble biomarkers of inflammation (e.g., IL-6 or C-Reactive Proteins) or hormonal status (such as fasting insulin and dehydroepiandrosterone sulfate). Early ‘first generation’ epigenetic biomarkers were also used to predict chronological age. However, cross-sectional age-associations can be biased by secular trends and selective attrition to study participation, which can preclude assessment of the predictive value of the marker in relation to future age-relevant outcomes. Furthermore, cross-sectional studies do not allow assessment of within-individual changes in response to interventions (sensitivity to change), a key requirement for the use of biomarkers of aging in clinical trials.
Figure 1. Different approaches to cohort study design in the context of biomarkers of aging.
Biomarkers of aging are commonly validated using cross-sectional or longitudinal study designs. Cross-sectional studies involve measurement of biomarkers and chronological age or aging-related outcome data at a single time point. These data can only support association of these measures at that time point. Longitudinal designs, on the other hand, allow for assessment of predictive validity of biomarkers measured at one time point and future aging-related outcomes.
In contrast to cross-sectional studies, longitudinal studies collect biological measures (omics or other biomarkers), phenotypes (clinical characteristics), and adverse age-related health outcomes serially over time in the same individuals (Figure 1b). Most longitudinal studies also include data on genetic variants and through Mendelian randomization studies, they may help determine whether specific biomarkers are causally related to health outcomes or rather reflect the activation of mechanisms aimed at counteracting the pathologic processes that lead to those adverse health outcomes (generally defined as “resilience” mechanisms). Most studies collect longitudinal information on participant demographics (e.g., age, sex), physiological measurements (e.g., body mass index, blood pressure), and routine laboratory results (e.g., complete blood count/hemograms or blood biochemistry) — and may additionally collect data on mortality and cause of death, as well as other aging-associated outcomes including multimorbidity, performance-based measures of physical and cognitive function, and frailty. Measures of disability in activities of daily living and instrumental activities of daily living provide information on a participant’s level of independence but also health deterioration over time.
Analytically, biomarkers are often considered at one point in time and related prospectively to future outcomes, such as disease onset, change in physical and cognitive function over time, or mortality. A more informative approach is to consider repeated measures obtained from the same participants at regular intervals. This approach allows the study of the relationship between biomarkers and the time-trajectories of clinical outcomes, which provide the best approximation of the ‘pace of aging’19. Therefore, longitudinal cohort data can uniquely support the development and validation of biomarkers of aging, such as prospective validation against multiple different outcomes and across independent populations. Additional approaches focus on resilience, healthy aging 20,21, or other aging-related outcomes 22-24. Moreover, outcomes related to healthcare resource utilization, such as the rate of hospital admissions and use of emergency rooms, may also be highly relevant. The prioritization of aging-associated outcomes and information on (functional) aging trajectories separate from mortality could make such biomarkers even more appealing for translation to clinical studies.
Many cohort studies establish biobanks that safely store biospecimens that can then be accessed in the future to test new hypotheses or employ newly available technology for analysis. Biobanks are invaluable resources for biomarkers research, especially if associated clinical and/or omic data and follow-up samples/data are available, particularly when it comes to testing and validation. In addition to the samples collected as part of a standard cohort study with specific research questions, other large-scale, general-purpose biobanks exist that can be used for biomarker development. For example, the UK Biobank contains in-depth genetic and health information and holds biological samples from half a million UK participants. Multiple studies have already evaluated omic-based predictors of various aging-related outcomes in the UK Biobank25-27. With the decreasing costs of measuring biomarkers, this and other biobanks are currently expanding their range of available omics data 28. The Finnish FinnGen cohort (n=~500,000 29), BioBank Japan (n=~260,000,30), and the Mass General Brigham Biobank (n=~135,000 31) have also recently generated large multi-omic datasets, which are expected to be used to validate multiple biomarkers for various aging-related outcomes. Some repositories are taking steps to organize their data in well documented and accessible databases: for instance, the US National Institute on Aging has launched complementary translational longevity initiatives to generate large-scale, cross-species, multi-omic datasets.
The current state of cross-population validation studies
Even with existing cohort studies and biobanks, systematic cross-population validation remains limited. Nevertheless, several biomarkers of aging have been tested across multiple cohorts, with the most commonly-examined outcome being all-cause mortality. Although there are issues surrounding mortality as an endpoint, it has the advantage of being clearly defined. We discuss endpoints beyond mortality below.
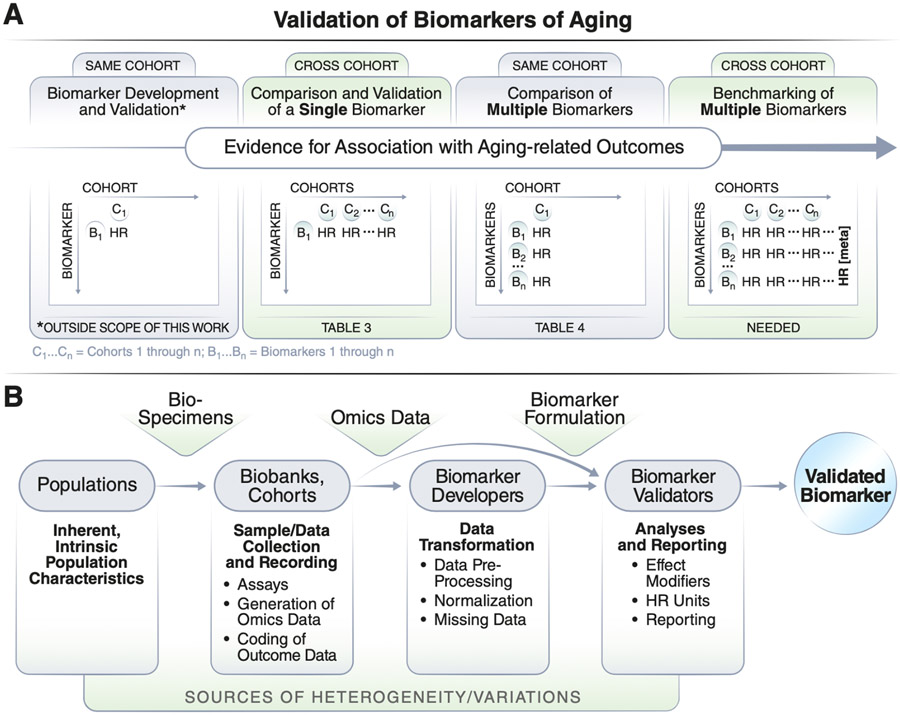
A representative list of studies validating blood-based composite biomarkers for prediction of future mortality is listed in Table 1. Many of these studies were conducted by researchers who developed the biomarker and validated that specific biomarker across multiple cohorts, or researchers who used biomarkers developed by others and compared multiple biomarkers within one cohort, with differences between the two approaches illustrated in Figure 2. In addition, a representative list of biomarkers of aging that have been tested across multiple cohorts is shown in Extended Data Table 1 (cohorts are described in Extended Data Table 2). Our intention here is not to systematically review previous studies or perform a meta-analysis; rather, the studies considered were selected to illustrate the challenges of validating biomarkers of aging in a reliable, comparable, and generalizable manner.
Table 1.
Validation studies of blood-based composite biomarkers based on future mortality*
| Study | Biomarker | Validation set | Validation sample size |
Events (%) | Adjusted HR† | HR adjusted for (in addition to age) |
HR unit/grouping |
|---|---|---|---|---|---|---|---|
| Levine et al. 2018 76 | "Phenotypic aging" | NHANES IV | 11,432 | 1,052 (9.2%) | 1.09 | - | per unit increase (year) |
| Sebastiani et al. 2017 77 | "Agglomerative algorithm" | FHS | 2,734 | 656 (24.0%) | 0.68 to 1.13 [varying] | sex | cluster 1 vs cluster 2-26 |
| Mamoshina et al. 2018 78 | BloodAge | NHANES | 2,768 | 873 (31%) | 1.66 | sex | AgeDev <−5 vs >+5 years |
| Levine et al. 2018 76 | PhenoAge | WHI, FHS, JHS, NAS | 8,965 | 2,074 (22.8%) | 1.05 [1.04-1.05] | ethnicity | per unit increase (year) |
| Lu et al. 2019 24 | Grim Age | FHS, WHI, JHS, InChianti | 7,375 | 1,848 (25.1%) | 1.10 [1.08-1.12] | ethnicity | per unit increase (year) |
| Lu et al. 2022 4 | GrimAge2 | FHS, WHI, JHS, InCHIANTI, NAS | 10,065 | 3900 § (39%) | 1.10 [1.09-1.10] | sex, ethnicity | per unit increase (year) |
| Zhang et al. 2017 79 | DNAmRS | KORA | 1,727 | 61 (3.5%) | 10.95 [3.09-38.84] | sex | scores 0-1 vs 5+ |
| Belsky et al. 2022 19 | DunedinPACE | NAS | 771 | 354 (45.9%) | 1.26 [1.14-1.40] | sex | per SD increase |
| FHS | 2,471 | 575 (23.3%) | 1.65 [1.51-1.79] | sex | per SD increase | ||
| Bernabeu et al. 2023 80 | bAge | LBC, FHS, WHI | 4,125 | 1,653 (40.1%) | 1.52 [1.44-1.59] | sex | per SD increase |
| Deelen et al. 2019 34 | MetaboHealth score | FINRISK 1997 | 7,603 | 1,213 (16.0%) | 2.73 [2.60-2.83] | sex | per unit increase (scores −2 to 3) |
| van den Akker et al. 2020 81 | Meta bo Age score | LLS_SIBS | 811 | 793 (97.7%) | 1.25 [1.14-1.37] | sex | per unit increase (year) |
| Balasubramanian et al. 2020 6 | M-metabo-score | WHI-HT | 1,355 | 685 (50.6%) | 1.95 [1.46-2.62] | clinical and lifestyle risk factors | highest vs lowest quartile |
| Tanaka et al. 2020 5 | "Proteomic signature" | InCHIANTI | 997 | 504 (50.6) | 1.03 [1.02-1.04] | sex, study site | per unit increase (year) |
| Huan et al. 2022 32 | "Integrative biomarker" | ARIC | 969 | 331 (34.1%) | 1.85 [1.44-2.37] | sex, clinical factors | per SD increase |
| Li et al. 2020 82 | DNAmAge (Horvath) | SATSA | 387 | 240 (62%) | 1.17 [1.01-1.36] | sex, education, lifestyle risk factors | per SD increase |
| DNAmAge (Hannum) | 1.17 [0.98-1.40] | ||||||
| PhenoAge | 1.26 [1.08-1.47] | ||||||
| Grim Age | 1.39 [1.11-1.75] | ||||||
| Hillary et al. 2020 83 | EEAA | GS | 2,578 | 57 (2.2%) | 1.39 [1.12-1.72] | sex | per SD increase |
| PhenoAge | 1.38 [1.11-1.73] | ||||||
| Grim Age | 1.70 [1.35-2.14] | ||||||
| DunedinPoAm | 1.69 [1.30-2.18] | ||||||
| DNAm estimate of telomere length | 0.81 [0.63-1.04] | ||||||
| McCrory et al. 2020 84 | DNAmAge (Horvath) | TILDA | 490 | 45 (7.2%) | 1.03 [0.74-1.44] | sex, lifestyle risk factors | per SD increase |
| DNAmAge (Hannum) | 0.92 [0.67-1.28] | ||||||
| PhenoAge | 1.13 [0.81-1.57] | ||||||
| Föhr et al. 2021 85 | DNAmAge (Horvath) | FITSA | 413 | 156 (35.2%) | 1.05 [0.89-1.23] | family relatedness, lifestyle risk factors | per SD increase |
| Grim Age | 1.31 [1.08-1.59] | ||||||
| Wang et al. 2021 86 | IEAA | NAS | 737 | 337 (45.7%) | 1.08 [0.92-1.28] | clinical and lifestyle risk factors | per SD increase |
| EEAA | 1.10 [0.93-1.3] | ||||||
| PhenoAge | 1.17 [0.98-1.41] | ||||||
| Grim Age | 1.56 [1.24-1.96] | ||||||
| DNAmRS | 1.37 [1.06-1.78] |
Studies with ≥ 10-year follow-up are listed.
Hazard ratios (HRs) derived from Cox proportional hazards regression for several biomarkers of aging against all-cause mortality are shown. We aimed to include the most representative adjustment model for studies that report multiple models and report the hazard ratio (HR) as the most frequently used metric for assessing performance of a biomarker with regards to time-to-event analysis. Note that reported HRs are not directly comparable because they are referred to different units of measure of the predictor. In addition, many factors (e.g. population characteristics and data preprocessing) may influence predictive performances.
Abbreviations: AgeDev, age deviation; SD, standard deviation
Cohorts: NHANES, National Health and Nutrition Examination Survey; FHS, Framingham Heart Study; WHI, Women’s Health Initiative; JHS, Jackson Heart Study; NAS, Normative Aging Study; InCHIANTI, Invecchiare in Chianti; KORA, Cooperative Health Research in the Region Augsburg; LBS, Lothian Birth Cohort; LLS_SIBS, Leiden Longevity Sibling Study; ARIC, Atherosclerosis Risk in Communities Study; SATSA, Swedish Adoption/Twin Study of Aging; GS, Generation Scotland; TILDA,
The Irish Longitudinal Study on Ageing; FITSA, The Finnish Twin Study on Ageing;
Figure 2. Validation of biomarkers of aging with different numbers of cohorts or biomarkers.
a) Most existing biomarkers have been developed using data from a single cohort and some have been validated in a second external cohort. Analysis of multiple biomarkers across multiple cohorts allows for a meta-analysis comparison across multiple cohorts. b) Biomarker validation studies need to consider different sources of variation, such as heterogeneity in population characteristics, sample collections, data preprocessing, analyses, and reporting.
In studies validating blood-based biomarkers, most reported HRs for prediction of mortality risk are in the moderate range; however, a few studies have reported impressive metrics that render those biomarkers good potential candidates for use in preclinical and clinical studies. For example, Huan et. al 32,33 and Deelen et al 34 reported increased mortality risk (HRs of 1.85 and 2.73) for their epigenetic and metabolomic biomarkers, respectively. These values should be considered with caution because they rely on different units of measure, need to be further substantiated by independent validation in a different cohort, and their performance should be compared with other biomarkers using consistent reporting measures. So far, relatively few studies have compared individual (composite) biomarkers across multiple cohorts, or multiple biomarkers across the same cohort using standardized and equivalent measurement units that make them fully comparable 32,35,36. We argue that studies featuring systematic and comprehensive benchmarking of diverse biomarkers of aging across many large cohorts with extended follow-ups (>10 years) are needed to substantially advance the field (Figure 2a).
Challenges for validation of biomarkers of aging
Despite ongoing progress, comparing the predictive strength of biomarkers of aging remains challenging. Even for a well-defined outcome such as mortality, studies evaluating predictive performance of omic biomarkers have provided heterogeneous results. Potential reasons for this inconsistency include different study populations with different characteristics; differences in recording, formatting, and coding of molecular and outcome data; differences in preprocessing and biomarker formulation; and different approaches to validation analyses and reporting (Figure 2b, Table 2). In the following sections, we focus on each one of these problems.
Table 2.
Challenges and associated recommendations for validation of biomarkers of aging.
| Challenge | Target stakeholder | Recommendation | Example |
|---|---|---|---|
| 1. Population-specific characteristics | Data maintainers | 1. Adopt data-sharing mechanisms that enable timely and broader access to enable validations using many populations. | Provide transparent information on available data as well as data access review processes including expected review time. |
| Validation study teams | 2. Include multiple diverse populations. | Validate biomarkers across multiple diverse cohorts and report stratified analyses. | |
| 2. Molecular and outcome data | Data maintainers | 3. Follow Findable, Accessible, Interoperable, and Reusable (FAIR) 66 data principles and provide detailed metadata and data dictionary and use standard data formats. | Ensure data is FAIR 66 by providing appropriate documentation and guidance: e.g., use GEO data format for expression (transcriptomics) and epigenetic data. |
| Biomarker developers | 4. Verify assumptions of statistical/machine learning models used to identify/learn the relation between biomarkers and aging outcomes | Consider non-linear or piecewise models for biomarkers with established non-linear relation with aging outcomes | |
| Validation study teams | 5. Standardization and harmonization of individual biomarker measurements and aging outcomes in different datasets. | Extend biomarkers standardization programs (e.g. RefMet) and consortium data harmonization efforts by CHARGE, TOPMed, UBiLim, BBMRI, BioSHaRE-EU toward developing assay-agnostic and generalizable biomarkers. | |
| 6. Consider aging outcomes beyond mortality | Consider multimorbidity, frailty, disability, quality of life measures, or health-focused metrics, such as vitality, resilience, and healthspan | ||
| 3. Biomarker procedures and formulations | Biomarker developers | 7. Improve transparency of sample preparation, data processing, and biomarker formulation. | Provide fully specified computational procedures, including details on normalization method and treatment of missing measurements. |
| Validation study teams | 8. Post-hoc harmonization of composite biomarker formulations. | Develop/extend packages or solutions for computation of multiple biomarkers, e.g., methylCIPHER, BioAge, ClockBase, MiMIR. | |
| 4. Comparability of validation studies | Biomarker developers | 9. Improve interpretability, generalizability, and robustness | Incorporate various omic data from diverse populations during biomarker development. |
| Validation study teams | 10. Account for potential effect modifiers by controlling, adjusting for, or stratifying based on them | Report hazard ratios for chronological age- and sex-adjusted models. | |
11. Standardize the validation process including:
|
“Lock down” predefined biomarker formulation and conduct validation studies according to defined standards, e.g., minimum follow up time for aging outcome, Cox proportional hazards regression, and hazard ratios per standard deviation and absolute unit increase. | ||
| 12. Report the results comprehensively and appropriately | Follow established guidelines for reporting of observational studies, such as STROBE. |
Population-specific characteristics.
Predictive performance of a biomarker of aging may vary by characteristics of the underlying population, including age demographics, ethnicity, health and disease status, or physical and cognitive function. For instance, in a population with high exposure to pollution or environmental contaminants, cancer biomarkers will appear to be highly predictive of all-cause mortality even if they are not in the general population. A related challenge is the lack of participant diversity in many large cohort studies and biobanks that suffer from heavy overrepresentation of European ancestry and predominantly white participants. Results from these studies may not apply to nonwhite, ethnically diverse individuals, which limits their external validity. Exceptions featuring more diverse populations exist, such as the Jackson Heart Study (https://www.jacksonheartstudy.org/) or Healthy Aging in Neighborhoods of Diversity across the Life Span 37, but many more studies are needed to understand similarities and difference in biomarkers of aging across diverse individuals. Notably, aging biomarkers that are reproducible across population groups likely reflect fundamental mechanisms of aging biology. Such biomarkers would be broadly useful for both clinical and basic research applications.
Molecular and outcome data.
Cohort studies are generally designed to address specific sets of scientific questions. Therefore, each cohort or biobank features unique content, collected, and recorded in a unique manner to address these questions. Even studies carrying out similar analyses may use different approaches. For example, epigenetic data could be collected using different microarray assays (27K, 450K, 850K) or isolating DNA with different methods, which produce slightly different estimates even for (epi)genetic targets shared between platforms 38. Similarly, metabolomic or proteomic data could be collected from plasma or serum, leading to different data distributions 34; measured using different technologies (e.g. mass spectrometry or aptamer-based assays); or tagged using different nomenclature39. Unfortunately, no harmonization standards currently exist for molecular data and aging-associated outcomes for the purpose of validating biomarkers of aging. Existing programs and consortia, such as RefMet (Reference Set of Metabolite Name 40, COMETS (Consortium of Metabolomics Studies 41), CHARGE (Cohorts for Heart and Aging Research in Genomic Epidemiology42), CINECA (Common Infrastructure for National Cohorts in Europe, Canada, and Africa 43), TOPMed (Trans Omics for Precision Medicine program 44), UK LLC (UK Longitudinal Linkage Collaboration, https://ukllc.ac.uk/), UBiLim (University Biobank Limburg 45), BioSHaRE-EU (Biobank Standardisation and Harmonisation for Research Excellence in the European Union 46), and BBMRI-NL (Biobanking and Biomolecular Resources Research Infrastructure Netherlands, https://www.bbmri.nl/), are developing standards for organization of specific data types to facilitate large-scale collaborations in other fields, but no such efforts have been initiated in geroscience.
Mortality is the most frequently used outcome for biomarker validation. Biomarkers constructed with time to mortality as a reference outcome also tend to predict chronic diseases as well as functional and cognitive outcomes independent of chronological age — suggesting that they capture a dimension related to overall health 1,35. However, the direct use of non-mortality aging-related outcomes, such as multimorbidity, poor mobility, and frailty, may better capture information on the pace of aging and may be more useful for clinical applications. These could include internationally recognized scoring systems multimorbidity, frailty, disability, cognition 47 or quality of life measures, as well as more health-focused metrics, such as vitality, resilience, and healthspan, although no consensus yet exists on how to quantify the latter two48. Beyond cohort-specific challenges, access to cohort data remains a general ongoing issue: applying for access to many government-funded datasets often requires lengthy paperwork and review processes, and can often span several months or years.
Biomarker procedures and formulations.
Statistical and machine learning models used to identify or learn the relation between biomarkers and aging outcomes are still in early stages of development and validation, and many modeling challenges remain to be addressed. For example, many existing models assume a linear relation between biomarkers of aging and the likelihood of aging outcomes throughout the lifespan, while recent studies have discovered multiple examples of non-linearity5,12,49. Technical considerations surrounding data preparation also pose challenges. For instance, recent work has demonstrated that calculating principal components from CpG-level data as input for biological age prediction can improve test-retest reliability of epigenetic biomarkers. 50. These and other unique transformations of individual measurements make cross-comparison of composite biomarkers challenging. Moreover, biomarkers or their components may be sensitive to underlying sample composition. For example, there is evidence that age-related methylation varies across different circulating immune cells 51. Therefore, comparative or validation studies should always carefully adjust for the proportions of different types of circulating cells. Studies may also treat missing data or repeated measurements for biomarkers or outcomes differently, potentially influencing power or skewing performance estimates. This issue is particularly important for proteomic assays that tend to generate many values ‘below the threshold of detection’, which may not be random but rather convey important information. Finally, there is currently no guidance on how to best integrate longitudinal repeated measures from the same individual, and whether trajectories or unique values should be considered.
Study design and reporting.
Several aspects of study design, such as follow-up time, number of events, and bias in mortality reporting may introduce variability across studies. Differences in statistical approaches are also a notable source of variation. For example, different validation studies often account for distinct potential effect modifiers by controlling, adjusting, or stratifying for them. These factors are expected to affect the magnitude of the relationship between the omic biomarkers and aging-associated outcomes, representing another challenge for comparison of biomarkers in (and across) validation studies. Additionally, studies can report performance metrics such as HRs in different ways (e.g., per standard deviation, compared to a reference group, or per unit increase), using different adjustment strategies for covariates (see Table 1). For example, in Cox proportional hazards regression, biomarkers can be coded as continuous variables (standardized or not), or as ordinal variables that capture quantiles of biomarker level or even as time-dependent covariates. The former approach provides information on risk estimates per one unit difference biomarker level (e.g., per standard deviation), while the second considers one level (typically the lowest quantile) as a reference group. These inconsistencies, which also plague other fields, have hindered reliable cross-comparison, benchmarking, and meta-analysis of evaluated biomarkers of aging.
Recommendations for validation of biomarkers of aging
Cross-population validation of multiple biomarkers across several cohorts on a larger scale is necessary but challenging and will require considerable coordinated effort and increased funding. Based on the current state of the field and the challenges outlined above, we provide the following recommendations (summarized in Table 2) for benchmarking and reporting of validation studies, grouped by target stakeholders.
Recommendations for biomarker developers
Before composite or algorithmic biomarkers are validated across populations, the underlying statistical or machine learning models capturing the biological relation between biomarkers and outcomes need to be verified. It is important to examine the extent to which an association could be reasonably attributed to the underlying biology. We recommend that biomarker developers verify that the statistical assumptions of their models reflect the expected biological phenomena, to the extent of our current knowledge. For example, as recent studies reveal unique age-dependent epigenetic changes during different phases of life, it is becoming clear that non-linear or piecewise epigenetic biomarkers might represent the whole human lifespan more accurately than those that assume a linear relationship with age across the life course 52,53.
Successful validation of biomarkers requires full transparency of the methods used for their development, computational pre-processing and analysis, and verification of their predictive validity in multiple independent populations. Hence, preprocessing pipelines should follow best practice guidelines that ultimately enable data harmonization 54. For example, the treatment of missing or repeated measurements (e.g., using imputation or machine learning methods 55,56), data normalization, and quality control influence predictive performance results, so it is important to establish and follow standards and best practices for these steps 57. Similarly, fully specified computational procedures (formulations) for composite biomarkers should be made available publicly (as recommended for all omic tests by US National Academy of Science, Institute of Medicine 17) to allow for computation of biomarker scores independently by other researchers (without the need to upload or transfer data to biomarker developers). In addition, biomarker formulation should allow for simple implementation across new datasets. For example, most omic biomarkers could be formulated in standardized mathematical terms (see harmonization efforts by e.g., ClockBase19 epigenetic biomarkers, and MiMIR metabolomic biomarkers 58) and standardized software packages, which enable streamlined calculation of various biomarkers including blood biochemistry (e.g., BioAge 59) and epigenetics (e.g., Biolearn at https://bio-learn.github.io and methylCIPHER 60). We believe that such a process of validation and implementation would provide even stronger results if it was undertaken according to guidelines that are widely discussed and adopted by the scientific community.
To support future validation studies, we recommend that developers consider methods and data sources that improve the likelihood of future generalizability, cross-population validity, and potential clinical validity of their biomarkers. Currently, epigenetic markers are the most commonly proposed and investigated type of composite biomarkers 1. We recommend developing methods to address many widely acknowledged challenges with these and other biomarkers, including interpretability 2,61 and technical robustness 19,50. Studies involving longitudinal sample collection may be particularly useful for this purpose, as the resulting data can enhance our understanding of the dynamic properties of these biomarkers. In addition, we recommend the use of other complementary omic data, including metabolomics, proteomics, transcriptomics, and lipidomics, whose biological interpretation is often less complex, to develop biomarkers capable of capturing aspects of aging that may not be best reflected by epigenetic data 62. As the costs of many omic assays are decreasing and cohorts/biobanks are increasingly incorporating multiple data modalities (Table 3), we expect multi-omic biomarkers to become common in the near future., emphasizing the need for accessible and standardized approaches.
Table 3.
Cohorts or biobanks with existing multi-omic data.
| Study characteristics | Omics | Follow-up outcomes | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Abbreviation | Cohort | Location | Description/Note | Total N | Methylation | Metabolome | Prateome | Other | Time to death |
Cause of death |
Multi-morbidity | Other |
| UKB | UK Biobank | UK | Largest multi-omic dataset to date | > 500,000 | ✓ | ✓ | Transcriptome | ✓ | ✓ | ✓ | Cognitive function, Accelerometer | |
| LIFELINES | Lifelines | NL | Multigenerational European Cohort | >167,000 | ✓ | ✓ | ✓ | Microbiome | ✓ | ✓ | ✓ | Cognitive function |
| MGB | Mass General Biobank | USA | Leading hospital biobank | > 135,000 | ✓ | ✓ | ✓ | Lipidome | ✓ | ✓ | ✓ | |
| RS | Rotterdam Study | NL | Three cohorts focusing on mid- to late life | >15,000 | ✓ | ✓ | ✓ | Microbiome, Lipidome | ✓ | ✓ | ✓ | |
| MESA | Multi-Ethnic Study of Atheroscler osis | USA | Multi-ethnic population followed since 2000 | > 6,000 | ✓ | ✓ | ✓ | Transcriptome | ✓ | ✓ | ||
| CARDIA | Coronary Artery Risk Developme nt in Young Adults | USA | Population of mixed ethnicities followed from young adulthood (18-30) | > 5,000 | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | Cognitive function | |
| HABC | Health, Aging and Body Composition Study | USA | Focus on functional decline and body composition | > 3,000 | ✓ | ✓ | ✓ | ✓ | ✓ | Cognitive function, Physical function, MRI | ||
| BLSA | Baltimore Longitudinal Study of Aging | USA | Comprehensive characterization of aging phenotypes | > 3,000 | ✓ | ✓ | ✓ | Microbiome | ✓ | ✓ | ✓ | Cognitive function, Physical function |
| AASK | African American Study of Kidney Disease and Hypertension | USA | African American population followed over time | 691 | ✓ | ✓ | ✓ | ✓ | ||||
| iPOP | Integrated Personal Omics Profiling | USA | Deep longitudinal profiling | >100 | ✓ | ✓ | ✓ | Microbiome, Lipidome | ✓ | ✓ | ✓ | |
Recommendations for data maintainers
Successful validation of biomarkers depends on access to data and harmonization of aging-related phenotypic and molecular data across relevant cohorts. Procedures for easy data-sharing that enable more timely and broader access, while maintaining privacy of individual human data (such as NHLBI BioData Catalyst 63), should be widely adopted. Data repositories can and should provide transparent information on available data and data formats, as well as the data access criteria and review processes — including expected review time based on historical statistics. In addition, synthetic datasets (with the same data structures and distributions), data safe havens (that is, secure storage and computing for sensitive data), and federated access (unified central access) would ideally be provided to facilitate broader access to the data. Providing one or more of the above should be incentivized by funding agencies and other financial supporters. For sensitive data with controlled-access, federated analysis – whereby data remains decentralized on host institution servers but is made available for analysis in a privacy-preserving manner64 – may offer a suitable compromise, especially using cloud-based methods. Rather than requesting transfer of sensitive data, individuals aiming to validate a biomarker could provide the formulation of the biomarker to data owners and/or conduct their analysis in a secure environment, with access to only summary or synthetic data.
Many initiatives (e.g., RefMet, CHARGE, TOPMed, UBiLim, BBMRI, BioSHaRE-EU) have taken steps to standardize biomarker nomenclatures or cohort or biobank data to facilitate crosspopulation studies, often following rigorous guidelines for retrospective data harmonization (e.g., Maelstrom65). While these post-hoc efforts are needed to improve existing data, cohort or biobank data maintainers may facilitate this process by following best practices in recording and reporting biomarkers and aging outcome measures from the inception of their biobanks or cohort studies. In particular, data owners should aim for alignment with FAIR data principles (ensuring data is Findable, Accessible, Interoperable and Reusable 66), provide machine-readable metadata and data dictionaries that allow for harmonization, and make available records of data structure in data description publications. Especially for older or ongoing longitudinal studies with long followup, the above steps represent a considerable challenge that requires increased support from the aging research community. Success with the above efforts would increase data utility, particularly for federated learning and analysis across populations, which require standardized data.
Recommendations for cross-population validation study teams
Biomarkers of aging should be evaluated across multiple diverse populations to account for differences across genetic ancestries, sex, geographic contexts, environmental or lifestyle factors, life stages, and health or disease states. This step is critical because even seemingly established biomarkers may not be valid in all human populations. For example, the ApoE4 allele is the strongest risk factor for Alzheimer’s disease risk in white populations, but the association is substantially weaker in African American and Hispanic populations 67. Moreover, in Tsimane horticulturalists (a subsistence population in Brazil), ApoE4 appears to be protective against cognitive decline 68. As mentioned above, many existing composite biomarkers of aging have been trained in cohorts of predominantly white, European ancestry. A similar bias in the design of genetic studies has resulted in the development of polygenic risk scores that have diminished predictive accuracy in populations with non-European ancestry69.
While many composite biomarkers of aging have shown some evidence of comparable predictive accuracy across genetic ancestry populations 24 70 71, establishing diverse cohorts with non-European ancestries to validate new composite biomarkers of aging remains a priority. Other key axes for cross-validation could include climate zones, country or continent, and exposure levels to various chemical or biological risk factors. This will require efforts to establish resources and research capacity in various geographical regions and minority populations. In addition to commonly used cohorts, many other cohort studies or biobanks (many of which are focused on recruiting diverse or minority populations) may be suitable for validation studies of biomarkers of aging. Some of these have already added or are in the process of adding (multi)omic data (Table 3), which will help to further improve development or validation of biomarkers of aging.
Efforts by developers to standardize various aspects of biomarker validation, including biomarker formulation and statistical analyses (described above), will allow for a reliable comparison across studies. For instance, biomarker formulations should be established “a priori” and not be further modified during validation (i.e. formulation “lock down” 17). Additionally, the result of statistical analysis, such as HRs, should be reported for unadjusted, chronological age- and sex-adjusted, and fully adjusted models, permitting a broader cross-comparison of studies. Studies may additionally account for other factors mentioned above, including sample composition. To ensure comparability of performance, the community needs to take steps to agree on the minimal set of covariates to be included in the analysis and the use of stratified analyses by subgroups, such as age, sex, and/or race. Finally, reporting HRs per standard deviation and the absolute unit differences in biomarker levels (e.g., one standard deviation and one unit of increase in the biomarker) allows for easier comparison of different biomarkers and meta-analysis. While perfect standardization may not be realistically achievable, moving in the direction of standardization will at least enable the qualitative assessment of the extent to which results in different populations converge.
Correct reporting of study results is vital to enable cross-population validation. We recommend investigators follow established guidelines for reporting of observational studies, such as STROBE 72, to enhance transparency and reproducibility of findings. All populations should be sufficiently described, either in summary or individually, when including multiple cohorts in one study. When focusing on mortality as a key aging-associated outcome, studies should report allcause mortality based on reliable information, and where possible, cause-specific mortality, which may differ based on underlying population characteristics. Several multi-omic datasets offer information on aging-related outcomes separate from mortality (Table 3) that may be utilized instead of, or in addition to, mortality. Analysis in subgroups with certain (chronic) conditions that lead to accelerated aging (e.g., HIV infection 73) will inform whether these changes in aging biomarker levels are associated with increased morbidity or mortality, or whether bespoke biomarkers may be required in those groups of individuals to predict clinical outcomes. At minimum, results should be stratified and reported separately by age group and sex, given the clear sexual dimorphism in aging. Additionally, extended reporting of stratified analyses by various demographics (e.g., ethnicity, country, or pre-existing health status) is recommended to evaluate generalizability, since models that perform well across distinct strata are more likely to have good external validity 74. Reporting highly stratified results will also facilitate meta-analyses.
Outlook
The past decade has seen substantial progress in the development of new blood-based biomarkers of aging. Despite the tremendous promise of these tools for use in trials for longevity interventions, major roadblocks persist in translating them to clinical use. Our article highlights challenges encountered in the validation of these biomarkers and proposes efforts to overcome these barriers. Addressing these relatively simple challenges, such as standardization of effect size reporting, stands to greatly benefit the comparison and validation of biomarkers of aging. We anticipate that studies benchmarking multiple biomarkers of aging, especially those using different technologies (e.g., metabolomics, proteomics, epigenetics, and multi-omic approaches), across multiple populations will provide a more comprehensive understanding of their performance and robustness. Performing such large-scale comparative studies is a key priority to progress in this field but will require increasing cooperation between research groups and creation of incentives for transparent sharing of biomarker formulations and data. Efforts toward harmonization will require endorsement across diverse stakeholder groups, including biomarker developers, data owners, and epidemiological researchers, and may ultimately enable the goal of identifying the most promising biomarker candidates for clinical prioritization.
We further recommend that future work should aim to incorporate more clinically relevant and potentially actionable outcomes instead of, or in addition to, mortality. Many cohort studies provide alternative health outcome data (Table 3) which may support this goal, including data on specific chronic diseases, multi-morbidity, organ specific physiological integrity as well as physical and cognitive function. However, agreement of standard definition and operationalization of these outcomes would be highly desirable and ensure true comparability. Prospective studies that develop individual, longitudinal profiles of biomarkers will add a dimension that is strongly needed in the field. Such studies will be critical particularly with respect to assessing whether biomarkers are sensitive to physiological changes such as those induced by longevity interventions and gerotherapeutics, or other preventive measures.
We anticipate that the ideal biomarkers of aging shall have moderate to strong associations with chronological age and predict multiple aging-related outcomes beyond mortality, such as functional decline, frailty, chronic diseases and disability, and (multi)morbidity. They should be sensitive to upstream factors thought to influence aging such as stress, adverse events, environment, genetics, and lifestyle, and they should mediate the relationship between these factors and aging outcomes. They should do so in many, diverse populations, and should do so relatively similarly across populations. Ideal biomarkers of aging meeting these requirements should be prioritized for validation as screening and diagnostic biomarkers, and eventually as surrogate endpoints in clinical trials. While a clear roadmap to realize this long-term goal does not yet exist, harmonization and standardization of biomarkers and population data across the field will greatly enhance our ability to identify, characterize, and validate the most promising biomarker candidates.
As biomarkers of aging move toward clinical implementation, several key questions remain to be addressed. First, there is no widespread agreement on the extent to which biological age may be captured by a single biomarker. Further validation of aging biomarkers through their use in clinical and epidemiological studies will help establish whether a single biomarker or multiple complementary biomarkers may be most useful. A looming question is whether biomarkers of aging should be integrated into the current disease-centric and disease-specific approach to healthcare. A shift towards holistic prevention, in line with the geroscience hypothesis, has the potential to substantially change public health and expand the portion of life free of diseases and disability, but will require endorsement across diverse stakeholder groups, particularly in the clinical realm. Next, the clinical utility of biomarkers of aging remains to be validated using prospective clinical trials to demonstrate that they can indeed improve how patients feel, function, and survive. Finally, while we focused on blood-based biomarkers of aging, more studies investigating aging across diverse organ systems and individuals are warranted to enhance our understanding and clinical potential of biomarkers of aging.
Conclusion
The translation of the science of aging to clinical applications holds substantial promises for the improvement of healthcare and the expansion of health expectancy, with the potential to both reduce health care expenditure and improve population health 75. An important prerequisite to accomplish this goal is the availability of solid and validated biomarkers of aging, which requires a process of validation to advance them into clinically valuable and actionable tools. It is our hope that the challenges we highlight and the recommendations we offer will aid in advancing biomarkers of aging to more robust tools that empower the action of health planners and health care providers.
Extended Data
Extended Data Table 1.
Blood-based* biomarkers of aging validated in cross-population studies.
| Biomarker | Type | Assay | Variable selection method |
Biomarker components | |
|---|---|---|---|---|---|
| Panel | “Phenotypic aging”11 | Blood biomarkers | Blood biochemistry and hematology | Penalized Cox regression model, 42 variables | 9 blood measures |
| “Agglomerative algorithm”12 | Blood biomarkers | Blood biochemistry and hematology | Hierarchical clustering, 40 variables | 19 blood measures | |
| BloodAge 13 | Blood biomarkers | Blood biochemistry and hematology | Deep Neural Networks | 45 blood measures | |
| Omic | DNAmRS 14 | Epigenetics | DNAm microarray | EWAS on mortality, ∼450K CpGs | Weighted DNAm levels at 10 CpGs |
| PhenoAge11 | Epigenetics | DNAm microarray | EN regression model with 20169 CpGs | Weighted DNAm levels at 513 CpGs | |
| GrimAge 15 | Epigenetics | DNAm microarray | EN Cox regression model with ∼450K CpGs | Weighted DNAm levels at 1030 CpGs | |
| GrimAge2 16 | Epigenetics | DNAm microarray | EN Cox regression model of time-to-death, 1030 CpGs | Weighted DNAm levels at 1030 CpGs | |
| DunedinPACE 17 | Epigenetics | DNAm microarray | EN regression model on 81239 CpGs with high test-retest reliability | Weighted DNAm levels at 173 CpGs | |
| bAge 18 | Epigenetics | DNAm microarray | EN Cox regression model of time-to-death, ~ 370K CpGs | 35 EpiScores based on weighted DNAm levels | |
| MetaboHealth score 19 | Metabolomic | Nuclear magnetic resonance | Stepwise Cox proportional hazards model, 226 variables | Concentration of 14 metabolites | |
| MetaboAge score 20 | Metabolomic | Nuclear magnetic resonance | Linear model trained on chronological age, 56 variables | Concentration of 56 metabolites | |
| M-metabo-score 21 | Metabolomic | Liquid chromatography-mass spectrometry | Cox proportional hazards model, 470 variables | Concentration of 17 metabolites | |
| “Proteomic signature” 22 | Proteomic | SOMAscan 1.3K | Cox proportional hazard model, 1301 variables | Concentration of 76 proteins | |
| Integrative | “Integrative biomarker” 23 | Clinical + Epigenetics | clinical measurements and DNAm microarray | EWAS and Elastic-coxph | 12 clinical measurements and DNAm levels at 76 CpGs |
Biomarker selection: Biomarkers were selected based on the following criteria 1) we reviewed biomarkers compiled by Justice et al. 14 10, Rutledge et al.24, and Kudryashova et al.25 which, to the best of our knowledge, collectively provide the most comprehensive list of blood-based and omic biomarkers of aging in the geroscience field; 2) we consulted a recent systematic ranking and scoring of biomarkers of aging performed by Hartmann et al. in 2021 to select the most cited biomarkers 26; and 3) we queried the literature using search terms [“epigenetic”, “metabolomic”, “proteomic”, “transcriptomic”], [“mortality”, “death”], and [“validation”] to identify additional omic biomarkers that have been validated in cohort studies. Genomic biomarkers of aging (e.g., genetic variants) were not included in this work, as we focused on biomarkers potentially responsive to interventions. All included biomarkers/studies met the following inclusion criteria: 1) studies that propose new ways to quantify biological aging (predicting aging outcomes better than chronological age 27); 2) biomarkers measured entirely or primarily in blood; 3) biomarkers that are applicable to the general population and not specific to certain subpopulations with specific diseases; 4) biomarkers that underwent validation in an independent cohort (cross-population validation) with at least 10 years of follow-up data on aging outcomes, including mortality (e.g., time to death); and 5) biomarkers whose predictive performance is reported in terms of hazard ratio (HR). As these strict inclusion criteria resulted in omission of numerous studies and biomarkers (see Extended Data Table 3 below for examples).
Abbreviations: SBP: Systolic blood pressure; FEV1: forced expiratory volume at 1 second; FVC: forced vital capacity; CRP: C-reactive protein; ALP: Alkaline phosphatase; WBC: White blood cell count, CN: Creatinine; ALB: Albumin; EWAS: epigenome-wide association studies; EN: Elastic Net; 10F CV: 10 fold cross validation; RFU: relative fluorescence units
Extended Data Table 2.
Cohorts for validation studies of biomarkers of aging described in this study.
| Abbreviation | Cohort | Location | Focus | Website |
|---|---|---|---|---|
| ARIC | Atherosclerosis Risk in Communities Study | USA | Risk for atherosclerosis | https://sites.cscc.unc.edu/aric/Cohort_Description |
| FHS | Framingham Heart Study | USA | Cardiovascular, lung, and blood diseases | https://www.framinahamheartstudy.org |
| FINRISK | FINRISK | Finland | Risk factors on chronic, noncommunicable diseases | https://thl.fi/en/web/thlfi-en/research-and-development/research-and-proiects/the-national-finrisk-study |
| FITSA | Finnish Twin Study on Aging | Finland | Genetic and environmental influence on aging | https://www.gerec.fi/en/research/health/the-finnish-twin-study-on-ageing-fitsa-taina-rantanen/ |
| GS | Generation Scotland | UK | Health and wellbeing across lifetime | https://www.ed.ac.uk/generation-scotland/for-researchers/generation-scotland |
| InCHIANTI | InCHIANTI | Italy | Older persons | https://www.nia.nih.gov/inchianti-study |
| JHS | Jackson Heart Study | USA | Cardiovascular disease in African Americans | https://www.jacksonheartstudy.org/ |
| KORA | Cooperative Health Research in the Augsburg Region | Germany | Risk factors for cardiovascular disease | https://www.maelstrom-research.org/study/kora |
| LBC | Lothian Birth Cohorts | UK | Longitudinal changes in brain aging | https://www.ed.ac.uk/lothian-birth-cohorts/history |
| LLS | Leiden Longevity Study | NL | Long-lived family members and controls | https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6132126/ |
| NAS | Normative Aging Study | USA | Aging and disease development | https://www.vacsp.research.va.gov/CSPEC/Studies/INVESTD-R/CSP-710-VA-Normative-Aging-Study.asp |
| NHANES | National Health and Nutrition Examination Survey | USA | Health and nutritional status | https://www.cdc.gov/nchs/nhanes/index.htm |
| SATSA | Swedish Adoption/Twin Study of Aging | Sweden | Genetic and environmental influence on aging | httDs://www.icpsr.umich.edu/web/NACDA/studies/3843/summary |
| TILDA | The Irish LongituDinal Study on Ageing | Ireland | Aging | https://tilda.tcd.je/participants/ |
| WHI | Women’s Health Initiative | USA | Heart disease, breast and colorectal cancer, and osteoporosis | https://www.whi.org |
Extended Data Table 3.
Summary of additional studies not included in Table 1.
| Biomarker | Reason for exclusion | Cohort | Cross-validation | Study |
|---|---|---|---|---|
| Adiponectin | Single molecule, follow-up < 10 years, no cross-population validation | Health ABC | n/a | Poehls et al. 2009 28 |
| Cystatin C | Single molecule, follow-up < 10 years, no cross-population validation | Health ABC | n/a | Shlipak et al. 2009 29 |
| Telomere length | Single molecule, no cross-population validation | UKB | n/a | Schneider et al. 2022 30 |
| “Protein Biomarker” | no cross-population validation | FHS | n/a | Ho et al. 2018 31 |
| "Biological Age" | no cross-population validation | FHS | n/a | Murabito et al. 2018 32 |
| DM17 | follow-up <10 years | BLSA, InChianti, NHANES | WHAS | Li et al. 2022 1 |
| Wang et al. | follow-up <10 years | n/a | KORA F4 cohort excluded | Wang et al. 2021 33 |
| Plasma proteomic biomarker | no cross-population validation, follow-up <10 years | LonGenity | LonGenity (∼50%) | Sathyan et al. 2020 8 |
| Proteomics | no cross-population validation | ICP+VSP1 | ICP+VSP1 (30%) | Eiriksdottir et al. 2021 34 |
| DNAm clocks | follow-up <10 years | n/a | 4 cohorts | Marioni et al. 2015 2 |
| Integrated proteomic and metabolomic biomarker | follow-up <10 years | ARIC | AASK | Zhou et al. 2022 35 |
| Multi-domain phenotypic panel | no cross-population validation, follow-up <10 years | BLSA | n/a | Kuo et al. 2022 9 |
| Heterogenous organ aging | no cross-population validation | UKB | n/a | Tian et al. 2023 4 |
| Integrative clinical and DNAm model | follow up <10 years | FHS | ARIC | Huan et al. 2022 23 |
| "Proteomic Biomarker" | no cross-population validation | Health ABC | n/a | Orwoll et al. 2020 7 |
| DNAm clocks | weighted median follow-up <10 | n/a | 13 cohorts | Chen et al. 2016 36 |
| DNAm Skin and Blood Clock | follow-up ? y, details not clear | n/a | WHI, FHS, ... | Horvath et al. 2018 37 |
Summary of additional studies not included in Table 1.
The criteria we defined for our overview resulted in exclusion of several studies. Exclusion was primarily due to follow-up times below 10 years (Mahalanobis distance, 1; Cross-cohort evaluation of DNAm biomarkers,2), a lack of cross-validation in independent cohorts (miRNA biomarkers, 3; advanced biological aging predictor,4), a lack of evaluation of association with mortality (GlycanAge, 5; transcriptomic clock, 6), or a combination of the above (Plasma proteomic biomarker panels, 7,8; Multi-domain phenotypic panel, 9). We provide an overview of example studies not included in Extended Data Table 3 below. We also note that single molecule biomarkers are not covered in this study as the focus is on composite biomarkers. For a detailed overview and comparison of blood-based single molecule biomarkers of aging please refer to 10.
Acknowledgments:
Supported in part by the Intramural Research Program of the National Institute on Aging, NIH, Baltimore, MD. The authors are very grateful to David M. Wilson III for the many suggestions during the writing of this manuscript.
Footnotes
Conflict of Interest statement: M.M., V.S., M.P.S, and V.N.G. have filed a patent on measuring cellular aging. C.H. is also affiliated with the Institute for Biomedical Aging Research, Universitat Innsbruck, Austria and an honorary research fellow at the Department of Women’s Cancer, EGA Institute for Women’s Health, University College London, United Kingdom. C.H. is a shareholder of Sola Diagnostics GmbH, and named as inventor on a patent on an epigenetic clock indicative of breast cancer risk. J.N.J. is also affiliated with the Sticht Center for Healthy Aging and Alzheimer’s Prevention, Wake Forest University School of Medicine, Winston-Salem, NC, USA and the XPRIZE Foundation, Culver City, CA, USA. J.N.J. serves on the advisory board for the American Federation for Aging Research (AFAR)’s Finding Aging biomarkers by Searching existing Trials (FAST) Initiative and the editorial board of Journals of Gerontology Series A Biological Sciences, eLife, and Experimental Gerontology. D.B. is also affiliated with the Child Brain Development Network, Canadian Institute for Advanced Research and SocioMed Research Nucleus, Universidad Mayor, Santiago, Chile. D.B. is an inventor of DunedinPACE, a Duke University and University of Otago invention licensed to TruDiagnostic. A.H.-C. has built epigenetic aging metrics that are licensed by Elysium Health through Yale University. B.H.C. owns stock in Illumina Inc., the manufacturer of the DNA methylation arrays used in epigenetic biomarkers of aging, and is listed as a co-inventor in filed patents on commercial applications of epigenetic prediction models. A.A.C. is a founder, president, and majority shareholder at Oken Health. R.E.M has received a speaker fee from Illumina and is an advisor to the Epigenetic Clock Development Foundation and Optima Partners. M.W. is also affiliated with the Institute for Biomedical Aging Research, Universität Innsbruck, Austria. M.W. is a shareholder of Sola Diagnostics GmbH, and named as inventor on a patent on an epigenetic clock indicative of breast cancer risk. K.F. is CEO of BioAge Labs P.O.F. is an employee and stakeholder in Gero PTE. A.Z. is the founder and CEO of Insilico Medicine, a clinical-stage generative AI and robotics biotechnology company specializing in aging research N.B. is the Scientific Director of the American Federation for Aging Research (AFAR), on the board of the executive committee of the Longevity Biotech Association, and advisor on the Board of the Academy for Health and Lifespan Research. D.P.K has received a grant from Solarea Bio and royalties from Wolters Kluwer. D.P.K sits on the Scientific Advisory Boards of Solarea Bio, Pfizer and Reneo and has participated on the Data Safety Monitoring Board for the AgNovos Healthcare osteoporosis treatment trial. E.V. is a scientific co-founder of Napa Therapeutics and BHB Therapeutics, serves on the scientific advisory board of Seneque, and is named co-inventor on a patent relating to an epigenetic clock robust to cell composition changes. A.B.M. declares to be Chief Medical Officer of NU. V.S is Cofounder, SAB Chair and Head of research of Turn Biotechnologies. M.P.S. is a cofounder and scientific advisor of Personalis, SensOmics, Qbio, January AI, Fodsel, Filtricine, Protos, RTHM, Iollo, Marble Therapeutics, Crosshair Therapeutics and Mirvie. He is a scientific advisor of Jupiter, Neuvivo, Swaza, Mitrix. S.H. is a founder of the nonprofit Epigenetic Clock Development Foundation that licenses patents surrounding epigenetic clocks. The Regents of the University of California is the sole owner of a patent application directed at GrimAge and other epigenetic clocks for which S.H. is a named inventor.
REFERENCES
- 1.Moqri M. et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758–3775 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Moqri M. et al. PRC2 clock: a universal epigenetic biomarker of aging and rejuvenation. 2022.06.03.494609 Preprint at 10.1101/2022.06.03.494609 (2022). [DOI] [Google Scholar]
- 3.Poganik JR et al. Biological age is increased by stress and restored upon recovery. Cell Metab. 35, 807–820.e5 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu AT et al. DNA methylation GrimAge version 2. Aging 14, 9484–9549 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tanaka T. et al. Plasma proteomic biomarker signature of age predicts health and life span eLife 9, e61073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balasubramanian R. et al. Metabolomic profiles associated with all-cause mortality in the Women’s Health Initiative. Int. J. Epidemiol 49, 289–300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sarkar TJ et al. Transient non-integrative expression of nuclear reprogramming factors promotes multifaceted amelioration of aging in human cells. Nat. Commun 11, 1545 (2020) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee JW et al. Fit-for-purpose method development and validation for successful biomarker measurement. Pharm. Res 23, 312–328 (2006). [DOI] [PubMed] [Google Scholar]
- 9.Wagner JA Overview of Biomarkers and Surrogate Endpoints in Drug Development. D/s. Markers 18, 41–6 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hunter DJ et al. A Pathway and Approach to Biomarker Validation and Qualification for Osteoarthritis Clinical Trials. Curr. Drug Targets 11, 536–545 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bortz J. et al. Biological age estimation using circulating blood biomarkers. Commun. Biol 6, 1–10 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lehallier B. et al. Undulating changes in human plasma proteome profiles across the lifespan. Nat. Med 25, 1843–1850 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ahadi S. et al. Personal aging markers and ageotypes revealed by deep longitudinal profiling. Nat. Med 26, 83–90 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Alpert A. et al. A clinically meaningful metric of immune age derived from high-dimensional longitudinal monitoring. Nat. Med 25, 487–495 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rutledge J, Oh H & Wyss-Coray T Measuring biological age using omics data. Nat. Rev. Genet 23, 715–727 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kudryashova KS, Burka K, Kulaga AY, Vorobyeva NS & Kennedy BK Aging Biomarkers: From Functional Tests to Multi-Omics Approaches. PROTEOMICS 20, 1900408 (2020). [DOI] [PubMed] [Google Scholar]
- 17.Committee on the Review of Omics-Based Tests for Predicting Patient Outcomes in Clinical Trials et al. Omics-Based Clinical Discovery: Science, Technology, and Applications. (2012). [Google Scholar]
- 18.Ying K. et al. ClockBase: a comprehensive platform for biological age profiling in human and mouse. 2023.02.28.530532 Preprint at 10.1101/2023.02.28.530532 (2023). [DOI] [Google Scholar]
- 19.Belsky DW et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 11, e73420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bautmans I. et al. WHO working definition of vitality capacity for healthy longevity monitoring. Lancet Healthy Longev. 3, e789–e796 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lara J. et al. A proposed panel of biomarkers of healthy ageing. BMC Med. 13, 222 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Crimmins EM, Thyagarajan B, Levine ME, Weir DR & Faul J Associations of Age, Sex, Race/Ethnicity, and Education With 13 Epigenetic Clocks in a Nationally Representative U.S. Sample: The Health and Retirement Study. J. Gerontol. Ser. A 76, 1117–1123 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Faul JD et al. Epigenetic-based age acceleration in a representative sample of older Americans: Associations with aging-related morbidity and mortality. Proc. Natl. Acad. Sci 120, e2215840120 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lu AT et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gadd DA et al. Blood protein levels predict leading incident diseases and mortality in UK Biobank. 2023.05.01.23288879 Preprint at 10.1101/2023.05.01.23288879 (2023). [DOI] [Google Scholar]
- 26.Tian YE et al. Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nat. Med 29, 1221–1231 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Buergel T. et al. Metabolomic profiles predict individual multidisease outcomes. Nat. Med 28, 2309–2320 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Raghavachari N, Wilmot B & Dutta C Optimizing Translational Research for Exceptional Health and Life Span: A Systematic Narrative of Studies to Identify Translatable Therapeutic Target(s) for Exceptional Health Span in Humans. J. Gerontol. Ser. A 77, 2272–2280 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kurki MI et al. FinnGen provides genetic insights from a well-phenotyped isolated population. Nature 613, 508–518 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nagai A. et al. Overview of the BioBank Japan Project: Study design and profile. J. Epidemiol 27, S2–S8 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Boutin NT et al. The Evolution of a Large Biobank at Mass General Brigham. J. Pers. Med 12, 1323 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huan T. et al. Integrative analysis of clinical and epigenetic biomarkers of mortality. Aging Cell 21, e 13608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eiriksdottir T. et al. Predicting the probability of death using proteomics. Commun. Biol 4, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Deelen J. et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuiper LM et al. Epigenetic and Metabolomic Biomarkers for Biological Age: A Comparative Analysis of Mortality and Frailty Risk. J. Gerontol. Ser. A glad 137 (2023) doi: 10.1093/gerona/glad137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang C. et al. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: The NAS, and KORA F4. eBioMedicine 63, (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Evans MK et al. Healthy aging in neighborhoods of diversity across the life span (HANDLS): overcoming barriers to implementing a longitudinal, epidemiologic, urban study of health, race, and socioeconomic status. Ethn. Dis 20, 267–275 (2010). [PMC free article] [PubMed] [Google Scholar]
- 38.Solomon Olivia et al. Comparison of DNA methylation measured by lllumina 450K and EPIC BeadChips in blood of newborns and 14-year-old children. 13, 655–664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koistinen V. et al. Towards a Rosetta stone for metabolomics: recommendations to overcome inconsistent metabolite nomenclature. Nat. Metab 5, 351–354 (2023). [DOI] [PubMed] [Google Scholar]
- 40.Fahy E & Subramaniam S RefMet: a reference nomenclature for metabolomics. Nat. Methods 17, 1173–1174 (2020). [DOI] [PubMed] [Google Scholar]
- 41.Yu B. et al. The Consortium of Metabolomics Studies (COMETS): Metabolomics in 47 Prospective Cohort Studies. Am. J. Epidemiol 188, 991–1012 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Psaty BM et al. Cohorts for Heart and Aging Research in Genomic Epidemiology (CHARGE) Consortium: Design of prospective meta-analyses of genome-wide association studies from 5 cohorts. Circ. Cardiovasc. Genet 2, 73–80 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keane T & Glass L CINECA: Common Infrastructure for National Cohorts in Europe, Canada, and Africa - Kick Off Report. (2019). [Google Scholar]
- 44.Taliun D. et al. Sequencing of 53,831 diverse genomes from the NHLBI TOPMed Program. Nature 590, 290–299 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Linsen L. et al. Raising to the Challenge: Building a Federated Biobank to Accelerate Translational Research—The University Biobank Limburg. Front. Med 6, 224 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kaye J. et al. Access Governance for Biobanks: The Case of the BioSHaRE-EU Cohorts. Biopreservation Biobanking 14, 201–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cummings SR & Kritchevsky SB Endpoints for geroscience clinical trials: health outcomes, biomarkers, and biologic age. GeroScience 44, 2925–2931 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kaeberlein M. How healthy is the healthspan concept? GeroScience 40, 361–364 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Okada D, Cheng JH, Zheng C, Kumaki T & Yamada R Data-driven identification and classification of nonlinear aging patterns reveals the landscape of associations between DNA methylation and aging. Hum. Genomics 17, 8 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Higgins-Chen AT et al. A computational solution for bolstering reliability of epigenetic clocks: Implications for clinical trials and longitudinal tracking. Nat. Aging 2, 644–661 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tomusiak A. et al. Development of a novel epigenetic clock resistant to changes in immune cell composition. http://biorxiv.Org/lookup/doi/10.1101/2023.03.01.530561 (2023) doi: 10.1101/2023.03.01.530561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fang F. et al. Evaluation of pediatric epigenetic clocks across multiple tissues. Clin. Epigenetics 15, 142 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson ND et al. Non-linear patterns in age-related DNA methylation may reflect CD4+T cell differentiation. Epigenetics 12, 492–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Methylation Array Harmonization Workflow. Natl. Cancer Inst. [Google Scholar]
- 55.Emmanuel T. et al. A survey on missing data in machine learning. J. Big Data 8, 140 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Li P, Stuart EA & Allison DB Multiple Imputation: A Flexible Tool for Handling Missing Data. JAMA 314, 1966–1967 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yousefi Paul et al. Considerations for Normalization of DNA Methylation Data by lllumina 450K BeadChip Assay in Population Studies. 8, 1141–1152 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bizzarri D, Reinders MJT, Beekman M, Slagboom PE & van den Akker EB MiMIR: R-shiny application to infer risk factors and endpoints from Nightingale Health’s 1H- NMR metabolomics data. Bioinformatics 38, 3847–3849 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kwon D & Belsky DW A toolkit for quantification of biological age from blood chemistry and organ function test data: BioAge. GeroScience 43, 2795–2808 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Thrush KL, Higgins-Chen AT, Liu Z & Levine ME R methylCIPHER: A Methylation Clock Investigational Package for Hypothesis-Driven Evaluation & Research. http://biorxiv.org/lookup/doi/10.1101/2022.07.13.499978 (2022) doi: 10.1101/2022.07.13.499978. [DOI] [Google Scholar]
- 61.Ying K. et al. Causal Epigenetic Age Uncouples Damage and Adaptation. 2022.10.07.511382 Preprint at 10.1101/2022.10.07.511382 (2022). [DOI] [Google Scholar]
- 62.Pun FW et al. Hallmarks of aging-based dual-purpose disease and age-associated targets predicted using PandaOmics Al-powered discovery engine. Aging 14, 2475–2506 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manning AK et al. NHLBI biodata catalyst and the future of cloud computing. 45, 774–775 (2021). [Google Scholar]
- 64.Li T, Sahu AK, Talwalkar A & Smith V Federated Learning: Challenges, Methods, and Future Directions. IEEE Signal Process. Mag 37, 50–60 (2020). [Google Scholar]
- 65.Fortier I. et al. Maelstrom Research guidelines for rigorous retrospective data harmonization. Int. J. Epidemiol 46, 103–105 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.FAIR Principles. GO FAIR https://www.go-fair.org/fair-principles/. [Google Scholar]
- 67.Tang MX et al. The APOE-epsilon4 allele and the risk of Alzheimer disease among African Americans, whites, and Hispanics. JAMA 279, 751–755 (1998). [DOI] [PubMed] [Google Scholar]
- 68.Benjamin C. Trumble et al. Apolipoprotein E4 is associated with improved cognitive function in Amazonian forager-horticulturalists with a high parasite burden. 31, 1251–1767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Prive F. et al. Portability of 245 polygenic scores when derived from the UK Biobank and applied to 9 ancestry groups from the same cohort. Am. J. Hum. Genet 109, 12–23 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Graf GH et al. Testing Black-White Disparities in Biological Aging Among Older Adults in the United States: Analysis of DNA-Methylation and Blood-Chemistry Methods. Am. J. Epidemiol 191, 613–625 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu Z. et al. A new aging measure captures morbidity and mortality risk across diverse subpopulations from NHANES IV: A cohort study. PLOS Med. 15, e1002718 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vandenbroucke JP et al. Strengthening the Reporting of Observational Studies in Epidemiology (STROBE): Explanation and Elaboration. PLoS Med. 4, e297 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rodés B, Cadiñanos J, Esteban-Cantos A, Rodríguez-Centeno J & Arribas JR Ageing with HIV: Challenges and biomarkers. eBioMedicine 77, 103896 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Cohen AA et al. Detection of a Novel, Integrative Aging Process Suggests Complex Physiological Integration. PLOS ONE 10, e0116489 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ferrucci L & Kohanski R Better care for older patients with complex multimorbidity and frailty: a call to action. Lancet Healthy Longev. 3, e581–e583 (2022). [DOI] [PubMed] [Google Scholar]
- 76.Levine ME et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sebastiani P. et al. Biomarker signatures of aging. Aging Cell 16, 329–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Mamoshina P. et al. Machine Learning on Human Muscle Transcriptomic Data for Biomarker Discovery and Tissue-Specific Drug Target Identification. Front. Genet 9, 242 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Zhang Y. et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun 8, 14617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Bernabeu E. et al. Refining epigenetic prediction of chronological and biological age. Genome Med. 15, 12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.van den Akker EB et al. Metabolic Age Based on the BBMRI-NL 1 H-NMR Metabolomics Repository as Biomarker of Age-related Disease. Circ. Genomic Precis. Med 13, 541–547 (2020). [DOI] [PubMed] [Google Scholar]
- 82.Li X. et al. Longitudinal trajectories, correlations and mortality associations of nine biological ages across 20-years follow-up. eLife 9, (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hillary RF et al. An epigenetic predictor of death captures multi-modal measures of brain health. Mol. Psychiatry 26, 3806–3816 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.McCrory C. et al. GrimAge Outperforms Other Epigenetic Clocks in the Prediction of Age-Related Clinical Phenotypes and All-Cause Mortality. J. Gerontol. Ser. A 76, 741–749 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tiina F. et al. Does the epigenetic clock GrimAge predict mortality independent of genetic influences: an 18 year follow-up study in older female twin pairs. Clin. Epigenetics 13, 128 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Wang C. et al. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: The NAS, and KORA F4. EBioMedicine 63, 103151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Baker GT & Sprott RL Biomarkers of aging. Exp. Gerontol 23, 223–239 (1988). [DOI] [PubMed] [Google Scholar]
Extended Data References
- 1.Li Q. et al. An objective metric of individual health and aging for population surveys. Popul. Health Metr 20, 11 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Marioni RE et al. DNA methylation age of blood predicts all-cause mortality in later life. Genome Biol. 16, 1–12 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huan T. et al. Age-associated microRNA expression in human peripheral blood is associated with all-cause mortality and age-related traits. Aging Cell 17, e12687 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tian YE et al. Heterogeneous aging across multiple organ systems and prediction of chronic disease and mortality. Nat. Med 29, 1221–1231 (2023). [DOI] [PubMed] [Google Scholar]
- 5.Kristie J. et al. Glycans Are a Novel Biomarker of Chronological and Biological Ages. J. Gerontol. A. Biol. Sci. Med. Sci 69, 779–789 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Peters MJ et al. The transcriptional landscape of age in human peripheral blood. Nat. Commun 6, 8570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Orwoll ES et al. Proteomic assessment of serum biomarkers of longevity in older men. Aging Cell 19, e13253 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sathyan S. et al. Plasma proteomic profile of age, health span, and all-cause mortality in older adults. Aging Cell 19, e13250 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kuo P-L et al. Longitudinal phenotypic aging metrics in the Baltimore Longitudinal Study of Aging. Nat. Aging 2, 635–643 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Justice JN et al. A framework for selection of blood-based biomarkers for geroscience-guided clinical trials: report from the TAME Biomarkers Workgroup. GeroScience 40, 419–436 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Levine ME et al. An epigenetic biomarker of aging for lifespan and healthspan. Aging 10, 573–591 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sebastiani P. et al. Biomarker signatures of aging. Aging Cell 16, 329–338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mamoshina P. et al. Population Specific Biomarkers of Human Aging: A Big Data Study Using South Korean, Canadian, and Eastern European Patient Populations. J. Gerontol. Ser. A 73, 1482–1490 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Y. et al. DNA methylation signatures in peripheral blood strongly predict all-cause mortality. Nat. Commun 8, 14617 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lu AT et al. DNA methylation GrimAge strongly predicts lifespan and healthspan. Aging 11, 303–327 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lu AT et al. DNA methylation GrimAge version 2. Aging 14, 9484–9549 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Belsky DW et al. DunedinPACE, a DNA methylation biomarker of the pace of aging. eLife 11, e73420 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bernabeu E. et al. Refining epigenetic prediction of chronological and biological age. Genome Med. 15, 12 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Deelen J. et al. A metabolic profile of all-cause mortality risk identified in an observational study of 44,168 individuals. Nat. Commun 10, (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Akker EB et al. Metabolic Age Based on the BBMRI-NL 1 H-NMR Metabolomics Repository as Biomarker of Age-related Disease. Circ. Genomic Precis. Med 13, 541–547 (2020). [DOI] [PubMed] [Google Scholar]
- 21.Balasubramanian R. et al. Metabolomic profiles associated with all-cause mortality in the Women’s Health Initiative. Int. J. Epidemiol 49, 289–300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tanaka T. et al. Plasma proteomic biomarker signature of age predicts health and life span. eLife 9, e61073 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huan T. et al. Integrative analysis of clinical and epigenetic biomarkers of mortality. Aging Cell 21, e13608 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rutledge J, Oh H & Wyss-Coray T Measuring biological age using omics data. Nat. Rev. Genet 23, 715–727 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kudryashova KS, Burka K, Kulaga AY, Vorobyeva NS & Kennedy BK Aging Biomarkers: From Functional Tests to Multi-Omics Approaches. PROTEOMICS 20, 1900408 (2020). [DOI] [PubMed] [Google Scholar]
- 26.Hartmann A. et al. Ranking Biomarkers of Aging by Citation Profiling and Effort Scoring. Front. Genet 12, 686320 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moqri M. et al. Biomarkers of aging for the identification and evaluation of longevity interventions. Cell 186, 3758–3775 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Poehls J. et al. Association of adiponectin with mortality in older adults: the Health, Aging, and Body Composition Study. Diabetologia 52, 591–595 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shlipak MG et al. Cystatin C and Mortality Risk in the Elderly: The Health, Aging, and Body Composition Study. J. Am. Soc. Nephrol 17, 254–261 (2006). [DOI] [PubMed] [Google Scholar]
- 30.Schneider CV et al. Association of Telomere Length With Risk of Disease and Mortality. JAMA Intern. Med 182, 291–300 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ho JE et al. Protein Biomarkers of Cardiovascular Disease and Mortality in the Community. J. Am. Heart Assoc. Cardiovasc. Cerebrovasc. Dis 7, e008108 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Murabito JM et al. Measures of Biologic Age in a Community Sample Predict Mortality and Age-Related Disease: The Framingham Offspring Study. J. Gerontol. A. Biol. Sci. Med. Sci 73, 757–762 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wang C. et al. DNA methylation-based biomarkers of age acceleration and all-cause death, myocardial infarction, stroke, and cancer in two cohorts: The NAS, and KORA F4. EBioMedicine 63, 103151 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eiriksdottir T. et al. Predicting the probability of death using proteomics. Commun. Biol 4, 758 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou L. et al. Integrated proteomic and metabolomic modules identified as biomarkers of mortality in the Atherosclerosis Risk in Communities study and the African American Study of Kidney Disease and Hypertension. Hum. Genomics 16, 53 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen BH et al. DNA methylation-based measures of biological age: meta-analysis predicting time to death. Aging 8, 1844–1865 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvath S. et al. Epigenetic clock for skin and blood cells applied to Hutchinson Gilford Progeria Syndrome and ex vivo studies. Aging 10, 1758–1775 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]